Abstract
The mammalian retina contains more than 40 retinal ganglion cell (RGC) subtypes based on their unique morphologies, functions, and molecular profiles. Among them, intrinsically photosensitive RGCs (ipRGCs) are the first specified RGC type emerging from a common retinal progenitor pool during development. Previous work has shown that T-box transcription factor T-brain 2 (Tbr2) is essential for the formation and maintenance of ipRGCs, and that Tbr2-expressing RGCs activate Opn4 expression upon native ipRGC ablation, suggesting that Tbr2+ RGCs contain a reservoir for ipRGCs. However, the identity of Tbr2+ RGCs has not been fully vetted. Here, using genetic sparse labeling and single cell recording, we showed that Tbr2-expressing retinal neurons include RGCs and a subset of GABAergic displaced amacrine cells (dACs). Most Tbr2+ RGCs are intrinsically photosensitive and morphologically resemble native ipRGCs with identical retinofugal projections. Tbr2+ RGCs also include a unique and rare Pou4f1-expressing OFF RGC subtype. Using a loss-of-function strategy, we have further demonstrated that Tbr2 is essential for the survival of these RGCs and dACs, as well as maintaining the expression of Opn4. These data set a strong foundation to study how Tbr2 regulates ipRGC development and survival, as well as the expression of molecular machinery regulating intrinsic photosensitivity.
Keywords: Intrinsically photosensitive retinal ganglion cells, displaced amacrine cells, Tbr2, Opn4
Graphical Abstract
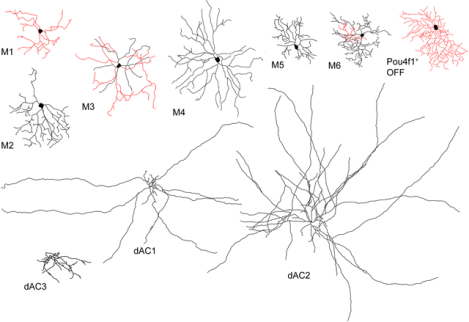
Using genetic sparse labeling and recording on a newly engineered Tbr2CreERT2 mouse line paired with various reporter lines, we identified seven types of Tbr2-expressing retina ganglion cells, including M1 to M6 types of ipRGCs and a Pou4f1+ RGC with bushy and small dendritic arbors stratifying into the OFF sub-laminae (in red). Additionally, we identified three types of Tbr2-expressing amacrine cells, including two widefield and a mediumfield displaced AC types, all with dendritic arbors stratifying into the ON sub-laminae (in black). Furthermore, we have demonstrated that Tbr2 is essential for the survival of these RGCs and dACs, as well as maintaining the expression of Opn4.
1. Introduction
Intrinsically photosensitive RGCs (ipRGCs), marked by the expression of opsin 4 (Opn4, aka melanopsin), are the first specified RGC types in developing mouse retina starting from embryonic day 14 (E14) (Mao et al., 2014; McNeill et al., 2011). IpRGCs play essential roles in non-image-forming vision, such as pupillary light reflex and circadian photo-entrainment (Lazzerini Ospri, Prusky, & Hattar, 2017). They also participate in image-forming vision by regulating RGC excitability as well as eye development (Brown et al., 2010; Rao et al., 2013; Schmidt et al., 2014; Sonoda, Lee, Birnbaumer, & Schmidt, 2018).
We and others have identified T-brain 2 (Tbr2 or Eomes) as the key regulator of ipRGC formation (Mao et al., 2008; Mao et al., 2014; Sweeney, Tierney, & Feldheim, 2014). The observation that many Tbr2-expressing RGCs activate Opn4 expression upon genetic ablation of native ipRGCs suggests that Tbr2+ RGCs contain a reservoir for native ipRGCs (Mao et al., 2014). However, the identity of Tbr2+ RGCs and the role Tbr2 plays in these processes remain unanswered. First, an estimated 17.8% of all RGCs were Tbr2 positive but only ~40% of Tbr2+ RGCs were thought to be reservoir for ipRGCs (Mao et al., 2014), leaving ~60% of Tbr2+ RGCs unaccountable. Second, it was shown that all supra-chiasmatic nucleus (SCN)-afferent RGCs expressed melanopsin (Baver, Pickard, Sollars, & Pickard, 2008), inconsistent with the observation that many cells in the Tbr2+ ipRGC reservoir do not express melanopsin or at levels below the sensitivity of immunohistochemical detection. Third, our previous conclusion that Tbr2 is essential for ipRGC survival was drawn from the near-complete loss of melanopsin-positive cells in adult Opn4Cre:Tbr2f/f retinas. This observation may alternatively be interpreted by the requirement of Tbr2 for ipRGC development and/or maturation because Opn4 expression is clearly detectable at early developmental stages. We shall tackle these unanswered questions here.
To examine the diversity of Tbr2+ retinal cells and Tbr2’s roles in ipRGC development, we generated a new knock-in mouse line containing the engineered Tbr2TauGFP-IRESCreERT2 allele (as shown in Fig. 1a). Using this line in conjunction with RosaiAP, Pou4f1CKOAP, and Ai9 reporter lines, we conducted genetic sparse labeling to investigate the diversity of Tbr2-expressing cells. We found that the vast majority of Tbr2+ RGCs are morphologically indistinguishable from the known M1-M6 ipRGCs. Evidently, these Tbr2+ RGCs project to SCN, ventral lateral geniculate nuclei (vLGN), inter-geniculate leaflet (IGL), and olivary pretectal nuclei (OPN), which constitute the primary targets of currently known ipRGC retinofugal projections (Delwig et al., 2016; Ecker et al., 2010; Hattar et al., 2006). We also found a small fraction of Pou4f1-expressing Tbr2+ RGCs and identified them morphologically as a unique bushy OFF RGC type with dendritic stratification distinctively different from and below those of the known M1 type. In addition to RGCs which constitute ~46% of Tbr2+ retinal neurons, surprisingly we found that ~54% of Tbr2+ retinal cells contain two types of GABAergic widefield displaced amacrine cells (dACs) and a mediumfield dAC type. These findings explains the mystery in our previous report where only ~40% of Tbr2+ cells were depleted in Opn4Cre:Tbr2f/f retinas (Mao et al., 2014). Moreover, by using a loss-of-function approach, we demonstrate that Tbr2 is indeed essential for survival of adult Tbr2+ RGCs and dACs. Most importantly, we found that Opn4 expression is down-regulated prior to the death of Tbr2-deleted RGCs in adult retinas, lending stronger support for the essential role of Tbr2 in maintaining Opn4 expression.
Figure 1. Tracing Tbr2-expressing retinal neurons.
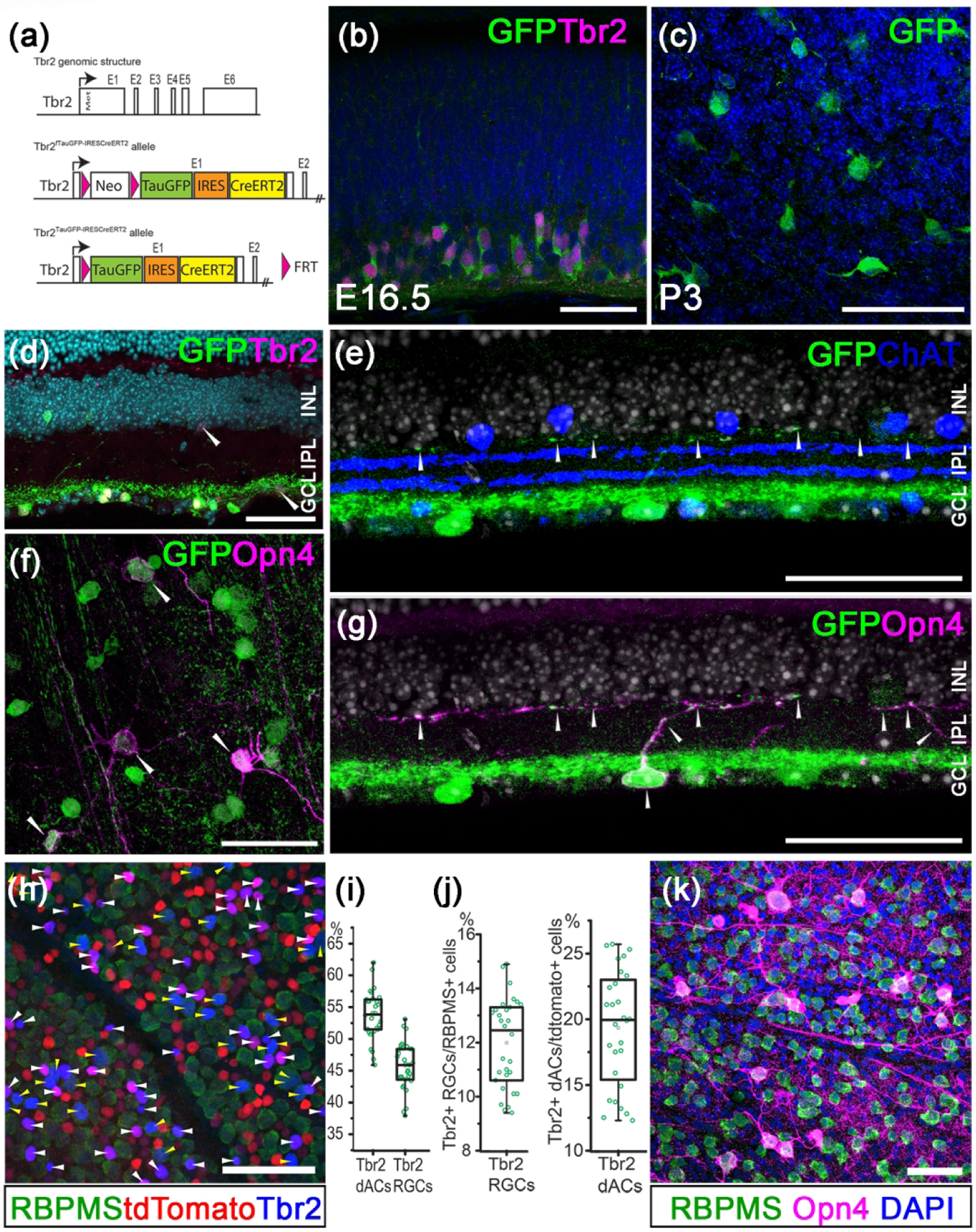
(a) Genomic structure of Tbr2 locus and the engineered allele in the Tbr2TauGFP-IRESCreERT2 mouse. (b) Double immunofluorescent staining showing Tbr2 expression (magenta) in Tbr2-driven GFP+ cells (green) in an E16.5 Tbr2TauGFP retinal section. (c) Representative immunofluorescent image showing Tbr2-driven GFP+ cells (green) in a P3 Tbr2TauGFP retinal flatmount. (d) Immunofluorescent image showing Tbr2 expression (magenta) in Tbr2-driven GFP+ cells (green) in a Tbr2TauGFP retinal section. (e) Fluorescent image of a Tbr2TauGFP retinal section showing dense Tbr2-driven GFP signal (green) in ON sub-laminae below the cholinergic ChAT bands (blue) in IPL. Note that GFP signal is separated from the ON ChAT band. (f) Representative co-immunofluorescent image showing melanopsin expression (magenta) in Tbr2-driven GFP+ cells (green) in a P30 Tbr2TauGFP retinal flatmount. (g) Double immunofluorescent staining showing melanopsin expression (magenta) in Tbr2-driven GFP+ cells (green). (h) Co-immunofluorescent staining on a Slc32a1Cre:Ai9 retinal flatmount showing Tbr2 expression (blue) with RBPMS+ RGCs (yellow arrowheads) or tdTomato+ RBPMS− dACs (white arrowheads). (i) Relative abundance of Tbr2+ RGCs and dACs described in G. (j) Relative abundance of Tbr2+ RGCs within the entire RGC population (left) and Tbr2+ dACs within the entire dAC population. (k) Co-immunofluorescent staining on a WT retinal flatmount showing melanopsin expression (magenta) with RBPMS+ RGCs (green). ChAT: cholinergic acetyltransferase. GCL: ganglion cell layer. INL: inner nuclear layer. IPL: inner plexiform layer. Scale bars: 50 μm (b-k).
2. Materials and Methods
2.1. Animals
Tbr2TauGFP-IRESCreERT2 mice (described in Fig. 1a) were generated by inserting a FrtPGKNeopA-Frt cassette and a synthetic TauGFP-IRESCreERT2 DNA fragment in the first exon behind the translation initiation codon of Tbr2. Gene targeting was conducted as described previously (Mao et al., 2008). Two targeted embryonic stem (ES) cell lines were used to generate Tbr2fTauGFPIRESCreERT2 mice. The Tbr2fTauGFPIRESCreERT2 mice were subsequently bred to a R26-FLPeR line to delete the Frt-flanked PGKNeopA cassette to generate the Tbr2TauGFPIRESCreERT2 mice. PCR primers Tbr2f1 (5’AAGGGGCACCTACAATCCCGA-3’) and Taur1 (5’-TTCGTTCCAGGCTTACCATCC-3’) were used for genotyping the Tbr2TauGFPIRESCreERT2 allele. Tbr2TauGFPIRESCreERT2 mice were bred to RosaiAP, Pou4f1CKOAP, or R26-tdTomato (Ai9) mice to produce double-transgenic mice for sparse labeling, dye filling and patch clamp recording. Tamoxifen (1–5 consecutive daily injections of 100 μg/g body weight; Sigma, St. Louis, MO) was injected intraperitoneally for Cre induction. Alternatively, 1 μl of 4-hydroxytamoxifen (0.1 mg/ml; Sigma) was injected into the vitreous space of the right eye to activate cells locally. The generation and genotyping of RosaiAP, Pou4f1CKOAP, Ai9, R26FLPeR, Slc32a1Cre, and Opn4TauLacZ mice were described previously (Badea, Cahill, Ecker, Hattar, & Nathans, 2009; Farley, Soriano, Steffen, & Dymecki, 2000; Hattar, Liao, Takao, Berson, & Yau, 2002; Madisen et al., 2010; Vong et al., 2011). All mice were maintained on C57BL6/129 mixed backgrounds. Mouse lines of either sex at various ages between P21 to 6 months were used. Pre-weaned animals were housed with their mother; weaned animals were housed in groups of no more than 5 in individually ventilated cages. All animal procedures followed the US Public Health Service Policy on Humane Care and Use of Laboratory Animals and were approved by the Animal Welfare Committee at The University of Texas Health Science Center at Houston and Institutional Animal Care and Use Committee at Baylor College of Medicine.
2.2. Immunohistochemical analysis
Retinal sections or flat-mounted retinas were fixed with 4% paraformaldehyde (PFA; Electron Microscopy Sciences, Hatfield, PA), then incubated with the primary antibodies. Alexa Fluor-conjugated secondary antibodies were obtained from Life Technologies, Carlsbad, CA (1:600 dilution). DAPI (2.5 μg/ml) was used to stain the nuclei. Images were acquired on a Zeiss 780 confocal laser scanning microscope (Carl Zeiss, Thornwood, NY) and exported as TIFF files into Adobe Photoshop (Adobe Systems, San Jose, CA). Cell counting was conducted using the cell counter plugin of NIH ImageJ.
