Abstract
Over the past two decades, evo-devo (evolution of development) studies have elucidated genetic mechanisms underlying novel dipteran body color patterns. Here we review the most recent developments, which show some departure from the model organism Drosophila melanogaster, leading the field into the investigation of more complex color patterns. We also discuss how the robust application of transgenic techniques has facilitated the study of many non-model pest species. Furthermore, we see that subtle pigmentation differences guide the discovery and description of new dipterans. Therefore, we argue that the existence of new field guides and the prevalence of pigmentation studies in non-model flies will enable scientists to adopt uninvestigated species into the lab, allowing them to study novel morphologies.
Keywords: Diptera, non-model species, color patterns, pigmentation, pigmentation genes, pest control, species discovery, wing patterns, head patterns, thorax patterns, abdominal patterns, Evo-Devo, drosophilids, tephritids, field guides
Graphical abstract
Introduction
Diptera, or the “true flies”, is an order of holometabolous insects set apart by their single pair of wings and set of flight-stabilizing halteres [1,2]. With regards to investigating body coloration, the most well-studied dipterans belong to the family Drosophilidae, which includes the “common fruit fly” (Figure 1). Attempts to understand the evolution of color pattern development in Drosophila (D.) have persisted for decades. In this review, we will discuss the ongoing research in not only “common fruit flies”, but the order Diptera as a whole. One early focus of dipteran evo-devo pertained to simple color patterns, like that of D. melanogaster. Today, we investigate body coloration across an array of genera and species. Recently, there appears to be a major trend in the use of the CRISPR/Cas9 system to disrupt pigmentation to develop pest control methods. Furthermore, the critical analysis of color patterns has led to the identification of new species. Here we review the recent literature on dipteran coloration and provide our opinion on how the field should move forward.
Figure 1: Drosophilid body coloration.

Here, we see just a fraction of the diverse body color patterning exhibited within the order Diptera. Some of these unique morphologies are the subjects of recent investigations detailed within this review. However, many other traits shown here are not mentioned in the literature, and their developments deserve to be investigated. Row 1: (A, B) D. guttifera, (C, D) D. deflecta, and (E, F) Leucophenga varia. Row 2: (G) D. palustris, (H) D. subpalustris, (I) D. busckii, (J) D. hydei, (K) the thoracic trident of D. melanogaster, and (L) the striped thorax of Zaprionus indianus. Row 3: (M) D. melanogaster (female), (N) D. melanogaster (male), (O) Hirtodrosophila duncani (female), (P) Hirtodrosophila duncani (male), (Q) Samoaia leonensis, and (R) Chymomyza amoena. Row 4: (S) D. americana, (T) Mycodrosophila claytonae, (U) Mycodrosophila dimidiata, and (V) Phortica variegata. Row 5: (W) D. macrospina, (X) D. borealis, (Y) D. peninsularis, and (Z) D. ananassae. Images are from [53*, 54*].
The evolution of dipteran color patterns
Dipterans exhibit a multitude of color patterns; however, the bulk of recent literature pertains to the genus Drosophila. Recent studies in these “common fruit flies” examined complex abdominal, wing, and thoracic color patterning. We also note the exhibition of new methods and tools with the potential to impact the field of evo-devo and a paradigm shift in our understanding of pigment biosynthesis. Finally, we see that a complex color pattern outside of the genus Drosophila is thoroughly analyzed, a progression welcome in the field.
Abdominal color patterns
The myriad of abdominal color patterns seen in Drosophila species, ranging from full-body pigmentation to intricate combinations of spots and stripes, have inspired investigators to study how these novel morphologies emerged [3,4]. The evolutionary divergence of coloration between the pale-yellow colored D. novamexicana and the brown D. americana was shown to have occurred, at least in part, from differences in the ebony alleles between the species [5*]. Sramkoski et al. (2020) examined the intraspecific pigmentation of D. americana. They noted that populations found in the eastern United States display a darker body coloration than the western populations, and that this progressively lighter body coloration seen in western D. americana results in a pigmentation phenotype that resembles D. novamexicana. Sramkoski et al. (2020) suggested that allelic similarities in ebony and tan between lightly colored D. americana morphs and D. novamexicana might underly the pigmentation cline seen in D. americana. However, this hypothesis was not supported [6].
