Abstract
We investigated changes in the predicted functions of the rumen bacterial community in Japanese Black beef cattle during fattening. Nine cattle were fed a high-concentrate diet during the early, middle, and late fattening stages consecutively (10–14, 15–22, and 23–30 months of age, respectively). The rumen fluid and solid samples collected at each stage were subjected to sequencing analyses. The sequencing results were clustered and classified into operational taxonomic units (OTUs). Representative sequences and a raw counting table for each OTU were submitted to the Piphillin website. The predicted functions were revealed by the Kyoto Encyclopedia of Genes and Genomes database as the ratio of the total sequence. In the early stage, “Biosynthesis of secondary metabolites” was significantly higher in the fluid fraction than in the solid fraction. “Two-component system” in the middle stage was significantly lower and “Purine metabolism” in the late stage was significantly higher in the fluid fraction than those in the solid fraction. The fluid fraction was significantly correlated with acetic acid, propionic acid, and bacterial metabolism, such as “Biosynthesis of secondary metabolites” and “Sugar metabolism.” Moreover, the solid fraction was correlated with “Purine metabolism” and “Biosynthesis of secondary metabolism”. These results suggest that the rumen bacterial community in Japanese Black beef cattle adapts to changes in rumen conditions by altering their functions in response to a long-term high-grain diet.
Keywords: bacterial function, Japanese Black beef cattle, Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, rumen fluid and solid bacteria
Japanese Black cattle are characterized by the ability to deposit a large amount of intramuscular fat and typically consume a large amount of concentrated diet from the age of 10–30 months [22, 23]. This, in addition to the lack of roughage, can lead to excessive generation and accumulation of volatile fatty acids (VFAs), decreasing ruminal pH, such as subacute ruminal acidosis (SARA) [10, 18, 22, 23]. Long-term high-grain diet feeding does not acutely alter bacterial diversity but causes bacteria to adapt and remain stable against the decrease in pH and changes in fermentation to enhance lactic acid production in Japanese Black beef cattle [23]. Furthermore, acute hepatitis related to SARA is a major problem in beef farms [18]. Meanwhile, the ruminal bacterial community and ruminal pH can adapt to and influence each other [19], and the effects of short- (days) and mid-term (weeks) SARA and RA challenges have been explored previously [12, 19, 30]. In general, grain-based SARA challenge induces death or lysis of gram-negative bacteria, such as Bacteroidetes and Proteobacteria, and eventually the proportion of Firmicutes increases with severely low ruminal pH [12, 19].
Large-scale sequencing has been used to detect large amounts of genomic DNA or RNA sequences, and the number of gene expression studies has increased. Ruminal pH, VFAs, and bacterial community structure change dramatically during the fattening stage in Japanese Black beef cattle [23]. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses enable the prediction of the meaning of screened DNA or RNA sequences from large-scale sequencing results. A previous KEGG pathway analysis revealed that live yeast contributes to the rumen microbiota of beef cattle [24]. Furthermore, the rumen microbiome, as a reservoir of antimicrobial resistance, is directly affected by diet [1] and by dietary energy sources and levels that shift the multi-kingdom microbiota and functions in the rumen of dairy cows [26]. However, no study to date has conducted predicted functional analyses of the rumen bacterial community in Japanese Black beef cattle during the fattening stages.
MATERIALS AND METHODS
We re-analyzed our previously published data regarding rumen fermentation parameters and sequencing data [22] to evaluate the relationship between rumen measurements and the predicted functions of the bacterial community in Japanese Black beef cattle. All experimental procedures were approved by the Iwate University Laboratory Animal Care and Use Committee (A201720) and the Hyogo Prefectural Technology Center for Agriculture, Forestry, and Fisheries.
Animals, experimental design, sampling, and measurements
Experimental animals, design, and measurements have been previously described [23]. Briefly, Japanese Black beef cattle (n=9) were castrated and equipped with a rumen cannula. The fattening period was subdivided into early, middle, and late stages (10–14, 15–22, and 23–30 months of age, respectively) according to a general agreement in Japan [25]. The cattle were fed concentrate and forage (rice straw) diets during all three stages, and the amount of the concentrate diet was increased gradually throughout the experimental period. The forage diet was supplied daily at 0930 and 1530 hr in two equal portions, and the concentrate was supplied 1-hr after rice straw. A calculated amount of roughage and the concentrate diet was given for a daily gain of 0.8 kg/day during all stages. The forage-to-concentrate ratios were 26:74, 13:87, and 14:86 during the early, middle, and late stages, respectively. The adequacy rate of the diet was calculated based on the nutrient requirements of the Japanese feeding standard for beef cattle [21].
Samples were collected and measurements were made as previously described [23]. Briefly, ruminal pH was measured continuously every 10 min during the final 7 days of the early, middle, and late fattening stages using a radio transmission system (YCOW-S; DKK-TOA, Yamagata, Japan), as previously described [31]. Rumen samples were collected through the rumen cannula every fourth day during the final 7 days of the pH measurements at each stage to analyze the rumen bacterial community, VFAs, lactic acid concentration, and lipopolysaccharide (LPS) activity. The rumen samples were filtered through two layers of cheesecloth, the fluid was treated as the fluid fraction, and the remaining sample on the cheesecloth was collected as the solid fraction.
The ruminal concentrations of VFAs (i.e., acetic acid, propionic acid, and butyric acid) were quantified by gas chromatography (GC-2014; Shimadzu, Kyoto, Japan). The lactic acid concentration was determined using a commercially available kit (F-kit [d-lactate/l-lactate], J.K. International Co., Tokyo, Japan). Ruminal LPS activity was assayed using a kinetic Limulus amebocyte lysate assay (Pyrochrome with Glucashield; Seikagaku Corp., Tokyo, Japan).
DNA extraction and sequencing
DNA extraction and Illumina sequencing analyses were performed as previously described [22]. Briefly, total bacterial DNA was extracted from the fluid and solid portions, as described by Kim et al. [15]. Sequencing libraries were prepared according to the Illumina 16S Metagenomic Sequencing Library preparation guide (2013). The bacterial 16S rRNA gene was amplified using the barcoded universal primers 515F (5-TGYCAGCMGCCGCGGTAA-3) and 805R (5-GACTACHVGGGTATCTAATCC-3) spanning the V4 hypervariable region. The sequence data were deposited into the Sequence Read Archive (SRA) of the National Center for Biotechnology Information database and can be accessed via SRA accession number PRJNA548210 (https://submit.ncbi.nlm.nih.gov/subs/sra/).
