Abstract
Injured and orphaned wildlife are often brought to Wildlife Rehabilitation Centers (WRC) to be cared for by professionals to ultimately be released back to their natural habitats. In these centers, animals may spend months and frequently receive prolonged antibiotic therapy. Therefore, WRC may play a role in the emergence and dissemination of antimicrobial resistance (AMR). The goal of this study was to investigate the presence and antibiotic resistance profiles of Gram-negative bacteria with reduced susceptibility to cephalosporins in both the wildlife admitted to a WRC and in the WRC built environment in Chile. A cross-sectional study was conducted sampling animals undergoing rehabilitation (n = 64) and the WRC environment (n = 160). Isolated bacterial species were identified with MALDI-TOF, and antimicrobial susceptibility determined using the disk diffusion method. Enterobacteriaceae and Pseudomonadaceae were the dominant bacterial families among the environmental (n = 78) and animal (n = 31) isolates. For Enterobacteriaceae, isolates of the most abundant species (E. coli) were classified into 20 antibiotic resistance profiles, with eight of those isolates being resistant to more than nine antibiotics, including imipenem. Isolates of the Pseudomonadaceae family identified 11 isolates with resistance to antibiotics such as carbapenems and quinolones. Even though a cluster analysis based on antibiotic resistance patterns did not show a clear overlap between environmental and animal isolates, it is important to highlight the identification of isolates resistant to carbapenems, which is very relevant from a public health perspective. Further, numerous antibiotic resistance profiles were observed in different bacterial species, indicating not only environmental contamination with a wide diversity of bacteria, but also a wide diversity of resistant bacteria in animals at the WRC. The approach taken by sampling animals and their hospital environment can be useful in understanding AMR dynamics in wildlife rehabilitation settings, as well as the potential dissemination of AMR into the natural environment.
Keywords: Wildlife, Antimicrobial resistance, Antibiotic, Cluster analysis, Latin America, Chile
Highlights
-
•
The wildlife center was contaminated with wide diversity of resistant bacteria.
-
•
There was wide diversity of resistant bacteria in wildlife at the center.
-
•
Resistant isolates to carbapenems were present, which has public health relevance.
-
•
No clear overlap between wildlife and the center antibiotic resistance patterns.
-
•
Wildlife rehabilitation should be considered in antimicrobial resistance dynamics.
1. Introduction
The interconnectedness between humans, animals, and the natural environment (otherwise known as One Health) is key in understanding and mitigating antimicrobial resistance (AMR) given that resistant bacteria and resistance genes have the ability to move between these three compartments [[1], [2], [3]]. Of these three compartments, the role of the natural environment (e.g., soil, water, air, and wildlife) in the ecology and dissemination of AMR has received increased attention and has been reviewed in several recent publications [1,[4], [5], [6]]. Waste from anthropogenic sources, such as hospitals, wastewater treatment plants, pharmaceutical industries, and agricultural activities are ultimately released into natural environments. This waste may contain antibiotics, their metabolites, antibiotic resistant bacteria, and resistance genes. Thus, the natural environment may act as a reservoir and as a pathway of AMR spread to humans, animals, and the natural ecosystem [4,7].
AMR is a phenomenon that has existed for eons, well before the ‘antibiotics era’. This has been shown in studies where antibiotic resistant bacteria and/or antibiotic resistance genes usually found in clinical settings have been detected in areas far-removed from human contact [8,9]. Despite it being a natural phenomenon, anthropogenic pressures, such as human wastewater systems or animal husbandry facilities, may increase the occurrence, diversity, and quantity of antibiotic resistant bacteria and genes in the environment [10,11].
Wildlife species are part of the natural environmental compartment and can also naturally harbor antibiotic resistant bacteria. However, selective and anthropogenic pressures may also increase the potential for free-ranging wildlife to carry emerging resistant bacteria and genes, as well as facilitate their dissemination [[12], [13], [14]]. Injured and orphaned wildlife are often brought to wildlife rehabilitation centers (WRC) so that they can be cared for by professionals to ultimately be released back to their natural habitats. In these centers, animals may spend months and frequently receive prolonged antibiotic therapy [15]. There are studies that have reported the presence of antibiotic resistant bacteria and resistance genes, including those of public health concern, in wild animals admitted to WRC in different parts of the world. Giacopello et al. (2016) found multi-drug resistant Enterobacteriaceae (resistant to three or more antibiotics) in wild birds admitted to a rehabilitation center in Italy [16]. In another study, Darwich et al. (2019) detected bacterial isolates resistant to fluoroquinolones, tetracyclines and aminoglycosides (among others), and cephalosporin resistant genes from wildlife admitted to a rehabilitation center in Spain [17]. Within Chile, antibiotic resistant bacteria and genes, including those of public health relevance, have been found in wildlife admitted to WRC. Specifically, extended spectrum beta-lactamases (ESBL)-producing Escherichia coli and Salmonella enterica serovar Infantis were found in wild owls [13], and mecA and blaCTX-M genes were found in Andean foxes (Lycalopex culpaeus). These studies however sampled only the animals but not the hospital environment where they were housed. These studies illustrate the importance of not only evaluating the role of free-ranging wildlife but also the role that WRC have in the epidemiology of AMR emergence and spread, especially as the number of injured wild animals continues to increase due to a growing number of human-wildlife interactions [18,19].
The goal of this study was to investigate antibiotic resistance profiles of Gram-negative bacteria with reduced susceptibility to cephalosporins in both the wildlife species admitted to a WRC in Chile and in the WRC hospital built environment [20,21]. We hypothesized that Gram negative antibiotic resistant bacteria are widespread in the WRC built environment and that antibiotic resistance profiles recovered from animals hospitalized at the center would be similar to those observed in the WRC built environment.
2. Materials and methods
2.1. Sampling design
A cross-sectional study was conducted at the Wildlife Rehabilitation Center at the Universidad Andrés Bello (UFAS), located in the city of Santiago, Metropolitan Region of Chile. The center receives an average of 600 animals per year of different species of mammals, birds, reptiles, and amphibians. The main causes of admission to the WRC are wildlife attacked by domestic carnivores, vehicle collisions, illegal hunting, illegal wildlife trafficking and/or possession, and intoxication. Animals are mostly received from the Metropolitan Region of Chile, but a smaller number of them are admitted from other regions of the country as well.
The WRC is comprised of the following sectors (and subdivisions): reception, kitchen, quarantine, exam room, hospital (hospital 1 and hospital 2), indoor (indoor 1, 2, and 3), outdoor (outdoor 1, 2, and 3), and soft release (small animal enclosure, semi-aquatic bird enclosure, small bird enclosure, carnivore enclosure, flight room, owl enclosure, and parrot aviary). The specific number and type of samples taken per sector and subdivision can be found in Table 1. In total, 160 samples at the WRC environment were collected with a gauze previously enriched in peptonized water in 100 mL sterile containers and passed through a 30 cm2 sampled surface.
Table 1.