The quinaria group’s 26 species [7] display an impressive array of pattern elements. Dion et al. (2020) demonstrated that the co-expression patterns of three key pigmentation genes, Dopa decarboxylase, tan, and yellow, prefigure the spot patterning of three quinaria species group members: D. guttifera, D. palustris, and D. subpalustris [8]. D. guttifera was further examined by KKB Raja et al. (bioRxiv doi: 10.1101/2020.04.09.034900), who showed that a cis-regulatory element in the yellow locus drove the expression of a reporter construct in a pattern that resembled the adult abdominal pigmentation. They also showed correlational in situ hybridization data indicating that the toolkit genes wingless, decapentaplegic, hedgehog, abdominal-A, and zerknullt may play a role in the formation of the abdominal color pattern. Kalay et al. (2019) focused on the cis-regulatory capabilities of sequences derived from the 5’ intergenic and intronic regions of the yellow gene in D. melanogaster, D. pseudoobscura, and D. willistoni. They observed reporter gene expression patterns driven by redundant and cryptic cis-regulatory elements, the latter being a sequence with the ability to regulate gene expression alone but not in its naturally occurring position in the genome. They suggested that the observed cis-regulatory complexity may underlie the yellow gene’s wide range of expression patterns within the genus Drosophila [9].
Sexually dimorphic pigmentation is widespread across Drosophila, a phenomenon that Hughes et al. (2020) proposed to have emerged through parallel evolution [10]. Roeske et al. (2018) demonstrated that in female D. melanogaster, bab represses posterior coloration by binding to a cis-regulatory element (body element) of yellow required for the male-specific pigmentation [11]. The development of this male-specific coloration was also shown to require the activity of the gene grainy head [12]. The female-specific pigmentation of this fly was shown to be controlled by a gene-regulatory network that is thermally sensitive. Abdominal B and (female-specific) doublesex initiate the expression of bab, a gene with a role in the development of sexually dimorphic body coloration [13], which dials down the activity of three pigmentation genes: Dopa decarboxylase, tan, and yellow. De Castro et al. (2018) demonstrated that higher temperatures yielded increased levels of bab, which resulted in reduced pigmentation. They further suggested that Abdominal B, a gene whose product has an established role in Drosophila pigmentation [14,15], may turn on tan expression at lower temperatures [16]. Abdominal B was further investigated by Liu et al. (2019), who demonstrated that untangling the evolution of a gene-regulatory network is not straightforward. Abdominal B facilitates dark body coloration in D. yakuba, and modifying its expression was shown to not alter the body pigmentation of the sister species D. santomea (whose absence of abdominal pigmentation resulted from an evolutionary loss of tan and yellow expression [17]) [18]. The morphological diversity of Drosophila abdominal coloration continues to be a fruitful resource for investigating the development of novel traits.
Wing pigmentation
Attempts to understand the development of dipteran wing coloration has been ongoing for more than 20 years [19]. Recent studies of Drosophila and Samoaia progressed our understanding of how wing color patterns evolved. The gene wingless is an established coordinator in the development of wing pigmentation in D. guttifera [20,21]. Fukutomi et al. (2020) further demonstrated that wingless is a key regulator of 151 genes associated with the color pattern on the wings of this polka-dotted fruit fly [22*]. Massey et al. (2020) suggested that the X-chromosomal gene optomotor-blind possibly underlies the disparity of pigmentation between two sibling species, D. elegans (which has a male-specific wing spot) and D. gunungcola (which lacks a wing spot) [23]. The male-specific spot seen in D. biarmipes was also the subject of a recent study. CC Galouzis et al. (bioRxiv doi: 10.1101/2020.03.23.003103) showed that homologous alleles for the X-chromosomal yellow gene interact with each other to inactivate the regulatory element (spot enhancer) responsible for this trait in females. They further noted that the intron of yellow and the protein Mod(mdg4) are required for this function to occur.
