The development of high-throughput sequencing technologies has provided the opportunity to platform genome-wide surveys. Recent applications of next generation sequencing (NGS) have revealed that the hepatitis B virus (HBV) genome is randomly integrated throughout the human genome in patients with HBV infection. Besides the induction of chronic infection of the liver, HBV integration of the host genome contributes to hepatocarcinogenesis, especially in young patients without cirrhosis (1,2). In addition to the oncogenic potency of inserted HBV itself due to mutational changes such as truncated HBx protein, viral DNA integration induces chromosomal instability and aberrant expression of driver genes (3). The burden of HBV infection is currently the highest in the Southeast Asia and sub-Saharan Africa; therefore, most HBV integrations in hepatocellular carcinoma (HCC) have been reported in Asian populations where HBV genotypes B and C are prevalent.
Péneau et al. (4) have elucidated the direct oncogenic consequences of HBV integrations in the host genome using NGS from HCC and non-cancer adjacent liver tissues obtained from HBV-positive patients in a French cohort with ancestry mainly from Europe and Africa. In 84% of the 170 adjacent non-cancer liver tissues, 6,610 HBV integration breakpoints in the HBV/human junction points in the genome were identified, most of which were unique (82%). Recurrent clonal (1%) and subclonal (17%) insertions were identified in very large genes (>100 kb) including FN1 (14 cases), CPS1 (4 cases), KCNT2 (3 cases), and ADH1B (3 cases). As described previously (5,6), FN1 was the most frequent target gene of HBV in the liver, and there were no hotspots of HBV insertions in this gene. There were two major types of fusion transcripts identified, including the out-of-frame HBx-FN1 and in-frame HBs-FN1 fusions, which did not have a functional impact.
Localization of HBV integration breakpoints was different between the HCC and adjacent liver tissues, and the number of HBV integrations was significantly associated with the HBV copy number per cell. In 88% of the 177 HCC samples, 31% and 9% of 2,199 break points were clonal and subclonal events, respectively, suggesting strong functional selection of cells harboring such events and that the hepatocytes with HBV integrations in driver genes clonally expanded during cancer progression. Only four known driver genes for recurrent HBV integration have been reported to date (1,5-11); TERT, KMT2B, CCNE1, and CCNA2. Indeed, TERT (48 cases), CCNE1 (4 cases), and KMT2B (3 cases) were recurrently targeted for HBV integration (Table 1). Other genes such as AHRR (2 cases), NRG1 (2 cases), TRIM16L (2 cases), and ST18 (2 cases) were also found to be recurrently integrated, which have not been previously reported as HBV integration targets. Except for TERT, KMT2B, CCNE1, and CCNA2, other targets of HBV integration vary among reports (1,6,7,10,11), and most of these have never been reported as cancer-driver genes. Therefore, most HBV integration events are considered to be passenger events. For example, our previous report (6) showed that several passenger genes harbor repetitive sequences, such as the poly (G) site in LINCO00486, the GGGTTA repeat in DEBT, and the TTGGGG repeat in DDX11L family members. These repetitive sequences exist in other sites of the host genome, suggesting that these genes might be counted as HBV-integrated genes by a capture sequencing approach.
Table 1. Recurrent HBV target genes.
