Abstract
The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), results in life-threatening disease in a minority of patients, especially elderly people and those with co-morbidities such as obesity and diabetes. Severe disease is characterized by dysregulated cytokine release, pneumonia and acute lung injury, which can rapidly progress to acute respiratory distress syndrome, disseminated intravascular coagulation, multisystem failure and death. However, a mechanistic understanding of COVID-19 progression remains unclear. Here we review evidence that SARS-CoV-2 directly or indirectly activates inflammasomes, which are large multiprotein assemblies that are broadly responsive to pathogen-associated and stress-associated cellular insults, leading to secretion of the pleiotropic IL-1 family cytokines (IL-1β and IL-18), and pyroptosis, an inflammatory form of cell death. We further discuss potential mechanisms of inflammasome activation and clinical efforts currently under way to suppress inflammation to prevent or ameliorate severe COVID-19.
Subject terms: Inflammasome, Cell death and immune response
This Progress article brings us up to date on the role of inflammasomes in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and COVID-19, describing how they may be activated during infection and contribute to the overexuberant inflammatory response in severe disease, and the efforts being taken to target them therapeutically.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus responsible for COVID-19, has so far infected more than 190 million people and caused death of more than 4.1 million people worldwide. The virus primarily infects the respiratory tract, causing fever, sore throat, anosmia and dyspnoea, but its tissue tropism still remains to be fully understood. As many as 10–15% of patients develop severe pneumonia, with some cases progressing to hypoxia and acute respiratory distress syndrome (ARDS), which requires mechanical ventilation in a critical care setting and has high mortality. Patients can also develop multi-organ failure, acute kidney injury and disseminated intravascular coagulation, among a host of other disorders1–11. Aside from supportive care, only a few treatments have been approved for COVID-19, and their reduction of mortality has been limited12–14. Although several vaccines against SARS-CoV-2 have been approved and are being administered internationally, there will still be a significant number of infections owing to people who are not vaccinated in regions with inadequate access or acceptance of vaccination. In addition, while global vaccination efforts strive to meet the challenge of ending the pandemic, the appearance of immune-evasive viral variants and the unlikelihood of reaching immediate herd immunity underscore the continued need for additional treatments mitigating disease progression15–19.
Most researchers agree that an inappropriate hyperinflammatory response lies at the root of many severe cases of COVID-19, driven by overexuberant inflammatory cytokine release. Consistently, co-morbidities, such as obesity, diabetes, heart disease, hypertension and ageing, which are prognostic of poor outcome, are associated with high basal inflammation7,11,20,21. It has been proposed since the beginning of the pandemic that these co-morbidities and the ensuing hyperinflammatory response may be aetiologically linked through overactive inflammasome signalling, which may account for the association of these co-morbidities with severe COVID-19 in the context of chronic inflammation as well as for COVID-19 progression in the context of a robust acute inflammatory response to infection22–29. However, many of the studies that seek to understand the immune response to SARS-CoV-2 are based on RNA sequencing, often of thawed cells, and infected, activated or dying cells do not survive freeze–thaw well, which could skew results. Moreover, inflammasome activation does not directly induce transcriptional responses, and its detection is less straightforward than that of most other signalling pathways. Nonetheless, several studies are now accumulating that support direct (infection-induced) and indirect inflammasome activation and the critical role of inflammasomes in severe COVID-19. Here we discuss the available evidence, potential mechanisms and the implications for therapy.
Inflammasomes
Key to inflammation and innate immunity, inflammasomes are large, micrometre-scale multiprotein cytosolic complexes that assemble in response to pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) and trigger proinflammatory cytokine release as well as pyroptosis, a proinflammatory lytic cell death30,31 (Fig. 1). Upon activation by PAMPs or DAMPs, canonical inflammasome sensors — mainly in monocytes, macrophages and barrier epithelial cells — oligomerize and recruit the adaptor apoptosis-associated speck-like protein containing a CARD (ASC) to form inflammasome specks, within which the inflammatory caspase 1 is recruited and activated. Inflammasome sensors are activated in response to different triggers and differ in their overall specificities to PAMPs or DAMPs. NLRP3, the most broadly activated inflammasome sensor and a member of the nucleotide-binding domain- and leucine-rich repeat-containing protein (NLR) family, responds to an array of insults to the cell that cause cytosolic K+ efflux, Ca2+ cytosolic influx or release of mitochondrial reactive oxygen species (ROS)31,32. These insults include extracellular ATP, membrane permeabilization by pore-forming toxins and large extracellular aggregates such as uric acid crystals, cholesterol crystals and amyloids30. Other sensors, such as AIM2 and NLRC4, are tuned to recognize specific PAMPs and DAMPs, such as cytosolic double-stranded DNA and bacterial proteins, respectively31. In a parallel pathway, the mouse inflammatory caspase 11 and human caspase 4 and caspase 5 sense PAMPs and DAMPs such as bacterial lipopolysaccharide (LPS) that gain cytosolic access and endogenous oxidized phospholipids, leading directly to membrane damage or pyroptosis, and secondary K+ efflux followed by noncanonical NLRP3 inflammasome activation33–36.
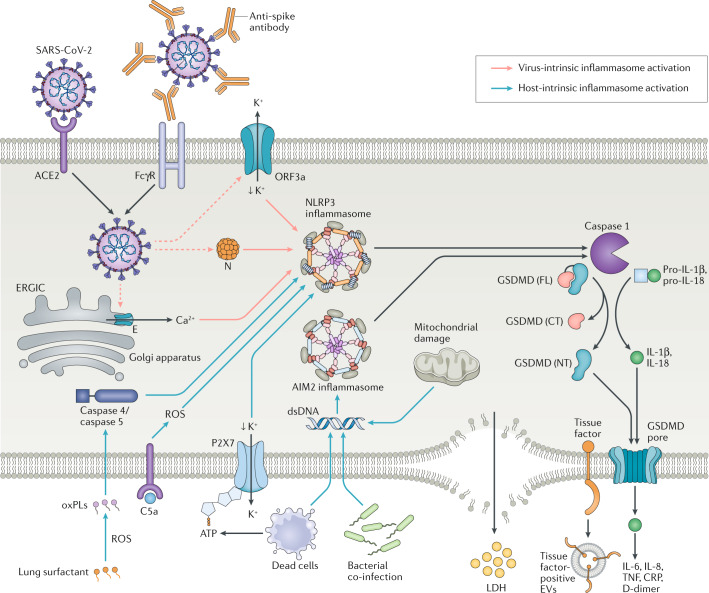
Fig. 1. Virus-intrinsic and host-intrinsic mechanisms of inflammasome activation.
Virus intrinsic mechanisms (red arrows): severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virions enter epithelial cells via angiotensin-converting enzyme 2 (ACE2) and can enter monocytes by binding to anti-spike antibodies followed by Fc receptor for IgG (FcγR)-mediated internalization. Upon translation of the viral genome, the viroporins ORF3a and E can trigger K+ efflux or Ca2+ influx to promote NLRP3 activation. Viral N protein can bind directly to NLRP3, resulting in its activation. Host-intrinsic mechanisms (blue arrows): oxidation of lung surfactant phospholipids results in oxidized phospholipids (oxPLs), which can activate caspase 4 and/or caspase 5 to promote noncanonical inflammasome activation. Complement products such as C5a can activate NLRP3 by promoting accumulation of reactive oxygen species (ROS). ATP released by dead cells binds to P2X7 receptor, which causes K+ efflux and NLRP3 activation. Dead cells, bacterial co-infection or damaged mitochondria can result in cytosolic double-stranded DNA (dsDNA), which activates the AIM2 inflammasome. NLRP3 and AIM2 inflammasome assembly activates caspase 1, which cleaves full-length (FL) gasdermin D (GSDMD) into amino-terminal (NT) and carboxy-terminal (CT) fragments. The GSDMD NT fragment binds to the plasma membrane, oligomerizes and inserts itself as a pore. Caspase 1 also cleaves pro-IL-1β and pro-IL-18 into their mature forms, which are released through the GSDMD pore. IL-1β can activate macrophages to secrete additional proinflammatory cytokines such as IL-6. Pyroptosis results after further membrane damage, which releases lactate dehydrogenase (LDH) and is associated with the formation of tissue factor-enriched extracellular vesicles (EVs). CRP, C-reactive protein; ERGIC, endoplasmic reticulum–Golgi intermediate compartment; TNF, tumour necrosis factor.
Upon activation, caspase 1 processes pro-IL-1β and pro-ΙL-18 into their functional cytokine forms, and all inflammatory caspases cleave the pyroptotic executor protein gasdermin D (GSDMD) into amino-terminal and carboxy-terminal fragments. The GSDMD amino-terminal fragment binds to acidic lipids, oligomerizes and inserts itself into cell and organelle membranes to form sizable pores37–41. These pores are large enough to directly release IL-1β and ΙL-18, strong pleiotropic inducers of downstream immune responses, as well as various alarmins, such as ATP and high mobility group protein B1 (HMGB1). Pyroptosis occurs when the plasma membrane is compromised, resulting in the release of larger alarmins such as lactate dehydrogenase (LDH) tetramer (144 kDa), which is a feature pathognomonic for pyroptosis and other necrotic cell deaths that cause cell membranes to rupture42–44. Although plasma membrane rupture during pyroptosis was originally thought to be a passive mechanism driven by oncotic pressure and osmotic lysis following GSDMD pore formation, a recent discovery reveals that it is an active process that depends on ninjurin 1, a small transmembrane protein that oligomerizes following GSDMD activation44.
