Abstract
Background
The emergence of SARS-Cov2 variants has highlighted the need to implement sequencing-based surveillance in developing countries for early response to mutant viruses of concern. However, limited information on how to implement sequencing-based surveillance is available, and the feasibility and performance of this new type of surveillance are still in question.
Methods
To understand the challenges with the implementation and to promote sequencing-based surveillance, we reported findings from a pilot for hepatitis A (HepA) in five sentinel provinces in China as an example of sequencing-based surveillance implementation. The performance of the surveillance system was evaluated by indicators related to acceptability, data quality, simplicity, utility, and timeliness. We use a scale from 1 to 3 was used to provide a score for each aspect.
Results
During the pilot, 306 cases of HepA were reported, and 49.79% of samples were available for sequencing. Eleven genomic clusters were found, of which seven clusters were potentially related to a foodborne outbreak oyster based on identical viral sequence and epidemiologic investigations. The greatest strength of the system was its simplicity (Score: 2.63). The acceptability (Score: 2.0) and utility (Score: 2.33) were modest, but data quality (Score: 1.75) and timeliness (Score: 1.75) were the main challenges.
Conclusions
Overall, the system performed satisfactorily and proved to be useful for virological characterization of cases and early outbreak detection, with a great potential for scale-up. Further efforts are required to address financial and human resource constraints and inadequate support among physicians. Education should be given to health care professionals to improve the data quality. The establishment of decentralized surveillance networks can be an approach to improve timeliness for emerging infections.
Keywords: Sequencing, surveillance evaluation, hepatitis A (HepA), emerging infections
Introduction
The frequency of outbreaks and the threat of infectious disease pathogens have been increasing significantly (1,2). The coronavirus diseases 2019 (COVID-19) pandemic is the latest one but will not be the last one. It has painfully proven that our world is under-prepared for large outbreaks of emerging infectious diseases, especially in developing countries where the public health and clinical infrastructure are often inadequate. To prepare for the next epidemic and to provide a timely and effective response, the World Health Organization (WHO) recommended scaling up sequencing-based surveillance, which depositing pathogens’ sequences in public databases, and sharing data among scientific and public health communities (3). However, developing countries have more challenges in speeding up the pace of establishing sequencing-based surveillance.
After SARS, the Chinese government has launched a national internet-based surveillance system named the National Notifiable Disease Report System (NNDRS) (4). Thirty-nine legally notifiable infectious diseases, including hepatitis A, must be reported electronically by physicians to the NNDRS within 2 to 24 hours after diagnosis, but this system does not collect data on the epidemiological details of the cases and the molecular information of pathogens (5). Although China is technologically more capable now and sequencing costs continue to fall sharply, limited efforts have been made to add sequencing into NNDRS, and sequencing-based surveillance at a large scale has not been implemented. Generating know-how guidance for the implementation of sequencing-based surveillance is urgent to mitigate the impact of the pandemics now and in the future.
Beginning in March 2019, we conducted a pilot of sequencing-based surveillance for hepatitis A (HepA) in 5 sentinel provinces in China. HepA was chosen because reported cases are primarily based on laboratory diagnosis, the number of cases is manageable and sequencing methods are widely used. Therefore, sequencing-based surveillance for HepA was technically and resource-wise among the “low hanging fruits”. In the meanwhile, the potential utility of sequence-based surveillance for early outbreak detection was rising because of China’s globalization and food imports
In this study, we evaluated the performance of the pilot surveillance using guidelines from the United States Centers for Disease Control and Prevention (CDC) and previous studies (6-8). The primary goal is to formally assess the feasibility and its performance, with the hope of improving future implementation efforts and identifying the key factors contributing to successful implementation.
We present the following article in accordance with the StaRI reporting checklist (available at https://dx.doi.org/10.21037/atm-21-1193).
Methods
National Notifiable Disease Report System (NNDRS)
NNDRS is a national internet-based reporting system for infectious diseases in China. Based on NNDRS, the incidence of notifiable infectious diseases and demographic characteristics of reported cases are monitored at different levels (9). However, the molecular information of pathogens from sequencing is not part of NNDRS. To be better prepared for the next epidemic, researchers from the Chinese CDC and Zhejiang University launched this pilot study. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). This study was approved by the Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (Number [2019]17). Written or oral consents were obtained from the patients.
Piloting sequencing-based surveillance for HepA
The sequencing-based enhanced surveillance for HepA was first initiated in Zhejiang province in March 2019, followed by Shandong province in June, and Shaanxi, Hainan, and Hubei provinces in September 2019, and continued through December 2020. Public health infrastructure, geographic representation, and financial resources needed were considered in the selection and duration of the pilot project. These five provinces (Zhejiang, Shandong, Shaanxi, Hainan, and Hubei) represented all three social-economical tiers of China with different HepA morbidity and healthcare resource levels (10). The incidence of HAV infection varied and was highest in Hubei at 1.47/100,000, and lowest in Shandong at 0.53/100,000 in 2015 (11). Government funding for healthcare also varied across the three regions, with the eastern region (Zhejiang, Shandong, and Hainan) having the highest budget (6,194 thousand Yuan), the western region (Shaanxi) having the lowest budget (2,643 thousand Yuan) (12).
