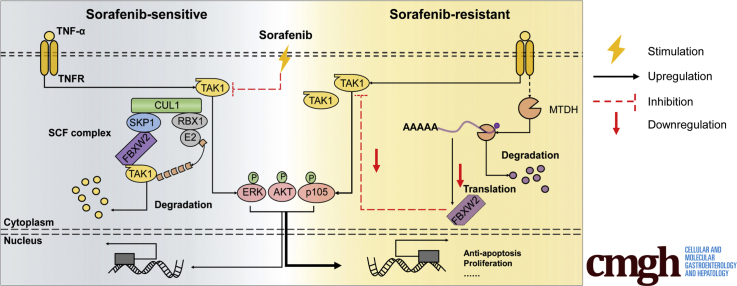
Abstract
Background & Aims
Identifying novel and actionable targets in hepatocellular carcinoma (HCC) remains an unmet medical need. TAK1 was originally identified as a transforming growth factor-β–activated kinase and was further proved to phosphorylate and activate numerous downstream targets and promote cancer progression. However, the role of TAK1 in developed HCC progression and targeted therapy resistance is poorly understood.
Methods
The expression of TAK1 or MTDH in HCC cell lines, tumor tissues, and sorafenib-resistant models was analyzed by in silico analysis, quantitative real-time polymerase chain reaction, Western blotting, and immunohistochemistry. In vivo and in vitro experiments were introduced to examine the function of TAK1 or MTDH in HCC and sorafenib resistance using small interfering RNA and pharmacologic inhibitors in combination with or without sorafenib. Co-immunoprecipitation and RNA immunoprecipitation were carried out to determine the binding between TAK1 and FBXW2 or between MTDH and FBXW2 mRNA. Protein half-life and in vitro ubiquitination experiment was performed to validate whether FBXW2 regulates TAK1 degradation.
Results
Our findings unraveled the clinical significance of TAK1 in promoting HCC and sorafenib resistance. We identified a novel E3 ubiquitin ligase, FBXW2, targeting TAK1 for K48-linked polyubiquitylation and subsequent degradation. We also found that MTDH contributes to TAK1 up-regulation in HCC and sorafenib resistance through binding to FBXW2 mRNA and accelerates its degradation. Moreover, combination of TAK1 inhibitor and sorafenib suppressed the growth of sorafenib-resistant HCCLM3 xenograft in mouse models.
Conclusions
These results revealed novel mechanism underlying TAK1 protein degradation and highlighted the therapeutic value of targeting TAK1 in suppressing HCC and overcoming sorafenib resistance.
Keywords: Hepatocellular Carcinoma, Sorafenib, Drug Resistance, TAK1, Ubiquitin-Mediated Proteolysis
Abbreviations used in this paper: CHX, cycloheximide; DAPI, 4′,6-diamidino-2-phenylindole; DFS, disease-free survival; DMSO, dimethyl sulfoxide; EMT, epithelial-mesenchymal transition; GEO, Gene Expression Omnibus; GSEA, gene set enrichment analysis; HCC, hepatocellular carcinoma; IC50, half maximal inhibitory concentration; IHC, immunohistochemistry; IP, immunoprecipitation; NAFLD, nonalcoholic fatty liver disease; NASH, nonalcoholic steatohepatitis; NF-κB, nuclear factor kappa B; OXO, 5z-7-oxozeaenol; qPCR, quantitative polymerase chain reaction; RIP, RNA immunoprecipitation; RT, real-time; siRNA, small interfering RNA; SR, sorafenib-resistant; SRRSH, Sir Run Run Shaw Hospital; TAK1, transforming growth factor-β–activated kinase 1; TCGA, The Cancer Genome Atlas; TNF-α, tumor necrosis factor-alpha
Graphical abstract
Summary.
The study unravels the clinical significance of TAK1 in HCC and sorafenib resistance. We identified a novel E3 ubiquitin ligase, FBXW2, targeting TAK1 for degradation, and MTDH contributes to TAK1 up-regulation through binding to FBXW2 mRNA and accelerates its degradation.
Liver cancer is among the most commonly diagnosed cancers and the leading cause of cancer deaths. Hepatocellular carcinoma (HCC) comprises 75%–85% of liver cancer cases.1 HCC is of great tumor heterogeneity, which is the major cause of tumor progression and treatment failures and could be the result of diverse mutations that have been identified by studies through multiomics approaches.2 Sorafenib is the first targeted drug but provides limited improvement in patient survival because of the low frequency of sorafenib-targeting mutations and the early occurrence of secondary drug resistance in HCC patients.3 As we previously reviewed, the underlying mechanisms of sorafenib resistance involve various cellular processes, complex regulatory signaling network, genetic and epigenetic regulations, and tumor microenvironment.4 On the basis of these researches, numerous drugs including small molecule agonists or inhibitors, epigenetic, metabolic, or microenvironmental modulators, stemness inhibitors, oxidative stress inducers, and even nucleic acid therapies were being explored in combination with sorafenib to improve HCC patient outcomes.4, 5, 6 However, none of them had been applied in clinical use. Hence, identifying targets especially those with multiple signals convergence or divergence to overcome sorafenib resistance is of great clinical significance.
Transforming growth factor-β–activated kinase 1 (TAK1, also known as MAP3K7) can be activated by a diverse set of intracellular and extracellular stressors and is known to regulate nuclear factor kappa B (NF-κB), transforming growth factor-β, and mitogen-activated protein kinase signaling pathways. Accumulating studies demonstrated that TAK1 mediates pro-survival activities, tumor progression, and chemoresistance in cancers.7 However, as a hub of cellular homeostasis, the activity of TAK1 is context-dependent and cancer type-dependent, which is mainly attributed to the dual effect of TAK1-mediated NF-κB and transforming growth factor-β signaling in cancers.8 The role of TAK1 in liver is diverse. Early studies presented that TAK1 deletion in hepatocytes caused severe cell death, compensatory proliferation, hepatic inflammation, liver fibrosis, and early-onset of hepatocarcinogenesis.9,10 However, TAK1 deletion is rarely found in HCC patients according to The Cancer Genome Atlas (TCGA) database. Moreover, recent studies also proved that TAK1 was associated with poor survival of HCC patients and promoted hepatic steatosis, epithelial-mesenchymal transition (EMT) phenotypes, drug resistance, and cancer metastasis.11, 12, 13 These studies indicated that TAK1 might exert pro-survival function in transformed malignant hepatocytes. In this context, the role of TAK1 in HCC progression and therapy resistance requires further elucidation.
TAK1 activity is regulated by its binding partners TAB1 and TAB2/TAB3 and relies on post-translational modifications including ubiquitination and phosphorylation.14 Several E3 ligases, deubiquitinating enzymes, and phosphatases had been previously identified mediating K63-linked polyubiquitylation or deubiquitylation and dephosphorylation of TAK1, leading to TAK1 activation or inactivation.7,11,15,16 However, the E3 candidates targeting TAK1 for K48-linked polyubiquitylation and degradation are largely unknown. MTDH has already been proved to be a master regulator in several crucial aspects of tumor progression, including transformation, evasion of apoptosis, invasion, metastasis, and chemoresistance. TAK1 has a large overlap with MTDH in terms of downstream pathways and biological functions.17,18 However, whether MTDH and TAK1 regulate with each other remains unknown, and their roles in sorafenib resistance have not been well-studied.
Here we demonstrated that both MTDH and TAK1 contribute to HCC progression and sorafenib resistance, with the combination of the in vitro and in vivo studies and clinical data analysis from HCC patients. We identified FBXW2 as a novel E3 ubiquitin ligase targeting TAK1 for K48-linked polyubiquitination and degradation, and MTDH functions as an upstream effector of TAK1 at post-translational level through binding to FBXW2 mRNA and promoting its degradation. Our results highlighted the key roles and the molecular mechanisms of MTDH/TAK1 axis in HCC and sorafenib resistance, thus providing novel targets for clinical intervention.
Results
TAK1 Expression Is Up-Regulated in Developed HCC
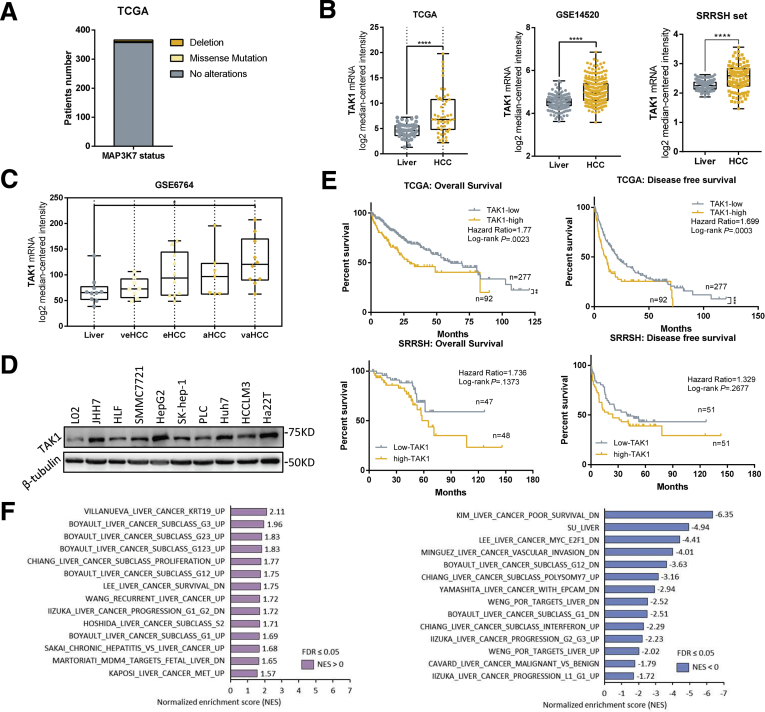
We first analyzed the mutation status and expression of TAK1 in silico using TCGA database, Gene Expression Omnibus (GEO) dataset (GSE14520), and our own dataset (Sir Run-Run Shaw Hospital [SRRSH]). We observed that TAK1 deletion is rarely found in HCC patients (6/366), not to mention missense mutation (3/366) with unknown function in TCGA dataset (Figure 1A). But mRNA expression of TAK1 was significantly elevated in human HCC tissues as compared with non-tumorous livers (Figure 1B). Moreover, analysis of GSE6764 dataset revealed that TAK1 expression was already up-regulated in very early HCC and tended to increase with HCC progression (Figure 1C). We also observed ranging TAK1 protein level among different HCC cell lines, almost all of which exhibited higher TAK1 expression than that in normal liver cells (Figure 1D). Survival analysis of TCGA dataset showed that TAK1 expression was negatively correlated with overall survival (OS) and disease-free survival (DFS) of HCC patients (Figure 1E). Moreover, SRRSH dataset analysis revealed similar results (Figure 1E). To address the role of TAK1 in HCC patients, we carried out gene set enrichment analysis (GSEA) using TCGA data and revealed that high TAK1 expression was positively correlated with a series of up-regulated gene sets in HCC and was negatively correlated with those down-regulated in HCC (Figure 1F). Taken together, these data suggested that although artificial TAK1 deletion leads to HCC initially, TAK1 functions as a tumor promoter in developed HCC and is a potential prognostic marker of HCC patients.
Figure 1.
TAK1 expression in HCC. (A) MAP3K7 (encoding TAK1 protein) mutation status in TCGA database; (B) mRNA expression of TAK1 in human HCC tissues as compared with non-tumorous livers from TCGA, GSE14520, and SRRSH datasets. (C) mRNA expression of TAK1 in different stages of HCC from GSE6764 dataset. (D) Protein level of TAK1 among different HCC cell lines compared with normal liver cell lines (L02 and Chang). (E) Kaplan-Meier survival curves of patients with low or high TAK1 mRNA expression based on TCGA and SRRSH datasets. Significance was determined using Kaplan-Meier analysis. Survival analysis was performed using log-rank test. (F) Gene sets associated with TAK1 expression using TCGA data and GSEA analysis.
