Abstract
Genomic analysis of Pancreatic Neuroendocrine Tumors (PanNETs) has revealed that these tumors often lack mutations in typical cancer-related genes such as the tumor suppressor gene p53. Instead, PanNET tumorigenesis usually involves mutations in specific PanNET-related genes, such as tumor suppressor gene MEN1. Using a PanNET mouse model, human tissues and human cell lines, we studied the cross-talk among MEN1, p53 and Notch signaling pathways and their role in PanNETs. Here, we show that reactivation of the early developmental program of islet cells underlies PanNET tumorigenesis by restoring the proliferative capacity of PanNET cells. We investigated the role of INSM1, a transcriptional regulator of islet cells’ development, and revealed that its expression and subcellular localization is regulated by MEN1 and p53. Both human and mouse data show that loss of MEN1 in a p53 wild-type genetic background results in increased nuclear INSM1 expression and cell proliferation. Additionally, inhibition of Notch signaling in a p53 wild-type background reduces the proliferation of PanNET cells, due to repression of INSM1 transcription and nuclear localization. Our study elucidates the molecular mechanisms governing the interactions of INSM1 with MEN1, p53 and Notch and their roles in PanNET tumorigenesis, suggesting INSM1 as a key transcriptional regulator of PanNET cell proliferation.
Keywords: Pancreatic neuroendocrine tumors, Men1, Notch, INSM1, p53, Diagnostic marker
Introduction
Pancreatic neuroendocrine tumors (PanNETs) comprise a heterogeneous group of endocrine tumors arising in the pancreas [1,2]. Studies of Asian and European populations show that these are uncommon neoplasms, with an incidence lower than 1 per 100,000 persons per year 2, 3, 4. Still, they are among the most common neuroendocrine tumors (NETs) and their diagnosis has been increasing, most likely owing to more sensitive detection methods, thus creating challenges for clinical management [5].
Numerous studies have demonstrated the utility of several neuroendocrine biomarkers, such as chromogranin A (CGA), synaptophysin (SYP), and CD56 for the diagnosis of PanNETs [6,7]. Based on such proliferative markers as mitotic count and Ki67 index, the current World Health Organization (WHO) classification categorizes PanNETs into well-differentiated PanNETs, which consist of low- (NET G1), intermediate- (NET G2), and high-grade (NET G3), and poorly-differentiated pancreatic neuroendocrine carcinoma (PanNEC), referred as PanNEC [4,5]. Still, our current understanding of the molecular pathology of PanNETs is inadequate for informed clinical management.
Previous studies have shown that driver mutations can influence the prognosis of patients diagnosed with PanNET. For instance, loss of heterozygosity (LOH) of the tumor suppressor gene MEN1 occurs in 35% of PanNETs [5]. In contrast, mutations in p53, a tumor suppressor gene commonly inactivated in various cancers, are rare in PanNETs [8]. Nonetheless, LOH of the p53 target gene PHLDA3 [8] has been reported in 72% of human PanNETs, and this mutation is especially related to high tumor grade and poor prognosis [9]. Inactivation of either MEN1 or PHLDA3 in mouse models causes islet hyperplasia [8,10, 11, 12]. However, hyperplastic islets in each of these mouse models retain their differentiated β-cell characteristics and lack the PanNET tumorigenic phenotype. Thus, mutations of these tumor suppressor genes does not adequately explain the molecular pathogenesis of PanNET tumor formation [6].
The insulinoma associated 1 (INSM1) protein has been described as a diagnostic marker for a wide range of NETs, including small cell lung cancers (SCLCs), pituitary tumors, medullary thyroid carcinomas, and small intestinal and colorectal NETs 13, 14, 15, 16. INSM1 is a zinc-finger transcription factor that plays a critical role in the development of several neuroendocrine tissues [13,14]. Still, its role and functions in PanNET tumorigenesis have yet to be fully elucidated. In this study, we used human PanNET specimens, genetically modified mouse models and PanNET cell lines carrying characteristic PanNET genomic abnormalities to investigate the role of INSM1 in PanNET development. Our results show that INSM1 is a key regulator of PanNET cell proliferation, its expression and function are correlated with the genomic status of MEN1 and p53, and it is regulated by Notch signaling. These data reveal a novel system of crosstalk among signaling pathways involved in early PanNET tumorigenesis.
Materials and methods
Tumor samples
Tissue samples were provided by the National Cancer Center Biobank, Japan. The tumor samples used in this study were surgically resected at the National Cancer Center Hospital between 1993 and 2012 after receiving written informed consent from each patient and had been previously characterized for genomic mutations [8]. All procedures performed in studies involving human samples were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Histological grading of the tumors was based on World Health Organization 2004 classification. We used surgically resected non-functioning well-differentiated PanNET primary tissues that had been previously found to be either normal or positive for MEN1 or PHLDA3 LOH [8]. This study was approved by the Institutional Review Board of the National Cancer Center, Tokyo. Clinical and pathological data were obtained through a retrospective review of the medical records and they are described in Table S1.
Genetically engineered mice
Generation and genotyping of mice with a floxed Men1 gene was previously reported [17]. Briefly, mice carrying floxed Men1 alleles (Men1f/f) were crossbred with mice expressing the Cre-recombinase from the rat insulin promoter (Rip-Cre) to selectively inactivate both copies of the endogenous Men1 gene in islet β-cells.
Generation and genotyping of Phlda3-deficient mice was previously reported [8,18]. Mouse experiments were performed in a specific pathogen-free environment at the National Cancer Center animal facility according to institutional guidelines. All the animal experiments were approved by the Committee for Ethics in Animal Experimentation at the National Cancer Center, Tokyo.
Cell lines, cell culture and siRNA transfection
The authenticated pancreatic neuroendocrine tumor cell lines BON-1, QGP1 [19] and NT-3 [20] were cultured in Roswell Park Media (RPMI) supplemented with 10% FCS, penicillin andstreptomycin, HEPES, EGF (20 ng/mL), and FGF2 (10 ng/mL; PeproTech, UK). NT3 tumor cells were cultivated on human collagen-coated culture plates.
H1299 (human lung NET) and Β-TC-06 (mouse pancreatic β cell) lines were cultivated in DMEM supplemented with 10% FCS and penicillin andstreptomycin. Cells regularly tested negative for mycoplasma contamination. Cells from passage 15 to 30 were used for the experiments. siRNAs were introduced into cells using RNAiMAX (Invitrogen, UK). ON-TARGET plus control and ON-TARGETplus human INSM1-SMARTpools targeting siRNAs were purchased from Dharmacon Research. Genomic features of the cell lines are described in Table S2.
Immunohistochemistry
Fixation of tissues was performed in 4% paraformaldehyde for 24 hours. Immunohistochemistry (IHC) of paraffin-embedded tissues was performed basically according to the manufacturer's instructions. Briefly, after deparaffinization, tissues sections were processed for antigen retrieval by autoclaving slides 15 min in 10 mm citrate buffer (pH 6.0). Sections were pre-treated with 0.3% H2O2 for inactivation of endogenous peroxidase. Nonspecific interactions were blocked for 1 hour using a 5% horse serum solution. The following primary antibodies were applied to separate slides and incubated overnight at 4 °C: mouse anti-INSM1 monoclonal antibody (Santa-Cruz; diluted 1:100), mouse anti-Cyclin D1 monoclonal antibody (Santa-Cruz; diluted 1:100), or mouse anti-Men1 (B-9) monoclonal antibody (Santa-Cruz; diluted 1:200). Dilution was with Signal Enhancer HIKARI (Nacalai Tesque, Japan). Biotinylated anti-mouse IgG antibody (VECTOR Laboratories) was used as the secondary antibody. We used 3,3′-diaminobenzidine tetrahydrochloride (DAB; Muto Pure Chemicals) as the substrate chromogen. The sections were counterstained with hematoxylin Staining.
Co-immunoprecipitation and Western blotting
Primary antibodies were used at a concentration of approximately 1 ug primary antibody per 200 μl protein extract for immunoprecipitation and Western blotting as described elsewhere [21]. Antibodies used were INSM1 (A-8) (1:1000), Menin (C-19) (1:1000), Actin (1:2000), Cyclin D1 (HD11) (1:300), p27 (F-8) (1:500), p53 (FL-393) (1:2000), Hes1 (E:5) (1:500), Notch 3 (A-6) (1:800); all purchased from Santa-Cruz Biotechnology (USA). Antibodies against phosphor-ERK (1:1000), phosphor-Akt (S473) (D-96) (1:1000), Akt (1:1000) (1:1000) were purchased from Cell Signaling (Japan). Secondary antibodies were obtained from GE Healthcare (UK).
Growth inhibition assays
Adherent cells were trypsinized into single cell suspensions and aliquots of 500 cells per well were seeded in triplicate in opaque 96-well plates (Corning) in 50μL medium and incubated overnight at 37 in 5% CO2. After 24 hours, serial dilutions of gamma-secretase inhibitor (GSI) N-[N-(3,5-Difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester (DAPT) (Sigma-Aldrich, USA) were added to the appropriate wells. Equal volumes of vehicle was used as controls. After incubation for 48 hours, cell viability was measured using the CellTiter-Glo Luminescent Assay (Promega, USA). Data from triplicate wells were averaged and normalized against the average signal of control samples, and dose-response curves were generated.
