Summary
Ultraviolet (UV) light and incompletely understood genetic and epigenetic variations determine skin color. Here, we describe an UV- and MITF-independent mechanism of skin pigmentation. Targeting the mitochondrial redox-regulating enzyme nicotinamide nucleotide transhydrogenase (NNT) resulted in cellular redox changes, which impact tyrosinase degradation. These changes regulate melanosome maturation and consequently eumelanin levels and pigmentation. Topical application of small-molecule inhibitors yielded skin darkening in human skin, and mice with decreased NNT function displayed increased pigmentation. Additionally, genetic modification of NNT in zebrafish alters melanocytic pigmentation. Analysis of four diverse human cohorts revealed significant associations of skin color, tanning, and sun protection use with various single nucleotide polymorphisms within NNT. NNT levels were independent of UVB irradiation and redox modulation. Patients with postinflammatory hyperpigmentation or lentigines displayed decreased skin NNT levels, suggesting an NNT-driven, redox-dependent pigmentation mechanism which can be targeted with NNT-modifying topical drugs for medical and cosmetic purposes.
Keywords: Pigmentation, Redox regulation, Nicotinamide nucleotide transhydrogenase, Melanosome, UVB, MITF
Graphical Abstract
In brief:
Nicotinamide nucleotide transhydrogenase (NNT) is a mitochondrial redox-regulating enzyme that mediates pigmentation via a UVB- and MITF-independent mechanism
Introduction
Pigmentation of human skin, which confers protection against skin cancer, evolved over one million years ago in the setting of evolutionary loss of body hair (Jablonski and Chaplin, 2017). Human skin color results from the relative amounts of yellow-orange pheomelanin and black-brown eumelanin (Del Bino et al., 2015). Darker pigmented individuals are more protected from, oncogenic UV radiation by the light scattering and antioxidant properties of eumelanin (Jablonski and Chaplin, 2012).
Pigment dictates how light is absorbed and disseminated in skin (Pathak et al., 1962). UV can interact photochemically with DNA to form cyclobutane pyrimidine dimers (CPD) and 6,4-photoproducts and causes production of reactive oxygen species (ROS) through multiple mechanisms, increasing the risk of skin cancer (Premi et al., 2015). Whereas eumelanin has antioxidant activity, ROS-mediated oxidation of DNA bases and lipid peroxidation are elevated in mice that produce pheomelanin only (Mitra et al., 2012).
Melanocytes produce melanin within subcellular organelles called melanosomes which mature from early, unpigmented (stages I-II) towards late, pigmented states (stages III-IV). Early-stage melanosomes are recognized by proteinaceous fibrils within the melanosomal lumen. In the late stages melanin is gradually deposited on the fibrils (Raposo and Marks, 2007). These mature melanosomes are ultimately transferred to keratinocytes (Park et al., 2009) where they coalesce in a supranuclear location on the sun-facing side. UV radiation triggers tanning through p53-mediated induction of POMC peptides in keratinocytes, leading to MC1R activation on melanocytes and cAMP-mediated induction of the microphthalmia-associated transcription factor (MITF), that induces expression of tyrosinase-related protein 1 and 2 (TYRP1 and DCT) (Lo and Fisher, 2014) and tyrosinase, which drive melanosome maturation (Paterson et al., 2015) and increased production of eumelanin (Iozumi et al., 1993).
The enzyme nicotinamide nucleotide transhydrogenase (NNT) is located in the inner mitochondrial membrane. It regulates mitochondrial redox levels by coupling hydride transfer between β-nicotinamide adenine dinucleotide NAD(H) and β-nicotinamide adenine dinucleotide 2’-phosphate NADP (+) to proton translocation across the inner mitochondrial membrane (Earle and Fisher, 1980; Rydstrom et al., 1970; Zhang et al., 2017). Even though The Human Protein Atlas (Uhlen et al., 2015) showed expression of NNT in human melanocytes, fibroblasts, keratinocytes, and other epidermal cells, so far, NNT has not been described to be involved in mechanisms of direct regulation of skin pigment. Here, we report a role for NNT in modulating melanosome maturation and pigmentation.
Results
NNT enables regulation of pigmentation via changing intracellular redox levels
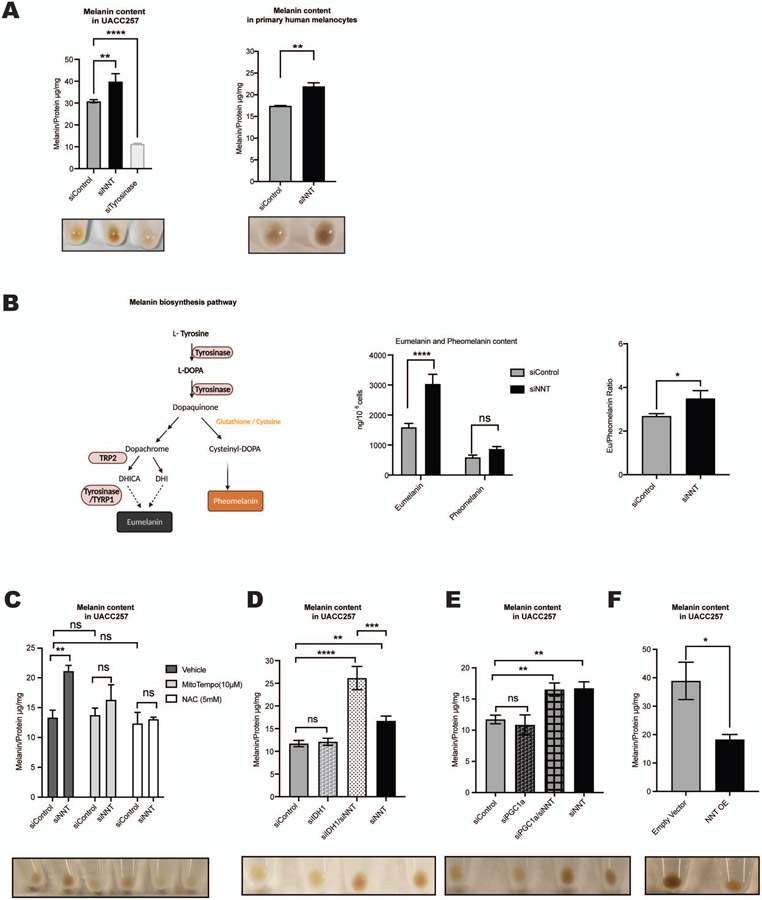
NNT was depleted using a pool of siRNAs (siNNT) in human melanoma cell lines UACC257 and SK-MEL-30, and in primary human melanocytes. In all three cell models knockdown of NNT led to a significant increase in melanin content (Figures 1A, S1A-D). The increase of pigmentation following siNNT was blocked by simultaneous knockdown of tyrosinase demonstrating the dependence of siNNT-mediated pigmentation on tyrosinase (Figure S1A).
Figure 1. Nicotinamide Nucleotide Transhydrogenase (NNT) regulates in vitro pigmentation via a redox-dependent mechanism.
(A) siNNT increases pigmentation. Quantification of intracellular melanin content of UACC257 cells treated with siControl, siNNT, or siTyrosinase for 72 hours (Left Panel) and human primary melanocytes treated with siControl or siNNT for 96 hours (Right Panel); n = 3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test (Left Panel) and unpaired, two-sided t-test (Right Panel). Below the graphs, representative cell pellets of the indicated treatment (1x106 cells). (B) Schema: Pathways of pheomelanin and eumelanin biosynthesis. DHICA, 5,6-dihydroxyindole-2-carboxylic acid; DHI, 5,6-dihydroxyindole. Graphs: UACC257 melanoma cells were treated with siControl, or siNNT for 5 days and eumelanin and pheomelanin were measured using HPLC techniques (n = 3). Absolute pigment levels (Left graph) were analyzed by ordinary two-way ANOVA. The eumelanin/pheomelanin ratio (Right graph) was analyzed by unpaired Student t test. (C) siNNT-induced increased pigmentation of human UACC257 melanoma cells is blocked by NAC (5 mM) or MitoTEMPO (20 μM) (daily treatment for 72 h); n = 3, analyzed by ordinary two-way ANOVA with Šídák’s post-test. (D-E) Quantification of intracellular melanin content of UACC257 cells treated for 72 hours with siControl, siNNT, siIDH1, or siIDH1 + siNNT (D), or with siControl, siNNT, siPGC1 a, or siNNT + siPGC1 a (E); n = 3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test. Below the graphs, representative cell pellets (1 x 106 cells) of the indicated treatments. (F) Overexpression of NNT reduced pigmentation. Melanin content in UACC257 cells that overexpressed NNT (NNT OE) or the corresponding control (Empty Vector) for 12 days; n = 3, analyzed by unpaired, two-sided t-test.
All data are expressed as mean ± SEM; *p<0.05, **p<0.01, ****p<0.0001.
NNT has been described to increase GSH in Nnt wild type versus Nnt mutant C57BL/6J mice (Ronchi et al., 2013), as well as in human myocardium (Sheeran et al., 2010). In line with this, silencing NNT caused a decrease of the GSH/GSSG ratio in UACC257 human melanoma cells (Figure S1E). Cysteine or reduced glutathione is a required component for pheomelanin synthesis (Ito and Ifpcs, 2003; Jara et al., 1988) (Schema, Figure 1B), suggesting that NNT may modulate pigmentation via its role in regenerating GSH and thereby affecting the pheomelanin to eumelanin ratio. To investigate this possibility, high-performance liquid chromatography (HPLC) was utilized and demonstrated significantly increased absolute levels of eumelanin, but not pheomelanin, upon NNT knockdown (Figure 1B, Left graph). The eumelanin to pheomelanin ratio also showed a significant increase, (Figure 1B, Right graph). Tyrosinase silencing was used as a positive control showing efficient and quick depigmentation five days after transfection (Figure 1A), resulting in decreased levels of both eumelanin and pheomelanin, and as suspected, no significant change in the eumelanin to pheomelanin ratio (Figure S1F). This data suggests that NNT modulates melanin synthesis towards a eumelanin phenotype.
Due to NNT’s essential role as an antioxidant enzyme against ROS by controlling the NADPH conversion, we hypothesized that the increase in pigmentation following silencing of NNT is driven by an oxidative stress-dependent mechanism. As expected, knockdown of NNT caused a significant increase in the NADP/NADPH ratio (Figure S1E) and induced cytosolic ROS (Figure S1G) in UACC257 cells. Adding thiol antioxidant N-acetylcysteine (NAC), mitochondria-targeted antioxidant MitoTEMPO, or NADPH to siNNT, inhibited the siNNT-mediated increase in pigmentation (Figures 1C, S1A and S1H), demonstrating the dependence of siNNT-mediated pigmentation on oxidative stress.
To understand how cytosolic and mitochondrial oxidative stress levels are connected, isocitrate dehydrogenase 1 (IDH1), a source of cytosolic NADPH (Zhao and McAlister-Henn, 1996) was depleted in UACC257 cells (Figures 1D, S1I and S1J). Interestingly, while siNNT alone increased pigmentation, siIDH1 alone had no significant effect on pigmentation (Figure 1D). However, the double knockdown of NNT and IDH1 increased the intracellular melanin content further, exceeding the siNNT-induction of pigmentation (Figure 1D). To exclude the possibility that siIDH1 or siIDH1-induced oxidative stress may increase NNT levels, NNT mRNA levels were measured (Figure S1I-J), which showed no changes. To understand if cytosolic ROS may be the driver of the observed pigmentation change, cytosolic oxidative stress was measured upon silencing of siNNT and siIDH1 (Figure S1G), showing similar effects of the different siRNAs, emphasizing the crucial role of NNT in human pigmentation.
In order to clarify the role of mitochondrial oxidative stress, we investigated the participation of peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC1α). As shown previously, intramitochondrial concentrations of ROS were significantly increased in PGC1α-depleted melanoma cells, associated with decreased levels of reduced glutathione (GSH), cystathionine, and 5-adenosylhomocysteine (Vazquez et al., 2013). However, no change of pigmentation was detected in PGC1α-depleted human UACC257 melanoma cells (Figures 1E and S1J), thus emphasizing the specific role of NNT and especially NNT-induced cytosolic oxidative stress for the pigmentation response. Finally, overexpression of NNT in UACC257 cells (Figure S1K) increased GSH/GSSG ratios and deceased NADP/NADPH ratios (Figure S1L). As opposed to the increase in pigmentation observed with silencing of NNT, overexpression of NNT induced a significant decrease in pigmentation (Figure 1F), confirming the relationship between NNT and pigmentation in both directions.
Taken together our data suggest that NNT affects pigmentation via a redox-dependent mechanism.
NNT depletion enhances pigmentation independently of the classic cAMP-MITF-pigmentation pathway
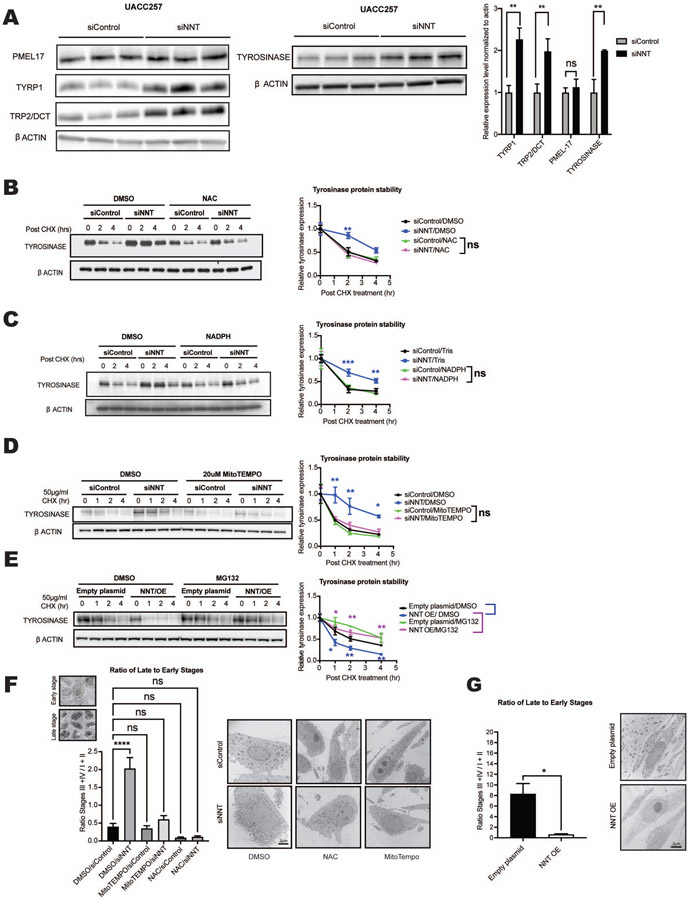
In order to elucidate the mechanism underlying hyperpigmentation after NNT knockdown, we investigated its effects on key melanin biosynthesis factors in UACC257 cells (Figure 2A). NNT knockdown revealed a significant increase in the levels of the melanin biosynthesis enzymes, tyrosinase, TYRP1 and TRP2/DCT (Figure 2A). In addition, tyrosinase activity was increased upon silencing of siNNT (Figure S2A). Since MITF is the main regulator of these enzymes and the master regulator of melanogenesis (Figures S2B-G), we measured MITF protein levels and its transcriptional activity. Upon silencing of NNT, neither MITF protein levels, nor mRNA levels were significantly changed (Figures S2C-D). Furthermore, MITF promoter activity was modestly decreased following siNNT (Figures S2E-F), while no significant change in the mRNA level of TYRP1, TRP2/DCT or tyrosinase was observed (Figures S2G). This suggests that NNT can impact tyrosinase, TRP2/DCT and TYRP1 protein levels without affecting their mRNA levels. As cAMP is a crucial messenger in UV-induced skin pigmentation (“classic cAMP-MITF-pigmentation pathway”) (Figure S2B), baseline cAMP levels in siControl-vs. siNNT-transfected UACC257 cells were assayed and found to be unaffected by siNNT (Figure S2H). Treatment of primary human melanocytes with forskolin, an activator of adenylate cyclase, which increases cAMP levels, did not affect NNT expression levels (Figure S2I), nor did UVB irradiation of human skin (Figure S2J). In addition, no increase in POMC (Figure S2G) or p53 (Figure S2K) was observed in UACC257 cells upon siNNT treatment. Further, modulating the general redox system by adding NAC, MitoTEMPO or H2O2 did not impact NNT protein levels (Figure S2L).
Figure 2. Inhibition of NNT enhances melanosome maturation and tyrosinase protein stability via a redox-dependent mechanism.
(A) Immunoblot analysis of whole cell lysates from UACC257 melanoma cells 72 hours post-treatment with either siControl or siNNT, showing increased tyrosinase, DCT/TRP2, and TYRP1, but not PMEL17 protein levels. Band intensities were quantified by ImageJ, normalized to β-actin, plotted relative to siControl (n = 3), and analyzed by multiple t-tests with the Holm-Šídák post-test. (B-D) siNNT-mediated increased protein stability is blocked by antioxidants. UACC257 cells transfected with siControl or siNNT were treated 24 hours post-transfection with 5 mM NAC (B), 0.1 mM NADPH (C), 20 μM MitoTEMPO (D), or control vehicle for 48 h, followed by CHX treatment. Cells were harvested 0, 1, 2 and 4 h post-CHX treatment for immunoblotting. Band intensities were quantified by ImageJ, normalized to β-actin, and plotted relative to t=0; n = 3, analyzed by repeated measures two-way ANOVA with Šídák’s post-test (Asterisks indicate significance of siControl/vehicle vs. each of the other three groups). (E) Proteasome inhibitor MG132 inhibits tyrosinase protein degradation upon CHX treatment of NNT-overexpressing UACC257 cells. The cells were treated with DMSO or MG132 (10 μM) for 6 h, followed by CHX treatment for 0, 1, 2 and 4 h and immunoblotting. Band intensities were quantified by ImageJ, normalized to β-actin and plotted relative to t = 0; n = 3, analyzed by repeated measures two-way ANOVA with Šídák’s post-test. (F) Enhanced melanosome maturation induced by siNNT in primary human melanocyte cells is blocked by NAC (5 mM) or MitoTEMPO (20 μM) (daily treatment for 96 h). The ratios of late stages (III + IV) to early stages (I + II) are presented. n = 4-5, analyzed by ordinary two-way ANOVA with Šídák’s post-test. (G) Inhibition of melanosome maturation induced by NNT overexpression in primary human melanocytes for 7 days. The ratios of late- to early-stage melanosomes were compared by unpaired, two-sided t-test, n=4 (NNT OE) and n=8 (Empty plasmid).
All data are expressed as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001
Finally, overexpression of NNT in UACC257 showed a significant decrease of tyrosinase protein levels (Figure S2M) but not its mRNA levels (Figure (S2N).
Together, these data suggest the existence of an NNT-dependent pigmentation mechanism, independent of the previously established cAMP-MITF-dependent pigmentation pathway.
NNT promotes ubiquitin-proteasome-dependent tyrosinase degradation and modulates melanosome maturation
Since altering NNT was found to impact the protein levels of tyrosinase and related key melanogenic enzymes (Figure 2A) without impacting their mRNA levels (Figure S2G), we hypothesized that NNT can affect the stability of certain melanosomal proteins. The impact of NNT-mediated redox changes on tyrosinase protein stability was investigated by knockdown of NNT mRNA in the presence or absence of an antioxidant, followed by inhibition of protein synthesis with cycloheximide (CHX) and measurements of the rate of decay of tyrosinase protein. Silencing of NNT increased tyrosinase protein stability significantly, and this effect was prevented by antioxidant treatment with either NAC, NADPH or Mito-Tempo (Figures 2B-D).
The mechanism of tyrosinase degradation is not fully understood, although it has been shown that tyrosinase is degraded via the ubiquitin-proteasome system (Bellei et al., 2010). Addition of carbobenzoxy-L-leucyl-L-leucyl-L-leucinal (MG132), a cell-permeable, reversible proteasome inhibitor prevented an NNT overexpression-induced decrease in Tyrosinase protein stability in UACC257 cells (Figure 2E), suggesting that NNT induces changes in melanin levels is through proteasome-mediated degradation of Tyrosinase protein.
Due to siNNT-induced increases in melanogenesis enzymes, NNT’s role in NADPH and GSH generation and its location in the inner mitochondrial membrane, we hypothesized that NNT function might be connected to the maturation of melanosomes. The effects of modulating NNT expression on the ultrastructure of melanosomes was assessed by electron microscopy in primary human melanocytes. Knockdown of NNT resulted in a striking increase in late-stage/pigmentated melanosomes (stages III and IV) (Figures 2F and S3A), while overexpression of NNT resulted in a switch towards early-stage/unpigmented melanosomes (stages I and II) (Figure 2G), establishing a role for NNT in regulating melanosome maturation. In line with the pigmentation data (Figure 1C), cotreatment with either NAC or MitoTEMPO prevented the siNNT-induced phenotype (Figures 2F and S3A). The absolute number of melanosomes per cytosolic area was not affected by NNT knockdown or overexpression (Figure S3B), which is in line with the observation that the pre-melanosome protein Pmel17, a marker for early melanosome development, did not change upon depletion of NNT (Figure 2A). Together, our data suggest that inhibition of NNT drives pigmentation via stabilizing tyrosinase and possibly other tyrosinase-related proteins (TYRP1 and TRP2/DCT) associated with increased melanosome maturation.
