Abstract
The bacterial genus, Borrelia, is comprised of vector-borne spirochete species that infect and are transmitted from multiple host species. Some Borrelia species cause highly-prevalent diseases in humans and domestic animals. Evolutionary, ecological, and molecular research on many Borrelia species have resulted in tremendous progress toward understanding the biology and natural history of these species. Yet, many outstanding questions, such as how Borrelia populations will be impacted by climate and land-use change, will require an interdisciplinary approach. The evolutionary ecology research framework incorporates theory and data from evolutionary, ecological, and molecular studies while overcoming common assumptions within each field that can hinder integration across these disciplines. Evolutionary ecology offers a framework to evaluate the ecological consequences of evolved traits and to predict how present-day ecological processes may result in further evolutionary change. Studies of microbes with complex transmission cycles, like Borrelia, which interact with multiple vertebrate hosts and arthropod vectors, are poised to leverage the power of the evolutionary ecology framework to identify the molecular interactions involved in ecological processes that result in evolutionary change. Using existing data, we outline how evolutionary ecology theory can delineate how interactions with other species and the physical environment create selective forces or impact migration of Borrelia populations and result in micro-evolutionary changes. We further discuss the ecological and molecular consequences of those micro-evolutionary changes. While many of the currently outstanding questions will necessitate new experimental designs and additional empirical data, many others can be addressed immediately by integrating existing molecular and ecological data within an evolutionary ecology framework.
Keywords: Evolutionary ecology, Borrelia, transmission, ecological interactions
1. Introduction
Zoonotic pathogens – those that transmit among wildlife and infect humans (Box 1)–are some of the most common causes of emerging and re-emerging infectious diseases in the world1,2. Many zoonotic bacterial and viral pathogens have complex transmission cycles during which they must interact with multiple natural host species and arthropod vectors2–4(Box 1). The bacterial genus Borrelia is comprised of vector-borne pathogens which infect and are transmitted from multiple vertebrate host species and can cause several highly-prevalent diseases in humans and domestic animals5–7. While not fully resolved, the genus has three major evolutionary groups now proposed as separate genera8–10; the most well-studied of these is the Lyme borreliosis clade (referred to here as the LB group), the species of which are vectored exclusively by hard-bodied ticks in the Ixodes genus. Here, we review research results on the LB group within an evolutionary ecology framework to explore the molecular interactions involved in ecological processes that result in evolutionary change within Borrelia and the ecological consequences of that evolutionary change (Box 1).
Box 1: Glossary.
Complex transmission: transmission that occurs through multiple hosts or vectors
Evolutionary ecology: A research framework that explicitly considers the evolutionary histories and the ecological interactions driving evolutionary change
Generalist pathogen: Pathogens that can successfully infect and transmit from a wide range of host species
Micro-evolution: the change in allele frequencies within populations or species caused by natural selection, gene flow, mutation or drift, including changes caused by horizontal gene or plasmid transfer within and between species81.
Negative frequency-dependent selection: When the strength and direction of natural selection is a function of the relative abundance a trait variant in a population. The fitness of a trait variant increases as the relative abundance, or frequency, of the variant decreases.
Niche: set of environmental conditions in which the members of a species can survive.
Multiple niche polymorphism: Diversity within a population that is maintained because the strength and direction of natural selection on each trait variant is a function of its ability to exploit different environmental features in a heterogeneous habitat153,154. This type of multi-niche selection can maintain multiple trait variants in a population if each variant has a selective advantage in some available habitats while other variants are superior in other habitats.
Specialist pathogen: Pathogens that can successfully infect and transmit from only one or a small subset of possible species
Zoonotic pathogens: pathogens naturally transmitted between animals and humans
The evolutionary history and ecology of Borrelia have been investigated extensively. Evolutionary studies of Borrelia have characterized within and between species diversity, genetic and phenotypic variation across space and among host species, and the intergenerational processes underlying the observed variation11–14. These studies have resulted in the identification of molecular factors associated with host and vector specialization, diversification rates and processes, mechanisms of immune escape, and the importance of virulence factors to the life cycle4,15–32. Similarly, ecological studies have provided critical advances in our understanding of the processes and interactions that affect the abundance of some Borrelia species on short time scales, identifying environmental conditions and host communities that directly influence year-to-year variations in Borrelia populations33–35. While both evolutionary and ecological studies have been instrumental to our understanding of Borrelia, many outstanding questions can be addressed only by transcending the assumption that ecological and evolutionary processes operate on different timescales (Table 1).
Table 1:
Evolutionary Ecology Framework
| ECOLOGY | EVOLUTIONARY BIOLOGY | EVOLUTIONARY ECOLOGY | |
|---|---|---|---|
| EXAMPLE QUESTIONS | • How does intraspecific diversity contribute to host-vector-pathogen interactions? | • How do pathogens co-evolve with their vectors and hosts? | • How do ecological interactions create selective forces or impact migration rates to cause micro-evolutionary changes? |
| • How important are multiple infections in driving disease dynamics? | • By what molecular mechanisms do pathogens replicate and how does that impact pathogen evolution? | • What are the ecological consequences – such as changes in geographic, host or vector range, abundance, or virulence – of these micro-evolutionary changes? | |
| • How do species interactions explain the distribution and abundance of different species? | |||
| ASSUMPTIONS | • Individuals within a species or group considered identical | • Constant/irrelevant population densities (Hartl and Clark 1989) | • Distribution and variance of genetic variation is constant (Holt 2005) |
| • No evolutionary change (short timescales) | • Fitness in light of ecological interactions considered as constant | ||
| • Most evolution is neutral | |||
| NECESSARY DATA & EXAMPLE APPROACHES |
Necessary Data: Measures of diversity (e.g. phenotypes, genotypes, species counts, functional traits); Measures of disease progression and transmission Examples: Devevey et al. 2015155, Walter et al. 2016117 |
Necessary Data: Sequencing (e.g. multi-locus markers, whole genome sequencing, reduced representation sequencing) Examples: Brisson et al. 2010156, Becker et al. 201618 |
Necessary Data: Sequencing (e.g. multi-locus markers, whole genome sequencing, reduced representation sequencing); Measures of diversity (e.g. phenotypes, genotypes, species counts, functional traits); Examples: MacDonald et al. 2019 bioRxiv157; Becker and Han 2020 bioRxiv152 |
An evolutionary ecology research framework integrates ecological and evolutionary timescales, incorporating feedbacks between ecological interactions and evolutionary change36 (Box 1). Ecological interactions, like those that Borrelia species experience throughout their transmission cycle, create evolutionary pressures that select for traits that enhance specific components of the life cycles of Borrelia species37. These evolutionary changes, in turn, effect subsequent ecological interactions with hosts or vectors that create additional evolutionary pressures. Investigating Borrelia within an evolutionary ecology framework provides a foundation to evaluate how ecological interactions result in micro-evolutionary change within Borrelia populations and the ecological consequences of those changes. This evolutionary ecology framework can be used address questions like why some Borrelia species are more host-specific than others or why rates of gene flow are different among populations. While some of these questions have been addressed, further investigations within an evolutionary ecology framework would provide new insights given an evolutionary context. Such evaluations are critical for understanding, predicting, and managing disease epidemics.
Central to the evolutionary ecology of Borrelia species are two questions:
How do intra- and interspecies interactions, or interactions with the physical environment, create selective forces or impact migration rates to cause micro-evolutionary changes?
What are the ecological consequences – such as changes in geographic, host or vector range, abundance, or virulence – of these micro-evolutionary changes?
Evolutionary ecology provides a particularly powerful investigatory framework for infectious disease systems38–41. For example, an investigation of a plant pathogen system within an evolutionary ecology framework resulted in accurate identification of novel host species to which plant pathogens are likely to adapt42. These predictions were made by considering relatedness among hosts, their ecological traits, and their geographic distributions. Research into microbes with complex transmission cycles, like those within the Borrelia genus, which interact with multiple hosts and environmental conditions, is particularly poised to leverage the power derived from an evolutionary ecology framework to identify ecological processes affecting evolutionary change.
