Abstract
Background
The Northern Territory (NT) has the highest tuberculosis (TB) rate of all Australian jurisdictions. We combined TB public health surveillance data with genomic sequencing of Mycobacterium tuberculosis isolates in the tropical ‘Top End’ of the NT to investigate trends in TB incidence and transmission.
Methods
This retrospective observational study included all 741 culture-confirmed cases of TB in the Top End over three decades from 1989–2020. All 497 available M. tuberculosis isolates were sequenced. We used contact tracing data to define a threshold pairwise SNP distance for hierarchical single linkage clustering, and examined putative transmission clusters in the context of epidemiologic information.
Findings
There were 359 (48%) cases born overseas, 329 (44%) cases among Australian First Nations peoples, and 52 (7%) cases were Australian-born and non-Indigenous. The annual incidence in First Nations peoples from 1989-2019 fell from average 50.4 to 11.0 per 100,000 (P<0·001). First Nations cases were more likely to die from TB (41/329, 12·5%) than overseas-born cases (11/359, 3·1%; P<0·001). Using a threshold of ≤12 SNPs, 28 clusters of between 2–64 individuals were identified, totalling 250 cases; 214 (86%) were First Nations cases and 189 (76%) were from a remote region. The time between cases and past epidemiologically- and genomically-linked contacts ranged from 4·5 months to 24 years.
Interpretation
Our findings support prioritisation of timely case detection, contact tracing augmented by genomic sequencing, and latent TB treatment to break transmission chains in Top End remote hotspot regions.
Research in context.
Evidence before this study
The Northern Territory (NT) has the highest rate of TB in Australia, with most cases occurring in those born overseas and in Australian First Nations peoples. The NT lies in close geographic proximity to Southeast Asia, and TB cases have increased with sporadic influxes of arrivals from neighbouring countries. Approximately a third of NT residents are Aboriginal peoples, three quarters of whom live in remote regions. Public health surveillance data suggest that TB rates in NT Aboriginal peoples have dropped markedly but remain consistently higher than in non-Indigenous Australian-born people.
A PubMed search using the terms “tuberculosis,” “Australia,” “genomic,” and “sequencing” revealed no previous TB genomic sequencing studies within the NT, and few studies reporting use of genomics to understand TB transmission in other jurisdictions of Australia.
Added value of this study
In this study we combined public health surveillance data with genomic sequencing of Mycobacterium tuberculosis isolates to understand trends in TB incidence and transmission in the tropical ‘Top End’ of the NT during three decades from 1989–2020. The annual TB incidence in Australian First Nations peoples dropped on average by 5% per year. First Nations TB cases were more likely to have smear-positive sputum at diagnosis and to die from TB than overseas-born cases, suggesting diagnosis later in the course of illness. Genomic clusters predominantly involved First Nations cases in remote hotspot regions, with evidence that both reactivation from latency and recent transmission are contributing to incident TB. Genomic sequencing identified putative transmission links which had not been evident during contact tracing. There were few overseas-born cases within transmission clusters, and there was no evidence of transmission of drug-resistant TB.
Implications of all the available evidence
This study provides support for focussing on timely case detection, contact tracing augmented by genomic sequencing, and effective latent TB treatment to break transmission chains in NT remote hotspot regions. Future work includes investigation of the factors contributing to late TB presentations among Australian First Nations peoples, incorporation of prospective genomic sequencing into TB surveillance and outbreak investigations, and evaluation of short, effective regimens for latent TB treatment in this setting.
Alt-text: Unlabelled box
1. Introduction
Australia has one of the lowest rates of tuberculosis (TB) in the world with an annual incidence of 5·2–7·0 per 100,000 population since the 1980s and with 86–89% of TB notifications being in people born overseas.1 First Nations populations are disproportionately affected by TB globally including in Australia.2,3 Here, the terms ‘First Nations’ or ‘Indigenous’ are respectfully used to refer to Australian Aboriginal and Torres Strait Islander peoples. Where appropriate in this study we refer to Aboriginal peoples, the main Indigenous population of the study setting. The TB rate in Australian First Nations populations is 3·8–7·5 per 100,000 population, consistently 5–6 times higher than in the Australian-born non-Indigenous population (<1 case per 100,000 population).1 The Northern Territory (NT) has the highest overall and childhood TB rates of Australian jurisdictions.1,4 Multidrug-resistant TB (MDR-TB) remains uncommon accounting for 1.5% of NT TB notifications.5
The NT is sparsely populated but culturally rich; 20% of the population were born overseas and 30% are Aboriginal people among whom ~30 different languages are spoken.6 The capital city Darwin is closer to Dili in Timor-Leste (686km) than to the nearest Australian capital city Adelaide over 3,000km away. Approximately three quarters of NT Aboriginal people live in remote areas, a partial legacy of forced relocation to missions and reserves associated with the 1869 Aboriginal Protection Act. Overcrowded housing persists with up to 16 people per household, with high rates of homelessness, food insecurity and malnutrition.7,8 There are high rates of smoking, hazardous alcohol consumption, diabetes, and chronic kidney disease.8 Together these factors contribute to increased TB risk in the NT.9,10
Following the disbanding of the NT TB control program in 1982, TB incidence in NT Aboriginal peoples rose and was 114 per 100,000 population annually by 1989.11 Consequently the program was reinstituted in 1989 with an increase in TB control officers, standardisation and supervision of treatment, recording of treatment compliance, extension of contact tracing and screening of high-risk groups, and promotion of latent TB treatment.12 TB rates in NT Aboriginal peoples have subsequently dropped, and in 2015-2018 the annual rates were 5·4–12·1 per 100,000 population.1 The number of overseas-born NT TB cases has fluctuated under the influence of regional policies and politics. TB cases increased in association with evacuation of >1,800 people to Darwin from Timor-Leste during conflict associated with transition to independence in 1999,13 with the on-shore detention of illegal fishers in northern Australian waters since 2005,14 and detention in the NT of people arriving by boat to seek asylum in Australia predominantly from 2007–2013.15
In this study, we combine TB public health surveillance data with genomic sequencing of M. tuberculosis isolates from the tropical ‘Top End’ of the NT during three decades from 1989–2020, to understand the changing epidemiology and identify priority groups and regions for public health initiatives.
2. Methods
2.1. Data sources and case definitions
Data were extracted from the NT Notifiable Diseases System (NDS), a Northern Territory Government database for managing notifiable disease data maintained by the NT Centre for Disease Control (CDC). All cases of culture-confirmed TB in the Top End of the NT between 1st January 1989 and 6th August 2020 were included. It is a legal requirement for diagnostic laboratories to notify the NT CDC when Mycobacterium tuberculosis is isolated, and for the NT CDC to notify the Australian Government of tuberculosis cases; it is therefore unlikely that cases were missed. We determined the demographics, risk factors, clinical presentations, and outcomes of Top End NT TB cases using NT NDS data. Demographic information included age, sex, ethnicity, country of birth, Top End NT region, whether the case was an illegal fisher, whether the case was a maritime arrival in immigration detention under other circumstances (for example, asylum seekers and crew), and whether the case had been evacuated from Timor-Leste as a result of the 1999 conflict. Risk factors included human immunodeficiency virus (HIV) infection and previous identification as a contact during past NT TB contact tracing investigation. Outcomes included TB treatment completion, incomplete TB treatment, death prior to TB treatment completion, and unknown outcome. We placed focus on the final 5 study years because past TB contact data were most complete for recently diagnosed cases and our main aim was to determine current TB control priorities.
