Abstract
Sensing physical forces is a critical first step in mechano-transduction of cells. Zyxin, a LIM domain-containing protein, is recruited to force-bearing actin filaments and is thought to repair and strengthen them. Yet, the precise force-induced protein interactions surrounding zyxin remain unclear. Using BioID analysis, we identified proximal proteins surrounding zyxin under normal and force-bearing conditions by label-free mass spectrometry analysis. Under force-bearing conditions, increased biotinylation of α-actinin 1, α-actinin 4, and AFAP1 were detected, and these proteins accumulated along force-bearing actin fibers independently from zyxin, albeit at a lower intensity than zyxin. VASP also accumulated along force-bearing actin fibers in a zyxin-dependent manner, but the biotinylation of VASP remained constant regardless of force, supporting the model of a free zyxin–VASP complex in the cytoplasm being corecruited to tensed actin fibers. In addition, ARHGAP42, a RhoA GAP, was also identified as a proximal protein of zyxin and colocalized with zyxin along contractile actin bundles. The overexpression of ARHGAP42 reduced the rate of small wound closure, a zyxin-dependent process. These results demonstrate that the application of proximal biotinylation can resolve the proximity and composition of protein complexes as a function of force, which had not been possible with traditional biochemical analysis.
INTRODUCTION
Mechanical cues are crucial for regulating cellular homeostasis, initiating a vast array of signaling pathways. Force-sensing modules such as adhesive receptors transmit external forces to cytoplasmic interacting partners, converting mechanical cues into biochemical signaling. Besides adhesive complexes, actin filaments are also thought of as tension-sensors with force-dependent protein-binding partners (Galkin et al., 2012; Harris et al., 2018). For example, zyxin, a LIM domain-containing protein, is recruited to focal adhesions and actin stress fibers in a force-sensitive manner either by uniaxial stretching of cells on a compliant substrate (Yoshigi et al., 2005; Hoffman et al., 2012), through the application of external force on the dorsal cell surface (Colombelli et al., 2009), or with local RhoA activation (Oakes et al., 2017). At focal adhesions, zyxin dynamics is tension-dependent (Lele et al., 2006), and zyxin mediates stretch-induced actin polymerization via its interaction with Ena/VASP (Hirata et al., 2008). Furthermore, the LIM domain of zyxin alone is sufficient for its recruitment to force-bearing actin filaments (Smith et al., 2013; Watanabe-Nakayama et al., 2013). Through these findings, zyxin emerged as a force-sensitive protein that promotes actin assembly at force-bearing sites; however, a complete list of force-dependent protein interactions surrounding zyxin remains largely unclear.
Force-dependent protein interactions are short-lived and weak. Consequently, they are difficult to isolate using traditional biochemical analyses that rely on stable protein–protein interactions during purification. This inherent nature of traditional biochemical assays prevents detection of force-dependent protein interactions. Currently, identification and analysis of mechano-responsive proteins largely depend on methods that often focus on single proteins (e.g., the use of conformation-sensitive antibodies, FRET, or single molecule force spectroscopy; see reviews by LaCroix et al., 2015; Harris et al., 2018) rather than a comprehensive analysis of protein compositions. Alternatively, the contractility-dependent composition of focal adhesions was identified with proteomic analysis (Kuo et al., 2011; Schiller et al., 2011) by extracting the cytoplasm and other organelles from adherent cells while preserving focal adhesions along the substrates for enrichment of focal adhesion proteins. However, these techniques have yet to be adapted to probe structures beyond adhesive complexes.
An alternative method more amenable to identify weak and transient protein interactions is BIOtin IDentification, BioID, using promiscuous biotin ligases (Roux et al., 2012). Proximal biotinylation has been applied to adhesion complexes (see review by Kim and Roux, 2016). In particular, stretch-induced proximal biotinylation surrounding α-catenin demonstrated that force-dependent proximity of myosin IIA can be detected by Western blot analysis (Ueda et al., 2015). Unfortunately, the detection of biotinylated proteins from the original biotin ligase typically requires overnight biotin incubation, while force-induced recruitment of zyxin occurs in the order of minutes (e.g., Supplemental Figure S1, B and C, and Supplemental Movie S1). The improved versions of biotin ligases, BASU–isolated from Bacillus subtilis (Ramanathan et al., 2018), TurboID and miniTurbo–engineered BirA* using directed evolution (Branon et al., 2018), have much faster biotinylation kinetics, and biotinylated proteins can be detected within 10 min after biotin addition (Branon et al., 2018; Ramanathan et al., 2018), a time scale more comparable to zyxin’s force-induced recruitment. Using zyxin as a model force-sensing protein, we demonstrate the use of proximal biotinylation combined with proteomic analysis to identify proximal proteins surrounding zyxin under force-bearing conditions.