As Borrelia species are not free-living microbes, all molecular and ecological interactions occur with or within a vector or a vertebrate host. The web of interactions among Borrelia, vectors, and hosts create selection pressures that drive Borrelia evolution43,44, impact migration and geographic distributions of Borrelia31,32,45–49, and shape mutation rates and genetic drift50, although the latter two evolutionary forces are difficult to discern in natural populations. Here, we describe the impacts of ecological interactions on micro-evolutionary change (Box 1) with Borrelia that can be deduced from independent studies of ecological interactions and molecular interactions. We then discuss future directions that can maximize the utility of an evolutionary ecology framework to identify additional ecological or molecular mechanisms that are key to the Borrelia life-cycle.
2. Borrelia-Host Interactions
2.1. Generalism to specialism
Ecological interactions between Borrelia and its vertebrate hosts select for evolutionarily-tuned molecular mechanisms within Borrelia species to enhance their ability to colonize, disseminate to distal tissues, evade host immune responses, and transmit from the host to feeding ticks15,25,26. Sequence variation among Borrelia species in the proteins that mediate these molecular mechanisms is shaped by natural selection to enhance the efficacy of interactions with host proteins that vary among vertebrate host species15,51,52. For example, variation in a complement regulator acquiring surface protein (CspA) among Borrelia species results in variation in their ability to bind complement molecules in different vertebrate hosts and thus limits the host range of each Borrelia species52–58. Ecological interactions with different host species cause each Borrelia species to experience unique selective pressures that result in divergent micro-evolutionary trajectories (Table 1). This evolutionary divergence has ecological consequences for Borrelia as it limits the subset of vertebrate hosts in which each Borrelia species can successfully complete its infectious cycle. A core component of evolutionary ecology is to assess these types of eco-evolutionary feedback loops. Investigating these eco-evolutionary feedback loops could address outstanding questions such as how the frequency and strength of selection on other Borrelia-host interactions are impacted by host specialization or the mechanisms by which Borrelia species evolve to become ever more specialized.
Host associations, sometimes referred to as host specialization59, determine the frequency and importance of ecological interactions between each Borrelia species and each vertebrate species60,61. The frequency of ecological interactions with each host species, in turn, determines the strength of natural selection imposed by each host species on Borrelia15,62. For example, variation among host species in immune system components imposes different selective regimes on invading Borrelia species60,61. The strength of these selective regimes is determined by how frequently that Borrelia species must evade the immune system of each host species and the host-to-tick transmission rate given a successful infection of that host15,62. Further, adaptations that maximize survival and transmission from one host species often limit survival and transmission from other hosts, leading to selection favoring host specialism15,63–70. This eco-evolutionary feedback loop favors evolution towards greater host specialization (Box 1, Figure 1). Host associations can even drive speciation events. For example, the divergence between the sister species B. garinii and B. bavariensis resulted from differential host species use; B. garinii specialized on bird species while B. bavariensis specialized on small mammals18,71. Subsequent genetic differentiation among these specialized Borrelia species arose in response to the different selective regimes imposed by the different vertebrate hosts with which they interact most frequently. An evolutionary ecology research framework can facilitate investigations into the molecular changes within B. bavariensis that enabled it to use a different host species by considering both how ecological interactions can cause micro-evolutionary change and how that evolutionary change impacts ecological interactions.
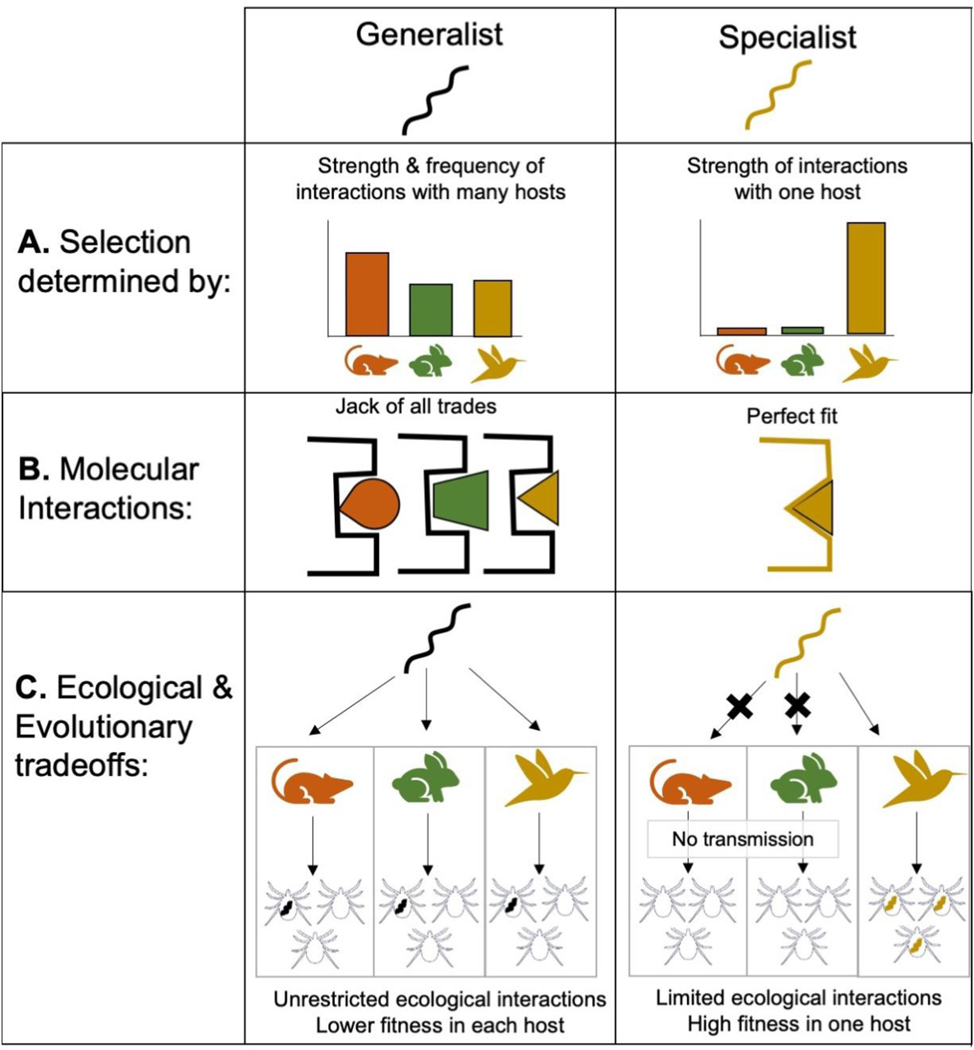
Figure 1: Why are some Borrelia species more host-specific than others?
Along the generalist-specialist continuum, the more generalist Borrelia species infect many vertebrate host species while the more specialist species infect only one or a few host species. Considering the ecological interactions each Borrelia species experiences elucidates the selective pressures that led to the differences in host species specialization. A. The strength of natural selection is derived, in part, from the frequency of ecological interactions that a Borrelia species experiences with each possible host species. That is, selection on generalist species is impacted by the frequency of interactions with the wider vertebrate host community whereas selection on specialist species populations is determined primarily by interactions with the one species it can infect and from which it is regularly transmitted. B. The molecular mechanisms underlying successful infection vary between generalist and specialist species due to differing selective pressures. For example, the surface proteins of generalist species interact successfully but not ideally with the corresponding receptors of multiple host species, C. resulting in sub-optimal infection or transmission success from many species; B. the surface proteins of specialist Borrelia species interact more effectively with the corresponding receptor of its one competent host C. resulting in optimal infection and transmission success from only one or a small number of host species. Thus, generalist species have less restricted ecological interactions with the vertebrate community but lower fitness in each host while specialist species are limited in their interactions to the vertebrate host species it can successfully infect and be transmitted from, but experience high fitness in that host.
Adaptations that cause Borrelia to complete their life cycles in only a specialized subset of host species subsequently limit the frequency of interactions with other vertebrate species, thus limiting the selective pressure they experience from these non-competent vertebrate species. Further, opportunities for genetic exchange between Borrelia species decrease when the species are less likely to occupy the same host. Thus, genetic mutations that fix in Borrelia species that specialize on different host species are rarely shared through horizontal gene transfer due to limited opportunities for genetic exchange72–74 leading to continuing evolutionary divergence among the Borrelia species. By contrast, horizontal gene transfer rates among genotypes within the same Borrelia species are 50 times greater than rates of inter-species horizontal gene transfer, at least partially due to the increased opportunity for transfer within co-infected hosts74. Evolutionary ecology provides a framework to investigate how eco-evolutionary feedback loops drive the high proportion of Borrelia species currently displaying host associations.
