Summary
Lymphoma is a group of blood cancers that develop from the immune system, and one of the main risk factors is associated with exposure to environmental chemicals. Bisphenol A (BPA) is a common chemical used in the manufacture of materials in polycarbonate and epoxy plastic products and can interfere with the immune system. BPA is considered to possibly induce lymphoma development by affecting the immune system, but its potential mechanisms have not been well established. This study performed a gene-network analysis of microarray data sets in human lymphoma tissues as well as in human cells with BPA exposure to explore module genes and construct the potential pathway for lymphomagenesis in response to BPA. This study provided evidence that BPA exposure resulted in disrupted cell cycle and DNA damage by activating CTNNB1, the initiator of the aberrant constructed CTNNB1-NFKB1-AR-IGF1-TWIST1 pathway, which may potentially lead to lymphomagenesis.
Subject areas: molecular biology, cancer
Graphical abstract
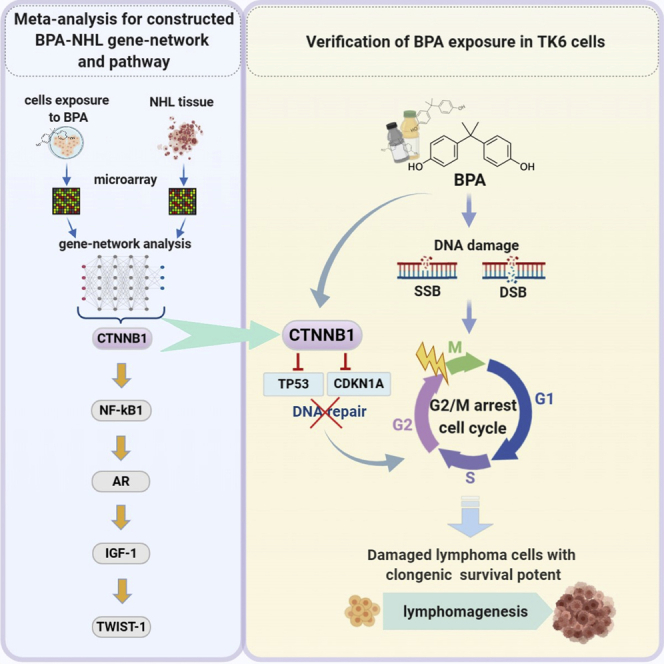
Highlights
-
•
CTNNB1 is an initiator of BPA-mediated lymphomagenesis by gene-network analysis
-
•
BPA exposure promotes clonogenic survival of damaged TK6 lymphoblastoid cells
-
•
BPA/CTNNB1 dysregulates DNA-repair-associated genes TP53 and CDKN1A
-
•
BPA-/CTNNB1-mediated lymphomagenesis is attributable to DNA breaks and G2/M arrest
Molecular biology; Cancer
Introduction
Lymphoma is a group of blood cancers developed from lymphocytes in the immune system, which is categorized into Hodgkin lymphoma (HL) and non-Hodgkin lymphoma (NHL). NHL is ranked as the fifth to ninth most common cancer in most countries worldwide, with almost 510,000 new cases estimated in 2018 (Miranda-Filho et al., 2019). It has been reported that the age-adjusted incidence rates of NHL per 100,000 between 2008 and 2012 were generally much higher in countries with high Human Development Index (HDI) (the highest 17.6/13, respectively, versus the lowest 1.6/1.0 per 100,000 males/females). Altered immune systems have an increased risk of NHL (Grulich et al., 2007; Purdue et al., 2011). Besides viral and bacterial infections as well as use of immunosuppressive drugs, previous epidemiologic studies have revealed that exposure to chemicals and changes in diet and lifestyle contributed to modifiable risk factors of NHL (Alexander et al., 2007; Chiu and Hou, 2015; Morton et al., 2006). Although descriptive epidemiology of NHL risk factors has been well characterized using population-based cancer registry data, the key effector and its downstream pathway for lymphomagenesis underlying such risk factors is not yet well established.
Bisphenol A (BPA) is an industrial chemical used to create polycarbonate plastic and epoxy resins. Owing to its hardness and strength over a wide range of temperatures, global production of BPA was predicted to exceed 5 million metric tons by 2015 (Gore et al., 2015). Most people are exposed to BPA leached from polycarbonate containers of consuming food and beverages and resin-based dental sealants and bonding agents (Konieczna et al., 2015). It has been reported that estimates of higher daily BPA intake between 2000 and 2016 were more common in economically developed countries (Huang et al., 2017b). BPA has a long-known ability to interfere with immune and inflammation responses through several signaling pathways and alteration of various immune cells (Clayton et al., 2011; Murata and Kang, 2018). Altered immune systems have an increased risk of NHL (Grulich et al., 2007; Purdue et al., 2011). In addition, DNA damage has been reported following BPA exposure to nonmalignant human breast cell lines (Pfeifer et al., 2015) and human bronchial cell lines (George and Rupasinghe, 2018). Cumulating evidence from solid tumors and acute myeloid leukemia highlights the roles of BPA exposure in promoting tumor progression through a variety of genes involved in cancer cell survival advantages (Nomiri et al., 2019). Exposure to BPA is associated with increased aggressiveness of a variety of cancers, such as lung cancer (Zhang et al., 2014), ovarian cancer (Ptak et al., 2014), colorectal cancer (Chen et al., 2015), breast cancer (Song et al., 2015), cervical cancer (Ma et al., 2015), and nasopharyngeal cancer (Zeng, 2020).
DNA damage accumulates in hematopoietic stem cells (HSCs) during aging (Beerman et al., 2014; Rübe et al., 2011). DNA double-strand breaks (DSBs) are considered the most lethal DNA damage and have been found increased in CD19-positive cells of chronic lymphocytic leukemia compared with those of monoclonal B cell lymphocytosis and healthy ones (Popp et al., 2019). Since BPA exposure contributes to DNA damage and dysregulated immune responses, DNA-damaged lymphoid cells would render lymphoma development and progression in a compromised immune microenvironment. However, it is currently unknown how human lymphoid cells survive DNA damage induced by BPA during the progression of lymphoma. Therefore, we proposed a novel and applicable pathway construction approach for potential lymphomagenesis in response to BPA exposure by performing a gene-network analysis of microarray data sets in human lymphoma tissues and in human cells exposed to BPA for exploring module genes and their potential interconnection. Furthermore, the identified key gene CTNNB1 dysregulated by BPA in the constructed pathway CTNNB1-NFKB1-AR-IGF1-TWIST1 underlying the lymphoma pathogenesis and progression was validated in human lymphoblastoid TK6 cells with BPA-induced DNA damage.
Results
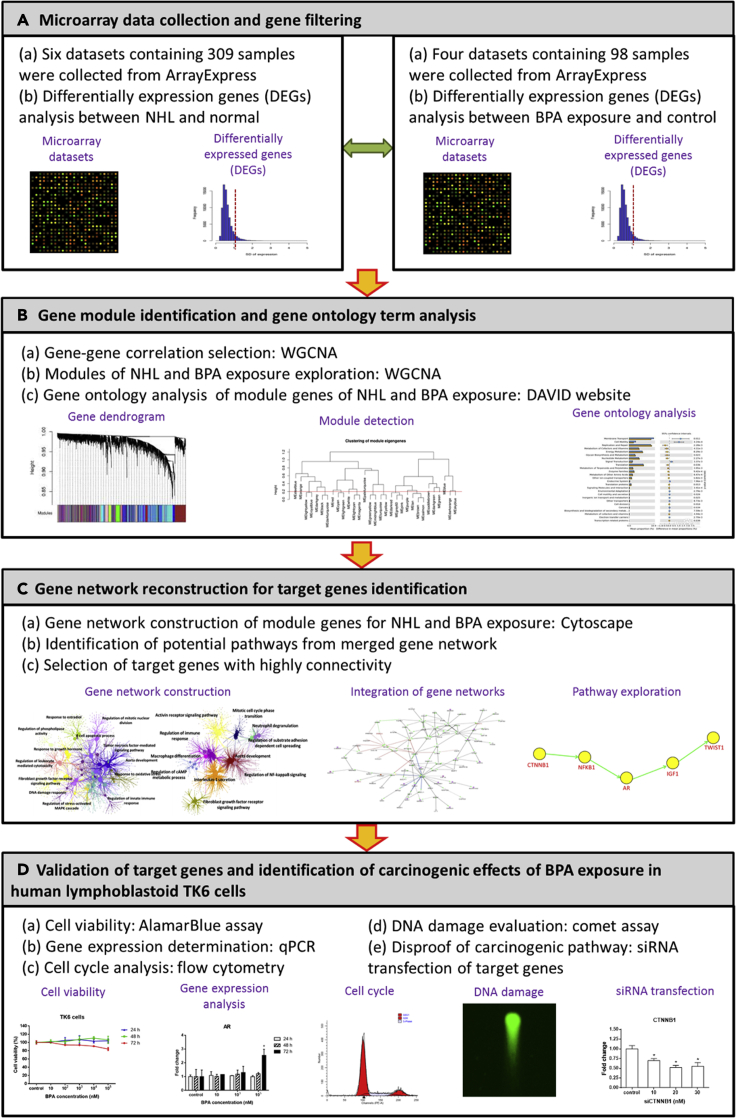
The framework of the present study is divided into four main parts (Figure 1): (A) collection of microarray data and gene filtering, (B) identification of gene modules of NHL and BPA exposure and analysis of their gene ontology term, (C) gene-network reconstruction for a potential pathway of the NHL progression corresponding to BPA exposure, and (D) validation of potential target genes and examination of carcinogenic effect in TK6 cells exposure to BPA.
Figure 1.
