Abstract
Peptide and peptidomimetic cyclization by copper-catalyzed alkyne–azide cycloaddition (CuAAC) reaction have been used to mimic disulfide bonds, alpha helices, amide bonds, and for one-bead-one-compound (OBOC) library development. A limited number of solid-supported CuAAC cyclization methods resulting in monomeric cyclic peptide formation have been reported for specific peptide sequences, but there exists no general study on monocyclic peptide formation using CuAAC cyclization. Since several cyclic peptides identified from an OBOC CuAAC cyclized library has been shown to have important biological applications, we discuss here an efficient method of alkyne–azide ‘click’ catalyzed monomeric cyclic peptide formation on a solid support. The reason behind the efficiency of the method is explored. CuAAC cyclization of a peptide sequence with azidolysine and propargylglycine is performed under various reaction conditions, with different catalysts, in the presence or absence of an organic base. The results indicate that piperidine plays a critical role in the reaction yield and monomeric cycle formation by coordinating to Cu and forming Cu–ligand clusters. A previously synthesized copper compound containing piperidine, [Cu4I4(pip)4], is found to catalyze the CuAAC cyclization of monomeric peptide effectively. The use of 1.5 equivalents of CuI and the use of DMF as solvent is found to give optimal CuAAC cyclized monomer yields. The effect of the peptide sequence and peptide length on monomer formation are also investigated by varying either parameter systemically. Peptide length is identified as the determining factor for whether the monomeric or dimeric cyclic peptide is the major product. For peptides with six, seven, or eight amino acids, the monomer is the major product from CuAAC cyclization. Longer and shorter peptides on cyclization show less monomer formation. CuAAC peptide cyclization of non-optimal peptide lengths such as pentamers is affected significantly by the amino acid sequence and give lower yields.
We report the heterogeneous controlled formation of monomeric cyclic peptides by CuAAC reaction using cooper–piperidine complexes.
Introduction
Cyclic peptides span the structural and molecular space between proteins and small molecules, with intermediate size, molecular weight, stability, and affinity characteristics.1 These desirable features are reflected in the increasing interest in research about naturally occurring peptides like cyclosporin2 and synthesized peptides such as octreotide.3 Peptide cyclization can be achieved with different orientations4 – head to tail, side-chain to side-chain, head to side-chain, and side-chain to tail. Lactamisation,5 disulfide bridge formation,6 olefin metathesis,7 thioether bridge formation,8–10 oxime formation,11 or SNAr reaction12 have been successfully used to cyclize peptides in the different orientations. ‘Click’ reaction or copper-catalyzed alkyne–azide cycloaddition (CuAAC) reaction is one such reaction successfully used for peptide cyclization. The 1,2,3-triazole formed during the CuAAC reaction can act as a disulfide mimic13 or as an amide bond mimic.14,15 Peptides cyclized by CuAAC have been used as alpha-helix mimics,16,17 and as disulfide-linked small proteins.18–20 While macrocyclization of peptides introducing constraints such as 1,3,4-oxadiazole and a tertiary amine induce peptide conformational rigidity and allow them to permeate into cells,21 we have previously demonstrated CuAAC cyclized heptameric peptides can have important biological properties. CuAAC cyclized peptides have been shown to bind active phosphorylated kinases like protein kinase B,22 inhibit in-cell the botulinum neurotoxin,23 bind malarial biomarker Plasmodium falciparum histidine-rich protein II (PfHRPII),22 inhibit heme sequestration by PfHRPII24 and distinguish interleukin IL-17F from its close isoform IL17A.25
In CuAAC cyclization, a peptide with alkyne and azide functionalities undergoes Huisgen 1,3 cycloaddition to form 1,4-disubstituted triazole linked cyclic peptide. The cyclic peptide may contain one26 or two triazoles,27 which will be referred to as ‘monomer’ (M) and ‘dimer’ (D), respectively (Fig. 1, ESI Fig. S.1†). The nature of the cyclized product may depend on several parameters such as peptide sequence,28 peptide length,26,28 and the choice of solvent.27 CuAAC cyclization reaction of peptides is done either in solution or on a solid support. The advantage of an in-solution ‘click’ is that in some instances, the monomeric cyclic peptide can form as the major product, even if the monomer is not formed or is minimally formed using on-resin cyclization.29,30 On the other hand, CuAAC of a peptide on a solid support is more convenient in terms of purification and reagents removal. But CuAAC on solid support has been reported to yield mainly dimeric cyclic products for peptides longer than hexamers,27,28 for small trimeric peptides,31 and in the cyclization of carbohydrates32 and carbohydrate/amino acid hybrids.33 Finn et al. did a systematic study of head-to-tail CuAAC cyclization of peptides on resin.27,28 They found that the process was independent of the peptide sequence, sensitive to the proximity of the alkyne to the resin, and sensitive to solvent composition.28 While monomeric peptide formation was found to be prevalent for peptides shorter than hexamers, the authors observed cyclodimerization for long peptides. Long peptides containing nine amino acids cyclized on solid support yielded dimers as the only product. It was hypothesized that the solvent (20% DMSO in acetonitrile) and the resin swelling allowed interstrand hydrogen bonding. For monomer formation, Meldal et al. explored CuAAC cyclization of a peptide on PEGA resin using CuI and DIEA, under side-chain protected and side-chain deprotected conditions. In both cases, a yield of 79% of the cyclic monomer was obtained.34 It was postulated that the low local substrate concentration of the PEGA resin was advantageous in maximizing monomer formation and minimizing dimerization. In 2009, Lokey et al.26 and Goncalves et al.35 reported monomer peptide formation using CuBr or CuI and sodium ascorbate in the presence of weak bases like DIEA and 2,6 lutidine, for specific peptide sequences. However, no specific guidelines for controlling the cyclization of peptides as monomers have been reported in the literature.
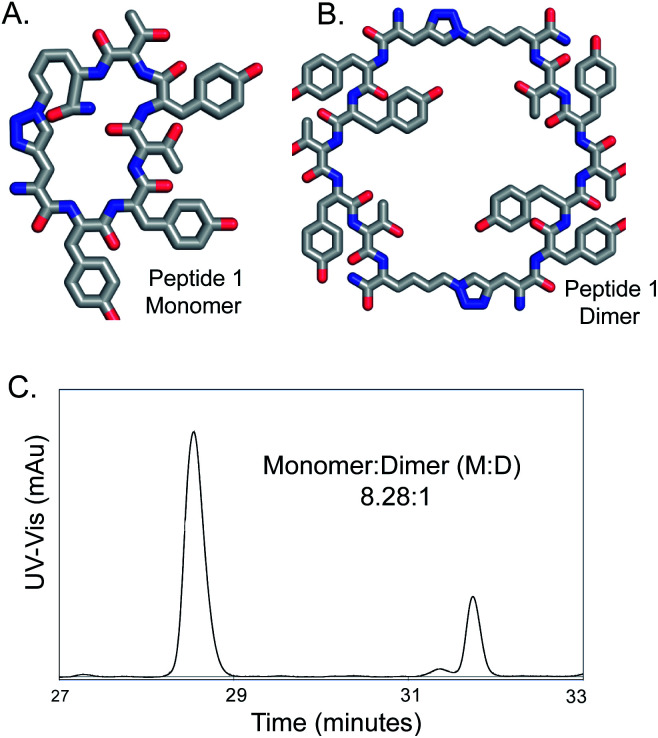
Fig. 1. On-bead CuAAC cyclization of a linear peptide yields peptide 1, predominantly in monomer form. (A) Structure of peptide monomer (M). (B) Structure of peptide dimer (D). (C) Analytical reverse phase HPLC trace of cyclized peptide 1 cleaved from resin shows M is formed as major product. Absorbance trace at 280 nm is shown.
