Abstract
The development of resistance to therapy is a significant obstacle to effective therapeutic regimens. Evaluating the effects of oncology drugs in the laboratory setting is limited by the lack of translational models that accurately recapitulate cell–microenvironment interactions present in tumors. Acquisition of resistance to therapy is facilitated, in part, by the composition of the tumor extracellular matrix (ECM), with the primary current in vitro model using collagen I (COL I). Here we seek to identify the prevalence of COL I-enhanced expression in the triple-negative breast cancer (TNBC) subtype. Furthermore, we identify if methods of response to therapy are altered depending on matrix composition. We demonstrated that collagen content varies in patient tumor samples across subtypes, with COL I expression dramatically increased in typically less aggressive estrogen receptor (ER)-positive(ER+)/progesterone receptor (PGR)-positive (PGR+) cancers irrespective of patient age or race. These findings are of significance considering how frequently COL I is implicated in tumor progression. In vitro analyses of ER+ and ER-negative (ER–) cell lines were used to determine the effects of ECM content (collagen I, collagen IV, fibronectin, and laminin) on proliferation, cellular phenotype, and survival. Neither ER+ nor ER– cells demonstrated significant increases in proliferation when cultured on these ECM substrates. ER– cells cultured on these substrates were sensitized to both chemotherapy and targeted therapy. In addition, MDA-MB-231 cells expressed different morphologies, binding affinities, and stiffness across these substrates. We also demonstrated that ECM composition significantly alters transcription of senescence-associated pathways across ER+ and ER– cell lines. Together, these results suggest that complex matrix composites should be incorporated into in vitro tumor models, especially for the drug-resistant TNBC subtype.
Impact statement
The importance of tumor extracellular matrix (ECM) in disease progression is often inadequately represented in models of breast cancer that rely heavily on collagen I and Matrigel. Through immunohistochemistry analysis of patient breast tumors, we show a wide variation in collagen content based on subtype, specifically a repression of fibril collagens in the receptor negative subtype, irrespective of age and race. We also demonstrated that tumor ECM composition alters cellular elasticity and oncogenic pathway activation demonstrating that physiologically relevant three-dimensional models of breast cancer should include an ECM that is subtype specific.
Keywords: breast cancer, extracellular matrix, optical tweezers, tumor microenvironment
Introduction
Molecular characteristics, such as receptor status, are used to classify breast cancer subtypes.1 Despite known molecular markers for intervention, many patients still develop resistance to therapies.2,3 One mechanism by which cancer cells develop drug resistance is tumor molecular heterogeneity. Breast cancer is a heterogeneous disease exhibiting diversity between patients (inter-tumor heterogeneity) and within the same individual (intra-tumor heterogeneity).4 Although not fully distinguished, inter-tumor heterogeneity may be driven in part by the tumor microenvironment and extracellular matrix (ECM).5 Cellular binding to ECM components promote prosurvival pathways that enhance the viability of cancer cells and build resistance to therapies.6 The role that the mechanical integrity of the ECM plays in activating survival and proliferation pathways to induce drug resistance is vital for the progression of breast cancer.7 Despite these known associations between ECM and cancer survival, to date, the effect of molecular composition in breast cancer progression is less understood.
Specific components of ECM, such as collagen I (COL I), are well studied because of their association with poorer prognosis by increasing overall tumor stiffness, which facilitates cancer progression.8–10 To date, the characterization of matrix stiffness has been well identified as a mechanism for drug resistance and breast cancer proliferation.11–14 As tumors expand, the native ECM is remodeled, altering its molecular composition to favor tumor progression.
A recent proteomic study evaluating tumor matrix elevated in breast tumors compared with proximal nondiseased breast tissue demonstrated that COL I was not specifically elevated in triple-negative breast cancer (TNBC). Furthermore, fibronectin (FN), fibril-associated COL (COL12A1), and tenascin C were observed to be elevated in all breast cancers compared with proximal tissue.15 An immunohistochemistry (IHC) analysis of 134 tumors isolated from breast cancer patients revealed that FN expression was found to positively correlate with an increased mortality risk, lymph node involvement, and Ki67 proliferation-associated index, but did not correlate with estrogen receptor (ER) or progesterone receptor (PGR) receptor status.16 Collagen IV (COL IV) expression positively correlated with tumor size and negatively correlated with ER and PGR status. Laminin (LAM) expression was inversely correlated with ER and PGR status and positively correlated with Ki67 expression.16 This suggests that ECM protein composition may correlate with breast cancer subtypes and patient outcomes.
Despite the observed relevance of subtype-specific expression of ECM in breast cancer and associated patient survival, much of breast cancer data are generated in artificial systems that rely primarily on COL I owing to the relatively low cost and high availability of COL I. In addition, there is no subtype-specific matrix for use in breast cancer research. Tumor-specific ECM is not yet an integral component of in vitro drug studies where two-dimensional tissue culture plastic (TCP) is the gold standard. What has not yet been identified is the role matrix composition has on altering cellular physiology and response to therapy.17 The goal of this study was to determine the need for subtype-specific matrix.
Materials and Methods
Cell culture
ER-positive (ER+; MCF-7 ZR-75) and TNBC (BT-549, MDA-MB-157, MDA-MB-231) were maintained in 5% carbon dioxide at 37°C and cultured in Dulbecco's modified Eagle's medium supplemented with 10% Hyclone Cosmic Calf Serum (HyClone Laboratories), 50 ng/mL insulin (Sigma-Aldrich), and 1% minimal essential medium amino acids, non-essential amino acids, sodium pyruvate, and antibiotic-antimycotic (Gibco). Research was done under approved IRB exemption number E10882.
Matrix-induced breast cancer cell viability
ER+ breast cancer lines and TNBC lines were cultured on TCP control or Corning® BioCoat™ plates (Corning) coated with either COL IV, LAM, or FN. Cell density was 2000 cells/well. Cells were allowed to expand on coated dishes for 3 days then treated with paclitaxel (10 nM), PI3K inhibitor idelalisib (100 nM), or dimethyl sulfoxide (DMSO) control. At day 5, plates were analyzed with alamarBlue (Thermo Fisher) and read at 595 nm. Cells cultured on matrix and treated with therapies were normalized to results of cells cultured on TCP and treated with the same therapies.
Cell senescence assay
Breast cancer cell lines were seeded on TCP, COL IV, FN, and LAM Corning BioCoat 96-well plates at 1000 cells/well in culture media. At day 7, SA-β-gal was used to quantify senescence (ENZ-KIT129-0120; Enzo Life Sciences, Inc.). Results were normalized to TCP.
Immunofluorescence staining and imaging
Breast cancer cell lines were seeded on TCP, COL IV, FN, and LAM coated 96-well plates (Corning BioCoat) at 2500 cells/well. After 3 days, cells were fixed and stained with Alexa Fluor® 488 Mouse anti-Ki-67 (BD Biosciences) and 4′,6-diamidino-2-phenylindole following manufacturer's protocol. Three representative images were taken of each well at 20 × magnification. Percent Ki67-positive cells was calculated by dividing the number of Ki-67 positive cells by the total number of DAPI stain (n = 3). Actin filament stain was performed on cells seeded on TCP, COL IV, FN, and LAM Corning BioCoat 96-well plates at 2500 cells/well. After 3 days of culture, cells were fixed and stained with Alexa Fluor 488 Phalloidin (Invitrogen) following manufacturer's protocol.