Total number of environmental samples (n = 160) and number of ceph-resistant isolates that were taken from each sector, subdivision, and equipment (when applicable) at the wildlife rehabilitation center. The numbers represent the sample size and the percentage (%) from the total.
| Sector | Number of samples (%) a |
Number of ceph-resistant isolates collected | Number of samples from subdivision(s) (%) b |
Location or Equipment where samples were obtained |
|---|---|---|---|---|
| Reception | 10 (6.2%) | 1 | NA | Wall, Floor, Computer, Remote control, Telephone, Knob, Light switch, and Table |
| Kitchen | 11 (6.9%) | 5 | NA | Wall, Floor, Remote control, Knob, Light switch, Table, and Microwave |
| Quarantine | 38 (23.8%) | 23 | NA | Wall, Floor, Remote control, Light switch, Table, Stethoscope, and Handling gloves |
| Exam room | 9 (5.6%) | 1 | NA | Wall, Floor, Remote control Knob, Light switch, Table, Stethoscope, Handling gloves, and Anesthesia machine |
| Hospital | 52 (32.5%) | 28 | Hospital 1: 23 (14.4%) Hospital 2: 29 (18.1%) |
Wall, Floor, Remote control, Knob, Light switch, and Table |
| Indoor | 6 (3.8%) | 2 | Indoor 1: 2 (1.2%) Indoor 2: 2 (1.2%) Indoor 3: 2 (1.2%) |
Wall and Floor |
| Outdoor | 11 (6.9%) | 3 | Outdoor 1: 2 (1.2%) Outdoor 2: 5 (3.1%) Outdoor 3: 4 (2.5%) |
Wall and Floor |
| Soft release | 23 (14.4%) | 15 | Small animal enclosure: 4 (2.5%) Semi-aquatic bird enclosure: 4 (2.5%) Small bird enclosure: 3 (1.9%) Carnivore enclosure: 3 (1.9%) Flight room: 3 (1.9%) Owl enclosure: 4 (2.5%) Parrot aviary: 2 (1.2%) |
Wall and Floor |
NA: Not applicable.
Percentage of total number of samples was calculated based on 160 samples.
Percentage of samples per subdivision was calculated based on 160 samples.
A random sample of animals from each sector of the rehabilitation center that were hospitalized on the day of the study were selected for sampling (n = 64). This not only included animals from each sector, but also undergoing different stages of the rehabilitation process, as well as different taxa, to have a good representative cross-sectional sample of the animals at the WRC (Table 2). Experienced veterinarians and trained volunteers collected rectal and/or cloacal swabs using a Cary Blair transport medium (Deltalab, Spain). In addition, data about the animals sampled (species, gender, age, animal admission date, origin, cause of admission, and previous antimicrobial therapy consisting of antibiotics used and length of treatment) were collected when available. All samples (environmental and animal) were kept at 4 °C until further analysis at the Universidad Andrés Bello research laboratory, where they were processed within 8 h of collection. The study was approved by the Universidad Andrés Bello bioethics committee (Act. 019/2014).
Table 2.
Summary table for the animal samples (n = 64) with numbers and percentage (%) for each category, and number of ceph-resistant isolates.
| Taxa: n(%) | Species: n(%) | Number of ceph-resistant isolates | Enclosure: n(%) |
|---|---|---|---|
| Birds: 55 (86.0%) |
Athene cunicularia: 2(3.6%) Bubo magellanicus: 3(5.5%) Cyanoliseus patagonus: 2(3.6%) Enicognathus ferrugineus: 1(1.8%) Enicognathus leptorhynchus: 2(3.6%) Falco peregrinus: 2(3.6%) Falco sparverius: 7(12.7%) Geranoaetus melanoleucus: 3(5.5%) Glaucidium nana: 3(5.5%) Geranoaetus polyosoma: 1(1.8%) Milvago chimango: 7(12.7%) Parabuteo unicinctus: 4(7.3%) Phrygilus fruticeti: 1(1.8%) Spatula platalea: 1(1.8%) Spinus barbatus: 1(1.8%) Turdus falcklandii: 3(5.5%) Tyto alba: 6(10.9%) Vanellus chilensis: 2(3.6%) Veniliornis lignarius: 1(1.8%) Zenaida auriculata: 3(5.5%) |
0 0 0 0 0 0 0 0 0 0 0 0 3 2 0 5 4 1 2 0 |
Flight Room: 2(3.6%) Hospital 1: 16(29.1%) Hospital 2: 14(25.5%) Outdoor 1: 1(1.8%) Parrot Aviary: 1(1.8%) Quarantine: 16(29.1%) Semi-aquatic birds: 2(3.6%) Small birds: 3(5.5%) |
| Mammals: 2 (3.0%) |
Lycalopex culpaeus: 2(100.0%) | 3 | Carnivores: 1(50.0%) Indoor 3: 1(50.0%) |
| Reptiles: 7 (11.0%) |
Chelonoidis chilensis: 5(71.4%) Philodryas chamissonis: 2(28.6%) |
9 2 |
Hospital 2: 2(28.6%) Indoor 1: 5(71.4%) |
2.2. Laboratory methods
For environmental samples, sterile containers with peptone water and gauzes were subjected to mixing by pulse vortexing for 15 s; this was followed by streaking 50 μL onto MacConkey agar (Becton Dickinson GmbH, Germany) supplemented with 1 mg/L of cefotaxime (Merck, Germany), as previously described [20,21]. For the animal samples, swabs were directly streaked into MacConkey agar, supplemented with cefotaxime as described above. All plates were incubated for 24–48 h at 37 °C, as previously described [22]. After incubation, distinct morphotypes were further isolated with at least three passages, and then isolated colonies were stored at −80 °C with 20% of glycerol.
Species identification was performed using a Vitek MS MALDI-TOF (bioMerieux, San Louis, MO, USA) following manufacturer's instructions as previously described [23]. Their antibiotic susceptibility profile was assessed using the disk diffusion method as per The Clinical & Laboratory Standards Institute (CLSI) recommendations [24]. Briefly, isolates were grown overnight in Tryptic Soy Broth (Becton Dickinson GmbH, Germany), then cultures were adjusted to a MacFarland 0.5 [25] and streaked in Muller Hilton agar (Becton Dickinson GmbH, Germany). All colonies representing different morphotypes that were grown on cephalosporin supplemented MacConkey Agar were further species identified and classified into families: Enterobacteriaceae/Yersiniaceae (order Enterobacteriales), Pseudomonadaceae, Comamonadaceae, Moraxellaceae, Xanthomonadaceae, and Alcaligenaceae. The combination of antibiotics tested varied according to bacterial families: Enterobacteriaceae/Yersiniaceae (order Enterobacteriales), Pseudomonadaceae, Comamonadaceae, Moraxellaceae, Xanthomonadaceae, and Alcaligenaceae. CLSI breakpoints were used to characterize the antibiotic resistance patterns [24]. For Enterobacteriales, 19 antibiotics were tested: amikacin (AMK), gentamicin (GEN), ampicillin (AMP), amoxicillin/clavulanic acid (AMC), ampicillin/sulbactam (SAM), piperacillin/tazobactam (TZP), cefazolin (CFZ), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), ertapenem (ETP), imipenem (IPM), meropenem (MEM), chloramphenicol (CHL), ciprofloxacin (CIP), fosfomycin (FOS), tetracycline (TET), and trimethoprim/sulfamethoxazole (SXT). For Pseudomonadaceae, eight antibiotics were tested: AMK, GEN, CAZ, FEP, IPM, MEM, CIP, and TZP. For Moraxellaceae, 10 antibiotics were tested: AMK, GEN, SAM, TZP, CAZ, FEP, IPM, MEM, CIP, and SXT. For Xanthomonadaceae, three antibiotics were tested: CAZ, levofloxacin (LEV), and SXT. Finally, CAZ, MEM, and SXT were tested for Comamonadaceae and Alcaligenaceae. All disks were obtained from OXOID, United Kingdom. The control strain Escherichia coli ATCC25922 was used. The inhibition zone diameters were interpreted following the Susceptible, Intermediate, Resistant (SIR) status from the Clinical and Laboratory Standards Institute guidelines [24], which differed depending on the bacterial family and species (Tables A.1-A.5).
