Abstract
Objective:
To determine the institutional diagnostic accuracy of an AI DSS, Aidoc, in diagnosing intracranial hemorrhage (ICH) on non-contrast head CTs and to assess the potential generalizability of an AI DSS.
Methods:
This retrospective study included 3605 consecutive, emergent, adult non-contrast head CT scans performed between 7/1/2019 and 12/30/2019 at our institution (51% female, mean age of 61 ± 21 years). Each scan was evaluated for ICH by both a certificate of added qualification certified neuroradiologist and Aidoc. We determined the diagnostic accuracy of the AI model and performed a failure mode analysis with quantitative CT radiomic image characterization.
Results:
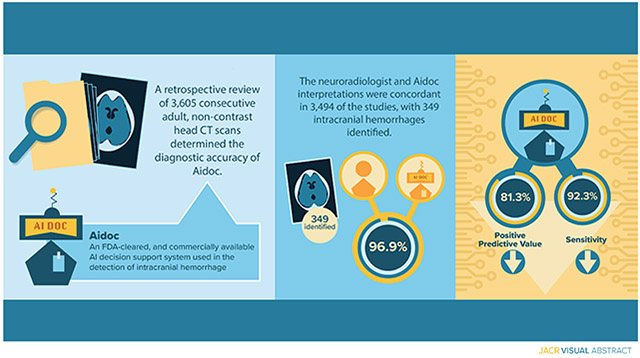
Of the 3605 scans, 349 cases of ICH (9.7% of studies) were identified. The neuroradiologist and Aidoc interpretations were concordant in 96.9% of cases and the overall sensitivity, specificity, positive predictive value, and negative predictive value were 92.3%, 97.7%, 81.3% and 99.2%, respectively, with sensitivity and positive predictive values unexpectedly lower than in previously reported studies. Prior neurosurgery, type of ICH, and number of ICH were significantly associated with decreased model performance. Quantitative image characterization with CT radiomics failed to reveal significant differences between concordant and discordant studies.
Discussion.
This study revealed decreased diagnostic accuracy of an AI DSS at our institution. Despite extensive evaluation, we were unable to identify the source of this discrepancy, raising concerns about the generalizability of these tools with indeterminate failure modes. These results further highlight the need for standardized study design to allow for rigorous and reproducible site-to-site comparison of emerging deep learning technologies.
Keywords: Artificial intelligence, decision support systems, intracranial hemorrhage, generalizability, non-contrast head CT
Graphical Abstract
Summary statement:
Unexpected lower sensitivity and positive predictive values were observed for an artificial intelligence decision support system for intracranial hemorrhage detection, raising concerns about the generalizability of deep learning tools.
Introduction.
The use of diagnostic imaging has dramatically increased over the last several decades.1-2 In the acute care setting, CT imaging is a critical diagnostic tool for numerous emergent medical conditions such as intracranial hemorrhage (ICH). Timely interpretation is required to guide clinical interventions, especially for ICHs, with half of resulting mortality being reported to occur in the first 24 hours.3-4 However, increased imaging volumes place a significant burden on radiologists who must maintain diagnostic accuracy and efficiency.5 While efforts have been made to reduce the number of unnecessary scans ordered, their effectiveness appears to be modest.6-7
To help radiologists maintain diagnostic performance in the face of increasing clinical volumes, artificial intelligence (AI) decision support systems (DSS) have been developed. In their typical implementation, a DSS analyzes studies immediately after acquisition and flags those with emergent findings. In theory, a DSS can assist radiologists by directing them to prioritize flagged studies,8 thereby reducing the risk of missing or delaying the communication of a critical finding. Despite the promise of AI and the significant potential gains to be had with a DSS, radiologists are faced with choosing from hundreds of independently developed deep learning algorithms9 and 76 FDA-cleared AI algorithms10 with considerable variation in the quality of the evidence supporting each one. AI algorithms also have many known limitations, such as the necessity for large and diverse training datasets,11 biases in dataset compilation,12 poor generalizability,13 overfitting,14 limited number of clinical validation studies,15-16 and the inability to interpret or analyze the underlying mechanisms. Additionally, a poorly performing DSS can hinder a clinician by highlighting false positive studies and promoting premature closure in falsely negative studies. Therefore, it is crucial to fully understand the role, performance, and generalizability of a DSS prior to widespread clinical implementation.
To these ends, we sought to determine and validate the performance and diagnostic accuracy of Aidoc, a widely used, FDA-cleared, and commercially available AI DSS used in the detection of ICH to examine the generalizability and reproducibility of deep learning tools. Early studies of this DSS have reported exceptional diagnostic accuracies.17-19 However, these studies have been limited by either small sample sizes or biased data processing, while more recent smaller validation studies report more modest operating characteristics.20 The aim of this study was to rigorously assess the performance of Aidoc for the detection of ICHs in its implementation at our institution.
Materials and methods.
This Health Insurance Portability and Accountability Act-compliant retrospective study was approved by our local institutional review board (IRB). The requirement for informed consent was waived by the IRB. The data was analyzed and controlled by the authors exclusively, none of whom are employees or consultants to Aidoc or its competitors.
Study population, data collection, and AI system.