Host-species associations are even apparent among genetically distinct strains, or genotypes, within some generalist Borrelia species59,75,76 (Box 1). Genotypes of B. burgdorferi sensu stricto (Bbss) appear to differ in their ability to complete their infectious cycles in different vertebrate hosts in nature64–69, although these statistical associations have not been experimentally verified and are distinct, in both mechanism and strength, from those confirmed among European Borrelia species. Nevertheless, Bbss genotypes differ in their binding efficacy to host plasminogen and complement regulator molecules from different host species due to molecular adaptations (i.e. OspC and OspE) 15,77,78. Genotypes also differ in their abilities to survive and disseminate to distal tissues in a range of vertebrate hosts63–69, both of which are critical to the ability of Borrelia to transmit to ticks. Evolutionary ecology provides the research foundation to experimentally evaluate the mechanisms driving any existing host association differences among Bbss genotypes by focusing on eco-evolutionary feedbacks between interactions with host species and the impacts of these interactions on micro-evolution.
Variation among host species in plasminogen and complement regulator molecules may require Bbss genotypes to adapt to only a subset of host species, resulting in interactions with molecules from other species that prevent successful infections62. For example, each Bbss genotype maintains a single OspC variant that interacts effectively with plasminogen from only a limited number of host species79–81. While expressing multiple OspC variants may increase the range of host species that a Bbss genotype can infect, it may come at the cost of increasing immune targeting82. OspC is targeted by a fast and lethal immune response such that more targets could increase the probability of immune clearance before the bacterium can establish a long-lasting infection. The evolutionary ecology research framework can be used to investigate if a tradeoff between host-range breadth and immune targeting imposes a barrier to the long-term maintenance of host generalism in Borrelia genotypes. That being said, there is growing evidence that the arrival of Borrelia in North America pre-dates the last Ice Age17,24, and such a long evolutionary history may suggest that generalism is being maintained. Future work within evolutionary ecology could elucidate the apparent discord.
2.2. Interactions with the vertebrate immune system
Interactions between Borrelia species and the vertebrate adaptive immune system impose strong selective pressures on Borrelia. These interactions result in an evolutionary arms race between host immune responses and molecular mechanisms that enable immune evasion within Borrelia species15. The vlsE locus in all Borrelia species encodes an immunodominant surface protein that undergoes extensive and rapid antigenic variation during vertebrate infection in order to evade antibodies11,83–85. Novel vlsE protein sequences that are unrecognized by circulating antibodies are generated by recombination between the vlsE expression site and one or more of the unexpressed, highly-variable vls cassettes. The conservation of highly-mutable tandem repeat structures across the otherwise highly-diverged cassettes suggests that the host immune system imposes a strong selective pressure to maintain evolvability at this locus and results in ongoing micro-evolutionary change in the vls antigenic variation system11.
Acquired immunity can also create negative frequency dependent selection processes that maintain genetic variation within Borrelia populations (Box 1). Differences among host individuals in their history of antigen exposure can result in acquired immunity to specific Borrelia genotypes, thus reducing the proportion of hosts susceptible to the more common Borrelia genotypes. Therefore, rare Borrelia genotypes to which few hosts have been exposed would have a selective advantage over the more common genotypes to which many hosts have mounted an immune response. Experimental studies have also shown that genotype-specific antibodies are maternally inherited such that selection against common genotypes may last across generations86. Although fluctuating frequencies of genotypes are often associated with negative frequency dependent selection, ospC-major groups do not appear to fluctuate cyclically in natural populations87. Contrary to most textbook examples, however, negative frequency dependent selection on multi-strain pathogens can produce a variety of dynamics in addition to cyclical frequency fluctuations. For example, as discussed in Durand et al. 2017, the host immune system could cause pathogen populations to organize into communities of serotypes that minimize cross-reactive acquired immunity whose frequencies remain stable over long periods of time88–90. The exact mechanisms underlying this somewhat counterintuitive population dynamic under negative frequency dependent selection can be further explored within an evolutionary ecology research framework.
2.3. Borrelia distribution and genetic structure is shaped by host associations
The geographic distribution and population genetic structure of Borrelia species is shaped, in part, by the migration patterns of infected vertebrate hosts. Borrelia species associated with highly-mobile hosts (i.e. birds) tend to have limited population genetic structure as hosts maintain genetic cohesion over large geographic ranges31,32,47–49. By contrast, species associated with less mobile hosts (i.e. small mammals) tend to show much stronger population genetic structure, with different selective environments in different geographic areas and few opportunities for horizontal gene transfer among isolated populations31,47,91. For example, population genetic structure can be detected only at very large (inter-continental) scales in bird-associated species like B. garinii and B. valaisiana while the rodent-associated species like B. afzelii shows extensive spatial structuring even at fine spatial scales31,32,92. Highly disconnected populations limit horizontal gene transfer and reduces effective population sizes, which in turn, amplifies the impact of genetic drift leading to further differentiation among subpopulations.
Host species diversity can select for the maintenance of genetic diversity within Borrelia populations. Multiple genotypes of Bbss coexist within geographic locations when the environment is heterogeneous (i.e. a diverse vertebrate community) and none of the genotypes have the highest fitness in all host species60,93. Molecular variation among genotypes within Borrelia species determines the transmission success of antigenically distinct genotypes in each host species resulting in niche separation33,59,60,75,94 (Box 1). The presence of multiple host species at individual locations permits an array of genetically distinct genotypes to be simultaneously maintained. As vertebrate communities have been and continue to be impacted by land use change95, evolutionary ecology will provide a framework to predict how changes in vertebrate host communities will impact the diversity of Borrelia populations.
Borrelia-Vector Interactions
3.1. Vector specialization, vector switches, and Borrelia distribution
All Borrelia species require at least one tick species to complete their life cycle. Similar to the molecular interactions between Borrelia and its vertebrate hosts, interactions between Borrelia and its vectors select for molecules that enable successful uptake by feeding ticks, persistence within the tick midgut throughout the tick molt and subsequent questing, and migration to the salivary glands and transmission to a subsequent host25,27–30. For example, Outer Surface Protein A (OspA) in Borrelia must bind to the tick midgut receptor, TROSPA, in order to colonize and persist within the tick96,97. Genes involved in interactions with ticks generally have little variation within Borrelia species98, but there is substantial variation between Borrelia species99. The among-species variation observed at these genes is likely caused by differences in binding efficiency to tick proteins (i.e. TROSPA receptors) that likely differ among tick species99. Past natural selection has resulted in molecular specialization to maximize efficacy in one or a few tick species resulting in the observed differences in vector competence and vector specialization among Borrelia species100. Future evolutionary ecology research can determine how the frequency and strength of selection on other molecular interactions is impacted by vector specialization and evaluate the ecological consequences caused by the resulting micro-evolution.
Specialization to one or a few vector species impacts the geographic range and migration rates of Borrelia species101,102, primarily through the mobility and range of the vertebrate species commonly parasitized by the vector. Some vectors, like I. dentatus, prefer mobile hosts like birds103, while others, like I. spinipalpis, feed primarily on rodents with smaller geographic ranges104,105. As expected, Borrelia associated with vectors that feed on more mobile hosts have less population genetic structure due to the high rates of migration among locations31,47,101. In contrast, Borrelia species in vectors parasitizing less-mobile hosts tend to have greater genetic divergence among geographically separated populations due to both neutral evolution and natural selection favoring specialization, despite experiencing limited ecological interactions within their restricted location24,106. Differences in Borrelia population structure can stem from both direct associations with vertebrates, as discussed in the Borrelia-host section, or indirectly through associations with vectors that differently associate with different vertebrate species. Evolutionary ecology investigations can differentiate among potential host- or vector-associated mechanisms underlying these patterns.