The regime of inflammasome activation followed by pyroptotic cell death can differ depending on cell type and stimulus. For example, in response to oxidized phospholipids produced in inflamed lung tissue, the IL-1 family cytokines IL-1β and IL-18 can be secreted continuously without pyroptotic cell death, a phenomenon called ‘hyperactivation’ that has been observed in macrophages, dendritic cells and neutrophils33,42,43. Sustained IL-1β and IL-18 production in surviving cells enhances proinflammatory cytokine release and may contribute to macrophage activation syndrome42.
Demonstration of inflammasome activation
LDH release and pyroptosis
Early suggestions of inflammasome activation in COVID-19 came from studies that revealed serum LDH concentration as the strongest single predictor among multiple serum factors of severe disease regardless of the criteria (acute physiology and chronic health evaluation II (APACHE II) mortality prediction score, sequential organ failure assessment score or pneumonia severity index)45 (Fig. 1). This correlation was later validated in other patient cohorts46–52.
Elevated LDH concentration is a general indicator of tissue damage, which is supported by widespread cell death observed among monocytes, alveolar epithelial cells and endothelial cells of the lungs and kidneys53,54. All of these cell types are competent to activate inflammasomes and undergo pyroptosis55–57. Interestingly, monocytes from patients with COVID-19 showed no increase in staining for annexin V, yet about ~10% took up small dyes, a sign of disrupted plasma membranes, which is a pattern indicative of pyroptosis or other forms of programmed necrosis58.
Inflammasome-directed cytokine profile
Extensive characterization of serum cytokines in COVID-19 has highlighted an overabundance of chemokines such as CXC-chemokine ligand 8 (CXCL8; also known as IL-8) and proinflammatory cytokines such as IL-6 and tumour necrosis factor (TNF) throughout the disease course59–61. This proinflammatory cytokine response is accompanied by serum markers of inflammation such as the IL-6-inducible hepatic factors C-reactive protein and ferritin, and the associated elevation of the concentration of the coagulation product D-dimer, which are all associated with poor prognosis62,63. These cytokines are common serum markers of severe inflammation that can be strongly induced by acute-phase IL-1β (Fig. 1), which is also consistent with the previously recognized role of IL-1β in IL-6 production64. However, they are not pathognomonic for inflammasome activation and can be induced by other inflammatory pathways, including those stimulated by nuclear factor-κB. Support for the hypothesis that IL-1β acts as a pleiotropic proinflammatory cytokine to unleash the inflammatory response is provided by the in vitro observation that exogenous IL-1 receptor antagonist (IL-1RA) completely abolishes IL-6 and TNF secretion in SARS-CoV-2-infected primary monocytes65. Another strong indication of inflammasome activation in COVID-19 is the fact that IL-18, the processing and secretion of which depend on inflammasome and GSDMD activation, is associated with severe COVID-19 and has emerged as a highly predictive biomarker of death52,58,61,66,67.
Direct measurements of serum IL-1β concentration have been more equivocal. An initial report on a small cohort of patients exposed in the Wuhan Huanan Seafood Market, the site of putative early SARS-CoV-2 transmission, measured markedly increased serum IL-1β and IL-1RA levels compared with levels in healthy adults8. However, IL-1β level was not shown to be further elevated in severe versus moderate cases, and concurrent studies differed in measuring a statistically significant increase in serum IL-1β level58–61,68,69. This is perhaps not surprising given the extremely short half-life of IL-1β68,70, which is often not detected in other diseases in humans that are clearly driven by IL-1 (refs71,72). Because of this problem, plasma levels of IL-1RA are often used as a surrogate for elevated IL-1β levels, and the level of IL-1β-inducible IL-1RA is consistently associated with severe COVID-19 (refs61,66,67). Moreover, the immunopathological activity of IL-1β in COVID-19 could also be restricted to local secretion and paracrine signalling20,59,70. Indeed, measurements in bronchoalveolar lavage fluid (BALF) — a readout more reflective of the lung microenvironment with massive monocyte, macrophage and neutrophil infiltration — showed a significant IL-1β level increase in patients with moderate to severe COVID-19 (refs73,74). Post-mortem histological sections from lung parenchyma also showed broadly elevated staining of IL-1β compared with control sections75,76, although one study attributed the elevated IL-1β staining to processing by caspase 8 and release by necroptosis rather than by inflammasome activation75. Interestingly, apoptotic caspases such as caspase 3 or caspase 8 can convert immunologically silent apoptotic cell death to immunogenic pyroptosis involving cytokine release by cleaving and activating gasdermin E (GSDME) and GSDMD, respectively77–80.
Detection of inflammasome specks and GSDMD amino-terminal processing and release
Recently, direct evidence of inflammasome activation in COVID-19 was provided by multiple studies that showed SARS-CoV-2 infection (detected by the presence of SARS-CoV-2 nucleocapsid protein or double-stranded RNA), inflammasome activation and pyroptosis in about 10% of blood monocytes from patients, but the percentage differs between patients and studies52,58,65,81. Most of these infected monocytes displayed a characteristic inflammasome speck, which stained for NLRP3, ASC and active caspase 1, and was accompanied by punctate relocalization of GSDMD to the membrane and pyroptotic cellular morphology52,58. Cleaved GSDMD was also detected in lung tissue sections as well as in BALF and plasma of patients with COVID-19, suggesting active pore formation and subsequent release of GSDMD fragments by pyroptotic cells58,76. Thus, pyroptotic monocytes fully activate all inflammasome pathway components. Interestingly, one preprint shows that monocytes harboured specks that were positive for both NLRP3 and AIM2 and thus activated multiple inflammasome sensors simultaneously58, a phenomenon that has been observed but is incompletely understood82. The involvement of AIM2 is also unexpected given its characterization as a double-stranded DNA sensor; however, AIM2 has been shown to participate in the lung injury and death caused by another RNA virus, influenza virus83. Secondary infection by bacteria is common in respiratory viral infections and is implicated in COVID-19. Plasma samples from patients with severe COVID-19 contain LPS and bacterial DNA; LPS boosts the activation of the NLRP3 and AIM2 inflammasomes84, and bacterial, host mitochondrial or nuclear DNA that gained access to the cytosol could trigger AIM2 activation84–87. Future studies should address the potential involvement of other inflammasome sensors in COVID-19 and, more broadly, elucidate the mechanisms behind their co-activation and combinatorial function. A role for GSDMD in COVID-19 pathogenesis is also supported by a preprint showing significant association of expression quantitative trait loci (eQTL) for increased GSDMD expression with severe COVID-19 (ref.58).
Other cell types in the lung
The fact that monocytes release proinflammatory cytokines and undergo pyroptosis by SARS-CoV-2 infection suggests they may be important drivers of cytokine release and severe COVID-19. It is less clear, however, whether other cell types in lung tissue such as alveolar epithelium, tissue macrophages and endothelial cells undergo pyroptosis. Histological analysis of COVID-19 lung samples has shown GSDMD expression in alveolar epithelium as well as NLRP3 and caspase 1 expression in lung vascular tissue88–90. Induction of inflammasome pathway components is also supported by in vitro experiments showing that SARS-CoV-2 infection of primary human bronchiolar epithelial cells upregulated IL1B expression91. Whether these components are actually activated in the lung or other involved tissues and how strongly they contribute to local and systemic inflammation remain to be carefully shown. Single-cell RNA analysis of BALF showed depletion of alveolar macrophages in COVID-19. These cells were shown to be infected by SARS-CoV-2 in vitro and in autopsy samples, although their expression of the virus entry receptor angiotensin-converting enzyme 2 (ACE2) and the mechanism of viral entry are unclear92–96. As alveolar macrophages activate inflammasomes during acute lung injury in mice, it will be important to determine whether this also occurs after SARS-CoV-2 infection through direct or indirect mechanisms. Mouse studies that deplete alveolar macrophages before SARS-CoV-2 infection could address their relevance to lung hyperinflammation in COVID-19 (ref.97).
IL-1β amplification of inflammation
Inflammasome activation can be massively amplified by positive feedback loops to result in uncontrolled overactivation and cytokine storm. IL-1β binding to IL-1R triggers a nuclear factor-κB response to increase transcription of pro-IL-1β, which can occur in myeloid cells recruited to the lung28. Single-cell RNA sequencing studies of patients with severe COVID-19 detected expanded peripheral CD14+IL-1β+ monocytes/macrophages and increased IL1B-expressing monocyte/macrophage populations in BALF, which also express IL-1-inducible chemokines69,73,98–100. Thus, cytokine release and subsequent immune cell recruitment can be serially repeated through positive feedback loops, culminating in the formation of a hyperinflammatory microenvironment carrying a high burden of cytokine-secreting monocytes and neutrophils associated with severe disease. This dramatic immune cell infiltration and cytokine release in lung tissue is a hallmark of ARDS and is thought to contribute to severe lung damage in patients with COVID-19.
Proinflammatory cytokines whose levels are elevated in COVID-19 may also participate in positive feedforward loops to exacerbate lung injury. For example, IL-1β-mediated activation of endothelial cells can downregulate vascular endothelial cadherin (VE-cadherin) transcription, resulting in loss of adherens junctions that are critical to barrier integrity101. Meanwhile, IL-1β-induced IL-6 secretion can increase production of vascular endothelial growth factor, which in turn weakens the pulmonary endothelium through VE-cadherin internalization102. These events can promote accumulation of interstitial and alveolar fluid that compromises gas exchange10,103. Alveolar fluid accumulation — which can be especially devastating given the loss of resorptive activity from infected alveolar epithelium — can disrupt pulmonary surfactant, leading to increased alveolar surface tension and collapse10,104,105. Thus, feedback-amplified immune cell recruitment and cascading feedforward tissue damage could synergistically exacerbate lung injury in response to early unrestrained IL-1-directed proinflammatory cytokine release.
