Abstract
The year 2020 was characterized by the COVID-19 pandemic that has caused, by the end of March 2021, more than 2.5 million deaths worldwide. Since the beginning, besides the laboratory test, used as the gold standard, many applications have been applying deep learning algorithms to chest X-ray images to recognize COVID-19 infected patients. In this context, we found out that convolutional neural networks perform well on a single dataset but struggle to generalize to other data sources. To overcome this limitation, we propose a late fusion approach where we combine the outputs of several state-of-the-art CNNs, introducing a novel method that allows us to construct an optimum ensemble determining which and how many base learners should be aggregated. This choice is driven by a two-objective function that maximizes, on a validation set, the accuracy and the diversity of the ensemble itself. A wide set of experiments on several publicly available datasets, accounting for more than 92,000 images, shows that the proposed approach provides average recognition rates up to 93.54% when tested on external datasets.
Keywords: COVID-19, X-ray, Deep-learning, Multi-expert systems, Optimization, Convolutional neural networks
Notation
model
set of all models
subset of models
average
with max accuracy
with max diversity
with max accuracy and diversity
# of available models
# of models in
set of all
# of all
# of labels
input scan
classification output
correlation coefficient
# of scans
dataset
set of objective functions
objective function
# of objective functions
set of conditions
condition
# of conditions
pair
point
weights
1. Introduction
The eruption of the severe acute respiratory syndrome COVID-19 in early 2020 developed into a pandemic. By the end of March 2021, more than 100 million coronavirus cases and more than 2.5 million deaths were reported worldwide. As a first reaction, the scientific community tried to cut the transmission of the virus by finding different diagnostic modalities of COVID-19. Further to the reverse-transcription polymerase chain reaction (RT-PCR) currently performed to diagnose the COVID-19 presence [1], computed tomography (CT) and chest X-ray (CXR) offer insights into lung involvement. Indeed, chest imaging findings of COVID-19 were discovered in patients with false-negative RT-PCR tests and asymptomatic patients [2], suggesting that the use of chest images may give an important contribution to the diagnosis. Moreover, in situations where RT-PCR results were delayed or initially negative, or if clinical symptoms were present, chest imaging has been used in the diagnostic path for patients with suspected COVID-19 [3]. Moreover, the World Health Organization guidelines suggest using X-rays to assess the clinical therapy of suspected or confirmed COVID-19 patients [4].
Among the different imaging modalities used for the COVID-19 diagnosis, CT and CXR images were used in most of the publications [5]. The authors of such a survey suggest that CT is the main diagnostic modality, due to its high specificity and the likelihood of identifying the various degrees of the disorder at early stages. However, CT scans have a poor specificity to differentiate between COVID-19 pneumonia and other pneumonia [5]. Besides, the difficulty in the cleaning process of the CT scanners results in a high risk of contamination for both patients and healthcare staff. On the contrary, as opposed to CT, CXR is the most widely used modality even though it has limited sensitivity at an early stage of infection. The value of the approach is the option of using compact CXR units, which have universal availability and reduce the chance of cross-infection. This offers the possibility to use portable units which are cost-effective and offers the chance to use such solution directly in emergency or intensive care units, or even directly in the patients houses.
Due to the aforementioned limitations of CT images, in this work we analyze CXR images, where the application of modern methodologies for image processing can empower the technique. In this respect, the literature provides several studies on deep learning (DL) applied to CXR scans to diagnose COVID-19 [6]. Nonetheless, these studies account two main limitations mostly due to the initial difficulty of retrieving CXR COVID-19 datasets: first, most of the works employ the Cohen dataset [7] which contains images retrieved from publications and, hence they are no true clinical images. Second, to the best of our knowledge, all the papers published ignore external testing of the approaches they propose, therefore using, in a hold-out or cross-validation scheme, the same data source for both training and testing. As presented in [8], external validation is essential to inspect the robustness of the model. Despite the high performance obtained, such approaches may result unusable in clinical practice since no real-life images were used to train the algorithms and no external validation was performed. We empirically confirm such an observation by showing that the performance of several state-of-the-art convolutional neural networks (CNNs), when applied to an external cohort of data, drops. To overcome this issue, this manuscript presents a multi-expert system (MES) exploiting a late fusion approach where the classifications provided by a set of individual CNNs are combined by the majority voting rule. The method presents a strategy to choose the best combination of learners for the MES, which is based on a Pareto multi-objective optimization problem with a unique optimum solution by maximizing the accuracy and the diversity on a validation set. Indeed, the literature agrees that the learning models included in an ensemble should be individually accurate but, at the same time, they should be diverse, i.e. they should make errors on different samples [9]. To show the effectiveness of our proposal we consider four publicly available datasets accounting for more than 92,000 CXR images, and we designed two classification tasks of interest. The first is the binary classification between COVID-19 patients and non-COVID-19 patients, whilst the second consists of a three-class classification task where we aim to recognize healthy subjects, patients suffering from COVID-19 and other pneumonia. Furthermore, we perform the experiments in two modalities: in one case we use the standard cross-validation, in the other case we train the MES on a repository and test it on the other three, which can be considered as external sources of data.
The rest of this manuscript is organized as follows: next section first summarizes the background on DL applications analyzing CXR images for COVID-19 diagnosis and then it presents a short overview of classifiers fusion methods. Section 3 introduces the datasets used in this work, whereas Section 4 presents our method. Section 5 describes the experimental configuration; Section 6 reports and discusses the results. Finally, Section 7 provides concluding remarks.
2. Background
In this section we first summarize the literature on AI-based models for the diagnosis of COVID-19 pneumonia in CXR images (Section 2.1), and then we overview the ways to combine several classifiers (Section 2.2).
2.1. State of the art on COVID-19 CXR applications
Nowadays, the power of DL of extrapolating the necessary features from data for a specific task has been widely explored and it has proven to be one of the best solutions in many fields [10]. In particular, COVID-19 diagnosis on medical images using DL models has been investigated during the pandemic. The review presented by Wynants et.al. [6] shows that the majority of the research efforts have been directed towards CT or X-ray images. Focusing our attention on work applying DL to X-ray images, as we present hereinafter, Table 1 summarizes the main features of papers published in peer-review journals that use publicly available CXR datasets. All of them develop DL algorithms on X-ray images for a binary classification task, i.e. recognizing COVID-19 from non-COVID-19 patients, or for a three-class task, i.e. distinguishing COVID-19 patients from patients with other forms of pneumonia and healthy patients.
Table 1.
Main features of peer-reviewed papers applying DL to CXR images for COVID-19 diagnosis. The papers are listed in chronological order.
| Paper | Datasets | # of cases | Task | DL Network | Validation | Accuracy |
|---|---|---|---|---|---|---|
| I.D. Apostolopoulos[11] | Cohen [7] Kermany [12] | 224 COVID-19 700 Pneumonia 504 Healthy | 2 classes 3 classes | VGG-19 | 10-fold CV | 98.75% 93.48% |
| M. Loey[13] | Cohen [7] Kermany [12] | 69 COVID-19 158 Pneumonia 79 Healthy | 2 classes 3 classes | GAN+ ALexNet | train-val-test split 90%-20%-10% | 100.00% 85.19% |
| M. Rahimzadeh[14] | Cohen [7] RSNA [15] | 180 COVID-19 6054 Pneumonia 8851 Healthy | 3 classes | Xception + ResNet50V2 | 5-fold CV | 91.40% |
| S. Vaid[16] | Cohen [7] RSNA [15] | 243 COVID-19 121 Healthy | 2 classes | VGG-19 | train-val-test split 80%-20%-20% | 96.30% |
| T. Ozturk[17] | Cohen [7] RSNA [15] | 127 COVID-19 500 Pneumonia 500 Healthy | 2 classes 3 classes | DarkCovidNet | 5-fold CV | 98.08% 87.02% |
| M. Togacar[18] | Cohen [7] Chowdhury [19] | 295 COVID-19 98 Pneumonia 65 Healthy | 3 classes | MobileNetV2 + SqueezeNet + SVM | train-test split 70%-30% | 99.27% |
| E.H. Chowdhury[19] | Cohen [7] RSNA [15] Kermany [12] | 423 COVID-19 1485 Pneumonia 1579 Healthy | 2 classes 3 classes | CheXNet | 5-fold CV | 99.70% 97.94% |
| Z. Wang[20] | Cohen [7] RSNA [15] | 225 COVID-19 2024 Pneumonia 1314 Healthy | 3 classes | VGG + ResNet + Residual Attention Network | train-test split 80%-20% | 94.00% |
| L. Brunese[21] | Cohen [7] Ozturk [17] RSNA [15] | 250 COVID-19 2753 non-COVID-19 | 2 classes | VGG-16 | train-val-test split 50%-20%-30% | 98.00% |
| A.I. Khan[22] | Cohen [7] Kermany [12] | 284 COVID-19 657 Pneumonia 310 Healthy | 3 classes | CoroNet | 4-fold CV | 95.00% |
| Z. Wang[23] | Cohen [7] RSNA [15] | 204 COVID-19 2004 Pneumonia 1314 Healthy | 3 classes | ResNet50 + FPN | train-test split 80%-20% | 94.00% |
| P. Afshar[24] | Cohen [7] RSNA [15] Chowdhury [19] | 358 COVID-19 13,604 non-COVID-19 | 2 classes | COVID-CAPS | train-test split 80%-10%-10% | 98.30% |
| S. Mohammad[25] | Cohen [7] Kermany [12] | 226 COVID-19 226 Pneumonia 226 Healthy | 2 classes 3 classes | MetaCOVID | train-test split 70%-30% | 96.50% 95.60% |
| L. Wang[26] | Cohen [7] RSNA [15] Chowdhury [19] | 358 COVID-19 5538 Pneumonia 8066 Healthy | 3 classes | COVID-Net | train-test split 98%-2% | 93.33% |
| L. Jinpeng[27] | Cohen [7] Kermany [12] | 231 COVID-19 4007 Pneumonia 1583 Healthy | 3 classes | CMT-CNN | 5-fold CV | 93.49% |
| F. Yuqi[28] | Cohen [7] RSNA [15] Chowdhury [19] | 500 COVID-19 500 non-COVID-19 | 2 classes | MKSC | 10-fold CV | 98.17% |
| V. Pablo[29] | Cohen [7] Desai [30] RSNA [15] Kermany [12] | 717 COVID-19 5617 Pneumonia 61,995 Healthy | 3 classes | NASNetLarge | 10-fold CV | 97.60% |
Due to the initial difficulty in retrieving COVID-19 datasets, the work in the literature try to face the problem of the lack of COVID-19 positive available data by joining the Cohen dataset [7] with other available non-COVID-19 datasets [12], [15], [17], [19], [30]. Despite the high and promising performance reported in Table 1, the use of different datasets, the combination of different methods, and the use of different validation techniques yield results that are not directly comparable. It is worth noticing that all the papers used the simple hold-out or the cross-validation and, hence, they did not externally validate the methodology proposed. This could be expected as they were published in response to the pandemic when a reduced amount of data were available. As a further consideration, note that the Cohen dataset [7] contains only low-quality images in jpg or png format obtained from publications and articles (Table 2 ). Nevertheless, recent works have made publicly available image repositories containing CXRs of COVID-19 patients collected in true clinical environments [31], [32]. These observations emphasize the need for further studies exploiting the past and new repositories also to externally validate the proposed systems to fasten their successful application in clinical practice.