| Sample No | Sequencing | Cancer related genes | Cancer non-related genes | Reference |
|---|---|---|---|---|
| 40 | MAPS | TERT [6], CCNA2 [1], KMT2B [1] | SMAD5 [2] | (7) |
| 81 | WGS | TERT [18], KMT2B [9], CCNE1 [4] | SEMP5 [3], ROCK1 [2] | (1) |
| 28 | Capture | TERT [7], KMT2B [5], CCNE1 [2] | (8) | |
| 79 | WES | TERT [17], KMT2B [6], CCNE1 [1], CCNA2 [1] | ALOX5 [2], ZFPM [2], SENP [2], MYO19 [2], RGS [2] | (10) |
| 22 | WGS | TERT [2], KMT2B [5] | (9) | |
| 426 | Capture | TERT [101], KMT2B [15], CCNE1 [7], CCNA2 [8] | ANKRD30BL [7], FAM157A [7], DDX11L1 [23] DDX11L5 [8], DDX11L9 [8] | (11) |
| 42 | Capture | TERT [8], KMT2B [6], CCNA2 [1] | (5) | |
| 154* | Capture | TERT [19], KMT2B [7], CCNE1 [1], CCNA2 [1] | DDX11L1 [2], DDX11L2 [2], DDX11L16 [2] CWH43 [2], DEBT [2], EYA1 [2], MACROD2 [2] PTPRD [2], ZNF595 [2] | (6) |
| 177 | Capture | TERT [48], KMT2B [3], CCNE1 [4], CCNA2 [1] | AHRR [2], NRG1 [2], TRIM161 [2], ST18 [2] | (4) |
*, including both patients with chronic and prior HBV infection. Data sets in which ≥20 cases were analyzed are chronologically listed. The numbers in square brackets correspond to the patient numbers. HBV, hepatitis B virus; MAPS, massive anchored parallel sequencing; WGS, whole-genome sequencing; WES, whole-exome sequencing.
Whole-exome or whole-genome sequencings showed that recurrent HBV integration into cancer-related genes including TERT, KMT2B, and CCNE1 in the liver cancer tissues and mutations of known driver genes such as TP53, CTNNB1, and the TERT promoter region in HCC were exclusive (5). Therefore, HBV integration into these genes followed by increased gene expression is considered to be one of the driver events in hepatocarcinogenesis, especially in patients without driver gene mutations.
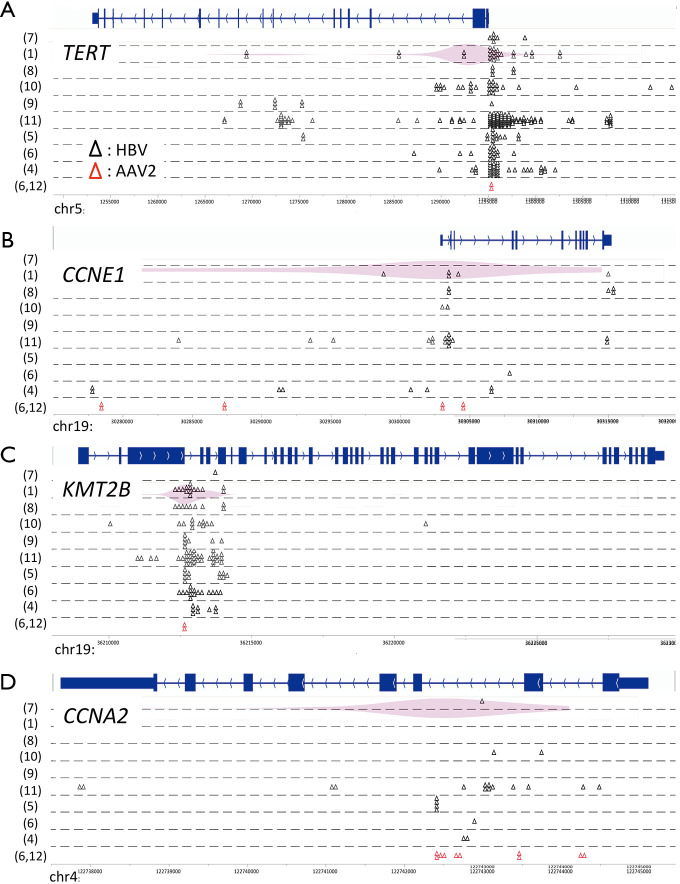
Potential preferential sites of virus integration have also been identified using NGS (Figure 1). HBV integration sites in the human genome were concentrated in the promoter region of TERT and CCNE1, intron 3 of KMT2B, and intron 2 of CCNA2 (6). Of note, these recurrent integration sites of HBV were similar to those of adeno-associated virus 2 in HCC (6,12), and are not reported as fragile sites in the human genome (13). Therefore, preferential virus integration regions may exist in driver genes, but not in passenger genes. Intriguingly, HBV insertion sites in the promoter region of TERT were exclusive to mutations or structural variations in the same region, and they induced higher gene expression than promoter mutations at the –124 and –146 hotspots of TERT and copy number aberrations near 5p (6). A luciferase reporter gene assay showed that such strong TERT activation induced by HBV insertion was conferred by one of two viral enhancer regions.