TAK1 Suppression Inhibits Clonogenicity, Proliferation, and Oncogenic Signaling in HCC
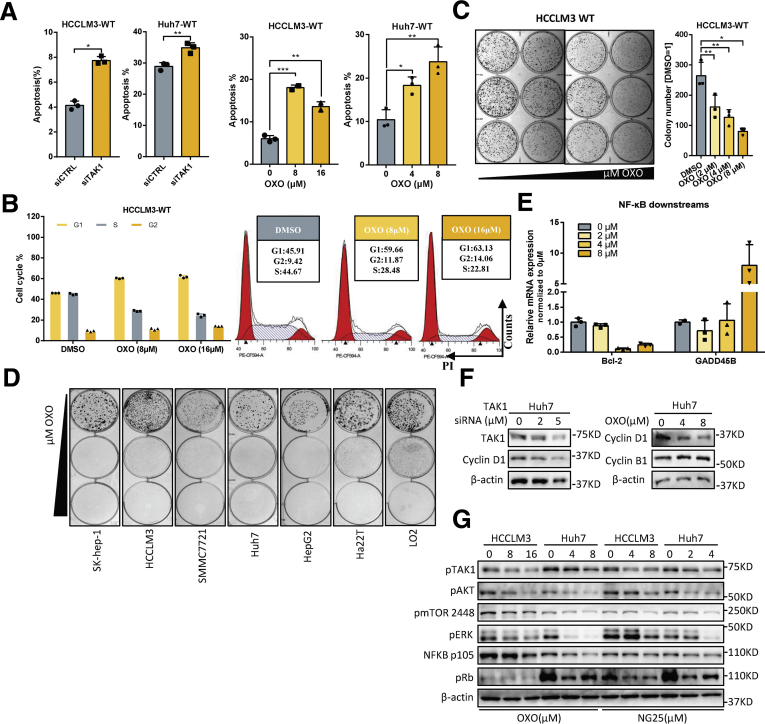
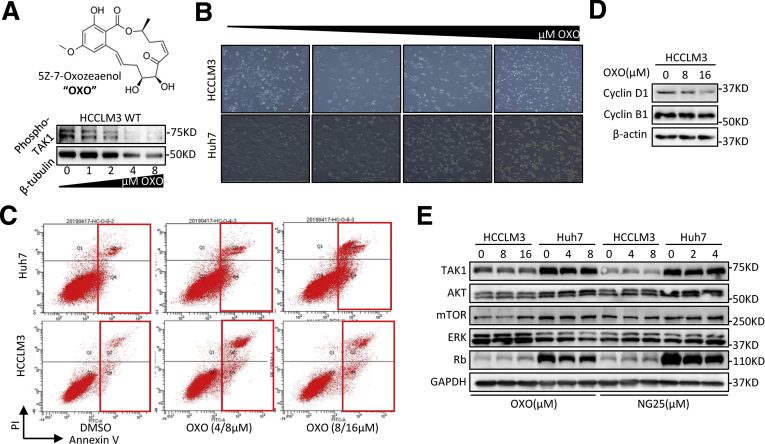
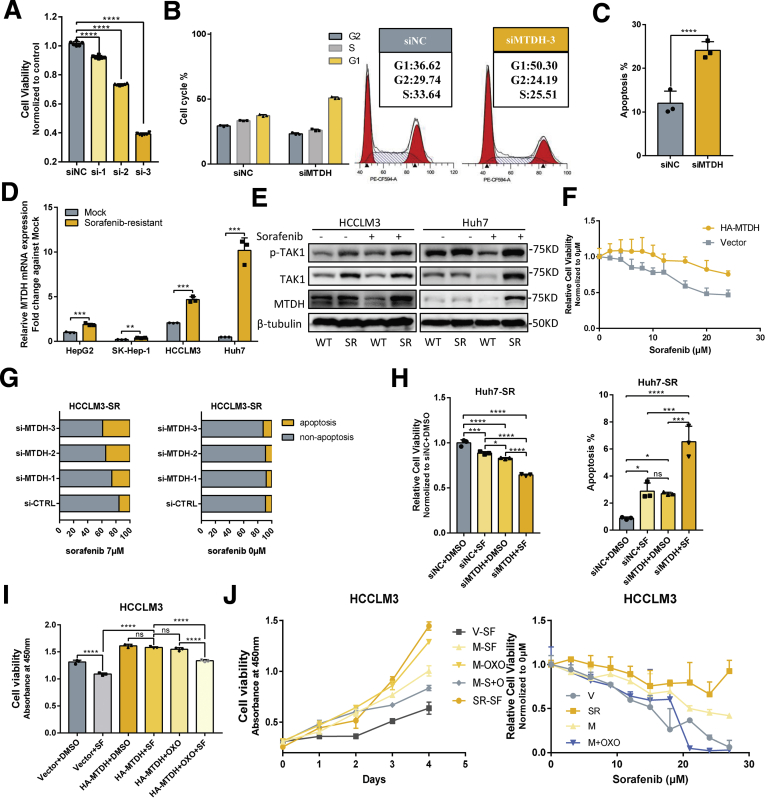
To better address the function of TAK1 in HCC, we carried out experiments to determine the functional impact of TAK1 in Huh7 and HCCLM3 cell lines. Genetic knockdown of TAK1 by specific small interfering RNA (siRNA) and pharmacologic inhibition of phospho-TAK1 by 5z-7-oxozeaenol (OXO)19 both strongly induced apoptosis in these 2 cell lines, with increased annexin V–stained apoptotic cells and accumulated floating cell fragments (Figure 2A, Figure 3A–C). In addition, TAK1 suppression also caused cell cycle arrest in G1 phase and inhibited clonogenicity of HCC cells (Figure 2B and C). Moreover, TAK1 suppression inhibited cell proliferation in most HCC cell lines (Figure 2D). TAK1 was known to boost NF-κB phosphorylation and its transcriptional activity,12 which mediates the transcription of numerous pro-survival and anti-apoptosis signals in HCC.20 Therefore, we tested the mRNA expression of NF-κB targets through quantitative polymerase chain reaction (qPCR), and the results showed lower expression of Bcl-2, an anti-apoptosis regulator, and higher expression of GADD45B, a DNA damage inducer, on OXO treatment (Figure 2E). TAK1 activates multiple oncogenic signals, which were known to be critical players in HCC tumorigenesis. Indeed, OXO treatment or TAK1 knockdown significantly reduced the protein expression of phosphorylated ERK, phosphorylated AKT, phosphorylated mTOR, phosphorylated Rb, and cyclin D1 but not cyclin B1, whereas total protein levels of above genes were not altered (Figure 2F and G, Figure 3D and E). Altogether, TAK1 suppression induced cell cycle arrest and cell apoptosis in HCC cells and inhibited clonogenicity, proliferation, and oncogenic signaling in vitro, indicating TAK1 as a potential target for developed HCC.
Figure 2.
Function of TAK1 in HCC. (A) Proportion of annexin V-stained apoptotic cells analyzed by flow cytometry in Huh7 or HCCLM3 cells transfected with TAK1 siRNA or OXO treatment, a TAK1 inhibitor. Three biological repeats per group. (B) Cell cycle analysis and representative results of HCCLM3 cells using flow cytometry on OXO treatment. Three biological repeats per group. (C) Colony formation of HCCLM3 cells treated with different concentration of OXO. Colony numbers were measured by Image J software. Three biological repeats per group. (D) Colony formation of different HCC cell lines on OXO treatment with different concentrations. (E) Real-time (RT) qPCR quantification of Bcl-2 and GADD45β mRNA in HCCLM3 cells treated with OXO or not. (F and G) Western blotting analysis of TAK1 downstream proteins levels on genetic knockdown or pharmacologic inhibition of TAK1 in Huh7 and HCCLM3 cells.
Figure 3.
Supplementary data for TAK1 inhibition in HCC cell lines. (A) Western blotting analysis of phosphorylated TAK1 protein at ser439 site in HCCLM3 cells on treatment of OXO. (B) Representative images of HCCLM3 treated with OXO of increasing concentrations. (C) Representative results of flow cytometry analysis of HCCLM3 and Huh7 cells treated with OXO or not. (D) Western blotting analysis of cyclin D1 and cyclin B1 in HCCLM3 on OXO treatment. (E) Western blotting analysis of total TAK1 downstream proteins in HCCLM3 and Huh7 on OXO or NG-25 treatment.
TAK1 Expression Can Be Reduced by Targeted Drugs and Is Retained in Sorafenib-Resistant Cell Lines
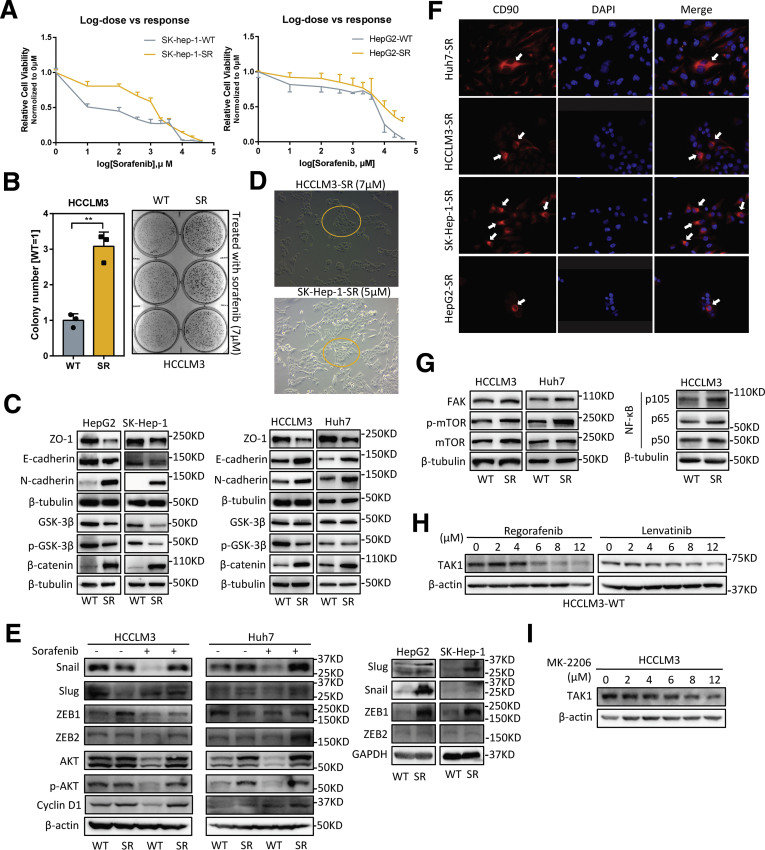
To unravel the role of TAK1 in sorafenib resistance, we established stable sorafenib-resistant (SR) models of normal HCC cell lines (Huh7, SK-hep-1, HepG2, and HCCLM3), following the instructions in previous study.6,21 The SR was determined by the half maximal inhibitory concentration (IC50) shift toward a higher concentration in all the resistant cell lines compared with their parental cell lines (Figure 4A, Figure 5A). The colony formation assays also showed that SR cells are more proliferative under sorafenib treatment (Figure 5B).
Figure 4.
Features of SR cells. (A) Relative cell viability of SK-hep-1-SR and HepG2-SR cells on sorafenib treatment with increasing dosage compared with their parental cells. (B) Colony formation assay was performed in HCCLM3 wild-type (WT) and SR cells treated with sorafenib (7 μmol/L). Colony numbers were measured by Image J software. Three biological repeats per group. (C) Western blotting analysis of EMT-related markers and proteins in SR cells compared with their parental cells. (D) Representative images of HCCLM3-SR and SK-hep-1-SR cells treated with OXO of indicated concentrations. (E) Western blotting analysis of EMT-related transcriptional factors in SR cells compared with their parental cells on sorafenib treatment or not. (F) Representative images of CD90 staining in 4 SR cells. Red, CD90; blue, DAPI. (G) Western blotting analysis of TAK1 downstream proteins in SR cells compared with their parental cells. (H and I) Western blotting analysis of TAK1 protein in HCCLM3 cells on treatment of regorafenib, lenvatinib, or MK-2206.
Figure 5.
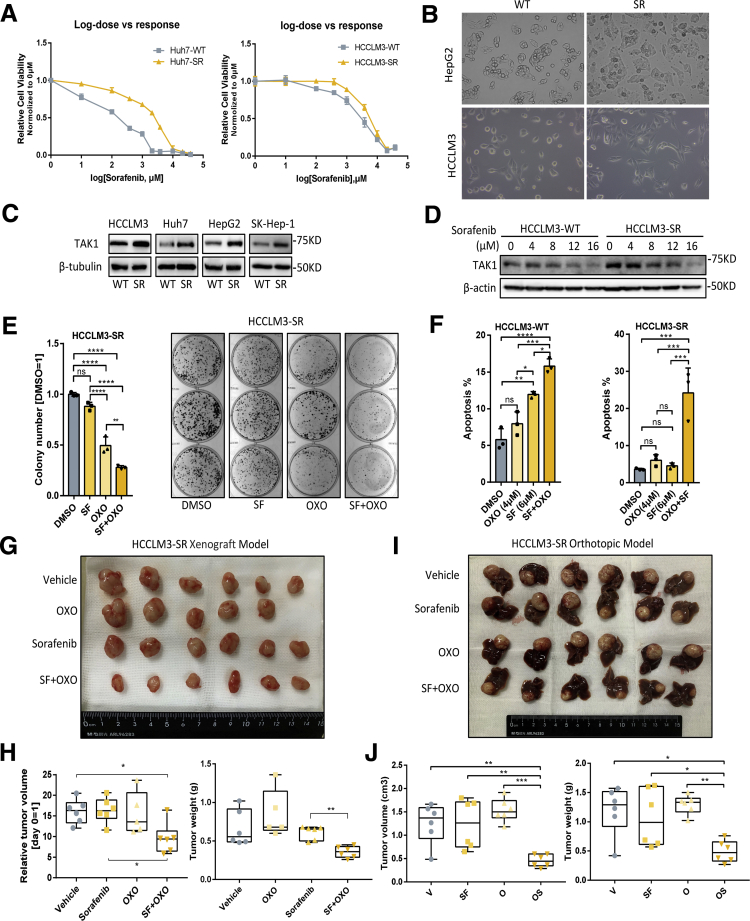
TAK1 expression and function in sorafenib resistance. (A) Relative cell viability of Huh7-SR and HCCLM3-SR cells on sorafenib treatment with increasing dosage compared with their parental cells. Relative cell viability was measured by cell counting kit 8 (CCK8) reagent and normalized to 0 μml/L. (B) Morphologic changes of Hepg2-SR and HCCLM3-SR cells compared with their parental cells. (C) Western blotting analysis of TAK1 protein level in 4 SR cells compared with their parental cells. (D) Western blotting analysis of TAK1 protein level in HCCLM3-SR cells compared with their parental cells in response to sorafenib treatment. (E) Colony formation assay was performed in HCCLM3-SR cells treated with sorafenib alone or in combination with OXO. Colony numbers were measured by Image J software. Three biological repeats per group. (F) Proportion of annexin V-stained apoptotic cells analyzed by flow cytometry in HCCLM3-SR or its parental cells treated with sorafenib alone or in combination with OXO. Three biological repeats per group. (G) Gross view of tumors from xenograft model treated with sorafenib alone or in combination with OXO. (H) Total volume or weight of tumor burden in mice treated with sorafenib alone or in combination with OXO from xenograft model. (I) Gross view of tumors attached with liver from orthotopic model treated with sorafenib alone or in combination with OXO. (J) Total volume or weight of tumor burden in mice treated with sorafenib alone or in combination with OXO from orthotopic model.