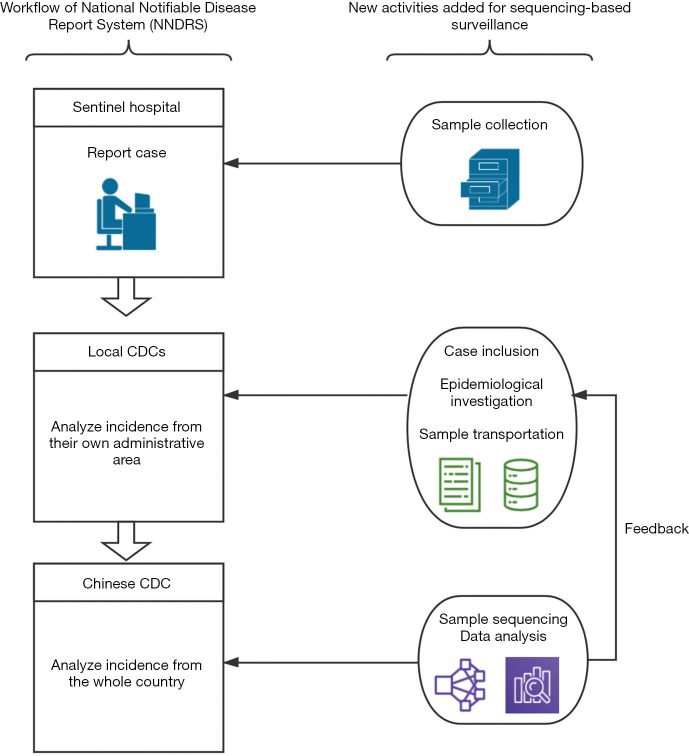
The pilot was built on case reporting to CDCs via NNDRS and adding viral sequencing and more detailed epidemiologic investigation of food and travel exposures, as shown in Figure 1:
Figure 1.
Core activities in the surveillance network. This figure indicates different work schemes of NNDRS in each stakeholder and new activities that we added for implementing sequencing-based surveillance. NNDRS, National Notifiable Disease Report System; CDC, Center for Disease Control and Prevention.
Step 1: Specimen collection and transportation to local CDCs: HepA cases were routinely reported by physicians from hospitals at the town level and above to NNDRS within 24 hours after diagnosis (5,13). Case definition of HepA is uniform nationwide and a case must be symptomatic patient with a discrete onset of symptoms consistent with acute viral hepatitis, and jaundice or elevated serum aminotransferase levels, and a positive anti-HAV IgM test. To acquire specimens for sequencing, we asked the clinical laboratories in the sentinel hospitals to preserve their leftover serum samples and to request that the public health practitioners (PHPs) perform an epidemiological investigation within 3 days.
Step 2: Transportation of specimens and collection of epidemiologic data by local CDCs: information on demographic, clinical, and epidemiological exposures, including food habits and work-related exposure, were collected. A web-based real-time reporting system for data entry using smartphones or computers was developed. Data collection by PHP was remotely supervised by our research team and data collection must be completed within 3 days after diagnosis. Before the project was launched, PHPs from each sentinel province were trained during a one-day training session and were provided with handy problem-solving support by the study team during the first month.
Step 3: Genomic sequencing and analysis at a central CDC laboratory: samples were preserved below −20 °C before they were shipped to a central CDC lab. According to the manufacturer's instructions, RNA was extracted using the QIAamp Viral RNA Mini Kit (Qiagen Inc., USA). The RNA of positive samples was then amplified by nested reverse transcription-polymerase chain reaction (RT-PCR), using the primers that were previously described (14,15). The first round of amplification was performed with the PrimeScript™ One-Step RT-PCR Kit (TaKaRa, Dalian, China). The PCR Premix TaqTM DNA Polymerase (Ex TaqTM Version 2.0 plus dye) from TaKaRa Biotechnology (Dalian, China) was then used for the second round of amplification. Eventually, the region of the VP1-P2A junction region, which is a 391-base-pair fragment of the HAV genome was amplified. Mega-X was used for the sequence alignment. A final phylogenetic tree was constructed based on Neighbor-Joining algorithms and using the Kimura2-parameter method. The molecular information of each case was integrated with the clinical and demographic data using a unique identification number. Data were cleaned and analyzed by personnel from different technical backgrounds (e.g., epidemiology, microbiology, public health, etc.). Local CDCs would be suggested to perform further epidemiological investigations if we detected genomic clustered cases. A genomic cluster was defined as a group of 1 or more laboratory-confirmed HepA cases that shared an identical VP1–2A junction region.
Evaluation of the sequencing-based surveillance
According to the CDC guideline and previous studies (6-8,16), we evaluated the performance of the current surveillance network using the following five aspects: (I) data quality, (II) acceptability, (III) simplicity, (IV) timeliness, and (V) utility. Data for evaluation were retrieved from the surveillance databases, laboratory documents, and a questionnaire survey given to PHPs at the sentinel provinces and central laboratory. The score for each indicator was defined as follows: Score 1 (<60%, weak performance); Score 2 (60–79%, moderate performance); Score 3 (≥80%, good performance).
Statistical analysis
Data analysis was conducted using SPSS software version 16.0 (SPSS Inc., Chicago, IL, USA). Continuous variables were expressed as means and categorical variables as percentages, and they were compared using the χ2 test, Fisher’s exact test, or Wilcoxon rank-sum test, where appropriate. P<0.05 was considered statistically significant. Univariate regression models were used to calculate the OR values and 95% confidence intervals (CI) for the risk factors that were associated with genomic clusters.
