Abstract
Surveillance of DNA damage and maintenance of lipid metabolism are critical factors for general cellular homeostasis. We discovered that in response to DNA damage–inducing UV light exposure, intact Caenorhabditis elegans accumulate intracellular lipids in a dose-dependent manner. The increase in intracellular lipids in response to exposure to UV light utilizes mafr-1, a negative regulator of RNA polymerase III and the apical kinases atm-1 and atl-1 of the DNA damage response (DDR) pathway. In the absence of exposure to UV light, the genetic ablation of mafr-1 results in the activation of the DDR, including increased intracellular lipid accumulation, phosphorylation of ATM/ATR target proteins, and expression of the Bcl-2 homology region genes, egl-1 and ced-13. Taken together, our results reveal mafr-1 as a component the DDR pathway response to regulating lipid homeostasis following exposure to UV genotoxic stress.
INTRODUCTION
Maf1 is a negative regulator of RNA polymerase (pol) III (Boguta et al., 1997; Pluta et al., 2001; Reina et al., 2006; Goodfellow et al., 2008; Khanna et al., 2014; Palian et al., 2014). It is activated in response to various cellular and environmental sources of stress (Upadhya et al., 2002; Willis and Moir, 2007; Willis, 2018) to conserve metabolic energy and translational capacity. The roles of Maf1 in regulating biosynthetic capacity and lipid homeostasis are key to its impact on cell physiology and metabolism. Dysregulation of these functions is associated with the molecular underpinnings of various disease states, which can lead to obesity, metabolic disease, or cancer (Willis and Moir, 2007; Khanna et al., 2015; Johnson and Stiles, 2016). Maf1 physically associates with TFIIIB and RNA pol III and prevents its transcription of rRNA, noncoding RNA (ncRNA), and (primarily) tRNA (Vannini et al., 2010; Vorlander et al., 2020). Study on glioblastoma cells reveals that Maf1 also regulates a subset of RNA pol I and pol II gene targets (Johnson et al., 2007). By reducing tRNA availability, Maf1 decreases overall biosynthetic potential, thereby acting as a regulator of translational capacity.
Maf1 is activated in response to DNA damage. In human cells, transcription of ncRNA molecules is reduced following DNA damage, as both RNA pol I and III activity are decreased following DNA damage induced by both methane methylsulfonate (MMS) and UV light exposure (Ghavidel and Schultz, 2001). While other transcriptional control mechanisms are likely employed, Maf1 appears to play a key role in the DNA damage–dependent inhibition of RNA pol III. In Saccharomyces cerevisiae, exposure to MMS results in reduced RNA pol III transcription, which is not present in strains lacking Maf1 (Upadhya et al., 2002). Furthermore, human MAF1 is dephosphorylated, a modification correlated with RNA pol III repression (Michels et al., 2010), in response to MMS exposure (Reina et al., 2006).
In Caenorhabditis elegans, deregulation of translation capacity and changes in lipid accumulation are seen in animals with altered MAFR-1 levels (Khanna et al., 2014; Pradhan et al., 2017). In mammalian cells, Maf1 is implicated in oncogenic metabolism (Palian et al., 2014; Li et al., 2016), while the lack of MAF1 in mice is associated with metabolic reprograming that results in futile RNA cycling, whereby reallocation of substrates is funneled toward energy generation and nucleotide synthesis (Bonhoure et al., 2015; Willis, 2018). Because metabolic reprogramming has the potential to increase the availability of substrates required for survival during a crisis, it could be advantageous for an organism facing genotoxic stress (Shetty et al., 2020). However, this process could also be hijacked by oncogenic metabolism to fuel tumorigenesis (Palian et al., 2014; Li et al., 2016). Given its known roles in regulating programmed resource allocation, we asked whether MAFR-1–mediated metabolic changes are implicated in the response to genotoxic stress.