The expression of engrailed is normally located only in the rear section of the early developing fly wing to dictate posterior wing identity. However, Engrailed also plays a role in the development of the wing spot seen in D. biarmipes [24]. Dufour et al. (2020) suggested that engrailed was co-opted into the development of wing pigmentation in Samoaia leonensis, a process that occurs after the development of the basic wing morphology. This co-option results in a complex wing color pattern; engrailed represses pigmentation in specific areas of an ancestrally black wing, resulting in many irregular light areas spread across the wing surface. Dufour et al. (2020) further proposed that engrailed gained new temporal and spatial domains of expression with an onset after its vital developmental role in posterior wing identity [25**]. This study provided valuable insights into how toolkit genes can gain novel domains of expression without interrupting their other, essential roles in organismal development.
Thoracic trident of D. melanogaster
One other specific pigmentation feature, the thoracic trident of D. melanogaster, inspired recent studies. Gibert et al. (2018) suggested that the expression pattern of stripe - a gene involved in flight muscle attachment and also the repression of pigmentation - caused the emergence of this unique color pattern as a consequence of its co-option into a pigmentation gene-regulatory network [26]. Endler et al. (2018) showed that single-nucleotide polymorphisms within the tan male-specific enhancer and a missense mutation in CG15370 (a gene upstream of tan) have a high impact on trident pigmentation [27]. Based on the discovery that alterations to the regulation of ebony impacted the abdominal coloration of D. melanogaster [28], Telonis-Scott and Hoffmann (2018) suggested that ebony enhancer variation, in part, underlies the diversity in color intensity of the thoracic trident pattern [29].
Methods and pigment biosynthesis
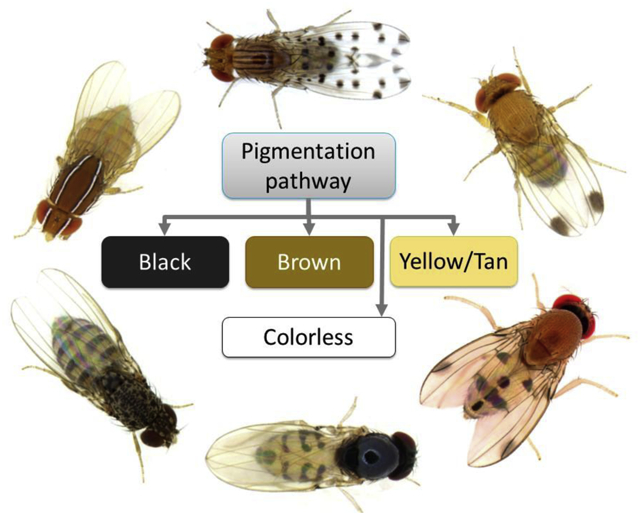
Besides “common fruit flies” being at the center of experiments that progressed our understanding of the molecular mechanisms underlying the evolution and development of body color patterns, we see techniques relevant to evo-devo designed and refined through the use of both model and non-model Drosophila species, such as a method to investigate enhancer-promoter interactions [30], a method to produce transgenics in a much more delicate “common fruit fly”, D. guttifera [31*], and the robust application of the Oxford Nanopore sequencing technology [32]. Also, it is now widely accepted that black pigment is produced by Yellow from dopamine, as opposed to dopa (Figure 2) [8,10,33,34]. These studies show that our evo-devo toolkit and our understanding of pigmentation continue to develop.
Figure 2: The pigmentation biosynthesis pathway.

The previously understood role of Yellow (Yellow*) is shown by a dashed line. POs are phenol oxidases, aaNATs is arylalkylamine N-acetyl transferases, NADA is N-acetyl dopamine, and NBAD is N-β-alanyl dopamine. Figure adapted from [8,10,33,34].
Head pigmentation in a tephritid species
One interesting study, notably outside of the genus Drosophila, investigated a complex spot pattern crowning the head of Bactrocera dorsalis. Bai et al. (2019) demonstrated that the knockout of the white gene partially erased this dark pigmentation pattern. These white mutants had lower expression levels of yellow (Bd-yellow1 specifically), prompting the authors to speculate that a regulatory relationship between white and yellow may exist, where White possibly facilitates the transport of secondary messengers that affect transcription factor activity and gene expression [35**]. This study clearly demonstrated the value of incorporating diverse fly species into our efforts to understand both the architecture of gene-regulatory networks and the evolution and development of complex color patterns.