Results
Implementation of the sequencing-based surveillance system and questionnaire survey
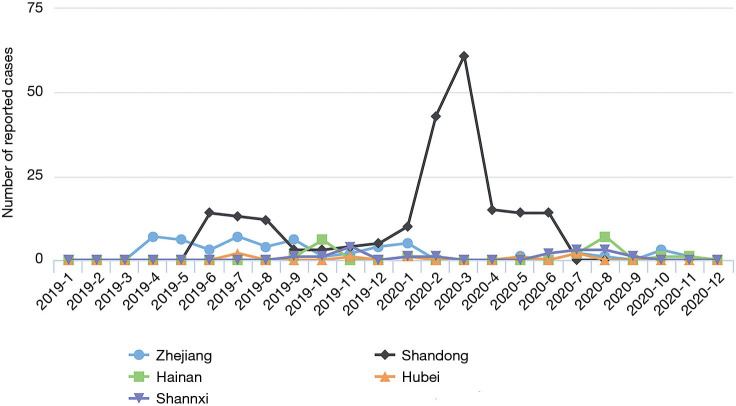
A total of 306 cases were reported by sentinel hospitals (Figure 2). As shown in Table 1, the majority were aged 40–64 years. About 20% of cases were asymptomatic cases that were detected during medical checkups or preoperative testing that included a HepA anti-IgM test. Only 237 (77%) patients had serum samples for further analysis. In total, there were 118 (50%) patients who had detectable HAV RNA. One hundred and sixteen (98%) specimens were typed as genotype IA by sequencing and only 2 were typed as IB. Sequencing indicated that 86% (102/118) of the patients shared an identical VP1-2A junction region sequence to one other, while the remaining 16 cases were sporadic with one or more nucleotide differences; eleven genomic clusters were found (Figure 3). The questionnaire survey was completed by 40/103 (39%) of the PHPs involved in the HAV surveillance.
Figure 2.
Total number of reported cases per sentinel province from 2019 to 2020 in China.
Table 1. characteristics of hepatitis A cases reported in sentinel provinces targeted for sequencing, 2019–2020.
| Characteristics | Reported cases (N=306) |
|---|---|
| Male patients | 186 (61%) |
| Median age, years (range) | 48 [22–97] |
| 22–39 | 104 (34%) |
| 40–64 | 161 (53%) |
| ≥65 | 41 (13%) |
| Patients without ALT elevated (%) | 63 (20%) |
| Median ALT, U/L | 1,232.60 (range: 8–8,576) |
| Median TBIL, μmol/L | 105.28 (range: 2–1,615) |
Figure 3.
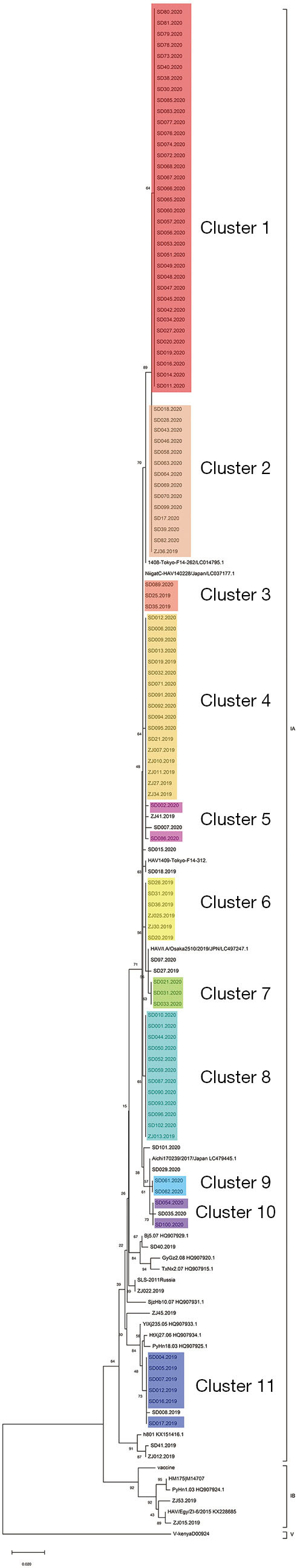
Phylogenetic analysis of all HAV isolates obtained from 2019 to 2020 in sentinel provinces. Nucleotide (nt) sequences from the VP1/2A junction region (391 bases) of reference strains retrieved from GenBank (*) and 118 sequences from this study were analyzed by the neighbor-joining (NJ) method. Branches with colors indicate different clusters. The numbers at the nodes indicate bootstrap percentages over 1,000 replicates (only values >70% are shown). The bar length indicates genetic distance. HAV, hepatitis A virus.
Evaluation of the surveillance system
Data quality
The mean score for data quality was 1.75. The three evaluated indicators had a moderate performance (Table 2). Of 306 enrolled patients who had positive tests for anti-HAV IgM, 243 patients had symptoms consistent with a HepA case definition (79.41%). Information regarding the history of close contact with recent acute hepA cases, travel history, food exposure, and other clinical characteristics was collected for 178 patients who had serum sample (75.10%). The proportion of eligible samples for sequencing analysis scored the lowest (49.79%).
Table 2. The evaluation of the HAV sequencing-based surveillance in sentinel provinces, 2019–2020.
