Abstract
Introduction
We performed a multicentre evaluation of the Elecsys® Anti-SARS-CoV-2 immunoassay (Roche Diagnostics), an assay utilising a recombinant protein representing the nucleocapsid (N) antigen, for the in vitro qualitative detection of antibodies to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
Methods
Specificity was evaluated using serum/plasma samples from blood donors and routine diagnostic specimens collected before September 2019 (i.e., presumed negative for SARS-CoV-2-specific antibodies); sensitivity was evaluated using samples from patients with polymerase chain reaction (PCR)-confirmed SARS-CoV-2 infection. Point estimates and 95% confidence intervals (CIs) were calculated. Method comparison was performed versus commercially available assays.
Results
Overall specificity for the Elecsys Anti-SARS-CoV-2 immunoassay (n = 9575) was 99.85% (95% CI 99.75–99.92): blood donors (n = 6714; 99.82%), routine diagnostic specimens (n = 2861; 99.93%), pregnant women (n = 2256; 99.91%), paediatric samples (n = 205; 100.00%). The Elecsys Anti-SARS-CoV-2 immunoassay demonstrated significantly higher specificity versus LIAISON SARS-CoV-2 S1/S2 IgG (99.71% vs. 98.48%), EUROIMMUN Anti-SARS-CoV-2 IgG (100.00% vs. 94.87%), ADVIA Centaur SARS-CoV-2 Total (100.00% vs. 87.32%) and iFlash SARS-CoV-2 IgM (100.00% vs. 99.58%) assays, and comparable specificity to ARCHITECT SARS-CoV-2 IgG (99.75% vs. 99.65%) and iFlash SARS-CoV-2 IgG (100.00% vs. 100.00%) assays. Overall sensitivity for Elecsys Anti-SARS-CoV-2 immunoassay samples drawn at least 14 days post-PCR confirmation (n = 219) was 93.61% (95% CI 89.51–96.46). No statistically significant differences in sensitivity were observed between the Elecsys Anti-SARS-CoV-2 immunoassay versus EUROIMMUN Anti-SARS-CoV-2 IgG (90.32% vs. 95.16%) and ARCHITECT SARS-CoV-2 IgG (84.81% vs. 87.34%) assays. The Elecsys Anti-SARS-CoV-2 immunoassay showed significantly lower sensitivity versus ADVIA Centaur SARS-CoV-2 Total (85.19% vs. 95.06%) and iFlash SARS-CoV-2 IgG (86.25% vs. 93.75%) assays, but significantly higher sensitivity versus the iFlash SARS-CoV-2 IgM assay (86.25% vs. 33.75%).
Conclusion
The Elecsys Anti-SARS-CoV-2 immunoassay demonstrated very high specificity and high sensitivity in samples collected at least 14 days post-PCR confirmation of SARS-CoV-2 infection, supporting its use to aid in determination of previous exposure to SARS-CoV-2.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40121-021-00504-9.
Keywords: SARS-CoV-2, COVID-19, Coronavirus, Serologic tests, Immunoassay, Clinical performance, Specificity, Sensitivity, Method comparison
Key Summary Points
| Why carry out this study? |
| The coronavirus disease 2019 (COVID-19) pandemic has prompted the rapid development of serological assays to detect prior exposure to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2); however, reliable information on the relative performance of these assays in a wide range of settings is urgently needed to better inform morbidity rates and virus containment measures. |
| The Elecsys® Anti-SARS-CoV-2 immunoassay (Roche Diagnostics), which utilises a recombinant protein representing the nucleocapsid (N) antigen, was developed to provide an accurate and reliable method for the detection of antibodies to SARS-CoV-2. |
| The aims of this multicentre study were to provide detailed evidence on the clinical performance of the Elecsys Anti-SARS-CoV-2 immunoassay and compare the results to other commercially available assays. |
| What was learned from the study? |
| The Elecsys Anti-SARS-CoV-2 immunoassay demonstrated a very high specificity (99.85% [95% CI 99.75–99.92]) in blood donor and routine diagnostic specimens, and high sensitivity (93.61% [95% CI 89.51–96.46]) in samples collected at least 14 days post-PCR confirmation of SARS-CoV-2 infection; specificity and sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay were comparable to several other commercially available assays. |
| These results support use of the Elecsys Anti-SARS-CoV-2 immunoassay as a tool to aid in determination of an immune response following previous exposure to SARS-CoV-2 in a wide variety of settings. |
Introduction
As of 16 March 2021, there have been more than 119 million confirmed cases worldwide of coronavirus disease 2019 (COVID-19), including over 2.6 million deaths, caused by infection with the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1–3]. SARS-CoV-2 is an enveloped, non-segmented, single-stranded RNA virus that shares similarities with other coronaviruses in the expression of its genome, which encodes 16 non-structural proteins and four structural proteins, known as the spike (S), envelope (E), membrane (M) and nucleocapsid (N) antigens [4, 5]. Individuals infected with SARS-CoV-2 may exhibit a range of respiratory symptoms, including a persistent cough, shortness of breath, fever and fatigue [2, 6]. Although up to 80% of infections are mild or asymptomatic, 15% are severe, requiring oxygen, and 5% are critical, requiring ventilation [6]. Symptomatic and pre-symptomatic transmission of SARS-CoV-2, occurring via contact with infected respiratory droplets and contaminated surfaces, is thought to play a greater role in the spread of the virus than asymptomatic transmission [7, 8].
Reverse transcriptase polymerase chain reaction (RT-PCR) is the current gold standard for detection of SARS-CoV-2 RNA in sputum gathered from patient nasopharyngeal or oropharyngeal swabs, which typically have high viral titres during the first few days of infection [9]. Recent evidence suggests that whilst the clinical sensitivity of PCR remains very high during the first few days after initial onset of symptoms, it then decreases over time, dropping from more than 90% over the first 5 days, to 70–71% between days 9–11, and 30% on day 21 following onset of symptoms [10]. Conversely, the clinical sensitivity of serological tests has been shown to increase over time following initial onset of symptoms, from more than 50% at day 7, to more than 80% at day 12, and finally 100% at day 21, using an in-house enzyme-linked immunosorbent assay (Massachusetts General Hospital, Boston, MA, USA and the Ragon Institute of MGH, MIT and Harvard, Cambridge, MA, USA) [10]. Therefore, it is possible that complementary, time-dependent use of PCR and serological testing will increase reliability when determining prior exposure to SARS-CoV-2 infection, which could better inform morbidity rates and virus containment measures.
The Elecsys® Anti-SARS-CoV-2 electrochemiluminescence immunoassay (Roche Diagnostics International Ltd, Rotkreuz, Switzerland) was developed to provide an accurate method for the in vitro qualitative detection of high-affinity antibodies to SARS-CoV-2 [11]. The clinical performance of the newly launched Elecsys Anti-SARS-CoV-2 serological assay is undergoing evaluation; however, high specificity (99.8%) and sensitivity (99.5%) were previously observed in samples with prior PCR-confirmed SARS-CoV-2 infection, supporting its use as a tool for identification of past SARS-CoV-2 infection [12].
