Summary
APOE is the strongest genetic risk factor for late-onset Alzheimer’s disease. ApoE exacerbates tau-associated neurodegeneration by driving microglial activation. However, how apoE regulates microglial activation and whether targeting apoE is therapeutically beneficial in tauopathy is unclear. Here we show that overexpressing an apoE metabolic receptor LDLR (low-density lipoprotein receptor) in P301S tauopathy mice markedly reduces brain apoE, and ameliorates tau pathology and neurodegeneration. LDLR overexpression (OX) in microglia cell-autonomously downregulates microglial Apoe expression, and is associated with suppressed microglial activation as in apoE-deficient microglia. Both apoE-deficiency and LDLR-OX strongly drive microglial immunometabolism towards enhanced catabolism over anabolism, whereas LDLR-overexpressing microglia also uniquely upregulate specific ion channels and neurotransmitter receptors upon activation. ApoE-deficient and LDLR-overexpressing mice harbor enlarged pools of oligodendrocyte progenitor cells (OPCs), and show greater preservation of myelin integrity under neurodegenerative conditions. They also show less reactive astrocyte activation in the setting of tauopathy.
Introduction
Apolipoprotein E (ApoE) is a lipid-binding protein that carries lipids and cholesterol for their transport and metabolism. Humans harbor three major APOE alleles: APOE2, APOE3 and APOE4. APOE4 strongly increases the risk of developing late-onset Alzheimer’s disease (AD), whereas APOE2 reduces AD risk relative to APOE3 (Bu, 2009). ApoE strongly regulates brain amyloid-β (Aβ) deposition in a dose- and isoform-dependent manner (E4>E3>E2) (Bales et al., 1997; Holtzman et al., 2000; Schmechel et al., 1993). We recently uncovered a prominent role of apoE in regulating neurodegeneration in the setting of tauopathy independent of its effect on Aβ (Shi et al., 2017). ApoE ablation in P301S tau transgenic (Tg) mice markedly reduced brain atrophy and tau pathology compared to the presence of apoE, whereas apoE4 significantly exacerbated neurodegeneration compared to apoE2 and apoE3 (Shi et al., 2017). We and others subsequently identified microglia as the principal driving force of neurodegeneration and tau pathogenesis in tauopathy mouse models (Mancuso et al., 2019; Shi et al., 2019). Microglial depletion in P301S mice fully blocked neurodegeneration and stopped tau pathology progression. ApoE had a strong immunomodulatory function and promoted neurodegeneration and tau pathogenesis predominantly by driving microglial activation (Shi et al., 2019). However, how apoE mechanistically regulates microglial activation and whether targeting apoE is therapeutically beneficial in the setting of tauopathy is unclear.
Low-density lipoprotein receptor (LDLR) is one of the two primary metabolic receptors (the other is low-density lipoprotein receptor-related protein 1, LRP1) mediating the clearance of apoE lipoproteins (Holtzman et al., 2012; Ishibashi et al., 1994). Upon binding to the metabolic receptors, apoE lipoproteins are endocytosed, with the core cargo lipids and a portion of apoE delivered to lysosomes for degradation and the remaining part of apoE being recycled (Heeren et al., 2006). In the brain, LDLR is expressed by most cell types (Rapp et al., 2006; William Rebeck et al., 1993; Zeisel et al., 2015). In an LDLR transgenic (Tg) mouse line, a PrP-driven Ldlr transgene was shown to be overexpressed in neurons and astrocytes by 10-fold, resulting in ~90% reduction of brain soluble apoE (Kim et al., 2009), suggesting that overexpressing LDLR is an effective approach to reduce extracellular brain apoE level via increased uptake and degradation. Here we crossed LDLR Tg mice to P301S mice, and assessed the effects of LDLR-mediated apoE reduction on tauopathy.
Results
LDLR OX in P301S mice attenuates neurodegeneration, synaptic loss and tau pathology
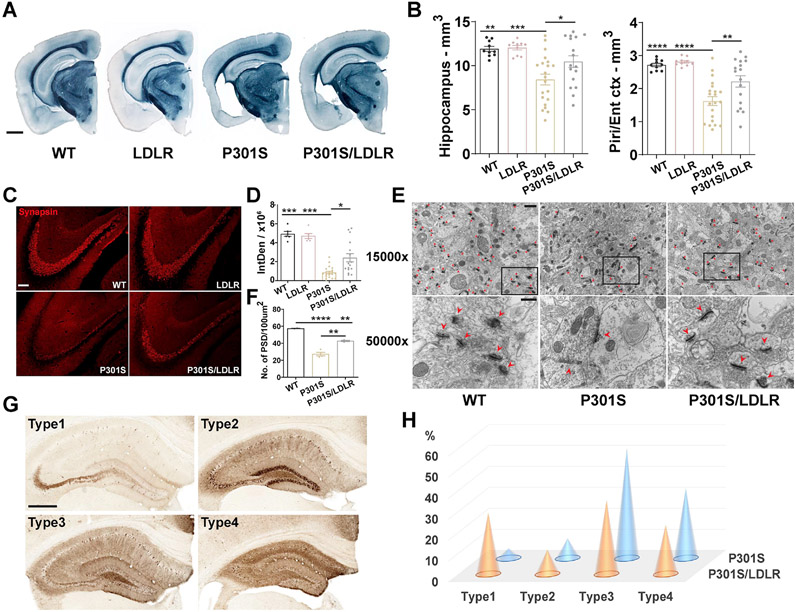
Littermates of male WT, LDLR, P301S, and P301S/LDLR mice were compared at 9 months of age, a time point when notable neurodegeneration occurs in the P301S mouse model. Compared to WT and LDLR mice that had a normal brain volume, P301S mice showed severe brain atrophy in the hippocampus and piriform/entorhinal cortex, whereas P301S/LDLR mice had significantly less brain volume loss than P301S mice (Figure 1A, B). Synapsin staining revealed a marked synaptic loss in P301S mice compared to WT and LDLR mice, particularly in the stratum lucidum of hippocampal CA3 region, which was significantly attenuated in P301S/LDLR mice (Figure 1C, D). Electron microscopy also revealed severe loss of postsynaptic densities (PSDs) in the stratum radiatum of hippocampal CA1 region in P301S mice, which was partially rescued in P301S/LDLR mice (Figure 1E, F). Phosphorylated tau – a pathological form of tau, was detected by immunostaining using a phospho-tau (p-tau) antibody, AT8. We previously identified four types of p-tau staining patterns (type1-type4) in P301S mice that represent progressively advancing disease stages (Shi et al., 2017) (Figure 1G). LDLR OX in P301S mice attenuated tau pathology, shifting p-tau staining towards an early-disease pattern (type1) (Figure 1H).
Figure 1. LDLR OX in P301S mice attenuates neurodegeneration, synaptic loss and tau histopathology.
(A, B) Sudan Black staining and brain volume quantification of 9-month WT, LDLR, P301S and P301S/LDLR mice (n=10-21). Scale bar: 1mm.
(C, D) Synapsin staining and quantification of the integrated density (IntDen, area × mean gray value) in the stratum lucidum of hippocampal CA3 region in 9-month mice (n=6-21). Scale bar: 100μm. Kruskal-Wallis test with Dunn’s multiple comparisons test.
(E, F) EM images of PSDs and quantification of PSD density in the stratum radiatum of hippocampal CA1 region in 9-month mice (n=2-4). Top (E): 15000x magnification, scale bar: 1μm; Bottom (E): 50000x magnification, scale bar: 400nm. Red asterisks or arrow heads point to PSDs.
(G) Four distinct p-tau patterns stained by AT8 in 9-month P301S and P301S/LDLR mice. Scale bar: 500μm.
(H) Distribution of p-tau staining patterns in 9-month P301S and P301S/LDLR mice (P301S: n=21, P301S/LDLR: n=17), p= 3.624e-05 (fisher’s exact test, two-sided). No p-tau immunoreactivity in WT and LDLR mice.
Data expressed as mean ± SEM, One-way ANOVA with Tukey’s post hoc test, two-sided in (B) and (F), *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Piri: piriform cortex, Ent: entorhinal cortex. See also Figure S2.
LDLR OX in P301S mice leads to a marked reduction of apoE and p-tau levels that strongly correlate with each other
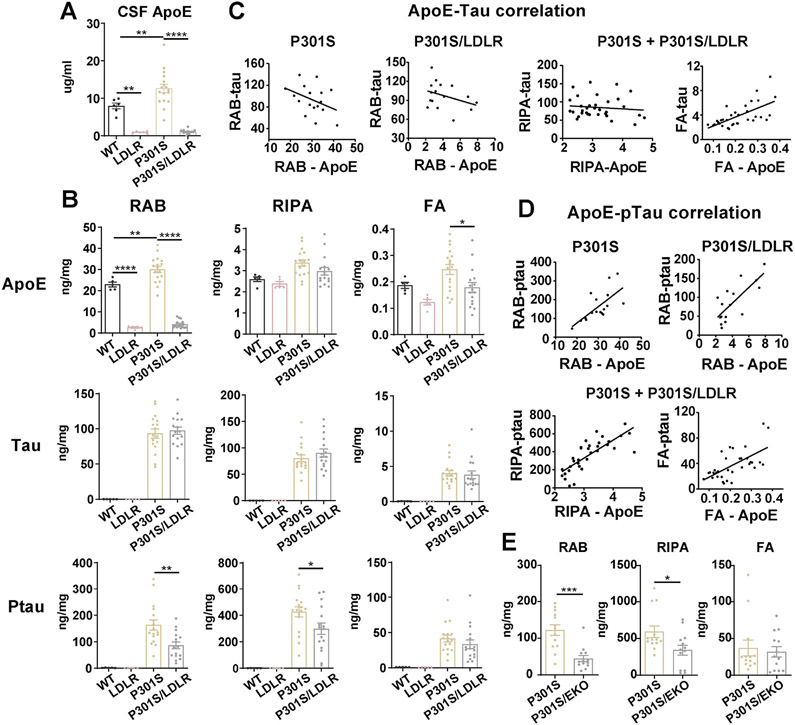
Corresponding to LDLR OX, apoE levels in the CSF of LDLR and P301S/LDLR mice were markedly reduced by ~90% compared to WT and P301S mice respectively (Figure 2A). Mouse cortical tissue was analyzed biochemically via quantitative ELISA following sequential tissue extraction in RAB (salt buffer), RIPA (detergent buffer) and 70% formic acid (FA), which contains soluble, less soluble, and insoluble proteins respectively. Both LDLR and P301S/LDLR mice showed an ~90% reduction of soluble apoE similar to that seen in the CSF (Figure 2B). Significantly reduced insoluble apoE was also observed in P301S/LDLR mice compared to P301S mice (Figure 2B). No human tau or p-tau signal was observed in WT and LDLR mice. P301S/LDLR mice showed no alteration in total human tau levels in any fractions compared to P301S mice (Figure 2B). However, apoE level had a strong positive correlation with insoluble tau level, and trended toward a negative correlation with soluble tau levels in both P301S and P301S/LDLR mice (Figure 2C). In contrast to no change of total human tau levels, LDLR OX in P301S mice significantly reduced p-tau levels in the RAB and RIPA fractions (Figure 2B). In addition, apoE levels positively correlated with p-tau levels in all fractions (Figure 2D). In a separate cohort of mice, we crossed P301S mice onto an apoE knockout (EKO) background, and found that 9-month P301S/EKO mice displayed the same pattern of p-tau reduction in the RAB and RIPA fractions compared to P301S mice (Figure 2E), supporting a key role of apoE in p-tau pathogenesis.
Figure 2. LDLR OX in P301S mice markedly reduces apoE and p-tau levels that highly correlate with each other.
(A) ApoE level in the CSF of 9-month WT, LDLR, P301S and P301S/LDLR mice measured by ELISA (n=6-17).
(B) ELISA measurement of apoE, human tau, and p-tau levels in RAB, RIPA and 70% FA fractions respectively for 9-month mouse posterior cortical lysates. Student’s t-test (two-sided) for human tau and ptau, no signal in WT and LDLR mice.
(C) Correlation between apoE and human tau level in RAB (non-significant), RIPA (no correlation) and FA (r2=0.4114, p<0.0001) fractions respectively in P301S and P301S/LDLR mice.
(D) Correlation between apoE level and p-tau level in RAB (P301S: r2=0.4649, p=0.0026; P301S/LDLR: r2=0.5690, p=0.0007), RIPA (r2=0.6124, p<0.0001), and FA (r2=0.3948, p<0.0001) fractions respectively in P301S and P301S/LDLR mice.
(E) ELISA measurement of p-tau levels in RAB, RIPA and 70% FA fractions of mouse brain lysates for a separate cohort of 9-month P301S and P301S/EKO mice. (n=13). Mann-Whitney test (two-sided).