2.3. Data analyses
Antibiotic resistance patterns for both environmental and animal samples were described for each bacterial family. Further analyses focused on Enterobacteriaceae/Yersiniaceae (order Enterobacteriales) and Pseudomonadaceae as most isolates belonged to these families. Fisher exact test was used to compare the frequency of isolates from animals with a history of previous antimicrobial exposure (yes/no) and their antibiotic resistance outcome (susceptible/intermediate/resistant) for Enterobacteriales order and for Pseudomonadaceae family separately across all the antibiotics tested. Statistical significance was defined with an alpha level of 5%.
A cluster analysis was performed to describe the resistance patterns obtained from Enterobacteriales and Pseudomonadaceae. The goal of the cluster analysis was to determine if isolates from the animals and the WRC environment were similar in their resistance profiles, as evidenced by isolates from both sources clustering together. To perform the cluster analysis, the zone of inhibition obtained for each isolate/antibiotic combination was used. Isolates susceptible to all antibiotics were removed prior to the analysis. Agglomerative Hierarchical Clustering (HC) was used, which is based on a dissimilarity matrix and has the advantage of not having the number of clusters chosen a priori [26]. The Gower distance was used to calculate the distance matrix, and Ward's method was used as the HC algorithm [27]. The functions ‘hclust’ and ‘daisy’ from the package ‘cluster’ in R were used to conduct the HC and the Gower distance respectively [28]. The optimal number of clusters was validated using the optimum average silhouette width with the ‘pamk’ function from the ‘fpc’ package in R [29]. All statistical analyses were performed in R software 3.6.3 [30].
3. Results
3.1. Presence of different families of resistant bacteria in the environmental samples
A total of 160 samples were collected from the WRC environment and a total of 78 bacterial isolates were recovered (Table 1). While isolates were obtained from all sampled sectors, most isolates were retrieved from the hospitals (n = 28), quarantine (n = 23), and soft release (n = 15) (Table 1). Further identification demonstrated that these isolates belonged to six bacterial families: 62.3% (n = 48) Pseudomonadaceae, 21.8% (n = 17) Enterobacteriaceae, 11.5% (n = 9) Yersiniaceae, 1.3% (n = 1) Alcaligenaceae, 2.6% (n = 1) Moraxellaceae, and 1.3% (n = 1) Xanthomonadaceae. For Enterobacteriaceae, species identified were Citrobacter braakii, Escherichia coli, E. vulneris, and Enterobacter cloacae. For Yersiniaceae, Rahnella aquatilis. For Pseudomonadaceae, species identified were P. aeruginosa, P. fluorescens, P. oryzihabitans, P. putida, P. stutzeri, and P. viridiflava. For Alcaligenaceae, Achromobacter xylosoxidans was identified. For Moraxellaceae, Acinetobacter baumannii, and for Xanthomonadaceae, Stenotrophomonas maltophilia was identified.
3.2. Presence of different families of resistant bacteria in the animal samples
A total of 64 animal samples were collected. Of those, 86.0% (n = 55) were avian species, 3.0% (n = 2) mammals, and 11.0% (n = 7) reptiles. There was a total of 25 different animal species, with the most common being Falco sparverius (n = 7), Milvago chimango (n = 7), and Tyto alba (n = 6) for birds, Chelonoidis chilensis (n = 5) for reptiles, and Lycalopex culpaeus (n = 2) for mammals (Table 2). Across all taxa, 54.7% (n = 35) were adults, 28.1% (n = 18) were juveniles, 15.6% (n = 10) were nestlings/pups, and in 1.6% (n = 1) age was not determined. In 64.1% (n = 41) of animals, gender was not determined, and for those where gender was determined, 18.6% (n = 12) were female and 17.2% (n = 11) were male. There was no information about the geographical origin of the animals for most animals sampled (60%, n = 38). For those with information about recovery location, the most frequent were counties 40-50 km from the city of Santiago. The average length of stay at the WRC among animals sampled was 6.7 months (range: 1 week - 3 years).
A total of 31 bacterial isolates were recovered from animal samples. These isolates were obtained from six different bird species: Turdus falcklandii (n = 5), Tyto alba (n = 4), Phrygilus fruticeti (n = 3), Spatula platalea (n = 2), Veniliornis lignarius (n = 2), and Vanellus chilensis (n = 1); one mammal species (three isolates from Lycalopex culpaeus), and two reptile species (Chelonoidis chilensis [n = 9] and Philodryas chamissonis [n = 2]).
These 31 isolates were further classified into five bacterial families: 51.6% (n = 16) Enterobacteriaceae, 29.0% (n = 9) Pseudomonadaceae, 9.7% (n = 3) Xanthomonadaceae, 6.5% (n = 2) Comamonadaceae, and 3.2% (n = 1) Moraxellaceae. For Enterobacteriaceae, Citrobacter braakii, Escherichia coli, E. vulneris, and Enterobacter cloacae were identified. For Pseudomonadaceae, Pseudomonas aeruginosa, P. deovorans, P. fluorescens, P. oryzihabitans, P. putida, P. stutzeri, and P. viridiflava. Stenotrophomonas maltophilia was the species identified for Xanthomonadaceae, Comamonas aquatica for Comamonadaceae, and Acinetobacter baumanni complex for Moraxellaceae.
3.3. Antimicrobial resistance in isolated bacteria from the environment and animals
For the order Enterobacteriales, resistance to antibiotics of different classes was found. Isolates obtained from environmental samples were resistant to penicillins (100%), cephalosporines (92.3%), aminoglycosides (42.3%), quinolones (42.3%), tetracyclines (38.4%), sulfonamides (30.8%), Fosfomycin (30.8%), chloramphenicol (23.1%), and carbapenems (11.5%). Isolates obtained from animal samples were resistant to penicillins (100%), cephalosporines (100%), tetracyclines (75.0%), quinolones (62.5%), sulfonamides (50%), chloramphenicol (31.3%), carbapenems (6.2%), and aminoglycosides (6.2%).