Figure 1.
Virus integration sites in the human genome. Virus integration sites are indicated by triangles (black, HBV; red, adeno-associated virus 2). The frequency of virus integration is indicated by a violin plot. The numbers in parentheses correspond to the reference numbers.
Viral dynamics and selection processes were also shown to differ between the HCC and adjacent liver tissues. Consistent with previous data (1,6,11), the C-terminal regions of HBx with structural variants such as deletions, duplications, and inversions were found to be the most frequently integrated in the host genome in the HCC samples. In addition, the localization and orientation of HBV/human junctions in the host genome showed frequent chromosomal rearrangements. The hotspot of HBV integration in the adjacent non-cancer liver samples was also the C-terminal region of HBx, but without structural variants, and RNA sequencing showed high expression of the HBs gene (in-frame) whereas the HBx gene (out-of-frame) did not show such high expression for the frequency of integration.
Integrated analysis of the capture sequence and whole-genome or whole-exome sequences suggested that viral insertions could drive liver cancer via another mechanism. We and others previously reported that non-genic HBV integrations were recurrently observed near the centromere, especially on chromosomes 1p, 8p, and 10q, which are associated with genome instability (1,10). In this study, 41% of clonal HBV integrations identified in tumors were involved in large rearrangements of the human genome, 48% of which were associated with copy number alterations. In addition to altering the expression of the closest gene along with inducing copy number alteration, HBV integration can cause aberrations in cancer-driver genes located at distance through integration-associated chromosomal structural rearrangements. Long-read sequencing showed that centromeric insertions of HBV are related to large deletions of chromosome 17p, including TP53 (translocation-like event), focal gain of chromosome 5p including TERT, and a large gain of chromosome 8q, including MYC (duplication-inverted-like event). This is the first report on the association between HBV integration and chromosome rearrangements; however, most HCC patients harbor loss of heterozygosity of 17p, more than 10% of patients have focal gain of 5p, and amplification of MYC is frequently observed (14). Given that the frequency of copy number alterations does not appear to different among the viral subtypes (10), further detailed study is needed to determine whether HBV integrations near the centromere actually cause such chromosomal rearrangements and functionally alter cancer driver genes at distance.
Clinically, the survival of patients with a higher number of HBV integrations in tumors was reported to be shorter than that of patients with a lower number of viral integrations despite their younger age (1). Moreover, other clinical parameters such as tumor size, serum alpha-fetoprotein level, and grade of adjacent liver fibrosis were not associated with the number of HBV integrations in this study, in contrast to a previous report using whole-genome sequencing in an Asian population (1). The negative effects of a higher number of HBV integrations in liver cancer could be attributed to the induction of chromosomal structural rearrangements and activation of oncogenes. HBV integrations could be considered one of the major mechanisms of HCC development considering the different clinicopathological features in younger and older patients, especially in the non-cirrhotic livers of younger patients.
Through the hundreds of cases analyzed by capture sequencing with or without whole-exome or whole-genome sequencings, the following common characteristics are clarified regardless of patient ethnicity and the HBV genotype; recurrent target cancer-related genes with their preferential integration regions, recurrent regions of the inserted HBV genome, genome instability associated with HBV integrations, and poor survival of patients with viral integrations. In the future, the detection of HBV integrations in addition to clinical sequencing may be available to predict the patient survival or HBV reactivation. However, elucidating the mechanism and effect of HBV integrations in non-cancerous tissues on fibrosis or carcinogenesis remains a future challenge in this field.
Acknowledgments
Funding: This work was mainly supported by a Grant-in-Aid for Scientific Research (C) 21K08807 (Y.M.) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan. The funding body has no role in the design of the study and analysis and interpretation of data and in writing the manuscript.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
Footnotes
Provenance and Peer Review: This article was commissioned by the editorial office of Hepatobiliary Surgery and Nutrition. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://dx.doi.org/10.21037/hbsn-21-228). The authors have no conflicts of interest to declare.