Immunohistochemistry
Frozen tissue sections (5 μm) of diagnosed breast cancer (Origene) were stained for Masson Trichrome through Tulane University Histology Core (New Orleans, LA, USA). Patient information (receptor status, age, and race) when known is given in Supplementary Table S1.
Cell elasticity and cell binding force assays
Three-dimensional (3D) printed blocks measuring 0.5 cm on all sides (0.125 cm3) were cast in polydimethylsiloxane and used to make molds (Supplementary Fig. S1A, B) for human COL I, COL IV, FN, and LAM (Corning). Molds were used as described hereunder to coat glass slides. Matrix solutions (0.25 mg/mL) were poured into the molds and incubated overnight at 37°C. Molds were removed after three coatings and cells were seeded. Optical tweezing was adapted from previously described methods.18 Elasticity measurements were performed after 3 h. Polystyrene beads were trapped by the infrared laser beam. The motorized stage of the scope (Nikon TE2000) was programmed to move the cell against the bead to make an indentation in the cell (Supplementary Fig. S1C–E). From the indentation and optical force measurements,19 the cell elasticity was obtained as described elsewhere.20
Cell binding force was measured using coated microscope slides prepared as detailed previously. Concentrations of ECM proteins for this assay were 1 mg/mL of COL type IV, LAM, and FN. After drying, a razor blade was used to make a straight cut at one edge of the coating. The cells were collected from the previously mentioned culture and added to the coated slide. Cell binding force was measured as previously described.18 Cells suspended in culture media were added to the coated microscope slide and a single cell was trapped with the optics and moved against the coating (Supplementary Fig. S1F–H). Cells were held for 10 s and then moved away at 5 μm/s. The experiment was repeated decreasing the laser power, until the cell remained attached to the coating. Finally, the optical force applied to detach the cells was determined using trapping force calibration curves generated before experimentation. The obtained optical force corresponds to the initial binding force. The Stokes' Force created by the movement of the cell was very low compared with the applied optical force and can be neglected. For each sample, the experiment was repeated with 15–20 different cells at several places on the material.
Quantitative Real-Time-polymerase chain reaction
MDA-MB-231 cell line was cultured on matrix Corning BioCoat flasks COL IV, LAM, and FN-coated T25 flasks and TCP at a density of 5 × 105 cells, cultured for 3 days and collected for RNA extraction. Total RNA was extracted with Zymo RNA extraction kit (Zymo Research, CA, USA). One microgram RNA was used to generate cDNA (qScript cDNA SuperMix; Quantabio, MA, USA) per manufacturer's protocol. PerfeCTa SYBR® Green SuperMix (Quantabio) was used and total gene expression analysis was through the ΔΔCt method. Data were normalized to housekeeping gene CYCB with biological triplicates (n = 3) ± standard error of the mean (SEM). Complete primer sequences are given in Supplementary Table S2.
Kaplan–Meier plots and gene expression correlation
Kaplan–Meier (KM) survival plots were made using KM plotter21 for relapse-free survival. The KM plotter mRNA gene chip breast cancer data set was used. Patient splits were median and restricted analysis for subtype was selected for ER+ (n = 2565 samples), ER-negative (ER−; n = 1214 samples), and no restrictions (n = 3951 samples). Significant difference was p < 0.05. For protein correlations based on race, the Tang-2018 data set was used, no receptor status was selected. Correlation of COL1A1 and COL1A2 gene expression and receptor status was evaluated through the Breast Cancer Gene-Expression Miner v4.4 (n = 4319).22 RNA sequencing TCGA (n = 1034) and The Sweden Cancerome Analysis Network - Breast data sets (n = 3678) were used. Population query was receptor status (ER/PGR) using IHC.
Statistical analysis
All data were analyzed for statistical significance using a t-test in Prism GraphPad. Survival and polymerase chain reaction (PCR) experiments were carried out in triplicate with error bars representing SEM. Error bars for binding force and elasticity measurements represent standard deviation (SD). Binding force measurements were carried out with 15–20 cells/condition. For elasticity of cells, nine different cells on each substrate were measured at four or five different places on the cell. The large SDs of binding force and elasticity measurements is owing to several experimental parameters, like the laser stability that distort the trapping force, external vibrations, the surface roughness of the polymer, the contact point between cell and polymer or the cell itself that can be more or less damage by the laser.
Results
Matrix fibrillar composition is subtype specific
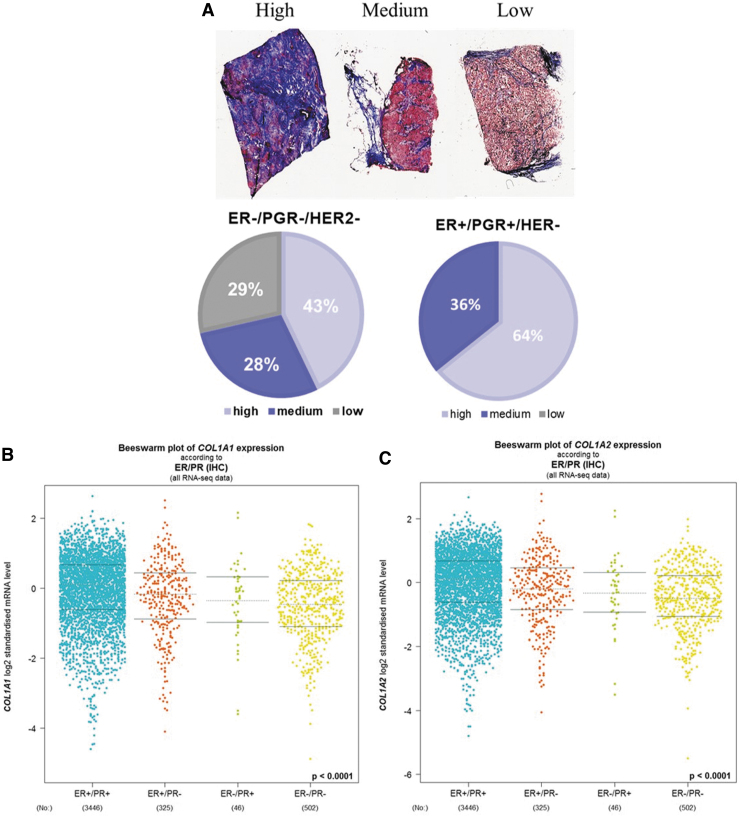
To assess the correlation of fibril COL content and breast cancer subtype, tumor slides from 35 patients were stained with Masson's Trichrome and total fibrillar COL content was evaluated across breast cancer subtypes: ER+/PGR+/human epidermal growth factor receptor 2 (HER2−), ER+/PGR−/HER2−, HER2+, and TNBC (ER–/PGR−/HER2−). Tumors were classified as having high, medium, or low COL content based on the intensity and coverage of stain (Fig. 1A). ER-positive tumors (ER+/PGR+/HER2− and ER+/PGR−/HER2−) had high fibrillar COL content (Table 1 and Fig. 1A). In contrast, the ER– tumor subtypes TNBC and HER2+ had only lower levels of COL (Table 1 and Fig. 1A).
