Abstract
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) has infected over 200 million people throughout the world as of August 2021. There are currently no approved treatments providing high chance of recovery from a severe case of coronavirus disease 2019 (COVID-19) caused by SARS-CoV-2, and the beneficial effect of Remdesivir and passive immunization therapies may only be seen when administered early on disease onset. The emergence of variants is also raising concerns regarding the efficacy of antibody therapies, antivirals, and vaccines. Therefore, there is still a need to develop new antivirals. Here, we investigated the suitability of primary human epithelial cells from the trachea/bronchia (NHBE) and small airway (SAEC) as lung models of SARS-CoV-2 infection to determine, whether the microbicide polyphenylene carboxymethylene (PPCM) has antiviral activity against SARS-CoV-2. Both NHBE and SAEC expressed proteins required for virus entry in lung epithelial cells. However, these cells were only low to moderately permissive to SARS-CoV-2 as titers increased at best by 2.5 log10 during an 8-day kinetic. Levels of replication in SAEC, unlike in NHBE, were consistent with data from other studies using human normal tissues or air-liquid interface cultures, suggesting that SAEC may be more relevant to use than NHBE for drug screening. PPCM EC50 against SARS-CoV-2 was between 32 and 132 μg/ml with a selectivity index between 12 and 41, depending on the cell type and the infective dose used. PPCM doses were consistent with those previously showing effect against other human viruses. Finally, PPCM antiviral effect observed in SAEC was in line with reduction of inflammatory markers observed overly expressed in severe COVID-19 patients. Altogether, our data support the fact that PPCM should be further evaluated in vivo for toxicity and antiviral activity against SARS-CoV-2.
Keywords: SARS-CoV-2, Coronavirus, Respiratory infections, Polyphenylene carboxymethylene, PPCM
1. Introduction
Mankind is currently amid the worst pandemic in 100 years that is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for coronavirus disease 2019 (COVID-19). The first identification of SARS-CoV-2 dates back to December 2019 (Zhou et al., 2020), and as of August 2021, there has been more than 200 million confirmed cases and 4.2 million deaths worldwide. Until recently the US was affected the hardest and 35 million cases with 614,000 deaths have been reported. The number of cases is still growing worldwide (Dong et al., 2020), despite the availability of several vaccine platforms (Nature, 2021), and the well-documented beneficial effects of taking social and behavioral measures (Chu et al., 2020; Prather et al., 2020; Studdert and Hall, 2020). COVID-19 symptoms can range from none to a mild or moderate respiratory disease, and progress into severe disease accompanied by respiratory distress, multiple organ failure, and death (Carvalho et al., 2021; To et al., 2021). Survivors may exhibit permanent respiratory injuries and lifelong cognitive impairments (Candan et al., 2020; Holmes et al., 2021; Moreno-Perez et al., 2021).
Unfortunately, in this rapidly evolving situation, a decrease in efficacy of some SARS-CoV-2 vaccines, and monoclonal antibody therapies has already been observed with emergence of SARS-CoV-2 variants (Chen et al., 2021; Diamond et al., 2021). Other antivirals and anti-inflammatory treatments are available to treat COVID-19 including the broad-spectrum nucleoside-analogue inhibitor Remdesivir (Veklury®, FDA-approved), and immunomodulators (under emergency use authorization) but they only have a modest reduction in mortality (Beigel et al., 2020; Calabrese and Calabrese, 2021; Paladugu and Donato, 2020; Ranjbar et al., 2021). Some of these treatments come at high cost as well. Therefore, there is still a need for identifying antivirals with a broad-spectrum efficacy to counteract the limitation of virus-specific vaccines and passive immunization treatments, as well as to improve the efficacy of other antivirals through potential combinatorial treatment.
Polyphenylene Carboxymethylene (PPCM) is a mandelic acid condensation polymer under development as a multipurpose prevention technology, and has a low cost of manufacturing under different formulations (Weitzel et al., 2020). PPCM has previously demonstrated microbicidal broad-spectrum activity against sexually transmitted infections in vitro and in animal models. Focusing on viruses, PPCM specifically prevented HIV, HSV, BPV and ebolavirus infection by binding to both the virus envelope glycoproteins and target cells (Chang et al., 2007; Escaffre et al., 2019; Herold et al., 2002; Mesquita et al., 2008; Weitzel et al., 2020; Zaneveld et al., 2002) while having low cell cytotoxicity in the vaginal flora (Zaneveld et al., 2002). These characteristics suggest that PPCM is a small molecule candidate to be tested as an antiviral against a broad range of human infectious diseases.
Here, we investigated in vitro antiviral activity of PPCM against SARS-CoV-2 using undifferentiated primary human respiratory epithelial cells from the trachea/bronchia (NHBE) and small airways (SAEC). NHBE and SAEC were rather low to moderately permissive to infection despite detecting in each of them expression of proteins responsible in virus attachment and entry process. High PPCM doses were not cytotoxic and interfered with both SARS-CoV-2 replication and virus-induced inflammatory response in a dose dependent manner.
2. Material and methods
2.1. Cells, PPCM, and virus
Normal NHBE derived from the bronchial tube and normal SAEC derived from the distal airspace were respectively obtained from the Clonetics® Bronchial/Tracheal or Human Small Airway Epithelial Cell System (Lonza) and maintained at 37 °C, 5% CO2 in appropriate media per manufacturer's recommendation. The NHBE donor was a 70-year-old tobacco-user Caucasian male. The SAEC donor was a tobacco-free 62-year-old Asian female. None of the donors had a history of diabetes, heart disease or hypertension. Both cell types were used at passage number 4 to 7 from original vials. Optimal seeding density of each cell type (biological quadruplicates) for up to 8 days of culture was carried out using tetrazolium dye (MTT), as previously described (Escaffre et al., 2019). Vero E6 (provided by R. Baric), A549 (ATCC, CCL-185), Vero E6 (ATCC, CRL1586), and Vero E6 stably expressing angiotensin-converting enzyme 2 (ACE2) (Oguntuyo et al., 2021) were cultured at 37 °C and 5% CO2 in Dulbecco's modified Eagle's medium (Gibco) containing 10% fetal bovine serum, and 2 μg/ml puromycin for selection where appropriate. All cells were mycoplasma-free.
Sterile PPCM polymer stock solution was prepared in phosphate-buffered saline (PBS) by Alera Labs under GMP and provided by Yaso Therapeutics Inc. PPCM stock was then diluted in the appropriate media to obtain concentrations ranging from 1 mg/ml to 10 μg/ml. The cell viability assay using MTT (Sigma) was also used to determine the 50% Cytotoxic Concentration (CC50) of PPCM for each cell type (biological quadruplicates). PPCM concentrations used to illustrate virus replication and antiviral effect were arbitrarily chosen, while being below the corresponding CC50 for each cell type at different time points. Additional concentrations within the same range were tested to allow for the half maximal effective concentration (EC50) calculation at peak virus titer in SAEC and NHBE cells. CC50 and EC50 were calculated by nonlinear regression analysis after normalization of data. The selectivity index (SI) was calculated by dividing the CC50 value by the EC50 value.
SARS-CoV-2 stock was derived from the first COVID-19 patient identified in the US in Washington state (GenBank Accession#: MT020880). The isolate (2019-nCoV/USA_WA1/2020) (Holshue et al., 2020) was provided by the World Reference Center for Emerging Viruses and Arboviruses (WRCEVA) at the University of Texas Medical Branch at Galveston (UTMB) and propagated on Vero E6 cells. Titration of the virus stock and SAEC/NHBE samples was performed by plaque assay using Vero E6 cells, 300 μl inoculum, and neutral red counter staining after 3 days incubation. Titers were reported as log10 pfu/ml with a limit of detection set a 1.52. All infectious work was performed in a biosafety level 4 laboratory (BSL4) at the Galveston National Laboratory, UTMB.