Framework of this study
Module genes and ontological network of NHL and BPA exposure
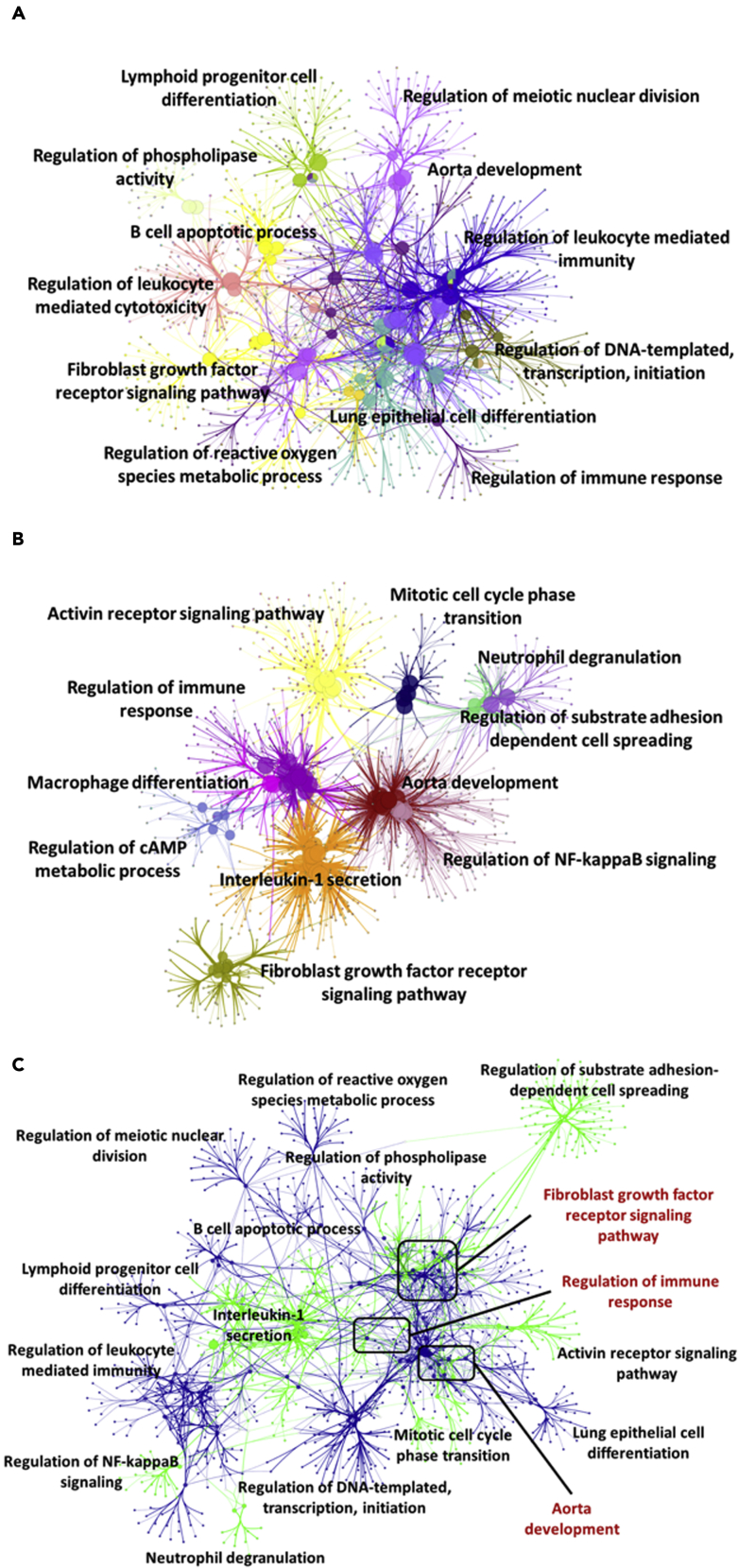
First of all, we utilized six NHL data sets containing 309 samples and four BPA data sets containing 98 samples from ArrayExpress. The DEGs of NHL (n = 4,593) and BPA exposure (n = 4,028) were constructed into gene modules by weighted gene correlation network analysis (WGCNA) to construct two subnetworks. In the gene ontological subnetworks (Figure 2), the functions of NHL module genes were clustered into regulation of phospholipase activity, lymphoid progenitor cell differentiation, B cell apoptotic process, regulation of leukocyte-mediated cytotoxicity, fibroblast growth factor receptor signaling pathway, regulation of reactive oxygen species metabolic process, regulation of meiotic nuclear division, aorta development, regulation of leukocyte-mediated immunity, regulation of DNA-templated transcription and initiation, lung epithelial cell differentiation, and regulation of immune response (Figure 2A). On the other hand, the functions of BPA module genes were categorized into activin receptor signaling pathway, regulation of immune response, macrophage differentiation, regulation of cyclic adenosine monophosphate metabolic process, interleukin-1 secretion, fibroblast growth factor receptor signaling pathway, mitotic cell cycle phase transition, neutrophil degranulation, regulation of substrate adhesion dependent cell spreading, aorta development, and regulation of nuclear factor kappa B (NF-κB) signals (Figure 2B). Attractively, there were three overlapping functions involved in regulation of immune response, aorta development, and fibroblast growth factor receptor signaling pathway between module genes of NHL and BPA exposure (Figure 2C).
Figure 2.
Ontological gene network of module genes
(A–C) (A) NHL, (B) BPA exposure, and (C) integration of NHL and BPA exposure. The ontological gene network was visualized by Cytoscape plug-in ClueGO.
Subnetwork and potential pathway of NHL and BPA exposure
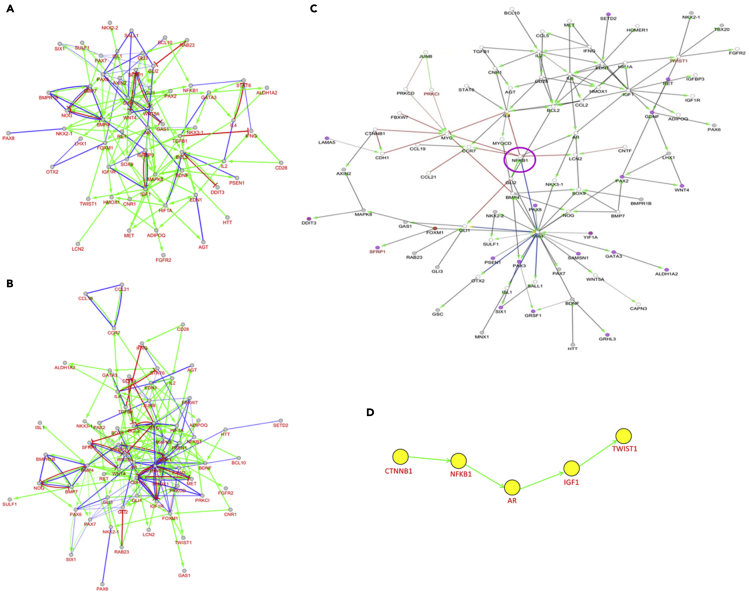
Genes in the aforementioned ontological subnetworks were further selected by high gene connectivity to predict potential regulatory pathways for development of NHL (Figure 3A), BPA exposure (Figure 3B), and progression of NHL in response to BPA exposure (Figure 3C). In Figure 3C, red lines represent the regulatory pathway of BPA exposure only. Blue lines display the regulatory pathway of NHL, while black lines indicate the collaborative regulatory pathway of BPA exposure and NHL. NFKB1 (purple circle) is the only one gene that interacts with red, blue, and black lines. As a result, NFKB1 was chosen as a middle point to explore its upstream (BPA only) and downstream (intersection of BPA and NHL) genes. The upstream gene of NFKB1, CTNNB1, was considered to be an initiator gene of BPA exposure. Previous studies have presented that Twist expression is higher in diffuse large B cell lymphoma (DLBCL) (Lemma et al., 2013), and Twist1 is related to epithelial-mesenchymal transition (Huang et al., 2018). Sequentially, Twist1 was regarded as the downstream end in the proposed pathway, and the downstream genes following NFKB1 were AR, IGF1, and TWIST1 and postulated to be carcinogenic genes of NHL progression after BPA exposure. The predicted potential pathway related to NHL progression attributed to BPA exposure is shown in Figure 3D. Eventually, the proposed pathway of NHL progression in response to BPA exposure was concentrated on dysregulated CTNNB1-NFKB1-AR-IGF1-TWIST1 pathway, which was validated with BPA-treated human lymphoblastoid TK6 cell line, one of the standard cell lines for in vitro mammalian cell genotoxicity tests.
Figure 3.
The collaborative regulatory pathway for progression of NHL in response to BPA exposure
(A–D) (A) NHL subnetwork, (B) BPA subnetwork, and (C) regulatory pathway from the intersection of NHL and BPA modules were illustrated; (D) the potential regulatory pathway of NHL progression in response to BPA exposure. Color lines reflect pathway differences between NHL and BPA (red: BPA exposure, blue: NHL, black: interactions of NHL and BPA exposure).
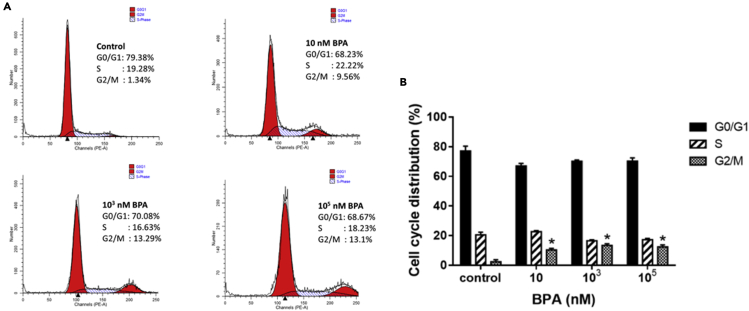
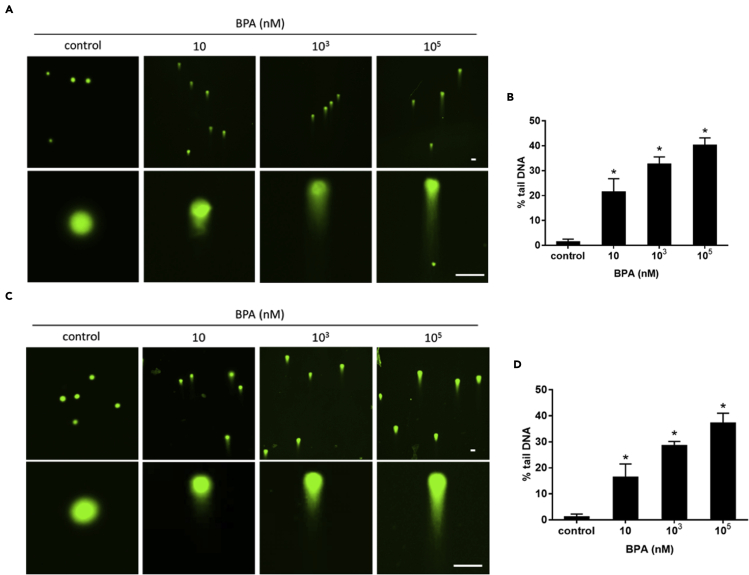
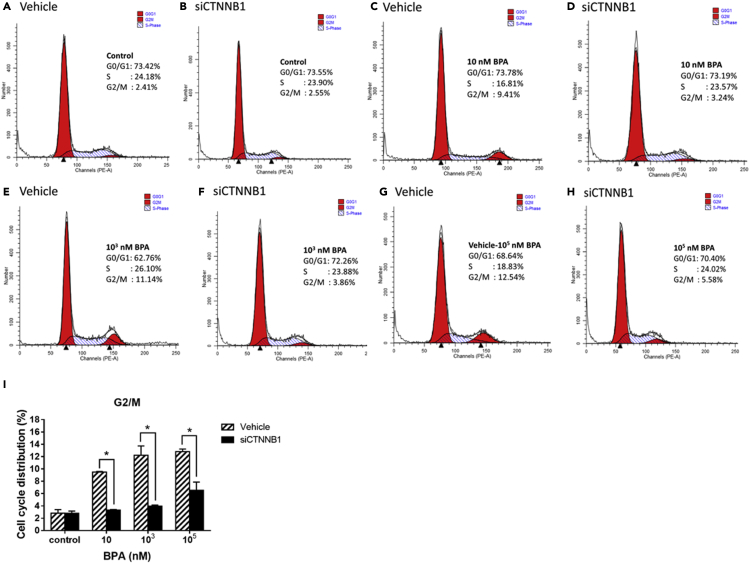
Target gene expression, cell cycle distribution, and DNA strand break in exposure to BPA
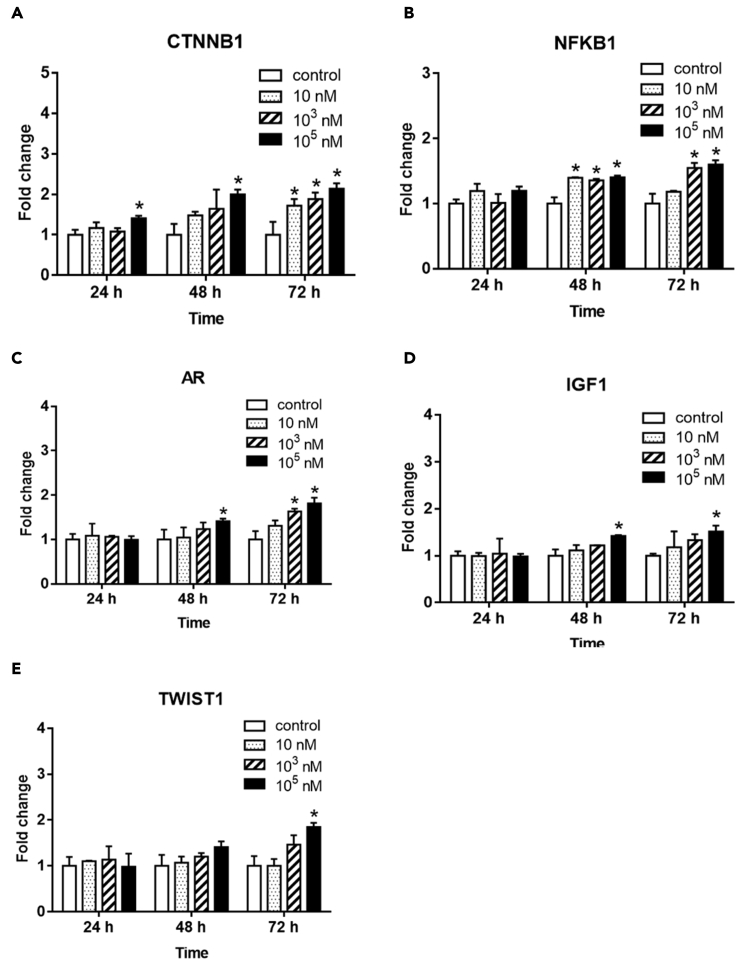
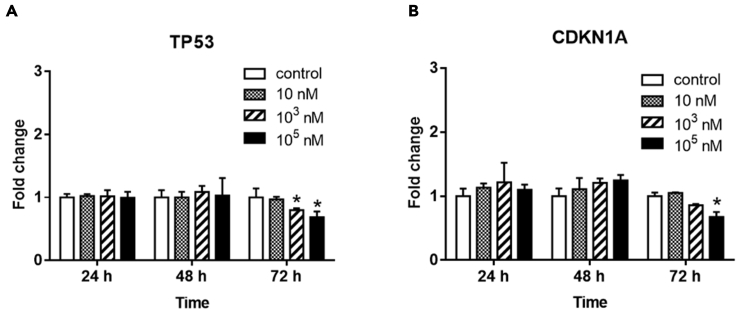
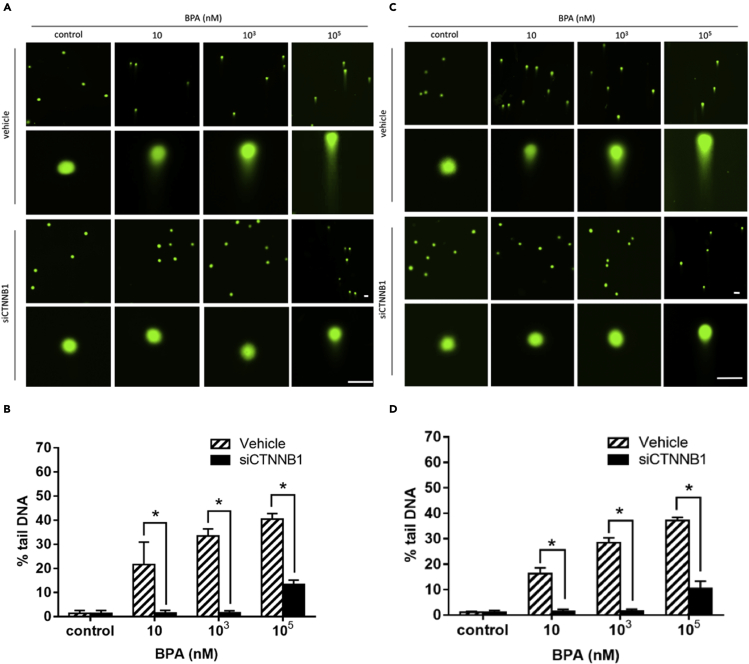
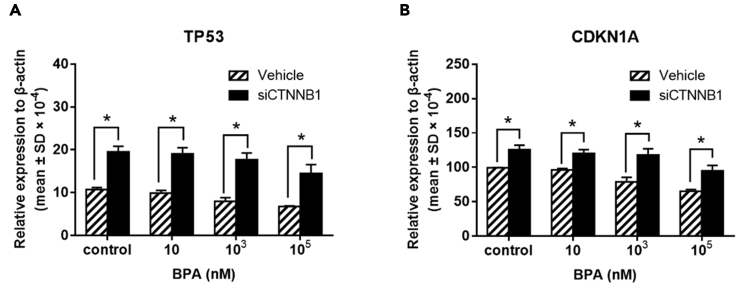
Increased mRNA expressions of CTNNB1, NFKB1, AR, IGF1, and TWIST1 in TK6 cells were shown in a dose-response and time-dependent manner following BPA treatment (Figure 4). Moreover, markedly increased cell percentage arrested in G2/M phase cell cycle was observed by flow cytometry after 72 hr exposure to 10-nM BPA compared with control (Figure 5). Comparable percentage increments of cells arrested were found between 103 and 105 nM BPA. Furthermore, the tail DNA% of a mixture of DNA damage (single-strand breaks [SSBs], double-strand breaks [DSBs], and alkali-labile sites) (Figures 6A and 6B) and DSBs (Figures 6C and 6D) were significantly increased in TK6 cells treated with 10-105 nM BPA for 72 hr, which indicated BPA-induced DNA SSBs and DSBs in TK6 cells. As to the BPA-associated DNA damage, we further determined the status of DNA-repair-associated genes in BPA-treated TK6 cells. TP53 and CDKN1A mRNA expression were significantly decreased after 72 hr of exposure to BPA (Figures 7A and 7B).