Both primary and second drug resistance are mainly attributed to tumor heterogeneity.22 In many tumor entities including HCC, therapy-induced evolving is achieved through epithelial-to-mesenchymal transition (EMT) and cancer stemness acquisition.23, 24, 25 Therefore, we first characterized the mesenchymal state and stemness features in SR cell line models. As expected, we observed profound morphologic changes in SR cells transforming into spindle-like shape under an electron microscope (Figure 5B), indicating that SR cells were undergoing EMT process, which was one of the well-known underlying mechanisms of SR.26 The EMT status in SR cells was further characterized by loss of ZO1 and gain of mesenchymal markers such as N-cadherin, although the expression of another epithelial marker E-cadherin was not consistently changed among 4 SR cell lines (Figure 4C). Unlike other studies,21 the SR cells we build tend to cluster together rather than being separated from each other, especially under sorafenib treatment, and this might be attributed to the retention of E-cadherin to maintain cell-to-cell contacts (Figure 4D). These features might also be explained by the existence of intermediate EMT states and multifunction of E-cadherin according to recent advances in research.27,28 EMT-related transcriptional factors are key regulators of EMT process and could also promote HCC progression.29 We further checked the expression of EMT- related transcriptional factors in SR cells compared with their parental cells on sorafenib treatment or not. The immunoblotting results showed that not all EMT-related transcriptional factors were changed accordingly, further indicating the heterogeneity at transcriptional level (Figure 4E). Because that gain of stemness improves drug tolerance of normal tumor cells,4 we examined the stem features of SR cells by CD90 staining, a stemness marker. Expectedly, CD90 positive SR cells were enriched in the center of normal SR cell clusters (Figure 4F), suggesting that some of SR cells acquired stemness traits and became the origin of sorafenib resistance. Oncogenic pathway activation and intracellular signaling compensation on drug treatment are important layers of HCC progression and sorafenib resistance.30 Indeed, we revealed higher cyclin D1, p-AKT, FAK, p-mTOR, p105/p50, GSK-3β, and β-catenin expression in SR cells, indicating activation of MAPK, PI3K/AKT, NF-κB, and Wnt pathways in sorafenib resistance (Figure 4C, E, and G).
Next we detected the TAK1 expression in SR cells compared with the parental cells and observed significantly higher protein level of TAK1 in 4 SR cells (Figure 5C). Interestingly, on sorafenib treatment, the protein level of TAK1 decreases as the concentration increases in the parental HCC cells but decreases more slowly in SR cells (Figure 5D). Such TAK1 protein decreases could also be observed on regorafenib or lenvatinib treatment (Figure 4H). These suggested TAK1 is a direct or indirect target of targeted drugs. Sorafenib as well as other tyrosine kinase inhibitors are well-known to inhibit Ras/MEK/ERK and PI3K/Akt/mTOR pathways. In this context, we treated HCC cells with increasing concentration of AKT inhibitor, MK-2206. We found that AKT inhibitor decreased total protein of TAK1 as well, suggesting that sorafenib might regulate TAK1 level through AKT pathway (Figure 4I). We also believed that there are other factors involved. More importantly, the regulatory mechanism of TAK1 expression in SR cells on long-term sorafenib treatment might be totally different from short-term inhibitory effect on TAK1 expression.
Targeting TAK1 Overcomes Sorafenib Resistance in vitro and in vivo
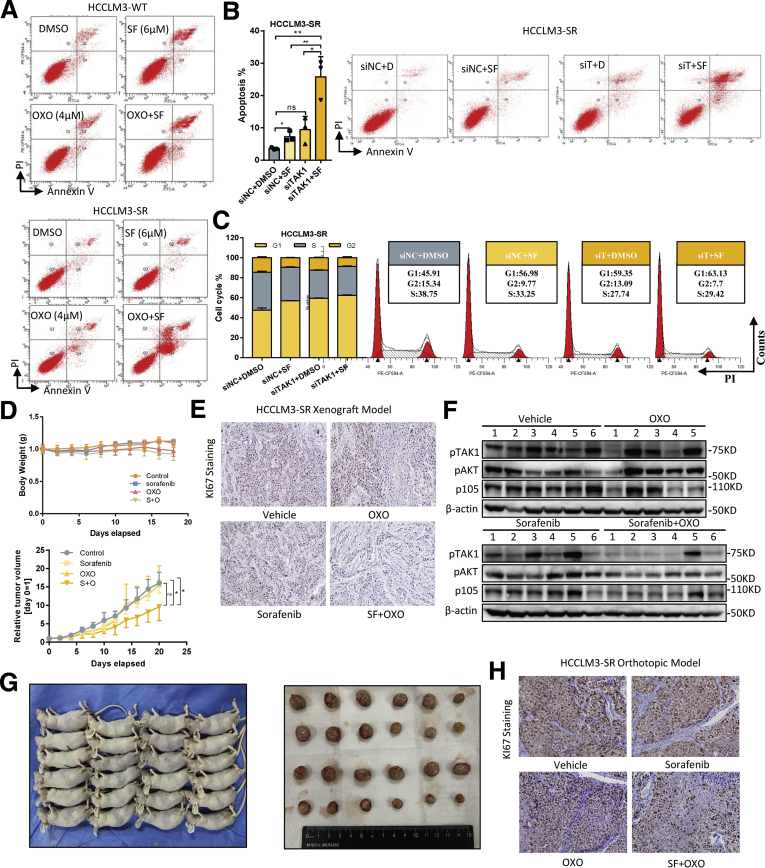
High expression of TAK1 in SR cells also prompted us to explore whether targeting TAK1 pharmacologically or via genetic knockdown reverses sorafenib resistance of SR cells. Indeed, combination of sorafenib and OXO strongly inhibited its clonogenicity and proliferation compared with sorafenib or OXO treatment alone in SR cell models (Figure 5E). Flow cytometry assays showed that OXO or TAK1 knockdown in combination with sorafenib largely increased proportion of apoptotic HCC cells and re-sensitized SR cells to sorafenib-induced apoptosis, whereas both sorafenib and OXO alone induced apoptosis in parental cells (Figure 5F, Figure 6A and B). Particularly, we used an in vivo SR cells xenograft and tumor model to evaluate the combined anticancer effect of sorafenib and OXO by inoculating subcutaneously the HCCLM3-SR cells into the flank sides of nude mice, followed by treatment with vehicle control, OXO, or sorafenib alone or in combination. When nontoxic concentrations were used (Figure 6D), sorafenib or OXO alone did not suppress the tumor growth in vivo; however, significant suppression was observed in combination group in terms of tumor volumes, tumor weights, and Ki67-staining, as well as downstream oncogenic proteins (Figure 5G and H, Figure 6D–F). Because liver microenvironment has a crucial impact on sorafenib resistance, we also adopted in vivo orthotopic model, and consistent results were observed (Figure 5I and J, Figure 6G and H). Thus, TAK1 inhibitors sensitized HCC SR cells to sorafenib as tested in both in vitro cell culture and in vivo xenograft models.
Figure 6.
Supplementary data for TAK1 inhibition in combination with sorafenib in vivo. (A) Representative results of apoptosis analysis in HCCLM3-SR and its parental cells treated with sorafenib and OXO alone or in combination. (B) Proportion of annexin V-stained apoptotic cells analyzed by flow cytometry, and (C) cell cycle analysis and representative results of HCCLM3-SR cells transfected with TAK1 siRNA or scramble control. Three biological repeats per group. (D) Body weights of mice and total volume tumor burden in mice treated sorafenib alone or in combination with OXO in indicated time period. (E) Representative images of Ki67 staining in tumors from 4 groups. (F) Western blotting analysis of downstream proteins in tumors from mice treated with sorafenib and OXO alone or in combination. (G) Gross view of mice and tumors from orthotopic model. (H) Representative images of Ki67 staining of tumors from orthotopic model.
FBXW2 E3 Ubiquitin Ligase Targets TAK1 for Ubiquitination and Degradation
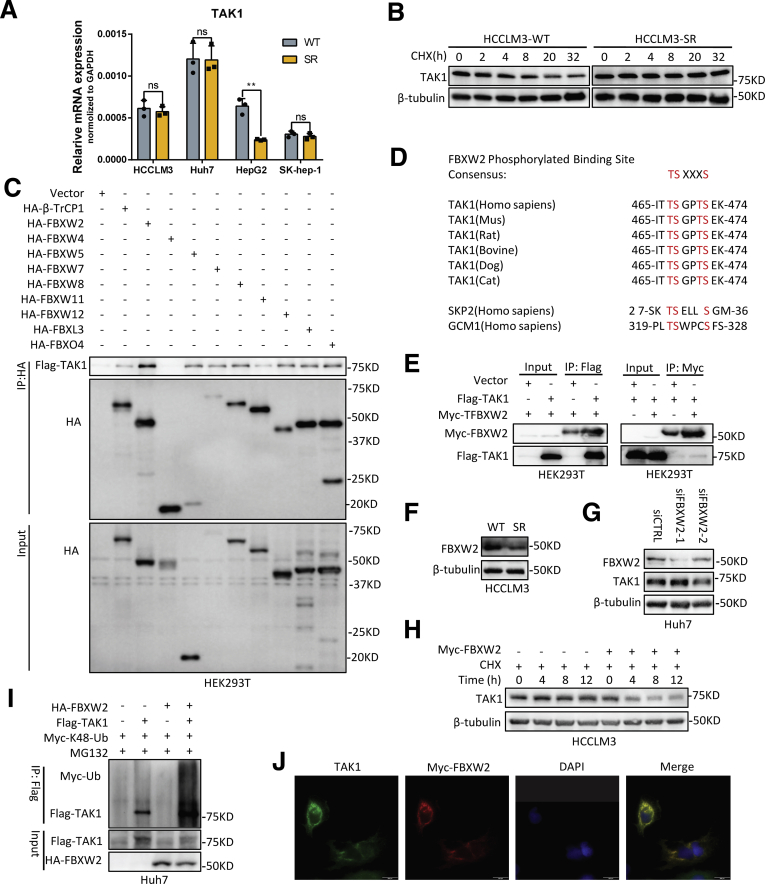
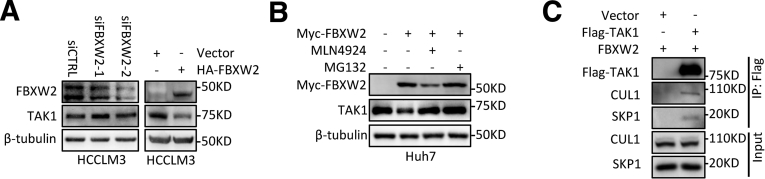
To uncover the regulatory mechanism of TAK1 expression in SR cells, we first detected mRNA level of TAK1 and observed no consistently significant changes between SR cell lines and their parental cells (Figure 7A). Furthermore, chlorhexidine (CHX) assay indicated that the TAK1 protein degradation was retarded in SR cells (Figure 7B). K48-linked polyubiquitination and degradation of TAK1 are largely unknown. F-box proteins are components of SKP1–cullin 1–F-box protein E3 ligase complexes and have pivotal roles in multiple cellular activities.31 However, none of F-box proteins have been proved to mediate TAK1 degradation. Following this lead, we first ectopically expressed a variety of F-box proteins including FBWX2 followed by immunoprecipitation (IP) pull-down and found in vitro interaction between FBXW2 and TAK1, although FBXW2 is not the only F-box protein that could bind to TAK1 but has the highest IP affinity of TAK1 (Figure 7C). F-box proteins bind short, defined degradation motifs in substrates.32 Regarding FBXW2 of which the substrates remain largely unknown, the consensus degron sequences defined as TSXXXS were required for its binding to substrates such as SKP2.33 Indeed, we also identified such motif in TAK1, which is evolutionarily conserved (Figure 7D), suggesting TAK1 is a potential substrate of FBXW2. The binding of FBXW2 and TAK1 was then specifically confirmed by IP pull-down of Flag-TAK1 or Myc-FBXW2 (Figure 7E).
Figure 7.