2.3. Alkaline phosphatase (AP) staining
Tamoxifen-treated Tbr2TauGFP/+:RosaiAP/+ mice were used for alkaline phosphatase (AP) staining, which was conducted as previously described with minor modifications (Badea, Wang, & Nathans, 2003; Jamal, Kiyama, & Mao, 2020). Whole eyeballs were fixed with 10% neutrally buffered formalin for 5 min. The retinas were removed and flat mounted on a piece of nitrocellulose membrane, post-fixed for 10 min at room temperature, washed twice in phosphate-buffered saline (PBS), and heated in PBS for 30 min in 65°C water bath to inactivate endogenous AP activity. AP staining was performed in 0.1 M Tris (pH 9.5), 0.1 M NaCl, 50 mM MgCl2, 0.34 g/ml p-nitroblue tetrazolium chloride (NBT), and 0.175 g/ml 5-bromo-4-chloro-3-indolyl phosphate (BCIP) solution for 24–48 hours at room temperature with gentle shaking. After staining, retinas were washed 3 times for 5 min in PBS, post-fixed in PBS with 4% PFA briefly, dehydrated through an ethanol series, and then cleared with 2:1 benzyl benzoate/benzyl alcohol.
Montages of the whole retina were acquired on a Zeiss Axio Imager 2 microscope equipped with a motorized xyz drive (Carl Zeiss, White Plains, NY). Individual RGCs were imaged under transmitted light, and z-stacks were collected with a 20X objective (3.1 pixels/μm) at 0.58 μm vertical (z-dimension) intervals. For each RGC, the areas of dendritic arbors and somata, the branching, and the stratification depth within the inner plexiform layer (IPL) were analyzed using Zen lite (Carl Zeiss). Diameters of dendritic arbors and cell bodies were measured for each cell by calculating the diameter of a circle having the same area as a convex polygon, minimally enclosing the dendritic or somatic profile. Individual examples of distinct morphological types for M2, M4, and M5-like cells were traced using Adobe Photoshop or the NeuronJ plugin of ImageJ (Meijering et al., 2004).
2.4. Tracing RGC axon targets
For inspecting Tbr2CreERT2-driven RosaiAP expression in brains, tamoxifen-treated mice were perfused with 4% cold PFA, and then the whole brains were dissected and post-fixed with 4% PFA. Cryopreserved brains were sectioned into consecutive 100 μm coronal sections for AP staining.
2.5. Dye filling
Dye filling was conducted under room light as previously described with minor modifications (Mao et al., 2014). Briefly, retinas were flat-mounted on black membrane filter paper, and perfused with oxygenated Ames’ medium (US Biological, Salem, MA) in a recording chamber. The glass microelectrodes were tip-filled with a mixture of 0.5% Lucifer yellow-CH and 3.5% NBT and then backfilled with 3 M LiCl. The tdTomato+ RGCs were viewed under an epifluorescence microscope with a 40X water immersion objective and randomly targeted. Cell penetration was verified within the first minute of iontophoresis of Lucifer yellow with negative current (−0.5 nA at 3 Hz). NBT was then injected into cells with a positive current (+0.5 nA at 3Hz) for 5–10 min. Tissue was fixed with 4% PFA in 0.1 M PBS for 1 hr at room temperature. Filled cells were visualized by incubating with Alexa Fluor 488-conjugated streptavidin (1:1000 dilution) at 4°C overnight. The injected retinas were stained by an anti-GFP antibody to verify the targeted RGCs and an anti-ChAT antibody to determine the depth of dendritic stratification of injected cells.
2.6. Intrinsic membrane property and photosensitivity
Whole cell patch clamp recording was used as described in Kiyama et al. (2019) to test intrinsic photosensitivity of Tbr2+ cells. Retinas from tamoxifen-treated Tbr2CreERT2/+:Ai9 mice were flat-mounted with ganglion cell side up under dim red light in a perfusion chamber on an Olympus BX51WI fixed-stage microscope (Olympus USA, Central Valley, PA, USA) and supplied (3 ml/minute) with a warm (~34°C) carbogenated mammalian Ringer solution containing (in mM) 120 NaCl, 5 KCl, 25 NaHCO3, 0.8 Na2HPO4, 0.1 NaH2PO4, 2 CaCl2, 1 MgSO4, 10 D-glucose, 0.01 L-AP4, 0.01 AP5 and 0.04 NBQX. Retinal neurons were visualized by differential interference contrast under infrared illumination (> 900 nm) and Tbr2+ cells marked by tdTomato expression were identified by brief 50 msec light pulses (35000 R*/rod/sec) delivered and imaged through the mCherry filter set (Olympus USA). The cumulative total exposure of the retina was less than 1 second to preserve light sensitivity. Once a cell was selected, a 20-minute wait time was used to allow dark adaptation before recording. Patch pipettes made from borosilicate glass tubes (8–12 MΩ) were filled with a potassium-based internal solution containing (in mM) 125 K-gluconate, 8 NaCl, 4 ATP-Mg, 0.5 Na-GTP, 5 EGTA, 10 HEPES and 0.2% biocytin (w/v) with pH adjusted to 7.3 using KOH. The liquid junction potential was 15 mV and not corrected. Current clamp recording was conducted using the AM2400 amplifier (A-M Systems, Sequim, WA, USA) driven by the WinWCP program provided by John Dempster (University of Strathclyde, Glasgow, UK). Signals were filtered at 5 kHz and sampled at 10 kHz. To obtain intrinsic membrane property (IMP), a small negative current was injected immediately after break-in to adjust membrane potential from rest to −64 ± 2 mV. This was followed by five 600-msec serial negative current injections for hyperpolarization and four serial positive current injections for depolarization. The current that hyperpolarizes a cell to 100 ± 5 mV was empirically determined and used to adjust the amplitudes of subsequent serial current steps. Under these conditions, Tbr2+ cells exhibit characteristic IMP profiles that correspond to cells’ dendritic morphologies including IPL stratification levels. Intrinsic photoresponse was initiated by a 1-sec 470 nm light stimulation (~430 μm in diameter) at intensities of 2.67 to 6.14 log R*/rod/sec. Dendritic morphology of M1-like and other cells were recovered post hoc by staining for biocytin. IPL stratification level was determined in reference to the distance between the two VaChT or ChAT bands with the ON band designated as 0 and the OFF band as −1. Dendritic structure was traced in the Neurolucida 360 and quantified in the Neurolucida Explorer programs (MBF Bioscience, Boston, MA).
2.7. Antibody Characterization
The Pou4f1 antibody (Millipore: Cat# MAB1585; RRID:AB_94166, used at 1:500) is a mouse monoclonal IgG1 antibody made against aa 186–224 of Pou4f1/Brn-3a fused to the T7 gene 10 protein. Immunostaining using this antibody detects RGCs in wild-type mouse retinas but not in Pou4f1-mutant retinas (Xiang et al., 1995). The Tbr2 antibody (Abcam: Cat# ab23345; RRID: AB_778267, used at 1:500) recognizes a synthetic peptide corresponding to mouse Tbr2 aa 650 to the C-terminus. Immunostaining using this antibody detects RGCs in wild-type mouse retina but not in Tbr2-mutant retinas (Mao et al., 2014). The ChAT antibody (Millipore: Cat #AB144p; RRID: AB_2079751, used at 1:400) is a goat polyclonal antibody made against choline acetyltransferase. It has been widely used on immunostaining and western blotting. The GFP antibody (Thermo Fisher, Cat# A10262, RRID:AB_2534023, used at 1:1000) is a chicken IgY polyclonal antibody that recognizes jellyfish Aequorea Victoria green fluorescent protein. Its specificity was demonstrated by detection of different targets fused to GFP tag in transiently transfected cells. This antibody is very specific for detecting GFP-tagged proteins. The RBPMS antibody (Phosphosolutions, Cat# 1832-RBPMS, RRID:AB_2492226, used at 1:500) recognizes a synthetic peptide corresponding to aa residues from the N-terminal region of the rat RBPMS. The antibody is prepared from guinea pig serum by affinity purification. It has been widely used by immunofluorescence to detect RGCs (Kwong, Caprioli, & Piri, 2010). The melanopsin antibody (Advanced Targeting Systems, Cat#AB-N38, RRID:AB_1608077, used at 1:1000) recognizes a 15 aa N-terminal peptide of the mouse melanopsin. This antibody is specific for mouse, and is routinely tested by immunofluorescence (Berson, Castrucci, & Provencio, 2010).
2.8. EXPERIMENTAL DESIGN AND STATISTICAL ANALYSIS
Mouse lines (Tbr2TauGFP-IRESCreERT2/+: RosaiAP; Tbr2TauGFP-IRESCreERT2/+:Pou4f1CKOAP/+ and Tbr2TauGFP-IRESCreERT2/+: Ai9) of either sex at various ages between P21 to 6 months were used for sparse labeling, dye filling and patch clamp recording experiments. Cell counting experiments described in Fig.1 were conducted on 3 Slc32aCre/+:Ai9 mice. All data are presented as mean ± standard deviation for each genotype. Morphometric data of Table 1 are presented as mean ± sem.
Table 1:
Morphometric data and input resistance of Tbr2+ RGCs
| Primary dendrites | Branching no. | Dendritic area (mm2) | Dendritic length (mm) | Branch order | Segment size (μm) | Input Resistance (MΩ) | |
|---|---|---|---|---|---|---|---|
| M1n (8*) | 3.6 ± 0.5 | 21.3 ± 6.2 | 0.14 ± 0.02 | 2.31 ± 0.34 | 7.9 ± 1.1 | 32.8 ± 2.8 | 1239 ± 70 (26%) |
| M1r (5) | 3.2 ± 0.4 | 18.4 ± 3.7 | 0.15 ± 0.03 | 2.39 ± 0.30 | 7.6 ± 1.4 | 33.0 ± 2.2 | 1439 ± 91 (16) |
| M1s (9) | 3.4± 0.4 | 17.2 ± 2.5 | 0.12 ± 0.02 | 2.07 ± 0.25 | 8.0 ± 1.1 | 37.4 ± 4.8 | 604 ± 75 (16) |
| M2-like (3) | 5 ± 0.6 | 32 ± 9.5 | 0.19 ± 0.05 | 3.34 ± 0.46 | 8.3 ± 1.4 | 27.9 ± 6.5 | 317 ± 52 (7) |
| Bistratified (7) | 3.6 ± 0.2 | 37 ± 12.1 | 0.16 ± 0.05 | 2.72 ± 0.61 | 10 ± 2.4 | 34.7 ± 8.1 | 473 ± 83 (12) |
| M4-like (4) | 4.5 ± 0.3 | 31 ± 3.5 | 0.26 ± 0.07 | 4.01 ± 0.69 | 10.8 ± 1.7 | 34.5 ± 7.5 | 170 ± 25 (13) |
| M5-like (9) | 3.5 ± 0.4 | 52 ± 13.9 | 0.08 ± 0.01 | 2.22 ± 0.28 | 12.7 ± 1.8 | 24 ± 4.8 | 376 ± 28 (26) |
The number of cells traced and analyzed.
The number of cells analyzed including those identified by IMP and dendritic morphology.
3. Results
3.1. Tbr2-driven TauGFP expression recapitulates endogenous Tbr2 expression
In order to trace and sparsely label retinal neurons that express Tbr2, we have generated a new knock-in mouse line Tbr2TauGFP-IRESCreERT2 (labeled as Tbr2TauGFP or Tbr2CreERT2 interchangeably in the text according to its usage relevancy) in which a TauGFPIRESCreERT2 dual reporter cassette was inserted in-frame behind the translational start codon of Tbr2 (Met, Fig. 1a). Immunostaining with an anti-GFP antibody detected GFP in E16.5 and P3 developing Tbr2TauGFP/+ retinas (Fig. 1b, 1c). In adult Tbr2TauGFP/+ retinas, GFP signals were clearly present in Tbr2-expressing cells and the signal weakened when Tbr2 expression levels were barely detectable (white arrowheads in Fig. 1d). These data validated the usefulness of this mouse line to track endogenous Tbr2 expression. We then found in adult Tbr2TauGFP retinas that GFP signal was conspicuously present at the lower part of IPL below the ON cholinergic acetyltransferase (ChAT) band (Fig. 1e). A noticeable though weaker signal was also found in a narrow band lining the border of IPL and inner nuclear layer (INL) (arrowheads in Fig. 1e, 1g) similar to that of Opn4/melanopsin staining pattern. By comparing Opn4 and GFP expression on Tbr2TauGFP retinal flatmounts and sections, we found that all Opn4-expressing cells express GFP (Fig. 1f, 1g) and weak but detectable GFP signal can be seen on melanopsin-marked dendrites (arrowheads in Fig. 1g), further affirming the utility of this mouse line for studying the relationship between Tbr2 and melanopsin-expressing ipRGCs.
Previous estimate put Tbr2-expressing retinal cells at approximately 8.9% of all DAPI+ nuclei in the ganglion cell layer (GCL) (Mao et al., 2014). The previous conclusion that Tbr2+ RGCs must account for 17.8% of all RGCs was based on two assumptions: 1) All Tbr2+ cells are RGCs, and 2) RGCs account for ~50% of all GCL cells. To test these assumptions, we co-stained GFP and a pan-RGC marker RBPMS (Kwong et al., 2010). Surprisingly, we found that less than half of the GFP+ cells expressed RBPMS (data not shown), indicating that only a fraction of Tbr2+ cells are RGCs and that the Tbr2+RBPMS− cells may be displaced amacrine cells (dACs). To verify this, we examined Tbr2 signal with tdTomato-labeled inhibitory neurons in a Slc32a1Cre:Ai9 mouse (Vong et al., 2011), and found that ~54% of the Tbr2+ cells in the GCL are Ai9+RBPMS− dACs (53.6 ± 1.5%, white arrowheads in Fig. 1h, 1i), and that the remaining ~46% Tbr2+ cells are RBPMS+ RGCs (46.4 ± 1.5%, yellow arrowheads in Fig. 1h, 1i). Using RBPMS and Slc32a1Cre:Ai9 as pan-RGC and dAC readouts, respectively, we conclude that Tbr2+ RGCs account for 11.6 ± 1.0% of all RGCs, and that Tbr2+ dACs account for 18.1 ± 1.3% of all dACs (Fig. 1j). Moreover, co-immunostaining of melanopsin and RBPMS revealed that ~6.3% (6.26 ± 1.23%) of RBPMS+ RGCs express melanopsin (Fig. 1k), suggesting that ~50% of Tbr2+ RGCs express detectable level of Opn4 under our experimental conditions.
3.2. Characterization of Tbr2-expressing retinal neurons
To determine the identities of Tbr2-expressing retinal neurons, we bred Tbr2CreERT2 with the RosaiAP mouse line to produce Tbr2CreERT2/+:RosaiAP/+ mice, then injected tamoxifen through the intraperitoneal (IP) route for genetic sparse AP labeling in retinal flatmounts (Fig. 2a) (Badea, Hua, et al., 2009; Badea & Nathans, 2004; Jamal et al., 2020). In agreement with the above finding that Tbr2 is expressed in ~18% of dACs, AP-marked retinal neurons were primarily comprised of widefield dACs (dendritic field diameter > 500 μm) amongst diverse types of RGCs with dendrites entangled within a dense and narrow band in IPL (Fig. 2b and arrowheads in Fig. 2b’). By breeding Tbr2CreERT2 into the Ai9 background (Fig. 2c), we found that tdTomato+ cells from tamoxifen-treated Tbr2CreERT2:Ai9 retinas co-labeled with Tbr2 and Tbr2-driven GFP expression (Figure 2d, 2e). Using neurobiotin (NBT) dye-filling to label the tdTomato+ cells in tamoxifen-treated Tbr2CreERT2:Ai9 retinas, we found that these dACs have long dendritic processes that ramify at the ON laminae of the IPL below the ON-ChAT band (Fig. 2f, f’ and Fig. 3 insets). This stratification pattern is also consistent with the location of GFP signals in crosssections of Tbr2TauGFP/+ retinas shown above (Fig. 1d–1g).
Figure 2. Identification of Tbr2+ RGCs and dACs.