2.2. Microbiological testing and genomic sequencing
Mycobacterial culture of samples from patients with suspected TB was done at Territory Pathology (Royal Darwin Hospital, Darwin), and positive cultures were referred to the Victorian Infectious Diseases Reference Laboratory (Doherty Institute, Melbourne) for identification and antimicrobial susceptibility testing. From 1989–2007 antimicrobial susceptibility testing was done using the radiometric BACTEC 460TB system (Becton Dickinson), and from 2007 onwards the BACTEC Mycobacterial Growth Indicator Tube 960 system (Becton Dickinson) was used. Between 1989–2007 DNA was extracted using phenol and chloroform,16 and from 2010 by ethanol precipitation.17 There were no samples available between 2008–2009. Whole genome sequencing was done at the Microbiological Diagnostic Unit Public Health Laboratory (Doherty Institute, Melbourne). Unique dual indexed libraries were prepared using the Nextera XT DNA sample preparation kit (Illumina) and libraries were sequenced on the Illumina NextSeq 500/550 with 150-cycle paired end chemistry as outlined in the manufacturer's protocols.
2.3. Bioinformatic analyses
The Sequence Read Archive (SRA) accessions for Top End NT M. tuberculosis isolates sequenced as part of this study are listed in Supplementary Table 1, and SRA accessions for publicly available global genomes included in the analyses are listed in Supplementary Table 2. Sequence reads were mapped to the H37Rv genome (GenBank accession NC_000962.3) and variants were called using Snippy v4.4.5 (https://github.com/tseemann/snippy) with minimum coverage 10 reads and minimum fraction of variant bases 90%. Repetitive regions as defined by Sekizuka et al were masked from the alignment.18 TBProfiler v2.8.12 was used to assign sublineages.19,20 To determine a pairwise single nucleotide polymorphism (SNP) distance threshold for defining genomic clusters, we determined the distribution of pairwise SNP distances between isolates from cases and their previous pulmonary TB contacts (if belonging to the same sublineage). Based on this, a cut-off of ≤12 SNPs was selected for hierarchical single-linkage clustering, consistent with previous studies.21,22 Maximum likelihood phylogenetic analyses were inferred using IQ-TREE v1.6.12 using a generalised time reversible model with 4 gamma categories, 1,000 ultrafast bootstrap approximation replicates and 1,000 bootstrap approximate likelihood ratio test replicates.23 Trees were visualised and annotated using ggtree.24 TempEst v1.5.1 was used to undertake regression of root-to-tip distance against sampling time to determine whether there was a temporal signal in the sublineage 1.2.1 and 1.2.2 trees.25 We did not proceed with molecular dating due to poor temporal signal within the data.
2.4. Statistical analyses
Statistical analyses were done using R v4.0.2.26 Comparison of demographics, clinical features and outcomes between groups was done using Fisher's exact test. The annual incidence rate ratio for culture-confirmed TB in Australian First Nations peoples was estimated using a negative binomial model, with the number of First Nations cases as outcome, year as predictor, and the log First Nations population as offset. Population numbers were extrapolated from Australian census data. The odds of a sequenced case belonging to a cluster was estimated using a quasi-binomial generalised linear model with annual proportion of sequenced cases belonging to a cluster as outcome, year and number of cases associated with the Timor-Leste evacuation as predictors, and total annual sequenced cases as weights. Years where fewer than 50% of cases were sequenced were excluded from this analysis. For both models, residuals were checked for random distribution across predictors and fitted outcome; there was no temporal autocorrelation and there were no overly influential outliers.
2.5. Ethics and governance
The study was approved by the Human Research Ethics Committee of Menzies School of Health Research (2017-2747). Feedback on the study design and reporting was sought from the Menzies Global and Tropical Health Indigenous Reference Group.
2.6. Role of the funding source
The study funders had no role in study design, data collection, data analysis, interpretation, or writing of the report
3. Results
3.1. Epidemiology
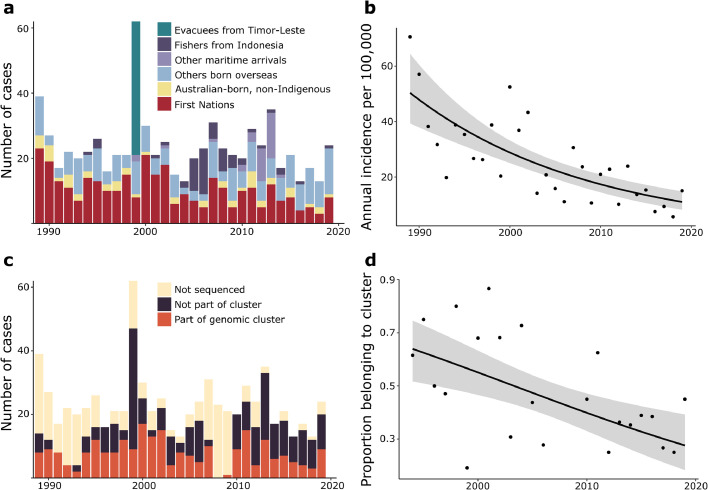
Between 1st January, 1989, and 6th August, 2020, there were 741 first-episode culture-confirmed TB cases in the Top End of the NT; 329 were Australian First Nations people (328 Aboriginal people and 1 Torres Strait Islander person), 52 were Australian-born and non-Indigenous, 359 were born overseas, and one had unknown country of birth (Table 1). The annual incidence of culture-confirmed TB in First Nations peoples in the Top End decreased significantly from 1989–2019 with an estimated annual incidence rate ratio of 0·95 (95% confidence interval [CI] 0·94–0·97, P<0·001); that is, on average the incidence rate decreased by 5% every year (Figure 1a and b). The annual incidence fell from 70.6 per 100,000 population (modeled average 50.4 per 100,000 population) in 1989 to 15.0 per 100,000 population (modeled average 11.0 per 100,000 population) in 2019 (Figure 1b). Among 329 First Nations people, 275 (84%) lived in a remote area outside urban Darwin, there were no cases with HIV infection, there were no cases with a multidrug-resistant M. tuberculosis isolate, and 41 (13%) died prior to completion of TB treatment (Table 1). 31/39 (79%) cases in children ≤14 years of age were First Nations children.
Table 1.