Sequencing data analyses
We used the sequencing data presented by Ogata et al. [22], analyzed all sequencing results using the MOTHUR program (version 1.41.1; University of Michigan; http://www.mothur.org/wiki/; Schloss) [32], and mapped sequence data with the SILVA reference database (SSURef release 128; [27]). Then, candidate sequences were screened and filtered, as follows: unique sequences were determined; candidate sequences were pre-clustered to eliminate outliers; chimeras were identified using the “chimera.vsearch” command; and sequence comparison was performed using the Mothur Ribosomal Database Project training set (version 16). Sequences were clustered and classified into operational taxonomic units (OTUs) using a cutoff value of 97% similarity. All samples were standardized by random subsampling (6,741 sequences/sample) using the “sub.sample” command.
Representative sequences for each OTU were set using the BLASTn program (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and compared to the 16S ribosomal RNA sequence database (Bacteria and Archaea; May 2019). Furthermore, representative sequences and tabulated raw count data were submitted to the Piphillin website (http://piphillin.secondgenome.com/; [13]) and analyzed. A sequence identity cutoff of 97% was applied to analyze the functional categories, and metagenomic functions were assigned using the KEGG database (November 2019).
Statistical analyses
The normality of the data distribution was assessed using the Shapiro-Wilk test. Significant differences (between groups) in variables during the early, middle, and late stages were detected using Student’s t-test for normal variables and Welch’s t-test for non-normal variables. Significant changes (within groups) in ruminal pH, VFAs, lactic acid concentration, and ruminal LPS activity during the early, middle, and late stages were assessed using the Friedman test, and the relative abundances of bacteria and their functions were assessed using the Kruskal-Wallis test. Correlation coefficients between the predicted function of rumen bacteria and rumen measurements were calculated. Pearson’s correlation coefficient (r) and significance levels (P-values) were used to determine the relationships between rumen measurements and the predicted functions of the bacterial community. All numerical data were analyzed using Prism version 8.10 (GraphPad Software Inc., La Jolla, CA, USA). Statistical significance was set at P<0.05.
RESULTS
pH and volatile fatty acids
The 24-hr mean pH, total VFAs, and acetic acid concentration decreased as the fattening stages proceeded. Propionic acid concentration increased during the middle stage. LPS activity increased throughout the study period (Table 1).
Table 1. Twenty four-hr mean pH, total volatile fatty acid (VFA), individual VFA proportions, acetic acid-to-propionic acid (A/P) ratio, lactic acid concentrations, and lipopolysaccharide (LPS) activity in Japanese Black beef cattle (n=9) during the fattening stages.
| Item | Stage |
P-valuea | |||
|---|---|---|---|---|---|
| Early | Middle | Late | |||
| 24-hr mean pH | 6.22 ± 0.06 | 6.06 ± 0.08 | 5.73 ± 0.13 | 0.031 | |
| Total VFA (mmol/dl) | 13.1 ± 0.59 | 12.3 ± 0.80 | 9.77 ± 0.76 | 0.057 | |
| Acetate (%) | 62.6 ± 2.04 | 57.1 ± 1.53 | 58.6 ± 1.50 | 0.329 | |
| Propionate (%) | 21.4 ± 1.81 | 27.1 ± 2.23 | 27.1 ± 2.21 | 0.187 | |
| Butyrate (%) | 11.9 ± 1.04 | 12.7 ± 0.86 | 11.2 ± 0.85 | 0.154 | |
| A/P ratio | 3.05 ± 0.22 | 2.24 ± 0.21 | 2.34 ± 0.28 | 0.154 | |
| Lactic acid (mg/l) | 67.9 ± 0.01 | 25.0 ± 0.00 | 141.4 ± 0.01 | <0.001 | |
| LPS (EU×104/ml) | 1.34 ± 0.39 | 4.29 ± 1.82 | 6.62 ± 2.24 | 0.048 | |
aFriedman test was used to determine within-group differences.
Bacterial abundance
The top 10 bacterial families comprised more than 80% of all sequences during each stage and in each fraction (fluid or solid) (Table 2). Four families (Lachnospiraceae, Ruminococcaceae, Coriobacteriaceae, and Prevotella) accounted for more than 60% of all bacteria in the fluid fraction throughout the fattening stages. Four of the five families (the above four families and Firmicutes-unclassified family) were the same in the solid fraction as in the fluid fraction, which included more than 60% of all bacteria during the early and middle stages, and six families (the above five families and the Acidaminocoddaceae family) accounted for more than 60% of all sequences during the late stage.
Table 2. Relative abundance of major bacterial families during each fattening stage (early, middle, and late stages) in the fluid and solid fractions of Japanese Black beef cattle (n=9).
| Item | Stage |
|||||
|---|---|---|---|---|---|---|
| Early |
Middle |
Late |
||||
| Fluid | Solid | Fluid | Solid | Fluid | Solid | |
| Lachnospiraceae | 19.6 ± 0.53 | 21.3 ± 0.51 | 15.4 ± 0.57 | 22.8 ± 0.50 | 20.8 ± 0.57 | 23.7 ± 0.56 |
| Ruminococcaceae | 25.4 ± 0.56 | 10.8 ± 0.54a | 14.9 ± 0.60 | 6.55 ± 0.52 | 16.6 ± 0.59 | 8.74 ± 0.58 |
| Coriobacteriaceae | 6.68 ± 0.94 | 11.8 ± 0.93 | 9.41 ± 1.00 | 16.2 ± 0.90a | 13.8 ± 1.02 | 11.1 ± 1.01 |
| Prevotellaceae | 8.72 ± 0.84 | 4.49 ± 0.84 | 23.7 ± 0.93 | 10.4 ± 0.81 | 9.36 ± 0.91 | 4.33 ± 0.90 |
| Firmicutes_unclassified | 9.39 ± 0.52 | 18.0 ± 0.51a | 4.66 ± 0.56 | 7.76 ± 0.49 | 5.38 ± 0.56 | 10.5 ± 0.56 |
| Acidaminococcaceae | 3.46 ± 0.18 | 1.04 ± 0.17 | 4.26 ± 0.21 | 5.14 ± 0.16 | 5.27 ± 0.19 | 14.8 ± 0.19a |
| Clostridiales_unclassified | 4.10 ± 0.48 | 8.15 ± 0.47a | 2.55 ± 0.52 | 7.46 ± 0.45a | 2.33 ± 0.51 | 4.48 ± 0.51a |
| Clostridiales_Incertae_Sedis_XIII | 1.72 ± 0.48 | 7.95 ± 0.47a | 2.10 ± 0.52 | 7.64 ± 0.45a | 1.61 ± 0.51 | 3.90 ± 0.51a |
| Veillonellaceae | 1.50 ± 0.67 | 2.23 ± 0.66 | 2.14 ± 0.74 | 4.88 ± 0.65a | 2.04 ± 0.73 | 4.02 ± 0.72 |
| Bacteria_unclassified | 3.00 ± 1.21 | 2.51 ± 1.21 | 3.98 ± 1.30 | 1.97 ± 1.16 | 2.77 ± 1.30 | 1.97 ± 1.30 |
aDenotes a significant difference (P<0.05) between the solid and fluid fractions.
