Abstract
Antibody dependent enhancement (ADE) of infection is a safety concern for vaccine strategies. In a recent publication, Li et al. (Cell 184 :4203–4219, 2021) have reported that infection-enhancing antibodies directed against the N-terminal domain (NTD) of the SARS-CoV-2 spike protein facilitate virus infection in vitro, but not in vivo. However, this study was performed with the original Wuhan/D614G strain. Since the Covid-19 pandemic is now dominated with Delta variants, we analyzed the interaction of facilitating antibodies with the NTD of these variants. Using molecular modeling approaches, we show that enhancing antibodies have a higher affinity for Delta variants than for Wuhan/D614G NTDs. We show that enhancing antibodies reinforce the binding of the spike trimer to the host cell membrane by clamping the NTD to lipid raft microdomains. This stabilizing mechanism may facilitate the conformational change that induces the demasking of the receptor binding domain. As the NTD is also targeted by neutralizing antibodies, our data suggest that the balance between neutralizing and facilitating antibodies in vaccinated individuals is in favor of neutralization for the original Wuhan/D614G strain. However, in the case of the Delta variant, neutralizing antibodies have a decreased affinity for the spike protein, whereas facilitating antibodies display a strikingly increased affinity. Thus, ADE may be a concern for people receiving vaccines based on the original Wuhan strain spike sequence (either mRNA or viral vectors). Under these circumstances, second generation vaccines with spike protein formulations lacking structurally-conserved ADE-related epitopes should be considered.
The aim of the present study was to evaluate the recognition of SARS-CoV-2 Delta variants by infection enhancing antibodies directed against the NTD. The antibody studied is 1052 (pdb file #7LAB) which has been isolated from a symptomatic Covid-19 patient1. Molecular modeling simulations were performed as previously described2. Two currently circulating Delta variants were investigated, with the following mutational patterns in the NTD :
- G142D/E154K (B.1.617.1)
- T19R/E156G/del157/del158/A222V (B.1.617.2)
Each mutational pattern was introduced in the original Wuhan/D614G strain, submitted to energy minimization, and then tested for antibody binding. The energy of interaction (ΔG) of the reference pdb file #7LAB (Wuhan/D614G strain) in the NTD region was estimated to −229 kJ/mol−1. In the case of Delta variants, the energy of interaction was raised to −272 kJ.mol−1 (B.1.617.1) and −246 kJ.mol−1 (B.1.617.2). Thus, these infection enhancing antibodies not only still recognize Delta variants but even display a higher affinity for those variants than for the original SARS-CoV-2 strain.
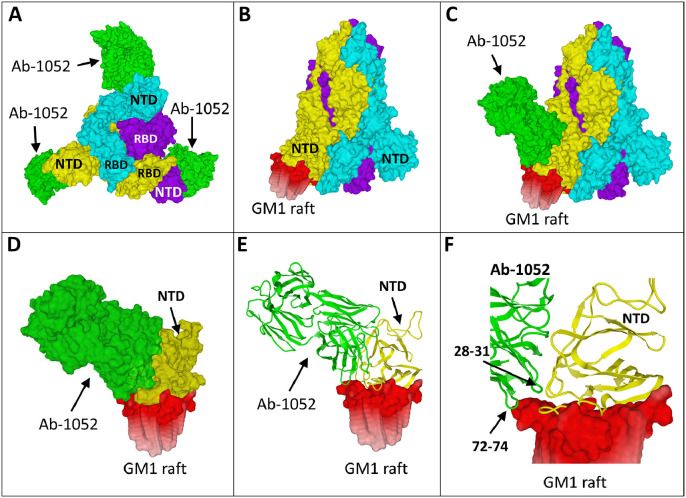
The global structure of the trimeric spike of the B.1.617.1 variant in the cell-facing view is shown in Fig. 1 A. As expected, the facilitating antibody bound to the NTD (in green) is located behind the contact surface so that it does not interfere with virus-cell attachment. Indeed, a preformed antibody-NTD complex could perfectly bind to the host cell membrane. The interaction between the NTD and a lipid raft is shown in Fig. 1 B, and a whole raft-spike-antibody complex in Fig. 1 C. Interestingly, a small part of the antibody was found to interact with the lipid raft, as further illustrated in Figs. 1 D-E. More precisely, two distinct loops of the heavy chain of the antibody encompassing amino acid residues 28–31 and 72–74, stabilize the complex through a direct interaction with the edge of the raft (Fig. 1 F). Overall, the energy of interaction of the NTD-raft complex was raised from −399 kJ.mol−1 in absence of the antibody to −457 kJ.mol−1 with the antibody. By clamping the NTD and the lipid raft, the antibody reinforces the attachment of the spike protein to the cell surface and thus facilitates the conformational change of the RBD which is the next step of the virus infection process2.
Fig. 1.