Movie S1.
Stretch‐induced accumulation of zyxin using a microneedle. See also Figure 1A.
RESULTS AND DISCUSSION
To identify force-sensitive protein interactions surrounding zyxin, zyxin was tagged with promiscuous biotin ligase, TurboID (Branon et al., 2018), and stably expressed in Madin-Darby Canine Kidney (MDCK) cells (Figure 1A). TurboID-tagged zyxin localized to focal adhesions and along actin fibers, and in the presence of biotin, biotinylated proteins colocalized with TurboID-tagged zyxin (Figure 1A). Using a small-scale PDMS substrate to apply a mechanical strain to adherent cells (60% static uniaxial substrate stretch for 10 min), TurboID-tagged zyxin was recruited to actin fibers and biotinylated proximal proteins along the actin fibers (Figure 1A); therefore, isolation of these biotinylated proteins should yield the identities of zyxin’s force-dependent proximal proteins.
FIGURE 1:
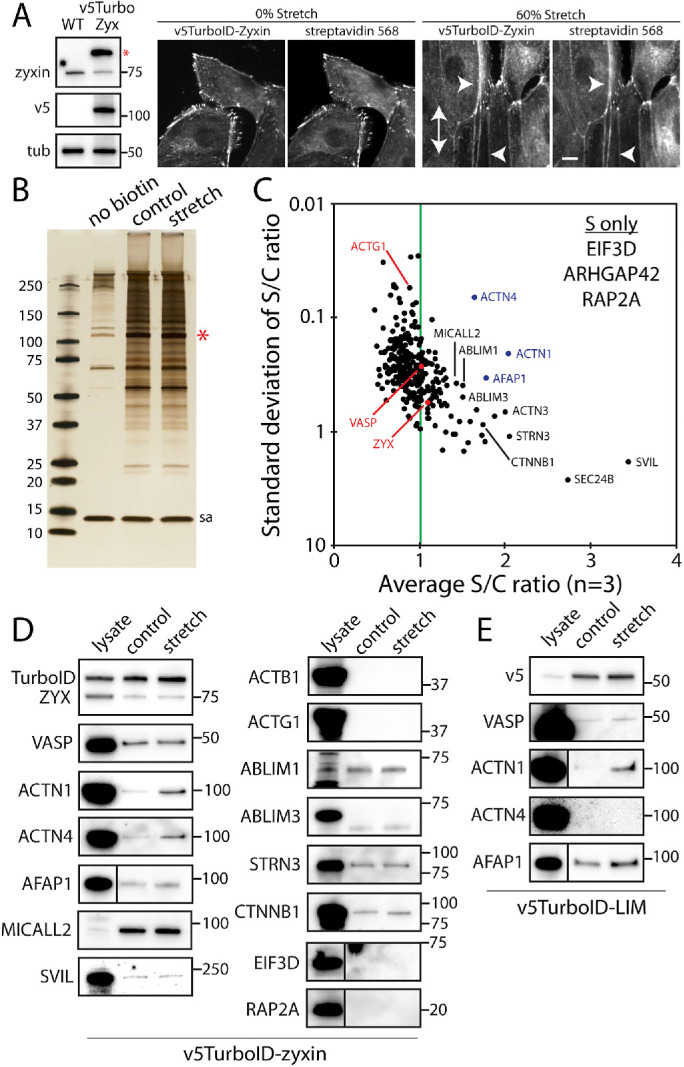
Stretch-dependent proximal biotinylation analysis using TurboID-tagged zyxin. (A) Characterization of MDCK cells stably expressing v5-TurboID-tagged zyxin using Western blot analysis (left) and immunofluorescent labeling (right). Tubulin (tub) was used as a loading control. The double-headed arrow indicates the stretch direction, and arrowheads indicate the zyxin positive fibers with streptavidin labeling. Scale bar, 10 μm. (B) Silver-stained SDS–PAGE analysis of purified biotinylated proteins from no biotin, control and stretch conditions. The red star indicates a band for self-biotinylated TurboID-tagged zyxin, and sa denotes streptavidin bands. (C) Mass spectrometry analysis of TurboID-tagged zyxin. The average and SD of ratios of sum intensities from control and stretch samples are plotted (n = 3). The green line indicates an equal level of biotinylation in control and stretch samples. Only top 320 hits based on the sum intensity (>107) were plotted. Top hits present in stretch condition only (S only) are also listed. (D) Western blot analysis of biotinylated proteins with control lysates as positive controls. (E) Western blot analysis of biotinylated proteins from TurboID-tagged LIM domain of zyxin.