Adapting to novel vector species exposes Borrelia populations to previously unencountered ecological interactions with both the novel vector and with the vertebrate community within the host range of the novel vector. For example, populations of Bbss that adapted to a new vector species, I. pacificus, diverged phenotypically from the ancestral populations carried by I. scapularis such that Bbss acquisition efficiency is higher in sympatric pairings of ticks and Bbss populations than allopatric pairings100. I. pacificus-associated Bbss populations also expanded geographically into the range of its new vector. While the evolutionary divergence is currently insufficient to observe geographically-based clusters when neutrally evolving loci are used to build phylogenies 107, the geographic differences in selection pressures at one or several genes have resulted in observable phenotypic differences. Adaptation to a novel vector, I. ricinus, also split two populations of B. bavariensis; one population was likely able to invade Europe by adapting to I.ricinus 18,108 (Figure 2). Through an evolutionary ecology lens, future research could consider the genetic mechanisms underlying this divergence and how the evolved changes influenced ecological interactions.
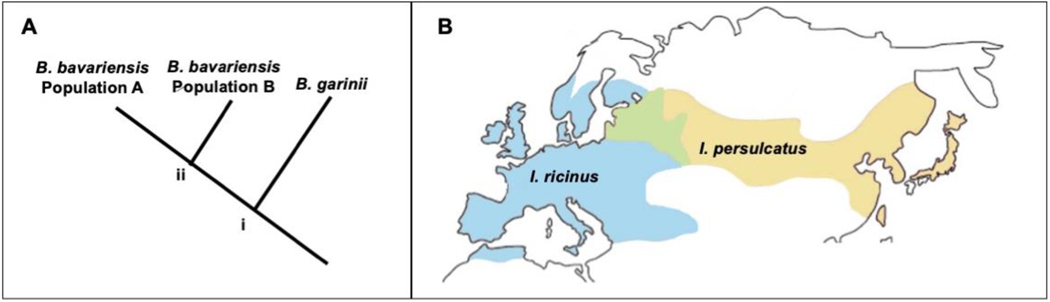
Figure 2: Diversification of Borrelia garinii and Borrelia bavariensis in Eurasia demonstrates how evolutionary changes impact the ecological interactions.
A. The sister species B. garinii and B. bavariensis diverged at node i as a result of specialization on different sets of host species with B. garinii specializing on bird species and B. bavariensis specializing on small mammals. Subsequent B. bavariensis divergence into two populations at node ii occurred due to adaptation to a novel vector, Ixodes ricinus, allowing B. bavariensis to invaded western Europe. B. The distributions of I. ricinus (blue) and I. persulcatus (yellow) overlap in eastern Europe (green) where the eastern B. bavariensis population predominates. The host and vector switches that have occurred during the evolutionary history of these Borrelia species has facilitated range expansion and increased the diversity of their ecological interactions. (Species ranges as described in Stanek et al. 2012158. Adapted with permission from the European Concerted Action on Lyme Borreliosis. Available at: http://www.eucalb.com/.)
Although Borrelia species can disperse within vectors and vertebrate hosts, the direction and rate of Borrelia migration (gene flow) and that of their primary vector are often only weakly correlated at fine geographic and temporal scales17. This discord may be explained by Borrelia dispersing predominantly within infected vertebrates. For example, migrating birds infected with Borrelia may seed local tick populations in nonendemic areas22. Borrelia that do disperse within ticks may still have different rates and direction of gene flow from the tick vector if the bacteria and vectors colonize areas at different rates. That is, while the colonization efficiency of Borrelia is expected to be positively correlated with the local density of vectors needed to transmit the bacterium, the colonization efficiency of ticks may be inversely proportional to local vector densities due to competition with resident ticks. While the mechanisms underlying competitive interactions between ticks within or between species – as well as the impact this competition may have on colonization efficiency – require further research, recent evidence suggests that ticks do compete for rodent hosts109. By considering how ecological interactions impact migration rates (micro-evolution), an evolutionary ecology framework could identify the cause of the observed differences in gene flow between Borrelia and tick.
3.2. Interactions within ticks
Borrelia interact with other microbes within tick vectors, including other Borrelia species or strains, other pathogens, and non-pathogenic components of the tick microbiome110–112. As Borrelia density is positively correlated with the probability of tick-to-host transmission113,114, the competition between Borrelia strains within individual ticks that reduces the density of each strain114–117 decreases the evolutionary fitness of Borrelia strains. Evolutionary theory predicts that inter-strain competition that negatively impacts evolutionary fitness should select for traits that increase growth rates within ticks or for molecules that suppress competing strains118–121. Although Borrelia is often found in multi-strain infections in ticks that reduce evolutionary fitness by reducing transmission probabilities114, there is no evidence that traits enhancing competitive ability have evolved. This may indicate that there are other important ecological interactions that are imposing selective pressures on Borrelia populations. Future evolutionary ecology research can experimentally or statistically investigate the selective pressures created by competitive interactions between coinfecting Borrelia strains, as well as other ecological interactions, to determine their relative evolutionary impact.
Borrelia can also interact with other human pathogens vectored by ticks such as Babesia microti, the protozoan pathogen that causes human babesiosis111. The number of ticks coinfected with B. microti and Bbss is higher than expected by random chance alone111,122 suggesting that Bbss may be facilitating infection with Babesia. This facilitation may be responsible for the increased prevalence and range expansion of Babesia in the northeastern United States111,123. While facilitation of Babesia may occur as a byproduct of natural selection Bbss otherwise experiences, future evolutionary ecology research can determine the traits of Bbss that are key to this ecological interaction and their impacts on the evolutionary trajectories of both species.
Experimentally detecting and quantifying biological interactions between pathogens is a long-standing challenge124–126. Longitudinal sampling, the current gold standard for inferring ecological interactions127,128, requires resource-intensive efforts over many years. Collecting cross-sectional data is less burdensome and can be used to identify deviations from the expected probability of coinfection given the prevalence of each pathogen129–131. A departure from the random expectation may indicate interactions between pathogens but may also result from environmental heterogeneity and spatial or ecological clustering132. Further, the assumption that the prevalence of noninteracting pathogens should be statistically independent has been challenged124. It is critical that ecological interactions are correctly inferred to consider them as selective forces within an evolutionary ecology framework. Appropriate analytical methods, such as those developed by Alizon et al 2019, can accurately infer species interactions from cross-sectional data while accounting for environmental heterogeneity but, to date, are rarely used132.
Borrelia species encounter limited diversity and densities of bacteria within Ixodid ticks overall such that selective pressures from interactions with the tick microbiome as a whole should be minimal. Although early studies found diverse microbiomes in multiple Ixodes species and life stages133–135, more recent studies that controlled for bacterial biomass found that the internal microbiome diversity is actually quite low110,136. Consistent with limited microbiome diversity and density, Borrelia has lost genes that encode known interbacterial interaction pathways110 that have not been under selective pressure by resident microbes137. As Borrelia geographic ranges expand, so too does the diversity of interactions Borrelia experiences, as the composition of tick microbiomes varies geographically134. Further, there is growing evidence that the ranges of other tick-associated microbes like Babesia and Anaplasma are expanding into areas already inhabited by Borrelia species138,139. The evolutionary loss of genes encoding inter-microbial competition pathways may make Borrelia particularly susceptible to inhibition or exclusion by competing tick-associated microbes. Evolutionary ecology provides the foundation to determine if the evolutionarily reduced Borrelia genome constrains the ability of Borrelia to outcompete novel microbial competitors.
Borrelia populations are also shaped by the abiotic factors that affect the populations of their non-homeothermic vector (e.g. temperature, climate, landscape connectivity). Survival within unfed or intermolt ticks poses significant challenges owing to temperature extremes caused by daily and seasonal fluctuations as well as nutritional stress140. Environmental stress can create directional selection pressures for tolerance to environmental variation141. In fact, experimental evidence suggests that fluctuating environments select for bacteria that can tolerate a range of conditions142,143. Although no genes have been identified in Borrelia to facilitate tolerance of extreme cold during overwintering or extreme heat during summer, the regulation of many genes is temperature-sensitive144–146. For example, a large fraction of novel Borrelia sRNAs (43%) are temperature-dependent both in function and expression levels147. Future evolutionary ecology research into how Borrelia has evolved to withstand year-round fluctuations in temperature is critical to determine how Borrelia persists in ticks and to predict how Borrelia distributions may shift with climate change.