Predicted functions of rumen bacteria
The predicted functions of the sequence data were revealed against the KEGG database (November 2019) as the ratio of the total sequences. Comparisons between the fluid and solid fractions during each stage are presented in Table 3. “Metabolic pathway” (KEGG pathway object identifier; ko01100) and “Biosynthesis of secondary metabolites” (ko01110) were significantly higher (P<0.05) in the fluid fraction than in the solid fraction during the early stage. “Microbial metabolism in diverse environments” (ko01120) was significantly higher (P<0.05) in the solid fraction than in the fluid fraction. “Ribosome” (ko03010) was significantly higher (P<0.05) in the fluid fraction than in the solid fraction during the middle stage, and “Microbial metabolism in diverse environments” (ko01120) and “Two-component system” (ko02020) were higher in the solid fraction than in the fluid fraction. “Ribosome” (ko03010) and “Purine metabolism” (ko00230) were significantly higher (P<0.05) in the fluid fraction than in the solid fraction during the late stage.
Table 3. Relative abundance of the bacterial predicted functions during each fattening stage (early, middle, and late stages) in the fluid and solid fractions of Japanese Black beef cattle (n=9).
| Item | Stage |
|||||
|---|---|---|---|---|---|---|
| Early |
Middle |
Late |
||||
| Fluid | Solid | Fluid | Solid | Fluid | Solid | |
| Metabolic pathways | 16.3 ± 0.15 | 15.7 ± 0.14a | 16.5 ± 0.22 | 16.0 ± 0.03 | 16.1 ± 0.11 | 16.0 ± 0.10 |
| Biosynthesis of secondary metabolites | 7.44 ± 0.06 | 7.12 ± 0.04a | 7.57 ± 0.07 | 7.32 ± 0.04 | 7.38 ± 0.06 | 7.31 ± 0.03 |
| Biosynthesis of antibiotics | 5.65 ± 0.03 | 5.61 ± 0.04 | 5.77 ± 0.04 | 5.77 ± 0.04 | 5.78 ± 0.04 | 5.94 ± 0.05 |
| Biosynthesis of amino acids | 4.20 ± 0.05 | 4.08 ± 0.07 | 4.34 ± 0.08 | 4.24 ± 0.06 | 4.29 ± 0.05 | 4.30 ± 0.05 |
| Microbial metabolism in diverse environments | 4.05 ± 0.05 | 4.29 ± 0.07a | 4.00 ± 0.04 | 4.19 ± 0.07a | 4.13 ± 0.04 | 4.18 ± 0.04 |
| Aminoacyl-tRNA biosynthesis | 2.85 ± 0.15 | 2.69 ± 0.13 | 3.10 ± 0.19 | 2.63 ± 0.13 | 2.65 ± 0.10 | 2.55 ± 0.13 |
| ABC transporters | 2.58 ± 0.18 | 3.07 ± 0.11 | 2.26 ± 0.29 | 2.86 ± 0.11 | 2.81 ± 0.14 | 2.73 ± 0.20 |
| Ribosome | 2.67 ± 0.05 | 2.53 ± 0.04 | 2.78 ± 0.06 | 2.53 ± 0.04a | 2.62 ± 0.03 | 2.53 ± 0.02a |
| Carbon metabolism | 2.33 ± 0.01 | 2.35 ± 0.02 | 2.31 ± 0.02 | 2.34 ± 0.02 | 2.33 ± 0.01 | 2.36 ± 0.01 |
| Purine metabolism | 2.07 ± 0.02 | 2.04 ± 0.02 | 2.07 ± 0.03 | 1.99 ± 0.02 | 2.03 ± 0.02 | 2.00 ± 0.01a |
| Pyrimidine metabolism | 1.72 ± 0.01 | 1.70 ± 0.02 | 1.69 ± 0.02 | 1.64 ± 0.02 | 1.68 ± 0.02 | 1.69 ± 0.02 |
| Quorum sensing | 1.51 ± 0.10 | 1.70 ± 0.02 | 1.55 ± 0.16 | 1.62 ± 0.02 | 1.55 ± 0.06 | 1.60 ± 0.07 |
| Amino sugar and nucleotide sugar metabolism | 1.55 ± 0.04 | 1.61 ± 0.03 | 1.36 ± 0.06 | 1.44 ± 0.03 | 1.46 ± 0.04 | 1.43 ± 0.04 |
| Starch and sucrose metabolism | 1.34 ± 0.04 | 1.41 ± 0.04 | 1.27 ± 0.06 | 1.39 ± 0.04 | 1.42 ± 0.04 | 1.46 ± 0.06 |
| Fructose and mannose metabolism | 1.02 ± 0.07 | 1.27 ± 0.08a | 0.83 ± 0.09 | 1.10 ± 0.08 | 1.07 ± 0.07 | 1.11 ± 0.06 |
| Two-component system | 1.05 ± 0.08 | 1.14 ± 0.11 | 0.90 ± 0.03 | 1.07 ± 0.10a | 0.97 ± 0.06 | 0.96 ± 0.02 |
| Glycolysis/Gluconeogenesis | 1.04 ± 0.02 | 1.07 ± 0.03 | 0.99 ± 0.03 | 1.02 ± 0.03 | 1.03 ± 0.02 | 1.04 ± 0.01 |
| Phosphotransferase system (PTS) | 0.85 ± 0.17 | 1.44 ± 0.18 | 0.64 ± 0.17 | 1.16 ± 0.18 | 1.09 ± 0.16 | 1.14 ± 0.15 |
| Homologous recombination | 0.94 ± 0.03 | 0.94 ± 0.04 | 0.99 ± 0.04 | 1.02 ± 0.04 | 1.04 ± 0.03 | 1.08 ± 0.04 |
| Pyruvate metabolism | 0.92 ± 0.03 | 1.00 ± 0.02 | 0.88 ± 0.05 | 1.01 ± 0.02 | 1.01 ± 0.02 | 1.07 ± 0.03 |
aDenotes a significant difference (P<0.05) between the solid and fluid fractions.
Comparisons of the fluid and solid fractions among the fattening stages are shown in Table 4. In the solid fraction during the late fattening stage, “Biosynthesis of secondary metabolites” (ko01110) and “Biosynthesis of antibiotics” (ko01130) significantly increased (P<0.05), and “Amino sugar and nucleotide sugar metabolism” (ko00520) significantly decreased (P<0.05).