Numerous resistance profiles were found that further characterized the collected isolates. Isolates of the most abundant species (E. coli) were found in the environment and animal samples, and these isolates were classified into 20 antibiotic resistance profiles (Table 3). The majority of E. coli isolates were resistant to more than 9/19 antibiotics tested, including highly resistant isolates, with one E. coli isolate from a sample obtained in the quarantine room that was resistant to 11/19 antibiotics (AMP-SAM-CFZ-FEP-CRO-FOX-ETP-IPM-MEM-GEN-FOS). One E. coli isolate from a sample obtained from a L. culpaeus also showed resistance to 11/19 antibiotics (AMP-SAM-AMC-CFZ-FEP-CRO-FOX-CAZ-TET-CIP-SXT).
Table 3.
Antimicrobial resistance profiles identified in isolates of the order Enterobacteriales and of the Pseudomonadaceae and Xanthomonadaceae families.
| Bacterial species | Antimicrobial resistance profile | Number of isolates | Origin |
|---|---|---|---|
| Enterobacteriales | |||
| Escherichia coli | AMP-SAM-CFZ | 1 | Environment |
| AMP-SAM-CFZ-CIP | 1 | Environment | |
| AMP-CFZ-CRO-GEN-TET | 1 | Environment | |
| AMP-AMC-CFZ-CRO-FOX | 1 | Animal | |
| AMP-SAM-AMC-CFZ-GEN-TET | 1 | Environment | |
| AMP-SAM-AMC-CFZ-CRO-FOX | 1 | Animal | |
| AMP-SAM-AMC-CFZ-CRO-GEN-TET | 1 | Environment | |
| AMP-SAM-AMC-CFZ-ETP-CIP-SXT-FOS | 1 | Environment | |
| AMP-SAM-AMC-CFZ-FOX-CAZ-TET-CIP | 1 | Animal | |
| AMP-SAM-AMC-CFZ-CRO-FOX-CAZ-TET | 1 | Animal | |
| AMP-TZP-CFZ-FEP-CRO-FOX-CAZ-FOS | 1 | Environment | |
| AMP-CFZ-FEP-CRO-FOX-TET-CIP-SXT-CHL | 4 | Animal | |
| AMP-SAM-AMC-CFZ-CRO-FOX-CAZ-TET-CHL | 1 | Animal | |
| AMP-AMC-CFZ-GEN-TET-CIP-SXT-CHL-FOS | 1 | Environment | |
| AMP-CFZ-FEP-CRO-MEM-AMK-TET-CIP-SXT-CHL | 1 | Environment | |
| AMP-SAM-AMC-CFZ-CRO-FOX-CAZ-TET-CIP-SXT | 1 | Animal | |
| AMP-SAM-AMC-CFZ-CRO-FOX-CAZ-TET-CIP-SXT | 1 | Environment | |
| AMP-CFZ-FEP-CRO-FOX-GEN-TET-CIP-SXT-CHL | 3 | Environment | |
| AMP-SAM-CFZ-FEP-CRO-FOX-ETP-IPM-MEM-GEN-FOS | 1 | Environment | |
| AMP-SAM-AMC-CFZ-FEP-CRO-FOX-CAZ-TET-CIP-SXT | 1 | Animal | |
| Citrobacter braakii | AMP-SAM-AMC-CFZ-FOX-TET | 1 | Animal |
| AMP-SAM-AMC-CFZ-CRO-GEN-CIP | 1 | Environment | |
| AMP-SAM-AMC-CFZ-FOX-AMK-CIP | 1 | Animal | |
| AMP-SAM-AMC-CFZ-FOX-IPM-TET-CIP | 1 | Animal | |
| Enterobacter cloacae | AMP-SAM-AMC-CFZ-FOX-TET-CIP-SXT | 1 | Animal |
| AMP-SAM-AMC-CFZ-GEN-TET-CIP-SXT-CHL | 1 | Environment | |
| AMP-SAM-AMC-CFZ-CRO-FOX-CAZ-SXT | 1 | Animal | |
| E. vulneris | AMP-AMC-CFZ-CRO-FOX-CAZ-CIP-FOS | 1 | Environment |
| Rahnella aquatilis | AMP-CFZ-CRO-FOS | 2 | Environment |
| AMP-CFZ-CRO | 4 | Environment | |
| AMP-CFZ-FOS | 1 | Environment | |
| SAM | 1 | Environment | |
| AMP | 1 | Environment | |
| Pseudomonadaceae | |||
| Pseudomonas aeruginosa | TZP-GEN-CIP | 1 | Animal |
| IPM-MEM | 1 | Environment | |
| Pseudomonas fluorescens | IPM | 1 | Environment |
| Pseudomonas putida | CIP | 5 | Environment |
| Pseudomonas stutzeri | MEM | 1 | Environment |
| IPM-MEM | 1 | Animal | |
| Pseudomonas viridiflava | IPM-MEM | 1 | Animal |
| Xanthomonadaceae | |||
| Stenotrophomonas maltophilia | CAZ | 1 | Environment |
| Stenotrophomonas maltophilia | CAZ-LEV-SXT | 1 | Animal |
Abbreviations: amikacin (AMK), gentamicin (GEN), ampicillin (AMP), amoxicillin/clavulanic acid (AMC), ampicillin/sulbactam (SAM), piperacillin/tazobactam (TZP), cefazolin (CFZ), cefoxitin (FOX), ceftazidime (CAZ), ceftriaxone (CRO), cefepime (FEP), ertapenem (ETP), imipenem (IPM), meropenem (MEM), chloramphenicol (CHL), ciprofloxacin (CIP), fosfomycin (FOS), tetracycline (TET), levofloxacin (LEV), and trimethoprim/sulfamethoxazole (SXT).
Isolates of the Pseudomonadaceae family were tested using 8 antibiotics, which further identified 11 isolates with resistance to antibiotics such as carbapenems and quinolones, including four isolates collected from the environment and animals (Table 3). For instance, one isolate of Pseudomonas aeruginosa obtained from an animal possessed resistance to TZP-GEN-CIP and another from the environment possessed resistance to IPM-MEM. Other species of Pseudomonas also possessed resistance to IPM-MEM (Table 3).
In addition, one isolate from an environmental sample from the family Moraxellaceae, was identified as Acinetobacter baumannii and was susceptible to all 10 antibiotics. For Xanthomonadaceae, one environmental isolate and one animal isolate of Stenotrophomonas maltophilia were resistant to one of the three antibiotics tested and to the three antibiotics respectively (Table 3). Lastly, for Alcaligenaceae there was one species identified, Achomobacter xylosoxidans, which was susceptible to all three antibiotics tested.