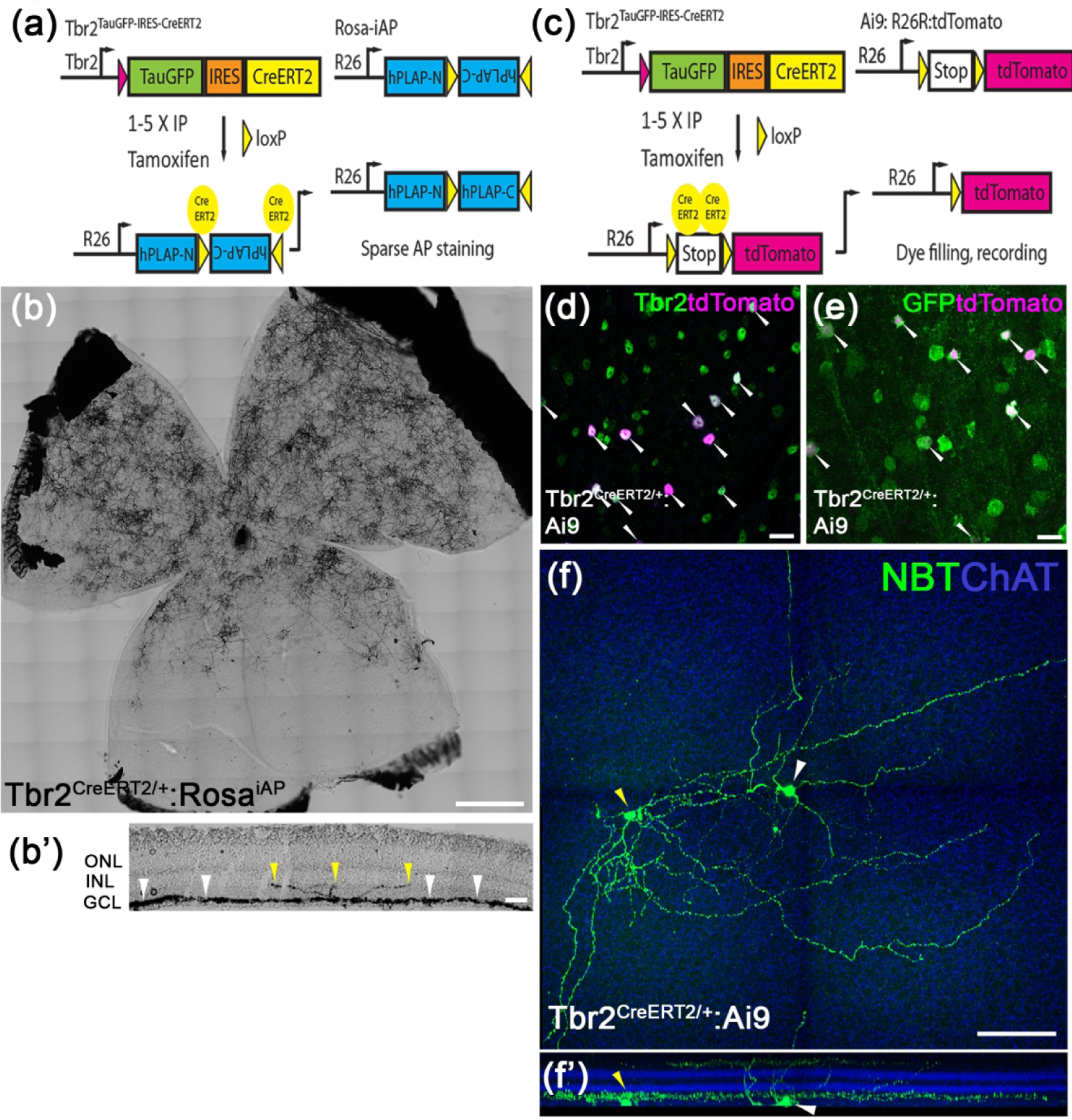
(a) The genetic sparse labeling system in the Tbr2CreERT2:RosaiAP mouse line. (b, b’) Representative AP staining from a flat-mounted P30 Tbr2CreERT2:RosaiAP retina showing intense AP+ Tbr2-expressing retinal neurons (b) and the location where their terminal dendrites stratify in IPL (b’). (c) The fluorescent labeling system of Tbr2+ neurons in the Tbr2CreERT2:Ai9 mouse line. (d, e) Co-immunofluorescent staining on a Tbr2CreERT2/+:Ai9 retinal flatmount showing tdTomato expression (magenta, white arrowheads) with Tbr2 (green) (d) or Tbr2-driven GFP (e). (f, f’) Representative images showing the morphology of a Tbr2-expressing displaced amacrine cell (yellow arrowhead) and a nearby RGC (white arrowhead) revealed by filled neurobiotin (f) and the location where their terminal dendrites stratify in reference to ChAT bands in IPL (f’). NBT: neurobiotin. ChAT: cholinergic acetyltransferase. Scale bars: 500 μm (b), 20 μm (d, e), 100 μm (f).
Figure 3. Characterization of Tbr2+ dACs.
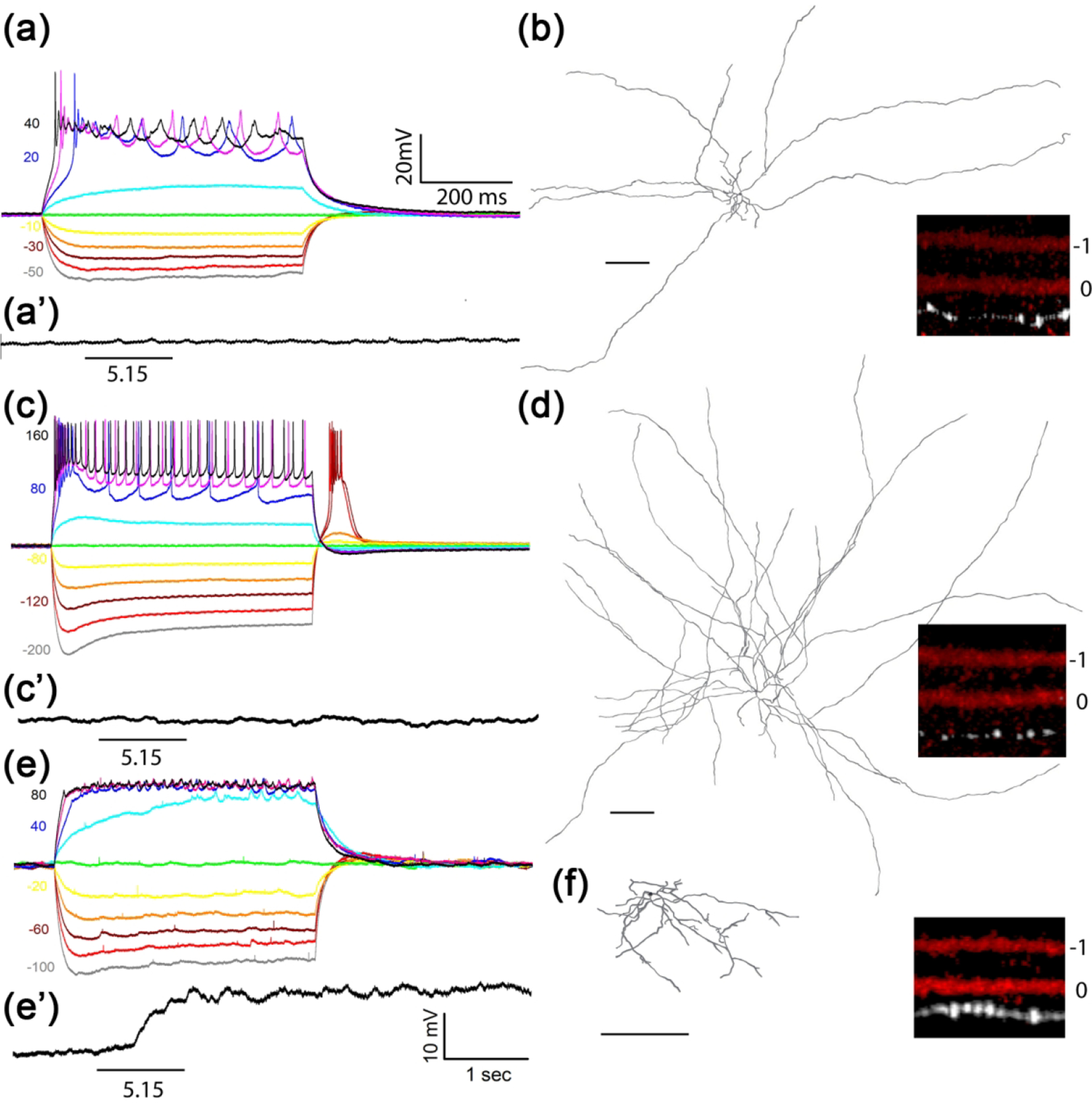
Representative intrinsic membrane property (IMP) profiles (a, c, e), intrinsic light sensitivity (a’, c’, e’), dendritic structure (b, d, f) and IPL stratification levels (insects) of three Tbr2+-dACs. Membrane potential changes in response to 600 msec current injections of varying amplitude and polarity are shown at left. (a, b) Tbr2+-dAC1, not intrinsically photosensitive (a’), is the most frequently encountered type (65%) with a relatively high input resistance (a) and characterized by asymmetric short and long dendrites (b) stratifying to 0.75 ± 0.04 IPL position in reference to designated ON-ChAT position at 0 and OFF-ChAT position at −1 (inset). (c, d) Tbr2+dAC2 (31%) has the lowest input resistance (c) and is not intrinsically photosensitive (c’) with many radiating long dendrites that span the retina (d) and stratify to 0.77 ± 0.07 IPL position (inset). Note the larger currents required to elicit similar amplitude changes in the Tbr2+-dAC2. (e, f) Tbr2+-dAC3 is a rarely encountered (4%) mediumfield dAC with discernable IMP (e) and apparent intrinsic photosensitivity (e’) with closer stratification level to ON-ChAT band at 0.50 ± 0.12 (inset). Intrinsic photosensitivity was tested under 1-sec light step of 5.15 log R*/rod/sec. Scale bar: 200 μm.
Whole-cell current clamp recording of 230 Tbr2+ neurons from 36 adult Tbr2CreERT2:Ai9 retinas found 156 dACs and among them three distinguishable IMP profiles. The most abundant type (65%) has a very high input resistance (1094 ± 38 MΩ, n = 75) and asymmetric dendritic morphology similar to the one identified by NBT dye filling (Fig. 2f), with dendrites stratifying to IPL level of 0.75 ± 0.04 (n = 6, Fig. 3a, 3b and inset, in reference to designated OFF-ChAT level of −1 and ON-ChAT at 0). The second dAC type (31%) has a much lower input resistance (300 ± 18 MΩ, n = 52) but a larger dendritic area and density whose dendrites stratify to 0.77 ± 0.07 IPL level (n = 6, Fig. 3c, 3d and inset). These two dAC types exhibited no intrinsic photosensitivity. The third type is rarely encountered and is a mediumfield dAC type (4%) with a medium input resistance (748 ± 87 MΩ, n = 6) and a statistically significant closer stratification level to the ON-ChAT band at 0.50 ± 0.12 (n = 6, P < 0.0001, Fig. 3e, 3f and inset). One cell out of six recorded in this mediumfield dAC type exhibited light sensitivity under glutamatergic synaptic blockade (Fig. 3e’), suggesting that it may be connected through electrical synapse to ipRGCs.
It is worth noting that by delivering 4-hydroxytamoxifen (4-OHT) directly into the vitreous space (1μl, 0.1 mg/ml), we could label more RGCs than dACs to reduce the interference from dAC-derived dendritic AP signals while characterizing RGC dendritic morphology. In subsequent studies, we shall focus on describing the Tbr2+ RGCs.
3.3. M1-like Tbr2+ RGCs
We have examined 291 cells by AP staining from 37 adult retinas and found 50 “M1-like” cells based on stratification of their terminal dendrites to the boundary between IPL and INL, a signature feature of conventional M1 ipRGCs (Fig. 4a–4d). These cells had medium-sized somata (16.16 ± 3.0 μm in diameter) but displayed large variations in dendritic arbor sizes (see green polygons in Fig. 4a–4d; arbor diameter: 264.08 ± 48.75 μm) and degree of dendritic branching (23.69 ± 6.20). The heterogeneity reflects discordant observations of M1-ipRGC sizes in the literature (Berson et al., 2010; Ecker et al., 2010; Emanuel, Kapur, & Do, 2017; Lee & Schmidt, 2018; Muller, Do, Yau, He, & Baldridge, 2010; Schmidt & Kofuji, 2011). Most of these M1-like Tbr2+ RGCs have 2 or 3 primary dendrites, although one cell with a single primary dendrite (red arrowheads in Fig. 4e, 4e’) and a smaller soma was also encountered (11.3 μm vs. 16.16 μm). We also detected several Tbr2+ RGCs whose somata were in the INL (red arrowheads in Fig. 4f–4h). These cells have variable dendritic arbor sizes as well but all stratify exclusively to the OFF sub-laminae of the IPL. In agreement with our previous finding that all displaced ipRGCs express Tbr2 (Mao et al., 2014), these cells appeared to be displaced M1-ipRGCs.
Figure 4. M1-like Tbr2+ RGCs.
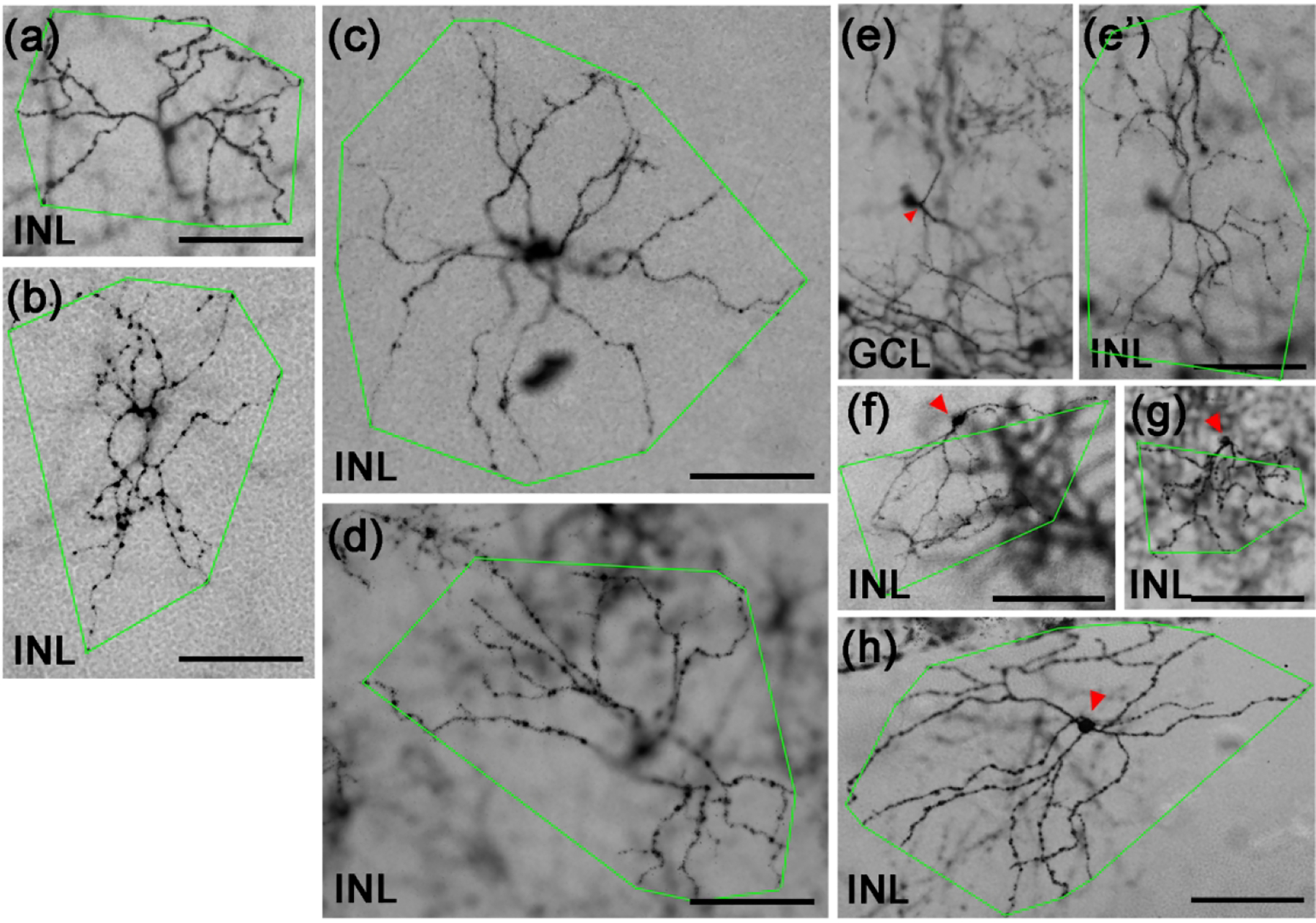
(a-h) Representative brightfield images of AP-stained Tbr2CreERT2/+:RosaiAP/+ retinal flatmounts. Images were focused on the terminal dendrites located in between INL/IPL boundary (labeled as INL) except in (e) where the focus was on the soma in the ganglion cell layer. The green polygons represent the terminal dendritic arbor. The red arrowhead in E indicates the single primary dendrite of this RGC. Three representative displaced M1-like RGCs (f-h) with somata located in INL (pointed by red arrowheads) were also shown. GCL: ganglion cell layer. INL: inner nuclear layer. IPL: inner plexiform layer. Scale bars: 100 μm (a-h).