FBXW2 E3 ubiquitin ligase targets TAK1 for ubiquitination. (A) RT-qPCR quantification of TAK1 mRNA expression in 4 SR cells compared with their parental cells. (B) CHX assay was performed in HCCLM3-SR cells and their parental cells for indicated time periods, and TAK1 protein was measured by Western blotting analysis. (C) Immunoblotting analysis of HA and Flag-TAK1 in HA tag pull-down derived from HEK293T cells transfected with indicated constructs. (D) Evolutionary conservation of FBXW2 degron motif on TAK1. (E) HEK293 cells were transfected with indicated plasmids, followed by IP with anti-Flag or anti-Myc (F) Western blotting analysis of FBXW2 expression in HCCLM3-SR and its parental cells. (H) Half-life of TAK1 protein in HCCLM3 cells transfected with Myc-FBXW2 or vector plasmid as control. (I) Immunoblotting analysis of Myc-Ub in Flag tag pull-down and input derived from HEK293T cells transfected with indicated constructs. (J) Immunofluorescence analysis of TAK1 and Myc-FBXW2 location in Huh7 cells. Green, TAK1; red, Myc-FBXW2; blue, DAPI.
Notably, it was found that FBXW2 protein expression was down-regulated in SR cells compared with parental HCC cells, indicating that it might be responsible for retarded degradation of TAK1 in SR cells (Figure 7F). Hence, we assessed the effect of FBXW2 on TAK1 level. Indeed, FBXW2 knockdown increased the levels of total TAK1 protein, whereas FBXW2 overexpression reduced them, which could be reversed by MG132 or MLN4924, a NEDD8-activating enzyme inhibitor that inhibits the neddylation of the Cullin subunits of Cullin RING E3 ligases (Figure 7G, Figure 8A and B). These data together with the existence of SKP1 and CUL1 protein in TAK1 immunoprecipitants (Figure 8C) suggested that FBXW2 regulated TAK1 protein level through SKP1-CUL1-FBXW2 complex. Indeed, FBXW2 promoted TAK1 K48-linked polyubiquitylation and shortened its protein half-life (Figure 7H and I). Immunofluorescence analysis suggested that TAK1 and FBXW2 were co-localized in cytoplasm (Figure 7J). Taken together, we identified FBXW2 as a new E3 ubiquitin ligase targeting TAK1 for K48-linked polyubiquitylation and degradation.
Figure 8.
Supplementary data for TAK1 and FBXW2. (A and B) Western blotting analysis of TAK1 expression in HCCLM3 cells or Huh7 cells with FBXW2 knockdown or overexpression, treated with MG132 or MLN4924. (C) Immunoblotting analysis of CUL1 and SKP1 in Flag tag pull-down and input derived from HEK293T cells transfected with indicated constructs.
MTDH Regulates TAK1 at Protein Level Through Promoting FBXW2 mRNA Degradation
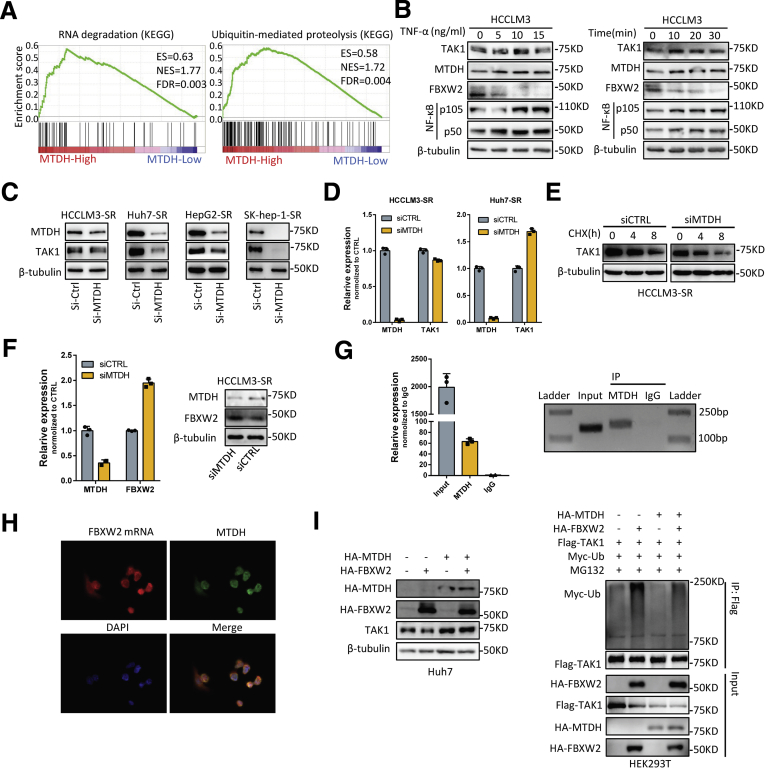
The upstream regulators of TAK1 remain largely unknown, whereas its downstream effectors have been widely explored. As a master regulator of cell death, TAK1 is well-known to regulate NK-κB pathway. Tumor necrosis factor-alpha (TNF-α)/NF-κB pathway plays important role in the development of inflammation-driven HCC including viral hepatitis-related and nonalcoholic steatohepatitis (NASH)- or nonalcoholic fatty liver disease (NAFLD)-related HCC.34 Recent study also indicated that TNF-α might serve as a predictor of sorafenib response in HCC patients.35 Following this clue, it is quite possible that TAK1 upstream regulators might also regulate NF-κB signaling and contribute to drug resistance. We reviewed relative articles in PubMed and found that MTDH functions similarly with TAK1 in tumor progression and therapy resistance, activates common downstream pathways, and responds to similar intracellular or extracellular stimuli.7,17 Moreover, GSEA analysis of defined gene sets revealed that MTDH was highly involved in RNA degradation and ubiquitin-mediated proteolysis (Figure 9A). In line with this, several researches had implied that MTDH acted as an RNA-binding protein.36, 37, 38 Also illustrated was the decreased mRNAs that could bind to MTDH including a large amount of E2 and E3 ubiquitin ligases such as FBXW2 on a dual PI3K/mTOR inhibitor treatment.36 This indicates that MTDH could accelerate the degradation of these mRNAs in response to drug stimuli. However, the role of MTDH in RNA stability or translation is not known. These findings suggested that MTDH might regulate TAK1 expression through inhibiting FBXW2 mediated TAK1 degradation. We first treated HCC cells with TNF-α. Results showed that TNF-α treatment increased MTDH and TAK1 protein expression but decreased FBXW2 protein level in a time course and in a dose-dependent manner (Figure 9B).
Figure 9.
MTDH functions as an upstream regulator of TAK1. (A) KEGG pathways significantly enriched in HCC patients with high expression of MTDH using TCGA data. (B) Western blotting analysis of TAK1, MTDH, and FBXW2 expression in HCCLM3 cells in response to TNF-α treatment. (C and D) RT-qPCR quantification and Western blotting analysis of TAK1 and MTDH expression in SR cells transfected with MTDH siRNA or scramble control. (E) Half-life of TAK1 protein in HCCLM3 cells transfected with MTDH siRNA or scramble control. (F) RT-qPCR quantification and Western blotting analysis of MTDH and FBXW2 protein expression in HCCLM3 cells transfected with MTDH siRNA or scramble control. (G) RIP and RT-qPCR analysis of FBXW2 mRNA binding to MTDH protein compared with immunoglobulin G. (H) Immunofluorescence analysis of MTDH and FBXW2 mRNA location in HCCLM3 cells. Green, MTDH; red, FBXW2 mRNA; blue, DAPI. (I) Immunoblotting analysis of TAK1 protein and Myc-Ub expression in Huh7 cell lysates transfected with indicated constructs. ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score.
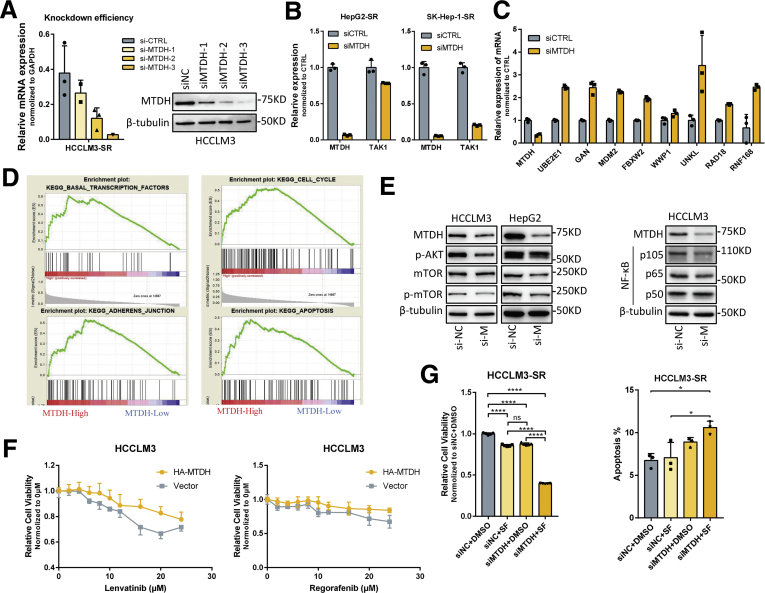
We then altered the MTDH levels to examine its effect on TAK1 level. MTDH knockdown significantly reduced TAK1 protein expression, whereas the mRNA levels of TAK1 were not consistent among 4 HCC cell lines (Figure 9C and D, Figure 10A and B), indicating that post-translational regulations were involved. Indeed, further mechanistic dissection demonstrated that MTDH knockdown could accelerate the protein degradation of TAK1 (Figure 9E). We thus detected the mRNA expression of those E2 and E3 ligases on MTDH knockdown and found that the decrease of MTDH expression increased their mRNA level (Figure 10C). Regarding FBXW2, MTDH knockdown also increased its protein level in HCC cells (Figure 9F).
Figure 10.
Supplementary data for regulation of TAK1 by MTDH. (A) RT-qPCR analysis and Western blotting analysis of MTDH expression in HCCLM3 cells on MTDH knockdown using 3 siRNAs. (B) RT-qPCR quantification of TAK1 and MTDH expression in SK-hep-1 and HepG2 cells transfected with MTDH siRNA or scramble control. (C) RT-qPCR quantification of E3 mRNAs expression in HCCLM3-SR cells transfected with MTDH siRNA or scramble control. (D) KEGG pathways significantly enriched in HCC patients with high expression of MTDH using TCGA data. (E) Western blotting analysis of MTDH and NF-κB pathway proteins in HCCLM3-SR and its parental cells; Western blotting analysis of MTDH and AKT pathway proteins in HCCLM3-SR and HepG2-SRs on MTDH knockdown. (F) Cell viability of HCCLM3 with stable expression MTDH treated with lenvatinib or regorafenib. (G) Relative cell viability and proportion of apoptotic cells in HCCLM3 cells transfected with MTDH siRNA or scramble control in combination with sorafenib or DMSO. Three biological repeats per group.
Altogether, we hypothesized that MTDH regulates K48-linked polyubiquitination and degradation of TAK1 through down-regulating its E3 ubiquitin ligase FBXW2. We first used RNA-IP (RIP) and revealed much higher MTDH-bound FBXW2 mRNA compared with immunoglobulin G, suggesting the specific association of FBXW2 mRNA and MTDH (Figure 9G). Immunofluorescence and fluorescence in situ hybridization assay for MTDH protein and FBXW2 mRNA also showed that FBXW2 mRNA overlapped with those MTDH localized at cytoplasm (Figure 9H). Moreover, we found that MTDH overexpression partially reduced TAK1 protein level and reduced K48-polyubiquitylation on FBXW2 transfection (Figure 9I), suggesting FBXW2-mediated TAK1 degradation could be mitigated by MTDH. In summary, MTDH regulates TAK1 protein level through binding to and subsequently degrading FBXW2 mRNA.
TAK1 Mediates MTDH-Induced Sorafenib Resistance
To further demonstrate the relationship between MTDH and TAK1, we then aimed to elucidate the function of MTDH in HCC and sorafenib resistance, and whether TAK1 mediates these functions. MTDH has been regarded as a driver oncogene in HCC.39 Although MTDH has been widely proved to be involved in chemotherapeutics resistance,17 its role in targeted therapy sensitivity remains unknown. We first used knockdown MTDH by 3 specific siRNAs and found that MTDH knockdown inhibited proliferation of HCC cells, and the third siRNA performed best according to the decrease in cell viability (Figure 11A). Knockdown of MTDH also induced apoptosis and cell cycle arrest in G1 phase of HCC cells (Figure 11B and C). GSEA using TCGA data revealed that MTDH expression was closely related to the regulation of transcription factor activity, cell cycle, apoptosis, mTOR pathways, and TGF-β pathway (Table 1, Figure 10D), as indicated by previous studies.18 Indeed, MTDH suppression reduced classic oncogenic signaling such as p-AKT, p-mTOR, and NF-κB in HCC (Figure 10E). We then detected the expression of MTDH in our SR cell models and observed significantly higher protein and mRNA levels of MTDH in SR cells, especially under sorafenib treatment, consistent with the expression alteration of both TAK1 and p-TAK1 (Figure 11D and E). To verify the role of MTDH in targeted-therapy resistance, we carried out functional experiments to assess the relationship between MTDH and the IC50 of several targeted drugs including sorafenib, regorafenib, and lenvatinib. The results showed that IC50 of these 3 drugs, especially sorafenib, was higher in HCC cells that stably expressed MTDH (Figure 10F, Figure 11F). We then combined MTDH knockdown and sorafenib treatment in SR cells to further evaluate the function of MTDH in sorafenib resistance. Results showed that MTDH knockdown enhanced sensitivity of SR cells to sorafenib-induced apoptosis and inhibitory of proliferation (Figure 10G, Figure 11G and H). Sorafenib or MTDH knockdown alone also induced apoptosis in SR cells, although the proportion of apoptotic cells remains low. This may be due to the heterogeneity of SR cells and the essential function of MTDH in HCC cell proliferation.