Virus-intrinsic mechanisms
The NLRP3 inflammasome is known to be activated by highly pathogenic coronaviruses, such as severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and mouse hepatitis virus27,106,107. NLRP3 is expressed in immune cells of both myeloid and lymphoid origin, but also in alveolar epithelial and pulmonary endothelial cells, where its overactivation can contribute to lung injury30,108–110. One mechanism of NLRP3 inflammasome activation involves K+ efflux or Ca2+ influx induced by ion-conductive transmembrane viral proteins or ‘viroporins’ (for example, ORF3a and envelope protein E) (Fig. 1). The SARS-CoV ORF3a, which is not encoded by avirulent coronaviruses, activates NLRP3 when overexpressed in monocytic cells or macrophages and has been linked to viral pathogenesis111,112. The SARS-CoV E protein embeds in the endoplasmic reticulum–Golgi intermediate compartment (ERGIC) membranes and causes cytosolic Ca2+ influx to activate NLRP3 in reconstituted Vero epithelial cells113,114. Another mechanism of NLRP3 inflammasome activation involves direct interactions with viral proteins. ORF3a can interact with the inflammasome adaptor ASC, resulting in its polyubiquitylation and speck formation112,115. The SARS-CoV protein ORF8b, generated de novo by a 29-nucleotide deletion in the mutation hotspot of ORF8 during the SARS-CoV outbreak116, can also directly activate the NLRP3 inflammasome by binding the leucine-rich repeat region of NLRP3 to promote oligomerization and form insoluble aggregates that colocalize with NLRP3 and ASC117. Importantly, a study in vivo using recombinant mouse-adapted SARS-CoVs showed that single amino acid mutations of the E protein that suppressed ion conductivity reduced proinflammatory activity of the virus, resulting in disease recovery and mouse survival113,114.
SARS-CoV-2 E protein and ORF3a have 95% and 72% amino acid identity, respectively, with E protein and ORF3a of SARS-CoV, suggesting functional conservation27, while ORF8b was likely not conserved. Indeed, one preprint reported overexpression of SARS-CoV-2 ORF3a induced K+ efflux, NLRP3 activation and IL-1β release in the lung epithelial cell line A549 (ref.118). SARS-CoV-2 E protein functions as a K+-permeable cation channel, which lyses cells when overexpressed and induces ARDS-like pathology in mice119. Beyond these mechanisms, it is conceivable that SARS-CoV-2 activates inflammasomes by novel processes not known to occur by SARS-CoV. This possibility is supported by a recent preprint showing SARS-CoV-2 nucleocapsid protein can directly bind NLRP3 to induce inflammasome assembly and IL-1β release in macrophages and dendritic cells120 (Fig. 1). Many of these reports relied on overexpression of SARS-CoV-2 proteins in cell lines, and in vivo infection models are needed to confirm direct SARS-CoV-2-induced inflammasome activation in COVID-19.
Host-intrinsic mechanisms
Infection of blood monocytes with SARS-CoV-2 is surprising given the lack of or weak ACE2 expression by monocytes121, and suggests involvement of alternative viral entry mechanisms20,58. Infection of healthy donor monocytes in vitro was markedly enhanced by anti-spike antibodies or patient convalescent plasma, suggesting uptake of opsonized viral particles by monocytes through antibody-dependent phagocytosis58. Indeed, antibody depletion of patient plasma and blocking antibodies to the monocyte Fc receptor FcγRIIIa (also known as CD16) largely abrogated monocyte infection. While this finding contradicts the finding of another study122, it is supported by the observation of increased afucosylated antiviral antibodies seen in patients with severe COVID-19 (refs123,124). The Fc domain of IgG antibodies contains a conserved N-linked glycan, and IgG lacking core fucosylation at this glycan has higher affinity for FcγRIIIa. Thus, afucosylation may initiate enhanced antibody-dependent engulfment, resulting in monocyte infection and inflammasome activation. Regardless of the viral entry route, the fact that monocytes are major contributors of proinflammatory cytokines and undergo pyroptosis following SARS-CoV-2 infection suggests that they may be important drivers of severe inflammation in COVID-19.
In addition to direct infection-mediated inflammasome activation by SARS-CoV-2, it is also plausible that inflammasomes are indirectly activated in COVID-19 (Fig. 1). Through various mechanisms, including host translation inhibition, SARS-CoV-2 suppresses an early type I interferon response to allow uncontrolled viral replication125–127. Lysis of infected lung cells could secrete alarmins to activate inflammatory macrophages and promote their recruitment to the lung128. Supernatants from Vero E6 cells lysed by overexpression of SARS-CoV-2 E protein can induce IL-1β secretion by recipient macrophages119, likely through NLRP3, but also possibly other inflammasome sensors, such as AIM2 and NLRC4 (ref.58). Interestingly, eQTL associated with increased expression of NLRC4 and NLRP3 were correlated with severe COVID-19, suggesting possible involvement of multiple inflammasome sensors in the disease58.
Host-intrinsic mechanisms activating inflammasomes in COVID-19 may also involve innate or adaptive lymphocytes. Cytotoxic T lymphocytes and natural killer cells can induce pyroptosis in infected cells through the delivery of granzyme A, which cleaves gasdermin B (GSDMB) or granzyme B, which cleaves GSDME, resulting in pore formation77,129. Given the high expression of GSDMB in the airway epithelium, cytotoxic lymphocyte-mediated killing of infected cells could contribute to cytokine and DAMP release during lung infection129,130. In addition, the pathogen-induced cleaved complement product C5a, which is highly abundant in BALF of patients with severe COVID-19, can facilitate IL-1β release in MERS-CoV-infected mice and can also trigger NLRP3 activation via a ROS-dependent mechanism in CD4+ T cells109,131,132.
Another, perhaps more pathologically relevant, ROS-dependent mechanism involves oxidation of the phospholipid-rich surfactant normally secreted by alveolar epithelium, which aberrantly generates oxidized phospholipids, potent inducers of ARDS in mouse infection models and of the noncanonical inflammasome (human caspase 4/5 or mouse caspase 11) in dendritic cells and macrophages33,133,134. Together, these phenomena indicate a complex response to DAMPs, complement products and local alveolar fluid to produce a lung microenvironment favouring inflammasome activation, resulting in inflammatory cytokine and alarmin release, hyperactivation of lung epithelial cells and tissue-resident macrophages, tissue insult via pyroptosis and subsequent proinflammatory recruitment of peripheral immune cells.
Thus, through direct, cell-autonomous activation by SARS-CoV-2 infection and/or indirect activation by DAMPs, inflammasome sensors such as NLRP3 could serve as key pleiotropic drivers of COVID-19 pathogenesis. Compelling evidence supporting this idea also comes from bats, which asymptomatically coexist with coronaviruses that are highly pathogenic to humans. Bat peripheral blood mononuclear cells express a hypomorphic isoform of NLRP3 and thus fail to release IL-1β after MERS-CoV infection; however, this function is restored by expression of human NLRP3, suggesting that NLRP3 suppression could be a potent strategy for reducing coronavirus pathogenicity106.
GSDMD, NETosis and coagulopathy
As in SARS-CoV and H1N1 influenza virus infections, SARS-CoV-2 can cause inflammation-directed coagulopathy, which may contribute to the high rates of venous and arterial thrombosis and the high mortality in COVID-19 (Fig. 2). Coagulopathy is marked by increased levels of plasma clotting markers, such as D-dimer, factor VIII and von Willebrand factor, and by abnormal prothrombin time9,135–137. Although proinflammatory cytokines are known to promote various procoagulation factors, they may not be sufficient for the dramatic systemic clot formation seen in some patients with severe COVID-19. GSDMD activation by inflammasomes in monocytes or neutrophils could also promote coagulation through multiple mechanisms. Inflammasome activation and pyroptosis can trigger massive coagulopathy in a cytokine-independent manner. In a mouse model of endotoxaemia, activation of inflammasome signalling via caspase 1 or caspase 11 resulted in systemic fibrin deposition reminiscent of disseminated intravascular coagulation, a phenomenon that depended on GSDMD but not IL-1β or IL-18 (refs138,139). GSDMD-dependent lysis of pyroptotic macrophages releases extracellular vesicles enriched in membrane-bound tissue factor, a transmembrane glycoprotein that circulates in the bloodstream and promotes coagulation by activating prothrombin138,140. Plasma extracellular vesicles from patients with COVID-19 showed increased tissue factor activity, which also correlated with disease severity140. Inflammasome-induced coagulopathy can also involve phosphatidylserine-mediated activation of tissue factor. Phosphatidylserine is ordinarily confined to the inner membrane leaflet; however, Ca2+ entry through GSDMD pores activates the phospholipid scramblase transmembrane protein 16F (TMEM16F), which externalizes phosphatidylserine to enhance tissue factor activity141.
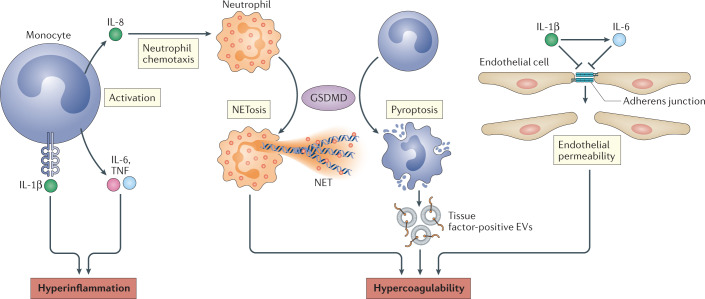
Fig. 2. Mechanisms of inflammasome-driven COVID-19.
IL-1β released by inflammasome signalling activates monocytes, which secrete IL-6, tumour necrosis factor (TNF) and IL-8. These cytokines cause inflammation through various mechanisms, including recruitment of neutrophils to the lung. Gasdermin D (GSDMD) activation in neutrophils leads to the formation of neutrophil extracellular traps (NETs), which can recruit platelets and promote hypercoagulability. IL-1β and IL-6 can downregulate adherens junctions in endothelial cells, which increases their permeability and could contribute to coagulation in the lung vasculature. Tissue factor-positive extracellular vesicles (EVs) released by pyroptotic monocytes can also directly activate the clotting cascade and promote coagulation in COVID-19.
