Abstract
Circulating tumor cells (CTCs) have a great potential for noninvasive diagnosis and real-time monitoring of cancer. A comprehensive evaluation of four whole genome amplification (WGA)/next-generation sequencing workflows for genomic analysis of single CTCs, including PCR-based (GenomePlex and Ampli1), multiple displacement amplification (Repli-g), and hybrid PCR- and multiple displacement amplification–based [multiple annealing and loop-based amplification cycling (MALBAC)] is reported herein. To demonstrate clinical utilities, copy number variations (CNVs) in single CTCs isolated from four patients with squamous non–small-cell lung cancer were profiled. Results indicate that MALBAC and Repli-g WGA have significantly broader genomic coverage compared with GenomePlex and Ampli1. Furthermore, MALBAC coupled with low-pass whole genome sequencing has better coverage breadth, uniformity, and reproducibility and is superior to Repli-g for genome-wide CNV profiling and detecting focal oncogenic amplifications. For mutation analysis, none of the WGA methods were found to achieve sufficient sensitivity and specificity by whole exome sequencing. Finally, profiling of single CTCs from patients with non–small-cell lung cancer revealed potentially clinically relevant CNVs. In conclusion, MALBAC WGA coupled with low-pass whole genome sequencing is a robust workflow for genome-wide CNV profiling at single-cell level and has great potential to be applied in clinical investigations. Nevertheless, data suggest that none of the evaluated single-cell sequencing workflows can reach sufficient sensitivity or specificity for mutation detection required for clinical applications.
Circulating tumor cells (CTCs) are a population of cells that are shed from a primary or metastatic tumor into the bloodstream. As a new type of liquid biopsy, CTCs bear tremendous potential for noninvasive diagnosis and real-time monitoring of cancer. Genomic analysis of CTCs using single-cell sequencing may help to reveal the underlying mechanisms of tumor metastasis and intratumor heterogeneity and to identify gene mutations that potentially contribute to disease relapse or drug resistance.1, 2, 3, 4, 5
To achieve accurate genomic analysis of CTCs at the single-cell level, whole genome amplification (WGA) of genomic DNA from a single cell must be performed with sufficient breadth and precision. Depending on the mode of downstream genomic analysis [ie, single-nucleotide variants (SNVs), insertions/deletions (indels), or copy number variations (CNVs)], specific performance metrics may be required for different applications. For example, a robust CNV analysis requires wide genome coverage (breadths) as well as high coverage uniformity and reproducibility. On the other hand, to achieve the high sensitivity and specificity required for the analysis of SNVs and indels, high genome coverage/low allele dropout (ADO) rate, and low amplification errors would be critical.6,7
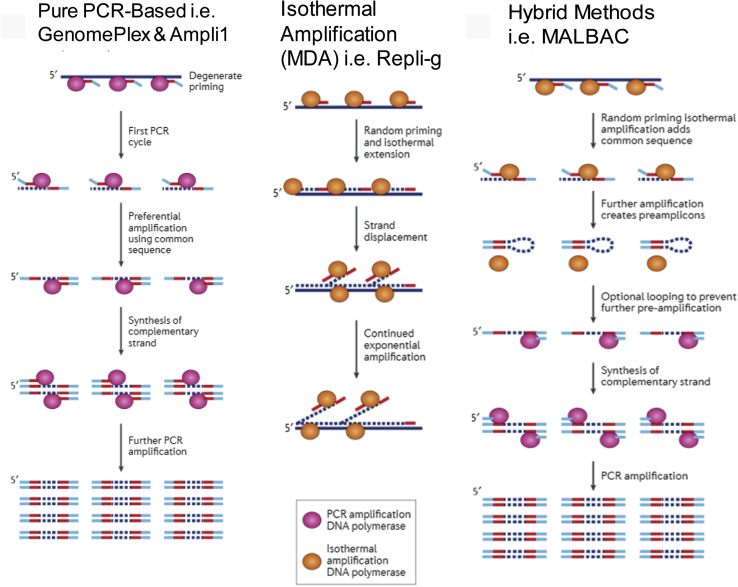
Currently, the most commonly implemented WGA methods for single-cell genomic analyses are PCR or isothermal multiple displacement amplification (MDA) based.8 PCR-based WGA methods typically conduct PCR amplification using degenerate-oligos as primers (ie, PicoPlex)9,10 or linker adaptors with universal sequences ligated to the DNA fragments (ie, GenomePlex and Ampli1).10 In general, PCR-based WGA methods are believed to generate higher coverage and uniformity but at the expense of introducing sequence-dependent coverage bias and significantly more single-nucleotide errors than MDA-based methods. On the other hand, the isothermal MDA-based methods (ie, Repli-g)11 use high-fidelity ф29 DNA polymerase for linear amplification of larger DNA fragments across the genome with higher fidelity than PCR-based approaches, making it potentially better suited for identification of point mutations. However, MDA-based methods are also known to suffer from amplification biases and nonuniformity that can prevent applications in CNV analysis. WGA methods that hybridize the principles of PCR- and MDA-based approaches have also been reported [ie, multiple annealing and loop-based amplification cycling (MALBAC)].12 MALBAC generates looped DNA molecules during the initial multiple rounds of displacement preamplification using a specific oligo design, which is intended to reduce the bias often observed with nonlinear amplification. These DNA loops are then further amplified using PCR amplification. Although such a hybrid approach has broader genomic coverage while maintaining uniformity sufficient for CNV analysis, it can still result in >30% base dropout,13 a potential significant sacrifice in sensitivity of detecting single-nucleotide mutations. Despite intensive efforts in method development in recent years, a comprehensive evaluation of methods and workflows for accurate single-cell genomic analysis has been lacking.
In this study, the performance of four different WGA methods for single CTC analysis, including two PCR-based methods (GenomePlex and Ampli1), an MDA-based method (Repli-g), and a hybrid approach (MALBAC), were systematically evaluated. Results indicate that MALBAC and Repli-g WGA have significantly higher genome coverage compared with GenomPlex and Ampli1, the two PCR-based WGA methods. Furthermore, MALBAC coupled with low-pass whole genome sequencing (LP-WGS) was found to be superior to Repli-g for genome-wide CNV profiling and detecting focal oncogenic amplifications. When coupled with whole exome sequencing (WES), MALBAC WGA had higher sensitivity but lower specificity in SNV/indel detection compared with Repli-g. Nevertheless, none of the WGA methods can achieve sufficient sensitivity and specificity for genome-wide point mutation analysis at the single-cell level. Finally, the clinical utilities of genetic analysis of single CTCs were found by profiling CNVs in single CTCs isolated from four patients with squamous non–small cell lung cancer (NSCLC) enrolled in a phase 2 trial (NCT01493843) treated with pictilisib in combination with chemotherapy.
Materials and Methods
Synthetic CTC Samples with Spiked-In Tumor Cells
To mimic CTCs in blood, synthetic CTC samples were created by spiking viable tumor cells from three tumor cell lines, EBC-1, KPL-4, and PC-3 (see cell line information in Supplemental Table S1), into 3.75-mL normal human donor blood samples. The spiked-in CTCs were selected with the CellSearch platform (Menarini Silicon Biosystems Inc., Huntington Valley, PA), a semiautomated system that enriches for cells expressing epithelial cell adhesion molecules (EpCAMs) but lacking the leukocyte-specific molecule CD45. Cells are further immunostained with fluorescent-labeled anti-keratin antibodies identifying, among others, cytokeratin (CK) 8, CK18, and CK19, and individual single cells were then isolated using DEPArray System (Menarini Silicon Biosystems).
CTCs from Patients With Squamous NSCLC
Four patients with squamous NSCLC treated with the phosphatidylinositol 3-kinase inhibitor pictilisib in combination with carboplatin and paclitaxel in a phase 2 trial (FIGARO, GO27912, NCT01493843) were selected for clinical application of single CTC analysis. All patient blood samples were obtained from the institutional review board or ethics committee at each site. Informed consent was obtained from all patients.
Patients' blood (7.5 mL) was collected in Streck tubes (Streck Inc., La Vista, NE) and shipped to Epic Sciences within 48 hours and processed immediately on arrival. Erythrocytes were lysed, and approximately 3 million nucleated blood cells were dispensed onto each of 10 to 16 glass microscope slides and placed at −80°C for long-term storage according to methods previously described.14,15 Prepared slides were thawed and subjected to automated immunofluorescent staining for CK, DAPI (DNA marker), and CD45 (blood lineage marker). Automated scanning identified candidate cells of interest among nucleated cell populations based on size and morphologic features of the cells, nuclear features, and CK expression in the absence of blood-lineage CD45 expression. Candidate cells were then reviewed by California-licensed clinical laboratory scientists to confirm immunohistochemical staining profile as well as to assess the cytomorphometric features of the cell (size, shape, nucleus/cytoplasm ratio, and so on as they relate to the features associated with CTCs). Candidate cells were given histologic classification of single cells, clusters (more than one sharing cytoplasmic boundaries), or apoptotic cells (nuclear features consistent with apoptosis).