Using a large-scale cell stretch device (Renner et al., 2017), biotinylated proteins were then biochemically isolated from cells plated on control and stretched membranes (60% uniaxial stretch for 30 min in the presence of 50 μM biotin) and analyzed using label-free mass spectrometry (Figure 1, B and C; see the complete list of proteins in the Supplemental Data). To identify highly biotinylated proteins under force-bearing conditions with minimal deviations among the triplicate samples, the ratios of relative abundance of identified biotinylated proteins in control versus stretch conditions were plotted against the SD of the ratios (Figure 1C and Supplemental Data, “ratio” tab). The majority of the proteins were equally biotinylated in control and stretch conditions (e.g., ACTG1, ZYX, and VASP, a known binding partner of zyxin; Drees et al., 2000), suggesting the proteins’ close proximity to zyxin regardless of stretch application. However, there were notable exceptions that had high stretch-to-control ratios (labeled in black or blue and appearing in the top right quadrant of Figure 1C). Note that all proteins examined had at least a 10-fold increase in the relative abundance compared with the samples collected in the absence of free biotin (except for ACTG1, which had a sevenfold increase). In addition, EIF3D, RAP2A, and ARHGAP42 were not detected in control samples but were present in all three stretch samples, albeit at low levels (see Supplemental Data, “S only” tab).
Selected candidates for force-dependent proximal proteins of zyxin were further analyzed using Western blots for verification (Figure 1D). Some proteins were detected by mass spectrometry, but not detected by Western blots (ACTB1, ACTG1, and ABLIM3 – Figure 1D). This was possibly caused by contamination in the mass spectrometry samples or by the protein level in the eluted fraction being below the detection level of Western blot. Interestingly, some proteins were only detected in eluted samples (e.g., MICALL2), suggesting that these proteins were expressed at low levels and were concentrated during purification of the biotinylated proteins (Figure 1D). While some biotinylated proteins were highly enriched in stretch conditions according to mass spectrometry analysis (e.g., supervillain—SVIL), only α-actinins (ACTN1, ACTN4), and AFAP1 were also enriched in stretch samples according to Western blot analysis (Figure 1D). Of these proteins, only α-actinin 1 has been previously identified as a binding partner of zyxin (Crawford et al., 1992; Li and Trueb, 2001).
Since the zyxin LIM domain is solely responsible for force-induced recruitment along actin fibers (Smith et al., 2013), the proximity between zyxin’s LIM domain and the candidate proteins were tested using TurboID-tagged zyxin LIM domain (TurboID-LIM) and Western blot analysis (Figure 1E). TurboID-LIM was stably expressed in MDCK cells, albeit at a lower level than full-length zyxin (Supplemental Figure S1A). Similar to the full-length zyxin, TurboID-LIM localized to focal adhesions, actin fibers and cell–cell contacts, and biotinylated proximal proteins (Supplemental Figure S1A). In Western blots, both α-actinin 1 and AFAP1 were enriched in stretch condition; however, α-actinin 4 was barely detectable (Figure 1E). Interestingly, α-actinin 1 binds to the N-terminus of zyxin (Li and Trueb, 2001), yet close proximity of α-actinin 1 and the LIM domain suggests that the direct binding may not be required to bring these proteins close to each other, and that these proteins may be independently recruited to force-bearing fibers (see below). In fact, zyxin does not bind to α-actinin 4 in vitro (Fukumoto et al., 2015) despite highly biotinylated α-actinin 4 by TurboID-zyxin being observed in stretch samples for both mass spectrometry analysis (Figure 1C) and Western blot (Figure 1D). It remains possible that in vitro biochemical analysis failed to recapitulate the zyxin-α-actinin 4 interaction observed in live cells. Further studies are needed to better define the molecular details of this interaction. Again, our finding highlights the advantage of proximal biotinylation approach to identify force-induced association in live cells as they are actively under force-bearing conditions versus a force-free in vitro biochemical analysis.