In neutrophils, GSDMD is required to form neutrophil extracellular traps (NETs), which are extruded fibrous assemblages of DNA decorated with antimicrobial proteins142–144. NETs promote coagulability when dysregulated during viral infections, which is of particular interest given massive neutrophil infiltration of pulmonary capillaries in COVID-19 (refs145,146). The circulating neutrophils in the blood of patients with COVID-19 show increased basal activation of NETs, and NETs are also highly abundant in the lungs147–149. NETs recruit platelets and incorporate them into microthrombi. Thus, GSDMD activation of coagulation and NETosis could become dysregulated during inflammation in COVID-19 to contribute to the disseminated intravascular coagulation seen in severe disease. The possibility that inflammasome activation drives both severe inflammation and coagulation in COVID-19 suggests that inhibiting it could be beneficial.
Clinical interventions
Inflammasome activation and IL-1β activity are dysregulated in many chronic inflammatory conditions and have been successfully targeted by drugs, many of which are currently being repurposed as treatments for COVID-19 (refs28,150). Anakinra, a recombinant and slightly modified version of human IL-1RA, demonstrated early promise in small cohort studies, but clinical trials have met with mixed results28,150–156. One trial was stopped early due to lack of a significant reduction in ventilation requirement and mortality155. It is worth noting that this study recruited only patients with mild to moderate disease with a marginally elevated C-reactive protein level cut-off and not requiring admission to the intensive care unit, and may have excluded individuals who would be most responsive to anti-IL-1 therapy155. Similar considerations have been raised for other immunomodulators, such as dexamethasone, for which reduction in mortality was restricted to individuals receiving respiratory support14,157. Another trial used levels of soluble urokinase plasminogen activator receptor, a strong early predictor of severe COVID-19, for eligibility, and found that daily administration of anakinra significantly reduced progression to severe respiratory failure (hazard ratio 0.3) and 30-day mortality (hazard ratio 0.49)156,158. However, as this was a single-arm study, placebo-controlled randomized trials are needed. The mixed success of anti-IL-1 intervention could indicate that IL-1-independent effects of pyroptosis contribute to disease progression, which are beyond the reach of single cytokine modulation.
Multiple phase II clinical trials are testing direct NLRP3 inhibition in patients with either mild or severe COVID-19 (Novartis, NCT04382053; Olatec Therapeutics, NCT04540120) (Table 1). In animal models, NLRP3 inhibition reduces cytokine release and lung inflammation in influenza A virus infection159, and the NLRP3 inhibitor MCC950 reduced caspase 1 activation and secretion of IL-1β in primary human monocytes infected with SARS-CoV-2 in vitro52. Another inhibitor of NLRP3, the sulfonylurea diabetes drug glyburide, reduced IL-6 secretion by SARS-CoV-2-infected human monocytes65. These data suggest that NLRP3 inhibition may be a superior strategy against COVID-19 in comparison with IL-1 inhibition. Indirect NLRP3 inhibition is also being tested by administration of the microtubule-depolymerizing drug colchicine, which among its other effects is a well-tolerated inhibitor of inflammasome formation160,161. A small randomized controlled trial in patients with moderate to severe disease demonstrated its ability to reduce oxygen requirement and hospitalization; in this study, low overall mortality precluded a determination of its effect on death162. Another study showed that patients with COVID-19 treated with colchicine had a significantly decreased rate of hospitalization or death combined, although neither the rate of hospitalization nor the rate of death was significantly reduced on its own and the study has not yet been peer reviewed163. The diabetes drug metformin indirectly inhibits NLRP3 by regulating mTOR, and its use is significantly associated with reduced mortality among patients with COVID-19 and type 2 diabetes. Metformin was also recently shown to inhibit NLRP3 activation and lung inflammation in SARS-CoV-2-infected mice164,165.
Table 1.
Ongoing and planned clinical trials of inflammasome pathway inhibition in COVID-19
| Trial | Type, size and inclusion | Intervention and end points |
|---|---|---|
| Targeting NLRP3 | ||
| Novartis DFV890 (NCT04382053) | Multicentre, randomized, controlled; 143 participants | Administered for 14 days in addition to standard of care; day 15 APACHE II severity or day of discharge |
| Olatec Therapeutics Dapansutrile (NCT04540120) | Randomized, double blind, placebo controlled; 80 participants | Initial dose of 8 × 250 mg on first day; 4 × 250-mg capsules for 14 days; proportion of participants with clinical deterioration at day 15, defined as COVID-19-related hospitalization or both worsening shortness of breath and oxygen saturation less than 92% on room air |
| Targeting gasdermin D | ||
| University of California, San Francisco Disulfiram for COVID-19 (DISCO) trial (NCT04485130) | Randomized, dose escalation, placebo controlled, double blind; 60 participants | 1,000 mg per day for 5 days or 2,000 mg per day for 5 days; plasma levels of IL-1β, IL-6 and other cytokines at 31 days |
| ETICA Disulfiram (NCT04594343) | Randomized, double blind, placebo controlled; 200 participants | 500 mg per day for 14 days; plasma IL-18 level at day 7 |
| University of Oxford RECOVERY trial Dimethyl fumarate (NCT04381936) | Randomized, open label | 120 mg every 12 hours for 2 days and 240 mg every 12 hours for 8 days; mortality 28 days after randomization |
APACHE II, acute physiology and chronic health evaluation II.
GSDMD is another attractive target as pore formation occurs downstream of all inflammasome sensors but upstream of IL-1β and DAMP release166. Two known small-molecule drugs have recently been described to inhibit GSDMD. Disulfiram (DSF), a drug approved by the US Food and Drug Administration (FDA) for alcohol dependence used for 70 years, inhibits GSDMD pore formation by covalently modifying Cys191 and protects against LPS-induced sepsis and inflammatory cytokine secretion in mice167. Dimethyl fumarate (DMF), an FDA-approved therapeutic for multiple sclerosis, as well as fumarate, the endogenous Krebs cycle intermediate related to DMF, was recently shown to inhibit GSDMD168. Interestingly, a case series of several patients with multiple sclerosis being treated with DMF showed that their SARS-CoV-2 infection was self-limiting without their receiving any specific treatment169, and an observational study using a large health-care database revealed a significantly reduced risk of SARS-CoV-2 infection and no death in the group using DSF for alcoholism compared with a control group, but this study has not been fully peer reviewed170. DSF is currently being studied in two controlled phase II clinical trials that are testing efficacy in mild to moderate disease (NCT04485130) as well as in patients who are hospitalized (NCT04594343) (Table 1). Similarly, DMF is being tested in a randomized open-label trial for its effect in preventing death in patients with COVID-19 (NCT04381936) (Table 1). Both DSF and DMF may also confer beneficial effects in treating COVID-19 through their antiviral activities. DSF can weakly inhibit the SARS-CoV-2 MPro polyprotein protease171. Patients with COVID-19 showed suppressed nuclear factor erythroid 2-related factor 2 (NRF2) antioxidant gene expression. The NRF2 agonists 4-octyl itaconate (a cell-permeable itaconate derivative) and DMF both activate an NRF2-dependent antiviral programme to inhibit replication of SARS-CoV-2 in lung epithelial cell lines172,173.
An important issue when one is considering anti-inflammatory drugs is when is the best time to intervene. Early on, inflammation promotes the development of protective adaptive immunity by recruiting immune cells to the site of infection and activating their expansion and protective functions, and intervening too early may interfere with the development of effective immunity. Later on, the feedforward mechanisms take over, and interventions may not be able to control the explosive amplification of inflammation and its dangerous consequences. Therefore, the ‘sweet spot’ may be when signs of pulmonary distress are just beginning to become apparent (for example, a slight decrease in blood oxygenation, mild dyspnoea and early evidence of pneumonia on chest X-ray). The caveat is that most patients with mild symptoms do not go on to develop severe disease, and detecting an improvement in severe disease end points would require large numbers of study participants. Overall, the timing of treatment and proper patient stratification will be important parameters for identifying whether inflammasome-inhibiting interventions and anti-inflammatory drugs in general are beneficial.
Concluding remarks
Recent data support inflammasome involvement in severe COVID-19, through either direct infection-mediated activation or indirect DAMP-mediated activation. A protective role of inflammasome signalling and IL-1β release has been demonstrated against multiple pathogens, particularly in the acute phase of the infection174–176. However, its prolonged late-stage activation may underlie immunopathology, including overexuberant cytokine release, damage to pulmonary endothelium accompanied by infiltration of immune cells and systemic hypercoagulability. Whether inflammasome signalling is protective in the context of early SARS-CoV-2 infection awaits further study using in vivo disease models, and the timing of treatment with inflammasome inhibitors may be critical.
It is also worth cautioning that NLRP3 is a promiscuously activated sensor of infection and stress and its chronic activation does not generally lead to the severe inflammation seen in some patients with COVID-19 (ref.30). The variability in disease course could be related to the extent or dissemination of inflammasome-activating stimuli, the site of inflammation or the cell type that is inflamed. Indeed, classic experiments in rats show that overexpression of IL-1β specifically in the lungs is sufficient to recapitulate many of the phenotypes of ARDS177. Differences in inflammasome-induced pathology could also be explained by negative regulatory mechanisms that limit feedback amplification downstream of chronic inflammasome signalling. Simultaneous activation of inflammasomes and inhibition of negative regulatory mechanisms that suppress inflammasomes could produce severe uncontrolled inflammation. Of note, the feedback mechanisms that suppress and terminate inflammasome activation are still poorly understood.