Table 2.
Overview of the dataset’s features.
| Dataset | AIforCOVID[31] | COVIDx +[26] | Brixia[32] | RSNA[15] | |
|---|---|---|---|---|---|
| #of images | 820 | 1770 | 4696 | 85,374 | |
| Prior class distribution (# of images) | COVID-19 (820) | COVID-19 (1770) | COVID-19 (4696) | Pneumonia (1062) | |
| Normal (84312) | |||||
| True Scan or Article Image | true | both | true | true | |
| Format | dicom | jpg, png | dicom | dicom | |
| Bit Depth (# of images) | 12 (394), 16 (426) | 8 (1770) | 12 (2583), 16 (2113) | 8 (85374) | |
| Resolution | Height | ||||
| Width | |||||
| Standardized dynamic rangea | |||||
| Contrastb | |||||
, where and denote the maximum and minimum gray level intensity in the image, and stands for the number of bits representing the gray scale intensity.
2.2. Fusion of classifiers
The combination or fusion of learners in an ensemble can be performed at different levels using different techniques. Indeed, joining multiple models allows to extract complementary and more powerful data representation, with the goal of improving the performance with respect to those of each stand-alone classifier considered. Although there is not any formal proof, this intuition has brought interesting results in many applications, medical imaging included [33].
Turning the attention to how learners can be combined in a MES, there exists three main types of fusion: early, joint, and late fusion [34]. Early fusion is the process of combining multiple inputs into a single feature vector, which is then passed to one single model. Also known as representation learning, it integrates different types of data to feed one expert and, hence, it falls out from the scope of this work since we have only CXR images.
For what concerns joint and late fusion, these two combination strategies can be used to aggregate different learners. Joint fusion combines extracted intermediate feature vectors from trained neural networks. This combined feature vector is then given to a final model that will output the classification, which constitutes a new sample representation that is the input of a new classifier [35].
Let us now delve into late fusion approaches as our proposal works at this level. In the past, late fusion has attracted considerable attention due to its potential to improve the performance of learners. The basic idea is to solve any classification problem by designing a set of independent systems, which are then combined by an aggregation function to reduce individual error rates. The aggregation rule takes as input the predictions provided by the different models that are combined according to a function [35] (e.g. minimum, maximum, average, median, majority vote). There is a consensus that the key to the success of late fusion is that it builds a mixture of diverse classifiers [9], providing different and complementary points of view to the ensemble. Diversity is a measure, applied to a set of classifiers, which expresses how much the classifications on a set of data vary, as we further detail in Section 4.2.1.
3. Materials
During the pandemic, the scientific community has made publicly available several COVID-19 imaging datasets necessary to better understand how the virus is working and how it affects the lungs. According to the focus of our work, i.e. to predict if a patient is affected by COVID-19 by analyzing CXR scans, this section summarizes the four public CXR datasets that we use hereinafter. The use of several repositories including images collected in different hospitals under diverse parameters and conditions make the data as general as possible, as summarized in Table 2, which shows the large variability among the images in the different repositories in terms of bits, resolution, standardized dynamic range and contrast. This variability is the desired value for our application, helping setting up a learning system able to correctly classify a CXR of any sort, fastening its adoption in clinical practice.
3.1. AIfor COVID
This dataset includes 820 X-ray images of COVID-19 positive patients [31] collected in six Italian hospitals from March to June 2020 with an anteroposterior and posteroanterior view. The images were acquired at the time of the patient’s hospitalization if he/she resulted positive to the RT-PCR test. Each patient was assigned to the mild or severe group. The former contains the patients sent back to domiciliary isolation or hospitalized without ventilatory support, whereas the latter is composed of patients who required non-invasive vcsed patients. All of the collected X-ray scans were in DICOM format.
3.2. COVIDx +
The second used dataset is COVIDx + , composed of only the COVID-19 positive cases of the COVIDx dataset introduced by L. Wang et.al. [26]. To generate this dataset the authors combined open source COVID-19 patient datasets: (i) COVID-19 images data collection [7]; (ii) COVID-19 radiography dataset [19]. The final dataset contains 1770 X-ray images of COVID-19 positive patients. Images are available in png and jpeg format and were partially retrieved from published articles.
3.3. Brixia
The Brixia dataset, introduced by A. Signoroni et.al. [32], contains 4696 X-ray images of positive COVID-19 patients collected in an Italian hospital between March and April 2020. Images belong to hospitalized patients, older than 20 years, with confirmed SARS-CoV-2 infection by using the RT-PCR test. Images are in DICOM format, they were acquired using anteroposterior or posteroanterior views and they were reviewed by radiologists with 15 years of experience.
3.4. RSNA
The non-COVID-19 dataset used here is necessary to complement the images of COVID-19 positive patients with those of healthy subjects as well as with those of patients suffering from other pathologies. The RSNA dataset [15] contains 108,948 X-ray images in DICOM format of 32,717 patients in anteroposterior view collected from NIH clinical center’s internal PACS systems. The dataset is composed of images of patients having one or more of the following pathologies: atelectasis, cardiomegaly, effusion, infiltration, mass, nodule, pneumonia and pneumothorax. For our application, we selected all of the scans labeled as healthy or pneumonia, resulting in 85,374 images.
4. Methods
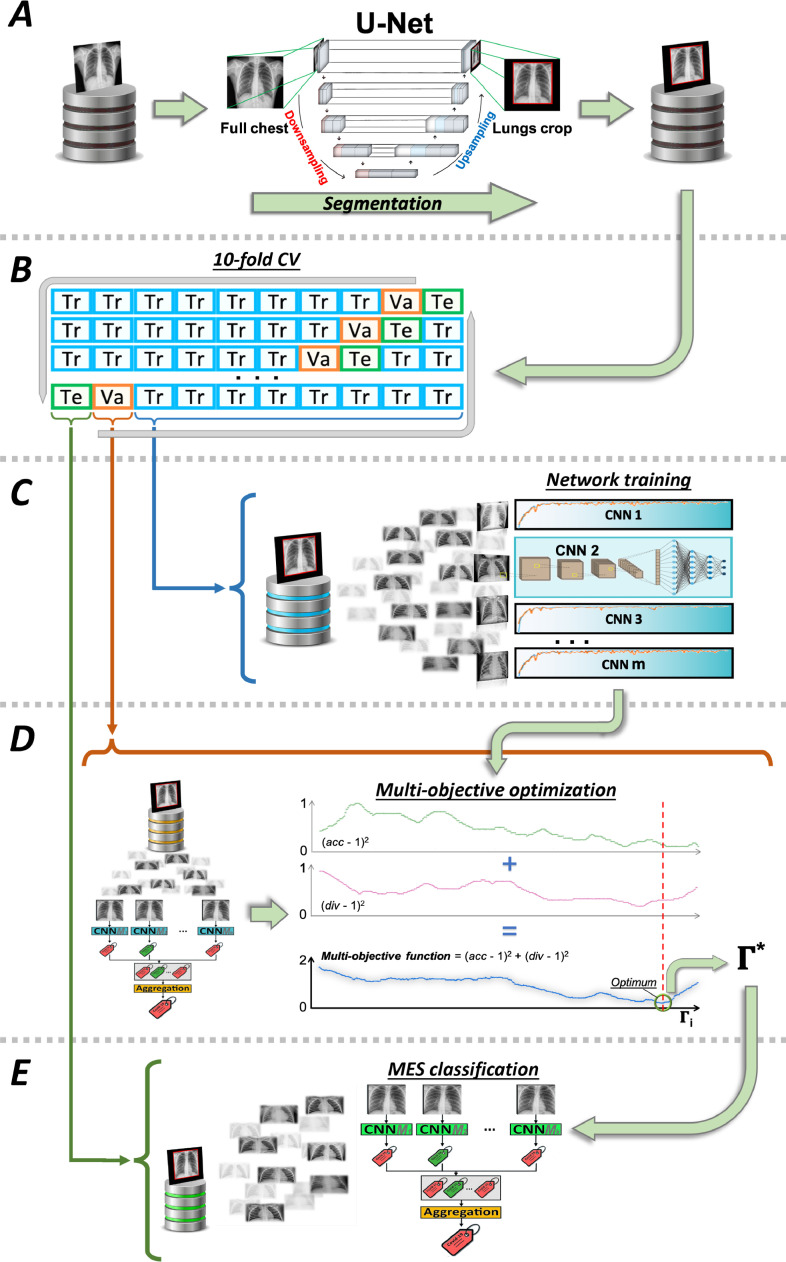
As described in Section 1, our work copes with two tasks: the first is the binary classification between COVID-19 patients and non-COVID-19 patients, whilst the second consists of a three-class classification task where we aim to recognize healthy subjects, patients suffering from COVID-19 and other pneumonia. To tackle the two different tasks, we adopt a common DL approach shown in Fig. 1 . First, panel A shows that each scan is segmented using a U-Net extracting a bounding box containing the patient’s lungs. Second, the cropped image is used to train CNNs (panel B and C), also referred to as base learners, models or classifiers, collected in
| (1) |
Third, still in the training stage, we construct an optimum ensemble finding which, and consequently how many, are the base learners to be aggregated (panel D). Indeed, the fusion of the decisions of all the models in could not be the best approach since some of them may degrade the performance of the MES: this happens since there exists a trade-off between accuracy and diversity [36]. Indeed, the literature shows that the base learners should be accurate and diverse, an issue further discussed in Section 4.2.1. Formally, , and is defined as
| (2) |
where . Hence, denotes the set of all the possible combinations of . To determine we maximize on a validation set a two-objective function based on the accuracy and the diversity of the ensemble. Fourth, in the test phase, the MES classifies the samples by applying the aggregation function on the outputs provided by each , as shown in panel E of Fig. 1. In the following subsections, we now deepen each of the previous steps. To simplify the reading, a summary of the notation adopted is presented after the conclusions.