Data expressed as mean ± SEM, One-way ANOVA with Tukey’s post hoc test, two-sided in (A) and (B), Pearson correlation analysis (two-sided) in (C) and (D). *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Microglial overexpression of the LDLR transgene reduces microglial Apoe expression and intracellular apoE level and is associated with less microglial activation
We previously showed that microglia are the principal driving force of tau pathogenesis and neurodegeneration, and apoE promotes tau pathogenesis and neurodegeneration predominantly by driving microglial activation (Shi et al., 2019). Thus, the reduced tau pathology and neurodegeneration upon LDLR OX may mainly result from attenuated microglial activation due to apoE reduction. How apoE activates microglia is still unclear. While the vastly reduced pool of extracellular apoE (primarily produced by astrocytes) may reduce microglial activation via non-cell autonomous mechanisms, microglia-derived apoE was shown to cell-autonomously promote microglial activation towards the neurodegenerative “MGnD/DAM” phenotype (Krasemann et al., 2017).
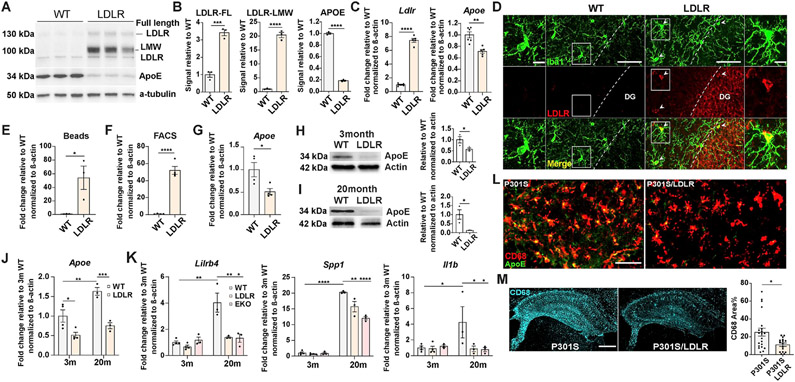
When we cultured primary microglia from the cortex of LDLR Tg and WT mouse littermates, we surprisingly found that in addition to the reported LDLR OX in neurons and astrocytes (Kim et al., 2009), microglia from LDLR Tg mice also overexpressed LDLR, particularly a low molecular weight form of LDLR (Figure 3A, B). Microglial overexpression of LDLR in LDLR Tg mice was subsequently confirmed in vivo by immunofluorescence staining (Figure 3D). Intriguingly, corresponding to increased Ldlr expression (Figure 3C), a significant reduction of Apoe gene expression (Figure 3C) as well as a marked reduction of intracellular apoE protein level (Figure 3A, B) was observed in cultured LDLR-overexpressing microglia. Similarly, when microglia were acutely isolated from adult mouse brain using CD11b-conjugated magnetic beads (Figure 3E), or via fluorescence-activated cell sorting (FACS) that yields highly pure microglia by using multiple cell type markers (Figure 3F, Figure S1), we consistently observed Ldlr overexpression in LDLR Tg microglia compared to WT microglia. Reduction of microglial Apoe expression (Figure 3G) and intracellular apoE protein levels was also observed in acutely-isolated LDLR Tg microglia (Figure 3H, I). These results suggest that Apoe downregulation was inherently associated with LDLR OX in microglia. Interestingly, the difference of intracellular apoE level between WT and LDLR Tg microglia was increased in aged mice (20-month) compared to young mice (3-month) (Figure 3H, I). As Apoe gene expression is upregulated in activated microglia under neurodegenerative conditions (Krasemann et al., 2017; Mathys et al., 2019; Zhou et al., 2020), the increased difference in apoE level is likely due to aging-induced microglial activation that is stronger in WT microglia (with a higher degree of Apoe upregulation) than in LDLR microglia. Indeed, we found microglial Apoe expression was significantly elevated in 20-month WT mice, but not in 20-month LDLR Tg mice compared to their 3-month counterparts (Figure 3J). This could suggest that LDLR OX and associated Apoe downregulation in microglia suppressed microglial activation. Supporting this idea, in microglia isolated from 20-month mice, LDLR Tg microglia, like EKO microglia, showed significantly reduced “DAM” and proinflammatory gene expression compared to WT microglia, whereas minimal levels of these gene expression changes and no differences in their expression level were observed in microglia isolated from 3-month mice (Figure 3K). In the setting of tauopathy, activated microglia (CD68+) in 9-month P301S mice almost invariably showed large punctate intracellular apoE staining; whereas in P301S/LDLR mice, minimal intracellular apoE, or only tiny apoE puncta were observed in activated microglia (Figure 3L). In association with the reduction of apoE levels in microglia, P301S/LDLR mice showed significantly attenuated microglial activation relative to P301S mice by CD68 staining (Figure 3M).
Figure 3. Microglial overexpression of the LDLR transgene reduces its Apoe expression and intracellular apoE level, and is associated with suppressed microglial activation.
(A, B) Western blot and quantification of apoE and LDLR in primarily cultured microglia isolated from P2 pups of WT and LDLR Tg mice (n=3, mixed sex).
(C) qPCR for Ldlr and Apoe in cultured microglia (n=5-6, mixed sex).
(D) Immunofluorescent staining of LDLR and Iba1 in 9-month WT and LDLR Tg mice. Column 1 and 4 (scale bar: 10μm): zoom-in images of square areas in column 2 and 3 (scale bar: 50μm) respectively. Arrow heads point to microglia overexpressing LDLR.
(E, F) qPCR for Ldlr in microglia acutely isolated from 3-month WT and LDLR Tg mice by magnetic beads (E) or by FACS (F) (n=3-4).
(G) qPCR for Apoe in FACS-sorted microglia from 3-month WT and LDLR Tg mice (n=4).
(H, I) Western blot and quantification of apoE level in acutely isolated microglia from 3-month (H) and 20-month (I) WT and LDLR Tg mice (n=3).
(J, K) qPCR for Apoe (J) and DAM/proinflammatory genes (K) in FACS-sorted microglia from 3-month and 20-month WT and LDLR Tg mice (n=3-4), two-way ANOVA with Tukey’s post hoc test, two-sided.
(L) Co-staining of apoE and CD68 in 9-month P301S mice and P301S/LDLR mice (scale bar: 50μm).
(M) Staining and quantification of CD68 in the hippocampus of 9-month P301S and P301S/LDLR mice (n=16-21), scale bar: 500μm. Mann-Whitney test (two-sided).
Data expressed as mean ± SEM. Student’s t-test (two-sided) in (B), (C), (E-I), *p<0.05, **p<0.01 ***p<0.001, ****p<0.0001. See also Figure S1.
LDLR OX does not affect tau pathology and brain volume at an early age prior to microglial activation
At 3-months of age, an early time point before activation of DAM microglia (Figure S2A), LDLR OX did not affect early ptau histopathology that is present in the mossy fibers of the hippocampus (type1 ptau, not present in WT mice) (Figure S2B). Biochemically, no differences in tau or ptau levels were detected despite a similar level of apoE reduction in P301S/LDLR mice (Figure S2C). The brains were generally intact at this stage, and no volume differences were observed between P301S and P301S/LDLR mice (Figure S2D).
Single-nuclei RNAseq identifies changes in distinct brain cell populations and in cell-type-specific DEGs under neurodegenerative, apoE-deficient and LDLR-OX conditions
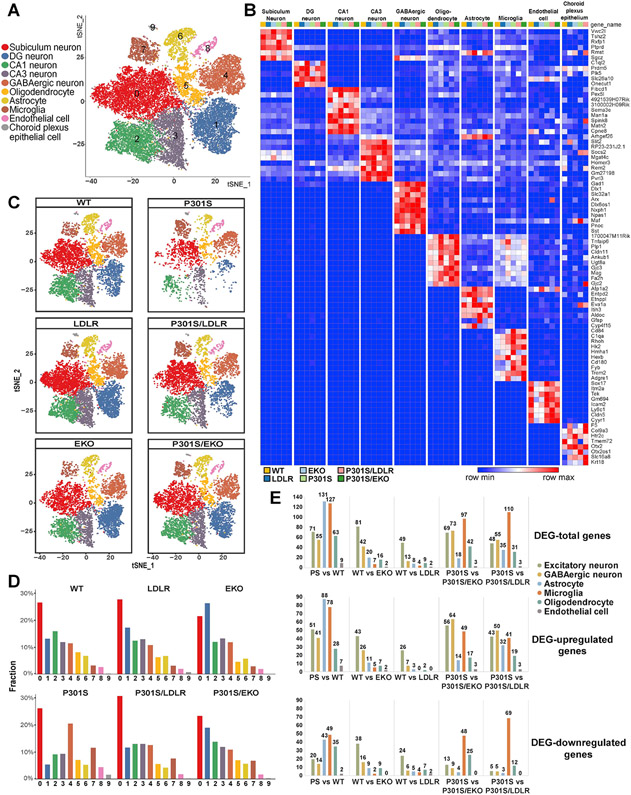
To further understand how apoE and LDLR-OX regulate neurodegeneration, we performed single nuclei RNAseq (snRNAseq) in 9-month old P301S, P301S/LDLR, P301S/EKO mice as well as their aged-matched non-tau Tg WT, LDLR and EKO littermates. Hippocampi from 7-8 mice per genotype were pooled together as a single sample. snRNAseq was performed using the 10x Genomics platform. We obtained sequencing data from a total of 31785 nuclei, and identified 10 distinct cell clusters across all samples based on the expression of known cell-type-specific markers, including neurons (cluster 0, 1, 2, 3, 4), oligodendrocytes (cluster 5), astrocytes (cluster 6), microglia (cluster 7), endothelial cells (cluster 8), and choroid plexus epithelial cells (cluster 9) (Figure 4A, B, Figure S3). Cell population analysis revealed that neurodegeneration in P301S mice featured loss of specific subtypes of excitatory hippocampal neurons, including dentate gyrus (DG), CA1 and CA3 neurons, particularly DG neurons, but had little impact on subiculum neurons (Figure 4C, D). The proportion of inhibitory neurons, in contrast, was increased in P301S mice compared to other groups (Figure 4C, D). Notably, microglia were markedly increased in P301S mice, and were only mildly increased in P301S/LDLR and P301S/EKO mice, whereas the astrocyte population remained stable across groups (Figure 4C, D).
Figure 4. Single-nucleus RNAseq identifies changes in major brain cell types and cell type-specific DEGs under neurodegenerative, apoE-deficient and LDLR-overexpressing conditions.
(A) t-SNE plot showing 10 clusters (0–9) identified in all nuclei from 9-month P301S, P301S/LDLR, P301S/EKO, WT, LDLR, and EKO mouse hippocampus.
(B) Heatmap showing distinct cell types identified by cell type-specific markers.
(C) Overview of all cell populations in six groups of mice by t-SNE plots.
(D) Relative frequency of all cell clusters in each genotype.
(E) The number of DEGs compared between different genotypes in major brain cell types. Top: total number of DEGs; middle: number of upregulated DEGs; bottom: number of downregulated DEGs (former versus latter genotype).
Differentially expressed genes (DEGs) analysis (Figure 4E, Table S1) showed that neurodegeneration-associated gene expression change (P301S vs. WT) mainly occurred in microglia and astrocytes. In the absence of the tau transgene, apoE- and LDLR-associated gene expression changes (WT vs. EKO or WT vs. LDLR) were primarily enriched in neurons, particularly in excitatory neurons. However, in the setting of tauopathy, the highest level of gene expression change shifted from neurons to microglia in both conditions (P301S vs. P301S/EKO, P301S vs. P301S/LDLR), suggesting a key role of microglia in mediating the effect of apoE and LDLR on neurodegeneration.
Apoe expression is upregulated uniquely in microglia over other brain cell types in tau-associated neurodegeneration
In a normal brain, apoE is produced predominantly by astrocytes, and to a much less extent, by microglia (Boyles et al., 1985; Uchihara et al., 1995). ApoE expression in neurons is minimal under physiological conditions, but can be upregulated upon neuronal injury in certain conditions (Xu et al., 2006). Here we found that compared to WT mice, P301S mice showed a marked increase of Apoe expression that occurred uniquely in microglia over other cell types (Figure S4A). Correspondingly, in P301S mice, apoE immunostaining revealed intracellular apoE accumulation as large bright puncta in activated microglia; whereas in WT mice, apoE signal was primarily present in astrocytes (Figure S4B). The distinctive upregulation of apoE in microglia in P301S mice highlights the potential role of microglia-derived apoE in neurodegeneration. Compared to P301S mice, Apoe expression in P301S/LDLR mice was notably lower in microglia, but was only mildly reduced in astrocytes (Figure S4A), further supporting a key role of microglial Apoe in mediating LDLR’s protective effect.
