Abstract
Kidney renal clear cell carcinoma (KIRC) is one of the most malignant diseases with poor survival rate over the world. The tumor microenvironment (TME) is highly related to the oncogenesis, development, and prognosis of KIRC. Thus, making the identification of KIRC biomarkers and immune infiltrates critically important. Microtubule Interacting and Trafficking Domain containing 1(MITD1) was reported to participate in cytokinesis of cell division. In the present study, multiple bioinformatics tools and databases were applied to investigate the expression level and clinical value of MITD1 in KIRC. We found that the expression of MITD1 was significantly increased in KIRC tissues. Further, the KIRC patients with high MITD1 levels showed a worse overall survival (OS) rate and disease free survival (DFS) rate. Otherwise, we found a significant correlation MITD1 expression and the abundance of CD8+ T cells. Functional enrichment analyses revealed that immune response and cytokine-cytokine receptor are very critical signaling pathways which associated with MITD1 in KIRC. In conclusion, our findings indicated that MITD1 may be a potential biomarker and associated with immune infiltration in KIRC.
Keywords: MITD1, KIRC, TCGA, bioinformatics, CD8+ T cells
Introduction
As one of the most common malignant cancers, renal cell carcinoma has been increasing during the last 2 decades, which accounts for 2%-3% of all new cancer patients. 1 Among kidney cancer, kidney renal clear cell carcinoma (KIRC) is the most dominant subtype, accounting for approximately 75% of all renal cell carcinoma patients 2,3 and the majority of cancer-associated deaths. 4 It was reported that the 5-year disease specific survival rate for kidney cancer less than 10% at stage IV. 5 Moreover, KIRC patients remain a worse prognosis compared to other kidney cancer subtypes patients. Nowadays, more and more therapies, including surgical operation, chemotherapy, radiotherapy and targeted therapies, has been used in the treatment of renal cancer, showing to be effective in the survival of patients. 6,7 However, these treatments methods are still far from enough to cure the KIRC patients. Thus, it is urgent to find new and effective biomarkers for the prognosis and therapy of KIRC. For example, by comprehensive analysis of RCC clinical datasets, RGS5, TOP2A andMYC showed to play a significant role in RCC. 8,9 What’s more, studies have shown that immune response is closely associated with clinical outcome in kidney cancer. 10,11 Tumor-infiltrating immune cells (TIIC) play an important role in the TME to regulate tumor development and are appealing therapeutic targets. 10 In multiple clinical and genomic investigations, KIRC has been shown to be significantly associated with immune infiltration. 12,13
MITD1 is a protein coding gene, which located in 2q11.2. And through the ESCRT-III, MITD1 is recruited to midbodies and then participate in cytokinesis abscission. 14 ESCRT-III was shown to be disorder in several cancers, such as ovarian cancer, 15 prostate cancer 16 and pancreatic tumors. 17 Moreover, cytokinesis disorders of abscission could enhance oncogene-induced mitotic stress to promote genomic instability and tumorigenesis. 18 This may indicate that there is a certain connection between MITD1 and cytokinesis in the process of tumorigenesis. Otherwise, MITD1 has been studied to be correlated with immune infiltrates of liver cancer. 19
In this study, we analyzed the expression of MITD1 in KIRC by Onconmine, TIMER and the Human Protein Atlas. Then, we performed the relationship between MITD1 and clinical information by the TCGA KIRC data. Furthermore, to further investigate the biological processes and pathways associated with the regulatory network of MITD1 that could promote KIRC occurrence and development. The Gene Set Enrichment Analysis (GSEA) was performed to elevate Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. Finally, the data on immune system infiltrates of KIRC from TCGA database was analyzed through the ImmuneCellAI and TIMER. The identification of prognostic MITD1 suggests the potential role of MITD1 in KIRC pathogenesis and progression.
Materials and Methods
Data Acquisition
The RNA-seq data of KIRC and normal tissue were obtained from the The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). Four kidney cancer datasets were obtained from Oncomine (http://www.oncomine.org) database, 20 a web-based data mining platform with RNA-seq and microarray data of human tumors. Some samples with incomplete clinical information were deleted when KIRC samples were analyzed by comprehensive bioinformatics. The RNA-seqdata and clinical information of 534 patients were downloaded from TCGA database. Subsequently, by chi-square test, the statistical difference of these characteristics with MITD1 expression was analyzed.
TIMER Database Analysis
TIMER (http://cistrome.dfci.harvard.edu/TIMER/) is a comprehensive database for systematic analysis of immune infiltrates with different kinds of cancer. 21 It used a previously published statistical method to analyze RNA-seq expression profile data to detect the infiltration of immune cells in tumor tissues. 22 The current release of TIMER incorporated 10,009 samples across 23 cancer types from TCGA dataset to estimate the abundance of immune infiltrates.