Table 4. Relative abundance of the bacterial predicted functions in each fraction (fluid and sold fractions) of Japanese Black beef cattle (n=9) during the fattening stages.
| Items | Fluid |
P-valuea | Solid |
P-value | ||||
|---|---|---|---|---|---|---|---|---|
| Early | Middle | Late | Early | Middle | Late | |||
| Metabolic pathways | 16.3 ± 0.15 | 16.5 ± 0.22 | 16.1 ± 0.11 | 0.382 | 15.7 ± 0.14 | 16.0 ± 0.03 | 16.0 ± 0.10 | 0.362 |
| Biosynthesis of secondary metabolites | 7.44 ± 0.06 | 7.57 ± 0.07 | 7.38 ± 0.06 | 0.290 | 7.12 ± 0.04 | 7.32 ± 0.04 | 7.31 ± 0.03 | 0.049 |
| Biosynthesis of antibiotics | 5.65 ± 0.03 | 5.77 ± 0.04 | 5.78 ± 0.04 | 0.154 | 5.61 ± 0.04 | 5.77 ± 0.04 | 5.94 ± 0.05 | 0.007 |
| Biosynthesis of amino acids | 4.20 ± 0.05 | 4.34 ± 0.08 | 4.29 ± 0.05 | 0.884 | 4.08 ± 0.07 | 4.24 ± 0.06 | 4.30 ± 0.05 | 0.213 |
| Microbial metabolism in diverse environments | 4.05 ± 0.05 | 4.00 ± 0.04 | 4.13 ± 0.04 | 0.220 | 4.29 ± 0.07 | 4.19 ± 0.07 | 4.18 ± 0.04 | 0.629 |
| Aminoacyl-tRNA biosynthesis | 2.85 ± 0.15 | 3.10 ± 0.19 | 2.65 ± 0.10 | 0.359 | 2.69 ± 0.13 | 2.63 ± 0.13 | 2.55 ± 0.13 | 0.711 |
| ABC transporters | 2.58 ± 0.18 | 2.26 ± 0.29 | 2.81 ± 0.14 | 0.299 | 3.07 ± 0.11 | 2.86 ± 0.11 | 2.73 ± 0.20 | 0.715 |
| Ribosome | 2.67 ± 0.05 | 2.78 ± 0.06 | 2.62 ± 0.03 | 0.469 | 2.53 ± 0.04 | 2.53 ± 0.04 | 2.53 ± 0.02 | 0.801 |
| Carbon metabolism | 2.33 ± 0.01 | 2.31 ± 0.02 | 2.33 ± 0.01 | 0.333 | 2.35 ± 0.02 | 2.34 ± 0.02 | 2.36 ± 0.01 | 0.717 |
| Purine metabolism | 2.07 ± 0.02 | 2.07 ± 0.03 | 2.03 ± 0.02 | 0.348 | 2.04 ± 0.02 | 1.99 ± 0.02 | 2.00 ± 0.01 | 0.071 |
| Pyrimidine metabolism | 1.72 ± 0.01 | 1.69 ± 0.02 | 1.68 ± 0.02 | 0.260 | 1.70 ± 0.02 | 1.64 ± 0.02 | 1.69 ± 0.02 | 0.211 |
| Quorum sensing | 1.51 ± 0.10 | 1.55 ± 0.16 | 1.55 ± 0.06 | 0.854 | 1.70 ± 0.02 | 1.62 ± 0.02 | 1.60 ± 0.07 | 0.778 |
| Amino sugar and nucleotide sugar metabolism | 1.55 ± 0.04 | 1.36 ± 0.06 | 1.46 ± 0.04 | 0.148 | 1.61 ± 0.03 | 1.44 ± 0.03 | 1.43 ± 0.04 | 0.003 |
| Starch and sucrose metabolism | 1.34 ± 0.04 | 1.27 ± 0.06 | 1.42 ± 0.04 | 0.308 | 1.41 ± 0.04 | 1.39 ± 0.04 | 1.46 ± 0.06 | 0.919 |
| Fructose and mannose metabolism | 1.02 ± 0.07 | 0.83 ± 0.09 | 1.07 ± 0.07 | 0.470 | 1.27 ± 0.08 | 1.10 ± 0.08 | 1.11 ± 0.06 | 0.326 |
| Two-component system | 1.05 ± 0.08 | 0.90 ± 0.03 | 0.97 ± 0.06 | 0.389 | 1.14 ± 0.11 | 1.07 ± 0.10 | 0.96 ± 0.02 | 0.206 |
| Glycolysis/Gluconeogenesis | 1.04 ± 0.02 | 0.99 ± 0.03 | 1.03 ± 0.02 | 0.841 | 1.07 ± 0.03 | 1.02 ± 0.03 | 1.04 ± 0.01 | 0.330 |
| Phosphotransferase system (PTS) | 0.85 ± 0.17 | 0.64 ± 0.17 | 1.09 ± 0.16 | 0.463 | 1.44 ± 0.18 | 1.16 ± 0.18 | 1.14 ± 0.15 | 0.422 |
| Homologous recombination | 0.94 ± 0.03 | 0.99 ± 0.04 | 1.04 ± 0.03 | 0.333 | 0.94 ± 0.04 | 1.02 ± 0.04 | 1.08 ± 0.04 | 0.292 |
| Pyruvate metabolism | 0.92 ± 0.03 | 0.88 ± 0.05 | 1.01 ± 0.02 | 0.126 | 1.00 ± 0.02 | 1.01 ± 0.02 | 1.07 ± 0.03 | 0.418 |
aKruskal-Wallis test was used to determine within-group differences.
Predicted functional analyses of the Lachnospiraceae and Ruminococcaceae families
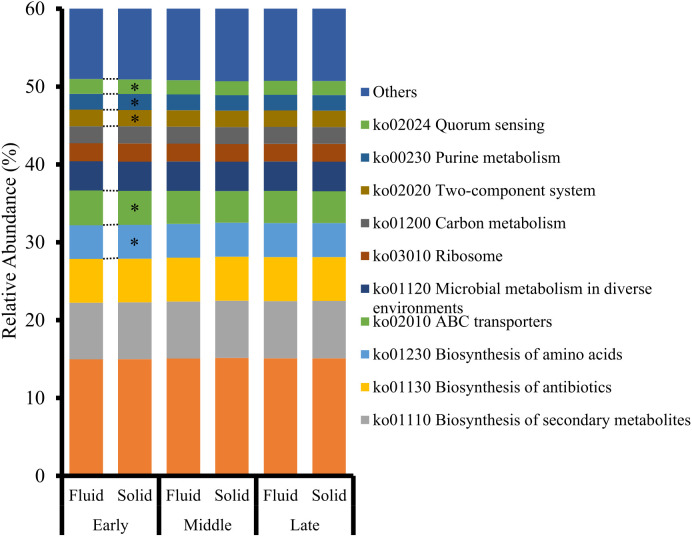
The predicted functions of the Lachnospiraceae family are shown in Fig. 1. The Lachnospiraceae family was present in the highest proportion in the solid fraction than in the fluid fraction during the early stage. “ABC transporters” (ko02010), “Two-component system” (ko02020), and “Quorum sensing” (ko02024) were significantly higher in the fluid fraction than in the solid fraction during the early stage (P<0.05). In contrast, “Biosynthesis of amino acids” (ko01230) was significantly higher in the solid fraction than in the fluid fraction (P<0.05). No significant differences were detected between the middle and late stages.