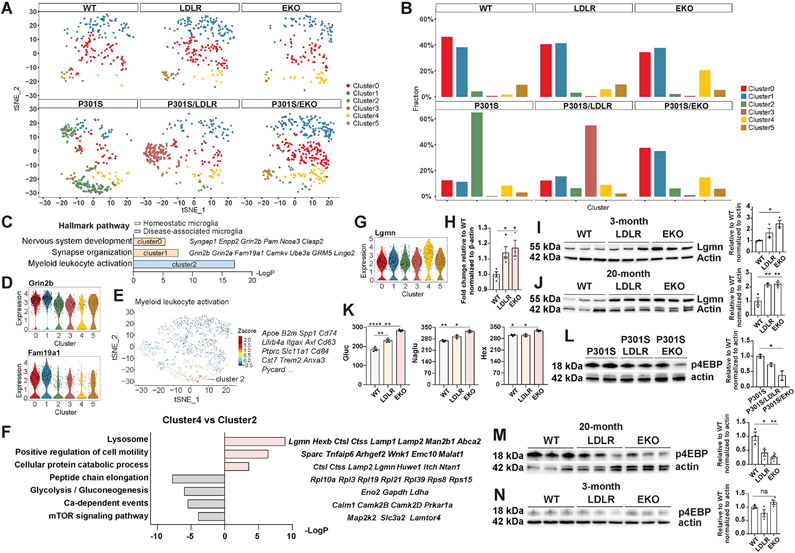
Microglia sub-clustering reveals distinct microglial subpopulations associated with neurodegeneration, apoE deficiency, or LDLR-OX
Given the central role of microglia in neurodegeneration, we assessed the microglia population (pooled from all samples, a total of 1633 nuclei) with sub-clustering. Unsupervised clustering yielded six subclusters among all samples (Figure 5A). We identified two subpopulations of homeostatic microglia (cluster 0 and 1) that were enriched in non-tau Tg mice, but were drastically reduced in P301S mice (Figure 5A, B). These subsets of microglia showed high expression of genes involved in regulating synaptic functions and neuronal development, such as Grin2b, Grin2a, Fam19a1, Camkv and others (Figure 5C, D). In contrast, cluster 2 was specifically enriched in P301S mice (Figure 5A, B), and featured hallmark “DAM/MGnD” genes such as Apoe, Spp1, Cd74, Lilrb4a, Itgax, Axl, Trem2 and B2m (Figure 5C, E), representing a population of activated microglia in neurodegeneration. Notably, cluster 3 was uniquely enriched in P301S/LDLR mice (Figure 5A, B), but not in LDLR mice, indicating a specific population of microglia associated with disease but whose property was regulated by LDLR OX. Cluster 4 was specifically associated with apoE deficiency, as it was enriched in apoE-deficient mice, regardless of the presence or absence of tau transgene (Figure 5A, B). Interestingly, this subset of microglia also appeared to be a type of activated microglia in the setting of apoE deficiency, as it clustered together with cluster 2 DAM microglia when using a lower resolution for cell clustering. Understanding how these “activated” classes of microglia (cluster 2, 3, 4) differ from each other could provide key insights into the role of apoE and LDLR in regulating microglial activation.
Figure 5. Microglia sub-clustering identifies distinct microglial subpopulations and uncovers a subset of microglia associated with apoE deficiency that show enhanced catabolism and suppressed mTOR activation.
(A) Microglia subclusters in each genotype.
(B) Relative frequency of microglia subclusters in different genotypes.
(C-E) Hallmark genes and pathways in homeostatic microglia (cluster 0, 1) (C, D) and disease-associated microglia (cluster 2) (C, E).
(F) Top upregulated and downregulated pathways with featured genes in apoE deficiency-associated microglia (cluster 4) compared to DAM microglia (cluster 2).
(G) Violin plot showing upregulation of the lysosomal gene Lgmn in apoE deficiency-associated microglia (cluster 4).
(H) qPCR for Lgmn in FACS-sorted microglia from 3-month WT, LDLR and EKO mice (n=3-4).
(I, J) Western blot and quantification of LGMN in microglia acutely isolated from 3-month (I) and 20-month (J) WT, LDLR and EKO mice (n=3).
(K) Enzyme activities of β-hexosaminidase (Hex), β-glucuronidase (Gluc) and N-Acetyl-Alpha-Glucosaminidase (Naglu) in cortices of 9-month WT, LDLR, EKO mice (n=3).
(L) Western blot and quantification of phospho-4EBP in microglia acutely isolated from 11-month female P301S, P301S/LDLR, and P301S/EKO mice (n=2).
(M, N) Western blot and quantification of phospho-4EBP in microglia acutely isolated from 20-month (M) and 3-month (N) WT, LDLR, and EKO mice (n=3).
Data expressed as mean ± SEM, One-way ANOVA with Tukey’s post hoc test, two-sided for all quantifications. *p<0.05, **p<0.01, ****p<0.0001. Expression level on the Y-axis of violin plots is normalized, natural log-transformed gene count.
See also Figure S5.
ApoE deficiency-associated microglia downregulate anabolic pathways and show robust upregulation of catabolic genes
Compared to cluster 2 “DAM” microglia, a prominent feature of cluster 4 microglia was upregulation of catabolic genes, and downregulation of anabolic genes as well as microglial activation pathways (Figure 5F). The largest group of upregulated genes in cluster 4 encode lysosomal enzymes and proteins involved in cellular degradation, including Lgmn, Ctss, Ctsl, Hexb, Man2b1, Lamp1, Lamp2, Abca2 and others (Figure 5F, G). We verified upregulation of Lgmn in acutely isolated microglia from EKO mice compared to that from WT mice by qPCR (Figure 5H), and found increased LGMN protein level in acutely isolated microglia from both young (3-month) and aged (20-month) EKO mice compared to WT mice by western blot (Figure 5I, J). LDLR microglia showed a similarly increased level of LGMN compared to WT microglia, indicating its potentially shared mechanisms with apoE-deficient microglia in elevating cell catabolism that was likely due to apoE reduction. Downregulated genes in cluster 4 microglia primarily fell into anabolic pathways, such as “peptide chain elongation” characterized by ribosome protein genes; “glycolysis”, including genes encoding glycolytic enzymes such as Eno2, Gapdh, and Ldha; and “mTOR signaling”, including genes directly involved in or regulating mTOR activation, such as Map2k2, Slc3a2, and Lamtor4 (Figure 5F). Cell metabolism is a key mechanism linked to microglial activation. Activated immune cells increase anabolism and rewire their metabolism towards enhanced glycolysis (Bories and Leitinger, 2017). ApoE deficiency-associated activation of cell catabolism and suppression of cell anabolism likely constitute a crucial mechanism preventing microglial activation.
ApoE deficiency boosts microglial lysosomal enzyme activities under physiological conditions, and reduces microglial mTOR activation and lipid/cholesterol accumulation upon microglial activation
Because metabolic changes are crucial in regulating microglial activation, we decided to verify whether lysosome-associated catabolic capacity in microglia is altered by apoE. We assessed the activity of three lysosomal enzymes - β-hexosaminidase (Hex), β-glucuronidase (Gluc) and N-Acetyl-Alpha-Glucosaminidase (Naglu) in mouse cortices from 9-month old WT, LDLR Tg and EKO mice. These enzymes are predominantly expressed by microglia. Therefore, the results largely reflect microglial lysosomal enzyme activity. We observed significantly increased activity of all three enzymes in EKO mice compared to WT mice (Figure 5K), indicating that apoE deficiency resulted in enhanced lysosomal functions under physiological conditions. LDLR mice also showed a moderate increase in enzyme activities compared to WT mice (Figure 5K), which may be related to apoE reduction.
In addition to enhanced lysosomal catabolic functions, immunoblotting of acutely isolated microglia from 11-month P301S, P301S/LDLR and P301S/EKO female mice showed reduced phosphorylation of 4EBP (p4EBP) - an mTORC1 effector, in P301S/EKO microglia compared to P301S microglia (Figure 5L), indicating reduced mTOR activation and mTOR-mediated anabolism in apoE-deficient microglia in the setting of tauopathy. Interestingly, even in the absence of the tau transgene, microglia isolated from aged (20-month) EKO mice showed a similar reduction of p4EBP compared to microglia from WT mice (Figure 5M). This effect was not observed in 3-month old mice (Figure 5N). As aging represents a mild neurodegenerative state that activates microglia, but doesn’t contain other confounding factors such as tau pathology, these results indicate that apoE-deficiency inherently suppresses mTOR activation upon microglial activation, likely by enhancing microglial catabolism. Interestingly, LDLR microglia showed an intermediate level of p4EBP compared to WT and EKO microglia; a similar pattern of p4EBP levels was also observed in P301S/LDLR microglia compared to P301S and P301S/EKO microglia (Figure 5L-N), suggesting that LDLR OX shares certain common mechanisms with apoE deficiency in controlling microglial metabolism, likely via apoE reduction.
LDLR-associated microglia upregulate genes encoding specific subsets of ion channels and neurotransmitter receptors and downregulate microglial activation pathways
In addition to similarities to EKO microglia, LDLR-associated microglia (cluster 3) also harbored unique features. Compared to cluster 2, top downregulated pathways in cluster 3 include “myeloid leukocyte activation”, “antigen processing and presentation”, and “myeloid leukocyte differentiation”, all featuring genes involved in microglial activation, including “DAM” genes such as Apoe, Cd74, Gpnmb, Lilrb4; MHC-related genes such as H2-K1 and B2m; the complement gene C1qc, and others (Figure S5A). In contrast, upregulated pathways featured “regulation of synaptic plasticity”, “regulation of ion transport”, and “regulation of membrane potential”. These pathways highlighted genes encoding ion channels and neurotransmitter receptors, such as voltage-gated K+ and Na+ channels (Kcnc1, Kcnc3, Scn2a, Scn2b), Na+/K+-ATPase (Atp1a1), NMDA receptor (Grin2c), GABA-A receptor (Gabra6), and metabotropic glutamate receptor (Grm4) (Figure S5A, B). We confirmed higher expression of top upregulated genes such as Gabra6 and Grm4 in acutely isolated microglia from 20-month LDLR Tg mice compared to WT mice, whereas no differences were observed in 3-month mice (Figure S5C, D). Microglia express major classes of neurotransmitter receptors and ion channels that critically regulate microglial physiology, which inherently controls microglial function and activation (Domercq et al., 2013; Izquierdo et al., 2019). The unique alteration in gene expression of these ion channels and receptors likely constitutes an important aspect of LDLR’s function in regulating microglial activation.
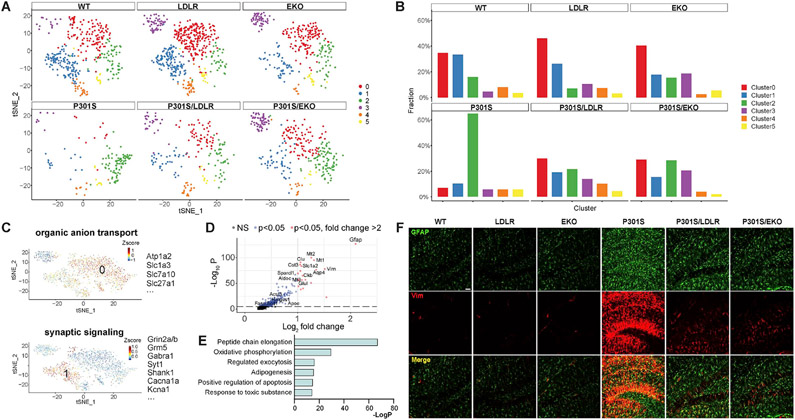
Astrocyte sub-clustering identifies features of homeostatic and reactive astrocytes
Astrocytes are another major immune-competent cell population in the brain, and have long been proposed to play a role in neurodegeneration (Ben Haim et al., 2015; Liddelow et al., 2017). However, the detailed transcriptional profile of astrocytes in tauopathy and in relation to apoE status has not been fully characterized. We performed sub-clustering of astrocytes (pooled from all samples, a total of 1863 nuclei), and identified homeostatic astrocyte populations and a reactive astrocyte subset, as well as a subpopulation of astrocytes enriched in apoE-deficient mice (EKO and P301S/EKO) (Figure 6A, B).
Figure 6. Astrocyte sub-clustering identifies homeostatic and reactive astrocytes and a subpopulation enriched in apoE-deficient and LDLR-overexpressing mice.
(A) Astrocyte subclusters in each genotype.
(B) Relative frequency of astrocyte subclusters in different genotypes.