Fig. 1.
Pipeline followed for the classification tasks.
4.1. Preprocessing
Given the variety of CXR scans, there is the need to automatically align the images by detecting the region of interest including the bounding box segmenting the lungs. To this goal, we use a U-Net, which has obtained very good performance in many biomedical applications [37]. It is composed of two parts: the encoder, used to capture the context in the image via convolutional and max-pooling layers, and the decoder, used to enable localization using transposed convolutions. We trained the network combining two lung CXR datasets: the Montgomery County CXR set (MC) [38], with 7470 scans, and the Japanese Society of Radiological Technology (JSRT) repository [39], with 247 scans. We used such repositories since they already contain the ground truth masks and, therefore, no manual segmentation is needed. It is worth noting that the use of a training set composed of non-COVID-19 images is not a limitation since lung detection does not depend upon the presence of any COVID-19 related signs. As input, the U-Net receives 256 256 normalized images and it was trained with an Adam optimizer and with a binary cross-entropy loss function. During training, we applied a random augmentation phase composed of: rotation (), horizontal and vertical shift ( pixels), and zoom (0-0.2). The batch size was set to 8 and the number of epochs was equal to 100. We also assessed the performance of this network in 5-fold cross-validation obtaining a Dice score equal to 0.972 and 0.956 on the MC and the JSRT datasets, respectively.
Once the U-Net is trained, we extract the bounding box containing both lungs. To this end, we do not use the minimum rectangular box containing the lungs, because in the further steps it has to be re-sized into a square to input the CNN. Indeed, this would cause the lungs to be deformed and the images would have an artificial source of variability that could make the CNN training harder as the classification models would be distracted by the different positions of the lungs. Therefore, we extract a squared box whose side is equal to the longest side of the minimum rectangle, while keeping fixed the barycenter of the box. Moreover, we disregard the regions smaller than 2% of the image.
4.2. Ensemble
The literature agrees that different CNNs have different ways of interpreting data [40]; to exploit this issue we set-up a hard ensemble method aggregating the predictions provided by several networks in via a late fusion approach that, in our case, consists in the well-known majority voting rule. Let us recall that and the related quantities are defined at the beginning of Section 4 Eqs. (1) and (2). Formally, given models in and labels, the output of the classifier on the class for a scan is , where and . The output given by majority voting is determined as
| (3) |
Straightforwardly, if applied to , Eq. (3) holds by replacing with .
The use of this simple rule to combine classifiers is also supported by observing that the choice of the fusion method is less important than to have diversity among the models [35], a topic deepened in the next section before presenting the approach we use to optimize the ensemble.
4.2.1. Ensemble diversity
In the case of non-ordinal outputs for classification, i.e. the case of majority voting rule, the literature reports that a necessary and sufficient condition for a MES to be more accurate than any of its base learners is if the models are accurate and diverse [41]. On new data points, a classifier is accurate if its error rate is lower than random guessing, whilst two classifiers are diverse if they make different errors. In this respect, a number of empirical investigations derived heuristic expressions try to estimate the diversity [41], which are divided into pairwise and non-pairwise measures [9]. The former consider the diversity of a pair of classifiers at a time by computing a metric of distance between all possible pairings of classifiers’ outputs in the MES, whilst the latter measures the diversity of the classifiers all at once calculating the correlation of each ensemble member with the averaged output. In the following, we focus on the pairwise measures. There exists four pairwise measures [9]: Q-statistics (), correlation coefficient (), disagreement measure () and double fault (). Since, the diversity should not be a replacement for the estimate of the accuracy of the ensemble we disregard as the smaller its values (i.e. higher diversity), the more accurate the MES [9]. Still according to [9], , or are correlated between themselves, a claim confirmed also in our preliminary experiments showing that the performance achieved is similar (or almost identical) regardless of the choice of , or . On these premises, in this manuscript we report the results achieved measuring the diversity between and by the correlation coefficient, defined as follows:
| (4) |
where is the number of wrong classifications made by both models, is the number of correct classifications made by the models, and are the number of images on which the classifications of the two models differ. To obtain the diversity between the models in we calculate:
| (5) |
Straightforwardly, if applied to , Eq. 5 holds by replacing with .
4.2.2. How to optimize the ensemble
Let us now turn back the attention to our multi-objective optimization task that finds out , i.e. the best combination of base learners to be aggregated. It consists in four steps:
-
(i)
the CNNs are trained and tested in 10-fold cross-validation on a dataset ;
-
(ii)
still in cross-validation on , we set-up the ensembles in , with the caution to have being an odd integer in , thus avoiding any tie in the majority voting rule;
-
(iii)
using the test set folders in , we measure the average performance of each ensemble in terms of accuracy and diversity ;
-
(iv)
among the different ensembles built, we find out the one () which maximizes both accuracy and diversity. This means that we look for the MES that, on the one side, returns the best classification performance and, on the other side, reduces the incidence and effect of coincident errors amongst its members. This step, therefore, returns an ensemble composed of a subset of all the available base classifiers.
The steps reported so far describe the application of our method when we have a single dataset where we employ the 10-fold cross-validation. Nevertheless, if we have also an external validation dataset, namely , we first perform steps (i)-(iv) setting equal to the ensemble selected in the majority of the cross-validation runs. Then we retrain the models in on the whole , still using a validation set for early stopping and, finally, we test on .
We now show how this intuitive four-step process corresponds to solve a multi-objective optimization problem. Given the pair , where is our set of objective functions and is a subset of defined by constraints, any point is said to be an admissible point for iff . Given 2 admissible points and for , we say that (dominates) according to Pareto when: , and for at least one . An admissible point for is said to be a Pareto optimum if there exists no other admissible points for such that . In other words, a Pareto optimum is an admissible point trying to minimize all functions in under the constraints , thus solving a multi-objective minimization problem over . Following the Fritz-John Theorem [42], the point is a Pareto optimum if there exist and s.t. the following system is satisfied:
| (6) |
On this basis, referring to step (iii) of our procedure, we seek for the MES maximizing both accuracy () and diversity (): hence, in this specific case we have two objective functions that straightforwardly are the accuracy and the diversity of the MES.
This makes reasonable to search for which is a Pareto optimum for the following minimization problem:
| (7) |
where denotes the cardinality of this set, i.e. the number of base learners in . The first constraint means that the number of chosen models must be equal or smaller than the total number of available models, whilst the second implies that the minimum number of models is 3. Following the Fritz-John Theorem [42], is a Pareto optimum if the quintuple satisfies:
| (8) |
As already stated, we would find the combination providing largest and . As a practical remark, both accuracy and diversity were normalized to the same range via a Min-Max scaler, in a way to give equal importance to both measures, and were evaluated as the average across the folds used in cross-validation. By observing that both and range in [0,1], we can write the following minimization problem:
| (9) |
It is worth noting that the first line of this system represents the optimization function, which simultaneously maximizes the accuracy and the diversity to obtain the best . According to the Kun-Tucker Theorem [43], we can solve this system:
| (10) |
This can be rewritten as:
| (11) |
Let the triple be the solution to this system. When and , where and since and ), satisfies the system and is a Pareto optimum for our optimization problem.
5. Experimental configuration
Here we first describe how the data was divided and used for the classification tasks, the single CNNs considered, and the competitors tested.
5.1. Train and test configurations
As described above, there were multiple sources for COVID-19 CXR images and our experiments would show that the proposed approach will generalize on unseen external data. For this reason, we keep the COVID-19 sets described in Section 3 separated and we do not integrate any images of the Brixia dataset in the training phase.