All consecutive adult, non-contrast head CT (NCCT) scans performed at two emergency departments of an academic medical center between 7/1/2019 and 12/30/2019 were analyzed. A total of 3605 consecutive studies were identified (60.6 ± 20.7 years, 1843 women), across seven General Electric (GE, Boston, MA) CT scanners. Our clinical site utilizes highly standardized GE CT protocols. Due to hardware differences, these protocols do vary slightly scanner to scanner; however, in general, our non-contrast head CT for adults is protocoled as a helical acquisition at a pitch of 0.531 (32x0.625 detector configuration) with a 0.4 s rotation at 120 kV using GE’s smart mA to achieve a noise index of 3.7. No smoothing algorithms are applied. The thin axial reconstruction is 1.25 mm slices at 0.625 mm intervals using the “Soft” kernel; similarly, sagittal and coronal reconstructions are contemporaneously generated and available to the interpreting radiologist at the time of study interpretation. Interpreting neuroradiologists also have access to advanced visualization tools to aid in study interpretation (Vitrea Advanced Visualization, Vital Images, Inc., Minnetonka, Minnesota, USA). The thin source images are also saved to and available in PACS and for Aidoc (Aidoc, Tel Aviv, Israel) analysis. The software only accepts and interprets thin (interval 0.5-1 mm) axial CT images from modern (>64 slice) CT scanners. Clinical instances of Aidoc flag studies in real-time that are determined to have intracranial bleeding. However, to avoid potential bias influencing the final interpretation of the neuroradiologist, our study period only encompassed NCCTs that were performed prior to the clinical implementation of Aidoc at our institution. These NCCTs were then retrospectively analyzed by Aidoc (using the same FDA-cleared algorithm) and subsequently matched to the final imaging report.
Data processing and analysis.
Following study inclusion, the presence of an ICH, type of ICH, and study indication were manually determined from the attending neuroradiologist imaging report of each study. All studies were also analyzed by Aidoc, an FDA-cleared neural network algorithm, without technical exclusions and classified as positive for ICH (ICH+) or negative for ICH (ICH−). Because of the difficultly in assessing the presence of multiple ICHs in the algorithm output, scans were scored as ICH + irrespective of the number of ICHs detected. Key images highlighting the pathology identified by the AI model were obtained for each of the discordant, ICH+ studies. To establish the ground truth of the presence or absence of an ICH, the interpretation of the neuroradiologist and Aidoc were compared and the final certificate of added qualification (CAQ)-neuroradiologist attending interpretation was regarded as the ground truth. Studies with concordant interpretations (i.e. both positive or both negative) were assumed to be correct, while those with discordant interpretations were adjudicated after evaluation by both a second-year radiology resident and a second CAQ-certified Neuroradiologist with 6 years of experience. Even if multiple ICHs were noted on the neuroradiologist interpretation, the presence of any ICH + indicated by the AI model was deemed concordant.
CT textural and quantitative image characterization.
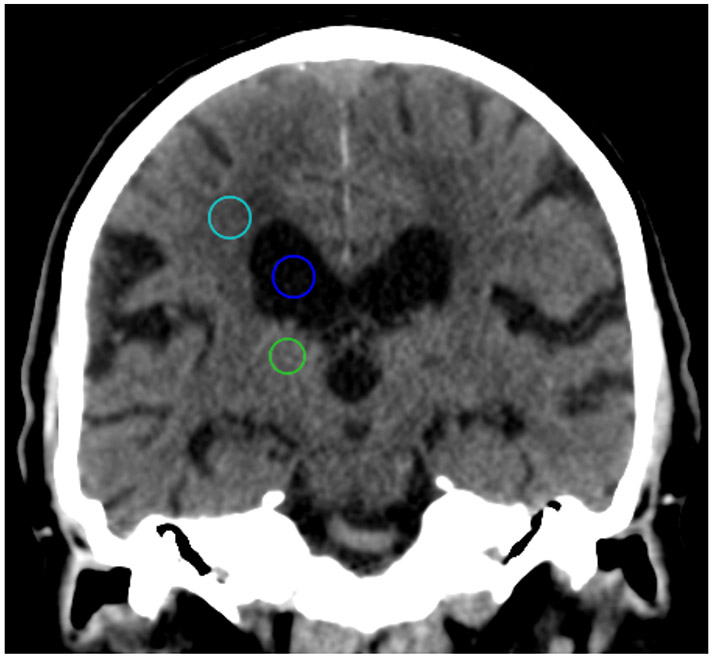
All ICH+ and all discordant studies were uploaded to the HealthMyne server (HealthMyne Inc, Madison, WI), a platform for segmentation and computation of CT radiomic and textural features. Standardized spherical regions of interest (ROI) were drawn in the centrum semiovale, the thalamus, and lateral ventricle to broadly and quantitatively characterize image texture and appearance in multiple tissue types and locations in the central nervous system (white matter, gray matter, cerebrospinal fluid). ROIs were drawn on the right side, with a diameter of 10 mm. ROIs were either moved to the left side or drawn with a reduced diameter when anatomical distortion precluded drawing our standard ROI. All ROIs were drawn by a medical student with assistance from an attending neuroradiologist as required. CT textural features of each ROI were calculated by HealthMyne. In this work, the use of radiomic features in well-defined anatomic locations is not intended to describe underlying pathologies or physiological features21-23, but rather to quantitatively analyze both overall image quality as well as quantify image features that while not observable to radiologists, might nonetheless be image features driving image analysis in the AI algorithm . Some of the selected features such as variance are very well established as image quality metrics on their own; however, other higher order features such as gray-level non-uniformity, kurtosis, and homogeneity describe other important characteristics of the image quality and texture which impact visual perception of the regions of interest.24-25
Statistical analysis.