Single-Cell WGA
Captured single CTCs were stored in a 0.2-mL PCR tube stored at −80°C. All single cells were washed with phosphate-buffered saline buffer; phosphate-buffered saline volume carried over with the cell sample into the amplification protocol should not exceed 1 μL. To avoid DNA contamination from external sources or from the amplified DNA product, sample preparation steps before amplification were performed in pre-PCR hood and room. Diluted control human genomic DNA in concentrations of 30 pg/μL and 1 μL (30 pg) was used for positive control for single-cell WGA. Four different WGA workflows were performed with commercial available kits (MALBAC Single Cell WGA Kit, catalog number YK001B, Yikon Genomics, Shanghai, China; Repli-g Single Cell Kit, catalog number 150345, Qigen, Venlo, the Netherlands; GenomePlex Single Cell Whole Genome Amplification Kit, product number WGA4, Sigma-Aldrich, St. Louis, MO; and Ampli1 WGA Kit, reference number WG 001 050 R02, Silicon Biosystems) and strictly followed by manufacturer's manual. Supplemental Figure S1 shows the schemes of these four different WGA methods.
PCR-Based Targeted Sequencing by MMP-Seq
Targeted sequencing was performed using a matrix metalloproteinase sequencing (MMP-seq) panel (963 amplicons that targeted 88 oncogenic and tumor suppresser genes) and workflow that developed and reported previously.16 The experiments were performed according to the Multiplex Amplicon Tagging Protocol from the manufacturer (Fluidigm, South San Francisco, CA). The resulting sequencing-ready amplicon libraries were sequenced on MiSeq using Illumina MiSeq version 2 chemistry [2 × 108-bp paired-end (PE) reads; Illumina Inc., San Diego, CA]. The mean yield is 18 million per run.
LP-WGS
TruSeq PCR-free libraries (Illumina) were generated from 2 μg of Repli-g amplified sample DNA. Libraries from 150 ng of MALBAC amplified DNA for WGS were prepared from the adaptor-ligated DNA before the pooling step in exome library preparation. Eight-cycle enrichment PCR was performed on an aliquot of adaptor-ligated DNA to complete the adaptor for Illumina PE sequencing. Both libraries were checked for quality (TapeStation, Agilent Technologies Inc., Santa Clara, CA) and quantity (KAPA Biosciences Library Quantification, Kapa Biosystems Inc., Wilmington, MA) and sequenced to 0.1× using Illumina MiSeq version chemistry (2 × 100 PE reads). Libraries were sequenced to 0.1× using Illumina MiSeq version chemistry (2 × 100 PE reads).
Whole Exome Sequencing
SureSelectXT (Agilent Technologies Inc.) next-generation sequencing libraries were prepared using Repli-g and MALBAC amplified samples. Repli-g (500 ng) and MALBAC (150 ng) amplified DNA was sheared to approximately 150-bp fragments using the Covaris E220 Focused ultrasonicator system (Covaris Inc., Woburn, MA). Fragmented DNA was processed according to manufacturer's protocol with slight modifications to generate partial adaptor ligated DNA suitable for target enrichment using the SureSelectXT Exome Target Enrichment System for Illumina Sequencing version 5 DNA baits (Agilent Technologies Inc.). Exome-enriched libraries were PCR amplified to complete the Illumina adaptor and then sequenced to 100× coverage (2 × 100 PE) using Illumina HiSeq 2500 Rapid Run with on-board cluster generation version 1 chemistry (Agilent Technologies Inc.). All sequencing data (including MMP-Seq, LP-WGS, and whole exome sequencing) in this study have been submitted to the Sequence Read Archive (https://trace.ncbi.nlm.nih.gov/Traces/sra; SRA number SRP256948).
Next-Generation Sequencing Statistical Analysis
Alignment
All the paired FASTQ files were mapped to the GRCh37 human reference genome with BWA-MEM version 0.7.15,17 and the BAM alignment files were sorted and indexed with SAMtools version 1.3.1.18 For the exome sequencing and WGS, BAM files (biobambam219) were used to mark duplicates. For each WGA platform, two of the 12 single cells (four replicas for each of the three cell lines) were removed from further analyses because of their low depth of coverage.
Coverage
For MMP-seq, the locations of 963 amplicons were merged to 416 amplified regions according to the overlapping of amplicons using Bedtools.20 The amplified regions cover approximately 100 Kb of the human genome. For WES, the targeted 230,417 exonic regions from the Agilent SureSelect platform (Agilent Technologies Inc.) cover approximately 50 Mb of the genome.
The mean depth of coverage of the amplified regions and the exonic regions were calculated for the MMP-seq and WES sequencing data, respectively, using Sambamba21 with the default setting. For WGS, the genome was split into nonoverlapping windows of fixed length. The breadth of coverage is defined as the number of windows covered by at least one read divided by the total number of windows. The number of covering reads is calculated by the program readCounter in HMMCopy software version 1.28.22 R package ineq software version 0.2-1323 was used to generate Lorenz curves to represent inequality of the depths of coverage across windows or targeted regions.
To calculate the mean depth of coverage for the three genes ERBB2, MET, and PTEN, Sambamba was used for the WGS and WES data. For WES, the depth of coverage was further normalized according to the total length of the exonic regions in the gene.
Variant Calling and Quality Analysis
The same strategy was applied for the WES and MMP-seq alignment data. With the use of the alignment data, the variants of four MALBAC replicates, four Repli-g replicates, and the bulk cell line DNA were jointly called with FreeBayes24 for the three cell lines PC-3, EBC-1, and KPL4, respectively. Only the variants in the exonic regions and in the amplified regions were reported for WES and for MMP, respectively. A variant was marked as known if it was in dbSNP Human Build 150 release25 and unknown if otherwise. The joint calling distinguished reference calls from noncovered sites for all the sites called in at least one tested sample. For each cell line, the sites where the depth of coverage is <10 in bulk were filtered, and the genotypes of bulk at these sites were used as the ground truth. The variants were further annotated with SnpEff software version 4.3g.26
Sensitivity
For each cancer cell line, first, the sites that were not genotyped as homologous reference and had depth of coverage ≥10× in the bulk DNA sample were identified as the ground truth. Supplemental Table S2 gives the number of these sites for the three cell lines using MMP-seq and WES. The sensitivity of the variant calls for the single cell is defined as the percentage of these sites having the same genotypes as in bulk DNA sequencing.
FDR
The false discovery rate (FDR) was calculated as previously reported.13 Briefly, high-confidence homozygous reference sites where the coverage is at least 20× and there was no evidence of a nonreference allele (checked with samtools mpileup) were identified. The numbers of these sites are listed in Supplemental Table S3. Next, the frequency of nonreference alleles detected at these sites in single cells was measured.
ADO Rates
The ADO rates27 were accessed using a similar approach as previously described.13 High-confidence heterozygous sites where the coverage is at least 10× and genotypes are heterozygous in bulk were identified. The numbers of these sites are listed in Supplemental Table S4. The frequency of homozygous alleles detected at these sites in single cells was then measured.
CNV Profiling by LP-WGS of Single Cells
For WGS data, HMMCopy was used to correct the number of reads across the windows based on the guanine-cytosine content and mapability. The corrected numbers of reads were then used for visualization and to identify the copy numbers with the HMM model where the parameter E20 was set as 0.9999999 and the window size was set as 200 Kb. For WES data, for each 200-Kb window, the number of reads by the total length of the exonic regions in the window was normalized, and windows without mapping reads or with a total length of the exonic regions <1 Kb were removed. HMMCopy was again used to identify the CNV for the windows.
Results
Benchmarking Performance of Four Different WGA Methods by Targeted Sequencing of Single Tumor Cells
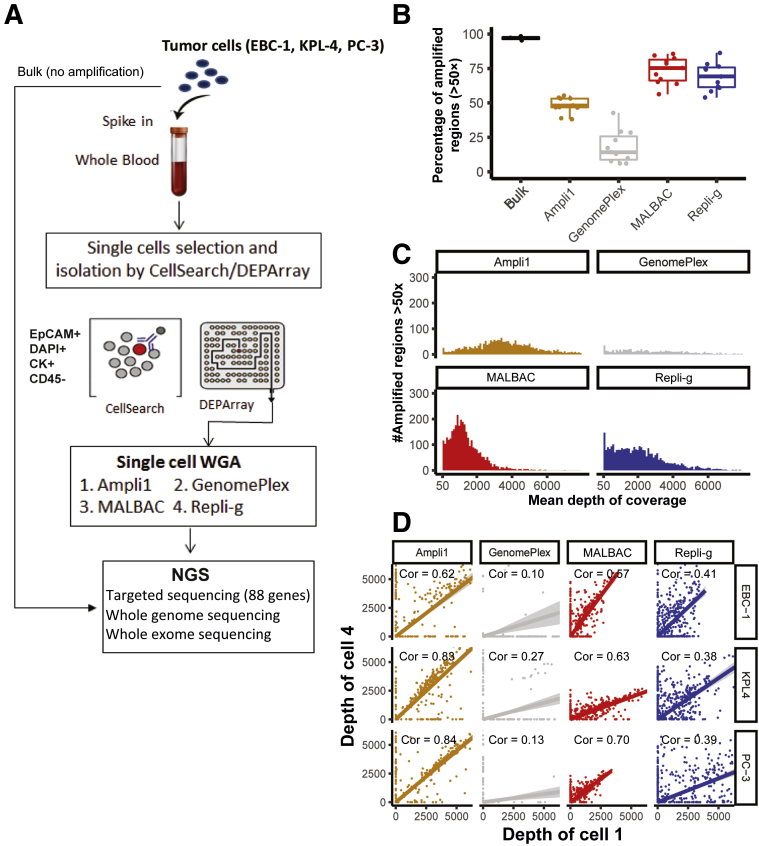
First, the performance of four existing WGA methods (Supplemental Figure S1), including PCR-based (GenomePlex and Ampli1), MDA (Repli-g), and hybrid PCR- and MDA-based (MALBAC), was systematically evaluated. Figure 1A summarizes the experimental design and analyzes the workflow. For this evaluation, three synthetic CTC samples were created by spiking viable tumor cells from three tumor cell lines, EBC-1, KPL-4, and PC-3 (Table 1), into normal human donor blood samples. The CTCs were then captured with the CellSearch platform (EpCAM+, CK+, and CD45−), and individual single cells were isolated using the DEPArray system (Materials and Methods). Four single tumor cells and nine white blood cells from each of the three synthetic CTC samples were subjected to different WGA methods followed by targeted sequencing, LP-WGS, and WES. As controls, 50 ng of bulk genomic DNA from each of the three cancer cell lines and a normal peripheral blood mononuclear cell sample were also sequenced without WGA (Figure 1A).
