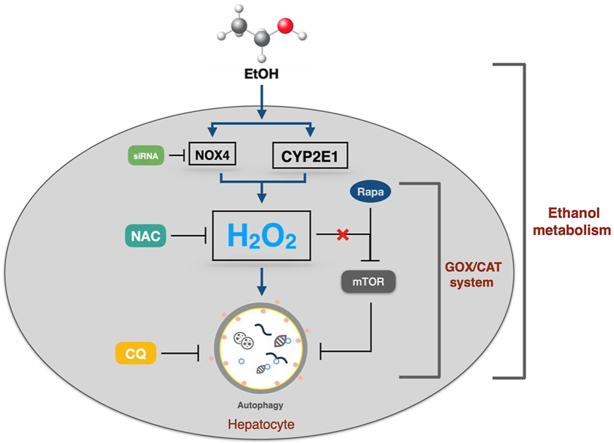
Abstract
Background
Alcoholic liver disease (ALD) is the most common liver disease worldwide and its underlying molecular mechanisms are still poorly understood. Moreover, conflicting data have been reported on potentially protective autophagy, the exact role of ethanol-metabolizing enzymes and ROS.
Methods
Expression of LC3B, CYP2E1, and NOX4 was studied in a mouse model of acute ethanol exposure by immunoblotting and immunohistochemistry. Autophagy was further studied in primary mouse hepatocytes and huh7 cells in response to ethanol and its major intermediator acetaldehyde. Experiments were carried out in cells overexpressing CYP2E1 and knock down of NOX4 using siRNA. The response to external H2O2 was studied by using the GOX/CAT system. Autophagic flux was monitored using the mRFP-GFP-LC3 plasmid, while rapamycin and chloroquine served as positive and negative controls.
Results
Acute ethanol exposure of mice over 24 h significantly induced autophagy as measured by LC3B expression but also induced the ROS-generating CYP2E1 and NOX4 enzymes. Notably, ethanol but not its downstream metabolite acetaldehyde induced autophagy in primary mouse hepatocytes. In contrast, autophagy could only be induced in huh7 cells in the presence of overexpressed CYP2E1. In addition, overexpression of NOX4 also significantly increased autophagy, which could be blocked by siRNA mediated knock down. The antioxidant N-acetylcysteine (NAC) also efficiently blocked CYP2E1-and NOX4-mediated induction of autophagy. Finally, specific and non-toxic production of H2O2 by the GOX/CAT system as evidenced by elevated peroxiredoxin (Prx-2) also induced LC3B which was efficiently blocked by NAC. H2O2 strongly increased the autophagic flux as measured by mRFP-GFP-LC3 plasmid.
Conclusion
We here provide evidence that short-term ethanol exposure induces autophagy in hepatocytes both in vivo and in vitro through the generation of ROS. These data suggest that suppression of autophagy by ethanol is most likely due to longer alcohol exposure during chronic alcohol consumption with the accumulation of e.g. misfolded proteins.
Keywords: Alcohol liver disease (ALD), Ethanol metabolism, Cytochrome P450 2E1(CYP2E1), NADPH oxidase (NOX), Hydrogen peroxide (H2O2), Reactive oxygen species (ROS)
Abbreviations: LC3B, microtubule-associated protein light chain 3B; ALD, alcoholic liver disease; ethanol, ethanol; ROS, reactive oxygen species; H2O2, hydrogen peroxide; NAC, N-acetyl cysteine; Prx2, peroxiredoxin 2; GOX, glucose oxidase; CAT, catalase; CQ, chloroquine; Rapa, rapamycin; mRFP, monomeric red fluorescent protein; GFP, green fluorescent protein; CYP2E1, cytochrome P450 2E1; NOX, NADPH oxidase; mTOR, mammalian target of rapamycin
Graphical abstract
Highlights
-
•
Generation of ROS is the major mechanism of autophagy activation during ethanol exposure in acute alcoholic liver disease (ALD).
-
•
Non-toxic H2O2 significantly induces autophagy without the engagement of mTOR suppression.
-
•
Important ROS-generating enzymes CYP2E1 and NOX4 are strongly induced in a mice model of acute alcohol exposure.
1. Introduction
Alcoholic liver disease (ALD), due to excess alcohol consumption is a major health problem worldwide [1]. ALD spans from hepatic steatosis over inflammation to fibrosis and typically develops over 10–15 years of heavy drinking. Most critical and life-threatening end points are decompensated alcoholic liver cirrhosis and the rare and clinically defined alcoholic hepatitis (AH) which typically affects younger patients with a shorter drinking history [1]. The exact underlying mechanisms for ALD and specifically for AH, however, are still incompletely understood, and alcohol abstinence remains the most effective treatment option in drinkers during all disease stages. Many pathophysiological pathways have been unraveled in the past four decades and the formation of acetaldehyde and reactive oxygen species (ROS) are considered a cornerstone for ethanol-mediated liver damage that early on causes mitochondrial damage [2]. The role of ROS is further underlined by the fact that, apart from steroids [3], the antioxidant N-acetylcysteine (NAC) has proven effectivity for the treatment of AH in a randomized controlled trial [4].
Recently, evidence has been provided that autophagy plays an important role in the pathology of ALD [5], next to well established hallmarks such as liver necrosis and apoptosis [1]. It is long known that liver enlargement in ALD arises both from the accumulation of proteins and lipids [6]. Similar to hepatic steatosis, protein accumulation during alcohol metabolism is due to protein modifications by reactive molecules generated from ethanol metabolism such as ROS [2]. Autophagy (formerly also known as macro-autophagy) is an evolutionarily conserved catabolic process in response to stress to eliminate proteins and cytoplasmic debris through lysosomal degradation [7]. Autophagy is vital in developmental biology [8] and in immunity against microbial infections [9]. It is also associated with a variety of pathological conditions in humans but starvation or ischemia are among the best understood conditions that lead to autophagy [10].
Based on studies on starvation-mediated autophagy, it has been shown that the mechanistic target of rapamycin (mTOR), a serine/threonine-specific protein kinase that belongs to the family of phosphatidylinositol-3 kinase (PI3K) related kinases (PIKKs) plays a central role in suppressing autophagy. Autophagosome generation, autophagosome-lysosomal fusion (autolysosome) and degradation are controlled by the evolutionary conserved autophagy-related (Atg) gene family [11,12]. To date, among known Atg-encoded proteins, only microtubule-associated protein light chain 3 (LC3) is known to exist in all types of autophagic membranes [13,14]. Nascent LC3 (proLC3) is cleaved by Atg4-family proteins into LC3-Ⅰ immediately after synthesis, and cleaved LC3-Ⅰ is further conjugated to phosphatidylethanolamine to become autophagosomal-membrane bound LC3-Ⅱ [14]. The lipid modified form of LC3, referred to as LC3-II, is believed to be involved in autophagosome membrane expansion and fusion events. To some extent, the expression of LC3 (LC3-Ⅱ/LC3-Ⅰ) turnover ratio can be used to precisely reflect the activated autophagy. Among the four isoforms of LC3, LC3B protein is widely used as biomarker for monitoring autophagy [[15], [16], [17]]. Nevertheless, the exact role of LC3 in the autophagic pathway still requires further clarification [18].
Similar to ethanol-mediated suppression of apoptosis and regeneration [19], it is generally believed that ethanol blocks autophagy [20] and numerous studies indicate that pharmacological or genetic inhibition of autophagy greatly enhances cell death [[21], [22], [23]]. Thus, in the absence of autophagy, cell death is significantly increased. Several mechanisms have been proposed to mediate the suppression of autophagy by ethanol including elevation of lysosomal pH with subsequent decreased proteolytic capacity or lowered cathepsin content by disrupted trafficking of these enzymes to lysosomes [10]. It has been generally thought that ethanol oxidation enhances autophagosome content by suppressed mTOR activity through CYP2E1 metabolites such as acetaldehyde or other reactive metabolites [24]. Moreover, ethanol is not able to induce autophagy in HepG2 cells due to the absence of functional CYP2E1 [25]. Ethanol is also known to inhibit the AMP kinase, an important mTOR suppressor and regulator of catabolic pathways. Finally, ethanol disrupts functional microtubule cytoskeleton in hepatocytes which is required for intracellular movement of autophagic cargo [26].
Conflicting results, however, have been reported from various laboratories regarding the role of autophagy during ethanol exposure [24,[27], [28], [29], [30], [31], [32]]. This may be due to the rather heterogenous molecular and cellular response of liver tissue to ethanol and its modulation by genetic and non-genetic factors. Nevertheless, it remains poorly understood at which step and by which ethanol modulates autophagy. In mice, acute gavage of ethanol caused ca. 35% increase of LC3-II protein after 12 h and this increased autophagosome content is found in parallel with indirect signs of enhanced oxidative stress such as reduced glutathione levels and increased lipid peroxides [27]. In addition, in livers of transgenic GFP-LC3 mice, an increased number of GFP-LC3 puncta as indicator for an enrichment of autophagosomes in the LAMP1-positive heavy membrane, lysosome containing fraction has been observed [5,33] and, likewise, electron microscopic studies also demonstrated an accumulation of autophagosomes in response to ethanol which could be blocked by knockdown of Atg7 [24]. Ding et al. demonstrated that autophagy protected cells from the toxic effects of ethanol in livers of mice [24]. Findings from another detailed study suggest that the effect of acute ethanol gavage on hepatic autophagy differs significantly from that after chronic ethanol feeding [27]. Similar, seemingly paradox findings have been recently reported on the central iron master switch hepcidin [34].