Assessment of Prognosis Clinical Characteristics and MITD1 in KIRC Cohort
Based on the expression level of MITD1, we divided the 534 KIRC samples into 2 groups: high MITD1 expression group and low MITD1 expression group. Then we performed Kaplan-Meier analysis on 534 KIRC patients through SPSS software (version 24.0, Chicago, IL, United States). A univariate Cox proportional hazard regression analysis was used to evaluate the association between overall survival (OS) time and clinical characteristics in the trial cohort. P-value < 0.05 was considered to have statistically significant prognostic value.
The Human Protein Atlas
The Human Protein Atlas database (HPA) (https://www.proteinatlas.org) was used to analyze protein expression of MITD1 between normal and KIRC tissues. HPA portal is an open database with millions of high-resolution images showing the spatial distribution of proteins in 44 different normal human tissues and 20 different cancer types. 23,24
Gene Set Enrichment Analysis
Gene Set Enrichment Analysis (GSEA) is a computational method used to determine whether a set of priori defined genes show statistically significant, concordant differences between 2 biological states. 25 GSEA preliminarily classified the genes according to their correlation with the MITD1 expression. This computational method illustrates the significant difference in survival observed between the high and low MITD1 groups. For each analysis, we ran 1000 repetitions of gene set permutations. In addition, nominal P values and standardized enrichment scores (NES) were used to classify enrichment pathways in each phenotype. 26
Correlation Analysis Between MITD1 and Immune Infiltration in KIRC
The composition and abundance of immune cells in the tumor microenvironment have a great influence on tumor progression and the effect of immunotherapy. GraphPad Prism software (version 8.4.2, https://www.graphpad.com/updates/) was used to analyze the correlation between MITD1 expression and TIICs, the cutoff was P-value < 0.01. The Immune Cell Abundance Identifier (ImmuCellAI) detected the abundance of immune cells, and the differences in the infiltration immune cells of 534 KIRC were analyzed. ImmuneCellAI is a tool for estimating the abundance of 24 immune cells from gene expression datasets, including RNA-Seq and microarray data, of which 24 immune cells are composed of 6 immune cell types and 18 T-cell subtypes (http://bioinfo.life.hust.edu.cn/web/ImmuCellAI/). 27
Statistical Analysis
All statistical analyses were enforced with corresponding packages of R (Version 3.6.3). Kaplan-Meier survival analysis and chi square test was conducted using SPSS (version 24.0). To calculate the 95% CI and HR, univariate Cox regression analyses were used to evaluate MITD1 expression between clinical factors and OS. For all analysis, the threshold P-value < 0.05 was considered significant statistically.
Results
MITD1 Was Upregulated in KIRC
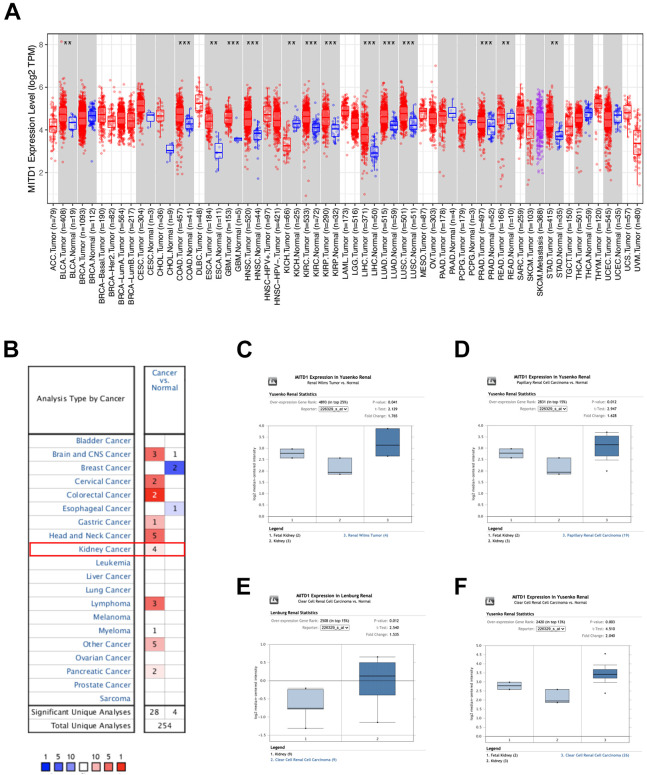
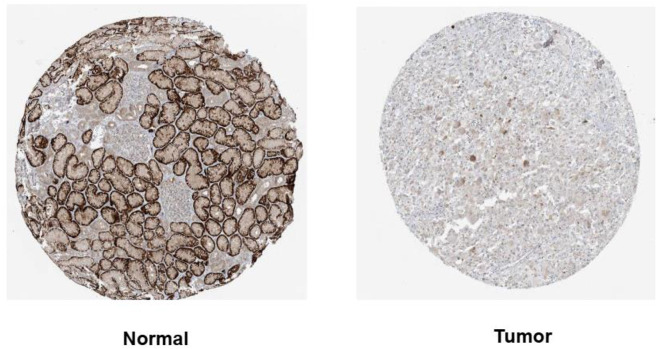
The mRNA-seq data from TCGA datasets were analyzed by the TIMER tools, which showed the expression levels of MITD1 in different kinds of tumors. MITD1 with a higher expression level existed in most cancers, including BLCA, ESCA, COAD, GBM, HNSC, KIRP, LIHC, LUAD, LUSC, PRAD, STAD, READ as well as KIRC (Figure 1A). Then, the expression level of MITD1 in tumors and normal tissues of kidney were analyzed by Oncomine database. As shown in Figure 1B, higher expression level of MITD1 was presented in kidney cancers which included 4 studies. Four studies on MITD1 mRNA levels in Renal cancer vs normal kidney tissue was shown in Figure 1C to F. Further, immunohistochemistry (IHC) results available from the HPA database showed that MITD1 has a higher expression in KIRC tumor tissues compared to nontumor tissues (Figure 2).