Sensitivity, specificity, PPV, and NPV calculations were all performed in Microsoft Excel (Excel 365, Microsoft, Redmond, WA). The logistical regression analysis was performed in R (version 4.0.2, R Foundation for Statistical Computing, Vienna, Austria) with a 0.05 threshold for statistical significance. Wald tests were used to test the effect of entire sets of categorical features. For CT textural analyses, for each feature, a two-sided t-test was used at each anatomical site to compare the set of concordant, ICH+ studies with 1) set of discordant studies and 2) all false negative studies. The threshold for statistical significance was corrected for multiple comparisons using the Holm-Bonferroni method with a familywise error rate of 0.05.
Results.
Aidoc diagnostic accuracy and failure mode analysis
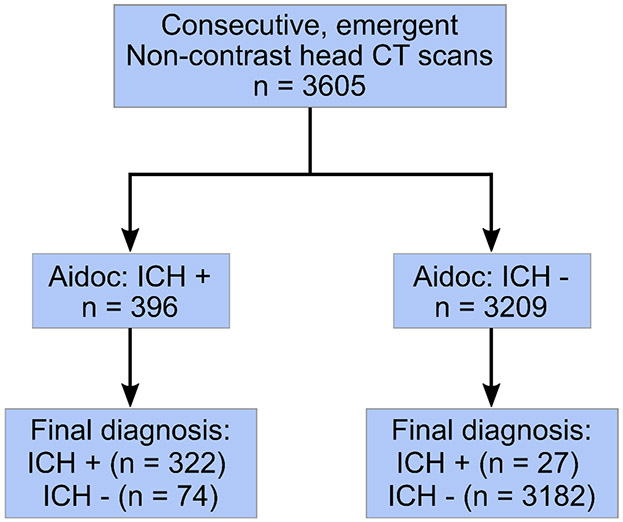
A total of 3605 eligible NCCTs were analyzed and 349 (9.7%) ICHs were identified (Fig 1). Patient characteristics are summarized in table 1. The neuroradiologist and Aidoc interpretations were concordant in 3494 (96.9%) of the studies. After establishing a ground truth in discordant studies, 74 (2.1%) false positive and 27 (0.75%) false negative readings were reported by the AI model and 4 (0.11%) false positive and 6 (0.17%) false negatives were observed for the neuroradiologist interpretation (Fig 2A). The overall sensitivity and specificity of the DSS was 92.3% (95% confidence interval, 88.9 – 94.8%) and 97.7% (97.2 – 98.2%) respectively with a positive predictive value of 81.3% (77.6 – 84.5%) and a negative predictive value of 99.2% (98.8 – 99.4%). We conducted a failure mode analysis to identify factors that could be contributing to the incorrect Aidoc interpretations. Factors were first analyzed using a univariable logistical regression model and age, sex, prior neurosurgery, the type of ICH and number of ICHs (a single type vs. mixed ICH) were all found to be significantly correlated with decreased diagnostic accuracy (Table 2). These factors were included in a multivariable logistic regression and all factors except for sex were found to significantly contribute to DSS performance (Table 2).
Figure 1.
STARD patient flow diagram.
Table 1.
Patient characteristics.
| All | (%) | ICH + | (%) | Aidoc Incorrect |
(%) | |
|---|---|---|---|---|---|---|
| Total | 3605 | 100 | 349 | 100 | 101 | 100 |
| Sex | ||||||
| Male | 1762 | 48.9 | 205 | 58.7 | 60 | 59.4 |
| Female | 1843 | 51.1 | 144 | 41.3 | 41 | 40.6 |
| Age (mean, SD) | (SD) | (SD) | (SD) | |||
| All | 60.6 | 20.7 | 63 | 19.7 | 67.5 | 19.1 |
| Male | 58.1 | 20.1 | 59 | 19.2 | 64.4 | 19.3 |
| Female | 62.9 | 21.0 | 69 | 18.8 | 72 | 18.1 |
| Prior neurosurgery | ||||||
| yes | 267 | 7.4 | 38 | 11 | 81 | 80.2 |
| No | 3338 | 92.6 | 311 | 89 | 20 | 19.8 |
| CT Scanner | 0.0 | |||||
| 1 | 82 | 2.3 | 5 | 1.4 | 3 | 3.0 |
| 2 | 72 | 2.0 | 8 | 2.3 | 3 | 3.0 |
| 3 | 252 | 7.0 | 22 | 6.3 | 6 | 5.9 |
| 4 | 346 | 9.6 | 48 | 13.8 | 10 | 9.9 |
| 5 | 2085 | 57.8 | 232 | 66.5 | 62 | 61.4 |
| 6 | 723 | 20.1 | 29 | 8.3 | 17 | 16.8 |
| 7 | 45 | 1.2 | 5 | 1.4 | 0 | 0.0 |
| NCCT indication | ||||||
| Trauma | 2064 | 57.3 | 173 | 49.6 | 60 | 59.4 |
| Altered mental status | 525 | 14.6 | 25 | 7.2 | 12 | 11.9 |
| Headache | 389 | 10.8 | 15 | 4.3 | 9 | 8.9 |
| Neurologic | 241 | 6.7 | 16 | 4.6 | 5 | 5.0 |
| Repeat study | 137 | 3.8 | 113 | 32.4 | 6 | 5.9 |
| Seizure | 104 | 2.9 | 3 | 0.9 | 4 | 4.0 |
| Loss of consciousness | 69 | 1.9 | 2 | 0.6 | 2 | 2.0 |
| Infection | 20 | 0.6 | 2 | 0.6 | 2 | 2.0 |
| Other | 56 | 1.6 | 0 | 0.0 | 1 | 1.0 |
| ICH type | ||||||
| No ICH | 3256 | 90.3 | 0 | 0.0 | 74 | 73.3 |
| Any ICH | 349 | 9.7 | 349 | 100.0 | 27 | 26.7 |
| Single ICH | 226 | 6.3 | 226 | 64.8 | 26 | 25.7 |
| SDH | 91 | 2.5 | 91 | 26.1 | 14 | 13.9 |
| SAH | 72 | 2.0 | 72 | 20.6 | 5 | 5.0 |
| IPH | 33 | 0.9 | 33 | 9.5 | 5 | 5.0 |
| Other | 30 | 0.8 | 30 | 8.6 | 2 | 2.0 |
| Univariable analysis |
Multivariable Analysis |
|||||
| IVH | 10 | 0.3 | 10 | 2.9 | 0 | 0.0 |
| Hem. Met. | 9 | 0.2 | 9 | 2.6 | 1 | 1.0 |
| Extra-axial (not specified) | 8 | 0.2 | 8 | 2.3 | 1 | 1.0 |
| EDH | 2 | 0.1 | 2 | 0.6 | 0 | 0.0 |
| Subependymal | 1 | 0.0 | 1 | 0.3 | 0 | 0.0 |
| Mixed ICH | 123 | 3.4 | 123 | 35.2 | 1 | 1.0 |
SD: standard deviation, NCCT: non-contrast head CT, ICH: intracranial hemorrhage, ICH+: positive intracranial hemorrhage, SDH: subdural hemorrhage, SAH: subarachnoid hemorrhage, IPH: intraparenchymal hemorrhage, IVH: intraventricular hemorrhage, Hem. Met: Hemorrhagic metastases, EDH: Epidural hematoma.