To determine whether morphological heterogeneity of these M1-like Tbr2+ RGCs further reflects the heterogeneity in biophysical properties reported for conventional M1 ipRGCs identified in transgenic lines or by retrograde labeling, we performed targeted patch clamp recording of 56 additional Tbr2+ cells under glutamatergic synaptic blockade in 13 tamoxifen-treated Tbr2CreERT2:Ai9 retinas. We found 33 so-called M1-like cells, all with robust intrinsic photosensitivity (Fig. 5). Together with cells identified by dendritic morphology only but without testing for their intrinsic photosensitivity, we have in total analyzed 67 cells and placed them in three subgroups based on their IMP profiles, namely, the presence of rebound spiking following current injection, the level of input resistance, and the appearance of a sag upon hyperpolarization (Table 1 and Fig. 5). The first and most abundant subgroup, called M1n, is characterized by the presence of a hyperpolarization sag but no rebound spiking (M1n: n=31, 46%) (Fig. 5a). The second subgroup has both a hyperpolarization sag and rebound spiking (M1r; n=19, 28%) (Fig. 5b). The third subgroup has no hyperpolarization sag and with or without rebound spiking (M1s; n=17, 26%) (Fig. 5C). By tracing representative cells randomly selected from each subgroups and performed quantitative morphometric analysis, we found no significant differences among them in primary dendrite numbers, branching numbers, dendritic lengths, dendritic field sizes, branching orders and segment sizes (Table 1). However, cells in the M1s subgroup have a significantly lower input resistance at 604 ± 75 MΩ (n=16) than those of M1n and M1r at 1239 ± 70 MΩ (n=26, p <0.00001) and 1439 ± 91 MΩ (n=16, P<0.00001), respectively. Because conventional M1-ipRGCs exhibit a broad range of intrinsic photosensitivity (Milner & Do, 2017); therefore, it will be interesting to determine whether input resistance difference observed here may correlate with intrinsic photosensitivity among the M1-like Tbr2+ cells in future studies.
Figure 5. IMP and intrinsic photosensitivity of M1-like Tbr2+ RGCs.
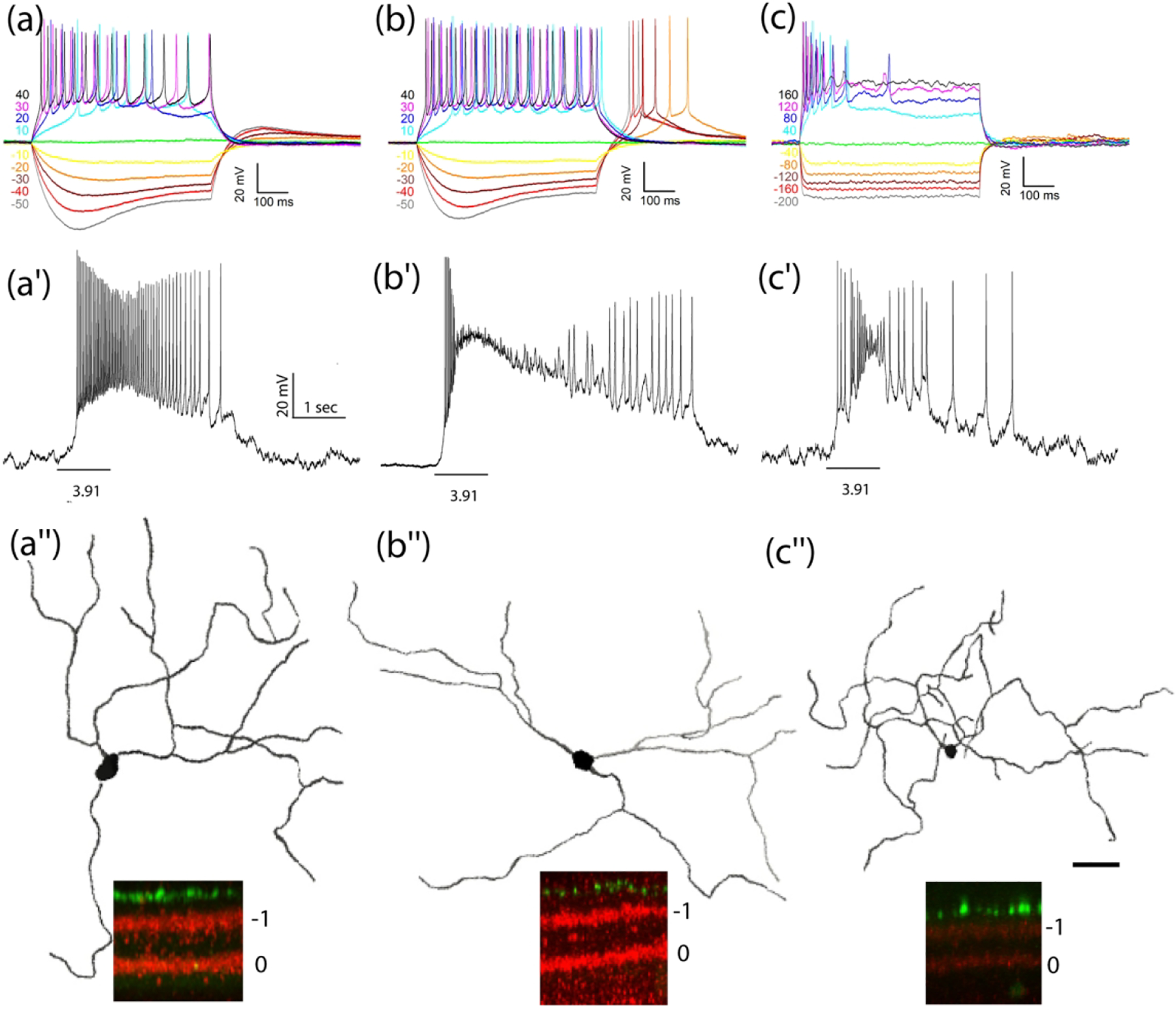
(a-c) Representative IMP profiles: M1n (a), M1r (b), M1s (c), intrinsic photosensitivity (a’-c’) to 1-sec light step of indicated intensity in log R*/rod/sec, and dendritic morphology and stratification patterns (a“-c”) from three M1-like Tbr2+ RGC subtypes. See Table 1 for detailed morphometric analysis which reveals no significant difference between the three except that the M1s subtype has a significantly lower input resistance than the other two. Scale bar: 50 μm.
3.4. Bistratified Tbr2+ RGCs
We identified 73 AP-labeled bistratified RGCs with diverse morphological characteristics (Fig. 6). The first type (n=14) had smaller dendritic arbor sizes and extensive branching patterns (Fig. 6a, 6b). Their 2 dendritic arbors stratified next to the inner (OFF) and outer (ON) margins in the IPL, and the dendritic arbors in the ON sub-laminae (red polygons) were larger than that in the OFF sub-laminae (green polygons) (Fig. 6a–6b’). This type of RGC is similar to the recently described M6 ipRGCs from a Cdh3-GFP mouse line (Quattrochi et al., 2019). The other 59 bistratified Tbr2+ RGCs had relatively larger arbors and less dendritic branching. Twenty-five of these cells had dendrites confined almost entirely to the ON sub-laminae (Fig. 6c, 6d) with only a few straying ones ramifying up into the OFF sub-laminae (black arrowheads in Fig. 6c’, 6d’). Eighteen cells had similar dendritic arbors in ON and OFF sub-lamina (Fig. 6e, 6e’), while the remaining 16 cells had dendrites stratified mainly to the OFF-layer (Fig. 6f, 6f’). These larger bistratified RGCs are similar to the M3 ipRGCs described previously (Schmidt & Kofuji, 2011). We have recorded from a total of 14 bistratified Tbr2+ RGCs by current clamp recording. Dendritic morphology were obtained from eight and analyzed as one bi-stratified group shown in Table 1. We found varying intrinsic photosensitivity, including six cells (43%) without detectable intrinsic photosensitivity under our brightest light stimulation (Fig. 6g). The IMP profiles of these bistratified Tbr2+ RGCs are not uniform. Those with larger OFF layer arbors have no hyperpolarization sag (n = 4, example in Fig. 6h), while those with larger ON layer arbors (n = 3) have a unique profile marked by a hyperpolarization sag with or without rebound spiking (Fig. 6i–k). These bistratified Tbr2+ RGCs therefore are diverse not only in dendritic morphology, but also in IMP, and intrinsic photosensitivity.
Figure 6. Bistratified Tbr2+ RGCs.
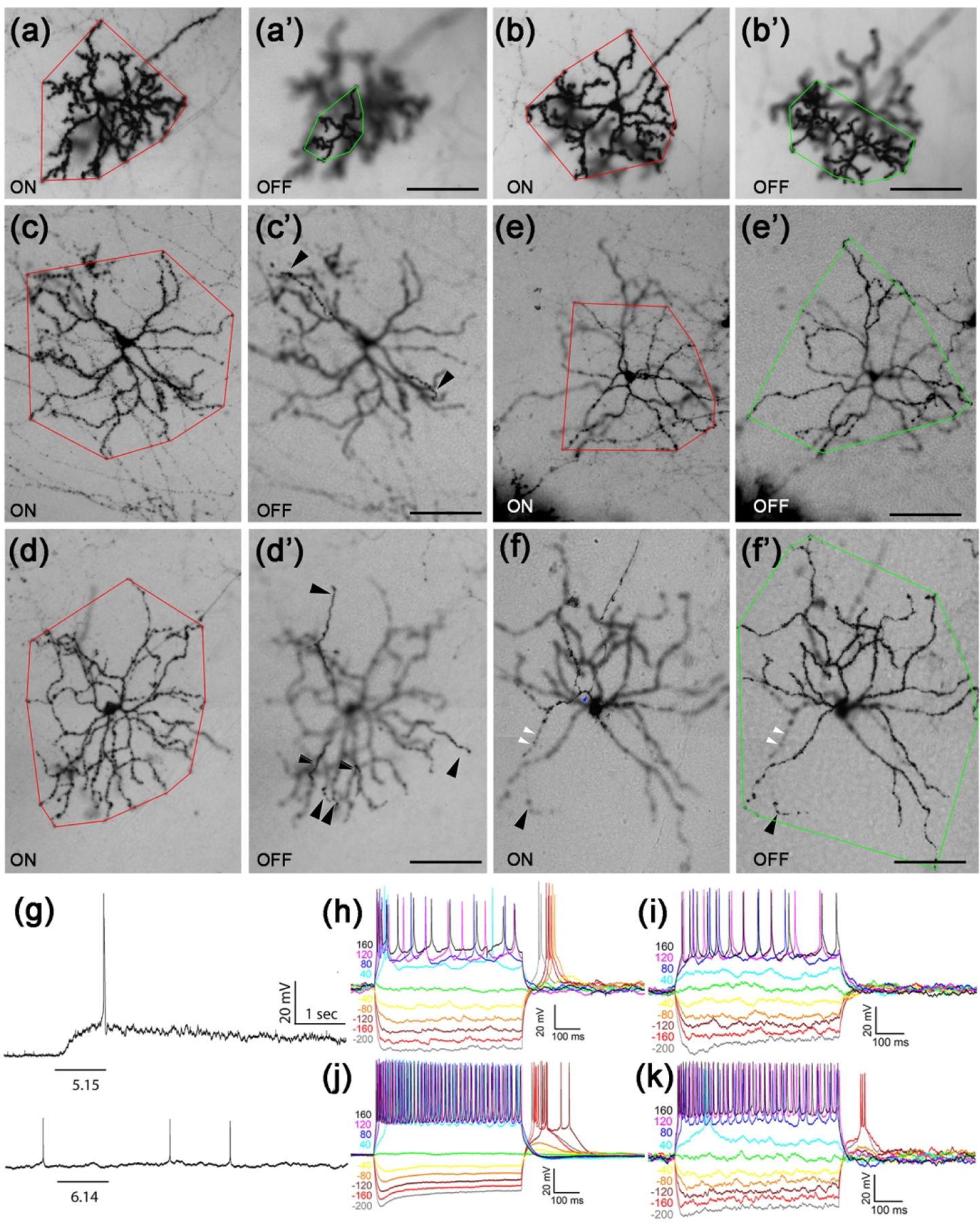
(a-f’) Representative AP-stained images of M6-like (a-b’) and various types of M3-like (c-f’) Tbr2+ RGCs from Tbr2CreERT2/+:RosaiAP/+ retinal flatmounts. Images were focused on the ON (a-f) or OFF (a’-f’) IPL sub-lamina.. Red polygons represent the terminal dendritic arbor in the ON-layer, while the green polygons represent those in the OFF-layer. The white arrowheads indicate the sparse terminal dendrites in the ON layer, and the black arrowheads indicat those in the OFF-layer. (g) Representative M3- (upper) and M6-like (lower) Tbr2+ RGC instrinsic photoresponses to 1-sec light step of indicated intensity in log R*/rod/sec. (h-k) Representative and diverse IMP profiles associated with the bistratified Tbr2+ RGCs. The M6 subtype has a distinguishable IMP profile (k) from the M3 subtypes (h-j). Scale bars: 100 μm (a’-f’).
3.5. ON-layer stratified Tbr2+ RGCs
Because the two widefield dAC types described above have long and narrowly stratified and massive dendritic network in the ON lamellae, detailed analysis of Tbr2+ RGCs with dendritic stratification to or near this layer was done on 165 cells whose dendrites were not entrenched with others. Their dendritic characteristics were similar to published ON layer stratified M2, M4, and M5 ipRGC subtypes. Below we separate them by the sizes of their dendritic arbors, somata, branching patterns and IMPs.