Immunocytochemistry
After seeding on cover glasses in 12 wells plates, cells were fixed with 1% paraformaldehyde for 10 min. After washes with PBST, cells were incubated overnight at 4 °C with the primary antibody: mouse anti-INSM1 monoclonal antibody (Santa-Cruz) diluted 1:100 in PBS-BSA 0.01%. The secondary antibody, Alexa Fluor 594 goat anti-mouse IgG antibody (Invitrogen), diluted 1:1,000 with PBST-BSA, was added to the slides and incubated 1 h at room temperature.
Image processing
Image analyses were performed using ImageJ software (imageJ.net), version 1.52k with Java 1.8.0_172 (64-bit) engine. Additionally, we used the color thresholding image adjustment employing the Color Deconvolution plugin (imagej.net/Color_Deconvolution). We used an optimized immunohistochemistry image processing protocol for DAB staining to semi-quantitatively analyze protein expression, as previously described [22,23]. Color deconvolution was performed by standard protocol [22,23]. Using fixed parameters of basal contrast, brightness and color threshold, we calibrated our baselines and recorded the integrated density for all images. The integrated density represents the sum of the values of the pixels in the area selected. The DAB staining intensity was calculated as the integrated density of the islet area covered by DAB stained profiles. This protocol corrects for staining variations among samples and standardizes the analyses.
RNA extraction and Quantitative real time PCR
Total cellular RNA was extracted using the RNeasy kit (Qiagen, UK) and was reverse transcribed using the Reverse TraAce Kit (Toyobo, Japan) according to the manufacturer's instructions. Quantitative real-time PCR (qRT-PCR) was performed for genes of interest using the CFX96 Real-time PCR (Biorad, UK), and the SYBRH Green Promix ExTaq TliRNaseHPlus reagent (Takara Bio Inc, Japan) according to manufacturer's instructions. Relative gene expression levels were calculated by normalization to the expression levels of housekeeping genes (GAPDH) using the Delta Delta Ct Method. Fold changes over 1.5 were considered statistically significant. The average of the 3 normal samples was used for relative expression. Additional information on the primers are included in Table S3.
Cell cycle analysis and flow cytometry
For cell cycle analysis, BrdU Flow Kits (BD Pharmigen, UK) were used according to the manufacturer's instructions. Briefly, BrdU was incorporated in seeded cells for an hour. Cell were harvested in PBS, fixed, permeabilized and stained with anti-BrdU antibody (1:50). Total DNA was then stained with 7AAD for 5 min and data were recorded using an EC800 Flow Cytometry Analyzer (Sony Biotechnologies, Japan). For DNA content analysis, cells were suspended in PBS, to which 70% ice-cold ethanol was added while vortexing, followed by incubation on ice for at least 30 min. Cells were washed with PBS, mixed with 10 mg/ml RNase A, and then incubated at 37°C for 20 min. Following addition of PI solution (0.1% trisodium citrate, 50 μg/ml PI) and incubation for 10 min at 4°C, the cells were analyzed by flow cytometry.
Statistical Analysis
All experiments were repeated at least on two separate occasions. Quantitative analysis was based on a minimum of 3 replicates. Data were analyzed using GraphPad Prism 6 (GraphPad Software) and all graphs and diagrams were generated using Microsoft Office 2017 software (Microsoft Corporation, USA). When data followed a normal distribution, two sample t-tests were used to compare the differences between two samples or one-sample t-tests to determine whether the sample mean was statistically different from a known or hypothesized mean. P values < 0.05 were considered statistically significant.
Results
INSM1 is highly expressed in PanNET human tissues having MEN1 LOH
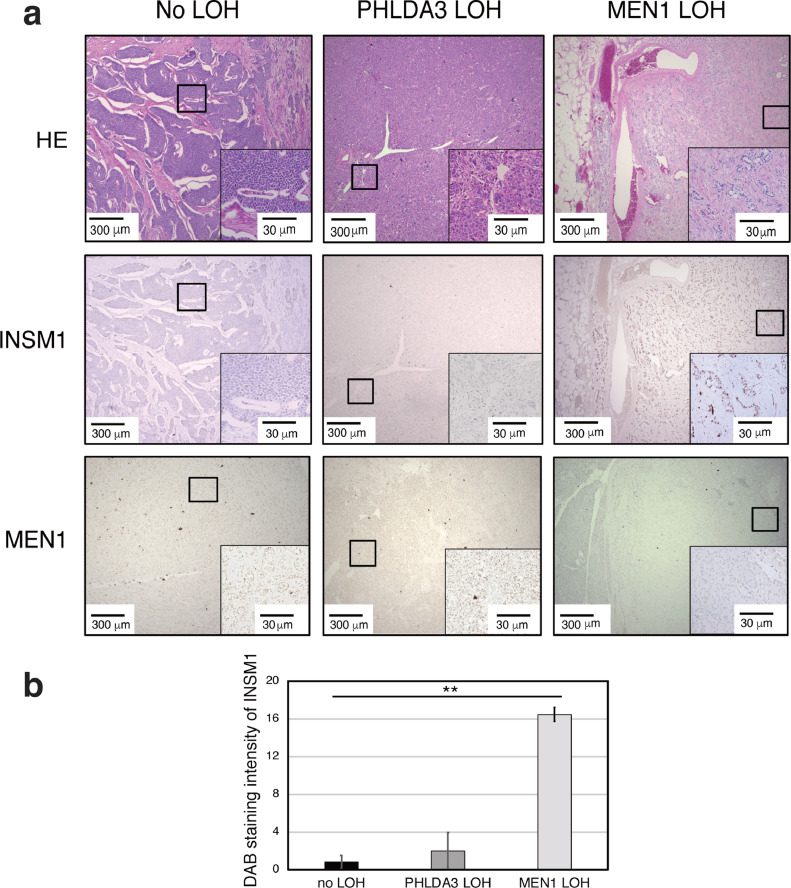
Since INSM1 protein can be detected in PanNETs [6], we analyzed INSM1 protein expression as a function of PanNET genomic status and specifically of MEN1 and PHLDA3 LOH status. We found that INSM1 was highly expressed in PanNET specimens positive for MEN1 LOH (Fig. 1A, B), independent of the tumor grade (Fig. S1). Immunohistochemical staining confirmed that expression of the MEN1 gene product, Menin, was absent only in the MEN1 LOH PanNET samples (Fig. 1A).
Fig. 1.
Relationship between INSM1 and Menin protein expression in PanNET human tissues with different genomic abnormalities. (A) Representative images of immunohistochemical staining of INSM1 and Menin of grade 3 surgically resected PanNET tumor tissues negative for PHLDA3 and MEN1 LOH, single PHLDA3 LOH and single MEN1 LOH. (B) INSM1 staining intensity in Human PanNET quantified by ImageJ. Values represents means of data obtained from five patient samples ± SD. **P-value < 0.01 as determined by t test.
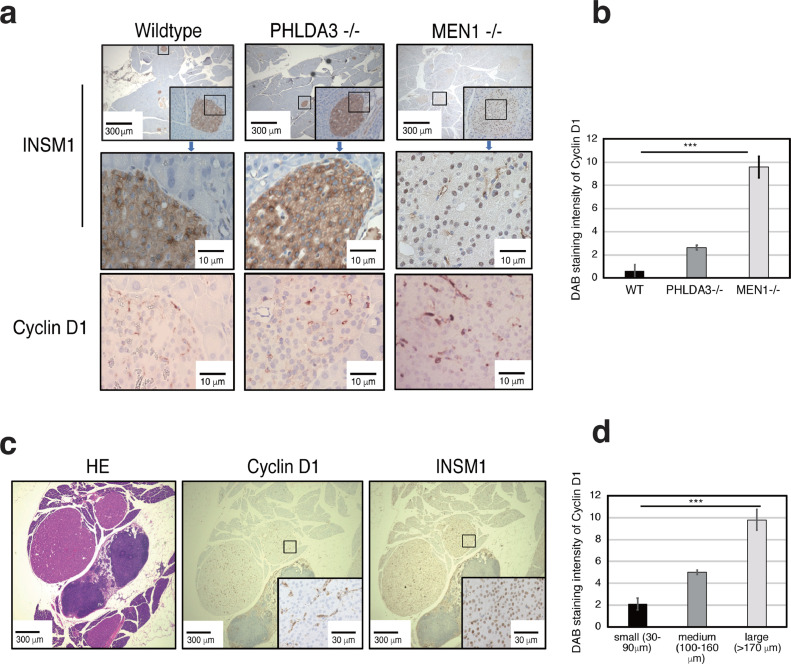
Nuclear localization of INSM1 in mice islets is directly correlated with loss of MEN1 and an increase in islet cells positively staining to cytoplasmic Cyclin D1
Previous studies have shown that the ability of INSM1 to transcriptionally regulate neuroendocrine cell fate depends on its subcellular localization [14,24, 25, 26]. To investigate the correlation between loss of MEN1 expression and INSM1 subcellular localization, we used genetically engineered mice having either wild-type, Phlda3−/− or Men1−/- genotypes. Our results show that in wild-type and Phlda3−/− mice, INSM1 localizes in the cytoplasm, whereas in Men1-/- mice, INSM1 localizes in the nucleus (Fig. 2A). In addition to its activity in the nucleus as a transcriptional regulator, it has been shown that cytoplasmic INSM1 can cause cell cycle arrest through binding to cellular regulators such as Cyclin D1 [27]. Considering that one phenotype of both Phlda3−/− and Men1−/- adult mice is islet hyperplasia [8,10], we examined Cyclin D1 expression in mice defective for MEN1 or PHLDA3. Immunohistochemical staining revealed a significant increase in Cyclin D1 cytoplasmic expression in Men1−/- mice, concomitant with INSM1 nuclear localization (Fig. 2A, B). Specifically, when we analyzed small vs large islets in Men1-/- mice, we observed a significant correlation between increased Cyclin D1 expression and increased islet size (Fig. 2C, D).