To investigate the force-dependent dynamics of zyxin and other proximal proteins in live cells, MDCK cells were transiently transfected with tandem dimer DsRed (tdDsRed)-tagged zyxin and stretched using a micron-sized needle tip (Cheah et al., 2019). The microneedle was gently placed on neighboring cells and moved away from the tdDsRed-zyxin expressing cell (Supplemental Figure S1B), which in turn stretched the tdDsRed-zyxin expressing cell via cell–cell contacts. Zyxin accumulated along fibers as the microneedle moved farther away, and on the release of the microneedle, therefore the release of tension, zyxin intensity at actin fibers decreased (Supplemental Figure S1B).
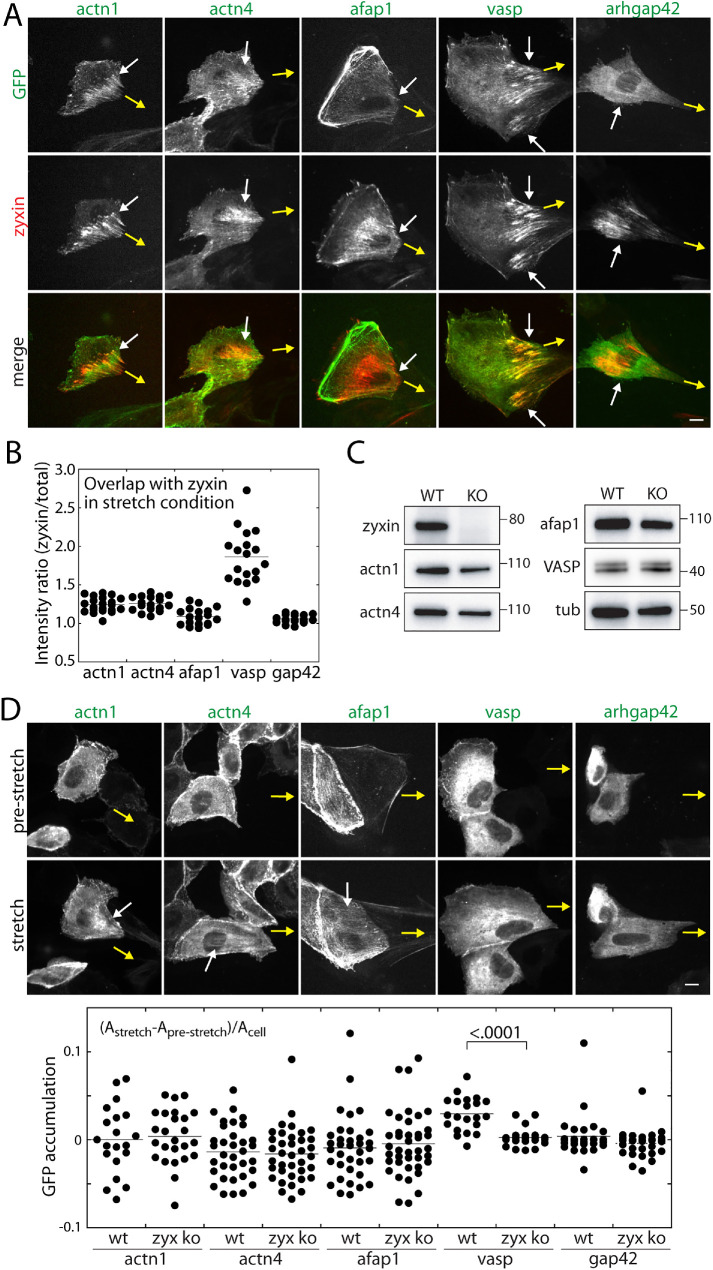
Since α-actinins and AFAP1 are in close proximity to zyxin under force-bearing conditions (Figure 1, C and D), α-actinins and AFAP1 dynamics were examined using the microneedle assay. As force is applied by a microneedle, α-actinin 1 and 4 occasionally accumulated along zyxin-positive actin fibers, while AFAP1 accumulated to a lesser extent than α-actinin 1 and 4 (Figure 2A). In addition, VASP accumulation along the actin fibers resembled that of zyxin, while ARHGAP42 (biotinylated only under stretch conditions and previously shown to localize to actin filaments; Luo et al., 2017) was barely detectable along the fibers (Figure 2A). This is also evident when the degree of overlap between α-actinins, AFAP1, VASP, and zyxin was quantified under stretch conditions (Figure 2B). The ratios of the candidate protein’s average intensity at the site of zyxin accumulation to average cell intensity were greater than 1 for both α-actinins and AFAP1, that is, the intensity of α-actinins or AFAP1 colocalized with zyxin was often higher than the average intensity per cell (Figure 2B and Supplemental Figure S1D). In contrast, VASP accumulated on force-bearing actin filaments (Figure 2, A and B), despite the relative biotinylation of VASP being force independent (Figure 1, C and D).