Figure 1.
MITD1 mRNA expression levels in normal and cancer tissues. A, Human MITD1 expression levels in different tumor types from TCGA database were determined by TIMER database (*P < 0.05, **P < 0.01, ***P < 0.001). B, MITD1 in datasets of different cancers compared with normal tissues in the oncomine database. Red color indicates upregulated expression in cancer, and blue color indicates downregulated expression in cancer. C-F, Meta-analysis of the 4 datasets from 4 studies on MITD1 mRNA levels in renal cancer vs normal kidney tissue searched by oncomine database.
Figure 2.
MITD1 protein expression levels in normal and KIRC tissues. The expression of MITD1 protein was visualized using immunohistochemistry via the human protein atlas database.
Connection Between the Expression of MITD1 and Clinicopathology
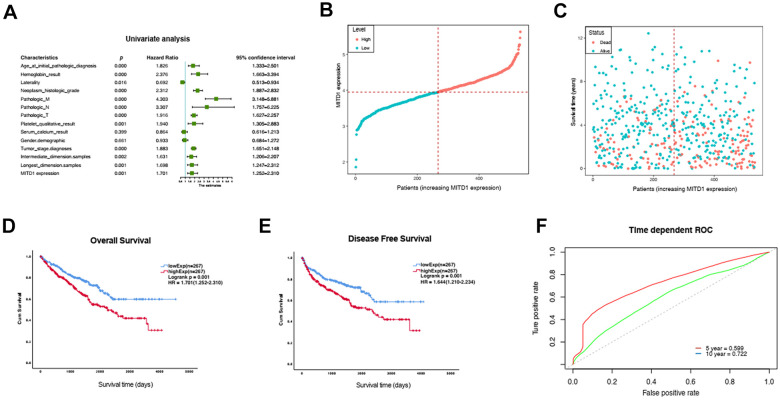
Then, univariate analyses were used to evaluate whether the MITD1 expression level and various clinic pathological characteristics were independent prognostic parameters of KIRC patient overall survival. We found that high expression of the MITD1 gene, age, hemoglobin, laterality, neoplasm, pathologic M, pathologic N, pathologic T, platelet, tumor stage, intermediate dimension and longest dimension were independent prognostic factors in patients with KIRC (Figure 3A). Next, the potentially prognostic significance of MITD1 expression levels was investigated by TCGA KIRC data. The patients’ clinical classification information is shown in Table 1, as a result, high MITD1 expression found to be closely correlated with pathologic T stage (P = 0), platelet qualitative (P = 0.009), live status (P = 0) and tumor stage (P = 0.001). These suggested that MITD1 could be an independent prognostic indicator for KIRC. The distribution of MITD1 expression and survival status of KIRC patients were shown in Figure 3B and C. Kaplan-Meier survival analysis of the KIRC dataset showed that MITD1 expression was significantly with OS (P = 0.001) and DFS (P = 0.001). Low MITD1 expression was associated with better prognosis, and high expression of MITD1 was associated with poor prognosis in KIRC patients (Figure 3D and E). As the time dependent ROC curve showed that the 5-year and 10-year AUC of MITD1 expression were 0.599 and 0.722 for predicting survival, respectively (Figure 3F), which indicated that MITD1 has great prognostic value.
Figure 3.
Univariate Cox analysis and survival curve of differential M1TD1 expression. A, Univariate Cox analysis of MITD1 expression and other clinicopathological variables. B-C, MITD1 expression distribution and survival status. D, Overall survival curve of differential MITD1 expression. E, Disease free survival curve of differential MITD1expression. F, ROC curves of MITD1 of 5 year and 10 year.