RESULTS AND DISCUSSION
Lipid accumulation of C. elegans in response to UV light exposure
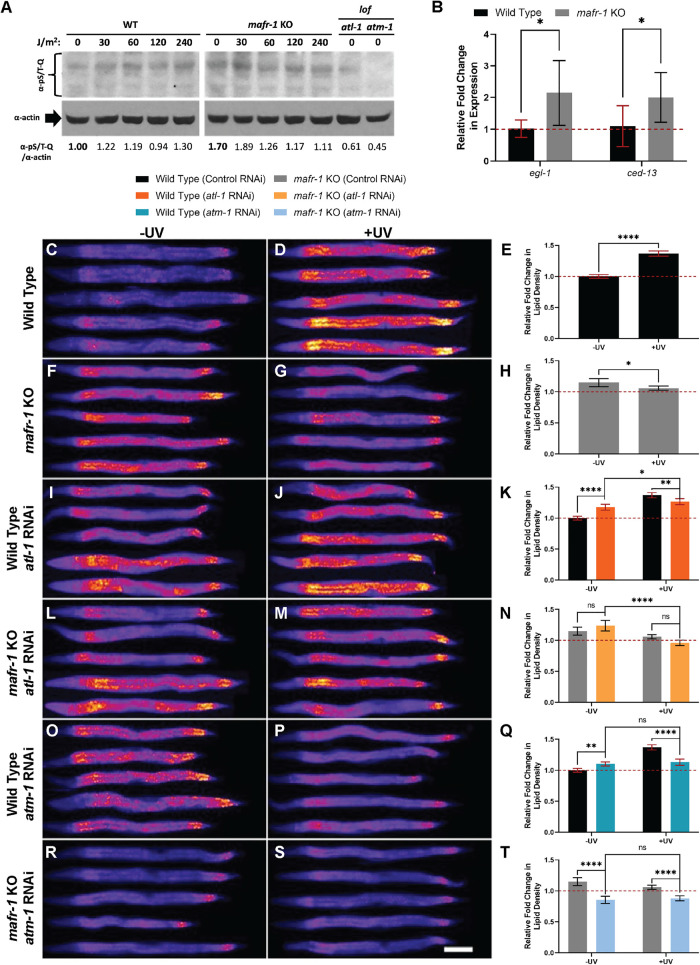
Although Maf1 function, and its role in the DNA damage response (DDR), has been widely researched in cultured yeast and mammalian cell models (Ghavidel and Schultz, 2001; Pluta et al., 2001; Upadhya et al., 2002; Reina et al., 2006; Johnson et al., 2007; Goodfellow et al., 2008; Vannini et al., 2010; Palian et al., 2014; Lee et al., 2015; Li et al., 2016; Shetty et al., 2020; Vorlander et al., 2020), fewer studies have been carried out in whole animals (Marshall et al., 2012; Rideout et al., 2012; Khanna et al., 2014; Bonhoure et al., 2015; Pradhan et al., 2017). To test whether C. elegans mafr-1 mediates organismal responses to UV light exposure, we analyzed animals carrying the CRISPR/Cas9-mediated deletion of the entire mafr-1 locus, hereafter referred to as mafr-1 knockout (KO), as compared with wild-type (WT) control animals. After exposing WT and mafr-1 KO C. elegans to increasing doses of UV light, we employed fixed Nile Red staining (Escorcia et al., 2018; Nhan and Curran, 2020) to measure the abundance of intracellular lipids. We observed increased accumulation of intracellular lipids when WT animals were exposed to increasing doses of UV irradiation (Figure 1, A–E; Supplemental Figure S1A). In contrast, although mafr-1 KO animals store more intracellular lipids in the absence of UV exposure, the dose-dependent accumulation of lipids was not observed in animals lacking mafr-1 (Figure 1, A and F–I; Supplemental Figure S1B). Interestingly, we observed a lack of lipid accumulation in both WT and mafr-1 KO animals following exposure to 240 J/m2 UV light, but at this higher dose of UV exposure we noted a marked increase of death in mafr-1 KO animals. These results suggest that MAFR-1 is involved in the modulation of lipid levels in response to UV phototoxicity. Because lipid homeostasis is governed by synthesis, utilization, and transport, it will be of great future interest to define how each, and perhaps all, are altered in order to survive DNA-damaging events.
FIGURE 1:
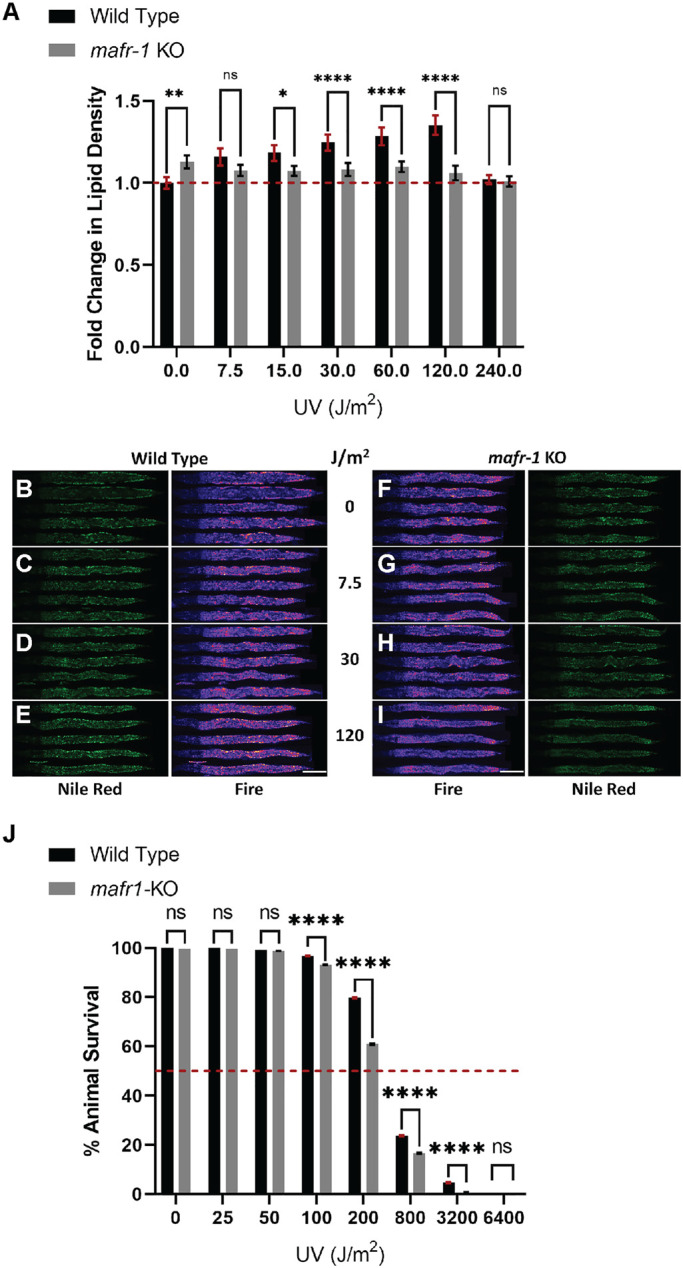
Loss of MAFR-1 results in failure to elevate lipid density in response to UV phototoxicity. (A) mafr-1 KO animals fail to increase lipid density following increasing levels of UV light exposure. Red dashed line indicates the mean lipid density in untreated WT animals. (B–I) Representative images of animals stained with Nile Red after exposure to UV light, quantified in panel A. (J) mafr-1 KO animals are more sensitive to UV light exposure. Red dashed line indicates 50% survival mark. Fisher’s exact test was used for pair-wise comparisons in survival data. One-way ANOVA test with Tukey’s post hoc test was used for multiple sample comparisons in lipid density data. See Descriptive Statistics Table for sample sizes used in each experiment. *, p < 0.05; **, p < 0.01; ****, p < 0.0001; n.s., no significance. Error bars show 95% C.I. of the mean. Scale bar is 100 μm.
Survival of C. elegans in response to UV light exposure
On the basis of the increased mortality observed at 240 J/m2 UV exposure, we asked how WT and mafr-1 KO worms respond to increasing doses of UV toxicity (Figure 1J; Supplemental Figure S1, C and D). After exposing worms to increasing doses of UV light, we allowed these animals to recover for 24 h and then assessed survival. We observed that doses of 100 J/m2 and higher significantly reduce the survival of WT animals. mafr-1 KO animals were more sensitive than WT animals at all UV doses greater than 50 J/m2, suggesting that MAFR-1 contributes to the survival response to UV-induced toxicity.