Future studies need to address the mutual inhibition of type I interferon signalling and inflammasome activation and how the interplay between the two innate immune signalling systems could contribute to COVID-19 severity (Fig. 3). These two principal innate immune responses often antagonize each other by mechanisms that are not completely understood, and what determines which response dominates in many situations is not completely clear178–180. SARS-CoV-2 infection inhibits early interferon signalling, while activating inflammasome signalling52,58,91. Loss-of-function mutations in type I interferon pathway genes and autoantibodies to type I interferons are associated with severe COVID-19 (refs181,182), and age further dampens type I interferon responses183. The virus-intrinsic and host-intrinsic dampening of the interferon response in SARS-CoV-2 infection should compromise antiviral immunity but may also contribute to unrestrained cytokine release by removing this negative regulator of inflammasome activity. Unbridled inflammasome activation can drive severe disease complications. For example, type I interferon-deficient mice that lacked inflammasome activity were less susceptible to influenza virus infection-associated complications despite having similar viral titres184,185. Early administered recombinant type I interferon might not only restore an innate antiviral immune response to better control SARS-CoV-2 but might also simultaneously restrain subsequent hyperinflammation. However, clinical trials have not yet demonstrated a clear benefit from type I interferons. While effective and specific antiviral drugs should do even better than immunomodulating drugs, only one antiviral drug has been approved186. Nonetheless, overactivation of inflammasome signalling and pyroptosis are increasingly likely drivers of COVID-19 pathogenesis, and controlled clinical studies to evaluate drug repurposing to suppress inflammation are needed.
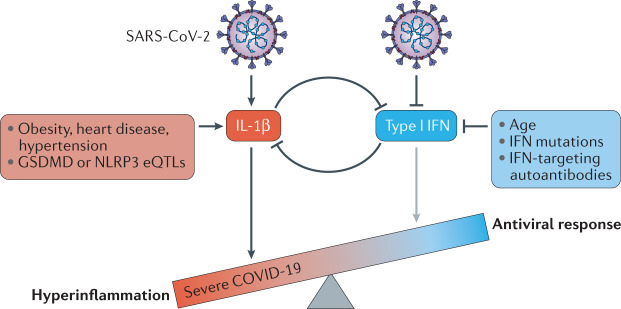
Fig. 3. Mutual inhibition of early type I interferon and IL-1β in COVID-19.
Early stages of infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) promote the secretion of IL-1β while inhibiting type I interferons. Various predispositions to severe disease are associated with increased basal IL-1β production (such as obesity, heart disease, hypertension and expression quantitative trait loci (eQTLs) associated with increased gasdermin D (GSDMD) or NLRP3 levels) or decreased type I interferon production (such as age, interferon pathway mutations and autoantibodies), which could offset the balance between these two signalling systems and push the innate immune response towards a hyperinflammatory state, while suppressing the antiviral response. IFN, interferon.
Acknowledgements
This work was supported by the US National Institutes of Health (1R01 Al124491 to H.W. and J.L.) and a postdoctoral fellowship from the American Cancer Society (133083-PF-19-034-01-LIB to S.M.V.). The authors apologize for incomplete citations due to space limitations and oversights in this fast-moving field.
Author contributions
The authors contributed equally to all aspects of the article.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information
Nature Reviews Immunology thanks M. Merad, D. Zamboni and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Judy Lieberman, Email: judy.lieberman@childrens.harvard.edu.
Hao Wu, Email: wu@crystal.harvard.edu.
References
- 1.Berlin DA, Gulick RM, Martinez FJ. Severe Covid-19. N. Engl. J. Med. 2020;383:2451–2460. doi: 10.1056/NEJMcp2009575. [DOI] [PubMed] [Google Scholar]
- 2.Jurado A, et al. COVID-19: age, interleukin-6, C-reactive protein, and lymphocytes as key clues from a multicentre retrospective study. Immun. Ageing. 2020;17:22. doi: 10.1186/s12979-020-00194-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argenziano MG, et al. Characterization and clinical course of 1000 patients with coronavirus disease 2019 in New York: retrospective case series. BMJ. 2020;369:m1996. doi: 10.1136/bmj.m1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrilli CM, et al. Factors associated with hospital admission and critical illness among 5279 people with coronavirus disease 2019 in New York City: prospective cohort study. BMJ. 2020;369:m1966. doi: 10.1136/bmj.m1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Price-Haywood EG, Burton J, Fort D, Seoane L. Hospitalization and mortality among black patients and white patients with Covid-19. N. Engl. J. Med. 2020;382:2534–2543. doi: 10.1056/NEJMsa2011686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bonafe M, et al. Inflamm-aging: why older men are the most susceptible to SARS-CoV-2 complicated outcomes. Cytokine Growth Factor. Rev. 2020;53:33–37. doi: 10.1016/j.cytogfr.2020.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng Z, et al. Risk factors of critical and mortal COVID-19 cases: a systematic literature review and meta-analysis. J. Infect. 2020;81:e16–e25. doi: 10.1016/j.jinf.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang C, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Samkari H, et al. COVID-19 and coagulation: bleeding and thrombotic manifestations of SARS-CoV-2 infection. Blood. 2020;136:489–500. doi: 10.1182/blood.2020006520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matthay MA, et al. Acute respiratory distress syndrome. Nat. Rev. Dis. Prim. 2019;5:18. doi: 10.1038/s41572-019-0069-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao X. COVID-19: immunopathology and its implications for therapy. Nat. Rev. Immunol. 2020;20:269–270. doi: 10.1038/s41577-020-0308-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomazini BM, et al. Effect of dexamethasone on days alive and ventilator-free in patients with moderate or severe acute respiratory distress syndrome and COVID-19: the CoDEX randomized clinical trial. JAMA. 2020;324:1307–1316. doi: 10.1001/jama.2020.17021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WHO Solidarity Trial Consortium et al. Repurposed antiviral drugs for Covid-19 - interim WHO solidarity trial results. N. Engl. J. Med. 2021;384:497–511. doi: 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.RECOVERY Collaborative Group et al. Dexamethasone in hospitalized patients with Covid-19. N. Engl. J. Med. 2021;384:693–704. doi: 10.1056/NEJMoa2021436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thomson EC, et al. Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity. Cell. 2021;184:1171–1187.e1120. doi: 10.1016/j.cell.2021.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Veldhoen M, Simas JP. Endemic SARS-CoV-2 will maintain post-pandemic immunity. Nat. Rev. Immunol. 2021;21:131–132. doi: 10.1038/s41577-020-00493-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lavine JS, Bjornstad ON, Antia R. Immunological characteristics govern the transition of COVID-19 to endemicity. Science. 2021;371:741–745. doi: 10.1126/science.abe6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou D, et al. Evidence of escape of SARS-CoV-2 variant B.1.351 from natural and vaccine-induced sera. Cell. 2021;184:2348–2361. doi: 10.1016/j.cell.2021.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Supasa P, et al. Reduced neutralization of SARS-CoV-2 B.1.1.7 variant by convalescent and vaccine sera. Cell. 2021;184:2201–2211. doi: 10.1016/j.cell.2021.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Merad M, Martin JC. Pathological inflammation in patients with COVID-19: a key role for monocytes and macrophages. Nat. Rev. Immunol. 2020;20:355–362. doi: 10.1038/s41577-020-0331-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay MZ, Poh CM, Renia L, MacAry PA, Ng LFP. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding S, et al. Modulatory mechanisms of the NLRP3 inflammasomes in diabetes. Biomolecules. 2019;9:850. doi: 10.3390/biom9120850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasqua T, Pagliaro P, Rocca C, Angelone T, Penna C. Role of NLRP-3 Inflammasome in hypertension: a potential therapeutic target. Curr. Pharm. Biotechnol. 2018;19:708–714. doi: 10.2174/1389201019666180808162011. [DOI] [PubMed] [Google Scholar]
- 24.Vandanmagsar B, et al. The NLRP3 inflammasome instigates obesity-induced inflammation and insulin resistance. Nat. Med. 2011;17:179–188. doi: 10.1038/nm.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.An N, et al. Regulatory mechanisms of the NLRP3 inflammasome, a novel immune-inflammatory marker in cardiovascular diseases. Front. Immunol. 2019;10:1592. doi: 10.3389/fimmu.2019.01592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brodin P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021;27:28–33. doi: 10.1038/s41591-020-01202-8. [DOI] [PubMed] [Google Scholar]
- 27.Yap JKY, Moriyama M, Iwasaki A. Inflammasomes and pyroptosis as therapeutic targets for COVID-19. J. Immunol. 2020;205:307–312. doi: 10.4049/jimmunol.2000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van de Veerdonk FL, Netea MG. Blocking IL-1 to prevent respiratory failure in COVID-19. Crit. Care. 2020;24:445. doi: 10.1186/s13054-020-03166-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Channappanavar R, Kanneganti T-D. Coronaviruses: Innate immunity, inflammasome activation, inflammatory cell death, and cytokines. Trends Immunol. 2020;41:1083–1099. doi: 10.1016/j.it.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swanson KV, Deng M, Ting JP. The NLRP3 inflammasome: molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019;19:477–489. doi: 10.1038/s41577-019-0165-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat. Rev. Immunol. 2016;16:407–420. doi: 10.1038/nri.2016.58. [DOI] [PubMed] [Google Scholar]
- 32.Munoz-Planillo R, et al. K+ efflux is the common trigger of NLRP3 inflammasome activation by bacterial toxins and particulate matter. Immunity. 2013;38:1142–1153. doi: 10.1016/j.immuni.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zanoni I, et al. An endogenous caspase-11 ligand elicits interleukin-1 release from living dendritic cells. Science. 2016;352:1232–1236. doi: 10.1126/science.aaf3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kayagaki N, et al. Noncanonical inflammasome activation by intracellular LPS independent of TLR4. Science. 2013;341:1246. doi: 10.1126/science.1240248. [DOI] [PubMed] [Google Scholar]