FIG. 1.
TNBC has collagen I expression. (A) Percent of tumors that stained for high, medium, or low fibril collagen expression evaluated with Masson Trichrome stain across breast cancer subtypes based on receptor status: ER+/PGR+/HER– or ER–/PGR–/HER–, n = 21 tumors. Data mining of RNA sequencing of breast cancer tumors for ER and PGR receptor expression of (B) COL1A1 and (C) COL1A2. N = 4319 tumor samples, Database used Breast Cancer Gene-Expression Miner v4.4. Statistically different p < 0.0001. ER, estrogen receptor; HER, human epidermal growth factor receptor; PGR, progesterone receptor, TNBC, triple-negative breast cancer.
Table 1.
Percentage of Tumors That Stained for High, Medium, and Low Collagen Content Grouped by Receptor Status, Origin of Tissue, Age, and Race
| High (%) | Medium (%) | Low (%) | |
|---|---|---|---|
| Receptor status | |||
| ER+/PGR+/HER2– | 64.29 | 35.71 | 0 |
| ER+/PGR–/HER2– | 57.14 | 28.57 | 14.28 |
| HER2+ | 20 | 80 | 0 |
| TNBC (ER–/PGR–/HER2–) | 42.86 | 28.57 | 28.57 |
| Origin | |||
| Lobular | 66.67 | 33.33 | 0 |
| Ductal | 44.44 | 44.44 | 11.11 |
| Lobular/ductal | 50 | 50 | 0 |
| Age | |||
| <45 Years of age | 31.25 | 62.5 | 6.25 |
| >60 Years of age | 69.23 | 23.08 | 7.69 |
| Race | |||
| African American | 57.14 | 28.57 | 14.28 |
| Caucasian | 53.85 | 38.46 | 7.63 |
ER, estrogen receptor; HER; PGR, progesterone receptor, TNBC, triple-negative breast cancer.
To determine if additional patient-specific factors regulated total fibrillar COL content, tumors (irrespective of receptor status) were evaluated for correlations of COL content with patient race (Black and White) and age. Black patients scored as having high fibril COL content were all ER+ and comprised 57.14% of the tumors. All TNBC tumors derived from Black patient samples had either medium (28.57%) or low (14.28%) total fibrillar COL. White patient samples had 53% scored as high COL content and all samples except one were ER+ (Table 1). The median patient age for all tumor slides, irrespective of tumor subtype, was 46 years of age and the age range was between 20 and 84 years of age. Tumor samples below the median age (≤45 years of age) were evaluated in comparison with tumor slides derived from tumor patients above the median age (age ≥60 years). For tissue slides obtained from patients who were ≤45 years of age, 31.25% of these tumors had high fibrillar COL content with four from the HER2+ cohort displaying no elevated COL. For tumor slides obtained from patients over ≥60 years of age, 69% of samples scored as having high COL content; all samples were ER+ with three samples designated as ER+/PGR− (Table 1). These data suggest that the TNBC and HER2+ subtypes have lower levels of total fibrillary COL compared with the ER+ subtypes, irrespective of additional factors such as patient age and race.
To quantifiably demonstrate that receptor status correlated with fibrillar COL expression, RNA sequencing from TCGA and SCANB data sets was used to evaluate COL1A1 and COL1A2 gene expression in ER+, PGR+, and HER2+ tumors. COL1A1 and COL1A2 had significantly (p < 0.0001) decreased expression in ER–/PGR− tumors compared with ER+ tumors (Fig. 1B, C). HER2 status did not correlate with the expression of either COL1A1 or COL1A2 expression (data not shown). Patient age inversely correlated with COL1A1 and COL1A2 gene expression (Supplementary Fig. S2). KM plotter demonstrated no correlation with COL1A1 or COL1A2 expression and survival in ER+/PGR+/HER2−, ER–/PGR−/HER−, or ER–/PGR−/HER2− data sets (data not shown). When segregated by ER status only, ER– tumors had high COL1A1 and COL1A2 expression correlated with poor patient survival (Table 2). To determine if other matrix components correlated with breast cancer overall survival, KM plots were generated for all COL, LAM, and integrin gene expression (Table 2). The hazard ratio varied between ER+ and ER– tumors (Table 2). In summary, 46 genes were found to correlate with patient survival: 16 collagens, 1 elastin, 9 integrins, and 10 laminins. Of those genes, only five were seen in both ER+ and ER– tumors: COL4A2, COL13A1, ITGA5, ITGAE, and ITGAL. There were no observed fibrillary COLs (I, III, V) or FN to be associated with patient survival uniformly across both ER+ and ER– tumors.
Table 2.
Upregulated Extracellular Matrix Genes Correlate with Hazard Ratio Using Kaplan–Meier Survival Analysis for Estrogen Receptor Positive and Estrogen Receptor Negative Patients
| Gene | HR | p | Gene | HR | p |
|---|---|---|---|---|---|
| ER+ | |||||
| COL4A1 | 1.46 | 6.80E–06 | ITGAL | 0.79 | 5.00E–03 |
| COL4A2 | 1.34 | 4.70E–04 | ITGB1 | 1.15 | 9.10E–02 |
| COL6A6 | 0.61 | 9.10E–04 | ITGB4 | 0.78 | 3.60E–03 |
| COL13A1 | 1.29 | 2.00E–03 | ITGB8 | 0.6 | 6.40E–04 |
| COL14A1 | 0.7 | 1.50E–02 | LAMA1 | 0.77 | 1.50E–03 |
| COL17A1 | 0.76 | 1.10E–03 | LAMA2 | 0.76 | 9.80E–04 |
| ELN | 0.74 | 2.50E–04 | LAMA3 | 0.74 | 4.10E–02 |
| ITGA5 | 1.19 | 4.20E–02 | LAMB3 | 0.73 | 2.10E–04 |
| ITGA8 | 0.74 | 4.00E–02 | LAMB4 | 0.83 | 2.50E–02 |
| ITGAE | 1.25 | 7.60E–03 | LAMC2 | 0.76 | 1.20E–03 |
| ER– | |||||
| COL1A1 | 1.56 | 1.3E–04 | COL18A1 | 1.6 | 5.4E–05 |
| COL1A2 | 1.52 | 3.0E–04 | ITGA4 | 0.71 | 3.4E–03 |
| COL3A1 | 1.4 | 3.9E–03 | ITGA5 | 1.46 | 1.1E–03 |
| COL4A1 | 1.48 | 6.6E–04 | ITGAE | 0.79 | 3.7E–02 |
| COL4A2 | 1.28 | 3.4E–02 | ITGAL | 0.76 | 1.7E–02 |
| COL4A5 | 1.3 | 2.2E–02 | ITGAM | 0.77 | 2.2E–02 |
| COL5A1 | 1.54 | 1.8E–04 | ITGAV | 1.4 | 3.3E–03 |
| COL5A2 | 1.51 | 3.6E–04 | ITGB2 | 0.75 | 1.3E–02 |
| COL5A3 | 1.36 | 7.3E–03 | ITGBL1 | 0.77 | 3.8E–06 |
| COL6A1 | 1.28 | 3.2E–02 | LAMA4 | 1.58 | 8.8E–05 |
| COL6A3 | 1.3 | 2.1E–02 | LAMB1 | 1.65 | 1.7E–05 |
| COL10A1 | 1.47 | 9.6E–04 | LAMB2 | 1.33 | 1.2E–02 |
| COL13A1 | 1.49 | 5.9E–04 | LAMC1 | 1.33 | 1.2E–02 |
HR, hazard ratio.