3.5.1. M4-like Tbr2+ RGCs
M4 ipRGCs have been characterized as having the largest dendritic arbor area and soma diameter among all known ipRGCs (Estevez et al., 2012) and a recent study identified them intersectionally in Opn4Cre:Z/EG mice using coincidental GFP and SMI32 signals (Sonoda, Okabe, & Schmidt, 2019). In 37 AP-stained retinas, we analyzed 15 these M4-like Tbr2+ RGCs with large dendritic arbors and somata (arbor diameter: 327.95 ± 48.51 μm; soma diameter: 19.28 ± 3.49 μm; N=15) and relatively loose dendritic arbors (Fig. 7a–7b’). They displayed less complex branching (branch points: 29.14 ± 3.46) when compared with other ON-layer stratified Tbr2+ RGCs. We encountered 13 of these M4-like cells by current clamp recording and in contrast to M1-like and bistratified RGCs, all exhibited a characteristic IMP profile as previously described (Fig. 7c) (Kiyama et al., 2019). We tested and found 2 cells with intrinsic photosensitivity under our brightest stimulation (Fig. 7d). We also randomly traced and analyzed 4 of these M4-like cells and found them to have the largest dendritic arbors among all RGCs in Tbr2CreERT2:Ai9 retinas (Table 1)
Figure 7. Large M4-like Tbr2+ RGCs.
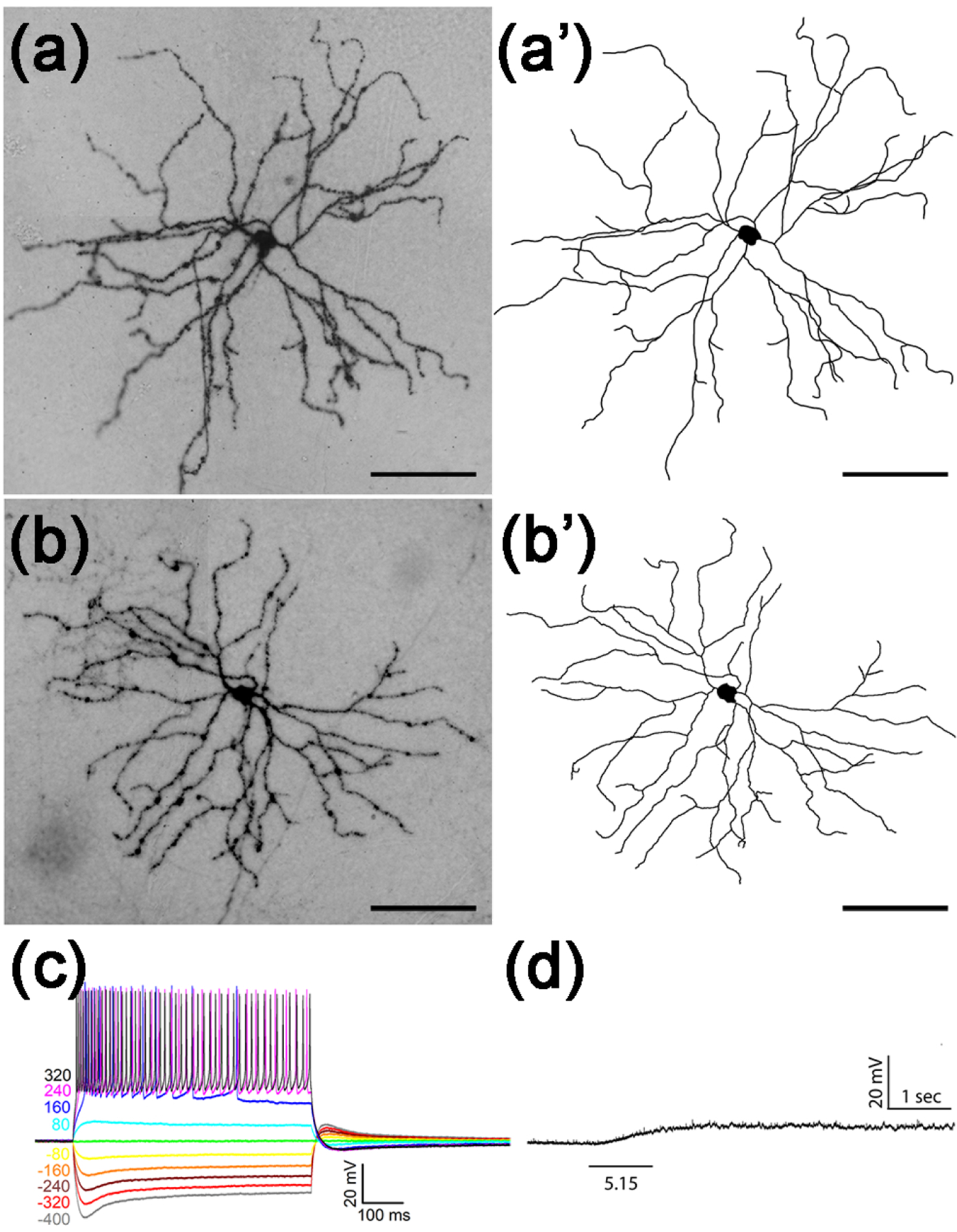
(a-b) Representative images of AP-stained M4-like Tbr2+ RGCs from Tbr2CreERT2/+:RosaiAP/+ retinal flatmounts. (a’-b’) Neuronal tracing of the ministacked images shown in A-B. (c-d) Stereotypic IMP profile (c) and representative intrinsic photoresponse of a M4-like Tbr2+ RGC to 1-sec light step of indicated intensity in log R*/rod/sec. (d). Scale bars: 100 μm (a-b’).
3.5.2. M5-like Tbr2+ RGCs
The native M5 ipRGC subtype has dense, bushy dendritic trees with smaller dendritic arbors when compared to M2 and M4 types and stratifies below the ON ChAT band in the IPL (Ecker et al., 2010; Stabio et al., 2018). We found 40 AP-stained Tbr2+ RGCs with the M5 morphological characteristics (arbor diameter: 158.32 ± 24.03 μm; soma diameter: 14.38 ± 1.86 μm, N=35, Fig. 8a, b). Notably, their dendrites emanated from 3 to 5 primary dendrites (3.75 ± 0.7, N=29) and stratified slightly below the ON ChAT band (depth in IPL: 85.73% ± 1.48%, N=20), forming more extensive branching than other ipRGCs (49.5 ± 9.58, N=20). We frequently encountered these M5-like Tbr2+ cells by current clamp recording (N=28). Of 10 cells where intrinsic photosensitivity was tested, 3 (30%) had no detectable intrinsic photosensitivity (Fig. 8c). The IMP profiles of these M5-like Tbr2+ cells appeared diverse but all contained a unique characteristic delayed return to baseline following positive current injections (Fig. 8d–f, arrows). We randomly traced and analyzed nine such M5-like cells and found them to have the smallest dendritic arbors but the highest number of dendritic branching (Table 1).
Figure 8. Small M5-like Tbr2+ RGCs.
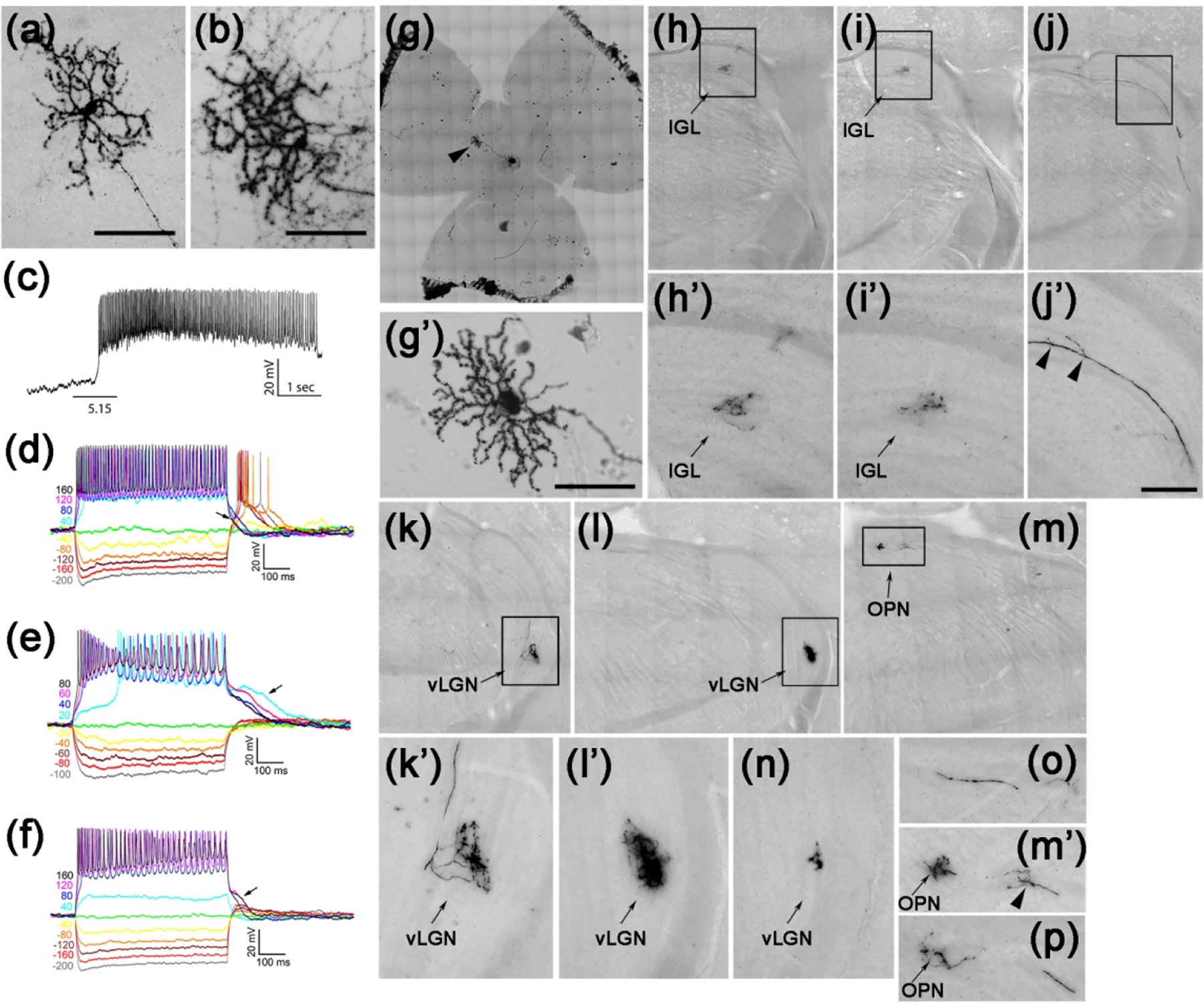
(a-b) Representative AP-stained images of M5-like Tbr2+ RGCs from Tbr2CreERT2/+:RosaiAP/+ retinal flatmounts. (c) Representative intrinsic photoresponse to 1-sec 5.15 log R*/rod/sec light step. (d-f) Representative and diverse IMP profiles of M5-like Tbr2+ RGCs. Note the characteristic return delay to the baseline following positive current injections (arrows). (g-g’) Retina with a single M5-like Tbr2+ RGC. (h-p) AP-stained brain sections showing multiple axonal projections of the M5-like RGC shown in g’ in several different regions in the brain. (h’-m’) Enlarged images of the bracketed regions in H to M, respectively. IGL: inter-geniculate leaflet. OPN: olivary pretectal nuclei. vLGN: ventral lateral geniculate nuclei. INL: inner nuclear layer. Scale bars: 100 μm (a, b, g’).
Fortuitously, in one animal whose right eye was injected with a low dose of 4-OHT (1 μg/μl), we found a single M5-like RGC in the entire retina (Fig. 8g, 8g’). AP staining on brain sections revealed its retinofugal projection to the IGL (Fig. 8h–8i’), vLGN (Fig. 8k–8l’, 8n), and OPN (Fig. 8m, 8m’, 8o). Notably, the size and complexity of its axonal arbors were different in different brain regions. In addition, the axon bifurcated and gave rise to several telodendria along its path (arrowheads in Fig. 8j’, 8m’). Such projection patterns are similar to those obtained from a single M1-ipRGC, which sends collateral axonal inputs to SCN, IGL, and other brain regions (Fernandez, Chang, Hattar, & Chen, 2016). Whether this is a common feature of all ipRGCs is an interesting topic for future investigations.
3.5.3. M2-like Tbr2+ RGCs
M2 ipRGCs have an intermediate dendritic arbor size between M4 and M5. We analyzed 24 such “M2-like” AP-stained Tbr2+ RGCs (Fig. 9a–9c’). They had relative medium dendritic arbors (arbor diameter: 223.16 ± 27.82 μm; N=21) and somata (soma diameter: 16.95 ± 2.22 μm; N=22). Their dendrites emanated from 3 to 5 primary dendrites (3.96 ± 0.61, N=23) and branched more variably than other ipRGCs (40.25 ± 14.03, N=20). These M2-like cells possessed an IMP profile different from those of M4-like and M5-like cells, characterized by the lack of rebound depolarization and immediate return to baseline following current injections (Fig. 9d). We encountered nine M2-like Tbr2+ cells and of three where intrinsic light response was examined, two were intrinsically photosensitive (66%) (Fig 9e). Three randomly selected M2-like cells were traced and analyzed with data obtained support the notion that dendritic arbor of these cells lied in between the bigger M4-like and the smaller M5-like cells. IMP profile of these M2-like RGC appears stereotypic (Fig. 9d).
Figure 9. M2-like Tbr2+ RGCs.