In this article, we explore the effect of the catalyst composition and ratio, peptide sequence and peptide length on CuAAC cyclized peptide monomer formation on a polystyrene high-loading solid support. We observe that for specific peptide lengths, irrespective of sequence, the monomeric cyclic peptide is the major product under the current reaction conditions. Monomeric peptides are the major product, when peptides containing six, seven, or eight amino acids are cyclized using this on-resin CuAAC reaction condition. Cyclization of smaller and longer peptides lead to a decrease in the formation of monomers and an increase in the formation of dimers. In addition to peptide length, the composition and ratio of catalyst are critical parameters for getting high yield and getting monomer formation. We demonstrate, for the first time, that the CuAAC reaction of peptides can be catalyzed with a high yield (approximately 89%) by a polynuclear Cu catalyst [Cu4I4(pip)4]. We also demonstrate CuI is as efficient as a catalyst as [Cu4I4(pip)4] under certain reaction conditions, giving a high yield of the cyclized monomer peptide. We hypothesize that this high yield may possibly be occurring due to Cu(i) coordination to piperidine and DMF to form catalytic Cu–ligand clusters. Our experimental data suggest that piperidine stabilizes Cu(i) and forms smaller Cu(i)–piperidine catalyst complexes which favor monomeric cyclic peptide formation. This research explores the effect of in situ catalyst complexes on CuAAC cyclic product formation and therefore allows the controlled synthesis of monomeric or dimeric cyclic peptides.
Results and discussion
We previously reported the synthesis and use of a CuAAC cyclized comprehensive One-Bead-One-Compound (OBOC) peptide library for screening against proteins and ligand identification.22–25,36–38 For this library, we noticed that for most cyclized peptides, the monomer was the main product, as evident from the Edman Peptide Sequencing of the on-bead peptides.22 The size of the peptide library (heptameric) were based on screening requirements, and the orientation and the identity of the reacting azide and the alkyne were serendipitous choices.39 Given the literature information available, obtaining high yields (80–90%) of cleanly synthesized monomer cyclic peptides through on-bead cyclization in this library was fortunate. Such high yields of cyclic monomers have rarely been reported for CuAAC cyclization reactions on-bead.40 In this article, the reason behind clean monomer formation was investigated by varying the different reaction parameters. First, the role of the catalyst was examined. Different catalysis conditions for synthesis of a peptide sequence 1 (Fig. 1, Figure S.1), which is a ligand for protein kinase B or Akt2 and binds Akt2 with an EC50 of 122 nM,22 were explored.
Effect of catalyst composition and ratio on solid-supported CuAAC peptide cyclization
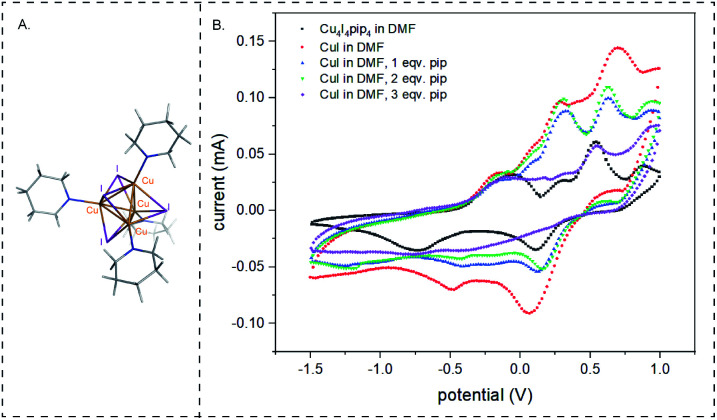
High yields of cyclized peptide 1 were obtained using CuI, ascorbic acid, and piperidine for on-bead CuAAC cyclization of linear peptide Fmoc-L-Pra-YYTYT-Az4-Rink (Fig. S.2†) (Table 1A, F, G), (Pra: l-propargylglycine; Az4: azidolysine; Rink: rink amide resin). Computational41 and kinetic studies42 indicate that more than one Cu center is involved in CuAAC reaction, as at intermediate concentrations, the reaction is second-order with respect to Cu(i) and alkyne concentrations. It had been previously hypothesized that a dynamically changing family of different Cu(i)–acetylide species might exist in solution.43,44 Recently, such a π,σ-bis(copper) acetylide complex was isolated and it was demonstrated that binuclear copper complexes are kinetically favored over mononuclear copper complexes for the click reaction.45 In terms of catalyst source, dinuclear46 and polynuclear47 copper acetate complexes are well-established as excellent click reaction catalysts. We hypothesize that under the described reaction conditions, polynuclear Cu(i)–piperidine clusters might be forming and that these clusters formed in situ are acting as an efficient catalyst. To test this hypothesis, we investigated the catalytic performance of a polynuclear complex of Cu(i), [Cu4I4(pip)4]48,49 for CuAAC peptide cyclization. The crystal structure of [Cu4I4(pip)4]49 has been previously published, but the CCDC data is not available. The crystal system is tetragonal and space group is P42/n (no. 86), with a = 14.7715(2) Å, c = 7.6073(2) Å. As a corresponding pyridine analog [Cu4I4(py4)] (CCDC 1529204) also has a stretched cubic structure, its crystal system being orthorhombic and space group being P212121 (no. 19), with the third axis (c 11.756(3) Å) being shorter than the other two axes (a 16.032(6) Å, b 15.510(2) Å),48 we modified the three-dimensional structure of the latter in Fig. 2A to depict the crystal structure of [Cu4I4(pip)4]. The cyclization of peptide 1 through on-bead catalysis by CuI and piperidine, or [Cu4I4(pip)4], were compared. Excess ascorbic acid was added to both reactions to maintain the oxidation state (I) of Cu. Both Cu(i) compounds catalyzed the CuAAC reaction with high cyclization percentages and yielded high monomer : dimer (M : D) ratios. [Cu4I4(pip)4] was the most efficient catalyst, converting 89.2% linear peptide to corresponding cyclic products (Table 1B). However, the most significant M : D ratio of 8.6 : 1 was obtained for CuI and 20% piperidine, with excess ascorbic acid (Table 1A). The yield using CuI and 20% piperidine was comparable but slightly less (87.4%) than when using [Cu4I4(pip)4].
Cyclic monomer and dimer formation with different catalyst compositionsa.
| Entry | Catalyst | Cu(i) | Ascorbic acid | Piperidine (% vol) | Monomer : dimer | % yield |
|---|---|---|---|---|---|---|
| A | CuI | 1.5 | 5 | 20 | 8.6 : 1 | 87.4 |
| B | [Cu4I4(pip)4] | 1.5 | 30 | 0 | 5.04 : 1 | 89.2 |
| C | [Cu4I4(pip)4] | 1.5 | 5 | 20 | 7.14 : 1 | 84.3 |
| D | CuI | 1.5 | 5 | 0 | 5.90 : 1 | 73.8 |
| E | CuI | 1.5 | 30 | 0 | 3.74 : 1 | 74.1 |
| F | CuI | 1.5 | 30 | 1 | 4.24 : 1 | 84.1 |
| G | CuI | 1.5 | 30 | 3 | 6.04 : 1 | 84.0 |
| H | CuI | 1 | 5 | 20 | 9.53 : 1 | 66.8 |
| I | CuI | 0.5 | 5 | 20 | 8.04 : 1 | 66.0 |
Cu(i) and ascorbic acid given in equivalents.
Fig. 2. Study of copper complexes formed during CuAAC reaction. (A) Structure of [Cu4I4(pip)4] complex. Structure modified from CCDC 1529204 using Pymol. (B) Comparing cyclic voltammetry of 1.25 mM [Cu4I4(pip)4] to 5 mM CuI with different concentrations of piperidine in DMF under inert atmosphere, with 0.25 mM supporting electrolyte tetrabutylammonium perchlorate (TBAP). Potential measured vs. Ag+/Ag electrode (reference electrode). Scan rate of 50 mV s−1 was used.