2.2. Western blot
Cell lysates were prepared from pelleted cells resuspended in a 2X Laemmli buffer (4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.125 M Tris-HCl), clarified, quantified on Nanodrop, boiled at 95 °C for 5min, and resolved on hand cast 4–8% SDS-PAGE gels. Assays were performed with technical duplicates. Note that 60 μg control protein lysates (Vero E6, and Vero E6 ACE2) per well were used, whereas 100 μg were used for all other samples. Proteins were transferred onto an Immuno-Blot PVDF membrane (Bio-Rad), and nonspecific binding sites were blocked with Tris-buffered saline–0.1% Tween 20 (TBST) containing 5% Bovine Serum Albumin powder (Thermo Fisher Scientific). The membranes were then incubated in TBST 2% BSA with the primary antibodies overnight at 4 °C, followed by incubation with the goat anti-rabbit IgG horseradish peroxidase (HRP)-conjugated secondary antibody (Abcam, #7090, dilution by 1000 or 2000) for 60 min. Primary antibodies used in this study were rabbit anti-ACE2 (Invitrogen, #MA5-32307, d/500), anti-TMPRSS2 (Invitrogen, #PA5-14264, d/5000) and anti-actin (Cell signaling, clone 13E5, d/5000). Stripping (2% SDS, 0.05 M Tris-HCl, 1% 2-mercaptoethanol) was performed on blots targeting tmprss2 (about 55 KDa) to then probe for actin (45 KDa) and avoid signal overlapping for densitometry analysis. Proteins of interest were visualized with a SuperSignal West Pico chemi-luminescent reagent (Thermo Fisher Scientific) on a C-DiGit Blot Scanner imaging system (LI-COR Biosciences). Time of exposure was up to 12 min. For densitometry analysis from the digital images, background was calculated using the median intensity of pixels at the top and bottom border of a band of interest. Normalization of protein amount was performed using the internal actin loading control.
2.3. Virus replication kinetics
SAEC and NHBE were seeded in 24-well plates the day prior infection at 0.35 × 106 cells/well in a 1 ml volume (biological duplicates or triplicates) for assessment of virus replication, nucleocapsid (N) gene expression, as well as cytokine and chemokine quantification. Cells were first treated with PPCM for 1 h (h) prior infection. Medium was then removed but no washes were performed between PPCM pretreatment and 1h virus contact. Multiplicity of infection (MOI) of 5 or 0.1 was used. Cells were then washed 3 times (1 ml) with warm 1 × phosphate-buffered saline (PBS), and 1 ml medium per well supplemented with PPCM was added, where appropriate. PPCM treatment was maintained from day 1–8 post infection (pi) with daily addition of media with PPCM (200 μl), where appropriate, for the purpose of maintaining treatment with freshly prepared PPCM and also to compensate for volume loss (200 μl) by sample collection.
2.4. Real-time (RT)-PCR assay
Total cellular RNA from SAEC and NHBE cells (biological duplicates or triplicates) was extracted using TRIzol reagent (Invitrogen), and then purified using Direct-zol RNA miniprep (Zymo Research), following manufacturer's recommendations. Quantitative reverse transcription (RT)-PCR assays were performed with a One-step QuantiFast Probe RT-PCR kit (Qiagen) using a single FAM-BHQ1 probe with two unlabeled oligonucleotide primers (Integrated DNA Technologies) that recognize the gene sequence corresponding to SARS-CoV-2 N gene. Quantification of GAPDH rRNA level was performed with the 20X GAPDH Pre-Developed TaqMan assay reagent (Thermo Fisher Scientific) and used as an endogenous control. Cut-off for SARS-CoV-2 positive samples was determined from mock-infected SAEC and NHBE cells and set at ΔCt = 40 (PCR run of 40 cycles total). The N gene expression fold change in PPCM-treated cells was calculated using samples from vehicle-treated infected cells as reference, after normalization of values using the housekeeping GAPDH rRNA gene data. Duplicate threshold cycle (Ct) values of each sample were analyzed using the comparative delta-delta Ct method (2–ΔΔCt). Primer and probe sequences were those used by the CDC as following: 2019-nCoV_N1-F: GACCCCAAAATCAGCGAAAT, 2019-nCoV_N1-R: TCTGGTTACTGCCAGTTGAATCTG, and 2019-nCoV_N1-P: ACCCCGCATTACGTTTGGTGGACC.
2.5. Bio-plex assay
Cytokine and chemokine concentrations in cell-free supernatants of SAEC (biological triplicates) were determined using a combination of Bio-Plex Pro Human inflammation panel 1 37-plex and cytokine group 1 panel 27-plex Immunoassay kits (Bio-Rad). The concentration of 59 analytes were quantified. Non-inactivated samples from mock- and virus-infected SAEC cells were analyzed on a BioRad BioPlex 2200 platform at BSL4.
2.6. Statistical analysis
Comparison of nonlinear fits of PPCM concentration versus cytotoxicity (CC50) or virus titer reduction (EC50) sets was performed through the least squares regression fitting method and then the extra sum-of-squares F test. One-way analysis of variance (ANOVA) with Tukey's post-hoc analysis was used in multiple comparisons of data from the virus replication kinetics and the immune response multiplex assays. Null hypotheses were rejected at p-values less than 0.05. All data presented in figures represent means ± standard error or standard deviation (*p < 0.05, **p < 0.01, ***p < 0.001). Statistical analysis was performed with GraphPad Prism 9 software (GraphPad Software, Inc.).
3. Results
3.1. Suitability of primary human respiratory epithelial cells to study SARS-CoV-2 replication
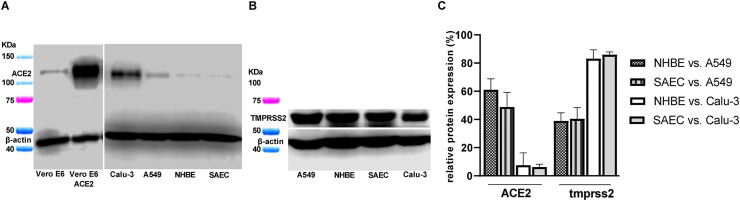
NHBE and SAEC were used in this study to assess PPCM potential antiviral activity against SARS-CoV-2. Both cell types equally expressed amounts (p > 0.05) of the angiotensin-converting enzyme 2 (ACE2) cell receptor and the type II transmembrane serine protease 2 (TMPRSS2) (Fig. 1 A and B) involved in SARS-CoV-2 entry into host cells (Hoffmann et al., 2020a; Walls et al., 2020). Interestingly, there was a 2- and 13.5-fold decrease of ACE2 expression in both primary cell types (Fig. 1A-C) when compared to the low-level virus replicating A549 or the highly permissive Calu-3 cell line (Blanco-Melo et al., 2020; Harcourt et al., 2020; Hoffmann et al., 2020a). Besides, TMPRSS2 expression level in NHBE and SAEC was 2.5 and 1.2-fold lower (Fig. 1B and C) than the counterpart in A549 or Calu-3 cell line, respectively. Altogether these results suggests that both NHBE and SAEC cells are susceptible to SARS-CoV-2 infection.
Fig. 1.