Previously, it has been shown that mitochondria are connected with melanosomes via physical contacts, requiring Mitofusin-2 (MFN2) (Daniele et al., 2014). The connection between these two organelles may enable localized interorganellar exchange (Daniele et al., 2014); (Wu and Hammer, 2014). To understand if siNNT-induced pigmentation may rely on an equivalent mechanism, we performed simultaneous knockdown of NNT and MFN2 in UACC257 cells (Figure S3G) and in human primary melanocytes (Figure S3H). Consistent with previous findings (Daniele et al., 2014) evaluation of mitochondria-melanosome proximities by electron microscopy confirmed that knockdown of MFN2, resulted in a strong decrease in close appositions (<20nm) compared to control (Figure S3C). By contrast, silencing of NNT alone lead to a relative increase of organelle contiguities, possibly related to the stimulation of melanogenesis (Figure S3C), and double knockdown prevented this increase (Figure S3C) while melanosome and mitochondria numbers remained unchanged (Figures S3D-E). Similar to the melanosome-mitochondria proximity, silencing of NNT in UACC257 human melanoma cells significantly increased the intracellular melanin content which was reversed by the simultaneous knockdown of NNT and MFN2 (Figures S3F). Finally, overexpression of NNT resulted in decrease in close appositions (<20nm) compared to control (Figure S3C), while no change was observed in both melanosome and mitochondria numbers (Figures S3B and S3E).
While these findings suggest that MFN2 and melanosome-mitochondria proximity may contribute to NNT regulation of pigmentation changes, the role of MFN2 in melanogenesis is complex. In addition to interorganellar connections, MFN2 regulates many functions in cells, including mitochondrial fusion, ATP production, and autophagy, which may impact pigmentation (Filadi et al., 2018). In particular, MFN2 deficiency has been associated with impaired autophagic degradation and the accumulation of autophagosomes (Zhao et al., 2012); (Sebastian et al., 2016). Consistent with those findings, knockdown of MFN2 in human primary melanocytes and UACC257 cells resulted in the presence of large autophagosome-like structures containing numerous and partly intact melanosomes (Figure S3I), as well as increased LCB3 Type II (S3J), which can be associated with either enhanced autophagosome synthesis or reduced autophagosome degradation (Barth et al., 2010). Since defects of autophagosome formation and/or turnover interfere with melanosome biogenesis and are associated with pigmentary defects (Ho and Ganesan, 2011), we conclude that MFN2 can regulate pigmentation via distinct – incompletely understood – pathways.
Topical NNT inhibitors increase pigmentation
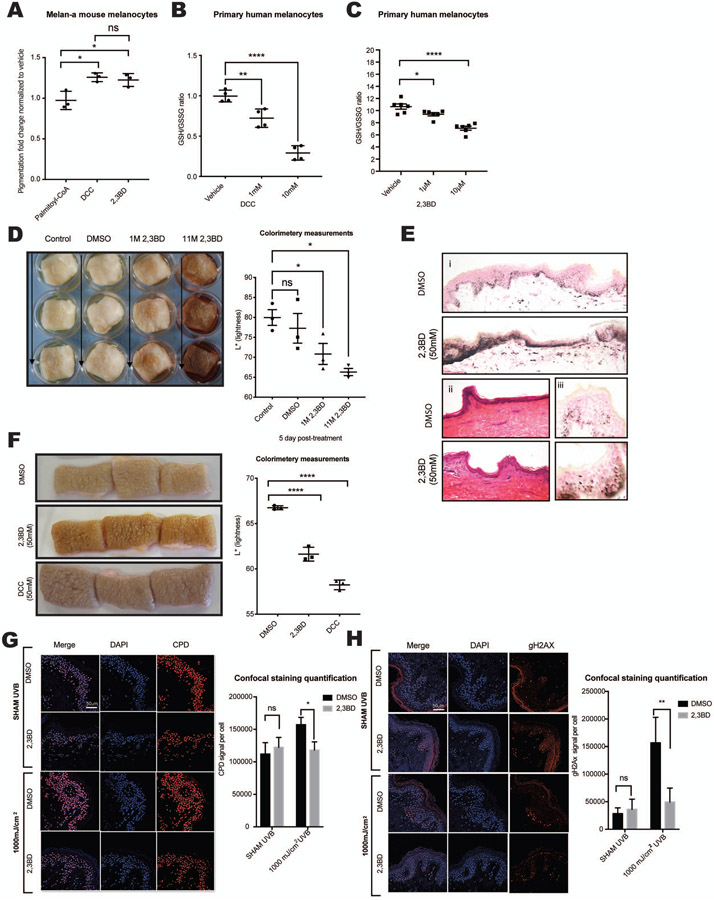
Currently, only a limited number of topical drugs are capable of altering pigmentation in human skin (Rendon and Gaviria, 2005). No topical skin darkeners are available for clinical use. Systemic administration of peptides such as α-MSH analogs (e.g., Melanotan) has been used to successfully increase skin pigmentation (Ugwu et al., 1997). Three NNT inhibitors (N,N’-Dicyclohexylcarbodiimide [DCC], 2,3-Butanedione [2,3BD], Palmitoyl-CoA) have been described previously (Figure S4A) (Rydstrom, 1972). DCC is commonly used as a peptide-coupling reagent and 2,3BD is used as a flavoring agent (Rigler and Longo, 2010). Both are low molecular weight compounds (DCC: 206.33 g/mol; 2,3BD: 86.09 g/mol) potentially capable of penetrating human epidermis. Palmitoyl-CoA, like 2,3BD, is a natural product, but has a high molecular weight (1005.94 g/mol), making skin penetration challenging. The effects of all three compounds on pigmentation of intermediately pigmented murine Melan-A cells (Figure 3A) were assessed. Both 2,3BD and DCC significantly increased the melanin content in intermediately pigmented murine Melan-A cells (Figure 3A) and in human primary melanocyte (Figure S4D). In vitro toxicity was assessed in primary human melanocytes, dermal fibroblasts and keratinocytes (Figure S4B) showing no significant toxicity in doses up to 10μM respectively, 100μM for 2,3BD in primary melanocytes (Figure S4C). To verify the effects of the small molecular weight compounds on NNT function, the GSH/GSSG ratio, an indirect endpoint of NNT enzyme activity, was measured, revealing decreased GSH/GSSG ratios induced by DCC and 2,3 BD in primary melanocytes (Figures 3B and 3C) and by DCC in UACC257 melanoma cells (Figure S4E), without significant toxicity (Figures S4C and S4E). Treatment of primary human melanocytes with either siNNT or 2,3BD significantly increased the intracellular melanin content, however simultaneous treatment with siNNT and 2,3 BD did not further increase the melanin (Figure S4D), suggesting that enhancement of pigmentation by 2,3 BD may be mediated by inhibition of NNT.
Figure 3. NNT inhibitors are non-toxic and induce pigmentation of primary melanocytes in vitro and in human skin explants.
(A) Murine melanocytes (Melan-A) showed increased melanin content after incubation with 2 mM 2,3BD or DCC, but not after incubation with palmitoyl-CoA; n =3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (B-C) Treatment of primary human melanocytes with different doses of DCC (B, n = 4) or 2,3BD (C, n = 6) for 24 hours yielded decreased GSH/GSSG ratios; analyzed by ordinary one-way ANOVA with Tukey’s (B) or Dunnett’s (C) post-test. (D) A single, one-time topical treatment with 2,3BD (1M or 11M) induces human skin pigmentation after 5 days. Left Panel: Representative images of at least three individual experiments are displayed. Right panel: Reflective colorimetry measurements of skin treated with 2,3BD (higher L* values represent lighter skin tones); n = 3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (E) Fontana-Masson staining of melanin in human skin after 2,3BD (50 mM) (i) and hematoxylin & eosin staining (ii) compared with vehicle control (DMSO). (iii) Supranuclear capping in human keratinocytes of 2,3BD- and vehicle control-treated skin displayed by Fontana-Masson staining. (F) NNT inhibitors, 2,3BD or DCC, applied daily at a 50 mM dose resulted in skin darkening after 5 days. Left Pane: Representative images of three individual experiments are displayed. Right panel: Reflective colorimetry measurements of human skin treated with 2,3BD, DCC, or DMSO vehicle (higher L* values represent lighter skin tones;) n = 3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (G) Immunofluorescence staining for CPD formation (Red) in human skin treated with 50 mM 2,3BD for 5 consecutive days. On the last day, skin was irradiated with 1000 mJ/cm2 UVB. The results show a protective role for 2,3BD from UVB-induced CPD damage. Representative images of three individual experiments are displayed. Scale bar 50μM. Quantified results were normalized to the total number of cells; n = 3, analyzed by ordinary two-way ANOVA with Šídák’s post-test. (H) Measurement of γ-H2AX (Red) in human skin revealed no significant toxicity of 2,3BD, while 2,3-BD-induced pigmentation protected from UVB-induced γ-H2AX formation. Representative images of three individual experiments are displayed Scale bar 50μM. Quantified results were normalized to the total number of cells; n = 3, analyzed by ordinary two-way ANOVA with Šídák’s post-test.
All data are expressed as mean ± SEM. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001
Next, we tested the compounds on human skin explants from different skin types. As suggested above, palmitoyl-CoA did not penetrate the epidermis and had no effect on pigmentation (data not shown). In abdominal skin from individuals of fair skin phototype 1-2, 2,3BD yielded a strong induction of pigmentation at relatively high doses (Figure 3D). Histology with Fontana-Masson staining showed increased melanin in the 2,3BD treated skin (Figures 3Ei and S4F) and no obvious cell damage or inflammation by H&E staining (Figure 3Eii), although the volatility of 2,3BD produces a strong butter-like aroma, potentially limiting its future clinical use. Importantly, keratinocytic supranuclear caps (Figures 3Eiii and Figure S4F) were present, suggesting the formation of functional melanosome/melanin transfer to keratinocytes, which allows cells to protect their nuclei from UV radiation. Daily application of 50 mM 2,3BD or DCC on skin from intermediately pigmented skin type 3-4 individuals yielded significantly increased pigmentation after 5 days (Figure 3F). Due to the activity of DCC as a coupling agent and its corresponding unclear toxicity risks, only 2,3BD was used in subsequent experiments.
2,3BD-induced skin pigmentation can prevent UVB-induced DNA damage
UV radiation interacting with DNA can directly produce cyclobutane pyrimidine dimers (CPD) and 6-4 photoproducts, whereas ROS-mediated DNA modifications produce alternative nucleotide adducts including 8,5-cyclo-2-deoxyadenosine, 8,5-cyclo-2-deoxyguanosine, and 8-oxo-deoxyguanine (Jaruga and Dizdaroglu, 2008; Wang, 2008).
While superficial epidermal cells containing modified proteins, lipids and DNA are continuously shed through corneocyte desquamation, durable basal cells require active DNA repair machinery for their maintenance. Melanomas have been found to contain high frequencies of somatic mutations with characteristic UV-induced signatures of C to T and G to A transitions (Berger et al., 2012). Protecting human skin from these intermediates is a major goal of skin cancer prevention strategies. As shown in previous studies, increased pigmentation can help to protect against CPD formation (D'Orazio et al., 2006; Mujahid et al., 2017). We tested if 2,3BD-induced pigmentation can protect skin from UVB-induced CPD formation. After inducing a visible increase in pigmentation of human skin by application of 50 mM 2,3BD to skin type 2-3 for 5 days (Figure 3G), UVB was applied and CPD formation was detected by immunofluorescence staining and normalized to the total number of cells. It was observed that 2,3BD treatment protected against formation of UVB-induced CPD (Figure 3G). We then measured γ-H2AX, a marker of DNA double-stranded breaks, in order to investigate potential 2,3BD-mediated toxicity as well as whether 2,3BD-mediated skin pigmentation could protect from UVB-induced γ-H2AX induction (Figure 3H). 2,3BD was observed to be non-toxic and the pigmentation it produced could protect human skin from UVB-induced γ-H2AX induction.
NNT regulates pigmentation in mice, zebrafish and human pigmentation disorders
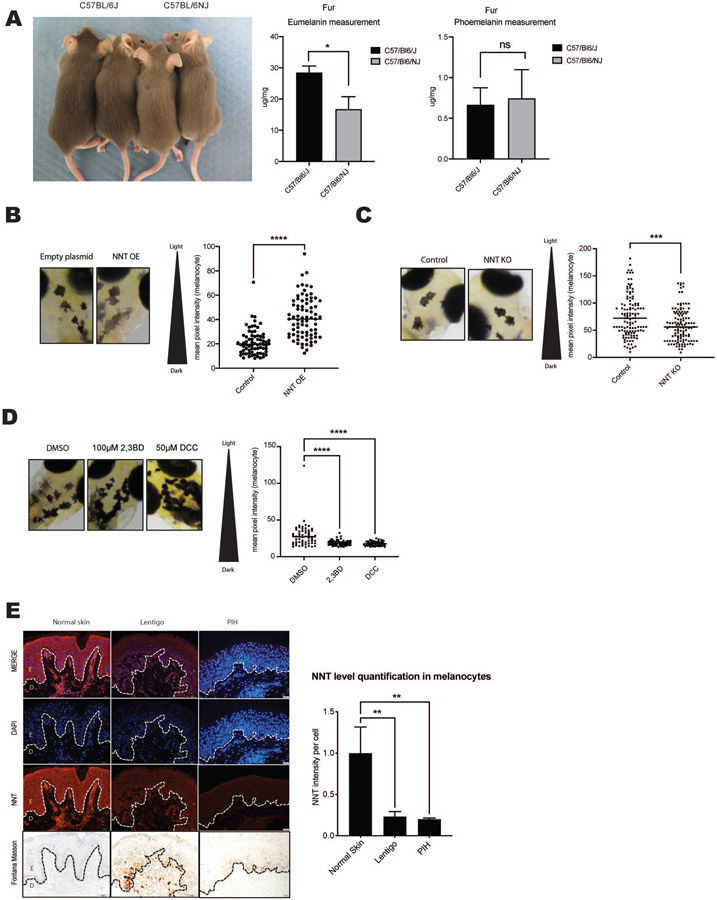
C57BL/6J and C57BL/6NJ mice are substrains of the C57BL/6 mouse with known genetic differences. While C57BL/6NJ mice are homozygous for the Nnt wild type allele, C57BL/6J mice are homozygous for the NntC57BL/6J mutation. This mutant allele is missing a stretch of 17,814 bp between exons 6 and 12, resulting in a lack of mature protein in these mutants (Toye et al., 2005) (Huang et al., 2006). In our experiments, C57BL/6J mice that are homozygous for the Nnt mutation (Figure S5A) showed increased fur pigmentation compared with C57BL6/NJ control (wild type Nnt) mice (Figure 4A, Left panel). Quantification of pheomelanin and eumelanin levels in mouse hair by HPLC shows higher eumelanin, but not pheomelanin, in C57BL/6J mice compared with C57BL/6NJ mice (Figure 4A).
Figure 4. NNT regulates pigmentation in mice, zebrafish and human pigmentation disorders.
(A) Left panel: C57BL/6J mice carrying a 5-exon deletion in the Nnt gene resulting in homozygous loss of NNT activity display increased fur pigmentation compared with C57BL/6NJ wild-type Nnt animals. Right graphs: Mouse fur samples were analyzed for pheomelanin and eumelanin levels by HPLC. n = 3, analyzed by multiple t-tests with the Holm-Šídák post-test. (B) Left panel: Zebrafish overexpressing NNT (NNT OE) display decreased pigmentation in individual melanocytes after 5 days. A representative image has been displayed. Results of mean melanocytic brightness, quantified by pixel-based analysis are shown in the graph at right; Empty plasmid (n = 11 fish; 72 melanocytes), NNT OE (n = 12 fish; 78 melanocytes), analyzed by unpaired, two-sided t-test. (C) Zebrafish with the nnt gene edited using CRISPR/Cas9 (NNT KO) display increased pigmentation after 4 days. A representative image has been displayed. Results of mean melanocytic brightness, quantified by pixel-based analysis are shown in the graph at right; Control (n = 42 fish; 120 melanocytes), NNT KO (n = 50 fish; 96 melanocytes). (D) Zebrafish treated for 24 hours with either 100 μM 2,3BD or 50 μM DCC display increased darkening after 4 days. A representative image has been displayed. Results of mean melanocytic brightness, quantified by pixel-based analysis are shown in the graph at right; DMSO (n = 21 fish; 97 melanocytes), 2,3BD (n = 20 fish; 59 melanocytes), DCC (n = 18 fish; 57 melanocytes), analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (E) Left panel: Human skin specimens from Asian individuals with lentigo or post inflammatory hyperpigmentation were compared to normal skin after staining for NNT, DAPI and Fontana Masson. Representative images of at least 3 samples are displayed (epidermis, E; dermis, D) Graph shows NNT signal intensities normalized to absolute cell numbers (DAPI); n = 3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test.
All data are expressed as mean ± SEM; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Next, a zebrafish (Danio rerio) model that overexpresses NNT selectively in melanocytes was engineered. Similar to humans and mice, zebrafish melanocytes originate from the neural crest, and the pathways leading to melanocyte differentiation and pigment production are conserved. Many human pigmentation genes and disorders have been successfully modeled in the zebrafish, highlighting the striking similarity between zebrafish and human melanocytes. Unlike humans, zebrafish have xanthophore and iridophore pigmentation cells, however in this manuscript we restrict our studies to melanocytes (van Rooijen et al., 2017). Five days after NNT overexpression, a decrease in intramelanocytic pigmentation was observed in NNT-overexpressing zebrafish compared with empty plasmid Zebrafish embryos (Figure 4B). This observation was confirmed by pixel-based brightness quantification analysis. Deletion of nnt using CRISPR-Cas9 (Figure S5B) resulted in darkened melanocytes (Figure 4C). Similar to the genetic deletion of nnt, treatment of zebrafish embryos for 24 hours with the chemical NNT-inhibitors (DCC and 2,3BD), resulted in a significant darkening (Figure 4D). However, subsequent treatment of NNT overexpressing fish with 2,3 BD prevented the NNT OE-induced decrease in melanocytic pigmentation (Figure S5C). This finding is in line with previous publications confirming an inhibitory role 2,3BD and DCC on NNT enzyme activity (Phelps and Hatefi, 1981) (Moody and Reid, 1983). Next, we examined the status of NNT in human hyperpigmentation disorders including post inflammatory hyperpigmentation (PIH) and lentigo. Skin biopsies of nine Asian patients were co-stained for NNT and 4’,6-diamidino-2-phenylindole (DAPI) immunofluorescence. NNT intensity was normalized to the sample’s DAPI intensity and cell count. Both epidermal and upper dermal skin were investigated. In line with the Human Protein Atlas, NNT is expressed in different epidermal cells including keratinocytes, fibroblasts, and melanocytes (Uhlen et al., 2015), were moderate levels of NNT expression (red) detected throughout the epidermis and upper dermis (Figure 4E, Left panels). While non-inflammatory skin disorders, such as ABNOM (Acquired, bilateral nevus of Ota-like macules, also known as Hori nevus), displayed NNT expression levels similar to those of healthy skin (data not shown), skin of patients with inflammation-induced disorders displayed decreased NNT expression levels. Disorders where intrinsic inflammation was present, such as post-inflammatory hyperpigmentation, or where extrinsic inflammation was present, such as UV-induced lentigo, NNT expression was significantly lower compared with healthy skin (Figure 4E, middle and right panels). Interestingly, this trend was further enhanced in areas of hyperpigmentation (Figure S5D).
Thus, NNT levels appear to be associated with murine and zebrafish pigmentation, as well as human disorders of hyperpigmentation.
Statistical associations between genetic variants of NNT and human skin pigmentation variation in diverse population cohorts.
Genetic associations
To investigate whether NNT plays a role in normal skin pigmentation variation in humans, we examined associations between pigmentation and genetic variants within the ~1.1 Mb NNT gene region. A meta-analysis was performed to combine P-values from Genome-Wide Association Studies (GWAS) conducted in 4 diverse population cohorts with a total of 462,885 individuals: two Western European cohorts (Rotterdam Study (Jacobs et al., 2015), UK Biobank (Hysi et al., 2018; Loh et al., 2018)), a multi-ethnic Latin American cohort (CANDELA (Adhikari et al., 2019)), and a multi-ethnic cohort from Eastern and Southern Africa (Crawford et al., 2017). In these studies skin pigmentation was measured either quantitatively by reflectometry or by an ordinal system (see Methods). UK Biobank summary statistics were also available for ease of skin tanning (sunburn) and use of sun protection.
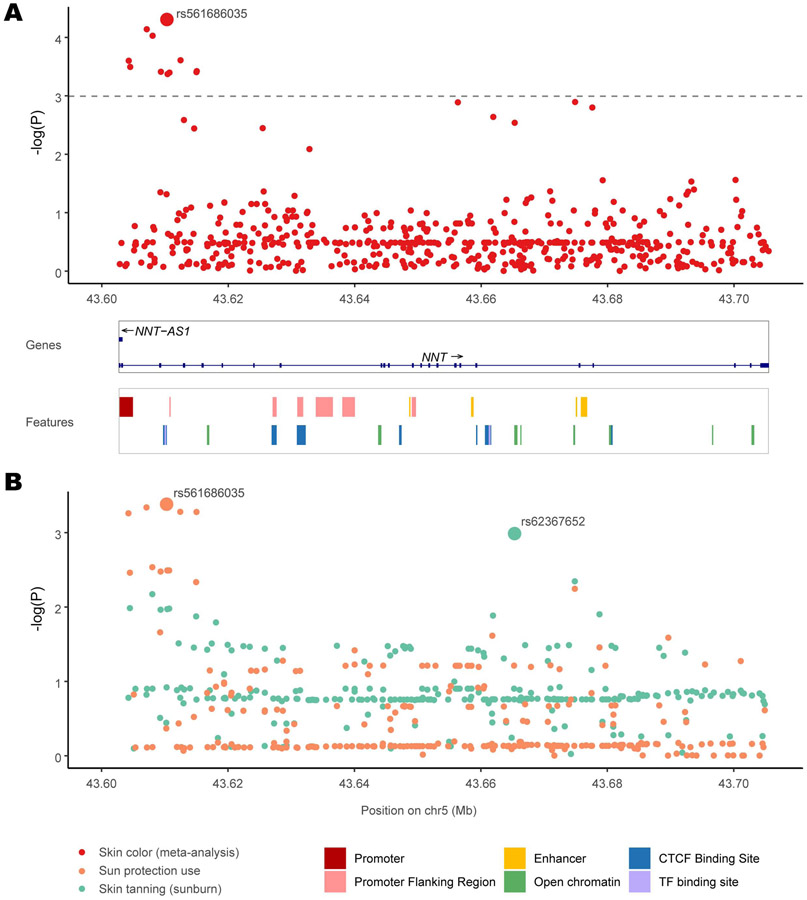
332 variants were available in the combined dataset; using a P-value significance threshold of 1.01E-3 (adjusted for multiple testing, see STAR Methods), 11 variants were significantly associated with skin pigmentation in the meta-analysis (Figure 5A and Supplementary Table 1). The variants were present in all worldwide populations, with the alternative alleles having the highest frequency in Africans (Supplementary Table 1 and Figure 6A) and associated with darker skin color. The strongest association (P = 4.94E-05) was observed for an intronic variant rs561686035.