- 35.Shi J, et al. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 2014;514:187–192. doi: 10.1038/nature13683. [DOI] [PubMed] [Google Scholar]
- 36.Yeon SH, Yang G, Lee HE, Lee JY. Oxidized phosphatidylcholine induces the activation of NLRP3 inflammasome in macrophages. J. Leukoc. Biol. 2017;101:205–215. doi: 10.1189/jlb.3VMA1215-579RR. [DOI] [PubMed] [Google Scholar]
- 37.Shi J, et al. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–665. doi: 10.1038/nature15514. [DOI] [PubMed] [Google Scholar]
- 38.Ding J, et al. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature. 2016;535:111–116. doi: 10.1038/nature18590. [DOI] [PubMed] [Google Scholar]
- 39.Liu X, et al. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–158. doi: 10.1038/nature18629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lieberman J, Wu H, Kagan JC. Gasdermin D activity in inflammation and host defense. Sci. Immunol. 2019;4:eaav1447. doi: 10.1126/sciimmunol.aav1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruan J, Xia S, Liu X, Lieberman J, Wu H. Cryo-EM structure of the gasdermin A3 membrane pore. Nature. 2018;557:62–676. doi: 10.1038/s41586-018-0058-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evavold CL, et al. The pore-forming protein gasdermin D regulates interleukin-1 secretion from living macrophages. Immunity. 2018;48:35–44 e36. doi: 10.1016/j.immuni.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heilig R, et al. The gasdermin-D pore acts as a conduit for IL-1β secretion in mice. Eur. J. Immunol. 2018;48:584–592. doi: 10.1002/eji.201747404. [DOI] [PubMed] [Google Scholar]
- 44.Kayagaki N, et al. NINJ1 mediates plasma membrane rupture during lytic cell death. Nature. 2021;591:131–136. doi: 10.1038/s41586-021-03218-7. [DOI] [PubMed] [Google Scholar]
- 45.Han Y, et al. Lactate dehydrogenase, an independent risk factor of severe COVID-19 patients: a retrospective and observational study. Aging. 2020;12:11245–11258. doi: 10.18632/aging.103372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Y, et al. Serum lactate dehydrogenase level may predict acute respiratory distress syndrome of patients with fever infected by SARS-CoV-2. Ann. Transl. Med. 2020;8:1118. doi: 10.21037/atm-20-2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu M-y, et al. Clinical evaluation of potential usefulness of serum lactate dehydrogenase (LDH) in 2019 novel coronavirus (COVID-19) pneumonia. Respir. Res. 2020;21:171. doi: 10.1186/s12931-020-01427-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang Y, et al. Risk factors associated with 28-day all-cause mortality in older severe COVID-19 patients in Wuhan, China: a retrospective observational study. Sci. Rep. 2020;10:22369. doi: 10.1038/s41598-020-79508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Henry BM, et al. Lactate dehydrogenase levels predict coronavirus disease 2019 (COVID-19) severity and mortality: a pooled analysis. Am. J. Emerg. Med. 2020;38:1722–1726. doi: 10.1016/j.ajem.2020.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Szarpak L, et al. Lactate dehydrogenase level as a COVID-19 severity marker. Am. J. Emerg. Med. 2020 doi: 10.1016/j.ajem.2020.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yan L, et al. An interpretable mortality prediction model for COVID-19 patients. Nat. Mach. Intell. 2020;2:283–288. doi: 10.1038/s42256-020-0180-7. [DOI] [Google Scholar]
- 52.Rodrigues TS, et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 2021;218:e20201707. doi: 10.1084/jem.20201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schurink B, et al. Viral presence and immunopathology in patients with lethal COVID-19: a prospective autopsy cohort study. Lancet Microbe. 2020;1:e290–e299. doi: 10.1016/S2666-5247(20)30144-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Varga Z, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu X, Lieberman J. A mechanistic understanding of pyroptosis: the fiery death triggered by invasive infection. Adv. Immunol. 2017;135:81–117. doi: 10.1016/bs.ai.2017.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palazon-Riquelme P, Lopez-Castejon G. The inflammasomes, immune guardians at defence barriers. Immunology. 2018;155:320–330. doi: 10.1111/imm.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bai B, et al. NLRP3 inflammasome in endothelial dysfunction. Cell Death Dis. 2020;11:776. doi: 10.1038/s41419-020-02985-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Junqueira C, et al. SARS-CoV-2 infects blood monocytes to activate NLRP3 and AIM2 inflammasomes, pyroptosis and cytokine release. medRxiv. 2021 doi: 10.1101/2021.03.06.21252796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Del Valle DM, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat. Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Laing AG, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat. Med. 2020;26:1623–1635. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 61.Lucas C, et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature. 2020;584:463–469. doi: 10.1038/s41586-020-2588-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manson JJ, et al. COVID-19-associated hyperinflammation and escalation of patient care: a retrospective longitudinal cohort study. Lancet Rheumatol. 2020;2:e594–e602. doi: 10.1016/S2665-9913(20)30275-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen W, et al. Plasma CRP level is positively associated with the severity of COVID-19. Ann. Clin. Microbiol. Antimicrob. 2020;19:18. doi: 10.1186/s12941-020-00362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tosato G, Jones KD. Interleukin-1 induces interleukin-6 production in peripheral blood monocytes. Blood. 1990;75:1305–1310. doi: 10.1182/blood.V75.6.1305.1305. [DOI] [PubMed] [Google Scholar]
- 65.Ferreira AC, et al. SARS-CoV-2 engages inflammasome and pyroptosis in human primary monocytes. Cell Death Discov. 2021;7:43. doi: 10.1038/s41420-021-00428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Satis H, et al. Prognostic value of interleukin-18 and its association with other inflammatory markers and disease severity in COVID-19. Cytokine. 2021;137:155302. doi: 10.1016/j.cyto.2020.155302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhao Y, et al. Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight. 2020;5:e139834. doi: 10.1172/jci.insight.139834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Buszko M, et al. The dynamic changes in cytokine responses in COVID-19: a snapshot of the current state of knowledge. Nat. Immunol. 2020;21:1146–1151. doi: 10.1038/s41590-020-0779-1. [DOI] [PubMed] [Google Scholar]
- 69.Vabret N, et al. Immunology of COVID-19: current state of the science. Immunity. 2020;52:910–941. doi: 10.1016/j.immuni.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kudo S, Mizuno K, Hirai Y, Shimizu T. Clearance and tissue distribution of recombinant human interleukin 1 beta in rats. Cancer Res. 1990;50:5751–5755. [PubMed] [Google Scholar]
- 71.Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pascual V, Allantaz F, Arce E, Punaro M, Banchereau J. Role of interleukin-1 (IL-1) in the pathogenesis of systemic onset juvenile idiopathic arthritis and clinical response to IL-1 blockade. J. Exp. Med. 2005;201:1479–1486. doi: 10.1084/jem.20050473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liao M, et al. Single-cell landscape of bronchoalveolar immune cells in patients with COVID-19. Nat. Med. 2020;26:842–844. doi: 10.1038/s41591-020-0901-9. [DOI] [PubMed] [Google Scholar]
- 74.Xu G, et al. The differential immune responses to COVID-19 in peripheral and lung revealed by single-cell RNA sequencing. Cell Discov. 2020;6:73. doi: 10.1038/s41421-020-00225-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li S, et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal. Transduct. Target. Ther. 2020;5:235. doi: 10.1038/s41392-020-00334-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J, et al. Pyroptotic macrophages stimulate the SARS-CoV-2-associated cytokine storm. Cell. Mol. Immunol. 2021;18:1305–1307. doi: 10.1038/s41423-021-00665-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang Z, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579:415–420. doi: 10.1038/s41586-020-2071-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rogers C, et al. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat. Commun. 2019;10:1689. doi: 10.1038/s41467-019-09397-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang Y, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547:99–103. doi: 10.1038/nature22393. [DOI] [PubMed] [Google Scholar]
- 80.Orning P, et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science. 2018;362:1064–1069. doi: 10.1126/science.aau2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zheng J, et al. Severe acute respiratory syndrome coronavirus 2-induced immune activation and death of monocyte-derived human macrophages and dendritic cells. J. Infect. Dis. 2021;223:785–795. doi: 10.1093/infdis/jiaa753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Man SM, et al. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl Acad. Sci. USA. 2014;111:7403–7408. doi: 10.1073/pnas.1402911111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang H, et al. AIM2 inflammasome is critical for influenza-induced lung injury and mortality. J. Immunol. 2017;198:4383–4393. doi: 10.4049/jimmunol.1600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schroder K, et al. Acute lipopolysaccharide priming boosts inflammasome activation independently of inflammasome sensor induction. Immunobiology. 2012;217:1325–1329. doi: 10.1016/j.imbio.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 85.Arunachalam PS, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–1220. doi: 10.1126/science.abc6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hornung V, et al. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hament J-M, Kimpen JLL, Fleer A, Wolfs TFW. Respiratory viral infection predisposing for bacterial disease: a concise review. FEMS Immunol. Med. Microbiol. 1999;26:189–195. doi: 10.1111/j.1574-695X.1999.tb01389.x. [DOI] [PubMed] [Google Scholar]