Demographics, clinical features, antimicrobial susceptibility patterns and outcomes of patients with culture-confirmed TB in the Top End of the NT, 1989-2020. P<0.01 for all comparisons between First Nations, overseas-born, and Australian-born non-Indigenous groups, with the exception of P=0.04 for multidrug-resistance.
| First Nations, number (%) | Born overseas*, number (%) | Australian-born, non-Indigenous, number (%) | |
|---|---|---|---|
| Median age in years (range) | 41 (1–79) | 36 (5–90) | 59 (17–80) |
| Age ≤14 years | 31/329 (9·4%) | 8/358 (2·2%) | 0/52 (0%) |
| Male sex | 167/329 (50·8%) | 249/359 (69·4%) | 44/52 (84·6%) |
| Outside urban Darwin area | 275/329 (83·6%) | 76/359 (21·2%) | 10/52 (19·2%) |
| HIV infection | 0/288 (0%) | 6/317 (1·9%) | 3/49 (6·1%) |
| Pulmonary TB | 245/329 (74·5%) | 298/359 (83·0%) | 47/52 (90·4%) |
| Sputum smear positive (if pulmonary TB) | 154/198 (78·3%) | 132/214 (61·7%) | 36/40 (90·0%) |
| Extra-pulmonary TB | 123/329 (37·4%) | 91/359 (25·3%) | 9/52 (17·3%) |
| Lymph node TB | 71/329 (21·6%) | 40/359 (11·1%) | 4/52 (7·7%) |
| Isoniazid resistance without rifampicin resistance | 5/326 (1·5%) | 29/330 (8·8%) | 3/52 (5·8%) |
| Multidrug-resistance | 0/326 (0%) | 5/330 (1·5%) | 1/52 (1·9%) |
| Completed treatment | 265/329 (80·5%) | 191/359 (53·2%) | 43/52 (82·7%) |
| Incomplete treatment | 20/329 (6·1%) | 5/359 (1·4%) | 1/52 (1·9%) |
| Died during treatment | 41/329 (12·5%) | 11/359 (3·1%) | 6/52 (11·5%) |
| Unknown treatment outcome | 3/329 (0·9%) | 152/359 (42·3%) | 2/52 (3·8%) |
| Part of genomic cluster | 209/236 (89%) | 19/229 (8%) | 16/31 (52%) |
| Lineage 1 | 54/236 (22·9%) | 93/229 (40·6%) | 5/31 (16·1%) |
| Lineage 2 | 15/236 (6·4%) | 41/229 (17·9%) | 5/31 (16·1%) |
| Lineage 3 | 0/236 (0%) | 22/229 (9·6%) | 1/31 (3·2%) |
| Lineage 4 | 167/236 (70·8%) | 73/229 (31·9%) | 20/31 (64·5%) |
*Southeast Asia 235; South Asia 32; Europe 25; Pacific 18; East Asia 14; Eurasia 14; South America 2; North America 1.
Fig. 1.
(a) Demographics of culture-confirmed TB cases in the Top End of the NT, 1989–2019. (b) Annual incidence of culture-confirmed TB in Australian First Nations peoples in the Top End. Line and shaded area indicate fitted mean and 95% confidence intervals. (c) Number of culture-confirmed cases of TB sequenced and belonging to clusters, 1989–2019. (d) Proportion of sequenced isolates belonging to a genomic cluster over time. Line and shaded area indicate fitted mean and 95% confidence intervals.
Of the 359 cases born overseas, 235 (66%) were from Southeast Asia with the majority from Indonesia (87 cases), Timor-Leste (61 cases), the Philippines (49 cases) and Vietnam (20 cases). In 1999 there were 41 cases in evacuees from Timor-Leste. There were 57 cases among illegal fishers from Indonesia predominantly between 2004–2009, and 34 cases among other maritime arrivals predominantly asylum seekers and crew between 2012–2013 (Figure 1a). Among people born overseas there were 5 cases of MDR-TB and 29 cases with isoniazid resistance without rifampicin resistance. Treatment outcome was unknown in 152 (42%) overseas-born cases (Table 1), the majority of whom had left the NT. Some fishers, asylum seekers and crew were deported prior to treatment completion, but no cases travelled while sputum was smear-positive for acid-fast bacilli.
Among pulmonary TB cases with a sputum smear microscopy result, greater proportions of First Nations cases (154/198, 78%) and Australian-born non-Indigenous cases (36/40, 90%) were smear-positive at diagnosis than overseas-born cases (132/214, 62%; P<0.001). First Nations cases (41/329, 12%) and Australian-born non-Indigenous cases (6/52, 12%) were more likely to die prior to treatment completion than overseas-born cases (11/359, 3%, P<0.001). However the median age in those who died was lower in First Nations cases (49 years, range 21–79 years) than in Australian-born non-Indigenous cases (63 years, range 53–71 years) or overseas-born cases (median 77 years, range 55–90 years; P<0.001).
3.2. Genomic clusters
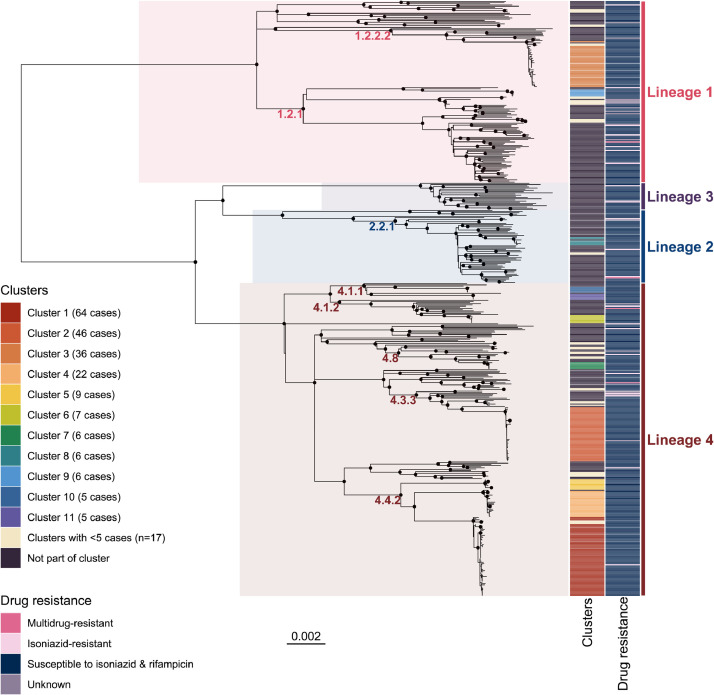
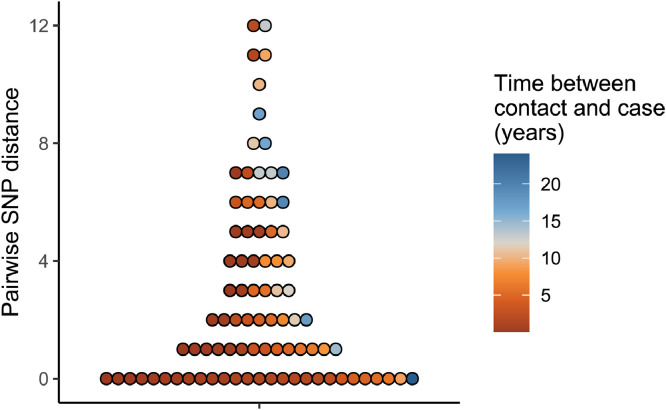
There were 497/741 (67%) first-episode M. tuberculosis isolates successfully sequenced (Figure 1c). Lineage 4 was most common (260 cases, 52·4%), followed by lineage 1 (152 cases, 30·6%), lineage 2 (61 cases, 12·3%) and lineage 3 (23 cases, 4·6%) (Figure 2, Table 1). There were 93 cases that had documented previous contact with a case of pulmonary TB with an M. tuberculosis isolate belonging to the same sublineage. In 85/93 (91%) of such instances the pairwise SNP distance was ≤12 SNPs; these 85 isolate pairs were separated by a median of 2 SNPs (interquartile range 0–5 SNPs) (Figure 3). The remaining 8 isolate pairs were separated by 71, 121, 178, 199, 201, 222, and 225 SNPs. Of 10 cases in the final 5 years of the study with a contact isolate separated by ≤12 SNPs, the contact was diagnosed a median of 6 years earlier (range 4.5 months–24 years earlier), suggesting that both reactivation from latency and recent transmission are contributing to incident TB.