Figure 11.
Role of MTDH in HCC and sorafenib resistance. (A) Relative cell viability measured by CCK8, (B) cell cycle analysis and representative results, and (C) proportion of annexin V-stained apoptotic cells analyzed by flow cytometry of HCCLM3 cells transfected with MTDH siRNA or scramble control. Three biological repeats per group. (D and E) RT-qPCR analysis of MTDH mRNA level and Western blotting analysis of MTDH, TAK1, and p-TAK1 protein level in 4 SR cells compared with their parental cells on sorafenib treatment or not. (F) Relative cell viability of HCCLM3 with stable expression MTDH or not treated with increasing dosage of sorafenib. (G and H) Relative cell viability and proportion of apoptotic cells in HCCLM3 or Huh7 cells transfected with MTDH siRNA or scramble control in combination with sorafenib or DMSO. Three biological repeats per group. (I) Relative cell viability of HCCLM3 with stable expression MTDH or not treated with sorafenib and OXO alone or in combination for 48 hours. (J) Relative cell viability of HCCLM3 with stable expression MTDH or not treated with OXO or NG25 alone or in combination sorafenib of increasing dosage or for indicated time periods. Three biological repeats per group.
Table 1.
KEGG gene sets enriched in MTDH -high HCC patients in TCGA dataset
| Name | Size | ES | NES | NOM P value | FDR q value |
|---|---|---|---|---|---|
| KEGG_AMINOACYL_TRNA_BIOSYNTHESIS | 41 | 0.646 | 1.774 | <.001 | 0.005 |
| KEGG_RNA_DEGRADATION | 57 | 0.626 | 1.769 | <.001 | 0.003 |
| KEGG_UBIQUITIN_MEDIATED_PROTEOLYSIS | 133 | 0.579 | 1.724 | <.001 | 0.004 |
| KEGG_SPLICEOSOME | 124 | 0.572 | 1.687 | <.001 | 0.005 |
| KEGG_BASAL_TRANSCRIPTION_FACTORS | 35 | 0.606 | 1.637 | .001 | 0.010 |
| KEGG_NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY | 61 | 0.551 | 1.571 | <.001 | 0.028 |
| KEGG_CELL_CYCLE | 124 | 0.518 | 1.536 | <.001 | 0.043 |
| KEGG_ADHERENS_JUNCTION | 73 | 0.534 | 1.531 | <.001 | 0.041 |
| KEGG_EPITHELIAL_CELL_SIGNALING_IN_HELICOBACTER_PYLORI_INFECTION | 67 | 0.530 | 1.520 | <.001 | 0.042 |
| KEGG_HOMOLOGOUS_RECOMBINATION | 28 | 0.569 | 1.516 | .009 | 0.040 |
| KEGG_CHRONIC_MYELOID_LEUKEMIA | 73 | 0.511 | 1.476 | <.001 | 0.064 |
| KEGG_INOSITOL_PHOSPHATE_METABOLISM | 54 | 0.519 | 1.467 | .005 | 0.067 |
| KEGG_MTOR_SIGNALING_PATHWAY | 52 | 0.518 | 1.460 | .002 | 0.067 |
| KEGG_PATHOGENIC_ESCHERICHIA_COLI_INFECTION | 55 | 0.506 | 1.440 | .004 | 0.082 |
| KEGG_APOPTOSIS | 86 | 0.485 | 1.415 | .001 | 0.093 |
| KEGG_NEUROTROPHIN_SIGNALING_PATHWAY | 126 | 0.476 | 1.412 | <.001 | 0.091 |
| KEGG_ENDOCYTOSIS | 178 | 0.469 | 1.412 | <.001 | 0.087 |
| KEGG_PANCREATIC_CANCER | 70 | 0.493 | 1.409 | .002 | 0.087 |
| KEGG_SMALL_CELL_LUNG_CANCER | 84 | 0.484 | 1.407 | .002 | 0.084 |
| KEGG_ACUTE_MYELOID_LEUKEMIA | 57 | 0.491 | 1.403 | .009 | 0.077 |
| KEGG_RENAL_CELL_CARCINOMA | 70 | 0.482 | 1.394 | .005 | 0.081 |
| KEGG_OOCYTE_MEIOSIS | 111 | 0.468 | 1.393 | .002 | 0.079 |
| KEGG_TGF_BETA_SIGNALING_PATHWAY | 85 | 0.476 | 1.380 | .004 | 0.089 |
ES, enrichment score; FDR, false discovery rate; NES, normalized enrichment score; NOM, nominal.
Next we aimed to explore whether TAK1 is involved in MTDH-mediated sorafenib resistance. Consistent with previous findings, HCC cells with ectopically expressed MTDH became more resistant to sorafenib, and such resistance could be reversed by sorafenib in combination with OXO or NG-25, another TAK1 inhibitor (Figure 11I and J). These data suggested the functional relationship between MTDH and TAK1 in sorafenib resistance.
MTDH and TAK1 Are Both Correlated With Poor Survival of HCC Receiving Sorafenib
To unravel the clinical significance of TAK1 and MTDH, we used immunohistochemistry (IHC) to stain the TAK1 and MTDH protein in tumor samples from 59 HCC patients who underwent surgery before sorafenib treatment. Representative images indicated that HCC patients with high MTDH expression might also have higher TAK1 protein level (Figure 12A). Further IHC scoring and statistical analysis revealed that high MTDH expression significantly correlates with high TAK1 protein level (Figure 12B). Those patients were then stratified into 2 groups on the basis of TAK1 or MTDH protein level via IHC scoring. Further Kaplan–Meier survival analysis revealed that HCC patients with higher level of TAK1 or MTDH expression had shorter overall survival and DFS (Figure 12C). Taken together, these data indicated that TAK1 and MTDH were involved in the development of sorafenib resistance, and their expression positively correlates with each other and predicts sorafenib response in HCC patients.
Figure 12.
MTDH/TAK1 expression in HCC patients receiving sorafenib. (A) Representative images of MTDH and TAK1 immunohistochemical staining in HCC patients receiving sorafenib before surgery. (B) Correlation between MTDH and TAK1 staining. Significance was determined using χ2 (and Fisher exact) test. (C) Kaplan-Meier survival curves of these patients with low or high TAK1 or MTDH staining. Significance was determined using Kaplan-Meier analyses. Survival analysis was performed using log-rank test.
Discussion
Most HCC patients are diagnosed at an advanced stage when surgical approaches and locoregional therapies are no longer feasible. Improvements in patient outcomes from most systematic therapies have been modest, although several targeted drugs and immune checkpoint inhibitors have been proved effective in the last decade,40 and more combinational strategies are being explored.41 The underlying mechanisms of sorafenib resistance remain complex and largely unknown. To better address this critical problem, we had established acquired SR HCC cell models by treating HCC cells with increasing concentrations of sorafenib in culture media over time.6,21 On the basis of these models, our previous studies had proved the crucial role of androgen receptor signals, EMT-related transcriptional factors, and several non-coding RNAs in sorafenib resistance, highlighting the epigenetic regulation of driver oncogenes in sorafenib resistance.5,6,42, 43, 44, 45 However, therapy-induced tumor cell evolving is comprehensive and systematic in terms of cellular and molecular alteration. Thus, identifying other novel targets still remains an unmet medical need, requiring new insights into underlying molecular mechanisms that support hepatocarcinogenesis, HCC progression, and drug resistance based on genomic, transcriptomic, and epigenomic studies.
In our study, some issues had been raised about SR cell models. We previously reviewed that EMT and cancer stem cells are common mechanisms underlying acquired sorafenib resistance. However, SR cells in our study underwent atypical EMT process that not all EMT markers and EMT-related transcriptional factors altered accordingly especially E-cadherin, although SR cells exhibited mesenchymal-like morphologic changes. Recent studies revealed that EMT is a processive program, and there are tumor cells undergoing intermediate EMT process. Actually, such EMT heterogeneity has already been proposed and remains controversial in recent years.46 Cells express CDH1 (encodes E-cadherin) but do not display E-cadherin at the cell surface, cells with partially mesenchymal features but without evident expression of EMT-related transcriptional factors, and so on had been reported.47,48 Another possible reason is that E-cadherin might be asymmetrically distributed toward the cell surfaces.49 Padmanaban et al28 found that E-cadherin promotes metastasis through limiting reactive oxygen-mediated apoptosis in breast ductal carcinomas. To this point, E-cadherin itself might have uncovered roles in our SR model. We found that SR cells tend to be clustered under sorafenib treatment, and stemness marker staining also indicated that the stem cells are located in the core of such “cluster”. It is possible that E-cadherin mediates cell-to-cell connection during SR cell clustering, but further experiments are required to demonstrate this hypothesis. Not all SR cells exhibited stemness features, which is consistent with EMT heterogeneity mentioned above. Such heterogeneity may also lead to functional heterogeneity of SR cells, that is, during cell proliferation and division, some SR cell subpopulation may lose sorafenib resistance. This may explain why sorafenib alone could still induce apoptosis in SR cells, although it remains a very low ratio.
Recently in many other tumor entities, TAK1 has been considered a robust therapeutic target especially in lung, colon, and pancreatic cancers, most of which exhibit frequent KRAS mutations.50,51 Although artificial TAK1-deletion leads to early onset of hepatocarcinogenesis in liver, HCC patients rarely harbor TAK1-deletion mutations according to TCGA database. Moreover, we observed that HCC specimens harbor higher TAK1 mRNA expression, which was increasing from early-stage to advanced-stage HCC patients. Metabolic syndrome together with alcohol abuse and hepatitis C infection contribute to the increasing incidence and death rate of liver cancers including HCC.52 From an etiologic perspective, recent studies indicated that TAK1 promoted NASH and NAFLD.15,16,53 The role of TAK1 in inflammation-driven HCC including hepatic steatosis and virus infection might be attributed to TNF-α stimulation, which was also indicated to be potential target to overcome sorafenib resistance recently.35 These clues prompted us to explore the role of TAK1 in developed HCC, especially in HCC progression and therapy resistance. Indeed, we found elevated protein levels of TAK1 in HCC patients, HCC cell lines, and SR cell models, and its expression predicts poor patient outcomes. In vivo and in vitro assay also suggested that genetic knockdown and pharmacologic inhibition of TAK1 using inhibitors OXO or NG-25 significantly suppressed cell proliferation and tumor growth and overcame sorafenib resistance. But it is never too cautious to make the conclusion that TAK1 is a therapeutic target for HCC.
Because TAK1 deletion could cause spontaneous hepatocarcinogenesis, it is worthy of discussion about whether pharmacologic TAK1 inhibition could induce detrimental effect. First, we believed that TAK1 deletion and pharmacologic TAK1 inhibition are totally different conditions. The latter one only inhibits the phosphorylation of TAK1 in HCC cells and will not completely eliminate all TAK1 protein, which could continue to play essential functions. In addition, TAK1 inhibitors have been applied in other tumor models by many studies, but none reported spontaneous hepatocarcinogenesis in mice liver. Of course, it is also possible that the authors were not aware of this issue, or the duration of TAK1 inhibitors treatment is too short to induce hepatocarcinogenesis. Because of role of TAK1 in steatohepatitis, TAK1 inhibitor was also used to treat HFD (High fat diet)-induced NAFLD in DUSP14-ko mice, and hepatocarcinogenesis was not reported in this model.54 Moreover, TAK1 deletion induced hepatocarcinogenesis was not attributed to the inhibition of antiproliferative role of TAK1 but cell death-induced inflammation and compensatory mechanisms, for instance, promoting macrophage infiltration.55 Last, genetic TAK1 deletion in liver is artificial, and TAK1-deficiency induced hepatocarcinogenesis in natural conditions had not been reported. All in all, the function of TAK1 is context-dependent. In our study, we focused on the status of TAK1 in transformed malignant HCC cells and its role in subsequent tumor progression and treatment resistance, which may not be in conflict with the role of TAK1 deletion in nonmalignant hepatocytes.
This study also explored the regulatory mechanism of TAK1 expression. We observed that sorafenib treatment reduced total protein of TAK1 in HCC cells, suggesting TAK1 is a direct or indirect target of sorafenib. Sorafenib as well as other tyrosine kinase inhibitors are well-known to inhibit Ras/MEK/ERK and PI3K/Akt/mTOR pathways. Previous research demonstrated that TAK1 functions in a KRAS-dependent way in other tumor entities especially in pancreatic adenocarcinoma. Sorafenib inhibits non-small-cell lung cancer cell growth by targeting B-RAF in cells with wild-type KRAS and C-RAF in those with mutant KRAS.56 We knocked down KRAS expression using siRNA and inhibited AKT function using selective inhibitor in HCC cells. The Western blotting results showed that KRAS knockdown and AKT inhibitor both decreased the total TAK1 expression (data not shown). Hence, sorafenib might down-regulate TAK1 expression through Ras/MEK/ERK and PI3K/Akt/mTOR pathways. One study also proved that sorafenib inhibited ATP-binding of TAK1 using in situ kinome.57 Although the function of TAK1 and FBXW2 binding to TAK1 depends on specific site phosphorylation or dephosphorylation, it is possible that sorafenib regulates TAK1 level through inhibiting corresponding phosphorylation kinases. Altogether, the mechanistic reason for the decreased levels of TAK1 on sorafenib treatment might be more diverse and is in need for further study.