Table 1.
MITD1 Expression Level and Clinical Pathological Factors in 534 KIRC Patient.a
| Characteristics | Case (No.) | No. of patients (%) | Expression level | P value | |
|---|---|---|---|---|---|
| Low (No. cases) | High (No. cases) | ||||
| Age | 534 | 0.193 | |||
| <60 | 247 (46.3) | 131 | 116 | ||
| ≥60 | 287 (53.7) | 136 | 151 | ||
| Hemoglobin_result | 454 | 0.191 | |||
| Normal | 187 (41.2) | 102 | 85 | ||
| Non-normal | 267 (58.8) | 129 | 138 | ||
| Laterality | 533 | 0.83 | |||
| Left | 250 (46.9) | 124 | 126 | ||
| Right | 283 (53.1) | 143 | 140 | ||
| Neoplasm_histologic_grade | 526 | 0.387 | |||
| G1 | 14 (2.7) | 7 | 7 | ||
| G2 | 231 (43.9) | 122 | 109 | ||
| G3 | 206 (39.2) | 101 | 105 | ||
| G4 | 75 (14.3) | 31 | 44 | ||
| Pathologic_M | 502 | 0.081 | |||
| M0 | 424 (84.5) | 225 | 199 | ||
| M1 | 78 (15.5) | 33 | 45 | ||
| Pathologic_N | 256 | 0.771 | |||
| N0 | 240 (93.8) | 114 | 126 | ||
| N1 | 16 (6.3) | 7 | 9 | ||
| Pathologic_T | 534 | 0 | |||
| T1 | 274 (51.3) | 162 | 112 | ||
| T2 | 70 (13.1) | 25 | 45 | ||
| T3 | 179 (33.5) | 75 | 104 | ||
| T4 | 11 (2.1) | 5 | 6 | ||
| Platelet_qualitative_result | 444 | 0.009 | |||
| Low | 47 (10.6) | 20 | 27 | ||
| Normal | 360 (81.1) | 195 | 165 | ||
| Elevated | 37 (8.3) | 11 | 26 | ||
| Serum_calcium_result | 364 | 0.957 | |||
| Normal | 151 (41.5) | 74 | 77 | ||
| Non-normal | 213 (58.5) | 105 | 108 | ||
| Gender.demographic | 534 | 0.586 | |||
| female | 186 (34.8) | 96 | 90 | ||
| male | 348 (65.2) | 171 | 177 | ||
| status | 534 | 0 | |||
| Alive | 361 (67.6) | 200 | 161 | ||
| Dead | 173 (32.4) | 67 | 106 | ||
| Tumor_stage | 531 | 0.001 | |||
| Stage I | 268 (50.5) | 158 | 110 | ||
| Stage II | 58 (10.9) | 23 | 35 | ||
| Stage III | 123 (23.2) | 50 | 73 | ||
| Stage IV | 82 (15.4) | 35 | 47 | ||
| Intermediate_dimension | 495 | 0.4 | |||
| Low | 290 (58.6) | 154 | 136 | ||
| High | 205 (41.4) | 101 | 104 | ||
| Tumor dimension | 495 | 0.059 | |||
| Low | 267 (53.9) | 148 | 119 | ||
| High | 228 (46.1) | 107 | 121 | ||
aThe boldface values indicate significance with P value < 0.05.
Biological Function Analysis of M1TD1
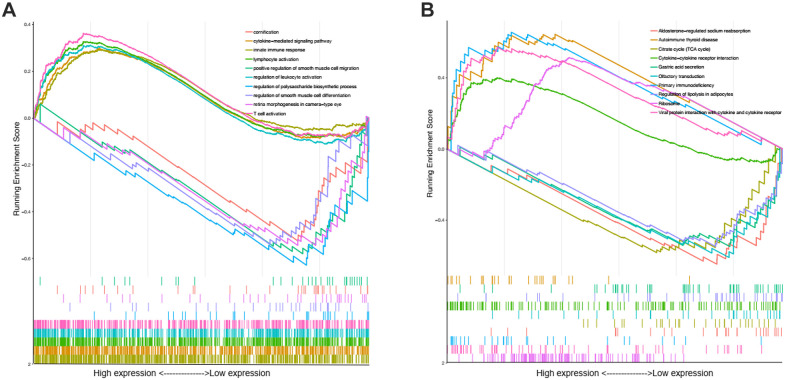
In order to investigate the potential biological functions of MITD1 in KIRC, the GO term and KEGG pathway enrichment analyses were used to address our needs. We used the GSEA to reveal the significantly enriched (FDR < 0.05, P-value < 0.05) GO terms and KEGG pathways with high MITD1 expression. The top 5 GO terms positively correlated with high expression of MITD1were innate immune response, cytokine-mediated signaling pathway, lymphocyte activation, regulation of leukocyte activation and T cell activation. The top 5 5 negatively correlated categories: regulation of polysaccharide biosynthetic process, regulation of smooth muscle cell differentiation, retina morphogenesis in camera-type eye, cornification and positive regulation of smooth muscle cell migration. As shown in Table 2 and Figure 4A, the biological processes (BPs) strongly associated with high expression of MITD1 were immune response and cell life. And the most significantly positively changed 5 pathways were Ribosome, Viral protein interaction with cytokine and cytokine receptor, Primary immunodeficiency, Cytokine-cytokine receptor interaction and Autoimmune thyroid disease. Also, the top 5 negative correlated pathways were Aldosterone-regulated sodium reabsorption Citrate cycle (TCA cycle), Olfactory transduction, Regulation of lipolysis in adipocytes, Gastric acid secretion, as shown in Table 3 and Figure 4B. These results indicated that cytokine receptor and citrate cycle are very critical pathways which associated with MITD1 in KIRC.