References
- 1.Sung WK, Zheng H, Li S, et al. Genome-wide survey of recurrent HBV integration in hepatocellular carcinoma. Nat Genet 2012;44:765-9. 10.1038/ng.2295 [DOI] [PubMed] [Google Scholar]
- 2.Toyoda H, Kumada T, Kaneoka Y, et al. Impact of hepatitis B virus (HBV) X gene integration in liver tissue on hepatocellular carcinoma development in serologically HBV-negative chronic hepatitis C patients. J Hepatol 2008;48:43-50. 10.1016/j.jhep.2007.08.016 [DOI] [PubMed] [Google Scholar]
- 3.Tu T, Budzinska MA, Shackel NA, et al. HBV DNA Integration: Molecular Mechanisms and Clinical Implications. Viruses 2017;9:75. 10.3390/v9040075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Péneau C, Imbeaud S, La Bella T, et al. Hepatitis B virus integrations promote local and distant oncogenic driver alterations in hepatocellular carcinoma. Gut 2021. [Epub ahead of print]. doi: . 10.1136/gutjnl-2020-323153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furuta M, Tanaka H, Shiraishi Y, et al. Characterization of HBV integration patterns and timing in liver cancer and HBV-infected livers. Oncotarget 2018;9:25075-88. Erratum in: Oncotarget 2018;9:31789. 10.18632/oncotarget.25308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatsuno K, Midorikawa Y, Takayama T, et al. Impact of AAV2 and Hepatitis B Virus Integration Into Genome on Development of Hepatocellular Carcinoma in Patients with Prior Hepatitis B Virus Infection. Clin Cancer Res 2019;25:6217-27. 10.1158/1078-0432.CCR-18-4041 [DOI] [PubMed] [Google Scholar]
- 7.Ding D, Lou X, Hua D, et al. Recurrent targeted genes of hepatitis B virus in the liver cancer genomes identified by a next-generation sequencing-based approach. PLoS Genet 2012;8:e1003065. 10.1371/journal.pgen.1003065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Zeng X, Lee NP, et al. HIVID: an efficient method to detect HBV integration using low coverage sequencing. Genomics 2013;102:338-44. 10.1016/j.ygeno.2013.07.002 [DOI] [PubMed] [Google Scholar]
- 9.Shiraishi Y, Fujimoto A, Furuta M, et al. Integrated analysis of whole genome and transcriptome sequencing reveals diverse transcriptomic aberrations driven by somatic genomic changes in liver cancers. PLoS One 2014;9:e114263. 10.1371/journal.pone.0114263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Totoki Y, Tatsuno K, Covington KR, et al. Trans-ancestry mutational landscape of hepatocellular carcinoma genomes. Nat Genet 2014;46:1267-73. 10.1038/ng.3126 [DOI] [PubMed] [Google Scholar]
- 11.Zhao LH, Liu X, Yan HX, et al. Genomic and oncogenic preference of HBV integration in hepatocellular carcinoma. Nat Commun. Nat Commun 2016;7:12992. Erratum in: Nat Commun 2016;7:13591. 10.1038/ncomms12992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nault JC, Datta S, Imbeaud S, et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat Genet 2015;47:1187-93. 10.1038/ng.3389 [DOI] [PubMed] [Google Scholar]
- 13.Fungtammasan A, Walsh E, Chiaromonte F, et al. A genome-wide analysis of common fragile sites: what features determine chromosomal instability in the human genome? Genome Res 2012;22:993-1005. Erratum in: Genome Res 2016;26:1451. 10.1101/gr.134395.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Midorikawa Y, Yamamoto S, Tatsuno K, et al. Accumulation of Molecular Aberrations Distinctive to Hepatocellular Carcinoma Progression. Cancer Res 2020;80:3810-9. 10.1158/0008-5472.CAN-20-0225 [DOI] [PubMed] [Google Scholar]