Figure 1.
Study design and performance of four different whole genome amplification (WGA) methods by targeted matrix metalloproteinase sequencing (MMP-seq) of single circulating tumor cells (CTCs). A: A schematic illustration of the experimental design and workflow for genetic analysis of single CTCs. B: Comparison of the coverage breadth of four WGA methods evaluated by targeted MMP-seq. Percentage of coverage of amplified regions (detectable amplicons with read depth >50×) was displayed for the bulk sequencing (control) and each of the four WGA methods evaluated. The lines in the boxes are the median values across 12 single cells derived from the three synthetic CTC samples. C: Uniformity of amplicon coverage. The distribution of amplicon coverage depth (dropout amplicons with read depth ≤50× were excluded) was presented for each WGA method. D: Reproducibility of amplicon coverage. Scatterplots of the read depth across all detectable amplicons between two single cells independently analyzed through a given WGA MMP-seq workflow. Cor, correlation; MALBAC, multiple annealing and loop-based amplification cycling; NGS, next-generation sequencing.
Table1.
Cell Lines Used in This Study
| Cancer cell line | Indication | Copy number variation | No. of copies | Ploidy |
|---|---|---|---|---|
| PC-3 | Prostate cancer | PTEN homozygous deletion | 0 | 2.9 |
| EBC-1 | Lung cancer | MET amplification | 9 | 2.8 |
| KPL-4 | Breast cancer | ERBB2 amplification | 6 | 2.4 |
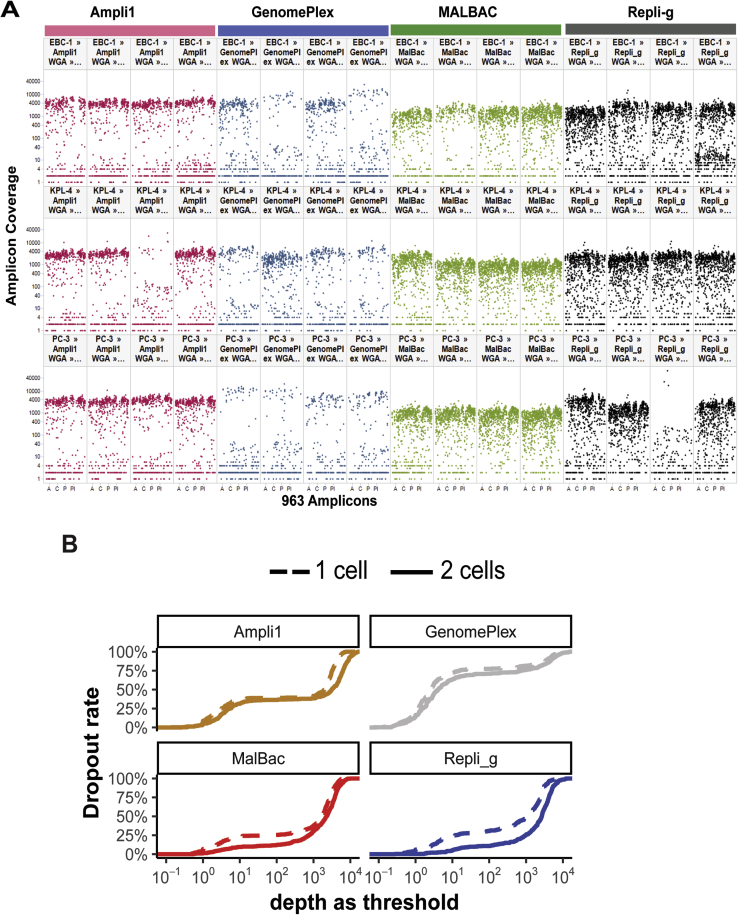
To have an initial assessment of the performance of the four WGA methods, first, targeted deep sequencing was performed by MMP-seq, which targets 88 clinically relevant oncogenes and tumor suppressor genes.16 The first performance metric evaluated was the coverage breadth of each WGA method, which was assessed as the fraction of the 416 amplified regions with >50× depth of coverage. MALBAC and Repli-g WGA had significantly broader coverage breadth (73.6% ± 9.6% and 69.0% ± 9.3%, respectively) than Ampli1 and GenomePlex (48.1% ± 5.7% and 17.9% ± 11.7%, respectively), whereas the breadth of the bulk was 97.0% ± 1.4%, indicating a vast difference in amplicon dropout rates among these WGA methods (Figure 1B and Supplemental Figure 2A). To further investigate whether the different amplicon dropout rates observed in different WGA methods are random or systematic events, a two-cell strategy was applied that calculates the number of amplicons covered by at least two of the four single cells of the same synthetic CTC sample. Use of the two-cell strategy was found to slightly improve the genomic coverage for each WGA method (Supplemental Table S2 and Supplemental Figure S2B), suggesting that the amplicon dropout is likely a result of systematic amplification bias intrinsic to each of the WGA methods.
The second performance metric evaluated was the coverage uniformity, which assesses whether the amplifications are biased toward some regions more than others. Among the four WGA methods evaluated, MALBAC showed the best coverage uniformity with a sharp peak in distribution of amplicon depth, whereas Repli-g and GenomePlex showed a wide range of amplicon coverage distribution, indicating unevenness in coverage across amplicons (Figure 1C). Interestingly, Ampli1 showed a unique binary distribution in amplicon coverage. This finding may be due to the bias introduced by the restriction enzyme digestion step to fragment genomic DNA before PCR amplification in this workflow (ie, genomic DNA regions that were digested into too small or too big fragments by the restriction enzyme may result in no or less amplification products).
Last, the reproducibility of each of the WGA methods was evaluated. For this, coverage depth of the same amplicons among different single cells isolated from the same synthetic CTC sample was compared. MALBAC and Ampli1 had higher cell-to-cell reproducibility (mean R = 0.63 and 0.76, respectively), whereas the Repli-g and GenomePlex had poor coverage reproducibility (mean R = 0.39 and 0.16, respectively) (Figure 1D).
Comparison of Performance of MALBAC and Repli-g WGA Methods Coupled with LP-WGS and WES
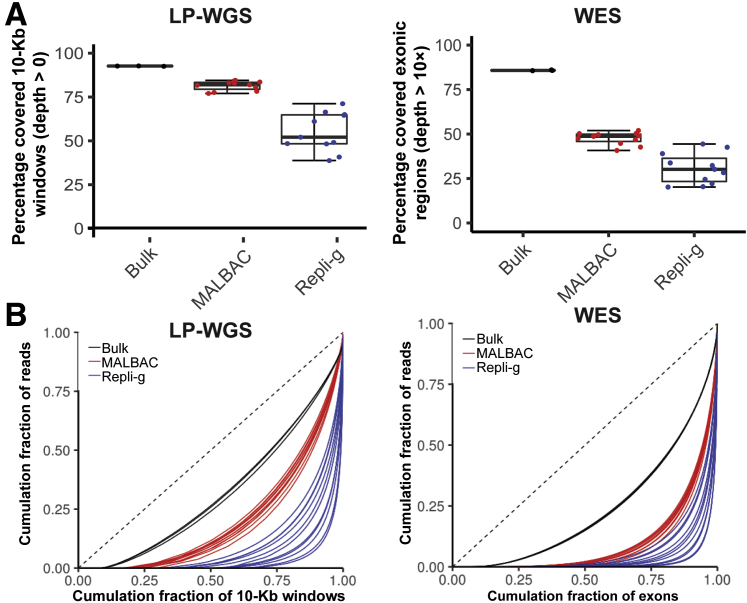
To further confirm findings from the targeted sequencing evaluation, MALBAC and Repli-g, the two WGA methods with better performance in the targeted sequencing evaluation, were the next focus. LP-WGS (approximately 0.1× mean sequencing depth) and WES (approximately 100× mean sequencing depth) were performed on the two WGA products. With the LP-WGS approach, MALBAC showed better genome-wide coverage (79.0% ± 3.8%) compared with that of Repli-g (52.2% ± 11.5%) (Figure 2A). A similar conclusion was derived from the WES data. Among exons with >10× coverage in the bulk DNA sequencing, the mean exonic coverages of MALBAC was 46% ± 6.2%, which was consistently higher than Repli-g (30.7% ± 8.6%) (Figure 2A).
Figure 2.
Performance of multiple annealing and loop-based amplification cycling (MALBAC) and Repli-g whole genome amplification (WGA) methods coupled with low-pass whole genome sequencing (LP-WGS) and whole exome sequencing (WES). A: Genome-wide coverage was displayed for bulk DNA sequencing and single cells subjected to MALBAC or Repli-g WGA. Left panel, LP-WGS (read depth 0.1×), 10-Kb window, coverage defined by read depth ≥1×. Right panel: WES (100×), coverage defined by read depth ≥10×. B: Genome-wide coverage uniformity represented as Lorenz curves for single cells amplified by MALBAC (solid red) and single cells amplified by Repli-g (solid blue), along with bulk genomic DNA controls (solid black). The ideal black diagonal line represents perfect uniformity. Left panel: LP-WGS; right panel: WES. n = 12 (B, MALBAC and Repli-g); n = 3 (B, genomic DNA).