3.4. Antibiotic use, antibiotic resistance, and clustering
Out of the animals sampled, 31.2% (n = 20) had received antibiotics (at least one dose of one antibiotic at some point in time) during their stay at the rehabilitation center, and 68.8% (n = 44) had not. Clindamycin (n = 10) followed by enrofloxacin (n = 8) were the most commonly used antibiotics. The longest antibiotic treatment was 3.9 months for enrofloxacin in a Patagonian land turtle (C. chilensis), and the shortest antibiotic treatment was a seven-day course of enrofloxacin in an Andean fox (L. culpaeus).
The 16 Enterobacteriaceae isolates that were recovered from animal samples belonged to 10 different animal species of which five had received antibiotic treatment and five had not. The nine Pseudomonadaceae isolates belonged to eight different animals of which one had received antibiotics and the remaining seven had not. There was no difference in the frequency of resistant isolates regardless of whether they had received antibiotics or not for Enterobacteriaceae (p = 0.35) and for Pseudomonadaceae (p = 0.56).
For the cluster analyses, 42 isolates for the order Enterobacteriales (26 from environmental samples and 16 from animal samples) and 11 for Pseudomonadaceae (8 for environmental samples and 3 for animal samples) were analyzed. The optimal number of clusters was two for Enterobacteriales (cluster I with 30 isolates and cluster II 12 isolates), and four for Pseudomonadaceae (cluster I with 4 isolates, cluster II with 4 isolates, cluster III with 2 isolates, and cluster IV with one isolate). In Enterobacteriales, cluster I isolates were resistant to 42.1% (8/19) of antibiotics, while isolates in cluster II were resistant to 15.8% (3/19) of antibiotics (Table 4). Cluster I was dominated by small groupings of isolates obtained from C. chilensis, Tyto alba, and hospital 1 isolates, while cluster II only contained environmental isolates that belonged mostly to the kitchen, owl enclosure, and hospital 2 (Fig. 1). For Pseudomonadaceae, Clusters II and III only contained environmental isolates and were dominated by hospital 2 and quarantine isolates, Cluster I had a mixed of animal and environmental isolates, and Cluster IV was made of an isolate of Vanelus chilensis (Table 5, Fig. 2).
Table 4.
Cluster results for Enterobacteriales isolates. The numbers represent the mean inhibition zone diameters in mm for each antibiotic that was tested. The number of isolates for each cluster is divided between environmental and animal isolates. Cluster I: 14 environmental and 16 animal isolates; Cluster II: 12 environmental isolates. The greyed-out fields represent those that are resistant according to the CLSI Susceptible Intermediate Resistant (SIR) status [24].
| Cluster |
Antibiotics |
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AMK | GEN | AMP | AMC | SAM | TZP | CFZ | FOX | CAZ | CRO | FEP | ETP | IPM | MEM | CHL | CIP | FOS | TET | SXT | |
| I | 19.3 | 15.4 | 0.0 | 9.8 | 7.7 | 24.3 | 0.3 | 5.8 | 19.7 | 14.9 | 22.3 | 26.3 | 25.0 | 28.3 | 16.7 | 10.2 | 23.9 | 5.3 | 11.1 |
| II | 26.0 | 26.0 | 1.1 | 24.0 | 20.8 | 25.0 | 6.3 | 23.5 | 27.0 | 19.0 | 25.6 | 32.1 | 29.8 | 31.5 | 27.6 | 25.4 | 21.8 | 24.4 | 25.2 |
Abbreviations: AMK: amikacin; GEN: gentamicin; AMP: ampicillin; AMC: amoxicillin/clavulanic acid; SAM: ampicillin/sulbactam; TZP: piperacillin/tazobactam; CFZ: cefazolin; FOX: cefoxitin; CAZ: ceftazidime; CRO: ceftriaxone; FEP: cefepime; ETP: ertapenem; IPM: imipenem; MEM: meropenem; CHL: chloramphenicol; CIP: ciprofloxacin; FOS: fosfomycin; TET: tetracycline; SXT: sulphamethoxazole/trimethoprim.
Fig. 1.

Dendrogram for Enterobacteriales that resulted from the cluster analysis. The y-axis (height) represents how close together observations were when they were merged into clusters. gower_distR refers to Gower distance which was used to calculate the distance matrix, and Ward's refers to the method used as the hierarchical clustering algorithm. The rectangular boxes represent each one of the two clusters (I and II).
Table 5.
Cluster results for Pseudomonadaceae isolates. The numbers represent the mean inhibition zone diameters in mm for each antibiotic that was tested. The number of isolates for each cluster is divided between environmental (Env.) and animal (An.) isolates. The greyed-out fields represent those that are resistant according to the CLSI Susceptible Intermediate Resistant (SIR) status [24].
| Cluster |
Antibiotics |
Env. |
An. |
||||
|---|---|---|---|---|---|---|---|
| GEN | TZP | IPM | MEM | CIP | |||
| I | 0.0 | 0.0 | 7.5 | 13.8 | 0.0 | 2 | 2 |
| II | 0.0 | 0.0 | 0.0 | 0.0 | 12.8 | 4 | |
| III | 0.0 | 0.0 | 7.5 | 7.5 | 0.0 | 2 | |
| IV | 7.0 | NA | NA | NA | NA | 1 | |
Abbreviations: GEN: gentamicin; TZP: piperacillin/tazobactam.
IPM: imipenem; MEM: meropenem; CIP: ciprofloxacin.
NA: Not applicable.
Fig. 2.
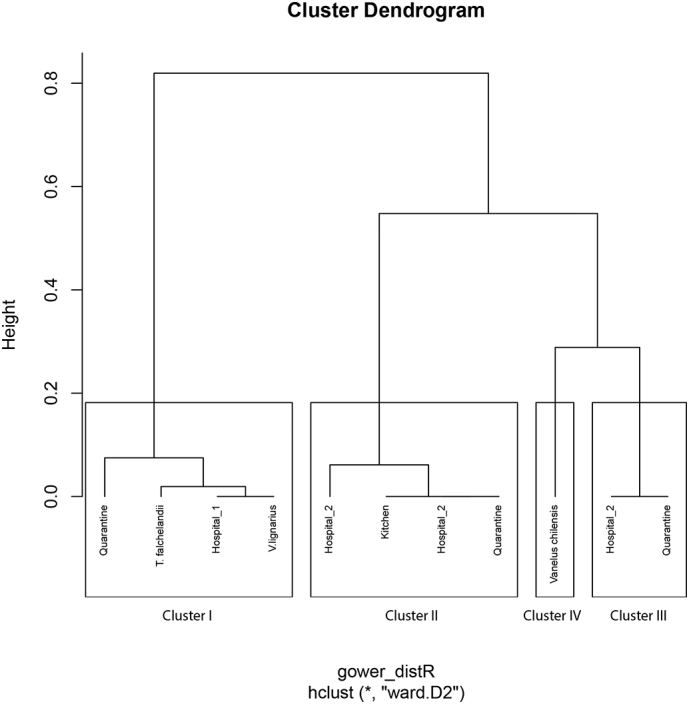
Dendrogram for Pseudomonadaceae that resulted from the cluster analysis. The y-axis (height) represents how close together observations were when they were merged into clusters. gower_distR refers to Gower distance which was used to calculate the distance matrix, and Ward's refers to the method used as the hierarchical clustering algorithm. The rectangular boxes represent each one of the four clusters (I, II, III, and IV).