Infection enhancing antibodies recognize the NTD of Delta variants. A. Molecular model of the Delta B.1.617.1 spike trimer as viewed from the host cell surface (chains A, B and C in cyan, yellow and purple, respectively), with the NTD and RBD of each chain indicated. The 1052 antibody is in green. B. Spike trimer with the B subunit bound to a lipid raft (with 6 ganglioside GM1 molecules). C. Trimolecular [spike-antibody-raft] complex. D. Focus on the NTD-antibody complex bound to the lipid raft. E. Secondary structures of the NTD (yellow) and the antibody (green) bound to lipid raft gangliosides. F. The 1052 antibody clamps the NTD and the edge of the lipid raft.
This notion of a dual NTD-raft recognition by an infection enhancing antibody may represent a new type of ADE that could be operative with other viruses. Incidentally, our data provide a mechanistic explanation of the FcR-independent enhancement of infection induced by anti-NTD antibodies1. The model we propose, which links for the first time lipid rafts to ADE of SARS-CoV-2, is in line with previous data showing that intact lipid rafts are required for ADE of dengue virus infection3.
Neutralizing antibodies directed against the NTD have also been detected in Covid-19 patients4, 5. The 4A8 antibody is a major representant of such antibodies5. The epitope recognized by this antibody on the flat NTD surface is dramatically affected in the NTD of Delta variants2, suggesting a significant loss of activity in vaccinated people exposed to Delta variants. More generally, it can be reasonably assumed that the balance between neutralizing and facilitating antibodies may greatly differ according to the virus strain (Fig. 2 ).
Fig. 2.
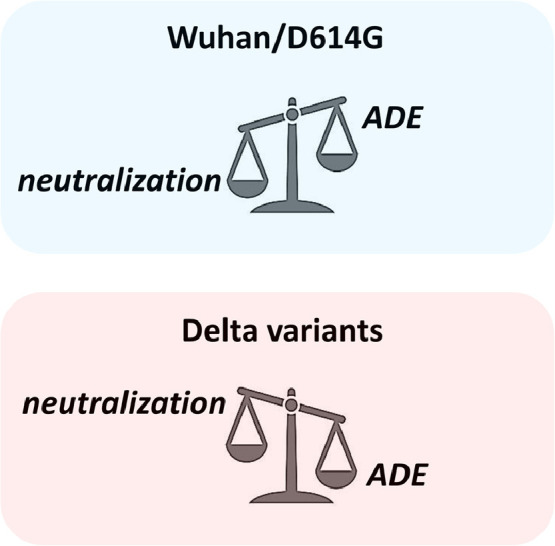
Neutralization vs ADE balance according to SARS-CoV-2 strains.
Current Covid-19 vaccines (either mRNA or viral vectors) are based on the original Wuhan spike sequence. Inasmuch as neutralizing antibodies overwhelm facilitating antibodies, ADE is not a concern. However, the emergence of SARS-CoV-2 variants may tip the scales in favor of infection enhancement. Our structural and modeling data suggest that it might be indeed the case for Delta variants.
In conclusion, ADE may occur in people receiving vaccines based on the original Wuhan strain spike sequence (either mRNA or viral vectors) and then exposed to a Delta variant. Although this potential risk has been cleverly anticipated before the massive use of Covid-19 vaccines6, the ability of SARS-CoV-2 antibodies to mediate infection enhancement in vivo has never been formally demonstrated. However, although the results obtained so far have been rather reassuring1, to the best of our knowledge ADE of Delta variants has not been specifically assessed. Since our data indicate that Delta variants are especially well recognized by infection enhancing antibodies targeting the NTD, the possibility of ADE should be further investigated as it may represent a potential risk for mass vaccination during the current Delta variant pandemic. In this respect, second generation vaccines7 with spike protein formulations lacking structurally-conserved ADE-related epitopes should be considered.
References
- 1.Li D., et al. In vitro and in vivo functions of SARS-CoV-2 infection-enhancing and neutralizing antibodies. Cell. 2021;184:4203–4219. doi: 10.1016/j.cell.2021.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fantini J., Yahi N., Azzaz F., Chahinian H. Structural dynamics of SARS-CoV-2 variants: a health monitoring strategy for anticipating Covid-19 outbreaks. J Infect. 2021;83:197–206. doi: 10.1016/j.jinf.2021.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Puerta-Guardo H., Mosso C., Medina F., Liprandi F., Ludert J.E., del Angel R.M. Antibody-dependent enhancement of dengue virus infection in U937 cells requires cholesterol-rich membrane microdomains. J Gen Virol. 2010;91:394–403. doi: 10.1099/vir.0.015420-0. [DOI] [PubMed] [Google Scholar]
- 4.Chi X., et al. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu L., et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- 6.Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat Rev Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fantini J., Chahinian H., Yahi N. Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19. Biochem Biophys Res Commun. 2021;538:132–136. doi: 10.1016/j.bbrc.2020.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]