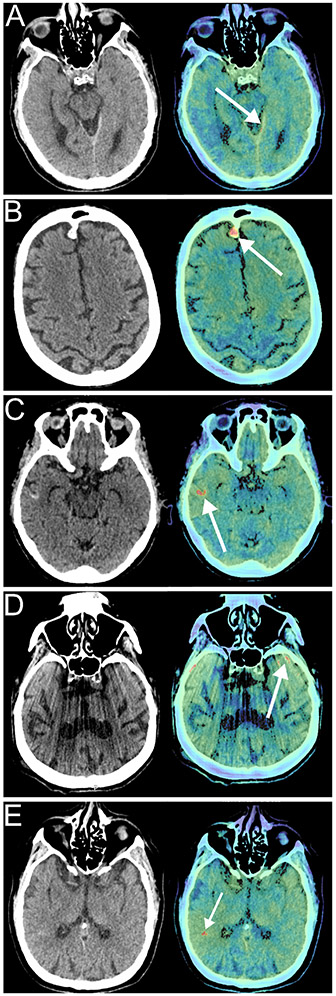
Figure 2. Examples of Aidoc reads and failure modes.
Each panel shows the non-contrast head CT (left) and the key image indicating the pathology identified by Aidoc (right). A) True positive, intracranial hemorrhage (ICH) missed by interpreting neuroradiologist. B) Benign finding (meningioma) misidentified as ICH by Aidoc. C) Pathology (cortical laminar necrosis) misidentified as ICH by Aidoc. D) Aidoc misidentification of an imaging artifact as an IC. E) Unclear failure mode without obvious pathology.
Table 2.
Results of univariable and multivariable logistical regression models for evaluating factors associated with AI DSS errors.
| Parameter | OR | 95% CI | P Value | OR | 95% CI | P value |
|---|---|---|---|---|---|---|
| Age | 1.018 | 1.008 - 1.029 | <0.001* | 1.018 | 1.007-1.030 | 0.002* |
| Sex | ||||||
| Male† | ||||||
| Female | 0.6 | 0.4-1.0 | 0.03* | 0.7 | 0.4-1.0 | 0.06 |
| Prior Neurosurgery | ||||||
| No† | ||||||
| Yes | 3.3 | 1.9 - 5.3 | <0.001* | 3.1 | 1.8-5.1 | <0.001* |
| NCCT Indication | ||||||
| Trauma† | ||||||
| Altered mental status | 0.78 | 0.40-1.41 | 0.44 | |||
| Headache | 0.79 | 0.37-1.53 | 0.52 | |||
| Neurologic | 0.71 | 0.25-1.61 | 0.46 | |||
| Repeat study | 1.5 | 0.58-3.3 | 0.33 | |||
| Seizure | 1.3 | 0.40-3.3 | 0.58 | |||
| Loss of consciousness | 1.0 | 0.16-3.28 | >0.99 | |||
| Infection | 3.7 | 0.58-13.3 | 0.08 | |||
| Other | 0.60 | 0.03-2.8 | 0.62 | |||
| Overall (χ2) | 0.60 | |||||
| CT scanner | ||||||
| 1† | ||||||
| 2 | 1.1 | 0.2-6.4 | 0.87 | |||
| 3 | 0.64 | 0.17-3.1 | 0.54 | |||
| 4 | 0.78 | 0.23-3.6 | 0.72 | |||
| 5 | 0.81 | 0.29-3.4 | 0.72 | |||
| 6 | 0.63 | 0.21-2.8 | 0.48 | |||
| 7 | 0 | 0-362 | 0.98 | |||
| Overall (χ2) | 0.95 | |||||
| Number of ICH types | ||||||
| None† | ||||||
| Single | 5.59 | 3.44-8.83 | <0.001* | |||
| Multiple | 0.35 | 0.02-1.61 | 0.3 | |||
| Overall (χ2) | <0.001* | |||||
| ICH type | ||||||
| No Bleed† | - | |||||
| SDH | 7.8 | 4.1-14.1 | <0.001* | 6 | 3.0-10.9 | <0.001* |
| SAH | 3.21 | 1.1-7.46 | 0.015* | 3.26 | 1.11-7.63 | 0.014* |
| IPH | 7.67 | 2.55-18.8 | <0.001* | 7.49 | 2.44-18.9 | <0.001* |
| other | 3.07 | 0.49-10.5 | 0.13 | 2.44 | 0.38-8.58 | 0.24 |
| mixed | 0.35 | 0.02-1.61 | 0.3 | 0.34 | 0.02-1.55 | 0.28 |
| Overall (χ2) | <0.001* | <0.001* |
OR: odds ratio, CI: confidence interval
Used as reference category
Statistically significant results (p <0.05)
We next explored the etiologies of the false positive studies. Each study flagged by the DSS is accompanied by a key image highlighting the abnormality identified by the algorithm, allowing us to identify the etiology in each false positive case (Table 3). The most common false positive etiology was misidentification of a benign hyperdense imaging finding, accounting for 77% of the false positives results (Fig 2B), with non-hemorrhage pathologies and imaging artifacts comprising another 12% and 4% respectively (Fig 2C, D).