Table 2.
Top 5 Significantly Correlated GO Term With MITD1 Expression Based on P-Value.
| Regulation | GO term | NES | P-value |
|---|---|---|---|
| Positive | Innate immune response | 1.416384989 | 0.001124859 |
| Cytokine-mediated signaling pathway | 1.435599139 | 0.001128668 | |
| Lymphocyte activation | 1.562070926 | 0.001148106 | |
| Regulation of leukocyte activation | 1.465387692 | 0.001157407 | |
| T cell activation | 1.695728707 | 0.001187648 | |
| Negative | Regulation of polysaccharide biosynthetic process | −2.17911054 | 0.002304147 |
| Regulation of smooth muscle cell differentiation | −1.87121347 | 0.002304147 | |
| Retina morphogenesis in camera-type eye | −1.88561074 | 0.002304147 | |
| Cornification | −1.82220123 | 0.002304147 | |
| Positive regulation of smooth muscle cell migration | −2.01859177 | 0.002336449 |
Figure 4.
Enrichment plots from gene set enrichment analysis. A, GSEA results showing differential enrichment of genes in GO biological process term with high MITD1 expression. B, GSEA results showing differential enrichment of genes in KEGG pathway with high MITD1 expression.
Table 3.
Top 5 Significantly Correlated KEGG Pathway With MITD1 Expression Based on P-Value.
| Regulation | KEGG pathway | NES | P-value |
|---|---|---|---|
| Positive | Ribosome | 2.111089495 | 0.000140095 |
| Viral protein interaction with cytokine and cytokine receptor | 2.123368383 | 0.000152975 | |
| Primary immunodeficiency | 2.055747293 | 0.000166667 | |
| Cytokine-cytokine receptor interaction | 1.704887989 | 0.000268132 | |
| Autoimmune thyroid disease | 2.040880331 | 0.000332557 | |
| Negative | Aldosterone-regulated sodium reabsorption | −2.17485499 | 0.000249377 |
| Citrate cycle (TCA cycle) | −1.99797266 | 0.000249875 | |
| Olfactory transduction | −2.063311 | 0.000249875 | |
| Regulation of lipolysis in adipocytes | −2.07478781 | 0.000273373 | |
| Gastric acid secretion | −2.11046891 | 0.000277855 |
Immune Infiltration of MITD1 in KIRC
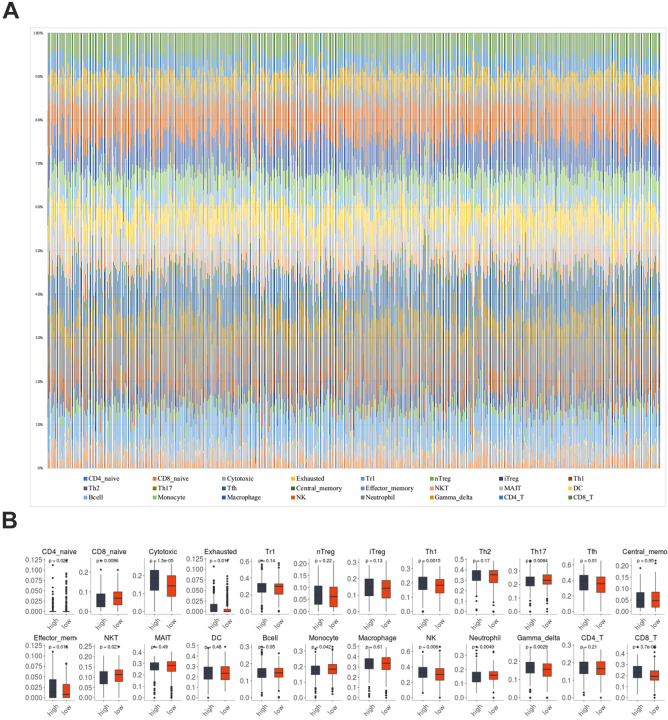
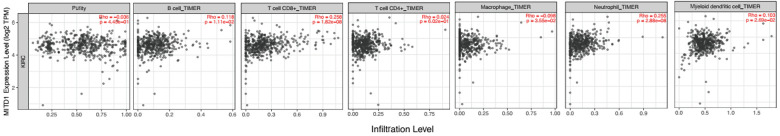
In order to investigate the possible effect of MITD1 on different immune cell types in the KIRC microenvironment, the ImmuCellAI database was utilized to calculate the ingredients of 24 immune cells in 534 KIRC sample. As was shown in Figure 5A, the immune cells varied significantly among different samples. Therefore, we compared the level of immune cells in the high expression MITD1 group and low expression group in the above study (Figure 5B). Most of the immune cells were significantly correlated with MITD1 expression. We found that CD8 navie, Th17, NKT, Monocyte and Neutrophil cells were negative correlation with MITD1 expression. Cytotoxic, Th1, Thf, effector memory, NK, Gamma delta and CD8 T cells were positively correlated with MITD1 expression. Further, the TIMER web tool was performed to investigate the association between MITD1 and tumor immune infiltrating levels (Figure 6A). Results indicated that MITD1 was not significantly correlated with tumor purity (r = −0.036, P > 0.05), CD4+ T cells (r = 0.024, P > 0.05), DC cells (r = 0.103, P > 0.05), B cell (r = 0.118, P > 0.05) and Macrophage (r = −0.098, P > 0.05) but positively correlated with infiltrating levels of CD8+ T cells (r = 0.258, P = 1.82e−8) and neutrophil (r = 0.255, P = 2.88e−8). All the analyses above may indicate that MITD1 expression is correlated with immune infiltration level in KIRC, especially by CD8+ T cells.