Figure 5. Association results for SNPs in the NNT gene with skin color in multiple cohorts.
(A) P-values of SNPs from a meta-analysis of skin color (red) combining association results from 4 worldwide cohorts across 462,885 individuals. For each of the 332 SNPs, its location in the NNT gene is shown in the X axis and the negative logarithm of the P-value is shown in the Y-axis. The SNP with the strongest association, rs574878126, is labeled. The adjusted significance threshold is shown with a dashed line. The NNT gene track and a track of regulatory regions obtained from the Ensembl genome browser are shown below. (B) P-values of SNPs from the UK Biobank for sun protection use (orange) and ease of skin tanning (green). For each SNP, its genomic location is shown in the X axis and negative logarithm of the P-value is shown in the Y-axis. The SNPs with the strongest association for each trait, rs574878126 for sun protection use and rs62367652 for skin tanning, are labeled.
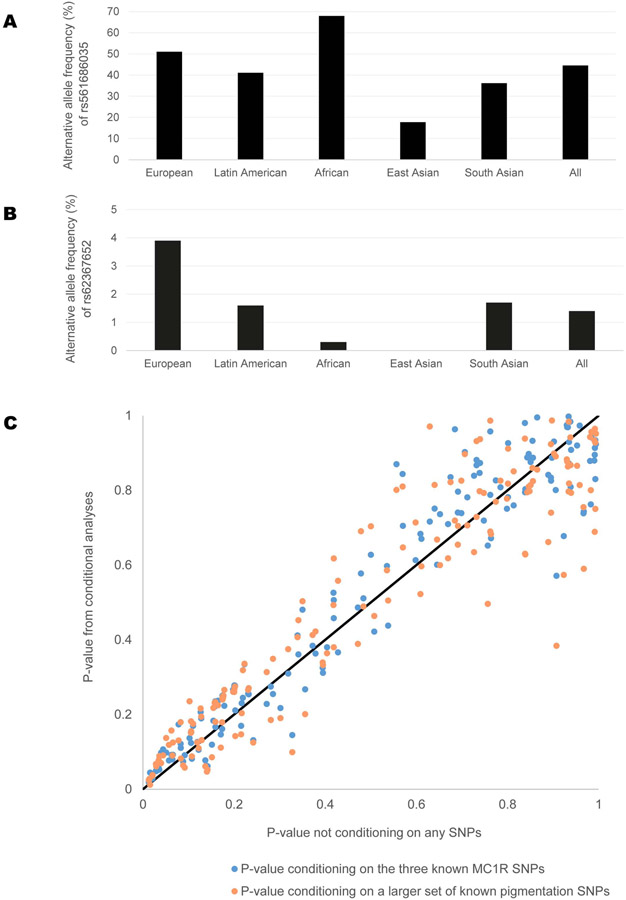
Figure 6. Association results and properties of SNPs from various human genetic association analyses.
(A-B): Allele frequencies for SNPs in the NNT gene showing most significant associations.
(A) Alternative allele frequencies of rs561686035 in various worldwide continental populations, obtained from 1000 Genomes Phase 3. This SNP showed the strongest association in the meta-analysis of skin color and for sun protection use. (B) Alternative allele frequencies of rs62367652 in various worldwide continental populations, obtained from 1000 Genomes Phase 3, are shown. This SNP showed the strongest association for ease of skin tanning (sunburn).
(C) Association results for SNPs in the NNT gene with or without conditioning on known pigmentation loci. P-values of SNPs from the Rotterdam Study are shown in this scatterplot. The X-axis represent P-values of SNPs from the standard GWAS analysis of skin pigmentation (not conditioned on any other SNP). P-values from two conditional analyses are plotted on the Y-axis: in blue, P-values conditioning on the three known MC1R SNPs; in orange, P-value conditioning on a larger set of known pigmentation SNPs. A diagonal line in black is shown for reference.
It was also the strongest associated variant for sun protection use in the UK Biobank cohort (P = 4.15E-04, Figure 5B), the minor allele being associated with increased use. The UK Biobank cohort also showed a significant association with ease of skin tanning (sunburn), the lowest P-value being 1E-3 for the intronic SNP rs62367652, the minor allele being associated with increased tanning (Figure5B, S6B).
In silico expression analysis of NNT variants
All the 11 variants that were significant in the meta-analysis of pigmentation are in linkage disequilibrium (LD) (r2 > 0.7), and they span a 11 KB region at the beginning of the NNT gene overlapping its promoter (ENSR00000180214) (Figure 5A), which shows regulatory activity in melanocytes and keratinocytes (according to the Ensembl database; Supplementary Table 1). Furthermore, several of these variants are highly significant eQTLs for the NNT gene in both sun exposed and unexposed skin tissues (according to the GTEx database, Supplementary Table 1). For these variants, the alternative alleles correlated with darker skin color and have negative effect sizes as eQTLs for NNT expression (Supplementary Table 1), indicating lower levels of expression of the NNT transcript.
Subsequently, we sought to understand the direction of effect of the NNT genetic variants on these traits and on the expression of NNT. We calculated the correlation between the GWAS effect sizes of the alternative allele of each genetic variant within the NNT region with their effect sizes as eQTLs on the expression of the NNT transcript according to GTEx in the two skin tissues (see Methods). The results are consistent with the direction of association between the NNT transcript expression and skin color as described earlier: expression levels of the NNT transcript in both tissues was negatively correlated (Supplementary Table 2) with darker skin color (especially in sun unexposed skin tissue, where the effect of external factors such as sunlight is less prominent), and sun protection use (especially in sun exposed skin tissue) as well as sunburn (especially in sun exposed skin tissue).
Therefore, several intronic SNPs within the NNT genomic region were associated with skin pigmentation, tanning, and sun protection use in 4 diverse cohorts including 462,885 individuals. Using eQTL expression data for NNT, we observe that lower expression of the NNT transcript in skin tissues correlates with darker skin color, and consequently less sunburn and less sun protection use.
Conditioning on known pigmentation SNPs
As MC1R is a major determinant of pigmentation, with known genetic variants associated with lighter skin color, red hair, and freckles in European populations (Quillen et al., 2019), we checked whether MC1R can be a confounder in the observed association of NNT with skin pigmentation. In the Western European cohort of the Rotterdam Study, conditioning on the three known MC1R SNPs in the GWAS did not significantly alter the P-values of the NNT variants ((P = 0.869, Figure 6B). Conditioning on a larger set of known pigmentation variants (see STAR Methods) in the GWAS does not significantly alter the P-values of the NNT variants (P = 0.191, Figure 6C) either.
Discussion
This study addresses the question of how redox metabolism interplays with skin pigmentation. It identifies (i) existence of a distinct redox-dependent, UV- and MITF-independent skin pigmentation mechanism; (ii) a role for the mitochondrial redox-regulating enzyme NNT in altering pigmentation by regulating tyrosinase protein stability and melanosome maturation via a redox-dependent mechanism; (iii) a class of topical NNT inhibitors that yield skin darkening (Martin et al., 2017)(Martin et al., 2017)(Martin et al., 2017).
SLC24A5 was the first gene to be identified as associated with light skin color in Europeans (Lamason et al., 2005). More recent genome-wide association studies (GWAS) in non-Europeans (Arjinpathana and Asawanonda, 2012; Crawford et al., 2017; Hysi et al., 2018; Lin et al., 2018; Martin et al., 2017) emphasized the complex nature of human skin pigmentation. In addition to certain key regulators such as TYR and MITF, many other genes may impact skin pigmentation and an individual’s skin color. It is thus plausible that factors involved in redox metabolism, such as NNT, may be responsive to environmental changes such as UV exposure or inflammation. Increasing eumelanin levels as a response to ROS-inducing events might have been beneficial during evolution by maintaining cutaneous redox equilibrium. An interplay between oxidative stress and skin pigmentation has been suspected (Arjinpathana and Asawanonda, 2012), while the exact mechanism and ways to potentially target this pathway have been incompletely elucidated.
From a clinical perspective, our findings are relevant due to the prevalence of pigmentary disorders, which are among the most common reasons for dermatological consultations (Cestari et al., 2014). In addition, lightly pigmented individuals have increased risk of melanoma, a life-threatening disease.
As shown previously (D'Orazio et al., 2006; Mujahid et al., 2017), skin pigmentation and, especially, high eumelanin levels can protect human skin from UVB-induced CPD formation. Most probably, this effect relates to both eumelanin-mediated absorbance of UV and buffering function of eumelanin towards oxidative stress radicals. Different approaches for increasing pigmentation have been tried so far, including the topical use of the cyclic AMP agonist forskolin (D'Orazio et al., 2006), which worked well in mice but does not penetrate human skin sufficiently. Afamelanotide, an α-MSH analog, has been used for treating erythropoietic protoporphyria by producing a hyperpigmentation that is able to protect skin against UV-induced photosensitivity (Langendonk et al., 2015). Topical administration of SIK inhibitors preclinically also induced pigmentation (Horike et al., 2010; Mujahid et al., 2017) As MITF is a transcription factor involved in numerous melanocyte functions, transiently targeting pigmentation via NNT inhibition provides a distinct and potentially complementary approach that might offer applications in contexts such as pigmentation disorders and skin cancer prevention.
In this report, we present evidence for the existence of a redox-dependent skin pigmentation pathway. In contrast to the established classic UVB-cAMP-MITF-dependent tanning pathway, this mechanism is independent of UV irradiation, MITF, and MITF signaling effects. Instead, a distinct pigmentation mechanism dependent on ROS is described, demonstrating how oxidative stress impacts pigmentation in cells of melanocytic origin via modifying GSH, NADPH, increasing tyrosinase protein stability, tyrosinase-related proteins and melanosome maturation. Evaluating the interplay between other pigmentation mechanisms, such as immediate and persistent pigment darkening, as well as understanding safety, penetration and efficacy of topical NNT modifiers may be worthy of study in future clinical settings.
Limitations of the study.
In this proof-of-principle study the effect of NNT on pigmentation has been shown. However, there are several limitations:
While this study used previously identified NNT inhibiting compounds, further research is needed to identify more specific compounds for modulating NNT enzyme activity. We believe caution is warranted when modulating cutaneous redox biology and skin pigmentation, requiring careful attention before human application.
We used the NntC57BL/6J mouse model, which are homozygous for the NntC57BL/6J mutation and lack a stretch of 17,814 bp between exons 6 and 12 resulting in a lack of mature NNT protein. As this mutation also affects the function of other proteins, the use of this model alone cannot confirm NNT’s role on pigmentation.
The statistical association analyses identify a correlation between SNPs within NNT and human pigmentation variation. While some of the associated SNPs are located within the promoter region of the NNT gene and are eQTLs for NNT expression, further biological experiments are needed to confirm causality.
Although zebrafish melanocyte function and melanin biosynthesis are similar to humans as many genes are shared, the contribution of other fish-specific cell types to melanocyte biology is incompletely understood. The MiniCoopR rescue system employed in this study overexpresses human NNT potentially at above physiological levels. This seems to cause slight toxicity to the melanocytes, which resulted in lower melanocyte rescue. Additionally, due to technical variability in single cell injection efficiency, variation in the number and location of melanocytes per fish was observed.
STAR★METHODS
RESOURCE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Elisabeth Roider (eroider@cbrc2.mgh.harvard.edu).
Material Availability
Plasmids, mouse and zebrafish lines generated in this study will be distributed upon request to other investigators under a Material Transfer Agreement. All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
DATA AND CODE AVAILABILITY
All the Software packages and methods used in this study have been properly detailed and referenced under the Software and Algorithms listed in KEY RESOURCES TABLE. All Human data (SNP analysis) generated in this study are available under the Deposited Data listed in KEY RESOURCES TABLE.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-MITF monoclonal antibody C5 | Made in the lab of Dr. David E. Fisher | (King et al., 1999) C5 |
| Mouse monoclonal anti-tyrosinase antibody, clone T311 | Sigma-Adlrich | Cat# 05-647: RRID:AB_309873 |
| Donkey anti-rabbit IgG-HRP | ThermoFisher Scientific | Cat# 45-000-683: RRID: AB_2721111 |
| Amersham ECL mouse IgG, HRP | ThermoFisher Scientific | Cat#45000680;RRID: AB_2721110 |
| Monoclonal anti-β-actin-peroxidase | Sigma Aldrich | Cat# A3854;RRID:AB_262011 |
| Alexa Fluor 555 goat anti-rabbit IgG (H+L), secondary antibody | ThermoFisher Scientific | Cat# A-21428;RRID: AB_2535849 |
| Alexa Fluor 647 goat anti mouse IgG (G+L); fluorescence conjugated secondary antibody | ThermoFisher Scientific | Cat# A-21236;RRID: AB_2535805 |
| Alexa Fluor 594 F(ab)2 fragment of goat anti-rabbit IgG (G+L); fluorescence conjugated secondary antibody | ThermoFisher Scientific | Cat# A-11072;RRID: AB_2534116 |
| Alexa Fluor 488-conjugated donkey anti-mouse secondary antibodies | ThermoFisher Scientific | Cat# A-21202;RRID: AB_141607 |
| Anti-Cyclobutane Pyrimidine Dimers (CPDs) mAb antibody (Clone TDM-2) | Cosmo Bio USA | Cat# CAC-NM-DND-001; RRID: AB_1962813 |
| Rabbit anti-γ-H2AX (P-ser139) polyclonal antibody | NOVUS Biologicals | Cat# NB100-384;RRID:AB_10002815 |
| Mouse monoclonal anti-Mitofusin 2 antibody [6A8] | Abcam | Cat# ab56889;RRID: AB_2142629 |
| Rabbit polyclonal anti TRP2/DCT antibody | Abcam | Cat# ab74073;RRID:AB_1524517 |
| Mouse monoclonal anti-NNT antibody [8B4BB10] | Abcam | Cat# ab110352;RRID:AB_10887748 |
| Rabbit anti-NNT (C-terminal) polyclonal antibody | Abcam | Cat# ab214212; RRID:AB_2889980 |
| Mouse anti-8-oxo-dG monoclonal antibody | Trevigen | Cat# 4354-MC-050; RRID:AB_1857195 |
| IDH1 (D2H1) Rabbit mAb | Cell Signaling Technology | Cat# 8137;RRID: AB_10950504 |
| Mouse monoclonal p53 antibody [PAb 240] | Abcam | Cat# ab26;RRID:AB_303198 |
| Rabbit monoclonal TRP1 antibody [EPR21960] | Abcam | Cat# ab235447; RRID:AB_2889980 |
| Mouse monoclonal antibody Pmel17 (E-7) | Santa Cruz Biotechnology | Cat# sc-377325; RRID:AB_2889982 |
| LC3B (D11) rabbit monoclonal antibody | Cell Signaling Technology | Cat#38668S RRID:AB_2137707 |
| Biological Samples | ||
| Full thickness human breast and abdominal skin explants | Massachusetts General Hospital | IRB# 2013P000093 |
| Paraffin-embedded formalin fixed slides, prepared from breast and abdominal biopsy samples | Massachusetts General Hospital | IRB# 2013P000093 |
| Human skin samples for genome wide association study (GWAS) | Massachusetts General Hospital or the Cooperative Human Tissue Network | IRB# 2013P000093 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 3-isobutyl-1-methylxanthine (IBMX) | Sigma-Aldrich | Cat# I5879 |
| 12-O-tetradecanoylphorbol-13-acetate (TPA) | Sigma-Aldrich | Cat# 16561-29-8 |
| Ham's F10 | Thermo Fisher Scientific | Cat# MT10070CV |
| N6,2′-O-Dibutyryladenosine 3′,5′-cyclic monophosphate sodium salt | Sigma-Aldrich | Cat# D0627 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat# 15140163 |
| Na3VO4 | Sigma-Aldrich | Cat# 450243 |
| Medium 254 | Life Technologies | Cat# M254500 |
| 0.05% Trypsin-EDTA w/ phenol red | Life Technologies | Cat# 25300120 |
| Human Melanocyte Growth Supplement (HMGS) | Life Technologies | Cat# S0025 |
| Bovine Serum Albumin | Sigma | Cat#A7030 |
| Goat serum | Sigma-Aldrich | Cat# G9023 |
| RPMI (Roswell Park Memorial Institute 1640 Medium) | Life Technologies | Cat# 11875119 |
| RIPA lysis buffer | Sigma-Aldrich | Cat# R0278 |
| FetalPlex Animal Serum Complex | Gemini Bio-Products | Cat# 100-602 |
| Western Lightning Plus-ECL | PerkinElmer | Cat # NEL105001EA |
| Non-fat milk powder | Boston BioProducts | Cat# P-1400 |
| Protein Block | Agilent | Cat# X090930-2 |
| Antibody Diluent | DAKO | Cat# S3022 |
| VECTASHIELD® HardSet™ Antifade Mounting Medium with DAPI | Vector Laboratories | Cat# H-1500 |
| synthetic melanin | Sigma Aldrich | Cat# M8631 |
| N,N-Dicyclohexylcarbodiimide [DCC] | Sigma Aldrich | Cat# D80002 |
| 2,3-Butanedione [2,3BD] | Sigma Aldrich | Cat# B85307 |
| Palmitoyl coenzyme A lithium salt | Sigma Aldrich | Cat# #P9716 |
| cycloheximide (CHX) | Sigma Aldrich | Cat# C7698 |
| NADPH | Sigma Aldrich | Cat# N7505 |
| N-Acetyl-L-cysteine (NAC) | Sigma Aldrich | Cat# A7250 |
| MitoTEMPO | ThermoFisher Scientific | Cat# 501872447 |
| Hydrogen peroxide solution | Sigma Aldrich | Cat# 216763 |
| SYBR FAST qPCR master mix | Kapa Biosystems | Cat# KK4600; |
| Protease and Phosphatase Inhibitor | ThermoFisher Scientific | Cat# PI78445 |
| Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate | Perkin Elmer | Cat# NEL105001EA |
| MitoSOX Red | ThermoFisher Scientific | Cat# M36008 |
| CM-H2DCFDA | ThermoFisher Scientific | Cat# C6827 |
| NucBlue | ThermoFisher Scientific | Cat# R37605 |
| Polybrene | Sigma-Aldrich | Cat# TR-1003 |
| Paraformaldehyde 16% | ThermoFisher Scientific | Cat# 50980487 |
| Ethanol | Thermo Fisher Scientific | Cat# 04355226 |
| Triton X-100 | Sigma Aldrich | Cat# T8787 |
| TWEEN® 20 | Sigma Aldrich | Cat# P7949 |
| Forskolin from Coleus forskohlii, ≥98% | Sigma Aldrich | Cat# F6886 |
| Lipofectamine RNAiMAX Transfection Reagent | Life Technologies | Cat# 13778150 |
| IQ5 High-fidelity DNA Polymerase | New England Biolabs | Cat# M0491S |
| Critical Commercial Assays | ||
| Direct cAMP ELISA Kit | Enzo Life Sciences | Cat# ADI-901-066 |
| GSH/GSSG-Glo Assay | Promega | Cat# V6611 |
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat# G7570 |
| Pierce BCA protein assay | ThermoFisher Scientific | Cat# 23225 |
| KAPA Library Quantification Kits | Roche | Cat# 7960140001 |
| MiSeq Reagent Kits v2 (300 cycles) | Illumina | Cat# MS-102-2002 |
| MaxBlock Autofluorescence Reducing Reagent Kit | MaxVision Biosciences | Cat# MB-L |
| Fontana-Masson Stain Kit (Melanin Stain) | Abcam | Cat# ab150669 |
| Dual Reporter System | GeneCopoeia | Cat# HPRM39435-LvPM02 |
| Secrete-Pair Gaussia Luciferase Assay Kit | GeneCopoeia | Cat# LF062 |
| NADP/NADPH-Glo Assay | Promega | Cat# G9082 |
| QUANTI-Blue™ Solution | InvivoGen | Cat# rep-qbs |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74136 |
| Deposited Data | ||
| Raw data supporting the human genetics association analyses | https://www.dropbox.com/scl/fi/ahdfnjo4puwzdz8ayw2ix/supporting-data-human-genetic-associations-2.xlsx?dl=0&rlkey=gvbpat4tjb6bnekgnxuu1a6yh | |
| Publicly Available Data | ||
| Ensembl database information for promoter ENSR00000180214 of the NNT gene | Ensembl | http://www.ensembl.org/Homo_sapiens/Regulation/Summary?fdb=funcgen;r=5:43600000- |
| GTEx expression database | GTEx | www.gtexportal.org |
| GWAS summary statistics from the CANDELA cohort | GWAS Central | http://www.gwascentral.org/study/HGVST3308 |
| GWAS summary statistics from the UK Biobank cohort | https://cnsgenomics.com/software/gcta/#DataResource | |
| Experimental Models: Cell Lines | ||
| Primary human melanocytes (isolated from neonatal foreskins) | Massachusetts General Hospital | IRB# 2013P000093 |
| Human melanoma cell line UACC257 | National Cancer Institute Division of Cancer Treatment and Diagnosis (DCTD) Tumor Repository | |
| Human melanoma cell line SK-MEL-30 | Memorial Sloan Kettering Cancer Center | https://www.mskcc.org/ |
| Mouse Melan-A cell line | Wellcome Trust Functional Genomics Cell Bank | Bennett et al., 1987 |
| Primary human keratinocytes isolated from discarded surgical human skin tissue (e.g., foreskins) | Massachusetts General Hospital | IRB# 2013P000093 |
| Primary human fibroblasts isolated from discarded surgical human skin tissue (e.g., foreskins) | Massachusetts General Hospital | IRB# 2013P000093 |
| Lenti-X™ 293T cells | Clontech | Cat# 632180 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice | Jackson Laboratory | Stock No: 000664 |
| C57BL/6NJ mice | Jackson Laboratory | Stock No: 005304 |
| Casper zebrafish (mitfa−/−;roy−/−) embryos | Laboratory of Dr. Leonard I. Zon | |
| Oligonucleotides | ||
| nheIkozakHAhNNT_f1: forward, 5’-ctagctagcCCGCCACCATGTACCCATACGATGTTCCAGATTACGCTGCAAACCTATTGAAAACAGTGGTGACTG-3’ | eurofins Genomics | For PLMJ1- HA-NNT |
| hNNTnheI_r1: reverse, 5’-ctagctagcTTACTTCTGATAGGATTCTCTAACTTTCGC-3’ | eurofins Genomics | For PLMJ1- HA-NNT |
| nheIkozakhMFN2_f1: forward, 5’-ctagctagcGCCACCATGTCCCTGCTCTTCTCTCGATGC-3’ | eurofins Genomics | For PLMJ1- HA MFN2 |
| hMFN2(HA)nheI_r1: reverse, 5’-ctagctagcTTAGGATCCAGCAGCGTAATCTGGAAC-3’ | eurofins Genomics | For PLMJ1- HA MFN2 |
| RT-Primers for NNT, IDH1, MFN2, TYRP1, DCT/TRP2, MITF, POMC, PPARGC1A, Tyrosinase: See Table in STAR Methods | ||
| Additional oligonucleotides that were used for nnt knockout in zebrafish are in Supplementary Table 3 | ||
| Recombinant DNA | ||
| CRISPR MiniCoopR-U6:gRNA-mitfa:Cas9 plasmid | Addgene | Cloned from Addgene plasmid ID 118840 |
| pMiniCoopR-mitfa:NNT expression plasmid | Addgene | Cloned from Addgene plasmid ID 118850 |
| pLMJ1-NNT-HA | This manuscript | Based on Addgene plasmid, # 19319 |
| pLMJ1-EGFP plasmid | Laboratory of Dr. David Sabatini | Addgene plasmid, # 19319; http://n2t.net/addgene:19319; RRID:Addgene_19319, (Sancak et al., 2008) |
| pcDNA3.1 Mfn2HA | Laboratory of Dr. Allan Weissman | Addgene plasmid, # 139192; http://n2t.net/addgene:139192; RRID:Addgene_139192 ((Leboucher et al., 2012) |
| PLMJ1-MFN2-HA | This manuscript | Based on Addgene plasmid, # 139192 and Addgene plasmid, # 19319 |
| Software and Algorithms | ||
| GraphPad Prism 8.4.3 (471) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ (v1.8.0) | National Institutes of Health | https://imagej.nih.gov/ij/ |
| FIJI software for pixel-based color quantification | FIJI | https://imagej.net/Fiji |
| Off-target prediction software (for design of guide RNAs) | (Bae et al., 2014) | http://www.rgenome.net/cas-offinder/ |
| On-target prediction software (for design of guide RNAs); CRISPRscan and CHOPCHOP | (Moreno-Mateoset al., 2015) (Labun et al., 2019) |
https://www.crisprscan.org/ https://chopchop.cbu.uib.no/ |
| Axiovision REL 4.7 software | Zeiss | https://carl-zeiss-axiovision-rel.software.informer.com/4.7/ |
| CRISPResso2 software (for genome editing) | Kendell Clement et al | Nat Biotechnol. 2019 March; 37(3):224–226. |
| MACH software | http://csg.sph.umich.edu/abecasis/mach/index.html | |
| GCTA program | Yang et al., 2011 | https://cnsgenomics.com/software/gcta/ |
| PLINK program | https://www.cog-genomics.org/plink/1.9/ | |
| BioRender | BioRender.com | |
| Fiji | Saalfeld, S., Schmid, B., et al. (2012) | https://imagej.net/Fiji |
| NDP.view2 Viewing software | HAMAMATSU | https://www.hamamatsu.com/us/en/product/type/U12388-01/index.html |
| Other | ||
| siGENOME Human MITF siRNA SMARTpool | Dharmacon | Cat# M-008674-00-0005 |
| ON-TARGETplus Human IDH1 siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-008294-01-0005 |
| ON-TARGETplus Human MFN2 siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-012961-00-0005 |
| ON-TARGETplus Human NNT siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-009809-00-0005 |
| ON-TARGETplus Human PPRGC1A siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-005111-00-0005 |
| ON-TARGETplus Human Tyrosinase siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-012555-00-0005 |
| ON-TARGETplus non-targeting siRNA control pool | Horizon Inspired Cell Solutions | Cat# D-001810-10-05 |
| 4-15% Criterion TGX Precast Midi Protein gels | Bio-Rad Laboratories | Cat# 5671084 |
| Chamber slides | ThermoFisher Scientific | Cat# 125657 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Ethics Statement
Mice studies and procedures were approved by the Institutional Animal Care and Use Committee of Massachusetts General Hospital and were conducted strictly in accordance with the approved animal handling protocol. Zebrafish experiments performed in this study were in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The animal research protocol, including zebrafish maintenance and euthanasia was approved by the Institutional Animal Care and Use Committee of Boston Children’s Hospital.