This multicentre study was conducted to further evaluate the specificity and sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay and compare its performance against other commercially available assays.
Methods
Study Design
This retrospective, non-interventional study to evaluate the performance of the Elecsys Anti-SARS-CoV-2 immunoassay was conducted at six European sites: four in Germany (Augsburg, Berlin, Hagen and Heidelberg), one in Switzerland (Bern) and one in Austria (Innsbruck). The Elecsys Anti-SARS-CoV-2 immunoassay was evaluated using serum/plasma samples from blood donors and routine diagnostic specimens that were presumed negative for SARS-CoV-2-specific antibodies (specificity) and samples from patients with PCR-confirmed SARS-CoV-2 infection (sensitivity). The first sample was tested on 20 May 2020 and the last sample was tested on 2 September 2020.
This study was conducted in accordance with the study protocol provided by Roche Diagnostics and in accordance with the principles of the Declaration of Helsinki. All human samples utilised were anonymised, frozen, residual samples for which no ethical approval was required or waiver was required in accordance with local legislation from ZEKO (Central Ethics Commission at the German Medical Association). A statement was obtained from the Ethics Committee of the Landesärztekammer Bayern confirming that there are no objections to the coherent use of anonymised residual samples. The study protocol was submitted to institutional review boards at study sites in Innsbruck (Austria) and Bern (Switzerland) prior to study initiation; ethical approval was granted for Innsbruck and a waiver granted for Bern.
Assay
The Elecsys Anti-SARS-CoV-2 electrochemiluminescence immunoassay was developed for the in vitro qualitative detection of antibodies to SARS-CoV-2 in human serum and plasma. It utilises a recombinant protein representing the nucleocapsid (N) antigen of the virus in a double-antigen sandwich test format, which favours detection of mature, high-affinity antibodies to SARS-CoV-2 [11, 13]. The Elecsys Anti-SARS-CoV-2 immunoassay is intended for use on cobas e analysers; total duration of the immunoassay is 18 min. The analyser automatically calculates a cut-off based on the measurement of two calibrators, one negative (ACOV2 Cal1) and one positive (ACOV2 Cal2). The result of a sample is given as either ‘reactive’ or ‘non-reactive’ in the form of a cut-off index (COI).
In this study, measurements determined using the Elecsys Anti-SARS-CoV-2 immunoassay were interpreted according to the manufacturer’s instructions. Samples with a COI < 1.0 were considered non-reactive and deemed negative for anti-SARS-CoV-2 antibodies, while those with a COI ≥ 1.0 were considered reactive and deemed positive for anti-SARS-CoV-2 antibodies [13].
Specificity Analysis
The specificity of the Elecsys Anti-SARS-CoV-2 immunoassay was evaluated at five sites using anonymised, frozen, residual samples from blood donor screening and routine diagnostic testing, including pregnancy screening and paediatric samples. Four sites (Augsburg, Hagen, Heidelberg and Bern) provided serum and/or plasma samples and performed testing using cobas e 601 and 801 analysers (Roche Diagnostics International Ltd, Rotkreuz, Switzerland); samples from Innsbruck were sent to Augsburg for measurement. All samples were obtained before September 2019 and were presumed negative for SARS-CoV-2-specific antibodies. Details of the cohorts enrolled and tested across the five sites are summarised in the Supplementary Material (Supplementary Table 1).
Sensitivity Analysis
Evaluation of the sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay was conducted at three sites in Germany (Augsburg, Heidelberg and Berlin) using anonymised, frozen, residual serum or plasma samples. All samples were single or sequential and confirmed positive for SARS-CoV-2 using PCR. Augsburg and Heidelberg included samples drawn from outpatients. Heidelberg also included samples from employees of MVZ Labor Limbach and hospitalised patients, including a subset from patients receiving dialysis. All samples provided by the study site in Berlin were collected from hospitalised patients, including a subset from patients monitored in the intensive care unit (ICU).
The sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay was evaluated on cobas e 601 and 801 analysers (Roche Diagnostics International Ltd, Rotkreuz, Switzerland). All samples were categorised by the week in which they were drawn following a positive PCR result, and grouped as follows: 0–6 days, 7–13 days, or at least 14 days post-PCR confirmation. For sequential samples, blood draws were performed over a period of minimum 2 to maximum 64 days, up to day 78. If more than one sample per patient was collected per time interval, only the last specimen per patient was included in the sensitivity calculation. For example, if blood was drawn from a patient on day 3, day 7, day 10, day 14 and day 21, only values from day 3, day 10 and day 21 would be included in the 0–6, 7–13 and at least 14 days post-PCR confirmation groups, respectively.
Method Comparison
Specificity and sensitivity results determined using the Elecsys Anti-SARS-CoV-2 immunoassay were compared with those calculated for other commercially available SARS-CoV-2 assays, as available at each study site. Comparator platforms included LIAISON SARS-CoV-2 S1/S2 IgG assay (DiaSorin, Saluggia, Italy); EUROIMMUN Anti-SARS-CoV-2 IgG assay (EUROIMMUN Medizinische Labordiagnostika AG, Luebeck, Germany); ARCHITECT SARS-CoV-2 IgG assay (Abbott Laboratories, Abbott Park, IL, USA); ADVIA Centaur SARS-CoV-2 Total assay (Siemens Healthcare GmbH, Erlangen, Germany); and iFlash SARS-CoV-2 IgG and IgM assays (Shenzhen YHLO Biotech Co., Ltd, Shenzhen, China). Results for each comparator assay were interpreted using cut-off values provided in the respective manufacturer’s package insert. For a breakdown of comparator assays by study site, please refer to the Supplementary Material (Supplementary Table 2). For method comparison of sensitivity between the Elecsys Anti-SARS-CoV-2 immunoassay and the LIAISON SARS-CoV-2 S1/S2 IgG assay, results were excluded because of the small sample size.
For all comparator assays, results considered ‘equivocal’ or ‘borderline’ per the cut-off values provided in the manufacturer’s package insert were grouped into a ‘grey zone’. For assays with results that were included in the grey zone, two calculations were performed: in the first, all samples with grey zone results were excluded from the analysis; in the second, all grey zone results were interpreted as reactive.
Overall percentage agreement (OPA) between the Elecsys Anti-SARS-CoV-2 immunoassay and the comparator assays was also calculated. For presumed negative samples, the same samples were used for specificity analysis and comparison of those results between methods and calculation of OPA. For confirmed positive samples, all samples with a valid measurement from both the Elecsys Anti-SARS-CoV-2 immunoassay and the comparator assay were included in the analysis; the OPA calculation was performed independently of the time point of blood draw.