- 88.Li S, et al. Clinical and pathological investigation of patients with severe COVID-19. JCI Insight. 2020;5:138070. doi: 10.1172/jci.insight.138070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paul O, et al. Vascular inflammation in lungs of patients with fatal coronavirus disease 2019 (COVID-19) infection: possible role for the NLRP3 inflammasome. medRxiv. 2021 doi: 10.1101/2021.03.19.21253815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nagashima S, et al. Endothelial dysfunction and thrombosis in patients with COVID-19-brief report. Arterioscler. Thromb. Vasc. Biol. 2020;40:2404–2407. doi: 10.1161/ATVBAHA.120.314860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Blanco-Melo D, et al. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell. 2020;181:1036–1045 e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lv J, et al. Distinct uptake, amplification, and release of SARS-CoV-2 by M1 and M2 alveolar macrophages. Cell Discov. 2021;7:24. doi: 10.1038/s41421-021-00258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Song X, et al. High expression of angiotensin-converting enzyme-2 (ACE2) on tissue macrophages that may be targeted by virus SARS-CoV-2 in COVID-19 patients. bioRxiv. 2020 doi: 10.1101/2020.07.18.210120. [DOI] [Google Scholar]
- 94.Grant RA, et al. Circuits between infected macrophages and T cells in SARS-CoV-2 pneumonia. Nature. 2021;590:635–641. doi: 10.1038/s41586-020-03148-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang C, et al. Alveolar macrophage dysfunction and cytokine storm in the pathogenesis of two severe COVID-19 patients. EBioMedicine. 2020;57:102833. doi: 10.1016/j.ebiom.2020.102833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bost P, et al. Host-viral infection maps reveal signatures of severe COVID-19 patients. Cell. 2020;181:1475–1488.e1412. doi: 10.1016/j.cell.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Fan EKY, Fan J. Regulation of alveolar macrophage death in acute lung inflammation. Respir. Res. 2018;19:50. doi: 10.1186/s12931-018-0756-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wen W, et al. Immune cell profiling of COVID-19 patients in the recovery stage by single-cell sequencing. Cell Discov. 2020;6:31. doi: 10.1038/s41421-020-0168-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ren X, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021;184:1895–1913 e1819. doi: 10.1016/j.cell.2021.01.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Szabo PA, et al. Longitudinal profiling of respiratory and systemic immune responses reveals myeloid cell-driven lung inflammation in severe COVID-19. Immunity. 2021;54:797–814.e796. doi: 10.1016/j.immuni.2021.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xiong S, et al. IL-1beta suppression of VE-cadherin transcription underlies sepsis-induced inflammatory lung injury. J. Clin. Invest. 2020;130:3684–3698. doi: 10.1172/JCI136908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Simons M, Gordon E, Claesson-Welsh L. Mechanisms and regulation of endothelial VEGF receptor signalling. Nat. Rev. Mol. Cell Biol. 2016;17:611–625. doi: 10.1038/nrm.2016.87. [DOI] [PubMed] [Google Scholar]
- 103.Costela-Ruiz VJ, Illescas-Montes R, Puerta-Puerta JM, Ruiz C, Melguizo-Rodríguez L. SARS-CoV-2 infection: the role of cytokines in COVID-19 disease. Cytokine Growth Factor. Rev. 2020;54:62–75. doi: 10.1016/j.cytogfr.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mason RJ. Thoughts on the alveolar phase of COVID-19. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319:L115–L120. doi: 10.1152/ajplung.00126.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Günther A, et al. Surfactant alteration and replacement in acute respiratory distress syndrome. Respir. Res. 2001;2:353–364. doi: 10.1186/rr86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ahn M, et al. Dampened NLRP3-mediated inflammation in bats and implications for a special viral reservoir host. Nat. Microbiol. 2019;4:789–799. doi: 10.1038/s41564-019-0371-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Guo S, et al. The NLRP3 inflammasome and IL-1beta accelerate immunologically mediated pathology in experimental viral fulminant hepatitis. PLoS Pathog. 2015;11:e1005155. doi: 10.1371/journal.ppat.1005155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wu J, et al. Activation of NLRP3 inflammasome in alveolar macrophages contributes to mechanical stretch-induced lung inflammation and injury. J. Immunol. 2013;190:3590–3599. doi: 10.4049/jimmunol.1200860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Arbore G, et al. T helper 1 immunity requires complement-driven NLRP3 inflammasome activity in CD4+ T cells. Science. 2016;352:aad1210. doi: 10.1126/science.aad1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ito H, et al. NLRP3 inflammasome activation in lung vascular endothelial cells contributes to intestinal ischemia/reperfusion-induced acute lung injury. J. Immunol. 2020;205:1393–1405. doi: 10.4049/jimmunol.2000217. [DOI] [PubMed] [Google Scholar]
- 111.Lu W, et al. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl Acad. Sci. USA. 2006;103:12540–12545. doi: 10.1073/pnas.0605402103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Chen IY, Moriyama M, Chang MF, Ichinohe T. Severe acute respiratory syndrome coronavirus viroporin 3a activates the NLRP3 inflammasome. Front. Microbiol. 2019;10:50. doi: 10.3389/fmicb.2019.00050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nieto-Torres JL, et al. Severe acute respiratory syndrome coronavirus envelope protein ion channel activity promotes virus fitness and pathogenesis. PLoS Pathog. 2014;10:e1004077. doi: 10.1371/journal.ppat.1004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nieto-Torres JL, et al. Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 2015;485:330–339. doi: 10.1016/j.virol.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Siu KL, et al. Severe acute respiratory syndrome coronavirus ORF3a protein activates the NLRP3 inflammasome by promoting TRAF3-dependent ubiquitination of ASC. FASEB J. 2019;33:8865–8877. doi: 10.1096/fj.201802418R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Oostra M, de Haan CAM, Rottier PJM. The 29-nucleotide deletion present in human but not in animal severe acute respiratory syndrome coronaviruses disrupts the functional expression of open reading frame 8. J. Virol. 2007;81:13876–13888. doi: 10.1128/JVI.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Shi C-S, Nabar NR, Huang N-N, Kehrl JH. SARS-coronavirus open reading Frame-8b triggers intracellular stress pathways and activates NLRP3 inflammasomes. Cell Death Discov. 2019;5:101. doi: 10.1038/s41420-019-0181-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Xu H, et al. SARS-CoV-2 viroporin triggers the NLRP3 inflammatory pathway. bioRxiv. 2020 doi: 10.1101/2020.10.27.357731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xia B, et al. SARS-CoV-2 envelope protein causes acute respiratory distress syndrome (ARDS)-like pathological damages and constitutes an antiviral target. Cell Res. 2021 doi: 10.1038/s41422-021-00519-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pan P, et al. SARS-CoV-2 N promotes the NLRP3 inflammasome activation to induce hyperinflammation. Review. 2020 doi: 10.21203/rs.3.rs-101224/v1. [DOI] [Google Scholar]
- 121.Ziegler CGK, et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035 e1019. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.García-Nicolás O, et al. No evidence for human monocyte-derived macrophage infection and antibody-mediated enhancement of SARS-CoV-2 infection. Front. Cell Infect. Microbiol. 2021;11:644574. doi: 10.3389/fcimb.2021.644574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Larsen MD, et al. Afucosylated IgG characterizes enveloped viral responses and correlates with COVID-19 severity. Science. 2021;371:eabc8378. doi: 10.1126/science.abc8378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chakraborty S, et al. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat. Immunol. 2021;22:67–73. doi: 10.1038/s41590-020-00828-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Shi M, et al. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. bioRxiv. 2020 doi: 10.1101/2020.09.18.302901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Schubert K, et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:959–966. doi: 10.1038/s41594-020-0511-8. [DOI] [PubMed] [Google Scholar]
- 127.Thoms M, et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020;369:1249–1255. doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Yang HZ, Oppenheim JJ. Alarmins and immunity. Immunol. Rev. 2017;280:41–56. doi: 10.1111/imr.12577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zhou Z, et al. Granzyme A from cytotoxic lymphocytes cleaves GSDMB to trigger pyroptosis in target cells. Science. 2020;368:eaaz7548. doi: 10.1126/science.aaz7548. [DOI] [PubMed] [Google Scholar]
- 130.Das S, et al. GSDMB induces an asthma phenotype characterized by increased airway responsiveness and remodeling without lung inflammation. Proc. Natl Acad. Sci. USA. 2016;113:13132–13137. doi: 10.1073/pnas.1610433113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Carvelli J, et al. Association of COVID-19 inflammation with activation of the C5a-C5aR1 axis. Nature. 2020;588:146–150. doi: 10.1038/s41586-020-2600-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Jiang Y, et al. Complement receptor C5aR1 inhibition reduces pyroptosis in hDPP4-transgenic mice infected with MERS-CoV. Viruses. 2019;11:39. doi: 10.3390/v11010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Imai Y, et al. Identification of oxidative stress and Toll-like receptor 4 signaling as a key pathway of acute lung injury. Cell. 2008;133:235–249. doi: 10.1016/j.cell.2008.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Fessler MB, Summer RS. Surfactant lipids at the host-environment interface. metabolic sensors, suppressors, and effectors of inflammatory lung disease. Am. J. Respir. Cell Mol. Biol. 2016;54:624–635. doi: 10.1165/rcmb.2016-0011PS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Chan NC, Weitz JI. COVID-19 coagulopathy, thrombosis, and bleeding. Blood. 2020;136:381–383. doi: 10.1182/blood.2020007335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Teuwen L-A, Geldhof V, Pasut A, Carmeliet P. COVID-19: the vasculature unleashed. Nat. Rev. Immunol. 2020;20:389–391. doi: 10.1038/s41577-020-0343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Deshpande C. Thromboembolic findings in COVID-19 autopsies: pulmonary thrombosis or embolism? Ann. Intern. Med. 2020;173:394–395. doi: 10.7326/M20-3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Wu C, et al. Inflammasome activation triggers blood clotting and host death through pyroptosis. Immunity. 2019;50:1401–1411.e1404. doi: 10.1016/j.immuni.2019.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Burzynski LC, Clarke MCH. Death is coming and the clot thickens, as pyroptosis feeds the fire. Immunity. 2019;50:1339–1341. doi: 10.1016/j.immuni.2019.05.015. [DOI] [PubMed] [Google Scholar]