Table 3.
Etiology of false positive findings
| False positive etiology | Count | % |
|---|---|---|
| Hyperdense structure | 24 | 32.4 |
| Calcification | 10 | 13.5 |
| Meningioma | 5 | 6.8 |
| Hyperdense mass | 3 | 4.1 |
| Choroid plexus | 2 | 2.7 |
| Calcified oligodendroma | 1 | 1.4 |
| Pineal calcification | 1 | 1.4 |
| Colloid cyst | 1 | 1.4 |
| Sellar mass | 1 | 1.4 |
| Thick dura | 19 | 25.7 |
| Dura | 3 | 4.1 |
| Falx | 8 | 10.8 |
| Tentorium | 8 | 10.8 |
| Vessel | 8 | 10.8 |
| Vessel | 6 | 8.1 |
| Aneurysm | 2 | 2.7 |
| Dural Sinus | 5 | 6.8 |
| Non-specific | 5 | 6.8 |
| Imaging artifact | 3 | 4.1 |
| High intrinsic gyral density | 3 | 4.1 |
| Post-surgical | 3 | 4.1 |
| Cortical laminar necrosis | 2 | 2.7 |
| Osseous lytic lesion | 2 | 2.7 |
Image quality assessment using radiomic and textural analysis
During analysis, we noticed extensive noise or artifact on several of the discordant studies (e.g., Fig. 2D). Therefore, we sought to see if systematic differences between our concordant and discordant studies might be uncovered in CT radiomics and CT textural analysis.26,27 A diverse set of 33 first order and higher ordered textural features were calculated in the centrum semiovale, the thalamus and ventricle (Fig. 3) and were compared between the ICH+, concordant studies and both the false negative studies and all discordant studies. After correction for multiple comparisons, no significant textural differences were observed for the either the false negative studies or the discordant studies (Supplemental Table 1).
Figure 3: Example regions of interest (ROI) for CT radiomics and quantitative image characterization.
Spherical ROIs with a 10 mm diameter were drawn in the centrum semiovale (teal), the thalamus (green) and ventricle (blue), to represent white matter, gray matter, and CSF, respectively.
Discussion.
We performed a retrospective analysis of the performance of a deep learning DSS for the detection of ICH at our clinical site. We measured the diagnostic performance of Aidoc, identified common etiologies of false positive findings and find that age, surgical history, ICH type and isolated ICH (versus multiple foci of ICH) are associated with decreased algorithm performance. Furthermore, we demonstrate that diagnostic performance is not related to quantitative differences in image quality when assessed by CT textural analyses. We observed that most of the incorrect Aidoc flags were false positives and the etiologies of these errors are consistent with the sources of false positives noted in prior studies, typically hyperdense structures or imaging artifacts.17,20,28
We and others17, 20 have noted a small number of false positives that are directly attributable to post-surgical changes. However, these cases cannot fully account for the impaired performance in post-surgical patients. Even in cases where surgical changes are not directly flagged, the error rate is three-fold higher in patients with a history of neurosurgery (2.4% vs 7.5%) and this compromised performance is seen with elevations in the rates of both the false negative and false positive studies. Because the mechanisms of neural networks are not readily interrogated, we cannot definitively determine the mechanisms for this observation, but in light of these changes in diagnostic performance, future deep learning models for detection of ICHs should more robustly include post-surgical patients to improve overall performance.
We also observed decreased diagnostic performance in patients with only a single ICH type relative to those with mixed ICHs. This could be explained in part by Aidoc only requiring a single bleed to score the study as positive for ICH. The presence of additional bleeds on a single study increases the probability that any bleed is detected. There was also decreased performance of the DSS in cases of a single ICH type compared to the ICH− studies, possibly due to the relatively lower number of ICH+ scans in our sample. Within the single ICH types, there were slight difference in the DSS performance, although the small number of bleeds within each subgroup increases the probability of spurious findings. Male sex was also associated with worsened performance of the algorithm in the univariable analysis, although this effect was not significant in the multivariable logistic regression and may simply be due to chance or confounding.
Next, we examined if image quality to might be responsible for the false negative findings. Poor image quality or artifacts can complicate or preclude interpretation by human readers, and we hypothesized that the performance of AI tools may also depend on image characteristics. Specifically, differences in diagnostic performance could stem from poor image quality or significant differences in image quality and that incorrectly flagged studies might have worse image quality compared to the correctly flagged studies. As examples, beam hardening artifacts29 and excessive noise30 alter the textural features of CT datasets. However, we did not observe any systematic differences between our concordant and discordant studies in our CT radiomics and CT textural analysis. Therefore, it seems unlikely that image quality per se is a meaningful failure mode in our data, although it remains to be seen if AI tools are insensitive to image quality as image quality may have different meanings for machine vision applications versus human visual interpretation.