Interestingly, when excess piperidine was added to [Cu4I4(pip)4] (Table 1C), the M : D ratio was significantly higher (7.14 : 1) than that for conditions in Table 1B (M : D 5.14 : 1), though the percentage yield was lower (84.3%). The presence of excess piperidine (Table 1C) might allow the exchange of the weak-field ligand iodide with strong-field ligand piperidine. Since piperidine is not a bridging ligand, piperidine co-ordination might encourage the formation of smaller Cu–piperidine complexes, shifting the equilibrium of CunIm clusters. These dynamic Cu–piperidine complexes could favor intramolecular reactions to form the cyclic monomer peptide. During the conversion of the catalyst from tetranuclear [Cu4I4(pip)4] to smaller complexes on piperidine addition, possibly some complexes are created that do not have similar catalytic activity.50 This explains why piperidine addition to [Cu4I4(pip)4] reduces the yield of reaction from 89.2% to 84.3%. These results indicate that the identity of the catalyst and the presence of piperidine play essential roles in obtaining a large yield and a high ratio of monomeric cyclic peptides. This set of results led to the next set of experiments in which piperidine concentrations were varied.
Effect of varying concentrations of piperidine on peptide cyclization for solid-supported CuAAC reaction
CuAAC reactions with CuI and excess ascorbic acid, without addition of piperidine, showed significantly decreased cyclization (Tables 1D and 1E). Unexpectedly, there was 73–74% cyclization yield from the CuAAC reaction, even in the absence of an added base. When CuI is used as Cu(i) source, one typically needs to add an amine base.34,51,52 As CuI has a tetrahedrally coordinated Wurtzite lattice structure,53 a certain amount of acetylide anion is required before the reactive complex can form. In the absence of the base, elevated temperatures54,55 or ultrasonication conditions,56 can allow a CuAAC reaction with CuI catalyst to proceed successfully. Though this reaction was performed at room temperature without any external source of energy like sonication, good cyclization yield was obtained.
We hypothesize that the high local acetylene concentration on solid support facilitates the formation of copper acetylide and hence the CuAAC reaction. As CuI has a well-defined lattice structure, with multiple Cu ions bridged by I− ions, CunIm clusters can form in solution on dissolving CuI.57 DMF, the solvent of choice, can also coordinate with Cu(i) as a ligand. When DMF coordinates to Cu(i), forming Cu–DMF complexes58 that favor intermolecular reactions, dimeric cyclic peptides are formed as the major product. The dimeric peptides undergo cyclization head to tail. IR spectroscopy was used to verify that both cyclization reactions occur (ESI Fig. S.3B and S3.C†). When one or more equivalents of piperidine are added in the reaction (Tables 1F and 1G), the cyclic product yield increased, as expected, but more importantly, the M : D value increased. Adding piperidine promoted the formation of complexes that favour intramolecular reaction of each peptide to form monomeric cycles.
To understand the nature of the catalytic complexes formed by copper during the reaction, CuAAC reactions catalysed by [Cu4I4(pip)4] and CuI, with and without piperidine, were monitored using cyclic voltammetry (CV). Instead of using ascorbic acid as the chemical reductant, the potential was held lower than 1 volt vs. Ag/AgCl electrode. DMF solvent and inert atmosphere were used for the CV studies. For [Cu4I4(pip)4] three quasi-reversible peaks were observed at −0.75 V, −0.056 V, and 0.5 V respectively and one irreversible peak was observed at 0.3 V. For CuI in DMF, two quasi-reversible peaks were observed at 0.3 V and 0.7 V and two irreversible peaks were observed at −0.2 V and −0.5 V. On the addition of piperidine, the peak at 0.7 V shifted to lower potential. Increasing equivalents of piperidine made this shift more prominent, and with three equivalents of piperidine, the peak was closest to the 0.5 V peak in [Cu4I4(pip)4]. Therefore, CuI with three equivalents of piperidine acts similarly to the [Cu4I4(pip)4] complex. On further addition of piperidine (4 equivalents), the peak at 0.5 V vanished. Further piperidine addition, up to 20 equivalents (ESI Fig. S.4†), showed the same cyclic voltammetry signature. The quasi-reversible peak at 0.3 V of CuI in piperidine and DMF slightly shifted to 0.2 V. The irreversible peak at −0.2 V increased.
In terms of the number of equivalents of Cu used in the CuAAC cyclization reaction, previously different authors have used different amounts of Cu(i) for CuAAC reactions – all the way from 0.5 equivalents to 2 equivalents. Finn et al. used 0.5 equivalents for resin-based cyclodimerization of a 9 amino acid containing oligopeptide. The authors observed that the use of higher equivalents like 10 equivalents had deleterious effects for synthesis of peptide cyclodimer synthesis by CuAAC.27 On addition of 1.5 equivalents of CuI, a lower yield of the cyclic dimer was observed, along with some of the unconverted linear peptide. In another paper by the same group in 2008, it was observed that use of 0.5 to 4 equivalents of CuI for resin-based cyclodimerization of the same oligopeptide had no effect on yield.28 The effect on varying the CuI concentration for CuAAC cyclization, therefore, seems unresolved.
Other researchers have used 1 equivalent40 or 2 equivalents59 of CuI for CuAAC reaction of peptides. For cyclization of the current heptameric peptides with specific azide and alkyne we observe that the use of 1.5 equivalents CuI is optimal. Using 0.5 equivalents or 1 equivalents of CuI gives similar results of the monomeric cyclic peptide being the predominant product, but the efficiency of the reaction decreases from 87.4% to approximately 66% (Tables 1H and 1I). During CuAAC cyclization, CuI is adsorbed to a large extent by the resin. This is evident as during the reaction clean-up, during washes with sodium diethyl dithiocarbamate, the resin changes colour from deep brown to light yellow, while the sodium diethyl dithiocarbamate solution changes colour from light yellow to brown as it reacts with copper salts.60 Because of the adsorption of Cu by the resin during the CuAAC reaction, only a certain fraction of the Cu is available to coordinate to the peptide in the solution. Use of 0.5 or 1 equivalent of CuI leads to lower CuI equivalents present in solution and leads to non-optimal CuAAC cyclization. We observe much less clean HPLC traces and more side products when 0.5 or 1 equivalents of CuI is used, as has been previously observed for non-optimal conditions for CuAAC cyclization.27
Effect of using a different organic base
Peptide 1 was cyclized using three different conditions, to compare piperidine with two standard bases- 2,6-lutidine and DIEA (N,N-diisopropylethylamine) (Table 2).11,26,27 A significantly higher conversion (87.4%) to cyclic products was obtained for reaction conditions 2A than previously reported reaction conditions (Table 2, ESI Fig. S.5†). Reaction conditions 2A also gave the most significant yield of monomeric cyclic peptides, with the best monomer to dimer ratio (M : D) of 8.28 : 1. If we compare the entries Tables 1A and 1D, lack of addition of the base decreases the yield of the reaction from 87.4 to 73%. These results lead us to conclude that the base piperidine has a significant effect on the reaction yield. Other researchers have previously found addition of a large excess of piperidine, such as 20% piperidine in DMF, was crucial for obtaining the quantitative yields required for repetitive CuAAC reactions to introduce four triazoles, whereas use of DIEA, pyridine or 2,6 lutidine gave lower yields.61
Reaction conditions, product formation and conversion for on-bead CuAAC cyclized peptides.
| Entry | Cu(i) (eq.) | Reagents (eq.)/solvent | Time (hrs) (inert gas) | Monomer % | Dimer | % conversiona |
|---|---|---|---|---|---|---|
| A | CuI (1.5) | Ascorbic acid (5), 20% piperidine/DMF | 15 | 51.1 | 13.0 | 87.4 |
| B | CuBr (1.0) | Sodium ascorbate (3)/2,6-lutidine (10)/DIEA (10)/MeCN : DMF (9.6 : 90.4) | 6 (argon) | 22.85 | 0e | 33.0 |
| C | CuI (0.5) | 2,6-Lutidine (2), DMSO : MeCN (20 : 80) | 16 (argon) | 3.8 | n.d.b,c | n.d.b,d |
With respect to linear starting material, the percentage of CuAAC products.
Could not be determined.
Mass corresponding to dimer could not be found.
Total yield could not be determined because of lack of dimer mass.