Matrix molecular composition regulates response to therapy
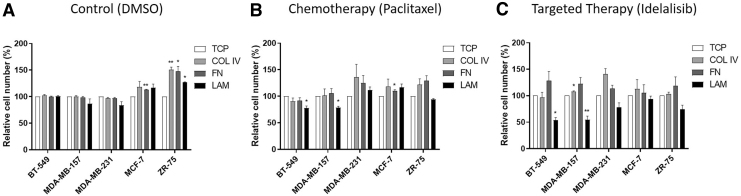
Breast cancers commonly present with heterogeneity between patients, even with common receptor status. Because of the observed differences in matrix fibrillar COL expression and patient outcome, we next sought to determine the effects of matrix composition on the proliferation and survival of a panel of ER+ (MCF-7 and ZR-75) and ER− (BT-549, MDA-MB-157, and MDA-MB-231) cell lines (Fig. 2). Paclitaxel, a commonly used chemotherapy in breast cancer, and idelalisib, a PI3K inhibitor, were chosen for this study.23–25 Idelalisib was chosen because the PI3K oncogenic pathway is suggested to be enhanced in breast cancer and with matrix adhesion.26 Cancer cell lines were seeded on TCP, COL I, COL IV, FN, and LAM-coated plates. These coatings were chosen as COL I is commonly used for breast cancer tumor models; COL IV, FN, and LAM have all been shown to influence patient survival and disease progression. However, a subtype-specific response has not been determined16,27 (Table 2). Cells seeded on TCP control or matrix-coated dishes were treated with DMSO control (Fig. 2A), paclitaxel (Fig. 2B), or idelalisib (Fig. 2C). The results from cells seeded on matrix and treated with DMSO control were normalized to cells seeded on TCP and treated with DMSO control (Fig. 2A). Evaluation of basal differences in cellular response to matrix composition demonstrated that the ER+ cells had increased cell numbers when cultured on matrix coatings, whereas ER− cells displayed no change (Fig. 2A).
FIG. 2.
Matrix composition and receptor status regulate response to therapy. ER– (BT-549, MDA-MB-157, and MDA-MB-231) and ER+ (MCF-7 and ZR-75) cell lines were cultured on TCP, COL IV, FN, and LAM then treated with (A) DMSO, (B) paclitaxel, and (C) idelalisib. Cells were cultured for 24 h and then treated. Results were analyzed using alamarBlue viability assay. Error bars represent SEM, n = 3, p < 0.05. Results were analyzed using alamarBlue viability assay and normalized to TCP for each treatment. Error bars represent SEM, *p ≤ 0.05, **p ≤ 0.01. COL IV, collagen IV; DMSO, dimethyl sulfoxide; FN, fibronectin; LAM, laminin; SEM, standard error of the mean; TCP, tissue culture plastic.
Results from cells seeded on matrix and treated with paclitaxel and idelalisib were normalized to cells cultured on TCP and treated with those therapies (Fig. 2B, C). Following drug treatment, ER− cell lines (BT-549, MDA-MB-157) were significantly sensitized to both chemotherapy and PI3K inhibition when seeded on LAM coatings compared with treatment response on TCP. The BT-549 cell line had an observed decrease in cell numbers of 77.7% ± 4.0% (p < 0.05) and 53.8 ± 4.9 (p < 0.05) on LAM when treated with paclitaxel and idelalisib, respectively, compared with treatment of cells on TCP (Fig. 2B, C). Similarly, the MDA-MB-157 cell line had decreased cell numbers, 78.8 ± 2.5 (p < 0.05) and 54.8 ± 6.8 (p < 0.01) on LAM when treated with paclitaxel and idelalisib, respectively (Fig. 2B, C). MDA-MB-231 did not yield significant differences in paclitaxel or idelalisib treatment. When treated with paclitaxel or idelalisib, the ER+ cell lines were less sensitive to therapy when cultured on all matrix types, MCF-7 cells cultured on FN had a significant increase in the percentage of cells (110.6 ± 1.9, p < 0.05) (Fig. 2B, C). TNBC cell lines cultured on FN also showed less sensitivity to both therapies (Fig. 2B, C). These data suggest that both matrix type and receptor status alter the response to therapy.
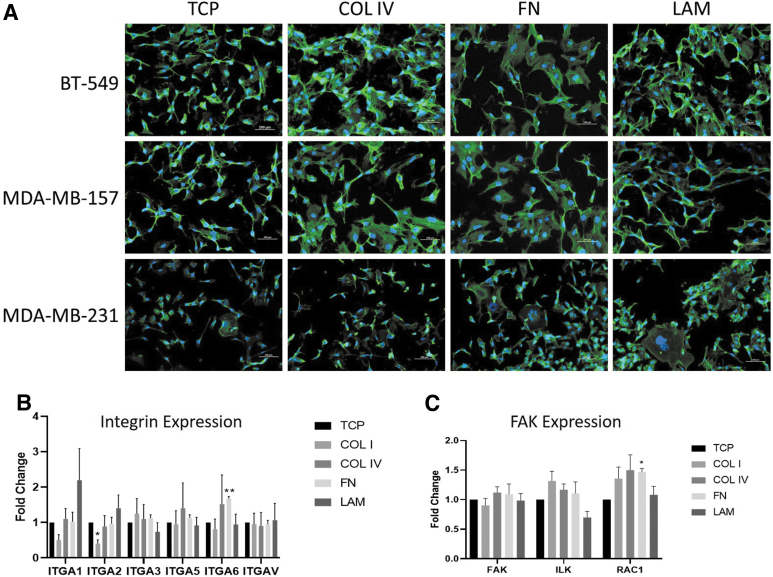
Matrix composition alters morphology, cell adhesion, and signaling
Earlier studies suggest changes in response to therapy on matrix composites are regulated by alterations in cell–matrix adhesions.11,28 Because of the observed differences in the response of the TNBC cell lines on matrix, we sought to determine if matrix composition altered the cellular actin cytoskeleton arrangements in TNBC. TNBC cell lines were seeded on COL IV, FN, and LAM coatings. Results demonstrated that all cell lines expressed different morphologies depending on the ECM substrate they were cultured on (Fig. 3A). BT-549 and MDA-MB-157 appear to be more polarized when cultured on LAM and have a morphology similar to that as TCP. Furthermore, BT-549 and MDA-MB-157 cell lines had more focal adhesions on COL IV and FN-coated dishes (Fig. 3A). MDA-MB-231 cells did not display the same morphological changes as MDA-MB-157 and BT-549 cells; specifically, cells maintained an overall polarized morphology in addition to areas of very large cytoskeletal arrangement. We next investigated alterations to genes associated with adhesion pathways and cancer progression. We looked at integrins associated with different ECMs in cancer: COL (ITGA1/2),29,30 LAM (ITGA3/6),31 and FN binding (ITGAV/5)32 (Fig. 3B). We also looked at focal adhesion kinase (FAK) pathway genes Ras-related C3 botulinum toxin substrate (RAC), integrin linked kinase (ILK), and FAK (Fig. 3C). Analysis of integrin gene expression in MDA-MB-231 showed significant changes in ITGA2 (0.41 ± 0.10-fold change, p < 0.05) when cultured on COL I and ITGA6 (1.68 ± 0.05-fold change, p < 0.01) when cultured on FN (Fig. 3B). RAC1 was significantly upregulated (1.47 ± 0.11-fold change, p < 0.05) when cells were cultured on FN; however, other core intracellular signaling genes (ILK, FAK) were unchanged (Fig. 3C).