4. Discussion
To fully understand and mitigate AMR, it is important to consider the role of the natural environment as part of the One Health approach that has been advocated towards this end. Wildlife species may be exposed to antibiotics and antimicrobial resistant organisms, and they may contribute to their dissemination. From a public health perspective, wildlife admitted to WRC have been mostly evaluated for their potential to carry and transmit zoonotic pathogens such as Salmonella spp. including raptors in Chile [[31], [32], [33], [34]]; however, the role of WRC in the emergence and dissemination of AMR has been overlooked [35]. In this study, antibiotic resistant Gram-negative bacteria with reduced susceptibility to cefotaxime were characterized in both animal and environmental samples at a WRC in central Chile.
The results showed a high proportion of the cef-resistant bacterial subpopulation to also be resistant to three or more antibiotics (90% of animal isolates and 66.7% of environmental isolates). This finding is consistent with other studies that have also found remarkable percentages of resistant bacteria in wildlife undergoing rehabilitation. In one study, samples taken from injured wildlife admitted to a WRC in Spain revealed that 71% of all E. coli isolates recovered from animals were resistant to more than three individual antibiotics [36]. In a wildlife rescue center in Italy, resistance to 15/16 of antibiotics tested occurred among isolates from raptors and waterbirds, while there was resistance to 10/16 of antibiotics tested in isolates from passerine species [16]. Furthermore, another study found that 77.8% of northern elephant seals (Mirounga angustirostris) had antimicrobial resistant E. coli prior to release, compared to 38.4% of the seals at admission to a WRC [37]. These findings are compatible with others that have reported that wild animals either in captivity or closer to anthropogenic pressures tend to a higher prevalence of antibiotic resistant bacteria compared to those that are free-ranging or further from human influence [[38], [39], [40]].
Animals sampled in this study had been at the WRC an average of six months, and from those where retrieval information was known, they had been found near the city of Santiago, the capital of Chile, a large urban center. These two factors (time in captivity and proximity to human activities) could have a large influence on the antibiotic resistance outcome. In fact, there were two animals with a higher proportion of Enterobacteriaceae resistant isolates than others that had been at the WRC for a considerable time. One was a Patagonian land turtle (C. chilensis), admitted to the WRC after being confiscated from the illegal wildlife trade, that had spent over four months at the WRC and had received enrofloxacin treatment for 12 weeks. The prolonged antibiotic treatment and/or the prolonged captivity could have led to increased antibiotic resistance. The other case was an Andean fox cub (L. culpaeus) that had been at the WRC for almost 4 months and had not received antibiotic treatment. Age has been reported as a strong predictor of antibiotic resistance, with younger animals shedding a higher prevalence of resistant bacteria [41,42]. However, in a recent study evaluating antimicrobial resistance gene occurrence in Andean foxes, the authors did not find significant differences related to age [43].
There was no association between antibiotic treatment and frequency of resistant isolates. This could be explained by the small sample size (n = 20), by a short time of exposure to the antibiotics, and by other factors that could not be accounted for in this study, such as location where the animal was originally found, as well as other components of the complexity of AMR epidemiology, and the wide presence of resistant bacteria in the built environment. Alternatively, the effect of the antibiotic therapy may have been short-lived, and the animals became repopulated with resident bacteria when the pressure of the antibiotics were off. This effect has been observed in other animal settings, with the duration of the effect being related to the fraction of the animal population that received antibiotic therapy [44].
Among the resistance patterns found in this study, it is important to highlight the identification of both Enterobacteriales and Pseudomonadaceae isolates resistant to carbapenems in the WRC environment and in the animals. This is very relevant from a public health perspective since these microorganisms were classified as critical priority by the World Health Organization (WHO) priority pathogens list for research and development of new antibiotics [45]. Another remarkable finding was the percentage (30.8%) of Enterobacteriales environmental isolates resistant to fosfomycin. This antibiotic with antibacterial activity against a wide range of gram-negative pathogens and some gram-positive pathogens, has been increasingly used worldwide in the last few years to treat uncomplicated urinary tract infections in humans when strains are resistant to other most commonly used drugs such as ciprofloxacin [46,47]. Antibiotic resistant bacteria in rehabilitated wildlife can be seeing from different perspectives. For instance, one aspect is the potential dispersal of antibiotic resistant bacteria from released wildlife to livestock and humans; another aspect is the environmental acquisition of antibiotic resistant bacteria by rescued wildlife, especially when this wildlife is found at or near urban areas or near livestock. However, a recent study conducted in the same geographical area as our study, found ESBL-producing E. coli in 24% of dogs, 3% of cows, but only in 0.5% of wildlife [48], values much lower than our results in the built environment. The different ways by which wildlife may play a role in the acquisition and in the dissemination of antibiotic resistant bacteria require further investigation.
A high percentage of Enterobacteriales (30.8% of the environmental isolates and 62.5% of the animal isolates) and Pseudomonadaceae (62.5% of environmental isolates and 33.3% of animal isolates) were resistant to ciprofloxacin, an antibiotic of the fluoroquinolone class. The wide use of enrofloxacin, another fluoroquinolone, at the WRC may have contributed to these results, as it has been noted in other studies [49]. In addition, commonly used disinfectants in hospital environments including this WRC such as quaternary ammoniums (QACs) could have contributed to an increase in fluoroquinolone resistant isolates. Even though there was no evidence to address this hypothesis at the genetic level in this study, there are documented interactions between the use of QACs and the emergence of fluoroquinolone resistance in bacteria [50,51].
In this study, the WRC built environment was an important reservoir of bacteria with reduced susceptibility to cephalosporins. While it was hypothesized that resistant bacteria from both types of samples would cluster together based on their antibiotic resistance patterns, the results did not support this hypothesis. Numerous antibiotic resistance profiles were observed in different bacterial species isolated here, indicating not only environmental contamination with a wide diversity of bacteria, but also a wide diversity of resistant bacteria in animals at the WRC. In our study, even though transfer of antibiotic resistant bacteria to WRC personnel was not investigated, we identified antibiotic resistant bacteria in human-touch surfaces, such as doorknobs, light switches, and areas within the WRC such as the reception and the kitchen. All these represent potential sites for dissemination of resistant bacteria to humans. Furthermore, the diversity of bacteria could be further analyzed using culture-independent methods, which would provide a broader perspective on the antibiotic resistance dynamics at the WRC and help overcome the inherent culture bias of culture-based methods [52].
The study design was cross-sectional, with samples only collected at one point in time. This means that results could have differed if samples had been collected at a different time. Furthermore, cross-sectional studies cannot provide an indication of the sequence of events, and thus it would not be possible to identify if the animals were admitted carrying resistant bacteria or instead they acquired the resistant bacteria during their stay at the WRC. Improved study designs consisting of longitudinal sampling of the animals from admission to their final outcome (release/euthanasia/transfer) would add valuable information about the potential emergence and/or acquisition of AMR at WRC.