Figure 4.
Gene expression of target genes of carcinogenesis in TK6 cells after BPA exposure
TK6 cells were treated with various concentrations (10, 103, 105 nM) of BPA for 24, 48, and 72 hr. Fold changes of gene expression of (A) CTNNB1, (B) NFKB1, (C) AR, (D) IGF1, and (E) TWIST1 between BPA exposure and control. ∗p < 0.05 indicated a significant difference between BPA exposure and control.
Figure 5.
Cell cycle distribution of TK6 cells exposed to BPA
(A) The graphs showed cell cycle profiles in TK6 cells exposed to various concentrations (10, 103, 105 nM) of BPA for 72 hr. The peaks in the illustration correspond to the G0/G1, S, and G2/M phases of the cell cycle.
(B) Histogram showed the percentages of cells in each phase of the cell cycle after BPA exposure for 72 hr.
Figure 6.
DNA single-strand breaks and double-strand breaks of TK6 cells exposed to BPA
DNA strand breaks were determined using comet assay in TK6 cells exposed to BPA for 72 hr.
(A and B) (A) The fluorescent images were taken and (B) the percentage of tail of a mixture of DNA DSBs and SSBs was measured under alkaline conditions.
(C and D) (C) The fluorescent images were taken and (D) the percentage of tail DNA DSBs was measured under neutral conditions. Scale bar = 50 μm; ∗p < 0.05 indicated a significant difference between BPA exposure and control.
Figure 7.
Expression of DNA-repair-associated genes TP53 and CDKN1A in TK6 cells after BPA exposure
TK6 cells were treated with various concentrations (10, 103 and 105 nM) of BPA for 24, 48, and 72 hr. Fold changes of gene expression of (A) TP53 and (B) CDKN1A between BPA exposure and control. ∗p < 0.05 indicated a significant difference between BPA exposure and control.
Expression of downstream genes of CTNNB1 in exposure to BPA after CTNNB1 siRNA transfection
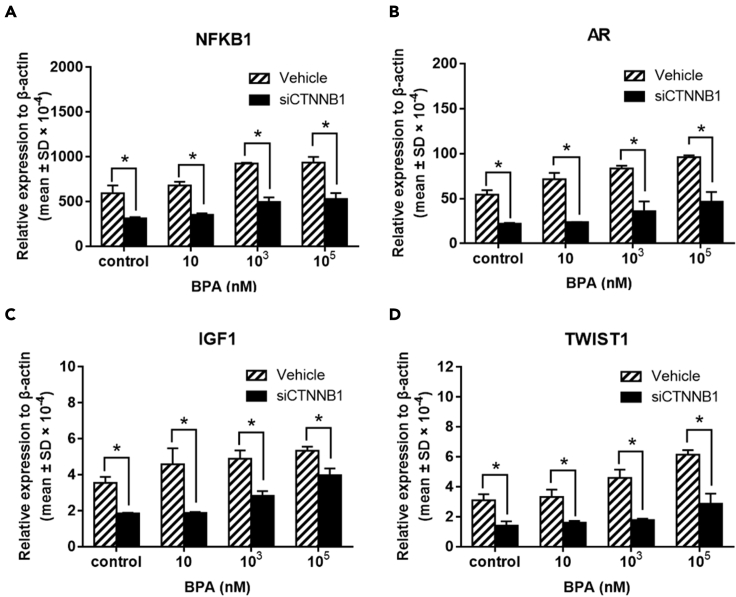
For validation, siRNA transfection was used to knockdown the upstream target gene CTNNB1 expression for evaluation of subsequent alterations of the regulatory pathway NFKB1-AR-IGF1-TWIST1, cell cycle distribution, DNA-repair-associated genes, and DNA damage in TK6 cells following BPA exposure. Compared with vehicles in each group of increasing BPA doses, the BPA upregulated mRNA expressions of NFKB1, AR, IGF1, and TWIST1 were reciprocally downregulated in CTNNBA siRNA-transfected TK6 cells (Figure 8). These results substantiated that CTNNB1 was the initiator gene of the constructed CTNNB1-NFKB1-AR-IGF1-TWIST1 pathway regulated by BPA.
Figure 8.
Gene expression of downstream genes of CTNNB1 in TK6 cells exposed to BPA after siCTNNB1 transfection
Relative gene expression of (A) NFKB1, (B) AR, (C) IGF1, and (D) TWIST1 in TK6 cells exposed to BPA for 72 hr after siCTNNB1 transfection (homo-960). ∗p < 0.05 indicated a significant difference between siCTNNB1 transfection and vehicle.
Cell cycle distribution and DNA strand break in exposure to BPA after CTNNB1 siRNA transfection
With respect to BPA-associated cell-cycle arrest, flow cytometric results were compared between TK6 cells under BPA treatment of increasing doses and with/without siCTNNB1 transfection (Figure 9). Compared with vehicles in each dose group, BPA-associated increased cell groups arrested in G2/M phase were reciprocally shifted in CTNNBA siRNA-transfected TK6 cells. These results further supported that BPA exposure played a role in cell-cycle arrest at G2/M phase through activation of the CTNNB1-NFKB1-AR-IGF1-TWIST1 pathway. Additionally, to understand the role of CTNNB1 playing in DNA strand break, the siCTNNB1-transfected TK6 cells were introduced in the comet assay under alkaline and neutral conditions for investigating the effects of CTNNB1 gene on BPA-induced DNA strand break. Increased tail DNA% of a mixture of DNA SSBs and DSBs by BPA in each dose group was significantly reduced in the presence of siCTNNB1 transfection (Figures 10A and 10B). Similar reciprocal reduction patterns in the tail DNA% of DSBs were demonstrated in siCTNNB1-transfected cells compared with vehicles in each BPA dose group (Figures 10C and 10D). These results verified that CTNNB1 participated in the initial event of BPA-mediated DNA breaks.
Figure 9.
Cell cycle distribution of TK6 cells exposed to BPA after siCTNNB1 transfection
(A–H) The graphs showed cell cycle profiles in TK6 cells exposed to BPA for 72 hr after siCTNNB1 transfection (homo-960). The peaks in the illustration correspond to the G0/G1, S, and G2/M phases of the cell cycle.
(I) Histogram showed the percentages of cells in G2/M phase of the cell cycle after BPA exposure for 72 hr ∗p < 0.05 indicated a significant difference between siCTNNB1 transfection and vehicle.
Figure 10.
DNA single-strand breaks and double-strand breaks in TK6 cells exposed to BPA after siCTNNB1 transfection
DNA strand breaks were determined using comet assay in TK6 cells exposed to BPA for 72 hr after siCTNNB1 transfection (homo-960).
(A and B) (A) The fluorescent images were taken and (B) the percentage of tail of a mixture of DNA DSBs and SSBs was measured under alkaline conditions.
(C and D) (C) The fluorescent images were taken and (D) the percentage of tail DNA DSBs was measured under neutral conditions. Scale bar = 50 μm; ∗p < 0.05 indicates a significant difference between siCTNNB1 transfection and vehicle.