FIGURE 2:
Dynamics of zyxin proximal proteins in the presence or absence of zyxin. (A) Relative localization of zyxin (tdDsRed) and α-actinin 1, α-actinin 4, AFAP1, VASP, and ARHGAP42 (all GFP-tagged) in cells stretched by microneedle. Yellow arrows point to the direction of microneedle movement and white arrows indicate the force-induced zyxin accumulation. (B) Colocalization of zyxin and α-actinin 1 (n = 21), α-actinin 4 (n = 17), AFAP1 (n = 18), and VASP (n = 18) in cells stretched by microneedle. See Materials and Methods for colocalization analysis. (C) Western blot analysis of zyxin knockout cell line. (D) Force-induced recruitment of α-actinin 1, α-actinin 4, AFAP1, VASP, and ARHGAP42 in zyxin knockout cells. Quantification of α-actinin 1 (n = 19 for WT and n = 24 for KO), α-actinin 4 (n = 34 for WT and n = 36 for KO), AFAP1 (n = 34 for WT and n = 39 for KO), and VASP (n = 20 for WT and n = 20 for KO) ARHGAP42 (n = 26 for WT and n = 28 for KO) accumulation in the presence (WT) or absence of zyxin (KO). The p value is calculated using one-way ANOVA. All scale bars, 10 μm.
To test whether the force-induced accumulation of α-actinins, AFAP1, and VASP depend on zyxin, we generated zyxin knockout MDCK cells using CRISPR/Cas9 (Figure 2C). In this knockout cell line, the protein levels of α-actinins, AFAP1, and VASP remain similar to that of wild-type cells (Figure 2C). Based on the fiber areas defined by relative intensity (see Materials and Methods), α-actinins and AFAP1 accumulated along force-bearing actin filaments in some cells (Figure 2D). In contrast, VASP accumulation was clearly diminished in the zyxin knockout cells (Figure 2, A and D), suggesting that zyxin is required for VASP recruitment to force-bearing actin filaments. Since the proximal biotinylation of VASP by TurboID-tagged zyxin remained similar between control and stretch samples, these results suggest that VASP is cotransported by zyxin to sites of force-bearing actin filaments. On the other hand, α-actinins and AFAP1 are recruited to force-bearing actin filaments independently from zyxin, but positioned in close proximity to zyxin.