At lower physiological levels, ROS and namely H2O2 can act as a classical intracellular signaling molecule regulating kinase-driven targets involved in proliferation, migration, survival, and autophagy [35,36]. The specific role of H2O2 in autophagy, however, is poorly understood. There are recent observations of spontaneous H2O2 release during autophagy induced by rapamycin and H2O2 signaling may be part of the autophagic process [37]. More complex, H2O2 can induce upstream proteins of autophagy but inhibited the autophagic flux [38]. The important but still poorly understood role of H2O2 in cell death or regeneration further complicates autophagy studies [39]. Moreover, ROS studies are often prone to artificial conditions. For instance, to explore the physiological role of H2O2, bolus treatments are commonly used which do not mimic the typical continuous release during (patho)physiological processes [40]. In addition, far too high H2O2 concentrations (>50 μM) are generally used as compared to the in vivo situation [40,41]. Finally, the exact role of how ROS are generated during ethanol metabolism is also complex and partly not clear. Thus, hepatic CYP2E1 which is strongly induced during the ethanol metabolism in liver, has been shown to generate ROS and enhance the progression of liver disease and cancer in ALD [42,43]. Likewise, NADPH-dependent oxidases (NOX), namely liver-expressed NOX4 also play a crucial role in the process of liver injury [1,44] and preliminary immunochemistry data indicate that NOX4 expression is increased in hepatocytes [34]. Moreover, in cardiomyocytes, NOX4 which directly produces H2O2 was shown to trigger autophagy [45].
Therefore, we here study in detail the role of ROS, namely H2O2, during ethanol metabolism and its effect on hepatocellular autophagy. We first confirm that in vivo markers of autophagy and the two important ROS-generating enzymes CYP2E1 and NOX4 are induced in a mice model of acute ethanol exposure. We also demonstrate that ethanol only induces autophagy in hepatocytes in the presence of functional CYP2E1. In addition, both overexpression of NOX4 or exposure to external H2O2 using the enzymatic glucose oxidase/catalase (GOX/CAT) system induce autophagy. In conclusion, our data indicate that ethanol metabolism efficiently modulates autophagy through the generation of ROS and that low nontoxic levels of H2O2 both from intra- and extracellular sources can induce hepatocyte autophagy.
2. Materials and methods
2.1. Animal experiment
All animals received humane care according to the rules of the local committee for Animal Welfare of the Regierungspräsidium Baden-Württemberg. 14 male C57BL/6 mice were randomly divided into control and ethanol treatment group. ethanol binge was conducted in the ethanol treatment group 12 h before the sacrificed, and each mouse was shortly given 33% (vol/vol) ethanol at a total accumulative dosage of 4.5 g/kg body weight by gavages. Control mice received the same energy contained maltodextrin solution. For autophagy inhibition, CQ (100 mg/kg) [46,47] was given intraperitoneally to one mouse in each group 30 min before the administration of ethanol.
2.2. Cell culture
Huh7 cells from the Japanese Cancer Research Resources Bank (JCRB, Tokyo, Japan) were grown under standard conditions using Dulbecco's modified Eagle medium (Sigma-Aldrich), 4500 mg/L glucose, and 10% fetal calf serum at 37 °C under 5% CO2 [48]. Murine primary hepatocytes kindly provided by Dr. Sai Wang (University of Heidelberg, Germany) were grown under standard conditions (1*105/well in 12-well plate) using Williams' medium (Sigma-Aldrich, Taufkirchen, Germany), 10% fetal bovine serum, 1% P/S (Penicillin and Streptomycin), 1% l-Glutamine, 0.5% ITS (Insulin-Transferrin-Selenium), 0.1% Dexamethasone. ethanol (50/100 mmol/L), acetaldehyde (100 μmol/L), and the following chemicals were applied as indicated in the figure legends: Rapa (20 nmol/L), CQ (20 μmol/L), NAC (10 mmol/L). Considering the high evaporation ability of acetaldehyde, we refreshed the medium every 6 h.
2.3. Reagents
The following reagents were used in this study: Glucose Oxidase (GOX, G0543), Catalase (CAT, C3155), Rapamycin (Rapa, R8781), Chloroquine (CQ, C6628), N-Acetyl-l-Cysteine (NAC, A8199), Methanethiosulfonate (MMTS, 64306), ethanol (ethanol, 32205-M), and Acetaldehyde solution (W200379) were purchased from Sigma-Aldrich.
2.4. Exposure of cells to steady-state H2O2
Steady-state H2O2 treatment was performed using the glucose oxidase and catalase system (GOX/CAT system) as described previously [40,49]. Briefly, to avoid starvation induced autophagy, only 5*104 Huh7 cells were seeded with 10% FCS medium in 12-well plate for 24 h, following with 24 h of H2O2 incubation controlled by GOX/CAT system. The GOX was always kept in dilution of 1:1600,000 in PBS, and CAT was diluted to 1: 300,000 in PBS.
2.5. Transfection experiments
Huh7 cells (5.5*104/well in 12-well plate) were transfected with mRFP-GFP-LC3 plasmid, CYP2E1 plasmid, or/and NOX4 plasmid constructs using 3 μl Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. For the construction of mRFP-GFP-LC3 plasmid, mRFP cDNA was exogenously added with restriction site of NheⅠ and AgeⅠto its 5′ end and 3′ end respectively, and removed of the termination codon by PCR. The PCR product was digested by each restriction enzyme and inserted into the NheⅠ-AgeⅠ site of pEGF-mRFP plasmid. CYP2E1 cDNA was cloned into the XhoⅠ and MluⅠ restriction sites of a pCI-neo vector, resulting in antisense orientation with respect to the cytomegalovirus (CMV) promoter. mRFP-GFP-LC3 or CYP2E1 or/and NOX4 plasmid [49] construct transfections were carried out for 24 h, after 6 h transfection, the medium was refreshed. GFP plasmid was performed as a control. For H2O2 and ethanol incubation, the transfected cells were kept under different treatments for additional 24 h.
2.6. RNA silencing
Huh7 cells (5.5*104/well in 12-well plate) were transfected using 3 μl Lipofectamine 2000 and 10 nM siRNA against NOX4 [49]. NOX4 was purchased from Ambion (Foster City, CA, USA). Equimolar concentrations of universal negative siRNA (Sigma-Aldrich, Taufkirchen, Germany) were used as control.
2.7. Immunoblotting
Cells were washed in ice-cold 1 × PBS and harvested in RIPA buffer plus 1 × Complete ® protease inhibitor with EDTA (Roche Applied Sciences) on ice. Frozen liver tissue (about 10 mg) was homogenized in 200 μl of RIPA buffer using a tight-fitting plastic homogenizer on ice. For Prx2 Western blotting, the cells were incubated with the thiol-blocking agent MMTS at 80 mM prepared in ice-cold PBS for 10 min before harvesting, to avoid lysis-induced oxidation of Prx2 [41]. Prx2 samples were subjected to non-reducing SDS-PAGE, and the Western Blotting was performed as described previously [48]. Equal protein loading was confirmed by protein staining with Ponceau-S solution as well as β-actin/GAPDH. Primary and secondary antibodies are listed in Suppl. Table 1. Immune-reactive bands were detected by chemiluminescence (Rotilumin, Carl Roth, Karlsruhe, Germany) and exposed to autoradiography film or after incubation with fluorescent secondary antibody. The membranes were scanned using an infrared imaging system (Odyssey CLx; LI-COR, Inc., Lincoln, NE, USA). Band intensities were quantified using ImageJ for further statistical analysis.
2.8. RNA isolation, cDNA synthesis, and real-time quantitative PCR analysis
RNA was isolated with Trifast (Peqlab Biotechnology GmbH, Erlangen, Germany) according to the manufacturer specifications. Reverse transcription and the real-time quantitative PCR reactions were performed as previously described [49]. Primers and probes were designed using the Probefinder software (Roche, Mannheim, Germany), and the sequences are provided in Suppl. Table 2
2.9. Fluorescence microscopy
Cells expressing fluorescent proteins were fixed for 15 min with 4% PFA, washed, and embedded directly with mountant solution (ThermoFisher, Lot: 2181004) containing DAPI for staining of nuclei. After covered with cover slips overnight, cells were imaged using a 60× oil immersion objective mounted on an Olympus B×41 equipped with a F-view II CCD camera controlled by the cell^D software.