Fig. 2.
INSM1 localization and Cyclin D1 expression varies depending on the islet genomic profile. (A) Representative images of INSM1 and Cyclin D1 immunohistochemical staining in wild-type, Phlda3−/− and Men1−/− male mice of 45 weeks. (B) Graph showing Cyclin D1 staining intensities in mice islets, quantified using ImageJ. Values represent means of data obtained from three mice ± SD. The relative intensity of the staining was calculated fractioning the integrated density by the area. (C) Representative images of hematoxylin & eosin, INSM1 and Cyclin D1 immunohistochemical staining in Men1−/- mice islets. (D) Graph showing Cyclin D1 staining intensities in mice islets, quantified using ImageJ. Values represent means of data obtained from three mice ± SD. ***P-value < 0.001 as determined by t test.
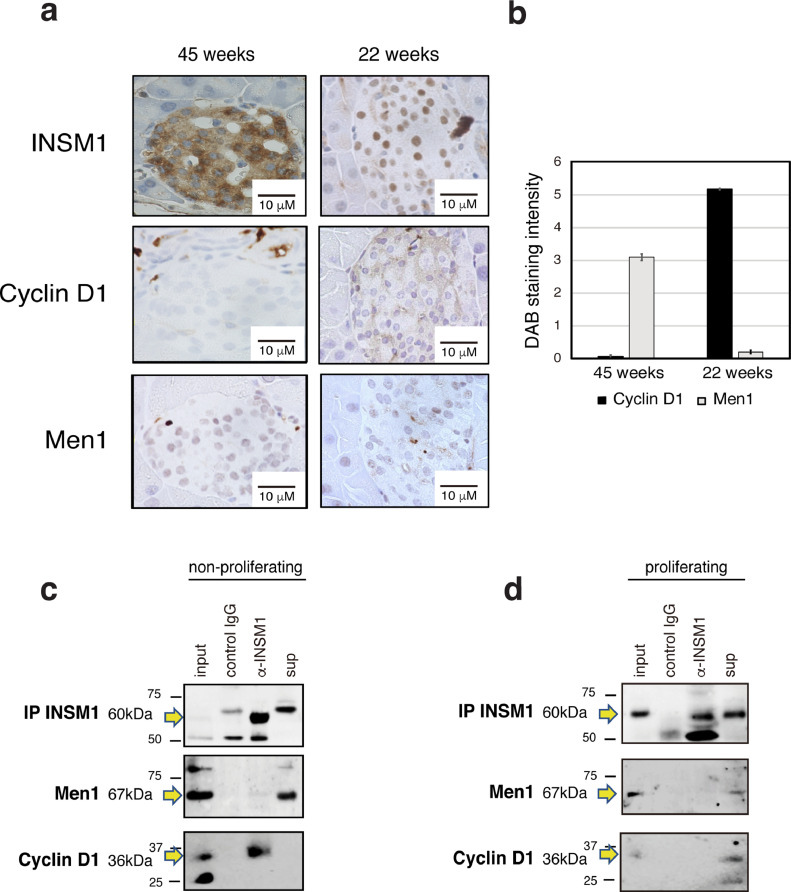
INSM1 subcellular localization influences islet proliferation and cell cycling
Islet endocrine cell mass usually increases up until young adulthood and gradually decreases thereafter [28]. Since INSM1 is closely associated with neuroendocrine embryonic development [24,27], we hypothesized that INSM1 localization might influence the proliferation of developing β cells. Immunohistochemical staining revealed that in islets from adult mice, INSM1 is localized in the cytoplasm of β cells, nuclear Menin expression is high and cytoplasmic Cyclin D1 expression is low (Fig. 3A, B). In contrast, in young mice, INSM1 is localized in the nucleus, nuclear Menin expression is concomitantly low, and cytoplasmic Cyclin D1 expression is high. These results reveal that Menin and Cyclin D1 expression are inversely correlated in mice β cells and their expression depends on the age of the mice (Fig. 3B). Additionally, INSM1 nuclear localization appears to be directly correlated with high cytoplasmic Cyclin D1 expression and low Menin nuclear expression. Previous studies have shown that INSM1 bound by Cyclin D1 acts as transcriptional co-repressor of the Cyclin D1 and INSM1 genes themselves, thereby inducing cell cycle arrest and inhibition of proliferation [26]. To test whether INSM1 and Cyclin D1 complex formation also occurs in mouse β cells, we used an immortalized mouse β-cell line (Β-TC-06) and compared INSM1 – Cyclin D1 interaction in both non-proliferating (Fig. 3C) and proliferating cells (Fig. 3D). Co-immunoprecipitation data show that INSM1 and Cyclin D1 form a complex when cells are arrested in the G1 phase (Fig. 3C), but do not interact when cells are in an active proliferative state (Fig. 3D). Menin does not bind to INSM1 in either case (Fig. 3C, D).
Fig. 3.
Variations in INSM1, Menin and Cyclin D1 expression with cell cycle and islet development. (A) Representative images of islets immunohistochemically stained for INSM1, Cyclin D1 and Menin in adult (45 weeks) and young (22 weeks) wild-type mice. (B) Graph showing Cyclin D1 and Menin expression in islets from young and adult mice, quantified from staining intensities by ImageJ. Values represent means of data obtained from three mice ± SD. Islets with a size above 50 μm (5 islets for each mouse) were used for the following calculation. (C, D) INSM1 protein was immunoprecipitated (IP) from non-proliferating (C) and proliferating (D) mouse β-cell lines (β-TC-06). Co-immunoprecipitation of cyclin D1 occurred only in non-proliferating β-TC-06 cells. Non-proliferative cells were synchronized in the G1 phase by double thymidine block. Normal IgG was used as control.
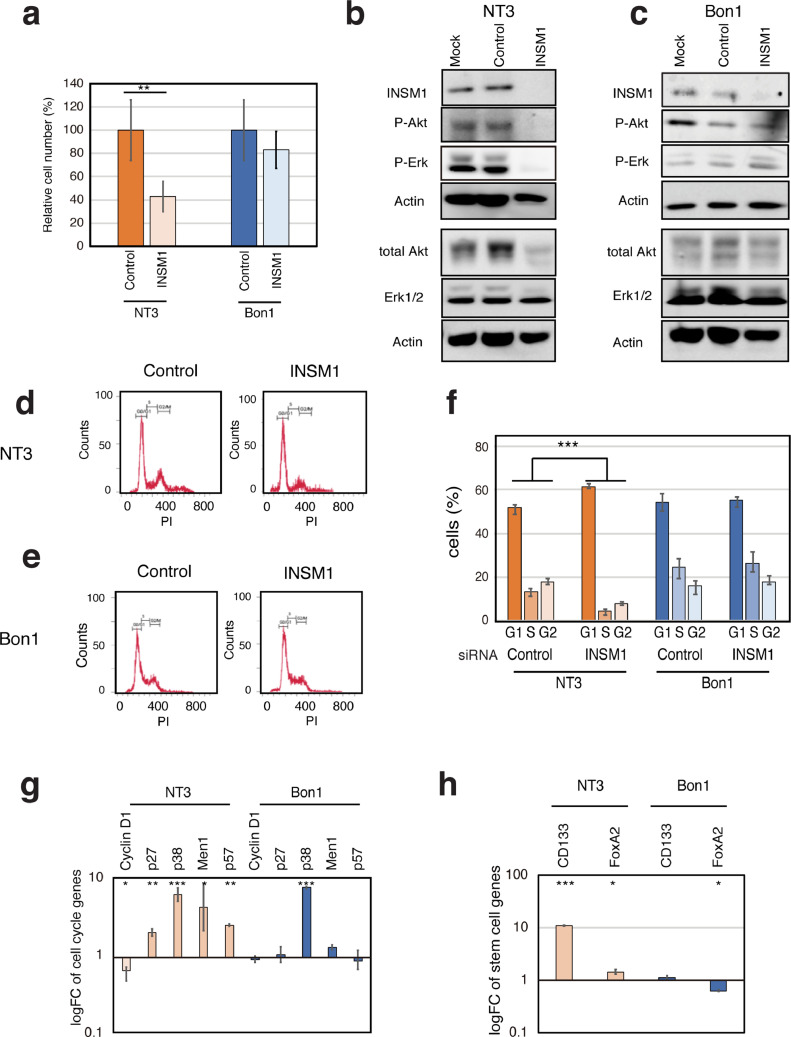
INSM1 knockdown inhibits PanNET cell proliferation and causes cell cycle arrest in p53 wild-type, MEN1 mutant cells
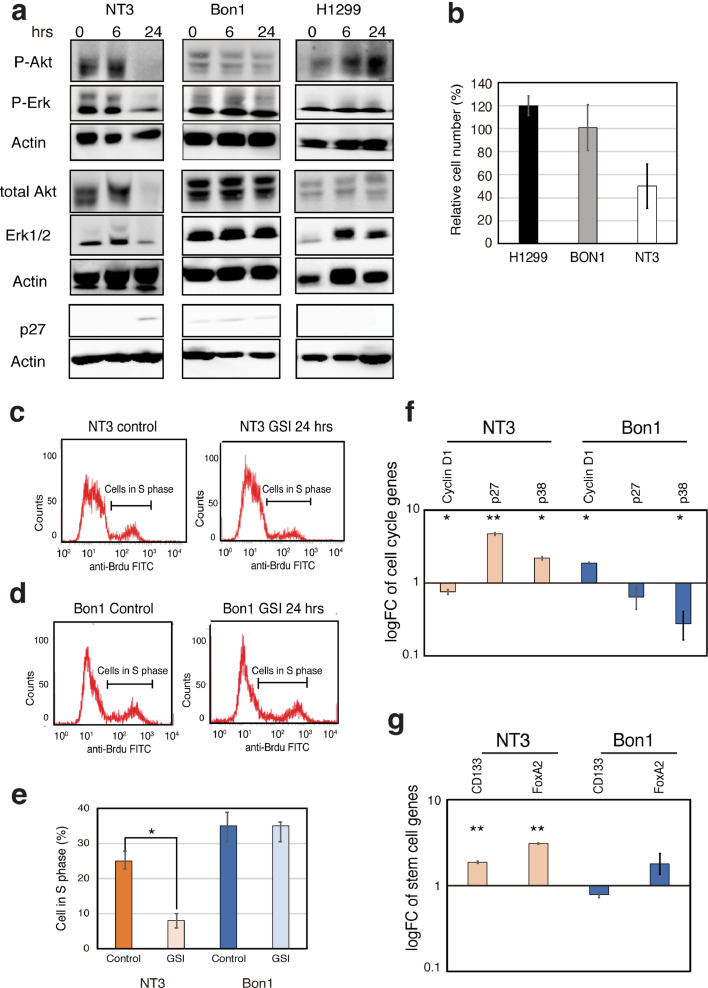
Our results so far show that interactions among INSM1, Cyclin D1 and Menin influence β cell proliferation. We have also shown that loss of MEN1 function causes β cells to switch from a non-proliferative to a proliferative state. To elucidate the role of INSM1 and its correlation with MEN1 and p53 during PanNET development, we examined a number of NET cell lines having various MEN1 and p53 genotypes (Table S2). Our data revealed that when Menin is present but p53 is null, INSM1 expression is abrogated (H1299). In contrast, when Menin is absent, INSM1 expression is observed in the nucleus, regardless of p53 status (NT3, Bon1 and QGP1) (Fig. S2). To examine the biological significance of these p53 and MEN1 genotypes on the regulation of INSM1 function, we used siRNA to knock down the expression of INSM1 in the PanNET cell lines NT3 and Bon1. In NT3 cells (p53 wild-type and MEN1 non-functional), knockdown of INSM1 resulted in a significant reduction in cell viability (Fig. 4A), together with a decrease in the expression and phosphorylation of ERK and AKT proteins, both of which are involved in cell proliferation (Fig. 4B). In contrast, in Bon1 cells (p53 mutated and MEN1 non-functional), no significant effects of INSM1 knockdown on cell viability (Fig. 4A) or the expression of proteins involved in proliferation (Fig. 4C) were observed. Accordingly, knockdown of INSM1 caused an increase in the fraction of G1 cells and a decrease in S and G2 phase cells in p53 wild-type NT3 cells (Fig. 4D, F). No significant changes were observed in Bon1 cells (Fig. 4E, F). Thus, we next tested whether reduced INSM1 expression can modulate the expression of master genes related to pancreatic progenitor cells and the slow-cycling signature. We analyzed mRNA expression both of genes related to cell cycle regulation [p38, MEN1, p57, cyclin D1 (CCND1), p27) and specific to pancreatic progenitor cells (CD133, FOXA2). Our results showed that p27, p38, p57, MEN1, CD133 and FOXA2 were upregulated in NT3 cells, whereas CCND1 was downregulated (Fig. 4G, H). The only significant changes observed in Bon1 cells were upregulation of p38 and downregulation of FOXA2 (Fig. 4G, H). Our results indicate that loss of INSM1 induces cell cycle arrest and decreases proliferation only in p53 wild-type cells, causing upregulation of slow-cycling progenitor cell markers.