In conclusion, an increased understanding on antibiotic use practices and AMR dynamics in wildlife rehabilitation is needed. It is critical to increase the knowledge about the influence of antibiotic and human exposure to wildlife populations, and when wild animals are placed in temporary captivity, to further understand the effects that hospitalization and reintroduction back into the natural environment can have on the potential emergence and spread of AMR, and thus on wildlife, human, and ecosystem health.
Funding
This work was supported by FONDECYT 1181167 and by the ANID Millennium Science Initiative/Millennium Initiative for Collaborative Research on Bacterial Resistance, MICROB-R, NCN17_81.
Credit author statement
Carla Baros Jorquera: Conceptualization, Methodology, Investigation, Writing; Andrea I. Moreno-Switt: Conceptualization, Methodology, Investigation, Writing, Supervision, Funding acquisition; Nicole Sallaberry-Pincheira: Resources, Conceptualization, Writing-Review & Editing; Jose M. Munita: Writing-Review & Editing, Funding acquisition; Camila Flores Navarro: Writing-Review & Editing; Rodolfo Tardone: Methodology, Writing-Review & Editing; Gerardo González-Rocha: Methodology, Writing-Review & Editing; Randall S. Singer: Methodology, Writing- Review & Editing; Irene Bueno: Methodology, Formal analysis, Writing, Supervision.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgments
We thank all the staff and volunteers at the UFAS UNAB/Buin Zoo wildlife rehabilitation center at the Universidad Andrés Bello (Santiago, Chile) that helped to make this study possible.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.onehlt.2021.100298.
Appendix A. Supplementary data
Clinical and Laboratory Standards Institute (CLSI) breakpoints for the isolates included in the study.
References
- 1.Larsson D.J. Critical knowledge gaps and research needs related to the environmental dimensions of antibiotic resistance. Environ. Int. 2018;117:132–138. doi: 10.1016/j.envint.2018.04.041. [DOI] [PubMed] [Google Scholar]
- 2.McEwen S.A., Collignon P.J. Antimicrobial resistance: a One Health perspective. Microbiol Spectr. 2018;6(2) doi: 10.1128/MICROBIOLSPEC.ARBA-0009-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rousham E.K., Unicomb L., Islam M.A. Human, animal and environmental contributors to antibiotic resistance in low-resource settings: integrating behavioural, epidemiological and one health approaches. Proc. Biol. Sci. 2018;285(1876):20180332. doi: 10.1098/rspb.2018.0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huijbers P.M. Role of the environment in the transmission of antimicrobial resistance to humans: a review. Environ. Sci. Technol. 2015;49(20):11993–12004. doi: 10.1021/acs.est.5b02566. [DOI] [PubMed] [Google Scholar]
- 5.Singer A.C. Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 2016;7:1728. doi: 10.3389/fmicb.2016.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bueno I. Systematic review: impact of point sources on antibiotic-resistant bacteria in the natural environment. Zoonoses Public Health. 2018;65(1):e162–e184. doi: 10.1111/zph.12426. [DOI] [PubMed] [Google Scholar]
- 7.Kraemer S.A., Ramachandran A., Perron G.G. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms. 2019;7(6):180. doi: 10.3390/microorganisms7060180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D’Costa V.M. Antibiotic resistance is ancient. Nature. 2011;477(7365):457. doi: 10.1038/nature10388. [DOI] [PubMed] [Google Scholar]
- 9.Bhullar K. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS One. 2012:7(4). doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rizzo L. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci. Total Environ. 2013;447:345–360. doi: 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 11.Berendonk T.U. Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 2015;13(5):310–317. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 12.Vittecoq M. Antimicrobial resistance in wildlife. J. Appl. Ecol. 2016;53(2):519–529. doi: 10.1111/1365-2664.12596. [DOI] [Google Scholar]
- 13.Fuentes-Castillo D. Wild owls colonized by international clones of extended-spectrum beta-lactamase (CTX-M)-producing Escherichia coli and Salmonella infantis in the southern cone of America. Sci. Total Environ. 2019;674:554–562. doi: 10.1016/j.scitotenv.2019.04.149. [DOI] [PubMed] [Google Scholar]
- 14.Dolejska M. Antibiotic-resistant bacteria in wildlife. In: Manaia C.M., editor. Antibiotic Resistance in the Environment. Springer; Switzerland: 2020. pp. 19–70. [DOI] [Google Scholar]
- 15.Marrow J. Prevalence and antibiotic-resistance characteristics of Enterococcus spp. isolated from free-living and captive raptors in Central Illinois. J. Wildl. Dis. 2009;45(2):302–313. doi: 10.7589/0090-3558-45.2.302. [DOI] [PubMed] [Google Scholar]
- 16.Giacopello C. Antimicrobial resistance patterns of Enterobacteriaceae in European wild bird species admitted in a wildlife rescue centre. Vet. Ital. 2016;52(2):139–144. doi: 10.12834/VetIt.327.1374.2. [DOI] [PubMed] [Google Scholar]
- 17.Darwich L. High prevalence and diversity of extended-spectrum beta-lactamase and emergence of OXA-48 producing Enterobacterales in wildlife in Catalonia. PLoS One. 2019;14(8) doi: 10.1371/journal.pone.0210686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soulsbury C.D., White P.C. Human–wildlife interactions in urban areas: a review of conflicts, benefits and opportunities. Wildl. Res. 2016;42(7):541–553. doi: 10.1071/WR14229. [DOI] [Google Scholar]
- 19.Heathcote G. Citizen reporting of wildlife interactions can improve impact-reduction programs and support wildlife carers. Wildl. Res. 2019;46(5):415–428. doi: 10.1071/WR18127. [DOI] [Google Scholar]
- 20.Hasman H. Abstract from 25th European Congress of Clinical Microbiology and Infectious Diseases, Copenhagen, Denmark. 2015. Validation of selective MacConkey agar plates supplemented with 1 mg/L cefotaxime for monitoring of ESBL- and AmpCproducing E. coli in meat and caecal samples.https://core.ac.uk/download/pdf/43250199.pdf [Google Scholar]
- 21.Worsley-Tonks K.E.L. Importance of anthropogenic sources at shaping the antimicrobial resistance profile of a peri-urban mesocarnivore. Sci. Total Environ. 2021;764:144166. doi: 10.1016/j.scitotenv.2020.144166. [DOI] [PubMed] [Google Scholar]
- 22.Veldman K. Characteristics of cefotaxime-resistant Escherichia coli from wild birds in the Netherlands. Appl. Environ. Microbiol. 2013;79(24):7556–7561. doi: 10.1128/AEM.01880-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porte L. Head-to-head comparison of Microflex LT and Vitek MS systems for routine identification of microorganisms by MALDI-TOF mass spectrometry in Chile. PLoS One. 2017;12(5) doi: 10.1371/journal.pone.0177929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.CLSI . CLSI; Wayne, PA: 2018. Performance Standards for Antimicrobial Susceptibility Testing, in CLSI supplement M100.https://clsi.org/standards/products/microbiology/documents/m100/ [Google Scholar]
- 25.Belbase A. Antibiotic resistance and biofilm production among the strains of Staphylococcus aureus isolated from pus/wound swab samples in a tertiary care hospital in Nepal. Ann Clin Microbiol. 2017;16(1):1–5. doi: 10.1186/s12941-017-0194-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hummel M., Edelmann D., Kopp-Schneider A. Clustering of samples and variables with mixed-type data. PLoS One. 2017;12(11) doi: 10.1371/journal.pone.0188274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Botrel M.A. Identifying antimicrobial multiresistance patterns of Escherichia coli sampled from diarrhoeic calves by cluster analysis techniques: a way to guide research on multiresistance mechanisms. Zoonoses Public Health. 2010;57(3):204–210. doi: 10.1111/j.1863-2378.2009.01262.x. [DOI] [PubMed] [Google Scholar]
- 28.Hubert M., Hornik K. 2019. Cluster: cluster analysis basics and extensions. (R package version 2.1.0) [Google Scholar]
- 29.Hennig C. Fpc: flexible procedures for clustering. R package version. 2010;2(2):0–3. [Google Scholar]
- 30.Team, R.C., R: a language and environment for statistical computing (3.5. 1)[computer software], 2020. R Foundation for Statistical Computing.