(C) Hallmark pathways and top enriched genes in homeostatic astrocytes (cluster 0, 1).
(D) Volcano plot showing genes enriched in reactive astrocytes (cluster 2).
(E) Top upregulated pathways in reactive astrocytes (cluster 2).
(F) Staining of reactive astrocytes with GFAP and Vimentin in 9-month old WT, LDLR, EKO, P301S, P301S/LDLR, and P301S/EKO mice. Scale bar: 50μm.
Among the six astrocyte subclusters, cluster 0 and cluster 1 shrunk drastically in P301S mice (Figure 6A, B), thus representing homeostatic states. Cluster 0 featured “organic anion transport”, and was enriched with genes involved in molecular transport, such as Atp1a2 and solute carrier genes Slc1a3, Slc7a10, Slc27a1 and Slco1c1 (Figure 6C). Cluster1 was featured by “synaptic signaling”, containing genes encoding various ion channels, glutamate and GABA receptors, and proteins essential for synaptic functions (Figure 6C).
Cluster 2 was highly enriched in P301S mice (Figure 6A, B), representing a population of reactive astrocytes. Top upregulated genes in cluster 2 (Figure 6D) included cytoskeleton genes (Gfap, Vim); the AD risk gene Clu; genes protecting against oxidative stress (Mt1, Mt2, Mt3, Prdx6, Prdx1); genes involved in glycolysis (Aldoc) and maintenance of ATP homeostasis (Ckb); and genes removing excess glutamate and K+ and maintaining extracellular volume (Slc1a2, Glul, Aqp4, Kcnj10, Gja1). In particular, a large group of genes involved in lipid synthesis were upregulated, such as Srebf1, Hmgcs1, Fasn, Acsl3, Acsbg, Acsl6, Scd1, Abca1, and Apoe. Genes encoding matricellular proteins, such as Sparcl1 and Cst3, were also highly upregulated.
Pathway analysis revealed multiple highly significantly upregulated pathways in cluster 2 astrocytes (Figure 6E), with the top pathway being “peptide chain elongation” that featured a large array of genes encoding ribosome proteins and translation elongation factors. Immediately following this was “oxidative phosphorylation”, highlighted by genes encoding proteins comprising the electron transport chain complexes and the ATP synthase, as well as glycolytic genes. Other highly upregulated pathways included “regulated exocytosis”, “adipogenesis”, “positive regulation of apoptotic process”, and “response to toxic substance”.
These gene and pathway alterations depicted the phenotype of reactive astrocytes in tauopathy as 1) highly active in protein synthesis and ATP production, generating large amounts of ROS and likely other toxic substances with concomitant enhanced detoxification capacity; 2) retaining and reinforcing certain neuroprotective features of homeostatic astrocytes, particularly to counteract neuronal excitotoxicity; 3) activated in lipid synthesis, likely to regulate cell signaling and immune response, to supply energy production through β-oxidation, and to assist in neuronal repair; and 4) boosted in exocytosis and extracellular matrix signaling that regulate neuronal function and immune response. We verified by immunofluorescent staining (GFAP, Vimentin) that reactive astrocytes were highly activated in 9-month P301S mice, but were only mildly increased in P301S/LDLR and P301S/EKO mice (Figure 6F). Functional characterization of reactive astrocytes in neurodegeneration need to be further addressed in future studies.
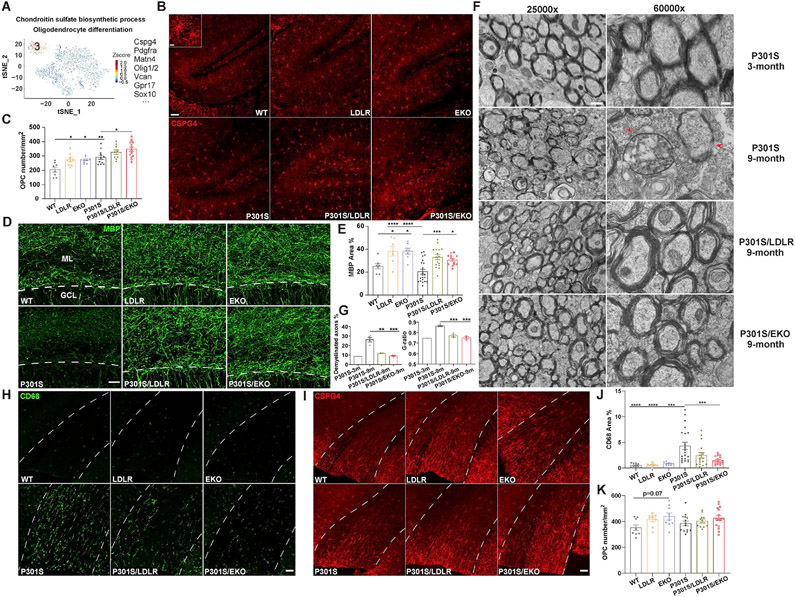
ApoE-deficient and LDLR-overexpressing mice harbor enlarged pools of oligodendrocyte progenitor cells in the brain and show greater preservation of myelin integrity under neurodegenerative conditions
Interestingly, in the astrocyte subclusters, cluster 3 was strongly enriched in EKO and P301S/EKO mice (~20%), and to a less extent in LDLR and P301S/LDLR mice (10-13%) compared to WT and P301S mice (3-5%) (Figure 6A, B). The uniquely high expression of Cspg4 and Pdgfra (Figure 7A) - markers of oligodendrocyte progenitor cells (OPCs, also known as NG2 glia) (Nishiyama et al., 2009), indicated that this cluster represented OPCs. Other conventional OPC genes such as Olig2, Olig1 and Sox10 were also among the top enriched genes in cluster 3. Pathway analysis highlighted “chondroitin sulfate biosynthetic process” and “oligodendrocyte differentiation” (Figure 7A), confirming their identity as OPCs.
Figure 7. ApoE-deficient and LDLR-overexpressing mice harbor an enlarged pool of oligodendrocyte progenitor cells, and show preserved myelin integrity in tauopathy.
(A) Astrocyte subcluster 3 in Figure 6A, B shows hallmark genes and pathways of OPCs.
(B) Staining of CSPG4 in all six groups of mice at 9-month of age (scale bar: 50μm). Inset shows enlarged image of OPCs (scale bar: 10μm).
(C) Quantification of OPC density in the molecular layer of DG in the hippocampus (n=8-14)
(D, E) MBP staining and quantification of MBP covered area in the molecular layer of DG (n=7-21). Above dash line: ML (molecular layer); below dash line: GCL (granule cell layer), scale bar: 25μm.
(F) EM images of corpus callosum in posterior brain regions of 3-month P301S, 9-month P301S, 9-month P301S/LDLR, and 9-month P301S/EKO mice. Left: 25000X magnification, scale bar: 600nm; Right: 60000X magnification, scale bar: 200nm. Arrowheads point to axons with myelin damage.
(G) Quantification of axonal G-ratio and the percentage of demyelinated axons (myelin layers ≤ 2) in EM images (n=1-4)
(H, J) Staining and quantification of CD68 in the corpus callosum (n=9-21). Corpus callosum located between the two dashed lines. Scale bar: 50μm.
(I, K) Staining and quantification of CSPG4 in the corpus callosum (n=9-16). Scale bar: 50μm.
Data expressed as mean ± SEM, One-way ANOVA with Turkey’s post hoc test, two-sided for all quantifications. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. See also Figure S6.
OPCs are a type of progenitor cells that mainly differentiate into mature oligodendrocytes. We confirmed increased density of OPCs in the molecular layer of dentate gyrus (DG) in the hippocampus of 9-month EKO and LDLR Tg mice compared to WT mice by immunofluorescent staining with the OPC marker CSPG4 (Figure 7B, C). Similar differences were also observed in tau Tg mice (Figure 7B, C). The density of OPCs was increased in P301S mice compared to WT mice, suggesting a potential role of proliferating OPCs in repairing damaged myelin in neurodegeneration. The enlarged OPC pools in EKO and LDLR Tg mice may be essential in promoting oligodendrocyte genesis and maintaining myelin integrity under neurodegenerative conditions. Supporting this idea, we found significantly more preserved myelin structure in the molecular layer of DG in P301S/LDLR and P301S/EKO mice compared to P301S mice by myelin basic protein (MBP) staining (Figure 7D, E). Surprisingly, even in the absence of tau pathology, LDLR and EKO mice showed higher myelin coverage in the molecular layer compared to WT mice (Figure 7D, E), suggesting intrinsic regulation of myelination by apoE/LDLR, likely in part through regulating OPCs.
Through electron microscopy, we also identified notable signs of myelin damage in the corpus callosum of 9-month P301S mice, which showed a significantly higher percentage of demyelinated axons as well as increased axonal G-ratio compared to 9-month P301S/LDLR and P301S/EKO mice, or 3-month P301S mice (Figure 7F, G). Corresponding to increased myelin damage, P301S mice showed marked microglial activation in the corpus callosum (Figure 7H, J), likely to clear damaged myelin. OPCs in the corpus callosum trended at a higher density in apoE-deficient and LDLR-overexpressing mice with or without tau transgene (Figure 7I, K), supporting their potential roles in maintaining myelin integrity.
Interestingly, a similar protective effect of apoE-deficiency and LDLR-OX on myelin integrity was also observed in aged mice in the absence of tauopathy. Axonal demyelination and altered G-ratio also occurred in the corpus callosum of 24-month old WT mice relative to 9-month WT mice, and this damage was significantly attenuated in 24-month LDLR Tg and EKO mice (Figure S6A-C). In the hippocampus, aged LDLR and EKO mice also showed a higher myelin density in the molecular layer compared to WT mice (Figure S6D, E). Regulation of the OPC pool and myelin homeostasis by apoE/LDLR may constitute a new mechanism underlying their effects on neurodegeneration.
Discussion
Here we found that LDLR OX protects against tau pathology and tau-associated neurodegeneration, suggesting LDLR as a new therapeutic target for treating tauopathy. A recent study shows that selective removal of astrocyte-derived apoE4 (but not apoE3) reduces tau-associated neurodegeneration (Wang et al., 2021). In our study, LDLR OX drastically reduces extracellular murine apoE that is mainly derived from astrocytes. Therefore, the protective effect may be partially due to reduction in this pool of apoE. However, it’s particularly interesting to see cell-autonomous downregulation of microglial Apoe by LDLR, which makes microglial LDLR a novel target to control microglial function. The other primary apoE metabolic receptor, LRP1, has been shown to mediate tau uptake in vitro and drastically increase the propagation of the P301L human tau protein expressed by an AAV virus in vivo (Rauch et al., 2020), which contrasts with our finding of a protective effect of LDLR on tau pathology in a tauopathy model. Although LDLR may affect tau propagation as a tau receptor similar to LRP1, here we didn’t observe exacerbation of early tau pathology in P301S/LDLR mice prior to microglial activation, suggesting that LDLR itself may not significantly impact pathological tau spreading. However, further study is required to carefully address this question. On the other hand, we find LDLR OX leads to a strong reduction in extracellular apoE levels and microglial Apoe downregulation that are directly linked with suppressed microglial activation. ApoE reduction doesn’t affect early tau pathology prior to microglial activation, suggesting that its protective effect on tau pathology is likely carried out at a later stage by regulating microglial activation under neurodegenerative conditions. Given that microglia are the principal driving force of tau pathogenesis and neurodegeneration in the tauopathy model, the effect of LDLR on suppressing microglial activation via apoE regulation may overwhelm any potential direct effects of LDLR on tau spreading. Modulating LRP1 has also been reported to affect brain apoE levels (Liu et al., 2007; Zerbinatti et al., 2006), yet its effects on microglial activation and tau-associated neurodegeneration have not been assessed. Therapies targeting these receptors should take both aspects into consideration, and should be carefully assessed to achieve therapeutic effects.
One key finding of our study is the role of apoE in regulating microglial metabolism. Cell metabolism plays a central role in controlling microglial activation. Proinflammatory “M1” microglia show a profound metabolic change characterized by increased glucose uptake and a switch from oxidative phosphorylation (OXPHOS) to glycolysis even in normoxic conditions (Warburg effect) (Bories and Leitinger, 2017). Disease-associated microglia shared many features with “M1” microglia and showed increased anabolism, whereas apoE deficiency-associated microglia showed enhanced catabolism and suppressed anabolism. Interestingly, TREM2-deficient microglia were reported to have increased autophagy and catabolism, impaired mTOR activation and anabolism, and reduced ATP production (Ulland et al., 2017). The defective anabolic and energetic metabolism in TREM2-deficient microglia was shown to underlie their ineffective activation. Interestingly, TREM2 deficiency also protects against tau-associated neurodegeneration in P301S mice (Leyns et al., 2017). APOE and TREM2 act in the same axis in regulating many aspects of microglial functions (Shi and Holtzman, 2018). The similar metabolic changes in apoE-deficient and TREM2-deficient microglia suggests that the inability to turn on the anabolic machinery in apoE-deficient microglia is likely a key mechanism preventing their activation. LDLR Tg microglia showed a similar metabolic pattern and reduced activation as apoE-deficient microglia, likely in part due to cell-autonomous reduction in Apoe expression. Lower extracellular apoE level may also contribute to suppressed microglial activation through reduced microglial signaling.