Effect of UV light exposure on mafr-1 expression and RNA pol III activity
Because complete loss of mafr-1 resulted in decreased survival following UV exposure, we asked whether mafr-1 expression was affected by UV light exposure. We used quantitative PCR (qPCR) to measure mafr-1 transcript levels in worms exposed to increasing doses of UV light. We observed in WT animals that there is a minor reduction in mafr-1 mRNA as UV light doses increase (Figure 2A; Supplemental Figure S2A). We next wanted to examine whether exposure to UV light leads to a reduction of MAFR-1 protein levels. We used worms harboring a single copy transgene encoding MAFR-1 fused to green fluorescent protein (GFP) at the C-terminus (MAFR-1::GFP), driven by the endogenous mafr-1 promoter, and exposed these worms to the same doses of UV light before measuring MAFR-1::GFP abundance by Western analysis. We observed that MAFR-1::GFP levels are diminished as UV light exposure increases (Figure 2B). This inverse relationship was also observed in animals carrying an integrated multicopy MAFR-1::GFP transgene (Supplemental Figure S2B). The decreased expression of mafr-1 mRNA implies that this observed reduction in MAFR-1 protein is due, at least in part, to reduced transcription in response to UV exposure, although we cannot exclude the possibility of additional posttranslational regulation of MAFR-1 protein expression.
FIGURE 2:
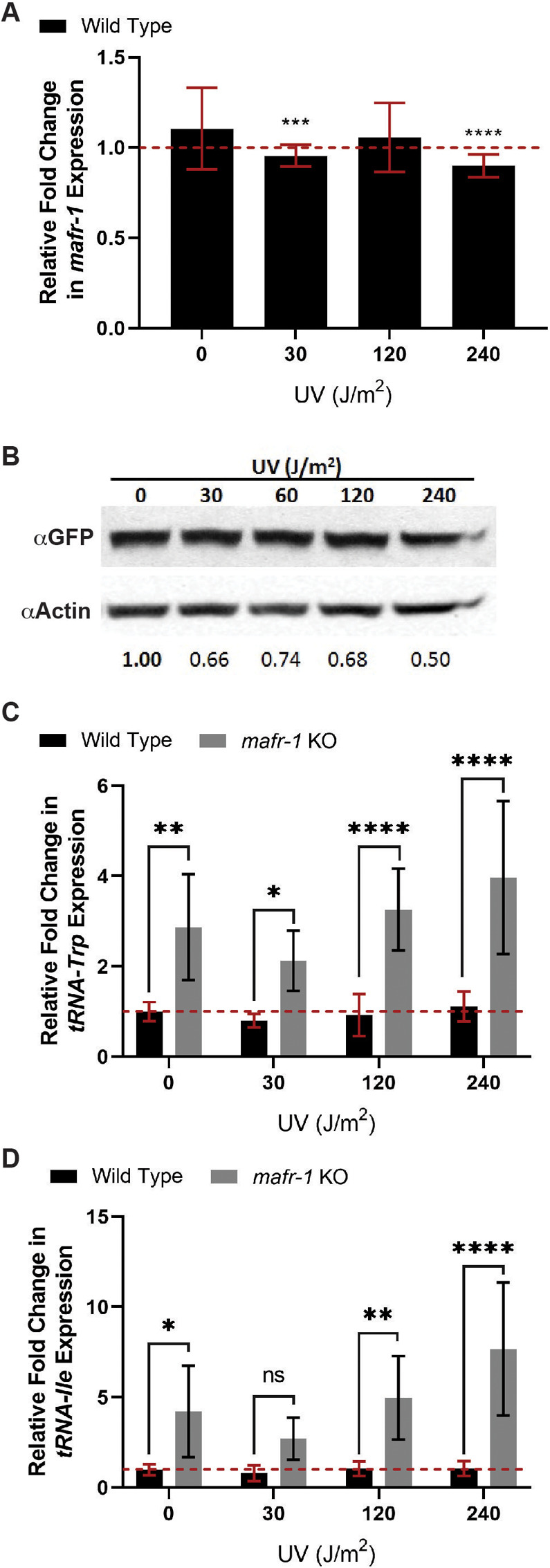
MAFR-1 expression is modestly reduced following UV exposure. (A) In WT animals, mafr-1 expression is reduced at 30 and 240 J/m2 relative to the untreated condition. (B) MAFR-1 stability decreases with increasing UV light exposure. (C, D) mafr-1 KO animals display increased abundance of tRNA-Trp (C) and tRNA-Ile (D), relative to WT animals. One-way ANOVA test with Tukey’s post hoc test was used for multiple sample comparisons in qPCR data. See Descriptive Statistics Table for sample sizes used in each experiment. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; n.s., no significance. Error bars show 95% C.I. of the mean.
Diminished MAFR-1 expression increases RNA pol III target expression (Khanna et al., 2014). As such, we measured the levels of two MAFR-1–responsive tRNA targets, tRNA-Trp and tRNA-Ile, by qPCR in UV-treated animals. Although several tRNA transcripts are regulated by mafr-1, previous studies identified these particular tRNAs as highly responsive to mafr-1 expression (Khanna et al., 2014). As expected, mafr-1 KO results in elevated tRNA-Trp and tRNA-Ile transcripts, as compared with WT animals. However, neither WT nor mafr-1 KO animals show significant changes in tRNA-Trp and tRNA-Ile expression with UV compared with untreated animals of the same genotype (Figure 2, C and D; Supplemental Figure S2, C and D). We noted a high degree of variability in the increased expression of these tRNAs in the absence of mafr-1, with and without UV exposure; perhaps due to the whole animal RNA extraction and the differential effects of MAFR-1 across tissues (Marshall et al., 2012; Rideout et al., 2012; Khanna et al., 2014; Bonhoure et al., 2015). Regardless, unlike lipid accumulation, RNA pol III activity does not appear to directly correlate with responses to UV.