Fig. 1.
Relative abundance of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways inferred from the Lachnospiraceae family in the rumen fluid and solid fractions of Japanese Black beef cattle (n=9) during the fattening stages. *Indicates a significant difference (P<0.05) between the fluid and solid fractions.
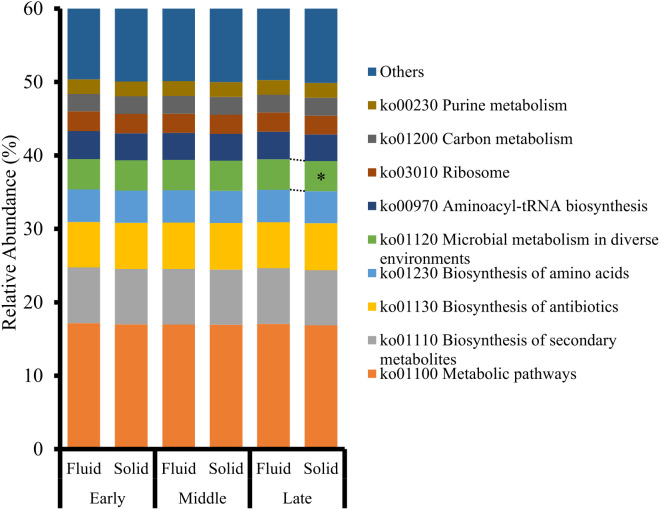
The predicted functions of the Ruminococcaceae family are shown in Fig. 2. The Ruminococcaceae family was more abundant in the solid fraction than in the fluid fraction during the early stage. “Microbial metabolism in diverse environments” (ko01120) was significantly higher in the fluid fraction than the solid fraction during the late stage (P<0.05). No significant differences were observed between the early and middle stages.
Fig. 2.
Relative abundance of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways inferred from the Ruminococcaceae family in the rumen fluid and solid fractions of Japanese Black beef cattle (n=9) during the fattening stages. *Indicates a significant difference (P<0.05) between the fluid and solid fractions.
Correlation coefficients between the predicted functions of the bacterial community and ruminal LPS activity and the proportion of acetic and propionic acids
Table 5 summarizes the significant correlation coefficients between the predicted functions of the bacterial community and ruminal LPS activity and the proportions of acetic and propionic acids. As a result, 9, 10, and 15 KEGG pathways related to LPS, acetic acid, and propionic acid, respectively, were identified in the fluid fraction (Table 5). Significant and strong correlations were observed in pathways related to LPS, including “Metabolism”, “Environmental Information Processing”, and “Cellular Processes”. Furthermore, among the pathways showing strong correlations with acetic acid, six pathways belonged to “Metabolism” or “Environmental Information Processing” and one belonged to “Genetic Information Processing”. The pathways strongly correlated with propionic acid and acetic acid mostly overlapped. Two propionic acid pathways belonged to “Metabolism” and one belonged to “Cellular Processes.”
Table 5. Pearson correlation coefficient analysis between inferred Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways in fluid and solid fractions and rumen lipopolysaccharide (LPS) activity and proportions of acetic and propionic acids (%) in Japanese Black beef cattle (n=9).
| KEGG pathways | KEGG identifier | Pearson’s correlation coefficient1 |
|||
|---|---|---|---|---|---|
| LPS | Acetic Acid | Propionic Acid | |||
| Fluid fraction | |||||
| AGE-RAGE signaling pathway in diabetic complications | ko00061 | 0.737 | |||
| ECM-receptor interaction | ko00524 | 0.738 | |||
| Focal adhesion | ko00950 | 0.738 | |||
| Isoflavonoid biosynthesis | ko02060 | 0.745 | |||
| Melanogenesis | ko00510 | 0.754 | |||
| Monoterpenoid biosynthesis | ko04964 | 0.733 | |||
| Platelet activation | ko00791 | 0.737 | |||
| Relaxin signaling pathway | ko05164 | 0.737 | |||
| Type I polyketide structures | ko01056 | 0.738 | |||
| Microbial metabolism in diverse environments | ko01120 | −0.545 | 0.650 | ||
| Pyruvate metabolism | ko00620 | −0.554 | 0.654 | ||
| Biosynthesis of secondary metabolites | ko01110 | −0.515 | 0.595 | ||
| Sphingolipid signaling pathway | ko04071 | −0.612 | 0.691 | ||
| Retinol metabolism | ko00830 | 0.536 | −0.506 | ||
| Taurine and hypotaurine metabolism | ko00430 | −0.522 | 0.530 | ||
| Acarbose and validamycin biosynthesis | ko00525 | −0.582 | 0.581 | ||
| RNA transport | ko03013 | −0.549 | 0.545 | ||
| Peptidoglycan biosynthesis | ko00550 | 0.569 | −0.627 | ||
| Carbon fixation pathways in prokaryotes | ko00720 | 0.543 | −0.623 | ||
| Bisphenol degradation | ko00363 | 0.556 | |||
| Renin-angiotensin system | ko04614 | 0.552 | |||
| Riboflavin metabolism | ko00740 | 0.528 | |||
| MicroRNAs in cancer | ko05206 | 0.518 | |||
| Meiosis −yeast | ko04113 | −0.530 | |||
| Solid fraction | |||||
| Hypertrophic cardiomyopathy (HCM) | ko05410 | 0.607 | |||
| AGE-RAGE signaling pathway in diabetic complications | ko04933 | 0.524 | |||
| Purine metabolism | ko00230 | 0.636 | |||
| Necroptosis | ko04217 | 0.608 | |||
| Tuberculosis | ko05152 | 0.580 | |||
| Sulfur metabolism | ko00920 | −0.599 | |||
| Vancomycin resistance | ko01502 | −0.524 | |||
| C5-Branched dibasic acid metabolism | ko00660 | −0.745 | 0.552 | ||
| Inositol phosphate metabolism | ko00562 | 0.564 | |||
| Flavone and flavonol biosynthesis | ko00944 | 0.551 | |||
| Amino sugar and nucleotide sugar metabolism | ko00520 | −0.542 | |||
| 2-Oxocarboxylic acid metabolism | ko01210 | −0.518 | |||
1Pathways showed significant correlation coefficients (>|0.5|) between the predicted functions of the bacterial community and ruminal LPS activity and the proportions of acetic and propionic acids.