FIG. 3.
Matrix composition alters morphology and gene expression of integrin and focal adhesion pathways. (A) ER+ and ER– cells were stained with phalloidin to illuminate differences in actin cytoskeletal arrangement. MDA-MB-231 cell line was cultured on TCP, COL I, COL IV, FN, and LAM for 3 days. Scale bar is 250 μm. Expression of (B) integrins and (C) FAK signaling for MDA-MB-231 cell line. Error bars represent SEM, n = 3, *p ≤ 0.05, **p ≤ 0.01. FAK, focal adhesion kinase.
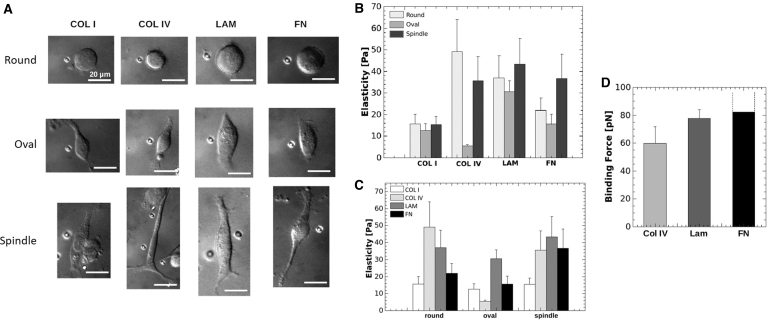
The effects of matrix composition on cellular elasticity and cell binding was next evaluated. Seeding of single-cell suspensions of MDA-MB-231 cells on matrix demonstrated three cell shapes (round, oval, or spindle) following 3 h of adhesion on different matrix types (Fig. 4A–C). There was no observed preference for a singular shape on different matrix types. Because cellular adhesions influence cell elasticity, we then quantified cellular elasticity for each defined cell shape on the different substrates. The effects of matrix on cell shape, elasticity, and binding were investigated using optical tweezers. Single cell suspensions were evaluated on each matrix composite with elasticity measurements performed after 3 h. Of interest, LAM and COL I did not appear to alter cellular elasticity when cells were grouped by shape (Fig. 4A–C).
FIG. 4.
Matrix alters cell shape and elasticity in MDA-MB-231 cell line. MDA-MB-231 cells were seeded on COL I, COL IV, FN, and LAM-coated microscope slides for 3 h. Optical tweezing was used to conduct elasticity measurements of cells corresponding to (A) round, oval, and spindle cell shapes. Each image of cells shows the polystyrene bead that was forced into the cell to measure elasticity. Elasticity results were grouped by (B) shape and (C) matrix coating. (D) Binding force assay results for COL IV, LAM, and FN. Binding force for FN was greater than the optical force generated.
The combined average elasticity of all cells cultured on substrates varied greatly with COL I measuring 14.5 Pa ±3.8 (SD) and LAM measuring LAM was 37 Pa ±9.1 (SD) (Fig. 4A). In contrast, FN and COL IV demonstrated changes in cellular elasticity that was dependent on cell shape and matrix (Fig. 4A). These differences suggest that cell shape may also be regulated by cellular affinity to matrix, particularly in COL IV where elasticities of 5.4 Pa ±0.71 (SD) were measured for oval-shaped cells and 49.1 Pa ±14.8 (SD) in round shape resulting in a nearly 10 × difference.
To determine if altered cell shape was a result of cellular affinity to different matrix composites, we again evaluated singular cell suspension of MDA-MB-231 cell line with optical tweezers. To determine cellular attachment forces, singular MDA-MB-231 cells were allowed to adhere to matrix for 10 s and then the force required to remove cells on each matrix was measured. Results demonstrated that MDA-MB-231 cells did not bind the matrix with equal affinity; FN (> 82.2 pN) induced the greatest cellular affinity and the binding force of the cells was greater than the optical force. LAM (77.8 pN ±6.3) was next followed by COL IV (59.9 pN ±11.8) (Fig. 4D).
Because of differences in response to therapy and observed changes in cell adhesion, we next sought to determine if there were alterations in cellular proliferation and survival pathways. Our initial evaluation of cell number did not demonstrate a significant change in TNBC cell lines (Fig. 3A). We next sough to precisely define if cellular proliferation was altered through the evaluation of cell proliferation marker Ki67. Ki67 expression was not significantly altered in TNBC cell lines when seeded on COL IV, FN, and LAM for 3 days (Supplementary Fig. S3A). In accordance with this finding, western blot analyses of MDA-MB-231 breast cancer cells cultured on TCP, COL IV, FN, and LAM-coated dishes for 3 days demonstrated no significant differences in activation of prosurvival and proliferation pathways AKT or ERK, as evident through western blot for p-AKT SER473 and p-ERK THR202/TYR204 (Supplementary Fig. S3B). Together, these results demonstrate that matrix composition did not significantly alter the long-term activation of AKT, ERK, or proliferation.