- 31.Steele C.M., Brown R.N., Botzler R.G. Prevalences of zoonotic bacteria among seabirds in rehabilitation centers along the Pacific coast of California and Washington. USA. J Wildl Dis. 2005;41(4):735–744. doi: 10.7589/0090-3558-41.4.735. [DOI] [PubMed] [Google Scholar]
- 32.Jijón S., Wetzel A., LeJeune J. Salmonella enterica isolated from wildlife at two Ohio rehabilitation centers. J Zoo Wildl Med. 2007;38(3):409–413. doi: 10.1638/1042-7260(2007)38[409:SEIFWA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 33.Molina-Lopez R.A. Multidrug-resistant Salmonella enterica serovar typhimurium monophasic variant 4,12:i:- isolated from asymptomatic wildlife in a Catalonian wildlife rehabilitation center. Spain. J Wildl Dis. 2015;51(3):759–763. doi: 10.7589/2015-01-019. [DOI] [PubMed] [Google Scholar]
- 34.Tardone R. Salmonella in raptors and aquatic wild birds in Chile. J. Wildl. Dis. 2020;56(3):707–712. doi: 10.7589/2019-08-198. [DOI] [PubMed] [Google Scholar]
- 35.Sellera F.P. Epidemiological implications of drug-resistant bacteria in wildlife rehabilitation centers. J Infect Public Health. 2019;12(5):748–749. doi: 10.1016/j.jiph.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 36.Vidal A. Microbiological diagnosis and antimicrobial sensitivity profiles in diseased free-living raptors. Avian Pathol. 2017;46(4):442–450. doi: 10.1080/03079457.2017.1304529. [DOI] [PubMed] [Google Scholar]
- 37.Stoddard R.A. The effect of rehabilitation of northern elephant seals (Mirounga angustirostris) on antimicrobial resistance of commensal Escherichia coli. Vet. Microbiol. 2009;133(3):264–271. doi: 10.1016/j.vetmic.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 38.Allen H.K. Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 2010;8(4):251–259. doi: 10.1038/nrmicro2312. [DOI] [PubMed] [Google Scholar]
- 39.Skurnik D. Effect of human vicinity on antimicrobial resistance and integrons in animal faecal Escherichia coli. J. Antimicrob. Chemother. 2006;57(6):1215–1219. doi: 10.1093/jac/dkl122. [DOI] [PubMed] [Google Scholar]
- 40.Guenther S. First insights into antimicrobial resistance among faecal Escherichia coli isolates from small wild mammals in rural areas. Sci. Total Environ. 2010;408(17):3519–3522. doi: 10.1016/j.scitotenv.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 41.Berge A., Atwill E.R., Sischo W. Animal and farm influences on the dynamics of antibiotic resistance in faecal Escherichia coli in young dairy calves. Prev Vet Med. 2005;69(1–2):25–38. doi: 10.1016/j.prevetmed.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 42.Langlois B.E. Effect of age and housing location on antibiotic resistance of fecal coliforms from pigs in a non-antibiotic-exposed herd. Appl. Environ. Microbiol. 1988;54(6):1341–1344. doi: 10.1128/aem.54.6.1341-1344.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cevidanes A. Antimicrobial resistance genes in Andean foxes inhabiting anthropized landscapes in Central Chile. Sci. Total Environ. 2020;724:138247. doi: 10.1016/j.scitotenv.2020.138247. [DOI] [PubMed] [Google Scholar]
- 44.Singer R.S., Patterson S.K., Wallace R.L. Effects of therapeutic ceftiofur administration to dairy cattle on Escherichia coli dynamics in the intestinal tract. Appl. Environ. Microbiol. 2008;74(22):6956–6962. doi: 10.1128/AEM.01241-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.World Health Organization . 2017. WHO publishes list of bacteria for which new antibiotics are urgently needed.https://www.who.int/news/item/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed [Google Scholar]
- 46.Falagas M.E. Fosfomycin. Clin. Microbiol. Rev. 2016;29(2):321–347. doi: 10.1128/CMR.00068-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alrowais H. Fosfomycin resistance in Escherichia coli, Pennsylvania. USA. Emerg Infect Dis. 2015;21(11):2045. doi: 10.3201/eid2111.150750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benavides J.A. ESBL-producing escherichia coli carrying CTX-M genes circulating among livestock, dogs, and wild mammals in small-scale farms of Central Chile. Antibiotics. 2021;10(5):510. doi: 10.3390/antibiotics10050510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fuentefria D.B., Ferreira A.E., Corção G. Antibiotic-resistant Pseudomonas aeruginosa from hospital wastewater and superficial water: are they genetically related? J. Environ. Manag. 2011;92(1):250–255. doi: 10.1016/j.jenvman.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 50.Wales A.D., Davies R.H. Co-selection of resistance to antibiotics, biocides and heavy metals, and its relevance to foodborne pathogens. Antibiotics. 2015;4(4):567–604. doi: 10.3390/antibiotics4040567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Buffet-Bataillon S. Efflux pump induction by quaternary ammonium compounds and fluoroquinolone resistance in bacteria. Future Microbiol. 2016;11(1):81–92. doi: 10.2217/fmb.15.131. [DOI] [PubMed] [Google Scholar]
- 52.McLain J.E. Culture-based methods for detection of antibiotic resistance in agroecosystems: advantages, challenges, and gaps in knowledge. J. Environ. Qual. 2016;45(2):432–440. doi: 10.2134/jeq2015.06.0317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical and Laboratory Standards Institute (CLSI) breakpoints for the isolates included in the study.