2.10. Immunohistochemistry and immunofluorescence staining
Paraffin-embedded liver tissues were prepared from sacrificed mice. Briefly, after dewaxing, liver sections were submitted to antigen retrieval at 99 °C for 25 min in a Tris-EDTA (10 mM Tris, 1 mM EDTA, pH 9) solution. After cooling and washing with PBS (3 × 5 min), the samples were blocked for 5 min using a blocking solution (ZUC007; Zytomed Systems), washed with PBS, and incubated overnight at 4 °C with the primary antibody diluted in antibody diluent (ZUC025) 1:200. Following washing in PBS (3 × 5 min), the samples were incubated with ZytoChem Plus (AP) Polymer anti-Rabbit (ZUC031) for 30 min and shortly washed with PBS after incubation. Liver sections were finally incubated with ImmPACT® Vector® Red AP Substrate (SK-5105; Vector Laboratories) for 10 min, washed with PBS to stop the reaction, and counterstained with hematoxylin for 5 min. After washing and clearing with xylene, the samples were mounted and further visualized using a Nikon Ni-E microscope equipped with a DS-Ri2 color camera (Nikon, Minato, Japan). Immunofluorescence staining was conducted to analyze the expression of NOX4. In brief, the frozen liver sections were incubated with NOX4 (1:100 dilution) antibody overnight at 4 °C. After washing three times with PBS, the slides were incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG secondary antibody for 1 h in the dark. Finally, the slides were rinsed with PBS and analyzed under a fluorescent microscope (Olympus FV500; Olympus, Tokyo, Japan). DAPI (Sigma, St Loise, MO, USA) was used as counterstain.
2.11. Statistical analysis
Statistical analysis was performed using GraphPad Prism version 5.0 (GraphPad Software, La Jolla, CA). Comparisons between two groups were performed by using Student's t-test. All the data are expressed as mean ± SD. A value of p < 0.05 was considered statistically significant.
3. Results
Ethanol induces CYP2E1, NOX4, and autophagy in a mouse model of acute ethanol exposure.
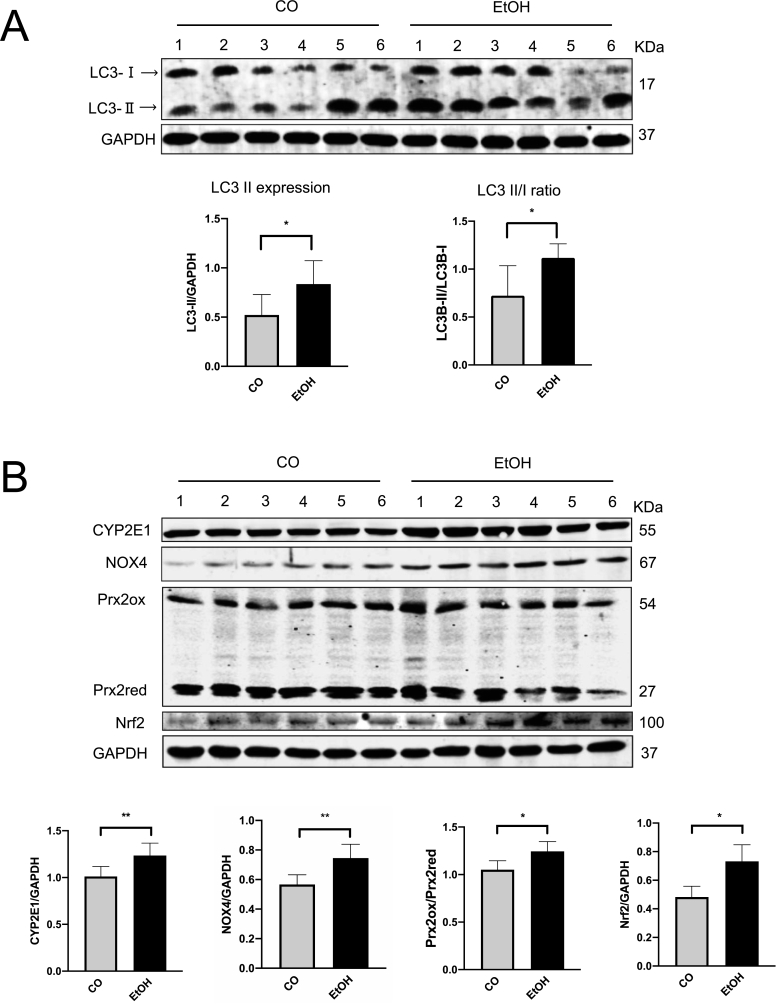
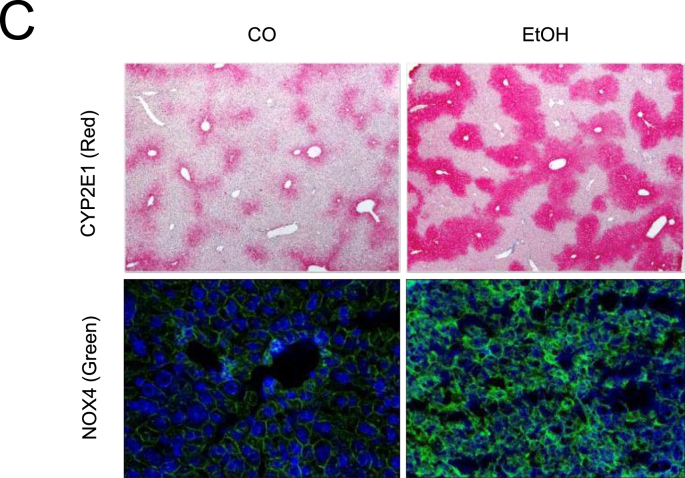
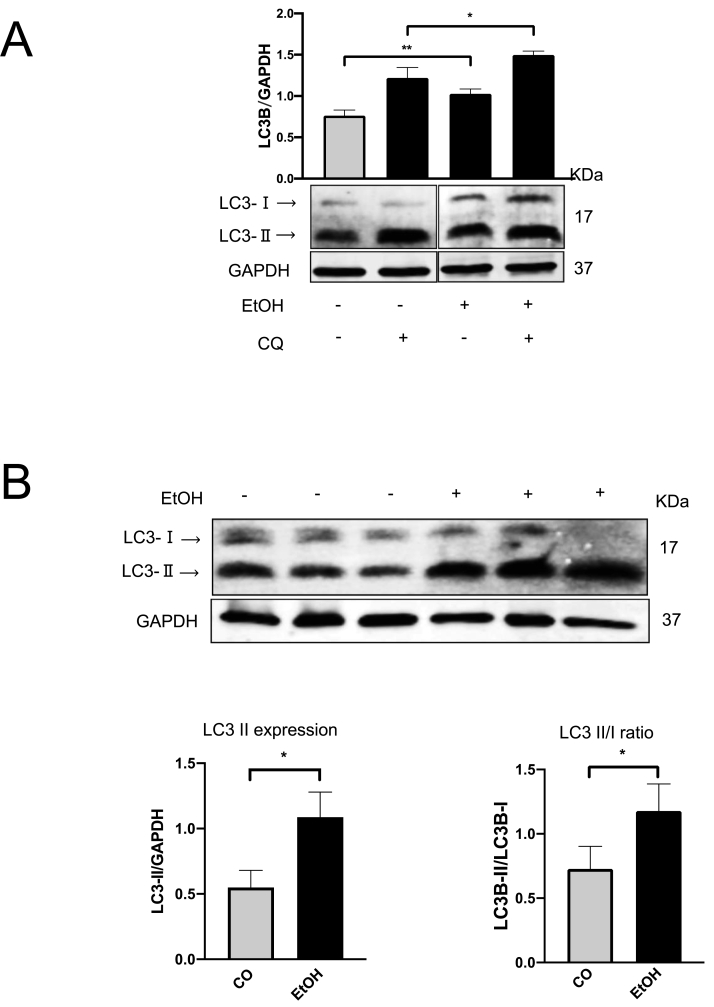
To confirm whether autophagy could be regulated during acute alcohol exposure, mice were treated by binge ethanol using gavage. Expression of LC3-Ⅱ and conversion to LC3-Ⅰ were used to assess autophagy. As shown in Fig. 1A, acute ethanol exposure significantly increased hepatic LC3-Ⅱ expression as well as LC3B turnover ratio (LC3-Ⅱ/LC3-Ⅰ). Ethanol also strongly induced CYP2E1 and NOX4 expression in ALD mice liver (Fig. 1B). Spearman Rho correlation between densitometric CYP2E1 and NOX4 expression levels showed a significant positive association (r = 0.692, p < 0.05) (not shown). Prx2 and nuclear factor erythroid-derived 2-like 2 (Nrf-2) [50,51], a transcription factor that plays a key role in the activation of cellular antioxidant enzymes in response to oxidative stress [52,53], were also significantly induced by ethanol suggesting elevated ROS, especially H2O2 generation existed in mice liver tissue (Fig. 1B). Nrf2 downstream target heme-oxygenase-1 (HO-1) was also upregulated (Suppl. Fig. 1A). Of note, both mTOR activation, related major upstream AMP kinase (Suppl. Fig. 1B) were not changed under these conditions. In addition, in line with previous data [54], CYP2E1 was drastically induced in the centrilobular area of liver tissues (Fig. 1C). NOX4 expression was also strongly induced (Fig. 1C). To confirm whether increased LC3B expression is due to activated autophagic flux and to monitor autophagic flux, Chloroquine (CQ) was intraperitoneally injected into one mouse 30 min before the ethanol gavage. Since autophagy is blocked by CQ, any elevated autophagy flux will result in further accumulation of total LC3B expression [24]. As shown in Fig. 2A, CQ caused further accumulation of LC3B, suggesting an increased flux. In line with in vivo data, similar results were observed in murine primary hepatocytes (Fig. 2B). Exposure of 100 mM ethanol for 24 h significantly increased LC3-Ⅱ expression as well as LC3B turnover ratio (LC3-Ⅱ/LC3-Ⅰ) in these cells.