| Aspects | Indicator definition | Indicator measurement | Indicator value | Score† | Performance |
|---|---|---|---|---|---|
| Data quality | 1.75 | Weak | |||
| (I) Met NRDSS case definition | % of HepA cases | 79.41 | 2 | ||
| (II) Serum samples for sequencing | % of cases obtained serum samples | 77.45 | 2 | ||
| (III) Samples sequenced | % of samples with detectable HAV RNA | 49.79 | 1 | ||
| (IV) Completeness of case information | % of forms with complete data | 75.10 | 2 | ||
| Timeliness | 1.75 | Weak | |||
| (I) Cases reported per NRDSS time requirement | % of cases reported within 24 hours | 50.98 | 1 | ||
| (II) Timely sample collection | % of samples were collected within 3 days | 75.49 | 2 | ||
| (III) Samples timely transferred to central CDC lab | % of samples transferred to central CDC within one month | 47.36 | 1 | ||
| (IV) Timely sequencing and sharing | <1 month: 83.331 to 2 months: 16.67 >2 months: 0.0 |
3‡ | |||
| Acceptability | 2 | Moderate | |||
| (I) Acceptable to patients | % of PHPs never be rejected by patients | 17.24 | 1 | ||
| (II) Acceptable to physicians from sentinel hospital | % of PHPs never fail to contact discharged patients due to delayed reporting | 72.41 | 2 | ||
| (III) Acceptable to PHP | % of PHPs within each category [not satisfied (NS), poorly satisfied (PS), satisfied (S), very satisfied (VS)] | VS: 66.52 S: 34.48 PS: 0.0 NS: 0.0 |
3§ | ||
| Utility | 2.33 | Moderate | |||
| (I) Detecting genomic clusters in lab | Eleven clusters were detected | 3 | |||
| (II) Sharing of HAV sequencing with CDCs | Sequence data were available in two provinces | 1 | |||
| (III) CDC investigation and outbreak control | One outbreak was detected and managed promptly | 3 |
†A scale from 1 to 3 was used to provide a score for indicators as follows: <60% scored 1 (weak performance); 60–79% scored 2 (moderate performance); ≥80% scored 3 (good performance), the score for each aspect was the mean score of its indicators. ‡The combined percentage of “one month” and “one to two months” was used to obtain the score. §The combined percentage of “very satisfied” and “satisfied” was used to obtain the score. HepA, hepatitis A; HAV, hepatitis A virus; RNA, ribonucleic acid; PHPs, public health practitioners; CDC, Centers for Disease Control and Prevention.
Timeliness
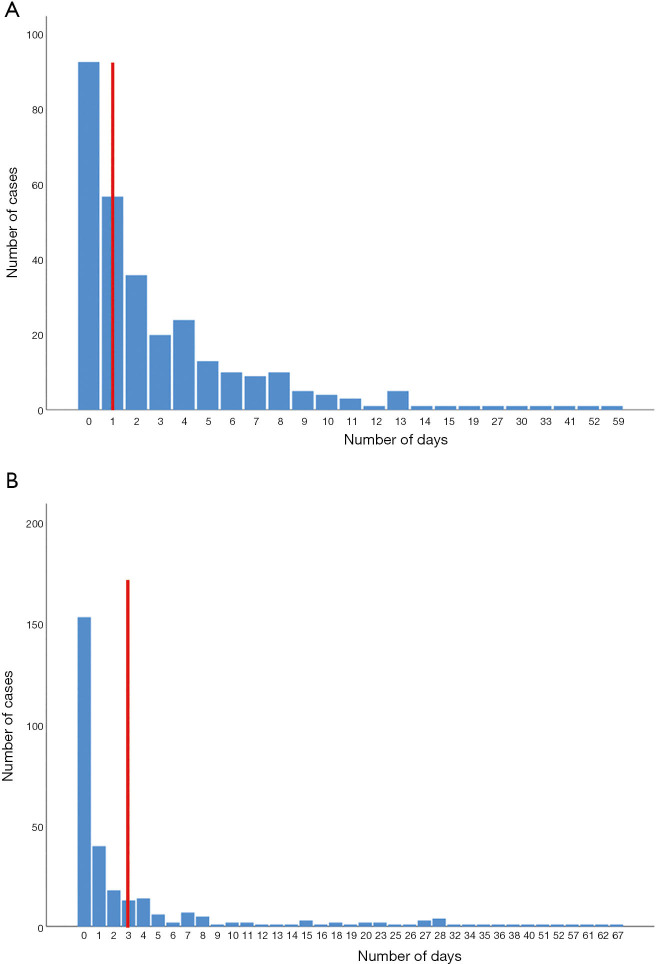
The mean score for timeliness was 1.75. Of the four indicators evaluated only one scored 3 points (Table 2). The time interval between sample collection and sequencing analysis reporting was too long. In China, HepA cases must be reported within 24 hours and this process is supervised by the public health department in each hospital. However, considerable reporting delays (49.02%) still occur when overworked physicians often forget their public health duty (Figure 4A). The average number of days between diagnosis and sample collection varied (Figure 4B), but only 75.49% of the samples were collected promptly, and the lengthiest period (67 days) was in Shaanxi province.
Figure 4.
Distribution of the time interval between first-time visit and case reporting to NNDRS, and the time interval between sample collection and first notification (N=306). (A) This interval represents the time between the patient coming into contact with the health care services and the physicians becoming aware of the case, the red vertical line indicated required time points of case reporting. (B) This interval represents the time between the patient was reported as a case and the PHPs collecting his serum sample, the red vertical line indicated required time points of sample collection. NNDRS, National Notifiable Disease Report System; PHPs, public health practitioners.
Acceptability
The mean score for acceptability was 2.0. Obtaining clinical samples is fairly complicated in China, and during this investigation, only 17.24% of the PHPs encountered no refusal to participate by patients. The most common reason for refusal was an unwillingness to give extra blood for viral sequencing for patients. Although we used the leftovers of serum samples when possible, it still leads to the loss of some specimens because the leftover amount was insufficient for further examination. Besides, around 28% of PHPs claimed that they were not get notified in time by physicians and then could not contact discharged patients, indicating the acceptability of sequencing-based surveillance among physicians was not ideal. All the PHPs gave positive feedback about the supervision and feedback provided by our surveillance system.