Pigmentation as a target for pest control
Applying our understanding of the underlying molecular pathways and adapted molecular techniques for model and non-model species has allowed for advancements in translational research. Mosquitoes are an established disease vector, and one method to manage their populations could be through manipulating their pigmentation development. Pigmentation studies in Aedes albopictus demonstrated that ovary-specific genes of the yellow family have been found to play roles in both melanization timing and proper development of the chorion [36]. CRISPR/Cas9 knockouts of kynurenine hydroxylase and yellow in Aedes albopictus demonstrated the feasibility of genetic manipulation in this mosquito [37], and RNAi knockdown of Laccase 2 in Anopheles sinensis identified a target that could facilitate the control of this mosquito population [38]. Juvenile hormone receptors are integral to insect development. Zhu et al. (2019) knocked out the gene methoprene-tolerant encoding one of these receptors in Aedes aegypti through the use of CRISPR/Cas9. This knockout led to black third-instar and fourth-instar larvae that eventually died before reaching the pupal stage [39].
In Cochliomyia hominivorax and Lucilia cuprina, two flesh-eating parasitic flies in livestock, yellow plays a key role in the development of dark adult body coloration, and the gene was targeted to prove the viability of the CRISPR/Cas9 system in these insects. Taking it one step further beyond pigmentation, Paulo et al. (2019) then knocked out the gene transformer in C. hominivorax, which disrupted reproductive organ development and demonstrated a possible avenue for pest control [40]. Additionally, the brown body locus in Musca domestica was shown to be orthologous to the yellow gene in Drosophila through the use of CRISPR/Cas9 [41].
The Tephritidae, or the “true fruit flies”, are a large and invasive dipteran family at the center of recent studies of genetic manipulation. CRISPR/Cas9 facilitated the first successful manipulation of the gene scarlet in Bactrocera oleae [42]. In Anastrepha ludens, a previously unidentified mutation affecting both pupal and adult pigmentation, slow larvae, was identified [43]. Chen et al. (2018) and Zhang et al. (2019) demonstrated that the pigmentation gene tyrosine hydroxylase (a.k.a. pale) plays an important role in the pupal pigmentation (tanning) of Bactrocera dorsalis [44] and Zeugodacus tau [45], respectively. These studies focused primarily on pest control; however, their contributions to refining transgenic techniques in non-model organisms and their data regarding the roles of key pigmentation genes are a boon to the field of dipteran color pattern evo-devo.
The identification and characterization of new species and traits
A new species, D. carrolli, isolated from Brunei has been recognized. Some defining characteristics that separated this species from the closely related D. rhopaloa included unique body and wing pigmentation traits [46*]. Additionally, wing pigmentation supported the identification of five previously unknown species of Culicoides [47], and differences in thoracic pigmentation helped to distinguish a new member of the genus Corethrella from an established species [48].
While body pigmentation is easily visible to the human eye, wing interference patterns require multispectral digital imaging to be properly visualized. Hawkes et al. (2019) suggested that these structural colors in the wings of D. simulans play a role in sexual selection [49], and NJ Butterworth et al. (bioRxiv doi: 10.1101/2020.02.18.948646) observed species-specific and sexually dimorphic wing interference patterns in species of the genus Chrysomya.
As a discipline, evo-devo is incomplete without field work. Field guides and references providing high-quality images and accurate descriptions of species across the order Diptera have recently become available [50–52, 53*, 54*]. We hope that these guides facilitate the identification and investigation of rarely studied flies.
Conclusion
The advancement in our understanding of dipteran body color evo-devo, though impressive, is mainly localized to well-characterized Drosophila species. We propose two directions for the field: First, we hope to see an even greater incorporation of non-model Drosophila species into evo-devo research. The availability of new, comprehensive field guides coupled with broadly applicable methods for genetic manipulation will facilitate the broader exploration of novel morphologies - a scenario that should excite every naturalist. Second, we propose that in-depth studies of body color patterns displayed by the family Tephritidae should be pursued. Recent literature demonstrated that genetic manipulation is practical in this dipteran family and that novel gene-regulatory architectures underlying pigmentation development may exist. We believe that the broader inclusion of non-model Drosophila and the “true fruit flies” into a field dominated by well-studied “common fruit flies” is a vital step towards understanding the evo-devo of very complex dipteran body color patterns.