Figure 5.
The correlation of MITD1 and immune infiltration in KIRC. (A) Immune cells estimated by ImmuCellAI in KIRC; (B) the comparison of immune cells with high and low expression of MITD1.
Figure 6.
Correlations between MITD1 expression and immune infiltration levels in KIRC by TIMER.
Discussion
KIRC is the main kind of renal cell carcinoma, which takes up for 60%-85% in all renal cell carcinoma cases. 28 Despite the development in diagnosis, surgery, drug therapy, radiotherapy and chemotherapy, the clinical outcome of KIRC still remains unsatisfactory. 29 -31 Although many studies have reported possible diagnostic or therapeutic targets for renal cancer, but remain mostly theoretical. As a well-known heterogeneous disease, KIRC still lacks specific diagnostic biomarkers for individualized therapy, especially in immunotherapy.
MITD1 protein is considered to associate with the midbody. A previous study reported that ESCRT-III subunits are very important for the recruitment of MITD1 to the midbody. Also, ESCRT-III complex helps MITD1 to participate in the abscission of cytokinesis. 14 ESCRT-III is in synergy with oncogene-induced mitotic stress, the abnormal regulation of abscission may lead to oncogenesis and genetic instability. During the process, MITD1 could coordinate ESCRT-III activity during abscission with early events in the final phase of cell division. 18 The genomic and clinical datasets on the public database have been used to mined to find potential novel biomarkers and underlying mechanism of kidney cancer. 8,32 In this study, we integrated a variety of bioinformatics approaches to prove that MITD1 could be a prognostic biomarker for KIRC. The TCGA RNA-seq data of the KIRC cases were used to analyze the prognostic value of MITD1. All the relationship of MITD1 with various tumor characteristics and immune cell responses support that high expression of MITD1 showed a poor survival of KIRC. Through our analyses, we found that high MITD1 expression of KIRC patients are more likely to present a more advanced pathologic T stage, platelet qualitative, live status and tumor stage than those patients with low expression of MITD1. We speculated that dysregulated expression of MITD1 could play an important role of tumorigenesis and tumor immunology in KIRC progression. Coincidentally, the founding was consistent with the expression model of MITD1 in liver cancer. 19
The functional enrichment analyses of MITD1 co-expressed related genes by GSEA were investigated to further figure out the function and associated signaling pathways in KIRC. Our work suggested that higher expression of MITD1 in KIRC patients could mainly alter immune response and cytokine-cytokine receptor interaction. Tumor immune infiltration by different kinds of immune cells is an active immune response against various tumors. 33 Cytokine-cytokine receptor interaction is usually enriched during tumorigenesis, such as liver cancer 34 and colorectal cancer. 35 All these above suggested that MITD1 may be a potential biomarker of prognosis and immune therapeutic target of KIRC.