Let us first recall that we investigate two classification tasks: the first is a dichotomy (i.e. COVID-19 vs non-COVID-19), whilst the second is a three-class polychotomy (i.e. COVID-19 vs Pneumonia vs Healthy). This choice permits us to assess the performance of our proposal in the two classification tasks reported in the literature. We include in the training set of each CNN samples representing all the classes designing the following four combinations, where the class priors are balanced:
-
#E1
AIforCOVID under-RSNA (binary task): in this case the training set contains COVID-19 images from the AIforCOVID dataset and non-COVID-19 images from RSNA, randomly extracted from healthy and pneumonia classes, collapsed into a single label, shortly denoted as under-RSNA. We first ran a stratified cross-validation to assess the performance of the base learners as well as of the ensembles determining according to the method presented in the previous section. Next, to study how each classifier and generalize to an independent test set, we train each learner on the whole AIforCOVID under-RSNA and test on RSNA, i.e. non-COVID-19 images from the RSNA repository not included in under-RSNA, as well as on COVID-19 samples from COVIDx + and Brixia.
-
#E2
COVIDx + under-RSNA (binary task): in this case the training set contains COVID-19 images from the COVIDx + dataset and non-COVID-19 images from RSNA, randomly extracted from healthy and pneumonia classes, collapsed into a single label, shortly denoted as under-RSNA. We first ran a stratified cross-validation to assess the performance of the base learners as well as of the ensembles, finding . As before, to study how the models and generalize to an independent test set, we train each learner on the whole COVIDx + under-RSNA and test on RSNA, AIforCOVID and Brixia.
-
#E3
AIforCOVID under-RSNA (3-class task): in this case we include COVID-19 images from AIforCOVID, as well as scans from RSNA from healthy and pneumonia groups, kept with different labels, shortly denoted as under-RSNA. As before, we first ran a stratified cross-validation to assess the performance of the base learners as well as of the ensembles, finding . Then, to study each classifier and generalize to an independent test set, we train each learner on the whole AIforCOVID under-RSNA and test on RSNA, i.e. healthy and pneumonia images from the RSNA repository non included in under-RSNA, as well as on COVIDx + and Brixia.
-
#E4
COVIDx + under-RSNA (3-class task): in this case we include COVID-19 images from COVIDx + , as well as scans from RSNA from healthy and pneumonia groups, kept with different labels, shortly denoted as under-RSNA. As before, we first ran a stratified cross-validation to assess the performance of the base learners as well as of the ensembles, looking for . Then, to study how the classifiers and generalize to an independent test set, we train each learner on the whole COVIDx + under-RSNA and test on RSNA, AIforCOVID and Brixia.
In all the experiments, we work in a 10-fold cross-validation fashion, using 7 folds for training, 2 for validation and 1 for testing.
5.2. Stand-alone models
To have the baseline performance we used several CNNs, which were individually trained and tested according to a transfer learning approach, for the two classification tasks and the four aforementioned experiments.
We considered 20 well-known CNNs that have obtained promising results in biomedical applications [10]. They belong to 9 main families:
-
•
AlexNet [44]: spatial exploitation-based CNN which uses Relu, dropout, and overlap pooling layers.
-
•
VGG [45]: a spatial exploitation-based CNN which has an homogeneous topology and uses small size kernels. We considered VGG11, VGG13, VGG16, and VGG19.
-
•
GoogLeNet [46]: the last spatial exploitation network that we tested; it was the first architecture using the block concept and the split transform, and the merge idea.
-
•
ResNet [47]: a depth and multi-path based CNN which uses residual learning and has identity mapping based skip connections. We used these versions: ResNet18, ResNet34, ResNet50, ResNet101 and ResNet152.
-
•
WideResNet [48]: a width-based multi-connection CNN that, compared to the networks presented before, has a greater width and a smaller depth. We adopted the WideResNet50(2).
-
•
ResNeXt [49]: a width-based multi-connection CNN whose main characteristics are cardinality, homogeneous topology, and grouped convolutions. We utilized the ResNeXt50(32x4d).
-
•
SqeezeNet [50]: a feature-map exploitation based CNN modeling interdependencies between feature-maps. We used the SqueezeNet1(0) and the SqueezeNet1(1).
-
•
DenseNet [51]: a multi-path based CNN exploiting cross-layer information flow. We tested DenseNet121, DenseNet161, DenseNet169 and DenseNet201.
-
•
MobileNet [52]: it has an inverted residual structure and it utilizes lightweight depth-wise convolutions. In particular, we adopted the MobileNetV2.
Such models have different architectures, layer organization and complexity, permitting us to explore how different models perform on the task at hand. To have a fair comparison, all the networks underwent the same process: we modified their last layer, where we set a number of neurons equal to the number of classes to be predicted. Then, as the input image is the output of the segmentation process, the two-dimensional matrix is normalized and stacked to a three-dimensional tensor with three identical channels, having a 224x224x3 tensor. As mentioned, we use a transfer learning approach: we started from the CNNs already trained on the ImageNet dataset [53] using the final weights as initialization weights for the modified networks working on CXR scans. We also trained the models from scratch but the performance was not satisfactory. Furthermore, we applied data augmentation with a 0.3 probability. The possible transformations are: horizontal and vertical shift ( pixels), flip along the vertical axis, rotation () and elastic transformation (, ). The networks were trained with a batch size of 32, a cross-entropy loss function, and stochastic gradient descent optimizer with an initial learning rate of 0.001 and a momentum of 0.9, a step learning rate scheduler with a step size of 7, and . We fixed to 300 the maximum number of epochs, introducing also an early stopping of 25 epochs on the validation accuracy.
5.3. Competitors
To deepen the analysis of the results, we compare our approach with five different CNNs that have reported high performance in classifying CXR scans. Such five models were selected among the peer-reviewed papers listed in Table 1, since their codes are publicly available (available by May 2021). They are:
-
•
COVID-Net [26]: proposed for the three-class task, it is characterized by a light-weight design pattern, selective long-range connectivity, and architectural diversity.
-
•
DarkCovidNet [17]: it was proposed for both the binary and three-class task. It consists of a smaller version of the DarkNet architecture, made up by convolutional layers with different filter numbers, sizes, and stride values.
-
•
CheXNet [19]: it is a variation of the DenseNet121 and, according to the experiments presented in [19] for the three-class task it is the most performing architectures among the eight tested.
-
•
COVID-CAPS [24]: proposed for the binary task non-COVID-19 vs COVID-19, it is capable of handling small datasets. Its architecture consists of four convolutional layers and three capsule layers [54]. Each capsule represents a specific image instance at a specific location, through several neurons.
-
•
ConcatenatedNet [14]: proposed for the three-class task, it consists of a joint fusion of the Xception and ResNet50V2 networks. It utilizes the concatenation of multiple features, which are then connected to a convolutional layer that is fed to a fully connected classifier.
All of these networks were tested following the four cases described before. When necessary, the output layer was modified according to the tasks studied here.
5.4. Other ensembles
We considered three other competitors based on an ensemble of classifiers. The first is denoted as , and its performance is equal to the average of the results obtained across all the . This corresponds to random guessing the ensemble among the 524,268 combinations and, in practice, it reflects the real situation where practitioners pick random models and simply try to fuse them in an ensemble. On the one hand, the down-side of this approach is the randomness of the selection of the models, which would reflect in a wrong image classification if the majority of the models in predict badly. On the other hand, if the models have a decent performance, their fusion may provide better results than using a single model, if there is a certain degree of diversity. The second competitor is referred to as and it builds the ensemble by simply maximizing only the accuracy following the steps described in Section 4.2.2. The third one is referred to as and, similarly, it builds the MES only maximizing the diversity. Hence, and permits us to compare the multi-objective optimization with the separate optimization of each of its target functions.
6. Results and discussion
The results of the four experimental combinations described in Section 5.1 are presented in Table 3 , 4 , 5 and 6 . By column, the tables are divided into three parts: the first lists the learning models adopted, the second reports the results achieved in cross-validation, and the third shows the performance attained when we validate each learner on external validation. We measure the global accuracy (column “Global”) as well as the recall per each class. On the one hand, in case of experiments #E1 and #E2 (Table 3 and 4) in the second group of columns (“10-fold CV”) we show the specificity (“non-COVID-19”) and the sensitivity (“COVID-19”), whilst the third group of columns reports the results observed in external validation on the three datasets. Straightforwardly, in both tables the results on RSNA correspond to the specificity, whereas those on COVIDx + in Table 3 (and AIforCOVID in Table 4) and Brixia to the sensitivity. On the other hand, in the case of experiments #E3 and #E4 (Table 5 and 6) accounting for a three-class learning task, the organization per column is almost the same, except for having an extra column for each vertical section: hence, now the recall per class refers to healthy samples, patients suffering from non-COVID-19 pneumonia (shortly “pneumonia” in the tables) and COVID-19 cases.
Table 3.