NHBE and SAEC express proteins involved in SARS-CoV-2 entry. (A) ACE2, and (B) TMPRSS2 protein expression in both NHBE, and SAEC as well as in several cell lines as controls (Vero E6, and Vero E6 ACE2) or for comparison purposes (Calu-3, and A549). (C) Relative ACE2 and TMPRSS2 protein expression in both NHBE and SAEC compared to A549 or Calu-3 cell line. (A–C) Assay performed using technical duplicates, and data are shown as mean with standard deviation.
3.2. SARS-CoV-2 replication in primary human respiratory epithelial cells
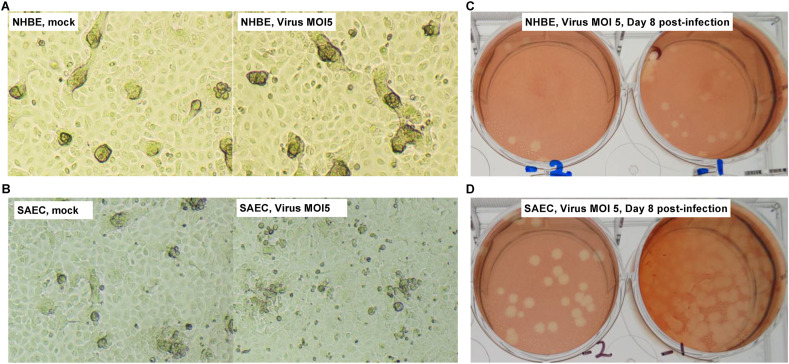
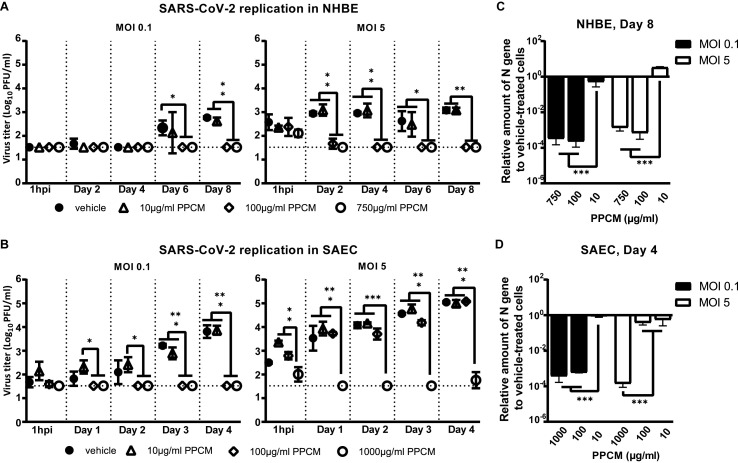
SARS-CoV-2 did not cause any CPE over 8 days of infection using a high infective dose (MOI) of 5, regardless of the cell type (Fig. 2 A and B). Therefore, virus titration of culture supernatants was only initially performed at day 8 post infection (pi). Consistent with the fact that a little amount of ACE2 was noted in both cell types compared to A549 or Calu-3 cell line, infectious virus was detected with low titers averaging 103 and 104 pfu/ml in respectively NHBE or SAEC culture supernatants (Fig. 2C and D), nevertheless suggesting that both cell types are moderately permissive to infection.
Fig. 2.
NHBE and SAEC are permissive to SARS-CoV-2 infection. Brightfield observation of (left) mock- and (right) virus-infected (A) NHBE or (B) SAEC cultures at day 8 post-infection (pi). Imaging at 10× magnification. Representative visualization of virus-induced plaques from 2 10-fold serial dilutions (−1 and −2) of (C) NHBE or (D) SAEC culture supernatants at day 8 pi. Here the virus titer from a NHBE or SAEC culture is respectively at 103.2 and 104.0 pfu/ml.
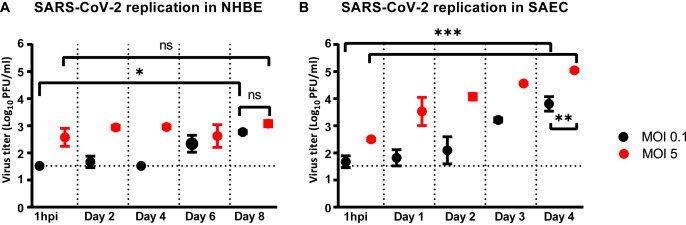
Further analysis of NHBE culture samples collected at earlier time points and from cells infected with a lower MOI showed similar peak titers (p > 0.05) that averaged 102.9 pfu/ml (Fig. 3 A). However, this was reached either on the last day or maintained to comparable levels throughout the course of infection using a MOI of 0.1 or 5, respectively (Fig. 3A). Specifically, virus titer only increased by 1.4 (p < 0.05) and 0.5 log(10) within 8 days of infection using respectively a MOI of 0.1 and 5 (Fig. 3A), altogether suggesting that virus replication in NHBE cells is sub-optimal. Likewise, virus replication in SAEC reached peak titers of 103.8 (MOI 0.1) and 105 (MOI 5) pfu/ml both by day 4 pi (p < 0.01) which respectively corresponded to an increase of 2.1 (p < 0.001) and 2.5 log(10) (p < 0.001) from the 1h pi time point (Fig. 3B). Starting day 5 pi, virus replication in SAEC then entered a plateau phase or progressively decreased by about 1 log(10) by the end of the study using a MOI of 0.1 or 5, respectively (data not shown). Interestingly, peak titers in SAEC at day 4 pi were 0.9 and 2.1 log(10) higher than in NHBE at day 8 pi using respectively a MOI of 0.1 or 5 (Fig. 3B), altogether suggesting that virus replication is more productive in SAEC than in NHBE.
Fig. 3.
SARS-CoV-2 replication kinetics in NHBE and SAEC. Virus replication kinetics in (A) NHBE or (B) SAEC following a low and high infective dose inoculation. The horizontal dotted lines correspond to the detection limit. ns, not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001 (ANOVA, Tukey's test).
3.3. Polyphenylene carboxymethylene (PPCM) dosing and antiviral activity against SARS-CoV-2
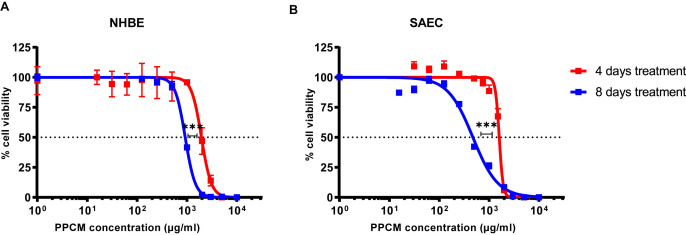
PPCM cytotoxic concentration (CC50) in NHBE and SAEC was determined after both 4 and 8 consecutive days of treatment, and were consistent with dose ranges at which PPCM previously demonstrated in vitro antiviral activity against sexually transmitted viruses (Chang et al., 2007; Escaffre et al., 2019; Herold et al., 2002). Note that experiments were not extended beyond an 8-day treatment as cytopathic effects (CPE) including cell clumping, rounding, and swelling started occurring past that time (data not shown). CC50 corresponding to the 2 different treatment regimens in NHBE or SAEC were significantly distinct (p < 0.001) (Fig. 4 A and B). Specifically, PPCM CC50 was 1951 (95% confidence interval (CI): 1171–2118) and 922 (95% CI: 899–944) μg/ml in NHBE at day 4 and 8, respectively. Likewise, PPCM CC50 was 1596 (95% CI: 1546–1647) and 473 (95% CI: 427–523) μg/ml in SAEC at day 4 and 8, respectively.
Fig. 4.