Fig. 2.
Midpoint-rooted maximum likelihood phylogenetic tree of Top End NT M. tuberculosis sequences between 1989–2020. Nodes with approximate likelihood ratio >95 and ultrafast bootstrap >95 are marked with a black circle. Scale bar indicates substitutions/site.
Fig. 3.
Distribution of pairwise SNP distances between cases and previous contacts with pulmonary TB due to M. tuberculosis belonging to the same sublineage. Each circle represents a pairwise SNP distance. Eight outlying datapoints were excluded from the figure; these were pairwise SNP distances of 25, 71, 121, 178, 199, 201, 222, and 225.
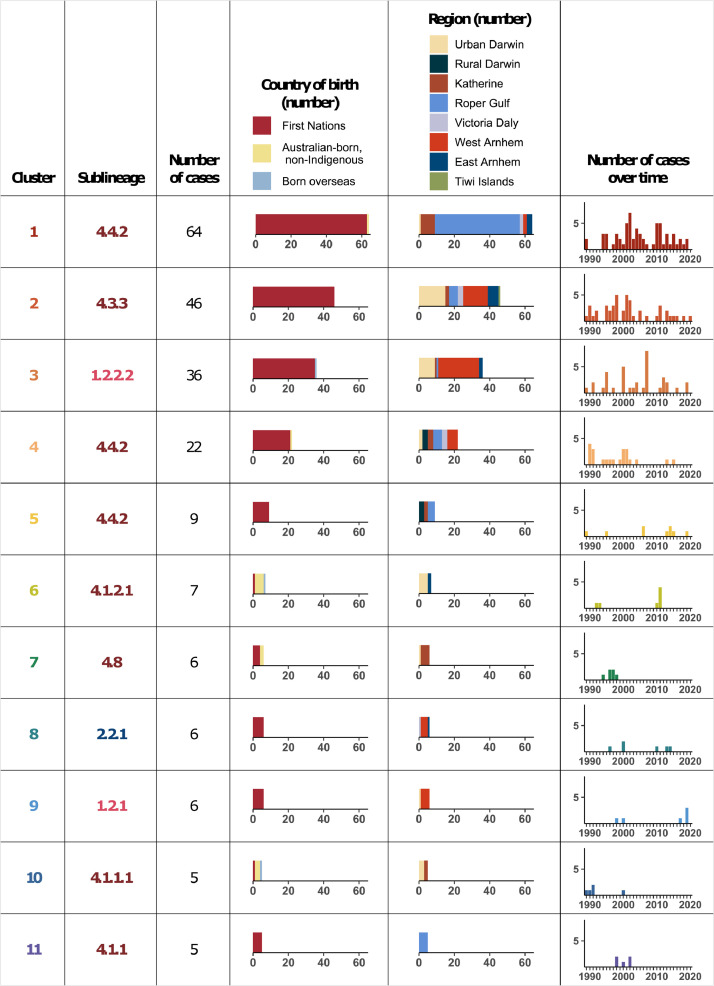
A cut-off of ≤12 SNPs was used for single-linkage hierarchical clustering; 28 genomic clusters including 250 cases were defined based on this, with 11 clusters having ≥5 cases (Figures 2 and 4). No multidrug-resistant or isoniazid-resistant isolates clustered together (Figure 2). The odds of a case belonging to a genomic cluster decreased on average by 6% every year (odds ratio 0·94, 95% CI 0·91–0·97, P=0·002) (Figure 1c and d). Of 250 clustered cases, 214 (86%) were First Nations people, 17 (7%) were Australian-born and non-Indigenous, and 19 (8%) were born overseas. Most clustered cases (189; 76%) were from a remote area. In 5/10 genomic clusters involving an overseas-born case, Australian-born cases within the cluster were diagnosed earlier. Genomic clusters with an overseas-born case as the first case were small; 1 cluster included 5 cases, 1 cluster included 4 cases, and 3 clusters each included 2 cases.
Fig. 4.
Clusters with ≥5 cases separated by ≤12 SNPs in the Top End NT, 1989–2020.
The largest genomic cluster (cluster 1, sublineage 4.4.2) spanned the duration of the study and included 64 cases, 48 (75%) of which were from the remote Roper Gulf region (Figure 4). The three next largest clusters (clusters 2–4) included 46, 36 and 22 cases (Figure 4). The Roper Gulf region had the greatest number of clustered cases (68 cases belonging to 6 clusters) followed by urban Darwin (61 cases belonging to 21 clusters [13 clusters with <5 cases]), and the remote West Arnhem region (56 cases belonging to 8 clusters). Of 19 cases belonging to genomic clusters in the final 5 study years, 12 belonged to the same cluster as a known previous TB contact, 1 belonged to a different genomic cluster to their previous TB contact, and 6 had not been identified as TB contacts as part of previous contact tracing.
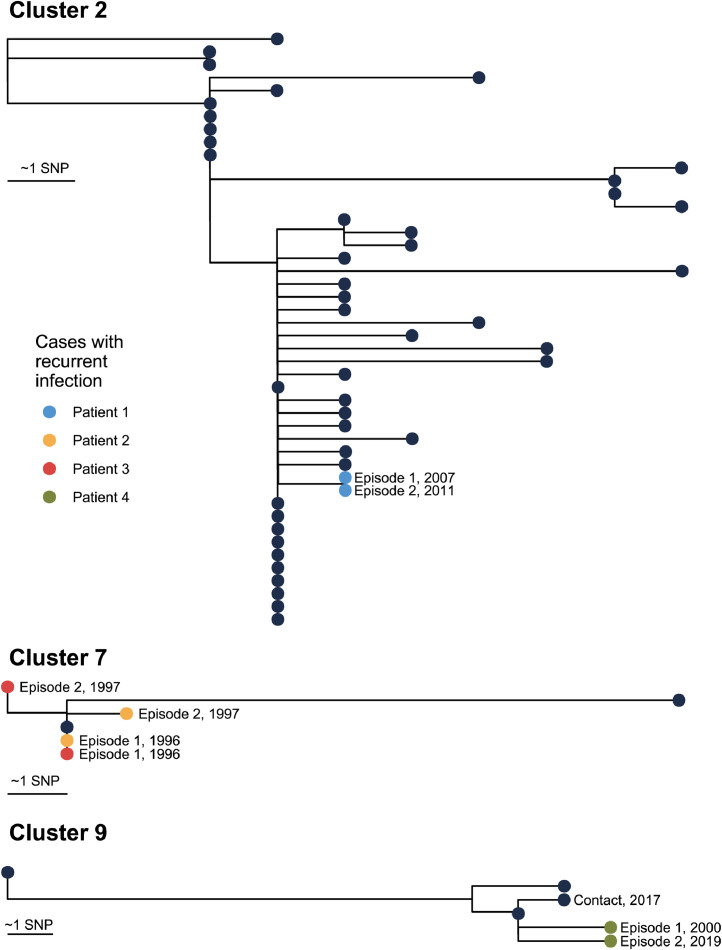
3.3. Recurrent TB cases
Seven cases had a second episode of culture-confirmed TB which occurred 1·2–19·3 years after initial diagnosis; All were prescribed directly observed treatment but two were lost to follow-up prior to treatment completion during their first episode. All initial and recurrent M. tuberculosis isolates were susceptible to all first-line antimicrobials (Table 2). Five recurrent episode isolate pairs were sequenced, and 4/5 pairs belonged to a larger cluster (Table 2, Figure 5). The pairwise SNP distance between initial and recurrent M. tuberculosis isolates was 0–4 SNPs. Based on phylogenetic analyses, in 3/5 cases it was unclear whether recurrent episodes were relapses due to inadequate treatment or reinfection from another case in a cluster (Table 2 and Figure 5). For instance, one case belonging to cluster 9 had an episode of cervical lymph node TB in 2000, exposure to a case of heavily smear-positive pulmonary TB in 2017, and recurrent lymph node TB at the original cervical site in 2019; the initial and recurrence isolates were separated from each other by 4 SNPs, and were both separated from the contact isolate by 3 SNPs (Figure 5).