3. Future Directions
Evolutionary ecology provides a particularly powerful framework to address outstanding questions on Borrelia species. This framework builds upon a strong research foundation in the biology of Borrelia constructed from the exceptional progress in molecular mechanistic, ecological, and evolutionary biological studies. Molecular studies have identified factors associated with host and vector specialization, diversification rates and processes, and mechanisms of immune escape which reveal ecological interactions with hosts or vectors that impact the evolution of these microbial species. Ecological investigations have characterized how environmental conditions, host communities, and host/vector associations directly influence the frequency and type of molecular interactions each Borrelia species experiences. Evolutionary ecology offers a framework by which we can evaluate the ecological consequences of evolved traits in Borrelia and predict how present-day ecological processes may result in further molecular evolutionary change.
Investigations into some outstanding questions about the biology of Borrelia within an evolutionary ecology framework may necessitate collecting new empirical data while others can be addressed by leveraging existing data. For example, more empirical data will be needed to characterize the frequency, strength, and type of interactions between Borrelia and pathogens coinfecting ticks or hosts and to determine the selective forces generated by those interactions. These data are necessary to determine how competitive interactions between coinfecting pathogens create selective forces that cause micro-evolutionary changes within Borrelia species. Similarly, how Borrelia has evolved to withstand year-round climatic fluctuations necessitates additional empirical data to predict how interactions with the environment under climate change scenarios will impact Borrelia transmission and evolution. In many cases, however, existing data from different disciplines can be integrated to address how ecological interactions – especially those between Borrelia and hosts and those between Borrelia and vectors – create evolutionary pressures that select for a diverse set of traits. For example, integrating ecological and molecular data can identify if and how ecological interactions drive population genetic structure in natural Borrelia populations.
The massive influx of Borrelia, tick, and vertebrate genomic informatione.g.,17,148–150 can be used to discern past evolutionary processes across multiple loci that can be used to generate tractable evolutionary ecology hypotheses which can subsequently be experimentally validated. If species under different selective regimes consistently evolve distinct trait combinations, comparative genomics can be used to draw conclusions about the evolutionary impact of ecological interactions. Comparative genomic approaches have already been used in an evolutionary ecology framework to determine the genetic underpinnings of host specialization in some infectious microbes151.
Understanding the evolutionary ecology of Borrelia has practical implications for predicting expansions in geographic, host, or vector ranges and the disease risk associated with these expansions. For example, predicting potential host species by their ecological and evolutionary similarities to known hosts can identify geographic areas in which populations are likely to establish152. Evolutionary ecology approaches are invaluable for predicting novel hosts of emerging diseases, especially those caused by pathogens that circulate among multiple hosts like many Borrelia species. By overcoming the assumption that ecology and evolution operate on distinct timescales, evolutionary ecology provides broad insight into factors regulating population and community dynamics, processes critical to understanding disease dynamics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References Cited
- 1.Jones CG, Lawton JH & Shachak M. Organisms as Ecosystem Engineers. Oikos 69, 373–386 (1994). [Google Scholar]
- 2.Taylor LH, Latham SM & Woolhouse ME Risk factors for human disease emergence. Philos. Trans. R. Soc. Lond. B. Biol. Sci 356, 983–989 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lloyd-Smith JO et al. Epidemic Dynamics at the Human-Animal Interface. Science. 326, 1362–1367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higgs S, Vanlandingham DL, Huang Y-JS & Vanlandingham DL Arbovirus-Mosquito Vector-Host Interactions and the Impact on Transmission and Disease Pathogenesis of Arboviruses. Frontiers in microbiology. 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parker JL & White KK Lyme borreliosis in cattle and horses: a review of the literature. Cornell Vet. 82, 253–274 (1992). [PubMed] [Google Scholar]
- 6.Burgdorfer W et al. Lyme disease-a tick-borne spirochetosis? Science (80-. ). 216, 1317 LP – 1319 (1982). [DOI] [PubMed] [Google Scholar]
- 7.Pritt BS et al. Identification of a novel pathogenic Borrelia species causing Lyme borreliosis with unusually high spirochaetaemia: a descriptive study. Lancet Infect. Dis 16, 556–564 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gofton AW et al. Genome-wide analysis of Borrelia turcica and ‘Candidatus Borrelia tachyglossi’shows relapsing fever-like genomes with unique genomic links to Lyme disease Borrelia. Infect. Genet. Evol 66, 72–81 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Loh S-M et al. Novel Borrelia species detected in echidna ticks, Bothriocroton concolor, in Australia. Parasit. Vectors 9, 339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takano A et al. Isolation and characterization of a novel Borrelia group of tick-borne borreliae from imported reptiles and their associated ticks. Environ. Microbiol 12, 134–146 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Graves CJ, Ros VID, Stevenson B, Sniegowski PD & Brisson D. Natural Selection Promotes Antigenic Evolvability. PLOS Pathog. 9, e1003766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dykhuizen DE et al. The propensity of different Borrelia burgdorferi sensu stricto genotypes to cause disseminated infections in humans. Am. J. Trop. Med. Hyg 78, 806–810 (2008). [PMC free article] [PubMed] [Google Scholar]
- 13.De Michelis S et al. Genetic Diversity of Borrelia burgdorferi Sensu Lato in Ticks from Mainland Portugal. J. Clin. Microbiol 38, 2128 LP – 2133 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coipan CE, van Duijvendijk GLA, Hofmeester TR, Takumi K & Sprong H. The genetic diversity of Borrelia afzelii is not maintained by the diversity of the rodent hosts. Parasit. Vectors 11, 454 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tufts DM et al. Outer surface protein polymorphisms linked to host spirochete association in Lyme borreliae. Mol. Microbiol 111, 868–882 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanincova K et al. Multilocus sequence typing of Borrelia burgdorferi suggests existence of lineages with differential pathogenic properties in humans. PLoS One 8, (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter KS, Carpi G, Caccone A & Diuk-wasser MA Genomic insights into the ancient spread of Lyme disease across North America. Nat. Ecol. Evol 1, 1569–1576 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker NS et al. Recurrent evolution of host and vector association in bacteria of the Borrelia burgdorferi sensu lato species complex. BMC Genomics 17, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Estrada-Peña A, Álvarez-Jarreta J & Cabezas-Cruz A. Reservoir and vector evolutionary pressures shaped the adaptation of Borrelia. Infect. Genet. Evol 66, 308–318 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Råberg L et al. Evolution of antigenic diversity in the tick transmitted bacterium Borrelia afzelii: a role for host specialization? J. Evol. Biol 30, 1034–1041 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Hanincová K et al. Association of Borrelia afzelii with rodents in Europe. Parasitology 126, 11–20 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Ogden NH et al. Active and passive surveillance and phylogenetic analysis of Borrelia burgdorferi elucidate the process of Lyme disease risk emergence in Canada. Environ. Health Perspect 118, 909–914 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhacheva TA & Kovalev SY Borrelia spirochetes in Russia: Genospecies differentiation by real-time PCR. Ticks and tick-borne diseases. 5, 722–726 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Hoen AG et al. Phylogeography of Borrelia burgdorferi in the eastern United States reflects multiple independent Lyme disease emergence events. Proc. Natl. Acad. Sci 106, 15013–15018 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kenedy MR, Lenhart TR & Akins DR The role of Borrelia burgdorferi outer surface proteins. FEMS Immunol. Med. Microbiol 66, 1–19 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ogden NH et al. Evolutionary Aspects of Emerging Lyme Disease in Canada. Appl. Environ. Microbiol 81, 7350–7359 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neelakanta G et al. Outer surface protein B is critical for Borrelia burgdorferi adherence and survival within Ixodes ticks. PLoS Pathog. 3, (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pal U et al. Inhibition of Borrelia burgdorferi-tick interactions in vivo by outer surface protein A antibody. J. Immunol 166, 7398–7403 (2001). [DOI] [PubMed] [Google Scholar]