Fig. 4.
INSM1 knockdown induces transcription of progenitor markers decreasing islet proliferation in wild-type p53 PanNET cells. (A) Graph showing cell viability of NT3 (p53 wild-type) and Bon1 (p53 mutated) cells. Values are mean of triplicates ± SD. (B, C) Western blotting analysis showing that INSM1 protein knockdown results in a decrease in ERK and AKT protein expression and phosphorylation in NT3 cells (B), but not in Bon1 cells (C). (D, E, F) NT3 (D) and Bon1 (E) cells were treated with INSM1 siRNA and analyzed by propidium iodide (PI) staining and flow cytometry to quantify cells in G1, S and G2 phase. Graphs shows cell cycle distribution in NT3 and Bon1 cells (F). Values represent mean of triplicates ± SD. ***P-value < 0.001 as determined by Fisher's exact test. (G, H) qRT-PCR analysis of cell cycle regulators (p38, MEN1, p57, p27, Cyclin D1) (G), and stem cell-related genes (CD133, FOXA2) (h) in NT3 and Bon1 cells treated with INSM1 or control siRNAs. Values were calculated using cDNA from untreated cells as the calibrator sample (logFC = 1). Values represent mean of triplicates ± SD. *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001 as determined by t test.
Inhibition of the Notch pathway causes cell cycle arrest and a decrease in proliferation only in p53 wild-type NET cells
These data reveal that loss of INSM1 expression in cells having functional p53 and nonfunctional MEN1 induces features characteristic of pancreatic slow-cycling progenitor cells. Notch signaling is essential to maintaining a progenitor pool in tissues of neuroendocrine origin [13,29]. Previous studies have indicated a prominent role of Notch signaling in NET tumorigenesis [30] and more specifically insulinoma tumorigenesis [31]. To investigate the significance of Notch inhibition on PanNET proliferation and cell cycle, p53 wild-type (NT3), mutated (Bon1) and null (H1299) NET cells were treated with a gamma secretase inhibitor (GSI), which inhibits Notch signaling, for up to 24 hours and ERK and AKT protein expression were analyzed. In p53 wild-type NT3 cells, GSI treatment caused a decrease in ERK and AKT protein expression and phosphorylation levels and caused an increase in the expression of the cell cycle arrest regulator p27 (Fig. 5A). In contrast, GSI treatment had no effect on p53-mutated Bon1 cells and caused an increase in ERK and AKT phosphorylation in p53-null H1299 cells (Fig. 5A). Concomitantly, cell viability decreased in p53 wild-type NT3 cells following GSI treatment, whereas viability increased in p53 null H1299 cells (Fig. 5B). We analyzed GSI-induced cell cycle changes by flow cytometry and demonstrated that Notch inhibition decreases the fraction of proliferating cells (S phase) in p53 wild-type NT3 cells (Fig. 5C, E). In contrast, no significant changes were observed in p53 mutated Bon1 cells (Fig. 5D, E). Finally, we tested whether suppression of the Notch pathway modulates the expression of master genes related to slow-cycling progenitor cells, i.e. cell cycle regulator genes (p38, p27, CCND1) (Fig. 5F) and pancreatic stem cell marker genes (CD133, FOXA2) (Fig. 5G). GSI treatment of p53 wild-type NT3 cells caused an increase in the mRNA expression of pancreatic stem cell markers (CD133, FOXA2) and slow-cycling cell markers (p38, p27), similar to what was seen when INSM1 was knocked down, whereas CCND1 mRNA expression decreased. The opposite occurred in p53-mutated Bon1 cells (Fig. 5F). These results indicate that Notch inhibition induces cell cycle arrest and decreased proliferation in p53 wild-type NT3 cells.
Fig. 5.
Notch inhibition in p53 wild-type cells causes cell cycle arrest and influences PanNET cell proliferation. (A) Western blotting analysis of NT3 (p53 wild-type), Bon1 (p53 mutated) and H1299 (p53 null) cells following 6 or 24 hrs of treatment by gamma secretase inhibitor (GSI; 10μg/ml). Results show that inhibition of the Notch pathway activity causes a decrease in ERK and AKT protein phosphorylation in NT3 cells, no change in Bon1 and an increase in H1299. Concomitantly, p27 expression increases in NT3, whereas no change is seen in Bon1 or H1299 cells. (B) Graph showing NT3, Bon1 and H1299 cell viabilities following treatment with 10 μg/ml GSI or DMSO for 48 hours. Values are means of triplicates ± SD. *P-value < 0.05 as determined by t test. (C, D, E) NT3 (C) and Bon1 (D) cells were treated with 10 μg/ml GSI or DMSO for 24 hours and then subjected to BrdU staining and flow cytometry to quantify cells in proliferating state (S phase). Graph shows cells in S phase (E). Values represent mean of triplicates ± SD. *P-value < 0.05 as determined by t test. (F, G) Graphs showing qRT-PCR analysis of cell cycle regulator (p38, p27, MEN1, p57, cyclin D1) (f), pancreatic stem cell-related genes (CD133, FOXA2) (g) comparing p53 wild-type (NT3) and mutated (Bon1) cells treated with GSI versus untreated cells. Expression values were calculated using cDNA from untreated cells as the calibrator sample (logFC=1). Values are means of triplicates ± SD. *P-value < 0.05, **P-value < 0.01, ***P-value < 0.001 as determined by t test.