Funding
TW was awarded a National Institutes of Health grant (grant number 1R15GM107801-01A1), which facilitated unpublished and published results discussed in this review. The funding source had no influence in the study design; collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest statement
Nothing declared.
References
References and author recommendations
Although every article included in this review contributed to our collective understanding of dipteran color pattern evo-devo, we would like to note literature of special interest (*) and of outstanding interest (**).
- 1.Courtney GW, Cranston PS: Chapter 40 - Order Diptera. In Thorp and Covich’s Freshwater Invertebrates (Fourth Edition). Edited by Thorp JH, Rogers DC. Academic Press; 2015:1043–1058. [Google Scholar]
- 2.Sarwar M: Typical flies: Natural history, lifestyle and diversity of Diptera. In Life Cycle and Development of Diptera. Edited by: Sarwar M IntechOpen; 2020: 1–50. [Google Scholar]
- 3.Wittkopp PJ, True JR, Carroll SB: Reciprocal functions of the Drosophila Yellow and Ebony proteins in the development and evolution of pigment patterns. Development 2002, 129:1849–1858. [DOI] [PubMed] [Google Scholar]
- 4.Wittkopp PJ, Vaccaro K, Carroll SB: Evolution of yellow gene regulation and pigmentation in Drosophila. Curr Biol 2002, 12:1547–1556. [DOI] [PubMed] [Google Scholar]
- 5.Lamb AM, Wang Z, Simmer P, Chung H, Wittkopp PJ: ebony Affects pigmentation divergence and cuticular hydrocarbons in Drosophila americana and D. novamexicana. Front Ecol Evol 2020, 8:184. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Here, the CRISPR/Cas9 system was used to knock out ebony in D. americana and D. novamexicana. The authors used these ebony knockout mutants to support the assumption that differences in ebony alleles impact the development of species-specific body coloration seen in these two fruit fly species.
- 6.Sramkoski LL, McLaughlin WN, Cooley AM, Yuan DC, John A, Wittkopp PJ: Genetic architecture of a body color cline in Drosophila americana. Mol Ecol 2020, 29:2840–2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scott Chialvo CH, White BE, Reed LK, Dyer KA: A phylogenetic examination of host use evolution in the quinaria and testacea groups of Drosophila. Mol Phylogenet Evol 2019, 130:233–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dion WA, Shittu MO, Steenwinkel TE, Raja KKB, Kokate PP, Werner T: The modular expression patterns of three pigmentation genes prefigure unique abdominal morphologies seen among three Drosophila species. Gene Expr Patterns 2020, 38:119132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalay G, Lachowiec J, Rosas U, Dome MR, Wittkopp P: Redundant and cryptic enhancer activities of the Drosophila yellow gene. Genetics 2019, 212:343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hughes JT, Williams ME, Johnson R, Grover S, Rebeiz M, Williams TM: Gene regulatory network homoplasy underlies recurrent sexually dimorphic fruit fly pigmentation. Front Ecol Evol 2020, 8:80. [Google Scholar]
- 11.Roeske MJ, Camino EM, Grover S, Rebeiz M, Williams TM: cis-Regulatory evolution integrated the Bric-à-brac transcription factors into a novel fruit fly gene regulatory network. Elife 2018, 7:e32273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grover S, Williams ME, Kaiser R, Hughes JT, Gresham L, Rebeiz M, Williams TM: Augmentation of a wound response element accompanies the origin of a Hox-regulated Drosophila abdominal pigmentation trait. Dev Biol 2018, 441:159–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopp A, Duncan I, Carroll SB: Genetic control and evolution of sexually dimorphic characters in Drosophila. Nature 2000, 408:553–559. [DOI] [PubMed] [Google Scholar]
- 14.Jeong S, Rokas A, Carroll SB: Regulation of body pigmentation by the Abdominal-B Hox protein and its gain and loss in Drosophila evolution. Cell 2006, 125:1387–1399. [DOI] [PubMed] [Google Scholar]