- 29.de Silva AM, Tyson KR & Pal U. Molecular characterization of the tick-Borrelia interface. Front. Biosci 14, 3051–3063 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fingerle V, Goettner G, Gern L, Wilske B & Schulte-Spechtel U. Complementation of a Borrelia afzelii OspC mutant highlights the crucial role of OspC for dissemination of Borrelia afzelii in Ixodes ricinus. Int. J. Med. Microbiol 297, 97–107 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Vollmer SA et al. Host migration impacts on the phylogeography of Lyme Borreliosis spirochaete species in Europe. Environ. Microbiol 13, 184–192 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Vollmer SA et al. Spatial spread and demographic expansion of Lyme borreliosis spirochaetes in Eurasia. Infect. Genet. Evol 14, 147–155 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Vuong HB et al. Influences of Host Community Characteristics on Borrelia burgdorferi Infection Prevalence in Blacklegged Ticks. PLoS One 12, e0167810–e0167810 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paul REL, Cote M, Le Naour E & Bonnet SI Environmental factors influencing tick densities over seven years in a French suburban forest. Parasit. Vectors 9, 309 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kilpatrick AM et al. Lyme disease ecology in a changing world : consensus, uncertainty and critical gaps for improving control. in Philosophical Transactions of the Royal Society B: Biological Sciences 372, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turcotte MM, Reznick DN & Hare JD The impact of rapid evolution on population dynamics in the wild: experimental test of eco evolutionary dynamics. Ecol. Lett 14, 1084–1092 (2011). [DOI] [PubMed] [Google Scholar]
- 37.Barraclough TG How do species interactions affect evolutionary dynamics across whole communities? Annu. Rev. Ecol. Evol. Syst 46, 25–48 (2015). [Google Scholar]
- 38.Gilbert GS & Webb CO Phylogenetic signal in plant pathogen-host range. Proc. Natl. Acad. Sci. U. S. A 104, 4979–83 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parker IM & Gilbert GS The Evolutionary Ecology of Novel Plant-Pathogen Interactions. Annu. Rev. Ecol. Evol. Syst 35, 675–700 (2004). [Google Scholar]
- 40.Jarosz AM & Davelos AL Effects of disease in wild plant populations and the evolution of pathogen aggressiveness. New Phytol. 129, 371–387 (1995). [Google Scholar]
- 41.Alexander HM, Thrall PH, Antonovics J, Jarosz AM & Oudemans PV Population dynamics and genetics of plant disease: a case study of anther smut disease. Ecology 77, 990–996 (1996). [Google Scholar]
- 42.Gilbert GS & Parker IM The Evolutionary Ecology of Plant Disease: A Phylogenetic Perspective. Annu. Rev. Phytopathol 54, 549–578 (2016). [DOI] [PubMed] [Google Scholar]
- 43.Haven J et al. Pervasive recombination and sympatric genome diversification driven by frequency-dependent selection in Borrelia burgdorferi, the Lyme disease bacterium. Genetics 189, 951–966 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tsao JI Reviewing molecular adaptations of lyme borreliosis spirochetes in the context of reproductive fitness in natural transmission cycles. Vet. Res 40, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seifert SN, Khatchikian CE, Zhou W & Brisson D. Evolution and population genomics of the Lyme borreliosis pathogen, Borrelia burgdorferi. Trends Genet. 31, 201–207 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khatchikian CE et al. Recent and rapid population growth and range expansion of the Lyme disease tick vector, Ixodes scapularis, in North America. Evolution (N. Y). 69, 1678–1689 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Norte AC et al. Host dispersal shapes the population structure of a tick-borne bacterial pathogen. Mol. Ecol 29, 485–501 (2020). [DOI] [PubMed] [Google Scholar]
- 48.Comstedt P, Jakobsson T & Bergström S. Global ecology and epidemiology of Borrelia garinii spirochetes. Infect. Ecol. Epidemiol 1, 10.3402/iee.v1i0.9545 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Munro HJ et al. Genetic diversity of Borrelia garinii from Ixodes uriae collected in seabird colonies of the northwestern Atlantic Ocean. Ticks Tick. Borne. Dis 10, 101255 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Brisson D, Drecktrah D, Eggers CH & Samuels DS Genetics of Borrelia burgdorferi. Annu Rev Genet. 46, 181–204 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roberts ED et al. Pathogenesis of Lyme neuroborreliosis in the rhesus monkey: the early disseminated and chronic phases of disease in the peripheral nervous system. J. Infect. Dis 178, 722–732 (1998). [DOI] [PubMed] [Google Scholar]
- 52.Hart T, Yang X, Pal U & Lin Y-P Identification of Lyme borreliae proteins promoting vertebrate host blood-specific spirochete survival in Ixodes scapularis nymphs using artificial feeding chambers. Ticks Tick. Borne. Dis 9, 1057–1063 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lin Y-P, Frye AM, Nowak TA & Kraiczy P. New Insights Into CRASP-Mediated Complement Evasion in the Lyme Disease Enzootic Cycle. Front. Cell. Infect. Microbiol 10, 1 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhide MR et al. Complement factor H binding by different Lyme disease and relapsing fever Borrelia in animals and human. BMC Res. Notes 2, 134 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kurtenbach K et al. Host association of Borrelia burgdorferi sensu lato--the key role of host complement. Trends Microbiol. 10, 74–79 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Wywial E et al. Fast, adaptive evolution at a bacterial host-resistance locus: the PFam54 gene array in Borrelia burgdorferi. Gene 445, 26–37 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hammerschmidt C et al. Versatile roles of CspA orthologs in complement inactivation of serum-resistant Lyme disease spirochetes. Infect. Immun 82, 380–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin Y-P, Diuk-Wasser MA, Stevenson B & Kraiczy P. Complement Evasion Contributes to Lyme Borreliae-Host Associations. Trends Parasitol. 36, 634–645 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mechai S et al. Evidence for host-genotype associations of Borrelia burgdorferi sensu stricto. PLoS One 11, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Brisson D & Dykhuizen DE ospC diversity in Borrelia burgdorferi: different hosts are different niches. Genetics 168, 713–722 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bäumler A & Fang FC Host specificity of bacterial pathogens. Cold Spring Harb. Perspect. Med 3, a010041 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ripoche J, Day AJ, Harris TJR & Sim RB The complete amino acid sequence of human complement factor H. Biochem. J 249, 593–602 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anderson JF, Barthold SW & Magnarelli LA Infectious but nonpathogenic isolate of Borrelia burgdorferi. J. Clin. Microbiol 28, 2693–2699 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Barthold SW, Persing DH, Armstrong AL & Peeples RA Kinetics of Borrelia burgdorferi dissemination and evolution of disease after intradermal inoculation of mice. Am. J. Pathol 139, 263 (1991). [PMC free article] [PubMed] [Google Scholar]
- 65.Norris SJ, Howell JK, Garza SA, Ferdows MS & Barbour AG High-and low-infectivity phenotypes of clonal populations of in vitro-cultured Borrelia burgdorferi. Infect. Immun 63, 2206–2212 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang G et al. Disease severity in a murine model of Lyme borreliosis is associated with the genotype of the infecting Borrelia burgdorferi sensu stricto strain. J. Infect. Dis 186, 782–791 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barbour AG et al. Niche partitioning of Borrelia burgdorferi and Borrelia miyamotoi in the same tick vector and mammalian reservoir species. Am. J. Trop. Med. Hyg 81, 1120–1131 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Baum E, Hue F & Barbour AG Experimental infections of the reservoir species Peromyscus leucopus with diverse strains of Borrelia burgdorferi, a Lyme disease agent. MBio 3, e00434–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chan K, Awan M, Barthold SW & Parveen N. Comparative molecular analyses of Borrelia burgdorferi sensu stricto strains B31 and N40D10/E9 and determination of their pathogenicity. BMC Microbiol. 12, 157 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang G et al. Impact of genotypic variation of Borrelia burgdorferi sensu stricto on kinetics of dissemination and severity of disease in C3H/HeJ mice. Infect. Immun 69, 4303–4312 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Margos G et al. A new Borrelia species defined by multilocus sequence analysis of housekeeping genes. Appl. Environ. Microbiol 75, 5410–5416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bunikis J et al. Sequence typing reveals extensive strain diversity of the Lyme borreliosis agents Borrelia burgdorferi in North America and Borrelia afzelii in Europe. Microbiology 150, 1741–1755 (2004). [DOI] [PubMed] [Google Scholar]