NHBE and SAEC tolerate high PPCM concentrations. PPCM cytotoxicity in (A) NHBE and (B) SAEC at both day 4 and 8 daily post-treatment. Cell viability is shown as a percentage of values obtained from PPCM-versus vehicle-treated cells. Assays performed using biological quadruplicates, and bars represent standard deviations. Horizontal dotted lines correspond to the 50% cell viability mark. ***, p < 0.001 (extra sum-of-squares F test).
Considering CC50 data, a set of low, medium, and high PPCM concentration was arbitrarily chosen to be used for each cell type to investigate the potential antiviral effect of PPCM against SARS-CoV-2. PPCM significantly decreased virus replication in both cell types in a dose-dependent manner (Fig. 5 A and B). Specifically, a 10 μg/ml dose did not interfere with virus replication (p > 0.5) at any time points, and irrespective of the MOI. However, a dose of at least 0.1 mg/ml in NHBE lowered virus titer by a minimum of 1.3 (p < 0.01) and 1.6 (p < 0.01) log10 using respectively a MOI of 0.1 or 5, as seen at day 8 pi (Fig. 5A). This was concomitant and correlated well with on average a 103.2 (p < 0.001) and 102.6 (p < 0.001) -fold nucleocapsid (N) gene expression decrease in cells, using respectively a MOI of 0.1 or 5 (Fig. 5C). Similarly, a 1 mg/ml dose in SAEC lowered virus titer by at least 2.3 (p < 0.001) and 3.2 (p < 0.001) log10 by day 4 pi using respectively a MOI of 0.1 or 5 (Fig. 5B), whereas a lower dose only had an effect following a MOI 0.1 challenge, as seen by titers below the detection level (p < 0.001). Results from N gene expression in SAEC also correlated well with virus titers at that time (Fig. 5D). Indeed, N gene expression was decreased on average by 103.4 -fold (p < 0.001) with a dose of at least 0.1 mg/ml compared to 10 μg/ml, using a MOI of 0.1. Likewise, a decrease by 103.5-fold (p < 0.001) was observed with a dose of 1 mg/ml compared to 0.1 mg/ml, using a MOI of 5. Overall, these in vitro data suggest that PPCM has antiviral properties against SARS-CoV-2.
Fig. 5.
PPCM antiviral effect against SARS-CoV-2 in NHBE and SAEC. Virus replication kinetics in (A) NHBE or (B) SAEC following a (left) low and (right) high infective dose inoculation. Assays performed with PPCM treatments (0–1000 μg/ml) starting 1h prior infection and maintained daily for up to 8 days, as described in materials and methods. Detection and relative quantification of SARS-CoV-2 nucleocapsid (N) gene expression in PPCM-versus vehicle-treated (C) NHBE or (D) SAEC cells at day 8 and 4 pi, respectively. (A–D) Results are expressed as the average of biological duplicates or triplicates, and error bars represent standard errors. (A, B) The horizontal dotted lines correspond to the detection limit. *, p < 0.05; **, p < 0.01; ***, p < 0.001 (ANOVA, Tukey's test).
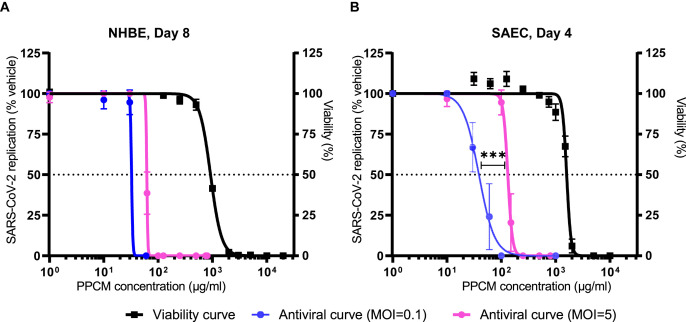
Additional PPCM concentrations were tested against SARS-CoV-2 in both cell types to determine the corresponding half maximal effective concentration (EC50) and selectivity index (SI) at time points matching peak virus titers. PPCM EC50 against SARS-CoV-2 MOI 0.1 and 5 was respectively 32 (SI of 29) or 62 μg/ml (SI of 15) in NHBE at day 8 pi (Fig. 6 ). Note that 95% confidence intervals calculation of EC50 and comparison of curves between MOI could not be performed due to the slope factor steepness for each PPCM dose-response combined to the narrow dynamic range between peak titer and limit of detection of the plaque assay. Likewise, PPCM EC50 against SARS-CoV-2 MOI 0.1 and 5 was respectively 39 (95% CI: 33–45; SI of 41) or 132 μg/ml (95% CI: 124–140; SI of 12) in SAEC at day 4 pi (Fig. 6). Comparison of fits also highlighted different PPCM-dose response data sets (p < 0.001) in SAEC between MOI 0.1 and 5 altogether suggesting that PPCM antiviral efficacy is non-surprisingly dependent of the infective dose.
Fig. 6.
PPCM efficacy parameters against SARS-CoV-2 in NHBE and SAEC. Determination of PPCM half maximal effective concentration (EC50) against SARS-CoV-2 in (A) NHBE and (B) SAEC at respectively day 8 and 4 pi. Values indicate the percentage of virus titer from PPCM-versus vehicle-treated cell cultures (left y-axis) using a low and high infective dose. Note that the corresponding viability curves from Fig. 1D were reported in these figures. Results are expressed as the average of biological duplicates or triplicates, and error bars represent standard deviations. The horizontal dotted lines correspond to either 50% of maximal virus replication level (left y-axis) or cell viability (right y-axis) mark. ***, p < 0.001 (extra sum-of-squares F test).
3.4. Inflammatory response of primary human small airway epithelial cells following SARS-CoV-2 infection
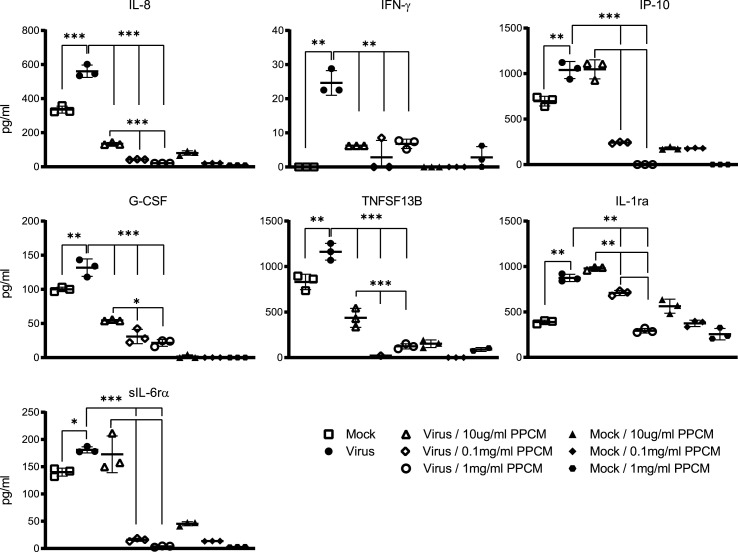
An exacerbated inflammatory response in the small airway is a key feature of COVID-19 that can lead to respiratory distress, and death. Virus-induced inflammation and the effect of PPCM at lowering it was therefore investigated in SAEC at day 4 pi. Samples from the experiment using MOI 0.1 were chosen as a lower infective dose than MOI 5 is likely more relevant in the context of a natural infection. However, this is still unknown.