To assess the genome-wide and exome-wide coverage uniformity of the two WGA methods, the Lorenz curves were plotted to benchmark their performance against the assumed perfect uniformity exhibited by bulk sequencing. Consistent with the MMP-seq targeted sequencing results, MALBAC WGA single-cell sequencing outperformed Repli-g in uniformity of both genome (10-Kb window) (Figure 2B) and exome (Figure 2B) coverage.
MALBAC WGA Coupled with LP-WGS Is Superior to Repli-g for CNV Profiling
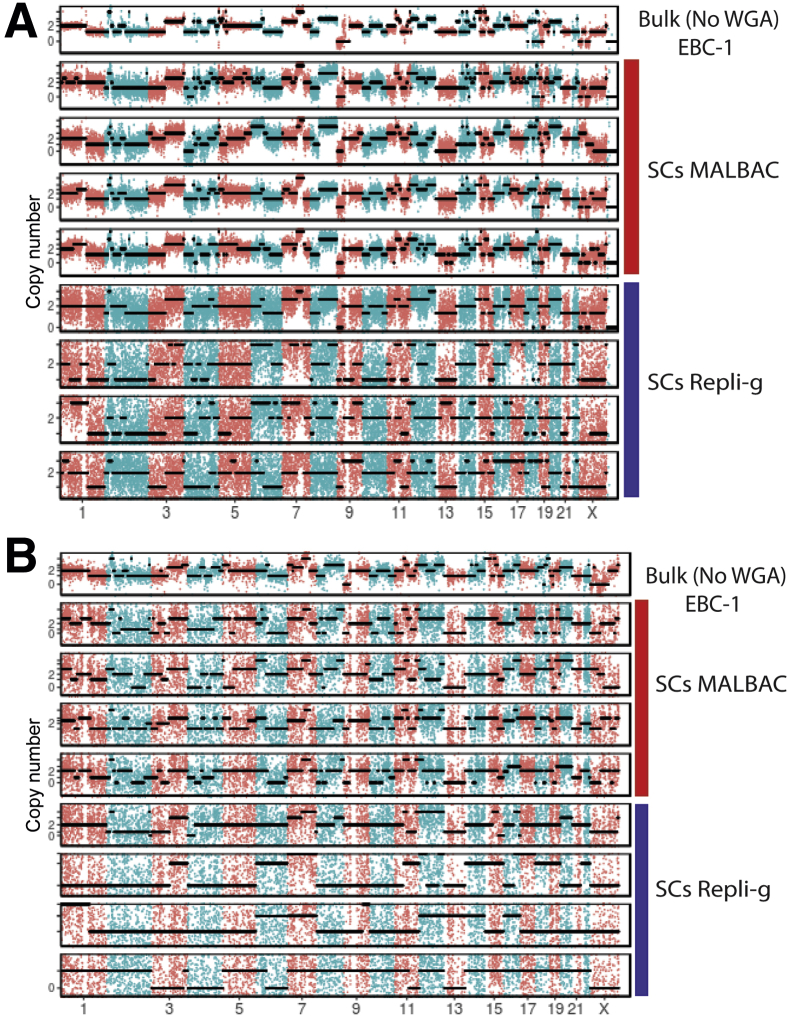
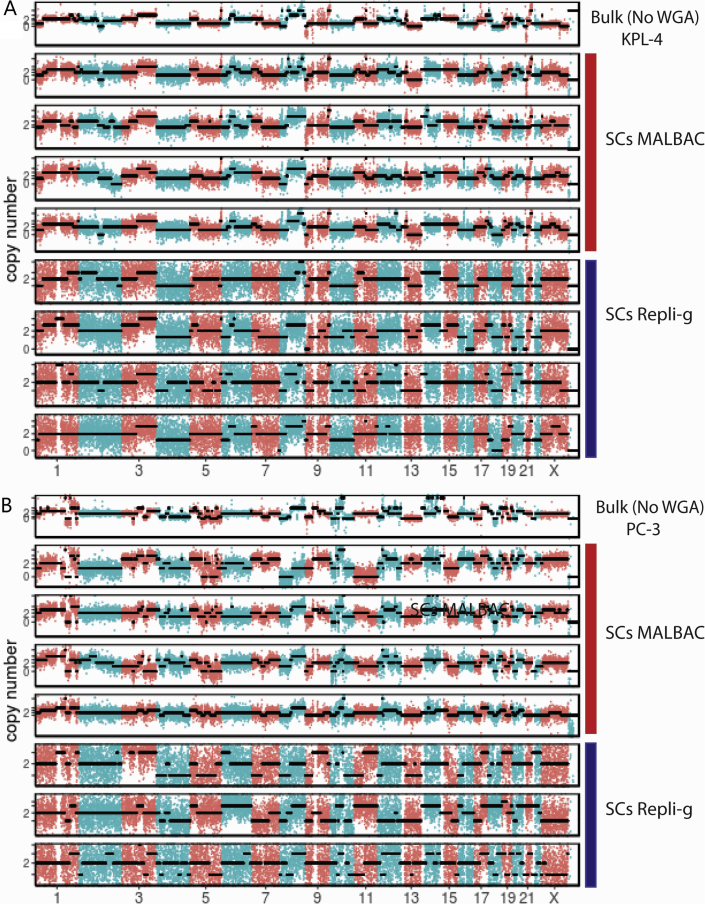
CNVs are one of the major forms of genetic variations in cancers. For noninvasive prognosis and diagnosis of cancer, theoretically it would be advantageous to analyze CTCs other than cell-free DNA fragments for CNV because it removes background contributed from the normal blood cells. Next, whether CNVs of single tumor cells can be precisely determined using MALBAC and Repli-g WGA products was examined. By LP-WGS, the CNVs across the whole genomes of four single cells from each of the three synthetic CTC samples were determined. Figure 3A shows the CNV patterns (segmented with a hidden Markov model) across all chromosomes for the synthetic CTCs derived from EBC-1 as a representative example. Results for additional synthetic CTCs (KPL-4 and PC-3) are included in the supplementary materials (Supplemental Figure S3). The results from the bulk DNA sequencing were used as the ground truth, assuming the cells derived from tumor cell lines were pure homogenous populations. The CNV profiles across different chromosomes of single cells by MALBAC WGA were consistent with those of the bulk DNA. Moreover, four single CTCs were found to exhibit highly reproducible CNV patterns using MALBAC workflow. On the other hand, Repli-g WGA found a much higher background noise level that resulted in less robust resolutions on CNV differences. The performance of CNV profiling based on the WES data was also evaluated. The resolution of CNVs by WES (mean depth of 100×) was significantly decreased compared with that of LP-WGS even for the bulk DNA sample (Figure 3B). Furthermore, the CNV pattern became too noisy to yield confident CNV profiles when using WES of single cells regardless of WGA methods. These results suggested that WES is not suitable for genome-scale CNV detections at the single-cell level.
Figure 3.
Performance of multiple annealing and loop-based amplification cycling (MALBAC) and Repli-g whole genome amplification (WGA) coupled with low-pass whole genome sequencing (LP-WGS) or whole exome sequencing (WES) for copy number variation (CNV) profiling. A: LP-WGS (read depth 0.1×); B: WES (read depth 100×). Digitized genome-wide CNV patterns across all chromosomes derived from single circulating tumor cells using EBC-1 as a representative example. Profiles of four individual single cells (SCs) are presented for each WGA method and for each sequencing method. The results from the bulk DNA sequencing (top panel) were used as the ground truth. Black dotted lines are fitted CNV numbers obtained from the hidden Markov model. The binning window is 200 Kb.