Linear peptide with one triazole formed instead of dimer; DMSO dimethyl sulfoxide, ACN acetonitrile, DMF dimethylformamide, Pip piperidine, DIEA diisopropylethylamine
Effect of solvent and reaction time
All three reactions in Table 2 were done in different polar aprotic solvents, and the solvent identity can play an important role, due to the different swelling properties of the resin in the three solvents.34 On-resin CuAAC cyclization of peptides can vary in efficiency with reaction solvent variation.27 High efficiency of 83% for triazole tethering of a helical peptide has been observed in 80 : 20 DMSO water mixture40 using CuBr as catalyst. Polar aprotic solvents like DMSO disrupt internal H-bonding and therefore cyclodimerization in the resin bound peptide,28 which favours monomer cyclic peptide formation. In fact, when Finn et al. changed the solvent system from 4 : 1 MeCN : DMSO to 100% DMSO, the disruption was enough that cyclomonomer was formed as the major product instead of cyclodimer with the same peptide sequence.28 Finn et al. noted28 that monocyclization of two azido–alkyne oligopeptides had been reported using the highly polar and protic combination of 20% H2O in N-methylpyrrolidinone (NMP) as the solvent.35 In this article, the use of the polar aprotic solvent DMF was explored. DMF, being a polar aprotic solvent, can disrupt internal hydrogen bonding,28 and provide high solubility for the catalytic Cu–piperidine complexes formed during the reaction. An efficiency of 87% was observed with DMF under these reaction conditions. While in Table 2A, long reaction times were used, the same reaction could be efficiently done in one hour (ESI Fig. S.6†). Therefore, the reaction is optimal for reaction times of one hour or more, when using the polar aprotic solvent DMF.
Effect of azide and alkyne orientation on CuAAC cyclization on-bead
It has been previously reported that dimeric cyclic peptides are formed as the major product after on-resin CuAAC reaction of peptide sequences that contain an alkyne on the carboxy terminal.27 Therefore, we wanted to look at the effect of alkyne/azide orientation on the M : D ratio. Peptide 2 containing C-terminal alkyne was cyclized under standard reaction conditions (Table 3A). While peptide 1 had an M : D ratio of 8.28 : 1, reversing the azide and alkyne orientations in peptide 2 causes the M : D ratio to change to 7.04 : 1. Thus, there was a small decrease in intramolecular cyclic monomer formation and an increase in cyclic dimer formation (Table 3). Switching the positions of azide and alkyne functionalities decreased the cyclization yield, from 87.4% to 78.8%, with the monomeric cyclic peptide remaining the overwhelmingly major product (ESI Fig. S.7†).
Effect of azide and alkyne orientation and presence of linker on CuAAC cyclization.
| Peptide | Cyclic peptide | Linear peptide precursor | % converteda | Monomer : dimer |
|---|---|---|---|---|
| 1 | (YYTYT-Tz4)cy | Fmoc-Pra-YYTYT-Az4 | 87.4 | 8.28 : 1 |
| 2 | (Tz4-YYTYT)cy | Fmoc-Az4-YYTYT-Pra | 78.8 | 7.02 : 1 |
| 3 | (YYTYT-Tz4)cy-(PEG)5 | Fmoc-Pra-YYTYT-Az4-(PEG)5 | 53.5 | 4.41 : 1 |
Compared to pure linear starting material.
Since on solid support there are high local concentrations of azide and alkyne, it was investigated if adding a polyethylene glycol (PEG) spacer between the peptide and the solid support increased or decreased CuAAC cyclization yield and affected M : D ratio. Since adding a spacer makes the environment less sterically crowded, it was expected the cyclization yield would increase. Interestingly, it was observed that the yield decreased significantly to 53.5% for peptide 3, and during product purification by HPLC, new peaks with molecular weights corresponding to polymeric peptide masses were observed. Adding a PEG spacer between the solid support and the peptide in peptide 3 (ESI Fig. S.8†) decreased the M : D ratio compared to peptide 1. Comparing the HPLC traces of peptide 1 and peptide 3, we observe that the monomer is formed to a much lower extent for peptide 3 (36.8%) than for peptide 1 (70.36%), while the extent of dimer formation remains more or less unchanged between peptide 3 (16.67%) and peptide 1 (17.00%). It has been previously established that interchain hydrogen bonding played a critical role in dimer formation and addition of PEG spacer did not disrupt cyclodimerization because it did not engage in competitive hydrogen bonding.28 This explains the consistency of dimer formation between peptides 1 and 3. For CuAAC cyclization of a peptide, the alkyne, the catalytic copper cluster, and the azide come together in the transition state. Whether the first cycloaddition is intermolecular or intramolecular is controlled mainly by structural probability.57 We believe that maintaining the optimal reaction conditions for on-resin side-chain protected heptameric sequence Fmoc-Pra-XXXXX-Az4 dictates the structural probability of monomer formation. However, the addition of a PEG spacer seems to reduce the structural probability somewhat and encourages the formation of polymers larger than dimers. We have seen this trend in HPLC traces of other peptides on incorporating PEG linker between resin and cyclic peptide.22 Difficulty in the isolation of the polymeric masses prevented us from determining whether they are cyclic polymers or linear polymers.
Effect of length of the peptide on CuAAC cyclization – exploring heptameric sequences
Monomeric cyclic peptides were formed as the major products on cyclization of heptameric peptides. In addition to peptide 1, cyclization of other heptameric peptide sequences (Table 4) yielded cyclic monomers as the major product (ESI Fig. S.9–S.13†). For some of the heptameric peptides, on cyclization there is no dimer formed at all (Table 4, peptides 4, 5, 6) while for some heptameric peptides, cyclic dimer is formed as minor product (peptides 1, 7, 8). Thus, the M : D ratio can vary, but the monomeric cycle is the major product for on-resin cyclization of side chain protected heptameric peptides of sequence Fmoc-Pra-XXXXX-Az4. An exception to this trend was observed for a polyalanine heptapeptide (ESI Fig. S.14†), which produced dimer as the major product (M : D 1 : 1.93). Polyalanine is known to form alpha helix homopolymers in solution,62 and also form alpha helices and beta sheets at different temperatures in the solid-state.63 Some secondary structures may develop in the crowded resin-bound environment that favor intermolecular reaction to form a dimeric cyclic peptide. We note that unusual behaviour of a polyalanine containing peptide on CuAAC cyclization was observed by another research group.27 The cyclization resulted in formation of a highly insoluble material whose composition could not be determined. The authors attributed this result to the β-sheet aggregation characteristic of long polyglycines.27 Apart from this exception, the cyclic monomer is the major product for most of the heptameric peptide sequences, though the exact yield of cyclic monomer and the M : D ratio depended on the peptide sequence. This result is similar to published reports for heptameric peptides, in which the monomer is identified as the major product (M : D 3.6 : 1).26
CuAAC on-bead cyclization of heptameric peptide sequences.
| Peptide | Cyclic peptide | Linear peptide precursor | M : Da |
|---|---|---|---|
| 1 | (YYTYT-Tz4)cy22 | Fmoc-Pra-YYTYT-Az4 | 8.28 : 1 |
| 4 | (NYRWLTz4)cy22,23 | Fmoc-Pra-NYRWL-Az4 | 43.8 : 0 |
| 5 | (RYKHYTz4)cy22,24 | Fmoc-Pra-RYKHY-Az4 | 40.9 : 0 |
| 6 | (YKYYR-Tz4)cy22,24 | Fmoc-Pra-YKYYR-Az4 | 52.3 : 0 |
| 7 | (DARNI-Tz4)cy22 | Fmoc-Pra-DARNI-Az4 | 6.52 : 1 |
| 8 | (RRATS-Tz4)cy22,25 | Fmoc-Pra-RRATS-Az4 | 5.71 : 1 |
Based on HPLC of crude sample of the cyclized product.
Effect of longer peptide length on cyclization
We synthesized and characterized peptides with eight, nine, and ten amino acids, and compared these results to the synthesis of heptameric sequence peptide 1 (Table 5). Increasing the peptide length by one amino acid had a drastic effect on the M : D ratio, reducing the cyclization of the peptide by 63% and the M : D ratio to 3.06 : 1 for peptide 9 (ESI Fig. S.15†).