Mice
All mice were bred on a heterozygous MiWhite background (Mitf white) (Steingrimsson et al., 2004). C57BL/6J mice (Jackson Laboratory, Stock No: 000664) displaying a 5-exon deletion in the Nnt gene resulting in a homozygous loss were compared to Nnt wild type C57BL/6NJ mice (Jackson Laboratory, Stock No: 005304). All mice were matched by gender and age (female, 6 weeks old). Mice were genotyped according to the protocol obtained from Jackson Laboratory (protocol 26539: Standard PCR Assay - Nnt<C57BL/6J>, Version 2.2).
Zebrafish
Overexpression of human NNT in Zebrafish
The human NNT gene was cloned into the MiniCoopR expression plasmid to allow melanocyte-specific overexpression of NNT (Ceol et al., 2011). The MiniCoopR plasmid contains an mitf mini-gene alongside mitfa driven NNT or an empty control. Casper zebrafish (mitfa−/−; roy−/−) embryos (Ablain et al., 2015) were injected at the single cell stage with plasmid DNA, which gets incorporated into the genome though Tol2 transgenesis. This results in the rescue of melanocytes via the mitfa minigene and melanocyte-specific overexpression of NNT. Larvae were raised for 5 days and imaged using a Nikon SMZ18 Stereomicroscope.
Deletion of Zebrafish nnt gene
SpCas9 guide RNAs (gRNAs) were designed to target the first two exons of the zebrafish nnt gene using on-target and off-target prediction software (Supplementary Table 3). gRNA expression plasmids were constructed by cloning oligonucleotides (Integrated DNA Technologies) into BseRI-digested pMiniCoopR-U6:gRNA-mitfa:Cas9 (Addgene plasmid ID 118840) (Ablain et al., Dev Cell 2015). A control CRISPR MiniCoopR plasmid was generated by cloning a scrambled gRNA into the CRISPR MiniCoopR vector. The CRISPR MiniCoopR plasmid contains an mitf mini-gene alongside mitfa:Cas9 and U6:gRNA. Casper zebrafish (mitfa−/−; roy−/−) embryos (Ablain et al., 2015) were injected at the single cell stage with plasmid DNA, which gets incorporated into the genome though Tol2 transgenesis. This results in the rescue of melanocytes via the mitfa minigene and melanocyte-specific knockout of nnt. Larvae were raised for 4 days and imaged using a Nikon SMZ18 Stereomicroscope.
DNA was extracted from the embryos at 4 days post fertilization using the Hot Shot method (Truett, et al, BioTechniques 2000), for analysis of genome editing. The efficiency of genome modification by SpCas9 was determined by next-generation sequencing using a 2-step PCR-based Illumina library construction method, as previously described (Walton et al., 2020). Briefly, genomic loci were amplified from gDNA extracted from pooled samples of 8-10 zebrafish embryos using Q5 High-fidelity DNA Polymerase (New England Biolabs, # M0491S) with the primers listed in Supplementary Table 3. PCR products were purified using paramagnetic beads prepared as previously described (Rohland and Reich, 2012) (Kleinstiver et al., 2019). Approximately 20 ng of purified PCR product was used as template for a second PCR to add Illumina barcodes and adapter sequences using Q5 and the primers (Supplementary Table 3). PCR products were purified prior to quantification via capillary electrophoresis (Qiagen QIAxcel), followed by normalization and pooling. Final libraries were quantified by qPCR using a KAPA Library Quantification Kit (Roche, #7960140001) and sequenced on a MiSeq sequencer using a 300-cycle v2 kit (Illumina, #MS-102-2002). Genome editing activities were determined from the sequencing data using CRISPResso2 (Clement et al., 2019) with default parameters.
Chemical treatment of Zebrafish
Wildtype Tübingen zebrafish (Figure 4D) or mcr:NNT or mcr:Empty rescued Casper Zebrafish (Figure S5C) were placed in a 24 well plate at 72 hours post-fertilization, with 10 larvae per well for a total twenty larvae per condition. Larvae were treated for 24 hours with either 2,3BD (1 μM, 10 μM, 100 μM, 1 mM; Sigma Aldrich, #B85307), DCC (1 μM, 10 μM, 50 μM, 100 μM; Sigma Aldrich, #D80002), or DMSO (1:500) in E3 embryo medium. At 4 days post fertilization, larvae were imaged using a Nikon SMZ18 Stereomicroscope. At least 57 melanocytes from 18 zebrafish embryos were analyzed using the FIJI software enabling pixel-based color quantification.
Quantification of pigmentation in the Zebrafish model
Pigmentation of free-standing melanocytes were identified at high magnification, making sure no overlapping signal was included into the analysis. The intra-melanocytic region was marked and the brightness was measured using the FIJI software. The measured output is the mean pixel intensity of the measured region (=melanocyte), which was plotted as one dot in the graph displayed.
Human skin explants
Skin samples considered surgical waste were obtained de-identified from healthy donors (IRB# 2013P000093) undergoing reconstructive surgery, according to institutional regulations. Full thickness human abdominal skin explants were cultured in petri dishes with a solid phase and liquid phase phenol red free DMEM medium containing 20% penicillin/streptomycin/glutamine, 5% fungizone (Gibco), and 10% fetal bovine serum. Explants were treated with vehicle (DMSO), 2,3BD (50 mM, 1 M, or 11 M;) or DCC (50 mM) as indicated in the figure legends. Compounds were applied strictly on top of the explants, making sure no drip occurred into the underlying media. For UV irradiation experiments, a UV lamp (UV Products) was used at 1000 mJ/cm2 UVB.
Cell lines
Primary human melanocytes were isolated from normal discarded foreskins and were established in TIVA medium as described previously (Khaled et al., 2010) or in Medium 254 (Life Technologies, #M254500) (Allouche et al., 2015). Human melanoma cell line UACC257 (sex unspecified) was obtained from the National Cancer Institute (NCI), Frederick Cancer Division of Cancer Treatment and Diagnosis (DCTD) Tumor Cell Line Repository. SK-MEL-30 (male) human melanoma cell line was from Memorial Sloan Kettering Cancer Center. Both melanoma cell lines have been authenticated by our lab using ATCC’s STR profiling service. UACC257 and SK-MEL-30 cells were cultured in DMEM and RPMI medium (Life Technologies, #11875119) respectively, supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin/L-glutamine in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
Murine Melan-A (Bennett et al., 1987)cells were obtained from the Wellcome Trust Functional Genomics Cell Bank. Melan-A cells were grown in RPMI 1640 supplemented with 10% FBS or FetalPlex (Gemini Bio-Products, #100-602), 100,000 U/L penicillin, 100 mg/L streptomycin sulphate, 100x Glutamax, and 200 nM TPA.
Primary human keratinocytes were cultured in EpiLife® medium supplemented with human keratinocyte growth supplement (HKGS, ThermoFisher Scientific). Primary human fibroblasts were cultured in medium 106 supplemented with low serum growth supplement (LSGS, ThermoFisher Scientific). 106 and 104 cells were plated per well of 6-well and 96-well plates, respectively. Drugs indicated in the figure legends were dissolved in DMSO and added 1:1000 to the culture media for 24 h at the concentrations indicated.
METHOD DETAILS
siRNA transfection
A single treatment of 10 nmol/L of siRNA was delivered to a 60% confluent culture by transfection with Lipofectamine RNAiMAX (Life Technologies, #13778150) according to the manufacturer’s recommendations. After 48-72 h of transfection, total RNA or protein was harvested.
Plasmid overexpression
Human NNT fused to a haemagglutinin (HA)-tag at the N-terminus was amplified from pEGFP-C1-hNNT (primer sequences are in the Key Resources Table) and was subcloned into the NheI restriction site of pLMJ1-EGFP [a gift from David Sabatini, Addgene plasmid #19319, http://n2t.net/addgene:19319, RRID:Addgene_19319 (Sancak et al., 2008)] using NheI (New England Biolabs, R3131S).
For human MFN2 overexpression, human MFN2 fused to three HA tags at the C-terminus was amplified from pcDNA3.1 Mfn2HA (a gift from Allan Weissman, Addgene plasmid 139192, http://n2t.net/addgene:139192, RRID:Addgene_139192 (Leboucher et al., 2012) (primer sequences are in the Key Resources Table) and was subcloned into the NheI restriction site of pLJM1-EGFP using NheI (New England Biolabs, #R3131S).
FLAG-tagged human NNT cDNA (NNT-FLAG) was purchased from Origene (RC224002). The NNT-FLAG cassette was re-cloned into pLJM1-EGFP (Addgene #19319) following NheI and EcoRI digestion.
Lentivirus generation and infection
Lentivirus was generated in Lenti-X™ 293T cells (Clontech, #632180). The Lenti-X cells were transfected using 250 ng pMD2.G, 1250 ng psPAX2, and 1250 ng lentiviral expression vector in the presence of PEI (MW:25K). For infection with lentivirus, 0.1–1 ml of lentivirus-containing medium was used in the presence of 8 μg/ml polybrene (Sigma, #TR-1003). Selection with puromycin (10 μg/ml) was performed the day after infection.
In vitro culture with NNT inhibitors
2,3-Butanedione 97% (2,3 BD) (Sigma Aldrich, #B85307) (1 μM, 10 μM, 100 μM, 2 mM), N,N-Dicyclohexylcarbodiimide (DCC) (Sigma Aldrich, #D80002) (1 mM, 2 mM, 10 mM), and Palmitoyl coenzyme A lithium salt (Sigma Aldrich, #P9716) (10 μM, 2 mM) were reconstituted with DMSO (American Type Culture Collection, 4-X).
Immunoblotting
Whole-cell protein lysates were prepared using RIPA lysis buffer (Sigma-Aldrich, #R0278) supplemented with Protease and Phosphatase Inhibitor (ThermoFisher Scientific, #PI78445). Protein concentrations were quantified using the Pierce BCA protein assay (ThermoFisher Scientific, #23225). Immunoblotting was performed by standard techniques using 4-15% Criterion TGX Precast Midi Protein gels (Bio-Rad Laboratories, #5671084) and transferring to 0.2 μm nitrocellulose membranes (Bio-Rad Laboratories, #1620112). Membranes were blocked with 5% non-fat milk (Boston BioProducts, #P-1400) in PBS containing 0.1% Tween 100 and incubated with one of the following primary antibodies at the indicated dilution (antibody sources are in the Key Resources Table): 1:20 dilution of anti-MITF monoclonal antibody C5, 1:1,000 dilution of anti-Tyrosinase clone T311, 1:1,000 dilution of anti- Mitofusin-2 antibody [6A8], 1:500 dilution of TRP2/DCT antibody, 1:1,000 dilution of anti-NNT antibody [8B4BB10], 1:1,000 dilution of anti-IDH1 (D2H1) antibody, 1:1,000 dilution of p53 antibody [PAb 240], 1:1,000 dilution of TYRP1 antibody [EPR21960], 1:1,000 dilution of mouse monoclonal antibody Pmel17 (E-7), or 1:1,000 dilution of LC3B (D11) rabbit monoclonal antibody. Incubation with the appropriate secondary antibody followed, either a 1:5,000 dilution of donkey anti-Rabbit IgG-HRP or a 1:3,000 dilution of Amersham ECL mouse IgG, HRP.
To verify equal loading of samples, membranes were re-probed with a 1:20,000 dilution of monoclonal anti-β-actin-peroxidase (Sigma Aldrich, #A3854). Protein bands were visualized using Western Lightning Plus ECL (PerkinElmer, #NEL105001EA) and quantified using ImageJ software (NIH).
RNA purification and quantitative RT-PCR
Total RNA was isolated from cultured primary melanocytes or melanoma cells at the indicated time points, using the RNeasy Plus Mini Kit (Qiagen, #74136). mRNA expression was determined using intron-spanning primers with SYBR FAST qPCR master mix (Kapa Biosystems, #KK4600).
Expression values were calculated using the comparative threshold cycle method (2−ΔΔCt) and normalized to human RPL11 mRNA. The primers used for quantitative RT-PCR (eurofins Genomics) and are listed below.
| Primer | Sequence |
|---|---|
| Human RPL11: forward | 5’-GTTGGGGAGAGTGGAGACAG-3’ |
| Human RPL11: reverse | 5’-TGCCAAAGGATCTGACAGTG-3’ |
| Human M isoform MITF: forward | 5’-CATTGTTATGCTGGAAATGCTAGAA-3’ |
| Human M isoform MITF: reverse | 5’-GGCTTGCTGTATGTGGTACTTGG-3’ |
| Human Tyrosinase: forward | 5’-ACCGGGAATCCTACATGGTTCCTT-3’ |
| Human Tyrosinase: reverse | 5’-ATGACCAGATCCGACTCGCTTGTT-3’ |
| Human NNT: forward | 5’-AGCTCAATACCCCATTGCTG-3’ |
| Human NNT: reverse | 5’-CACATTAAGCTGACCAGGCA-3’ |
| Human IDH1: forward | 5’-GTC GTCATGCTTATGGGG AT-3’ |
| Human IDH1 reverse | 5’-CTT TTGGGTTCCGTCACT TG-3’ |
| Huma MFN2: forward | 5’-CTG CTA AGG AGGTGCTCA A-3’ |
| Human MFN2: reverse | 5’-TCC TCA CTTGAAAGC CTT CTG C-3’ |
| Human PPARGC1A: forward | 5’-CTG CTA GCA AGTTTG CCT CA-3’ |
| Human PPARGC1A: reverse | 5’-AGTGGTGCAGTGACCAATCA-3’ |
| Human POMC: forward | 5’-AAGAGGCTAGAGGTCATCAG-3’ |
| Human POMC: reverse | 5’-AGAACGCCATCATCAAGAAC-3’ |
| Human TYRP1 forward | 5’-CCAGTCACCAACACAGAAATG-3’ |
| Human TYRP1 reverse | 5’-GTGCAACCAGTAACAAAGCG-3’ |
| Human TRP2/DCT forward | 5’-TTCTCACATCAAGGACCTGC-3’ |
| Human TRP2/DCT reverse | 5’-ACACATCACACTCGTTCCTC-3’ |
Cycloheximide chase assay
72 h after siRNA transfection (siControl or siNNT), UACC257 melanoma cells were treated with a protein synthesis inhibitor, cyclohexamide (CHX, Sigma Aldrich #C7698, 50 μg/ml), for the indicated times and then immediately subjected to immunoblotting for tyrosinase protein expression. The expression of tyrosinase was quantified using ImageJ software based on band intensities and normalized to the intensities of the corresponding β-actin bands. The normalized tyrosinase expression was then defined as relative tyrosinase expression by setting the mean values at t=0 in each experimental group to 1.0.
In the ROS rescue experiments, siRNA-containing medium was replaced with fresh culture medium containing either N-acetyl-L-cysteine (NAC; Sigma Aldrich #A7250, 5 mM), β-nicontinamide adenine dinucleotide 2’-phosphate (NADPH; Sigma Aldrich #N7505, 0.1 mM), MitoTEMPO (ThermoFisher #501872447, 20 μM) or control vehicle (DMSO or TrisHCl respectively) 24h after siRNA transfection. The siRNA-transfected cells were cultured for an additional 48 h in the presence of these agents and then examined by the CHX chase assay as described above.
pLJM-1-EGFP or pLJM1-NNT/FLAG was introduced into UACC257 cells using Lipofectamine 3000. 48 after transfection, the transfection medium was replaced with fresh medium containing DMSO or 10 μM MG132 (Sigma Aldrich #M8699) and pre-incubated for 6 h. Then, CHX was added to assess tyrosinase protein stability as described above.