Intersection of MAFR-1 with ATM-1 and ATL-1 activation
Although previous work implicates Maf1 activation following DNA damage (Ghavidel and Schultz, 2001; Upadhya et al., 2002; Reina et al., 2006), little is known about how Maf1 communicates with the DDR (Shetty et al., 2020), especially in intact animals. As is the case in other organisms, C. elegans depends on the phosphorylation of downstream targets by the apical kinases ATL-1 (homologue of mammalian ATR) and ATM-1 (homologue of mammalian ATM) to promote activation of the DDR (Lans et al., 2010; Vermezovic et al., 2012). We first asked whether MAFR-1 influences ATL-1 and ATM-1 activity in response to UV light exposure by measuring global phosphorylation of the serine/threonine-glutamine epitope (S/T-Q) found in ATL-1 and ATM-1 targets after exposing animals to increasing doses of UV light (Vermezovic et al., 2012). As expected, with increased exposure to UV treatment, we observed an increase in phosphorylated S/T-Q (pS/T-Q) products in WT animals, which was diminished in atl-1(ok1063) and atm-1(gk186) loss of function mutants (Figure 3A). We noted that mafr-1 KO mutants have increased pS/T-Q phosphorylation in the absence of UV treatment, which suggests that loss of mafr-1 induces the DDR and could explain the increased intracellular lipids observed in these animals in the absence of UV exposure. In light of this observation, we measured the expression egl-1 and ced-13, two well-established downstream transcriptional targets of DDR activation (Stergiou et al., 2007; Ye et al., 2014). EGL-1 and CED-13 both encode proteins with BH3 (Bcl-2 homology region 3) domains found in mammalian cell death activators (Schumacher et al., 2005). We found expression of both to be significantly increased in mafr-1 KO animals relative to WT in the absence of UV (Figure 3B). Responses to DNA damage–inducing events are dose dependent and culminate in lethality when the capacities of homeostatic response systems are exceeded (Lo et al., 2017; Rieckher et al., 2018) Intriguingly, as UV light increases to lethal levels, pS/T-Q phosphorylation decreased in mafr-1 KO worms (Figure 3A). These results correlate with the reduced survival observed in animals lacking MAFR-1, and suggest that the diminished capacity for ATM-1/ATL-1 signaling may be linked to elevated DNA damage and compromised survival in C. elegans (Lans et al., 2010; Vermezovic et al., 2012). This implies a connection between elevated lipid density and perceived genotoxic damage, and taken together, these data reveal that in the absence of mafr-1, ATM-1 and ATL-1 activity is deregulated with and without UV exposure.
FIGURE 3:
mafr-1 intersects with the DDR pathway to mediate lipid abundance in response to UV phototoxicity. (A) mafr-1 KO animals activate ATL-1/ATM-1 target phosphorylation in response to UV light. (B) Expression of egl-1 and ced-13 is elevated in mafr-1 KO animals relative to WT. (C-D, F-G, I-J, L-M, O-P, R-S) Images of WT and mafr-1 KO animals treated with RNAi against atl-1 and atm-1 and stained with Nile Red after exposure to 30 J/m2 UV light. White scale bar is 100 μm. (E, H, K, N, Q, T) Quantification of Nile Red staining. Red dashed line shows the mean lipid density of untreated, WT animals. Student’s t test was used to determine the significance of egl-1 and ced-13 expression. A one-way ANOVA test with Tukey’s post hoc test was used for multiple sample comparisons in lipid density data. See Descriptive Statistics Table for sample sizes used in each experiment. *, p < 0.05; **, p < 0.01; ****, p < 0.0001; n.s., no significance. Error bars show 95% C.I. of the mean.
We next tested whether MAFR-1 regulation of lipids, in response to sublethal exposure to UV light, involves the apical kinases ATM-1 and ATL-1. We exposed WT and mafr-1 KO animals raised on control, atm-1, and atl-1 RNA interference (RNAi) to 30 J/m2 of UV-C light and measured lipid density by Nile Red staining. Importantly, we observed that WT animals fed control RNAi (in the HT115/Escherichia coli K-12 host) displayed increased intracellular lipid density after UV light treatment (Figure 3, C–E), similar to the results observed when fed the standard OP50/E. coli B diet. Similarly, mafr-1 KO animals fed control RNAi bacteria displayed increased lipid density in the absence of UV exposure and failed to increase lipid density when exposed to UV (Figure 3, F–H). Although the magnitude of the lipid response is enhanced on the HT115 diet, the directionality of the response is diet-independent (Figure 1A; Supplemental Figure S3A). Several genetically regulated metabolic and health conditions can be suppressed by the nutritional quality of the diet in C. elegans (Pang and Curran, 2014; Lynn et al., 2015; Yen and Curran, 2016; Verma et al., 2018; Nhan et al., 2019), whereas mafr-1–dependent accumulation of intracellular lipids is similar on multiple diets, perhaps even enhanced on the HT115/K-12 diet as compared with the OP50/B food source. The diet-independent nature of this response (Figure 1A; Supplemental Figure S3A) suggests that increasing intracellular lipids is an important physiological response to DNA damage that occurs regardless of nutrient availability.
RNAi of atl-1 in WT animals results in increased lipid density in the absence of UV (Figure 3, I–K). Mutations in DDR pathway components, even in the absence of damage, can result in genotoxic stress (Ou and Schumacher, 2018), which further links DNA damage with intracellular lipid accumulation. However, atl-1 RNAi significantly attenuated the UV-dependent increase in lipid density observed in WT animals (Figure 3, I–K), suggesting a role for ATL-1 in moderating lipid homeostasis following UV exposure. mafr-1 KO animals showed no significant change in lipid density between control RNAi and atl-1 RNAi treatment, regardless of UV exposure (Figure 3, L–N). Intriguingly, mafr-1 KO animals on atl-1 RNAi exposed to UV showed a precipitous decrease in lipid density relative to mafr-1 KO animals on atl-1 RNAi that had not been exposed to UV (Figure 3, L–N), suggesting a synthetic interaction between ATL-1 and MAFR-1 in mediating lipid homeostasis in response to UV toxicity. Taken together, these results suggest a role for ATL-1 in the UV-induced accumulation of lipids.