Table 2.
Cases of recurrent of TB.
| Patient demographics# | First presentation | Time between episodes (years) | Second presentation | Pairwise SNP distance | Genomic cluster* | Relapse or reinfection |
|---|---|---|---|---|---|---|
| 1. 41M, Aboriginal | Pulmonary TB, fully susceptible, lost to follow-up during treatment | 3·9 | Pulmonary TB, fully susceptible, completed treatment | 0 | 2 | Relapse |
| 2. 36F, Aboriginal | Pulmonary TB, fully susceptible, completed treatment | 1·8 | Pulmonary TB, fully susceptible, died of other cause during 2nd treatment course | 1 | 7 | Uncertain |
| 3. 64M, Australian-born, non-Indigenous | Pulmonary TB, fully susceptible, completed treatment | 1·2 | Pulmonary TB, fully susceptible, completed treatment | 0 | 7 | Uncertain |
| 4. 3F, Aboriginal | Lymph node TB, fully susceptible, completed treatment | 19·3 | Lymph node TB, fully susceptible, lost to follow-up during treatment | 4 | 9 | Uncertain |
| 5. 42M, born overseas | Pulmonary TB, fully susceptible, completed treatment | 1·3 | Pulmonary TB, fully susceptible, completed treatment | 3 | Not part of cluster | Relapse |
| 6. 39M, Aboriginal | Pulmonary TB, fully susceptible, lost to follow-up during treatment | 8·8 | Pulmonary TB, fully susceptible, completed treatment | Not sequenced | Not sequenced | Uncertain |
| 7. 24M, Aboriginal | Pulmonary TB, fully susceptible, completed treatment | 10·0 | Pulmonary TB, fully susceptible, completed treatment | Not sequenced | Not sequenced | Uncertain |
Fig. 5.
Maximum likelihood phylogenetic trees of clusters with cases with recurrent TB infection.
3.4. Phylogeography of M. tuberculosis sublineages
Among cases born in Australia lineage 4 was most common (187/267 [70%] cases), while lineage 1 was most common in people born in Southeast Asia (82/145 [57%] cases) including among Indonesian fishers (18/28 [64%] cases) and evacuees from Timor-Leste (14/29 [48%] cases) (Table 1). The genomic clusters predominantly comprising Australian-born cases belonged to a diverse range of lineage 1 and lineage 4 sublineages (Figures 2 and 4).
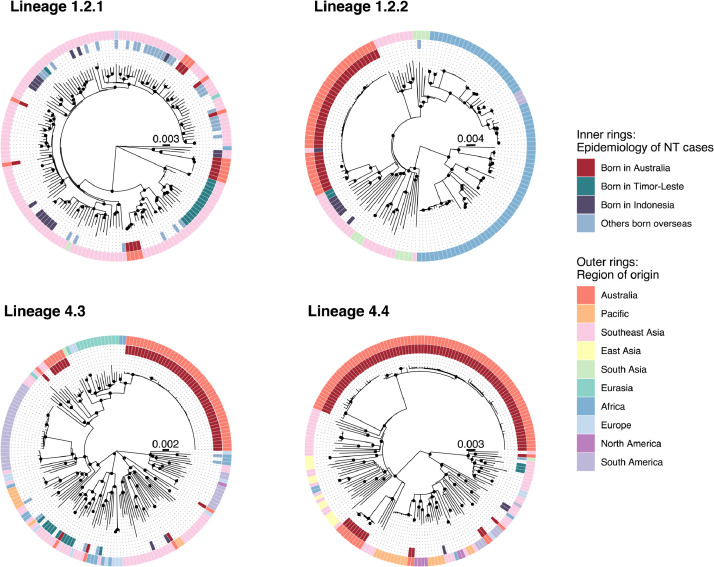
Phylogenetic analyses of the study M. tuberculosis genomes in the context of diverse publicly available global M. tuberculosis genomes demonstrated that two Australian lineage 1 clusters (clusters 3 and 9) belonged to the same clades as isolates from Indonesian fishers and evacuees from Timor-Leste (Figure 6). Cluster 9 (sublineage 1.2.1) was most closely related to two isolates from Indonesian fishers (AUSMDU00010247 and AUSMDU00030071). Cluster 3 (sublineage 1.2.2.2) was most closely related to two isolates from evacuees from Timor-Leste (AUSMDU00044352 and AUSMDU00045785), with isolates from Indonesian fishers also located on this clade (Figure 6). There was poor correlation between sampling time and genetic divergence in the sublineage 1.2.1 and 1.2.2 trees, with correlation coefficients 0·11 and 0·17 respectively; we therefore did not proceed with temporal phylogenetic analyses.
Fig. 6.
Maximum likelihood phylogenetic trees including Top End NT (inner rings) and publicly available global M. tuberculosis sequences (outer rings). “Region of origin” is based on country of birth if known. Nodes with approximate likelihood ratio >95 and ultrafast bootstrap >95 are marked with a black circle. Scale bars indicate substitutions/site.