- 73.Dykhuizen DE & Baranton G. The implications of a low rate of horizontal transfer in Borrelia. Trends Microbiol. 9, 344–350 (2001). [DOI] [PubMed] [Google Scholar]
- 74.Jacquot M et al. Comparative population genomics of the Borrelia burgdorferi species complex reveals high degree of genetic isolation among species and underscores benefits and constraints to studying intra-specific epidemiological processes. PLoS One 9, e94384 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hanincová K, Kurtenbach K, Diuk-Wasser M, Brei B & Fish D. Epidemic spread of Lyme borreliosis, northeastern United States. Emerg. Infect. Dis 12, 604 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kurtenbach K et al. Fundamental processes in the evolutionary ecology of Lyme borreliosis. Nat. Rev. Microbiol 4, 660–669 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Önder Ö et al. OspC is potent plasminogen receptor on surface of Borrelia burgdorferi. J. Biol. Chem 287, 16860–16868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lagal V, Portnoï D, Faure G, Postic D & Baranton G. Borrelia burgdorferi sensu stricto invasiveness is correlated with OspC–plasminogen affinity. Microbes Infect. 8, 645–652 (2006). [DOI] [PubMed] [Google Scholar]
- 79.Marconi RT, Samuels DS & Garon CF Transcriptional analyses and mapping of the ospC gene in Lyme disease spirochetes. J. Bacteriol 175, 926–932 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Livey I, Gibbs CP, Schuster R & Dorner F. Evidence for lateral transfer and recombination in OspC variation in Lyme disease Borrelia. Mol. Microbiol 18, 257–269 (1995). [DOI] [PubMed] [Google Scholar]
- 81.Casjens SR et al. Primordial origin and diversification of plasmids in Lyme disease agent bacteria. BMC Genomics 19, 218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tilly K et al. Borrelia burgdorferi OspC protein required exclusively in a crucial early stage of mammalian infection. Infect. Immun 74, 3554–3564 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang J-R & Norris SJ Kinetics and in vivo induction of genetic variation of vlsE in Borrelia burgdorferi. Infect. Immun 66, 3689–3697 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Coutte L, Botkin DJ, Gao L & Norris SJ Detailed analysis of sequence changes occurring during vlsE antigenic variation in the mouse model of Borrelia burgdorferi infection. PLoS Pathog. 5, (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Norris SJ vls antigenic variation systems of Lyme disease Borrelia: eluding host immunity through both random, segmental gene conversion and framework heterogeneity. Mob. DNA III 471–489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gomez-Chamorro A et al. Maternal Antibodies Provide Bank Voles with Strain-Specific Protection against Infection by the Lyme Disease Pathogen. Appl. Environ. Microbiol 85, e01887–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Durand J, Jacquet M, Rais O, Gern L & Voordouw MJ Fitness estimates from experimental infections predict the long-term strain structure of a vector-borne pathogen in the field. Sci. Rep 7, 1851 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gupta S & Anderson RM Population structure of pathogens: the role of immune selection. Parasitol. Today 15, 497–501 (1999). [DOI] [PubMed] [Google Scholar]
- 89.Gupta S, Ferguson N & Anderson R. Chaos, persistence, and evolution of strain structure in antigenically diverse infectious agents. Science (80-. ). 280, 912–915 (1998). [DOI] [PubMed] [Google Scholar]
- 90.Gupta S et al. The maintenance of strain structure in populations of recombining infectious agents. Nat. Med 2, 437–442 (1996). [DOI] [PubMed] [Google Scholar]
- 91.Vitorino LR et al. Fine-scale phylogeographic structure of Borrelia lusitaniae revealed by multilocus sequence typing. PLoS One 3, (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gómez-Díaz E, Jordà M, Peinado MA & Rivero A. Epigenetics of host-pathogen interactions: the road ahead and the road behind. PLoS Pathog. 8, e1003007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Cobey S & Lipsitch M. Niche and neutral effects of acquired immunity permit coexistence of pneumococcal serotypes. Science (80-. ). 335, 1376–1380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brisson D. Negative Frequency-Dependent Selection Is Frequently Confounding. Front. Ecol. Evol 6, 1–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kilpatrick AM et al. Lyme disease ecology in a changing world: consensus, uncertainty and critical gaps for improving control. Philos. Trans. R. Soc. Lond. B. Biol. Sci 372, 20160117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Battisti JM et al. Outer surface protein A protects Lyme disease spirochetes from acquired host immunity in the tick vector. Infect. Immun 76, 5228–5237 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pal U et al. TROSPA, an Ixodes scapularis receptor for Borrelia burgdorferi. Cell 119, 457–468 (2004). [DOI] [PubMed] [Google Scholar]
- 98.Wilske B et al. Recombinant immunoblot in the serodiagnosis of Lyme borreliosis. Med. Microbiol. Immunol 182, 255–270 (1993). [DOI] [PubMed] [Google Scholar]
- 99.Konnai S et al. Identification of TROSPA homologue in Ixodes persulcatus Schulze, the specific vector for human Lyme borreliosis in Japan. Ticks Tick. Borne. Dis 3, 75–77 (2012). [DOI] [PubMed] [Google Scholar]
- 100.Couper LI, Yang Y, Yang XF & Swei A. Comparative vector competence of North American Lyme disease vectors. Parasit. Vectors 13, 29 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Margos G, Vollmer SA, Ogden NH & Fish D. Population genetics, taxonomy, phylogeny and evolution of Borrelia burgdorferi sensu lato. Infect. Genet. Evol 11, 1545–1563 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Margos G et al. Core genome phylogenetic analysis of the avian associated Borrelia turdi indicates a close relationship to Borrelia garinii. Mol. Phylogenet. Evol 131, 93–98 (2019). [DOI] [PubMed] [Google Scholar]
- 103.Sonenshine DE Ticks of Virginia. Virginia Polytech. Inst. State Univ. Coll. Agric. Life Sci. Blacksburg, VA 42 (1979). [Google Scholar]
- 104.Maupin GO et al. Discovery of an enzootic cycle of Borrelia burgdorferi in Neotoma mexicana and Ixodes spinipalpis from northern Colorado, an area where Lyme disease is nonendemic. J. Infect. Dis 170, 636–643 (1994). [DOI] [PubMed] [Google Scholar]
- 105.Burkot TR et al. Babesia microti and Borrelia bissettii transmission by Ixodes spinipalpis ticks among prairie voles, Microtus ochrogaster, in Colorado. Parasitology 121, 595–599 (2000). [PubMed] [Google Scholar]
- 106.Humphrey PT, Caporale DA & Brisson D. Uncoordinated phylogeography of Borrelia burgdorferi and its tick vector, Ixodes scapularis. Evolution (N. Y). 64, 2653–2663 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tyler S et al. Whole genome sequencing and phylogenetic analysis of strains of the agent of Lyme disease Borrelia burgdorferi from Canadian emergence zones. Sci. Rep 1–12 (2018). doi: 10.1038/s41598-018-28908-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gatzmann F et al. NGS population genetics analyses reveal divergent evolution of a Lyme Borreliosis agent in Europe and Asia. Ticks Tick. Borne. Dis 6, 344–351 (2015). [DOI] [PubMed] [Google Scholar]
- 109.Karbowiak G et al. The Competition Between Immatures of Ixodes ricinus and Dermacentor reticulatus (Ixodida: Ixodidae) Ticks for Rodent Hosts. J. Med. Entomol 56, 448–452 (2019). [DOI] [PubMed] [Google Scholar]