SARS-CoV-2 infection resulted in an increased secretion of inflammatory markers including at least 1 chemokine (Interleukin 8: IL-8) and 6 cytokines/soluble cytokine receptors (Interferon gamma: IFN-γ; Interferon gamma-induced protein 10: IP-10; Granulocyte colony-stimulating factor: G-CSF; Tumor necrosis factor superfamily member 13b: TNFSF13B; Interleukin-1 receptor antagonist: IL-1Rα; Soluble IL-6 receptor alpha: sIL6-Rα) (p < 0.01 or <0.001) when compared to mock-infected cells at day 4 pi (Fig. 7 ). Interestingly, the secretion level of all these markers was significantly decreased in a PPCM dose-dependent manner (Fig. 7). For example, no changes in IP-10, IL-1Rα, and IL-6ra expression were noted between vehicle- and 10 μg/ml PPCM-treated infected cells which is consistent with the fact that virus titers were comparable between these 2 conditions. However, the expression level of all markers remained either at a similar or decreased to a significantly lower level compared to their counterparts in mock-infected cells using PPCM at 1 or 0.1 mg/ml. This is in line with the fact that no replication was detected at that time point (Fig. 7). It is also interesting to note that except for IFN-γ, and IL-1Rα, all other above-mentioned analytes were significantly decreased compared to their reciprocal in PPCM-versus vehicle-treated mock-infected cells in a dose dependent-manner. Altogether, these data suggest that PPCM does not cause any inflammatory response even at the highest tolerated dose tested in SAEC, and that it can maintain a level of inflammation comparable or even lower to what is seen in mock-infected cells even though infectious virus is detected.
Fig. 7.
Effect of PPCM on SARS-CoV-2-induced immune response in SAEC. Shown are cytokine/chemokine levels in mock- and virus (MOI 0.1)-infected SAEC cells, with or without PPCM treatment at day 4 pi. Results are expressed as averages of data from biological triplicates, and bars represent standard deviations. *, p < 0.05; **, p < 0.01; ***; p < 0.001 (ANOVA, Tukey's test). Note that statistical differences between cytokine/chemokine levels in mock-infected cells with and without PPCM treatment are not highlighted in this figure.
4. Discussion
Since the first reports of pneumonia of an unknown origin in December 2019 (Zhou et al., 2020), the causative agent, now identified as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has spread throughout the world and infected over 200 million people as of August 2021 (Dong et al., 2020). A worldwide vaccine distribution is ongoing, but the total number of new cases is still increasing (Dong et al., 2020). This scenario is all the more disturbing as the treatments of patients with antivirals and immunomodulators, that already have limited efficacy, could offer no beneficial effect due to emergence of variant strains (Chen et al., 2021; Diamond et al., 2021).
The goal of this study was to evaluate the antiviral efficacy of PPCM against SARS-CoV-2, which would support future studies on repurposing PPCM as a prophylaxis or therapeutic antiviral treatment method via aerosol delivery against respiratory viruses. PPCM has previously demonstrated in vitro broad-spectrum activity against several sexually transmitted infections including some of viral origin such as HIV-1, HSV-1 and 2, BPV, and Ebola virus (Chang et al., 2007; Escaffre et al., 2019; Herold et al., 2002; Mesquita et al., 2008; Weitzel et al., 2020; Zaneveld et al., 2002). In addition, PPCM toxicity studies have shown that a 4% gel formulation was safe in a vaginal irritation assay in New Zealand White rabbits (Zaneveld et al., 2002), and did not cause any apparent disease in BALB/c mice (Mesquita et al., 2008). A 33% solution delivered at 5.0 g per kilogram of body weight also had low acute oral toxicity in rats (Zaneveld et al., 2002).
In vitro testing of SARS-CoV-2 antiviral candidates has often been performed with cell lines originating from different species, and not necessarily from the human lungs (Hoffmann et al., 2020b; Schooley et al., 2020; Wang et al., 2020), which can slow down the drug-screening process as to whether a drug candidate is of interest. For example, SARS-CoV-2 entry mechanism in human respiratory epithelial cells, as opposed to monkey kidney epithelial cells, has been shown to occur without the need of endosomal acidification (Hoffmann et al., 2020b; Ou et al., 2021), which in turn caused the entry inhibitor hydroxychloroquine to be ineffective in the lung model. Therefore, PPCM antiviral efficacy against SARS-CoV-2 was chosen to be only tested in human respiratory epithelial cells. The reasoning for specifically using trachea/bronchia (NHBE) and small airways (SAEC) undifferentiated cells was that these cultures allow for PPCM treatment to be maintained throughout the infection as opposed to air-liquid interface models. Indeed, prolonged submersion of a pseudostratified respiratory epithelium on inserts will cause cell dedifferentiation and complicate data analysis. Lastly, NHBE and SAEC are primary cells which are more relevant than cell lines to study the inflammatory response to infection or antiviral treatment, regardless of the potential donor-to-donor variability. However, reproducibility of data could be affected.
ACE2 and TMPRSS2 are involved in virus attachment and entry process (Hoffmann et al., 2020a), and NHBE and SAEC equally expressed each of these proteins, consistent with the fact that they are found in the lower respiratory tract epithelium (Bertram et al., 2012; Sungnak et al., 2020), although mostly in alveolar epithelial type II cells (Qi et al., 2020; Zhao et al., 2020). A lower SARS-CoV-2 replication level was however anticipated in both cell types when compared to the highly permissive Calu-3 cell line since the endogenous level of ACE2 was reported to be a limiting factor in the susceptibility of cells to infection (Hoffmann et al., 2020a; Hou et al., 2020; Wu et al., 2021). Indeed, while TMPRSS2 expression in these primary cells and Calu-3 was similar, the amount of ACE2 significantly differed between them, and resulted in virus replication in Calu-3 at least 3 log10 higher by 48h pi using a comparable infective dose (Plante et al., 2020). Note that overexpressing ACE2 in a given cell line has demonstrated to increase virus permissiveness (Hoffmann et al., 2020a; Letko et al., 2020). The rather low virus titer increase of over the course of infection in NHBE and SAEC was also in line with the fact that these and the almost refractory to infection A549 cells (Hoffmann et al., 2020a) expressed similar ACE2 levels. Differences in replication kinetics between using NHBE and SAEC will be further investigated but could be due to non-matching donors. Another possibility is the distinct anatomical location these cells originate from, with their capabilities to produce ciliated, secretory, and basal cells in different proportions (Mercer et al., 1994; Rock et al., 2009). However, a recent study refuted that changes in ciliated cell distribution between epithelia correlated with differences in SARS-CoV-2 susceptibility (Hou et al., 2020). Interestingly, SARS-CoV-2 infection of the fully differentiated NHBE-derived epithelium or equivalent from freshly excised normal tissues yielded significant higher titers (Busnadiego et al., 2020; Hou et al., 2020; Plante et al., 2020) than in NHBE highlighting one limitation regarding the relevance of our model. This discrepancy was however not observed between SAEC and fresh tissues from three donors (Hou et al., 2020), altogether suggesting that SAEC is more relevant than NHBE as a model for drug-screening for SARS-CoV-2 in spite of allowing low replication rates compared to using cell lines.
PPCM CC50 in NHBE (922 μg/ml), and SAEC (1596 μg/ml) were comparable to those reported in MT-2 lymphoblastic cells (>1000 μg/ml), and VK2/E6E7 vaginal (927 μg/ml) and HeLa cervical (>300 μg/ml) epithelial cells (Chang et al., 2007; Escaffre et al., 2019; Herold et al., 2002; Mesquita et al., 2008; Zaneveld et al., 2002). Our results are also consistent with the fact that PPCM CC50 in human alveolar epithelial A549 cells was higher than 1250 μg/ml (unpublished data), altogether suggesting that high PPCM concentrations are not cytotoxic regardless of the human cell origin.