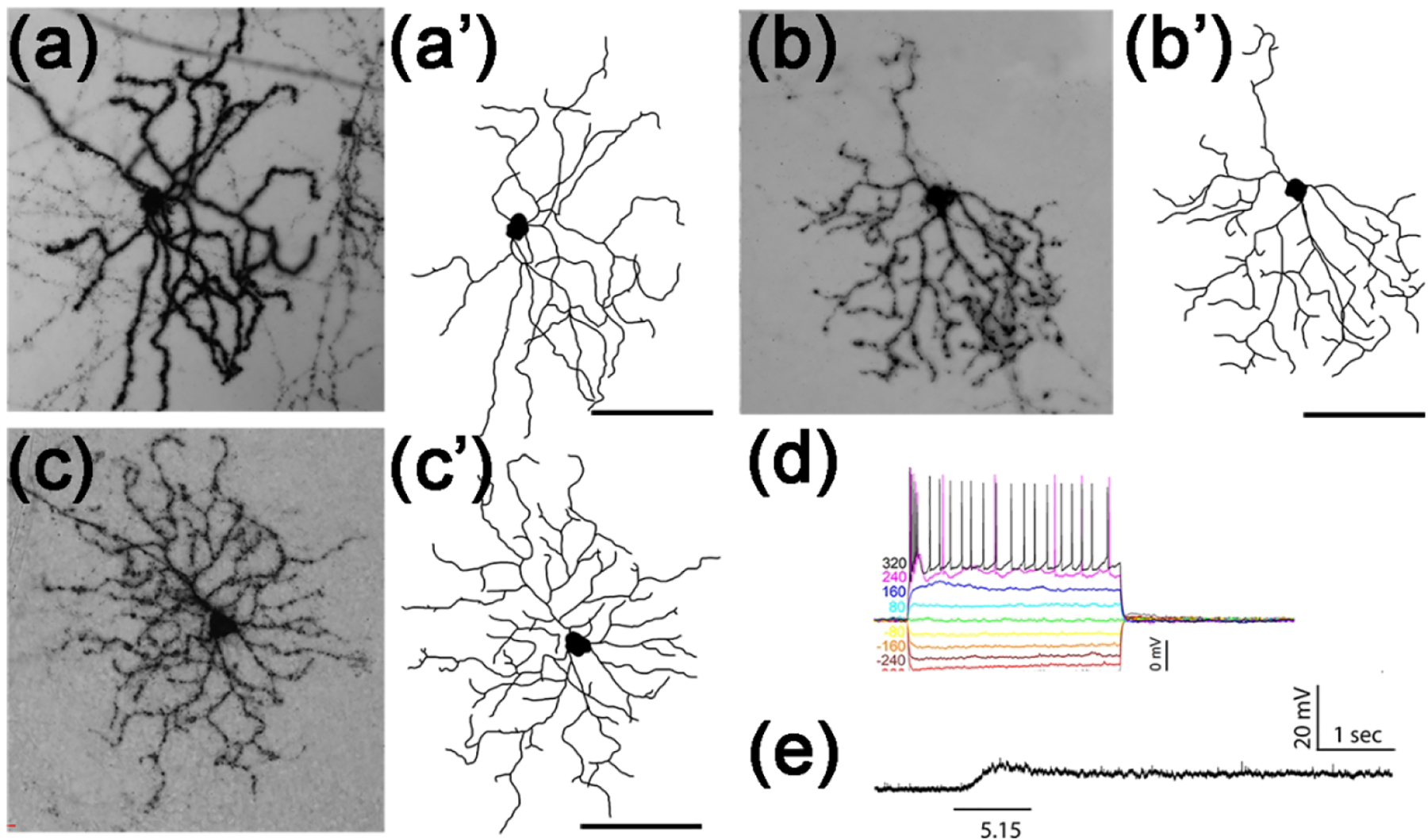
(a-c) Representative AP-stained images of 3 M2-like Tbr2+ RGCs from Tbr2CreERT2/+:RosaiAP/+ retinal flatmounts. (a’-c’) Corresponding tracing of ministacked images shown in A to C. (d) Representative stereotypic IMP profile of M2-like Tbr2+ RGCs. Note the quick return to baseline and the absence of regenerative changes following large current injections. (e) Representative intrinsic response to 1-sec light step of indicated intensity in log R*/rod/sec. Scale bars: 100 μm (q’-c’).
3.6. Pou4f1-expressing Tbr2+ RGCs
A small percentage of Tbr2-expressing cells expressed RGC marker Pou4f1 (Fig. 10a), which is not expressed, or at a very low level, in ipRGCs (Sweeney et al., 2014), suggesting that these cells are different from aforementioned reservoir ipRGCs. To track them, we bred Tbr2CreERT2 and Pou4f1CKO to produce a Tbr2CreERT2/+:Pou4f1CKO/+ mouse line to enable sparse AP staining (Fig. 10b). In all retinas examined thus far, we found low abundance of these cells with 0 to 2 AP-positive RGCs per retina. The most notable morphological characteristics was the dense and bushy dendrites (Fig. 10c–10h’) resulting from numerous branching with short terminal dendrites emanating from multiple primary dendrites (4.15 ± 0.36, N=13). Their dendritic arbors and soma sizes were relatively smaller (dendritic arbor diameter: 196.5 ± 26.8 μm; soma diameter: 15.54 ± 1.1 μm; N=14) and their terminal dendrites stratified to the OFF sub-laminae (depth in the IPL: 12.8% ± 1.5%, N=12). We classified this RGC type into a unique but rare Tbr2+Pou4f1+ RGC subtype because we could only characterize 3 of them in all Tbr2CreERT2/+:RosaiAP retinas examined. Despite increasing the sample size in current clamp recording, we have yet to encounter this rare Tbr2+ RGC type and whether it is intrinsically photosensitive remains to be determined. .
Figure 10. OFF-layer stratified Tbr2+Pou4f1+ RGCs.
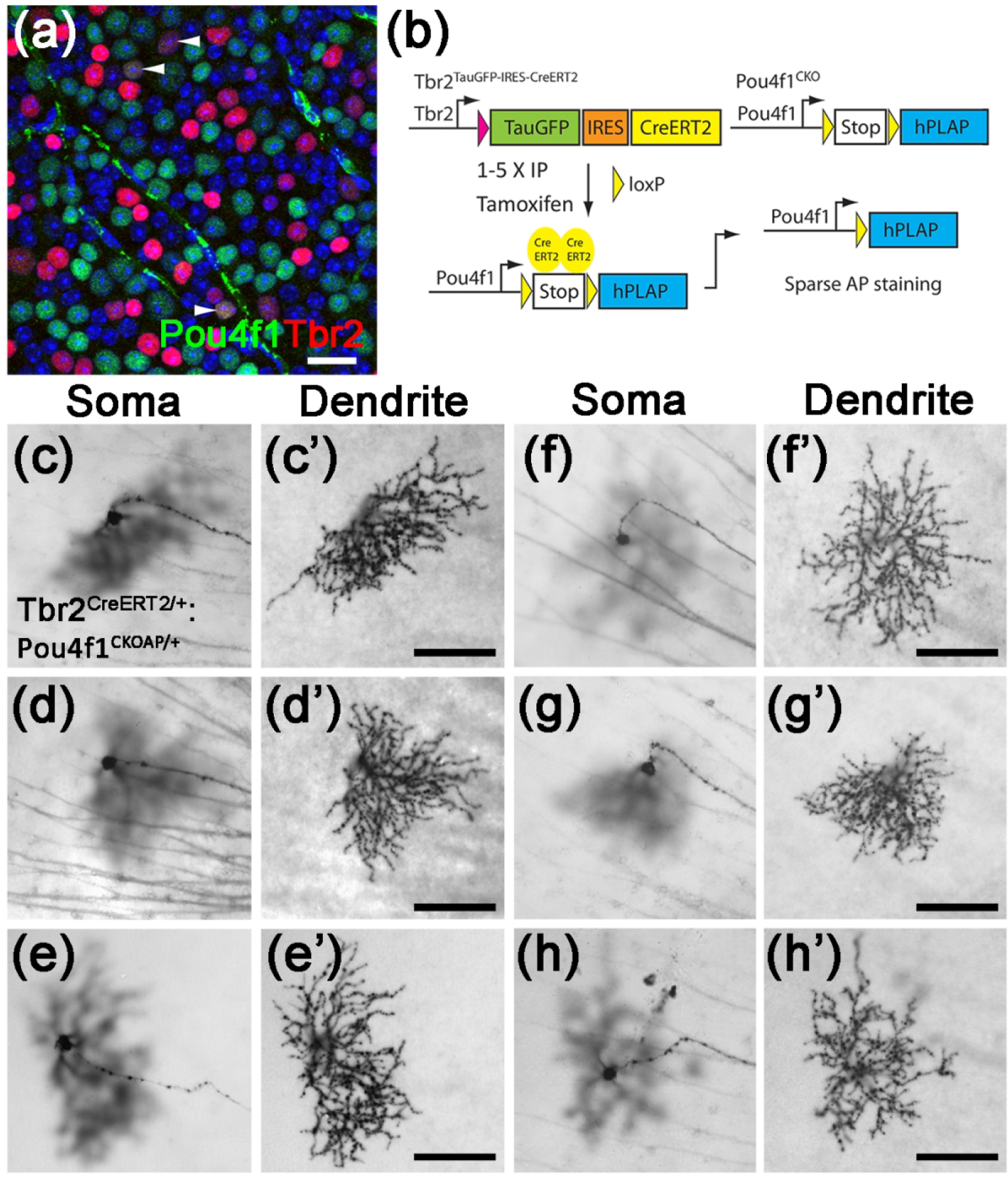
(a) Fluorescent image of a representative wild-type retinal flatmount showing Tbr2 expression (red) with Pou4f1-expressing (green) RGCs (white arrowheads). (b) The genetic sparse labeling system in the Tbr2CreERT2:Pou4f1CKO mouse line for brightfield identification of this cell type by AP staining. (c-h’) Representative brightfield AP-stained images of the Tbr2+Pou4f1+ RGCs. Scale bars: 100 μm (c’-h’). AP: alkaline phosphatase.
3.7. Axonal projection of Tbr2+ RGCs in brain
To determine the central projection of Tbr2+ RGCs, we examined AP staining patterns on brain sections from Tbr2CreERT2/+:RosaiAP/+ mice that received 5 consecutive tamoxifen doses through the IP route. As expected, intense AP staining was detected in SCN (Fig. 11a), vLGN, IGL, OPN, and medial pretectal nuclei (MPT) (Fig. 11b–11c). In addition, a few AP+ axonal arbors with relatively diffused signals were found in the dorsal lateral geniculate nuclei (dLGN) and lateral dorsal nucleus (LD) (Fig. 11b), superior colliculus (SC) (Fig. 11d–11d), peri-supraoptic nucleus (pSON) (Fig. 11f), lateral hypothalamus (LHA) (Fig. 11g1–g2), and lateral habenula (LHb) (Fig. 11h). These data confirm that Tbr2+ RGCs are not merely morphologically similar to ipRGCs, they also have very similar, if not identical, retinofugal projections (Delwig et al., 2016; Fernandez et al., 2016; Hattar et al., 2002; Li & Schmidt, 2018).
Fig. 11. Retinofugal projections of Tbr2+ RGCs in Tbr2CreERT2/+:RosaiAP mice.

(a-g) AP-stained brightfield images showing projections of Tbr2+ RGCs into SCN (a), IGL, vLGN, dLGN, LD (b), OPN and MPT (c), SC (d-e), pSON (f), LDH (g1-g2), and LHb (h). dLGN: dorsal lateral geniculate nuclei. IGL: inter-geniculate leaflet. LD: lateral dorsal nucleus. LDH: lateral hypothalamus. LHb: lateral habenula. OPN: olivary pretectal nuclei. pSON: peri-supraoptic nucleus SC: superior colliculus. SCN: suprachiasmatic nuclei. SO: stratum opticum. u/lSGS: upper/lower stratum griseum superficiale. vLGN: ventral lateral geniculate nuclei.
3.8. Tbr2 is essential for the survival of Tbr2+ RGCs and Opn4 expression in adult retina
Previous data obtained from Opn4Cre:Tbr2fx/fx mice showed that most Opn4-expressing ipRGCs had vanished, leading to the conclusion that Tbr2 is essential for ipRGC survival. However, since Opn4 is expressed as early as E14, a caveat lurks that the observed ipRGC loss might result from the requirement of Tbr2 for ipRGC survival and/or maturation during development. It is therefore necessary to determine whether Tbr2 is essential for ipRGC survival in adult retinas. To do this, we injected AAV2-Cre into the vitreous space of adult (P30) Tbr2TauGFP/+ (control) and Tbr2TauGFP/fx mice, isolated retinas 12 days post-injection, and examined co-immunostaining patterns of Tbr2, GFP, and RBPMS (Fig. 12a, 12b). We found that while Tbr2 expression was efficiently removed by AAV2-Cre in Tbr2TauGFP/fx retinas after 12 days (d12), many GFP+ Tbr2-deficient RGCs remained (double white arrowheads in Fig. 12b). However, at 21 days post-injection, we found that GFP+RBPMS+ cells in AAV2-Cre treated Tbr2TauGFP/fx retinas decreased by approximately 50% (data not shown). At 38 days post-injection (d38), we could barely find GFP+RBPMS+ RGCs in Tbr2-deleted regions (compared Fig. 12c and 12d. Ctrl: 55.2 ± 4.8; AAV2-Cre: 10.2 ±1.5, P = 2.2E-06, per unit area 1.0 × 105 μm2). Together, these data indicate that Tbr2 is indeed essential for the survival of adult Tbr2+ reservoir ipRGCs as well as dACs, ascertaining our previous conclusions.
Figure 12. Tbr2 is essential for maintaining ipRGC survival and Opn4 expression.
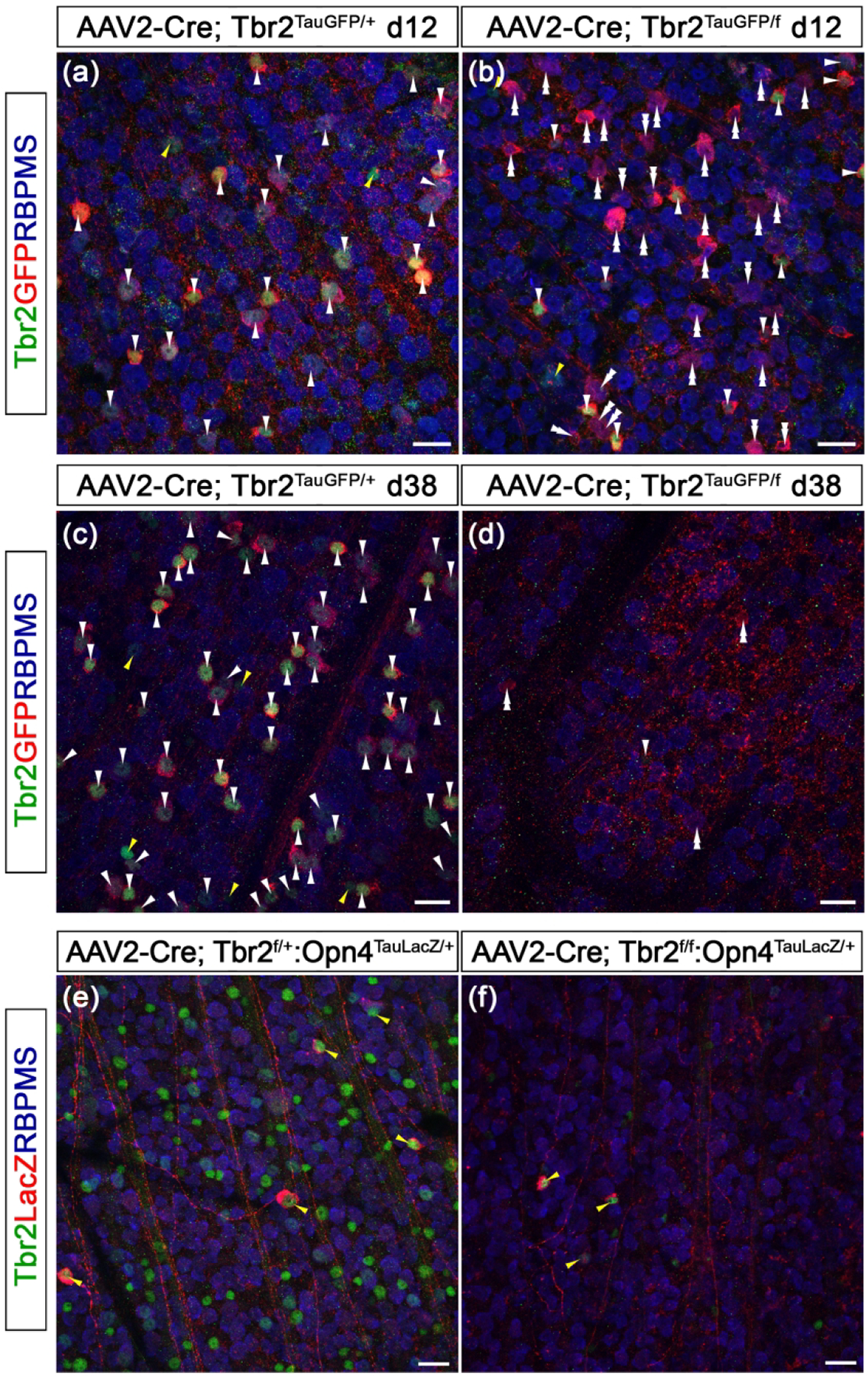
(a-d) Fluorescent images of Tbr2, GFP, and RBPMS on Tbr2TauGFP/+ retinas (a, c) and Tbr2TauGFP/fx (b, d) that was treated with AAV2-Cre. Retinas were isolated 12 days (a, b) or 38 days (c, d) post-injection. (e-f) Co-immunofluorescent staining of Tbr2, LacZ, and RBPMS on Tbr2fx/+:Opn4TaulacZ (e) or Tbr2fx/fx:Opn4TaulacZ (f) retinas. Retinas were isolated 14 days post-injection. Scale bars: 20 μm.