Australian sublineage 4.3.3 genomes belonging to cluster 2 were located on a clade with public genomes from South Africa and Russia, and Australian sublineage 4.4.2 genomes belonging to clusters 1, 4 and 5 resided on a clade with public genomes from China, Vietnam and Thailand (Figure 6). An Australian sublineage 4.8 cluster (cluster 7) was separated by 40–55 SNPs from an isolate (SRR6040105) from the Torres Strait Protected Zone located between the north-eastern tip of Australia and Papua New Guinea,27 and two Australian sublineage 4.4.1.1 genomes (AUSMDU00009954 and AUSMDU00043809) were separated by 63–71 SNPs from genomes belonging to the New Zealand ‘Rangipo’ strain.28
4. Discussion
In this study we have described the changing epidemiology of TB in the NT Top End over a 31-year period to identify priority areas for public health initiatives. From 1989–2019, the incidence of culture-confirmed TB in Australian First Nations people dropped markedly. Increases in overseas-born cases were noted to be associated with influxes of arrivals from neighbouring countries.[13], [14], [15] Pre-elimination of TB, <1 case per 100,000 population per year, was reached over a decade ago in non-Indigenous Australian-born people.2 However, if a 5% per year incidence reduction continues it is predicted that TB pre-elimination in Top End First Nations peoples will not be reached until ~2066.
Similar changes in TB epidemiology have been noted in Far North Queensland, Australia, where incidence in Aboriginal peoples has fallen and the proportion of cases born overseas has risen.29 Increased case numbers from neighbouring Papua New Guinea have also been noted, with high annual TB incidence (54 per 100,000 population) in Torres Strait Islander peoples. First Nations populations are disproportionately affected by TB in other countries with low TB incidence including New Zealand, Canada, and the United States of America.3 For example, the annual TB incidence in New Zealanders of European descent is 1 per 100,000 population but is 6 times higher in Māori.30 High rates of diabetes and cancers, overcrowded housing, and barriers to accessing healthcare have been highlighted as contributing factors, with genomic sequencing demonstrating ongoing transmission of the dominant ‘Rangipo’ strain over three decades in the Waikato region.31,32
Our analyses suggest that both reactivation from latency and recent transmission with progression are currently contributing to incident TB in the Top End; in the final 5 study years the time interval between cases and epidemiologically- and genomically-linked contacts ranged from 4·5 months to 24 years. 79% of child cases (versus 48% of total cases) were in Aboriginal people, with incident childhood TB also being an indicator of recent community transmission.4 Over three quarters of clustered cases occurred outside urban Darwin with the remote Roper Gulf and West Arnhem regions having the largest numbers of clustered cases. When the findings are considered together, the findings indicate that TB control resources should continue to be directed to TB hotspot regions with focus on timely and complete case detection, contact tracing, and latent TB treatment.
Australian First Nations cases with pulmonary TB were more likely to be smear-positive at diagnosis than overseas-born cases (78% versus 62%, Table 1), and mortality was higher in First Nations patients than overseas-born patients (13% versus 3%) with First Nations TB deaths occurring at a younger age (median 49 versus median 77 years). Late TB presentations resulting in intensive care unit admission and/or death continue in the NT Top End. Such cases may be heavily smear-positive and highly infectious, and transmission may occur over a prolonged period prior to diagnosis. We hypothesise that normalisation of cough in the context of high rates of smoking and chronic lung disease,33 under-recognition of TB by health staff due to relative infrequency of the condition and high staff turnover,34,35 lack of access to radiologic investigations in remote regions, and over-stretched health services with competing priorities could be contributing factors to late diagnoses. These could be addressed by further emphasising TB as an uncommon but serious cause of cough in remote treatment guidelines and as part of community and staff orientation and education sessions. TB patient educational materials are available in some Top End First Nations languages, however evaluation of their effectiveness, expansion to further language groups, and broader community public health messaging should be prioritised.
In our study, 6/19 clustered cases in the final 5 study years had no previous TB contact identified. Social networks in northern Australian regions can be complex with frequent movement between households and communities that can pose a challenge to contact tracing.36,37 Extended community screening is done when there are secondary cases identified through contact tracing or there are ≥2 cases within 12 months in a community, but it is labour-intensive.37 Prospective, timely M. tuberculosis genomic sequencing for tracking TB spread would augment contact tracing by identifying occult transmission.38 Treatment of contacts confirmed to have latent TB infection with short, effective regimens administered by remote clinics would likely improve completion rates.39
Genomic analyses suggested there were very few instances of community transmission arising from overseas-born cases. This supports the current practice of chest radiograph screening and treatment of active TB prior to immigration to Australia, and screening and treatment for latent TB after arrival in Australia.[2] Active TB treatment completion rates were low among overseas-born cases predominantly due to departure from the NT prior to treatment completion. Many irregular maritime arrivals from overseas were repatriated during treatment, and while efforts were made to encourage treatment completion on return home, this was not assured.14
There was no evidence of acquisition of antimicrobial-resistance during treatment and there were very few recurrent TB episodes. This suggests that active TB treatment for patients surviving and remaining in the NT was adequate in almost all cases and provides support for the current directly observed therapy program. It was not possible to differentiate relapse from reinfection in 3/5 sequenced recurrent TB episodes. Previous studies have differentiated relapse from reinfection based on pairwise SNP distances of ≤5–10 SNPs.40,41 However even with the addition of phylogenetic analysis it was not possible to differentiate relapse from reinfection within 3 genomic clusters in our study, reflecting a limitation of genomics when active transmission of a strain with little diversity is occurring in a community. Alignment to a closed reference genome closely related to the cluster genomes and sequencing to a greater depth of coverage could improve resolution.42
There is no evidence to suggest that TB was present in Australia prior to European colonisation in 1788, however it did spread rapidly among Aboriginal peoples in southern Australia soon thereafter.43,44 Lineages 4 and 1 were most common among Australian First Nations peoples in the NT Top End. Lineage 4 is the dominant M. tuberculosis lineage in Europe and is predicted to have expanded globally from there subsequent to colonisation of the Americas, Africa and the Asia-Pacific,19,45 with outbreaks reported in First Nations groups previously reported in New Zealand,28 and Canada.46 Lineage 1 is found predominantly in regions bordering the Indian Ocean and is predicted to have expanded from South Asia to Southeast Asia and East Africa.47,48 The diversity of endemic lineage 4 and lineage 1 M. tuberculosis sublineages in the NT Top End is consistent with multiple introductions from Europe and Asia, compatible with recent history of the region.
Permanent European settlement on Larrakia land at Port Darwin occurred in 1869, with expansion of the Chinese expatriate population soon thereafter.44 TB was likely introduced to the Top End at this time, but it is uncertain whether it was present prior to this. Makassans from present-day Indonesia visited the northern coastline since the 1700s and formed important social and economic ties with Aboriginal peoples. The Makassans travelled annually from the port of Makassar in South Sulawesi to fish and trade for sea cucumbers (trepang) which were sought after for aphrodisiac qualities and traded on return.49 Indonesian fishers from South Sulawesi and surrounding islands have continued to visit northern Australian waters into the present day, including during this study.14 The phylogenetic proximity between some M. tuberculosis isolates from the remote Top End and those from fishers from Indonesia could indicate introduction from Indonesia, however the timing is uncertain.
Our study has three main limitations. Only two thirds of M. tuberculosis isolates during the study period were sequenced and it is likely that there were many cases belonging to genomic clusters that were not sequenced. There was a 2-year period in which no samples were available. Some cultures were contaminated or did not grow on arrival at the reference laboratory, and a minority of cases did not have mycobacterial culture done by the study laboratories. Despite these limitations, it is likely that the subset of sequenced cases is representative. Secondly, we did not account for within-host diversity in our analyses of transmission clusters. Deep sequencing is expensive but can enable detection of mixed infection and onward transmission of M. tuberculosis subpopulations that may be missed using conventional methods and therefore confound analyses of transmission.42,50 Finally, we were unable to make definitive conclusions about the timing and source of TB introductions to the Top End due to inadequate temporal signal in the genomic data.