PPCM antiviral activity was demonstrated against SARS-CoV-2, with an EC50 ranging from 32 to 132 μg/ml and SI between 12 and 41, depending on the infective dose and cell type. These data compare well with PPCM efficacy against ebolavirus (EC50 < 5 μg/ml; SI > 185) (Escaffre et al., 2019), HIV-1 (EC50 = 6.5 μg/ml; SI > 154) (Zaneveld et al., 2002), and to a lower extend HSV (EC50 < 54 ng/ml; SI > 5,500) (Zaneveld et al., 2002) considering that the assays were set up differently, and used lower infective doses compared to our study. Mechanistically, PPCM was previously shown to bind to the envelope glycoprotein 120 of HIV, B of HSV, and GP of ebolavirus at unidentified sites, but also to directly interact with cells, altogether preventing virus attachment and entry (Chang et al., 2007; Escaffre et al., 2019; Mesquita et al., 2008; Zaneveld et al., 2002). Indeed, PPCM was proposed to mask the ubiquitous cell surface glycosaminoglycan heparan sulfate (HS) that is important in HIV, HSV and ebolavirus infection or virus binding (Mondor et al., 1998; O'Hearn et al., 2015; Shieh et al., 1992). Interestingly, recent studies also showed that HS has a role in SARS-CoV-2 infection by serving as an attachment cofactor with ACE2 (Clausen et al., 2020; Hu et al., 2021) and a site in the N-terminal domain of the spike protein has even been identified (Schuurs et al., 2021). Although the mechanism on how PPCM interferes with SARS-CoV-2 replication still needs to be investigated, it could therefore involve competition for HS binding on cells similar to its mechanism of action against other viruses. The immune response of SAEC rather than NHBE was investigated as SARS-CoV-2 can cause severe pneumonia and acute airspace disease with virus detected in ciliated airway cells among others (Harrison et al., 2020; Schaefer et al., 2020). SAEC demonstrated permissiveness to virus replication which is consistent with type II interferon (IFN) secretion, and upregulation of IFN-stimulated gene (ISG) effectors including IL-8 and IP-10 altogether suggesting inflammation of the respiratory epithelium. Interestingly, high levels of these two markers have been found in sera and lung tissues of severe COVID-19 patients (Del Valle et al., 2020; Huang et al., 2020), also indicating that SAEC is a relevant model to study virus-induced inflammation, and support screening of therapeutics. PPCM treatment of infected cells resulted in a significant reduction of expression of all inflammatory markers highlighted in this study in a dose dependent manner. In fact, these markers remained either at a similar or decreased to a significantly lower level when compared to the counterparts in mock-infected cells using an optimal PPCM dose. Altogether, these data suggest that PPCM can inhibit SARS-CoV-2-induced inflammation of the respiratory epithelium similar to our previous observations with ebolavirus in a vaginal epithelial model (Escaffre et al., 2019).
In conclusion, our study investigated in vitro PPCM antiviral efficacy against SARS-CoV-2. We first determined the suitability of using primary human undifferentiated respiratory epithelial cells as a platform to perform drug screening against SARS-CoV-2 and reported a low permissiveness of these cells to infection. We then showed that PPCM inhibited infection and virus-induced inflammation by a mechanism that remains to be identified although potentially through masking of the ubiquitous cell surface heparan sulfate. Our data support that PPCM, in the form of an aerosol, should be evaluated in vivo for toxicity and antiviral activity against SARS-CoV-2, and potentially other human pathogenic coronaviruses.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements and Funding
We thank Yaso Therapeutics Inc. (supported by NIH/NICHD R44 HD092206) for generously providing PPCM stock. This work was supported by a COVID-19 pilot project provided through the Institute for Human Infections & Immunity and Sealy & Smith Foundation (UTMB, awarded to A.N.F.).
References
- Beigel J.H., Tomashek K.M., Dodd L.E., Mehta A.K., Zingman B.S., Kalil A.C., Hohmann E., Chu H.Y., Luetkemeyer A., Kline S., Lopez de Castilla D., Finberg R.W., Dierberg K., Tapson V., Hsieh L., Patterson T.F., Paredes R., Sweeney D.A., Short W.R., Touloumi G., Lye D.C., Ohmagari N., Oh M.D., Ruiz-Palacios G.M., Benfield T., Fatkenheuer G., Kortepeter M.G., Atmar R.L., Creech C.B., Lundgren J., Babiker A.G., Pett S., Neaton J.D., Burgess T.H., Bonnett T., Green M., Makowski M., Osinusi A., Nayak S., Lane H.C., Members A.-S.G. Remdesivir for the treatment of covid-19 - final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram S., Heurich A., Lavender H., Gierer S., Danisch S., Perin P., Lucas J.M., Nelson P.S., Pohlmann S., Soilleux E.J. Influenza and SARS-coronavirus activating proteases TMPRSS2 and HAT are expressed at multiple sites in human respiratory and gastrointestinal tracts. PloS One. 2012;7 doi: 10.1371/journal.pone.0035876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D., Nilsson-Payant B.E., Liu W.C., Uhl S., Hoagland D., Moller R., Jordan T.X., Oishi K., Panis M., Sachs D., Wang T.T., Schwartz R.E., Lim J.K., Albrecht R.A., tenOever B.R. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busnadiego I., Fernbach S., Pohl M.O., Karakus U., Huber M., Trkola A., Stertz S., Hale B.G. 2020. Antiviral Activity of Type I, II, and III Interferons Counterbalances ACE2 Inducibility and Restricts SARS-CoV-2. mBio 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese L.H., Calabrese C. Baricitinib and dexamethasone for hospitalized patients with COVID-19. Cleve. Clin. J. Med. 2021 doi: 10.3949/ccjm.88a.ccc073. Accession number: 33526440. [DOI] [PubMed] [Google Scholar]
- Candan S.A., Elibol N., Abdullahi A. Consideration of prevention and management of long-term consequences of post-acute respiratory distress syndrome in patients with COVID-19. Physiother. Theory Pract. 2020;36:663–668. doi: 10.1080/09593985.2020.1766181. [DOI] [PubMed] [Google Scholar]