Colony formation in exposure to BPA after CTNNB1 siRNA transfection
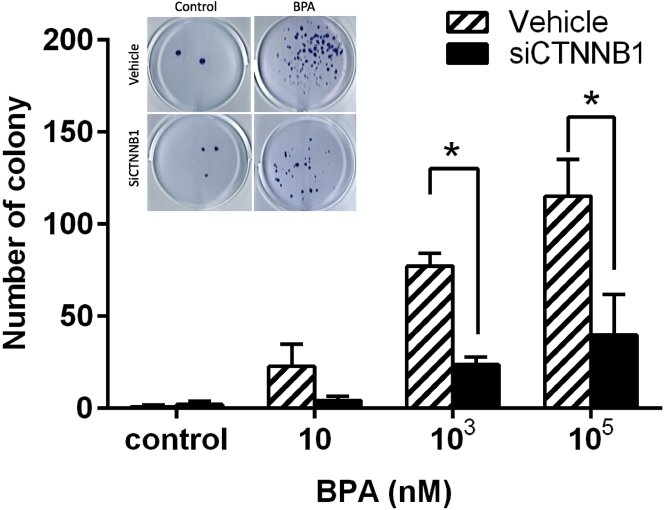
To further clarify interconnections between the constructed pathway and DNA-associated repair genes, compared with vehicles in each dose group, BPA-downregulated mRNA expressions of TP53 and CDKN1A were reciprocally upregulated in siCTNNB1-transfected TK6 cells (Figure 11). These results provided evidence that BPA exposure dysregulated DNA-repair-associated genes TP53 and CDKN1A through CTNNB1 activation. Most importantly, we performed TK6 cell colony-forming assay (Figure 12) to evaluate whether BPA exposure was involved in lymphomagenesis via activation of CTNNB1. The 72-hr dose-dependent colony formations of TK6 cells visibly promoted by BPA treatment were significantly suppressed in siCTNNB1-transfected TK6 cells in each dose group. Strikingly, these compelling results demonstrated that BPA-promoted colony formations, indicative of lymphomagenesis, were mediated by activation of CTNNB1, the initiator of the novel constructed CTNNB1-NFKB1-AR-IGF1-TWIST1 pathway.
Figure 11.
Gene expressions of TP53 and CDKN1A in TK6 cells exposed to BPA after siCTNNB1 transfection
Relative gene expression of (A) TP53 and (B) CDKN1A expression in TK6 cells exposed to BPA for 72 hr after siCTNNB1 transfection (homo-960). ∗p < 0.05 indicates a significant difference between siCTNNB1 transfection and vehicle.
Figure 12.
Colony formation of TK6 cells in exposed to BPA after siCTNNB1 transfection
The average number of colonies formed in TK6 cells exposed to BPA for 72 hr after siCTNNB1 transfection (homo-960) in six randomly sampled visual fields. ∗p < 0.05 indicates a significant difference between siCTNNB1 transfection and vehicle.
Discussion
Following rapid global economic and industrial development in recent years, cumulating modifiable cancer risk factors are identified mainly based on epidemiological studies (Islami et al., 2018; Stein and Colditz, 2004). A recently published proteogenomic analysis proves the perspective of environmental-carcinogens-driven sequential pathways initiated with DNA damage contributing to pathogenesis and progression of non-smoking lung cancer in East Asia (Chen et al., 2020). With regard to BPA, most epidemiological and laboratory studies focused on and indicated the carcinogenic risk of BPA for breast cancer (Seachrist et al., 2016). The environmentally relevant concentration of BPA is at ppb (μg/L) level, and BPA was routinely detected in blood and urine in the range of ng/mL (ppb) (Vandenberg et al., 2010). Based on the reported levels of BPA in body fluids and tissues, the theoretical internal dose of the general population has been estimated around 10–100 nM (Vandenberg et al., 2007). Genotoxicity of BPA has been demonstrated in BPA-treated human breast cancer cells (MCF-7) (Aghajanpour-Mir et al., 2016), human epithelial type 2 cells (Hep-2), and human lung fibroblasts (MRC-5) (Ramos et al., 2019).
This study combined the gene-network meta-analysis and in vitro validation to clarify the potential adverse effects of BPA on lymphomagenesis through CTNNB1-NFKB1-AR-IGF1-TWIST1 pathway. Human TK6 cell line, characterized by chromosome stability, high efficiency of cell proliferation, and colony formation (Lorge et al., 2016), used in the present study was one of the standard cell lines for in vitro mammalian cell genotoxicity tests. A second cell line, human lymphoblastoid SUP-B15 cells, was used to explore BPA effects on genes of interest as well (see Figures S1–S3). In spite of inevitable intrinsic differences between these two cell lines, we presented the results of BPA exposure in SUP-B15 cells to validate the certain key findings in TK6 cells. After BPA exposure, the expression of NFKB1 and TWIST1 in SUP-B15 cells was increased (see Figures S1B and S1E) with similar patterns observed in TK6 cells. An associated review showed that activated Wnt/CTNNB1 (β-catenin) pathway to intermediate to high levels lead to lymphopoiesis of T and B lineages, and dysregulated Wnt/CTNNB1 signaling is a frequent event in oncogenesis of T and B cell lymphoid leukemias (Chiarini et al., 2020). Another interesting review (Karimaian et al., 2017) provided a comprehensive molecular insight of cross talk between Wnt/CTNNB1 pathway and DNA damage response. In this study, BPA exposure increased the gene expression of CTNNB1, NFKB1, AR, IGF1, and TWIST1 and caused DNA SSB and DSB damage, accompanied by cell cycle arrest in the G2/M phase for DNA repair. BPA exposure also reduced gene expression of TP53 and CDKN1A and enhanced colony formation, which indicated that DNA repair dysfunction potentially promotes the occurrence of lymphomagenesis underlying the CTNNB1-mediated pathway. To the best of our knowledge, this study was the first to present the evidence that BPA exposure promoted lymphomagenesis through CTNNB1-activation-associated cell cycle arrest, DNA breaks, and reduced DNA repair potential.
Initial ontological gene-network analysis identified common pathways of BPA exposure and lymphomagenesis which were involved in regulation of fibroblast growth factor receptor, aorta development, and immune response. Uterine fibroblast growth factor signal was upregulated by chronic BPA exposure in a mouse model (Neff et al., 2019). Altered angiogenic activity of the human endometrial endothelial cell was observed following in vitro BPA treatment. Aortic atherosclerotic lesions have been demonstrated as a result of BPA exposure (Fang et al., 2014; Sui et al., 2014; Trusca et al., 2019) and associated NF-κB activation (Trusca et al., 2019). Basic fibroblast growth factor, a key angiogenic factor, was found to be a poor prognostic factor in NHL (Pazgal et al., 2002). Intravascular large B cell lymphoma, an aggressive type NHL, has been reported with aortic involvement (Aghajanpour-Mir et al., 2016). Emerging evidence indicated the importance of angiogenesis and angiogenic therapy to B cell NHL (Jiang and Li, 2020). Altered immune systems have an increased risk of NHL (Grulich et al., 2007; Purdue et al., 2011).
According to our gene-network analysis, the CTNNB1 gene encoding β-catenin protein was the key initiator gene of the novel constructed BPA-dysregulated CTNNB1-NFKB1-AR-IGF1-TWIST1 pathway involved in lymphomagenesis. The key role of CTNNB1 gene in the CTNNB1-NFKB1-AR-IGF1-TWIST1 pathway was well demonstrated in the BPA-treated human lymphoblastoid TK6 cell line. Activation of the canonical Wnt pathway was shown in BPA-treated human ovarian (Hui et al., 2018) and nasopharyngeal (Zeng, 2020) cancer cell lines. Compared with reactive hyperplasia of lymph nodes, β-catenin was overexpressed and nuclear accentuation in those of DLBCL, which were strongly correlated with clinical stages (Ge et al., 2012). Similar β-catenin expression patterns were also found in initiating cells of mantle cell lymphoma (MCL) (Mathur et al., 2015). There are several ways of cross talk between Wnt/β-catenin and NF-κB pathways during inflammation (Aghajanpour-Mir et al., 2016) and MCL lymphomagenesis (Lazarian et al., 2020). The NFKB1 gene encodes canonical NF-κB subunit p105, which is processed by the proteasome into p50 to form a p65/p50 heterodimer transcription factor, and is considered a regulator of innate immunity. Canonical NF-κB signals are connected to pathological pathways in low-grade Mucosa-associated lymphoid tissue (MALT) lymphoma (Hamoudi et al., 2010; Nagel et al., 2014), high-grade MCL (Aghajanpour-Mir et al., 2016; Lazarian et al., 2020), as well as DLBCL of ABC type (Compagno et al., 2009; Davis et al., 2001; Nagel et al., 2014). AR (androgen receptor) interplay with NF-κB has been linked in prostate cancer (Malinen et al., 2017; Zhang et al., 2009). BPA regulates the function of AR via multiple ways (Lee et al., 2003; Pelch et al., 2019; Teng et al., 2013). AR blockade has been investigated for MCL therapy (Chow et al., 2020; Mostaghel et al., 2017), a high-grade lymphoma with male predominance (Aghajanpour-Mir et al., 2016), and increased AR expression (Mostaghel et al., 2017). Androgens were demonstrated to promote mRNA expression of IGF1 (insulin-like growth factor 1) and its receptor in the primate ovary (Aghajanpour-Mir et al., 2016). Correlation between serum levels of IGF1 and androgens was reported in all women enrolled in an acne study (Cappel et al., 2005). It has been shown in adipose stromal/stem cells (Ohlstein et al., 2014) and human embryonic stem cells (Huang et al., 2017a) that IGF1 was regulated by BPA exposure. Variable levels of IGF1 receptor (IGF1-R) were found between 8 DLBCL cell lines (Stromberg et al., 2015). IGF1 signal was associated with proliferation of DLBCL (Zhou et al., 2017). TWIST is a transcription factor involved in mesoderm development (Miraoui and Marie, 2010). IGF1-induced Twist expression in mouse fibroblast cell lines was associated with antiapoptotic effects of IGF1-R (Dupont et al., 2001). Twist1 expression was upregulated, and epithelial-mesenchymal transition was induced in prostate tissue from aged mice exposed to oral BPA (Huang et al., 2018). Much higher percentages of nuclear Twist expression were found in DLBCL, compared with lack of expression in lymphocytes from lymph nodes with benign hyperplasia (Lemma et al., 2013).
DNA damage can change the DNA sequence of cells on tumor promotion and activate the DNA repair mechanism to correct the errors. Thus, when DNA repair is inefficient, such lesions can lead to mutations that ultimately cause cancer. This study revealed that BPA exposure suppressed gene expression of TP53 and CDKN1A in TK6 cells and SUP-B15 cells (see Figure S2). DNA SSBs and DSBs were observed in human peripheral blood mononuclear cells post 1 to 4 hr of BPA treatment in a dose-dependent manner (Mokra et al., 2017). DNA damage on peripheral blood mononuclear cells following 4 hr of BPA exposure was not completely repaired after 2 hr of BPA-free duration (Mokra et al., 2017). Such BPA dose-dependent DNA damage was reproduced by TK6 cells treated with BPA for 72 hr in the present study. It was reported with a model of nematode, Caenorhabditis elegans, that BPA exposure altered DNA DSBs repair process, checkpoint activation, as well as repair gene expression and eventually left severe chromosome abnormalities, maturation delay in late prophase (Allard and Colaiácovo, 2010). A variety of recurrent chromosome translocations are found in B cell lymphomas ranging from low-grade to high-grade subtypes (Vega and Medeiros, 2003). Such translocations usually result from DNA DSBs (Nambiar and Raghavan, 2011) and erroneous repair (Iarovaia et al., 2014). G2/M checkpoint prevents damaged cells from mitosis initiation. Cells inflicted with unimpaired damages during G1 or S phase accumulate in G2 phase and usually undergo repair and/or apoptosis. Cells with G2/M failure exposed to etoposide led to cell proliferation with fixed chromosome translocations (Nakada et al., 2006). Our flow cytometric results showed that TK6 cell percentage arrested in G2/M was markedly increased in 72-hr 10-nM BPA-treated group compared with the control. Such increments of cells arrested were comparable between groups with 72-hr 103 and 105 nM BPA exposure. However, colony formations of TK6 cells were significantly promoted by BPA exposure in a dose-dependent pattern from 10 to 105 nM. Both BPA-induced G2/M arrest and promoted colony formation were proved to be CTNNB1 gene regulated.