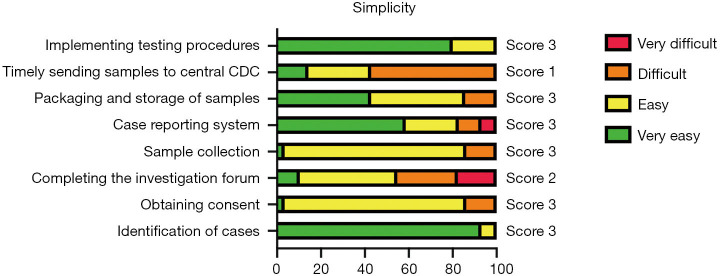
Simplicity
The mean score for simplicity was 2.63. Among all of the eight indicators evaluated, six had a good performance, one had moderate performance, and one had weak performance (Figure 5). For the indicators used to assess the implementation of the different surveillance activities, most PHPs had no trouble with implementation except for the timely transfer of samples to the central CDC laboratory. Also, laboratory personnel reported that it was easy to conduct the sample testing procedures.
Figure 5.
List of indicators and scores for simplicity, interviews with public health practitioners (N=40). The combined percentage of “very easy” and “easy” was used to obtain the score. A scale from 1 to 3 was used to provide a score for indicators as follows: <60% scored 1; 60–79% scored 2; ≥80% scored 3.
Utility
Based on the purpose of this study, we evaluated the utility of the current surveillance system by its ability to achieve the following three indicators: (I) to detect genomic clustered cases occurring at different times and in different places; (II) to provide information on the identification and characteristics of HAV strains in the five sentinel provinces; (III) to detect and manage potential HepA outbreak promptly. Of the three indicators evaluated, two showed a good performance and one showed a weak performance (Table 2).
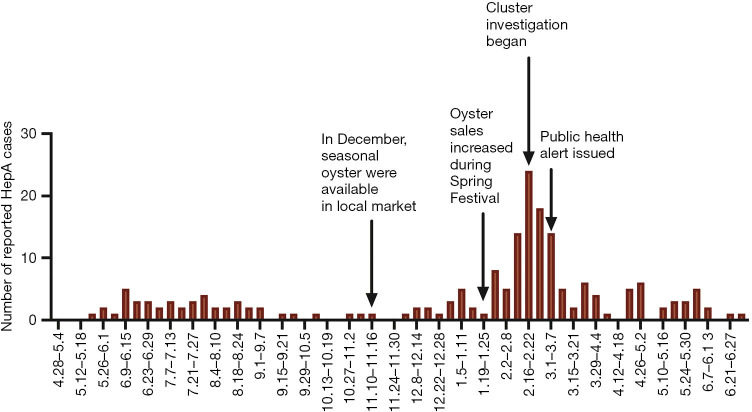
We found 11 clusters involving 102 cases from two provinces (Figure 3). Sixty-two cases (61%) were reported at different times or in different places, and the most common risk factor was raw seafood consumption (Table 3). Forty-two patients had raw seafood exposure; of these, 74% ate raw oysters. To identify localized epidemic events, local CDC used to base on a case count threshold per month at the catchment area level. An outbreak investigation would be initiated if two cases were occurring in the same area within one month. However, the link between those clustered cases would be unknown without sequencing data. The new information from sequencing-based surveillance may help the early detection and management of an outbreak. For instance, as shown in Figure 6, 40% of cases from Cluster 1 were dispersed over time and geographic locations, and sequencing-based surveillance revealed the virologic links among the sporadic cases, and this information could advance the detection of this outbreak by two months if the virologic data had been shared promptly. According to the abovementioned threshold, the first two cases of outbreak concerns (SD61 and SD60) were triggered in February 2020. But, the first two clustered cases (SD002 and SD086) actually occurred in Dec 2019. Sequencing-based surveillance could improve the efficiency of traceback investigation since patients with identical sequence patterns and similar epidemiologic characteristics usually suggest a common-source exposure. According to laboratory data, only 56% of cases that were reported after Feb 2020 were genetically identical with the outbreak strains. The combination of epidemiological and virologic information among clustered cases implicated raw oysters as the potential vehicle of the outbreak. Communication campaigns by CDC warned the general public about the risk of eating raw seafood. Although HAV was not detected in food samples, the local health authority still halted a potentially worsening outbreak based on epidemiological and sequencing evidence (Figure 7).
Table 3. Comparison of hepatitis A cases did and did not belong to genomic clusters (N=237).
| Characteristics | Non-cluster (N=140) | Genomic clusters by sequencing | Cluster‡ (N=97) | Odd ratio† (95% CI) | P value |
|---|---|---|---|---|---|
| Sex (%) | 0.050 | ||||
| Female | 64 (45.7) | 32 (33.0) | 1.0 | ||
| Male | 76 (54.3) | 65 (67.0) | 1.7 (1.0–2.9) | ||
| Median age, years (range) | 49 [22–97] | 40 [25–64] | 0.000 | ||
| Risk factors (%) | |||||
| Raw seafood exposure | 31 (22.1) | 42 (43.3) | 2.6 (1.4–4.5) | 0.001 | |
| Raw vegetable exposure | 44 (31.4) | 43 (44.3) | 1.6 (0.9–2.7) | 0.165 | |
| Recent travel history | 14 (10.0) | 5 (5.2) | 0.5 (0.2–1.4) | 0.122 | |
| Household contact | 0 (0.0) | 1 (1.0) | NA |
Data are n (%) unless otherwise specified. †Comparison between gnomically clustered and non-clustered hepatitis A cases. ‡4 individuals in cluster 3 and 1 individual in cluster 6 have no epidemiological information.
Figure 6.