We next tested RNAi against atm-1. Similar to atl-1 RNAi, RNAi of atm-1 resulted in a slight increase in lipid density in WT animals and attenuated accumulation of lipids following UV exposure (Figure 3, O–Q), suggesting a role similar to that of ATL-1 in moderating UV-dependent lipid homeostasis. Interestingly, mafr-1 KO animals exposed to atm-1 RNAi were lean, regardless of exposure to UV (Figure 3, R–T), indicating the necessity of ATM-1 for mafr-1–dependent lipid accumulation. These data suggest that ATM-1 is an integral part in both UV-dependent and MAFR-1-dependent lipid homeostasis. We noted that RNAi of atm-1, but not atl-1, reduced survival following UV exposure (Supplemental Figure S3B), although there was no measurable effect on survival at the doses used to measure lipid density (Supplemental Figure S3C). The synthetic relationship between mafr-1 KO animals and reduced atm-1 or atl-1 implies that DDR-dependent responses and MAFR-1 activity are coordinated to modulate lipid homeostasis when confronted with genotoxic stress.
Lipid accumulation of cultured MEF cells in response to UV light exposure
Recent work in fission yeast cells identifies a role for Maf1 in the maintenance of genomic stability (Shetty et al., 2020; Noguchi et al., 2021). Because changes in Maf1 levels are linked to alterations in lipid accumulation in mammalian cancer cell lines (Palian et al., 2014; Li et al., 2016; Pradhan et al., 2017), we employed mouse embryonic fibroblast (MEF) cells derived from WT or Maf1 KO mice (Bonhoure et al., 2015) to examine whether UV phototoxicity alters intracellular lipid homeostasis. The organization, size, and number of lipid droplets within a cell are important qualities that can indicate cellular health and metabolic state (Thiam and Beller, 2017). We examined the relationship between lipid droplets in WT and Maf1 KO MEF cells using Oil Red O (ORO), which allowed us to enumerate lipid droplets (Supplemental Figure S4, A–D). We noted that Maf1 KO MEFs contain a greater number of lipid droplets than WT cells (Supplemental Figure S4E), although the total area of ORO-stained lipid droplets was unchanged (Supplemental Figure S4F). This increase in lipid droplet number is consistent with our observations in C. elegans, while the maintenance of overall lipid droplet area may be due to compensatory physiological responses (Bostrom et al., 2005; Murphy et al., 2010) or methodological limitations (Fukumoto and Fujimoto, 2002). These observations are also consistent with previous studies, as loss of Maf1 has been shown to cause lipid accumulation in cultured mammalian cells (Pradhan et al., 2017; Chen et al., 2018) and across various model systems (Khanna et al., 2014; Mierzejewska and Chreptowicz, 2016; Pradhan et al., 2017). UV exposure increased the number (Supplemental Figure S4E) and area (Supplemental Figure S4F) of lipid droplets in WT MEFs, as well as in Maf1 KO cells. The magnitude of the UV-induced lipid accumulation was reduced in Maf1 KO cells relative to WT (Supplemental Figure S4, E and F). This response suggests that mammals may have evolved Maf1-independent mechanisms to increase lipid stores following DNA damage but supports the importance of intracellular lipid accumulation in response to DNA damage. Intriguingly, we noted that the distribution of lipid droplets is perinuclear in response to UV exposure (Supplemental Figure S4, G and H). Although the mechanisms that regulate the spatial dynamics of lipids are not well understood (Thiam and Beller, 2017), our work suggests a model where increased levels of intracellular lipids, and perhaps their organization, might be a protective measure in response to UV exposure. Our study suggests that increasing intracellular lipids is a conserved response to DNA damage. However, contrary to intact C. elegans, MEF cells lacking Maf1 in vitro do not exhibit defects in elevating lipids to meet genotoxic demands. These results indicate that some, but not all, aspects of Maf1 control of lipid homeostasis are evolutionarily conserved or that mammalian cells have acquired additional compensatory mechanisms to induce lipid accumulation independent of Maf1.
Recent work suggests a role for Maf1 driving adipogenesis in mesoderm (Chen et al., 2018), while studies of C. elegans lipid homeostasis pertain primarily to the intestine, an endoderm-derived tissue, in which MAFR-1 may function differently. The differences observed between C. elegans and mouse cells may also reflect evolutionary differences in lipid accumulation and utilization in response to genotoxic stress. It will be of interest to see whether in vivo exposure to genotoxic stress can elevate lipid storage in the absence of Maf1 in mice, which will reveal whether in vitro studies of isolated cells can fully encapsulate the in vivo responses in intact animals.
DNA damage has been linked to altered lipid homeostasis, as loss of DDR genes is associated with metabolic syndromes (Vose and Mitchell, 2011; Goldstein and Rotter, 2012), and UV-induced DNA damage causes accumulation of phosphoinositides (Wang et al., 2017). p53, a tumor suppressor gene and inhibitor of RNA pol III (Crighton et al., 2003), is activated by DNA damage sensors ATM and ATR (Yang et al., 2004) and has been shown to promote expression of genes involved in catabolism of lipids (Goldstein et al., 2012). Here, we show that in C. elegans, mafr-1 is integral in the accumulation of lipids as a homeostatic response to UV-induced DNA damage.