Because there was a lengthy delay between the deletion of Tbr2 (12 days postinjection) and the disappearance of Tbr2-deficient RGCs (38 days post-injection), we reasoned that if Opn4 expression requires Tbr2, Opn4 should be down-regulated while most of the Tbr2-deficient RGCs were still present between d12 and d38. To test this, we injected AAV2-Cre into the vitreous space of P21 Tbr2fx/+:Opn4TauLacZ/+ (control) and Tbr2fx/fx:Opn4TauLacZ/+ retinas, then isolated retinas 14 days post-injection and conducted co-immunostaining for Tbr2, RBPMS, and Opn4-driven LacZ. We detected fewer LacZexpressing ipRGCs in the Tbr2-deleted region in Tbr2fx/fx:Opn4TauLacZ/+ retinas (compared Fig. 12e and 12f. Ctrl: 6.3 ± 0.6; AAV2-Cre: 3 ± 0.7, P = 0.0003, per unit area 1.0 × 105 μm2). Additionally, most of the remaining LacZ+ cells expressed Tbr2, suggesting that these cells were not infected by AAV2-Cre. While the loss of Opn4 expression did not result in ipRGC death, taken together, these data strongly suggest that Tbr2 is essential for maintaining Opn4 expression. Alternatively, the loss of Opn4 expression is secondary to the eventual death of ipRGCs when Tbr2 is deleted.
4. Discussion
Tbr2 is an essential transcription factor for many developmental processes including trophoblast development, mesoderm formation, T cell differentiation, induction of anterior visceral endoderm, and neurogenesis of olfactory and cortical neurons. (Arnold et al., 2008; Mizuguchi et al., 2012; Nowotschin et al., 2013; Pearce et al., 2003; Russ et al., 2000; Sessa et al., 2010; Sessa, Mao, Hadjantonakis, Klein, & Broccoli, 2008). In retina, we were the first to show that it is a direct downstream target of Pou4f2, implicating its role as a regulator for RGC subtype development within the hierarchical transcription network controlling RGC development (Mao et al., 2008; Mu, Fu, Beremand, Thomas, & Klein, 2008). In follow-up studies, Tbr2 was independently shown to be essential for ipRGC formation (Mao et al., 2014; Sweeney et al., 2014). Using genetic loss-of-function strategies, we further demonstrated that Tbr2 is indispensable for ipRGC maintenance and hence proposed that Opn4 expression may be activated in Tbr2+ RGCs, allowing them to serve as a reservoir for ipRGCs (Mao et al., 2014). In this study, we developed new mouse resources to address a number of unanswered questions stemming from this theme.
4.1. Tbr2+ RGCs are ipRGCs
The first and foremost puzzle that we have solved here is why are there so many Tbr2+ cells but only a small fraction express detectable level of Opn4? Our current co-immunostaining and genetically sparse labeling studies reveal that approximately 54% of Tbr2-expressing cells in the GCL are dACs and that the Tbr2+ dACs account for approximately 18% of all dACs in the GCL. Based on their unique IMP profiles and dendritic structures, we have identifies two widefield types without intrinsic photosensitivity and one mediumfield type of Tbr2+ dACs with light responsiveness under glutamatergic synaptic blockade. They are all ON layer-stratified. The finding of two widefield Tbr2+ dACs and a mediumfield type is consistent with one recent scRNA-seq profiling study that revealed several AC subtypes with Tbr2 expression (Yan et al., 2020). While these dACs were not characterized further, the new Tbr2TauGFP-IRESCreERT2 mouse generated for this study should facilitate future studies for their functions. In the RGC population, we found that Tbr2+ RGCs account for approximately 12% of all RGCs, much less than previous estimates. The assumption that all Tbr2+ cells were RGCs must therefore be discarded. Moreover, using melanopsin expression together with a pan-RGC marker (RBPMS), we found that melanopsin-expressing cells account for approximately 6% of all RGCs. This number now approximates the number of GFP+ RGCs found in Opn4Cre:Z/EG retinas in our previous analysis (No. of GFP+: 2028 at 1-month-old vs. 2870 at 10 months) (Mao et al., 2014). However, it still represents only about half of all Tbr2+ RGCs. Based on our current findings that the majority of Tbr2+ RGCs are anatomically indistinguishable from known ipRGC types and with the same retinofugal projections and many possess intrinsic photosensitivity, we have further strengthened the notion that Tbr2+ RGCs indeed serve as a reservoir for native ipRGCs. The simplest reconciling explanation is that some Tbr2+ RGCs express Opn4 above detection by conventional immunostaining and those whose levels fall below were not accounted for in previous works. This explanation is supported by the fact that ~82% Tbr2+ RGCs we recorded exhibited intrinsic light sensitivity because whole-cell recording is more sensitive than immunostaining for Opn4 detection. An alternative and more sophisticated explanation, inspired by the literature, is that the expression of Opn4 within the entire Tbr2+ RGC population may be dynamically regulated by ambient light level. IpRGC number and Opn4 level can be modulated by light for the building of a more sensitive and broader photoreceptive system in rats (Hannibal, 2006; Hannibal, Georg, Hindersson, & Fahrenkrug, 2005). It is therefore plausible that the Tbr2+ ipRGC reservoir confirmed in this work allows Opn4 expression to be dynamically regulated to match the need to adapt to distinct lighting conditions in different seasons and/or geographic locations (Peirson, Halford, & Foster, 2009). Our data that Tbr2 is essential for Opn4 expression supports this notion. Together, these data strongly bolster our conclusion that Tbr2+ RGCs are bona fide ipRGCs and that it would be of interest in the future to study how Tbr2 regulates Opn4 expression.
4.2. Tbr2 expression marks all known ipRGC subtypes
Within Tbr2+ RGCs, we identified 2 types of OFF-layer stratified RGCs, several types of bistratified RGCs, and 3 types of ON-layer stratified RGCs. The predominant type of the OFF-layer Tbr2+ RGCs resembles previously described M1 ipRGCs, which branch sparsely with monostratified dendritic ramifications to the outermost IPL with variable dendritic field sizes and IMP profiles. This M1 type responds to light through a phototransduction mechanism involving members of the Gq family, Phospholipase C beta 4, and Trpc6 and trpc7 channels (Emanuel et al., 2017; Xue et al., 2011). We have classified the Tbr2+ M1-like ipRGCs here into three arbitrary subtypes based on their IMP profiles: M1n, M1r and M1s. Although quantitative morphometric analysis found no significant difference in dendritic structure, the M1s subtype has a significantly lower input resistance than M1n and M1r. While the exact relationship between IMP profiles, dendritic morphology, and instrinsic photosensitivity was not determined further here, these data suggest that IMP may be a useful “on the fly” cell identifier during recording and can therefore be used to complement post hoc morphological identification. The second and much rarer type of OFF-layer stratified RGCs was discovered in Tbr2CreERT2:Pou4f1CKO retinas. The co-expression of Tbr2 and Pou4f1 marks this unique subtype, which has bushy and small dendritic arbors stratifying into the OFF sub-laminae slightly below the IPL/INL boundary. The overall morphology of this RGC subtype is reminiscent of types “d” and “j” Pou4f1+ RGCs (Badea & Nathans, 2011). We have also encountered a few of this RGC subtype in Tbr2CreERT2/+: RosaiAP retinas, and it is likely that more of this type of RGC were buried in the dense dendritic AP signals from dACs. The IMP and intrinsic photosensitivity of this subtype was not determined, but can be aided in the future by generating new Pou4f1-driven reporter mouse lines allowing intersectional fluorescent labeling. We encountered various types of bistratified Tbr2+ RGCs, which matched well with the published M3 and M6 types. Finally, we found 3 ON-layer stratified Tbr2+ RGC types that morphologically correlated well with known M2, M4, and M5 ipRGCs. We have noted that among the three types, M2- and M4- types have stereotypic IMP profiles, while those of the M5 type appear more diverse. It is also noteworthy that phototransduction mechanism in ON-layer-stratified Tbr2+ RGCs is different from that in the M1-type and also known to be diverse as hyperpolarization activated channels and/or leaky potassium channels are involved (Jiang, Yue, Chen, Sheng, & Yau, 2018; Sonoda et al., 2018). The stereotypic IMP profiles of the M2 and the M4 types may aid future investigative efforts.Taken together, the data we have collected confirm that Tbr2+ RGCs are indeed ipRGCs. In agreement with our past and current findings, two recent profiling studies using singlecell RNA sequencing (scRNA-seq) found in P5 and adult mouse retinas that Tbr2 expression is enriched in diverse RGC clusters (Rheaume et al., 2018; Tran et al., 2019). In P5 retinas, Tbr2 was found in 7 (out of 40) RGC clusters (clusters 5, 6, 25, 26, 33, 37, 40), while Opn4 expression was expressed in 6 of the Tbr2+ clusters (higher levels in clusters 6, 25, and 33, medium to low levels in clusters 5, 26, and 37, and absent in 40) (Rheaume et al., 2018). In adult retinas, Tbr2 was detected in 8 (out of 46) RGC clusters (clusters 7, 8, 22, 29, 31, 33, 40, 43), while Opn4 expression is enriched in 5 of the Tbr2+ clusters (22, 31, 33, 40, 43, and not found in 7, 8, 29) (Tran et al., 2019). Furthermore, our birth-dating data suggest that the onset of ipRGC differentiation matches that of Tbr2+ RGCs (data not shown). Additionally, our unpublished RNA-seq dataset comparing E15.5 WT and Tbr2-mutant retinas has uncovered several Tbr2-dependent genes that are enriched in distinct ipRGC subtype clusters. Together, these data suggest that the terminal fate of ipRGC subtypes may have been segregated as early as E15.5, and thus Tbr2-mediated transcription network is involved in ipRGC subtype specification. Future research that compares the molecular profiles within each Tbr2+ cell type at different developmental stages will help elucidating the complex genetic network controlling the specification and differentiation of each of the ipRGC subtypes.
4.3. Role of Tbr2 in regulating Opn4 expression
A well known function of T-box transcription factors is to regulate molecular pathways involved in cell migration and cell-cell and cell-environment interactions (Ciruna & Rossant, 2001; Costello et al., 2011; Nowotschin et al., 2013; Pearce et al., 2003; Russ et al., 2000; Shen et al., 2014; Strumpf et al., 2005). For example, Tbr1, a paralogue of Tbr2, is necessary for neuronal migration, axon projection, establishment of regional and laminar identity in developing cortex and olfactory bulb, as well as RGC dendritic morphogenesis (Hevner et al., 2001; Huang et al., 2014; Kiyama et al., 2019; Liu et al., 2018). In mouse retinas, Tbr2 expression appears at ~E14 in a subset of RGCs that settled to the innermost region of GCL, suggesting that it plays a role in early steps of RGC development, such as cell fate determination and/or axon pathfinding. However, how Tbr2 achieves this and what are its downstream targets remain undetermined. Our study here suggests that deleting Tbr2 in mature retinas leads first to downregulation of Opn4 expression in ipRGCs, followed by their death, suggesting that in additional to its role in development, Tbr2 is also essential for maintaining Opn4 expression and ipRGC survival in adult. Alternatively but less likely, the loss of Opn4 expression reflects not only the requirement of Tbr2 presence but also the health of cells in which Tbr2 expression becomes undetectable. Nonetheless, we think that Tbr2 uses similar set of downstream targets to exert its functions during development and in the adult stage. In conjunction with our previous study in which Tbr2 is shown to be a direct downstream target of Pou4f2 and Isl1 (Mao et al., 2008; Wu et al., 2015), it now appears that the Pou4f2/Isl1-Tbr2-Opn4 regulatory hierarchy is necessary for ipRGC formation and survival. However, how the diverse ipRGC subtypes arise from this seemingly simple gene regulatory hierarchy to produce distinct dendritic morphologies and IPL stratification levels, retinofugal projections, intrinsic membrane properties, and molecular profiles remain poorly understood. Nevertheless, the close association of Tbr2 with all ipRGC subtypes suggests a pivotal role it plays in this process.
In summary, we have demonstrated that Tbr2-expressing RGCs are morphologically indistinguishable from known ipRGCs and that Tbr2 is essential for maintaining Opn4 expression and the survival of adult ipRGCs as well as three types of displaced amacrine cells. Future studies should investigate Tbr2’s co-regulators and downstream effectors in controlling the formation and maintenance of all ipRGC and dAC subtypes. The generation of the new Tbr2 reporter mouse line provides a unique opportunity to study in depth several biological questions regarding RGC specification, maturation, and terminal differentiation. One example we have shown here is the projection pattern of a single M5-like Tbr2+ RGC to multiple brain targets including IGL, vLGN, and OPN. This suggests that a single Tbr2+ RGC can relay photic information that requires different photo-response kinetics and threshold sensitivities, underlining the multiple roles of a single ipRGC in non-image forming vision. The comprehensive brain targeting pattern of a single RGC has been reported for M1-ipRGCs (Fernandez et al., 2016), but such information is still lacking for other ipRGC subtypes. It is of great interest to delineate retinofugal projections of all Tbr2+ RGC subtypes at the single cell level to fill the gap in the anatomical architecture and hence functional implications for ipRGCs.
Acknowledgments
This work was supported by grants from the National Institutes of Health-National Eye Institute to C.A.M. (EY024376) and C.K.C (EY026930), C.M.W. (EY021958), S.C.M. (EY006515), and in part by National Eye Institute Vision Core Grants P30EY028102 (UTHealth) and P30EY002520 (BCM). The Ophthalmology Department of Baylor College Medicine received an unrestricted grant from Research to Prevent Blindness, Inc. CKC is the Alice McPherson Retinal Research Foundation Chair at BCM. We acknowledge Dr. Jan Parker-Thornburg and Genetically Engineered Mouse Facility at The University of Texas MD Anderson Cancer Center in making Tbr2TauGFP-IRESCreERT2/+ mouse line, Dr. King-Wai Yau at Johns Hopkins University for sharing Opn4TauLacZ mouse, and Dr. Kimberly Mankiewicz at UTHealth for proofreading the manuscript.
Footnotes
Declaration of Interest
The authors declare no competing interest.