- 15.Williams TM, Selegue JE, Werner T, Gompel N, Kopp A, Carroll SB: The regulation and evolution of a genetic switch controlling sexually dimorphic traits in Drosophila. Cell 2008, 134:610–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Castro S, Peronnet F, Gilles J-F, Mouchel-Vielh E, Gibert J-M: bric à brac (bab), A central player in the gene regulatory network that mediates thermal plasticity of pigmentation in Drosophila melanogaster. PLoS Genet 2018, 14:e1007573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jeong S, Rebeiz M, Andolfatto P, Werner T, True J, Carroll SB: The evolution of gene regulation underlies a morphological difference between two Drosophila sister species. Cell 2008, 132:783–793. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Ramos-Womack M, Han C, Reilly P, Brackett KL, Rogers W, Williams TM, Andolfatto P, Stern DL, Rebeiz M: Changes throughout a genetic network mask the contribution of Hox gene evolution. Curr Biol 2019, 29:2157–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.True JR, Edwards KA, Yamamoto D, Carroll SB: Drosophila wing melanin patterns form by vein-dependent elaboration of enzymatic prepatterns. Curr Biol 1999, 9:1382–1391. [DOI] [PubMed] [Google Scholar]
- 20.Werner T, Koshikawa S, Williams TM, Carroll SB: Generation of a novel wing colour pattern by the Wingless morphogen. Nature 2010, 464:1143–1148. [DOI] [PubMed] [Google Scholar]
- 21.Koshikawa S, Giorgianni MW, Vaccaro K, Kassner VA, Yoder JH, Werner T, Carroll SB: Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc Natl Acad Sci U S A 2015, 112:7524–7529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fukutomi Y, Kondo S, Toyoda A, Shigenobu S, Koshikawa S: Transcriptome analysis reveals wingless regulates neural development and signaling genes in the region of wing pigmentation of a polka-dotted fruit fly. FEBS J 2020. DOI: 10.1111/febs.15338. [DOI] [PubMed] [Google Scholar]; *de novo Genome sequencing and transcriptomics identified wingless as a key regulator of 151 genes responsible for the wing pigmentation of D. guttifera. The authors suggested that this unique wing pattern arose through the incremental acquisition of different developmental mechanisms to produce the genetic architecture required to develop the complex wing color pattern of D. guttifera.
- 23.Massey JH, Rice GR, Firdaus AS, Chen C-Y, Yeh S-D, Stern DL, Wittkopp PJ: Co-evolving wing spots and mating displays are genetically separable traits in Drosophila. Evolution 2020, 74:1098–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gompel N, Prud’homme B, Wittkopp PJ, Kassner VA, Carroll SB: Chance caught on the wing: cis-Regulatory evolution and the origin of pigment patterns in Drosophila. Nature 2005, 433:481–487. [DOI] [PubMed] [Google Scholar]
- 25.Dufour HD, Koshikawa S, Finet C: Temporal flexibility of gene regulatory network underlies a novel wing pattern in flies. Proc Natl Acad Sci U S A 2020, 117:11589–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]; **By manipulating key genes over the course of D. melanogaster wing development, the authors demonstrated how toolkit genes’ essential activities are limited to specific timeframes. This conclusion allowed the authors to suggest how the sole co-option of engrailed, a key gene in posterior wing identity, occurred to create a novel wing color pattern in Samoaia leonensis without affecting its essential role in early wing development.
- 26.Gibert J-M, Mouchel-Vielh E, Peronnet F: Pigmentation pattern and developmental constraints: flight muscle attachment sites delimit the thoracic trident of Drosophila melanogaster. Sci Rep 2018, 8:5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Endler L, Gibert J-M, Nolte V, Schlötterer C: Pleiotropic effects of regulatory variation in tan result in correlation of two pigmentation traits in Drosophila melanogaster. Mol Ecol 2018, 27:3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rebeiz M, Pool JE, Kassner VA, Aquadro CF, Carroll SB: Stepwise modification of a modular enhancer underlies adaptation in a Drosophila population. Science 2009, 326:1663–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Telonis-Scott M, Hoffmann AA: Enhancing ebony? Common associations with a cis-regulatory haplotype for Drosophila melanogaster thoracic pigmentation in a Japanese population and Australian populations. Front Physiol 2018, 9:822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Camino EM, Weinstein ML, List MP, Vellky JE, Rebeiz M, Williams TM: Red Light/Green Light, a dual fluorescent protein reporter system to study enhancer-promoter specificity in Drosophila. G3 (Bethesda) 2020, 10:985–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shittu M, Steenwinkel T, Koshikawa S, Werner T: The making of transgenic Drosophila guttifera. Methods Protoc 2020, 3:31. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This well-detailed methods paper (video included) highlighted the additional considerations that must be taken when examining a delicate species. The authors suggested that their refined approach to D. guttifera could be translated to many other sensitive species.