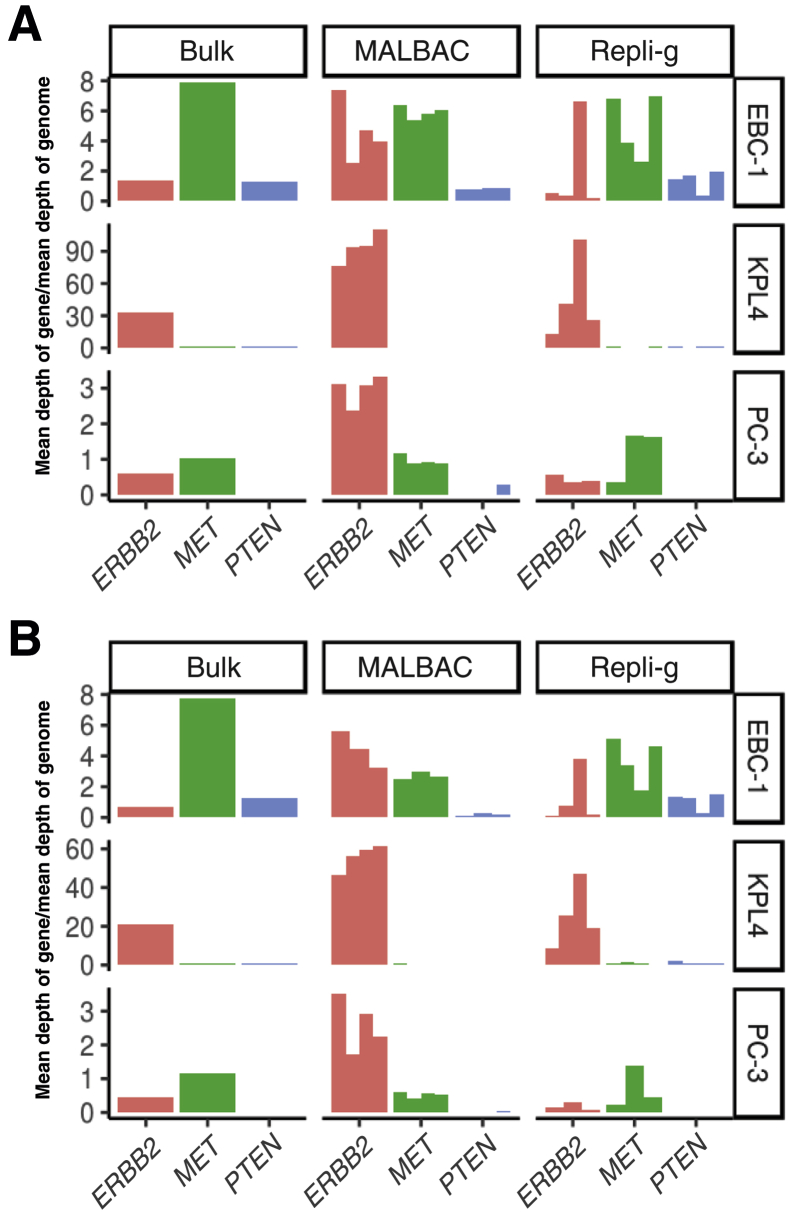
In addition to the genome-wide CNV patterns, the two WGA methods for detecting focal CNVs in oncogenic drivers were also evaluated. On the basis of bulk WES, it was established that each of the three cancer cell lines used for creating the synthetic CTC samples harbors specific oncogenic CNVs, including MET amplification in EBC-1, ERBB2 amplification in KPL-4, and PTEN deletion in PC-3 (Table 1). To assess the sensitivity and feasibility of focal CNV detection using low-pass WGS and WES, the copy number fold changes of MET, ERBB2, and PTEN genes in single cells were calculated and compared with bulk samples. Although LP-WGS (Figure 4A) and WES (Figure 4B) data indicate that MALBAC reproducibly detected the three known oncogenic CNVs among four single cells, significantly bigger variations were observed with the workflow using Repli-g WGA followed by LP-WGS or WES.
Figure 4.
Performance of multiple annealing and loop-based amplification cycling (MALBAC) and Repli-g for detecting focal copy number variations (CNVs) in oncogenic drivers. Each of the whole genome amplification methods were coupled with LP-WGS (read depth 0.1×) (A) or whole exome sequencing (read depth 100×) (B) to test whether they can detect known focal CNVs identified by bulk sequencing of the three cancer cell lines used for creating the synthetic circulating tumor cell samples in this study, including MET amplification in EBC-1, ERBB2 amplification in KPL-4, and PTEN deletion in PC-3 (Table 1). The calculated copy number fold changes of MET, ERBB2, and PTEN genes in single cells were compared with those determined by bulk DNA sequencing. Each bar represents one single cell.
Mutation Detection from Single CTCs by WES and Targeted Sequencing
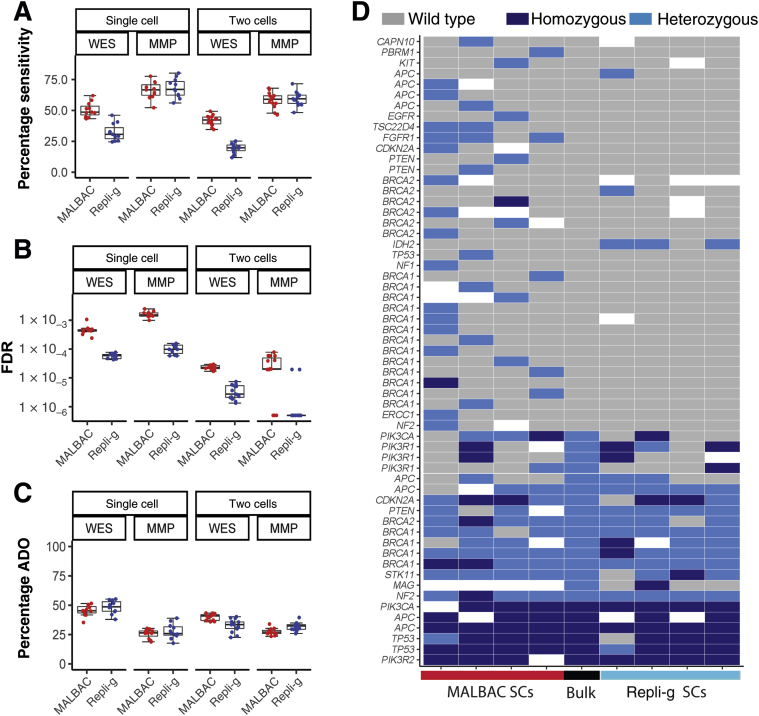
Another potential utility of genetic analysis of single CTCs is detecting mutations, such as SNVs and indels, including known oncogenic driver mutations and emerging acquired new somatic mutations during the disease evolution and treatment regimens. For assessing the sensitivity of SNV detection, the targeted exome regions of each cell were sequenced to a mean depth of 60×, and the high-confidence SNVs (with ≥10× read depth) from the WES data of the bulk DNA samples from the three cell lines as the ground truth (Materials and Methods). Both WGA and WES workflows had relatively low sensitivities (Figure 5A and Table 2). The MALBAC-WES single cell workflow had a 50.5% sensitivity in detecting all SNVs, whereas Repli-g's sensitivity was only 32.2%. Applying the two-cell strategy, which requires SNVs to be detected in at least two single cells from a given sample, the sensitivity of MALBAC and Repli-g workflows further decreased to 41.8% and 19.4%, respectively.
Figure 5.
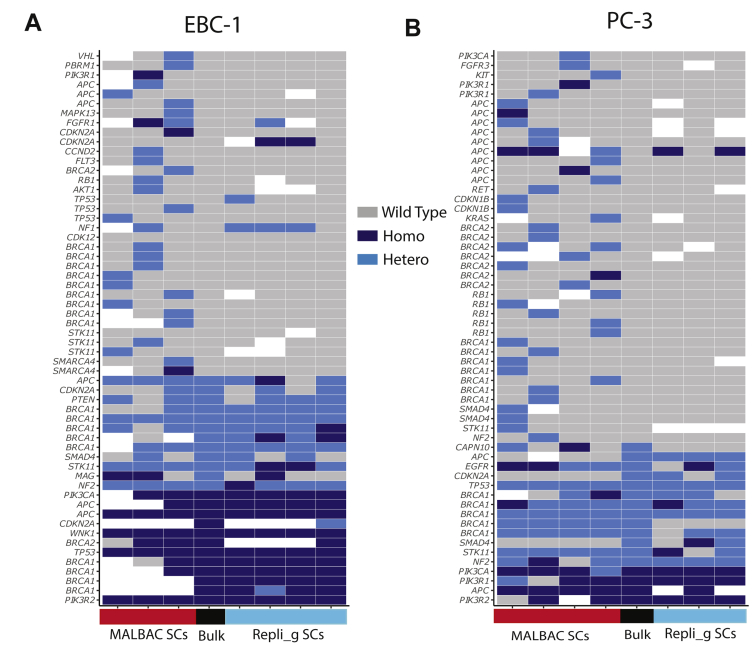
Mutation detection from single circulating tumor cells (CTCs) by whole exome sequencing (WES) or targeted sequencing. Performance of mutation detection was evaluated using the WES data of the bulk DNA samples (combination of EBC1, KPL-4, and PC-3 tumor cells) as the ground truth (Materials and Methods). Each performance metric was also evaluated with the single-cell (SC) [single nucleotide variations (SNVs) detected in any single cell from a given sample) or two-cell strategy (SNVs detected in at least two single cells from a given sample). A: The sensitivity of the variant calls for the SC was defined as the percentage of homozygous reference sites showing the same genotypes as in the bulk DNA samples. B: The false discovery rate (FDR) was calculated as previously reported.13 High-confidence homozygous sites (reference) where the coverage was at least 20× were identified in the bulk DNA samples and used as the ground truth, whereas high-confidence heterozygous sites where the coverage was at least 10× and genotypes were heterozygous in the bulk DNA samples. C: The allele dropout (ADO) rate27 was determined as the percentage of homozygous alleles detected at these sites in SCs. D: Heatmap of mutation detection by matrix metalloproteinase sequencing (MMP-seq) in synthetic single CTCs and bulk of KPL-4 cell line. Only the variants with high or moderate effects, annotated with SnpEff, are shown in this figure. The full list is given in Supplemental Table S4. Nonsynonymous heterozygous and homozygous mutations are in blue and wild-type mutations are in gray. Bulk (ground truth) is shown in the middle column, four SCs by the MALBAC whole genome amplification (WGA) method are shown in the left columns, and 4 SCs by the Repli-g WGA method are shown in the right columns. The blank regions represent no sequence coverage. The mutated genes are listed along the left side of the heatmap.
Table 2.