Statistical Analysis
According to a previously published formulae [14], sample sizes of n = 3435–20,964 for the specificity analyses (assuming a specificity of 0.998) and n = 32–50 for the sensitivity analyses (assuming a sensitivity of 0.999) would be required to ensure a significance level of 0.05 and power of 0.8. Point estimates and 95% confidence intervals (CIs; two sided) were calculated using the exact method for specificity and sensitivity. For the specificity analysis, samples were grouped into those obtained from archived blood donations (group A) and those from archived routine diagnostic specimens (group B). For the method comparison, two-sided Wald CIs were calculated for the differences between estimated specificities and sensitivities for the Elecsys Anti-SARS-CoV-2 immunoassay and comparator assays, as recommended by Wenzel and Zapf [15]. If these CIs did not include zero, differences were considered statistically significant.
Results
Specificity Analysis
Specificity results for the overall sample cohort (n = 9575) and by analysis group are summarised in Table 1. Using an assay COI of 1.0 resulted in an overall specificity of 99.85% (95% CI 99.75–99.92) in samples obtained across all five sites. Among 6714 serum and/or plasma samples from blood donors and 2861 serum and/or plasma samples from routine diagnostic samples, specificity was 99.82% (95% CI 99.69–99.91) and 99.93% (95% CI 99.75–99.99), respectively. Among 2256 samples from pregnant women, specificity was 99.91% (95% CI 99.68–99.99). Among 205 paediatric samples, specificity was 100.00% (95% CI 98.22–100.00). Across groups A (blood donors) and B (routine diagnostic specimens), a total of 14 reactive samples were detected (group A, n = 12; group B, n = 2).
Table 1.
Summary of specificity results for the Elecsys Anti-SARS-CoV-2 immunoassay in blood donor and routine diagnostic samples
| Group | Sample cohort | Samples tested, n | Reactive, n | Non-reactive, n | Specificity, % (95% CI) |
|---|---|---|---|---|---|
| Groups A and B | 9575 | 14 | 9561 | 99.85 (99.75–99.92) | |
| Group A (blood donors) | Austria (Innsbruck), influenza seasona | 1048 | 5 | 1043 | 99.52 (98.89–99.84) |
| Germany (Hagen), no seasonal selection | 2625 | 2 | 2623 | 99.92 (99.73–99.99) | |
| Switzerland (Bern), no seasonal selection | 3041 | 5 | 3036 | 99.84 (99.62–99.95) | |
| Switzerland (Bern), outside influenza season | 2003 | 2 | 2001 | 99.90 (99.64–99.99) | |
| Switzerland (Bern), influenza season | 1038 | 3 | 1035 | 99.71 (99.16–99.94) | |
| All | 6714 | 12 | 6702 | 99.82 (99.69–99.91) | |
| Group B (routine diagnostic testing) | Germany (Augsburg), routine diagnostic | 400 | 0 | 400 | 100.00 (99.08–100.00) |
| Germany (Augsburg), pregnancy | 1498 | 2 | 1496 | 99.87 (99.52–99.98) | |
| Germany (Heidelberg), pregnancy | 758 | 0 | 758 | 100.00 (99.51–100.00) | |
| Germany (Augsburg and Heidelberg), pregnancy | 2256 | 2 | 2254 | 99.91 (99.68–99.99) | |
| Germany (Heidelberg), paediatrics | 205 | 0 | 205 | 100.00 (98.22–100.00) | |
| All | 2861 | 2 | 2859 | 99.93 (99.75–99.99) |
CI confidence interval, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2
aSamples from Innsbruck were analysed at Augsburg
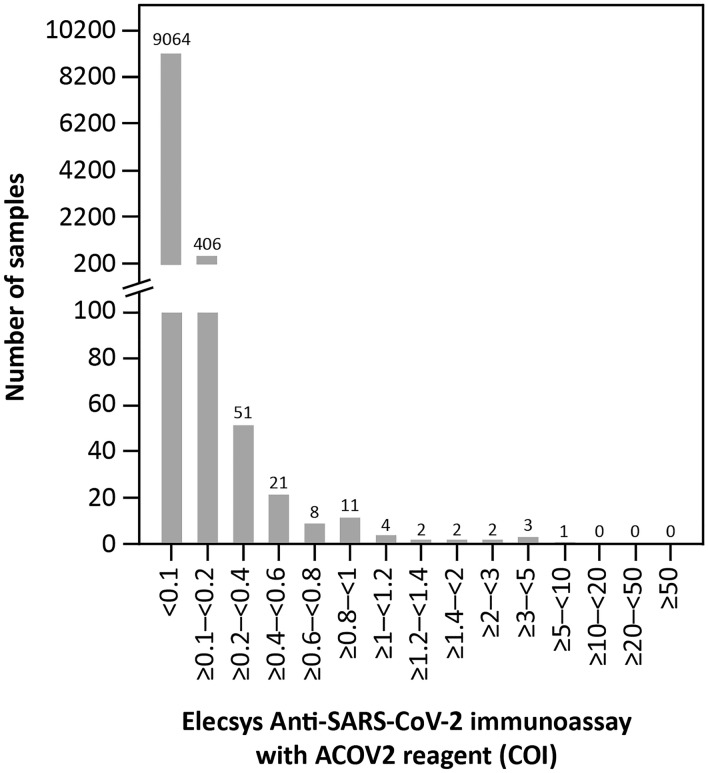
In total, 9561 of the presumed SARS-CoV-2 antibody negative samples tested had a COI < 1.0; the vast majority of these had COIs < 0.1 (n = 9064; Fig. 1). Only 14 samples had a COI ≥ 1.0 (pre-specified cut-off for reactivity).
Fig. 1.
COI distribution in patient samples presumed negative for SARS-CoV-2 antibodies (n = 9575). COI cut-off index, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2
Sensitivity Analysis
A total of 806 single and sequential SARS-CoV-2 PCR-confirmed positive samples from 255 patients across the three study sites were included in this analysis (Supplementary Material; Supplementary Table 3). Twenty samples were excluded because of an unclear or negative PCR result, missing Elecsys Anti-SARS-CoV-2 immunoassay result, or incorrect inclusion in the sample cohort. All samples from the study site in Augsburg were from outpatients. Of the samples from the study sites in Heidelberg, approximately half were drawn from dialysis patients. All samples from the study site in Berlin were drawn from hospitalised patients; approximately 60% of these samples were drawn from patients in the ICU.
Overall sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay was 93.61% (95% CI 89.51–96.46) in samples collected at least 14 days post-PCR confirmation of SARS-CoV-2 infection (n = 219; Table 2); sensitivity was 43.14% (95% CI 29.35–57.75) at 0–6 days (n = 51) and 84.00% (95% CI 70.89–92.83) at 7–13 days post-PCR confirmation (n = 50). Fourteen samples collected at least 14 days post-PCR confirmation were found to be non-reactive: 12 from Heidelberg, one from Augsburg, and one from Berlin. Site-specific calculations showed that sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay was highest at Augsburg (98.94% [95% CI 94.21–99.97]) in samples drawn at least 14 days post-PCR confirmation, and lowest at Augsburg in samples drawn 0–6 days post-PCR confirmation (33.33% [95% CI 0.84–90.57]). For all three sites, sensitivity was highest at at least 14 days post-PCR confirmation and lowest at 0–6 days post-PCR confirmation (Table 2).