- 32.Miller DE, Staber C, Zeitlinger J, Hawley RS: Highly contiguous genome assemblies of 15 Drosophila species generated using Nanopore sequencing. G3 (Bethesda) 2018, 8:3131–3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gibert J-M, Mouchel-Vielh E, Peronnet F: Modulation of yellow expression contributes to thermal plasticity of female abdominal pigmentation in Drosophila melanogaster. Sci Rep 2017, 7:43370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Massey JH, Akiyama N, Bien T, Dreisewerd K, Wittkopp PJ, Yew JY, Takahashi A: Pleiotropic effects of ebony and tan on pigmentation and cuticular hydrocarbon composition in Drosophila melanogaster. Front Physiol 2019, 10:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bai X, Zeng T, Ni X-Y, Su H-A, Huang J, Ye G-Y, Lu Y-Y, Qi Y-X: CRISPR/Cas9-mediated knockout of the eye pigmentation gene white leads to alterations in colour of head spots in the oriental fruit fly, Bactrocera dorsalis. Insect Mol Biol 2019, 28:837–849. [DOI] [PubMed] [Google Scholar]; **The authors used CRISPR/Cas9 to knockout white in Bactrocera dorsalis, altering a complex spot pattern on the head of this dipteran. This knockout resulted in a decrease in Bd-yellow1 expression, which demonstrated the possibility of a regulatory relationship between white and yellow.
- 36.Noh MY, Kim SH, Gorman MJ, Kramer KJ, Muthukrishnan S, Arakane Y: Yellow-g and Yellow-g2 proteins are required for egg desiccation resistance and temporal pigmentation in the Asian tiger mosquito, Aedes albopictus. Insect Biochem Mol Biol 2020, 122:103386. [DOI] [PubMed] [Google Scholar]
- 37.Liu T, Yang W-Q, Xie Y-G, Liu P-W, Xie L-H, Lin F, Li C-Y, Gu J-B, Wu K, Yan G-Y, Chen X-G: Construction of an efficient genomic editing system with CRISPR/Cas9 in the vector mosquito Aedes albopictus. Insect Sci 2019, 26:1045–1054. [DOI] [PubMed] [Google Scholar]
- 38.Du M-H, Yan Z-W, Hao Y-J, Yan Z-T, Si F-L, Chen B, Qiao L: Suppression of Laccase 2 severely impairs cuticle tanning and pathogen resistance during the pupal metamorphosis of Anopheles sinensis (Diptera: Culicidae). Parasit Vectors 2017, 10:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu G-H, Jiao Y, Chereddy SCRR, Noh MY, Palli SR: Knockout of juvenile hormone receptor, Methoprene-tolerant, induces black larval phenotype in the yellow fever mosquito, Aedes aegypti. Proc Natl Acad Sci U S A 2019, 116:21501–21507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paulo DF, Williamson ME, Arp AP, Li F, Sagel A, Skoda SR, Sanchez-Gallego J, Vasquez M, Quintero G, Pérez de León AA, et al. : Specific gene disruption in the major livestock pests Cochliomyia hominivorax and Lucilia cuprina using CRISPR/Cas9. G3 (Bethesda) 2019, 9:3045–3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heinze SD, Kohlbrenner T, Ippolito D, Meccariello A, Burger A, Mosimann C, Saccone G, Bopp D: CRISPR-Cas9 targeted disruption of the yellow ortholog in the housefly identifies the brown body locus. Sci Rep 2017, 7:4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koidou V, Denecke S, Ioannidis P, Vlatakis I, Livadaras I, Vontas J: Efficient genome editing in the olive fruit fly, Bactrocera oleae. Insect Mol Biol 2020, 29:363–372. [DOI] [PubMed] [Google Scholar]