Fig. 1.
Acute ethanol treatment activates autophagy, CYP2E1, and NOX4 in mice. (A) Six mice in the ethanol group were shortly given 33% (vol/vol) ethanol at a total accumulative dosage of 4.5 g/kg body weight by gavages. Western blot analysis of LC3B in mice liver total extracts (35 μg per lane, n = 6 mice per group). (B) Western blot analysis of CYP2E1, NOX4, Prx2, and Nrf2 in mice liver total extracts (35 μg per lane, n = 6 mice per group). (C) Immunohistochemistry staining of CYP2E1 in the paraffin-embedded liver section. Immunofluorescence staining of NOX4 in the frozen liver section. All target protein levels were quantified and normalized to those of GAPDH. *, P < 0.05, **, P < 0.01.
Fig. 2.
Ethanol induces autophagy flux in mice liver and triggers autophagy in primary murine hepatocytes. (A) Western blot analysis of LC3B in mice liver total extracts. One mouse in the control (CO) and ethanol group was injected with CQ (100 mg/kg) 0.5 h before getting the ethanol gavage. (B) The murine primary hepatocytes were incubated with 100 mM ethanol for 24 h, the LC3B expression was detected by Western blot. LC3-Ⅱ levels were quantified and normalized to those of GAPDH. *, P < 0.05.
In conclusion, acute ethanol significantly induces autophagy, CYP2E1, NOX4, and ROS in mice liver and autophagy as well as autophagic flux are induced by ethanol both in vivo but also in primary mouse hepatocytes.
CYP2E1 overexpression re-establishes ethanol mediated induction of autophagy in huh7 cells.
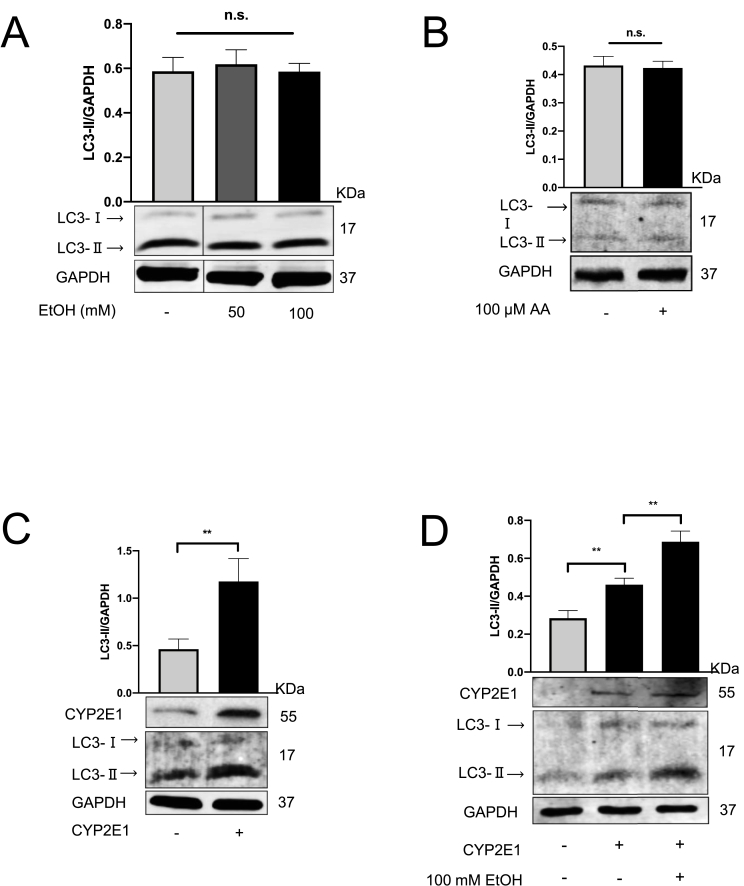
We next set up a series of experiments to study ethanol-mediated autophagy in hepatoma huh7 cells, a widely used in vitro model for liver studies. Notably, neither 100 mM ethanol nor 100 μM of its major metabolite acetaldehyde was able to induce autophagy in hepatoma huh7 cells (Fig. 3A and B). Detailed time response studies during 3–24 h of acetaldehyde exposure also did not show any effects on autophagy (not shown). In order to further validate the autophagic flux in vitro, we treated Huh7 cells with ethanol or acetaldehyde in the presence of CQ which did not result in further accumulation of LC3B (Suppl. Fig. 2A and 2B). In addition, a tandem mRFP-GFP-LC3 plasmid transfection experiment, widely used to monitor autophagy flux [55,56] was explored. Briefly, the increased formation of LC3 protein can be traced with increased mRFP (red) or GFP (green) -tagged LC3 dots [15,57]. Huh7 cells were transfected with two fluorescence proteins labeled LC3 plasmids and treated with ethanol. However, neither the number of RFP and RFP/GFP merged autophagosomes were significantly induced (Suppl. Fig. 2C). Since some hepatoma cells are known to lack functional CYP2E1 [25] and CYP2E1 overexpression is required to recapitulate ethanol metabolism [1,58,59], we repeated the studies with huh7 cells transiently transfected with CYP2E1-overexpressing plasmids. CYP2E1 overexpression was successfully confirmed by Western blotting (Fig. 3C) (Suppl. Fig. 3A) and treatment of these cells with 100 mM ethanol significantly induced autophagy (Fig. 3D). Transfection efficiency was confirmed by GFP (Suppl. Fig. 3B). These findings highly suggest that CYP2E1 and potentially CYP2E1-generated ROS seem to be essential for ethanol-induced autophagy.
Fig. 3.
Transfection of huh7 cells with CYP2E1 recapitulates ethanol-mediated induction of autophagy. (A) Huh7 cells were exposed to 50 and 100 mM ethanol for 24 h. The expression levels of LC3-Ⅱ were analyzed by the western bolt. (B) Huh7 cells were exposed to 100 μM acetaldehyde (AA) for 24 h. (C) Huh7 cells were transfected with the CYP2E1 plasmids for 24 h. The expression levels of CYP2E1 and LC3-Ⅱ were analyzed by the western bolt. (D) After 24 h of CYP2E1 transfection, the cells were further treated with 100 mM ethanol for another 24 h. LC3-Ⅱ levels were quantified and normalized to those of GAPDH. Representative data of at least three independent experiments are shown for protein data. **, P < 0.01, n.s.: not significant.
3.1. Induction of autophagy by CYP2E1-and NOX4-mediated generation of intracellular ROS
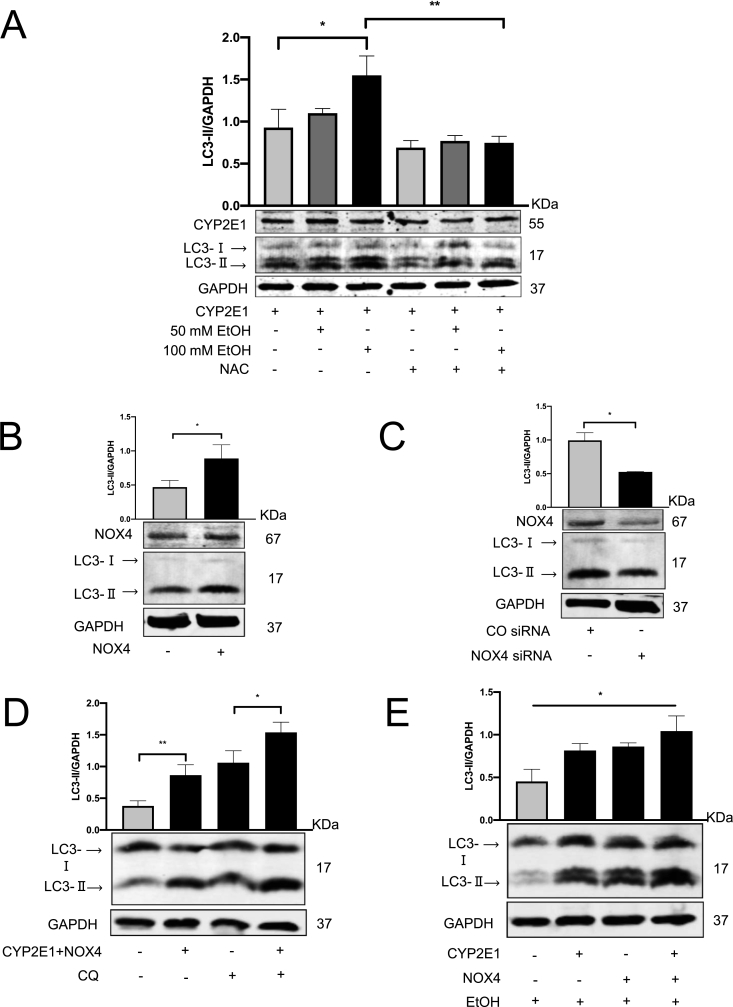
To further confirm whether known CYP2E1-mediated ROS production is involved in autophagy, we co-treated CYP2E1-overexpressing huh7 in addition to ethanol with 10 mM N-Acetyl-l-cysteine (NAC). As shown in Fig. 4A, NAC was able to efficiently block autophagy, suggesting the involvement of ROS in this model. We next explored NOX4, an additional important ROS source involved in hepatocyte signaling and induced during ethanol abuse [49] and autophagy [45,60]. Transfection NOX4 plasmid into Huh7 cells drastically increased NOX4 mRNA (Suppl. Fig. 4A) and markedly elevated NOX4 protein followed by LC3-Ⅱ induction (Fig. 4B) (Suppl. Fig. 4B). Moreover, and in line with CYP2E1 experiments, NOX4-mediated autophagy could be efficiently blocked by 10 mM NAC (Suppl. Fig. 4C). Vice versa, siRNA-mediated knockdown of NOX4 (Suppl. Fig. 4D and 4E) reduced LC3-Ⅱ expression in Huh7 cells (Fig. 4C). As demonstrated in Fig. 4D and E, co-transfection with CYP2E1 and NOX4 further enhanced autophagy. As compared with CQ treatment alone, the co-transfection caused further LC3-Ⅱ accumulation, when the degradation was blocked by CQ. Furthermore, Huh7 cells co-transfected with CYP2E1 and NOX4 plasmid showed stronger LC3B expression in the presence of ethanol, which attributes to a robust ROS generation.