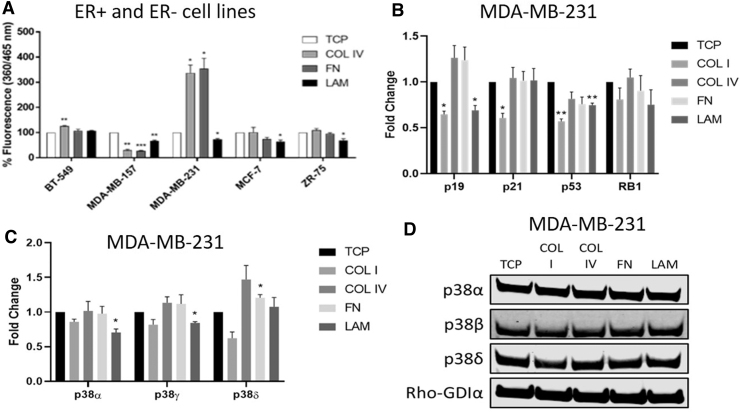
Cellular proliferation was not altered; we next investigated cellular senescence with the detection of the presence of SAβgal. TNBC and ER+ cell lines were seeded on matrix-coated plates (COL, FN, LAM) for 3 days and the amount of SAβgal present was measured using a plate reader (Fig. 5A). BT-549 and MDA-MB-231 cell lines on COL IV had increased SAβgal (126.4 ± 2.0, p < 0.01 and 337.1 ± 31.3, p < 0.05, respectively) compared with TCP. In contrast, MDA-MB-157 cells had significantly decreased levels of SAβgal (30.4 ± 3.6, p < 0.01). FN demonstrated a similar trend, MDA-MB-231 cells had significantly increased levels of SAβgal (354.2 ± 40.8, p < 0.05) and MDA-MB-157 had decreased SAβgal (27.6 ± 2.0 (p < 0.001). When cultured on LAM, MDA-MB-231 (72.9 ± 3.9, p < 0.05), MCF-7 (64.3 ± 6.4, p < 0.05), MDA-MB-157 (67.0 ± 3.3, p < 0.01), and ZR-75 (68.9 ± 6.8, p < 0.05) all demonstrated significant decreases in Saβgal; BT-549 remained unchanged. There was no change in ER+ cells on COL IV and FN.
FIG. 5.
Matrix alters senescence-associated signaling at the transcriptional level. MDA-MB-231 cell line was cultured on TCP, COL I, COL IV, FN, and LAM for 3 days. (A) Cellular senescence was measured using an assay for SAβgal for three ER– cell lines (BT-549, MDA-MB-157, and MDA-MB-231) and two ER+ cell lines (MCF-7 and ZR-75). Expression of (B) MAP kinases and (C) DNA damage pathways were analyzed using qPCR for MDA-MB-231 cell lines. (D) Western blot images of p38 MAP kinases for MDA-MB-231. Error bars represent SEM, *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001.
To determine if cell senescence pathways were altered with cellular adhesion, quantitative PCR for senescence-associated genes (p53, p19, p21, RB1) was performed on MDA-MB-231 cells seeded on matrix (Fig. 5B). MDA-MB-231 cells cultured on LAM had decreased p19 (0.69 ± 0.05-fold, p < 0.05) and p53 (0.75 ± 0.02-fold, p < 0.01) expression compared with TCP control. In comparison, MDA-MB-231 cells cultured on COL I showed significantly decreased expression of p19 (0.65 ± 0.04-fold, p < 0.05), p21 (0.61 ± 0.05-fold, p < 0.05), and p53 (0.57 ± 0.03-fold, p < 0.01) compared with TCP.
We next performed western blot for phosphorylation of p53 S15 adhesion of MDA-MB-231 cells on COL IV, FN, and LAM for 3 days. Results demonstrated no significant changes in p53 on any matrix composite (Supplementary Fig. S4). We next evaluated changes in p38 following adhesion of MDA-MB-231 cells on different matrix substrates. qPCR (Fig. 5C) and western blot (Fig. 5D) for p38 expression and activation in MDA-MB-231 cells cultured on COL I, COL IV, FN, and LAM after 3 days of adhesion demonstrated a significant reduction in expression in p38 gene isoforms but no change in total protein expression. These data demonstrate alterations in senescence-associated pathways at the transcriptional level; however, long-term studies may be needed to see associated protein changes.
Discussion
These data provide novel insight on mechanisms of inter- and intra-tumor heterogeneity in breast cancers. We highlighted the fact that collagen content in tumors can vary significantly from patient-to-patient with respect to receptor status (Table 1 and Fig. 1) and age of the patient (Table 1 and Supplementary Fig. S2). Of particular note are the findings that COL1A1 and COL1A2 have an overall medium to low expression in TNBC tumors, which is in contrast to ER+ tumors that have high collagen expression (Fig. 1B, C). These fundamental differences shine a light on the need for subtype-specific, physiologically relevant, translational models to understand the complexity created by breast cancer heterogeneity. With this insight, we can gain increased understanding of why individuals with similar subtypes respond differently to treatment.
Efforts by other researchers to improve the biological relevance of culture conditions include the use of 3D hydrogel cultures that can be found in microphysiological systems (MPSs). The cancer cells cultured in 3D hydrogels may be cocultured with stromal cells,33,34 cultured as tumor spheroids,35–37 and even as vascularized cocultured tumor spheroids.38,39 Often, rat-tail-derived COL I is chosen as the hydrogel in these cultures because of its wide commercial availability, low cost, and ability to crosslink simply by increasing the temperature from 4°C to 37°C. With the data presented here, we demonstrate the need for designing engineered, subtype-specific ECM to be used for in vitro therapeutic screening. Combining subtype-specific ECM with advanced hydrogel MPSs could be key in improving clinical translation of models.
Breast cancer progression and resistance to therapy, which require more physiologically relevant models to accurately study, is achieved through multiple mechanisms including increased survival and proliferation pathways, induction of cell dormancy/cellular senescence, immune evasion, and cancer stem cell phenotype. Here we demonstrate, for the first time, a novel role for tumor ECM as a mediator of morphology, adhesion, and potentially even cellular senescence. Preliminary results indicate that different matrix components elicit subtype-specific responses. Here, matrix induced a pro-survival phenotype in ER+ cancer cells (Fig. 2A). Matrix is a key factor in progression, drug resistance and enhanced proliferation through matrix stiffening in both ER+ and ER– cells.11–14
Survival and proliferation are increased by the upregulation of integrin signaling pathways. Here, we see no observed increase in proliferative markers (Ki67) or pathways in our TNBC cells on COL IV (Supplementary Fig. S3). These findings could be explained by the similarities in stiffness of TCP and thinly coated dishes and suggest that the proproliferative effect is mediated by substrate stiffness, not composition. One limit to this study was that we utilized a single 3-day time point for endpoint analysis. This time point was used to allow cells time to acclimate to the new culture condition; however, additional time points may be required to see translational effects of matrix adhesion as induced protein changes were not observed. Future work should include multiple time points.
Another limitation of this study was the use of monoculture, which is not representative of the heterogeneous cell populations found in vivo. However, our goal was not to recreate in vivo conditions, but instead to demonstrate that even changes in individual ECM proteins can dramatically alter cellular behavior and response. Future work could aim at addressing this limitation by including more complex culture conditions.
We identified dysregulation in cellular senescence pathways at the transcriptional level (Fig. 5). Cellular senescence is a potent mechanism to suppress proliferation. However, senescent cells can secrete molecules that promote tumorigenesis and transform cells in the microenvironment.40,41 This is observed in cancer cells in metastatic sites, during evasion of immune response, and in cells that fail to respond to chemotherapy. The TNBC cell lines demonstrated the largest increase in induction of cell senescence on different matrix substrates, providing new insight on drug resistance. Cells that undergo senescence as a defense mechanism can propagate tumorigenesis through their secretions.42 We did not see increased expression or activation of p53 in MDA-MB-231 cells that had increased Saβgal (Fig. 5A). p19 and p21 were downregulated, suggesting that other mechanisms may be governing this process (Fig. 5B). Future work should include extended time points to elucidate the discrepancy in data of transcriptional versus translational changes in senescence-associated pathways and a more thorough profiling of senescence-associated markers.