- 140.Rosell A, et al. Patients with COVID-19 have elevated levels of circulating extracellular vesicle tissue factor activity that is associated with severity and mortality. Arterioscler. Thromb. Vasc. Biol. 2020;41:878–882. doi: 10.1161/ATVBAHA.120.315547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Yang X, et al. Bacterial endotoxin activates the coagulation cascade through gasdermin D-dependent phosphatidylserine exposure. Immunity. 2019;51:983–996.e986. doi: 10.1016/j.immuni.2019.11.005. [DOI] [PubMed] [Google Scholar]
- 142.Chen KW, et al. Noncanonical inflammasome signaling elicits gasdermin D-dependent neutrophil extracellular traps. Sci. Immunol. 2018;3:eaar6676. doi: 10.1126/sciimmunol.aar6676. [DOI] [PubMed] [Google Scholar]
- 143.Sollberger G, et al. Gasdermin D plays a vital role in the generation of neutrophil extracellular traps. Sci. Immunol. 2018;3:aar6689. doi: 10.1126/sciimmunol.aar6689. [DOI] [PubMed] [Google Scholar]
- 144.Thiam HR, Wong SL, Wagner DD, Waterman CM. Cellular mechanisms of NETosis. Annu. Rev. Cell Dev. Biol. 2020;36:191–218. doi: 10.1146/annurev-cellbio-020520-111016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Bont CM, Boelens WC, Pruijn GJM. NETosis, complement, and coagulation: a triangular relationship. Cell. Mol. Immunol. 2019;16:19–27. doi: 10.1038/s41423-018-0024-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Laforge M, et al. Tissue damage from neutrophil-induced oxidative stress in COVID-19. Nat. Rev. Immunol. 2020;20:515–516. doi: 10.1038/s41577-020-0407-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Middleton EA, et al. Neutrophil extracellular traps contribute to immunothrombosis in COVID-19 acute respiratory distress syndrome. Blood. 2020;136:1169–1179. doi: 10.1182/blood.2020007008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Skendros P, et al. Complement and tissue factor–enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Invest. 2020;130:6151–6157. doi: 10.1172/JCI141374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Veras FP, et al. SARS-CoV-2–triggered neutrophil extracellular traps mediate COVID-19 pathology. J. Exp. Med. 2020;217:e20201129. doi: 10.1084/jem.20201129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Gupta R. Anakinra: a silver lining in COVID-19? Crit. Care. 2020;24:598. doi: 10.1186/s13054-020-03312-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Huet T, et al. Anakinra for severe forms of COVID-19: a cohort study. Lancet Rheumatol. 2020;2:e393–e400. doi: 10.1016/S2665-9913(20)30164-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Navarro-Millán I, et al. Use of anakinra to prevent mechanical ventilation in severe COVID-19: a case series. Arthritis Rheumatol. 2020;72:1990–1997. doi: 10.1002/art.41422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nemchand P, Tahir H, Mediwake R, Lee J. Cytokine storm and use of anakinra in a patient with COVID-19. BMJ Case Rep. 2020;13:e237525. doi: 10.1136/bcr-2020-237525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Cauchois R, et al. Early IL-1 receptor blockade in severe inflammatory respiratory failure complicating COVID-19. Proc. Natl Acad. Sci. USA. 2020;117:18951–18953. doi: 10.1073/pnas.2009017117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Tharaux P-L, et al. Effect of anakinra versus usual care in adults in hospital with COVID-19 and mild-to-moderate pneumonia (CORIMUNO-ANA-1): a randomised controlled trial. Lancet Respir. Med. 2021;9:295–304. doi: 10.1016/S2213-2600(20)30556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Kyriazopoulou E, et al. An open label trial of anakinra to prevent respiratory failure in COVID-19. eLife. 2021;10:e66125. doi: 10.7554/eLife.66125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Cavalli G, Dagna L. The right place for IL-1 inhibition in COVID-19. Lancet Respir. Med. 2021;9:223–224. doi: 10.1016/S2213-2600(21)00035-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Rovina N, et al. Soluble urokinase plasminogen activator receptor (suPAR) as an early predictor of severe respiratory failure in patients with COVID-19 pneumonia. Crit. Care. 2020;24:187. doi: 10.1186/s13054-020-02897-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tate MD, et al. Reassessing the role of the NLRP3 inflammasome during pathogenic influenza A virus infection via temporal inhibition. Sci. Rep. 2016;6:27912. doi: 10.1038/srep27912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Magupalli VG, et al. HDAC6 mediates an aggresome-like mechanism for NLRP3 and pyrin inflammasome activation. Science. 2020;369:eaas8995. doi: 10.1126/science.aas8995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Reyes AZ, et al. Anti-inflammatory therapy for COVID-19 infection: the case for colchicine. Ann. Rheum. Dis. 2020 doi: 10.1136/annrheumdis-2020-219174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Lopes MI, et al. Beneficial effects of colchicine for moderate to severe COVID-19: a randomised, double-blinded, placebo-controlled clinical trial. RMD Open. 2021;7:e001455. doi: 10.1136/rmdopen-2020-001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Tardif J-C, et al. Efficacy of colchicine in non-hospitalized patients with COVID-19. medRxiv. 2021 doi: 10.1101/2021.01.26.21250494. [DOI] [Google Scholar]
- 164.Li J, et al. Metformin use in diabetes prior to hospitalization: effects on mortality in Covid-19. Endocr. Pract. 2020;26:1166–1172. doi: 10.4158/EP-2020-0466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xian H, et al. Metformin inhibition of mitochondrial ATP and DNA synthesis abrogates NLRP3 inflammasome activation and pulmonary inflammation. Immunity. 2021;54:1463–1477. doi: 10.1016/j.immuni.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu X, Xia S, Zhang Z, Wu H, Lieberman J. Channelling inflammation: gasdermins in physiology and disease. Nat. Rev. Drug Discov. 2021;20:384–405. doi: 10.1038/s41573-021-00154-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hu JJ, et al. FDA-approved disulfiram inhibits pyroptosis by blocking gasdermin D pore formation. Nat. Immunol. 2020;21:736–745. doi: 10.1038/s41590-020-0669-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Humphries F, et al. Succination inactivates gasdermin D and blocks pyroptosis. Science. 2020;369:1633–1637. doi: 10.1126/science.abb9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Mantero V, et al. COVID-19 in dimethyl fumarate-treated patients with multiple sclerosis. J. Neurol. 2020;268:2023–2025. doi: 10.1007/s00415-020-10015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Fillmore N, et al. Disulfiram associated with lower risk of Covid-19: a retrospective cohort study. medRxiv. 2021 doi: 10.1101/2021.03.10.21253331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Jin Z, et al. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 172.Olagnier D, et al. SARS-CoV2-mediated suppression of NRF2-signaling reveals potent antiviral and anti-inflammatory activity of 4-octyl-itaconate and dimethyl fumarate. Nat. Commun. 2020;11:4938. doi: 10.1038/s41467-020-18764-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mills EL, et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Crowley SM, et al. Intestinal restriction of Salmonella Typhimurium requires caspase-1 and caspase-11 epithelial intrinsic inflammasomes. PLoS Pathog. 2020;16:e1008498. doi: 10.1371/journal.ppat.1008498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Cerqueira DM, et al. Guanylate-binding protein 5 licenses caspase-11 for gasdermin-D mediated host resistance to Brucella abortus infection. PLoS Pathog. 2018;14:e1007519. doi: 10.1371/journal.ppat.1007519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Yamada H, Mizumo S, Horai R, Iwakura Y, Sugawara I. Protective role of interleukin-1 in mycobacterial infection in IL-1 α/β double-knockout mice. Lab. Invest. 2000;80:759–767. doi: 10.1038/labinvest.3780079. [DOI] [PubMed] [Google Scholar]
- 177.Kolb M, Margetts PJ, Anthony DC, Pitossi F, Gauldie J. Transient expression of IL-1β induces acute lung injury and chronic repair leading to pulmonary fibrosis. J. Clin. Invest. 2001;107:1529–1536. doi: 10.1172/JCI12568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Kohase M, et al. Interleukin-1 can inhibit interferon-beta synthesis and its antiviral action: comparison with tumor necrosis factor. J. Interferon Res. 1988;8:559–570. doi: 10.1089/jir.1988.8.559. [DOI] [PubMed] [Google Scholar]
- 179.Mayer-Barber KD, et al. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 2014;511:99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Guarda G, et al. Type I interferon inhibits interleukin-1 production and inflammasome activation. Immunity. 2011;34:213–223. doi: 10.1016/j.immuni.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 181.Zhang Q, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370:eabd4570. doi: 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bastard P, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370:eabd4585. doi: 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Molony RD, et al. Aging impairs both primary and secondary RIG-I signaling for interferon induction in human monocytes. Sci. Signal. 2017;10:eaan2392. doi: 10.1126/scisignal.aan2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Pillai PS, et al. Mx1 reveals innate pathways to antiviral resistance and lethal influenza disease. Science. 2016;352:463–466. doi: 10.1126/science.aaf3926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Meffre E, Iwasaki A. Interferon deficiency can lead to severe COVID. Nature. 2020;587:374–376. doi: 10.1038/d41586-020-03070-1. [DOI] [PubMed] [Google Scholar]
- 186.Rubin D, Chan-Tack K, Farley J, Sherwat A. FDA approval of remdesivir - a step in the right direction. N. Engl. J. Med. 2020;383:2598–2600. doi: 10.1056/NEJMp2032369. [DOI] [PubMed] [Google Scholar]