Table 2.
Sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay by time interval post-PCR confirmation
| Time interval (days post-PCR confirmation) | Sensitivity, % (95% CI) n/N |
|||
|---|---|---|---|---|
| Augsburg | Heidelberg | Berlin | All sites combined | |
| 0–6 |
33.33 (0.84–90.57) 1/3 |
40.00 (21.13–61.33) 10/25 |
47.83 (26.82–69.41) 11/23 |
43.14 (29.35–57.75) 22/51 |
| 7–13 |
80.00 (28.36–99.49) 4/5 |
81.82 (48.22–97.72) 9/11 |
85.29 (68.94–95.05) 29/34 |
84.00 (70.89–92.83) 42/50 |
| ≥ 14 (up to day 78) |
98.94 (94.21–99.97) 93/94 |
85.71 (76.38–92.39) 72/84 |
97.56 (87.14–99.94) 40/41 |
93.61 (89.51–96.46) 205/219 |
CI confidence interval, PCR polymerase chain reaction, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2
Method Comparison
Specificity
Method comparison was performed using 934–2039 samples per comparison. The specificity of the Elecsys Anti-SARS-CoV-2 immunoassay was similar or higher than the specificity of all tested comparator assays (Table 3). Statistically significant differences were observed between the Elecsys Anti-SARS-CoV-2 immunoassay versus the following comparator assays: LIAISON SARS-CoV-2 S1/S2 IgG (n = 2039, 99.71% vs. 98.48%, difference 1.23%); EUROIMMUN Anti-SARS-CoV-2 IgG (n = 956, 100.00% vs. 94.87%, difference 5.13%); ADVIA Centaur SARS-CoV-2 Total (n = 962, 100.00% vs. 87.32%, difference 12.68%); and iFlash SARS-CoV-2 IgM (n = 962, 100.00% vs. 99.58%, difference 0.42%). No statistically significant differences were observed between the Elecsys Anti-SARS-CoV-2 immunoassay versus the following comparator assays: ARCHITECT SARS-CoV-2 IgG (n = 2006, 99.75% vs. 99.65%, difference 0.10%); and iFlash SARS-CoV-2 IgG (n = 962, 100.00% vs. 100.00%, difference 0%).
Table 3.
Summary of specificity values for the Elecsys Anti-SARS-CoV-2 immunoassay and comparator assays in presumed SARS-CoV-2 antibody negative samples
| Assay | Inclusion of grey zone values | Samples tested, n | Specificity, % (95% CI) | Difference between specificity values, % (95% Wald CI) | Significant difference |
|---|---|---|---|---|---|
| LIAISON SARS-CoV-2 S1/S2 IgG | Excluded | 2032 | 98.82 (98.25–99.24) | 0.89 (0.36–1.41) | Yes |
| 99.70 (99.36–99.89)a | |||||
| Included and considered reactive | 2039 | 98.48 (97.85–98.96) | 1.23 (0.64–1.81) | Yes | |
| 99.71 (99.36–99.89)a | |||||
| EUROIMMUN Anti-SARS-CoV-2 IgG | Excluded | 934 | 97.11 (95.82–98.09) | 2.89 (1.82–3.97) | Yes |
| 100.00 (99.61–100.00)a | |||||
| Included and considered reactive | 956 | 94.87 (93.28–96.18) | 5.13 (3.73–6.52) | Yes | |
| 100.00 (99.61–100.00)a | |||||
| ARCHITECT SARS-CoV-2 IgG | N/A | 2006 | 99.65 (99.28–99.86) | 0.10 (− 0.24 to 0.44) | No |
| 99.75 (99.42–99.92)a | |||||
| ADVIA Centaur SARS-CoV-2 Total | N/A | 962 | 87.32 (85.05–89.36) | 12.68 (10.58–14.78) | Yes |
| 100.00 (99.62–100.00)a | |||||
| iFlash SARS-CoV-2 IgG | N/A | 962 | 100.00 (99.62–100.00) | 0.00 (0.00–0.00) | No |
| 100.00 (99.62–100.00)a | |||||
| iFlash SARS-CoV-2 IgM | N/A | 962 | 99.58 (98.94–99.89) | 0.42 (0.01–0.82) | Yes |
| 100.00 (99.62–100.00)a |
CI confidence interval, IgG immunoglobulin G, IgM immunoglobulin M, N/A not applicable, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2
aMeasurements taken on the Elecsys Anti-SARS-CoV-2 immunoassay using the same samples
Sensitivity
Method comparison was performed using 79–124 samples drawn at least 14 days post-PCR confirmation per comparison across the three study sites. Across all assays evaluated, sensitivity was highest for the EUROIMMUN Anti-SARS-CoV-2 IgG assay for samples drawn at least 14 days post-PCR confirmation at 95.16% (95% CI 89.77–98.20; Table 4). No statistically significant differences in sensitivity for samples drawn at least 14 days post-PCR confirmation were observed between the Elecsys Anti-SARS-CoV-2 immunoassay and the EUROIMMUN Anti-SARS-CoV-2 IgG (n = 124, 90.32% vs. 95.16%, difference − 4.84%) and ARCHITECT SARS-CoV-2 IgG (n = 79, 84.81% vs. 87.34%, difference − 2.53%) assays. The Elecsys Anti-SARS-CoV-2 immunoassay showed significantly lower sensitivity versus the ADVIA Centaur SARS-CoV-2 Total (n = 81, 85.19% vs. 95.06%, difference − 9.88%) and iFlash SARS-CoV-2 IgG (n = 80, 86.25% vs. 93.75%, difference − 7.50%) assays, but significantly higher sensitivity versus the iFlash SARS-CoV-2 IgM assay (n = 80, 86.25% vs. 33.75%, difference 52.50%).
Table 4.