Performance of Single-Nucleotide Variation Detection by Single-Cell WES or Targeted Sequencing
| Workflow | Single cells |
At least two cells |
||
|---|---|---|---|---|
| WES | MMP-seq | WES | MMP-seq | |
| Sensitivity, means ± SD | ||||
| MALBAC | 50.5% ± 6% | 66.2% ± 7% | 41.8% ± 4% | 58.5% ± 6% |
| Repli-g | 32.2% ± 7% | 68.1% ± 8% | 19.4% ± 4% | 59.5% ± 6% |
| Mean false discovery rate | ||||
| MALBAC | 4.79 × 10−4 | 1.63 × 10−3 | 2.33 × 10−5 | 3.02 × 10−5 |
| Repli-g | 5.83 × 10−5 | 1.04 × 10−4 | 3.68 × 10−6 | 2.60 × 10−6 |
| Allele dropout, means ± SD | ||||
| MALBAC | 45.5% ± 5% | 25.6% ± 4% | 39.8% ± 3% | 27.5% ± 3% |
| Repli-g | 48.4% ± 6% | 27.8% ± 6% | 32.6% ± 5% | 31.8% ± 3% |
MALBAC, multiple annealing and loop-based amplification cycling; MMP-seq, matrix metalloproteinase sequencing; WES, whole exome sequencing.
On the basis of high-confidence homozygous reference sites identified from EBC-1, KPL4, and PC3 bulk DNA samples (Supplemental Table S3), the FDR in SNV detection were evaluated by single-cell sequencing. Repli-g outperforms MALBAC in specificity and has an approximately 10-fold lower FDR (mean, 5.83 ×10−5) in SNV detection compared with MALBAC (mean, 4.79 ×10−4) (Figure 5B and Table 2). Furthermore, applying the two-cell strategy can further improve the specificity of both methods by approximately 15- to 20-fold.
Because WGA amplification bias may also lead to amplification of one allele at heterozygous sites, the ADO27 rate was evaluated next by these two single-cell WES workflows. To quantify ADO rates, 14,366, 21,619, and 16,650 high-confidence heterozygous sites were identified (Supplemental Table S3) in the EBC-1, KPL4, and PC-3 bulk genomic DNA controls, respectively, and the frequency with which one allele or the other was lost in each of the single cells assayed was determined. Relatively high ADO rates were observed for the MALBAC (45.5%) and Repli-g (48.4%) methods (Figure 5C and Table 2). The two-cell strategy reduced the ADO rate for MALBAC (39.8%) and Repli-g (32.6%). Collectively, data indicate that neither of these WGA-WES single-cell sequencing workflows demonstrated sufficient sensitivity or specificity for SNV detection required for clinical implementation.
Last, single-cell WGA coupled with targeted sequencing (ie, MMP-seq), which focuses on 88 well-characterized oncogenes,16 was evaluated for performance in detecting clinically relevant mutations. Potential advantages of targeted sequencing also include significantly increased throughput and reduced cost compared with WES, two important considerations for clinical feasibility of single-cell mutation analysis. Among 207, 197, and 183 high-confidence variant calls identified in the EBC-1, KPL4, and PC-3 bulk genomic DNA, respectively, by MMP-seq (Supplemental Table S2), MALBAC, and Repli-g, single-cell sequencing had a similar SNV detection sensitivity (66.2% and 68.1%) and a similar ADO rate (25.6% and 27.8%) (Figure 5, A and C, Table 2), both a significant improvement compared with WGA-WES workflows. Figure 5D shows the total of 61 predicted high- and moderate-impact variant calls in bulk (23 mutations and 38 wild type) and single cells of KPL-4 as a representative (Supplemental Table S4). Although the mutation detection in single cells from both WGA methods are similar by visualization of the heatmap, the number of false-positive calls by Repli-g WGA is significantly fewer than those by MALBAC WGA, indicating a better specificity with the Repli-g method. Consistent findings were also observed from single cells of EBC-1 and PC-3 (Supplemental Figure S4). Repli-g coupled with targeted sequencing also had an order of magnitude lower FDR, which was consistent with the WES data, highlighting its superior specificity in SNV detection compared with MALBAC (Figure 5B and Table 2). Similar to the WES results, the two-cell strategy further significantly reduced the FDR of SNV detection in MMP-seq by >40- to 50-fold in both methods. The data suggest that, with further optimization, WGA coupled with targeted sequencing may have promising potential in detecting actionable mutations in single CTCs in the clinical settings.
CNV Profiling of Single CTCs in Patients with Squamous NSCLC
From the above analysis, it was concluded that MALBAC WGA coupled with LP-WGS is a robust workflow for CNV profiling at single-cell level and has great potential to be applied in clinical investigations. Next, clinical utilities of profiling CNVs of single CTCs isolated from patients with cancer were evaluated using this workflow. For this, blood samples collected from four patients with squamous NSCLC treated with chemotherapy in combination with pictilisib (a potent inhibitor of phosphatidylinositol 3-kinase α/δ) in a phase 2 clinical trial (NCT01493843, Materials and Methods) were analyzed. Of the four patients studied, a total of 80 circulating epithelial cells (CK+, CD45−, DAPI+) were collected to enrich CTCs using Epic Sciences platform (Materials and Methods, Supplemental Table S5). MALBAC WGA and LP-WGS (0.1×) were performed on each of the single cells from the enriched CTC population along with 11 white blood cells as normal controls. Of the 80 circulating epithelial cells profiled across the four patients, CNV changes were detected in 15 cells (Supplemental Table S5). The observed relatively low rate of CNV detection (mean, 19%; range, 13% to 40% across patients) may be due to the low CTC enrichment efficiency using the Epic Sciences platform (Discussion).
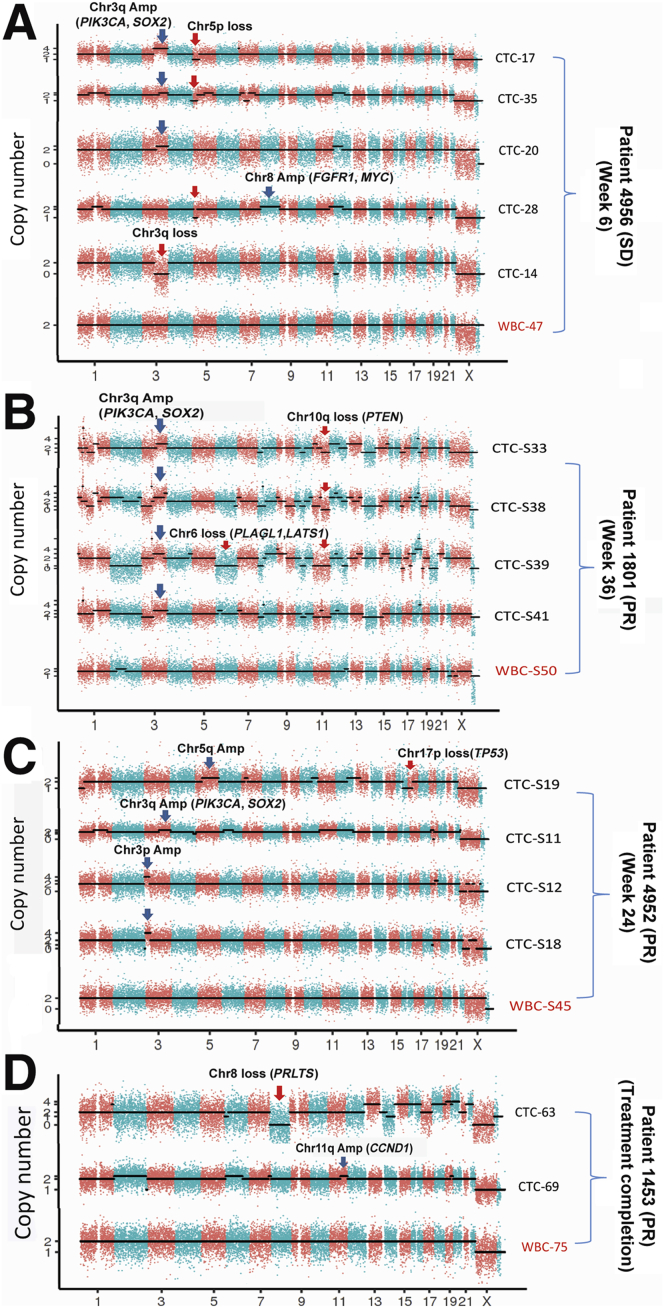
The genome-wide CNV profiles of the 15 single CTCs are outlined in Figure 6. For patient 4956, single-cell CNV profiling (6 weeks after treatment) detected Chr3q amplification (encompassing the PIK3CA and SOX2 genes) in three CTCs (Figure 6A). The PIK3CA gene amplification (PIK3CA/Chr3 ratio is 4.38) was further validated by PIK3CA chromogenic in situ hybridization assay on the tumor biopsy sample collected from the same patient before treatment. Interestingly, Chr3q loss was detected in one of the CTCs, indicating loss of PIC3CA may be one of the potential mechanisms of acquired resistance to combination treatment by PIK3CA inhibitor and chemotherapy. In addition, other oncogenic CNVs frequently found in patients with NSCLC, including Chr8 amplification (where the FGFR1 and MYC genes are located) and Chr5p loss were also detected in several of the CTCs from patient 4956.
Figure 6.
Copy number variation (CNV) profiling of single circulating tumor cells (CTCs) in patients with squamous non–small-cell lung cancer (NSCLC). Digitized copy numbers across the genome were plotted for selected single CTCs isolated from four patients with squamous NSCLC A: Patient 4956. B: Patient 1801. C: Patient 4952. D: Patient 1453. The CNV profiles of patient-matched white blood cells (WBCs) were used as normal controls. Black dotted lines are fitted CNV numbers obtained from the hidden Markov model. The single cells were sequenced at only 0.1× depth, and the binning window was 200 Kb. PR, partial response; SD, stable disease.