Two, six, and six KEGG pathways (P<0.05, correlation coefficients >0.5) were related to LPS, acetic acid, and propionic acid, respectively, in the solid fraction (Table 5). Among the pathways related to acetic acid, three belonged to “Metabolism”, and one belonged to “Cellular Processes”. “C5-Branched dibasic acid metabolism” (ko00660) overlapped with both acetic and propionic acid. The pathways strongly related to propionic acid included four that belonged to “Metabolism” and one that belonged to “Secondary metabolites”.
DISCUSSION
Bacterial adhesions for feeding particles are characterized by a complex and diverse population [16], and the free-living bacterial species in the fluid fraction do not contribute much to metabolic activity. Instead, species in the fluid fraction contribute to the development of solid-adherent biofilms to newly ingested feed [17]. Therefore, bacterial communities in the fluid and solid fractions continuously interact with each other [6], but with potentially different functions [30]. Hackmann et al. [10] discovered that the recognized pathways for metabolizing pentose and hexose sugars into short-chain fatty acids do not adequately explain the fermentation products generated by a wide range of rumen bacteria. The Piphillin algorithm used in the present study for bacterial community analyses strongly relies on the amount and quality of reference genomes and can be used to obtain accurate results in animals [1, 24, 26].
The predicted functions of the bacterial community changed during the fattening stages following the altered rumen fermentation measurements described above. Several cellulolytic bacteria were found in the solid fraction during the early stage, and the higher proportion of free-living species in the fluid plays a role in metabolizing the end products of carbohydrate fermentation [6]. For example, secondary metabolites are synthesized via multistep pathways, and Fusarium avenaceum, a widespread pathogen of important crops, can produce many bioactive secondary metabolites using different carbon sources [35]. In addition, bacterial secondary metabolites often contain carbohydrate attachments that play a significant role in conferring biological activity [37]. However, the investigation of “Biosynthesis of secondary metabolites” in the rumen bacterial community is still limited. In contrast, the most abundant family in the fluid fraction during the middle fattening stage, Prevotellaceae (accounting for 24%), was engaged in various types of glycation and proteolysis [2]; thus, “Two-component systems” may be responsible for the signaling mechanism to respond to diverse environmental changes [4]. Furthermore, “Purine metabolism”, which is a metabolic pathway of the purine bases in nucleic acids, was identified as a significantly enriched pathway during the late stage. This pathway has been reported in carnivore fecal gut bacteria that predominantly have purine metabolic function [42] and in lactic acid-producing species of goats (Lactococcus lactis) [14] and humans (Lactobacillus gaasseri PA-3) [40], which might be consistent with the increase in lactic acid concentration during the late stage (Table 1) in the present study. The solid fraction bacteria also changed to digest cellulose materials during the early and middle stages and to respond to ruminal environment changes during the late stage. These results suggest that adaptation by the bacteria was identified simultaneously with rumen measurements, such as pH, VFA, and lactic acid, as well as the predicted functions of the bacterial community.
When fermentable fibers are lacking, microorganisms switch their energy metabolism to less energetically desirable sources such as amino acids, endogenous proteins, or dietary fats [5, 39]. As a result, fermentation activity decreases and VFAs as end products decrease [29]. Bacterial amino acid fermentation compensates for the decrease in VFAs [34]. Purine metabolism significantly increased in the fluid fraction during the late fattening stage, as shown in Table 3, when VFA concentrations tended to be low. Therefore, the significantly lower proportion of “Purine metabolism” in the solid fraction than in the fluid fraction suggested that fermentation did not proceed at a high rate during the late fattening stage.
During the fattening stages, a significant change in the predicted bacterial function was identified in the solid fraction. The glycosylated secondary metabolites in bacteria continue to serve as an important source for antibiotic discovery [37], and “Biosynthesis of antibiotics” by Bacillus amyloliquefaciens play an important role in its bacterial activity to suppress Rhizoctonia solani and other fungal plant pathogens as biocontrol agents [41]. Furthermore, the “Amino sugar and nucleotide sugar metabolism” pathway may be involved in carbohydrate transport and metabolism [9], and altered nucleotide sugar metabolism influences exopolysaccharide-producing strains of lactic acid bacteria such as Streptococcus thermophilus LY03 [36]. Collectively, significant changes in the bacterial predicted function of the solid fraction might imply the adaptation of rumen bacteria to altered feeding management and rumen fermentation during the fattening stages. However, the investigation of the predicted bacterial function of rumen bacterial communities is still limited. Therefore, further studies are required to elucidate the relationships between changes in rumen fermentation and the relative abundance of bacteria and their metagenomic functions.
Lachnospiraceae and Ruminococcaceae had the highest and second highest relative abundances among bacterial families in the present study, respectively. The Lachnospiraceae family mainly hydrolyzes starch and other sugars to butyric acid and other VFAs [3]. Metagenomic analyses have revealed the ability of the Lachnospiraceae family to decompose complex plant structures and transport decomposed products of various sizes [7]. This is achieved by a secondary product of an “ABC transporter” protein encoded by the Lachnospiraceae family [38]. In addition, the Ruminococcaceae family made up a larger proportion of the fluid fraction than the solid fraction, which was related to the significantly higher proportion of “Microbial metabolic pathways in diverse environments” in the fluid compared to the solid fraction during the late fattening stage (Fig. 2); in contrast, there was no difference in the functional proportion between the fractions during the same stage (Table 4). Therefore, Lachnospiraceae had a relatively high abundance in the solid fraction, which was probably composed of species that use sugars from decomposed products to synthesize amino acids. Furthermore, the adaptation of Japanese Black beef cattle to long-term ruminal acidic conditions using a high-grain diet to increase intramuscular fat content [22] suggests that the Ruminococcaceae family in the fluid fraction adapted to the late ruminal environment under conditions of significantly higher “Microbial metabolic pathways in diverse environments”.
Pearson’s correlation coefficient analyses revealed that all pathways correlated with LPS were positively correlated, which was influenced by the increase in LPS during the fattening stages. Cell adhesion points and platelet activation were correlated with “ECM-receptor interactions” in the KEGG pathway analyses; ECM-receptors are important in neuronal information processing [8]. In the present study, LPS activity increased throughout the fattening period, and information processing in the ECM receptors and surrounding pathways may have been more active during the latter fattening stages due to an increase in ruminal LPS activity. Furthermore, acetic acid is produced from pyruvate by intestinal bacteria [28], and propionic acid is produced by the conversion of succinate to methylmalonyl-CoA in the TCA circuit from the acrylate pathway [11] and via the propanediol pathway [33]. Therefore, acetic and propionic acids were strongly correlated with pyruvate metabolism, the carbon fixation pathway, and secondary metabolites during glycolysis [20] to produce or decompose acetic and propionic acids in the present study.