It’s interesting to see upregulation of specific ion channels and neurotransmitter receptors in LDLR Tg microglia in tauopathy. Microglia express a broad spectrum of neurotransmitter receptors and ion channels that critically regulate microglial functions such as surveillance (Madry et al., 2018), cytokine release (Nguyen et al., 2017), chemotaxis, and phagocytosis (Haynes et al., 2006; Koizumi et al., 2007; Ohsawa et al., 2007). Development of outward K+ current in microglia is associated with an advanced microglial activation status (Boucsein et al., 2000; Wendt et al., 2017), and is largely mediated by voltage-gated K+ channels, particularly Kv1.3, which critically regulates proinflammatory cytokine generation (Di Lucente et al., 2018). LDLR OX upregulates Kv3.1 (Kcnc1) and Kv3.3 (Kcnc3). The Kv3 channels have a 10-50 mV higher activation threshold than any other known mammalian voltage-gated K+ channels (Coetzee et al., 1999). Hence, increased Kv3 channel expression in microglia may reduce proinflammatory microglial activation. In addition, the high expression of GABA receptor (Gabra6, second most enriched DEG in cluster3 microglia) may also play a role in inhibiting microglial cytokine release (Kuhn et al., 2004) and attenuating microglial activation.
The discovery that apoE deficiency promotes the expansion of OPCs is intriguing. The ability of apoE to inhibit cell proliferation has also been reported in other cell types, such as hippocampal neural stem/progenitor cells (NSC/NPC) (Yang et al., 2011) and hematopoietic cells (Murphy et al., 2011). ApoE’s effect appears to occur mainly in a cell-autonomous manner by regulating cellular cholesterol content, which critically regulates the level of membrane-associated signaling complexes that are essential for triggering cell proliferation (Murphy et al., 2011). LDLR OX also increases the OPC population, which is likely due to apoE reduction, or may also be contributed by a direct effect of LDLR on cellular lipid/cholesterol homeostasis. The discovery of apoE/LDLR’s effects on OPCs and myelination provides a novel perspective in understanding their roles in neurodegeneration.
STAR★Methods
RESOUECE AVAILABILITY
Lead Contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, David M. Holtzman (holtzman@wustl.edu).
Materials Availability
This study did not generate new unique reagents.
Data and Code Availability
Data that support the findings of this study are available from the corresponding author upon request. snRNAseq raw data has been deposited to GEO, accession number GSE166261.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
P301S mice overexpressing human P301S 1N4R tau driven by the PrP promoter were bred on a C57BL/6 background. Male P301S mice start developing overt tau pathology from ~5-6 months of age and show severe brain atrophy at 9 months of age. Female mice have a slightly delayed disease onset than male mice by 1~2month. LDLR Tg mice were generated in house previously on a B6/CBA background expressing murine LDLR under the PrP promoter (Kim et al., 2009). Briefly, murine LDLR was cloned into the pcDNA3.1 vector using mRNA isolated from mouse brain. LDLR cDNA was then excised by XhoI and inserted into the cloning site of the mouse prion promoter vector MoPrP.Xho (Borchelt et al., 1996), which is then used to generate LDLR Tg mice. LDLR Tg mice were crossed to P301S mice to generate P301S and P301S/LDLR littermates. WT mice were purchased from Charles River Laboratories (#027). ApoE KO mice (C57BL/6) were purchased from The Jackson Laboratory (#002052). P301S mice were crossed to ApoE KO mice to generate P301S and P301S/EKO littermates. Male mice were used for all experiments, except for a few cases where the sex of mice was specifically stated in the figure legends. The age of mice used in each experiment was stated in the main text and in the respective figure legends. Mice were maintained under 12:12 light/dark cycles in a pathogen-free condition, with food and water provided ad libitum. All animal procedures and experiments were performed under guidelines approved by the animal studies committee at Washington University School of Medicine.
Primary cell cultures
WT mice were bred to LDLR Tg mice to generate WT and LDLR littermates. Microglia derived from individual P2 pups (mixed sex) were cultured separately as independent samples.
METHOD DETAILS
Primary microglia culture
Cortices of P2 pups were dissected in calcium- and magnesium-free Hanks' Balanced Salt solution (HBSS) with careful strip of meninges. Tissue was digested in HBSS containing 0.25% trypsin (GIBCO, Cat# 15090-046) and 0.2mg/ml DNase (Sigma-Aldrich, Cat# DN25) at 37 °C for 10 min, and was dissociated in HBSS containing 0.4mg/ml DNase using a fire-polished Pasteur glass pipette and filtered through a 70-μm nylon mesh. Filtered material was pelleted at 1000 g for 5 min, washed with glia medium (DMEM + 10% heat-inactivated fetal bovine serum + 1x pen/strep + 1x GlutaMAX, all from Gibco) once, and plated onto 10cm plates that had been coated with 10 μg/ml Poly-L-lysine (PLL, Sigma-Aldrich, Cat# P2636) in glia medium. The cells were replaced with new glia medium the next day and were cultured at 37°C for a week to allow the formation of a confluent layer of astrocytes. Fresh medium was then added, and the cells were cultured for an additional week to allow microglia to grow on top of astrocytes. Loosely attached microglia were then washed off the astrocyte layer with gentle pipetting, and were seeded onto 24-well plates coated with PLL at a density of 2×105 cells per well in glia medium. The next day, half of the glia medium were replaced with serum free medium (SFM: DMEM/F12 [Gibco]+ 1x GlutaMAX [Gibco] + 1x NEAA [Gibco] + 1x ITS-G [Gibco] + 1x B-27 [Gibco] + 1x N2 [Gibco] + 200uM 1-Thioglycerol [Sigma-Aldrich] + 5ug/mL insulin [Sigma-Aldrich] + 50 ng/mL TGFβ [R&D Systems] + 100 ng/mL IL34 [R&D Systems] + 25 ng/mL MCSF [R&D Systems]) that was described previously (TCW et al., 2019). On the 3rd day, microglia were washed once with SFM and switched to SFM. Microglia were cultured in SFM for two days, and were then lysed in RIPA buffer supplemented with protease inhibitors (cOmplete, Roche, Cat# 04693132001) and phosphatase inhibitors (PhosSTOP, Roche, Cat# 4906837001). Cell lysates were spun down at 15000rpm for 10min to remove cell debris. The supernatants were used for western blotting.
Volumetric analysis
The left hemi-brain of each mouse was sectioned coronally at 50 μm using a freezing sliding microtome. Every sixth coronal brain section (300 um between sections) starting from the appearance of hippocampus to the dorsal end of the hippocampus were mounted for each mouse. All mounted sections were stained with 0.1% Sudan black (Sigma-Aldrich) in 70% ethanol at RT for 20 min, then washed in 70% ethanol for 1 min, 3 times. The sections were finally washed in Milli-Q water for 3 times and coverslipped with Fluoromount-G mounting medium (SouthenBiotech, Cat# 0100-01). The stained slices were imaged with the NanoZoomer and areas of interest were traced and measured in each slice using the NDP viewer. The volume was calculated using the following formula: volume = (sum of area) * 0.3 mm. The piriform/entorhinal cortex was traced from bregma −2.3 and to bregma −3.9.
Immunohistochemistry
Three sections from each mouse (300 um apart), corresponding approximately to bregma coordinates −1.4, −1.7, and −2.0 mm respectively, were used for p-tau staining. Brain sections were washed in Tris-buffered saline (TBS) buffer 3 times followed by incubation in 0.3% hydrogen peroxide in TBS for 10 min at RT. After 3 washes in TBS, sections were blocked with 3% milk in 0.25% TBS-X (Triton X-100) for 0.5 h followed by incubation at 4°C overnight with biotinylated AT8 antibody (Ser202/Thr205, Thermo Fisher Scientific, Cat# MN1020B). The next day, the slices were developed using the VECTASTAIN Elite ABC HRP Kit (Vector laboratories, Cat# PK-6100) following manufacturer’s instructions. Stained sections were imaged by the NanoZoomer digital pathology system and pathology was quantified using Image J. For immunofluorescent staining, sections were washed in TBS 3 times, and blocked in blocking buffer (3% BSA + 3% goat or donkey serum in 0.3% TBS-X) for 0.5h at RT, followed by overnight incubation at 4°C with primary antibodies (Synapsin 1: Synaptic Systems, Cat# 106103; CD68: AbD SeroTec, Cat# MCA1957; GFAP: Abcam, Cat# ab7260; ApoE: HJ6.3, in house; Iba1: Wako, Cat# 019-19741; LDLR: R&D Systems, Cat# AF2255; Vimentin: Santa Cruz, Ca# sc-373717, CSPG4: Sigma, Cat# AB5320; MBP: Abcam, Cat# ab40390) in stain buffer (1% BSA + 1% goat or donkey serum in 0.3% TBS-X). The next day, the sections were washed in TBS and incubated with fluorescence-labeled secondary antibodies (Invitrogen) for 2h at RT (For BODIPY staining, brain sections were directly incubated in 1ug/ml BODIPY in PBS for 30min at RT). The slices were then washed and mounted in prolong gold antifade mounting media (Invitrogen, Cat# P36931). Images in Figure 1C and Figure 3M were taken by an epifluorescence microscope and quantified using ImageJ. Images in Figure 3D, L, Figure 6F, Figure 7B, Figure S2A, and Figure S4B were taken by a Nikon A1Rsi Confocal Microscope. Images in Figure 7D, H, I and Figure S6D were taken by a Leica STELLARIS Confocal Microscope.
Brain extraction
Mouse posterior cortex was processed in RAB, RIPA and 70% FA buffer sequentially. Briefly, the tissue was weighed and homogenized using a pestle at 10ul buffer/1mg tissue in RAB buffer (100mM MES, 1mM EGTA, 0.5mM MgSO4, 750mM NaCl, 20mM NaF, 1mM Na3VO4, pH=7.0, supplemented by protease inhibitors (cOmplete, Roche) and phosphatase inhibitors (PhosSTOP, Roche). After centrifugation at 50,000 × g for 20 min, the supernatant was taken as the RAB-soluble fraction and the pellet was dissolved in RIPA buffer (150mM NaCl, 50mM Tris, 0.5% deoxycholic acid, 1% Triton X-100, 0.1% SDS, 5mM EDTA, 20mM NaF, 1mM Na3VO4, pH 8.0, supplemented by cOmplete and PhosSTOP) at 10ul buffer/1mg tissue by sonication. After centrifugation at 50,000 × g for 20 min, the supernatant was taken as the RIPA-soluble fraction. The pellet was sonicated in 70% formic acid at 10ul buffer/1mg tissue, and centrifuged at 50,000 × g for 20 min. The supernatant was taken as the FA-soluble fraction. All fractions were stored at −80°C until analyzed.
ELISA
The ELISA plates were coated with 20ug/ml coating antibody overnight at 4°. The next day, the plates were washed in PBS, and mouse brain lysates were diluted in sample buffer (human tau and p-tau sample buffer: 0.25% BSA in PBS with 300 uM Tris, pH 7.4; apoE sample buffer: 0.5% BSA in 0.025% PBS-tween20; both buffers were supplemented with protease inhibitors) and loaded into the plates in duplicates. The plates were incubated at 4° overnight. On the 3rd day, the plates were washed in PBS, followed by addition of the detection antibody and incubation at 37° for 90min. The plates were then washed with PBS and incubated with streptavidin-poly-horseradish peroxidase-40 for 90min at room temperature (RT). Plates were then washed and developed with Super Slow ELISA TMB (Sigma-Aldrich, Cat# T5569) and read at 650 nm. For FA fractions, samples were neutralized with 3M Tris buffer (pH 8.0) before loading into the plate. Tau5 (gift from L. Binder, Northwestern University, Chicago, IL), HJ14.5 (in house anti-p-tau antibody, Thr181) and HJ6.2 (in house anti-apoE antibody) were used as coating antibodies, and biotinylated HT7 (Thermo Fisher Scientific, Cat# MN1000B), biotinylated AT8 (Ser202/Thr205, Thermo Fisher Scientific), and biotinylated HJ6.8 (in house anti-murine apoE antibody) were used as detection antibodies for human tau, p-tau and murine apoE ELISAs respectively. ptau ELISA detects ptau species that have phosphorylation at both Thr181 and Ser202/Thr205 epitopes, which represent hyperphosphorylated ptau. Human tau ELISA uses 2N4R recombinant human tau (MW ~46 kDa) as the standard. The pTau ELISA uses synthetic human tau peptide (176-229) phosphorylated at Thr-181, Ser-198, Ser-199, Ser-202, and Thr-205 (MW ~5.4 kDa) as the standard. The absolute values between tau and ptau levels reported here cannot be directly compared to each other because 1) tau and ptau standards were generated by different techniques, and their concentration and purity were determined by different approaches that are not directly comparable and 2) the capacity of ptau peptide standard to bind the ELISA antibodies may be different from the capacity of ptau proteins in the tissue samples to bind the antibodies.