Over the past few years, the medical treatment of RCC has transited from nonspecific immune approaches to novel immunotherapy agents. 36 Targeted immunotherapy is considered to be a promising new strategy for hereditary and sporadic disease. 37 Subsequently, ImmuCellAI tools and TIMER database were used to reveal the relationship between MITD1 expression and immune cells infiltration levels in KIRC. By comprehensive analyses, we found that the relationship betweenMITD1 and CD8+ T cells is the strongest. CD8+ T cells are the preferred immune cells for targeting cancer cell therapy. 38 Macrophage type 2 (M2) cells, Cancer-associated fibroblasts (CAFs), and regulatory T cells (Tregs) could form immune barriers against CD8+ T cell-mediated anti-tumor immune responses. Thus, CD8+ T cells need to initiate and activate effector CTLs in a process known as the tumor immune cycle to produce a durable and effective anti-tumor immune response. 38 Also, a study has shown that infiltrating CD 8+ T cells was correlated with kidney injury in patients with anti-glomerular basement membrane disease. 39 In a word, these results indicated that MITD1 could regulate and recruit immune infiltrating cells in KIRC. However, in order to more accurately understand the relationship between MITD1 and CD8 + T cells in vivo and in vitro, controlled and multicenter clinical trials are needed.
Taken together, we found that MITD1 is highly expressed in KIRC through integrated bioinformatics analyses. Otherwise, increased MITD1 expression indicates poor prognosis and increased immune infiltration levels of CD8+ T cells. Therefore, MITD1 may be a potential prognostic biomarker for KIRC and play a vital role in immune cell infiltration.
Footnotes
Authors’ Note: Chujie Chen and Yiyu Sheng conceived, designed, and performed the project. All data will be provided by reasonable request from the corresponding author. This study did not require an ethical board approval because it was a study based on bioinformatics.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Yiyu Sheng, MD  https://orcid.org/0000-0002-9020-1548
https://orcid.org/0000-0002-9020-1548
References
- 1. Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shuch B, Amin A, Armstrong AJ, et al. Understanding pathologic variants of renal cell carcinoma: distilling therapeutic opportunities from biologic complexity. Eur Urol. 2015;67(1):85–97. [DOI] [PubMed] [Google Scholar]
- 3. Sjöberg E, Frödin M, Lövrot J, et al. A minority-group of renal cell cancer patients with high infiltration of CD20+ B-cells is associated with poor prognosis. Br J Cancer. 2018;119(7):840–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hsieh JJ, Purdue MP, Signoretti S, et al. Renal cell carcinoma. Nat Rev Dis Primers. 2017;3:17009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mekhail TM, Abou-Jawde RM, Boumerhi G, et al. Validation and extension of the Memorial Sloan-Kettering prognostic factors model for survival in patients with previously untreated metastatic renal cell carcinoma. J Clin Oncol. 2005;23(4):832–841. [DOI] [PubMed] [Google Scholar]
- 6. Fernández-Pello S, Hofmann F, Tahbaz R, et al. A systematic review and meta-analysis comparing the effectiveness and adverse effects of different systemic treatments for non-clear cell renal cell carcinoma. Eur Urol. 2017;71(3):426–436. [DOI] [PubMed] [Google Scholar]
- 7. Vera-Badillo FE, Templeton AJ, Duran I, et al. Systemic therapy for non-clear cell renal cell carcinomas: a systematic review and meta-analysis. Eur Urol. 2015;67(4):740–749. [DOI] [PubMed] [Google Scholar]
- 8. Su SL, Shahriyari. RGS5 plays a significant role in renal cell carcinoma. R Soc Open Sci. 2020;7(4):191422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuan L, Zeng G, Chen L, et al. Identification of key genes and pathways in human clear cell renal cell carcinoma (ccRCC) by co-expression analysis. Int J Biol Sci. 2018;14(3):266–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang S, Zhang E, Long J, et al. Immune infiltration in renal cell carcinoma. Cancer Sci. 2019;110(5):1564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xu WH, Xu Y, Wang J, et al. Prognostic value and immune infiltration of novel signatures in clear cell renal cell carcinoma microenvironment. Aging (Albany NY). 2019;11(17):6999–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yoshihara K, Shahmoradgoli M, Martínez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4:2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Thompson RH, Dong H, Lohse CM, et al. PD-1 is expressed by tumor-infiltrating immune cells and is associated with poor outcome for patients with renal cell carcinoma. Clin Cancer Res. 2007;13(6):1757–1761. [DOI] [PubMed] [Google Scholar]