Summary of sensitivity values for the Elecsys Anti-SARS-CoV-2 immunoassay and comparator assays determined in samples drawn at least 14 days post-PCR confirmation across all sites
| Assaya | Inclusion of grey zone values | Samples tested, n | Sensitivity, % (95% CI) | Difference between sensitivity values, % (95% Wald CI) | Significant difference |
|---|---|---|---|---|---|
| EUROIMMUN Anti-SARS-CoV-2 IgG | Excluded | 120 | 94.17 (88.35–97.62) | − 3.33 (− 8.96 to 2.29) | No |
| 90.83 (84.19–95.33)b | |||||
| Included and considered reactive | 124 | 95.16 (89.77–98.20) | − 4.84 (− 10.25 to 0.57) | No | |
| 90.32 (83.71–94.90)b | |||||
| ARCHITECT SARS-CoV-2 IgG | N/A | 79 | 87.34 (77.95–93.76) | − 2.53 (− 9.53 to 4.46) | No |
| 84.81 (74.97–91.90)b | |||||
| ADVIA Centaur SARS-CoV-2 Total | N/A | 81 | 95.06 (87.84–98.64) | − 9.88 (− 18.67 to − 1.08) | Yes |
| 85.19 (75.55–92.10)b | |||||
| iFlash SARS-CoV-2 IgG | N/A | 80 | 93.75 (86.01–97.94) | − 7.50 (− 14.23 to − 0.77) | Yes |
| 86.25 (76.73–92.93)b | |||||
| iFlash SARS-CoV-2 IgM | N/A | 80 | 33.75 (23.55–45.19) | 52.50 (40.51 to 64.49) | Yes |
| 86.25 (76.73–92.93)b |
CI confidence interval, IgG immunoglobulin G, IgM immunoglobulin M, N/A not applicable, PCR polymerase chain reaction, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2
aFor method comparison of sensitivity between the Elecsys Anti-SARS-CoV-2 immunoassay and the LIAISON SARS-CoV-2 S1/S2 IgG assay, results were excluded because of the small sample size
bMeasurements taken on the Elecsys Anti-SARS-CoV-2 immunoassay using the same samples
Agreement Rates
Agreement rates (OPA) for the Elecsys Anti-SARS-CoV-2 immunoassay versus comparator assays in presumed SARS-CoV-2 antibody negative samples ranged between 87.32% and 100.00%, and in SARS-CoV-2 PCR-confirmed positive samples ranged between 44.22% and 93.80% (Table 5).
Table 5.
Agreement rates for the Elecsys Anti-SARS-CoV-2 immunoassay and comparator assays in presumed SARS-CoV-2 antibody negative and PCR-confirmed positive samples
| Assay | Elecsys Anti-SARS-CoV-2 | ||
|---|---|---|---|
| Sample type | Samples tested, n | Overall agreement, % (95% CI) | |
| LIAISON SARS-CoV-2 S1/S2 IgG | Presumed SARS-CoV-2 negative | 2039 | 98.19 (97.51–98.72) |
| PCR-confirmed positive | N/A | N/A | |
| EUROIMMUN Anti-SARS-CoV-2 IgG | Presumed SARS-CoV-2 negative | 956 | 94.87 (93.28–96.18) |
| PCR-confirmed positive | 710 | 93.80 (91.77–95.46) | |
| ARCHITECT SARS-CoV-2 IgG | Presumed SARS-CoV-2 negative | 2006 | 99.40 (98.96–99.69) |
| PCR-confirmed positive | 147 | 91.16 (85.35–95.21) | |
| ADVIA Centaur SARS-CoV-2 Total | Presumed SARS-CoV-2 negative | 962 | 87.32 (85.05–89.36) |
| PCR-confirmed positive | 149 | 83.89 (76.99–89.40) | |
| iFlash SARS-CoV-2 IgG | Presumed SARS-CoV-2 negative | 962 | 100.00 (99.62–100.00) |
| PCR-confirmed positive | 147 | 90.48 (84.54–94.69) | |
| iFlash SARS-CoV-2 IgM | Presumed SARS-CoV-2 negative | 962 | 99.58 (98.94–99.89) |
| PCR-confirmed positive | 147 | 44.22 (36.04–52.63) | |
CI confidence interval, IgG immunoglobulin G, IgM immunoglobulin M, N/A not applicable, PCR polymerase chain reaction, SARS-CoV-2 severe acute respiratory syndrome coronavirus 2
Discussion
There is an urgent unmet need for highly sensitive serological assays for detecting antibodies to SARS-CoV-2 in order to aid identification of individuals previously exposed to the virus and inform containment procedures [16, 17]. In this multicentre study, the Elecsys Anti-SARS-CoV-2 immunoassay demonstrated a very high specificity of 99.85% across a large cohort of blood donor and routine diagnostic samples and a high overall sensitivity of 93.61% in samples collected at least 14 days post-PCR confirmation of SARS-CoV-2 infection. Specificity and sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay were comparable to several other commercially available SARS-CoV-2 assays tested. These findings provide broader evidence for the favourable performance of the assay across different diagnostic laboratories and support its use as a tool to aid the determination of previous exposure to SARS-CoV-2 infection.
The very high overall specificity reported here (99.85%) for the Elecsys Anti-SARS-CoV-2 immunoassay corresponds to that reported by the manufacturer (99.80%) [13] and in previous studies using samples drawn pre-COVID-19 (98.00–100%) [12, 18–21]. The overall sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay in samples collected at least 14 days post-PCR confirmation of SARS-CoV-2 infection in the present study (93.61%) was similar to that reported in a previous study in PCR-confirmed SARS-CoV-2 infected samples collected 14 days post-symptom onset (92.0%) [20], but lower than that reported in the manufacturer’s package insert (99.5%) [13]. A potential reason for this may be differences in assay sensitivity between the sites in the present study; in particular, the lower sensitivity calculated for samples analysed at Heidelberg (85.71%), compared with Augsburg (98.94%) and Berlin (97.56%). The samples analysed at Berlin were taken from hospitalised patients, including a subset of samples from patients monitored in the ICU, for whom blood draws were closely monitored. In contrast, the majority of samples tested at Augsburg and Heidelberg were from outpatients. Disease severity in ICU patients was likely far greater than that of non-hospitalised patients infected with SARS-CoV-2, which may exacerbate immune response and increase the likelihood of a positive test result from the Elecsys Anti-SARS-CoV-2 immunoassay [22–24]. It is possible that the observed difference in sensitivity performance of the Elecsys Anti-SARS-CoV-2 immunoassay between study sites was partly caused by the pre-characterisation and selection of cohorts applied at each site. However, given the unprecedented situation of the ongoing COVID-19 pandemic, the clinical heterogeneity included in this study should be considered a strength rather than a limitation. Moreover, approximately 50% of all samples tested at Heidelberg were from patients undergoing dialysis, in whom infections are one of the main causes of morbidity and mortality due to immunodeficiency associated with end-stage renal disease [25–27]. Therefore, it is less likely that these patients were able to mount a detectable antibody response to SARS-CoV-2 [28, 29]. In the present study, 10/12 non-reactive samples collected at least 14 days post-PCR confirmation at Heidelberg were from patients receiving dialysis. These findings support the use of PCR confirmation alongside serological testing for immunocompromised patients to ensure an accurate test result.
The Elecsys Anti-SARS-CoV-2 immunoassay demonstrated a very high specificity of 99.91% in samples from pregnant women. Accurate serological assays for SARS-CoV-2 are especially important for use in pregnant women, who may have increased susceptibility to viral respiratory infections because of altered immune status [30]. At present, there are limited data available on the performance of other commercially available serological assays for SARS-CoV-2 in pregnant women. Rushworth et al. conducted a performance evaluation of the Mount Sinai SARS-CoV-2 IgG antibody ELISA and observed no false positive results in a cohort of negative control samples (n = 50) drawn from presumed healthy pregnant women pre-COVID-19 [31]. In the present study, two reactive results were observed in pre-COVID-19 samples from pregnant women, which could be the result of an unknown cross-reactant. A very high specificity of 100% for the Elecsys Anti-SARS-CoV-2 immunoassay was also observed in the present study in a paediatric cohort. However, it should be noted that the statistical significance of these results is not very powerful because of the relatively small number of samples in this cohort (n = 205) and further investigation is warranted.