Western Blotting
Mouse cortical tissue or isolated microglia were lysed by sonication in RIPA buffer supplemented with cOmplete and PhosSTOP. Samples were separated by 4–12% NuPAGE gels (Invitrogen, Cat# NP0335BOX) in MOPS buffer (Invitrogen, Cat# NP0001) and transferred to nitrocellulose membranes using the iBlot™ 2 Gel Transfer Device (Thermo Fisher Scientific). The membranes were blocked with 5% milk in 0.125% PBSX for 30min at RT, followed by incubation of primary antibodies overnight at 4°. The membranes were then washed with 0.0125% PBSX and incubated with HRP-conjugated secondary antibodies (Santa Cruz) for 1.5h at RT. Membranes were then washed and developed using the ChemiDoc™ MP Imaging System (BIO-RAD). Primary antibodies: HJ6.8B (for murine apoE, in house), α-tubulin (Sigma-Aldrich, Cat# T5168), LDLR (Abcam, Cat# ab52818), Lgmn (Santa Cruz, Cat# sc-133234), p4EBP (Cell signaling, Cat# 2855), actin (Cell signaling, Cat# 8456).
Lysosomal enzyme activity assay
10 mg of mouse brain cortex was homogenized in buffer containing 10 mM Tris (pH 7.5), 150 mM NaCl, 1 mM dithiothreitol, and 0.2% Triton X-100 and centrifuged at 14,000 rpms for 1 min at 4°C. Following centrifugation, the supernatant was removed and used for Naglu, β-gluc, and β-Hexa enzyme assays using the 4-MU fluorometric assay and normalized to total protein as previously described(Benitez and Sands, 2017). In brief, 1-5 uL of supernatant was added to 100 μl of 5 mM 4-Methylumbelliferyl-β-D-glucuronide hydrate (β-Gluc substrate) (Sigma-Aldrich, Cat# M9130), 4-Methylumbelliferyl-6-sulfo-N-acetyl-β-D-glucosaminide (HexA substrate) (Sigma-Aldrich, Cat# 454428) or 4-Methylumbelliferyl N-acetyl-β-D-glucosaminide (Naglu Substrate) (Sigma-Aldrich, Cat# M2133) respectively, and incubated at 37°C for 1 h. The reactions were stopped by adding 1 ml of 0.1 M Na2CO3. Substrate cleavage was measured at 448 nm emission and 365 nm excitation in a Hitachi F-2000 fluorescence spectrophotometer (Hitachi) using a standard curve ranging from 0.5 to 5 mM of 4-methylumbelliferone (Sigma-Aldrich, Cat# M1381). The values were normalized to total protein measured using a Coomassie dye-binding assay (Bio-Rad Laboratories).
Acute microglial isolation from mouse brains using magnetic beads
Mice were perfused with perfusion buffer (DPBS + 1x Glutamax). Whole brain was removed, cut into small pieces, and dissociated into single cell suspension using the Neural Tissue Dissociation Kit (T) (Miltenyi Biotec, Cat# 130-093-231) with a gentleMACS Dissociator (Miltenyi Biotec, Cat# 130-093-235) following manufacturer’s instructions. Single cells were resuspended in 800ul cold MACS buffer (DPBS + 0.5% BSA + 1mM EDTA degas overnight), and incubated with 40ul CD11b MicroBeads (Miltenyi Biotec, Cat# 130-093-634) at 4° for 15min. The cells were washed with 15ml cold MACS buffer, and spun down at 1500rpm for 5min. LS columns (Miltenyi Biotec, Cat# 130-042-401) were put onto a QuadroMAC Separator (Miltenyi Biotec, Cat# 130-091-051) and pre-equilibrated with 5ml MACS buffer. Cell pellets were resuspended in 5ml MACS buffer, loaded onto LS columns through a MACS SmartStrainer (Miltenyi Biotec, Cat# 130-098-462), and allowed to flow through by gravity. The columns were washed 3 times with MACS buffer (6ml each), and were taken off the separator to sit on a 15ml tube. Microglia were eluted with MACS buffer (3ml eluted by gravity first, followed by 6ml eluted with gentle push of plunger). Eluted microglia were mixed with 100% Percoll (GE-healthcare: 17-0891-01, 9 volumes of Percoll were mixed with 1 volume of 10x DPBS, and the mixture was taken as 100% Percoll) to achieve a final concentration of 30% percoll. The mixture was spun down at 700g for 15min. Supernatant containing cell debris was removed. The clean cell pellets were washed twice with MACS buffer, and were used for downstream analysis.
Isolating microglia from mouse brains by fluorescence-activated cell sorting
Mouse brains were dissociated into single cell suspension using the Adult Brain Dissociation Kit (Miltenyi Biotec, Cat# 130-107-677) with cell debris removal following manufacturer’s instructions. Single cells were resuspended in cold MACS buffer supplemented with mouse Fcγ receptor block reagent (553141; BD Biosciences) and incubated on ice for 5 min, followed by staining with fluorescently tagged antibodies in the brilliant stain buffer (563794; BD Biosciences) on ice for 20min. The cells were washed twice with MACS buffer, and used for FACS sorting. Antibodies: APC-Cy7 rat anti-CD11b (clone M1/70, Cat# 557657; BD Biosciences), ACSA2-PE (Miltenyi Biotec, Cat# 130-123-284), O4-APC (Miltenyi Biotec, Cat# 130-118-978), BV421 Rat Anti-Mouse CD24 (clone M1/69, Cat# 562563; BD Biosciences).
RT-qPCR
RNA was isolated from acutely isolated microglia using RNeasy Mini Kit (QIAGEN, Cat# 74104) following manufacturer’s instructions, and reverse transcribed to cDNA using the high capacity RNA-to-cDNA kit (Applied Biosystems, #4387406). Quantitative PCR was performed with Taqman primers (Thermo Fisher Scientific) and Taqman universal PCR master mix (Applied Biosystems, #4304437) using the QuantStudio 12K Flex Real-Time PCR System (Applied Biosystems).
Staining of acutely isolated microglia and flow cytometry analysis
Acutely isolated microglia were fixed with 4% PFA, washed with MACS buffer, and then stained with 1ug/ml BODIPY (Invitrogen, Cat# D3922) or 12.5ug/ml Filipin3 (Tocris Bioscience, Cat# 62501NB) for 20min. Cells were washed, resuspended in MACS buffer, and run through flow cytometry using a BD LSRFortessa X-20 cell analyzer (BD Biosciences). Data were analyzed using FlowJo v10.
Isolation of single nuclei from frozen brain tissue
Frozen hippocampi from 7-8 mice of the same genotype were pooled as a single sample. Tissue was homogenized with a Dounce homogenizer in lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, and 0.005% NP-40 in nuclease free water), and incubated on ice for 15 min. The suspension was filtered through a 30-μm filter to remove debris and pelleted at 500g for 5 min at 4 °. Nuclei were washed and filtered twice with Nuclei Wash Buffer (1x PBS with 1.0% BSA and 0.2 U/uL RNasin Ribonuclease Inhibitors (Promega)). Nuclei pellets were resuspended in 500 μl Nuclei Wash Buffer, and mixed with 900 μl Sucrose Cushion Buffer (2.7 mL Nuclei PURE 2M Sucrose Cushion Solution (Sigma-Aldrich, Cat# S9308) + 300 uL Nuclei PURE Sucrose Cushion Solution (Sigma-Aldrich, Cat# S9058)). The 1,400 μl mixture was carefully layered on top of 500 μl Sucrose Cushion Buffer, and centrifuged at 13,000g for 45 min at 4 ° to separate the nuclei from cell debris. The nuclei pellet was resuspended in Nuclei Wash Buffer and filtered through a 40-μm FlowMi Cell Strainer (Sigma-Aldrich, Cat# BAH136800040). The nuclei were adjusted to a concentration of 1000 nuclei/uL and proceeded to the 10x Genomics protocol.
Single-nuclei RNA sequencing
Isolated cells were subjected to droplet-based 3’ end massively parallel single-cell RNA sequencing using Chromium Single Cell 3’ Reagent Kits (10x Genomics) following manufacturer’s instructions. The libraries were sequenced using a HiSeq3000 instrument (Illumina). Sample demultiplexing, barcode processing, and single-cell 3’ counting were performed using the Cell Ranger Single-Cell Software Suite (10x Genomics). Cellranger count was used to align samples to the reference genome (mm10), quantify reads, and filter reads with a quality score below 30.
Electron microscopic imaging
Mice were perfused with mammalian ringer’s solution (2.2 mM CaCl2, 5.6 mM KCl, 154 mM NaCl, 2.4 mM NaHCO3, 2 mM Tris-Cl, pH=7.4) and 4% PFA. Fixed brains were put in 4% PFA for 24h. Corpus callosum together with a portion of hippocampus was isolated from posterior brain regions, and was further fixed in a modified Karnovsky's fixative of 3% glutaraldehyde, 1% paraformaldehyde in 0.1M sodium cacodylate buffer for 5 days. Tissue was then post-fixed in 2% osmium tetroxide in 0.1M sodium cacodylate buffer for 1 hr, en bloc stained with 3% aqueous uranyl acetate for 30 min, dehydrated in graded ethanol and embedded in PolyBed 812 (Polysciences, Cat# 08792-1). Tissue blocks were sectioned at ninety nanometers thick, post stained with Venable's lead citrate and viewed with a JEOL model 1400EX transmission electron microscope (JEOL, Tokyo, Japan). Digital images were acquired using the AMT NanoSprint 12A-B (Advanced Microscopy Technology, Danvers MA) CMOS, 12 megapixel TEM camera.
Quantification of G-ratio and demyelinated axon ratio
G-ratio was calculated as the ratio between the axon diameter to the total diameter of a myelinated axon (i.e., axon + myelin). These measurements were performed by using the “line” tool in ImageJ. For each axon, two sets of the axon and myelin + axon diameters perpendicular to each other were measured, and the average value of the two G-ratios was used to represent the G-ratio of the axon. For each sample, 20 representative axons from 5 different images were measured and their averaged G-ratio was used to represent the axonal G-ratio of this sample. Demyelinated axons were defined as axons having comparable or larger diameters to adjacent myelinated axons, but had ≤ two layers of myelin. Over 200 axons were assessed for each sample, and the percentage of demyelinated axons was calculated by dividing the number of demyelinated axons by the total number of axons.
Processing data with Seurat Package
The Seurat package in R was used for subsequent analysis (Butler et al., 2018). For quality control, nuclei with mitochondrial content >5% were removed. A total of 31,785 individual nuclei across all genotypes remained, with a median of 1,620 UMIs and 871 genes per nucleus for downstream analysis. Data were normalized using a scaling factor of 10,000, and nUMI was regressed with a negative binomial model. The highly variable genes were selected using the FindVariableFeatures function with a mean value greater than 0.0125 or less than 3 and dispersion greater than 0.5. These genes were used in performing the linear dimensionality reduction. Principal component analysis was performed using the top 3000 most variable genes and t-SNE analysis was performed with the top 10 PCAs based on the ElbowPlot. Clustering was performed using the FindClusters function which works on the K-nearest neighbor (KNN) graph model with the granularity ranging from 0.1-0.9. Clustering was performed using a resolution of 0.5. Differential gene expression between each cell cluster and all other clusters was performed to identify marker genes for each individual cell cluster.
Gene differential expression and pathway analysis
Differential expression of genes between conditions was done using the MAST algorithm of the Seurat package in R, which implements a two-part hurdle model. Log2(fold change) of average expression and the percentage of cells (pct) expressing the genes in each condition were generated. The adjusted P-value was calculated using Bonferroni correction. Genes with an adjusted P < 0.05 were taken as differentially expressed genes (DEGs). Pathway analysis was performed using Metascape. For microglia and astrocytes, total nuclei of each cell type from all 6 genotypes were further sub-clustered using PC5 and resolution 0.5 & 0.4 respectively. DEGs and pathway analysis were performed for microglial and astrocyte sub-populations as described above. A portion of microglia from all 6 groups of mice show expression of certain oligodendrocyte genes, which was likely due to formation of doublets between a portion of microglia and oligodendrocyte nuclei instead of being real biological effects. These oligodendrocyte genes did not drive apoE-deficiency or LDLR-associated effects in microglia, as these effects were either not present or only weakly showed up in oligodendrocytes. Oligodendrocyte genes and pathways showing up in microglial analysis results were excluded.