Melanin quantification
Equal numbers of cells were plated in 6-well plates. The cells were then harvested 72 – 96 hours post siRNA or NNT inhibitors compounds, as indicated in the legends, pelleted, washed in PBS and counted. 106 cells were used for measurement of protein concentration with the Pierce BCA protein assay (Thermo Fisher Scientific, #23225) and 106 cells were resuspended in 60 μl of 1 N NaOH solution and incubated at 60°C for 2 h or until the melanin was completely dissolved. After cooling down to room temperature, samples were centrifuged at 500 × g for 10 min and the supernatants were loaded onto a 96-well plate. The melanin content was determined by measuring the absorbance at 405 nm on an Envision plate reader, compared with a melanin standard (0 to 50 μg/ml; Sigma Aldrich, #M8631). Melanin content was expressed as micrograms per milligram of protein.
Eumelanin and pheomelanin analysis
Lyophilized cells (1 x 106) from human abdominal full thickness skin explants were ultrasonicated in 400 μL of water and fur samples were homogenized at a concentration of 10 mg/mL in water in a Ten-Broeck homogenizer. Aliquots of 100 μL were subjected to alkaline hydrogen peroxide oxidation to yield the eumelanin marker, pyrrole-2,3,5-tricarboxylic acid (PTCA) (Ito et al., 2011), or to hydroiodic acid (HI) hydrolysis to yield the pheomelanin marker, 4-amino-3-hydroxyphenylalanine (4-AHP) (Wakamatsu et al., 2002), then the samples were analyzed by HPLC. Amounts of each marker are reported as ng of marker per 106 cells or mg fur. Pheomelanin and eumelanin contents were calculated by multiplying the 4-AHP and PTCA contents by factors of 7 and 25, respectively (d'Ischia et al., 2013).
Skin colorimeter measurements
Skin reflectance measurements were made using a CR-400 Colorimeter (Minolta Corporation, Japan). Before each measurement, the instrument was calibrated against the white standard background provided by the manufacturer. The degree of melanization (darkness) is defined as the colorimetric measurement on the *L axis (luminance, ranging from completely white to completely black) of the Centre Internationale d’Eclairage (CIE) L*a*b* color system (Park et al., 1999). Each data point is the mean of measurements performed in technical triplicate (three different locations within the same ear).
Determination of intracellular cAMP content
Cyclic adenosine monophosphate (cAMP) was measured directly using an enzyme-linked immunosorbent assay (ELISA) (Enzo Life Sciences, #ADI-901-066). cAMP was quantified in 100,000 cells based on a standard curve.
Cell viability assay
Human melanoma cell lines and isolated primary cultured human melanocytes were propagated and tested in early passage (Passages 7 to 9). The effects of NNT inhibitors (2,3BD, DCC, and Palmitoyl coenzyme A lithium salt) on cell viability were evaluated by the CellTiter-Glo Luminescent Cell Viability Assay (Promega, #G7570) and measurement of luminescence was performed on an EnVision 2104 Multilabel Reader (PerkinElmer). Human melanoma cell lines and primary melanocytes were plated on 96-well white plates (10,000 cells/well) and were treated with the NNT inhibitors at the indicated concentrations for 24 h.
Glutathione measurements
Cell lysates were prepared from equal numbers of cells after 24 h of DCC or 2,3BD treatment, following the manufacturer’s protocols. Seventy-two h post siRNA treatment or overexpression of NNT and their corresponding controls, glutathione levels were determined using the GSH/GSSG-Glo assay (Promega, #V6611) and luminescence was measured using an EnVision 2104 Multilabel Reader (PerkinElmer).
Determination of NADPH/NADP ratio
Cell lysates were prepared from equal numbers of UACC257 human melanoma cells 72 h post siRNA treatment or overexpression of NNT and their corresponding controls. NADPH/NADP+ ratios were determined using the NADP/NADPH-Glo Assay (Promega, #G9082) following the manufacturer’s protocol and luminescence was measured using an EnVision 2104 Multilabel Reader (PerkinElmer).
Luciferase reporter assay
To measure MITF transcriptional activity, UACC257 melanoma cell lines were infected with the dual-reporter system (GeneCopoeia, #HPRM39435-LvPM02), which expresses secreted Gaussia luciferase (GLuc) under the TRPM1 promoter and SEAP (secreted alkaline phosphatase) as an internal control for signal normalization. The cells were grown in complete RPMI medium containing 10% Fetal Plex. Medium was collected 24, 48, and 72 h post siRNA transfection. GLuc and SEAP activities were measured by Secrete-Pair Gaussia Luciferase Assay Kit (GeneCopoeia, #LF062) and QUANTI-Blue™ Solution (Invivogen, #rep-qbs), respectively, according to the manufacturers’ instructions.
Histology and Immunofluorescence
For histology, paraffin sections were prepared and stained with hematoxylin and eosin (H&E) using the ihisto service (https://www.ihisto.io/). For visualization of melanin, paraffin sections were stained using a Fontana-Masson Stain kit (abcam, #ab150669). Briefly, the samples were incubated in warmed Ammoniacal silver solution for 30 min, followed by a Nuclear Fast Red stain.
For immunofluorescence, paraffin sections were deparaffinized by xylene and rehydrated gradually with ethanol to distilled water. Sections were submerged in 0.01 M citrate buffer and boiled for 10 min for retrieval of antigen. The sections were washed with TBST (0.1% Tween 20) and blocked with protein blocking solution (Agilent, #X090930-2) for 1 h at room temperature before application of primary antibody [1:100 diluted in Antibody Diluent (DAKO, #S3022)] and incubation overnight at 4°C. The following day, sections were washed with TBST three times and incubated with secondary antibody Alexa Fluor 647 goat anti-mouse IgG (G+L) (ThermoFisher Scientific, #A-21236), Alexa Fluor 594 F(ab)2 fragment of goat anti-rabbit IgG (G+L) (ThermoFisher Scientific, #A-11072), or Alexa Fluor 555 goat anti-rabbit IgG (ThermoFisher Scientific, #A-21428). After washing, the tissue sections were cover-slipped with mounting medium (SlowFade® Gold Antifade Reagent with DAPI, ThermoFisher Scientific, #S36939). MaxBlock Autofluorescence Reducing Reagent Kit (MaxVision Biosciences, #MB-L) was used to quench skin tissue autofluorescence according to the reagent instructions.
The following primary antibodies were used at the indicated dilutions (antibody sources are in the Key Resources Table): anti-CPDs monoclonal antibody (1:1,500), rabbit anti-γ-H2AX (P-ser139) polyclonal antibody (1:5,000), rabbit anti-NNT (C-terminal) polyclonal antibody (1:100), rabbit anti-γ-H2AX [p Ser139] polyclonal antibody (1:100).
Primary human melanocytes (50,000 cells/well) were cultured on chamber slides (ThermoFisher Scientific, #125657). Seventy-two hours post siRNA transfection, the cells were fixed with 4% paraformaldehyde (PFA) (ThermoFisher Scientific, #50980487) for 20 min at room temperature, followed by treatment with 0.1% Triton X-100 (Sigma) for 5 min and blocking with 10% goat serum (Sigma Aldrich, #G9023) containing 5% BSA in PBS for 60 min atroom temperature. Mouse anti-NNT monoclonal antibody [8B4BB10] was diluted with the blocking solution to a final concentration of 5 μg/ml and incubated with the cells overnight at 4°C. The following day, the slides were washed with TBST three times and incubated with donkey anti-mouse Alexa Fluor 488 secondary antibody (1:500). Sections were washed with TBST three times and mounted in mounting medium (VECTASHIELD® HardSet™ Antifade Mounting Medium with DAPI, Vector Laboratories, #H-1500). Images were captured using confocal microscopy (Zeiss Axio Observer Z1 Inverted Phase Contrast Fluorescence microscope).
Detection of cellular reactive oxygen species (ROS)
The redox-sensitive fluorescent dye chloromethyl-2′, 7′-dichlorodihydrofluorescein diacetates (CM-H2DCFDA, ThermoFisher Scientific, #C6827) was used to measure intracellular ROS accumulation. UACC257 melanoma cells were cultured on a glass bottom dish and treated with the indicated siRNAs. Forty-eight h post siRNA treatment, 2 μM CM-H2DCFDA in PBS/5% FBS was added and the samples were incubated at 37°C for 30 min to assess overall ROS production. Subsequently, the cells were incubated with 5 μM MitoSOX Red (ThermoFisher Scientific, #M36008) in PBS/5% FBS at 37°C for 10 min, washed with HBSS, and analyzed by immunofluorescence imaging (Zeiss Axio Observer Z1 Inverted Phase Contrast Fluorescence microscope). The results were normalized to cell numbers, which were determined by nuclear staining with 1 drop per ml of NucBlue (ThermoFisher Scientific, #R37605) at 37°C for 15 min.
Transmission electron microscopy
Cultured primary human melanocytes were grown in Medium 254 in 6-well transwell plates. Ninety-six h post siRNA or overexpression treatment, the cells were fixed with a modified Karnovsky’s fixative (2% paraformaldehyde/2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer, pH 7.4) for at least 2 h on a gentle rotator, followed by rinsing several times with 0.1 M cacodylate buffer. Then, the cells were treated with 1% osmium tetroxide/0.1 M cacodylate buffer for 1 h, rinsed thoroughly in 0.1 M cacodylate buffer, scraped, and the cell suspensions were transferred into 15 ml centrifuge tubes and centrifuged (3,000 rpm) for 15 min at 4°C). Pelleted material was embedded in 2% agarose, dehydrated through an ethanol gradient (series of solutions from 30% to 100% ethanol), dehydrated briefly in 100% propylene oxide, then allowed to infiltrate overnight on a gentle rotator in a 1:1 mix of propylene oxide and Eponate resin (Ted Pella, Inc., kit with DMP30, #18010’). The following day, specimens were transferred into fresh 100% Eponate resin for 2-3 hours, then embedded in flat molds in 100% fresh Eponate resin, and embeddings were allowed to polymerize for 24-48 h at 60°C. Thin (70 nm) sections were cut using a Leica EM UC7 ultramicrotome, collected onto formvar-coated grids, stained with 2% uranyl acetate and Reynold's lead citrate, and examined in a JEOL JEM 1011 transmission electron microscope at 80 kV. Images were collected using an AMT digital imaging system with proprietary image capture software (Advanced Microscopy Techniques, Danvers, MA).
Melanosomes-mitochondria distance measurements
Measurements of distances between melanosomes and mitochondria were quantified in FIJI (ImageJ) (Schindelin et al., 2012) by applying a customized macro to TEM micrographs. Melanosomes (N = ~ 50) were randomly selected for each condition within the whole image data set. Thirty Euclidean distances from the melanosome surface to the closest mitochondria surface were measured in nm. From these 30 single measurements the mean was calculated to give a final single mean value per melanosome-mitochondria event. A total of ~50 events (N) were quantified per condition. Data were plotted and statistically analyzed using Prism 8 (Version 8.4.3). Melanosome-mitochondria distances closer than 20 nm were considered melanosome-mitochondria close appositions or contacts, consistent with (Daniele et al., 2014). Cell area (μm2), number of melanosome-mitochondria contacts, and number of mitochondria were quantified in FIJI (ImageJ) using polygon and multi-point selection tools.
Melanosome stage quantification
Melanosome identification and quantification were performed with images at 40,000 x magnification or higher. Stages were estimated based on morphological features previously noted, namely multivesicular endosomes (Stage I), unpigmented fibrils (Stage II), pigmented fibrils (stage III), and darkly pigmented filled melanosomes (Stage IV). All identifiable melanosomes in 4 cells per condition were quantified and classified, and the proportions of each stage were normalized to cell cytosolic area (determined by ImageJ).
Tyrosinase activity assay
UACC257 human melanoma cells were treated with human NNT siRNA or non-targeting siRNA control pool for 4 days. Cell lysates were prepared by adding 1% Trion X100 in PBS for 1 h at room temperature with shaking. Tyrosinase activity was measured as previously described (Iozumi et al., 1993). Briefly, freshly made 25 mM L-DOPA in PBS was heated and added to the cell lysates in a 96-well plate. L-DOPA levels were determined by measuring the absorbance at 490 nm with shaking for 30 cycles, compared with mushroom tyrosinase (Sigma-Aldrich #T3824, 0 to 50 μg/μl in PBS), using an Envision 2104 Multilabel plate reader (PerkinElmer).
Human genetic association studies
For all cohorts, the GRCh37/hg19 human genome build was used. SNPs with minor allele frequency less than 1% were excluded from each cohort.
Ethics Approval
Rotterdam study:
The Medical Ethics Committee of the Erasmus Medical Center and the review board of the Dutch Ministry of Health, Welfare and Sports have ratified the Rotterdam study. Written informed consent was obtained from all participants.
East and South Africa:
As detailed in Crawford et al. (Crawford et al., 2017), individuals used in the study were sampled from Ethiopia, Tanzania and Botswana. IRB approval for this project was obtained from the University of Pennsylvania. Written informed consent was obtained from all participants and research/ethics approval and permits were obtained from the following institutions prior to sample collection: the University of Addis Ababa and the Federal Democratic Republic of Ethiopia Ministry of Science and Technology National Health Research Ethics Review Committee; COSTECH, NIMR and Muhimbili University of Health and Allied Sciences in Dar es Salaam, Tanzania; the University of Botswana and the Ministry of Health in Gaborone, Botswana.
A. The Rotterdam Study:
Population:
The Rotterdam Study (RS) is a prospective population-based follow-up study of the determinants and prognosis of chronic diseases in middle age and elderly participants (aged 45 years and older) living in the Ommoord district (Rotterdam, the Netherlands) (Ikram et al., 2017). The RS consists of 4,694 people of predominantly North European ancestry.
Phenotyping:
As part of the dermatological investigation within the RS, participants from three cohorts (RSI, RSII and RSIII) were screened to assess their skin color. In brief, trained physicians scored the skin color of the participants using a scale from 1 to 6, with 1 for albino, 2 for white, 3 for white to olive color, 4 for light brown, 5 for brown, and 6 for dark brown to black. The reliability of the assessment has been validated before (Jacobs et al., 2015). Individuals with dark skin were excluded since they were likely to have a different genetic background than Europeans.
Genotyping and imputation:
The RS-I and RS-II cohorts were genotyped with the Infinium II HumanHap550K Genotyping BeadChip version 3 (Illumina, San Diego, California USA) and the RS-III cohort was genotyped using the Illumina Human 610 Quad BeadChip. The RS-I, RS-II and RS-III cohorts were imputed separately using 1000 Genomes phase 3 (Genomes Project et al., 2012) as the reference dataset. Quality control on the single nucleotide polymorphisms (SNPs) has been described before (Hofman et al., 2015). SNPs were filtered out if they had a minor allele frequency of less than 1% or an imputation quality (R2) of less than 0.3. We used MACH software for the imputation with parameter defaults. Best-guess genotypes were called using the GCTA program (Yang et al., 2011) with parameter defaults.
Statistical analysis:
We used a multivariate linear regression model to test for associations between SNPs within the NNT region and skin color in the RS using an additive model (Purcell et al., 2007). The model was adjusted for age, sex and four principal components (variables derived from principal component analysis that were added to correct for possible population stratification and hidden relatedness between participants). The PLINK program was used for conducting associations.
B. The CANDELA cohort:
A GWAS study of skin color in the CANDELA cohort has been published (Adhikari et al., 2019) and summary statistics are available at http://www.gwascentral.org/study/HGVST3308. Details of the cohort and analyses are in the published study, so only the cohort population and phenotyping are summarized here.
Population:
6,357 Latin American individuals were recruited in Brazil, Chile, Colombia, Mexico and Peru. Participants were mostly young, with an average age of 24.
Phenotyping:
A quantitative measure of constitutive skin pigmentation (the Melanin Index, MI) was obtained using a DermaSpectrometer DSMEII reflectometer (Cortex Technology, Hadsund, Denmark). The MI was recorded from both inner arms and the mean of the two readings was used in the analyses.
Statistical analysis:
P-values for SNPs in the NNT region were obtained from the published CANDELA summary statistics.
C. The East & South African cohort:
The summary statistics were obtained from a previous study of pigmentation evolution in Africans (Crawford et al., 2017). Details of the cohort and analyses are in the published study, so only the cohort population and phenotyping are summarized here.
Population:
A total of 1,570 ethnically and genetically diverse Africans living in Ethiopia, Tanzania, and Botswana were sampled in this cohort.
Phenotyping:
A DSM II ColorMeter was used to quantify reflectance from the inner underarm. Reflectance values were converted to a standard melanin index score.
Statistical analysis:
P-values for SNPs in the NNT region were obtained from the published summary statistics.
D. The UK Biobank cohort:
There have been many published studies on pigmentation phenotypes in the UK Biobank (Jiang et al., 2019) and the summary statistics are publicly available at https://cnsgenomics.com/software/gcta/#DataResource. Details of the cohort and analyses are in the published study, so only the cohort population and phenotyping are summarized here.
Population:
The UK Biobank includes more than 500,000 individuals from across the UK, with predominantly White British ancestry.
Phenotyping:
Self-reported categorical questions were used to record data on skin color and ease of skin tanning.
For skin color, 6 categories were used: very fair, fair, light olive, dark olive, brown, and black (https://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=1717). 450,264 responses were available.
For ease of skin tanning (https://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=1727), participants were asked “What would happen to your skin if it was repeatedly exposed to bright sunlight without any protection?” Four categories were used: very, moderately, mildly, and never tanned. 446,744 responses were available.
For sun protection use (https://biobank.ctsu.ox.ac.uk/crystal/field.cgi?id=2267), participants were asked “Do you wear sun protection (e.g., sunscreen lotion, hat) when you spend time outdoors in the summer?” Four categories were used: never/rarely, sometimes, most of the time, and always. 452,925 responses were available.
Statistical analysis:
P-values for SNPs in the NNT region were obtained from the published UK Biobank summary statistics.
Meta-analysis of the cohorts
Considering the huge variation in sample size among the 4 cohorts, Fisher’s method (Won et al., 2009) of combining p-values from independent studies was used, in which p-values for one marker across different cohorts were combined to provide an aggregate p-value for the meta-analysis of that marker.
Multiple testing adjustment
Since we tested 332 independent associations, we corrected the significance threshold for multiple testing. We used the false discovery rate (FDR) method of controlling the multiple testing error rate, following the Benjamini-Hochberg procedure (Benjamini and Cohen, 2017). Applying the FDR procedure on the set of p-values to achieve an overall false positive level of 5%, the adjusted significance threshold was p = 1.01E-3. As there is substantial LD (linkage disequilibrium) between the SNPs, a Bonferroni correction would have been overly conservative.
GWAS conditional on known pigmentation variants
MC1R is a major determinant of pigmentation, with known genetic variants associated with lighter skin color, red hair, and freckles in European populations (Quillen et al., 2019). Among the two European cohorts used in this study, individual-level data were only available for the Rotterdam Study, so the conditional GWAS analysis was conducted only in this cohort.
We retrieved the dose allele of major MC1R variants data from the Rotterdam studies and used them as covariates in the earlier used multiple linear regression model, in addition to the previously mentioned covariates. The association P-value of the NNT variant is thus conditioned on the known pigmentation variants in this analysis. These conditioned P-values were then compared to the original (unconditioned) P-values with a Wilcoxon rank-sum test to assess whether they have been significantly altered due to the conditioning on the known pigmentation variants.
Jacobs et al. 2015 examined three functional variants in MC1R for their relationship with pigmentation in the Rotterdam Study: rs1805007, rs1805008, rs1805009 (Jacobs et al., 2015). Therefore, the first conditional analysis was performed using these three MC1R variants.
Subsequently, an additional set of well-established genetic variants in other pigmentation genes (Adhikari et al., 2019) were also used for conditioning: rs28777 (SLC45A2), rs12203592 (IRF4), rs1042602 (TYR), rs1800404 (OCA2), rs12913832 (HERC2), rs1426654 (SLC24A5), and rs885479 (MC1R).
Correlation between trait effect sizes and eQTL expression data
eQTL expression data corresponding to expression levels of the NNT transcript were downloaded from the GTEx database. For each genetic variant in the NNT region, we obtained the normalized effect size (NES) and P-value for the alternative (non-reference) allele in each of the two skin tissues: “Skin - Not Sun Exposed (Suprapubic)” and “Skin - Sun Exposed (Lower leg)”.
Correlation values were calculated between the regression coefficients for the alternative alleles of each variant from the UK Biobank for each of the three traits and the NES values corresponding to the same alleles (to ensure consistency of effect direction) in each of the two skin tissues.
QUANTIFICATION AND STATISTICAL ANALYSIS
ImageJ v1.8.0 (https://imagej.nih.gov/ij/) was used to quantify the immunoblots. FIJI software enabling pixel-based color quantification was used for Zebrafish analysis.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8. In general. for comparisons of two groups, significance was determined by two-tailed, unpaired Student’s t tests, correcting for multiple t tests with the same two groups using the Holm-Šídák method. One-way and two-way ANOVA tests were used for comparisons of more than two groups involving effects of one or two factors, respectively, using the recommended post-tests for selected pairwise comparisons. The specific statistical tests used for experiments are described in the figure legends. P values less than 0.05 were considered statistically significant. Levels of significance are indicated by *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001; ns, not significant.