Because degrees of DNA damage and colony formation were increased under comparable percentage of G2/M arrest between 103 and 105 nM BPA exposure, p53 and CDKN1A genes, the two closely interconnected key players in cell cycle, were further investigated for β-catenin-mediated clonogenic survival potential. Both p53 and CDKN1A genes were upregulated upon CTNNB1 gene knockdown across BPA time and dose exposure groups. In 72-hr exposure, p53 and CDKN1A were both significantly downregulated in 105 nM BPA-treated groups compared with control. It is well known that p53, a central tumor suppressor gene, is important for cell cycle arrest and following repair to stabilize a variable forms of DNA damage (Williams and Schumacher, 2016) by activation of DNA repair machinery to preserve cells (Mahemuti et al., 2018) or by induction of apoptosis to eliminate cells from the irrevocable damage (Liu et al., 2017) DNA damage accelerates the mutagenesis of human cancer cells carrying TP53 mutation (Moulder et al., 2018). Disrupted p53 function was shown to be a predictor of treatment failure and poor prognosis in B and T cell NHL (Moller et al., 1999). Mutations of p53 also predicted poor prognosis of DLBCL and MCL in a meta-analysis (Xu et al., 2017). p21 protein encoded by CDKN1A gene is a potent cyclin-dependent kinase inhibitor with multiple functions in regulation of cell cycle, apoptosis, and transcription after DNA damage for repair (Karimian et al., 2016). This study found that downregulated CTNNB1 by its targeting siRNA can elevate the gene expression of BPA-reduced TP53 and CDKN1A to attenuate BPA-induced DNA damage. β-Catenin inhibited CDKN1A gene expression in breast cancer cell lines (Xu et al., 2016), which was consistent with our results. NF-κB activation following irradiation to human leukemic and breast cancer cell lines induced p21 expression and resultant G2/M cell cycle arrest (Wuerzberger-Davis et al., 2005). BPA exposure (10 and 103 nM) reduces CDKN1A gene expression and increases cell proliferation in human breast cancer MCF-7 cells (Lee et al., 2012) Eμ-Myc mice lacking both CDKN1A and TP53 significantly increase the incidence of lymphoma (Valente et al., 2016). Additionally, the excess CTNNB1 can reduce TP53 protein levels and restrain TP53 signaling in human colon cancer cells (Sadot et al., 2001). Knockdown of CTNNB1 increases the expression of CDKN1A to decrease cell viability and neurosphere formation in glioma stem cells (Yang et al., 2016). According to the aforementioned studies and our findings, BPA exposure would induce CTNNB1 to decrease TP53 and CDKN1A expressions and cause DNA damage and DNA repair dysregulation, which may lead to lymphomagenesis.
DNA DSBs were much more common in cells of diffuse large B cell lymphomas (DLBCL) than in chronic lymphocytic leukemia cells and cells of reactive follicles (Derenzini et al., 2015). In patients with DLBCL following rituximab plus cyclophosphamide, doxorubicin, vincristine and prednisone/cyclophosphamide, doxorubicin, vincristine and prednisone-like therapies, those with high DNA DSBs had worse overall survival than those with low DNA DSBs. β-Catenin protein encoded by CTNNB1 gene is a key downstream effector of the Wnt/β-catenin pathway. Wnt/β-catenin pathway induced aging of mesenchymal stem cells through DNA damage response (Zhang et al., 2011). β-Catenin is responsible for self-renewal activity and leukemic potential of CML progenitor cells (Jamieson et al., 2004; Zhao et al., 2007). Cat T cell lymphoma induced by β-catenin activation had recurrent translocations but survived under DNA damage not repaired (Dose et al., 2014).
To sum up, our present study provided evidence that BPA genotoxicities to p53 wild-type TK6 cells took place with CTNNB1-mediated DNA damage and G2M arrest. Downregulation of p53 and CDKN1A genes following CTNNB1 activation by BPA enhanced potent clonogenic survival in DNA-damaged TK6 cells. BPA-activated CTNNB1-NFKB1-AR-IGF1-TWIST1 pathway might be involved in proliferation of TK6 cells selected by BPA-induced DNA damage.
Limitations of the study
This research only focused on cell line testing. If we can verify in animal experiments and compare with actual patient samples, the results will increase clinical application value.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Chemicals, peptides, and recombinant proteins | ||
| BPA (bis-(4-hydroxyphenyl)-propane | Sigma-Aldrich, Whitehouse Station, NJ | Cat# 239658 |
| DMSO (dimethyl sulfoxide) | Sigma-Aldrich | Cat# D4540 |
| Horse serum (HS) | Gibco, Grand Island, NY | Cat# 16050-122 |
| Penicillin-streptomycin | Corning, Blacksburg, VA | Cat# 30-002-CI |
| RNA TRIzol | Invitrogen, Carlsbad, CA | Cat# 1559601 |
| Chloroform | Sigma-Aldrich | Cat# 67-66-3 |
| Isopropanol | Sigma-Aldrich | Cat# RD-34863 |
| SYBR Green PCR master mix | Applied Biosystems, Foster City, CA | Cat# 4368706 |
| Propidium iodide | Invitrogen | P3566 |
| Low-melting agarose | Amresco, Solon, OH | Cat# 9012-36-6 |
| RPMI-1640 medium | Gibco, Rockville, MD | Cat# 31800-022 |
| Lipofectamine | Invitrogen | N/A |
| Critical commercial assays | ||
| High-capacity cDNA reverse transcription kit | Applied Biosystems | Cat# 4368814 |
| Sybr green master mix | Applied Biosystems | Cat# 4368706 |
| Comet assay kit | Trevigen Inc, Gaithersburg, MD | Cat# 4250-050-K |
| Deposited data | ||
| Microarray data sets of BPA exposure | ArrayExpress | E-GEOD-10270, E-GEOD-17624, E-GEOD-32158, E-GEOD-32160 |
| Microarray data sets of non-Hodgkin lymphoma | ArrayExpress | E-GEOD-65135, E-GEOD-53820, E-GEOD-12195, E-GEOD-10524, E-GEOD-66384, E-GEOD-55267 |
| Experimental models: Cell lines | ||
| Human lymphoblastoid TK6 cell line | ATCC | CRL-8015™ |
| Human lymphoblastoid SUP-B15 cell line | ATCC | CRL-1929™ |
| Oligonucleotides | ||
| Primer sequences of β-actin (sense): 5’-TTGTTACAGGAAGTCCCTTGCC-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of β-actin (anti-sense): 5’-ATGCTATCACCTCCCCTGTGTG-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of CTNNB1 (sense): 5’-ACCAGCCGACACCAAGAAG-3 |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of CTNNB1 (anti-sense): 5’-TCGAATCAATCCAACAGTAGCCT-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of NFKB1 (sense): 5’-CAATCATCCACCTTCATTCTCAAC-3 |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of NFKB1 (anti-sense): 5’-TCCACCACATCTTCCTGCTTA -3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of AR (sense): 5’-TGCCCATACTCACTCAGATTCC-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of AR (anti-sense): 5’-CCACACCAACCAGACCTTTCC-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of IGF1 (sense): 5’-CACCACCTCCCTTCATAACCTT-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of IGF1 (anti-sense) 5’-TCTTGACGACTTGCTGCTGCT-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of TWIST1 (sense): 5’-CGGGAGTCCGCAGTCTTA-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of TWIST1 (anti-sense): 5’-GCTTGAGGGTCTGAATCTTG-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of TP53 (sense): 5’-CCTGAGGTGTAGACGCCAACT-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of TP53 (anti-sense): 5’-GTACTGTAGGAAGAGGAAGGAGAC-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of CDKN1A (sense): 5’-GGGACAGCAGAGGAAGACCAT-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| Primer sequences of CDKN1A (anti-sense): 5’-GGAGTGGTAGAAATCTGTCATGCT-3’ |
Genomics, Xizhi District, Taiwan | N/A |
| CTNNB1 siRNA homo-960: F-5’CCCAAGCUUUAGUAAAUAUTT-3’ R-5’AUA UUUACUAAAGCUUGGGTT-3’ |
Integrated DNA Technologies, Coralville, IA | N/A |
| CTNNB1 siRNA homo-1996: F-5’GCCACAAGAUUACAAGAAATT-3’ R-5’UUUCUUGUAAUCUUGUGGCTT-3’ |
Integrated DNA Technologies, Coralville, IA | N/A |
| Software and algorithms | ||
| ModFit LTTM software | Verity Software House, Topsham, ME | http://www.vsh.com |
| Robust Multi-array Average | Weighted gene correlation network analysis (WGCNA) | N/A |
| Differentially expressed genes (D56EG) | NetworkAnalyst | http://www.networkanalyst.ca/ |
| Gene ontology (GO) | DAVID | http://david.ncifcrf.gov |
| Cytoscape | Cytoscape Consortium | http://cytoscape.org |
| ClueGO | Cytoscape APP | N/A |
| CluePedia | Cytoscape APP | N/A |
| CaspLab | Comet assay software project (CASP) | N/A |
| Other | ||
| Affymetrix GeneChip | Thermo Fisher Scientific, Wilmington, MA | Human Genome U133 Plus 2.0 |
| CometSlideTM | Trevigen, Gaithersburg, MD | N/A |
| Nanodrop 2000c | Thermo Fisher Scientific, Wilmington, MA | N/A |
| G:Box ChemiXT16 system | Syngene, Frederick, MD | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be contacted directly to and will be fulfilled by the Lead Contact, Chun-Yu Chuang (cychuang@mx.nthu.edu.tw).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All bioinformatic data sets used in this manuscript were obtained from publicly accessible ArrayExpress as indicated in the Method details.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request
Experimental model and subject details
A human lymphoblastoid cell line TK6 (ATCC® CRL-8015™) derived from a 5-year-old male with hereditary spherocytosis was cultured with RPMI-1640 medium (31800-022, Gibco, Rockville, MD), 10% horse serum (HS; 16050-122, Gibco, Grand Island, NY), and 1% (v/v) penicillin-streptomycin (30-002-CI; Corning, Blacksburg, VA), in a 5% CO2 incubator at 37°C. A human B lymphoblast cell line SUP-B15 (ATCC® CRL-1929™) derived from a 8-year-old male with acute lymphoblastic leukemia was cultured with Iscove's modified Dulbecco's medium (IMDM; Gibco™), supplemented with 20% fetal bovine serum, 0.05 μM 2-β-mercaptoethanol, and 1% (v/v) penicillin-streptomycin, in a 5% CO2 incubator at 37°C. BPA (bis-(4-hydroxyphenyl)-propane; purity > 99%, 239658, Sigma-Aldrich, Whitehouse Station, NJ) was dissolved in dimethyl sulfoxide (DMSO; D4540, Sigma-Aldrich) to prepare a stock solution (100 mM) and diluted into different concentrations (10, 103, and 105 nM) by the culture medium. TK6 cells were treated with BPA at 10, 102, 103, 104, and 105 nM for 24, 48, and 72 h. SUP-B15 cells were treated with BPA at 10 and 103 nM for 24, 48, and 72 h, respectively.
Method details
Collection and normalization of microarray data sets
Microarray data sets of NHL collected in this study were systematically searched with the terms “Homo sapiens” and “non-Hodgkin lymphoma” from Jan 1, 2011 to Aug 31, 2015 on the ArrayExpress website. Six data sets containing raw data files of 309 samples were ultimately used in the present study (E-GEOD-65135; E-GEOD-53820; E-GEOD-12195; E-GEOD-10524; E-GEOD-66384; E-GEOD-55267). On the other hand, microarray data sets of BPA exposure were collected on the ArrayExpress website using the keyword “Homo sapiens” and “bisphenol A” from Jan 1, 2011 to Aug 31, 2015. Four data sets containing raw data files of 98 samples were ultimately used in the present study (E-GEOD-10270; E-GEOD-17624; E-GEOD-32158; E-GEOD-32160). All of these recruited samples were accessed in Affymetrix GeneChip (Human Genome U133 Plus 2.0) platform and normalized by processing background correction, expression estimation, and summarization using Robust Multi-array Average (RMA) methods to produce log2 expression measurements in Affy package.