Quantification and statistical analysis
Data were presented as mean ± SEM. Statistical analysis was performed using Prism 8 (GraphPad). For all experiments, data normality was analyzed using D’Agostino-Pearson omnibus normality test. For data following normal distribution, differences between groups were evaluated by student’s t-test or one-way analysis of variance tests with post hoc Tukey’s multiple comparisons tests. For data not following normal distribution, Mann-Whitney test or Kruskal-Wallis test with Dunn’s multiple comparisons test was performed. The method of statistical test for each figure was stated in the figure legend. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. In all figures, N represents the number of animals.
Supplementary Material
Table S1. DEGs between different genotypes in major brain cell types, Related to Figure 4E
Upregulated and downregulated DEGs (former versus latter genotype) compared between the following groups: P301S vs. WT, WT vs. EKO, WT vs. LDLR, P301S vs. P301S/EKO, P301S vs. P301S/LDLR. DEGs were defined as adjusted p-value < 0.05. Mitochondrial genes were excluded from DEG counts.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti- Phospho-tau (Ser202, Thr205) (AT8), biotinylated | Thermo Fisher Scientific | Cat# MN1020B, RRID: AB_223648 |
| Mouse monoclonal anti-human tau (HT7), biotinylated | Thermo Fisher Scientific | Cat# MN1000B, RRID: AB_223453 |
| Rabbit polyclonal anti-Synapsin 1 | Synaptic Systems | Cat# 106103, RRID: AB_11042000 |
| Rat monoclonal anti-CD68, clone FA-11 | AbD SeroTec | Cat# MCA1957 |
| Rabbit polyclonal anti-GFAP (GFAP) | Abcam | Cat# ab7260, RRID: AB_305808 |
| Mouse monoclonal anti-p-tau HJ14.5 (Thr181) | In house (Shi et al., 2019) | N/A |
| Mouse monoclonal anti-tau (TAU-5) | Gift from L. Binder, Northwestern University (LoPresti et al., 1995) | N/A |
| Mouse monoclonal anti-apoE HJ6.2 | In house (Kim et al., 2012) | N/A |
| Mouse monoclonal anti-murine apoE HJ6.8, biotinylated | In house (Liao et al., 2015) | N/A |
| Mouse monoclonal anti-murine apoE HJ6.3 | In house (Kim et al., 2012) | N/A |
| APC-Cy7 rat anti-CD11b (clone M1/70) | BD Biosciences | Cat# 557657, RRID: AB_396772 |
| BV421 Rat Anti-Mouse CD24 (clone M1/69) | BD Biosciences | Cat# 562563, RRID: AB_2737002 |
| ACSA2-PE | Miltenyi Biotec | Cat# 130-123-284, RRID: AB_2811488 |
| O4-APC | Miltenyi Biotec | Cat# 130-118-978, RRID: AB_2751598 |
| Rabbit polyclonal anti-NG2 (CSPG4) | Sigma-Aldrich | Cat# AB5320, RRID: AB_11213678 |
| Mouse monoclonal anti-α-tubulin | Sigma-Aldrich | Cat# T5168, RRID: AB_477579 |
| Rabbit monoclonal anti-LDLR (EP1553Y) | Abcam | Cat# ab52818, RRID: AB_881213 |
| Rabbit polyclonal anti Iba1 | Wako | Cat# 019-19741, RRID: AB_839504 |
| Goat Polyclonal anti-LDLR | R&D Systems | Cat# AF2255, RRID: AB_355203 |
| Mouse monoclonal anti- Legumain (B-8) | Santa Cruz Biotechnology | Cat# sc-133234, RRID: AB_2135017 |
| Mouse monoclonal anti Vimentin (E-5) | Santa Cruz Biotechnology | Cat# sc-373717, RRID: AB_10917747 |
| Rabbit monoclonal anti-Phospho-4E-BP1 (Thr37/46) (236B4) | Cell Signaling Technology | Cat# 2855, RRID: AB_560835 |
| Rabbit polyclonal anti- Myelin Basic Protein | Abcam | Cat# ab40390 RRID: AB_1141521 |
| Rabbit monoclonal anti-Pan-Actin (D18C11) | Cell Signaling Technology | Cat# 8456, RRID: AB_10998774 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Sudan Black B | Sigma-Aldrich | Cat# 199664 |
| DMEM, high glucose | Gibco | Cat# 11965084 |
| Fetal Bovine Serum, qualified, USDA-approved regions | Gibco | Cat# 10437028 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | Cat# 15140122 |
| GlutaMAX™ Supplement | Gibco | Cat# 35050061 |
| Poly-L-lysine hydrobromide | Sigma-Aldrich | Cat# P2636 |
| MEM Non-Essential Amino Acids Solution (100X) | Gibco | Cat# 11140050 |
| DMEM/F-12, HEPES, no phenol red | Gibco | Cat# 11039021 |
| Insulin-Transferrin-Selenium (ITS -G) (100X) | Gibco | Cat# 41400045 |
| B-27 Supplement (50X), minus vitamin A | Gibco | Cat# 12587010 |
| N-2 Supplement (100X) | Gibco | Cat# 17502048 |
| 1-Thioglycerol | Sigma-Aldrich | Cat# M1753 |
| Insulin solution human | Sigma-Aldrich | Cat# I9278 |
| Recombinant Human TGF-beta 1 Protein | R&D Systems | Cat# 240-B |
| Recombinant Human IL-34 Protein | R&D Systems | Cat# 5265-IL |
| Recombinant Human M-CSF Protein | R&D Systems | Cat# 216-MC |
| 4-Methylumbelliferyl-β-D-glucuronide hydrate (β-Gluc substrate) | Sigma-Aldrich | Cat# M9130 |
| 4-Methylumbelliferyl-6-sulfo-N-acetyl-β-D-glucosaminide (Hexa substrate) | Sigma-Aldrich | Cat# 454428 |
| 4-Methylumbelliferyl N-acetyl-β-D-glucosaminide (Naglu Substrate) | Sigma-Aldrich | Cat# M2133 |
| Mouse BD Fc Block | BD Biosciences | Cat# 553141 |
| Brilliant stain buffer | BD Biosciences | Cat# 563794 |
| 4-Methylumbelliferone | Sigma-Aldrich | Cat# M1381 |
| Critical Commercial Assays | ||
| VECTASTAIN Elite ABC-HRP Kit, Peroxidase (Standard) | Vector laboratories | Cat# PK-6100 |
| Micro BCA Protein Assay Kit | Thermo Fisher Scientific | Cat# 23235 |
| Neural Tissue Dissociation Kit (T) | Miltenyi Biotec | Cat# 130-093-231 |
| Adult Brain Dissociation Kit | Miltenyi Biotec | Cat# 130-107-677 |
| RNeasy Mini Kit | QIAGEN | Cat# 74104 |
| Nuclei Isolation Kit: Nuclei PURE Prep | Sigma-Aldrich | Cat# NUC201 |
| RNA-to-cDNA kit | Applied Biosystems | Cat# 4387406 |
| DC Protein Assay Kit I | BIO-RAD | Cat# 5000111 |
| Deposited Data | ||
| snRNAseq raw data | This paper | GEO accession number GSE166261 |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6/C3-Tg (Prnp-MAPT*P301S) PS19Vle/J, crossed to B6 background | The Jackson Laboratory | Cat# 008169 |
| Mouse: Ldlr Tg | (Kim et al., 2009) | N/A |
| Mouse: B6.129P2-Apoetm1Unc/J | The Jackson Laboratory | Cat# 002052 |
| Mouse: C57BL/6NCrl WT | Charles River Laboratories | Cat# 027 |
| Oligonucleotides | ||
| Taqman qPCR primers for Gabra6 | Thermo Fisher Scientific | ID: Mm01227754_m1 |
| Taqman qPCR primers for Grm4 | Thermo Fisher Scientific | ID: Mm01306128_m1 |
| Taqman qPCR primers for Apoe | Thermo Fisher Scientific | ID: Mm01307193_g1 |
| Taqman qPCR primers for Lilrb4 | Thermo Fisher Scientific | ID: Mm01614371_m1 |
| Taqman qPCR primers for Spp1 | Thermo Fisher Scientific | ID: Mm00436767_m1 |
| Taqman qPCR primers for Il1b | Thermo Fisher Scientific | ID: Mm00434228_m1 |
| Taqman qPCR primers for Ldlr | Thermo Fisher Scientific | ID: Mm01177349_m1 |
| Taqman qPCR primers for Lgmn | Thermo Fisher Scientific | ID: Mm01325350_m1 |
| Taqman qPCR primers for Actb | Thermo Fisher Scientific | ID: Mm01205647_g1 |
| Software and Algorithms | ||
| Prism 8 | GraphPad | https://www.graphpad.com/ |
| ImageJ 1.52p | ImageJ | https://imagej.net/Fiji |
| FlowJo v10 | FlowJo, LLC | https://www.flowjo.com/solutions/flowjo/downloads |
| Cell Ranger Single-Cell Software Suite | 10x Genomics | https://support.10xgenomics.com/single-cell-gene-expression/software/overview/welcome |
| Seurat v2 | (Butler et al., 2018) | https://satijalab.org/seurat/articles/install.html |
| R 2.4.0 (for snRNAseq analysis) | R foundation | https://cran-archive.r-project.org/bin/windows/base/old/2.4.0/ |
| Metascape | Metascape | https://metascape.org/gp/index.html#/main/step1 |
Acknowledgements
This study was funded by NIH grants NS090934 (D.M.H.), AG047644 (D.M.H.), the Tau consortium (D.M.H.), and the JPB Foundation (D.M.H.). The EM study was supported by the DRC grant.