However, it is the TAK1 protein level but not mRNA expression that was elevated in SR cells and was retained even under sorafenib treatment. Although most studies focus on K63-polyubiquitylation-dependent activation of TAK1,11,58 these data lead us to explore which E3 ligase mediates K48-linked degradation of TAK1. F-box proteins mediate the degradation of a large number of regulatory proteins involved in diverse processes and have emerging roles in cancer and drug resistance.31,59 Previous study identified FBXW5 as a functional suppressor rather than an expression regulator for TAK1.60 We used IP assays and identified FBXW2 as a novel E3 ligase targeting TAK1 for K48-linked ubiquitylation and subsequent degradation through specific degron sequences. Our results also revealed that FBXW2 is not the only F-box protein that could bind to TAK1, regardless of nonspecific binding to tag beads. It is not surprising that other F-box proteins also regulate TAK1 activation and protein degradation because TAK1 is a gatekeeper of cellular homeostasis and must be under tight regulation. However, how other F-box proteins regulate TAK1 needs further investigation. Moreover, some issues remain unexplored in this study including how FBXW2 recruits the substrate TAK1, whether such recruitments depend on degron phosphorylation, and which kinases are responsible for the degron phosphorylation.
MTDH has been identified as a master and actionable gene in HCC as well as other tumor entities. It functions as a downstream mediator of the transforming activity of oncogenic Ha-Ras and c-Myc and activates MAPK, PI3K/Akt, NF-κB, and Wnt/β-catenin signaling pathways.18 Several studies have demonstrated that MTDH is an RNA-binding protein and could bind to many E3 or E2 mRNAs including FBXW2 and regulate their stability or translation.36,38 Furthermore, MTDH and TAK1 function similarly in tumor progression and therapy resistance, respond to overlapped stimuli, and activate common downstream pathways.7,17 Our expression analysis and functional experiments also proved that MTDH is essential for HCC development and sorafenib resistance. These findings and clues suggested that MTDH might be a potential upstream regulator for TAK1 at post-translational level. Indeed, our findings suggested that MTDH regulates TAK1 protein level through binding to FBXW2 mRNA and promoting its degradation. However, pharmacologic suppression of MTDH that is based on neither druggable structure nor bioactive compounds remains achievable. This study also proved that targeting TAK1 reversed MTDH-dependent and -independent sorafenib resistance in vitro and in vivo. In summary, MTDH overexpression in SR cells retarded the degradation of TAK1 protein and thus mitigated the inhibitory effect from sorafenib on TAK1. However, how long-term sorafenib treatment resulted in up-regulation of MTDH is still unknown and has not been elucidated in this study. Taken together, present study identified the MTDH/FBXW2/TAK1 axis, which is important in HCC progression and sorafenib resistance, and provided evidence for TAK1 as a robust therapeutic target in HCC and MTDH-dependent or -independent sorafenib resistance, rendering further preclinical and clinical studies to explore.
Materials and Methods
Cell Culture and Transfections
Human embryonic kidney 293 (HEK293) cells, Huh7, HepG2, SK-hep-1, HCCLM3, HLF, SMMC7721, JHH7, PLC/PRF/5, and Ha22T cells, normal liver L02 and Chang cells were obtained from the American Type Culture Collection and were maintained in Dulbecco modified Eagle medium containing 10% (v/v) fetal bovine serum at 37°C in 5% CO2 condition. All cell lines were routinely tested to be negative for mycoplasma contamination. Transfection of siRNA and plasmids was performed using Lipofectamine 3000 Reagents (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions.
Bioinformatic Analysis and GSEA
HCC datasets were downloaded from TCGA data portal (http://www.tcga-data.nci.nih.gov). MAP3K7 (TAK1) and MTDH mRNA levels were analyzed from TCGA and NCBI GEO databases (https://www.ncbi.nlm.nih.gov). For TCGA data, 5 of 374 HCC patients were excluded because of the absence of follow-up data. Finally, 369 HCC patients were subjected to mRNA expression in our analysis. GSEA was used to analyze whether a series of defined KEGG gene sets show statistically significant, concordant differences between 2 groups stratified on the basis of MTDH expression of HCC cases in TCGA dataset.
Plasmids, SiRNAs, and Reagents
HA-MTDH, HA-β-TrCP1, HA-FBXW2, HA-FBXW4, HA-FBXW5, HA-FBXW7, HA-FBXW8, HA-FBXW11, HA-FBXW12, HA-FBXL3, and HA-FBXO4 expression plasmids were subcloned with N-terminal double HA tags into pXF4H expression vector between the BamHI and EcoRI restriction sites. Flag-TAK1 was constructed with N-terminal triple Flag tags into pXF6F expression vector between the BamHI and EcoRI restriction sites. Myc-FBXW2 was constructed with N-terminal Myc tag into Pxf4H expression vector between the ClaI and EcoRI sites. All vectors and Myc-Ub were gifts from Lidan Hou. PCR and plasmids construction were performed using 2× Phanta Max Master Mix (#P511) and ClonExpress II One Step Cloning Kit (#C112) purchased from Vazyme Biotech Co, Ltd, Nanjing, China according to the manufacturer’s instructions. The primers used to generate the above cDNA fragments were purchased from Tsingke, Beijing, China and are shown in Table 2. The pooled siRNA oligos targeting for MTDH, TAK1, and FBXW2 were purchased from Guangzhou RiboBio Co, Ltd (Guangzhou, China), and the sequences of siRNA oligos are shown in Table 3. Sorafenib and MG132 were purchased from MedChemExpress (Princeton, NJ). OXO was purchased from MedChemExpress. APExBio, Cayman, and CHX were purchased from Beyotime Biotechnology (Nanjing, China). Puromycin (Solarbio, Beijing, China; P8230) was purchased from Solarbio. MLN4924 was a gift from Lidan Hou (Zhejiang University). Apoptosis (70-AP101) and Cell Cycle Assay (70-CCS012) were purchased from Multi Science, Beijing, China. Cell Counting Kits (40203ES60) were purchased from Yeasen Biotechnology, Shanghai, China. DAPI solution (C0060) and Mounting Medium (S2110) were purchased from Solarbio.
Table 2.
Lists of Primers for Constructs
| Target gene | Forward primer | Reverse primer |
|---|---|---|
| HA-BTRC | GTTCCAGATTACGCTGAATTCATGGACCCGGCCGAGG | CAGGTCGACTCTAGAGGATCCTTATCTGGAGATGTAGGTGTATGTTCGAGAAG |
| HA-FBXW2 | GTTCCAGATTACGCTGAATTCATGGAGAGAAAGGACTTTGAGACATGG | CAGGTCGACTCTAGAGGATCCTCAGCCGTGCTCCTTCCAC |
| HA-FBXW4 | GTTCCAGATTACGCTGAATTCATGGCGGCGGCGGCCGGGGAGGAGG | CAGGTCGACTCTAGAGGATCCTCATGGGTTTTGAAAATCCAGGACG |
| HA-FBXW5 | GTTCCAGATTACGCTGAATTCATGGGCCTGTCGCCC | CAGGTCGACTCTAGAGGATCCTCAGCGCCTCTGGCTGG |
| HA-FBXW7 | GTTCCAGATTACGCTGAATTCATGTGTGTCCCGAGAAGCG | CAGGTCGACTCTAGAGGATCCTCACTTCATGTCCACATCAAAGTCCAG |
| HA-FBXW8 | GTTCCAGATTACGCTGAATTCATGGACGACTACAGCCTGGATG | CAGGTCGACTCTAGAGGATCCCTAAACATGGTTATAGGGAAAGGCCAGT |
| HA-FBXW11 | GTTCCAGATTACGCTGAATTCATGGAGCCCGACTCGGT | CAGGTCGACTCTAGAGGATCCTTATCTAGAGATGTAAGTGTATGTTCTGGAGGG |
| HA-FBXW12 | GTTCCAGATTACGCTGAATTCATGAGCCCCACTCACCAGAT | CAGGTCGACTCTAGAGGATCCTTACGTATTCAAAGAATACATCACCAGAATGCT |
| HA-FBXO4 | GTTCCAGATTACGCTGAATTCATGGCGGGAAGCGAGC | CAGGTCGACTCTAGAGGATCCTCATCTTGCACGCTTAGATTCCACT |
| HA-FBXL3 | GTTCCAGATTACGCTGAATTCATGAAACGAGGAGGAAGAGATAGTGACC | CAGGTCGACTCTAGAGGATCCTTACCAAGTGGGCATCATGTCG |
| HA-MTDH | GTTCCAGATTACGCTGAATTCATGGCTGCACGGAGCT | CAGGTCGACTCTAGAGGATCCTCACGTTTCTCGTCTGGC |
| Myc-FBXW2 | CTCGGTTCTATCGATGCCACCATGGAGCAGAAACTCATCTCTGAAGAGGATCTGATGGAGAGAAAGGACTTTGAGA | TCTGATATCGAATTCTCAGCCGTGCTCCTTCCA |
| Flag-TAK1 | GATGATGACGACAAGGAATTCATGTCTACAGCCTCTGCCGC | CAGGTCGACTCTAGAGGATCCTCATGAAGTGCCTTGTCGTTTC |
Table 3.
List of siRNAs
| Target | Sense | Anti-sense |
|---|---|---|
| siTAK1 | GCAGUGAUUCUUGGAUUGU | ACAAUCCAAGAAUCACUGC |
| siFBXW2-1 | GCAGCGGUGAAGUUUGAUGAA | UUCAUCAAACUUCACCGCUGC |
| siFBXW2-2 | GCCUUUGAAACCUCGUCAU | AUGACGAGGUUUCAAAGGC |
| siMTDH-1 | GGAGGAGGCUGGAAUGAAA | UUUCAUUCCAGCCUCCUCC |
| siMTDH-2 | CAGAUAAAUCCAAGUCAAA | UUUGACUUGGAUUUAUCUG |
| siMTDH-3 | CUUAGUGAAUUGUGAUAAAGAAA | UUUCUUUAUCACAAUUCACUAAGUGAU |
IP and Immunoblotting
Cells were lysed in an IP lysis buffer (150 mmol/L NaCl, 1% NP-40, 50 mmol/L Tris-HCl, pH 8.0, 1 mmol/L Na3VO4), supplemented with complete protease inhibitor cocktail (EDTA free, mini-Tablet; MedChemExpress). For IP, 1–2 mg lysates were incubated with bead-conjugated FLAG, MYC, or HA (Sigma-Aldrich) or the appropriate antibody (2 μg) in a rotating incubator overnight at 4°C, followed by 2-hour incubation with Protein-A Sepharose beads (Santa Cruz Technology, Santa Cruz, CA). Immuno-complexes were washed 4 times with TBST or IP lysis buffer before resolved by sodium dodecyl sulfulate-polyacrylamide gel electrophoresis and analyzed by immunoblotting. For direct IB analysis, cells were lysed in lysis buffer (50 mmol/L Tris-HCl, pH 8.0, 150 mmol/L NaCl, 1% TritonX-100, 1% sodium deoxycholate, and 0.1% sodium dodecyl sulfate, 1 mmol/L EDTA). Proteins were resolved on sodium dodecyl sulfate polyacrylamide gels and then transferred to a polyvinylidene difluoride membrane. After blocking with 5% (w/v) milk or bovine serum albumin, the membrane was stained with the corresponding primary antibodies that are shown in Table 4. All antibodies were diluted in Primary Antibody Dilution Buffer (Solarbio, A1810).
Table 4.
List of Antibodies for Indicated Uses
| Target gene | Catalog no. | Producer | Target gene | Catalog no. | Producer |
|---|---|---|---|---|---|
| Anti-TAK1 | Ab109526 | Abcam | Anti-TAK1 | #5206S | CST |
| Anti-TAK1 (p-S439) | Ab109404 | Abcam | Anti-LYRIC | Ab227981 | Abcam |
| Anti-FBXW2 | 11499-1-AP | ProteinTech | Anti-CUL1 | Ab75817 | Abcam |
| Anti-Flag | F3165 | Sigma-Aldrich | Anti-SKP1 | Ab76502 | Abcam |
| Anti-HA | H6908 | Sigma-Aldrich | EMT Kit | 9782 | CST |
| Anti-Myc | Ab32 | Abcam |
RNA Extraction and Real-Time PCR
Cells were transiently transfected with indicated plasmids or siRNA for 24–48 hours, and the total RNA was then isolated using TRIzol Reagent (15596-018; Invitrogen, Carlsbad, CA). The concentration of RNA was measured by NanoDrop 2000. Total RNA was then transcribed into complementary DNA using Hifair Ⅱ 1st Strand cDNA Synthesis Kit (11121ES60, Yeasen Biotechnology) according to the manufacturer’s protocol. The quantification of complementary DNA was performed using SYBR Green Master Mix (11201ES08-5, Yeasen Biotechnology) on LightCycler 480 (Roche, Basel, Switzerland). The housekeeping gene, glyceraldehyde-3-phosphate dehydrogenase, was used as a loading control. The sequences of primer sets are shown in Table 5.
Table 5.