Overall, our estimates of the specificity and NPV agree with prior studies of the performance of Aidoc. However, we observed lower sensitives and PPV than in prior reports (Table 4).8,17,18,20,28 The lower PPV could be explained in part by the lower prevalence of ICH in our study, as prior studies report higher rates of ICHs, with correspondingly higher PPVs. However, sensitivity is independent of the prevalence and we sought to identify explanations for our elevated rate of false positive findings. As none of the population or study-specific factors we examined account for the impaired diagnostic performance of the AI DSS at our site when compared to previous reports, we hypothesize that in addition to unaccountable model biases, these differences may be explained in large part by study design. Numerous prior studies have relied on small sample sizes or enriched their data set with ICH+ studies.8, 18, 28 The largest study to date used the presence of repeat NCCTs to indicate a presumed ICH.17 While simplifying analysis, this necessarily biases the data set with an increased prevalence of ICH and thus inflates measures of diagnostic accuracy including PPV. Indeed, a manual review of a subset of their studies revealed a 2% error rate.17 Our study design circumvents these limitations and to the best of our knowledge, is the largest to use manual review of the neuroradiologist interpretation to establish a ground truth and to include the true prevalence of ICH in our study population. There are numerous barriers to performing multi-institutional studies, meaning single site studies will play an outsized role in evaluating the performance of deep learning algorithms and AI DSS. Our work suggests that the development of a common set of rigorous study criteria will facilitate collaboration, helping safely grow the role of AI DSS in clinical practice.
Table 4.
Operating characteristics of Aidoc in this and similar studies. Values are reported as percentages with 95% confidence intervals in parenthesis.
| This study | Aidoc 510(k)18 |
Chodakiewitz28 | Ojeda17 | Ginat20 | Wismuller8 | |
|---|---|---|---|---|---|---|
| Sens | 92.3% (88.9-94.8) |
93.6% (86.6-97.6) |
96.2% (93.2-98.2) |
95% (93.9-96.1) |
88.7% (84.7-92.0) |
95 (88.6-98.0) |
| Spec | 97.7% (97.2-98.2) |
92.3% (85.4-96.6) |
93.3% (89.6-96.0) |
99% (98.4-99.0) |
94.2% (93.0-95.3) |
96.7 (94.7-98.0) |
| PPV | 81.3% (77.6-84.5) |
91.7%1 (84.9-95.6) |
93.4% (90.1-95.7) |
96% (94.6 – 96.6) |
73.7 (69.8-77.4) |
86.1%1 (79.4-90.8) |
| NPV | 99.2% (98.8-99.4) |
94.1%1 (88.4-97.2) |
96.2% (93.2-97.9) |
98% (98.2-98.8) |
97.7 (97.1-98.4) |
99.0%1 (97.4-99.4) |
| ICH+ (%) | 9.70% | 47.5%1 | 49.7% | 23.2% | 20.3% | 17.9% |
| Total N | 3605 | 198 | 533 | 7112 | 2011 | 620 |
ICH+: Intracranial hemorrhage positive
Values were inferred from reported sensitivity, specificity, and N.
AI DSSs are not limited to the detection of ICHs. Algorithms have been developed for breast cancer screening,31 detection of pneumonia,32 pulmonary nodules,33 and others. While the performance of these systems has improved in recent years, challenges have arisen in rigorously assessing the performance of algorithms34 and a failure of the algorithms to generalize to broad patient populations,13,35 similar to what we observed in our work here. The implementation of reporting standards (i.e. STARD) has facilitated the comparison between trials,36 although there can still be considerable differences even among STARD conforming studies.34,37,38 Generalizability can be improved by diversifying training sets, especially with the inclusion of local data.39 While site-specific training of DSSs is not currently permitted by the FDA, there is a movement towards relaxing these restrictions with the goal of improving algorithm performance.40
Our study has limitations. As with other studies examining diagnostic accuracy of deep learning technologies, our study is a retrospective single site study and the degree to which our findings are generalizable to other clinical sites remains unknown. However, our study design, which examined consecutive NCCT head examinations performed in our emergency department, provides an unfiltered review of imaging cases evaluated by our AI DSS and allows for transparent comparison to future studies investigating diagnostic accuracy of ICH deep learning algorithms. An addition limitation is our inability to probe the source and frequency of false negative exams in further detail due to the black box nature of the AI algorithm. Another possible limitation is the assumption of accuracy in cases with concordant Aidoc and radiologist findings. Because each may have identified separate, albeit coincident, findings, this could artificially inflate the sensitivity of the model. Similarly, each may have independently failed to identify the same ICH, as it was not feasible to reassess all concordant scans. However, it is unlikely that enough ICHs were missed to significantly change our findings. Another potential limitation to our study is the equipment used. All of our studies were performed on a suite of GE CT scanners and while we did not find quantitative differences in image quality or texture between correctly and incorrectly flagged studies, potential vendor differences were not assessed and should be a consideration in future studies. Lastly, non-emergent referrals and other patient factors such as socioeconomic status and race were not considered in this work and will serve as the basis of future studies.
Supplementary Material
Take-home points:
Unexpected lower sensitivity and positive predictive values were observed for an artificial intelligence decision support system for intracranial hemorrhage detection, raising concerns about the generalizability of deep learning tools.
Decreased diagnostic performance was associated with prior neurosurgery, type and number of hemorrhages, but is not associated with image quality.