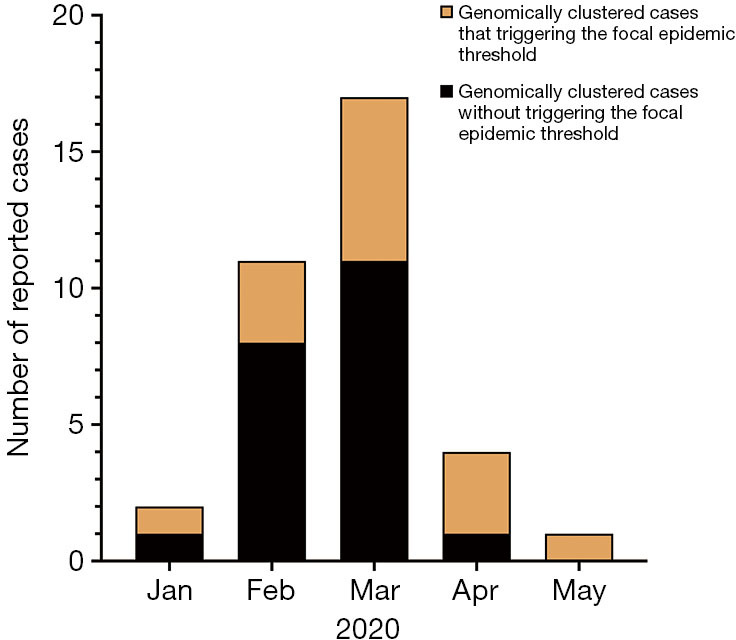
Spatial distribution and temporal distribution of cases from Cluster 1 (N=35). Different colors represent genomically clustered cases with or without triggering the focal epidemic threshold.
Figure 7.
Timeline of hepatitis A cases reported to local Centers for Disease Control and Prevention from April 2019 to June 2020 in one sentinel province (N=211).
Main challenges during the pilot
Low yield of RNA sequencing from samples
Less than 50% of the received samples were available for sequencing. The possible reason for such a low rate is that the case reporting criteria were more sensitive and not highly specific in identifying true HepA cases. We compared patients who had symptoms meeting the surveillance case definitions for hepatitis A in China and in the United States. Instead of elevated ALT, the case definition used in the United States requiring a high level of ALT of 200. As shown in Table 4, patients who met the case definition of the United States were more likely to have detectable HAV RNA (P=0.003), and those who met the Chinese HepA case definition and did not have ALT level at 200 or above, only 17% (8/47) had detectable RNA. Besides, only 79% of reported cases met China’s case definition Although most PHPs thought screening out reports that failed to meet the case definition should be easy, the implementation rate was low (10%).
Table 4. Demographic and clinical characteristics of cases met the surveillance case definition for Hepatitis A of China and United States.
| Characteristics | Patients met case definition of HepA in China (N=206) | Patients met case definition of HepA in the United states (N=159) | P value |
|---|---|---|---|
| Age, years (mean ± SD) | 46±13 | 43±12 | 0.113 |
| Gender (% male) | 62.1 | 67.9 | 0.251 |
| Median ALT, U/L | 1,014.8 (range: 16–8,586) | 1,789.1 (range: 202–8,586) | 0.000 |
| Median TBIL, μmol/L | 85.1 (range: 3.2–1,615) | 131.5 (range: 10.40–1,616) | 0.037 |
| Detection rate of HAV RNA in sample, N (%) | 116 (56.31%) | 114 (71.70%) | 0.003 |
HepA, hepatitis A; ALT, alanine transaminase; TBIL, total bilirubin; RNA, Ribonucleic Acid; CDC, Centers for Disease Control and Prevention.
Timely reporting
By case reporting regulations, HepA cases must be reported by physicians within 24 hours. However, from our panel discussion in the sentinel hospitals, many physicians thought reporting to NNDRS is not a priority. Considerable reporting delay occurs when overworked physicians often forget their public health responsibilities, only 50.98% of HepA cases were reported to NNDRS within 24 hours. Delayed reporting resulted in delayed serum sample collection and epidemiological investigation by the PHPs.
Financial and human resource burden
The main implementation challenges were additional burdens of human and financial resources, making timely sample collection and transportation difficult to sustain. Designated PHPs had to retrieve samples from every sentinel hospital and arranged transportation to the central laboratory. Due to time and cost restraints, the frequency of transport varied in different sentinel provinces, and less than half of the samples were sent to the central laboratory within one month. Secondly, around 38% of the PHPs we interviewed complained about work overload, which also impacted the sample collection and transportation.
Discussion
During 2019–2020, the sequencing-based surveillance in the sentinel provinces performed moderately well. The utility of the system was demonstrated by its ability to monitor the spatial and temporal trends of the HAV isolates circulating in sentinel provinces and to facilitate the timely identification and management of HAV outbreaks. The gaps and challenges affecting the surveillance performance were overcomable. For example, the quality of surveillance data can be improved by training. With more resources, the timeliness problem can be solved, so public health authorities can identify emerging problems and take early action. These challenges are similar in many developing countries (17-22).
Addressing the data quality problem should be the priority. In contrast to other developing countries where missing or inaccurate data may have affected the results of the reported trends (23), the data generated from the sentinel provinces was of relatively good quality regarding completeness. However, some key variables were missing due to the individual’s concern for privacy. We believe that credit for this improvement should go to using the electronic reporting system that uses smartphones or computers connected to the network service, which facilitates more accurate, convenient, and timely quality control of the data. With the increased popularity of smartphones and other information technologies, it provides a huge potential for scaling up surveillance in developing countries.