List of Primers for qPCR
| Target | Forward primer | Reverse primer |
|---|---|---|
| MTDH | AAGGAGATTCTACACTTCAGGTTTC | TTCCAGCCTCCTCCATTGAC |
| TAK1 | ATTGTAGAGCTTCGGCAGTT | TATATAAAGAGCCCCCTTCAGC |
| FBXW2 | CGGGGGCGGTGTTCAGT | AGCCCATACTTTCACAGCGA |
| GAPDH | GTCTTCACCACCATGGAGAAGG | ATGATCTTGAGGCTGTTGTCAT |
| Bcl-2 | CAGGATAACGGAGGCTGGGATG | AGAAATCAAACAGAGGCCGCA |
| GADD45B | CTGGTCACGAACCCTCACAC | CTTTCTTCGCAGTAGCTGGC |
| UBE2E1 | AGTTGCTGTTGCTGCACTTC | AACAGCCCCTCTCTTTGTGTC |
| GAN | AGCCCGTACATCAGGACAAAG | TGATCTGCCCACTGAAGATGT |
| RNF168 | AACGTGGAACTGTGGACGAT | GTCATCAGCCACTTCCTCTGA |
| RAD18 | TTTTGCACGGAATCATCTGCTG | TTAACCTGCTCCCCTGCTTT |
| MDM2 | CAGTAGCAGTGAATCTACAGGGA | CTGATCCAACCAATCACCTGAAT |
| UNKL | TGGCAAGATGCCAACTTCGT | GTGGACCTGTACTGGAACCG |
| WWP1 | ACTGCAGCTCATCTCCAACC | TTCAACAGCCAACCTGGCA |
Half-Life Analysis
After gene manipulation, 100 μg/ml CHX (Beyotime) was added to the cell medium. At the indicated time points, cells were harvested, lysed, and subjected to Western blotting analysis. The densitometry quantification was performed using Image Lab Processing software (Bio-Rad, Hercules, CA).
In Vivo Antitumor Study
Four- to five-week-old BALB/c athymic nude mice (nu/nu, male) were purchased from Shanghai SLAC Laboratory Animal Centre. Mice were fed with a regular diet and had free access to water and food. All mice procedures were approved by the Sir Run-Run Shaw Hospital Committee on Use and Care of Animals. The cell line–derived xenograft and orthotopic models were constructed as described below. Around 5 × 106 HCCLM3-SR cells were mixed with phosphate-buffered saline in a total volume of 0.2 mL and were then subcutaneously injected into both flanks of mice. When the tumors reached a volume of ~500 mm3, the mice were then killed, and the tumors were cut into small pieces evenly and replanted into 1 flank of mice. Those mice were then divided into 2 groups. To maintain the sorafenib resistance in HCCLM3-SR–derived tumors, sorafenib (25 mg/kg, oral) was given once a day in one group, whereas the other group received only vehicle as control. Two weeks later, the xenograft tumor volume was compared between 2 groups, and SR tumors were then chosen and replanted into the flank of mice or liver of mice to construct xenograft or orthotopic models. The abdomen of the nude mice was touched to determine the size of the tumor every week. When the tumors reached ~100 mm3, mice were then randomized into 4 groups (6 mice per group). Sorafenib (25 mg/kg, oral) was given once a day; OXO (20 mg/kg, intraperitoneal) was administered every 2 days; mice of the control group received only dimethyl sulfoxide (DMSO) as the vehicle control. The growth of tumors was measured at the indicated time points, and average tumor volumes were calculated according to the equation, Volume = (Length × Width × Width)/2. After 2 weeks, mice were killed, and all the tumor tissues were collected, fixed, and sectioned.
Human Specimens and Immunohistochemical Staining
Human HCC tumor tissue microarrays data were purchased from Shanghai Xinchao Biotech, China. For immunohistochemical staining, the sections were deparaffinized in xylene and rehydrated through graded ethanol. Antigen retrieval was performed for 20 minutes at 100°C with 0.1% sodium citrate buffer (pH 6.0). After quenching of endogenous peroxidase activity with 3% H2O2·dH2O and blocking of nonspecific binding with 5% goat serum albumin buffer, sections were incubated overnight at 4°C with indicated antibodies. After 3 times washes of phosphate-buffered saline, the sections were treated with horseradish peroxidase conjugated secondary antibody for 30 minutes at room temperature and stained with 0.05% 3, 3-diaminobenzidine tetrahydrochloride. Slides were photographed with virtual slide microscope. Quantification of the immunohistochemical staining was conducted on the basis of the ratio and intensity of the staining.
Statistical Analysis
Results are expressed as mean ± standard error of the mean. The Student t test was used for comparison between 2 groups. One-way analysis of variance was used for comparisons between the means of 3 or more groups. The Tukey test, Bonferroni test, or Dunnett test were used for post multiple comparisons between groups. For analysis of tissue microarrays, the Fisher exact test was used. The level of significance was P < .05 (∗P < .05, ∗∗P < .01, and ∗∗∗P < .001). The number of independent experiments was n ≥3 (if not depicted otherwise). Calculations were performed using the GraphPad Prism Software (San Diego, CA).
Acknowledgments
The authors thank Prof Yin Sun at University of Rochester Medical Centre for experimental skills and instrumental help; Dr Lidan Hou at Zhejiang University for providing plasmids (PXF6F and PXF4H) and other reagents and instrumental help; and Dr Hui Liu at Sir Run Run Shaw Hospital, Zhejiang University School of Medicine for her professional advice on statistical analysis.
CRediT Authorship Contributions
Shunjie Xia, PhD (Conceptualization: Lead; Data curation: Lead; Software: Lead; Validation: Equal; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead)
Lin Ji, PhD (Writing – review & editing: Supporting)
Liye Tao (Data curation: Supporting; Methodology: Supporting; Writing – original draft: Supporting; Writing – review & editing: Supporting)
Yu Pan, PhD (Conceptualization: Supporting; Data curation: Supporting; Methodology: Supporting; Software: Supporting; Writing – review & editing: Supporting)
Zhongjie Lin, PhD (Data curation: Supporting; Methodology: Supporting)
Zhe Wan, PhD (Writing – review & editing: Supporting)
Haoqi Pan (Methodology: Supporting; Writing – review & editing: Supporting)
Jie Zhao, PhD (Funding acquisition: Supporting)
Liuxin Cai, PhD (Funding acquisition: Supporting)
Junjie Xu, PhD (Funding acquisition: Equal; Supervision: Equal; Validation: Equal; Visualization: Supporting; Writing – original draft: Supporting; Writing – review & editing: Equal)
Xiujun Cai, MD, PhD (Funding acquisition: Lead; Validation: Lead; Writing – review & editing: Equal)
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding Supported by National Natural Science Foundation of China under grant no. 81827804 and no. 81772546 (X. Cai) and no. 81902367 (J. Xu); Zhejiang Provincial Natural Science Foundation of China under grant no. LQ18H160010 (J. Zhao) and no. LY15H160014 (L. Cai); China Postdoctoral Science Foundation under grant no. 2020M671755 (J. Xu); Hepatobiliary and Pancreatic Cancer Research of Hubei Chen Xiaoping Science and Technology Development Foundation under grant no. CXPJJH11900001-2019308 (J. Xu); Zhejiang Clinical Research Center of Minimally Invasive Diagnosis and Treatment of Abdominal Diseases grant no. 2018E50003 (X. Cai); Key Research and Development Project of Zhejiang Province under grant no. 2018C03083 (X. Cai); and Health Innovation Talent Support Project of Zhejiang Medical and Health Science and Technology Plan under grant no. 2021447581 (J. Xu).
Contributor Information
Junjie Xu, Email: walter235@zju.edu.cn.
Xiujun Cai, Email: srrsh_cxj@zju.edu.cn.
References
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Gao Q., Zhu H., Dong L., Shi W., Chen R., Song Z., Huang C., Li J., Dong X., Zhou Y., Liu Q., Ma L., Wang X., Zhou J., Liu Y., Boja E., Robles A.I., Ma W., Wang P., Li Y., Ding L., Wen B., Zhang B., Rodriguez H., Gao D., Zhou H., Fan J. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:1240. doi: 10.1016/j.cell.2019.10.038. [DOI] [PubMed] [Google Scholar]
- 3.Xu L.X., He M.H., Dai Z.H., Yu J., Wang J.G., Li X.C., Jiang B.B., Ke Z.F., Su T.H., Peng Z.W., Guo Y., Chen Z.B., Chen S.L., Peng S., Kuang M. Genomic and transcriptional heterogeneity of multifocal hepatocellular carcinoma. Ann Oncol. 2019 doi: 10.1093/annonc/mdz103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xia S., Pan Y., Liang Y., Xu J., Cai X. The microenvironmental and metabolic aspects of sorafenib resistance in hepatocellular carcinoma. EBioMedicine. 2020:51. doi: 10.1016/j.ebiom.2019.102610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J., Zheng L., Chen J., Sun Y., Lin H., Jin R.A., Tang M., Liang X., Cai X. Increasing AR by HIF-2alpha inhibitor (PT-2385) overcomes the side-effects of sorafenib by suppressing hepatocellular carcinoma invasion via alteration of pSTAT3, pAKT and pERK signals. Cell Death Disease. 2017;8 doi: 10.1038/cddis.2017.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin Z., Xia S., Liang Y., Ji L., Pan Y., Jiang S., Wan Z., Tao L., Chen J., Lin C., Liang X., Xu J., Cai X. LXR activation potentiates sorafenib sensitivity in HCC by activating microRNA-378a transcription. Theranostics. 2020;10:8834–8850. doi: 10.7150/thno.45158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santoro R., Carbone C., Piro G., Chiao P.J., Melisi D. TAK-ing aim at chemoresistance: the emerging role of MAP3K7 as a target for cancer therapy. Drug Resistance Updates. 2017;33–35:36–42. doi: 10.1016/j.drup.2017.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Mukhopadhyay H., Lee N.Y. Multifaceted roles of TAK1 signaling in cancer. Oncogene. 2020;39:1402–1413. doi: 10.1038/s41388-019-1088-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Inokuchi S., Aoyama T., Miura K., Österreicher C.H., Kodama Y., Miyai K., Akira S., Brenner D.A., Seki E. Disruption of TAK1 in hepatocytes causes hepatic injury, inflammation, fibrosis, and carcinogenesis. Proc Natl Acad Sci. 2010;107:844–849. doi: 10.1073/pnas.0909781107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang L., Inokuchi S., Roh Y.S., Song J., Loomba R., Park E.J., Seki E. Transforming growth factor-b2: signaling in hepatocytes promotes hepatic fibrosis and carcinogenesis in mice with hepatocyte-specific deletion of TAK1. Gastroenterology. 2013;144:1042–1054.e4. doi: 10.1053/j.gastro.2013.01.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan F.-J., Zhang X.-J., Wang W.-X., Ji Y.-X., Wang P.-X., Yang Y., Gong J., Shen L.-J., Zhu X.-Y., Huang Z., Li H. The E3 ligase tripartite motif 8 targets TAK1 to promote insulin resistance and steatohepatitis. Hepatology. 2017;65:1492–1511. doi: 10.1002/hep.28971. [DOI] [PubMed] [Google Scholar]
- 12.Mitra A., Yan J., Xia X., Zhou S., Chen J., Mishra L., Li S. IL6-mediated inflammatory loop reprograms normal to epithelial-mesenchymal transition+ metastatic cancer stem cells in preneoplastic liver of transforming growth factor beta–deficient β2-spectrin+/− mice. Hepatology. 2017;65:1222–1236. doi: 10.1002/hep.28951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao N., Wang R., Zhou L., Zhu Y., Gong J., Zhuang S.M. MicroRNA-26b suppresses the NF-κB signaling and enhances the chemosensitivity of hepatocellular carcinoma cells by targeting TAK1 and TAB3. Mol Cancer. 2014;13:35. doi: 10.1186/1476-4598-13-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roh Y.S., Song J., Seki E. TAK1 regulates hepatic cell survival and carcinogenesis. J Gastroenterol. 2014;49:185–194. doi: 10.1007/s00535-013-0931-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y., Wang F., Gao L., Xu L., Tong R., Lin N., Su Y., Yan Y., Gao Y., He J., Kong L., Yuan A., Zhuge Y., Pu J. Ubiquitin-specific protease 4 is an endogenous negative regulator of metabolic dysfunctions in nonalcoholic fatty liver disease in mice. Hepatology. 2018;68:897–917. doi: 10.1002/hep.29889. [DOI] [PubMed] [Google Scholar]
- 16.Ji Y.-X., Huang Z., Yang X., Wang X., Zhao L.-P., Wang P.-X., Zhang X.-J., Alves-Bezerra M., Cai L., Zhang P., Lu Y.-X., Bai L., Gao M.-M., Zhao H., Tian S., Wang Y., Huang Z.-X., Zhu X.-Y., Zhang Y., Gong J., She Z.-G., Li F., Cohen D.E., Li H. The deubiquitinating enzyme cylindromatosis mitigates nonalcoholic steatohepatitis. Nat Med. 2018;24:213–223. doi: 10.1038/nm.4461. [DOI] [PubMed] [Google Scholar]