Performance on experiment #E1 (AIforCOVID under-RSNA).
| Learning Model |
10-fold CV |
External Validation |
|||||
|---|---|---|---|---|---|---|---|
| Global | non-COVID-19 | COVID-19 | RSNA | COVIDx | Brixia | ||
| Single Model | AlexNet | 93.99.64 | 97.00.67 | 90.36.85 | 94.25 | 50.08 | 88.03 |
| DenseNet121 | 97.27.03 | 98.00.50 | 96.39.99 | 97.00 | 49.43 | 93.04 | |
| DenseNet161 | 96.72.11 | 96.00.70 | 97.59.70 | 95.71 | 62.07 | 93.82 | |
| DenseNet169 | 95.08.54 | 96.00.27 | 93.98.87 | 95.22 | 60.45 | 96.72 | |
| DenseNet201 | 93.99.58 | 96.00.47 | 91.57.38 | 96.19 | 51.54 | 81.00 | |
| GoogleNet | 96.17.31 | 99.00.77 | 92.77.60 | 95.14 | 47.16 | 73.53 | |
| MobileNetV2 | 95.63.92 | 95.00.70 | 96.39.80 | 96.03 | 52.35 | 78.91 | |
| ResNet18 | 95.08.87 | 96.00.77 | 93.98.94 | 96.03 | 54.78 | 90.18 | |
| ResNet34 | 95.63.58 | 96.00.81 | 95.18.96 | 95.47 | 48.14 | 87.16 | |
| ResNet50 | 97.27.61 | 99.00.03 | 95.18.82 | 96.43 | 58.67 | 83.34 | |
| ResNet101 | 96.17.07 | 97.00.17 | 95.18.83 | 95.22 | 63.37 | 95.85 | |
| ResNet152 | 96.17.09 | 96.00.90 | 96.39.02 | 96.03 | 59.00 | 95.48 | |
| ResNeXt50(32x4d) | 98.36.43 | 98.00.23 | 98.80.80 | 94.57 | 63.05 | 96.68 | |
| SqueezeNet1(0) | 91.80.60 | 95.00.78 | 87.95.91 | 95.71 | 38.74 | 79.74 | |
| SqueezeNet1(1) | 89.62.80 | 91.00.88 | 87.95.68 | 94.25 | 42.63 | 84.26 | |
| VGG11 | 93.99.36 | 95.00.88 | 92.77.43 | 94.73 | 45.38 | 92.35 | |
| VGG13 | 93.44.85 | 93.00.50 | 93.98.40 | 94.49 | 54.94 | 94.33 | |
| VGG16 | 93.44.65 | 96.00.79 | 90.36.69 | 95.95 | 51.70 | 85.13 | |
| VGG19 | 94.54.51 | 99.00.98 | 89.16.21 | 97.33 | 46.19 | 74.12 | |
| WideResNet50(2) | 96.17.58 | 98.00.65 | 93.98.83 | 96.03 | 61.26 | 88.07 | |
| Average | 95.032.03 | 96.302.00 | 93.503.11 | 95.59.86 | 53.057.21 | 87.597.38 | |
| Competitors | COVID-Net | 98.20.09 | 98.96.38 | 95.32.80 | 96.43 | 51.38 | 50.63 |
| DarkCovidNet | 96.06.44 | 96.80.54 | 95.18.71 | 95.84 | 56.79 | 90.40 | |
| CheXNet | 95.77.32 | 96.50.49 | 94.88.74 | 96.03 | 55.87 | 91.15 | |
| COVID-CAPS | 92.76.74 | 94.50.76 | 90.66.81 | 95.06 | 45.95 | 82.74 | |
| ConcatenatedNet | 97.27.51 | 98.00.44 | 96.39.82 | 95.30 | 62.16 | 92.38 | |
| MES | 96.152.78 | 97.272.72 | 94.802.82 | 96.011.90 | 67.829.89 | 93.172.84 | |
| 99.45.19 | 100.00.00 | 98.80.48 | 98.67 | 82.83 | 96.38 | ||
| 98.36.76 | 100.00.00 | 96.39.63 | 98.50 | 80.00 | 96.41 | ||
| 98.91.32 | 100.00.00 | 97.59.72 | 99.00 | 86.20 | 96.84 | ||
Table 4.
Performance on experiment #E2 (COVIDxunder-RSNA).
| Learning Model |
10-fold CV |
External Validation |
|||||
|---|---|---|---|---|---|---|---|
| Global | non-COVID-19 | COVID-19 | RSNA | AIforCOVID | Brixia | ||
| Single Model | AlexNet | 91.84.72 | 94.31.51 | 86.89.71 | 92.70 | 65.79 | 79.72 |
| DenseNet121 | 91.85.70 | 92.68.29 | 90.16.75 | 93.60 | 83.55 | 92.14 | |
| DenseNet161 | 92.93.79 | 93.50.44 | 91.80.73 | 94.10 | 81.51 | 91.52 | |
| DenseNet169 | 93.48.61 | 94.31.22 | 91.80.58 | 92.20 | 84.75 | 93.40 | |
| DenseNet201 | 91.30.56 | 91.87.01 | 90.16.84 | 93.60 | 84.51 | 91.82 | |
| GoogleNet | 92.93.40 | 93.50.60 | 91.80.92 | 92.40 | 75.03 | 87.39 | |
| MobileNetV2 | 92.93.74 | 95.12.95 | 88.52.92 | 93.70 | 77.19 | 87.07 | |
| ResNet18 | 92.93.67 | 95.12.10 | 88.52.82 | 93.60 | 72.51 | 86.99 | |
| ResNet34 | 94.02.94 | 95.94.81 | 90.16.64 | 93.60 | 78.15 | 88.22 | |
| ResNet50 | 92.93.76 | 95.12.54 | 88.52.12 | 93.80 | 79.23 | 89.71 | |
| ResNet101 | 90.76.67 | 94.31.95 | 83.61.91 | 94.20 | 74.91 | 86.77 | |
| ResNet152 | 90.22.40 | 93.50.78 | 83.61.08 | 94.00 | 79.35 | 87.58 | |
| ResNeXt50(32x4d) | 91.85.19 | 93.50.25 | 88.52.54 | 93.80 | 80.43 | 91.82 | |
| SqueezeNet1(0) | 90.76.17 | 91.87.78 | 88.52.90 | 91.00 | 64.23 | 72.03 | |
| SqueezeNet1(1) | 92.93.47 | 92.68.54 | 93.44.77 | 91.50 | 78.15 | 79.40 | |
| VGG11 | 92.39.96 | 91.87.37 | 93.44.59 | 92.80 | 71.31 | 84.22 | |
| VGG13 | 91.30.81 | 94.31.24 | 85.25.67 | 94.00 | 78.63 | 76.91 | |
| VGG16 | 91.85.94 | 94.31.66 | 86.89.54 | 93.50 | 74.19 | 85.81 | |
| VGG19 | 91.84.74 | 93.50.40 | 88.52.28 | 93.20 | 78.27 | 84.37 | |
| WideResNet50(2) | 91.85.88 | 92.68.54 | 90.16.37 | 92.40 | 80.43 | 92.46 | |
| Average | 92.14.99 | 93.701.18 | 89.022.82 | 93.19.89 | 77.115.51 | 86.475.71 | |
| Competitors | COVID-Net | 93.90.02 | 95.30.06 | 90.90.74 | 94.03 | 53.26 | 51.45 |
| DarkCovidNet | 92.17.69 | 94.80.64 | 86.88.51 | 93.84 | 76.83 | 87.85 | |
| CheXNet | 92.39.67 | 93.09.24 | 90.98.73 | 93.38 | 83.58 | 92.22 | |
| COVID-CAPS | 92.12.53 | 93.50.70 | 89.34.83 | 92.23 | 71.34 | 79.56 | |
| ConcatenatedNet | 91.85.54 | 93.09.40 | 89.34.46 | 93.10 | 80.43 | 92.14 | |
| MES | 93.633.49 | 94.642.46 | 91.594.63 | 94.533.75 | 83.035.63 | 92.653.24 | |
| 98.11.29 | 98.12.25 | 98.08.25 | 98.90 | 86.95 | 94.34 | ||
| 95.93.26 | 97.31.19 | 93.16.23 | 99.00 | 89.28 | 95.59 | ||
| 95.39.19 | 97.31.16 | 91.52.20 | 99.50 | 90.79 | 96.40 | ||
Table 5.
Performance on experiment #E3 (AIforCOVID under-RSNA).