We also observed changes in cellular elasticity (Fig. 4A–E), morphology (Figs. 3A and 4A–C), and altered senescence-associated pathway gene expression (Fig. 5B–E). Here we demonstrated significant changes in the expression of integrins in MDA-MB-231 cells (Fig. 3B). Loss of ITGA2 drives metastasis in multiple cancers, including breast.43–46 Upregulation of ITGA6 is implicated in metastasis and reduced sensitivity to radiotherapies.47–50 Altered expression of these integrins could explain altered cellular morphologies, elasticities, and cellular binding (Fig. 4). Although we only profiled a small subset of integrin proteins based on their association with LAM, COL, and FN, the role for integrin expression and activation of oncogenic pathways is undisputed.51 Preclinical success has been achieved for integrin inhibitors52; using ECM composition to select integrin inhibitors would create precision medicine for patients.
Conclusion
Although further studies are needed, these preliminary results suggest that novel matrix mechanisms are responsible for differences in the cellular response to therapy and that these differences are subtype specific. The informatics data presented in Figure 3 and Table 1 can guide future studies to identify those mechanisms that play a role in response to therapy and patient prognosis. The research presented here suggests that matrix composition plays a key role in breast cancer response to therapy. We demonstrate for the first time that matrix composition alters cell elasticity and morphology. Pinpointing the mechanism for matrix-induced drug resistance and the source of differential matrix remodeling has the potential to elucidate the mechanism for the heterogeneous response of tumors to therapy. In addition, novel neoadjuvant therapies designed to target integrin adhesion may prove useful in treating TNBC, which currently has no molecular targets, and provide better patient precision medicine.
Supplementary Material
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Disclosure Statement
No competing financial interests exist.
Funding Information
Supported in part by U54 GM104940 from the National Institute of General Medical Sciences of the National Institutes of Health, which funds the Louisiana Clinical and Translational Science Center.
Supplementary Material
References
- 1. Glass, A.G., Lacey, J.V., Carreon, J.D., and Hoover, R.N.. Breast cancer incidence, 1980–2006: combined roles of menopausal hormone therapy, screening mammography, and estrogen receptor status. J Natl Cancer Inst 99, 1152, 2007 [DOI] [PubMed] [Google Scholar]
- 2. Mancuso, M.R., and Massarweh, S.A.. Endocrine therapy and strategies to overcome therapeutic resistance in breast cancer. Curr Probl Cancer 40, 95, 2016 [DOI] [PubMed] [Google Scholar]
- 3. Samanta, D., Gilkes, D.M., Chaturvedi, P., Xiang, L., and Semenza, G.L.. Hypoxia-inducible factors are required for chemotherapy resistance of breast cancer stem cells. Proc Natl Acad Sci U S A 111, E5429, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 4. Barcus, C.E., O'Leary, K.A., Brockman, J.L., et al. Elevated collagen-I augments tumor progressive signals, intravasation and metastasis of prolactin-induced estrogen receptor alpha positive mammary tumor cells. Breast Cancer Res 19, 9, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Getzenberg, R.H., Pienta, K.J., Huang, E.Y., Murphy, B.C., and Coffey, D.S.. Modifications of the intermediate filament and nuclear matrix networks by the extracellular matrix. Biochem Biophys Res Commun 179, 340, 1991 [DOI] [PubMed] [Google Scholar]
- 6. Neophytou, C., Boutsikos, P., and Papageorgis, P.. Molecular mechanisms and emerging therapeutic targets of triple-negative breast cancer metastasis. Front Oncol 8, 31, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Insua-Rodríguez, J., and Oskarsson, T.. The extracellular matrix in breast cancer. Adv Drug Deliv Rev 97, 41, 2016 [DOI] [PubMed] [Google Scholar]
- 8. Xiong, G., Deng, L., Zhu, J., Rychahou, P.G., and Xu, R.. Prolyl-4-hydroxylase α subunit 2 promotes breast cancer progression and metastasis by regulating collagen deposition. BMC Cancer 14, 1, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Provenzano, P.P., Eliceiri, K.W., Campbell, J.M., Inman, D.R., White, J.G., and Keely, P.J.. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med 4, 38, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu, J., Shen, J.-X., Wu, H.-T., et al. Collagen 1A1 (COL1A1) promotes metastasis of breast cancer and is a potential therapeutic target. Discov Med 25, 211, 2018 [PubMed] [Google Scholar]
- 11. Schrader, J., Gordon-Walker, T.T., Aucott, R.L., et al. Matrix stiffness modulates proliferation, chemotherapeutic response, and dormancy in hepatocellular carcinoma cells. Hepatology 53, 1192, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Provenzano, P.P., Inman, D.R., Eliceiri, K.W., and Keely, P.J.. Matrix density-induced mechanoregulation of breast cell phenotype, signaling and gene expression through a FAK-ERK linkage. Oncogene 28, 4326, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Barcus, C.E., Holt, E.C., Keely, P.J., Eliceiri, K.W., and Schuler, L.A.. Dense collagen-I matrices enhance pro-tumorigenic estrogen-prolactin crosstalk in MCF-7 and T47D breast cancer cells. PLoS One 10, e0116891, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Boyd, N.F., Li, Q., Melnichouk, O., et al. Evidence that breast tissue stiffness is associated with risk of breast cancer. PLoS One 9, e100937, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang, W., Zhou, M., Dorsey, T.H., et al. Integrated proteotranscriptomics of breast cancer reveals globally increased protein-mRNA concordance associated with subtypes and survival. Genome Med 10, 94, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ioachim, E., Charchanti, A., Briasoulis, E., et al. Immunohistochemical expression of extracellular matrix components tenascin, fibronectin, collagen type IV and laminin in breast cancer: their prognostic value and role in tumour invasion and progression. Eur J Cancer 38, 2362, 2002 [DOI] [PubMed] [Google Scholar]
- 17. Pupa, S.M., Ménard, S., Forti, S., and Tagliabue, E.. New insights into the role of extracellular matrix during tumor onset and progression. J Cell Physiol 192, 259, 2002 [DOI] [PubMed] [Google Scholar]
- 18. Andersson, M., Madgavkar, A., Stjerndahl, M., et al. Using optical tweezers for measuring the interaction forces between human bone cells and implant surfaces: system design and force calibration. Rev Sci Instrum 78, 074302, 2007 [DOI] [PubMed] [Google Scholar]
- 19. Decombe, J.-B., Huant, S., and Fick, J.. Single and dual fiber nano-tip optical tweezers: trapping and analysis. Opt Express 21, 30521, 2013 [DOI] [PubMed] [Google Scholar]