This study reveals that in the absence of MAFR-1, C. elegans responds as if there was exposure to a genotoxic stress. Specifically, we observed that loss of mafr-1 without UV treatment results in 1) the increased accumulation of intracellular lipids, as observed in UV-treated WT animals, 2) increased phosphorylation of ATM/ATR targets, and 3) increased expression of egl-1 and ced-13 transcripts that are activated in response to DDR. Taken together, these data reveal a Maf1-dependent regulation of lipid metabolism in response to genotoxic stress that acts in concert with the canonical DDR pathways. This link is of importance to understanding how fat metabolism affects genome integrity, or vice versa, which is of increasing importance in cancer etiology and metabolic disorders.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Caenorhabditis elegans growth and maintenance
Growth and maintenance of C. elegans was performed with minor changes as previously reported (Yen et al., 2020). Worms were grown and maintained at 20°C on 6-cm nematode growth medium (NGM) plates supplemented with streptomycin and seeded with OP50 E. coli. For all experiments, animals were synchronized at the L1 development stage. For RNAi experiments, NGM plates were supplemented with 5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) and 100 μg/ml carbenicillin and were seeded with HT115 E. coli expressing control (L4440) or gene-specific double-stranded RNAi plasmids. L1 animals were dropped onto plates after RNAi was induced for 24 h. Strains used in this study include WT N2 Bristol, PHX580 (mafr-1(syb580) I) (6× backcrossed to N2), VC381 (atm-1(gk186) I) (C. elegans Deletion Mutant Consortium, 2012), VC728 (atl-1(ok1063) V), SPC328 (mafr-1p::mafr-1ORF:6XHis:TEV:GFP::mafr-1 3′UTR::unc-119(+) II), and COP394, a high copy integrated version of mafr-1p::mafr-1ORF:6XHis:TEV:GFP::mafr-1 3′UTR::unc-119(+) obtained by bombardment.
UV exposure
A UV cross-linker (Stratagene) was used to expose L4 worms to various UV-C light (254 nm) doses ranging from 0 to 6400 J/m2. For UV exposures, approximately 1500 animals were used per 6-cm NGM plate. Growth from the L1 to the L4 stage at this animal density depletes the E. coli (OP50) lawn low enough for sufficient UV light penetrance but leaves enough food to avoid starvation before harvest 4–8 h later. For RNAi experiments, E. coli (HT115) was concentrated fivefold to recapitulate the lawn thickness of OP50 bacteria before UV light treatment. For exposure of mouse cells to UV light, cultures were allowed to reach confluence (∼70%). Growth medium was removed, and cells were washed once with phosphate-buffered saline (PBS) solution and then exposed to either 0 or 30 J/m2 UV light doses. Fresh growth medium was subsequently used to let cells recover for 24 h before harvest.
Fat staining
Lipid staining in C. elegans was carried out as previously reported (Khanna et al., 2014; Pradhan et al., 2017; Escorcia et al., 2018) on animals 8 h after exposure to 0, 7.5, 15, 30, 60, 120, or 240 J/m2 UV light. Late L4 worms were harvested in 1.5 ml centrifuge tubes.
Lipid staining in mouse cell culture was performed as previously described (Pradhan et al., 2017) on samples 24 h after UV exposure, growth medium was removed, and cells were washed once in PBS and then fixed with 10% neutral buffered Formalin (Pradhan et al., 2017) for 1 h at room temperature. Lipid droplet size and number were detected using the Color Transformer 2 plug-in, and Threshold and Analyze Particles feature in ImageJ (National Institutes of Health). All lipid droplet counting was carried out with these tools.
Survival assay
After UV exposure, worms were allowed to recover for 3 h before further handling to avoid issues associated with UV exposure–induced immobility (DeBardeleben et al., 2017). Worms without apparent movement anomalies were transferred to fresh NGM plates and incubated at 20°C for 21 h and reassessed. For the 3200 J/m2 dose, worms were gently prodded to assess viability. Worm survivors developed into adult animals, while dead animals developed abnormalities.
Gene expression
RNA extraction and qPCR were carried out as described before (Khanna et al., 2014; Ly et al., 2015; Pradhan et al., 2017). The Livak method was used to calculate gene expression values. snb-1 was used as in previous reports (Khanna et al., 2014, 2015) to normalize gene expression values. Relative change in expression values originate from dividing all sample values by that of the WT, untreated sample. egl-1 and ced-13 primer sequences are from Schumacher et al. (2005).
Western blotting
Western blotting techniques were followed as recently described (Lo et al., 2017; Pradhan et al., 2017; Spatola et al., 2019). Images of scanned films were used along with ImageJ to quantify Western blot data. Actin signal intensity was used to normalize the values of all other proteins of interest. Antibodies and dilutions used were rabbit anti-GFP (abcam290) (1:10,000); rabbit anti-S/Q-T-p (Vermezovic et al., 2012) (1:1000); mouse anti-actin (1:10,000); goat anti-rabbit immunoglobulin G (IgG) horseradish peroxidase (HRP) (Pradhan et al., 2017) (1:10,000); and goat anti-mouse IgG HRP (Pradhan et al., 2017) (1:10,000).
Cell culture
MEF (C57B American Type Culture Collection [ATCC]) cells were grown and maintained on DMEM (Invitrogen) supplemented with 10% heat-inactivated fetal bovine serum (Life Technologies), antibiotic–antimycotic solution (Invitrogen), and Glutamax (Invitrogen). Multiwell plates (BD Biosciences) were seeded at 1 × 105 cells/plate and incubated for 24 h at 37°C in 95% humidity and 5% CO2 levels. Upon reaching 70–80% confluence, cells were utilized in experiments. For fat staining, cells were dispensed on top of poly-d-lysine–coated glass coverslips that were placed at the bottom of growth plates and were subsequently processed with Nile Red or ORO stains for image analysis.
Statistical analysis
A one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test was used in multiple comparisons among different samples. For counts data, a Fisher’s exact test was used (survival data). Data are presented as the mean values or fractions ±95% confidence intervals (C.I.s). In this study, replicates are designated as follows: a plate of at least 1000 worms constitutes a technical replicate; three technical replicates form a biological replicate; and three biological replicates harvested in at least two experiments were the minimum for data requiring pooled animals (staining) or whole-cell extracts (Western blotting). For lipid staining image data, more than 100 animals per sample were imaged from animals collected in at least two staining experiments. Survival data were collected from 600 animals harvested in three different experiments. GraphPad Prism 8 and Excel Data Analysis tools were used for all statistical analyses.