TB in the Top End of the NT has been shaped by local and regional political policies, and geographic proximity to Southeast Asia. The incidence in Australian First Nations peoples and the proportion of cases belonging to clusters dropped significantly during the study, however rates remain much higher than in Australian-born non-Indigenous people. In addition to TB screening and treatment of those born overseas, priorities in remote Top End hotspot regions comprise community and health staff education to improve the timely diagnosis of active TB, thorough contact tracing augmented by genomic sequencing to find and diagnose TB, and implementation of short, effective latent TB regimens.
Contributors
Ella Meumann, Vicki Krause, Bart Currie, Anna Ralph, Deborah Williamson, and Timothy Stinear conceived and designed the study. Ella Meumann, Kristy Horan, Torsten Seemann, and Koen Vandelannoote planned, performed, and interpreted genomic analyses. Ella Meumann and Mirjam Kaestli planned, performed, and interpreted statistical analyses. Vicki Krause, Belinda Farmer, Maria Globan, Elizabeth Stephenson, Tracy Popple, Rowena Boyd, Christopher Lowbridge, Robert Baird, and Ella Meumann were involved with data acquisition and curation. Ella Meumann prepared the manuscript with contributions from all authors. Ella Meumann, Kristy Horan, and Vicki Krause have accessed and verified the underlying data.
Declaration of Competing Interest
The authors have no conflicts of interest to declare.
Acknowledgments
Acknowledgements
We would like to thank Linda Viberg for assistance with culture and sequencing of M. tuberculosis isolates, Heather D'Antoine and Jane Davies for facilitating presentation to the Indigenous Reference Group, and Mark Mayo for reviewing the manuscript. We are grateful for the valuable feedback from the Menzies Global and Tropical Health Indigenous Reference Group.
This research was funded under the Australian National Health and Medical Research Council (NHMRC) grant number 1131932 (The HOT NORTH initiative), a NHMRC postgraduate scholarship to Ella Meumann (1114696), NHMRC investigator grants to Deborah Williamson (GNT1174555) and Timothy Stinear (GNT1194325), and a NHMRC fellowship to Anna Ralph (1142011). The funding source had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication
Data Availability
Accessions for genomes sequenced as part of this study are listed in Supplementary Table S1.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.lanwpc.2021.100229.
Appendix. Supplementary materials
References
- 1.Bright A, Denholm J, Coulter C, Waring J, Stapledon R. Tuberculosis notifications in Australia, 2015–2018. Commun Dis Intell (2018) 2020:44. doi: 10.33321/cdi.2020.44.88. [DOI] [PubMed] [Google Scholar]
- 2.The National Tuberculosis Advisory Committee for the Communicable Diseases Network Australia. The strategic plan for control of tuberculosis in Australia, 2016–2020: towards disease elimination. Commun Dis Intell. 2018;2019:43. [PubMed] [Google Scholar]
- 3.Tollefson D, Bloss E, Fanning A, Redd JT, Barker K, McCray E. Burden of tuberculosis in Indigenous peoples globally: a systematic review. Int J Tuberc Lung Dis. 2013;17:1139–1150. doi: 10.5588/ijtld.12.0385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teo SS, Tay EL, Douglas P, Krause VL, Graham SM. The epidemiology of tuberculosis in children in Australia, 2003–2012. Med J Aust. 2015;203:440. doi: 10.5694/mja15.00717. [DOI] [PubMed] [Google Scholar]
- 5.Judge D, Krause VL. Multidrug-resistant tuberculosis in the Northern Territory: a 10-year retrospective case series. Commun Dis Intell Q Rep. 2016;40:E334–E339. [PubMed] [Google Scholar]
- 6.The Northern Territory Government. Northern Territory Economy: Population. https://nteconomy.nt.gov.au/population (accessed 10th June, 2021).
- 7.Wyber R, Kelly A, Lee AM. Formative evaluation of a community-based approach to reduce the incidence of Strep A infections and acute rheumatic fever. Aust NZ J Public health. 2021 doi: 10.1111/1753-6405.13127. Online ahead of print. [DOI] [PubMed] [Google Scholar]
- 8.Quilty S, Wood L, Scrimgeour S. Addressing profound disadvantages to improve Indigenous health and reduce hospitalisation: a collaborative community program in remote Northern Territory. Int J Environ Res Public Health. 2019;16 doi: 10.3390/ijerph16224306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pai M, Behr MA, Dowdy D. Tuberculosis. Nat Rev Dis Primers. 2016;2:16076. doi: 10.1038/nrdp.2016.76. [DOI] [PubMed] [Google Scholar]
- 10.Cormier M, Schwartzman K, N'Diaye DS. Proximate determinants of tuberculosis in Indigenous peoples worldwide: a systematic review. Lancet Glob Health. 2019;7:e68–e80. doi: 10.1016/S2214-109X(18)30435-2. [DOI] [PubMed] [Google Scholar]
- 11.Krause VL, Britton WJ. Tuberculosis in the tropics. Med J Aust. 1993;159:412–415. doi: 10.5694/j.1326-5377.1993.tb137920.x. [DOI] [PubMed] [Google Scholar]
- 12.Krause V. Priorities for tuberculosis control in the Northern Territory. Intern Med J. 1991;21:595. [Google Scholar]
- 13.Kelly PM, Scott L, Krause VL. Tuberculosis in East Timorese refugees: implications for health care needs in East Timor. Int J Tuberc Lung Dis. 2002;6:980–987. [PubMed] [Google Scholar]
- 14.Gray NJ, Hansen-Knarhoi M, Krause VL. Tuberculosis in illegal foreign fishermen: whose public health are we protecting? Med J Aust. 2008;188:144–147. doi: 10.5694/j.1326-5377.2008.tb01556.x. [DOI] [PubMed] [Google Scholar]
- 15.Siegel M. Australia Adopts Tough Measures to Curb Asylum Seekers. The New York Times. July 19, 2013. https://www.nytimes.com/2013/07/20/world/asia/australia-adopts-tough-measures-to-curb-asylum-seekers.html (accessed 25th April, 2021).