In conclusion, although it was difficult to apply the KEGG pathway results directly to rumen bacterial functions, our results suggest that the overall functional structure of the rumen bacterial community is affected by long-term feeding of a high-grain diet. Combined with a dietary transition and changes in ruminal pH, our results suggest underlying differences in fermentation or functional adaptation of Japanese Black beef cattle during the fattening stages.
CONFLICT OF INTEREST
The authors have nothing to disclose.
REFERENCES
- 1.Auffret M. D., Dewhurst R. J., Duthie C. A., Rooke J. A., John Wallace R., Freeman T. C., Stewart R., Watson M., Roehe R.2017. The rumen microbiome as a reservoir of antimicrobial resistance and pathogenicity genes is directly affected by diet in beef cattle. Microbiome 5: 159. doi: 10.1186/s40168-017-0378-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avgustin G., Wallace R. J., Flint H. J.1997. Phenotypic diversity among ruminal isolates of Prevotella ruminicola: proposal of Prevotella brevis sp. nov., Prevotella bryantii sp. nov., and Prevotella albensis sp. nov. and redefinition of Prevotella ruminicola. Int. J. Syst. Bacteriol. 47: 284–288. doi: 10.1099/00207713-47-2-284 [DOI] [PubMed] [Google Scholar]
- 3.Biddle A., Stewart L., Blanchard J., Leschine S.2013. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity (Basel) 5: 627–640. doi: 10.3390/d5030627 [DOI] [Google Scholar]
- 4.Capra E. J., Laub M. T.2012. Evolution of two-component signal transduction systems. Annu. Rev. Microbiol. 66: 325–347. doi: 10.1146/annurev-micro-092611-150039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cummings J. H., Macfarlane G. T.1991. The control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70: 443–459. doi: 10.1111/j.1365-2672.1991.tb02739.x [DOI] [PubMed] [Google Scholar]
- 6.De Mulder T., Goossens K., Peiren N., Vandaele L., Haegeman A., De Tender C., Ruttink T., de Wiele T. V., De Campeneere S.2017. Exploring the methanogen and bacterial communities of rumen environments: solid adherent, fluid and epimural. FEMS Microbiol. Ecol. 93: fiw251. [DOI] [PubMed] [Google Scholar]
- 7.Di Iorio B. R., Rocchetti M. T., De Angelis M., Cosola C., Marzocco S., Di Micco L., di Bari I., Accetturo M., Vacca M., Gobbetti M., Di Iorio M., Bellasi A., Gesualdo L.2019. Nutritional therapy modulates intestinal microbiota and reduces serum levels of total and free indoxyl sulfate and P-Cresyl sulfate in chronic kidney disease (Medika Study). J. Clin. Med. 8: 1424. doi: 10.3390/jcm8091424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dityatev A., Schachner M., Sonderegger P.2010. The dual role of the extracellular matrix in synaptic plasticity and homeostasis. Nat. Rev. Neurosci. 11: 735–746. doi: 10.1038/nrn2898 [DOI] [PubMed] [Google Scholar]
- 9.Feng Y., Zhao Y., Guo Y., Liu S.2018. Microbial transcript and metabolome analysis uncover discrepant metabolic pathways in autotrophic and mixotrophic anammox consortia. Water Res. 128: 402–411. doi: 10.1016/j.watres.2017.10.069 [DOI] [PubMed] [Google Scholar]
- 10.Hackmann T. J., Ngugi D. K., Firkins J. L., Tao J.2017. Genomes of rumen bacteria encode atypical pathways for fermenting hexoses to short-chain fatty acids. Environ. Microbiol. 19: 4670–4683. doi: 10.1111/1462-2920.13929 [DOI] [PubMed] [Google Scholar]
- 11.Hetzel M., Brock M., Selmer T., Pierik A. J., Golding B. T., Buckel W.2003. Acryloyl-CoA reductase from Clostridium propionicum. An enzyme complex of propionyl-CoA dehydrogenase and electron-transferring flavoprotein. Eur. J. Biochem. 270: 902–910. doi: 10.1046/j.1432-1033.2003.03450.x [DOI] [PubMed] [Google Scholar]
- 12.Hook S. E., Steele M. A., Northwood K. S., Dijkstra J., France J., Wright A. D. G., McBride B. W.2011. Impact of subacute ruminal acidosis (SARA) adaptation and recovery on the density and diversity of bacteria in the rumen of dairy cows. FEMS Microbiol. Ecol. 78: 275–284. doi: 10.1111/j.1574-6941.2011.01154.x [DOI] [PubMed] [Google Scholar]
- 13.Iwai S., Weinmaier T., Schmidt B. L., Albertson D. G., Poloso N. J., Dabbagh K., DeSantis T. Z.2016. Piphillin: improved prediction of metagenomic content by direct inference from human microbiomes. PLoS One 11: e0166104. doi: 10.1371/journal.pone.0166104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kilstrup M., Hammer K., Ruhdal Jensen P., Martinussen J.2005. Nucleotide metabolism and its control in lactic acid bacteria. FEMS Microbiol. Rev. 29: 555–590. doi: 10.1016/j.fmrre.2005.04.006 [DOI] [PubMed] [Google Scholar]
- 15.Kim Y. H., Nagata R., Ohtani N., Ichijo T., Ikuta K., Sato S.2016. Effects of dietary forage and calf starter diet on ruminal pH and bacteria in Holstein calves during weaning transition. Front. Microbiol. 7: 1575. doi: 10.3389/fmicb.2016.01575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larue R., Yu Z., Parisi V. A., Egan A. R., Morrison M.2005. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ. Microbiol. 7: 530–543. doi: 10.1111/j.1462-2920.2005.00721.x [DOI] [PubMed] [Google Scholar]
- 17.Leng R. A.2011. The Rumen −a fermentation vat or a series of organized structured microbial consortia: implications for the mitigation of enteric methane production by feed additives. Livest. Res. Rural Dev. 23: 258. [Google Scholar]
- 18.Nagaraja T. G., Chengappa M. M.1998. Liver abscesses in feedlot cattle: a review. J. Anim. Sci. 76: 287–298. doi: 10.2527/1998.761287x [DOI] [PubMed] [Google Scholar]
- 19.Nagata R., Kim Y. H., Ohkubo A., Kushibiki S., Ichijo T., Sato S.2018. Effects of repeated subacute ruminal acidosis challenges on the adaptation of the rumen bacterial community in Holstein bulls. J. Dairy Sci. 101: 4424–4436. doi: 10.3168/jds.2017-13859 [DOI] [PubMed] [Google Scholar]