However, the main reason for the low yield rate of RNA sequencing is likely linked with the diagnostic quality and physicians’ attitudes toward the surveillance. Currently, the Chinese criterion for HepA case definition does not specify the cutoff value to define the term “elevated aminotransferase levels”. Meanwhile, HAV infection symptoms are often vague and may resemble symptoms of the flu and other common illnesses at the early stage of the infection, allowing some physicians to use their discretion liberally to identify patients. The overuse of detection measures can further fuel the situation mentioned above. Researchers have shown that positive anti-HAV IgM existed among individuals who did not have HAV infection (24-26), and patients who had other medical conditions could be misdiagnosed based on the anti-HAV IgM test. Moreover, convalescent persons could also be reported as new cases since it usually takes 3 to 6 months for IgM to become undetectable (27) and the persistence of IgM for years after the primary infection also existed (28). Since performing sequencing for these patients would be a waste of the limited human and financial resources, more accurate and specific criteria for HepA sequencing should be developed.
Traditional medical education is largely inadequate (29) and training programs are needed about sequencing-based surveillance and its value for individual and population health. Besides, similar to other countries (30), potential reasons for problems in data quality may have included the high volume of inpatients seen by the doctors. Integrating the reporting system with hospitals’ existing electronic health records could be a possible solution. The reporting efficiency will substantially improve if physicians can enter data directly into the electronic health record, allowing the system to extract data that already exists in the record automatically.
As a cornerstone of the laboratory-based surveillance network, the Central CDC laboratory should analyze the sequence data quickly to constructing an accurate picture of the transmission dynamics of pathogens to detecting outbreaks promptly. While the web-based reporting system allowed the timely transmission of data, some delays were experienced in the shipment of samples. Therefore, feedbacks were often delayed. Various sophisticated data-sharing platforms that have been established in developed countries have shown that decentralized surveillance networks will substantially shorten the time of result’ feedback and make the prompt detection of potential outbreaks possible (31). Samples are transported to a local laboratory for testing without long-haul transports. After being analyzed, the sequence data are uploaded to the central laboratory for further analysis (https://www.rivm.nl/en/havnet). This process may introduce several advantages, including reduced transportation cost, a decreased workload for PHPs, and a reduced dependency on skilled technicians at the local labs, as the central lab can perform further analysis, and then deliver the results to the local CDCs. A possible foodborne HAV outbreak occurred during the study. But samples could not be delivered to the central CDC due to the COVID-19 pandemic. Emergency actions were deployed, and the samples were transported to a local laboratory capable of sequencing. The experiment protocol from the central lab was shared with local personnel. Ultimately, the results turn out to be acceptable. This was a good example of utilizing a decentralized network. However, more investment in local laboratories utilities and advanced training programs for personnel is needed, especially for less economically developed countries.
The major limitation of this study is that we only selected five sentinel provinces based on voluntary participation and our findings may not be widely generalizable. However, we are confident that the provinces we selected were represented as the three social-economical areas of China with different HepA morbidity and healthcare resources. This study provides an informative overview of how to implement sequencing-based surveillance, how to operate at a large scale with sustainability, and the challenges that need to be addressed in China.
Conclusions
Overall, the system performed reasonably well and provided reliable data in the China context. Timeliness and data quality are among the main challenges, due to financial and human resource constraints and inadequate support among Chinese physicians regarding public health surveillance. Enhanced sequencing-based surveillance can be feasible with innovation and investments, including developing more suitable criteria for case detection, conducting training and education for physicians, and establishing decentralized surveillance networks.
Supplementary
The article’s supplementary files as
Acknowledgments
We would like to thank all the relevant public health workers who took part in the collection samples from CDCs of five provinces in this study.
Funding: This work was supported by the National Science and Technology Major Project of China (No. 2018ZX10201002), and Shandong Medical Health Science and Technology Development Program (No. 2018WS308).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Research Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University (Number [2019]17). Written or oral consents were obtained from the patients. The procedures performed involving human were in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the StaRI reporting checklist. Available at https://dx.doi.org/10.21037/atm-21-1193
Data Sharing Statement: Available at https://dx.doi.org/10.21037/atm-21-1193
Peer Review File: Available at https://dx.doi.org/10.21037/atm-21-1193
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/atm-21-1193). The authors have no conflicts of interest to declare.
(English Language Editors: D. Fitzgerald and J. Chapnickz)
References
- 1.The Global Preparedness Monitoring Board. A world in disorder: Global Preparedness Monitoring Board annual report 2020. Geneva: World Health Organization; 2020. [cited 2021 Mar 14]. Available online: https://apps.who.int/gpmb/annual_report.html
- 2.Smith KF, Goldberg M, Rosenthal S, et al. Global rise in human infectious disease outbreaks. J R Soc Interface 2014;11:20140950. 10.1098/rsif.2014.0950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. SARS-CoV-2 genomic sequencing for public health goals: Interim guidance, 8 January 2021. 2021. [cited 2021 Mar 14]. Available online: https://www.who.int/publications/i/item/WHO-2019-nCoV-genomic_sequencing-2021.1