- 16.Ross BC, Raios K, Jackson K, Sievers A, Dwyer B. Differentiation of Mycobacterium tuberculosis strains by use of a nonradioactive Southern blot hybridization method. J Infect Dis. 1991;163:904–907. doi: 10.1093/infdis/163.4.904. [DOI] [PubMed] [Google Scholar]
- 17.Votintseva AA, Pankhurst LJ, Anson LW. Mycobacterial DNA extraction for whole-genome sequencing from early positive liquid (MGIT) cultures. J Clin Microbiol. 2015;53:1137–1143. doi: 10.1128/JCM.03073-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sekizuka T, Yamashita A, Murase Y. TGS-TB: Total genotyping solution for Mycobacterium tuberculosis using short-read whole-genome sequencing. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Napier G, Campino S, Merid Y. Robust barcoding and identification of Mycobacterium tuberculosis lineages for epidemiological and clinical studies. Genome Med. 2020;12:114. doi: 10.1186/s13073-020-00817-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Phelan JE, O’Sullivan DM, Machado D. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med. 2019;11:41. doi: 10.1186/s13073-019-0650-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meehan CJ, Moris P, Kohl TA. The relationship between transmission time and clustering methods in Mycobacterium tuberculosis epidemiology. EBioMedicine. 2018;37:410–416. doi: 10.1016/j.ebiom.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Walker TM, Ip CL, Harrell RH. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis. 2013;13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen LT, Schmidt HA, von, Haeseler A. Minh BQ. IQ–TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu G, Lam TT, Zhu H, Guan Y. Two methods for mapping and visualizing associated data on phylogeny using ggtree. Mol Biol Evol. 2018;35:3041–3043. doi: 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rambaut A, Lam TT, Max Carvalho L, Pybus OG. Exploring the temporal structure of heterochronous sequences using TempEst (formerly Path-O-Gen) Virus Evol. 2016;2:vew007. doi: 10.1093/ve/vew007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R Core Team. R: A Language and Environment for Statistical Computing. https://www.R-project.org/ 2021
- 27.Bainomugisa A, Pandey S, Donnan E. Cross-border movement of highly drug-resistant Mycobacterium tuberculosis from Papua New Guinea to Australia through Torres Strait Protected Zone, 2010–2015. Emerg Infect Dis. 2019;25:406–415. doi: 10.3201/eid2503.181003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mulholland CV, Shockey AC, Aung HL. Dispersal of Mycobacterium tuberculosis driven by historical European trade in the South Pacific. Front Microbiol. 2019;10:2778. doi: 10.3389/fmicb.2019.02778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson M, Weston J, Mullen A, Knight T, Simpson G. Tuberculosis in Far North Queensland, Australia: a retrospective clinical audit. Intern Med J. 2019;49:333–338. doi: 10.1111/imj.13994. [DOI] [PubMed] [Google Scholar]
- 30.Aung HL, Devine TJ. Reducing the burden of tuberculosis in the Maori, the Indigenous people of New Zealand. Lancet Glob Health. 2019;7:e845. doi: 10.1016/S2214-109X(19)30236-0. [DOI] [PubMed] [Google Scholar]
- 31.Aung HL, Devine TJ, Mulholland CV, Arcus VL, Cook GM. Tackling tuberculosis in the Indigenous people of New Zealand. Lancet Public Health. 2019;4:e496. doi: 10.1016/S2468-2667(19)30180-X. [DOI] [PubMed] [Google Scholar]
- 32.Verrall AJ, Hill PC, Thorburn D. Towards elimination of tuberculosis in New Zealand. N Z Med J. 2020;133:89–96. [PubMed] [Google Scholar]
- 33.D'Sylva P, Walker R, Lane M, Chang AB, Schultz A. Chronic wet cough in Aboriginal children: it's not just a cough. J Paediatr Child Health. 2019;55:833–843. doi: 10.1111/jpc.14305. [DOI] [PubMed] [Google Scholar]
- 34.Wakerman J, Humphreys J, Russell D. Remote health workforce turnover and retention: what are the policy and practice priorities? Hum Resour Health. 2019;17:99. doi: 10.1186/s12960-019-0432-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosewarne C, Boffa J. An analysis of the Primary Health Care Access Program in the Northern Territory: a major Aboriginal health policy reform. Aust J Primary Health. 2004;10:12. [Google Scholar]
- 36.Devlin S, Passmore E. Ongoing transmission of tuberculosis in Aboriginal communities in NSW. N S W Public Health Bull. 2013;24:38–42. doi: 10.1071/NB12113. [DOI] [PubMed] [Google Scholar]
- 37.Krause VL, Cooper M, Bullen J, DiFrancesco M. TB Community Screening - a new TB threat promotes community action. The Northern Territory Disease Control Bulletin. 2002;9:3–5. [Google Scholar]
- 38.Gardy JL, Johnston JC, Ho Sui SJ. Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med. 2011;364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 39.Sterling TR, Villarino ME, Borisov AS. Three months of rifapentine and isoniazid for latent tuberculosis infection. N Engl J Med. 2011;365:2155–2166. doi: 10.1056/NEJMoa1104875. [DOI] [PubMed] [Google Scholar]
- 40.Bryant JM, Harris SR, Parkhill J. Whole-genome sequencing to establish relapse or re-infection with Mycobacterium tuberculosis: a retrospective observational study. Lancet Respir Med. 2013;1:786–792. doi: 10.1016/S2213-2600(13)70231-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guerra-Assuncao JA, Houben RM, Crampin AC. Recurrence due to relapse or reinfection with Mycobacterium tuberculosis: a whole-genome sequencing approach in a large, population-based cohort with a high HIV infection prevalence and active follow–up. J Infect Dis. 2015;211:1154–1163. doi: 10.1093/infdis/jiu574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee RS, Proulx JF, McIntosh F, Behr MA, Hanage WP. Previously undetected super-spreading of Mycobacterium tuberculosis revealed by deep sequencing. Elife. 2020;9 doi: 10.7554/eLife.53245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devlin S, MacLaren D, Massey PD, Widders R, Judd JA. The missing voices of Indigenous Australians in the social, cultural and historical experiences of tuberculosis: a systematic and integrative review. BMJ Glob Health. 2019;4 doi: 10.1136/bmjgh-2019-001794. e001794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Proust AJ. Brolga Press; New Zealand and Papua New Guinea. Curtin, Australia: 1991. History of tuberculosis in Australia. [Google Scholar]
- 45.Brynildsrud OB, Pepperell CS, Suffys P. Global expansion of Mycobacterium tuberculosis lineage 4 shaped by colonial migration and local adaptation. Sci Adv. 2018;4:eaat5869. doi: 10.1126/sciadv.aat5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee RS, Radomski N, Proulx JF. Reemergence and amplification of tuberculosis in the Canadian arctic. J Infect Dis. 2015;211:1905–1914. doi: 10.1093/infdis/jiv011. [DOI] [PubMed] [Google Scholar]
- 47.Menardo F, Rutaihwa LK, Zwyer M. Local adaptation in populations of Mycobacterium tuberculosis endemic to the Indian Ocean Rim. F1000Res. 2021;10:60. doi: 10.12688/f1000research.28318.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Neill MB, Shockey A, Zarley A. Lineage specific histories of Mycobacterium tuberculosis dispersal in Africa and Eurasia. Mol Ecol. 2019;28:3241–3256. doi: 10.1111/mec.15120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clark MA, May SK. ANU E Press; Canberra, Australia: 2013. Macassan history and heritage: journeys, encounters and influences. [Google Scholar]
- 50.Seraphin MN, Norman A, Rasmussen EM. Direct transmission of within-host Mycobacterium tuberculosis diversity to secondary cases can lead to variable between-host heterogeneity without de novo mutation: A genomic investigation. EBioMedicine. 2019;47:293–300. doi: 10.1016/j.ebiom.2019.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accessions for genomes sequenced as part of this study are listed in Supplementary Table S1.