The Notch pathway regulates p53 activation, INSM1 expression and INSM1 localization only in p53 wild-type PanNET cells via the Notch/ p53 axis
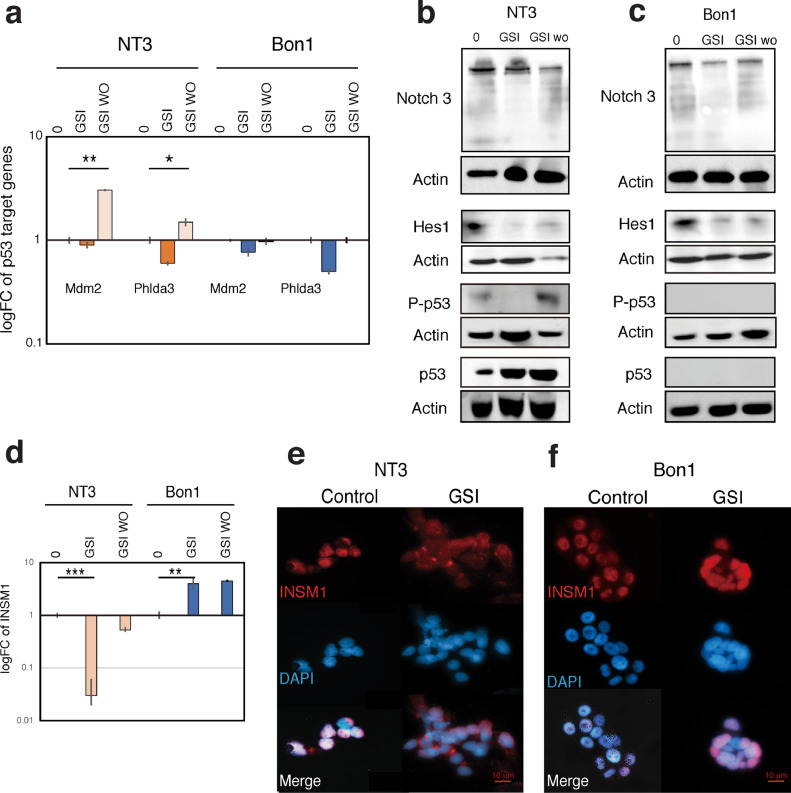
Since we found that p53 status determines the efficacy of Notch pathway inhibition, we investigated the effect of Notch inhibitor addition and removal on p53 activation. Specifically, we analyzed the induction of the p53 target genes MDM2 and PHLDA3 using a GSI washout assay as previously described [32] (Fig. 6A). First, we analyzed and compared Notch-related gene expression in p53 wild-type, mutated and null cells to identify Notch markers highly expressed in p53 wild-type cells. From this we selected NOTCH3, HES1, HEY2 and JAG2, as the top expressed Notch pathway genes expressed in p53 wild-type NT3 cells compared with p53 null H1299 cells (Fig. S3). Next, we examined Notch target gene mRNA expression to confirm the validity of this method, i.e. confirming an increase in the expression of these genes above those observed in both untreated p53 wild-type and p53 mutated cells following GSI washout (Fig. S4). Our results show that expression of these p53 target genes decreases after Notch inhibition and increases after Notch reactivation (Fig. 6A). Similarly, we observed that p53 phosphorylation decreases with GSI treatment and then increases after GSI washout (Fig. 6B), confirming a correlation between Notch signaling and p53 activation. Such changes were not observed in p53-mutated Bon1 cells (Fig. 6A, C). These data show that the Notch pathway influences p53 activation in p53 wild-type NT3 cells.
Fig. 6.
Notch signaling regulates p53 activation and INSM1 localization depending on p53 status in PanNET cells. (A) NT3(p53 wild-type) and Bon1 (p53 mutated) cells were treated with 10 μg/ml of gamma secretase inhibitor (GSI) or DMSO (control) for 48 hours to force cell surface accumulation of Notch, then washed out to activate Notch signaling. After 4 hours, RNA was isolated and cDNA prepared for real-time PCR analysis of p53 target genes (MDM2 and PHLDA3). Expression values were calculated using cDNA from untreated cells as the calibrator sample (logFC=1). Values are means of triplicates ± SD. (B, C) Western blotting analysis showing that changes in Notch pathway protein expression (Notch 3 and Hes1) directly influence p53 phosphorylation in p53 wild-type (B), but not p53 mutated cells (C). (D) NT3 and Bon1 cells were treated with 10 μg/ml GSI or DMSO for 48 hours. After 4 hours, RNA was isolated and cDNA prepared for real-time PCR analysis of INSM1. Expression values were calculated using cDNA from untreated cells as the calibrator sample (logFC=1). (E, F) Representative images of immunofluorescence analysis of NT3 (E) and Bon1 (F) cells after GSI treatment.
Our data so far has revealed that both INSM1 and the Notch pathway influence PanNET cell proliferation, depending on p53 status. To understand the p53-modulated feedback mechanisms operating between Notch signaling and INSM1 in PanNET cells, we tested the effect of Notch inhibition on INSM1 transcription by GSI washout. We found that inhibition of the Notch pathway caused a significant decrease in INSM1 gene expression only in p53 wild-type NT3 cells (Fig. 6D). In contrast, in p53-mutated Bon1 cells INSM1 expression increased with Notch inhibition (Fig. 6D). INSM1 can influence endocrine cell differentiation and proliferation depends on where it is localized [25] thus, we examined INSM1 localization following GSI treatment. Our results showed that when p53 wild-type NT3 cells were treated with GSI, INSM1 expression increased in the cytoplasm and decreased in the nucleus (Fig. 6E). In contrast, no changes in localization were observed in p53-mutated Bon1 cells (Fig. 6F).
Discussion
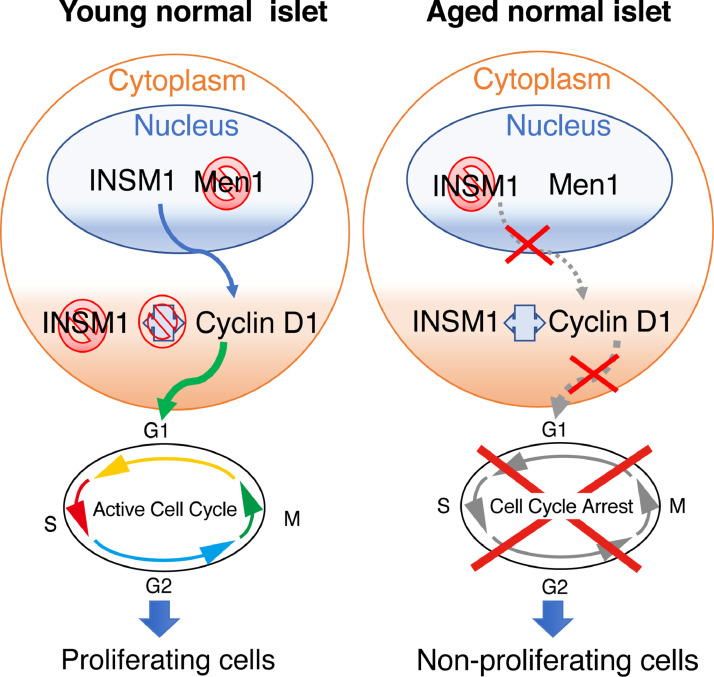
In the current study, we demonstrate that the crosstalk between p53, MEN1 and the NET diagnostic marker INSM1 regulates the proliferation and tumorigenesis of β cells. Our mice studies showed that the subcellular localization of INSM1 is inversely correlated to Menin expression: INSM1 is nuclear-localized and nuclear Menin expression is low in young mice, and this pattern is reversed in older mice. INSM1 usually localizes in the nucleus of neuroendocrine cells, where it regulates pancreatic differentiation by transcriptional repression of various β-cell markers including insulin and INSM1 itself [14,27,33]. Previous studies have revealed that INSM1 also functions in the cytoplasm to influence endocrine cell differentiation [25] and to regulate cell cycle arrest via an interaction with Cyclin D1 [26]. Cyclin D1 is a key regulator of cell cycle progression whose levels vary widely through the cell cycle 34, 35, 36. Immunohistochemical analysis of Cyclin D1 has been previously used for assessing cell proliferation in multiple types of cancer, including PanNETs [26,34,35]. We therefore assessed Cyclin D1 protein expression by semi-quantitative image analysis, and examined its correlation with MEN1 mutation and INSM1 expression. Our immunohistochemical analysis revealed scattered staining of Cyclin D1 in mice β cells. These findings are consistent with a Cyclin D1 staining pattern previously shown in multiple normal and carcinogenic tissues 34, 35, 36, 37. We revealed that Cyclin D1 staining increased concomitant with MEN1 loss and INSM1 nuclear staining. INSM1 nuclear localization is directly correlated with MEN1 loss and increased positivity in cytoplasmic Cyclin D1 staining in mice β cells. This data shows that changes in the subcellular localization of INSM1 significantly affects β cell proliferation in mouse islets. In young proliferative islets, INSM1 localizes in the nucleus causing low nuclear Menin expression and high Cyclin D1 cytoplasmic expression, thereby promoting islet cell proliferation (Fig. 7). When islets reach maturity, Menin expression in the nucleus increases, and INSM1 localizes in the cytoplasm and binds to Cyclin D1, thereby contributing to cell cycle arrest and the inhibition of islet proliferation (Fig. 7).