Fig. 4.
NOX4 cooperates with CYP2E1 and further up-regulated autophagy. NAC impaired the autophagy induction. (A) CYP2E1 transfected Huh7 cells were further incubated with 50/100 mM ethanol separately or in the presence of 10 mM NAC. LC3-II and CYP2E1 expression were analyzed by Western blot. (B) Huh7 cells were transfected with the NOX4 plasmids or (C) NOX4 siRNA for 48 h. LC3-II and NOX4 expression were analyzed by Western blot. (D) Huh7 cells were co-transfected with CYP2E1 and NOX4 and further incubated within the presence or absence of 20 μM CQ for 6 h. LC3-II expression was analyzed by Western blot. (E) CYP2E1, NOX4, or co-transfection were performed 24 h, followed by 24 h 100 mM ethanol incubation. LC3-II expression was analyzed by Western blot. *, P < 0.05, **, P < 0.01.
In summary, we here show that intracellular ROS generation by both CYP2E1 and NOX4 are able to induce autophagy in huh7 cells in an additive manner.
3.2. External non-toxic H2O2 significantly increases LC3-Ⅱ expression without mTOR suppression
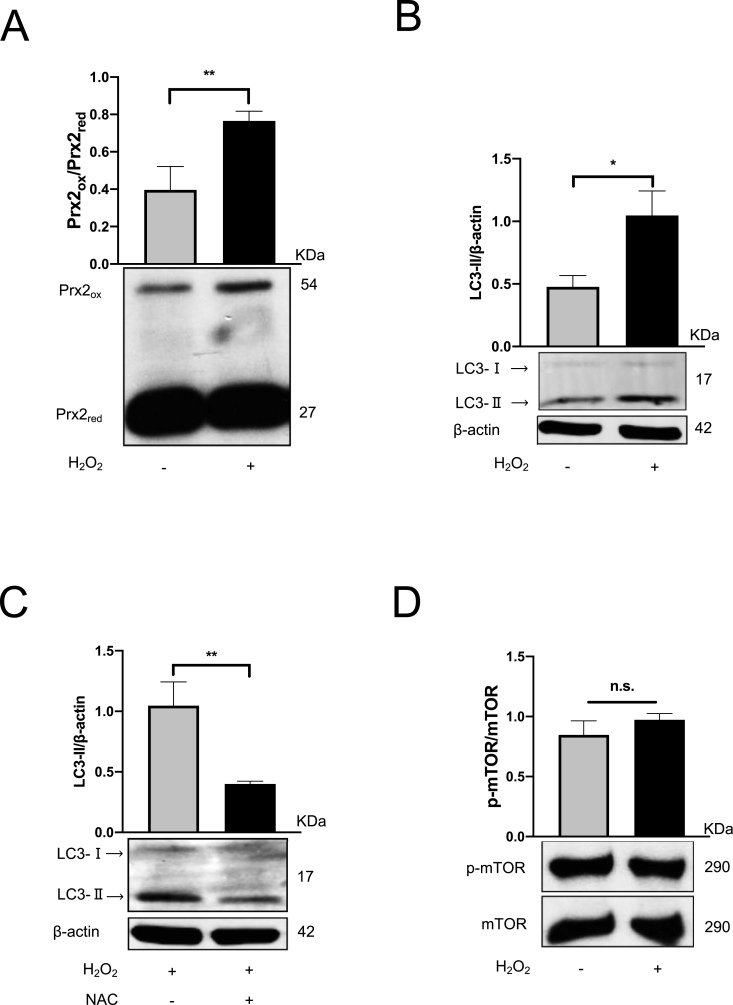
We next set up several experiments to analyze the role of the external release of H2O2 on autophagy using the enzymatic GOX/CAT system [40], which allows the specific formation of steady-state H2O2 (ssH2O2) level over a huge concentration range and time period [40]. Cell toxicity studies confirmed not toxic conditions (not shown) but effective elevation of cytosolic H2O2 levels were confirmed by Prx2 Western blotting [41,49]. Indeed and as shown in Fig. 5A, H2O2 induced oxidized Prx2 (Prx2ox). These low and non-toxic ssH2O2 levels (1–2 μM) were also able to induce LC3B (Fig. 5B), which could be efficiently blocked by 10 mM NAC (Fig. 5C). Although ROS have been shown to suppress p-mTOR [24,43,61], no suppression was observed under conditions of non-toxic ssH2O2 (Fig. 5D). In contrast, based on the dose-dependent disappearance of total LC3B with the co-incubation of H2O2 and rapamycin (Rapa), the specific inhibitor of mTOR, H2O2 highly contributed to autophagy degradation process, which is the final stage of autophagy flux. Notably, enhanced degradation was observed without a further mTOR impairment (Suppl. Fig. 5A and 5B). Although mTOR was completely blocked by rapamycin, H2O2 was still able to modulate autophagy ruling out any major mTOR involvement under these experimental. In line with unchanged mTOR expression, no changes of major upper-stream proteins of mTOR, such as PI3K/Akt and p-AMPK/AMPK, were observed (data not shown). Collectively, low level H2O2 induces LC3-Ⅱ expression in Huh7 cells in the absence of mTOR suppression.
Fig. 5.
H2O2activates autophagy in Huh7 cells.
Huh7 cells were exposed to H2O2, which was independently controlled by GOX/CAT system. (A) H2O2 generation was confirmed by Prx2ox expression. (B) The expression levels of LC3-Ⅱ were analyzed by the western bolt. LC3-Ⅱ levels were quantified and normalized to those of β-actin. (C) Huh7 cells were exposed to H2O2 in the presence or absence of 10 mM NAC for 24 h. The expression levels of LC3-II and (D) p-mTOR/mTOR were analyzed by Western blot. Representative data of at least three independent experiments are shown for protein data. *, P < 0.05, **, P < 0.01, n.s.: not significant.