Supplementary Material
Figure S1. Inhibition of NNT Increase pigmentation via redox dependent mechanism. Related to Figure 1
(A) siNNT-induced increased pigmentation in human SK-MEL-30 melanoma cells is dependent on tyrosinase and reactive oxygen species. Left panel: Representative lysates from SK-MEL-30 cells following treatment with siControl, siNNT, siNNT + siTyrosinase (siTYR), or siNNT + 5mM NAC. Right panel: Quantification of intracellular melanin content in SK-MEL-30 cells; n = 3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (B, C) qRT-PCR analysis of NNT (B) and immunofluorescence of NNT (C) in primary human melanocytes treated with siControl or siNNT for 96 hours. IF staining of human NNT (Green) and nuclei (DAPI, blue) are shown. Scale bar 50 μM. Relative NNT mRNA levels and fluorescent intensities (n = 3) were analyzed by unpaired, two-sided t-tests. (D) Immunoblot analysis of NNT expression in UACC257 human melanoma cells. Band intensities were quantified by ImageJ, normalized to β-actin and plotted relative to siControl; n = 3, analyzed by unpaired, two-sided t-test. (E) Treatment of UACC257 cells with siNNT for 24 hours resulted in increased NADPH/NADP (Left panel, n = 9) and decreased GSH/GSSG (Right panel, n = 6) ratios. The data were analyzed by multiple t tests with the Holm-Šídák post-test. (F) UACC257 melanoma cells were treated with siControl or siTyrosinase for 5 days and eumelanin and pheomelanin were measured using HPLC techniques (n = 3). Absolute pigment levels (Left graph) were analyzed by ordinary two-way ANOVA, separately for eumelanin and pheomelanin. The eumelanin/pheomelanin ratio (Right graph) was analyzed by unpaired Student t test (G). Increased ROS in UACC257 cells following 48 hours of siNNT or siIDH1 treatment, but not after 48 hours of siPGCa treatment. IF images of ROS indicator DCFDA (Green) and nuclei (DAPI, blue), representative of five experiments, are displayed. Quantified results were normalized to the total number of cells and analyzed by ordinary one-way ANOVA with Šídák’s post-test. (H) Increase of melanin content by siNNT is blocked by cotreatment with NADPH. Intracellular melanin content was quantified in UACC257 cells treated with siControl or siNNT for 72 hours, with 0.1 M NADPH or Vehicle (Tris-HCl, pH 8.0) added after the first 24 hours. n = 3, analyzed by ordinary two-way ANOVA with Šídák’s post-test. (I) Immunoblot analysis of IDH1 in UACC257 cells treated with siControl, siNNT, siIDH1, or siNNT + siIDH1 together for 72 hours. Band intensities were quantified by ImageJ, normalized to β-actin (n = 3), and analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (J) qRT-PCR analysis of NNT, IDH1, and PGC1α mRNAs in UACC257 cells treated with siRNA for one of those genes or siControl. qRT-PCR data are normalized to RPL11 RNA and RNA levels are presented as fold change relative to siControl; (n = 3), analyzed by ordinary one-way ANOVA with Dunnett’s post-test, followed by the Bonferroni correction for three ANOVA analyses. (K, L) Overexpression of NNT in the UACC257 cell line: (K) qRT-PCR analysis of NNT mRNA five days post transfection; n=3, analyzed by unpaired, two-sided t-test. (L) Overexpression of NNT resulted in decreased NADPH/NADP (Left panel, n = 8) and increased GSH/GSSG (Right panel, n = 4-6) ratios, analyzed by multiple t-tests with the Holm-Šídák post-test.
All data are expressed as mean ± SEM; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Figure S2. NNT does not impact TYR mRNA expression levels and acts independently of the cAMP pathway. Related to Figure 2
(A) Tyrosinase activity increase following siNNT in UACC257 melanoma cells; n = 4, analyzed by unpaired, two-sided t-test (B) Diagram of the “Tanning Pathway”. Briefly, UV exposure results in DNA damage and activation of P53 in keratinocytes. POMC is transcriptionally activated by P53 and the pro-protein is cleaved to α-MSH, which is secreted from the keratinocyte. α-MSH binds to MC1R in the melanocyte membrane, resulting in an increase in cAMP and activation of PKA. Active PKA results in an increase of MITF, activated transcriptionally by CREB. MITF transcriptionally regulates pigmentation enzymes such us TYRP1, TRP2 and tyrosinase. (C) Immunoblot analysis of MITF in UACC257 cells transfected with siNNT or siControl for 72 h. Band intensities were quantified by ImageJ, normalized to β-actin, plotted relative to siControl values (n = 3), and analyzed by unpaired, two-sided t-test. (D-F) Analyses of UACC257 cells stably expressing secreted luciferase under the TRPM1 promoter and SEAP under the CMV promoter. The cells were treated with either siControl, siNNT, or siMITF (n = 3): (D) qRT-PCR analysis of NNT, mMITF and TYRP1 72 hours post siRNA transfection. Data were normalized to RPL11 RNA, analyzed by unpaired, two-sided t-test (NNT) or ordinary one-way ANOVA with Dunnett’s post-test (mMITF and TYRP1). (E) Luciferase secretion normalized to secreted SEAP 72 hours post siRNA transfection, showing decreased luciferase activity following siMITF and siNNT, analyzed by ordinary one-way ANOVA with Dunnett’s post-test; representative cell pellets (1 x 106 cells) are below the graph. (F) Luciferase secretion normalized to secreted SEAP 24, 48 and 72 hours post siRNA transfection was analyzed by repeated measures two-way ANOVA with Šídák’s post-test. (G) qRT-PCR analysis of NNT, MITF, TYRP1, TRP2/DCT, NNT, tyrosinase, and POMC in UACC257 cells 72 hours post transfection of siNNT or siControl. Data were normalized to RPL11 RNA, presented as fold change relative to siControl (n = 3), and analyzed by multiple t-tests with the Holm-Šídák post-test. (H) cAMP content of UACC257 cells transfected with siNNT or siControl for 48 h, measured by cAMP ELISA and normalized to siControl cells; n = 3, analyzed by unpaired, two-sided t-test. (I) Primary human melanocytes were starved for 24 hours and Forskolin (FSK; 20 μM) was added to the medium for 2 hours. qRT-PCR analysis of NNT was performed with MITF as a positive control for the treatment. The data were normalized to RPL11 RNA (n = 3) and analyzed by multiple t-tests with the Holm-Šídák post-test. (J) No change in NNT mRNA upon UVB. Abdominal skin was irradiated with 1J/cm2 UVB, skin was collected at 0, 24, 48 and 72 hours post UVB, and qRT-PCR analysis of NNT was performed. The data were normalized to RPL11 RNA and presented as fold change relative to t = 0. n = 5-6 (two different donors), analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (K) Immunoblots of P53 and β-actin in UACC257 cells following siControl or siNNT treatment for 72 hours. (L) Immunoblot of NNT and β-actin in UACC257 melanoma cells (Left panel), daily treatment with NAC (5 mM), MitoTEMPO (20 μM) and H2O2 (100μM) for 72 hours (n = 3) analyzed by ordinary one-way ANOVA with Tukey post-test. (M) Immunoblots of tyrosinase and β-actin in UACC257 melanoma cells (Left panel), showing decreased tyrosinase protein levels following overexpression of NNT for 12 days. Band intensities were quantified by ImageJ, normalized to β-actin and plotted relative to siControl values (Right Panel). (n = 3), analyzed by unpaired, two-sided t-test. (N) qRT-PCR analysis of MITF, TYRP1 and tyrosinase mRNAs in UACC257 cells that overexpressed NNT (NNT OE), compared to control (Empty Vector). The data were normalized to RPL11 RNA (n = 3) and analyzed by ordinary one-way ANOVA with Dunnett’s post-test, followed by the Bonferroni correction for three ANOVA analyses.
Figure S3. NNT knockdown enhances melanosome maturation, melanosome-mitochondria proximity and pigmentation by NNT knockdown. Related to Figure 2
(A) Enhanced melanosome maturation induced by siNNT in human primary melanocyte cells is blocked by NAC (5 mM) or MitoTEMPO (20 μM) (daily treatment for 96 h). The number of melanosomes per um2 in the classified stages is represented. n = 4-5 cells, analyzed by ordinary two-way ANOVA with Šídák’s post-test (B) The total number of melanosomes per um2 in primary human melanocytes is not altered by siNNT and/or daily treatment with NAC (5 mM) or MitoTEMPO (20 μM) for 96 hours (Left graph, n = 8-10, analyzed by ordinary one-way ANOVA with Dunnett’s post-test) or by overexpression of NNT (Middle graph, n = 8-10), analyzed by unpaired, two-sided t-test. The total numbers of mitochondria per um2 by overexpression of NNT (Right graph, n=5) is not altered. (C) Measurements of proximities between melanosomes and mitochondria were quantified in FIJI (ImageJ) by applying a customized macro to TEM micrographs (n = 100 events per condition). Melanosome-mitochondria proximities closer than 20 nm are considered melanosome-mitochondria close appositions/contacts. Right panel: FIJI graphical user interface showing a TEM micrograph of mitochondria (m) and a melanosome (*) with a yellow line indicating the Euclidean distance between melanosome and mitochondrion surfaces, quantified with a customized macro to measure distances between two surfaces. Scale bar 400 nm. Table shows the percentages and, in parentheses, the fractions of melanosome-mitochondria proximities that were <20 nm. Denominators are the total number of measurements (events) performed in each group. Adjusted P values were determined by pairwise F-tests of the control group to each of the other groups, followed by the Bonferroni correction for three comparisons. (D-E) The total numbers of melanosomes (D) and mitochondria (E) per um2 in primary human melanocytes is not altered; n = 5 cells, analyzed by ordinary two-way ANOVA with Šídák’s post-test. (F) MFN2 enables siNNT-mediated pigmentation. Top panel: Quantification by spectrophotometry of intracellular melanin content of UACC257 human melanoma cells treated with siControl, siNNT, siMFN2 + siNNT, or siMFN2 for 72 hours. n = 3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test. Bottom panel: Representative cell pellets (106 cells). (G) Immunoblot analysis of MFN2 expression in UACC257 human melanoma cell lines. Band intensities (n = 3) were quantified by ImageJ, normalized to β-actin, and analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (H) qRT-PCR analysis of MFN2 in primary human melanocytes that were transfected with siMFN2. The data were normalized to RPL11 RNA, plotted relative to the control (n = 3), and analyzed by unpaired, two-sided t-tests (I) siMFN2 resulted in accumulation of large autophagosomes (white arrows), containing numerous melanosomes (arrowheads), in normal human melanocytes. Scale bar 2 μm. (J) Immunoblot analysis of LC3B in UACC257 cells treated with siMFN2, siNNT, siMFN2+siNNT, or siControl for 72 hours. Band intensities were quantified by ImageJ and normalized to β-actin. The ratios of LC3BII to LC3BI were plotted (n = 3) and analyzed by ordinary one-way ANOVA with Dunnett’s post-test.
All data are expressed as mean ± SEM; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Figure S4. NNT inhibitors are non-toxic in vitro. Related to Figure 3
(A) Chemical formulas of all three published NNT inhibitors. (B) Viability measurements showed no significant toxicity after treatment of human melanocytes, dermal fibroblasts, and keratinocytes with up to 10 μM of DCC, Palmitoyl-CoA, or 2,3BD. (C) Treatments with different doses of DCC (Left graph) or 2,3 BD (Right graph) had no impacts on cell viability. The data were plotted relative to vehicle treatment (0) and analyzed by ordinary one-way ANOVA with Dunnett’s post-test (n = 4). (D) Intracellular melanin content normalized to total protein levels in primary human melanocytes that were treated with siControl or siNNT for 24 hours, followed by incubation with 2,3 BD (2 mM) or DMSO vehicle for 72 hours. n = 3, analyzed by ordinary one-way ANOVA with Šídák’s post-test. (E) Treatments with different doses of DCC had no impact on cell viability (Right graph), but resulted in decreased GSH/GSSG ratios (Left graph) in the UACC257 human melanoma cell line. n = 4, analyzed by ordinary one-way ANOVA with Tukey’s (Left graph) or Šídák’s (Right graph) post-test. (F) Fontana-Masson staining of melanin in human abdominal skin 5 days after a single treatment of 2,3BD (1M), showing supranuclear capping (Black arrows) in keratinocytes of 2,3BD-treated skin. Scale bar 50 μM
All data are expressed as mean ± SEM; *p<0.05, **p<0.01, ****p<0.0001.
Figure S5. NNT regulates pigmentation in mice, zebrafish and human pigmentation disorders. Related to Figure 4
(A) Agarose gel showing PCR genotyping of DNA from C57BL/6J mice (single 743 bp product indicates homozygous 5-exon deletion in the Nnt gene) and C57BL/6NJ mice (single 570 bp product indicates homozygous wild type Nnt gene). (B) Modification of NNT sites in zebrafish using WT SpCas9. Editing was assessed by next-generation targeted amplicon sequencing. (C) Zebrafish overexpressing NNT (NNT OE) or empty plasmid were treated at 3 days post fertilization with 100 μM of 2,3BD or vehicle for 24 hours. A representative image has been displayed. Results of mean melanocytic brightness, quantified by pixel-based analysis are shown in the graph at right; Empty plasmid (n = 12 fish; 30 melanocytes), NNT OE (n = 10 fish; 24 melanocytes), Empty plasmid + 2,3 BD (n = 8 fish; 30 melanocytes), NNT OE + 2,3BD (n = 11 fish; 31 melanocytes), analyzed by ordinary one-way ANOVA with Dunnett’s post-test (D) Representative images of the specific areas of hyperpigmentation in human lentigo-affected skin after staining for NNT (left image, red) or Fontana Masson (right image). Graph at right shows NNT signal intensities in melanocytes of healthy and lesional skin. n = 9 (bars indicate means), analyzed by ordinary one-way ANOVA with Dunnett’s post-test.
Supplementary Table 1. Characteristics of the NNT genetic variants significant in the meta-analysis for skin color. Related to Figure 5 and Figure 6
Highlights.
Identification of a redox-dependent skin pigmentation mechanism
Modification of NNT impacts ubiquitin-proteasome-mediated tyrosinase degradation
Alteration of NNT levels affects skin pigmentation through melanosome maturation
Human NNT SNPs associate with skin pigmentation, tanning, and use of sun protection
Acknowledgments.
We thank C. Thomas Powell and Robert Liu for the help with statistical analysis and editing, Tiziana Daniele and Diane Capen for helping on electron microscopy-related questions, Sharon Germana for administrative help, Paolo Gameiro for providing plasmid constructs, Pawel Pelczar for his efforts on generating transgenic mice, and Ruth Halaban, Micky Marks, Doug Richardson, Desmond Tobin, Rizwan Haq, Pere Puigserver, Alfred Goldberg, Lajos Kemeny senior, and Steven Gygi for helpful discussions.
FUNDING
ER gratefully acknowledges support from the Mildred Scheel Grant of the German Cancer Society and the Filling the Gap Grant of the University of Zurich, Switzerland. DEF gratefully acknowledges support from NIH grants 5P01-CA163222, 5R01CA222871, 5R01AR043369, and 5R01AR072304 and the Dr. Miriam and Sheldon Adelson Medical Research Foundation. ST is funded by NIH grants R01AR076241 and R35 GM134957-01. This work was conducted with support from Harvard Catalyst ∣ The Harvard Clinical and Translational Science Center (National Center for Advancing Translational Sciences, National Institutes of Health Award UL 1TR002541) and financial contributions from Harvard University and its affiliated academic healthcare centers. The content is solely the responsibility of the authors and does not necessarily represent the official views of Harvard Catalyst, Harvard University and its affiliated academic healthcare centers, or the National Institutes of Health. P.P.N was supported by the Swiss National Science Foundation (SNSF) Early Postdoc. Mobility Fellowship CRSII3_154461 and Postdoc. Mobility Fellowship P400PB_199252. B.P.K. was supported by NCI R00 CA218870 and NHLBI P01 HL142494 K.A. was supported by the Santander Research and Scholarship Award, Bogue Fellowship from University College London. A.R.L. was supported by the Leverhulme Trust (F/07 134/DF), BBSRC (BB/I021213/1), the Excellence Initiative of Aix-Marseille University - A*MIDEX (a French “Investissements d′Avenir” programme, 2RUIZLRE/RHRE/ID18HRU201 and 20-07874), the National Natural Science Foundation of China (#31771393), the Scientific and Technology Committee of Shanghai Municipality (18490750300), Ministry of Science and Technology of China (2020YFE0201600), Shanghai Municipal Science and Technology Major Project (2017SHZDZX01) and the 111 Project (B13016). G.B. was supported by the Universidad de Antioquia (CODI sostenibilidad de grupos 2013–2014 and MASO 2013–2014). The Rotterdam Study is funded by Erasmus Medical Center and Erasmus University Rotterdam (The Netherlands); Netherlands Organization for the Health Research and Development (ZonMw); the Research Institute for Diseases in the Elderly; the Ministry of Education, Culture and Science; the Ministry for Health, Welfare and Sports; the European Commission (DG XII); and the Municipality of Rotterdam. The Microscopy Core of the MGH Program in Membrane Biology partially supported by an Inflammatory Bowel Disease Center Grant DK043351 and a Boston Area Diabetes and Endocrinology Research Center Grant DK057521.
Footnotes
Declaration of Interests. D.E.F and E.R. have a patent filed on “Methods and compositions for enhancing skin pigmentation” (publication number WO/2016/077817, May 19, 2016.). D.E.F. has a financial interest in Soltego, Inc., a company developing SIK inhibitors for topical skin darkening treatments that might be used for a broad set of human applications. D.E.F.’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict-of-interest policies. B.P.K is an inventor on patents and patent applications filed by Mass General Brigham that describe genome engineering technologies. B.P.K. consults for Avectas Inc., ElevateBio, and EcoR1 capital, and is an advisor to Acrigen Biosciences. Q.Y.W. is a shareholder in Mymiel Skincare. L.I.Z. is a founder and stockholder of Fate Therapeutics, CAMP4 Therapeutics, Amagma Therapeutics, and Scholar Rock. He is a consultant for Celularity and Cellarity. H.W. is an employee and shareholder of Johnson and Johnson. Funding has been decared in the supplemental data section.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Human Protein Atlas available from http://www.proteinatlas.org.