- Carvalho T., Krammer F., Iwasaki A. The first 12 months of COVID-19: a timeline of immunological insights. Nat. Rev. Immunol. 2021;21:245–256. doi: 10.1038/s41577-021-00522-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang T.L., Teleshova N., Rapista A., Paluch M., Anderson R.A., Waller D.P., Zaneveld L.J., Granelli-Piperno A., Klotman M.E. SAMMA, a mandelic acid condensation polymer, inhibits dendritic cell-mediated HIV transmission. FEBS Lett. 2007;581:4596–4602. doi: 10.1016/j.febslet.2007.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N., Gilchuk P., Zost S.J., Tahan S., Droit L., Turner J.S., Kim W., Schmitz A.J., Thapa M., Wang D., Boon A.C.M., Presti R.M., O'Halloran J.A., Kim A.H.J., Deepak P., Pinto D., Fremont D.H., Crowe J.E., Jr., Corti D., Virgin H.W., Ellebedy A.H., Shi P.Y., Diamond M.S. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K., Akl E.A., Duda S., Solo K., Yaacoub S., Schunemann H.J. Physical distancing, face masks, and eye protection to prevent person-to-person transmission of SARS-CoV-2 and COVID-19: a systematic review and meta-analysis. Lancet. 2020;395:1973–1987. doi: 10.1016/S0140-6736(20)31142-9. authors, C.-S.U.R.G.E.s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., Thacker B.E., Glass C.A., Yang Z., Torres J.L., Golden G.J., Bartels P.L., Porell R.N., Garretson A.F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B.M., Caradonna T.M., Kellman B.P., Martino C., Gordts P., Chanda S.K., Schmidt A.G., Godula K., Leibel S.L., Jose J., Corbett K.D., Ward A.B., Carlin A.F., Esko J.D. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183:1043–1057 e1015. doi: 10.1016/j.cell.2020.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Valle D.M., Kim-Schulze S., Hsin-Hui H., Beckmann N.D., Nirenberg S., Wang B., Lavin Y., Swartz T., Madduri D., Stock A., Marron T., Xie H., Patel M.K., van Oekelen O., Rahman A., Kovatch P., Aberg J., Schadt E., Jagannath S., Mazumdar M., Charney A., Firpo-Betancourt A., Mendu D.R., Jhang J., Reich D., Sigel K., Cordon-Cardo C., Feldmann M., Parekh S., Merad M., Gnjatic S. An inflammatory cytokine signature helps predict COVID-19 severity and death. medRxiv : Preprint Server Health Sci. 2020 doi: 10.1101/2020.05.28.20115758. Accession number: 32511562PMCID: PMC7274243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond M., Chen R., Xie X., Case J., Zhang X., VanBlargan L., Liu Y., Liu J., Errico J., Winkler E., Suryadevara N., Tahan S., Turner J., Kim W., Schmitz A., Thapa M., Wang D., Boon A., Pinto D., Presti R., O’Halloran J., Kim A., Deepak P., Fremont D., Corti D., Virgin H., Crowe J., Droit L., Ellebedy A., Shi P.Y., Gilchuk P. SARS-CoV-2 variants show resistance to neutralization by many monoclonal and serum-derived polyclonal antibodies. Res Sq. 2021 doi: 10.21203/rs.3.rs-228079/v1. PMCID: PMC7885928 ; Accession number: 33594356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E., Du H., Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis. 2020;20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escaffre O., Juelich T.L., Freiberg A.N. Polyphenylene carboxymethylene (PPCM) in vitro antiviral efficacy against Ebola virus in the context of a sexually transmitted infection. Antivir. Res. 2019;170:104567. doi: 10.1016/j.antiviral.2019.104567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harcourt J., Tamin A., Lu X., Kamili S., Sakthivel S.K., Murray J., Queen K., Tao Y., Paden C.R., Zhang J., Li Y., Uehara A., Wang H., Goldsmith C., Bullock H.A., Wang L., Whitaker B., Lynch B., Gautam R., Schindewolf C., Lokugamage K.G., Scharton D., Plante J.A., Mirchandani D., Widen S.G., Narayanan K., Makino S., Ksiazek T.G., Plante K.S., Weaver S.C., Lindstrom S., Tong S., Menachery V.D., Thornburg N.J. Severe acute respiratory syndrome coronavirus 2 from patient with coronavirus disease, United States. Emerg. Infect. Dis. 2020;26:1266–1273. doi: 10.3201/eid2606.200516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison A.G., Lin T., Wang P. Mechanisms of SARS-CoV-2 transmission and pathogenesis. Trends Immunol. 2020;41:1100–1115. doi: 10.1016/j.it.2020.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold B.C., Scordi-Bello I., Cheshenko N., Marcellino D., Dzuzelewski M., Francois F., Morin R., Casullo V.M., Anderson R.A., Chany C., 2nd, Waller D.P., Zaneveld L.J., Klotman M.E. Mandelic acid condensation polymer: novel candidate microbicide for prevention of human immunodeficiency virus and herpes simplex virus entry. J. Virol. 2002;76:11236–11244. doi: 10.1128/JVI.76.22.11236-11244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Muller M.A., Drosten C., Pohlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. e278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Mosbauer K., Hofmann-Winkler H., Kaul A., Kleine-Weber H., Kruger N., Gassen N.C., Muller M.A., Drosten C., Pohlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- Holmes E., Wist J., Masuda R., Lodge S., Nitschke P., Kimhofer T., Loo R.L., Begum S., Boughton B., Yang R., Morillon A.C., Chin S.T., Hall D., Ryan M., Bong S.H., Gay M., Edgar D.W., Lindon J.C., Richards T., Yeap B.B., Pettersson S., Spraul M., Schaefer H., Lawler N.G., Gray N., Whiley L., Nicholson J.K. Incomplete systemic recovery and metabolic phenoreversion in post-acute-phase nonhospitalized COVID-19 patients: implications for assessment of post-acute COVID-19 syndrome. J. Proteome Res. 2021;(6):3315–3329. doi: 10.1021/acs.jproteome.1c00224. PMCID: PMC8147448. Accession number:34009992. [DOI] [PubMed] [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., Spitters C., Ericson K., Wilkerson S., Tural A., Diaz G., Cohn A., Fox L., Patel A., Gerber S.I., Kim L., Tong S., Lu X., Lindstrom S., Pallansch M.A., Weldon W.C., Biggs H.M., Uyeki T.M., Pillai S.K., Washington State -nCo V.C.I.T. First case of 2019 novel coronavirus in the United States. N. Engl. J. Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Okuda K., Edwards C.E., Martinez D.R., Asakura T., Dinnon K.H., 3rd, Kato T., Lee R.E., Yount B.L., Mascenik T.M., Chen G., Olivier K.N., Ghio A., Tse L.V., Leist S.R., Gralinski L.E., Schafer A., Dang H., Gilmore R., Nakano S., Sun L., Fulcher M.L., Livraghi-Butrico A., Nicely N.I., Cameron M., Cameron C., Kelvin D.J., de Silva A., Margolis D.M., Markmann A., Bartelt L., Zumwalt R., Martinez F.J., Salvatore S.P., Borczuk A., Tata P.R., Sontake V., Kimple A., Jaspers I., O'Neal W.K., Randell S.H., Boucher R.C., Baric R.S. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446 e414. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Meng X., Zhang F., Xiang Y., Wang J. The in vitro antiviral activity of lactoferrin against common human coronaviruses and SARS-CoV-2 is mediated by targeting the heparan sulfate co-receptor. Emerg. Microb. Infect. 2021;10:317–330. doi: 10.1080/22221751.2021.1888660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko M., Marzi A., Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat Microbiol. 2020;5:562–569. doi: 10.1038/s41564-020-0688-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer R.R., Russell M.L., Roggli V.L., Crapo J.D. Cell number and distribution in human and rat airways. Am. J. Respir. Cell Mol. Biol. 1994;10:613–624. doi: 10.1165/ajrcmb.10.6.8003339. [DOI] [PubMed] [Google Scholar]
- Mesquita P.M., Wilson S.S., Manlow P., Fischetti L., Keller M.J., Herold B.C., Shattock R.J. Candidate microbicide PPCM blocks human immunodeficiency virus type 1 infection in cell and tissue cultures and prevents genital herpes in a murine model. J. Virol. 2008;82:6576–6584. doi: 10.1128/JVI.00335-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondor I., Ugolini S., Sattentau Q.J. Human immunodeficiency virus type 1 attachment to HeLa CD4 cells is CD4 independent and gp120 dependent and requires cell surface heparans. J. Virol. 1998;72:3623–3634. doi: 10.1128/jvi.72.5.3623-3634.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Perez O., Merino E., Leon-Ramirez J.M., Andres M., Ramos J.M., Arenas-Jimenez J., Asensio S., Sanchez R., Ruiz-Torregrosa P., Galan I., Scholz A., Amo A., Gonzalez-delaAleja P., Boix V., Gil J., group C.A.r. Post-acute COVID-19 syndrome. Incidence and risk factors: a Mediterranean cohort study. J. Infect. 2021;82:378–383. doi: 10.1016/j.jinf.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nature . 2021. COVID Research: a Year of Scientific Milestones, Nature, 2020/03/30. [DOI] [PubMed] [Google Scholar]