In the present study, the sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay was lowest 0–6 days post-PCR confirmation at 43.14%, before increasing to 93.61% for samples drawn at least 14 days post-PCR confirmation. This trend towards increased sensitivity over time following PCR confirmation was observed across all three sites and is in accordance with previous research [10, 12, 21]. However, in a similar cohort of SARS-CoV-2 PCR-confirmed positive samples, Oved et al. previously found that approximately 5% of patients remained seronegative at least 14 days post-PCR confirmation, and thus did not seroconvert [32]. These individuals would not have a positive result on a serological assay. Therefore, it is important to consider this phenomenon when evaluating these results, as it could affect the trends observed herein.
In the present method comparison analysis, the specificity of the Elecsys Anti-SARS-CoV-2 immunoassay was similar to or higher than that of the comparator assays tested. The sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay was also similar to the EUROIMMUN Anti-SARS-CoV-2 IgG and ARCHITECT SARS-CoV-2 IgG assays, but higher than that of the iFlash SARS-CoV-2 IgM assay, and lower than that of the ADVIA Centaur SARS-CoV-2 Total and iFlash SARS-CoV-2 IgG assays. It is important to note, however, that the available assays differ in terms of their assay design (e.g. antibody class) and the viral antigens they detect (e.g. spike protein or nucleocapsid protein). For example, the format of the Elecsys Anti-SARS-CoV-2 immunoassay requires binding of an antibody in the patient sample to two specific antigens, and thus favours preferential detection of mature, high-affinity antibodies characteristic of the late stages of SARS-CoV-2 infection [11, 13]. In addition, the clinical performance of serological assays is a compromise between sensitivity and specificity; whilst some assays are designed for higher sensitivity, others are designed for higher specificity.
Importantly, the overall specificity of the Elecsys Anti-SARS-CoV-2 immunoassay determined in the present study (99.85%) is comparable with the specificities of other commercially available assays reported in previous studies, including the ARCHITECT SARS-CoV-2 IgG ELISA (99.7%), LIAISON SARS-CoV-2 S1/S2 IgG ELISA (97.9–98.3%) and Elecsys Anti-SARS-CoV-2 S (99.8–100%) assays [18, 20, 32–34]. Higher specificity was observed for the Elecsys Anti-SARS-CoV-2 immunoassay in the present study than previously reported for the EUROIMMUN Anti-SARS-CoV-2 IgG (96–96.5%) ELISA [20, 35]. The relatively lower specificities measured for the Elecsys Anti-SARS-CoV-2 immunoassay and EUROIMMUN Anti-SARS-CoV-2 IgG ELISA could be attributed to cross-reactivity with antibodies to adenoviruses and other human coronaviruses [35]. A recent study reported no cross-reactivity with the Elecsys Anti-SARS-CoV-2 immunoassay in common cold or endemic coronavirus sample panels [12].
In general, the sensitivity of the comparator assays tested in the present study is consistent with previous findings [35–39]. The low sensitivity of the iFlash SARS-CoV-2 IgM assay in the present study is in accordance with that previously reported [40]. A potential explanation for this is the fact that detection of SARS-CoV-2-specific IgM is limited to very early on in the infection cascade; only 20% of SARS-CoV-2 infected individuals present IgM before IgG, and the majority will present both IgM and IgG in tandem [41]. Some studies have reported the detection of SARS-CoV-2-specific IgG even before IgM [42, 43]. Moreover, a 10-month follow-up study showed that assays designed to detect total antibodies had significantly higher rates of positive results compared with assays directed against IgG alone [44]. Taken together, the clinical value of IgM for diagnosis of COVID-19 remains unclear [45].
A major strength of this study is the large cohort of presumed SARS-CoV-2-negative (n = 9575) and SARS-CoV-2 PCR-confirmed positive (n = 806) serum and plasma samples used to evaluate specificity and sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay, respectively, across multiple sites in different countries and patient groups. This ensures that our data are reliable and can be used to form robust comparisons with immunoassay performance data from other studies. To our knowledge, the present study includes the largest cohort of samples from pregnant women used to date to evaluate the performance of a serological assay for SARS-CoV-2 with regard to specificity. Existing literature on the performance of commercially available assays for detection of antibodies to SARS-CoV-2 is limited in pregnant women and paediatric populations; thus, additional research in these groups is warranted to further inform clinical decision-making. This study was performed under accelerated timelines due to the high scientific value of the data in the ongoing COVID-19 pandemic, and so long-term stability data for frozen samples were not available prior to study initiation. However, the prolonged stability of IgG antibodies is well documented and so this should not impact the present data [46, 47]. For evaluation of sensitivity, the follow-up period for sequential samples was relatively short (maximum 64 days). Recent findings suggest that a lack of long-term follow-up samples may impact the sensitivity of SARS-CoV-2 serological assays because of the absence of seroconversion; therefore, inclusion of long-term samples may be beneficial in future studies [33, 34].
Conclusion
The performance of SARS-CoV-2 antibody assays in general is of high importance for public health and may affect political decision-making in pandemic management. This study generated additional data on the performance of the Elecsys Anti-SARS-CoV-2 immunoassay and provides broader evidence for the very high specificity of the assay, including in cohorts of pregnant women and paediatric populations. The Elecsys Anti-SARS-CoV-2 immunoassay also demonstrated a high overall sensitivity in samples collected at least 14 days post-PCR confirmation of SARS-CoV-2 infection, which was comparable to several other commercially available assays. Our findings support the use of the Elecsys Anti-SARS-CoV-2 immunoassay as a tool to aid in determination of previous exposure to SARS-CoV-2.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors thank: Peter Gowland (Interregionale Blood Transfusion Swiss Red Cross) for involvement in preparation of prepandemic samples and laboratory work; Sigrid Reichhuber, Janina Edion and Yvonne Knack (Roche Diagnostics) for management of investigation sites, data acquisition, and study monitoring. COBAS, COBAS E and ELECSYS are trademarks of Roche. All other product names and trademarks are the property of their respective owners.
Funding
This study was funded by Roche Diagnostics GmbH (Mannheim, Germany). The journal’s Rapid Service Fee was funded by Roche Diagnostics International Ltd (Rotkreuz, Switzerland).
Medical Writing and/or Editorial Assistance
Third-party medical writing support, under the direction of the authors, was provided by Chloe Fletcher, MSc, of Ashfield MedComms (Macclesfield, UK), an Ashfield Health Company, and was funded by Roche Diagnostics International Ltd (Rotkreuz, Switzerland).