| Learning Model |
10-fold CV |
External Validation |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
RSNA |
COVIDx | Brixia | |||||||
| Global | Healthy | Pneumonia | COVID-19 | Healthy | Pneumonia | ||||
| Single Model | AlexNet | 89.05.83 | 93.00.10 | 84.00.69 | 90.36.53 | 92.71 | 86.06 | 50.19 | 77.57 |
| DenseNet121 | 90.81.33 | 95.00.93 | 87.00.12 | 90.36.70 | 95.46 | 90.44 | 58.30 | 89.95 | |
| DenseNet161 | 93.29.53 | 96.00.44 | 89.00.32 | 95.18.96 | 94.98 | 89.47 | 61.86 | 90.59 | |
| DenseNet169 | 92.93.85 | 95.00.55 | 89.00.03 | 95.18.37 | 96.11 | 88.33 | 61.05 | 86.56 | |
| DenseNet201 | 92.23.54 | 93.00.12 | 91.00.53 | 92.77.06 | 94.81 | 89.47 | 58.95 | 68.95 | |
| GoogleNet | 89.05.32 | 95.00.99 | 84.00.54 | 87.95.99 | 93.19 | 86.22 | 55.22 | 89.01 | |
| MobileNetV2 | 92.93.49 | 95.00.70 | 91.00.33 | 92.77.25 | 94.33 | 89.14 | 56.60 | 87.41 | |
| ResNet18 | 91.87.67 | 93.00.65 | 91.00.29 | 91.57.02 | 94.33 | 89.63 | 60.41 | 92.23 | |
| ResNet34 | 90.81.57 | 94.00.99 | 88.00.12 | 90.36.26 | 95.14 | 89.30 | 59.92 | 88.56 | |
| ResNet50 | 92.93.17 | 92.00.06 | 91.00.63 | 96.39.96 | 93.52 | 87.52 | 66.73 | 82.02 | |
| ResNet101 | 93.64.02 | 96.00.06 | 90.00.72 | 95.18.6 | 94.33 | 89.14 | 59.92 | 93.21 | |
| ResNet152 | 94.35.77 | 95.00.87 | 91.00.46 | 97.59.48 | 96.43 | 86.71 | 73.21 | 92.01 | |
| ResNeXt50(32x4d) | 92.93.39 | 93.00.17 | 91.00.77 | 95.18.83 | 95.79 | 89.63 | 61.22 | 90.65 | |
| SqueezeNet1(0) | 91.17.74 | 94.00.05 | 89.00.10 | 90.36.47 | 93.19 | 87.52 | 49.87 | 82.47 | |
| SqueezeNet1(1) | 89.05.90 | 94.00.32 | 84.00.65 | 89.16.56 | 92.22 | 86.39 | 51.49 | 89.22 | |
| VGG11 | 91.87.85 | 95.00.65 | 90.00.77 | 90.36.51 | 93.35 | 88.87 | 56.52 | 92.20 | |
| VGG13 | 91.87.83 | 95.00.91 | 88.00.03 | 92.77.95 | 93.84 | 86.71 | 57.89 | 88.67 | |
| VGG16 | 91.17.38 | 94.00.78 | 90.00.74 | 89.16.70 | 92.22 | 88.49 | 50.52 | 72.10 | |
| VGG19 | 93.29.82 | 94.00.44 | 91.00.78 | 95.18.09 | 93.84 | 88.82 | 51.82 | 79.85 | |
| WideResNet50(2) | 92.23.74 | 94.00.90 | 90.00.53 | 92.77.57 | 95.14 | 88.01 | 57.65 | 83.01 | |
| Average | 91.871.54 | 94.251.07 | 88.952.44 | 92.532.75 | 94.251.24 | 88.291.32 | 59.518.97 | 85.816.84 | |
| Competitors | COVID-Net | 91.89.80 | 94.42.67 | 90.53.76 | 88.76.04 | 93.52 | 90.21 | 49.02 | 48.89 |
| DarkCovidNet | 92.72.44 | 94.00.53 | 90.20.44 | 94.22.46 | 94.75 | 88.46 | 64.04 | 89.61 | |
| CheXNet | 92.32.56 | 94.75.51 | 89.00.25 | 93.37.52 | 95.34 | 89.43 | 60.04 | 84.01 | |
| COVID-CAPS | 90.55.74 | 94.00.29 | 87.00.44 | 90.66.45 | 93.11 | 87.28 | 52.04 | 84.17 | |
| ConcatenatedNet | 92.58.57 | 93.50.54 | 90.50.65 | 93.98.70 | 95.47 | 88.82 | 59.44 | 86.83 | |
| MES | 93.012.28 | 95.182.49 | 90.192.42 | 93.782.19 | 95.47.62 | 93.962.69 | 64.639.96 | 88.823.56 | |
| 98.41.36 | 99.00.29 | 95.00.28 | 99.59.17 | 97.03 | 96.13 | 77.22 | 92.71 | ||
| 95.23.18 | 98.00.36 | 92.00.20 | 94.77.14 | 96.69 | 96.00 | 74.29 | 92.94 | ||
| 94.87.29 | 98.00.19 | 91.00.31 | 95.77.16 | 98.02 | 96.85 | 83.38 | 93.59 | ||
Table 6.
Performance on experiment #E4 (COVIDxunder-RSNA).
| Learning Model |
10-fold CV |
External Validation |
|||||||
|---|---|---|---|---|---|---|---|---|---|
|
RSNA |
AIforCOVID | Brixia | |||||||
| Global | Healthy | Pneumonia | COVID-19 | Healthy | Pneumonia | ||||
| Single Model | AlexNet | 86.89.66 | 88.52.33 | 86.89.47 | 85.25.84 | 88.50 | 82.40 | 49.46 | 60.68 |
| DenseNet121 | 89.62.76 | 93.44.50 | 86.89.39 | 88.52.61 | 91.70 | 83.60 | 82.35 | 90.71 | |
| DenseNet161 | 89.07.73 | 88.52.66 | 86.89.51 | 91.80.68 | 89.80 | 86.60 | 85.59 | 93.67 | |
| DenseNet169 | 87.98.85 | 91.80.58 | 85.25.70 | 86.89.62 | 92.50 | 81.60 | 75.75 | 89.63 | |
| DenseNet201 | 91.26.75 | 91.80.64 | 88.52.78 | 93.44.68 | 91.70 | 84.00 | 74.07 | 87.90 | |
| GoogleNet | 87.98.77 | 88.52.79 | 85.25.92 | 90.16.75 | 88.60 | 81.90 | 72.03 | 87.07 | |
| MobileNetV2 | 90.71.53 | 96.72.48 | 86.89.70 | 88.52.63 | 91.10 | 85.80 | 71.31 | 85.09 | |
| ResNet18 | 89.07.97 | 93.44.19 | 85.25.86 | 88.52.08 | 92.30 | 82.90 | 73.47 | 86.26 | |
| ResNet34 | 90.16.68 | 93.44.82 | 90.16.79 | 86.88.72 | 91.40 | 86.00 | 70.83 | 79.72 | |
| ResNet50 | 91.80.89 | 95.08.93 | 91.80.54 | 88.52.92 | 91.50 | 84.70 | 70.47 | 87.13 | |
| ResNet101 | 92.35.95 | 95.08.25 | 86.89.29 | 95.08.83 | 91.10 | 82.50 | 85.35 | 93.38 | |
| ResNet152 | 88.52.29 | 93.44.77 | 85.25.25 | 86.88.58 | 92.80 | 82.60 | 83.55 | 92.10 | |
| ResNeXt50(32x4d) | 90.16.17 | 93.44.48 | 86.89.60 | 90.16.61 | 92.80 | 83.80 | 85.11 | 92.14 | |
| SqueezeNet1(0) | 87.43.80 | 91.80.83 | 86.89.77 | 83.61.77 | 88.30 | 83.00 | 63.15 | 82.09 | |
| SqueezeNet1(1) | 89.07.01 | 91.80.39 | 86.89.85 | 88.52.67 | 88.40 | 84.10 | 66.03 | 89.71 | |
| VGG11 | 88.52.79 | 88.52.71 | 90.16.57 | 86.89.32 | 88.80 | 86.10 | 69.15 | 86.39 | |
| VGG13 | 89.62.42 | 93.44.94 | 83.61.82 | 91.80.51 | 88.20 | 84.50 | 76.83 | 77.06 | |
| VGG16 | 91.80.76 | 96.72.84 | 86.89.75 | 91.80.58 | 91.10 | 79.40 | 78.03 | 87.33 | |
| VGG19 | 89.62.23 | 91.80.21 | 85.25.50 | 91.80.67 | 89.60 | 83.60 | 77.43 | 90.07 | |
| WideResNet50(2) | 90.16.71 | 93.44.51 | 83.61.24 | 93.44.42 | 91.50 | 83.90 | 84.15 | 92.01 | |
| Average | 89.591.50 | 92.542.52 | 86.812.09 | 89.422.98 | 90.591.63 | 83.651.74 | 74.718.95 | 86.517.50 | |
| Competitors | COVID-Net | 92.08.27 | 91.70.66 | 95.90.46 | 90.90.26 | 88.67 | 85.77 | 47.54 | 46.26 |
| DarkCovidNet | 90.38.76 | 94.10.59 | 87.87.55 | 89.18.63 | 91.82 | 83.74 | 76.73 | 87.72 | |
| CheXNet | 89.48.77 | 91.39.60 | 86.89.60 | 90.16.65 | 91.43 | 83.95 | 79.44 | 90.48 | |
| COVID-CAPS | 88.53.50 | 92.21.51 | 86.89.70 | 86.48.73 | 89.08 | 83.83 | 62.49 | 79.39 | |
| ConcatenatedNet | 90.97.48 | 93.46.51 | 87.00.50 | 92.53.58 | 93.26 | 85.51 | 76.23 | 90.33 | |
| MES | 90.762.62 | 92.912.43 | 88.492.25 | 90.873.27 | 92.574.73 | 90.541.72 | 84.315.62 | 93.381.02 | |
| 96.08.17 | 97.72.21 | 92.80.16 | 97.72.18 | 98.07 | 93.22 | 90.73 | 95.18 | ||
| 95.44.14 | 98.72.23 | 93.80.19 | 93.80.15 | 98.05 | 92.69 | 90.35 | 94.99 | ||
| 92.47.08 | 93.16.08 | 91.52.13 | 91.52.15 | 98.16 | 93.68 | 91.70 | 97.48 | ||
By row, all the four tables are organized into three sections named as Single Model, Competitors and MES. The first reports the average results attained by each single CNN tested, and the average of such results (denoted as “Average”). The second section reports the performance yield by the competitors, which are single model too. The third section shows the results reported by the four ensembles of classifiers we tested, where is our proposal.
Let us now turn the attention to the results attained in the binary task aiming to discriminating between CXR images of non-COVID-19 and COVID-19 patients (Table 3 and 4). First, regarding the performance of the single CNNs, we notice that those attained in cross-validation are quite high, according to previous findings [13], [18], [55]. Nevertheless, such scores are larger than those attained in the external validation, showing that such models suffer to generalize to other images. Indeed, while the specificity (i.e. the recall on RSNA) in most of the cases decreases by a few percentage points, we find a severe drop in sensitivity. This holds for all the CNNs, competitors included, and it is observed using both COVID + and AIforCOVID datasets as external sources of validation. The average sensitivity decreases from 93.49% to 53.05% for the experiment #E1, and from 89.02% to 77.11% for experiment #E2. The same considerations apply also when Brixia is the external dataset, although the decrease is smaller, with the average sensitivity now being 87.59% and 86.47%. We deem that this happens since Brixia contains only images of hospitalized patients, having, therefore, a worse clinic picture than those positive returning home included in COVIDx + and AIforCOVID. We speculate that such a difference makes Brixia images simpler than the other two repositories.