References
- Arnold SJ, Huang GJ, Cheung AF, Era T, Nishikawa S, Bikoff EK, … Groszer M (2008). The T-box transcription factor Eomes/Tbr2 regulates neurogenesis in the cortical subventricular zone. Genes Dev, 22(18), 2479–2484. doi: 10.1101/gad.475408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Cahill H, Ecker J, Hattar S, & Nathans J (2009). Distinct roles of transcription factors brn3a and brn3b in controlling the development, morphology, and function of retinal ganglion cells. Neuron, 61(6), 852–864. doi: 10.1016/j.neuron.2009.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Hua ZL, Smallwood PM, Williams J, Rotolo T, Ye X, & Nathans J (2009). New mouse lines for the analysis of neuronal morphology using CreER(T)/loxP-directed sparse labeling. PLoS One, 4(11), e7859. doi: 10.1371/journal.pone.0007859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, & Nathans J (2004). Quantitative analysis of neuronal morphologies in the mouse retina visualized by using a genetically directed reporter. J Comp Neurol, 480(4), 331–351. doi: 10.1002/cne.20304 [DOI] [PubMed] [Google Scholar]
- Badea TC, & Nathans J (2011). Morphologies of mouse retinal ganglion cells expressing transcription factors Brn3a, Brn3b, and Brn3c: analysis of wild type and mutant cells using genetically-directed sparse labeling. Vision Res, 51(2), 269–279. doi: 10.1016/j.visres.2010.08.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badea TC, Wang Y, & Nathans J (2003). A noninvasive genetic/pharmacologic strategy for visualizing cell morphology and clonal relationships in the mouse. J Neurosci, 23(6), 2314–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baver SB, Pickard GE, Sollars PJ, & Pickard GE (2008). Two types of melanopsin retinal ganglion cell differentially innervate the hypothalamic suprachiasmatic nucleus and the olivary pretectal nucleus. Eur J Neurosci, 27(7), 1763–1770. doi: 10.1111/j.1460-9568.2008.06149.x [DOI] [PubMed] [Google Scholar]
- Berson DM, Castrucci AM, & Provencio I (2010). Morphology and mosaics of melanopsin-expressing retinal ganglion cell types in mice. J Comp Neurol, 518(13), 2405–2422. doi: 10.1002/cne.22381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown TM, Gias C, Hatori M, Keding SR, Semo M, Coffey PJ, … Lucas RJ (2010). Melanopsin contributions to irradiance coding in the thalamo-cortical visual system. PLoS Biol, 8(12), e1000558. doi: 10.1371/journal.pbio.1000558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciruna B, & Rossant J (2001). FGF signaling regulates mesoderm cell fate specification and morphogenetic movement at the primitive streak. Dev Cell, 1(1), 37–49. [DOI] [PubMed] [Google Scholar]
- Costello I, Pimeisl IM, Drager S, Bikoff EK, Robertson EJ, & Arnold SJ (2011). The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat Cell Biol, 13(9), 1084–1091. doi: 10.1038/ncb2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delwig A, Larsen DD, Yasumura D, Yang CF, Shah NM, & Copenhagen DR (2016). Retinofugal Projections from Melanopsin-Expressing Retinal Ganglion Cells Revealed by Intraocular Injections of Cre-Dependent Virus. PLoS One, 11(2), e0149501. doi: 10.1371/journal.pone.0149501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker JL, Dumitrescu ON, Wong KY, Alam NM, Chen SK, LeGates T, … Hattar S (2010). Melanopsin-expressing retinal ganglion-cell photoreceptors: cellular diversity and role in pattern vision. Neuron, 67(1), 49–60. doi: 10.1016/j.neuron.2010.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel AJ, Kapur K, & Do MTH (2017). Biophysical Variation within the M1 Type of Ganglion Cell Photoreceptor. Cell Rep, 21(4), 1048–1062. doi: 10.1016/j.celrep.2017.09.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estevez ME, Fogerson PM, Ilardi MC, Borghuis BG, Chan E, Weng S, … Berson DM (2012). Form and function of the M4 cell, an intrinsically photosensitive retinal ganglion cell type contributing to geniculocortical vision. J Neurosci, 32(39), 13608–13620. doi: 10.1523/JNEUROSCI.1422-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farley FW, Soriano P, Steffen LS, & Dymecki SM (2000). Widespread recombinase expression using FLPeR (flipper) mice. Genesis, 28(3–4), 106–110. [PubMed] [Google Scholar]
- Fernandez DC, Chang YT, Hattar S, & Chen SK (2016). Architecture of retinal projections to the central circadian pacemaker. Proc Natl Acad Sci U S A, 113(21), 6047–6052. doi: 10.1073/pnas.1523629113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannibal J (2006). Regulation of melanopsin expression. Chronobiol Int, 23(1–2), 159–166. doi: 10.1080/07420520500464544 [DOI] [PubMed] [Google Scholar]
- Hannibal J, Georg B, Hindersson P, & Fahrenkrug J (2005). Light and darkness regulate melanopsin in the retinal ganglion cells of the albino Wistar rat. J Mol Neurosci, 27(2), 147–155. doi: 10.1385/JMN:27:2:147 [DOI] [PubMed] [Google Scholar]
- Hattar S, Kumar M, Park A, Tong P, Tung J, Yau KW, & Berson DM (2006). Central projections of melanopsin-expressing retinal ganglion cells in the mouse. J Comp Neurol, 497(3), 326–349. doi: 10.1002/cne.20970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattar S, Liao HW, Takao M, Berson DM, & Yau KW (2002). Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science, 295(5557), 10651070. doi: 10.1126/science.1069609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevner RF, Shi L, Justice N, Hsueh Y, Sheng M, Smiga S, … Rubenstein JL (2001). Tbr1 regulates differentiation of the preplate and layer 6. Neuron, 29(2), 353–366. [DOI] [PubMed] [Google Scholar]
- Huang TN, Chuang HC, Chou WH, Chen CY, Wang HF, Chou SJ, & Hsueh YP (2014). Tbr1 haploinsufficiency impairs amygdalar axonal projections and results in cognitive abnormality. Nat Neurosci, 17(2), 240–247. doi: 10.1038/nn.3626 [DOI] [PubMed] [Google Scholar]
- Jamal L, Kiyama T, & Mao CA (2020). Genetically Directed Sparse Labeling System for Anatomical Studies of Retinal Ganglion Cells. Methods Mol Biol, 2092, 187–194. doi: 10.1007/978-1-07160175-4_13 [DOI] [PubMed] [Google Scholar]
- Jiang Z, Yue WWS, Chen L, Sheng Y, & Yau KW (2018). Cyclic-Nucleotide- and HCN-ChannelMediated Phototransduction in Intrinsically Photosensitive Retinal Ganglion Cells. Cell, 175(3), 652–664 e612. doi: 10.1016/j.cell.2018.08.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyama T, Long Y, Chen CK, Whitaker CM, Shay A, Wu H, … Mao CA (2019). Essential Roles of Tbr1 in the Formation and Maintenance of the Orientation-Selective J-RGCs and a Group of OFF-Sustained RGCs in Mouse. Cell Rep, 27(3), 900–915 e905. doi: 10.1016/j.celrep.2019.03.077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong JM, Caprioli J, & Piri N (2010). RNA binding protein with multiple splicing: a new marker for retinal ganglion cells. Invest Ophthalmol Vis Sci, 51(2), 1052–1058. doi: 10.1167/iovs.09-4098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzerini Ospri L, Prusky G, & Hattar S (2017). Mood, the Circadian System, and Melanopsin Retinal Ganglion Cells. Annu Rev Neurosci, 40, 539–556. doi: 10.1146/annurev-neuro-072116-031324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SK, & Schmidt TM (2018). Morphological Identification of Melanopsin-Expressing Retinal Ganglion Cell Subtypes in Mice. Methods Mol Biol, 1753, 275–287. doi: 10.1007/978-1-49397720-8_19 [DOI] [PubMed] [Google Scholar]
- Li JY, & Schmidt TM (2018). Divergent projection patterns of M1 ipRGC subtypes. J Comp Neurol, 526(13), 2010–2018. doi: 10.1002/cne.24469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Reggiani JDS, Laboulaye MA, Pandey S, Chen B, Rubenstein JLR, … Sanes JR (2018). Tbr1 instructs laminar patterning of retinal ganglion cell dendrites. Nat Neurosci, 21(5), 659–670.doi: 10.1038/s41593-018-0127-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, … Zeng H (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci, 13(1), 133–140. doi: 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Kiyama T, Pan P, Furuta Y, Hadjantonakis AK, & Klein WH (2008). Eomesodermin, a target gene of Pou4f2, is required for retinal ganglion cell and optic nerve development in the mouse. Development, 135(2), 271–280. doi: 10.1242/dev.009688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao CA, Li H, Zhang Z, Kiyama T, Panda S, Hattar S, … Wang SW (2014). T-box transcription regulator Tbr2 is essential for the formation and maintenance of Opn4/melanopsin-expressing intrinsically photosensitive retinal ganglion cells. J Neurosci, 34(39), 13083–13095. doi: 10.1523/JNEUROSCI.1027-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill DS, Sheely CJ, Ecker JL, Badea TC, Morhardt D, Guido W, & Hattar S (2011). Development of melanopsin-based irradiance detecting circuitry. Neural Dev, 6, 8. doi: 10.1186/1749-8104-6-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijering E, Jacob M, Sarria JC, Steiner P, Hirling H, & Unser M (2004). Design and validation of a tool for neurite tracing and analysis in fluorescence microscopy images. Cytometry A, 58(2), 167–176. doi: 10.1002/cyto.a.20022 [DOI] [PubMed] [Google Scholar]
- Milner ES, & Do MTH (2017). A Population Representation of Absolute Light Intensity in the Mammalian Retina. Cell, 171(4), 865–876 e816. doi: 10.1016/j.cell.2017.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Naritsuka H, Mori K, Mao CA, Klein WH, & Yoshihara Y (2012). Tbr2 deficiency in mitral and tufted cells disrupts excitatory-inhibitory balance of neural circuitry in the mouse olfactory bulb. J Neurosci, 32(26), 8831–8844. doi: 10.1523/JNEUROSCI.5746-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu X, Fu X, Beremand PD, Thomas TL, & Klein WH (2008). Gene regulation logic in retinal ganglion cell development: Isl1 defines a critical branch distinct from but overlapping with Pou4f2. Proc Natl Acad Sci U S A, 105(19), 6942–6947. doi: 10.1073/pnas.0802627105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller LP, Do MT, Yau KW, He S, & Baldridge WH (2010). Tracer coupling of intrinsically photosensitive retinal ganglion cells to amacrine cells in the mouse retina. J Comp Neurol, 518(23), 4813–4824. doi: 10.1002/cne.22490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S, Costello I, Piliszek A, Kwon GS, Mao CA, Klein WH, … Hadjantonakis AK (2013). The T-box transcription factor Eomesodermin is essential for AVE induction in the mouse embryo. Genes Dev, 27(9), 997–1002. doi: 10.1101/gad.215152.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Mullen AC, Martins GA, Krawczyk CM, Hutchins AS, Zediak VP, … Reiner SL (2003). Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science, 302(5647), 1041–1043. doi: 10.1126/science.1090148 [DOI] [PubMed] [Google Scholar]
- Peirson SN, Halford S, & Foster RG (2009). The evolution of irradiance detection: melanopsin and the non-visual opsins. Philos Trans R Soc Lond B Biol Sci, 364(1531), 2849–2865. doi: 10.1098/rstb.2009.0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quattrochi LE, Stabio ME, Kim I, Ilardi MC, Michelle Fogerson P, Leyrer ML, & Berson DM (2019). The M6 cell: A small-field bistratified photosensitive retinal ganglion cell. J Comp Neurol, 527(1), 297–311. doi: 10.1002/cne.24556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Chun C, Fan J, Kofron JM, Yang MB, Hegde RS, … Lang RA (2013). A direct and melanopsin-dependent fetal light response regulates mouse eye development. Nature, 494(7436), 243–246. doi: 10.1038/nature11823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rheaume BA, Jereen A, Bolisetty M, Sajid MS, Yang Y, Renna K, … Trakhtenberg EF (2018). Single cell transcriptome profiling of retinal ganglion cells identifies cellular subtypes. Nat Commun, 9(1), 2759. doi: 10.1038/s41467-018-05134-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ AP, Wattler S, Colledge WH, Aparicio SA, Carlton MB, Pearce JJ, … Evans MJ (2000). Eomesodermin is required for mouse trophoblast development and mesoderm formation. Nature, 404(6773), 95–99. doi: 10.1038/35003601 [DOI] [PubMed] [Google Scholar]
- Schmidt TM, Alam NM, Chen S, Kofuji P, Li W, Prusky GT, & Hattar S (2014). A role for melanopsin in alpha retinal ganglion cells and contrast detection. Neuron, 82(4), 781–788. doi: 10.1016/j.neuron.2014.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt TM, & Kofuji P (2011). Structure and function of bistratified intrinsically photosensitive retinal ganglion cells in the mouse. J Comp Neurol, 519(8), 1492–1504. doi: 10.1002/cne.22579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Colasante G, Nini A, Klein WH, & Broccoli V (2010). Tbr2-positive intermediate (basal) neuronal progenitors safeguard cerebral cortex expansion by controlling amplification of pallial glutamatergic neurons and attraction of subpallial GABAergic interneurons. Genes Dev, 24(16), 1816–1826. doi: 10.1101/gad.575410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa A, Mao CA, Hadjantonakis AK, Klein WH, & Broccoli V (2008). Tbr2 directs conversion of radial glia into basal precursors and guides neuronal amplification by indirect neurogenesis in the developing neocortex. Neuron, 60(1), 56–69. doi: 10.1016/j.neuron.2008.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Lu J, Sui L, Wang D, Yin M, Hoffmann I, … Pflugfelder GO (2014). The orthologous Tbx transcription factors Omb and TBX2 induce epithelial cell migration and extrusion in vivo without involvement of matrix metalloproteinases. Oncotarget, 5(23), 11998–12015. doi: 10.18632/oncotarget.2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda T, Lee SK, Birnbaumer L, & Schmidt TM (2018). Melanopsin Phototransduction Is Repurposed by ipRGC Subtypes to Shape the Function of Distinct Visual Circuits. Neuron, 99(4), 754–767 e754. doi: 10.1016/j.neuron.2018.06.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonoda T, Okabe Y, & Schmidt TM (2019). Overlapping morphological and functional properties between M4 and M5 intrinsically photosensitive retinal ganglion cells. J Comp Neurol. doi: 10.1002/cne.24806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stabio ME, Sabbah S, Quattrochi LE, Ilardi MC, Fogerson PM, Leyrer ML, … Berson DM (2018). The M5 Cell: A Color-Opponent Intrinsically Photosensitive Retinal Ganglion Cell. Neuron, 97(1), 251. doi: 10.1016/j.neuron.2017.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, & Rossant J (2005). Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development, 132(9), 2093–2102. doi: 10.1242/dev.01801 [DOI] [PubMed] [Google Scholar]
- Sweeney NT, Tierney H, & Feldheim DA (2014). Tbr2 is required to generate a neural circuit mediating the pupillary light reflex. J Neurosci, 34(16), 5447–5453. doi: 10.1523/JNEUROSCI.0035-14.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, … Sanes JR (2019). Single-Cell Profiles of Retinal Ganglion Cells Differing in Resilience to Injury Reveal Neuroprotective Genes. Neuron, 104(6), 1039–1055 e1012. doi: 10.1016/j.neuron.2019.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vong L, Ye C, Yang Z, Choi B, Chua S Jr., & Lowell BB (2011). Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron, 71(1), 142154. doi: 10.1016/j.neuron.2011.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Kaczynski TJ, Sethuramanujam S, Li R, Jain V, Slaughter M, & Mu X (2015). Two transcription factors, Pou4f2 and Isl1, are sufficient to specify the retinal ganglion cell fate. Proc Natl Acad Sci U S A, 112(13), E1559–1568. doi: 10.1073/pnas.1421535112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M, Zhou L, Macke JP, Yoshioka T, Hendry SH, Eddy RL, … Nathans J (1995). The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J Neurosci, 15(7 Pt 1), 4762–4785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue T, Do MT, Riccio A, Jiang Z, Hsieh J, Wang HC, … Yau KW (2011). Melanopsin signalling in mammalian iris and retina. Nature, 479(7371), 67–73. doi: 10.1038/nature10567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Laboulaye MA, Tran NM, Whitney IE, Benhar I, & Sanes JR (2020). Mouse Retinal Cell Atlas: Molecular Identification of over Sixty Amacrine Cell Types. J Neurosci. doi: 10.1523/JNEUROSCI.0471-20.2020 [DOI] [PMC free article] [PubMed] [Google Scholar]