Authorship
All named authors meet the International Committee of Medical Journal Editors (ICMJE) criteria for authorship for this article, take responsibility for the integrity of the work as a whole, and have given their approval for this version to be published.
Author Contributions
ML, CMR, KS, FL, TL: study conception and design. ER, MM, AM, CT, PF, JKH, CN: data acquisition. All authors contributed to data analysis and/or interpretation, provided critical review of the manuscript during development and final approval of the submitted version.
Prior Presentation
The manuscript includes previously published data from the following articles: Elena Riester, Beda Krieter, Peter Findeisen, Michael Laimighofer, Kathrin Schoenfeld, Tina Laengin, Christoph Niederhauser. Performance of an automated anti-SARS-CoV-2 immunoassay in prepandemic cohorts. medRxiv 2020.08.07.20169987. https://doi.org/10.1101/2020.08.07.20169987. Johannes Kolja Hegel, Elena Riester, Christopher M. Rank, Florina Langen, Tina Laengin, Peter Findeisen. Sensitivity of the Elecsys Anti-SARS-CoV-2 immunoassay as an aid in determining previous exposure to SARS-CoV-2. medRxiv 2021.02.11.21250290. https://doi.org/10.1101/2021.02.11.21250290.
Disclosures
Elena Riester received speaker’s honorarium from Roche. Kathrin Schönfeld is an employee of Roche Diagnostics GmbH and owner of shares in Roche Holding AG. Florina Langen is an employee of Roche Diagnostics GmbH. Tina Laengin is an employee of Roche Diagnostics GmbH. Christoph Niederhauser received speaker’s honorarium from Roche. Christopher M. Rank is an employee of Roche Diagnostics GmbH. Michael Laimighofer is an employee of Roche Diagnostics GmbH. Annelies Mühlbacher, Mario Majchrzak, Caroline Tinguely, Peter Findeisen and Johannes Kolja Hegel have no conflicts of interest to disclose.
Compliance with Ethics Guidelines
This study was conducted in accordance with the study protocol provided by Roche Diagnostics and in accordance with the principles of the Declaration of Helsinki. All human samples utilised were anonymised, frozen, residual samples for which no ethical approval was required or waiver was required in accordance with local legislation from ZEKO (Central Ethics Commission at the German Medical Association). A statement was obtained from the Ethics Committee of the Landesärztekammer Bayern confirming that there are no objections to the coherent use of anonymised residual samples. The study protocol was submitted to institutional review boards at study sites in Innsbruck (Austria) and Bern (Switzerland) prior to study initiation; ethical approval was granted for Innsbruck, and a waiver granted for Bern.
Data Availability
The datasets generated and analysed during the current study are available to qualified researchers on request via the clinical study data request platform (https://vivli.org/). Further details on Roche’s criteria for eligible studies are available here: https://vivli.org/members/ourmembers/. For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here: https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm.
Contributor Information
Elena Riester, Email: elena.riester@labkom.de.
Mario Majchrzak, Email: m.Majchrzak@bsdwest.de.
Annelies Mühlbacher, Email: annelies@heider.tirol.
Caroline Tinguely, Email: Caroline.Tinguely@itransfusion.ch.
Peter Findeisen, Email: peter.findeisen@labor-limbach.de.
Johannes Kolja Hegel, Email: kolja.hegel@laborberlin.com.
Michael Laimighofer, Email: michael.laimighofer@roche.com.
Christopher M. Rank, Email: christopher.rank@roche.com
Kathrin Schönfeld, Email: kathrin.schoenfeld@roche.com.
Florina Langen, Email: florina.langen@roche.com.
Tina Laengin, Email: tina.laengin@roche.com.
Christoph Niederhauser, Email: Christoph.Niederhauser@itransfusion.ch.
References
- 1.World Health Organization (WHO). WHO Coronavirus Disease (COVID-19) Dashboard. https://covid19.who.int/. Accessed 16 Mar 2021.
- 2.Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses. J Med Virol. 2020;92:424–432. doi: 10.1002/jmv.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khailany RA, Safdar M, Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Rep. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.World Health Organization (WHO). Coronavirus disease 2019 (COVID-19): Situation report—46. Data as reported by national authorities by 10AM CET 06 March 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200306-sitrep-46-covid-19.pdf?sfvrsn=96b04adf_4. Accessed 1 Dec 2020.
- 7.Cevik M, Kuppalli K, Kindrachuk J, Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. 2020;371:m3862. doi: 10.1136/bmj.m3862. [DOI] [PubMed] [Google Scholar]
- 8.Qiu X, Nergiz AI, Maraolo AE, Bogoch II, Low N, Cevik MJM. Defining the role of asymptomatic and pre-symptomatic SARS-CoV-2 transmission: a living systematic review. medRxiv. 2020 doi: 10.1101/2020.09.01.20135194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wölfel R, Corman VM, Guggemos W, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 10.Miller TE, Garcia Beltran WF, Bard AZ, et al. Clinical sensitivity and interpretation of PCR and serological COVID-19 diagnostics for patients presenting to the hospital. FASEB J. 2020;34:13877–13884. doi: 10.1096/fj.202001700RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egger M, Bundschuh C, Wiesinger K, et al. Comparison of the Elecsys® Anti-SARS-CoV-2 immunoassay with the EDI™ enzyme linked immunosorbent assays for the detection of SARS-CoV-2 antibodies in human plasma. Clin Chim Acta. 2020;509:18–21. doi: 10.1016/j.cca.2020.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muench P, Jochum S, Wenderoth V, et al. Development and validation of the Elecsys Anti-SARS-CoV-2 immunoassay as a highly specific tool for determining past exposure to SARS-CoV-2. J Clin Microbiol. 2020;58:e01694–e1720. doi: 10.1128/JCM.01694-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roche Diagnostics GmbH. Elecsys Anti-SARS-CoV-2 method sheet. 2020-10, V6.0 English.