Fig. 7.
Relationships among INSM1, Menin and Cyclin D1 during islet development. This diagram summarizes results from our data, which show that in young islets INSM1 is localized in the nucleus and Menin expression is low. Localization of INSM1 in the nucleus causes an increase in Cyclin D1 transcription, which promotes cell cycle activation and proliferation. In adult islets, Menin expression increases, causing INSM1 to localize in the cytoplasm and bind to Cyclin D1, arresting the cell cycle and halting proliferation. Thus, loss of MEN1 in mature islets reverses the cell cycle arrest caused by INSM1 translocation to the cytoplasm, resulting in islet proliferation.
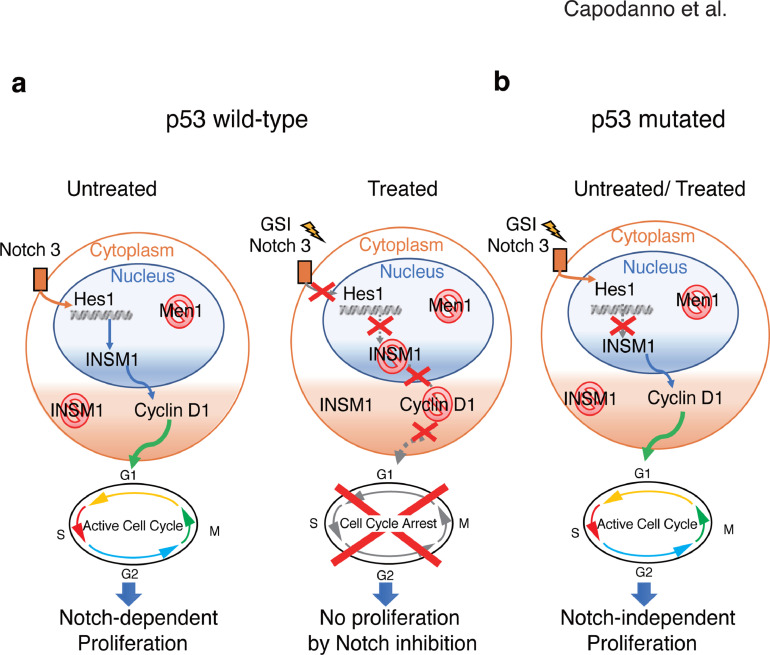
Our data suggest that dysregulation of these mechanisms may be involved in the onset of PanNETs (Fig. 8). Previous studies have described INSM1 as a highly specific NET immunohistochemical diagnostic marker [6,38]. Mice studies have revealed that INSM1 expression can directly influence PanNET metastatic behavior [39]. Still, its role in PanNET tumor formation in relation with common PanNET genomic abnormalities has not been fully elucidated. Our data show that INSM1 is highly expressed in human PanNET tissues when MEN1 LOH is present, revealing a correlation between MEN1 loss and INSM1 expression. Considering that INSM1 expression is usually prevalent in embryonic neuroendocrine tissues but restricted in adult tissues [27], the strong expression of the INSM1 protein when MEN1 is lost, suggests that tumors of neuroendocrine origin have undergone a de-differentiation event that mimics the reversal of normal islet development influencing β cell proliferation [40].
Fig. 8.
Hypothetical model of p53-dependent interactions among INSM1, Cyclin D1 and Notch during PanNET tumorigenesis. (A) In PanNET cells with wild-type p53 and loss of MEN1, Notch signaling controls the expression of INSM1 and its localization to the nucleus. When cells are treated with GSI, Notch signaling is inhibited, causing INSM1 to localize in the cytoplasm, thereby decreasing Cyclin D1 transcription and blocking the cell cycle. (B) In PanNET cells with mutated p53 and loss of MEN1, INSM1 localization in the nucleus and proliferation is not influenced by the Notch pathway.
It has been previously shown that altering or ablating expression of INSM1 in knockout mice and human cells significantly affects the terminal cellular differentiation and proliferation of NET cells including pancreatic endocrine cells [13,14,24,38]. Our data reveal that knockdown of INSM1 in PanNET cells having functional p53 and non-functional MEN1 results in significant changes in the transcription of pancreatic progenitor-related genes such as CD133 and FOXA2, cell cycle inhibitors such as p27, p57 and p38 and pancreatic-progenitor-related pathways such as the Notch signaling pathway. Pancreatic progenitor cells are multipotent stem cells that are usually arrested in the G1 phase and which can differentiate into the lineage-specific progenitors responsible for the developing pancreas [41,42]. p27 and p57 are cyclin-dependent kinase inhibitors (CDKIs) that play an important role in regulating the cell cycle exit in pancreatic progenitor cells [29,43]. p38 is an inhibitor of cell proliferation that regulates the cell cycle progression at the G1/S and G2/M transitions in multiple tissues, including the pancreas [44]. CD133 (prominin-1) and FOXA2 (forkhead box protein A2) are both highly expressed in pancreatic progenitor cells [41,45,46]. Notch signalling regulates progenitor self-renewal in early pancreatic development. Indeed, during pancreatic embryogenesis, INSM1 function is regulated by the Notch pathway [27]. Here, we show that Notch signaling significantly influences INSM1 transcription and subcellular localization in PanNET cells having functional p53 and non-functional MEN1. Specifically, we show that Notch inhibition decreases INSM1 transcription, induces cytoplasmic localization of INSM1 and causes cell cycle arrest, thereby reducing cell proliferation in p53 wild-type PanNET cells. It has been shown that Notch signaling regulates INSM1 expression and acts as an upstream regulator of INSM1 in SCLCs, influencing cell proliferation and differentiation [13,47]. Recent mice studies have shown that during the early stages of PanNET tumorigenesis, p53 is typically activated to higher levels compared to normal islets, revealing a potential role for p53 in PanNET tumorigenesis [48]. Additionally, the Notch and p53 pathways have been directly correlated in a variety of cancers, including late-stage cervical cancer and Ewing's sarcoma [49]. For instance, in cervical cancer, a positive feedback loop within the Notch pathway and p53 can counteract cervical carcinogenesis. In Erwing's sarcoma, the Notch pathway activates p53 through the transcriptional regulation of MDM2, resulting in cell cycle arrest [49]. To understand the interactions between Notch and p53 function in PanNET cells, we tested the effect of Notch activation on p53 activity. Our data indicate that the Notch pathway positively regulates p53 activation and induces transcription of the p53 target genes MDM2 and PHLDA3. These results reveal that the Notch pathway influences p53 activation in p53 wild-type PanNET cells, controls INSM1 localization and thereby regulates PanNET proliferation in a Notch-dependent manner. To the best of our knowledge, this is the first evidence showing that loss of MEN1 function in p53 wild-type in PanNET cells results in the creation of a positive feedback loop involving the Notch and p53 pathways, which in turn influences INSM1 localization and modulates cell cycle progression and PanNET proliferation (Fig. 8A). Specifically, activation of the Notch pathway regulates INSM1 nuclear localization and the consequent transcriptional regulation of Cyclin D1, thereby promoting cell cycle progression (Fig. 8A). In contrast, when p53 is mutated in PanNETs, cell cycle regulation becomes independent of Notch activation. In this case, inhibition of the Notch pathway does not result in INSM1 translocation to the cytoplasm and does not cause cell cycle arrest (Fig. 8B).
Conclusions
In summary, we have provided evidence that INSM1 is a key regulator of proliferation in β cells. INSM1 expression is strongly influenced by Menin expression and p53 activity during normal islet development and tumorigenesis. In PanNETs, inactivation of MEN1 in a p53 wild-type background causes an increase in INSM1 nuclear expression, and promotes cell cycle progression in a Notch-dependent manner. In these cells, when Notch signalling is inhibited, INSM1 localizes in the cytoplasm, inducing cell cycle arrest and a progenitor non-proliferating cell phenotype. It has been shown that different mechanisms influence early tumor formation and malignant progression in PanNETs [48,50]. For instance, it has been suggested that mutation of p53 may be involved in the late stages of PanNETs tumorigenesis [48]. Here, we hypothesize that wild-type p53 may play a role in early PanNET tumorigenesis influencing INSM1 transcription and subcellular localization depending on Notch activation. Although an infrequent event, when p53 mutations occur in late-stage of the disease, the Notch-p53 positive feedback loop is lost and Notch activation does not influence PanNET proliferation. Given the complicated nature of this intracellular signalling, further study is required to determine the precise molecular mechanisms regulating INSM1 function and early PanNET tumorigenesis. The shortage of PanNET cell lines represents one of the main limitations to this study as currently, NT3 represents the only available PanNET p53 wild-type cell line. NT3 cells proliferation index is lower compared to Bon1 cells [20], thus the Notch effect on INSM1 localization and proliferation might also be influenced by the slow-proliferating NT3 cell phenotype. Nevertheless, this study elucidates some of the mechanistic details underlying PanNET cell proliferation and suggests that INSM1 may serve both as a diagnostic biomarker and a potential target for improving the treatment of MEN1-deficient PanNETs. Future studies on a wider set of PanNET samples with either MEN1 mutations and/or additional PanNET-associated genomic abnormalities, may elucidate the significance of INSM1 as a biomarker for early disease and/or malignant progression in the MEN1-deficient PanNETs. Considering the current lack of a system to stratify all PanNET patients based on their genomic abnormalities, these findings may thereby provide an opportunity to improve future clinical decisions and improve the prognosis of PanNET patients.