Supplementary Material
Acknowledgments
We thank N. Hernandez for the Maf1 KO MEF cell line; J. Gonzalez and L. Thomas for technical assistance; and N. Stuhr and C.-A. Yen for critical reading of the manuscript. Some strains were provided by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health (NIH) Office of Research Infrastructure Programs (P40 OD010440). This work was funded by NIH grants R01GM109028 and R01AG058610 to S.P.C., T32AG000037 to A.M.H., and T32AG052374 to W.E.
Abbreviations used:
- DDR
DNA damage response
- GFP
green fluorescent protein
- KO
knockout
- MEF
mouse embryonic fibroblast
- MMS
methane methylsulfonate
- ncRNA
non-coding RNA
- pol
polymerase
- pS-T/Q
phospho-serine/threonine-glutamine
- qPCR
quantitative PCR
- RNAi
RNA interference
- S/T-Q
serine/threonine-glutamine motif
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-06-0378).
REFERENCES
- Boguta M, Czerska K, Zoladek T (1997). Mutation in a new gene MAF1 affects tRNA suppressor efficiency in Saccharomyces cerevisiae. Gene 185, 291–296. [DOI] [PubMed] [Google Scholar]
- Bonhoure N, Byrnes A, Moir RD, Hodroj W, Preitner F, Praz V, Marcelin G, Chua SC Jr, Martinez-Lopez N, Singh R, et al. (2015). Loss of the RNA polymerase III repressor MAF1 confers obesity resistance. Genes Dev 29, 934–947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom P, Rutberg M, Ericsson J, Holmdahl P, Andersson L, Frohman MA, Boren J, Olofsson SO (2005). Cytosolic lipid droplets increase in size by microtubule-dependent complex formation. Arterioscler Thromb Vasc Biol 25, 1945–1951. [DOI] [PubMed] [Google Scholar]
- C. elegans Deletion Mutant Consortium (2012). Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2, 1415–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Lanz RB, Walkey CJ, Chang WH, Lu W, Johnson DL (2018). Maf1 and repression of RNA polymerase III-mediated transcription drive adipocyte differentiation. Cell Rep 24, 1852–1864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D, Woiwode A, Zhang C, Mandavia N, Morton JP, Warnock LJ, Milner J, White RJ, Johnson DL (2003). p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J 22, 2810–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBardeleben HK, Lopes LE, Nessel MP, Raizen DM (2017). Stress-induced sleep after exposure to ultraviolet light is promoted by p53 in Caenorhabditis elegans. Genetics 207, 571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escorcia W, Ruter DL, Nhan J, Curran SP (2018). Quantification of lipid abundance and evaluation of lipid distribution in Caenorhabditis elegans by nile red and oil red O staining. J Vis Exp 2018, 57352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto S, Fujimoto T (2002). Deformation of lipid droplets in fixed samples. Histochem Cell Biol 118, 423–428. [DOI] [PubMed] [Google Scholar]
- Ghavidel A, Schultz MC (2001). TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell 106, 575–584. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Ezra O, Rivlin N, Molchadsky A, Madar S, Goldfinger N, Rotter V (2012). p53, a novel regulator of lipid metabolism pathways. J Hepatol 56, 656–662. [DOI] [PubMed] [Google Scholar]
- Goldstein I, Rotter V (2012). Regulation of lipid metabolism by p53—fighting two villains with one sword. Trends Endocrinol Metab 23, 567–575. [DOI] [PubMed] [Google Scholar]
- Goodfellow SJ, Graham EL, Kantidakis T, Marshall L, Coppins BA, Oficjalska-Pham D, Gerard M, Lefebvre O, White RJ (2008). Regulation of RNA polymerase III transcription by Maf1 in mammalian cells. J Mol Biol 378, 481–491. [DOI] [PubMed] [Google Scholar]
- Johnson DL, Stiles BL (2016). Maf1, a new PTEN target linking RNA and lipid metabolism. Trends Endocrinol Metab 27, 742–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SS, Zhang C, Fromm J, Willis IM, Johnson DL (2007). Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell 26, 367–379. [DOI] [PubMed] [Google Scholar]
- Khanna A, Johnson DL, Curran SP (2014). Physiological roles for mafr-1 in reproduction and lipid homeostasis. Cell Rep 9, 2180–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna A, Pradhan A, Curran SP (2015). Emerging roles for Maf1 beyond the regulation of RNA polymerase III activity. J Mol Biol 427, 2577–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lans H, Marteijn JA, Schumacher B, Hoeijmakers JH, Jansen G, Vermeulen W (2010). Involvement of global genome repair, transcription coupled repair, and chromatin remodeling in UV DNA damage response changes during development. PLoS Genet 6, e1000941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YL, Li YC, Su CH, Chiao CH, Lin IH, Hsu MT (2015). MAF1 represses CDKN1A through a Pol III-dependent mechanism. eLife 4, e06283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Tsang CK, Wang S, Li XX, Yang Y, Fu L, Huang W, Li M, Wang HY, Zheng XF (2016). MAF1 suppresses AKT-mTOR signaling and liver cancer through activation of PTEN transcription. Hepatology 63, 1928–1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo JY, Spatola BN, Curran SP (2017). WDR23 regulates NRF2 independently of KEAP1. PLoS Genet 13, e1006762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly K, Reid SJ, Snell RG (2015). Rapid RNA analysis of individual Caenorhabditis elegans. MethodsX 2, 59–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynn DA, Dalton HM, Sowa JN, Wang MC, Soukas AA, Curran SP (2015). Omega-3 and -6 fatty acids allocate somatic and germline lipids to ensure fitness during nutrient and oxidative stress in Caenorhabditis elegans. Proc Natl Acad Sci USA 112, 15378–15383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Rideout EJ, Grewal SS (2012). Nutrient/TOR-dependent regulation of RNA polymerase III controls tissue and organismal growth in Drosophila. EMBO J 31, 1916–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michels AA, Robitaille AM, Buczynski-Ruchonnet D, Hodroj W, Reina JH, Hall MN, Hernandez N (2010). mTORC1 directly phosphorylates and regulates human MAF1. Mol Cell Biol 30, 3749–3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzejewska J, Chreptowicz K (2016). Lack of Maf1 enhances pyruvate kinase activity and fermentative metabolism while influencing lipid homeostasis in Saccharomyces cerevisiae. FEBS Lett 590, 93–100. [DOI] [PubMed] [Google Scholar]