- 110.Ross BD et al. Ixodes scapularis does not harbor a stable midgut microbiome. ISME J. 12, 2596–2607 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Diuk-Wasser MA, Vannier E & Krause PJ Coinfection by Ixodes Tick-Borne Pathogens: Ecological, Epidemiological, and Clinical Consequences. Trends Parasitol. 32, 30–42 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Moutailler S et al. Co-infection of ticks: the rule rather than the exception. PLoS Negl. Trop. Dis 10, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rego ROM, Bestor A, Štefka J & Rosa PA Population bottlenecks during the infectious cycle of the Lyme disease spirochete Borrelia burgdorferi. PLoS One 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Durand J et al. Multistrain infections with Lyme borreliosis pathogens in the tick vector. Appl. Environ. Microbiol 83, e02552–16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Genne D et al. Competition between strains of Borrelia afzelii inside the rodent host and the tick vector. Proc. R. Soc. B Biol. Sci 285, 17–20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Genné D, Sarr A, Rais O & Voordouw MJ Competition Between Strains of Borrelia afzelii in Immature Ixodes ricinus Ticks Is Not Affected by Season. Front. Cell. Infect. Microbiol 9, 431 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Walter KS, Carpi G, Evans BR, Caccone A & Diuk-Wasser MA Vectors as epidemiological sentinels: patterns of within-tick Borrelia burgdorferi diversity. PLoS Pathog. 12, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Alizon S, de Roode JC & Michalakis Y. Multiple infections and the evolution of virulence. Ecol. Lett 16, 556–567 (2013). [DOI] [PubMed] [Google Scholar]
- 119.Cattadori IM, Boag B & Hudson PJ Parasite co-infection and interaction as drivers of host heterogeneity. Int. J. Parasitol 38, 371–380 (2008). [DOI] [PubMed] [Google Scholar]
- 120.Susi H, Barrès B, Vale PF & Laine A-L Co-infection alters population dynamics of infectious disease. Nat. Commun 6, 5975 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Telfer S et al. Species Interactions in a Parasite Community Drive Infection Risk in a Wildlife Population. Science 330, 243–246 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hersh MH et al. Co-infection of blacklegged ticks with Babesia microti and Borrelia burgdorferi is higher than expected and acquired from small mammal hosts. PLoS One 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Dunn JM et al. Borrelia burgdorferi promotes the establishment of Babesia microti in the northeastern United States. PLoS One 9, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hamelin FM et al. Coinfections by noninteracting pathogens are not independent and require new tests of interaction. PLoS Biol. 17, 1–25 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Johnson PTJ & Buller ID Parasite competition hidden by correlated coinfect: using surveys and experiments to understand parasite interactions. Ecology 92, 535–541 (2011). [DOI] [PubMed] [Google Scholar]
- 126.Hellard E, Fouchet D, Vavre F & Pontier D. Parasite-Parasite Interactions in the Wild: How To Detect Them? Trends Parasitol. 31, 640–652 (2015). [DOI] [PubMed] [Google Scholar]
- 127.Gerber GK The dynamic microbiome. FEBS Lett. 588, 4131–4139 (2014). [DOI] [PubMed] [Google Scholar]
- 128.Fenton A, Knowles SCL, Petchey OL & Pedersen AB The reliability of observational approaches for detecting interspecific parasite interactions: comparison with experimental results. Int. J. Parasitol 44, 437–445 (2014). [DOI] [PubMed] [Google Scholar]
- 129.Degarege A, Legesse M, Medhin G, Animut A & Erko B. Malaria and related outcomes in patients with intestinal helminths: a cross-sectional study. BMC Infect. Dis 12, 291 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Traub RJ et al. The prevalence and distribution of gastrointestinal parasites of stray and refuge dogs in four locations in India. Vet. Parasitol 205, 233–238 (2014). [DOI] [PubMed] [Google Scholar]
- 131.Gelaw A et al. Prevalence of intestinal parasitic infections and risk factors among schoolchildren at the University of Gondar Community School, Northwest Ethiopia: a cross-sectional study. BMC Public Health 13, 304 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Alizon S, Murall CL, Saulnier E & Sofonea MT Detecting within-host interactions from genotype combination prevalence data. Epidemics 29, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Abraham NM et al. Pathogen-mediated manipulation of arthropod microbiota to promote infection. Proc. Natl. Acad. Sci. U. S. A 114, E781–E790 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.van Treuren W et al. Variation in the microbiota of Ixodes ticks with regard to geography, species, and sex. Appl. Environ. Microbiol 81, 6200–6209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zolnik CP, Prill RJ, Falco RC, Daniels TJ & Kolokotronis S-O Microbiome changes through ontogeny of a tick pathogen vector. Mol. Ecol 25, 4963–4977 (2016). [DOI] [PubMed] [Google Scholar]
- 136.Couper LI, Kwan JY, Ma J & Swei A. Drivers and patterns of microbial community assembly in a Lyme disease vector. Ecol. Evol 7768–7779 (2019). doi: 10.1002/ece3.5361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zhang D, de Souza RF, Anantharaman V, Iyer LM & Aravind L. Polymorphic toxin systems: Comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct 7, 18 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ogden NH et al. Role of Migratory Birds in Introduction and Range Expansion of Ixodes scapularis Ticks and of Borrelia burgdorferi and Anaplasma phagocytophilum in Canada. Appl. Environ. Microbiol 74, 1780 LP – 1790 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Walter KS et al. Invasion of two tick-borne diseases across New England: harnessing human surveillance data to capture underlying ecological invasion processes. Proc. R. Soc. B Biol. Sci 283, 20160834 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kung F, Anguita J & Pal U. Borrelia burgdorferi and tick proteins supporting pathogen persistence in the vector. Future Microbiol. 8, 41–56 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Chevin L-M & Hoffmann AA Evolution of phenotypic plasticity in extreme environments. Philos. Trans. R. Soc. B Biol. Sci 372, 20160138 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Condon C, Cooper BS, Yeaman S & Angilletta MJ Jr Temporal variation favors the evolution of generalists in experimental populations of Drosophila melanogaster. Evolution (N. Y). 68, 720–728 (2014). [DOI] [PubMed] [Google Scholar]
- 143.Saarinen K, Laakso J, Lindström L & Ketola T. Adaptation to fluctuations in temperature by nine species of bacteria. Ecol. Evol 8, 2901–2910 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ojaimi C et al. Profiling of temperature-induced changes in Borrelia burgdorferi gene expression by using whole genome arrays. Infect. Immun 71, 1689–1705 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tokarz R, Anderton JM, Katona LI & Benach JL Combined effects of blood and temperature shift on Borrelia burgdorferi gene expression as determined by whole genome DNA array. Infect. Immun 72, 5419–5432 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Phelan JP et al. Genome-wide screen identifies novel genes required for Borrelia burgdorferi survival in its Ixodes tick vector. PLoS Pathog. 15, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Popitsch N, Bilusic I, Rescheneder P, Schroeder R & Lybecker M. Temperature-dependent sRNA transcriptome of the Lyme disease spirochete. BMC Genomics 18, 28 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mongodin EF et al. Inter- and intra-specific pan-genomes of Borrelia burgdorferi sensu lato: genome stability and adaptive radiation. BMC Genomics 14, 693 (2013).24112474 [Google Scholar]
- 149.Gulia-Nuss M et al. Genomic insights into the Ixodes scapularis tick vector of Lyme disease. Nat. Commun 7, 10507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kingry LC et al. Whole Genome Sequence and Comparative Genomics of the Novel Lyme Borreliosis Causing Pathogen, Borrelia mayonii. PLoS One 11, e0168994–e0168994 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Mourkas E et al. Agricultural intensification and the evolution of host specialism in the enteric pathogen Campylobacter jejuni Proc. Natl. Acad. Sci 117, 11018 LP – 11028 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Becker DJ & Han BA The macroecology and evolution of avian competence for Borrelia burgdorferi bioRxiv 2020.04.15.040352 (2020). doi: 10.1101/2020.04.15.040352 [DOI] [Google Scholar]
- 153.Levene H. Genetic equilibrium when more than one ecological niche is available. Am. Nat 87, 331–333 (1953). [Google Scholar]
- 154.Ravigné V, Olivieri I & Dieckmann U. Implications of habitat choice for protected polymorphisms. Evol. Ecol. Res 6, 125–145 (2004). [Google Scholar]
- 155.Devevey G, Dang T, Graves CJ, Murray S & Brisson D. First arrived takes all: Inhibitory priority effects dominate competition between co-infecting Borrelia burgdorferi strains Ecological and evolutionary microbiology. BMC Microbiol. 15, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Brisson D, Vandermause MF, Meece JK, Reed KD & Dykhuizen DE Evolution of northeastern and midwestern Borrelia burgdorferi, United States. Emerg. Infect. Dis 16, 911–917 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.MacDonald H, Akçay E & Brisson D. The role of host phenology for parasite transmission. bioRxiv 855031 (2019). doi: 10.1101/855031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stanek G, Wormser GP, Gray J & Strle F. Lyme borreliosis. Lancet 379, 461–473 (2012). [DOI] [PubMed] [Google Scholar]