- 43.Meza JS, Cáceres C, Bourtzis K: slow larvae Mutant and its potential to improve the pupal color-based genetic sexing system in Mexican fruit fly, (Diptera: Tephritidae). J Econ Entomol 2019, 112:1604–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen E-H, Hou Q-L, Wei D-D, Dou W, Liu Z, Yang P-J, Smagghe G, Wang J-J: Tyrosine hydroxylase coordinates larval-pupal tanning and immunity in oriental fruit fly (Bactrocera dorsalis). Pest Manag Sci 2018, 74:569–578. [DOI] [PubMed] [Google Scholar]
- 45.Zhang H-H, Zhang Q-W, Idrees A, Lin J, Song X-S, Ji Q-E, Du Y-G, Zheng M-L, Chen J-H: Tyrosine hydroxylase is crucial for pupal pigmentation in Zeugodacus tau (Walker) (Diptera: Tephritidae). Comp Biochem Physiol B Biochem Mol Biol 2019, 231:11–19. [DOI] [PubMed] [Google Scholar]
- 46.Gompel N, Kopp A: Drosophila (Sophophora) carrolli n. sp., a new species from Brunei, closely related to Drosophila (Sophophora) rhopaloa Bock & Wheeler, 1972 (Diptera: Drosophilidae). Zootaxa 2018, 4434:502–510. [DOI] [PMC free article] [PubMed] [Google Scholar]; *This article provided quality images and a clear description of a new Drosophila species. It is a testament to the importance of field work to evo-devo. The discovery of new species and novel morphologies are just as vital to our field as the invesitgation of the molecular underpinnings of novel traits.
- 47.Hakima B, Hwang H-S, Lee K-Y: Molecular identification of Culicoides (Diptera: Ceratopogonidae) species in Algeria. Acta Trop 2020, 202:105261. [DOI] [PubMed] [Google Scholar]
- 48.Kvifte GM, Bernal XE: A new species of frog-biting midge from Papua New Guinea with a key to the described Corethrellidae of the Australopapuan region (Diptera, Corethrellidae, Corethrella). Zookeys 2018, 795:39–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hawkes MF, Duffy E, Joag R, Skeats A, Radwan J, Wedell N, Sharma MD, Hosken DJ, Troscianko J: Sexual selection drives the evolution of male wing interference patterns. Proc Biol Sci 2019, 286:20182850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bourquia M, Garros C, Rakotoarivony I, Gardès L, Huber K, Boukhari I, Delécolle J-C, Baldet T, Mignotte A, Lhor Y, et al. : Update of the species checklist of Culicoides Latreille, 1809 biting midges (Diptera: Ceratopogonidae) of Morocco. Parasit Vectors 2019, 12:459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skevington JH, Locke MM, Young AD, Moran K, Crins WJ, Marshall SA: Field guide to the flower flies of northeastern North America. Princeton University Press; 2019. [Google Scholar]
- 52.Miller ME, Marshall SA, Grimaldi DA: A review of the species of Drosophila (Diptera: Drosophilidae) and genera of Drosophilidae of northeastern North America. Can J Arthropod Identif 2017, 31:1–282. [Google Scholar]
- 53.Werner T, Steenwinkel T, Jaenike J: The Encyclopedia of North American Drosophilids. Vol. 1: Drosophilids of the Midwest and Northeast (Version 3). Open Access Books; 2020. [Google Scholar]; *This open access, comprehensive field guide provided highly detailed photographs and accurate descriptions for drosophilids across the North American Midwest and Northeast.
- 54.Werner T, Steenwinkel T, Jaenike J: The Encyclopedia of North American Drosophilids. Vol. 2: Drosophilids of the Southeast. Open Access Books; 2020. [Google Scholar]; *This is volume two in a series of field guides that will encompass the entire North American mainland. Here, the authors focused on the Southeastern drosophilid species of North America.