- Murphy S, Martin S, Parton RG (2010). Quantitative analysis of lipid droplet fusion: inefficient steady state fusion but rapid stimulation by chemical fusogens. PLoS One 5, e15030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nhan JD, Curran SP (2020). Metabolic assessment of lipid abundance and distribution. Methods Mol Biol 2144, 103–110. [DOI] [PubMed] [Google Scholar]
- Nhan JD, Turner CD, Anderson SM, Yen CA, Dalton HM, Cheesman HK, Ruter DL, Uma Naresh N, Haynes CM, Soukas AA, et al. (2019). Redirection of SKN-1 abates the negative metabolic outcomes of a perceived pathogen infection. Proc Natl Acad Sci USA 116, 22322–22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi C, Wang L, Shetty M, Mell JC, Sell C, Noguchi E (2021). Maf1 limits RNA polymerase III-directed transcription to preserve genomic integrity and extend lifespan. Cell Cycle 20, 247–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ou HL, Schumacher B (2018). DNA damage responses and p53 in the aging process. Blood 131, 488–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palian BM, Rohira AD, Johnson SA, He L, Zheng N, Dubeau L, Stiles BL, Johnson DL (2014). Maf1 is a novel target of PTEN and PI3K signaling that negatively regulates oncogenesis and lipid metabolism. PLoS Genet 10, e1004789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pang S, Curran SP (2014). Adaptive Capacity to Bacterial Diet Modulates Aging in C. elegans. Cell Metab 19, 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta K, Lefebvre O, Martin NC, Smagowicz WJ, Stanford DR, Ellis SR, Hopper AK, Sentenac A, Boguta M (2001). Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol Cell Biol 21, 5031–5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan A, Hammerquist AM, Khanna A, Curran SP (2017). The C-box region of MAF1 regulates transcriptional activity and protein stability. J Mol Biol 429, 192–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reina JH, Azzouz TN, Hernandez N (2006). Maf1, a new player in the regulation of human RNA polymerase III transcription. PLoS One 1, e134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rideout EJ, Marshall L, Grewal SS (2012). Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc Natl Acad Sci USA 109, 1139–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckher M, Bujarrabal A, Doll MA, Soltanmohammadi N, Schumacher B (2018). A simple answer to complex questions: Caenorhabditis elegans as an experimental model for examining the DNA damage response and disease genes. J Cell Physiol 233, 2781–2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher B, Schertel C, Wittenburg N, Tuck S, Mitani S, Gartner A, Conradt B, Shaham S (2005). C. elegans ced-13 can promote apoptosis and is induced in response to DNA damage. Cell Death Differ 12, 153–161. [DOI] [PubMed] [Google Scholar]
- Shetty M, Noguchi C, Wilson S, Martinez E, Shiozaki K, Sell C, Mell JC, Noguchi E (2020). Maf1-dependent transcriptional regulation of tRNAs prevents genomic instability and is associated with extended lifespan. Aging Cell 19, e13068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatola BN, Lo JY, Wang B, Curran SP (2019). Nuclear and cytoplasmic WDR-23 isoforms mediate differential effects on GEN-1 and SKN-1 substrates. Sci Rep 9, 11783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stergiou L, Doukoumetzidis K, Sendoel A, Hengartner MO (2007). The nucleotide excision repair pathway is required for UV-C-induced apoptosis in Caenorhabditis elegans. Cell Death Differ 14, 1129–1138. [DOI] [PubMed] [Google Scholar]
- Thiam AR, Beller M (2017). The why, when and how of lipid droplet diversity. J Cell Sci 130, 315–324. [DOI] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM (2002). Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell 10, 1489–1494. [DOI] [PubMed] [Google Scholar]
- Vannini A, Ringel R, Kusser AG, Berninghausen O, Kassavetis GA, Cramer P (2010). Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 143, 59–70. [DOI] [PubMed] [Google Scholar]
- Verma S, Jagtap U, Goyala A, Mukhopadhyay A (2018). A novel gene-diet pair modulates C. elegans aging. PLoS Genet 14, e1007608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermezovic J, Stergiou L, Hengartner MO, d’Adda di Fagagna F (2012). Differential regulation of DNA damage response activation between somatic and germline cells in Caenorhabditis elegans. Cell Death Differ 19, 1847–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorlander MK, Baudin F, Moir RD, Wetzel R, Hagen WJH, Willis IM, Muller CW (2020). Structural basis for RNA polymerase III transcription repression by Maf1. Nat Struct Mol Biol 27, 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vose S, Mitchell J (2011). Relationship between DNA damage and energy metabolism: evidence from DNA repair deficiency syndromes. In: DNA Repair and Human Health, ed S. Vengrova. InTech Open, DOI: 10.5772/25053.
- Wang YH, Hariharan A, Bastianello G, Toyama Y, Shivashankar GV, Foiani M, Sheetz MP (2017). DNA damage causes rapid accumulation of phosphoinositides for ATR signaling. Nat Commun 8, 2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis IM (2018). Maf1 phenotypes and cell physiology. Biochim Biophys Acta 1861, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis IM, Moir RD (2007). Integration of nutritional and stress signaling pathways by Maf1. Trends Biochem Sci 32, 51–53. [DOI] [PubMed] [Google Scholar]
- Yang J, Xu ZP, Huang Y, Hamrick HE, Duerksen-Hughes PJ, Yu YN (2004). ATM and ATR: sensing DNA damage. World J Gastroenterol 10, 155–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye AL, Ragle JM, Conradt B, Bhalla N (2014). Differential regulation of germline apoptosis in response to meiotic checkpoint activation. Genetics 198, 995–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CA, Curran SP (2016). Gene-diet interactions and aging in C. elegans. Exp Gerontol 86, 106–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen CA, Ruter DL, Turner CD, Pang S, Curran SP (2020). Loss of flavin adenine dinucleotide (FAD) impairs sperm function and male reproductive advantage in C. elegans. eLife 9, e52899. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.