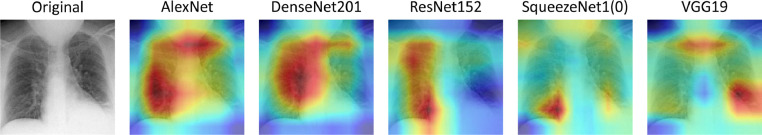
Still in the binary task (Table 3 and 4), we now focus on the results attained by using an ensemble of classifiers. In cross-validation experiments, we observe that the ensembles perform better than the average of the single CNNs, and than all the competitors. Nevertheless, there exist some single CNNs that in a few cases have a single performance score larger than one of the MES. Furthermore, within the four different ensembles, is the one with the lowest performance, although they are slightly better than the average of the CNNs. This can be expected since displays the average of the results obtained across all the possible MES (Section 5.2) whereas, on the contrary, both , and maximize an objective function. What is interesting is to look at the results of the external validation: the MESs still get a slightly larger specificity (i.e. RSNA columns in Table 3 and 4) than the single models, and they largely outperform the CNNs in terms of sensitivity on the COVID-19 datasets, showing that the fusion of several classifiers permits us to get satisfactory performance on external data sources. This result confirms our initial intuition about the loss of the discrimination ability of a single network when tested on an external dataset, which is a limitation overcome by the ensemble of classifiers. Appositely, an ensemble of classifiers exploits the fact that different networks focus on different features of the images: as a visual proof of this last assertion, Fig. 2 shows the CNNs’ Grad-CAM [56] activation maps for a single scan, where the largest activations are located in different areas of the chest.
Fig. 2.
Grad-CAM activation maps generated by different CNNs on a single scan from the AIforCOVID dataset.
Furthermore, this observation is also confirmed by the results attained by . To some extent, it can be considered a baseline for the ensemble-based approach since it shows what performance we can expect picking at random one MES, regardless of which and how many are the base learners used. The fact that this solution outperforms the single networks still confirms that a MES better generalizes to unseen data exploiting the diversity among the models. Such trend is more evident if we look at , , and where the maximization on a validation set of an objective function allows the MES to get better results. Furthermore, in both experiments #E1 and #E2, outperforms all the other solutions in terms of both sensitivity and specificity.
Let us now turn our attention to the results of experiments #E3 and #E4, i.e. those addressing the three-class task, which are reported in Table 5 and 6. It is worth noting that, as in the case of the binary task, outperforms all the other MESs as well as the single CNNs in the case of external validation. Furthermore, even if the task is now more difficult as the models have to discriminate images belonging from COVID-19, pneumonia and healthy subjects, we observe high recognition rates. Indeed, Table 5 shows that our proposal gets values of recall per class up to 98.02%, 96.85% for healthy and pneumonia patients, and up to 83.38% and 93.59% on COVID-19 images from COVIDx + and Brixia datasets. Similarly, in Table 6, gets values of recall per class up to 98.16%, 93.68% for healthy and pneumonia patients, and up to 91.70% and 97.48% on COVID-19 images from AIforCOVID and Brixia datasets. In the case of the experiments in cross-validation, we again find that an ensemble-based approach outperforms most of the single CNNs. Furthermore, among the MES alternatives, the reader can observe that in most of the cross-validation results yields the best performance. This finding can be expected since is the MES maximizing the accuracy on a validation set that belongs to the same dataset as the test set, so it is reasonable that the diversity has a smaller impact in this experimental setup. Nevertheless, the fact that diversity is a valuable property in an ensemble is demonstrated computing the performance on the external validation datasets, where it is that outperforms the other MES schema, as already discussed.
For the sake of completeness, Table 7 lists the base learners included in , and for the four experiments considered, showing that no network dominates with respect to others, thus corroborating the need for the proposed optimization stage.
Table 7.
Composition of , , and in all the four experiments.
| #EX1 | #EX2 | #EX3 | #EX4 | |
|---|---|---|---|---|
| DenseNet161 ResNet50 ResNeXt50(32x4d) | DenseNet121 ResNet50 ResNet101 ResNet152 VGG19 | DenseNet169 MobileNetV2 SqueezeNet1(1) | DenseNet201 ResNet101 VGG16 | |
| AlexNet DenseNet121 DenseNet161 | GoogleNet ResNet34 ResNet50 | DenseNet201 ResNet101 VGG11 | ResNet34 ResNet101 VGG16 | |
| DenseNet161 ResNeXt50(32x4d) WideResNet50(2) | ResNet34 ResNet50 VGG19 | MobileNetV2 VGG11 VGG13 | ResNet34 ResNet101 VGG16 |
Shifting the attention to the computation time, all the experiments were implemented in Python 3.7 using PyTorch as the main DL package, using an NVIDIA Tesla V100 16GB. We compute the average time needed to obtain a test prediction for the single models and for the MESs, with and without parallelization of each learner in the ensemble. The single models take , the MESs with no parallelization , and the MESs with parallelization . The Z-test pairwise applied to the three cases shows that the computation times are significantly different (0.05).
7. Conclusions
In this paper, we proposed an ensemble of CNNs that, combining multiple base learners by a late fusion approach, exploits the diversity of several state-of-the-art CNNs to maximize the recognition of chest X-rays. Although already explored in other medical and non-medical contexts, we confirm that an ensemble of classifiers can address challenging classification tasks: in our case, the proposed methodology not only performs at least as the best approaches currently available, but it also generalizes well to unseen data, differently from what happens for the other approaches. This happens since our method mixes different base learners by maximizing both the global accuracy and the diversity between the individual classifications by applying a criterion that provides an optimum solution according to Pareto’s multi-objective optimization theory. Indeed, our goal is not to find the absolute optimal combination of networks, but rather to define an algorithmic approach to decide which are the best networks to include in an ensemble, given a pool of classifiers and any available image dataset. This implies that, when a new external dataset becomes available, we will train the models on the data at hand, then we will find out and, using this optimized MES, we will classify such external dataset.
A large experimental campaign on more than 92,000 images publicly available, retrieved from four repositories, has validated the effectiveness of the algorithm used to set-up the MES, achieving recognition performance on the external validation sets that in most cases are larger than 95%. This could have a disruptive impact in the medical practice, supporting one or more of the following actions: (1) making it easier to carry out mass screening campaigns by pre-selecting the cases to be examined, enabling the medical doctors to focus the attention only on relevant cases, (2) serving as a second reader, empowering the physician capabilities and reducing errors, (3) working as a tool for training and education of specialized operators.
Future work are directed towards two different directions. First, to consolidate the robustness of the proposed method, we plan to explore other classification tasks such as the four-class problem proposed in [57], which aims to discriminate between healthy, pneumonia, other diseases (that may co-occur with pneumonia) and COVID-19. Second, we plan to investigate the applicability of our proposal to other classification tasks in medical imaging where CNNs have proven to be effective in cross-validation, but still fail when applied to external validation sets.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work is partially funded by POR CAMPANIA FESR 2014 - 2020, AP1-OS1.3 (DGR n.140/2020) for project “Protocolli TC del torace a bassissima dose e tecniche di intelligenza artificiale per la diagnosi precoce e quantificazione della malattia da COVID-19” CUP D54I20001410002. We acknowledge FSTechnology SpA which offered us GPU usage for the experiments. Our method won the AI against COVID-19 Competition, organized by IEEE SIGHT Montreal, Vision and Image Processing Research Group of the University of Waterloo, and DarwinAI Corp., and sponsored by Microsoft.
Biographies
Valerio Guarrasi received the B.C degree in Management Engineering, and the M.S. degree in Data Science from Sapienza University of Rome, Italy. He is persuing the PhD degree in Data Science at Sapienza University of Rome. His research interests include machine learning and deep learning, especially on bio-medical applications.
Natascha Claudia D’Amico graduated in Biomedical Engineer at Universitá of Stuttgart, Germany in 2016. Currently, she is a Data Scientist at Centro Diagnostico Italiano and is a Ph.D student in Biomedical Engineering of the University Campus Bio-medico di Roma. Her research interests include radiomics and deep learning applications on images.
Rosa Sicilia was born in 1993. In 2020 she finished her Ph.D. studies in Biomedical Engineering at University Campus Bio-Medico of Rome. As Post Doc researcher, her main interests are machine learning, data mining, radiomics, radiopathomics, eXplainable AI, automatic rumours detection in social networks and multivariate time series analysis.
Ermanno Cordelli graduated in Biomedical Engineering at the University Campus Bio-Medico di Roma in 2014 and in 2019 he obtained his PhD in Biomedical Engineering at the same University. Currently, Ermanno is in his second year of post-doctoral research and his main interests are machine learning and data mining.
Prof. Paolo Soda received a Ph.D Biomedical Engineering in 2008 at Universitá Campus Bio- Medico (UCBM). Currently he is Associate Professor of Computer Science and Computer Engineering at UCBM, and he is co-responsible of the Collaborative Laboratory of Precision Medicine & BioData Analytics between UCBM and Centro Diagnostico Italiano, Milan.
References
- 1.Tahamtan A., et al. Real-time RT-PCR in COVID-19 detection: issues affecting the results. Expert Rev. Mol. Diagn. 2020;20(5):453–454. doi: 10.1080/14737159.2020.1757437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fang Y., et al. Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology. 2020;296(2):E115–E117. doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manna S., et al. COVID-19: A multimodality review of radiologic techniques, clinical utility, and imaging features. Radiology: Cardiothoracic Imaging. 2020;2(3):e200210. doi: 10.1148/ryct.2020200210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.W.H. Organisation, Use of chest imaging in COVID-19, (file:///C:/Users/00020626/Desktop/WHO-2019-nCoV-Clinical-Radiology_imaging-2020.1-eng.pdf), Online; accessed 31 March 2021.