- 14.Hajian-Tilaki K. Sample size estimation in diagnostic test studies of biomedical informatics. J Biomed Inform. 2014;48:193–204. doi: 10.1016/j.jbi.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 15.Wenzel D, Zapf A. Difference of two dependent sensitivities and specificities: comparison of various approaches. Biom J. 2013;55:705–718. doi: 10.1002/bimj.201200186. [DOI] [PubMed] [Google Scholar]
- 16.Lou B, Li T-D, Zheng S-F, et al. Serology characteristics of SARS-CoV-2 infection since exposure and post symptom onset. Eur Respir J. 2020;52(2):2000763. doi: 10.1183/13993003.00763-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peeling RW, Wedderburn CJ, Garcia PJ, et al. Serology testing in the COVID-19 pandemic response. Lancet Infect Dis. 2020;20:e245–e249. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perkmann T, Perkmann-Nagele N, Breyer MK, et al. Side by side comparison of three fully automated SARS-CoV-2 antibody assays with a focus on specificity. medRxiv. 2020 doi: 10.1101/2020.06.04.20117911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ekelund O, Ekblom K, Somajo S, et al. High-throughput immunoassays for SARS-CoV-2—considerable differences in performance when comparing three methods. medRxiv. 2020 doi: 10.1101/2020.05.22.20106294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Favresse J, Cadrobbi J, Eucher C, et al. Clinical performance of three fully automated anti-SARS-CoV-2 immunoassays targeting the nucleocapsid or spike proteins. J Med Virol. 2021;93:2262–2269. doi: 10.1002/jmv.26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Favresse J, Eucher C, Elsen M, et al. Clinical performance of the Elecsys electrochemiluminescent immunoassay for the detection of SARS-CoV-2 total antibodies. Clin Chem. 2020;66:1104–1106. doi: 10.1093/clinchem/hvaa131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guthmiller JJ, Stovicek O, Wang J, et al. SARS-CoV-2 infection severity is linked to superior humoral immunity against the spike. bioRxiv. 2020 doi: 10.1101/2020.09.12.294066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long Q-X, Deng H-J, Chen J, et al. Antibody responses to SARS-CoV-2 in COVID-19 patients: the perspective application of serological tests in clinical practice. medRxiv. 2020 doi: 10.1101/2020.03.18.20038018. [DOI] [Google Scholar]
- 24.Wang Y, Zhang L, Sang L, et al. Kinetics of viral load and antibody response in relation to COVID-19 severity. J Clin Invest. 2020;130:5235–5244. doi: 10.1172/JCI138759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saran R, Robinson B, Abbott KC, et al. US renal data system 2017 annual data report: epidemiology of kidney disease in the United States. Am J Kidney Dis. 2018;71:A7. doi: 10.1053/j.ajkd.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Betjes MGH, Litjens NHR. Chronic kidney disease and premature ageing of the adaptive immune response. Curr Urol Rep. 2014;16:471. doi: 10.1007/s11934-014-0471-9. [DOI] [PubMed] [Google Scholar]
- 27.Lisowska KA, Pindel M, Pietruczuk K, et al. The influence of a single hemodialysis procedure on human T lymphocytes. Sci Rep. 2019;9:5041. doi: 10.1038/s41598-019-41619-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Edey M, Barraclough K, Johnson DW. Review article: hepatitis B and dialysis. Nephrology. 2010;15:137–145. doi: 10.1111/j.1440-1797.2009.01268.x. [DOI] [PubMed] [Google Scholar]
- 29.Flower B, Atchison C. SARS-CoV-2 antibody seroprevalence in patients receiving dialysis in the USA. Lancet. 2020;396:1308–1309. doi: 10.1016/S0140-6736(20)32006-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu H, Wang LL, Zhao SJ, et al. Why are pregnant women susceptible to COVID-19? An immunological viewpoint. J Reprod Immunol. 2020;139:103122. doi: 10.1016/j.jri.2020.103122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rushworth SA, Johnson BB, Ashurst K, et al. Performance and health economic evaluation of the Mount Sinai COVID-19 serological assay identifies modification of thresholding as necessary to maximise specificity of the assay. medRxiv. 2020 doi: 10.1101/2020.06.11.20128306. [DOI] [Google Scholar]
- 32.Oved K, Olmer L, Shemer-Avni Y, et al. Multi-center nationwide comparison of seven serology assays reveals a SARS-CoV-2 non-responding seronegative subpopulation. EClinicalMedicine. 2020;29:1006511. doi: 10.1016/j.eclinm.2020.100651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Higgins V, Fabros A, Kulasingam V. Quantitative measurement of anti-SARS-CoV-2 antibodies: analytical and clinical evaluation. J Clin Microbiol. 2021;59:e03149–e3220. doi: 10.1128/JCM.03149-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Favresse J, Douxfils J. Evaluations of SARS-CoV-2 serological assay performance need inclusion of long-term samples. J Clin Microbiol. 2021;59:e00487–e521. doi: 10.1128/JCM.00487-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lassaunière R, Frische A, Harboe ZB, et al. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv. 2020 doi: 10.1101/2020.04.09.20056325. [DOI] [Google Scholar]
- 36.Bryan A, Pepper G, Wener MH, et al. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise. Idaho J Clin Microbiol. 2020;58:e00941–e1020. doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parai D, Dash GC, Choudhary HR, et al. Diagnostic comparison of three fully automated chemiluminescent immunoassay platforms for the detection of SARS-CoV-2 antibodies. medRxiv. 2020 doi: 10.1101/2020.10.07.20207696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Public Health England. Evaluation of the Abbott SARS-CoV-2 IgG for the detection of anti-SARS-CoV-2 antibodies. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/890566/Evaluation_of_Abbott_SARS_CoV_2_IgG_PHE.pdf. Accessed 1 Dec 2020.
- 39.Harritshøj LH, Gybel-Brask M, Afzal S, et al. Comparison of sixteen serological SARS-CoV-2 immunoassays in sixteen clinical laboratories. medRxiv. 2020 doi: 10.1101/2020.07.30.20165373. [DOI] [Google Scholar]
- 40.Kittel M, Muth M, Zahn I, et al. Clinical evaluation of commercial automated SARS-CoV-2 immunoassays. Int J Infect Dis. 2020;103:590–596. doi: 10.1016/j.ijid.2020.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maine GN, Lao KM, Krishnan SM, et al. Longitudinal characterization of the IgM and IgG humoral response in symptomatic COVID-19 patients using the Abbott Architect. J Clin Virol. 2020;133:104663. doi: 10.1016/j.jcv.2020.104663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thevarajan I, Nguyen THO, Koutsakos M, et al. Breadth of concomitant immune responses prior to patient recovery: a case report of non-severe COVID-19. Nat Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.To KK-W, Tsang OT-Y, Leung W-S, et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Favresse J, Eucher C, Elsen M, et al. Persistence of anti-SARS-CoV-2 antibodies depends on the analytical kit: a report for up to 10 months after infection. Microorganisms. 2021;9:556. doi: 10.3390/microorganisms9030556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harley K, Gunsolus IL. Comparison of the clinical performances of the Abbott Alinity IgG, Abbott Architect IgM, and Roche Elecsys Total SARS-CoV-2 antibody assays. J Clin Microbiol. 2020;59:e02104–e2120. doi: 10.1128/JCM.02104-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gislefoss RE, Grimsrud TK, Mørkrid L. Stability of selected serum proteins after long-term storage in the Janus Serum Bank. Clin Chem Lab Med. 2009;47:596–603. doi: 10.1515/CCLM.2009.121. [DOI] [PubMed] [Google Scholar]
- 47.Siennicka J, Laskowska A, Trzcińska A. Evaluating of influence of repeated thaw/freeze cycles on IgG and IgM stability. Med Dosw Mikrobiol. 2010;62:281–283. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analysed during the current study are available to qualified researchers on request via the clinical study data request platform (https://vivli.org/). Further details on Roche’s criteria for eligible studies are available here: https://vivli.org/members/ourmembers/. For further details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here: https://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing.htm.