- 20.Nakagawa A., Minami H., Kim J. S., Koyanagi T., Katayama T., Sato F., Kumagai H.2011. A bacterial platform for fermentative production of plant alkaloids. Nat. Commun. 2: 326. doi: 10.1038/ncomms1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.National Agriculture and Food Research Organization (NARO). 2009. Japanese Feeding Standard for Beef Cattle. 2008 ed. Japan Livestock Industry Association (in Japanese). [Google Scholar]
- 22.Ogata T., Kim Y. H., Iwamoto E., Masaki T., Ikuta K., Sato S.2020. Comparison of pH and bacterial communities in the rumen and reticulum during fattening of Japanese Black beef cattle. Anim. Sci. J. 91: e13487. doi: 10.1111/asj.13487 [DOI] [PubMed] [Google Scholar]
- 23.Ogata T., Makino H., Ishizuka N., Iwamoto E., Masaki T., Ikuta K., Kim Y. H., Sato S.2019. Long-term high-grain diet altered the ruminal pH, fermentation, and composition and functions of the rumen bacterial community, leading to enhanced lactic acid production in Japanese Black beef cattle during fattening. PLoS One 14: e0225448. doi: 10.1371/journal.pone.0225448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogunade I. M., Lay J., Andries K., McManus C. J., Bebe F.2019. Effects of live yeast on differential genetic and functional attributes of rumen microbiota in beef cattle. J. Anim. Sci. Biotechnol. 10: 68. doi: 10.1186/s40104-019-0378-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oka A., Maruo Y., Miki T., Yamasaki T., Saito T.1998. Influence of vitamin A on the quality of beef from the Tajima strain of Japanese Black cattle. Meat Sci. 48: 159–167. doi: 10.1016/S0309-1740(97)00086-7 [DOI] [PubMed] [Google Scholar]
- 26.Park T., Ma L., Ma Y., Zhou X., Bu D., Yu Z.2020. Dietary energy sources and levels shift the multi-kingdom microbiota and functions in the rumen of lactating dairy cows. J. Anim. Sci. Biotechnol. 11: 66. doi: 10.1186/s40104-020-00461-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruesse E., Quast C., Knittel K., Fuchs B. M., Ludwig W., Peplies J., Glöckner F. O.2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35: 7188–7196. doi: 10.1093/nar/gkm864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ragsdale S. W., Pierce E.2008. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim. Biophys. Acta 1784: 1873–1898. doi: 10.1016/j.bbapap.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russell W. R., Gratz S. W., Duncan S. H., Holtrop G., Ince J., Scobbie L., Duncan G., Johnstone A. M., Lobley G. E., Wallace R. J., Duthie G. G., Flint H. J.2011. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am. J. Clin. Nutr. 93: 1062–1072. doi: 10.3945/ajcn.110.002188 [DOI] [PubMed] [Google Scholar]
- 30.Sato S.2016. Pathophysiological evaluation of subacute ruminal acidosis (SARA) by continuous ruminal pH monitoring. Anim. Sci. J. 87: 168–177. doi: 10.1111/asj.12415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato S., Kimura A., Anan T., Yamagishi N., Okada K., Mizuguchi H., Ito K.2012. A radio transmission pH measurement system for continuous evaluation of fluid pH in the rumen of cows. Vet. Res. Commun. 36: 85–89. doi: 10.1007/s11259-012-9518-x [DOI] [PubMed] [Google Scholar]
- 32.Schloss P. D., Westcott S. L., Ryabin T., Hall J. R., Hartmann M., Hollister E. B., Ryan A. L., Brian B. O., Donovan H. P., Courtney J. R., Jason W. S., Blaz S., Gerhard G. T., David J. V. H., Carolyn F. W.2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scott K. P., Martin J. C., Campbell G., Mayer C. D., Flint H. J.2006. Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium “Roseburia inulinivorans”. J. Bacteriol. 188: 4340–4349. doi: 10.1128/JB.00137-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith E. A., Macfarlane G. T.1997. Dissimilatory amino Acid metabolism in human colonic bacteria. Anaerobe 3: 327–337. doi: 10.1006/anae.1997.0121 [DOI] [PubMed] [Google Scholar]
- 35.Sørensen J. L., Giese H.2013. Influence of carbohydrates on secondary metabolism in Fusarium avenaceum. Toxins (Basel) 5: 1655–1663. doi: 10.3390/toxins5091655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Svensson M., Lohmeier-Vogel E., Waak E., Svensson U., Rådström P.2007. Altered nucleotide sugar metabolism in Streptococcus thermophilus interferes with nitrogen metabolism. Int. J. Food Microbiol. 113: 195–200. doi: 10.1016/j.ijfoodmicro.2006.06.032 [DOI] [PubMed] [Google Scholar]
- 37.Timmons S. C., Thorson J. S.2008. Increasing carbohydrate diversity via amine oxidation: aminosugar, hydroxyaminosugar, nitrososugar, and nitrosugar biosynthesis in bacteria. Curr. Opin. Chem. Biol. 12: 297–305. doi: 10.1016/j.cbpa.2008.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vacca M., Celano G., Calabrese F. M., Portincasa P., Gobbetti M., De Angelis M.2020. The controversial role of human gut Lachnospiraceae. Microorganisms 8: 573. doi: 10.3390/microorganisms8040573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wall R., Ross R. P., Shanahan F., O’Mahony L., O’Mahony C., Coakley M., Hart O., Lawlor P., Quigley E. M., Kiely B., Fitzgerald G. F., Stanton C.2009. Metabolic activity of the enteric microbiota influences the fatty acid composition of murine and porcine liver and adipose tissues. Am. J. Clin. Nutr. 89: 1393–1401. doi: 10.3945/ajcn.2008.27023 [DOI] [PubMed] [Google Scholar]
- 40.Yamanaka H., Taniguchi A., Tsuboi H., Kano H., Asami Y.2019. Hypouricaemic effects of yoghurt containing Lactobacillus gasseri PA-3 in patients with hyperuricaemia and/or gout: A randomised, double-blind, placebo-controlled study. Mod. Rheumatol. 29: 146–150. doi: 10.1080/14397595.2018.1442183 [DOI] [PubMed] [Google Scholar]
- 41.Yu G. Y., Sinclair J. B., Hartman G. L., Bertagnolli B. L.2002. Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol. Biochem. 34: 955–963. doi: 10.1016/S0038-0717(02)00027-5 [DOI] [Google Scholar]
- 42.Zhu L., Wu Q., Deng C., Zhang M., Zhang C., Chen H., Lu G., Wei F.2018. Adaptive evolution to a high purine and fat diet of carnivorans revealed by gut microbiomes and host genomes. Environ. Microbiol. 20: 1711–1722. doi: 10.1111/1462-2920.14096 [DOI] [PubMed] [Google Scholar]