CuAAC cyclization of peptides containing seven to ten amino acids.
| Peptide | Cyclic peptide | Linear peptide Precursor | Monomer : dimer | Number of amino acids |
|---|---|---|---|---|
| 1 | (YYTYTTz4)cy | Fmoc-Pra-YYTYT-Az4 | 8.28 : 1 | 7 |
| 9 | (AYYTYT-Tz4)cy | Fmoc-Pra-AYYTYT-Az4 | 3.06 : 1 | 8 |
| 10 | (AAYYTYT-Tz4)cy | Fmoc-Pra-AAYYTYT-Az4 | 1 : 3.98 | 9 |
| 11 | (AAAYYTYT-Tz4)cy | Fmoc-Pra-AAAYYTYT-Az4 | 1 : 4.5 | 10 |
This trend was also observed for the nonameric peptide 10 (ESI Fig. S.16†) and decameric peptide 11 (ESI Fig. S.17†), with the dimer being the major product in both cases.
Another long peptide series with eight, nine, and ten amino acids were synthesized (Table 6), in which serine and glycine were added to peptide 1. For the octameric peptide 12, an M : D ratio of 8.98 : 1 was observed (ESI Fig. S.18†), which meant the monomer formed at a higher proportion than for peptide 1. For the nonameric peptide 13, M : D drastically reduced by 38% to 5.54 : 1 (ESI Fig. S.19†). Therefore, the trend for this series was similar to the earlier series in Table 5, though the extent of reduction of the M : D ratio varies significantly for the two sets. Nonameric peptide 9 (Table 6) yielded monomer as the major cyclized product, while nonameric peptide 10 (Table 5) formed dimer as the major cyclized product.
Cyclization of Gly or Ser containing peptides with seven to ten amino acids.
| Peptide | Cyclic peptide | Linear peptide precursor | Monomer : dimer | Number of amino acids |
|---|---|---|---|---|
| 1 | (YYTYT-Tz4)cy | Fmoc-Pra-YYTYT-Az4 | 8.28 : 1 | 7 |
| 12 | (SYYTYT-Tz4)cy | Fmoc-Pra-SYYTYT-Az4 | 8.98 : 1 | 8 |
| 13 | (GSYYTYT-Tz4)cy | Fmoc-Pra-GSYYTYT-Az4 | 5.54 : 1 | 9 |
A dimeric cyclic peptide has been reported as the major product for a nonameric peptide cyclized by on-resin CuAAC.27 However, Goncalves et al. reported the formation of cyclic monomers for octameric peptides, which we find as the major product for both peptide series in Tables 5 and 6. We conclude that one can vary the peptide length from six amino acids to eight amino acids and get monomeric cyclic peptide as the major product for the on-resin CuAAC cyclization reaction.
Effect of shorter peptide length on cyclization
For investigating the effect of decreasing the peptide length on CuAAC cyclization, linear peptides containing lower than seven amino acids were synthesized by omitting amino acids from on the N-terminal end of peptide 1. On-resin CuAAC reaction was performed to cyclize the peptides. Lokey et al. previously reported monomeric cyclic peptide formation for polyleucine peptides containing four to seven amino acids. These polyleucine peptides included azidoglycine and propargylglycine as the azide and alkyne, respectively.26 They found that the pentameric peptide had the highest M : D ratio of 5.5 : 1, and the trimeric peptide had cyclic dimer as the major product. In this research, azidolysine, a long and flexible analog of azidoglycine was used instead of azidoglycine. It was expected that the flexible nature of the azide would enable higher rates of intramolecular reaction forming monomers so that the M : D ratio would be higher than 5.5 : 1. While the hexameric peptide 14 (ESI Fig. S.20†) had a monomer major product (M : D 6.68 : 1), a much lower M : D ratio of 1.90 : 1 was observed for the pentameric peptide 15 (Table 7, ESI Fig. S.21†). We next investigated if the observed M : D ratio was particular to the peptide sequence and if the identity and nature of the amino acids in pentamers could cause the M : D ratio to vary significantly.
Effect of short peptide length on CuAAC cyclization on-bead.
| Peptide | Cyclic peptide | Linear peptide precursor | M : D | Number of amino acids |
|---|---|---|---|---|
| 1 | (YYTYT-Tz4)cy | Fmoc-Pra-YYTYT-Az4 | 8.28 : 1 | 7 |
| 14 | (YTYT-Tz4)cy | Fmoc-Pra-YTYT-Az4 | 6.68 : 1 | 6 |
| 15 | (TYT-Tz4)cy | Fmoc-Pra-TYT-Az4 | 1.90 : 1 | 5 |
Effect of amino acid identity on the monomer–dimer ratio for pentameric cyclic peptides
To investigate the impact of sequence identity on cyclization of peptides with five amino acids, the amino acids in the 2nd, 3rd, and 4th positions from the N-terminal end were varied. The first residue, forming part of the cyclic triazole, was kept constant.
Two different peptides containing the same amino acids as peptide 15 were made, and their M : D ratios were determined (Table 8). The considerable variation in the M : D ratio observed between peptides 15 (ESI Fig. S.21†), 16 (ESI Fig. S.22†) and 17 (ESI Fig. S.23†) in Table 8 indicate that even if the same amino acids are used, the specific sequence identity significantly affects product formation.
CuAAC Cyclization of pentameric peptides with same amino acids but different sequences.
| Peptide | Cyclic peptide sequence | Linear peptide precursor | Monomer : dimer |
|---|---|---|---|
| 15 | (TYT-Tz4)cy | Fmoc-Pra-TYT-Az4 | 1.90 : 1 |
| 16 | (YTT-Tz4)cy | Fmoc-Pra-YTT-Az4 | 3.72 : 1 |
| 17 | (TTY-Tz4)cy | Fmoc-Pra-TTY-Az4 | 8.02 : 1 |
We next systemically investigated the effect of amino acid nature on the M : D ratio, by varying one amino acid in a particular position and keeping all other amino acids constant. Tables 9–11 list variable 2nd, 3rd and 4th residues from the N-terminal end, respectively. Table 9 lists pentameric cyclic sequences containing tyrosine (Y), valine (V), tryptophan (W), and phenylalanine (F) as the variable residue. Table 9 shows that peptide 18 containing V (ESI Fig. S.24†) and peptide 19 containing F (ESI Fig. S.25†) have similar M : D ratios, with peptide 19 containing the lowest M : D ratio. Also had a similar M : D ratio During cyclization by CuAAC on-resin, the reactive side-chains of amino acids were bonded to side-chain protecting groups. The phenolic group in Y in peptide 16 and the imidazole nitrogen in W in peptide 20 (ESI Fig. S.26†), therefore, had protecting groups while V (peptide 18) and F (peptide 19) did not have any protecting groups. Thus, in the presence of a sterically bulky functionality such as Tyr(OtBu) or Trp(Boc) in the 2nd position, the M : D ratio was higher.
Cyclization of pentameric peptides with variable 2nd residue from N-terminal.
| Peptide | Cyclic peptide | Linear peptide precursor | Monomer : dimer |
|---|---|---|---|
| 16 | (YTT-Tz4)cy | Fmoc-Pra-YTT-Az4 | 3.72 : 1 |
| 18 | (VTT-Tz4)cy | Fmoc-Pra-VTT-Az4 | 1.94 : 1 |
| 19 | (FTT-Tz4)cy | Fmoc-Pra-FTT-Az4 | 1.83 : 1 |
| 20 | (WTT-Tz4)cy | Fmoc-Pra-WTT-Az4 | 4.74 : 1 |
Cyclization of pentameric peptides with variable 3rd residue from N-terminal.
| Peptide | Cyclic peptide | Linear peptide precursor | Monomer : dimer |
|---|---|---|---|
| 15 | (TYT-Tz4)cy | Fmoc-Pra-TYT-Az4 | 1.90 : 1 |
| 21 | (TVT-Tz4)cy | Fmoc-Pra-TVT-Az4 | 3.68 : 1 |
| 22 | (TFT-Tz4)cy | Fmoc-Pra-TFT-Az4 | 2.82 : 1 |
| 23 | (TWT-Tz4)cy | Fmoc-Pra-TWT-Az4 | 2.90 : 1 |
Cyclization of pentameric peptides with different amino acids in the 4th residue from N-terminal.