3.3. H2O2 activates the autophagic flux in Huh7 cells
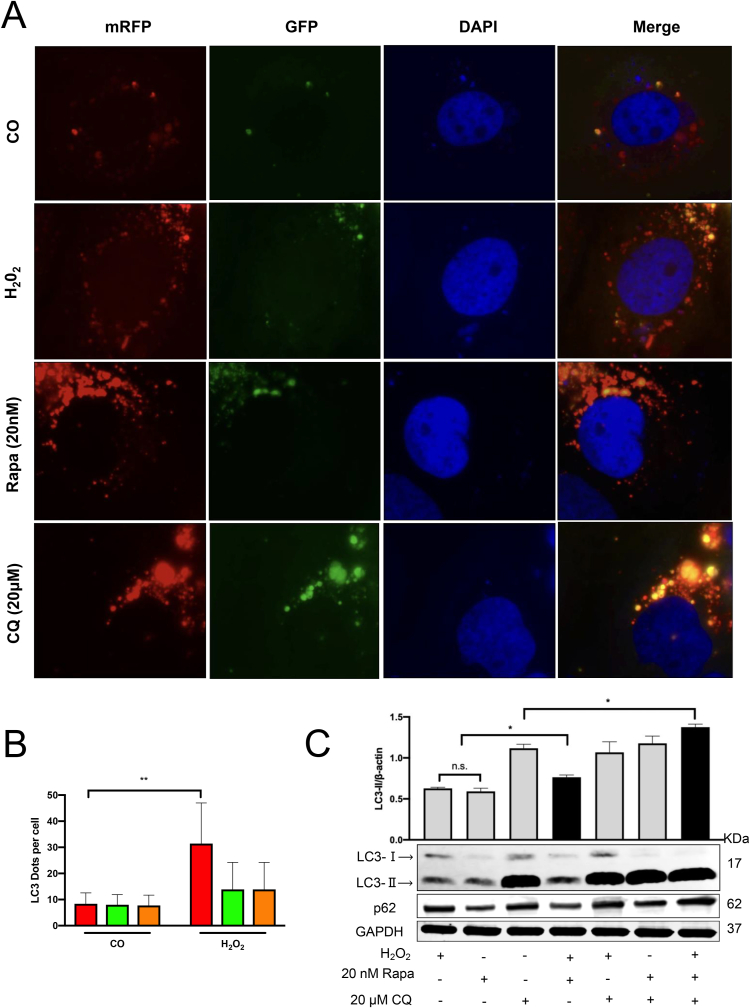
We now studied H2O2-mediated modulation of autophagic flux in Huh7 cells by tandem fluorescent-tagged mRFP-GFP-LC3 transfection. Rapamycin and CQ were used as the positive and negative control. In this assay, GFP fluorescence (green) is rapidly quenched in the acidic environment, e.g. in autolysosome, while RFP fluorescence (red) remains stable and serves as a more specific marker of LC3B expressed in autolysosome. After transfection, mRFP-LC3 visualizes autolysosomes while most GFP-LC3 signals do not colocalize with autolysosome [15,62]. As shown in Fig. 6A, H2O2 exposure led to significantly increased numbers of RFP tagged LC3 protein puncta, while the number of GFP puncta did not increase in line with positive control rapamycin expression. Compared with the control group, there are significantly increased RFP puncta, but no GFP puncta after H2O2 incubation (Fig. 6B). This observation highly suggests that H2O2 improves the fusion between the lysosome and the autophagosome to successfully form the autolysosome and activates the autophagic flux in Huh7 cells. On the contrary, after CQ treatment, more yellow labeled puncta (overlay between RFP and GFP puncta) was detected since the GFP fluorescence was not quenched due to impaired acidification of the autolysosome compartment. Subsequently, the induced autophagic flux was also confirmed using western blotting. The H2O2 treatment in the presence of CQ and rapamycin showed the most elevated LC3-Ⅱ levels as compared to any other treatment group (Fig. 6C). p62, a cargo protein responsible for delivering the damaged organelles to autophagosomes for further degradation [56], typically serving as a negative-related marker to study autophagic flux [63], was likewise induced in the three-treatment group. In addition, H2O2 and rapamycin co-treatment further increased LC3-Ⅱ accumulation in a synergistic effect, and a weaker p62 protein expression indicated a functional autophagic flux. Thus, these data suggest that H2O2 is an mTOR-independent inducer of autophagy that can activate autophagic flux in Huh7 cells (Fig. 7).
Fig. 6.
Effect of H2O2on autophagic flux in Huh7 cells by mRFP-GFP-LC3 and immunoblotting analysis. (A) Huh7 cells were transfected with the tandem mRFP-GFP-LC3 plasmids. 24 h after the transfection, the cells were treated with H2O2 for another 24 h 6 h CQ (20 μM) and Rapa (20 nM) treatment group for negative and positive control. (B) More than 9 cells were counted in the control and the H2O2 group, and data (mean ± SD) are representative of two independent experiments. (C) Huh7 cells were treated with H2O2 for 24 h in the presence or absence of 6 h CQ or/and Rapa treatment. The expression levels of LC3-II and p62 were analyzed by Western blot. *, P < 0.05, **, P < 0.01, n.s.: not significant.
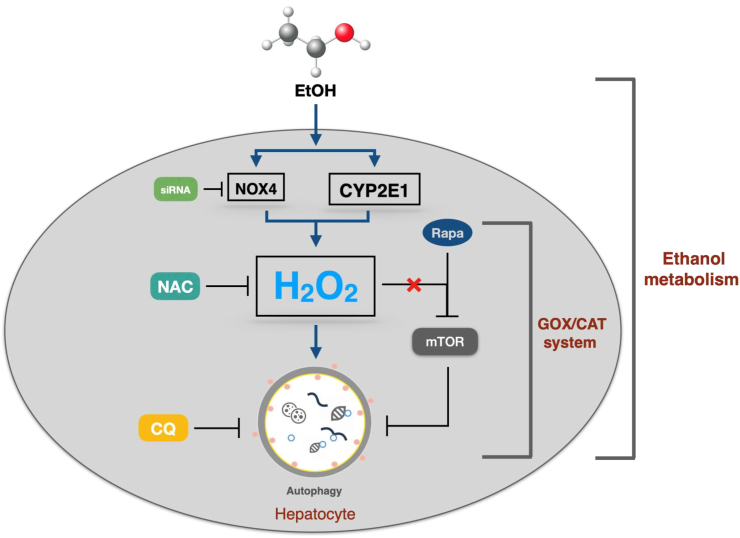
Fig. 7.
Scheme depictingthe role of ROS on autophagy. Especially H2O2 is generated by CYP2E1 and NOX4 during alcohol metabolism in the liver. Note that mTOR seems not to be involved.
4. Discussion
We here study the role of ethanol and its specific intermediate acetaldehyde on hepatocellular autophagy in vivo and in vitro, namely the role for important ALD-associated ROS-producing enzymes such as CYP2E1 and NOX4 under non-toxic conditions. We first confirm in a murine model of acute ethanol exposure that CYP2E and NOX4 are both induced, leading to enhanced autophagy as measured by LC3B levels. While these findings could be recapitulated in primary mouse hepatocytes, commonly used hepatoma cells huh7 required overexpression of CYP2E1 to restore ethanol-mediated autophagy. Moreover, overexpression and knock down of NOX4 could also strongly modulate autophagy in these cells and both CYP2E1-and NOX4-mediated autophagy was efficiently suppressed by the potent antioxidant NAC. Finally, using an enzymatic model of the specific, continuous and non-toxic release of steady-state H2O2 mimicking an extracellular e.g. inflammatory ROS source, we demonstrate that H2O2 is a potent modulator of autophagic flux and, surprisingly, without the involvement of mTOR under these non-toxic conditions.
Our data shed new light on the role of ROS in autophagy, especially H2O2. Oxidation of ethanol by the microsomal ethanol oxidizing system (MEOS) namely CYP2E1 has long been established as an important pathway of ethanol metabolism and it has been implicated in the progression of disease [64]. Deletion of CYP2E1 in mice has been shown to decrease ROS and DNA adduct formation [65] and also reduced diethylnitrosamine (DEN)-induced hepatic tumour formation [66]. However, the mechanisms by which CYP2E1 generates ROS are still complex and not completely understood although being studied using active-site mutants designed to alter the coupling of ethylbenzene hydroxylation in former reports [67]. In addition to CYP2E1, we therefore studied here two other enzymatic ROS sources: First, overexpression and knock down of NOX4, an important NADPH oxidase expressed in hepatocytes, and induced by ethanol [34] and, second, external and specific generation of non-toxic dosages of H2O2 by the GOX/CAT system [40] mimicking an extracellular H2O2 source in patients with ALD e.g., activated neutrophils.
In confirmation of previous data [27], we demonstrate the induction of LC3B and CYP2E1 in a mouse model of acute ethanol exposure. In addition, we also show an induction of NOX4 and indirect signs of oxidative stress through increased levels of the redox-sensitive transcription factor Nrf2 and peroxiredoxin 2 [41]. We were then able to demonstrate in various in vitro experiments using LC3B to monitor autophagy that both, the presence of CYP2E1 and NOX4, can induce autophagy. Notably, the important ethanol intermediate acetaldehyde was not able to induce autophagy while external and specific exposure to H2O2 at non-toxic concentrations or incubation with NAC potently modulated autophagy. We conclude that ROS such as H2O2 seems to be an important effector of ethanol metabolism rather than acetaldehyde which most likely enhances the need for autophagy through irreversible protein modifications. We also confirm that H2O2 at these conditions is indeed able to enhance the autophagic flux using well established tandem mRFP-GFP-LC3 plasmid transfection experiments. To our surprise, no direct impact of H2O2 on mTOR or upstream regulators such as AMP kinase was observed. This is in contrast to previous reports, where almost 500 times higher bolus levels of peroxide were used to observe mTOR changes [68]. Even the mTOR signaling pathway inhibited by ROS generation was reported as an underlying mechanism [24]. We have demonstrated in the past that even opposite effects can be observed with redox sensitive signaling molecules e.g. during hepcidin signal transdurction if non-physiological H2O2 concentrations are used [48]. While H2O2 levels in the 100 μM range were able to block hepcidin through unspecific inhibition of the transcription machinery, low continuous levels induced hepcidin and the effect was even potentiated under conditions of mild hypoxia [48,49]. However, it remains unclear how H2O2 induces autophagy. The recent in vivo observation of H2O2 release during autophagy could suggest that H2O2 actively participates in the autophagic process as a signaling molecule [37].
Moreover, our data clearly point to ROS-mediated induction of autophagy by ethanol at these non-toxic conditions. First, we demonstrate that CYP2E1 is required for autophagy induction in huh7 cells in the presence of ethanol and NAC completely blocked this induction. Second, although another mandatory end-product of CYP2E1-mediated ethanol metabolism, acetaldehyde in various concentrations and time exposure had no effect on autophagy. Third, autophagy was also induced by other ROS/H2O2 sources such as NOX4 [49] or H2O2 release by the GOX/CAT system. Thus, the induction of autophagy by ROS could explain some of the conflicting data on how alcohol affects autophagy [24,[27], [28], [29], [30], [31], [32]] namely whether exposed to acutely or chronically as discussed recently [27]. On the one side, ROS formation and degradation depends on the mode of ethanol application (chronic versus acute) and on the other side, we suggest to use more standardized ROS conditions in cell culture experiments. For instance, bolus addition of H2O2 requires the usage of unphysiologically high H2O2 dosages since H2O2 is rapidly removed within minutes from cultured cells [40]. In such settings, cells will be exposed for a too short time to too high H2O2 levels that may cause unspecific oxidation of various cellular proteins [69].