- 14. Lee S, Chang J, Renvoisé B, Tipirneni A, Yang S, Blackstone C. MITD1 is recruited to midbodies by ESCRT-III and participates in cytokinesis. Mol Biol Cell. 2012;23(22):4347–4361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nikolova DN, Doganov N, Dimitrov R, et al. Genome-wide gene expression profiles of ovarian carcinoma: identification of molecular targets for the treatment of ovarian carcinoma. Mol Med Rep. 2009;2(3):365–384. [DOI] [PubMed] [Google Scholar]
- 16. Hornung T, O’Neill HA, Logie SC, et al. ADAPT identifies an ESCRT complex composition that discriminates VCaP from LNCaP prostate cancer cell exosomes. Nucleic Acids Res. 2020;48(8):4013–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li J, Belogortseva N, Porter D, Park M. Chmp1A functions as a novel tumor suppressor gene in human embryonic kidney and ductal pancreatic tumor cells. Cell Cycle. 2008;7(18):2886–2893. [DOI] [PubMed] [Google Scholar]
- 18. Sadler JBA, Wenzel DM, Williams LK, et al. A cancer-associated polymorphism in ESCRT-III disrupts the abscission checkpoint and promotes genome instability. Proc Natl Acad Sci U S A. 2018;115(38):E8900–E8908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shen H, Wang Z, Ren S, et al. Prognostic biomarker MITD1 and its correlation with immune infiltrates in hepatocellular carcinoma (HCC). Int Immunopharmacol. 2020;81:106222. [DOI] [PubMed] [Google Scholar]
- 20. Wurmbach E, Chen YB, Khitrov G, et al. Genome-wide molecular profiles of HCV-induced dysplasia and hepatocellular carcinoma. Hepatology. 2007;45(4):938–947. [DOI] [PubMed] [Google Scholar]
- 21. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tebani A, Gummesson A, Zhong W, et al. Integration of molecular profiles in a longitudinal wellness profiling cohort. Nat Commun. 2020;11(1):4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Uhlén M, Fagerberg L, Hallström BM, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419. [DOI] [PubMed] [Google Scholar]
- 25. Subramanian A, Kuehn H, Gould J, Tamayo P, Mesirov JP. GSEA-P: a desktop application for gene set enrichment analysis. Bioinformatics. 2007;23(23):3251–3253. [DOI] [PubMed] [Google Scholar]
- 26. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Miao YR, Zhang Q, Lei Q, et al. ImmuCellAI: a unique method for comprehensive T-cell subsets abundance prediction and its application in cancer immunotherapy. Adv Sci (Weinh). 2020;7(7):1902880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Delahunt B, Eble JN. Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol. 1997;10(6):537–544. [PubMed] [Google Scholar]
- 29. Garcia JA, Rini BI. Recent progress in the management of advanced renal cell carcinoma. CA Cancer J Clin. 2007;57(2):112–125. [DOI] [PubMed] [Google Scholar]
- 30. Schmidinger M, Danesi R. Management of adverse events associated with cabozantinib therapy in renal cell carcinoma. Oncologist. 2018;23(3):306–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Atkins MB, Clark JI, Quinn DI. Immune checkpoint inhibitors in advanced renal cell carcinoma: experience to date and future directions. Ann Oncol. 2017;28(7):1484–1494. [DOI] [PubMed] [Google Scholar]
- 32. Sato Y, Yoshizato T, Shiraishi Y, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45(8):860–867. [DOI] [PubMed] [Google Scholar]
- 33. Sokratous G, Polyzoidis S, Ashkan K. Immune infiltration of tumor microenvironment following immunotherapy for glioblastoma multiforme. Hum Vaccin Immunother. 2017;13(11):2575–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li X, Lei Y, Wu M, Li N. Regulation of macrophage activation and polarization by HCC-derived exosomal lncRNA TUC339. Int J Mol Sci. 2018;19(10):2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu J, Li H, Sun L, Wang Z, Xing C, Yuan Y. Aberrantly methylated-differentially expressed genes and pathways in colorectal cancer. Cancer Cell Int. 2017;17:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67(6):507–524. [DOI] [PubMed] [Google Scholar]
- 37. Courthod G, Tucci M, Di Maio M, Scagliotti GV. Papillary renal cell carcinoma: a review of the current therapeutic landscape. Crit Rev Oncol Hematol. 2015;96(1):100–112. [DOI] [PubMed] [Google Scholar]
- 38. Farhood B, Najafi M, Mortezaee K. CD8+ cytotoxic T lymphocytes in cancer immunotherapy: a review. J Cell Physiol. 2019;234(6):8509–8521. [DOI] [PubMed] [Google Scholar]
- 39. Hu SY, Jia XY, Li JN, et al. T cell infiltration is associated with kidney injury in patients with anti-glomerular basement membrane disease. Sci China Life Sci. 2016;59(12):1282–1289. [DOI] [PubMed] [Google Scholar]