For patient 1801, Chr3q amplification (containing PIK3CA and SOX2) was detected in all four CTCs isolated at week 36 after treatment (Figure 6B). In addition, Chr10q loss (containing the tumor suppressor gene PTEN) was detected in three of the four CTCs. Chr6 deletion (containing the tumor suppressor genes PLAGL1 and LATS1) was also detected in one of the CTCs.
For patient 4952, Chr3q amplification was detected in one CTC, Chr3p amplification in two CTCs, and Chr5p amplification in one CTC. Chr17p deletion (containing TP53) was also detected in one CTC (Figure 6C). Lastly, for patient 1453 (Figure 6D), the two CTCs isolated at disease progression had Chr8 deletion (containing the tumor suppressor gene PRLTS) and Chr13p amplification (containing the oncogene CCND1).
Collectively, these data provided promising clinical utility of CNV profiling of single CTCs from patients with NSCLC. The identified potentially clinically relevant CNVs and intrapatient cell-to-cell heterogeneities in CTCs may inform mechanisms of response and resistance and novel therapeutic strategies.
Discussion
Genomic analysis of CTCs at the single-cell level holds great promise in clinical applications, such as uncovering the mechanism of tumor metastasis, intratumor heterogeneity, and genetic alterations conferring drug resistance. Despite great technical advancements in CTC selection and isolation,28 WGA method development, and downstream sequencing analysis, only a few studies have reported on their potential clinical utilities, such as mutational analysis of single CTCs in metastatic breast cancer29,30 and genetic profiling in small cell lung cancer31; a systematic assessment of performance and clinical applications of different workflows of single CTCs analysis has been lacking. The current study reports a comprehensive evaluation of multiple WGA/NGS workflows for single CTC genomic analysis. This study also highlights the strengths and limitations of these workflows in clinical applications of single CTC analysis. These findings are also applicable to the genetic analysis of single cells in general.
By spiking viable tumor cells from tumor cell lines into normal human donor blood samples, three synthetic CTC samples were created and single CTCs were isolated using CellSearch and DEPArray systems. First, a quick assessment of four commercially available WGA methods was performed by targeted sequencing. Compared with GenomPlex and Ampli1, the two PCR-based WGA methods, MALBAC and Repli-g were superior in several key performance metrics, including broader coverage breadth, coverage uniformity, and reproducibility.
MALBAC and Repli-g were analyzed next. Coupled with LP-WGS or WES, the performance of these workflows was evaluated in two different downstream genomic applications: CNV profiling and mutation analysis of single CTCs. MALBAC coupled with LP-WGS (0.1×) was identified as the most robust workflow for single CTC CNV analysis. Compared with Repli-g, MALBAC had better coverage breadth, uniformity, and reproducibility and accurately detected genome-wide CNV alterations and focal oncogenic amplifications. These findings are consistent with the known strength of MALBAC WGA, including good reproducibility and lack of large-scale bias coverage, both important requirements for robust CNV detection.32, 33, 34
Another important finding of this study is the limitations of current single CTC analysis workflows in mutation analysis. Results indicate that MALBAC and Repli-g coupled with WES exhibited different strengths and weaknesses in SNV/indel detection (ie, MALBAC had better overall sensitivity, whereas Repli-g had better specificity). Compared with the previous study by Hou et al,7 which reported a mean ADO rate of 43.09% and a mean FDR of 6.04 × 10−5 of WES from single CTCs by MDA, in this study, data from single cells by Repli-g (48.4% ± 6% and 5.83 × 10−5, respectively) are in the same range or magnitude. Nevertheless, it is concluded that none of the WGA methods can achieve sufficient sensitivity and specificity for genome-wide point mutation analysis at the single-cell level. Further optimization of the WGA methods (ie, increasing coverage breadth and uniformity and decreasing ADO rates and amplification errors) may improve the sensitivity and specificity of single CTC mutation analysis and enable robust clinical applications in the future.6
More importantly, the clinical utilities of CNV profiling of single CTCs derived from patients with squamous NSCLC treated with a combination of chemotherapy and a PIK3CA inhibitor (pictilisib) was established.35,36 MALBAC coupled with LP-WGS on single CTCs can identify frequent oncogenic CNV alterations found in squamous NSCLC, including amplifications in PIK3CA and SOX2 as well as loss of the tumor suppressor gene PTEN. Of interest, Chr3q loss (encompassing PIK3CA) from a CTC isolated from a patient at 6 weeks after treatment was identified, indicating loss of PIK3CA may be one of the potential mechanisms of acquired resistance to combination treatment by PIK3CA inhibitor and chemotherapy.
Lastly, further improvement of platforms for selecting and isolating single CTCs is also critical to improve the efficiency and cost-effectiveness of clinical application of single CTC analysis. This optimization may also need to be tailored to specific indications. For example, the lungs, as part of the respiratory system, are the front-line barrier to the outer world. In responding to environmental stimuli, such as allergen, virus, air pollution, and smoke, the process of epithelial cell repair and regeneration may result in the shedding of normal epithelial cells into the circulating system along with the CTCs. Some previous studies used cancer-specific markers combined with epithelial markers to better specifically select CTCs. For example, a panel of immunomagnetic nanoparticles against four markers, EpCAM, HER2/neu, epidermal growth factor receptor, and mucin-1, was applied to detect CTCs in patients with ovarian cancer.27,28 Miyamoto et al37 successfully measured androgen suppression treatment–induced signaling responses within CTCs in prostate cancer by analyzing prostate CTCs with prostate-specific antigen and prostate specific membrane antigen quantitative immunofluorescence assay.37 Therefore, developing tumor-specific biomarkers in conjunction with epithelial markers for CTC selection may enable further improvement in the sensitivity and the specificity of the CTC-capturing platforms.
Footnotes
Supported by a Genentech Inc. grant.
S.L. and C.-J.C. contributed equally to this work.
Disclosure: S.L., Y.G., E.P., A.D., B.S., J. S., M.R.L., and Y.W. are employees of Genentech Inc. and hold stock in Roche. R.G. holds equity in Expression Analysis. A.R and M.L. hold equity in Epic Sciences.
Supplemental material for this article can be found at https://doi.org/10.1016/j.jmoldx.2020.02.013
Contributor Information
Wenzhong Xiao, Email: wzxiao@stanford.edu.
Yulei Wang, Email: wang.yulei@gene.com.
Supplemental Data
Supplemental Figure S1.
The schemes of different whole genome amplification methods assessed in this study. MALBAC, multiple annealing and loop-based amplification cycling; MDA, multiple displacement amplification.
Supplemental Figure S2.
Amplicon coverages of all single cells by matrix metalloproteinase sequencing (MMP-seq). A: A total of 48 single cells. Each dot represents amplicon depth by MMP-seq for each single cell. Four cells per cell line and 12 single cells per whole genome amplification (WGA) protocol. One cell of KPL4 and the other cell of PC-3 are excluded for the further analysis because of significant amplicon dropout. B: Comparison of amplicon dropout rate between the single-cell and two-cell strategies. The solid lines represent the single-cell strategy and dotted lines the two-cell strategy. MALBAC, multiple annealing and loop-based amplification cycling.
Supplemental Figure S3.
Copy number variation (CNV) profiling for KPL4 and PC-3 synthetic circulating tumor cells by multiple annealing and loop-based amplification cycling (MALBAC) and Repli-g whole genome amplification (WGA) coupled with low-pass whole genome sequencing. A: CNV for KPL-4. B: CNV for PC-3. PC-3 single cell (SC) 3 by Repli-g was excluded because of failure of quality control. Black dotted lines are fitted CNV numbers obtained from the hidden Markov model. The single cells were sequenced at only 0.1× depth, and the binning window was 200 Kb.
Supplemental Figure S4.
Heatmap of mutation detections by matrix metalloproteinase sequencing in synthetic single circulating tumor cells and bulk of EBC-1 and PC-3 cell lines. A: EBC-1; EBC-1 single cell (SC) 2 by multiple annealing and loop-based amplification cycling (MALBAC) was excluded because of poor coverages. B:) PC-3; PC-3 SC 3 by Repli-g was excluded because of poor coverage.