Footnotes
Declaration of interests
D.M.H. and C.I. are listed as inventors on a patent licensed by Washington University to C2N Diagnostics on the therapeutic use of anti-tau antibodies. D.M.H. co-founded and is on the scientific advisory board of C2N Diagnostics. C2N Diagnostics has licensed certain anti-tau antibodies to AbbVie for therapeutic development. D.M.H. is on the scientific advisory board of Denali and consults for Genentech, Merck, and Cajal Neurosciences. All other authors have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bales KR, Verina T, Dodel RC, Du Y, Altstiel L, Bender M, Hyslop P, Johnstone EM, Little SP, Cummins DJ, et al. (1997). Lack of apolipoprotein E dramatically reduces amyloid beta-peptide deposition. Nat Genet 17, 263–264. 10.1038/ng1197-263. [DOI] [PubMed] [Google Scholar]
- Ben Haim L, Carrillo-de Sauvage MA, Ceyzeriat K, and Escartin C (2015). Elusive roles for reactive astrocytes in neurodegenerative diseases. Front Cell Neurosci 9, 278. 10.3389/fncel.2015.00278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez BA, and Sands MS (2017). Primary fibroblasts from CSPα mutation carriers recapitulate hallmarks of the adult onset neuronal ceroid lipofuscinosis. Sci Rep 7, 6332. 10.1038/s41598-017-06710-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borchelt DR, Davis J, Fischer M, Lee MK, Slunt HH, Ratovitsky T, Regard J, Copeland NG, Jenkins NA, Sisodia SS, and Price DL (1996). A vector for expressing foreign genes in the brains and hearts of transgenic mice. Genetic Analysis: Biomolecular Engineering 13, 159–163. [DOI] [PubMed] [Google Scholar]
- Bories GFP, and Leitinger N (2017). Macrophage metabolism in atherosclerosis. FEBS Lett 591, 3042–3060. 10.1002/1873-3468.12786. [DOI] [PubMed] [Google Scholar]
- Boucsein C, Kettenmann H, and Nolte C (2000). Electrophysiological properties of microglial cells in normal and pathologic rat brain slices. Eur J Neurosci 12, 2049–2058. 10.1046/j.1460-9568.2000.00100.x. [DOI] [PubMed] [Google Scholar]
- Boyles JK, Pitas RE, Wilson E, Mahley RW, and Taylor JM (1985). Apolipoprotein E associated with astrocytic glia of the central nervous system and with nonmyelinating glia of the peripheral nervous system. J Clin Invest 76, 1501–1513. 10.1172/jci112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu G (2009). Apolipoprotein E and its receptors in Alzheimer's disease: pathways, pathogenesis and therapy. Nature reviews. Neuroscience 10, 333–344. 10.1038/nrn2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, and Satija R (2018). Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36, 411–420. 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coetzee WA, Amarillo Y, Chiu J, Chow A, Lau D, McCormack T, Moreno H, Nadal MS, Ozaita A, Pountney D, et al. (1999). Molecular diversity of K+ channels. Ann N Y Acad Sci 868, 233–285. 10.1111/j.1749-6632.1999.tb11293.x. [DOI] [PubMed] [Google Scholar]
- Di Lucente J, Nguyen HM, Wulff H, Jin LW, and Maezawa I (2018). The voltage-gated potassium channel Kv1.3 is required for microglial pro-inflammatory activation in vivo. Glia 66, 1881–1895. 10.1002/glia.23457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domercq M, Vazquez-Villoldo N, and Matute C (2013). Neurotransmitter signaling in the pathophysiology of microglia. Front Cell Neurosci 7, 49. 10.3389/fncel.2013.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes SE, Hollopeter G, Yang G, Kurpius D, Dailey ME, Gan WB, and Julius D (2006). The P2Y12 receptor regulates microglial activation by extracellular nucleotides. Nat Neurosci 9, 1512–1519. 10.1038/nn1805. [DOI] [PubMed] [Google Scholar]
- Heeren J, Beisiegel U, and Grewal T (2006). Apolipoprotein E recycling: implications for dyslipidemia and atherosclerosis. Arterioscler Thromb Vasc Biol 26, 442–448. 10.1161/01.atv.0000201282.64751.47. [DOI] [PubMed] [Google Scholar]
- Holtzman DM, Bales KR, Tenkova T, Fagan AM, Parsadanian M, Sartorius LJ, Mackey B, Olney J, McKeel D, Wozniak D, and Paul SM (2000). Apolipoprotein E isoform-dependent amyloid deposition and neuritic degeneration in a mouse model of Alzheimer's disease. Proceedings of the National Academy of Sciences of the United States of America 97, 2892–2897. 10.1073/pnas.050004797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, Herz J, and Bu G (2012). Apolipoprotein E and apolipoprotein E receptors: normal biology and roles in Alzheimer disease. Cold Spring Harbor perspectives in medicine 2. 10.1101/cshperspect.a006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi S, Herz J, Maeda N, Goldstein JL, and Brown MS (1994). The two-receptor model of lipoprotein clearance: tests of the hypothesis in "knockout" mice lacking the low density lipoprotein receptor, apolipoprotein E, or both proteins. Proc Natl Acad Sci U S A 91, 4431–4435. 10.1073/pnas.91.10.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo P, Attwell D, and Madry C (2019). Ion Channels and Receptors as Determinants of Microglial Function. Trends Neurosci 42, 278–292. 10.1016/j.tins.2018.12.007. [DOI] [PubMed] [Google Scholar]
- Kim J, Castellano JM, Jiang H, Basak JM, Parsadanian M, Pham V, Mason SM, Paul SM, and Holtzman DM (2009). Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular A beta clearance. Neuron 64, 632–644. 10.1016/j.neuron.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Eltorai AEM, Jiang H, Liao F, Verghese PB, Kim J, Stewart FR, Basak JM, and Holtzman DM (2012). Anti-apoE immunotherapy inhibits amyloid accumulation in a transgenic mouse model of Aβ amyloidosis. The Journal of experimental medicine 209, 2149–2156. 10.1084/jem.20121274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koizumi S, Shigemoto-Mogami Y, Nasu-Tada K, Shinozaki Y, Ohsawa K, Tsuda M, Joshi BV, Jacobson KA, Kohsaka S, and Inoue K (2007). UDP acting at P2Y6 receptors is a mediator of microglial phagocytosis. Nature 446, 1091–1095. 10.1038/nature05704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, Beckers L, O'Loughlin E, Xu Y, Fanek Z, et al. (2017). The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581.e569. 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn SA, van Landeghem FK, Zacharias R, Farber K, Rappert A, Pavlovic S, Hoffmann A, Nolte C, and Kettenmann H (2004). Microglia express GABA(B) receptors to modulate interleukin release. Mol Cell Neurosci 25, 312–322. 10.1016/j.mcn.2003.10.023. [DOI] [PubMed] [Google Scholar]
- Leyns CEG, Ulrich JD, Finn MB, Stewart FR, Koscal LJ, Remolina Serrano J, Robinson GO, Anderson E, Colonna M, and Holtzman DM (2017). TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc Natl Acad Sci U S A 114, 11524–11529. 10.1073/pnas.1710311114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao F, Zhang TJ, Jiang H, Lefton KB, Robinson GO, Vassar R, Sullivan PM, and Holtzman DM (2015). Murine versus human apolipoprotein E4: differential facilitation of and co-localization in cerebral amyloid angiopathy and amyloid plaques in APP transgenic mouse models. Acta neuropathologica communications 3, 70–70. 10.1186/s40478-015-0250-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow SA, Guttenplan KA, Clarke LE, Bennett FC, Bohlen CJ, Schirmer L, Bennett ML, Munch AE, Chung WS, Peterson TC, et al. (2017). Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487. 10.1038/nature21029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, Herz J, Muglia L, and Bu G (2007). Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron 56, 66–78. 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LoPresti P, Szuchet S, Papasozomenos SC, Zinkowski RP, and Binder LI (1995). Functional implications for the microtubule-associated protein tau: localization in oligodendrocytes. Proceedings of the National Academy of Sciences of the United States of America 92, 10369–10373. 10.1073/pnas.92.22.10369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madry C, Kyrargyri V, Arancibia-Carcamo IL, Jolivet R, Kohsaka S, Bryan RM, and Attwell D (2018). Microglial Ramification, Surveillance, and Interleukin-1beta Release Are Regulated by the Two-Pore Domain K(+) Channel THIK-1. Neuron 97, 299–312.e296. 10.1016/j.neuron.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso R, Fryatt G, Cleal M, Obst J, Pipi E, Monzon-Sandoval J, Ribe E, Winchester L, Webber C, Nevado A, et al. (2019). CSF1R inhibitor JNJ-40346527 attenuates microglial proliferation and neurodegeneration in P301S mice. Brain 142, 3243–3264. 10.1093/brain/awz241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathys H, Davila-Velderrain J, Peng Z, Gao F, Mohammadi S, Young JZ, Menon M, He L, Abdurrob F, Jiang X, et al. (2019). Single-cell transcriptomic analysis of Alzheimer's disease. Nature 570, 332–337. 10.1038/s41586-019-1195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy AJ, Akhtari M, Tolani S, Pagler T, Bijl N, Kuo CL, Wang M, Sanson M, Abramowicz S, Welch C, et al. (2011). ApoE regulates hematopoietic stem cell proliferation, monocytosis, and monocyte accumulation in atherosclerotic lesions in mice. J Clin Invest 121, 4138–4149. 10.1172/jci57559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen HM, Grossinger EM, Horiuchi M, Davis KW, Jin LW, Maezawa I, and Wulff H (2017). Differential Kv1.3, KCa3.1, and Kir2.1 expression in "classically" and "alternatively" activated microglia. Glia 65, 106–121. 10.1002/glia.23078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama A, Komitova M, Suzuki R, and Zhu X (2009). Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci 10, 9–22. 10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- Ohsawa K, Irino Y, Nakamura Y, Akazawa C, Inoue K, and Kohsaka S (2007). Involvement of P2X4 and P2Y12 receptors in ATP-induced microglial chemotaxis. Glia 55, 604–616. 10.1002/glia.20489. [DOI] [PubMed] [Google Scholar]
- Rapp A, Gmeiner B, and Huttinger M (2006). Implication of apoE isoforms in cholesterol metabolism by primary rat hippocampal neurons and astrocytes. Biochimie 88, 473–483. 10.1016/j.biochi.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Rauch JN, Luna G, Guzman E, Audouard M, Challis C, Sibih YE, Leshuk C, Hernandez I, Wegmann S, Hyman BT, et al. (2020). LRP1 is a master regulator of tau uptake and spread. Nature 580, 381–385. 10.1038/s41586-020-2156-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmechel DE, Saunders AM, Strittmatter WJ, Crain BJ, Hulette CM, Joo SH, Pericak-Vance MA, Goldgaber D, and Roses AD (1993). Increased amyloid beta-peptide deposition in cerebral cortex as a consequence of apolipoprotein E genotype in late-onset Alzheimer disease. Proceedings of the National Academy of Sciences of the United States of America 90, 9649–9653. 10.1073/pnas.90.20.9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, and Holtzman DM (2018). Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol 18, 759–772. 10.1038/s41577-018-0051-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Manis M, Long J, Wang K, Sullivan PM, Remolina Serrano J, Hoyle R, and Holtzman DM (2019). Microglia drive APOE-dependent neurodegeneration in a tauopathy mouse model. J Exp Med. 10.1084/jem.20190980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Yamada K, Liddelow SA, Smith ST, Zhao L, Luo W, Tsai RM, Spina S, Grinberg LT, Rojas JC, et al. (2017). ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature 549, 523–527. 10.1038/nature24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCW J, Liang S, Qian L, Pipalia N, Chao M, Shi Y, Bertelsen SE, Kapoor M, Marcora E, et al. (2019). Cholesterol and matrisome pathways dysregulated in human APOE ε4 glia. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchihara T, Duyckaerts C, He Y, Kobayashi K, Seilhean D, Amouyel P, and Hauw JJ (1995). ApoE immunoreactivity and microglial cells in Alzheimer's disease brain. Neurosci Lett 195, 5–8. [DOI] [PubMed] [Google Scholar]
- Ulland TK, Song WM, Huang SC, Ulrich JD, Sergushichev A, Beatty WL, Loboda AA, Zhou Y, Cairns NJ, Kambal A, et al. (2017). TREM2 Maintains Microglial Metabolic Fitness in Alzheimer's Disease. Cell 170, 649–663.e613. 10.1016/j.cell.2017.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Xiong M, Gratuze M, Bao X, Shi Y, Andhey PS, Manis M, Schroeder C, Yin Z, Madore C, et al. (2021). Selective removal of astrocytic APOE4 strongly protects against tau-mediated neurodegeneration and decreases synaptic phagocytosis by microglia. Neuron. 10.1016/j.neuron.2021.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt S, Maricos M, Vana N, Meyer N, Guneykaya D, Semtner M, and Kettenmann H (2017). Changes in phagocytosis and potassium channel activity in microglia of 5xFAD mice indicate alterations in purinergic signaling in a mouse model of Alzheimer's disease. Neurobiol Aging 58, 41–53. 10.1016/j.neurobiolaging.2017.05.027. [DOI] [PubMed] [Google Scholar]
- William Rebeck G, Reiter JS, Strickland DK, and Hyman BT (1993). Apolipoprotein E in sporadic Alzheimer's disease: Allelic variation and receptor interactions. Neuron 11, 575–580. 10.1016/0896-6273(93)90070-8. [DOI] [PubMed] [Google Scholar]
- Xu Q, Bernardo A, Walker D, Kanegawa T, Mahley RW, and Huang Y (2006). Profile and regulation of apolipoprotein E (ApoE) expression in the CNS in mice with targeting of green fluorescent protein gene to the ApoE locus. J Neurosci 26, 4985–4994. 10.1523/jneurosci.5476-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CP, Gilley JA, Zhang G, and Kernie SG (2011). ApoE is required for maintenance of the dentate gyrus neural progenitor pool. Development 138, 4351–4362. 10.1242/dev.065540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A, Marques S, Munguba H, He L, Betsholtz C, et al. (2015). Brain structure. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 347, 1138–1142. 10.1126/science.aaa1934. [DOI] [PubMed] [Google Scholar]
- Zerbinatti CV, Wahrle SE, Kim H, Cam JA, Bales K, Paul SM, Holtzman DM, and Bu G (2006). Apolipoprotein E and low density lipoprotein receptor-related protein facilitate intraneuronal A??42 accumulation in amyloid model mice. Journal of Biological Chemistry 281, 36180–36186. 10.1074/jbc.M604436200. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, Poliani PL, Cominelli M, Grover S, Gilfillan S, et al. (2020). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer's disease. Nat Med 26, 131–142. 10.1038/s41591-019-0695-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. DEGs between different genotypes in major brain cell types, Related to Figure 4E
Upregulated and downregulated DEGs (former versus latter genotype) compared between the following groups: P301S vs. WT, WT vs. EKO, WT vs. LDLR, P301S vs. P301S/EKO, P301S vs. P301S/LDLR. DEGs were defined as adjusted p-value < 0.05. Mitochondrial genes were excluded from DEG counts.
Data Availability Statement
Data that support the findings of this study are available from the corresponding author upon request. snRNAseq raw data has been deposited to GEO, accession number GSE166261.