- 20. Yousafzai, M.S., Ndoye, F., Coceano, G., et al. Substrate-dependent cell elasticity measured by optical tweezers indentation. Opt Lasers Eng 76, 27, 2016 [DOI] [PubMed] [Google Scholar]
- 21. Nagy, Á., Lánczky, A., Menyhárt, O., and Győrffy, B.. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep 8, 9227, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jézéquel, P., Frénel, J.-S., Campion, L., et al. bc-GenExMiner 3.0: new mining module computes breast cancer gene expression correlation analyses. Database (Oxford) 2013, bas060, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Furman, R.R., Sharman, J.P., Coutre, S.E., et al. Idelalisib and rituximab in relapsed chronic lymphocytic leukemia. N Engl J Med 370, 997, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Adam, E., Kim, H.N., Gang, E.J., et al. The pi3kδ inhibitor idelalisib inhibits homing in an in vitro and in vivo model of B ALL. Cancers (Basel) 9, 121, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sparano, J.A., Wang, M., Martino, S., et al. Weekly paclitaxel in the adjuvant treatment of breast cancer. N Engl J Med 358, 1663, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mukohara, T. PI3K mutations in breast cancer: prognostic and therapeutic implications. Breast Cancer (Dove Med Press) 7, 111, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fernandez-Garcia, B., Eiró, N., Marín, L., et al. Expression and prognostic significance of fibronectin and matrix metalloproteases in breast cancer metastasis. Histopathology 64, 512, 2014 [DOI] [PubMed] [Google Scholar]
- 28. Wu, C., and Dedhar, S.. Integrin-linked kinase (ILK) and its interactors: a new paradigm for the coupling of extracellular matrix to actin cytoskeleton and signaling complexes. J Cell Biol 155, 505, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley, J., Knight, C.G., Farndale, R.W., Barnes, M.J., and Liddington, R.C.. Structural basis of collagen recognition by integrin alpha2beta1. Cell 101, 47, 2000 [DOI] [PubMed] [Google Scholar]
- 30. Cheli, Y., Kanaji, S., Jacquelin, B., Chang, M., Nugent, D.J., and Kunicki, T.J.. Transcriptional and epigenetic regulation of the integrin collagen receptor locus ITGA1-PELO-ITGA2. Biochim Biophys Acta 1769, 546, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ramovs, V., Te Molder, L., and Sonnenberg, A.. The opposing roles of laminin-binding integrins in cancer. Matrix Biol 57, 213, 2017 [DOI] [PubMed] [Google Scholar]
- 32. Akiyama, S.K., Aota, S., and Yamada, K.M.. Function and receptor specificity of a minimal 20 kilodalton cell adhesive fragment of fibronectin. Cell Adhes Commun 3, 13, 1995 [DOI] [PubMed] [Google Scholar]
- 33. Lan, S.-F., and Starly, B.. Alginate based 3D hydrogels as an in vitro co-culture model platform for the toxicity screening of new chemical entities. Toxicol Appl Pharmacol 256, 62, 2011 [DOI] [PubMed] [Google Scholar]
- 34. Shafran, Y., Deutsch, M., Afrimzon, E., et al. Co-culture hydrogel micro-chamber array-based plate for anti-tumor drug development at single-element resolution. Toxicol In Vitro 71, 105067, 2021 [DOI] [PubMed] [Google Scholar]
- 35. Liang, Y., Jeong, J., DeVolder, R.J., et al. A cell-instructive hydrogel to regulate malignancy of 3D tumor spheroids with matrix rigidity. Biomaterials 32, 9308, 2011 [DOI] [PubMed] [Google Scholar]
- 36. Kaur, P., Ward, B., Saha, B., et al. Human breast cancer histoid: an in vitro 3-dimensional co-culture model that mimics breast cancer tissue. J Histochem Cytochem 59, 1087, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yip, D., and Cho, C.H.. A multicellular 3D heterospheroid model of liver tumor and stromal cells in collagen gel for anti-cancer drug testing. Biochem Biophys Res Commun 433, 327, 2013 [DOI] [PubMed] [Google Scholar]
- 38. Nashimoto, Y., Okada, R., Hanada, S., et al. Vascularized cancer on a chip: the effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials 229, 119547, 2020 [DOI] [PubMed] [Google Scholar]
- 39. Paek, J., Park, S.E., Lu, Q., et al. Microphysiological engineering of self-assembled and perfusable microvascular beds for the production of vascularized three-dimensional human microtissues. ACS Nano 13, 7627, 2019 [DOI] [PubMed] [Google Scholar]
- 40. Coppé, J.-P., Patil, C.K., Rodier, F., et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol 6, 2853, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baker, D.J., Alimirah, F., van Deursen, J.M., Campisi, J., and Hildesheim, J.. Oncogenic senescence: a multi-functional perspective. Oncotarget 8, 27661, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chandeck, C., and Mooi, W.J.. Oncogene-induced cellular senescence. Adv Anat Pathol 17, 42, 2010 [DOI] [PubMed] [Google Scholar]
- 43. Ramirez, N.E., Zhang, Z., Madamanchi, A., et al. The α2β1 integrin is a metastasis suppressor in mouse models and human cancer. J Clin Invest 121, 226, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sawhney, R.S., Cookson, M.M., Omar, Y., Hauser, J., and Brattain, M.G.. Integrin alpha2-mediated ERK and calpain activation play a critical role in cell adhesion and motility via focal adhesion kinase signaling: identification of a novel signaling pathway. J Biol Chem 281, 8497, 2006 [DOI] [PubMed] [Google Scholar]
- 45. Robertson, J.H., Yang, S.Y., Winslet, M.C., and Seifalian, A.M.. Functional blocking of specific integrins inhibit colonic cancer migration. Clin Exp Metastasis 26, 769, 2009 [DOI] [PubMed] [Google Scholar]
- 46. Ding, W., Fan, X.-L., Xu, X., et al. Epigenetic silencing of ITGA2 by MiR-373 promotes cell migration in breast cancer. PLoS One 10, e0135128, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pawar, S.C., Dougherty, S., Pennington, M.E., et al. alpha6 integrin cleavage: sensitizing human prostate cancer to ionizing radiation. Int J Radiat Biol 83, 761, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim, H.I., Huang, H., Cheepala, S., Huang, S., and Chung, J.. Curcumin inhibition of integrin (alpha6beta4)-dependent breast cancer cell motility and invasion. Cancer Prev Res (Phila) 1, 385, 2008 [DOI] [PubMed] [Google Scholar]
- 49. Mukhopadhyay, R., Theriault, R.L., and Price, J.E.. Increased levels of alpha6 integrins are associated with the metastatic phenotype of human breast cancer cells. Clin Exp Metastasis 17, 325, 1999 [DOI] [PubMed] [Google Scholar]
- 50. Hu, T., Zhou, R., Zhao, Y., and Wu, G.. Integrin α6/Akt/Erk signaling is essential for human breast cancer resistance to radiotherapy. Sci Rep 6, 33376, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li, Z.-H., Zhou, Y., Ding, Y.-X., Guo, Q.-L., and Zhao, L.. Roles of integrin in tumor development and the target inhibitors. Chin J Nat Med 17, 241, 2019 [DOI] [PubMed] [Google Scholar]
- 52. Alday-Parejo, B., Stupp, R., and Rüegg, C.. Are integrins still practicable targets for anti-cancer therapy? Cancers (Basel) 11, 978, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.