Identification of differentially expressed genes
We merged microarray data sets using R software after preprocessing and normalizing gene expression data from different microarrays. The third quarter of standard deviation (SD) was used to filter differentially expressed genes (DEGs) between NHL versus normal of microarray data sets and BPA exposure versus nonexposure of microarray data sets. The filtering 4,593 DEGs of NHL (fold change ≥ 1.3) and 4,028 DEGs of BPA exposure (fold change ≥ 1.2) were imported into WGCNA for constructing gene modules. The consensus modules for NHL (NHL vs. normal) and BPA exposure (BPA exposure vs. nonexposure) were identified to further analyze the relationship among these modules. For each consensus module (NHL and BPA exposure), the exploration first created correlation matrices from DEGs (obtained by calculating the Pearson correlation coefficients (r) between all variable gene sets across all subjects in processed data sets) and then weighted DEGs based on the number of samples used in these data sets. The modules formed from the weighted correlation matrices were conducted using the WGCNA package in the R environment (data not shown). In the NHL group, the two modules (blue: r = 0.52, p = 0.03; turquoise: r = 0.49, p = 0.03) containing 699 genes revealed the higher correlation with cancer traits. In the BPA group, the turquoise (r = 0.36, p = 0.05) module containing 400 genes presented a representatively higher correlation.
Construction of gene network for identifying gene ontology and regulatory pathway
The module genes of NHL and BPA exposure outputted from R analysis were analyzed by Cytoscape to construct two individual gene subnetworks of NHL and BPA exposure, respectively, and plugged-in ClueGO and CluePedia to figure out potential biological functions and predict a potential pathway that presented NHL development along with BPA exposure. In these two subnetwork analyses, 699 module genes of NHL were extended to 1152 genes and 400 module genes of BPA exposure were expanded to 839 genes in the network analysis. These two gene subnetworks of NHL and BPA exposure were further merged to reconstruct a union gene-gene regulatory network corresponding to NHL and BPA exposure. Then, a regulatory pathway predicted by the relatively higher connectivity genes in the above merged network represented the potential progression of NHL in response to BPA exposure.
Total RNA extraction
The total RNA of TK6 cells was isolated using RNA TRIzol (15596018; Invitrogen, Carlsbad, CA). After the culture medium was removed, cells were dissolved in TRIzol reagent and mixed with chloroform (67-66-3; Sigma-Aldrich). The mixture was shaken vigorously and centrifuged at 12,000 g for 15 min at 4°C. The layer of RNA was in the upper supernatant and transferred to a fresh eppendorf tube. The supernatant was added to isopropanol (RD-34863, Sigma-Aldrich) for 10 min at room temperature. The RNA pellet was precipitated after centrifugation at 12,000 g for 10 min at 4°C and washed by 75% ethanol with centrifugation at 7,500 g for 5 min at 4°C to remove ethanol. The RNA pellet was dried up and dissolved in 30 μL of RNase-free water. The quantity and quality of purified RNA was measured using Nanodrop 2000c (Thermo, Wilmington, MA).
Reverse transcription polymerase chain reaction and quantitative real-time polymerase chain reaction for mRNA determination
cDNA was synthesized from total RNA by a high-capacity cDNA reverse transcription kit (4368814, Applied Biosystems, Foster City, CA). In brief, 2 μg of RNA was added into MultiScribeTM reverse transcriptase, 10X RT random primers, 25X dNTP mix, 10X RT buffer, and RNase-free water in a PCR tube and subsequently amplified using ABI 2720 thermal cycler (Applied Biosystems).
A quantitative real-time polymerase chain reaction (PCR) was performed to quantify the message expression pattern of synthesized cDNA products. cDNA was amplified by PCR using 2X power SYBR Green PCR master mix (4368706, Applied Biosystems), 10 μM forward and reverse primers, and RNase-free water. Quantitative determination of PCR products was carried out by ABI 7300 Sequence Detection System (Applied Biosystems) according to the manufacturer’s instruction.
Cell cycle analysis
TK6 cells were treated with 10, 103, and 105 nM concentrations of BPA for 72 h. All harvested cells were washed with ice-cold phosphate-buffered saline (PBS) and then fixed with ice-cold 70% ethanol overnight at -20°C. The cells were then stained with propidium iodide (PI; P3566; Invitrogen) solution containing a final concentration of 200 μg/mL RNase A and 20 μg/mL PI. After incubation at 37°C for 30 min in the dark, the cell cycle profiles were determined using the PE channel of a flow cytometer (Becton Dickinson BD FACSCanto, San Jose, CA) and analyzed using ModFit LTTM software (Verity Software House, Topsham, ME).
Comet assay
A comet assay kit was purchased from Trevigen Inc. (4250-050-K; Gaithersburg, MD) to determine the phenomenon of DNA damage. TK6 cells treated with BPA (10, 103, and 105 nM) for 72 h were detected for DNA damage at the level of single cells under neutral (DSBs) or alkaline (a mixture of DNA SSBs, DSBs, and alkali-labile sites) conditions. After 72 h of BPA exposure, TK6 cells were harvested and mixed with a low-melting-temperature agarose (LMA) at a ratio of 1:10 (v/v) onto TrevCometSlideTM and kept for 10 min at 4°C. The slides were then immersed in a 4°C lysis solution for at least 1 h. After removing from the lysis buffer, the slides were gently immersed in 50 mL of 4°C 1X neutral electrophoresis buffer for neutral comet assay or 4°C 1X alkaline unwinding solution for alkaline comet assay for 30 min. Electrophoresis was performed at 25 V for 30 min in the dark. After electrophoresis, the slides were immersed in a DNA precipitation solution for 30 min and quickly immersed in 70% ethanol for 30 min at room temperature. Then, the slides were then allowed to dry before being stained with SYBR Gold dye for 30 min in a dark refrigerator. Ten random fields per slide were photographed under fluorescence microscope (Ti-TIRF-E, Nikon, Tokyo, Japan), and each experimental condition group was carried out in triplicates for three time-independent experiments. Tail DNA% of 60 cells at least was analyzed using the comet assay software project (CASP). Tail DNA% was calculated following the equation: Tail DNA% = 100 × Tail DNA intensity/Cell DNA intensity.
Colony formation assay
TK6 cells were resuspended in 1.5 mL of 0.3% low-melting agarose (9012-36-6; Amresco, Solon, OH) containing 2X RPMI 1640 and 2X HS and plated on top of 1.5 mL bottom layer of 0.5% low-melting agarose in RPMI 1640. The cells in a 6-well plate were fed with 200 μL of medium per well twice a week, and the colonies were counted after 17 days of growth. The colonies were fixed with absolute methanol and stained with 10% aqueous Giemsa solution for 30 min. Colony formation images of each well were photographed under a G:Box ChemiXT16 system (Syngene, Frederick, MD), and the number of colonies was counted.
Transfection of small interfering RNA
The CTNNB1 siRNA was transfected into TK6 and SUP-B15 cells using lipofectamine transfection reagent (Invitrogen) in serum-free medium. The CTNNB1 siRNA is obtained from Integrated DNA Technologies (Coralville, IA; homo-960: F 5’CCCAAGCUUUAGUAAAUAUTT3’, R 5’AUA UUUACUAAAGCUUGGGTT3’; homo-1996: F 5’GCCACAAGAUUACAAGAAATT3’, R 5’UUUCUUGUAAUCUUGUGGCTT3’). The siRNAs at 10, 20, and 30 nM were diluted in 150 μL of RPMI-1640 medium without HS at 25°C for 5 min. The 5 μL of lipofectamine was added into 150 μL of RPMI-1640 medium without HS at 25°C for 5 min. The siRNA-lipofectamine transfection complex was formed by mixing the siRNA and lipofectamine and incubated at 25°C for 5 min. TK6 cells were cultured in RPMI-1640 medium with the transfection complex at 37°C in a 5% CO2 incubator for 24 h. Twenty-four hours after transfection, the transfection medium was changed with fresh culture medium supplemented with 10% HS. Then, the transfected cells were maintained for an additional 8 h. Total RNA was extracted from siRNA-transfected cells to determine mRNAs expression using quantitative real-time PCR (q-PCR).
Quantification and statistical analysis
Results were described as mean ± SD. The student t-test was used to analyze the differences of gene expression between BPA exposure groups and control (BPA nonexposure group), and one-way analysis of variance test was used to determine the difference between three or more groups. All statistical significances were determined at two-tailed p-value < 0.05.
Additional resources
This study has not generated or contributed to a new website/forum and is not part of a clinical trial.
Acknowledgments
This study was supported by the grant from the Ministry of Science and Technology of Taiwan (105-2221-E-007-080-MY3; 109-2221-E-007 -042 -MY3).
Author contributions
YKC, MY, and CYC participated and coordinated in the research design. YKC, YYT, and CYC wrote and edited the draft of the manuscript. MHT provided the cell line and instructed cell culture. YKC, YYT, HCL, YJH, and CYC conducted the experiments and performed the data analysis. CMC assisted the microscope experiments. YYL, TTH, and CWC assisted in illustrating the graphical abstract for the concepts and key findings of this research. All authors read and approved the final manuscript.
Declaration of interests
The authors declare that they have no competing interests.
Published: August 20, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.102888.