| Peptide number | Cyclic peptide sequence | Linear peptide precursor | Monomer : dimer |
|---|---|---|---|
| 17 | (TTY-Tz4)cy | Fmoc-Pra-TTY-Az4 | 8.02 : 1 |
| 24 | (TTW-Tz4)cy | Fmoc-Pra-TTW-Az4 | 59.1 : 0 |
| 25 | (TTF-Tz4)cy | Fmoc-Pra-TTF-Az4 | 6.28 : 1 |
| 26 | (TTV-Tz4)cy | Fmoc-Pra-TTV-Az4 | 6.34 : 1 |
A different trend was observed in Table 10, where the 3rd residue from the N-terminal end is varied. Peptide 21, containing the least bulky amino acid V in the variable position, had the largest M : D ratio (ESI Fig. S.27†). The 3rd residue in peptide 15, Tyr(OtBu), is bulkier than Phe in peptide 22 but less bulky than Trp(Boc) in peptide 23. But peptide 15 cyclization yielded a lower M : D ratio than either of peptide 22 (ESI Fig. S.28†) or peptide 23 (ESI Fig. S.29†). Therefore, we could not find any clear correlation between the bulk of the amino acids and the M : D ratios. However, small amino acids in the 3rd residue from the N-terminal seem to favor monomer formation.
High M : D ratios were observed for peptide sequences listed in Table 11, with variable 4th residue from the N-terminal. The trend observed is somewhat similar to the trend seen for variable 2nd residue (Table 9). Bulky functionalities like Tyr(OtBu) in peptide 17 or Trp(Boc) in peptide 24 (ESI Fig. S.30†) strongly favour monomer formation. Relatively smaller amino acids F (peptide 25, ESI Fig. S.31†) and V (peptide 26, ESI Fig. S.32†) in the 4th residue position also favour monomer formation, but not to the same extent.
It should be noted that a significant amount of side products was formed in the CuAAC reaction of pentameric sequences. On-resin CuAAC cyclization of Fmoc-Pra-XXX-Az4 is much less optimal compared to that of Fmoc-Pra-YYTYT-Az4 and therefore has lower yield. The M : D ratio was highly sequence-dependent for the pentameric cyclic peptides.
Conclusions
In this article, we explored how the various Cu(i) complexes formed in solution catalyze the CuAAC on-bead cyclization. We hypothesized that piperidine played a critical role, due to formation Cu(i)–ligand clusters during the CuAAC reaction. Looking for compounds with similar ratios of Cu(i) : piperidine led us to explore the catalytic properties of a previously synthetically and structurally characterized Cu(i) compound [Cu4I4(pip)4]. We demonstrated that [Cu4I4(pip)4] exhibits a high catalytic performance for CuAAC cyclization of peptides under reducing atmosphere. Since [Cu4I4(pip)4] contains piperidine base as co-ligands in addition to iodide, the catalyst can be used without the addition of a base for high yield, in applications where the addition of a base causes side-reactions. Electrochemical studies of [Cu4I4(pip)4] show that the reduction potential of Cu in the complex is similar to that in CuI in DMF with three equivalents of piperidine added, further supporting our hypothesis.
Surprisingly, on-resin peptide CuAAC cyclization reaction in DMF using 1.5 equivalents of CuI in the presence of excess ascorbic acid was found to be successful without a heteronuclear base, with a percentage yield between 70–80%. The addition of excess piperidine to CuI increased the cyclization yield by around 15%. Piperidine addition also increased the monomer–dimer ratio. DMF and piperidine were found to be the optimal solvent and base for this reaction, as cyclization of Fmoc-Pra-YYTYT-Az4 with other bases and solvents varied widely. Addition of 1.5 equivalents of CuI was found to be optimal, as opposed to lower equivalents.
Following the insight into active catalyst formation during the CuAAC reaction, a systematic study was done on the effect of location of azide and alkyne functionalities, peptide length, and peptide sequence on on-resin CuAAC cyclization. Sequence length was identified as a critical parameter for cyclization and determined whether cyclic monomer or cyclic dimer was formed as the major product. Peptides with six, seven, or eight amino acids mostly yielded the cyclic monomer as the major product. Longer peptides containing nine or ten amino acids produced cyclic dimer as the major product on CuAAC cyclization. Peptides shorter than hexamers had lower conversions to cyclic products compared to longer sequences, and the monomer to dimer ratio was highly dependent on the peptide sequence in that case.
In summary, this thorough investigation of on-resin CuAAC cyclization of peptides shall enable researchers to design the synthesis of monomeric or dimeric cyclic peptides through the reaction successfully. One can also use the CuAAC cyclization reaction condition described here for successfully synthesizing comprehensive on-bead cyclic peptide libraries with six, seven, or eight amino acids.
Experimental
Synthesis of peptides by solid phase peptide synthesis (SPPS)
All peptides were synthesized manually by solid phase peptide synthesis (SPPS) with rink amide resin (loading 0.67 mmol g−1) as solid support. After swelling the resin for 5–8 hours in NMP, N-terminal fluorenylmethyloxycarbonyl (Fmoc) deprotection was done in 20% piperidine in N-methyl-2-pyrrolidone (NMP) (v/v). The deprotection was performed 3 times for 10–15 min each time. The resin is washed 5 times for 1 minute with NMP, and Fmoc-amino acid is then coupled. Coupling reactions were performed in NMP with 3 equivalents (eq.) amino acid, 2.98 eq. HATU and 9 eq. of DIEA added. The coupling of the first amino acid to the resin was done for 10 hours at room temperature. All subsequent amino acids were coupled for 5 hours. The resin was washed 3 times with NMP after each coupling. After the last coupling the resin was washed with DCM and dried under vacuum. 5–10 mg resin was taken, swollen in NMP and deprotected and cleaved to get the linear peptide. An aliquot of the protected peptide sequence on resin was used for cyclization reactions.
CuAAC cyclization of Fmoc-protected peptides on resin
The cyclization was done following a previous published protocol.22 For the cyclization reaction, 25 mg or 50 mg of the resin was used, taking into account the weight of the peptide. The resin was swollen in NMP for 5 hours and treated with DMF overnight before cyclization. 1.5 eq. of CuI and 7.0 eq. of ascorbic acid were each dissolved in a 1 mL solution of 20% piperidine in DMF. The two solutions were added to the resin and the closed tube was wrapped in aluminium foil. The light yellow colour of the solution was checked 5, 15 and 30 min after initiation of the reaction. The reaction was performed at room temperature for 15 h. The excess copper catalyst was removed by washing the resin with DMF, followed by multiple washes with a 5% (w/v) sodium diethyldithiocarbamate, 5% (v/v) DIEA solution in DMF, methanol and DMF. Washing with the three solutions was repeated 4 to 5 times until the wash is light yellow in color. This was followed by three washes with DMF and two washes with NMP. The resin was treated with 20% piperidine in DMF for 15 minutes, then washed with NMP and DCM and dried.
Cleavage and precipitation of peptides
For the cleavage of peptides from the rink amide resin (100 mg, 50 mg, 25 mg, 10 mg) a TFA cocktail solution with 95 : 2.5 : 2.5 (v/v/v) TFA : TES : H2O was prepared. The resin with 0.7–1.5 mL of the TFA cocktail solution was taken in a glass round bottom flask and stirred for 2 hours. The solution was filtered into ice cold diethyl ether (35–40 mL), when the peptide precipitated. The precipitate was centrifuged at 4000 rpm for 15 min, and the resultant pellet dissolved in double distilled water with minimal acetonitrile and lyophilized.
Preparation of [Cu4I4(pip)4]
[Cu4I4(pip)4] was synthesized according to published procedure64 with some slight modifications. CuI was dissolved (78 mg, 0.41 mmol) in 3.5 M KI (2 mL) and transferred to an argon flushed SPE tube. An equimolar amount of piperidine (40.4 μL) was added. Shaking the tube results in a white precipitate that was washed with water (2×), methanol 1×, then hexane under argon atmosphere. The white precipitate was transferred to glass vials for storage and stored under argon.