- 5.Aljondi R., et al. Diagnostic value of imaging modalities for COVID-19: scoping review. J. Med. Internet Res. 2020;22(8):e19673. doi: 10.2196/19673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wynants L., et al. Prediction models for diagnosis and prognosis of COVID-19 infection: systematic review and critical appraisal. Br Med J. 2020;369 doi: 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J.P. Cohen, et al., COVID-19 image data collection: Prospective predictions are the future, arXiv preprint arXiv:2006.11988(2020).
- 8.Hryniewska W., et al. Checklist for responsible deep learning modeling of medical images based on COVID-19 detection studies. Pattern Recognit. 2021;118:108035. doi: 10.1016/j.patcog.2021.108035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavalcanti G.D.C., et al. Combining diversity measures for ensemble pruning. Pattern Recognit Lett. 2016;74:38–45. [Google Scholar]
- 10.Gu J., et al. Recent advances in convolutional neural networks. Pattern Recognit. 2018;77:354–377. [Google Scholar]
- 11.Apostolopoulos I.D., et al. COVID-19: Automatic detection from X-ray images utilizing transfer learning with convolutional neural networks. Physical and Engineering Sciences in Medicine. 2020:1. doi: 10.1007/s13246-020-00865-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kermany D.S., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 13.Loey M., et al. Within the lack of chest COVID-19 X-ray dataset: a novel detection model based on GAN and deep transfer learning. Symmetry (Basel) 2020;12(4):651. [Google Scholar]
- 14.Rahimzadeh M., et al. A modified deep convolutional neural network for detecting COVID-19 and pneumonia from chest X-ray images based on the concatenation of xception and resnet50v2. Informatics in Medicine Unlocked. 2020:100360. doi: 10.1016/j.imu.2020.100360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X., et al. Proceedings of the IEEE Conf. on computer vision and pattern recognition. 2017. ChestX-Ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases; pp. 2097–2106. [Google Scholar]
- 16.Vaid S., et al. Deep learning COVID-19 detection bias: accuracy through artificial intelligence. Int Orthop. 2020:1. doi: 10.1007/s00264-020-04609-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ozturk T., et al. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. 2020:103792. doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Toğaçar M., et al. COVID-19 Detection using deep learning models to exploit social mimic optimization and structured chest X-ray images using fuzzy color and stacking approaches. Comput. Biol. Med. 2020:103805. doi: 10.1016/j.compbiomed.2020.103805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chowdhury M.E.H., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 20.Wang Z., et al. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest X-rays. Pattern Recognit. 2021;110:107613. doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brunese L., et al. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput Methods Programs Biomed. 2020;196:105608. doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khan A.I., et al. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest X-ray images. Comput Methods Programs Biomed. 2020:105581. doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Z., et al. Automatically discriminating and localizing COVID-19 from community-acquired pneumonia on chest x-rays. Pattern Recognit. 2021;110:107613. doi: 10.1016/j.patcog.2020.107613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Afshar P., et al. COVID-CAPS: A capsule network-based framework for identification of COVID-19 cases from X-ray images. Pattern Recognit Lett. 2020;138:638–643. doi: 10.1016/j.patrec.2020.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shorfuzzaman M., et al. Metacovid: a siamese neural network framework with contrastive loss for n-shot diagnosis of COVID-19 patients. Pattern Recognit. 2021;113:107700. doi: 10.1016/j.patcog.2020.107700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L., et al. COVID-Net: A tailored deep convolutional neural network design for detection of COVID-19 cases from chest X-ray images. Sci Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J., et al. Multi-task contrastive learning for automatic CT and X-ray diagnosis of COVID-19. Pattern Recognit. 2021;114:107848. doi: 10.1016/j.patcog.2021.107848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan Y., et al. COVID-19 detection from X-ray images using multi-kernel-size spatial-channel attention network. Pattern Recognit. 2021:108055. doi: 10.1016/j.patcog.2021.108055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vieira P., et al. Detecting pulmonary diseases using deep features in X-ray images. Pattern Recognit. 2021:108081. doi: 10.1016/j.patcog.2021.108081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Desai S., et al. Chest imaging representing a COVID-19 positive rural US population. Sci Data. 2020;7(1):1–6. doi: 10.1038/s41597-020-00741-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.P. Soda, et al., AIforCOVID: predicting the clinical outcomes in patients with COVID-19 applying AI to chest-X-rays. an italian multicentre study, arXiv preprint arXiv:2012.06531(2020). [DOI] [PMC free article] [PubMed]
- 32.Signoroni A., et al. BS-Net: Learning COVID-19 pneumonia severity on a large chest X-ray dataset. Med Image Anal. 2021;71:102046. doi: 10.1016/j.media.2021.102046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang S.C., et al. Fusion of medical imaging and electronic health records using deep learning: a systematic review and implementation guidelines. NPJ digital medicine. 2020;3(1):1–9. doi: 10.1038/s41746-020-00341-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baltrušaitis T., et al. Multimodal machine learning: a survey and taxonomy. IEEE Trans Pattern Anal Mach Intell. 2018;41(2):423–443. doi: 10.1109/TPAMI.2018.2798607. [DOI] [PubMed] [Google Scholar]
- 35.Kuncheva L.I. A theoretical study on six classifier fusion strategies. IEEE Trans Pattern Anal Mach Intell. 2002;24(2):281–286. [Google Scholar]
- 36.Dietterich T.G. An experimental comparison of three methods for constructing ensembles of decision trees: bagging, boosting, and randomization. Mach Learn. 2000;40(2):139–157. [Google Scholar]
- 37.Ronneberger O., et al. International Conf. on Medical image computing and computer-assisted intervention. Springer; 2015. U-Net: Convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 38.Jaeger S., et al. Two public chest X-ray datasets for computer-aided screening of pulmonary diseases. Quant Imaging Med Surg. 2014;4(6):475. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shiraishi J., et al. Development of a digital image database for chest radiographs with and without a lung nodule: receiver operating characteristic analysis of radiologists’ detection of pulmonary nodules. American Journal of Roentgenology. 2000;174(1):71–74. doi: 10.2214/ajr.174.1.1740071. [DOI] [PubMed] [Google Scholar]
- 40.Rawat W., et al. Deep convolutional neural networks for image classification: a comprehensive review. Neural Comput. 2017;29(9):2352–2449. doi: 10.1162/NECO_a_00990. [DOI] [PubMed] [Google Scholar]
- 41.Brown G., et al. Diversity creation methods: a survey and categorisation. Information fusion. 2005;6(1):5–20. [Google Scholar]
- 42.John F. Traces and emergence of nonlinear programming. Springer; 2014. Extremum Problems with Inequalities as Subsidiary Conditions; pp. 197–215. [Google Scholar]
- 43.Kuhn H.W., et al. Traces and emergence of nonlinear programming. Springer; 2014. Nonlinear Programming; pp. 247–258. [Google Scholar]
- 44.A. Krizhevsky, One weird trick for parallelizing convolutional neural networks, arXiv preprint arXiv:1404.5997(2014).
- 45.K. Simonyan, et al., Very deep convolutional networks for large-scale image recognition, arXiv preprint arXiv:1409.1556(2014).
- 46.Szegedy C., et al. Proceedings of the IEEE Conf. on computer vision and pattern recognition. 2015. Going deeper with convolutions; pp. 1–9. [Google Scholar]
- 47.He K., et al. Proceedings of the IEEE Conf. on computer vision and pattern recognition. 2016. Deep residual learning for image recognition; pp. 770–778. [Google Scholar]
- 48.S. Zagoruyko, et al., Wide residual networks, arXiv preprint arXiv:1605.07146(2016).
- 49.Xie S., et al. Proceedings of the IEEE Conf. on computer vision and pattern recognition. 2017. Aggregated residual transformations for deep neural networks; pp. 1492–1500. [Google Scholar]
- 50.F.N. Iandola, et al., SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5 MB model size, arXiv preprint arXiv:1602.07360(2016).
- 51.Huang G., et al. Proceedings of the IEEE Conf. on computer vision and pattern recognition. 2017. Densely connected convolutional networks; pp. 4700–4708. [Google Scholar]
- 52.Sandler M., et al. Proceedings of the IEEE Conf. on computer vision and pattern recognition. 2018. MobileNetV2: Inverted residuals and linear bottlenecks; pp. 4510–4520. [Google Scholar]
- 53.Deng J., et al. 2009 IEEE Conf. on computer vision and pattern recognition. IEEE; 2009. ImageNet: A large-scale hierarchical image database; pp. 248–255. [Google Scholar]
- 54.Hinton G.E., et al. International conference on learning representations. 2018. Matrix capsules with EM routing. [Google Scholar]
- 55.Das D., et al. Truncated inception net: COVID-19 outbreak screening using chest X-rays. Physical and engineering sciences in medicine. 2020;43(3):915–925. doi: 10.1007/s13246-020-00888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Selvaraju R.R., et al. Proceedings of the IEEE international Conf. on computer vision. 2017. Grad-CAM: Visual explanations from deep networks via gradient-based localization; pp. 618–626. [Google Scholar]
- 57.Basu S., et al. 2020 IEEE Symposium Series on Computational Intelligence (SSCI) IEEE; 2020. Deep learning for screening covid-19 using chest x-ray images; pp. 2521–2527. [Google Scholar]