Supplemental information
References
- Aghajanpour-Mir S.M., Zabihi E., Akhavan-Niaki H., Keyhani E., Bagherizadeh I., Biglari S., Behjati F. The genotoxic and cytotoxic effects of bisphenol-A (BPA) in MCF-7 cell line and amniocytes. Int. J. Mol. Cell. Med. 2016;5:19–29. [PMC free article] [PubMed] [Google Scholar]
- Alexander D.D., Mink P.J., Adami H.O., Chang E.T., Cole P., Mandel J.S., Trichopoulos D. The non-Hodgkin lymphomas: a review of the epidemiologic literature. Int. J. Cancer. 2007;120:1–39. doi: 10.1002/ijc.22719. [DOI] [PubMed] [Google Scholar]
- Allard P., Colaiácovo M.P. Bisphenol A impairs the double-strand break repair machinery in the germline and causes chromosome abnormalities. Proc. Natl. Acad. Sci. U S A. 2010;107:20405–20410. doi: 10.1073/pnas.1010386107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beerman I., Seita J., Inlay M.A., Weissman I.L., Rossi D.J. Quiescent hematopoietic stem cells accumulate DNA damage during aging that is repaired upon entry into cell cycle. Cell Stem Cell. 2014;15:37–50. doi: 10.1016/j.stem.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappel M., Mauger D., Thiboutot D. Correlation between serum levels of insulin-like growth factor 1, dehydroepiandrosterone sulfate, and dihydrotestosterone and acne lesion counts in adult women. Arch. Dermatol. 2005;141:333–338. doi: 10.1001/archderm.141.3.333. [DOI] [PubMed] [Google Scholar]
- Chen Y.J., Roumeliotis T.I., Chang Y.H., Chen C.T., Han C.L., Lin M.H., Chen H.W., Chang G.C., Chang Y.L., Wu C.T. Proteogenomics of non-smoking lung cancer in east Asia delineates molecular signatures of pathogenesis and progression. Cell. 2020;182:226–244.e217. doi: 10.1016/j.cell.2020.06.012. [DOI] [PubMed] [Google Scholar]
- Chen Z.J., Yang X.L., Liu H., Wei W., Zhang K.S., Huang H.B., Giesy J.P., Liu H.L., Du J., Wang H.S. Bisphenol A modulates colorectal cancer protein profile and promotes the metastasis via induction of epithelial to mesenchymal transitions. Arch. Toxicol. 2015;89:1371–1381. doi: 10.1007/s00204-014-1301-z. [DOI] [PubMed] [Google Scholar]
- Chiarini F., Paganelli F., Martelli A.B., Evangelisti C. The role played by Wnt/β-catenin signaling pathway in acute lymphoblastic leukemia. Int. J. Mol. Sci. 2020;21:1098. doi: 10.3390/ijms21031098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu B.C., Hou N. Epidemiology and etiology of non-hodgkin lymphoma. Cancer Treat. Res. 2015;165:1–25. doi: 10.1007/978-3-319-13150-4_1. [DOI] [PubMed] [Google Scholar]
- Chow V.A., Martin P.S., Smith S.D., Till B.G., Graf S.A., Edlefsen K.L., Hannan L.M., Mostaghel E.A., Gopal A.K. Addressing the conundrum of male predominance in mantle cell lymphoma using androgen receptor blockade. Br. J. Haematol. 2020;190:e332–e335. doi: 10.1111/bjh.16902. [DOI] [PubMed] [Google Scholar]
- Clayton E.M., Todd M., Dowd J.B., Aiello A.E. The impact of bisphenol A and triclosan on immune parameters in the U.S. population, NHANES 2003-2006. Environ. Health Perspect. 2011;119:390–396. doi: 10.1289/ehp.1002883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compagno M., Lim W.K., Grunn A., Nandula S.V., Brahmachary M., Shen Q., Bertoni F., Ponzoni M., Scandurra M., Califano A. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.E., Brown K.D., Siebenlist U., Staudt L.M. Constitutive nuclear factor kappaB activity is required for survival of activated B cell-like diffuse large B cell lymphoma cells. J. Exp. Med. 2001;194:1861–1874. doi: 10.1084/jem.194.12.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derenzini E., Agostinelli C., Imbrogno E., Iacobucci I., Casadei B., Brighenti E., Righi S., Fuligni F., Ghelli Luserna Di Rorà A., Ferrari A. Constitutive activation of the DNA damage response pathway as a novel therapeutic target in diffuse large B-cell lymphoma. Oncotarget. 2015;6:6553–6569. doi: 10.18632/oncotarget.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dose M., Emmanuel A.O., Chaumeil J., Zhang J., Sun T., Germar K., Aghajani K., Davis E.M., Keerthivasan S., Bredemeyer A.L. beta-Catenin induces T-cell transformation by promoting genomic instability. Proc. Natl. Acad. Sci. U S A. 2014;111:391–396. doi: 10.1073/pnas.1315752111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont J., Fernandez A.M., Glackin C.A., Helman L., LeRoith D. Insulin-like growth factor 1 (IGF-1)-induced twist expression is involved in the anti-apoptotic effects of the IGF-1 receptor. J. Biol. Chem. 2001;276:26699–26707. doi: 10.1074/jbc.M102664200. [DOI] [PubMed] [Google Scholar]
- Fang C., Ning B., Waqar A.B., Niimi M., Li S., Satoh K., Shiomi M., Ye T., Dong S., Fan J. Bisphenol A exposure enhances atherosclerosis in WHHL rabbits. PLoS One. 2014;9:e110977. doi: 10.1371/journal.pone.0110977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Lv X., Feng L., Liu X., Wang X. High expression and nuclear localization of beta-catenin in diffuse large B-cell lymphoma. Mol. Med. Rep. 2012;5:1433–1437. doi: 10.3892/mmr.2012.835. [DOI] [PubMed] [Google Scholar]
- George V.C., Rupasinghe H.P.V. DNA damaging and apoptotic potentials of Bisphenol A and Bisphenol S in human bronchial epithelial cells. Environ. Toxicol. Pharmacol. 2018;60:52–57. doi: 10.1016/j.etap.2018.04.009. [DOI] [PubMed] [Google Scholar]
- Gore A.C., Chappell V.A., Fenton S.E., Flaws J.A., Nadal A., Prins G.S., Toppari J., Zoeller R.T. EDC-2: the endocrine society's second scientific statement on endocrine-disrupting chemicals. Endocr. Rev. 2015;36 doi: 10.1210/er.2015-1010. E1–e150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grulich A.E., Vajdic C.M., Cozen W. Altered immunity as a risk factor for non-Hodgkin lymphoma. Cancer Epidemiol. Biomarkers Prev. 2007;16:405–408. doi: 10.1158/1055-9965.EPI-06-1070. [DOI] [PubMed] [Google Scholar]
- Hamoudi R.A., Appert A., Ye H., Ruskone-Fourmestraux A., Streubel B., Chott A., Raderer M., Gong L., Wlodarska I., De Wolf-Peeters C. Differential expression of NF-kappaB target genes in MALT lymphoma with and without chromosome translocation: insights into molecular mechanism. Leukemia. 2010;24:1487–1497. doi: 10.1038/leu.2010.118. [DOI] [PubMed] [Google Scholar]
- Huang B., Ning S., Zhang Q., Chen A., Jiang C., Cui Y., Hu J., Li H., Fan G., Qin L. Bisphenol A represses dopaminergic neuron differentiation from human embryonic stem cells through downregulating the expression of insulin-like growth factor 1. Mol. Neurobiol. 2017;54:3798–3812. doi: 10.1007/s12035-016-9898-y. [DOI] [PubMed] [Google Scholar]
- Huang D.Y., Zheng C.C., Pan Q., Wu S.S., Su X., Li L., Wu J.H., Sun Z.Y. Oral exposure of low-dose bisphenol A promotes proliferation of dorsolateral prostate and induces epithelial-mesenchymal transition in aged rats. Sci. Rep. 2018;8:490. doi: 10.1038/s41598-017-18869-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.P., Liu Z.H., Yuan S.F., Yin H., Dang Z., Wu P.X. Worldwide human daily intakes of bisphenol A (BPA) estimated from global urinary concentration data (2000-2016) and its risk analysis. Environ. Pollut. 2017;230:143–152. doi: 10.1016/j.envpol.2017.06.026. [DOI] [PubMed] [Google Scholar]
- Hui L., Li H., Lu G., Chen Z., Sun W., Shi Y., Fu Z., Huang B., Zhu X., Lu W. Low dose of bisphenol A modulates ovarian cancer gene expression profile and promotes epithelial to mesenchymal transition via canonical Wnt pathway. Toxicol. Sci. 2018;164:527–538. doi: 10.1093/toxsci/kfy107. [DOI] [PubMed] [Google Scholar]
- Iarovaia O.V., Rubtsov M., Ioudinkova E., Tsfasman T., Razin S.V., Vassetzky Y.S. Dynamics of double strand breaks and chromosomal translocations. Mol. Cancer. 2014;13:249. doi: 10.1186/1476-4598-13-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islami F., Goding Sauer A., Miller K.D., Siegel R.L., Fedewa S.A., Jacobs E.J., McCullough M.L., Patel A.V., Ma J., Soerjomataram I. Proportion and number of cancer cases and deaths attributable to potentially modifiable risk factors in the United States. CA Cancer J. Clin. 2018;68:31–54. doi: 10.3322/caac.21440. [DOI] [PubMed] [Google Scholar]
- Jamieson C.H., Ailles L.E., Dylla S.J., Muijtjens M., Jones C., Zehnder J.L., Gotlib J., Li K., Manz M.G., Keating A. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N. Engl. J. Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- Jiang L., Li N. B-cell non-Hodgkin lymphoma: importance of angiogenesis and antiangiogenic therapy. Angiogenesis. 2020;23:515–529. doi: 10.1007/s10456-020-09729-7. [DOI] [PubMed] [Google Scholar]
- Karimian A., Ahmadi Y., Yousefi B. Multiple functions of p21 in cell cycle, apoptosis and transcriptional regulation after DNA damage. DNA Repair (Amst) 2016;42:63–71. doi: 10.1016/j.dnarep.2016.04.008. [DOI] [PubMed] [Google Scholar]
- Karimaian A., Majidinia M., Baghi H.B., Yousefi B. The crosstalk between Wnt/β-catenin signaling pathway with DNA damage response and oxidative stress: implications in cancer therapy. DNA Repair. 2017;51:14–19. doi: 10.1016/j.dnarep.2017.01.003. [DOI] [PubMed] [Google Scholar]
- Konieczna A., Rutkowska A., Rachoń D. Health risk of exposure to Bisphenol A (BPA) Rocz. Panstw. Zakl. Hig. 2015;66:5–11. [PubMed] [Google Scholar]
- Lazarian G., Friedrich C., Quinquenel A., Tran J., Ouriemmi S., Dondi E., Martin A., Mihoub I., Chiron D., Bellanger C. Stabilization of beta-catenin upon B-cell receptor signaling promotes NF-kB target genes transcription in mantle cell lymphoma. Oncogene. 2020;39:2934–2947. doi: 10.1038/s41388-020-1183-x. [DOI] [PubMed] [Google Scholar]
- Lee H.J., Chattopadhyay S., Gong E.Y., Ahn R.S., Lee K. Antiandrogenic effects of bisphenol A and nonylphenol on the function of androgen receptor. Toxicol. Sci. 2003;75:40–46. doi: 10.1093/toxsci/kfg150. [DOI] [PubMed] [Google Scholar]
- Lee H.R., Hwang K.A., Park M.A., Yi B.R., Jeung E.B., Choi K.C. Treatment with bisphenol A and methoxychlor results in the growth of human breast cancer cells and alteration of the expression of cell cycle-related genes, cyclin D1 and p21, via an estrogen receptor-dependent signaling pathway. Int. J. Mol. Med. 2012;29:883–890. doi: 10.3892/ijmm.2012.903. [DOI] [PubMed] [Google Scholar]
- Lemma S., Karihtala P., Haapasaari K.M., Jantunen E., Soini Y., Bloigu R., Pasanen A.K., Turpeenniemi-Hujanen T., Kuittinen O. Biological roles and prognostic values of the epithelial-mesenchymal transition-mediating transcription factors Twist, ZEB1 and Slug in diffuse large B-cell lymphoma. Histopathology. 2013;62:326–333. doi: 10.1111/his.12000. [DOI] [PubMed] [Google Scholar]
- Liu W., Li L., Ye H., Tu W. Weighted gene co-expression network analysis in biomedicine research. Sheng Wu Gong Cheng Xue Bao. 2017;33:1791–1801. doi: 10.13345/j.cjb.170006. [DOI] [PubMed] [Google Scholar]
- Lorge E., Moore M.M., Clements J., O'Donovan M., Fellows M.D., Honma M., Kohara A., Galloway S., Armstrong M.J., Thybaud V. Standardized cell sources and recommendations for good cell culture practices in genotoxicity testing. Mutat. Res. 2016;809:1–15. doi: 10.1016/j.mrgentox.2016.08.001. [DOI] [PubMed] [Google Scholar]
- Ma X.F., Zhang J., Shuai H.L., Guan B.Z., Luo X., Yan R.L. IKKbeta/NF-kappaB mediated the low doses of bisphenol A induced migration of cervical cancer cells. Arch. Biochem. Biophys. 2015;573:52–58. doi: 10.1016/j.abb.2015.03.010. [DOI] [PubMed] [Google Scholar]
- Mahemuti L., Chen Q., Coughlan M.C., Qiao C., Chepelev N.L., Florian M., Dong D., Woodworth R.G., Yan J., Cao X.L. Bisphenol A induces DSB-ATM-p53 signaling leading to cell cycle arrest, senescence, autophagy, stress response, and estrogen release in human fetal lung fibroblasts. Arch. Toxicol. 2018;92:1453–1469. doi: 10.1007/s00204-017-2150-3. [DOI] [PubMed] [Google Scholar]
- Malinen M., Niskanen E.A., Kaikkonen M.U., Palvimo J.J. Crosstalk between androgen and pro-inflammatory signaling remodels androgen receptor and NF-kappaB cistrome to reprogram the prostate cancer cell transcriptome. Nucleic Acids Res. 2017;45:619–630. doi: 10.1093/nar/gkw855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathur R., Sehgal L., Braun F.K., Berkova Z., Romaguerra J., Wang M., Rodriguez M.A., Fayad L., Neelapu S.S., Samaniego F. Targeting Wnt pathway in mantle cell lymphoma-initiating cells. J. Hematol. Oncol. 2015;8:63. doi: 10.1186/s13045-015-0161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Filho A., Piñeros M., Znaor A., Marcos-Gragera R., Steliarova-Foucher E., Bray F. Global patterns and trends in the incidence of non-Hodgkin lymphoma. Cancer Causes Control. 2019;30:489–499. doi: 10.1007/s10552-019-01155-5. [DOI] [PubMed] [Google Scholar]