Acknowledgments
We thank Dr. Marc Lamphier for critical reading of the manuscript. National Cancer Center Biobank is supported by the National Cancer Center Research and Development Fund, Japan.
Author contributions
Conceptualization of the study, Y. Capodanno. and R.O.; Methodology, Y. Capodanno, J.S, M.T, S.S, A.Y, N.H., Y. Chen. Formal analysis, Investigation, Data curation and Visualization, Y. Capodanno. Resources, J.S, M.T, S.S, A.Y, N.H. Writing—original draft preparation, Y. Capodanno and R.O. Writing-Review and editing, Y. Capodanno, Y. Chen, J.S, M.T, S.S, A.Y, N.H., R.O. All authors have read and agreed to the published version of the manuscript.
Footnotes
Abbreviations: MEN1, Multiple Endocrine Neoplasia 1; PanNET, pancreatic neuroendocrine tumors; INSM1, insulinoma associated 1; PHLDA3, Pleckstrin homology-like domain family A member 3
Funding: The authors (Y. Capodanno and R.O.) acknowledge the grant support of Grant-in-Aid for JSPS fellow (#18F18758). Y. Capodanno is a JSPS International Research fellow. This study was partly supported by a Grant-in-Aid for Scientific Research (B) (#20H03523) (R.O.), Grant-in-Aid for Scientific Research (C) (#20K07605) (R.O.) and Grant-in-Aid for Young Scientist (B) (#19K16732) (Y.Chen) from the Ministry of Education, Culture, Sports, Science and Technology of Japan; research grants from Project for Development of Innovative Research on Cancer Therapeutics (P-Direct) and Project for Cancer Research and Therapeutic Evolution (P-Create) from Japan Agency for Medical Research and Development (R.O.), research grant of the Ichiro Kanehara Foundation for the promotion of Medical Sciences and Medical Care (Y.Chen).
Conflicts of interest: The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neo.2021.07.008.
Appendix. Supplementary materials
References
- 1.Zhang J, Francois R, Iyer R, Seshadri M, Zajac-Kaye M, Hochwald SN. Current understanding of the molecular biology of pancreatic neuroendocrine tumors. J Natl Cancer Inst. 2013;105(14):1005–1017. doi: 10.1093/jnci/djt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu LM, Tang L, Qiao XW, Wolin E, Nissen NN, Dhall D, Chen J, Shen L, Chi Y, Yuan YZ. Differences and similarities in the clinicopathological features of pancreatic neuroendocrine tumors in China and the United States: A multicenter study. Medicine (Baltimore) 2016;95(7):e2836. doi: 10.1097/MD.0000000000002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Halfdanarson TR, Rubin J, Farnell MB, Grant CS, Petersen GM. Pancreatic endocrine neoplasms: Epidemiology and prognosis of pancreatic endocrine tumors. Endocr Relat Cancer. 2008;15(2):409–427. doi: 10.1677/ERC-07-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bu J, Youn S, Kwon W, Jang KT, Han S, Han S, You Y, Heo JS, Choi SH, Choi DW. Prognostic factors of non-functioning pancreatic neuroendocrine tumor revisited: The value of WHO 2010 classification. Ann Hepato-Biliary-Pancreatic Surg. 2018;22(1):66. doi: 10.14701/ahbps.2018.22.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Scarpa A. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature. 2017;543(7643):65–71. doi: 10.1038/nature21063. [DOI] [PubMed] [Google Scholar]
- 6.Fujino K, Yasufuku K, Kudoh S, Motooka Y, Sato Y, Wakimoto J. INSM1 is the best marker for the diagnosis of neuroendocrine tumors : Comparison with CGA, SYP and CD56. Int J Clin Exp Pathol. 2017;10(5):5393–5405. [Google Scholar]
- 7.Wang H, Wang S, Hu J, Kong Y, Chen S, Li L, Li L. Oct4 is expressed in Nestin-positive cells as a marker for pancreatic endocrine progenitor. Histochem Cell Biol. 2009;131(5):553–563. doi: 10.1007/s00418-009-0560-x. [DOI] [PubMed] [Google Scholar]
- 8.Ohki R, Saito K, Chen Y, Kawase T, Hiraoka N, Saigawa R, Minegishi M, Aita Y, Yanai G, Shimizu H. PHLDA3 is a novel tumor suppressor of pancreatic neuroendocrine tumors. Proc Natl Acad Sci. 2014;111(23):E2404–E2413. doi: 10.1073/pnas.1319962111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawase T, Ohki R, Shibata T, Tsutsumi S, Kamimura N, Inazawa J, Ohta T, Ichikawa H, Aburatani H, Tashiro F. PH domain-only protein PHLDA3 Is a p53-regulated repressor of Akt. Cell. 2009;136(3):535–550. doi: 10.1016/j.cell.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Fontanière S, Tost J, Wierinckx A, Lachuer J, Lu J, Hussein N, Busato F, Gut I, Wang ZQ, Zhang CX. Gene expression profiling in insulinomas of Men1 beta-cell mutant mice reveals early genetic and epigenetic events involved in pancreatic beta-cell tumorigenesis. Endocr Relat Cancer. 2006;13(4):1223–1236. doi: 10.1677/erc.1.01294. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, Gurung B, Wu T, Wang H, Stoffers DA, Hua X. Reversal of preexisting hyperglycemia in diabetic mice by acute deletion of the Men1 gene. Proc Natl Acad Sci. 2010;107(47):20358–20363. doi: 10.1073/pnas.1012257107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agarwal SK. Exploring the tumors of multiple endocrine neoplasia type 1 in mouse models for basic and preclinical studies. Int J Endocr Oncol. 2014;1(2):153–161. doi: 10.2217/ije.14.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujino K, Motooka Y, Hassan WA, Ali Abdalla MO, Sato Y, Kudoh S, Hasegawa K, Niimori-Kita K, Kobayashi H, Kubota I. Insulinoma-associated protein 1 is a crucial regulator of neuroendocrine differentiation in lung cancer. Am J Pathol. 2015;185(12):3164–3177. doi: 10.1016/j.ajpath.2015.08.018. [DOI] [PubMed] [Google Scholar]
- 14.Jia S, Wildner H, Birchmeier C. Insm1 controls the differentiation of pulmonary neuroendocrine cells by repressing Hes1. Dev Biol. 2015;408(1):90–98. doi: 10.1016/j.ydbio.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 15.Aldera A Pietro, Govender D, Locketz ML, Mukhopadhyay S, McHugh K, Allende D. Combined Use of INSM1 and synaptophysin is the most sensitive and specific panel to detect neuroendocrine neoplasms in the digestive tract. Am J Clin Pathol. 2020;154(6):870–871. doi: 10.1093/ajcp/aqaa164. [DOI] [PubMed] [Google Scholar]
- 16.Juhlin CC, Zedenius J, Höög A. Clinical Routine Application of the Second-generation Neuroendocrine Markers ISL1, INSM1, and Secretagogin in Neuroendocrine Neoplasia: Staining outcomes and potential clues for determining tumor origin. Endocr Pathol. 2020;31(4):401–410. doi: 10.1007/s12022-020-09645-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Shanmugam KS, Hayes DN, Bhattacharjee A, Biondi CA. Menin associates with a trithorax family histone methyltransferase complex and with the Hoxc8 locus. Mol Cell. 2004;13(4):587–597. doi: 10.1016/s1097-2765(04)00081-4. [DOI] [PubMed] [Google Scholar]
- 18.Frank D, Fortino W, Clark L, Musalo R, Wang W, Saxena A, Li CM, Reik W, Ludwig T, Tycko B. Placental overgrowth in mice lacking the imprinted gene lpl. Proc Natl Acad Sci U S A. 2002;99(11):7490–7495. doi: 10.1073/pnas.122039999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vandamme T, Peeters M, Dogan F, Pauwels P, Van Assche E, Beyens M, Mortier G, Vanderweyer G, De Herder W, Camp GV, Hofland LJ, De Beeck KP. Whole-exome characterization of pancreatic neuroendocrine tumor cell lines BON-1 and QGP-1. J Mol Endocrinol. 2015;54(2):137–147. doi: 10.1530/JME-14-0304. [DOI] [PubMed] [Google Scholar]
- 20.Benten D, Behrang Y, Unrau L, Weissmann V, Wolters-Eisfeld G, Burdak-Rothkamm S. Establishment of the first well-differentiated human pancreatic neuroendocrine tumor model. Mol Cancer Res. 2018;16(3):496–507. doi: 10.1158/1541-7786.MCR-17-0163. [DOI] [PubMed] [Google Scholar]
- 21.Yokoyama A, Somervaille TCP, Smith KS, Rozenblatt-Rosen O, Meyerson M, Cleary ML. The menin tumor suppressor protein is an essential oncogenic cofactor for MLL-associated leukemogenesis. Cell. 2005;123(2):207–218. doi: 10.1016/j.cell.2005.09.025. [DOI] [PubMed] [Google Scholar]