Peptide analysis by analytical HPLC
All the peptides were analyzed and purified by RP-HPLC (Agilent 1260). Agilent Pursuit XRs C18 (3 μm, 150 × 4.6 mm) analytical column or Phenomenex Luna C18 (5 μm, 100A, 250 × 4.6 mm) analytical column was used. The mobile phase composition was isocratic for 10 minutes with 5% ACN (0.1%TFA), then there was a gradient from 5% to 40% ACN from 10 to 60 min. This was followed by a steep gradient to 95% ACN (0.1% TFA) as a start of the column cleaning procedure. The flow rate was 1.5 mL min−1 for most samples. For certain strongly lipophilic samples, the flow rate was modified to 1.0 mL min−1. Collected fractions were lyophilized and either stored or analysed by LC-MS.
Determination of peptide masses by ESI LC-MS
Samples were either dissolved LC-MS grade water or methanol or acetonitrile after lyophilization. The dissolved samples were measured by ESI LC-MS (Agilent 1260 Infinity, 6120 Quadrupole) using an Eclipse plus C18 column (3 μm, 2.1 × 100 mm). The LC-traces were detected at 214 nm.
Cyclic voltammetry (CV) of copper catalysts CuI or [Cu4I4(pip)4] in DMF
All solvents and solutions were purged with argon prior to use. The supporting electrolyte tetrabutylammonium perchlorate (TBAP) (0.25 mM) was added to a solution of 5 mM CuI or 1.25 mM [Cu4I4(pip)4] in DMF. The solutions were flushed with argon and the measured on a VMP2 Multichannel Potentiostat (BioLogic, Princeton Applied Research) with a three electrode setup: a glassy carbon electrode (working electrode), Ag+/Ag electrode (reference electrode) and a Ni-mesh electrode (counter electrode). The current was measured for a scan rate of 50 mV s−1 and from −1.5 to 1.0 V.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
Research reported in this publication is supported by National Institute of General Medical Sciences of the National Institutes of Health under award number 1R15GM139155.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra07491h
References
- Dougherty P. G. Sahni A. Pei D. Chem. Rev. 2019;119:10241–10287. doi: 10.1021/acs.chemrev.9b00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenfeld E. Pflugfelder S. C. Surv. Ophthalmol. 2009;54:321–338. doi: 10.1016/j.survophthal.2009.02.002. [DOI] [PubMed] [Google Scholar]
- Biermasz N. R. Pituitary. 2017;20:149–153. doi: 10.1007/s11102-016-0785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wójcik P. Berlicki Ł. Bioorg. Med. Chem. Lett. 2016;26:707–713. doi: 10.1016/j.bmcl.2015.12.084. [DOI] [PubMed] [Google Scholar]
- Bracken C. Gulyas J. Taylor J. W. Baum J. J. Am. Chem. Soc. 1994;116:6431–6432. doi: 10.1021/ja00093a052. [DOI] [Google Scholar]
- Leduc A. M. Trent J. O. Wittliff J. L. Bramlett K. S. Briggs S. L. Chirgadze N. Y. Wang Y. Burris T. P. Spatola A. F. Proc. Natl. Acad. Sci. U. S. A. 2003;100:11273–11278. doi: 10.1073/pnas.1934759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackwell H. E. Sadowsky J. D. Howard R. J. Sampson J. N. Chao J. A. Steinmetz W. E. O'Leary D. J. Grubbs R. H. J. Org. Chem. 2001;66:5291–5302. doi: 10.1021/jo015533k. [DOI] [PubMed] [Google Scholar]
- Jo H. Meinhardt N. Wu Y. Kulkarni S. Hu X. Low K. E. Davies P. L. DeGrado W. F. Greenbaum D. C. J. Am. Chem. Soc. 2012;134:17704–17713. doi: 10.1021/ja307599z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assem N. Ferreira D. J. Wolan D. W. Dawson P. E. Angew. Chem., Int. Ed. Engl. 2015;54:8665–8668. doi: 10.1002/anie.201502607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diderich P. Bertoldo D. Dessen P. Khan M. M. Pizzitola I. Held W. Huelsken J. Heinis C. ACS Chem. Biol. 2016;11:1422–1427. doi: 10.1021/acschembio.5b00963. [DOI] [PubMed] [Google Scholar]
- Haney C. M. Loch M. T. Horne W. S. Chem. Commun. 2011;47:10915–10917. doi: 10.1039/C1CC12010G. [DOI] [PubMed] [Google Scholar]
- Brown S. P. Smith A. B. J. Am. Chem. Soc. 2015;137:4034–4037. doi: 10.1021/ja512880g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Empting M. Avrutina O. Meusinger R. Fabritz S. Reinwarth M. Biesalski M. Voigt S. Buntkowsky G. Kolmar H. Angew. Chem., Int. Ed. Engl. 2011;50:5207–5211. doi: 10.1002/anie.201008142. [DOI] [PubMed] [Google Scholar]
- Hou J. Liu X. Shen J. Zhao G. Wang P. G. Expert Opin. Drug Discovery. 2012;7:489–501. doi: 10.1517/17460441.2012.682725. [DOI] [PubMed] [Google Scholar]
- Tron G. C. Pirali T. Billington R. A. Canonico P. L. Sorba G. Genazzani A. A. Med. Res. Rev. 2008;28:278–308. doi: 10.1002/med.20107. [DOI] [PubMed] [Google Scholar]
- Lau Y. H. de Andrade P. Quah S.-T. Rossmann M. Laraia L. Sköld N. Sum T. J. Rowling P. J. E. Joseph T. L. Verma C. Hyvönen M. Itzhaki L. S. Venkitaraman A. R. Brown C. J. Lane D. P. Spring D. R. Chem. Sci. 2014;5:1804–1809. doi: 10.1039/C4SC00045E. [DOI] [Google Scholar]
- Kawamoto S. A. Coleska A. Ran X. Yi H. Yang C. Y. Wang S. J. Med. Chem. 2012;55:1137–1146. doi: 10.1021/jm201125d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tala S. R. Singh A. Lensing C. J. Schnell S. M. Freeman K. T. Rocca J. R. Haskell-Luevano C. ACS Chem. Neurosci. 2018;9:1001–1013. doi: 10.1021/acschemneuro.7b00422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams G. M. Lee K. Li X. Cooper G. J. Brimble M. A. Org. Biomol. Chem. 2015;13:4059–4063. doi: 10.1039/C5OB00160A. [DOI] [PubMed] [Google Scholar]
- Wright Z. V. F. McCarthy S. Dickman R. Reyes F. E. Sanchez-Martinez S. Cryar A. Kilford I. Hall A. Takle A. K. Topf M. Gonen T. Thalassinos K. Tabor A. B. J. Am. Chem. Soc. 2017;139:13063–13075. doi: 10.1021/jacs.7b06506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost J. R. Scully C. C. Yudin A. K. Nat. Chem. 2016;8:1105–1111. doi: 10.1038/nchem.2636. [DOI] [PubMed] [Google Scholar]
- Das S. Nag A. Liang J. Bunck D. N. Umeda A. Farrow B. Coppock M. B. Sarkes D. A. Finch A. S. Agnew H. D. Pitram S. Lai B. Yu M. B. Museth A. K. Deyle K. M. Lepe B. Rodriguez-Rivera F. P. McCarthy A. Alvarez-Villalonga B. Chen A. Heath J. Stratis-Cullum D. N. Heath J. R. Angew. Chem., Int. Ed. Engl. 2015;54:13219–13224. doi: 10.1002/anie.201505243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrow B. Wong M. Malette J. Lai B. Deyle K. M. Das S. Nag A. Agnew H. D. Heath J. R. Angew. Chem., Int. Ed. Engl. 2015;54:7114–7119. doi: 10.1002/anie.201502451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J. Bunck D. N. Mishra A. Hong S. Idso M. N. Heath J. R. J. Pept. Sci. 2019;25:e3203. doi: 10.1002/psc.3203. [DOI] [PubMed] [Google Scholar]