- Ablain J, Durand EM, Yang S, Zhou Y, and Zon LI (2015). A CRISPR/Cas9 vector system for tissue-specific gene disruption in zebrafish. Dev Cell 32, 756–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari K, Mendoza-Revilla J, Sohail A, Fuentes-Guajardo M, Lampert J, Chacon-Duque JC, Hurtado M, Villegas V, Granja V, Acuna-Alonzo V, et al. (2019). A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia. Nature communications 10, 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allouche J, Bellon N, Saidani M, Stanchina-Chatrousse L, Masson Y, Patwardhan A, Gilles-Marsens F, Delevoye C, Domingues S, Nissan X, et al. (2015). In vitro modeling of hyperpigmentation associated to neurofibromatosis type 1 using melanocytes derived from human embryonic stem cells. Proc Natl Acad Sci U S A 112, 9034–9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arjinpathana N, and Asawanonda P (2012). Glutathione as an oral whitening agent: a randomized, double-blind, placebo-controlled study. J Dermatolog Treat 23, 97–102. [DOI] [PubMed] [Google Scholar]
- Bae S, Park J, and Kim JS (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth S, Glick D, and Macleod KF (2010). Autophagy: assays and artifacts. J Pathol 221, 117–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellei B, Maresca V, Flori E, Pitisci A, Larue L, and Picardo M (2010). p38 regulates pigmentation via proteasomal degradation of tyrosinase. The Journal of biological chemistry 285, 7288–7299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, and Cohen R (2017). Weighted false discovery rate controlling procedures for clinical trials. Biostatistics 18, 91–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett DC, Cooper PJ, and Hart IR (1987). A line of non-tumorigenic mouse melanocytes, syngeneic with the B16 melanoma and requiring a tumour promoter for growth. Int J Cancer 39, 414–418. [DOI] [PubMed] [Google Scholar]
- Berger MF, Hodis E, Heffernan TP, Deribe YL, Lawrence MS, Protopopov A, Ivanova E, Watson IR, Nickerson E, Ghosh P, et al. (2012). Melanoma genome sequencing reveals frequent PREX2 mutations. Nature 485, 502–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceol CJ, Houvras Y, Jane-Valbuena J, Bilodeau S, Orlando DA, Battisti V, Fritsch L, Lin WM, Hollmann TJ, Ferre F, et al. (2011). The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cestari TF, Dantas LP, and Boza JC (2014). Acquired hyperpigmentations. Anais brasileiros de dermatologia 89, 11–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement K, Rees H, Canver MC, Gehrke JM, Farouni R, Hsu JY, Cole MA, Liu DR, Joung JK, Bauer DE, et al. (2019). CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NG, Kelly DE, Hansen MEB, Beltrame MH, Fan S, Bowman SL, Jewett E, Ranciaro A, Thompson S, Lo Y, et al. (2017). Loci associated with skin pigmentation identified in African populations. Science 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- d'Ischia M, Wakamatsu K, Napolitano A, Briganti S, Garcia-Borron JC, Kovacs D, Meredith P, Pezzella A, Picardo M, Sarna T, et al. (2013). Melanins and melanogenesis: methods, standards, protocols. Pigment Cell Melanoma Res 26, 616–633. [DOI] [PubMed] [Google Scholar]
- D'Orazio JA, Nobuhisa T, Cui R, Arya M, Spry M, Wakamatsu K, Igras V, Kunisada T, Granter SR, Nishimura EK, et al. (2006). Topical drug rescue strategy and skin protection based on the role of Mc1r in UV-induced tanning. Nature 443, 340–344. [DOI] [PubMed] [Google Scholar]
- Daniele T, Hurbain I, Vago R, Casari G, Raposo G, Tacchetti C, and Schiaffino MV (2014). Mitochondria and melanosomes establish physical contacts modulated by Mfn2 and involved in organelle biogenesis. Curr Biol 24, 393–403. [DOI] [PubMed] [Google Scholar]
- Del Bino S, Ito S, Sok J, Nakanishi Y, Bastien P, Wakamatsu K, and Bernerd F (2015). Chemical analysis of constitutive pigmentation of human epidermis reveals constant eumelanin to pheomelanin ratio. Pigment cell & melanoma research 28, 707–717. [DOI] [PubMed] [Google Scholar]
- Earle SR, and Fisher RR (1980). A direct demonstration of proton translocation coupled to transhydrogenation in reconstituted vesicles. The Journal of biological chemistry 255, 827–830. [PubMed] [Google Scholar]
- Filadi R, Pendin D, and Pizzo P (2018). Mitofusin 2: from functions to disease. Cell Death Dis 9, 330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project, C., Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, and McVean GA (2012). An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho H, and Ganesan AK (2011). The pleiotropic roles of autophagy regulators in melanogenesis. Pigment Cell Melanoma Res 24, 595–604. [DOI] [PubMed] [Google Scholar]
- Hofman A, Brusselle GG, Darwish Murad S, van Duijn CM, Franco OH, Goedegebure A, Ikram MA, Klaver CC, Nijsten TE, Peeters RP, et al. (2015). The Rotterdam Study: 2016 objectives and design update. Eur J Epidemiol 30, 661–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horike N, Kumagai A, Shimono Y, Onishi T, Itoh Y, Sasaki T, Kitagawa K, Hatano O, Takagi H, Susumu T, et al. (2010). Downregulation of SIK2 expression promotes the melanogenic program in mice. Pigment cell & melanoma research 23, 809–819. [DOI] [PubMed] [Google Scholar]
- Huang TT, Naeemuddin M, Elchuri S, Yamaguchi M, Kozy HM, Carlson EJ, and Epstein CJ (2006). Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum Mol Genet 15, 1187–1194. [DOI] [PubMed] [Google Scholar]
- Hysi PG, Valdes AM, Liu F, Furlotte NA, Evans DM, Bataille V, Visconti A, Hemani G, McMahon G, Ring SM, et al. (2018). Genome-wide association meta-analysis of individuals of European ancestry identifies new loci explaining a substantial fraction of hair color variation and heritability. Nature genetics 50, 652–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikram MA, Brusselle GGO, Murad SD, van Duijn CM, Franco OH, Goedegebure A, Klaver CCW, Nijsten TEC, Peeters RP, Stricker BH, et al. (2017). The Rotterdam Study: 2018 update on objectives, design and main results. Eur J Epidemiol 32, 807–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iozumi K, Hoganson GE, Pennella R, Everett MA, and Fuller BB (1993). Role of tyrosinase as the determinant of pigmentation in cultured human melanocytes. J Invest Dermatol 100, 806–811. [DOI] [PubMed] [Google Scholar]
- Ito S, and Ifpcs (2003). The IFPCS presidential lecture: a chemist's view of melanogenesis. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society 16, 230–236. [DOI] [PubMed] [Google Scholar]
- Ito S, Nakanishi Y, Valenzuela RK, Brilliant MH, Kolbe L, and Wakamatsu K (2011). Usefulness of alkaline hydrogen peroxide oxidation to analyze eumelanin and pheomelanin in various tissue samples: application to chemical analysis of human hair melanins. Pigment Cell Melanoma Res 24, 605–613. [DOI] [PubMed] [Google Scholar]
- Jablonski NG, and Chaplin G (2012). Human skin pigmentation, migration and disease susceptibility. Philos Trans R Soc Lond B Biol Sci 367, 785–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jablonski NG, and Chaplin G (2017). The colours of humanity: the evolution of pigmentation in the human lineage. Philos Trans R Soc Lond B Biol Sci 372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs LC, Hamer MA, Verkouteren JA, Pardo LM, Liu F, and Nijsten T (2015). Perceived skin colour seems a swift, valid and reliable measurement. Br J Dermatol 173, 1084–1086. [DOI] [PubMed] [Google Scholar]
- Jara JR, Aroca P, Solano F, Martinez JH, and Lozano JA (1988). The role of sulfhydryl compounds in mammalian melanogenesis: the effect of cysteine and glutathione upon tyrosinase and the intermediates of the pathway. Biochimica et biophysica acta 967, 296–303. [DOI] [PubMed] [Google Scholar]
- Jaruga P, and Dizdaroglu M (2008). 8,5'-Cyclopurine-2'-deoxynucleosides in DNA: mechanisms of formation, measurement, repair and biological effects. DNA Repair (Amst) 7, 1413–1425. [DOI] [PubMed] [Google Scholar]
- Jiang L, Zheng Z, Qi T, Kemper KE, Wray NR, Visscher PM, and Yang J (2019). A resource-efficient tool for mixed model association analysis of large-scale data. Nature genetics 51, 1749–1755. [DOI] [PubMed] [Google Scholar]
- Khaled M, Levy C, and Fisher DE (2010). Control of melanocyte differentiation by a MITF-PDE4D3 homeostatic circuit. Genes & development 24, 2276–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R, Weilbaecher KN, McGill G, Cooley E, Mihm M, and Fisher DE (1999). Microphthalmia transcription factor. A sensitive and specific melanocyte marker for MelanomaDiagnosis. Am J Pathol 155, 731–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfo I, et al. (2019). Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol 37, 276–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, and Valen E (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res 47, W171–W174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, Aros MC, Jurynec MJ, Mao X, Humphreville VR, Humbert JE, et al. (2005). SLC24A5, a putative cation exchanger, affects pigmentation in zebrafish and humans. Science 310, 1782–1786. [DOI] [PubMed] [Google Scholar]
- Langendonk JG, Balwani M, Anderson KE, Bonkovsky HL, Anstey AV, Bissell DM, Bloomer J, Edwards C, Neumann NJ, Parker C, et al. (2015). Afamelanotide for Erythropoietic Protoporphyria. The New England journal of medicine 373, 48–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leboucher GP, Tsai YC, Yang M, Shaw KC, Zhou M, Veenstra TD, Glickman MH, and Weissman AM (2012). Stress-induced phosphorylation and proteasomal degradation of mitofusin 2 facilitates mitochondrial fragmentation and apoptosis. Mol Cell 47, 547–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Siford RL, Martin AR, Nakagome S, Moller M, Hoal EG, Bustamante CD, Gignoux CR, and Henn BM (2018). Rapid evolution of a skin-lightening allele in southern African KhoeSan. Proceedings of the National Academy of Sciences of the United States of America 115, 13324–13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo JA, and Fisher DE (2014). The melanoma revolution: from UV carcinogenesis to a new era in therapeutics. Science 346, 945–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh PR, Kichaev G, Gazal S, Schoech AP, and Price AL (2018). Mixed-model association for biobank-scale datasets. Nature genetics 50, 906–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Lin M, Granka JM, Myrick JW, Liu X, Sockell A, Atkinson EG, Werely CJ, Moller M, Sandhu MS, et al. (2017). An Unexpectedly Complex Architecture for Skin Pigmentation in Africans. Cell 171, 1340–1353 e1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra D, Luo X, Morgan A, Wang J, Hoang MP, Lo J, Guerrero CR, Lennerz JK, Mihm MC, Wargo JA, et al. (2012). An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature 491, 449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody AJ, and Reid RA (1983). Inhibition of nicotinamide nucleotide transhydrogenase in rat liver submitochondrial particles by dicyclohexylcarbodiimide and butanedione. Biochem J 209, 889–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, and Giraldez AJ (2015). CRISPRscan: designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods 12, 982–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mujahid N, Liang Y, Murakami R, Choi HG, Dobry AS, Wang J, Suita Y, Weng QY, Allouche J, Kemeny LV, et al. (2017). A UV-Independent Topical Small-Molecule Approach for Melanin Production in Human Skin. Cell Rep 19, 2177–2184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HY, Kosmadaki M, Yaar M, and Gilchrest BA (2009). Cellular mechanisms regulating human melanogenesis. Cell Mol Life Sci 66, 1493–1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SB, Suh DH, and Youn JI (1999). A long-term time course of colorimetric evaluation of ultraviolet light-induced skin reactions. Clin Exp Dermatol 24, 315–320. [DOI] [PubMed] [Google Scholar]
- Paterson EK, Fielder TJ, MacGregor GR, Ito S, Wakamatsu K, Gillen DL, Eby V, Boissy RE, and Ganesan AK (2015). Tyrosinase Depletion Prevents the Maturation of Melanosomes in the Mouse Hair Follicle. PLoS One 10, e0143702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathak MA, Riley FJ, Fitzpatrick TB, and Curwen WL (1962). Melanin formation in human skin induced by long-wave ultra-violet and visible light. Nature 193, 148–150. [DOI] [PubMed] [Google Scholar]
- Phelps DC, and Hatefi Y (1981). Inhibition of the mitochondrial nicotinamide nucleotide transhydrogenase by dicyclohexylcarbodiimide and diethylpyrocarbonate. J Biol Chem 256, 8217–8221. [PubMed] [Google Scholar]
- Premi S, Wallisch S, Mano CM, Weiner AB, Bacchiocchi A, Wakamatsu K, Bechara EJ, Halaban R, Douki T, and Brash DE (2015). Photochemistry. Chemiexcitation of melanin derivatives induces DNA photoproducts long after UV exposure. Science 347, 842–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. (2007). PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quillen EE, Norton HL, Parra EJ, Lona-Durazo F, Ang KC, Illiescu FM, Pearson LN, Shriver MD, Lasisi T, Gokcumen O, et al. (2019). Shades of complexity: New perspectives on the evolution and genetic architecture of human skin. Am J Phys Anthropol 168 Suppl 67, 4–26. [DOI] [PubMed] [Google Scholar]
- Raposo G, and Marks MS (2007). Melanosomes--dark organelles enlighten endosomal membrane transport. Nat Rev Mol Cell Biol 8, 786–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rendon MI, and Gaviria JI (2005). Review of skin-lightening agents. Dermatologic surgery : official publication for American Society for Dermatologic Surgery [et al. ] 31, 886–889; discussion 889. [DOI] [PubMed] [Google Scholar]
- Rigler MW, and Longo WE (2010). Emission of diacetyl (2,3 butanedione) from natural butter, microwave popcorn butter flavor powder, paste, and liquid products. Int J Occup Environ Health 16, 291–302. [DOI] [PubMed] [Google Scholar]
- Rohland N, and Reich D (2012). Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome Res 22, 939–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronchi JA, Figueira TR, Ravagnani FG, Oliveira HC, Vercesi AE, and Castilho RF (2013). A spontaneous mutation in the nicotinamide nucleotide transhydrogenase gene of C57BL/6J mice results in mitochondrial redox abnormalities. Free Radic Biol Med 63, 446–456. [DOI] [PubMed] [Google Scholar]
- Rydstrom J (1972). Site-specific inhibitors of mitochondrial nicotinamide-nucleotide transhydrogenase. Eur J Biochem 31, 496–504. [DOI] [PubMed] [Google Scholar]
- Rydstrom J, Teixeira da Cruz A, and Ernster L (1970). Factors governing the steady state of the mitochondrial nicotinamide nucleotide transhydrogenase system. The Biochemical journal 116, 12P–13P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, and Sabatini DM (2008). The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320, 1496–1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian D, Sorianello E, Segales J, Irazoki A, Ruiz-Bonilla V, Sala D, Planet E, Berenguer-Llergo A, Munoz JP, Sanchez-Feutrie M, et al. (2016). Mfn2 deficiency links age-related sarcopenia and impaired autophagy to activation of an adaptive mitophagy pathway. EMBO J 35, 1677–1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheeran FL, Rydstrom J, Shakhparonov MI, Pestov NB, and Pepe S (2010). Diminished NADPH transhydrogenase activity and mitochondrial redox regulation in human failing myocardium. Biochimica et biophysica acta 1797, 1138–1148. [DOI] [PubMed] [Google Scholar]
- Steingrimsson E, Copeland NG, and Jenkins NA (2004). Melanocytes and the microphthalmia transcription factor network. Annu Rev Genet 38, 365–411. [DOI] [PubMed] [Google Scholar]
- Toye AA, Lippiat JD, Proks P, Shimomura K, Bentley L, Hugill A, Mijat V, Goldsworthy M, Moir L, Haynes A, et al. (2005). A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia 48, 675–686. [DOI] [PubMed] [Google Scholar]
- Ugwu SO, Blanchard J, Dorr RT, Levine N, Brooks C, Hadley ME, Aickin M, and Hruby VJ (1997). Skin pigmentation and pharmacokinetics of melanotan-I in humans. Biopharm Drug Dispos 18, 259–269. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, et al. (2015). Proteomics. Tissue-based map of the human proteome. Science 347, 1260419. [DOI] [PubMed] [Google Scholar]
- van Rooijen E, Fazio M, and Zon LI (2017). From fish bowl to bedside: The power of zebrafish to unravel melanoma pathogenesis and discover new therapeutics. Pigment Cell Melanoma Res 30, 402–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Lim JH, Chim H, Bhalla K, Girnun G, Pierce K, Clish CB, Granter SR, Widlund HR, Spiegelman BM, et al. (2013). PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell 23, 287–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamatsu K, Ito S, and Rees JL (2002). The usefulness of 4-amino-3-hydroxyphenylalanine as a specific marker of pheomelanin. Pigment cell research / sponsored by the European Society for Pigment Cell Research and the International Pigment Cell Society 15, 225–232. [DOI] [PubMed] [Google Scholar]
- Walton RT, Christie KA, Whittaker MN, and Kleinstiver BP (2020). Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y (2008). Bulky DNA lesions induced by reactive oxygen species. Chem Res Toxicol 21, 276–281. [DOI] [PubMed] [Google Scholar]
- Won S, Morris N, Lu Q, and Elston RC (2009). Choosing an optimal method to combine P-values. Stat Med 28, 1537–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, and Hammer JA (2014). Melanosome transfer: it is best to give and receive. Curr Opin Cell Biol 29, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Lee SH, Goddard ME, and Visscher PM (2011). GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 88, 76–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Padayatti PS, and Leung JH (2017). Proton-Translocating Nicotinamide Nucleotide Transhydrogenase: A Structural Perspective. Front Physiol 8, 1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Huang X, Han L, Wang X, Cheng H, Zhao Y, Chen Q, Chen J, Cheng H, Xiao R, et al. (2012). Central role of mitofusin 2 in autophagosome-lysosome fusion in cardiomyocytes. J Biol Chem 287, 23615–23625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao WN, and McAlister-Henn L (1996). Assembly and function of a cytosolic form of NADH-specific isocitrate dehydrogenase in yeast. The Journal of biological chemistry 271, 10347–10352. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Inhibition of NNT Increase pigmentation via redox dependent mechanism. Related to Figure 1
(A) siNNT-induced increased pigmentation in human SK-MEL-30 melanoma cells is dependent on tyrosinase and reactive oxygen species. Left panel: Representative lysates from SK-MEL-30 cells following treatment with siControl, siNNT, siNNT + siTyrosinase (siTYR), or siNNT + 5mM NAC. Right panel: Quantification of intracellular melanin content in SK-MEL-30 cells; n = 3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (B, C) qRT-PCR analysis of NNT (B) and immunofluorescence of NNT (C) in primary human melanocytes treated with siControl or siNNT for 96 hours. IF staining of human NNT (Green) and nuclei (DAPI, blue) are shown. Scale bar 50 μM. Relative NNT mRNA levels and fluorescent intensities (n = 3) were analyzed by unpaired, two-sided t-tests. (D) Immunoblot analysis of NNT expression in UACC257 human melanoma cells. Band intensities were quantified by ImageJ, normalized to β-actin and plotted relative to siControl; n = 3, analyzed by unpaired, two-sided t-test. (E) Treatment of UACC257 cells with siNNT for 24 hours resulted in increased NADPH/NADP (Left panel, n = 9) and decreased GSH/GSSG (Right panel, n = 6) ratios. The data were analyzed by multiple t tests with the Holm-Šídák post-test. (F) UACC257 melanoma cells were treated with siControl or siTyrosinase for 5 days and eumelanin and pheomelanin were measured using HPLC techniques (n = 3). Absolute pigment levels (Left graph) were analyzed by ordinary two-way ANOVA, separately for eumelanin and pheomelanin. The eumelanin/pheomelanin ratio (Right graph) was analyzed by unpaired Student t test (G). Increased ROS in UACC257 cells following 48 hours of siNNT or siIDH1 treatment, but not after 48 hours of siPGCa treatment. IF images of ROS indicator DCFDA (Green) and nuclei (DAPI, blue), representative of five experiments, are displayed. Quantified results were normalized to the total number of cells and analyzed by ordinary one-way ANOVA with Šídák’s post-test. (H) Increase of melanin content by siNNT is blocked by cotreatment with NADPH. Intracellular melanin content was quantified in UACC257 cells treated with siControl or siNNT for 72 hours, with 0.1 M NADPH or Vehicle (Tris-HCl, pH 8.0) added after the first 24 hours. n = 3, analyzed by ordinary two-way ANOVA with Šídák’s post-test. (I) Immunoblot analysis of IDH1 in UACC257 cells treated with siControl, siNNT, siIDH1, or siNNT + siIDH1 together for 72 hours. Band intensities were quantified by ImageJ, normalized to β-actin (n = 3), and analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (J) qRT-PCR analysis of NNT, IDH1, and PGC1α mRNAs in UACC257 cells treated with siRNA for one of those genes or siControl. qRT-PCR data are normalized to RPL11 RNA and RNA levels are presented as fold change relative to siControl; (n = 3), analyzed by ordinary one-way ANOVA with Dunnett’s post-test, followed by the Bonferroni correction for three ANOVA analyses. (K, L) Overexpression of NNT in the UACC257 cell line: (K) qRT-PCR analysis of NNT mRNA five days post transfection; n=3, analyzed by unpaired, two-sided t-test. (L) Overexpression of NNT resulted in decreased NADPH/NADP (Left panel, n = 8) and increased GSH/GSSG (Right panel, n = 4-6) ratios, analyzed by multiple t-tests with the Holm-Šídák post-test.
All data are expressed as mean ± SEM; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Figure S2. NNT does not impact TYR mRNA expression levels and acts independently of the cAMP pathway. Related to Figure 2
(A) Tyrosinase activity increase following siNNT in UACC257 melanoma cells; n = 4, analyzed by unpaired, two-sided t-test (B) Diagram of the “Tanning Pathway”. Briefly, UV exposure results in DNA damage and activation of P53 in keratinocytes. POMC is transcriptionally activated by P53 and the pro-protein is cleaved to α-MSH, which is secreted from the keratinocyte. α-MSH binds to MC1R in the melanocyte membrane, resulting in an increase in cAMP and activation of PKA. Active PKA results in an increase of MITF, activated transcriptionally by CREB. MITF transcriptionally regulates pigmentation enzymes such us TYRP1, TRP2 and tyrosinase. (C) Immunoblot analysis of MITF in UACC257 cells transfected with siNNT or siControl for 72 h. Band intensities were quantified by ImageJ, normalized to β-actin, plotted relative to siControl values (n = 3), and analyzed by unpaired, two-sided t-test. (D-F) Analyses of UACC257 cells stably expressing secreted luciferase under the TRPM1 promoter and SEAP under the CMV promoter. The cells were treated with either siControl, siNNT, or siMITF (n = 3): (D) qRT-PCR analysis of NNT, mMITF and TYRP1 72 hours post siRNA transfection. Data were normalized to RPL11 RNA, analyzed by unpaired, two-sided t-test (NNT) or ordinary one-way ANOVA with Dunnett’s post-test (mMITF and TYRP1). (E) Luciferase secretion normalized to secreted SEAP 72 hours post siRNA transfection, showing decreased luciferase activity following siMITF and siNNT, analyzed by ordinary one-way ANOVA with Dunnett’s post-test; representative cell pellets (1 x 106 cells) are below the graph. (F) Luciferase secretion normalized to secreted SEAP 24, 48 and 72 hours post siRNA transfection was analyzed by repeated measures two-way ANOVA with Šídák’s post-test. (G) qRT-PCR analysis of NNT, MITF, TYRP1, TRP2/DCT, NNT, tyrosinase, and POMC in UACC257 cells 72 hours post transfection of siNNT or siControl. Data were normalized to RPL11 RNA, presented as fold change relative to siControl (n = 3), and analyzed by multiple t-tests with the Holm-Šídák post-test. (H) cAMP content of UACC257 cells transfected with siNNT or siControl for 48 h, measured by cAMP ELISA and normalized to siControl cells; n = 3, analyzed by unpaired, two-sided t-test. (I) Primary human melanocytes were starved for 24 hours and Forskolin (FSK; 20 μM) was added to the medium for 2 hours. qRT-PCR analysis of NNT was performed with MITF as a positive control for the treatment. The data were normalized to RPL11 RNA (n = 3) and analyzed by multiple t-tests with the Holm-Šídák post-test. (J) No change in NNT mRNA upon UVB. Abdominal skin was irradiated with 1J/cm2 UVB, skin was collected at 0, 24, 48 and 72 hours post UVB, and qRT-PCR analysis of NNT was performed. The data were normalized to RPL11 RNA and presented as fold change relative to t = 0. n = 5-6 (two different donors), analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (K) Immunoblots of P53 and β-actin in UACC257 cells following siControl or siNNT treatment for 72 hours. (L) Immunoblot of NNT and β-actin in UACC257 melanoma cells (Left panel), daily treatment with NAC (5 mM), MitoTEMPO (20 μM) and H2O2 (100μM) for 72 hours (n = 3) analyzed by ordinary one-way ANOVA with Tukey post-test. (M) Immunoblots of tyrosinase and β-actin in UACC257 melanoma cells (Left panel), showing decreased tyrosinase protein levels following overexpression of NNT for 12 days. Band intensities were quantified by ImageJ, normalized to β-actin and plotted relative to siControl values (Right Panel). (n = 3), analyzed by unpaired, two-sided t-test. (N) qRT-PCR analysis of MITF, TYRP1 and tyrosinase mRNAs in UACC257 cells that overexpressed NNT (NNT OE), compared to control (Empty Vector). The data were normalized to RPL11 RNA (n = 3) and analyzed by ordinary one-way ANOVA with Dunnett’s post-test, followed by the Bonferroni correction for three ANOVA analyses.