- 17.Meng X., Thiel K.W., Leslie K.K. Drug resistance mediated by AEG-1/MTDH/LYRIC. Adv Cancer Res. 2013;120:135–157. doi: 10.1016/B978-0-12-401676-7.00005-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu G., Wei Y., Kang Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res. 2009;15:5615–5620. doi: 10.1158/1078-0432.CCR-09-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ninomiya-Tsuji J., Kajino T., Ono K., Ohtomo T., Matsumoto M., Shiina M., Mihara M., Tsuchiya M., Matsumoto K. A resorcylic acid lactone, 5z-7-oxozeaenol, prevents inflammation by inhibiting the catalytic activity of TAK1 MAPK kinase kinase. J Biol Chem. 2003;278:18485–18490. doi: 10.1074/jbc.M207453200. [DOI] [PubMed] [Google Scholar]
- 20.Luedde T., Schwabe R.F. NF-κB in the liver: linking injury, fibrosis and hepatocellular carcinoma. Nat Rev Gastroenterol Hepatol. 2011;8:108–118. doi: 10.1038/nrgastro.2010.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Malenstein H., Dekervel J., Verslype C., Van Cutsem E., Windmolders P., Nevens F., van Pelt J. Long-term exposure to sorafenib of liver cancer cells induces resistance with epithelial-to-mesenchymal transition, increased invasion and risk of rebound growth. Cancer Lett. 2013;329:74–83. doi: 10.1016/j.canlet.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Shibue T., Weinberg R.A. EMT, CSCs, and drug resistance: the mechanistic link and clinical implications. Nat Rev Clin Oncol. 2017;14:611. doi: 10.1038/nrclinonc.2017.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viswanathan V.S., Ryan M.J., Dhruv H.D., Gill S., Eichhoff O.M., Seashore-Ludlow B., Kaffenberger S.D., Eaton J.K., Shimada K., Aguirre A.J. Dependency of a therapy-resistant state of cancer cells on a lipid peroxidase pathway. Nature. 2017;547:453. doi: 10.1038/nature23007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Zhang Q., Lou Y., Fu Q., Chen Q., Wei T., Yang J., Tang J., Wang J., Chen Y., Zhang X., Zhang J., Bai X., Liang T. Hypoxia-inducible factor-1α/interleukin-1β signaling enhances hepatoma epithelial–mesenchymal transition through macrophages in a hypoxic-inflammatory microenvironment. Hepatology. 2018;67:1872–1889. doi: 10.1002/hep.29681. [DOI] [PubMed] [Google Scholar]
- 25.Zhou S.-L., Yin D., Hu Z.-Q., Luo C.-B., Zhou Z.-J., Xin H.-Y., Yang X.-R., Shi Y.-H., Wang Z., Huang X.-W., Cao Y., Fan J., Zhou J. A positive feedback loop between cancer stem-like cells and tumor-associated neutrophils controls hepatocellular carcinoma progression. Hepatology. 2019;70:1214–1230. doi: 10.1002/hep.30630. [DOI] [PubMed] [Google Scholar]
- 26.Lin X., Li A.M., Li Y.H., Luo R.C., Zou Y.J., Liu Y.Y., Liu C., Xie Y.Y., Zuo S., Liu Z., Liu Z., Fang W.Y. Silencing MYH9 blocks HBx-induced GSK3β ubiquitination and degradation to inhibit tumor stemness in hepatocellular carcinoma. Signal Transduct Target Ther. 2020;5:13. doi: 10.1038/s41392-020-0111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- 28.Padmanaban V., Krol I., Suhail Y., Szczerba B.M., Aceto N., Bader J.S., Ewald A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature. 2019;573:439–444. doi: 10.1038/s41586-019-1526-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng L., Xu M., Xu J., Wu K., Fang Q., Liang Y., Zhou S., Cen D., Ji L., Han W., Cai X. ELF3 promotes epithelial-mesenchymal transition by protecting ZEB1 from miR-141-3p-mediated silencing in hepatocellular carcinoma. Cell Death Disease. 2018;9:387. doi: 10.1038/s41419-018-0399-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harding J.J., Nandakumar S., Armenia J., Khalil D.N., Albano M., Ly M., Shia J., Hechtman J.F., Kundra R., El Dika I., Do R.K. Prospective genotyping of hepatocellular carcinoma: clinical implications of next generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res. 2019;25:2116–2126. doi: 10.1158/1078-0432.CCR-18-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z., Liu P., Inuzuka H., Wei W. Roles of F-box proteins in cancer. Nat Rev Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skaar J.R., Pagan J.K., Pagano M. Mechanisms and function of substrate recruitment by F-box proteins. Nat Rev Mol Cell Biol. 2013;14:369–381. doi: 10.1038/nrm3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang F., Xu J., Li H., Tan M., Xiong X., Sun Y. FBXW2 suppresses migration and invasion of lung cancer cells via promoting β-catenin ubiquitylation and degradation. Nat Commun. 2019;10:1382. doi: 10.1038/s41467-019-09289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang Y.M., Kim S.Y., Seki E. Inflammation and liver cancer: molecular mechanisms and therapeutic targets. Semin Liver Dis. 2019;39:26–42. doi: 10.1055/s-0038-1676806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tan W., Luo X., Li W., Zhong J., Cao J., Zhu S., Chen X., Zhou R., Shang C., Chen Y. TNF-α is a potential therapeutic target to overcome sorafenib resistance in hepatocellular carcinoma. EBioMedicine. 2019;40:446–456. doi: 10.1016/j.ebiom.2018.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meng X., Zhu D., Yang S., Wang X., Xiong Z., Zhang Y., Brachova P., Leslie K.K. Cytoplasmic Metadherin (MTDH) provides survival advantage under conditions of stress by acting as RNA-binding protein. J Biol Chem. 2012;287:4485–4491. doi: 10.1074/jbc.C111.291518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsu J.C., Reid D.W., Hoffman A.M., Sarkar D., Nicchitta C.V. Oncoprotein AEG-1 is an endoplasmic reticulum RNA-binding protein whose interactome is enriched in organelle resident protein-encoding mRNAs. RNA. 2018;24:688–703. doi: 10.1261/rna.063313.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo E.C., Nathanson J.L., Tan F.E., Schwartz J.L., Schmok J.C., Shankar A. Large-scale tethered function assays identify factors that regulate mRNA stability and translation. Nat Struct Mol Biol. 2020;27:989–1000. doi: 10.1038/s41594-020-0477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang K., Lim H.Y., Shi S., Lee J., Deng S., Xie T., Zhu Z., Wang Y., Pocalyko D., Yang W.J., Rejto P.A., Mao M., Park C.K., Xu J. Genomic landscape of copy number aberrations enables the identification of oncogenic drivers in hepatocellular carcinoma. Hepatology. 2013;58:706–717. doi: 10.1002/hep.26402. [DOI] [PubMed] [Google Scholar]
- 40.Llovet J.M., Montal R., Sia D., Finn R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat Rev Clin Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PubMed] [Google Scholar]
- 41.Cheng A.-L., Hsu C., Chan S.L., Choo S.-P., Kudo M. Challenges of combination therapy with immune checkpoint inhibitors for hepatocellular carcinoma. J Hepatol. 2020;72:307–319. doi: 10.1016/j.jhep.2019.09.025. [DOI] [PubMed] [Google Scholar]
- 42.Xu J., Lin H., Li G., Sun Y., Chen J., Shi L., Cai X., Chang C. The miR-367-3p increases sorafenib chemotherapy efficacy to suppress hepatocellular carcinoma metastasis through altering the androgen receptor signals. EBioMedicine. 2016;12:55–67. doi: 10.1016/j.ebiom.2016.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu J., Lin H., Li G., Sun Y., Shi L., Ma W.-L., Chen J., Cai X., Chang C. Sorafenib with ASC-J9(®) synergistically suppresses the HCC progression via altering the pSTAT3-CCL2/Bcl2 signals. Int J Cancer. 2017;140:705–717. doi: 10.1002/ijc.30446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu J., Wan Z., Tang M., Lin Z., Jiang S., Ji L., Gorshkov K., Mao Q., Xia S., Cen D., Zheng J., Liang X., Cai X. N(6)-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. 2020;19:163. doi: 10.1186/s12943-020-01281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu J., Ji L., Liang Y., Wan Z., Zheng W., Song X., Gorshkov K., Sun Q., Lin H., Zheng X., Chen J., Jin R.A., Liang X., Cai X. CircRNA-SORE mediates sorafenib resistance in hepatocellular carcinoma by stabilizing YBX1. Signal Transduct Target Ther. 2020;5:298. doi: 10.1038/s41392-020-00375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambert A.W., Weinberg R.A. Linking EMT programmes to normal and neoplastic epithelial stem cells. Nat Rev Cancer. 2021 doi: 10.1038/s41568-021-00332-6. [DOI] [PubMed] [Google Scholar]
- 47.Aiello N.M., Maddipati R., Norgard R.J., Balli D., Li J., Yuan S., Yamazoe T., Black T., Sahmoud A., Furth E.E., Bar-Sagi D., Stanger B.Z. EMT subtype influences epithelial plasticity and mode of cell migration. Dev Cell. 2018;45:681–695.e4. doi: 10.1016/j.devcel.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Puram S.V., Tirosh I., Parikh A.S., Patel A.P., Yizhak K., Gillespie S., Rodman C., Luo C.L., Mroz E.A., Emerick K.S., Deschler D.G., Varvares M.A., Mylvaganam R., Rozenblatt-Rosen O., Rocco J.W., Faquin W.C., Lin D.T., Regev A., Bernstein B.E. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171:1611–1624.e24. doi: 10.1016/j.cell.2017.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ye X., Tam W.L., Shibue T., Kaygusuz Y., Reinhardt F., Ng Eaton E., Weinberg R.A. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525:256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh A., Sweeney Michael F., Yu M., Burger A., Greninger P., Benes C., Haber Daniel A., Settleman J. TAK1 inhibition promotes apoptosis in KRAS-dependent colon cancers. Cell. 2012;148:639–650. doi: 10.1016/j.cell.2011.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bang D., Wilson W., Ryan M., Yeh J.J., Baldwin A.S. GSK-3α promotes oncogenic KRAS function in pancreatic cancer via TAK1–TAB stabilization and regulation of noncanonical NF-κB. Cancer Discovery. 2013;3:690–703. doi: 10.1158/2159-8290.CD-12-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Siegel R.L., Jemal A., Wender R.C., Gansler T., Ma J., Brawley O.W. An assessment of progress in cancer control. CA: Cancer J Clin. 2018;68:329–339. doi: 10.3322/caac.21460. [DOI] [PubMed] [Google Scholar]
- 53.An S., Zhao L.-P., Shen L.-J., Wang S., Zhang K., Qi Y., Zheng J., Zhang X.-J., Zhu X.-Y., Bao R., Yang L., Lu Y.-X., She Z.-G., Tang Y.-D. USP18 protects against hepatic steatosis and insulin resistance through its deubiquitinating activity. Hepatology. 2017;66:1866–1884. doi: 10.1002/hep.29375. [DOI] [PubMed] [Google Scholar]
- 54.Wang S., Yan Z.Z., Yang X., An S., Zhang K., Qi Y., Zheng J., Ji Y.X., Wang P.X., Fang C., Zhu X.Y., Shen L.J., Yan F.J., Bao R., Tian S., She Z.G., Tang Y.D. Hepatocyte DUSP14 maintains metabolic homeostasis and suppresses inflammation in the liver. Hepatology. 2018;67:1320–1338. doi: 10.1002/hep.29616. [DOI] [PubMed] [Google Scholar]
- 55.Tan S., Zhao J., Sun Z., Cao S., Niu K., Zhong Y., Wang H., Shi L., Pan H., Hu J., Qian L., Liu N., Yuan J. Hepatocyte-specific TAK1 deficiency drives RIPK1 kinase-dependent inflammation to promote liver fibrosis and hepatocellular carcinoma. Proc Natl Acad Sci U S A. 2020;117:14231–14242. doi: 10.1073/pnas.2005353117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takezawa K., Okamoto I., Yonesaka K., Hatashita E., Yamada Y., Fukuoka M., Nakagawa K. Sorafenib inhibits non-small cell lung cancer cell growth by targeting B-RAF in KRAS wild-type cells and C-RAF in KRAS mutant cells. Cancer Res. 2009;69:6515–6521. doi: 10.1158/0008-5472.CAN-09-1076. [DOI] [PubMed] [Google Scholar]
- 57.Zimmerman E.I., Gibson A.A., Hu S., Vasilyeva A., Orwick S.J., Du G., Mascara G.P., Ong S.S., Chen T., Vogel P., Inaba H., Maitland M.L., Sparreboom A., Baker S.D. Multikinase inhibitors induce cutaneous toxicity through OAT6-mediated uptake and MAP3K7-driven cell death. Cancer Res. 2016;76:117–126. doi: 10.1158/0008-5472.CAN-15-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang P.-X., Zhang X.-J., Luo P., Jiang X., Zhang P., Guo J., Zhao G.-N., Zhu X., Zhang Y., Yang S., Li H. Hepatocyte TRAF3 promotes liver steatosis and systemic insulin resistance through targeting TAK1-dependent signalling. Nature Communications. 2016;7:10592. doi: 10.1038/ncomms10592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan L., Lin M., Pan S., Assaraf Y.G., Wang Z.W., Zhu X. Emerging roles of F-box proteins in cancer drug resistance. Drug Resist Updat. 2020;49:100673. doi: 10.1016/j.drup.2019.100673. [DOI] [PubMed] [Google Scholar]
- 60.Minoda Y., Sakurai H., Kobayashi T., Yoshimura A., Takaesu G. An F-box protein, FBXW5, negatively regulates TAK1 MAP3K in the IL-1beta signaling pathway. Biochem Biophys Res Commun. 2009;381:412–417. doi: 10.1016/j.bbrc.2009.02.052. [DOI] [PubMed] [Google Scholar]