- Lai B. T. Wilson J. A. Malette Loredo J. Pitram S. M. LaBerge N. A. Heath J. R. Agnew H. D. Chem. - Eur. J. 2018;24:3760–3767. doi: 10.1002/chem.201704752. [DOI] [PubMed] [Google Scholar]
- Turner R. A. Oliver A. G. Lokey R. S. Org. Lett. 2007;9:5011–5014. doi: 10.1021/ol702228u. [DOI] [PubMed] [Google Scholar]
- Punna S. Kuzelka J. Wang Q. Finn M. G. Angew. Chem., Int. Ed. Engl. 2005;44:2215–2220. doi: 10.1002/anie.200461656. [DOI] [PubMed] [Google Scholar]
- Jagasia R. Holub J. M. Bollinger M. Kirshenbaum K. Finn M. G. J. Org. Chem. 2009;74:2964–2974. doi: 10.1021/jo802097m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantel S. Isaad Ale C. Scrima M. Levy J. J. DiMarchi R. D. Rovero P. Halperin J. A. D'Ursi A. M. Papini A. M. Chorev M. J. Org. Chem. 2008;73:5663–5674. doi: 10.1021/jo800142s. [DOI] [PubMed] [Google Scholar]
- Holland-Nell K. Meldal M. Angew. Chem., Int. Ed. Engl. 2011;50:5204–5206. doi: 10.1002/anie.201005846. [DOI] [PubMed] [Google Scholar]
- van Maarseveen J. H. Horne W. S. Ghadiri M. R. Org. Lett. 2005;7:4503–4506. doi: 10.1021/ol0518028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine K. D. Gin D. Y. Gin M. S. J. Am. Chem. Soc. 2004;126:1638–1639. doi: 10.1021/ja039374t. [DOI] [PubMed] [Google Scholar]
- Billing J. F. Nilsson U. J. J. Org. Chem. 2005;70:4847–4850. doi: 10.1021/jo050585l. [DOI] [PubMed] [Google Scholar]
- Roice M. Johannsen I. Meldal M. QSAR Comb. Sci. 2004;23:662–673. doi: 10.1002/qsar.200420021. [DOI] [Google Scholar]
- Goncalves V. Gautier B. Regazzetti A. Coric P. Bouaziz S. Garbay C. Vidal M. Inguimbert N. Bioorg. Med. Chem. Lett. 2007;17:5590–5594. doi: 10.1016/j.bmcl.2007.07.087. [DOI] [PubMed] [Google Scholar]
- Ul-Ain Q. Kandler R. Gillespie D. Nag A. Journal of Medicinal Chemistry and Drug Design. 2018;1:1–7. [Google Scholar]
- Deyle K. M. Farrow B. Qiao Hee Y. Work J. Wong M. Lai B. Umeda A. Millward S. W. Nag A. Das S. Heath J. R. Nat. Chem. 2015;7:455–462. doi: 10.1038/nchem.2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idso M. N. Akhade A. S. Arrieta-Ortiz M. L. Lai B. T. Srinivas V. Hopkins J. P. Gomes A. O. Subramanian N. Baliga N. Heath J. R. Chem. Sci. 2020;11:3054–3067. doi: 10.1039/C9SC04842A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag A., Doctor of Philosophy, Californai Institute of Technology, 2013 [Google Scholar]
- Ingale S. Dawson P. E. Org. Lett. 2011;13:2822–2825. doi: 10.1021/ol200775h. [DOI] [PubMed] [Google Scholar]
- Berg R. Straub B. F. Beilstein J. Org. Chem. 2013;9:2715–2750. doi: 10.3762/bjoc.9.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov V. O. Fokin V. V. Finn M. G. Angew. Chem., Int. Ed. Engl. 2005;44:2210–2215. doi: 10.1002/anie.200461496. [DOI] [PubMed] [Google Scholar]
- Bock V. D. Hiemstra H. van Maarseveen J. H. Eur. J. Org. Chem. 2006;2006:51–68. doi: 10.1002/ejoc.200500483. [DOI] [Google Scholar]
- Bastide J. and Henri-Rousseau O., in The Carbon–Carbon Triple Bond (1978), 1978, ch. 11, pp. 447–522, 10.1002/9780470771563.ch11 [DOI] [Google Scholar]
- Jin L. Tolentino D. R. Melaimi M. Bertrand G. Sci. Adv. 2015;1:e1500304. doi: 10.1126/sciadv.1500304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C. Cheng G. Su D. Xu J. Wang X. Hu Y. Adv. Synth. Catal. 2010;352:1587–1592. doi: 10.1002/adsc.200900768. [DOI] [Google Scholar]
- Lu B.-B. Yang J. Che G.-B. Pei W.-Y. Ma J.-F. ACS Appl. Mater. Interfaces. 2018;10:2628–2636. doi: 10.1021/acsami.7b17306. [DOI] [PubMed] [Google Scholar]
- Raston C. L. White A. H. J. Chem. Soc., Dalton Trans. 1976:2153–2156. doi: 10.1039/DT9760002153. doi: 10.1039/DT9760002153. [DOI] [Google Scholar]
- Schramm V. Inorg. Chem. 1978;17:714–718. doi: 10.1021/ic50181a043. [DOI] [Google Scholar]
- Zheng L. Wang Y. Meng X. Chen Y. Catal. Commun. 2021;148:106165. doi: 10.1016/j.catcom.2020.106165. [DOI] [Google Scholar]
- Giguère D. Patnam R. Bellefleur M.-A. St-Pierre C. Sato S. Roy R. Chem. Commun. 2006:2379–2381. doi: 10.1039/B517529A. doi: 10.1039/B517529A. [DOI] [PubMed] [Google Scholar]
- Cheshev P. Marra A. Dondoni A. Org. Biomol. Chem. 2006;4:3225–3227. doi: 10.1039/B609734K. [DOI] [PubMed] [Google Scholar]
- Wells A. F., Structural Inorganic Chemistry, Oxford University Press, 5th edn, 2012 [Google Scholar]
- Liang C.-H. Yao S. Chiu Y.-H. Leung P. Y. Robert N. Seddon J. Sears P. Hwang C.-K. Ichikawa Y. Romero A. Bioorg. Med. Chem. Lett. 2005;15:1307–1310. doi: 10.1016/j.bmcl.2005.01.027. [DOI] [PubMed] [Google Scholar]
- Romero A. Liang C.-H. Chiu Y.-H. Yao S. Duffield J. Sucheck S. J. Marby K. Rabuka D. Leung P. Y. Shue Y.-K. Ichikawa Y. Hwang C.-K. Tetrahedron Lett. 2005;46:1483–1487. doi: 10.1016/j.tetlet.2005.01.023. [DOI] [Google Scholar]
- Sreedhar B. Surendra Reddy P. Synth. Commun. 2007;37:805–812. doi: 10.1080/00397910601133599. [DOI] [Google Scholar]
- Meldal M. Tornoe C. W. Chem. Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- Chang W.-K. Lee G.-H. Wang Y. Oliver Su Y. Ho T.-I. Lin Y.-C. J. Chin. Chem. Soc. 1993;40:523–526. doi: 10.1002/jccs.199300084. [DOI] [Google Scholar]
- Tornoe C. W. and Meldal M., Peptides: The Wave of the Future, ed. M. Lebl and R. A. Houghten, American Peptide Symposia, 2001, pp. 263–264 [Google Scholar]
- Hendrickson A. R. Martin R. L. Rohde N. M. Inorg. Chem. 1976;15:2115–2119. doi: 10.1021/ic50163a021. [DOI] [Google Scholar]
- Zhang Z. Fan E. Tetrahedron Lett. 2006;47:665–669. doi: 10.1016/j.tetlet.2005.11.111. [DOI] [Google Scholar]
- Vila J. A. Ripoll D. R. Scheraga H. A. Proc. Natl. Acad. Sci. U.S.A. 2000;97:13075. doi: 10.1073/pnas.240455797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildman K. Lee D.-K. Ramamoorthy A. Biopolymers. 2002;64:246–254. doi: 10.1002/bip.10180. [DOI] [PubMed] [Google Scholar]
- Vitale M. Ford P. C. Coord. Chem. Rev. 2001;3:3–16. doi: 10.1016/S0010-8545(00)00414-8. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.