Figure S3. NNT knockdown enhances melanosome maturation, melanosome-mitochondria proximity and pigmentation by NNT knockdown. Related to Figure 2
(A) Enhanced melanosome maturation induced by siNNT in human primary melanocyte cells is blocked by NAC (5 mM) or MitoTEMPO (20 μM) (daily treatment for 96 h). The number of melanosomes per um2 in the classified stages is represented. n = 4-5 cells, analyzed by ordinary two-way ANOVA with Šídák’s post-test (B) The total number of melanosomes per um2 in primary human melanocytes is not altered by siNNT and/or daily treatment with NAC (5 mM) or MitoTEMPO (20 μM) for 96 hours (Left graph, n = 8-10, analyzed by ordinary one-way ANOVA with Dunnett’s post-test) or by overexpression of NNT (Middle graph, n = 8-10), analyzed by unpaired, two-sided t-test. The total numbers of mitochondria per um2 by overexpression of NNT (Right graph, n=5) is not altered. (C) Measurements of proximities between melanosomes and mitochondria were quantified in FIJI (ImageJ) by applying a customized macro to TEM micrographs (n = 100 events per condition). Melanosome-mitochondria proximities closer than 20 nm are considered melanosome-mitochondria close appositions/contacts. Right panel: FIJI graphical user interface showing a TEM micrograph of mitochondria (m) and a melanosome (*) with a yellow line indicating the Euclidean distance between melanosome and mitochondrion surfaces, quantified with a customized macro to measure distances between two surfaces. Scale bar 400 nm. Table shows the percentages and, in parentheses, the fractions of melanosome-mitochondria proximities that were <20 nm. Denominators are the total number of measurements (events) performed in each group. Adjusted P values were determined by pairwise F-tests of the control group to each of the other groups, followed by the Bonferroni correction for three comparisons. (D-E) The total numbers of melanosomes (D) and mitochondria (E) per um2 in primary human melanocytes is not altered; n = 5 cells, analyzed by ordinary two-way ANOVA with Šídák’s post-test. (F) MFN2 enables siNNT-mediated pigmentation. Top panel: Quantification by spectrophotometry of intracellular melanin content of UACC257 human melanoma cells treated with siControl, siNNT, siMFN2 + siNNT, or siMFN2 for 72 hours. n = 3, analyzed by ordinary one-way ANOVA with Dunnett’s post-test. Bottom panel: Representative cell pellets (106 cells). (G) Immunoblot analysis of MFN2 expression in UACC257 human melanoma cell lines. Band intensities (n = 3) were quantified by ImageJ, normalized to β-actin, and analyzed by ordinary one-way ANOVA with Dunnett’s post-test. (H) qRT-PCR analysis of MFN2 in primary human melanocytes that were transfected with siMFN2. The data were normalized to RPL11 RNA, plotted relative to the control (n = 3), and analyzed by unpaired, two-sided t-tests (I) siMFN2 resulted in accumulation of large autophagosomes (white arrows), containing numerous melanosomes (arrowheads), in normal human melanocytes. Scale bar 2 μm. (J) Immunoblot analysis of LC3B in UACC257 cells treated with siMFN2, siNNT, siMFN2+siNNT, or siControl for 72 hours. Band intensities were quantified by ImageJ and normalized to β-actin. The ratios of LC3BII to LC3BI were plotted (n = 3) and analyzed by ordinary one-way ANOVA with Dunnett’s post-test.
All data are expressed as mean ± SEM; *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Figure S4. NNT inhibitors are non-toxic in vitro. Related to Figure 3
(A) Chemical formulas of all three published NNT inhibitors. (B) Viability measurements showed no significant toxicity after treatment of human melanocytes, dermal fibroblasts, and keratinocytes with up to 10 μM of DCC, Palmitoyl-CoA, or 2,3BD. (C) Treatments with different doses of DCC (Left graph) or 2,3 BD (Right graph) had no impacts on cell viability. The data were plotted relative to vehicle treatment (0) and analyzed by ordinary one-way ANOVA with Dunnett’s post-test (n = 4). (D) Intracellular melanin content normalized to total protein levels in primary human melanocytes that were treated with siControl or siNNT for 24 hours, followed by incubation with 2,3 BD (2 mM) or DMSO vehicle for 72 hours. n = 3, analyzed by ordinary one-way ANOVA with Šídák’s post-test. (E) Treatments with different doses of DCC had no impact on cell viability (Right graph), but resulted in decreased GSH/GSSG ratios (Left graph) in the UACC257 human melanoma cell line. n = 4, analyzed by ordinary one-way ANOVA with Tukey’s (Left graph) or Šídák’s (Right graph) post-test. (F) Fontana-Masson staining of melanin in human abdominal skin 5 days after a single treatment of 2,3BD (1M), showing supranuclear capping (Black arrows) in keratinocytes of 2,3BD-treated skin. Scale bar 50 μM
All data are expressed as mean ± SEM; *p<0.05, **p<0.01, ****p<0.0001.
Figure S5. NNT regulates pigmentation in mice, zebrafish and human pigmentation disorders. Related to Figure 4
(A) Agarose gel showing PCR genotyping of DNA from C57BL/6J mice (single 743 bp product indicates homozygous 5-exon deletion in the Nnt gene) and C57BL/6NJ mice (single 570 bp product indicates homozygous wild type Nnt gene). (B) Modification of NNT sites in zebrafish using WT SpCas9. Editing was assessed by next-generation targeted amplicon sequencing. (C) Zebrafish overexpressing NNT (NNT OE) or empty plasmid were treated at 3 days post fertilization with 100 μM of 2,3BD or vehicle for 24 hours. A representative image has been displayed. Results of mean melanocytic brightness, quantified by pixel-based analysis are shown in the graph at right; Empty plasmid (n = 12 fish; 30 melanocytes), NNT OE (n = 10 fish; 24 melanocytes), Empty plasmid + 2,3 BD (n = 8 fish; 30 melanocytes), NNT OE + 2,3BD (n = 11 fish; 31 melanocytes), analyzed by ordinary one-way ANOVA with Dunnett’s post-test (D) Representative images of the specific areas of hyperpigmentation in human lentigo-affected skin after staining for NNT (left image, red) or Fontana Masson (right image). Graph at right shows NNT signal intensities in melanocytes of healthy and lesional skin. n = 9 (bars indicate means), analyzed by ordinary one-way ANOVA with Dunnett’s post-test.
Supplementary Table 1. Characteristics of the NNT genetic variants significant in the meta-analysis for skin color. Related to Figure 5 and Figure 6
Data Availability Statement
All the Software packages and methods used in this study have been properly detailed and referenced under the Software and Algorithms listed in KEY RESOURCES TABLE. All Human data (SNP analysis) generated in this study are available under the Deposited Data listed in KEY RESOURCES TABLE.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-MITF monoclonal antibody C5 | Made in the lab of Dr. David E. Fisher | (King et al., 1999) C5 |
| Mouse monoclonal anti-tyrosinase antibody, clone T311 | Sigma-Adlrich | Cat# 05-647: RRID:AB_309873 |
| Donkey anti-rabbit IgG-HRP | ThermoFisher Scientific | Cat# 45-000-683: RRID: AB_2721111 |
| Amersham ECL mouse IgG, HRP | ThermoFisher Scientific | Cat#45000680;RRID: AB_2721110 |
| Monoclonal anti-β-actin-peroxidase | Sigma Aldrich | Cat# A3854;RRID:AB_262011 |
| Alexa Fluor 555 goat anti-rabbit IgG (H+L), secondary antibody | ThermoFisher Scientific | Cat# A-21428;RRID: AB_2535849 |
| Alexa Fluor 647 goat anti mouse IgG (G+L); fluorescence conjugated secondary antibody | ThermoFisher Scientific | Cat# A-21236;RRID: AB_2535805 |
| Alexa Fluor 594 F(ab)2 fragment of goat anti-rabbit IgG (G+L); fluorescence conjugated secondary antibody | ThermoFisher Scientific | Cat# A-11072;RRID: AB_2534116 |
| Alexa Fluor 488-conjugated donkey anti-mouse secondary antibodies | ThermoFisher Scientific | Cat# A-21202;RRID: AB_141607 |
| Anti-Cyclobutane Pyrimidine Dimers (CPDs) mAb antibody (Clone TDM-2) | Cosmo Bio USA | Cat# CAC-NM-DND-001; RRID: AB_1962813 |
| Rabbit anti-γ-H2AX (P-ser139) polyclonal antibody | NOVUS Biologicals | Cat# NB100-384;RRID:AB_10002815 |
| Mouse monoclonal anti-Mitofusin 2 antibody [6A8] | Abcam | Cat# ab56889;RRID: AB_2142629 |
| Rabbit polyclonal anti TRP2/DCT antibody | Abcam | Cat# ab74073;RRID:AB_1524517 |
| Mouse monoclonal anti-NNT antibody [8B4BB10] | Abcam | Cat# ab110352;RRID:AB_10887748 |
| Rabbit anti-NNT (C-terminal) polyclonal antibody | Abcam | Cat# ab214212; RRID:AB_2889980 |
| Mouse anti-8-oxo-dG monoclonal antibody | Trevigen | Cat# 4354-MC-050; RRID:AB_1857195 |
| IDH1 (D2H1) Rabbit mAb | Cell Signaling Technology | Cat# 8137;RRID: AB_10950504 |
| Mouse monoclonal p53 antibody [PAb 240] | Abcam | Cat# ab26;RRID:AB_303198 |
| Rabbit monoclonal TRP1 antibody [EPR21960] | Abcam | Cat# ab235447; RRID:AB_2889980 |
| Mouse monoclonal antibody Pmel17 (E-7) | Santa Cruz Biotechnology | Cat# sc-377325; RRID:AB_2889982 |
| LC3B (D11) rabbit monoclonal antibody | Cell Signaling Technology | Cat#38668S RRID:AB_2137707 |
| Biological Samples | ||
| Full thickness human breast and abdominal skin explants | Massachusetts General Hospital | IRB# 2013P000093 |
| Paraffin-embedded formalin fixed slides, prepared from breast and abdominal biopsy samples | Massachusetts General Hospital | IRB# 2013P000093 |
| Human skin samples for genome wide association study (GWAS) | Massachusetts General Hospital or the Cooperative Human Tissue Network | IRB# 2013P000093 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| 3-isobutyl-1-methylxanthine (IBMX) | Sigma-Aldrich | Cat# I5879 |
| 12-O-tetradecanoylphorbol-13-acetate (TPA) | Sigma-Aldrich | Cat# 16561-29-8 |
| Ham's F10 | Thermo Fisher Scientific | Cat# MT10070CV |
| N6,2′-O-Dibutyryladenosine 3′,5′-cyclic monophosphate sodium salt | Sigma-Aldrich | Cat# D0627 |
| Penicillin-Streptomycin | Thermo Fisher Scientific | Cat# 15140163 |
| Na3VO4 | Sigma-Aldrich | Cat# 450243 |
| Medium 254 | Life Technologies | Cat# M254500 |
| 0.05% Trypsin-EDTA w/ phenol red | Life Technologies | Cat# 25300120 |
| Human Melanocyte Growth Supplement (HMGS) | Life Technologies | Cat# S0025 |
| Bovine Serum Albumin | Sigma | Cat#A7030 |
| Goat serum | Sigma-Aldrich | Cat# G9023 |
| RPMI (Roswell Park Memorial Institute 1640 Medium) | Life Technologies | Cat# 11875119 |
| RIPA lysis buffer | Sigma-Aldrich | Cat# R0278 |
| FetalPlex Animal Serum Complex | Gemini Bio-Products | Cat# 100-602 |
| Western Lightning Plus-ECL | PerkinElmer | Cat # NEL105001EA |
| Non-fat milk powder | Boston BioProducts | Cat# P-1400 |
| Protein Block | Agilent | Cat# X090930-2 |
| Antibody Diluent | DAKO | Cat# S3022 |
| VECTASHIELD® HardSet™ Antifade Mounting Medium with DAPI | Vector Laboratories | Cat# H-1500 |
| synthetic melanin | Sigma Aldrich | Cat# M8631 |
| N,N-Dicyclohexylcarbodiimide [DCC] | Sigma Aldrich | Cat# D80002 |
| 2,3-Butanedione [2,3BD] | Sigma Aldrich | Cat# B85307 |
| Palmitoyl coenzyme A lithium salt | Sigma Aldrich | Cat# #P9716 |
| cycloheximide (CHX) | Sigma Aldrich | Cat# C7698 |
| NADPH | Sigma Aldrich | Cat# N7505 |
| N-Acetyl-L-cysteine (NAC) | Sigma Aldrich | Cat# A7250 |
| MitoTEMPO | ThermoFisher Scientific | Cat# 501872447 |
| Hydrogen peroxide solution | Sigma Aldrich | Cat# 216763 |
| SYBR FAST qPCR master mix | Kapa Biosystems | Cat# KK4600; |
| Protease and Phosphatase Inhibitor | ThermoFisher Scientific | Cat# PI78445 |
| Western Lightning Plus-ECL, Enhanced Chemiluminescence Substrate | Perkin Elmer | Cat# NEL105001EA |
| MitoSOX Red | ThermoFisher Scientific | Cat# M36008 |
| CM-H2DCFDA | ThermoFisher Scientific | Cat# C6827 |
| NucBlue | ThermoFisher Scientific | Cat# R37605 |
| Polybrene | Sigma-Aldrich | Cat# TR-1003 |
| Paraformaldehyde 16% | ThermoFisher Scientific | Cat# 50980487 |
| Ethanol | Thermo Fisher Scientific | Cat# 04355226 |
| Triton X-100 | Sigma Aldrich | Cat# T8787 |
| TWEEN® 20 | Sigma Aldrich | Cat# P7949 |
| Forskolin from Coleus forskohlii, ≥98% | Sigma Aldrich | Cat# F6886 |
| Lipofectamine RNAiMAX Transfection Reagent | Life Technologies | Cat# 13778150 |
| IQ5 High-fidelity DNA Polymerase | New England Biolabs | Cat# M0491S |
| Critical Commercial Assays | ||
| Direct cAMP ELISA Kit | Enzo Life Sciences | Cat# ADI-901-066 |
| GSH/GSSG-Glo Assay | Promega | Cat# V6611 |
| CellTiter-Glo Luminescent Cell Viability Assay | Promega | Cat# G7570 |
| Pierce BCA protein assay | ThermoFisher Scientific | Cat# 23225 |
| KAPA Library Quantification Kits | Roche | Cat# 7960140001 |
| MiSeq Reagent Kits v2 (300 cycles) | Illumina | Cat# MS-102-2002 |
| MaxBlock Autofluorescence Reducing Reagent Kit | MaxVision Biosciences | Cat# MB-L |
| Fontana-Masson Stain Kit (Melanin Stain) | Abcam | Cat# ab150669 |
| Dual Reporter System | GeneCopoeia | Cat# HPRM39435-LvPM02 |
| Secrete-Pair Gaussia Luciferase Assay Kit | GeneCopoeia | Cat# LF062 |
| NADP/NADPH-Glo Assay | Promega | Cat# G9082 |
| QUANTI-Blue™ Solution | InvivoGen | Cat# rep-qbs |
| RNeasy Plus Mini Kit | Qiagen | Cat# 74136 |
| Deposited Data | ||
| Raw data supporting the human genetics association analyses | https://www.dropbox.com/scl/fi/ahdfnjo4puwzdz8ayw2ix/supporting-data-human-genetic-associations-2.xlsx?dl=0&rlkey=gvbpat4tjb6bnekgnxuu1a6yh | |
| Publicly Available Data | ||
| Ensembl database information for promoter ENSR00000180214 of the NNT gene | Ensembl | http://www.ensembl.org/Homo_sapiens/Regulation/Summary?fdb=funcgen;r=5:43600000- |
| GTEx expression database | GTEx | www.gtexportal.org |
| GWAS summary statistics from the CANDELA cohort | GWAS Central | http://www.gwascentral.org/study/HGVST3308 |
| GWAS summary statistics from the UK Biobank cohort | https://cnsgenomics.com/software/gcta/#DataResource | |
| Experimental Models: Cell Lines | ||
| Primary human melanocytes (isolated from neonatal foreskins) | Massachusetts General Hospital | IRB# 2013P000093 |
| Human melanoma cell line UACC257 | National Cancer Institute Division of Cancer Treatment and Diagnosis (DCTD) Tumor Repository | |
| Human melanoma cell line SK-MEL-30 | Memorial Sloan Kettering Cancer Center | https://www.mskcc.org/ |
| Mouse Melan-A cell line | Wellcome Trust Functional Genomics Cell Bank | Bennett et al., 1987 |
| Primary human keratinocytes isolated from discarded surgical human skin tissue (e.g., foreskins) | Massachusetts General Hospital | IRB# 2013P000093 |
| Primary human fibroblasts isolated from discarded surgical human skin tissue (e.g., foreskins) | Massachusetts General Hospital | IRB# 2013P000093 |
| Lenti-X™ 293T cells | Clontech | Cat# 632180 |
| Experimental Models: Organisms/Strains | ||
| C57BL/6J mice | Jackson Laboratory | Stock No: 000664 |
| C57BL/6NJ mice | Jackson Laboratory | Stock No: 005304 |
| Casper zebrafish (mitfa−/−;roy−/−) embryos | Laboratory of Dr. Leonard I. Zon | |
| Oligonucleotides | ||
| nheIkozakHAhNNT_f1: forward, 5’-ctagctagcCCGCCACCATGTACCCATACGATGTTCCAGATTACGCTGCAAACCTATTGAAAACAGTGGTGACTG-3’ | eurofins Genomics | For PLMJ1- HA-NNT |
| hNNTnheI_r1: reverse, 5’-ctagctagcTTACTTCTGATAGGATTCTCTAACTTTCGC-3’ | eurofins Genomics | For PLMJ1- HA-NNT |
| nheIkozakhMFN2_f1: forward, 5’-ctagctagcGCCACCATGTCCCTGCTCTTCTCTCGATGC-3’ | eurofins Genomics | For PLMJ1- HA MFN2 |
| hMFN2(HA)nheI_r1: reverse, 5’-ctagctagcTTAGGATCCAGCAGCGTAATCTGGAAC-3’ | eurofins Genomics | For PLMJ1- HA MFN2 |
| RT-Primers for NNT, IDH1, MFN2, TYRP1, DCT/TRP2, MITF, POMC, PPARGC1A, Tyrosinase: See Table in STAR Methods | ||
| Additional oligonucleotides that were used for nnt knockout in zebrafish are in Supplementary Table 3 | ||
| Recombinant DNA | ||
| CRISPR MiniCoopR-U6:gRNA-mitfa:Cas9 plasmid | Addgene | Cloned from Addgene plasmid ID 118840 |
| pMiniCoopR-mitfa:NNT expression plasmid | Addgene | Cloned from Addgene plasmid ID 118850 |
| pLMJ1-NNT-HA | This manuscript | Based on Addgene plasmid, # 19319 |
| pLMJ1-EGFP plasmid | Laboratory of Dr. David Sabatini | Addgene plasmid, # 19319; http://n2t.net/addgene:19319; RRID:Addgene_19319, (Sancak et al., 2008) |
| pcDNA3.1 Mfn2HA | Laboratory of Dr. Allan Weissman | Addgene plasmid, # 139192; http://n2t.net/addgene:139192; RRID:Addgene_139192 ((Leboucher et al., 2012) |
| PLMJ1-MFN2-HA | This manuscript | Based on Addgene plasmid, # 139192 and Addgene plasmid, # 19319 |
| Software and Algorithms | ||
| GraphPad Prism 8.4.3 (471) | GraphPad | https://www.graphpad.com/scientific-software/prism/ |
| ImageJ (v1.8.0) | National Institutes of Health | https://imagej.nih.gov/ij/ |
| FIJI software for pixel-based color quantification | FIJI | https://imagej.net/Fiji |
| Off-target prediction software (for design of guide RNAs) | (Bae et al., 2014) | http://www.rgenome.net/cas-offinder/ |
| On-target prediction software (for design of guide RNAs); CRISPRscan and CHOPCHOP | (Moreno-Mateoset al., 2015) (Labun et al., 2019) |
https://www.crisprscan.org/ https://chopchop.cbu.uib.no/ |
| Axiovision REL 4.7 software | Zeiss | https://carl-zeiss-axiovision-rel.software.informer.com/4.7/ |
| CRISPResso2 software (for genome editing) | Kendell Clement et al | Nat Biotechnol. 2019 March; 37(3):224–226. |
| MACH software | http://csg.sph.umich.edu/abecasis/mach/index.html | |
| GCTA program | Yang et al., 2011 | https://cnsgenomics.com/software/gcta/ |
| PLINK program | https://www.cog-genomics.org/plink/1.9/ | |
| BioRender | BioRender.com | |
| Fiji | Saalfeld, S., Schmid, B., et al. (2012) | https://imagej.net/Fiji |
| NDP.view2 Viewing software | HAMAMATSU | https://www.hamamatsu.com/us/en/product/type/U12388-01/index.html |
| Other | ||
| siGENOME Human MITF siRNA SMARTpool | Dharmacon | Cat# M-008674-00-0005 |
| ON-TARGETplus Human IDH1 siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-008294-01-0005 |
| ON-TARGETplus Human MFN2 siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-012961-00-0005 |
| ON-TARGETplus Human NNT siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-009809-00-0005 |
| ON-TARGETplus Human PPRGC1A siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-005111-00-0005 |
| ON-TARGETplus Human Tyrosinase siRNA SMARTpool | Horizon Inspired Cell Solutions | Cat# L-012555-00-0005 |
| ON-TARGETplus non-targeting siRNA control pool | Horizon Inspired Cell Solutions | Cat# D-001810-10-05 |
| 4-15% Criterion TGX Precast Midi Protein gels | Bio-Rad Laboratories | Cat# 5671084 |
| Chamber slides | ThermoFisher Scientific | Cat# 125657 |