- O'Hearn A., Wang M., Cheng H., Lear-Rooney C.M., Koning K., Rumschlag-Booms E., Varhegyi E., Olinger G., Rong L. Role of EXT1 and glycosaminoglycans in the early stage of filovirus entry. J. Virol. 2015;89:5441–5449. doi: 10.1128/JVI.03689-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oguntuyo K.Y., Stevens C.S., Hung C.T., Ikegame S., Acklin J.A., Kowdle S.S., Carmichael J.C., Chiu H.P., Azarm K.D., Haas G.D., Amanat F., Klingler J., Baine I., Arinsburg S., Bandres J.C., Siddiquey M.N.A., Schilke R.M., Woolard M.D., Zhang H., Consortium C.A., Duty A.J., Kraus T.A., Moran T.M., Tortorella D., Lim J.K., Gamarnik A.V., Hioe C.E., Zolla-Pazner S., Ivanov S.S., Kamil J.P., Krammer F., Lee B. Quantifying absolute neutralization titers against SARS-CoV-2 by a standardized virus neutralization assay allows for cross-cohort comparisons of COVID-19 sera. mBio. 2021;12 doi: 10.1128/mBio.02492-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou T., Mou H., Zhang L., Ojha A., Choe H., Farzan M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paladugu S., Donato A.A. Remdesivir improved time to recovery in adults hospitalized with COVID-19 and lower respiratory tract involvement. Ann. Intern. Med. 2020;173:JC4. doi: 10.7326/ACPJ202007210-005. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R., Mirchandani D., Scharton D., Bilello J.P., Ku Z., An Z., Kalveram B., Freiberg A.N., Menachery V.D., Xie X., Plante K.S., Weaver S.C., Shi P.Y. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2020;(7852):116–121. doi: 10.1038/s41586-020-2895-3. PMCID: PMC8158177. Accession number: 33106671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather K.A., Wang C.C., Schooley R.T. Reducing transmission of SARS-CoV-2. Science. 2020;368:1422–1424. doi: 10.1126/science.abc6197. [DOI] [PubMed] [Google Scholar]
- Qi F., Qian S., Zhang S., Zhang Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020;526:135–140. doi: 10.1016/j.bbrc.2020.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjbar K., Moghadami M., Mirahmadizadeh A., Fallahi M.J., Khaloo V., Shahriarirad R., Erfani A., Khodamoradi Z., Saadi M.H.G. Correction to: methylprednisolone or dexamethasone, which one is superior corticosteroid in the treatment of hospitalized COVID-19 patients: a triple-blinded randomized controlled trial. BMC Infect. Dis. 2021;21:436. doi: 10.1186/s12879-021-06130-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock J.R., Onaitis M.W., Rawlins E.L., Lu Y., Clark C.P., Xue Y., Randell S.H., Hogan B.L. Basal cells as stem cells of the mouse trachea and human airway epithelium. Proc. Natl. Acad. Sci. U.S.A. 2009;106:12771–12775. doi: 10.1073/pnas.0906850106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer I.M., Padera R.F., Solomon I.H., Kanjilal S., Hammer M.M., Hornick J.L., Sholl L.M. In situ detection of SARS-CoV-2 in lungs and airways of patients with COVID-19. Mod. Pathol. 2020;33:2104–2114. doi: 10.1038/s41379-020-0595-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schooley R.T., Carlin A.F., Beadle J.R., Valiaeva N., Zhang X.Q., Garretson A.F., Smith V.I., Murphy J., Hostetler K.Y. Rethinking Remdesivir: synthesis of lipid prodrugs that substantially enhance anti-coronavirus activity. bioRxiv : Preprint Server Biol. 2020 doi: 10.1101/2020.08.26.269159. PMCID: PMC7457622. Accession number: 32869033. [DOI] [Google Scholar]
- Schuurs Z.P., Hammond E., Elli S., Rudd T.R., Mycroft-West C.J., Lima M.A., Skidmore M.A., Karlsson R., Chen Y.H., Bagdonaite I., Yang Z., Ahmed Y.A., Richard D.J., Turnbull J., Ferro V., Coombe D.R., Gandhi N.S. Evidence of a putative glycosaminoglycan binding site on the glycosylated SARS-CoV-2 spike protein N-terminal domain. Comput. Struct. Biotechnol. J. 2021;19:2806–2818. doi: 10.1016/j.csbj.2021.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh M.T., WuDunn D., Montgomery R.I., Esko J.D., Spear P.G. Cell surface receptors for herpes simplex virus are heparan sulfate proteoglycans. J. Cell Biol. 1992;116:1273–1281. doi: 10.1083/jcb.116.5.1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert D.M., Hall M.A. Disease control, civil liberties, and mass testing - calibrating restrictions during the covid-19 pandemic. N. Engl. J. Med. 2020;383:102–104. doi: 10.1056/NEJMp2007637. [DOI] [PubMed] [Google Scholar]
- Sungnak W., Huang N., Becavin C., Berg M., Queen R., Litvinukova M., Talavera-Lopez C., Maatz H., Reichart D., Sampaziotis F., Worlock K.B., Yoshida M., Barnes J.L., Network H.C.A.L.B. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat. Med. 2020;26:681–687. doi: 10.1038/s41591-020-0868-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To K.K., Sridhar S., Chiu K.H., Hung D.L., Li X., Hung I.F., Tam A.R., Chung T.W., Chan J.F., Zhang A.J., Cheng V.C., Yuen K.Y. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg. Microb. Infect. 2021;10:507–535. doi: 10.1080/22221751.2021.1898291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292 e286. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Luo R., Zhang M., Wang Y., Song T., Tao T., Li Z., Jin L., Zheng H., Chen W., Zhao M., Zheng Y., Qin J. A cross-talk between epithelium and endothelium mediates human alveolar-capillary injury during SARS-CoV-2 infection. Cell Death Dis. 2020;11:1042. doi: 10.1038/s41419-020-03252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzel M., North B.B., Waller D. Development of multipurpose technologies products for pregnancy and STI prevention: update on polyphenylene carboxymethylene MPT gel development dagger. Biol. Reprod. 2020;103:299–309. doi: 10.1093/biolre/ioaa087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J., Deng W., Li S., Yang X. Advances in research on ACE2 as a receptor for 2019-nCoV. Cell. Mol. Life Sci. 2021;78:531–544. doi: 10.1007/s00018-020-03611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaneveld L.J., Anderson R.A., Diao X.H., Waller D.P., Chany C., Feathergill K., Doncel G., Cooper M.D., Herold B. Use of mandelic acid condensation polymer (SAMMA), a new antimicrobial contraceptive agent, for vaginal prophylaxis. Fertil. Steril. 2002;78:1107–1115. doi: 10.1016/s0015-0282(02)04210-3. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zhao Z., Wang Y., Zhou Y., Ma Y., Zuo W. Single-cell RNA expression profiling of ACE2, the receptor of SARS-CoV-2. Am. J. Respir. Crit. Care Med. 2020;202:756–759. doi: 10.1164/rccm.202001-0179LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]