Our data on the role of H2O2 on autophagic activity also point to the complex but essential relation between autophagy and programmed cell death. H2O2 is known to be a traditional inducer of apoptosis e.g. in lymphocytes [35] and it also stimulates proliferation and regeneration and low levels [36]. In the context with our findings, H2O2 at non-toxic low levels could stimulate both apoptosis and autophagy. On the other side, however, numerous studies indicate that pharmacological or genetic inhibition of autophagy greatly enhances cell death [[21], [22], [23]]. It is generally agreed that autophagy is a pro-survival mechanism by not only providing nutrients for cell survival during starvation but also selectively removing damaged organelles, including damaged mitochondria [70]. Liver-specific knockout of Atg7 leads to hepatomegaly and severe liver injury [23] and suppression of autophagy exacerbates alcohol-induced liver injury by increasing alcohol induced apoptosis [24]. A recent human study on heavy drinkers undergoing alcohol detoxification clearly demonstrated that ethanol primarily suppresses apoptosis and regeneration. In this study, it was shown that levels of caspase 3 cleaved CK18 fragments (M30) in serum were highly correlated with its expression in liver tissue and M30 levels could be used as rather specific marker of liver apoptosis [19]. In addition, detoxication from ethanol caused significant induction of M30 levels and markers of regeneration while signs of necrosis rapidly normalized [19]. We therefore feel that future studies on ethanol-mediated liver damage should better dissect the various biochemical effects of ethanol (e.g. ROS versus acetaldehyde) and should also take into consideration that some of the metabolites such as H2O2 could be part of the physiological signaling machinery e.g. autophagy or apoptosis. Moreover, as has been already conceived in human [1] and animal studies [27], the drinking pattern (acute versus chronic) is critical for the understanding of ethanol-mediated organ damage and may explain some of the seemingly contradictive findings in the literature. In this concept, a continuous exposure to ethanol may eventually block these physiological processes due to accumulation of misfolded or modified proteins and congested lysosomes as is typically seen in livers from ALD patients with the formation of Mallory-Denk bodies.
In summary, we here demonstrate that ethanol primarily stimulates autophagy by induction of enzyme systems such as CYP2E1 or NOX4 and enhanced ROS generation such as H2O2, a condition that is able to induce autophagy at non-toxic conditions. Of note, such non-toxic conditions are not able to suppress mTOR and it remains to be clarified in future studies whether H2O2 is an essential mediator of autophagy and what specific molecular steps are involved in this regulatory process.
Author contributions
CC, VR, and SM conceived the study, designed the experiments, and analyzed the data. CC, SJ performed the experiments. CC and SM wrote the manuscript. All authors critically discussed the experimental designs and data.
Declaration of competing interest
The authors declare no potential conflicts of interest.
Acknowledgements
This study was funded by a grant of the DFG to SM (RA 2677/1–2). CC, SJW, and LNY were supported by a fellowship from the China Scholarship Council (CSC). We would further like to thank Dr. Frederik Bergler (Medical Faculty of Heidelberg University) and Dr. Chaohui Lin (University Düsseldorf) for providing technical support and useful discussion.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2021.102081.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Seitz H.K. Alcoholic liver disease. Nat Rev Dis Primers. 2018;4(1):16. doi: 10.1038/s41572-018-0014-7. [DOI] [PubMed] [Google Scholar]
- 2.Bardag-Gorce F. The effect of ethanol-induced CYP2E1 on proteasome activity: the role of 4-hydroxynonenal. Exp. Mol. Pathol. 2005;78(2):109–115. doi: 10.1016/j.yexmp.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Thursz M.R. Prednisolone or pentoxifylline for alcoholic hepatitis. N. Engl. J. Med. 2015;372(17):1619–1628. doi: 10.1056/NEJMoa1412278. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen-Khac E. Glucocorticoids plus N-acetylcysteine in severe alcoholic hepatitis. N. Engl. J. Med. 2011;365(19):1781–1789. doi: 10.1056/NEJMoa1101214. [DOI] [PubMed] [Google Scholar]
- 5.Ding W.X., Manley S., Ni H.M. The emerging role of autophagy in alcoholic liver disease. Exp. Biol. Med. 2011;236(5):546–556. doi: 10.1258/ebm.2011.010360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baraona E. Alcoholic hepatomegaly - accumulation of protein in liver. Science. 1975;190(4216):794–795. doi: 10.1126/science.1198096. [DOI] [PubMed] [Google Scholar]
- 7.Shibutani S.T., Yoshimori T. A current perspective of autophagosome biogenesis. Cell Res. 2014;24(1):58–68. doi: 10.1038/cr.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang Z., Klionsky D.J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 2010;12(9):814–822. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirkegaard K., Taylor M.P., Jackson W.T. Cellular autophagy: surrender, avoidance and subversion by microorganisms. Nat. Rev. Microbiol. 2004;2(4):301–314. doi: 10.1038/nrmicro865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dolganiuc A. Autophagy in alcohol-induced liver diseases. Alcohol Clin. Exp. Res. 2012;36(8):1301–1308. doi: 10.1111/j.1530-0277.2012.01742.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feng Y. The machinery of macroautophagy. Cell Res. 2013;24(1):24–41. doi: 10.1038/cr.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lopez A., Fleming A., Rubinsztein D.C. Seeing is believing: methods to monitor vertebrate autophagy in vivo. Open Biol. 2018;8(10) doi: 10.1098/rsob.180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabeya Y. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. J. Cell Sci. 2004;117(Pt 13):2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 14.Kabeya Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. The EMBO Jouranl. 2000:5720–5728. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mizushima N., Yoshimori T., Levine B. Methods in mammalian autophagy research. Cell. 2010;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mizushima N., Yoshimori T. How to interpret LC3 immunoblotting. Autophagy. 2007;3(6):542–545. doi: 10.4161/auto.4600. [DOI] [PubMed] [Google Scholar]
- 17.Tanida I. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- 18.Weidberg H. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell. 2011;20(4):444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Mueller S. Caspase‐cleaved keratin‐18 fragments increase during alcohol withdrawal and predict liver‐related death in patients with alcoholic liver disease. Hepatology. 2017;66(1):96–107. doi: 10.1002/hep.29099. [DOI] [PubMed] [Google Scholar]
- 20.Donohue T.M., Jr. Autophagy and ethanol-induced liver injury. World J. Gastroenterol. 2009;15(10):1178–1185. doi: 10.3748/wjg.15.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding W.-X. Differential effects of endoplasmic reticulum stress-induced autophagy on cell survival. J. Biol. Chem. 2007;282(7):4702–4710. doi: 10.1074/jbc.M609267200. [DOI] [PubMed] [Google Scholar]
- 22.Ding W.-X. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am. J. Pathol. 2007;171(2):513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Komatsu M. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. JCB (J. Cell Biol.) 2005;169(3):425–434. doi: 10.1083/jcb.200412022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ding W.X. Autophagy reduces acute ethanol-induced hepatotoxicity and steatosis in mice. Gastroenterology. 2010;139(5):1740–1752. doi: 10.1053/j.gastro.2010.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu D., Cederbaum A.I. Inhibition of autophagy promotes CYP2E1-dependent toxicity in HepG2 cells via elevated oxidative stress, mitochondria dysfunction and activation of p38 and JNK MAPK. Redox Biol. 2013;1:552–565. doi: 10.1016/j.redox.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kharbanda K.K. Ethanol consumption alters trafficking of lysosomal enzymes and affects the processing of procathepsin L in rat liver. Biochim. Biophys. Acta Gen. Subj. 1996;1291(1):45–52. doi: 10.1016/0304-4165(96)00043-8. [DOI] [PubMed] [Google Scholar]
- 27.Thomes P.G. Acute and chronic ethanol administration differentially modulate hepatic autophagy and transcription factor EB. Alcohol Clin. Exp. Res. 2015;39(12):2354–2363. doi: 10.1111/acer.12904. [DOI] [PubMed] [Google Scholar]