- 4.Cyranoski D. SARS triggers biomedical shake-up in China. Nature 2003;425:333. [DOI] [PubMed] [Google Scholar]
- 5.Feng Z, Li W, Varma JK. Gaps remain in China's ability to detect emerging infectious diseases despite advances since the onset of SARS and avian flu. Health Aff (Millwood) 2011;30:127-35. 10.1377/hlthaff.2010.0606 [DOI] [PubMed] [Google Scholar]
- 6.German RR, Lee LM, Horan JM, et al. Updated guidelines for evaluating public health surveillance systems: recommendations from the Guidelines Working Group. MMWR Recomm Rep 2001;50:1-35; quiz CE1-7. [PubMed] [Google Scholar]
- 7.Babakazo P, Kabamba-Tshilobo J, Wemakoy EO, et al. Evaluation of the influenza sentinel surveillance system in the Democratic Republic of Congo, 2012-2015. BMC Public Health 2019;19:1652. 10.1186/s12889-019-8008-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rakotoarisoa A, Randrianasolo L, Tempia S, et al. Evaluation of the influenza sentinel surveillance system in Madagascar, 2009-2014. Bull World Health Organ 2017;95:375-81. 10.2471/BLT.16.171280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlieg WL, Fanoy EB, van Asten L, et al. Comparing national infectious disease surveillance systems: China and the Netherlands. BMC Public Health 2017;17:415. 10.1186/s12889-017-4319-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun XJ, Zhang GM, Zhou RJ, et al. Changes in the epidemiology of hepatitis A in three socio-economic regions of China, 1990-2017. Infect Dis Poverty 2019;8:80. 10.1186/s40249-019-0591-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu B, Liu J, Fu Y, et al. Spatio-Temporal Epidemiology of Viral Hepatitis in China (2003-2015): Implications for Prevention and Control Policies. Int J Environ Res Public Health 2018;15:661. 10.3390/ijerph15040661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li C, Sun M, Shen JJ, et al. Evaluation on the efficiencies of county-level Centers for Disease Control and Prevention in China: results from a national survey. Trop Med Int Health 2016;21:1106-14. 10.1111/tmi.12753 [DOI] [PubMed] [Google Scholar]
- 13.Ren X, Wu P, Wang L, et al. Changing Epidemiology of Hepatitis A and Hepatitis E Viruses in China, 1990-2014. Emerg Infect Dis 2017;23:276-9. 10.3201/eid2302.161095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hutin YJ, Pool V, Cramer EH, et al. A multistate, foodborne outbreak of hepatitis A. National Hepatitis A Investigation Team. N Engl J Med 1999;340:595-602. 10.1056/NEJM199902253400802 [DOI] [PubMed] [Google Scholar]
- 15.Houde A, Guevremont E, Poitras E, et al. Comparative evaluation of new TaqMan real-time assays for the detection of hepatitis A virus. J Virol Methods 2007;140:80-9. 10.1016/j.jviromet.2006.11.003 [DOI] [PubMed] [Google Scholar]
- 16.Budgell E, Cohen AL, McAnerney J, et al. Evaluation of two influenza surveillance systems in South Africa. PLoS One 2015;10:e0120226. 10.1371/journal.pone.0120226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibrahim NK. Epidemiologic surveillance for controlling Covid-19 pandemic: types, challenges and implications. J Infect Public Health 2020;13:1630-8. 10.1016/j.jiph.2020.07.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bawa SB, Olumide EA, Umar US. The knowledge, attitude and practices of the reporting of notifiable diseases among health workers in Yobe State, Nigeria. Afr J Med Med Sci 2003;32:49-53. [PubMed] [Google Scholar]
- 19.Benson FG, Levin J, Rispel LC. Health care providers' compliance with the notifiable diseases surveillance system in South Africa. PLoS One 2018;13:e0195194. 10.1371/journal.pone.0195194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ngwa MC, Liang S, Mbam LM, et al. Cholera public health surveillance in the Republic of Cameroon-opportunities and challenges. Pan Afr Med J 2016;24:222. 10.11604/pamj.2016.24.222.8045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler D. Flu surveillance lacking. Nature 2012;483:520-2. 10.1038/483520a [DOI] [PubMed] [Google Scholar]
- 22.Hoen AG, Keller M, Verma AD, et al. Electronic event-based surveillance for monitoring dengue, Latin America. Emerg Infect Dis 2012;18:1147-50. 10.3201/eid1808.120055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maponga BA, Chirundu D, Shambira G, et al. Evaluation of the Notifiable diseases surveillance system in Sanyati district, Zimbabwe, 2010-2011. Pan Afr Med J 2014;19:278. 10.11604/pamj.2014.19.278.5202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Centers for Disease C , Prevention. Positive test results for acute hepatitis A virus infection among persons with no recent history of acute hepatitis--United States, 2002-2004. MMWR Morb Mortal Wkly Rep 2005;54:453-6. [PubMed] [Google Scholar]
- 25.Castrodale L, Fiore A, Schmidt T. Detection of immunoglobulin M antibody to hepatitis A virus in Alaska residents without other evidence of hepatitis. Clin Infect Dis 2005;41:e86-8. 10.1086/497073 [DOI] [PubMed] [Google Scholar]
- 26.Huang W, Xue Y, Yang P, et al. Effects of Electrochemiluminescence Immunoassay and Enzyme - linked Immunosorbent Assay for Detection of Hepatitis A Virus IgM Antibody: a Comparative Study. Modern Hospital 2016;16:383-5. [Google Scholar]
- 27.Kao HW, Ashcavai M, Redeker AG. The persistence of hepatitis A IgM antibody after acute clinical hepatitis A. Hepatology 1984;4:933-6. 10.1002/hep.1840040525 [DOI] [PubMed] [Google Scholar]
- 28.Qiu F, Cao J, Su Q, et al. Multiplex hydrolysis probe real-time PCR for simultaneous detection of hepatitis A virus and hepatitis E virus. Int J Mol Sci 2014;15:9780-8. 10.3390/ijms15069780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li LM, Tang JL, Lv J, et al. The need for integration in health sciences sets the future direction for public health education. Public Health 2011;125:20-4. 10.1016/j.puhe.2010.10.008 [DOI] [PubMed] [Google Scholar]
- 30.Joseph Wu TS, Kagoli M, Kaasboll JJ, et al. Integrated Disease Surveillance and Response (IDSR) in Malawi: Implementation gaps and challenges for timely alert. PLoS One 2018;13:e0200858. 10.1371/journal.pone.0200858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu X, Collier MG, Xu F, Hepatitis A. Outbreaks in Developed Countries: Detection, Control, and Prevention. Foodborne Pathog Dis 2020;17:166-71. 10.1089/fpd.2019.2648 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as