- Miraoui H., Marie P.J. Pivotal role of Twist in skeletal biology and pathology. Gene. 2010;468:1–7. doi: 10.1016/j.gene.2010.07.013. [DOI] [PubMed] [Google Scholar]
- Mokra K., Kuźmińska-Surowaniec A., Woźniak K., Michałowicz J. Evaluation of DNA-damaging potential of bisphenol A and its selected analogs in human peripheral blood mononuclear cells (in vitro study) Food Chem. Toxicol. 2017;100:62–69. doi: 10.1016/j.fct.2016.12.003. [DOI] [PubMed] [Google Scholar]
- Moller M.B., Gerdes A.M., Skjodt K., Mortensen L.S., Pedersen N.T. Disrupted p53 function as predictor of treatment failure and poor prognosis in B- and T-cell non-Hodgkin's lymphoma. Clin. Cancer Res. 1999;5:1085–1091. [PubMed] [Google Scholar]
- Morton L.M., Wang S.S., Devesa S.S., Hartge P., Weisenburger D.D., Linet M.S. Lymphoma incidence patterns by WHO subtype in the United States, 1992-2001. Blood. 2006;107:265–276. doi: 10.1182/blood-2005-06-2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostaghel E.A., Martin P.S., Mongovin S., Frayo S., Zhang A., Edlefsen K.L., Press O.W., Gopal A.K. Androgen receptor expression in mantle cell lymphoma: potential novel therapeutic implications. Exp. Hematol. 2017;49:34–38 e32. doi: 10.1016/j.exphem.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulder D.E., Hatoum D., Tay E., Lin Y., McGowan E.M. The roles of p53 in mitochondrial dynamics and cancer metabolism: the pendulum between survival and death in breast cancer? Cancers (Basel) 2018;10:189. doi: 10.3390/cancers10060189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata M., Kang J.H. Bisphenol A (BPA) and cell signaling pathways. Biotechnol. Adv. 2018;36:311–327. doi: 10.1016/j.biotechadv.2017.12.002. [DOI] [PubMed] [Google Scholar]
- Nagel D., Vincendeau M., Eitelhuber A.C., Krappmann D. Mechanisms and consequences of constitutive NF-kappaB activation in B-cell lymphoid malignancies. Oncogene. 2014;33:5655–5665. doi: 10.1038/onc.2013.565. [DOI] [PubMed] [Google Scholar]
- Nakada S., Katsuki Y., Imoto I., Yokoyama T., Nagasawa M., Inazawa J., Mizutani S. Early G2/M checkpoint failure as a molecular mechanism underlying etoposide-induced chromosomal aberrations. J. Clin. Invest. 2006;116:80–89. doi: 10.1172/JCI25716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambiar M., Raghavan S.C. How does DNA break during chromosomal translocations? Nucleic Acids Res. 2011;39:5813–5825. doi: 10.1093/nar/gkr223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff A.M., Blanco S.C., Flaws J.A., Bagchi I.C., Bagchi M.K. Chronic exposure of mice to bisphenol-A alters uterine fibroblast growth factor signaling and leads to aberrant epithelial proliferation. Endocrinology. 2019;160:1234–1246. doi: 10.1210/en.2018-00872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomiri S., Hoshyar R., Ambrosino C., Tyler C.R., Mansouri B. A mini review of bisphenol A (BPA) effects on cancer-related cellular signaling pathways. Environ. Sci. Pollut. Res. Int. 2019;26:8459–8467. doi: 10.1007/s11356-019-04228-9. [DOI] [PubMed] [Google Scholar]
- Ohlstein J.F., Strong A.L., McLachlan J.A., Gimble J.M., Burow M.E., Bunnell B.A. Bisphenol A enhances adipogenic differentiation of human adipose stromal/stem cells. J. Mol. Endocrinol. 2014;53:345–353. doi: 10.1530/JME-14-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazgal I., Zimra Y., Tzabar C., Okon E., Rabizadeh E., Shaklai M., Bairey O. Expression of basic fibroblast growth factor is associated with poor outcome in non-Hodgkin's lymphoma. Br. J. Cancer. 2002;86:1770–1775. doi: 10.1038/sj.bjc.6600330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelch K.E., Li Y., Perera L., Thayer K.A., Korach K.S. Characterization of estrogenic and androgenic activities for bisphenol A-like chemicals (BPs): in vitro estrogen and androgen receptors transcriptional activation, gene regulation, and binding profiles. Toxicol. Sci. 2019;172:23–37. doi: 10.1093/toxsci/kfz173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer D., Chung Y.M., Hu M.C. Effects of low-dose bisphenol A on DNA damage and proliferation of breast cells: the role of c-myc. Environ. Health Perspect. 2015;123:1271–1279. doi: 10.1289/ehp.1409199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp H.D., Flach J., Brendel S., Ruppenthal S., Kleiner H., Seifarth W., Schneider S., Schulze T.J., Weiss C., Wenz F. Accumulation of DNA damage and alteration of the DNA damage response in monoclonal B-cell lymphocytosis and chronic lymphocytic leukemia. Leuk. Lymphoma. 2019;60:795–804. doi: 10.1080/10428194.2018.1498494. [DOI] [PubMed] [Google Scholar]
- Ptak A., Hoffmann M., Gruca I., Barć J. Bisphenol A induce ovarian cancer cell migration via the MAPK and PI3K/Akt signalling pathways. Toxicol. Lett. 2014;229:357–365. doi: 10.1016/j.toxlet.2014.07.001. [DOI] [PubMed] [Google Scholar]
- Purdue M.P., Lan Q., Bagni R., Hocking W.G., Baris D., Reding D.J., Rothman N. Prediagnostic serum levels of cytokines and other immune markers and risk of non-hodgkin lymphoma. Cancer Res. 2011;71:4898–4907. doi: 10.1158/0008-5472.CAN-11-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rübe C.E., Fricke A., Widmann T.A., Fürst T., Madry H., Pfreundschuh M., Rübe C. Accumulation of DNA damage in hematopoietic stem and progenitor cells during human aging. PLoS One. 2011;6:e17487. doi: 10.1371/journal.pone.0017487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos C., Ladeira C., Zeferino S., Dias A., Faria I., Cristovam E., Gomes M., Ribeiro E. Cytotoxic and genotoxic effects of environmental relevant concentrations of bisphenol A and interactions with doxorubicin. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2019;838:28–36. doi: 10.1016/j.mrgentox.2018.11.009. [DOI] [PubMed] [Google Scholar]
- Sadot E., Geiger B., Oren M., Ben-Ze'ev A. Down-regulation of beta-catenin by activated p53. Mol. Cell. Biol. 2001;21:6768–6781. doi: 10.1128/MCB.21.20.6768-6781.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seachrist D.D., Bonk K.W., Ho S.M., Prins G.S., Soto A.M., Keri R.A. A review of the carcinogenic potential of bisphenol A. Reprod. Toxicol. 2016;59:167–182. doi: 10.1016/j.reprotox.2015.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H., Zhang T., Yang P., Li M., Yang Y., Wang Y., Du J., Pan K., Zhang K. Low doses of bisphenol A stimulate the proliferation of breast cancer cells via ERK1/2/ERRgamma signals. Toxicol. Vitro. 2015;30:521–528. doi: 10.1016/j.tiv.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Stein C.J., Colditz G.A. Modifiable risk factors for cancer. Br. J. Cancer. 2004;90:299–303. doi: 10.1038/sj.bjc.6601509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stromberg T., Feng X., Delforoush M., Berglund M., Lin Y., Axelson M., Larsson O., Georgii-Hemming P., Lennartsson J., Enblad G. Picropodophyllin inhibits proliferation and survival of diffuse large B-cell lymphoma cells. Med. Oncol. 2015;32:188. doi: 10.1007/s12032-015-0630-y. [DOI] [PubMed] [Google Scholar]
- Sui Y., Park S.H., Helsley R.N., Sunkara M., Gonzalez F.J., Morris A.J., Zhou C. Bisphenol A increases atherosclerosis in pregnane X receptor-humanized ApoE deficient mice. J. Am. Heart Assoc. 2014;3:e000492. doi: 10.1161/JAHA.113.000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng C., Goodwin B., Shockley K., Xia M., Huang R., Norris J., Merrick B.A., Jetten A.M., Austin C.P., Tice R.R. Bisphenol A affects androgen receptor function via multiple mechanisms. Chem. Biol. Interact. 2013;203:556–564. doi: 10.1016/j.cbi.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusca V.G., Dumitrescu M., Fenyo I.M., Tudorache I.F., Simionescu M., Gafencu A.V. The mechanism of bisphenol A atherogenicity involves apolipoprotein A-I downregulation through NF-kappaB activation. Int. J. Mol. Sci. 2019;20:6281. doi: 10.3390/ijms20246281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L.J., Grabow S., Vandenberg C.J., Strasser A., Janic A. Combined loss of PUMA and p21 accelerates c-MYC-driven lymphoma development considerably less than loss of one allele of p53. Oncogene. 2016;35:3866–3871. doi: 10.1038/onc.2015.457. [DOI] [PubMed] [Google Scholar]
- Vandenberg L.N., Hauser R., Marcus M., Olea N., Welshons W.V. Human exposure to bisphenol A (BPA) Reprod. Toxicol. 2007;24:139–177. doi: 10.1016/j.reprotox.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Vandenberg L.N., Chahoud I., Heindel J.J., Padmanabhan V., Paumgartten F.J., Schoenfelder G. Urinary, circulating, and tissue biomonitoring studies indicate widespread exposure to bisphenol A. Environ. Health Perspect. 2010;118:1055–1070. doi: 10.1289/ehp.0901716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega F., Medeiros L.J. Chromosomal translocations involved in non-Hodgkin lymphomas. Arch. Pathol. Lab Med. 2003;127:1148–1160. doi: 10.5858/2003-127-1148-CTIINL. [DOI] [PubMed] [Google Scholar]
- Williams A.B., Schumacher B. p53 in the DNA-Damage-Repair Process. Cold Spring Harb. Perspect. Med. 2016;6:a026070. doi: 10.1101/cshperspect.a026070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerzberger-Davis S.M., Chang P.Y., Berchtold C., Miyamoto S. Enhanced G2-M arrest by nuclear factor-{kappa}B-dependent p21waf1/cip1 induction. Mol. Cancer Res. 2005;3:345–353. doi: 10.1158/1541-7786.MCR-05-0028. [DOI] [PubMed] [Google Scholar]
- Xu J., Chen Y., Huo D., Khramtsov A., Khramtsova G., Zhang C., Goss K.H., Olopade O.I. beta-catenin regulates c-Myc and CDKN1A expression in breast cancer cells. Mol. Carcinog. 2016;55:431–439. doi: 10.1002/mc.22292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Liu X., Ouyang J., Chen B. TP53 mutation predicts the poor prognosis of non-Hodgkin lymphomas: evidence from a meta-analysis. PLoS One. 2017;12:e0174809. doi: 10.1371/journal.pone.0174809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Yu H., Shen Y., Liu Y., Yang Z., Sun T. MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNA-p21/β-catenin pathway. Oncotarget. 2016;7:41505–41526. doi: 10.18632/oncotarget.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W. Bisphenol A triggers the malignancy of nasopharyngeal carcinoma cells via activation of Wnt/beta-catenin pathway. Toxicol. Vitro. 2020;66:104881. doi: 10.1016/j.tiv.2020.104881. [DOI] [PubMed] [Google Scholar]
- Zhang D.Y., Wang H.J., Tan Y.Z. Wnt/β-catenin signaling induces the aging of mesenchymal stem cells through the DNA damage response and the p53/p21 pathway. PLoS One. 2011;6:e21397. doi: 10.1371/journal.pone.0021397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K.S., Chen H.Q., Chen Y.S., Qiu K.F., Zheng X.B., Li G.C., Yang H.D., Wen C.J. Bisphenol A stimulates human lung cancer cell migration via upregulation of matrix metalloproteinases by GPER/EGFR/ERK1/2 signal pathway. Biomed. Pharmacother. 2014;68:1037–1043. doi: 10.1016/j.biopha.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Zhang L., Altuwaijri S., Deng F., Chen L., Lal P., Bhanot U.K., Korets R., Wenske S., Lilja H.G., Chang C. NF-kappaB regulates androgen receptor expression and prostate cancer growth. Am. J. Pathol. 2009;175:489–499. doi: 10.2353/ajpath.2009.080727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C., Blum J., Chen A., Kwon H.Y., Jung S.H., Cook J.M., Lagoo A., Reya T. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell. 2007;12:528–541. doi: 10.1016/j.ccr.2007.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Fang X., Jiang Y., Geng L., Li X., Li Y., Lu K., Li P., Lv X., Wang X. Klotho, an anti-aging gene, acts as a tumor suppressor and inhibitor of IGF-1R signaling in diffuse large B cell lymphoma. J. Hematol. Oncol. 2017;10:37. doi: 10.1186/s13045-017-0391-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All bioinformatic data sets used in this manuscript were obtained from publicly accessible ArrayExpress as indicated in the Method details.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request