Comparisons of decision support systems is complicated by variations in study design. Creation of standardized study parameters will facilitate the unbiased evaluation of these valuable tools.
Acknowledgements:
The authors would like to thank Aidoc for assistance in processing the NCCTs and HealthMyne for technical assistance with the textural analysis.
Funding information:
A.F.V. was supported by NIH F30 CA210465 and T32 GM008692; J.J.Y. was supported by the Clinical and Translational Science Award (CTSA) program, through the NIH National Center for Advancing Translational Sciences (NCATS), grant UL1TR002373. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
The authors declare no conflict of interest
The authors declare that they had full access to all of the data in this study and the authors take complete responsibility for the integrity of the data and the accuracy of the data analysis.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Hess EP, Haas LR, Shah ND, Stroebel RJ, Denham CR, Swensen SJ. Trends in computed tomography utilization rates: a longitudinal practice-based study. J Patient Saf 2014, 10(1), 52–8. [DOI] [PubMed] [Google Scholar]
- 2.Kocher KE, Meurer WJ, Fazel R, Scott PA, Krumholz HM, Nallamothu BK. National trends in use of computed tomography in the emergency department. Ann Emerg Med 2011, 58 (5), 452–62 e3. [DOI] [PubMed] [Google Scholar]
- 3.Elliott J, Smith M. The acute management of intracerebral hemorrhage: a clinical review. Anesth Analg 2010, 110 (5), 1419–27. [DOI] [PubMed] [Google Scholar]
- 4.Fujitsu K, Muramoto M, Ikeda Y, Inada Y, Kim I, Kuwabara T. Indications for surgical treatment of putaminal hemorrhage. Comparative study based on serial CT and time-course analysis. J Neurosurg 1990, 73 (4), 518–25. [DOI] [PubMed] [Google Scholar]
- 5.McDonald RJ, Schwartz KM, Eckel LJ, Diehn FE, Hunt CH, Bartholmai BJ, Erickson BJ, Kallmes DF. The effects of changes in utilization and technological advancements of cross-sectional imaging on radiologist workload. Acad radiol 2015, 22 (9), 1191–1198. [DOI] [PubMed] [Google Scholar]
- 6.Sharp AL, Huang BZ, Tang T, Shen E, Melnick ER, Venkatesh AK, Kanter MH, Gould MK. Implementation of the Canadian CT Head Rule and Its Association With Use of Computed Tomography Among Patients With Head Injury. Ann Emerg Med 2018, 71 (1), 54–63 e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mower WR, Gupta M, Rodriguez R, Hendey GW. Validation of the sensitivity of the National Emergency X-Radiography Utilization Study (NEXUS) Head computed tomographic (CT) decision instrument for selective imaging of blunt head injury patients: An observational study. PLoS Med 2017, 14 (7), e1002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wismuller A, Stockmaster L. A prospective randomized clinical trial for measuring radiology study reporting time on Artificial Intelligence-based detection of intracranial hemorrhage in emergent care head CT. Proc SPIE 2020, 11317. [Google Scholar]
- 9.Litjens G, Kooi T, Bejnordi BE, Setio AAA, Ciompi F, Ghafoorian M, van der Laak J, van Ginneken B, Sanchez CI. A survey on deep learning in medical image analysis. Med Image Anal 2017, 42, 60–88. [DOI] [PubMed] [Google Scholar]
- 10.Data Science Institute. FDA Cleared AI Algorithms. (accessed 9/15/2020).
- 11.Alwosheel A, van Cranenburgh S, Chorus CG. Is your dataset big enough? Sample size requirements when using artificial neural networks for discrete choice analysis. J Choice Model 2018, 28, 167–182. [Google Scholar]
- 12.Park SH, Han K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology 2018, 286 (3), 800–809. [DOI] [PubMed] [Google Scholar]
- 13.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: A cross-sectional study. PLoS Med 2018, 15 (11), e1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamashita R, Nishio M, Do RKG, Togashi K. Convolutional neural networks: an overview and application in radiology. Insights Imaging 2018, 9 (4), 611–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim DW, Jang HY, Kim KW, Shin Y, Park SH. Design Characteristics of Studies Reporting the Performance of Artificial Intelligence Algorithms for Diagnostic Analysis of Medical Images: Results from Recently Published Papers. Korean J Radiol 2019, 20 (3), 405–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu X, Faes L, Kale AU, Wagner SK, Fu DJ, Bruynseels A, Mahendiran T, Moraes G, Shamdas M, Kern C, Ledsam JR, Schmid MK, Balaskas K, Topol EJ, Bachmann LM, Keane PA, Denniston AK. A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. The Lancet Digital Health 2019, 1 (6), e271–e297. [DOI] [PubMed] [Google Scholar]
- 17.Ojeda P, Zawaideh M, Mossa-Basha M, Haynor D. The utility of deep learning: evaluation of a convolutional neural network for detection of intracranial bleeds on non-contrast head computed tomography studies. Proc SPIE 2019, 10949. [Google Scholar]
- 18.K180647. US Food and Drug Administration. Silver Spring, MD, 2018. [Google Scholar]
- 19.Rao B, Zohrabian V, Cedeno P, Saha A, Pahade J, Davis MA. Utility of artificial intellegence tool as a prospective radiology peer reviewer – detection of unreported intracranial hemorrhage. Acad Radiology 2020, 1–9. [DOI] [PubMed] [Google Scholar]