- 22.Mezei T, Szakacs M, Dénes L, Egyed-Zsigmond I. Semiautomated image analysis of high contrast tissue areas using Hue/Saturation/Brightness based color filtering. Acta Medica Marisiensis. 2011;57(6):679–684. [Google Scholar]
- 23.Crowe A, Yue W. Semi-quantitative determination of protein expression using immunohistochemistry staining and analysis: An integrated protocol. Bio-Protocol. 2019;9(24):1–15. doi: 10.21769/BioProtoc.3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Osipovich AB, Long Q, Manduchi E, Gangula R, Hipkens SB, Schneider J, Okubo T, Stoeckert CJ, Takada S, Magnuson MA. Insm1 promotes endocrine cell differentiation by modulating the expression of a network of genes that includes Neurog3 and Ripply3. Development. 2014;141(15):2939–2949. doi: 10.1242/dev.104810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang T, Chen C, Breslin MB, Song K, Lan MS. Extra-nuclear activity of INSM1 transcription factor enhances insulin receptor signaling pathway and Nkx6.1 expression through RACK1 interaction. Cell Signal. 2014;26(4):740–747. doi: 10.1016/j.cellsig.2013.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang T, Liu WD, Saunee NA, Breslin MB, Lan MS. Zinc finger transcription factor INSM1 interrupts cyclin D1 and CDK4 binding and induces cell cycle arrest. J Biol Chem. 2009;284(9):5574–5581. doi: 10.1074/jbc.M808843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lan MS, Breslin MB. Structure, expression, and biological function of INSM1 transcription factor in neuroendocrine differentiation. FASEB J. 2009;23(7):2024–2033. doi: 10.1096/fj.08-125971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mizukami H, Takahashi K, Inaba W, Osonoi S, Kamata K, Tsuboi K, Yagihashi S. Age-associated changes of islet endocrine cells and the effects of body mass index in Japanese. J Diabetes Investig. 2014;5(1):38–47. doi: 10.1111/jdi.12118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Georgia S, Soliz R, Li M, Zhang P, Bhushan A. p57 and Hes1 coordinate cell cycle exit with self-renewal of pancreatic progenitors. Dev Biol. 2006;298(1):22–31. doi: 10.1016/j.ydbio.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 30.Crabtree JS, Singleton CS, Miele L. Notch signaling in neuroendocrine tumors. Front Oncol. 2016;6(94):1–11. doi: 10.3389/fonc.2016.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Capodanno Y, Buishand FO, Pang LY, Kirpensteijn J, Mol JA, Argyle DJ. Notch pathway inhibition targets chemoresistant insulinoma cancer stem cells. Endocr Relat Cancer. 2017;25(2):1–14. doi: 10.1530/ERC-17-0415. [DOI] [PubMed] [Google Scholar]
- 32.Chadwick N, Zeef L, Portillo V, Fennessy C, Warrander F, Hoyle S, Buckle AM. Identification of novel Notch target genes in T cell leukaemia. Mol Cancer. 2009;16(8):1–16. doi: 10.1186/1476-4598-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W, Wang H, Muguira M, Breslin MB, Lan MS. INSM1 functions as a transcriptional repressor of the neuroD /β 2 gene through the recruitment of cyclin D1 and histone deacetylases. Biochem J. 2006;177:169–177. doi: 10.1042/BJ20051669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saawarn S, Astekar M, Saawarn N, Dhakar N. Gomateshwar Sagari S. Cyclin D1 expression and its correlation with histopathological differentiation in oral squamous cell carcinoma. Sci World J. 2012;2012:2–7. doi: 10.1100/2012/978327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guo SS, Wu X, Shimoide AT, Wong J, Moatamed F, Sawicki MP. Frequent overexpression of cyclin D1 in sporadic pancreatic endocrine tumours. J Endocrinol. 2003;179(1):73–79. doi: 10.1677/joe.0.1790073. [DOI] [PubMed] [Google Scholar]
- 36.Zhang X, Gaspard JP, Mizukami Y, Li J, Graeme-cook F, Chung DC. Overexpression of cyclin d1 in pancreatic beta cells in vivo results in islet hyperplasia without hypoglycemia. Diabetes. 2005;54:712–719. doi: 10.2337/diabetes.54.3.712. [DOI] [PubMed] [Google Scholar]
- 37.Assem M, Youssef EA, Rashad RM, Yahia MA-H. Immunohistochemical Expression of Cyclin D1 in Invasive Ductal Carcinoma of Human Breast. Oncomedicine. 2017;2:80–87. [Google Scholar]
- 38.Rosenbaum JN, Guo Z, Baus RM, Werner H, Rehrauer WM, Lloyd R.V. A novel immunohistochemical and molecular marker for neuroendocrine and neuroepithelial neoplasms. Am J Clin Pathol. 2015;144(4):579–591. doi: 10.1309/AJCPGZWXXBSNL4VD. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi S, Contractor T, Vosburgh E, Du YCN, Tang LH, Clausen R, Harris CR. Alleles of Insm1 determine whether RIP1-Tag2 mice produce insulinomas or nonfunctioning pancreatic neuroendocrine tumors. Oncogenesis. 2019;8(3):1–13. doi: 10.1038/s41389-019-0127-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Puri S, Roy N, Russ HA, Leonhardt L, French EK, Roy R, Bengtsson H, Scott DK, Stewart AF, Hebrok M. Replication confers β cell immaturity. Nat Commun. 2018;9(1):1–12. doi: 10.1038/s41467-018-02939-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang Y, Lanzoni G, Carpino G, Cui C, Dominguez-Bendala J, Wauthier E, Cardinale V, Oikawa T, Pileggi A, Gerber D. Biliary tree stem cells, precursors to pancreatic committed progenitors: evidence for possible life-long pancreatic organogenesis. Stem Cell. 2013;31(9):1966–1979. doi: 10.1002/stem.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venkatesan V, Gopurappilly R, Goteti SK, Dorisetty RK, Bhonde RR. Pancreatic progenitors: The shortest route to restore islet cell mass. Islets. 2011;3:295–301. doi: 10.4161/isl.3.6.17704. (February 2015) [DOI] [PubMed] [Google Scholar]
- 43.Nölting S, Rentsch J, Freitag H, Detjen K, Briest F, Möbs M, Weissman V, Siegmund B, Auernhammer CJ, Aristizabal Prada ET. The selective PI3Kα inhibitor BYL719 as a novel therapeutic option for neuroendocrine tumors: Results from multiple cell line models. PLoS One. 2017;12(8):1–29. doi: 10.1371/journal.pone.0182852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aasrum M, Thoresen GH, Christoffersen T, Brusevold IJ. p38 differentially regulates ERK, p21, and mitogenic signalling in two pancreatic carcinoma cell lines. J Cell Commun Signal. 2018;12(4):699–707. doi: 10.1007/s12079-017-0444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee K, Cho H, Rickert RW, Li Q.V., Pulecio J, Leslie CS, Huangfu D. FOXA2 Is required for enhancer priming during pancreatic differentiation. Cell Rep. 2019;28(2):382–393. doi: 10.1016/j.celrep.2019.06.034. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Immervoll H, Hoem D, Sakariassen P, Steffensen OJ, Molven A. Expression of the “stem cell marker” CD133 in pancreas and pancreatic ductal adenocarcinomas. BMC Cancer. 2008;8:1–14. doi: 10.1186/1471-2407-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ito T, Matsuo A, Hassan WA. Notch signaling and Tp53/RB1 pathway in pulmonary neuroendocrine tumorigenesis. Transl Cancer Res. 2016;5(2):213–219. [Google Scholar]
- 48.Yamauchi Y, Kodama Y, Shiokawa M, Kakiuchi N, Marui S, Kuwada T, Sogabe Y, Tomono T, Mima A, Morita T. Rb and p53 execute distinct roles in the development of pancreatic neuroendocrine tumors. Cancer Res. 2020;(16):3620–3630. doi: 10.1158/0008-5472.CAN-19-2232. [DOI] [PubMed] [Google Scholar]
- 49.Dotto GP. Crosstalk of Notch with p53 and p63 in cancer growth control. Nature. 2018;9(8):587–595. doi: 10.1038/nrc2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Capodanno Y, Buishand FO, Pang L, Kirpensteijn J, Mol J, Elders R, Argyle DJ. Transcriptomic analysis by RnA sequencing characterises malignant progression of canine insulinoma from normal tissue to metastatic disease. Sci Rep. 2020;10(11581):1–12. doi: 10.1038/s41598-020-68507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.