- 28.Han W. MiR-26a enhances autophagy to protect against ethanol-induced acute liver injury. J. Mol. Med. (Berl.) 2015;93(9):1045–1055. doi: 10.1007/s00109-015-1282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi X. Promotion of autophagosome–lysosome fusion via salvianolic acid A-mediated SIRT1 up-regulation ameliorates alcoholic liver disease. RSC Adv. 2018;8(36):20411–20422. doi: 10.1039/c8ra00798e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang L. Autophagy in alcoholic liver disease, self-eating triggered by drinking. Clin Res Hepatol Gastroenterol. 2015;39(Suppl 1):S2–S6. doi: 10.1016/j.clinre.2015.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lin C.W. Pharmacological promotion of autophagy alleviates steatosis and injury in alcoholic and non-alcoholic fatty liver conditions in mice. J. Hepatol. 2013;58(5):993–999. doi: 10.1016/j.jhep.2013.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khambu B. Autophagy in non-alcoholic fatty liver disease and alcoholic liver disease. Liver Res. 2018;2(3):112–119. doi: 10.1016/j.livres.2018.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ding W.-X., Li M., Yin X.-M. Selective taste of ethanol-induced autophagy for mitochondria and lipid droplets. Autophagy. 2011;7(2):248–249. doi: 10.4161/auto.7.2.14347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Silva I. Center for Alcohol Research. University of Heidelberg; Heidelberg: 2019. Mechanisms of carcinogenic iron accumulation in alcoholic liver disease. [Google Scholar]
- 35.Droge W. Free radicals in the physiological control of cell function. Physiol. Rev. 2002;82(1):47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 36.Gough D.R., Cotter T.G. Hydrogen peroxide: a Jekyll and Hyde signalling molecule. Cell Death Dis. 2011;2:e213. doi: 10.1038/cddis.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu F. A NIR fluorescent probe: imaging endogenous hydrogen peroxide during an autophagy process induced by rapamycin. J. Mater. Chem. B. 2016;4(46):7363–7367. doi: 10.1039/c6tb02463g. [DOI] [PubMed] [Google Scholar]
- 38.Jiang P. Hydrogen peroxide impairs autophagic flux in a cell model of nonalcoholic fatty liver disease. Biochem. Biophys. Res. Commun. 2013;433(4):408–414. doi: 10.1016/j.bbrc.2013.02.118. [DOI] [PubMed] [Google Scholar]
- 39.Stone J.R., Yang S. Hydrogen peroxide: a signaling messenger. Antioxidants Redox Signal. 2006;8(3–4):243–270. doi: 10.1089/ars.2006.8.243. [DOI] [PubMed] [Google Scholar]
- 40.Mueller S., Millonig G., Waite G.N. The GOX/CAT system: a novel enzymatic method to independently control hydrogen peroxide and hypoxia in cell culture. Adv. Med. Sci. 2009;54(2):121–135. doi: 10.2478/v10039-009-0042-3. [DOI] [PubMed] [Google Scholar]
- 41.Sobotta M.C. Exposing cells to H2O2: a quantitative comparison between continuous low-dose and one-time high-dose treatments. Free Radic. Biol. Med. 2013;60:325–335. doi: 10.1016/j.freeradbiomed.2013.02.017. [DOI] [PubMed] [Google Scholar]
- 42.Cederbaum Y.L.a.A.I. CYP2E1 and oxidative liver injury by alcohol. Free Radic. Biol. Med. 2018;44(5):723–738. doi: 10.1016/j.freeradbiomed.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen Y., Gibson S.B. Is mitochondrial generation of reactive oxygen species a trigger for autophagy? Autophagy. 2008;4(2):246–248. doi: 10.4161/auto.5432. [DOI] [PubMed] [Google Scholar]
- 44.De Minicis S., Brenner D.A. Oxidative stress in alcoholic liver disease: role of NADPH oxidase complex. J. Gastroenterol. Hepatol. 2008;23(Suppl 1):S98–S103. doi: 10.1111/j.1440-1746.2007.05277.x. [DOI] [PubMed] [Google Scholar]
- 45.Sciarretta S. Activation of NADPH oxidase 4 in the endoplasmic reticulum promotes cardiomyocyte autophagy and survival during energy stress through the protein kinase RNA-activated-like endoplasmic reticulum kinase/eukaryotic initiation factor 2alpha/activating transcription factor 4 pathway. Circ. Res. 2013;113(11):1253–1264. doi: 10.1161/CIRCRESAHA.113.301787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kurdi A. Continuous administration of the mTORC1 inhibitor everolimus induces tolerance and decreases autophagy in mice. Br. J. Pharmacol. 2016;173(23):3359–3371. doi: 10.1111/bph.13626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haspel J. Characterization of macroautophagic flux in vivo using a leupeptin-based assay. Autophagy. 2011;7(6):629–642. doi: 10.4161/auto.7.6.15100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Millonig G. Sustained submicromolar H2O2 levels induce hepcidin via signal transducer and activator of transcription 3 (STAT3) J. Biol. Chem. 2012;287(44):37472–37482. doi: 10.1074/jbc.M112.358911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silva I. Hypoxia enhances H2O2-mediated upregulation of hepcidin: evidence for NOX4-mediated iron regulation. Redox Biol. 2018;16:1–10. doi: 10.1016/j.redox.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao N. Targeting Nrf-2 is a promising intervention approach for the prevention of ethanol-induced liver disease. Cell. Mol. Life Sci. 2018;75(17):3143–3157. doi: 10.1007/s00018-018-2852-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.He Y. Dysregulated autophagy increased melanocyte sensitivity to H2O2-induced oxidative stress in vitiligo. Sci. Rep. 2017;7(1) doi: 10.1038/srep42394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tonelli C., Chio I.I.C., Tuveson D.A. Transcriptional regulation by Nrf2. Antioxidants Redox Signal. 2018;29(17):1727–1745. doi: 10.1089/ars.2017.7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loboda A. Role of Nrf2/HO-1 system in development, oxidative stress response and diseases: an evolutionarily conserved mechanism. Cell. Mol. Life Sci. 2016;73(17):3221–3247. doi: 10.1007/s00018-016-2223-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ingelman-Sundberg M., Johansson I P.K., Glaumann H L.K. Centrilobular expression of ethanolinducible cytochrome P450 (IIE1) in rat liver. Biochem. Biophys. Res. Commun. 1988;157(1):55–60. doi: 10.1016/s0006-291x(88)80010-x. [DOI] [PubMed] [Google Scholar]
- 55.Sarkar S. Methodological considerations for assessing autophagy modulators: a study with calcium phosphate precipitates. Autophagy. 2009;5(3):307–313. doi: 10.4161/auto.5.3.7664. [DOI] [PubMed] [Google Scholar]
- 56.Klionsky D.J. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2016;12(1):1–222. doi: 10.1080/15548627.2015.1100356. third ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klionsky D.J. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition) Autophagy. 2021:1–382. doi: 10.1080/15548627.2020.1797280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho Y.E. Increased ethanol-inducible cytochrome P450-2E1 and cytochrome P450 isoforms in exosomes of alcohol-exposed rodents and patients with alcoholism through oxidative and endoplasmic reticulum stress. Hepatol Commun. 2017;1(7):675–690. doi: 10.1002/hep4.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dey A., Cederbaum A.I. Alcohol and oxidative liver injury. Hepatology. 2006;43(2 Suppl 1):S63–S74. doi: 10.1002/hep.20957. [DOI] [PubMed] [Google Scholar]
- 60.Sciarretta S. Role of NADPH oxidase in the regulation of autophagy in cardiomyocytes. Clin. Sci. (Lond.) 2015;128(7):387–403. doi: 10.1042/CS20140336. [DOI] [PubMed] [Google Scholar]
- 61.Liu L. Hypoxic reactive oxygen species regulate the integrated stress response and cell survival. J. Biol. Chem. 2008;283(45):31153–31162. doi: 10.1074/jbc.M805056200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ni H.M. Dissecting the dynamic turnover of GFP-LC3 in the autolysosome. Autophagy. 2011;7(2):188–204. doi: 10.4161/auto.7.2.14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang P., Mizushima N. LC3- and p62-based biochemical methods for the analysis of autophagy progression in mammalian cells. Methods. 2015;75:13–18. doi: 10.1016/j.ymeth.2014.11.021. [DOI] [PubMed] [Google Scholar]
- 64.Lieber C.S. Cytochrome P-4502E1: its physiological and pathological role. Physiol. Rev. 1997;77(2):517–544. doi: 10.1152/physrev.1997.77.2.517. [DOI] [PubMed] [Google Scholar]
- 65.Lu Y. Chronic alcohol-induced liver injury and oxidant stress are decreased in cytochrome P4502E1 knockout mice and restored in humanized cytochrome P4502E1 knock-in mice. Free Radic. Biol. Med. 2010;49(9):1406–1416. doi: 10.1016/j.freeradbiomed.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kang J.S. Role of CYP2E1 in diethylnitrosamine-induced hepatocarcinogenesis in vivo. Canc. Res. 2007;67(23):11141–11146. doi: 10.1158/0008-5472.CAN-07-1369. [DOI] [PubMed] [Google Scholar]
- 67.Loida P.J., Sligar S.G. Molecular recognition in cytochrome P-450: mechanism for the control of uncoupling reactions. Biochemistry. 1993;32(43):11530–11538. doi: 10.1021/bi00094a009. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J. Autophagic lysosomal reformation depends on mTOR reactivation in H2O2-induced autophagy. Int. J. Biochem. Cell Biol. 2016;70:76–81. doi: 10.1016/j.biocel.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 69.Forman H.J. Use and abuse of exogenous H2O2 in studies of signal transduction. Free Radic. Biol. Med. 2007;42(7):926–932. doi: 10.1016/j.freeradbiomed.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim I., Lemasters J.J. Mitophagy selectively degrades individual damaged mitochondria after photoirradiation. Antioxidants Redox Signal. 2011;14(10):1919–1928. doi: 10.1089/ars.2010.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.