- 20.Ginat DT. Analysis of head CT scans flagged by deep learning software for acute intracranial hemorrhage. Neuroradiology 2020, 62 (3), 335–340. [DOI] [PubMed] [Google Scholar]
- 21.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology 2016, 278 (2), 563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lambin P, Leijenaar RT, Deist TM, Peerlings J, De Jong EE, Van Timmeren J, Sanduleanu S, Larue RT, Even AJ, Jochems A. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin oncol 2017, 14 (12), 749–762. [DOI] [PubMed] [Google Scholar]
- 23.Amadasun M, King R. Textural features corresponding to textural properties. IEEE T Syst Man Cyb 1989, 19 (5), 1264–1274. [Google Scholar]
- 24.Ganeshan B, Miles KA, Young RC, Chatwin CR. Texture analysis in non-contrast enhanced CT: impact of malignancy on texture in apparently disease-free areas of the liver. Eur J Radiol 2009, 70 (1), 101–10. [DOI] [PubMed] [Google Scholar]
- 25.Cruz-Bastida JP, Gomez-Cardona D, Garrett J, Szczykutowicz T, Chen GH, Li K. Modified ideal observer model (MIOM) for high-contrast and high-spatial resolution CT imaging tasks. Med Phys 2017, 44 (9), 4496–4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tao S, Rajendran K, Zhou W, Fletcher JG, McCollough CH, Leng S. Noise Reduction in CT Image Using Prior Knowledge Aware Iterative Denoising. Phys Med Biol 2020, Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kociolek M, Strzelecki M, Obuchowicz R. Does Image Normalization and Intensity Resolution impact Texture Classification? Comput Med Imaging Graph 2020, 81:101716. [DOI] [PubMed] [Google Scholar]
- 28.Chodakiewitz YG, Maya MM, Pressman BD. Prescreening for Intracranial Hemorrhage on CT Head Scans with an AI-Based Radiology Workflow Triage Tool: An Accuracy Study. J Med Diag Meth 2019, 8 (2), 1–5. [Google Scholar]
- 29.Ger RB, Craft DF, Mackin DS, Zhou S, Layman RR, Jones AK, Elhalawani H, Fuller CD, Howell RM, Li H, Stafford RJ, Court LE. Practical Guidelines for Handling Head and Neck Computed Tomography Artifacts for Quantitative Image Analysis. Comput Med Imaging Graph 2018, 69, 134–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bagher-Ebadian H, Siddiqui F, Liu C, Movsas B, Chetty IJ. On the Impact of Smoothing and Noise on Robustness of CT and CBCT Radiomics Features for Patients with Head and Neck Cancers. Med Phys 2017, 44 (5), 1755–1770. [DOI] [PubMed] [Google Scholar]
- 31.Yassin NIR, Omaran S, El Houby EMF, Allam H. Machine Learning Techniques for Breast Cancer Computer Aided Diagnosis Using Different Image Modalities: A Systematic Review. Comput. Meth. Prog. Bio 2018, 156:25–45. [DOI] [PubMed] [Google Scholar]
- 32.Li Y, Zhang Z, Dai C, Dong Q, Badrigilan S. Accuracy of Deep Learning for Automated Detection of Pneumonia Using Chest X-Ray Images: A Systematic Review and Meta-analysis. Comput. Biol. Med 2020, 123:103898. [DOI] [PubMed] [Google Scholar]
- 33.Ather S, Kadir T, Gleeson F. Artificial Intelligence and Radiomics in Pulmonary Nodule Management: Current Status and Future Applications. Clin. Radiol 2020, 75(1):13–19. [DOI] [PubMed] [Google Scholar]
- 34.Nagendran M, Chen Y, Lovejoy CA, Gordon AC, Komorowski M, Harvey H, Topol EJ, Ioannidis JPA, Collins GS, Maruthappu M. Artificial Intelligence Versus Clinicians: Systematic Review of Design, Reporting Standards and Claims of Deep Learning. BMJ 2020, 368:m689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X, Liang G, Zhang Y, Blanton H, Bessinger Z, Jacobs N. Inconsistent Performance of Deep Learning Models on Mammogram Classification. J. Am. Coll. Rad 2020, 17(6):796–803. [DOI] [PubMed] [Google Scholar]
- 36.Cohen JF, Korevaar DA, Altman DG, Bruns DE, Gatsonis CA, Hooft L, Irwig L, Levine D, Reitsma JB, de Vet HCW, Bossuyt PMM. STARD 2015 Guidelines for Reporting Diagnostic Accuracy Studies: Explanation and Elaboration. BMJ Open 2016, 6e012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Korevaar DA, Wang J, Van Est WA, Leeflang MM, Hooft L, Smidt N, Bossuyt PMM. Reporting Diagnostic Accuracy Studies: Some Improvements After 10 Years of STARD. Radiology 2015, 274(3), 781–789. [DOI] [PubMed] [Google Scholar]
- 38.Larson DB, Harvey H, Rubin DL, Irani N, Tse JR, Langlotz CP. Regulatory Frameworks for Development and Evaluation of Artificial Intelligence-Based Diagnostic Imaging Algorithms: Summary and Recommendations. J. Am. Coll. Rad 2020, in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.: McKinney SM, Sieniek M, Godbole V, Godwin J, Antropova N, Ashrafian H, et al. International Evaluation of an AI System for Breast Cancer Screening. Nature 2020, 577:89–94. [DOI] [PubMed] [Google Scholar]
- 40.US FDA. Proposed Regulatory Framework for Modifications to Artificial Intelligence/Machine Learning (AI/ML)-Based Software as a Medical Device (SaMD): Discussion Paper and Request for Feedback. Available at https://www.fda.gov/media/122535/download. Published April 2, 2019. Accessed January 28, 2021.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.