References
- 1.Wang Y., Waters J., Leung M.L., Unruh A., Roh W., Shi X., Chen K., Scheet P., Vattathil S., Liang H., Multani A., Zhang H., Zhao R., Michor F., Meric-Bernstam F., Navin N.E. Clonal evolution in breast cancer revealed by single nucleus genome sequencing. Nature. 2014;512:155–160. doi: 10.1038/nature13600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alderton G.K. Genomics: one cell at a time. Nat Rev Cancer. 2011;11:312. doi: 10.1038/nrc3060. [DOI] [PubMed] [Google Scholar]
- 3.Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., Cook K., Stepansky A., Levy D., Esposito D., Muthuswamy L., Krasnitz A., McCombie W.R., Hicks J., Wigler M. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maheswaran S., Sequist L.V., Nagrath S., Ulkus L., Brannigan B., Collura C.V., Inserra E., Diederichs S., Iafrate A.J., Bell D.W., Digumarthy S., Muzikansky A., Irimia D., Settleman J., Tompkins R.G., Lynch T.J., Toner M., Haber D.A. Detection of mutations in EGFR in circulating lung-cancer cells. N Engl J Med. 2008;359:366–377. doi: 10.1056/NEJMoa0800668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Polzer B., Medoro G., Pasch S., Fontana F., Zorzino L., Andergassen U., Meier-stiegen F., Czyz Z.T., Alberter B., Schamberger T., Sergio M., Bregola G., Doffini A., Gianni S., Calanca A., Signorini G., Bolognesi C., Hartmann A., Fasching P.A., Maria T. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol Med. 2014;6:1371–1387. doi: 10.15252/emmm.201404033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bourcy CFa, De Vlaminck I., Kanbar J.N., Wang J., Gawad C., Quake S.R. A quantitative comparison of single-cell whole genome amplification methods. PLoS One. 2014;9:e105585. doi: 10.1371/journal.pone.0105585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou Y., Song L., Zhu P., Zhang B., Tao Y., Xu X. Single-cell exome sequencing and monoclonal evolution of a JAK2-negative myeloproliferative neoplasm. Cell. 2012;148:873–885. doi: 10.1016/j.cell.2012.02.028. [DOI] [PubMed] [Google Scholar]
- 8.Gawad C., Koh W., Quake S.R. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–188. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]
- 9.Deleye L., Tilleman L., Vander Plaetsen A.S., Cornelis S., Deforce D., Van Nieuwerburgh F. Performance of four modern whole genome amplification methods for copy number variant detection in single cells. Sci Rep. 2017;7:3422. doi: 10.1038/s41598-017-03711-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L., Ma F., Chapman A., Lu S., Xie X.S. Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet. 2015;16:79–102. doi: 10.1146/annurev-genom-090413-025352. [DOI] [PubMed] [Google Scholar]
- 11.Deleye L., Gansemans Y., De Coninck D., Van Nieuwerburgh F., Deforce D. Massively parallel sequencing of micro-manipulated cells targeting a comprehensive panel of disease-causing genes: a comparative evaluation of upstream whole-genome amplification methods. PLoS One. 2018;13:e0196334. doi: 10.1371/journal.pone.0196334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zong C., Lu S., Chapman A., Xie X. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012:1622–1627. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Szulwach K.E., Chen P., Wang X., Wang J., Weaver L.S., Gonzales M.L., Sun G., Unger M.A., Ramakrishnan R. Single-cell genetic analysis using automated microfluidics to resolve somatic mosaicism. PLoS One. 2015;10:e0135007. doi: 10.1371/journal.pone.0135007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Werner S.L., Graf R.P., Landers M., Valenta D.T., Schroeder M., Greene S.B., Bales N., Dittamore R., Marrinucci D. Analytical validation and capabilities of the epic CTC platform: enrichment-free circulating tumour cell detection and characterization. J Circ Biomark. 2015;4:3. doi: 10.5772/60725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beltran H., Jendrisak A., Landers M., Mosquera J.M., Kossai M., Louw J., Krupa R., Graf R.P., Schreiber N.A., Nanus D.M., Tagawa S.T., Marrinucci D., Dittamore R., Scher H.I. The initial detection and partial characterization of circulating tumor cells in neuroendocrine prostate cancer. Clin Cancer Res. 2016;22:1510–1519. doi: 10.1158/1078-0432.CCR-15-0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bourgon R., Lu S., Yan Y., Lackner M.R., Wang W., Weigman V., Wang D., Guan Y., Ryner L., Koeppen H., Patel R., Hampton G.M., Amler L.C., Wang Y. High-throughput detection of clinically relevant mutations in archived tumor samples by multiplexed PCR and next-generation sequencing. Clin Cancer Res. 2014;20:2080–2091. doi: 10.1158/1078-0432.CCR-13-3114. [DOI] [PubMed] [Google Scholar]
- 17.Li H. Aligning Sequence Reads, Clone Sequences and Assembly Contigs with BWA-MEM. [Epub] arXiv. 2013:1303.3997. [Google Scholar]
- 18.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tischler G., Leonard S. biobambam: tools for read pair collation based algorithms on BAM files. Source Code Biol Med. 2014;9:13. [Google Scholar]
- 20.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tarasov A., Vilella A.J., Cuppen E., Nijman I.J., Prins P. Sambamba: fast processing of NGS alignment formats. Bioinformatics. 2015;31:2032–2034. doi: 10.1093/bioinformatics/btv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lai D., Ha G. R Foundation for Statistical Computing; Vienna, Austria: 2016. HMMcopy: A Package for Bias-Free Copy Number Estimation and Robust CNA Detection in Tumour Samples from WGS HTS Data; p. 14. [Google Scholar]
- 23.Zeileis A. R Foundation for Statistical Computing; Vienna, Austria: 2014. ineq: Measuring Inequality, Concentration, and Poverty. [Google Scholar]
- 24.Garrison E., Marth G. 2012. Haplotype-Based Variant Detection from Short-Read Sequencing. Preprint. Posted July 20, 2012. arXiv 12073907 [q-bio] [Google Scholar]
- 25.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cingolani P., Platts A., Wang leL., Coon M., Nguyen T., Wang L., Land S.J., Lu X., Ruden D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Issadore D., Chung J., Shao H., Liong M., Ghazani A.A., Castro C.M., Weissleder R., Lee H. Ultrasensitive clinical enumeration of rare cells ex vivo using a micro-hall detector. Sci Transl Med. 2012;4:141ra192. doi: 10.1126/scitranslmed.3003747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heitzer E., Auer M., Ulz P., Geigl J.B., Speicher M.R. Circulating tumor cells and DNA as liquid biopsies. Genome Med. 2013;5:73. doi: 10.1186/gm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.De Luca F., Rotunno G., Salvianti F., Galardi F., Pestrin M., Gabellini S., Simi L., Mancini I., Vannucchi A.M., Pazzagli M., Di Leo A., Pinzani P. Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer. Oncotarget. 2016;7:26107–26119. doi: 10.18632/oncotarget.8431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw J.A., Guttery D.S., Hills A., Fernandez-Garcia D., Page K., Rosales B.M., Goddard K.S., Hastings R.K., Luo J., Ogle O., Woodley L., Ali S., Stebbing J., Coombes R.C. Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high circulating tumor cell counts. Clin Cancer Res. 2017;23:88–96. doi: 10.1158/1078-0432.CCR-16-0825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodgkinson C.L., Morrow C.J., Li Y., Metcalf R.L., Rothwell D.G., Trapani F., Polanski R., Burt D.J., Simpson K.L., Morris K., Pepper S.D., Nonaka D., Greystoke A., Kelly P., Bola B., Krebs M.G., Antonello J., Ayub M., Faulkner S., Priest L., Carter L., Tate C., Miller C.J., Blackhall F., Brady G., Dive C. Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer. Nat Med. 2014;20:897–903. doi: 10.1038/nm.3600. [DOI] [PubMed] [Google Scholar]
- 32.Ni X., Zhuo M., Su Z., Duan J., Gao Y., Wang Z., Zong C., Bai H., Chapman A.R., Zhao J., Xu L., An T., Ma Q., Wang Y., Wu M., Sun Y., Wang S., Li Z., Yang X., Yong J., Su X.D., Lu Y., Bai F., Xie X.S., Wang J. Reproducible copy number variation patterns among single circulating tumor cells of lung cancer patients. Proc Natl Acad Sci U S A. 2013;110:21083–21088. doi: 10.1073/pnas.1320659110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao Y., Ni X., Guo H., Su Z., Ba Y., Tong Z., Guo Z., Yao X., Chen X., Yin J., Yan Z., Guo L., Liu Y., Bai F., Xie X.S., Zhang N. Single-cell sequencing deciphers a convergent evolution of copy number alterations from primary to circulating tumor cells. Genome Res. 2017;27:1312–1322. doi: 10.1101/gr.216788.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su Z., Wang Z., Ni X., Duan J., Gao Y., Zhuo M., Li R., Zhao J., Ma Q., Bai H., Chen H., Wang S., Chen X., An T., Wang Y., Tian Y., Yu J., Wang D., Xie X.S., Bai F., Wang J. Inferring the evolution and progression of small-cell lung cancer by single-cell sequencing of circulating tumor cells. Clin Cancer Res. 2019;25:5049–5060. doi: 10.1158/1078-0432.CCR-18-3571. [DOI] [PubMed] [Google Scholar]
- 35.Chen D., Zhen H., Qiu Y., Liu P., Zeng P., Xia J., Shi Q., Xie L., Zhu Z., Gao Y., Huang G., Wang J., Yang H., Chen F. Comparison of single cell sequencing data between two whole genome amplification methods on two sequencing platforms. Sci Rep. 2018;8:4963. doi: 10.1038/s41598-018-23325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X., Chang C.W., Spoerke J.M., Yoh K.E., Kapoor V., Baudo C., Aimi J., Yu M., Liang-Chu M.M.Y., Suttmann R., Huw L.Y., Gendreau S., Cummings C., Lackner M.R. Low-pass whole-genome sequencing of circulating cell-free DNA demonstrates dynamic changes in genomic copy number in a squamous lung cancer clinical cohort. Clin Cancer Res. 2019;25:2254–2263. doi: 10.1158/1078-0432.CCR-18-1593. [DOI] [PubMed] [Google Scholar]
- 37.Miyamoto D.T., Lee R.J., Stott S.L., Ting D.T., Wittner B.S., Ulman M., Smas M.E., Lord J.B., Brannigan B.W., Trautwein J., Bander N.H., Wu C.L., Sequist L.V., Smith M.R., Ramaswamy S., Toner M., Maheswaran S., Haber D.A. Androgen receptor signaling in circulating tumor cells as a marker of hormonally responsive prostate cancer. Cancer Discov. 2012;2:995–1003. doi: 10.1158/2159-8290.CD-12-0222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.