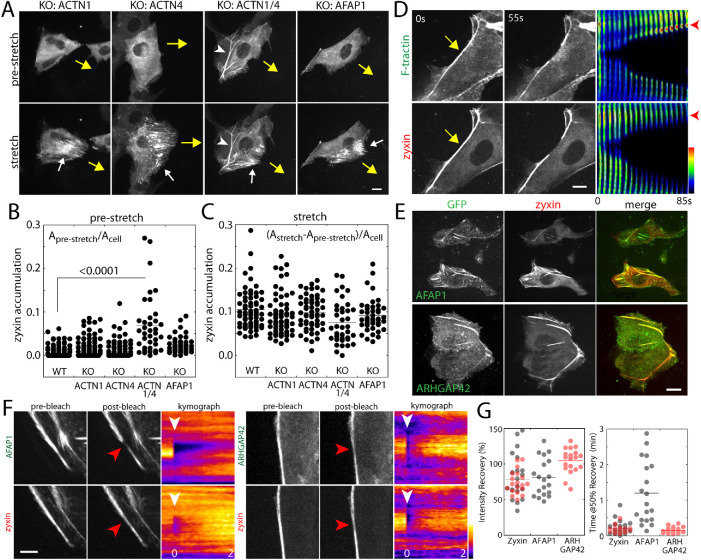
To test whether zyxin recruitment to actin filaments requires either α-actinins or AFAP1, we generated MDCK cells deficient in α-actinin 1, α-actinin 4, α-actinin 1/4, or AFAP1 using CRISPR/Cas9 (Supplemental Figure S1E). Note that MDCK cells express α-actinin 1 and 4, but not 2 (the expression of which is restricted to the heart) or 3 (detected in mass spectrometry results but not in Western blots). The protein level of zyxin in knockout cells remained similar to those of wild-type cells (Supplemental Figure S1E). Despite the absence of these proteins, zyxin organization remained similar to that of wild-type cells (Figure 3A), except in α-actinin 1/4 double knockout cells. In the absence of both α-actinin 1/4, zyxin was prominently recruited to actin bundles even prior to cell stretch (Figure 3, A and B). In the absence of either α-actinins or AFAP1, zyxin accumulated along actin filaments with the application of stretch by a microneedle at similar levels to that of wild-type cells (Figure 3C). These results suggest that zyxin recruitment to the force-bearing actin fibers does not require α-actinins or AFAP1. This is consistent with recent observations that zyxin binds directly to strained actin filaments (Sun et al., 2020; Winkelman et al., 2020).
FIGURE 3:
Force-induced zyxin accumulation is independent of α-actinin and AFAP1. (A) Stretch-induced accumulation of zyxin in α-actinin and AFAP1 knockout cell lines. Yellow arrows indicate the movement of microneedle and white arrows indicate the zyxin accumulation. White arrows indicate zyxin accumulation on actin fibers under tension. The white arrowhead indicates zyxin-positive fibers in α-actinin 1/4 double knockout cells. Scale bar, 10 μm. (B) Quantification of zyxin accumulation in prestretch cells. Zyxin accumulation was quantified using the thresholded area based on zyxin intensity and was normalized to total cell area. (C) Under force-bearing conditions, zyxin accumulation was quantified using the difference of the thresholded area of zyxin intensity in prestretch and stretch cells, and then the value was normalized to cell area. N = 63 (WT), 58 (KO: ACTN1), 54 (KO: ACTN4), 38 (KO: ACTN1/4), and 41 (KO: AFAP1). The p value is calculated using one-way ANOVA. (D) Severing the actin fibers in actinin 1/4 deficient cells using a microneedle. Using a microneedle to gently touch and sever actin fibers at the cell edge (yellow arrow), the fate of actin fibers was monitored as the montages of F-tractin and zyxin intensity profiles along actin fibers (right panel). Scale bar, 5 μm. (E) AFAP1 and ARHGAP42 colocalize with zyxin-positive actin fibers in α-actinin 1- and 4-deficient cells. Scale bar, 10 μm. (F) Turnover of AFAP1 and ARHGAP42 analyzed by fluorescence recovery after photobleaching (FRAP). The α-actinin 1- and 4-deficient cells transfected with zyxin-tdDsRed and GFP-tagged AFAP1 or ARHGAP42. A small region of fiber was photobleached (see red and white arrowheads) and the intensity recovery was monitored over time. The intensity line scan along the fiber was used to generate kymograph of the intensity recovery. Scale bar, 5 μm. (G) The intensity recovery curve was characterized based on the percentage of intensity recovery over the prebleached intensity during the first 5 min and the time it took to recover 50% of the initial intensity. Gray circles represent the analysis of AFAP1 and zyxin coexpressing cells and red circles represent the analysis of ARHGAP42 and zyxin coexpressing cells.
Since zyxin prominently accumulated along actin fibers in α-actinin 1/4 knockout cells, we examined the characteristics of these actin bundles and whether these bundles resemble force-induced zyxin-positive actin bundles in our microneedle analysis. Previously, similar actin bundles were observed in α-actinin 4-deficient MDCK cells (Kemp and Brieher, 2018). In our hand, however, these actin bundles were the most frequent in α-actinin 1/4 double knockout cells compared with the individual α-actinin knockout cells (Figure 3, A and D). Interestingly, in α–actinin-deficient fibroblasts, these actin bundles found to be under tension and contractile (Hu et al., 2017, 2019). While the actin bundles in fibroblasts are much more ordered structures exhibiting clear striation of actin binding proteins (Hu et al., 2017, 2019), these zyxin-positive actin bundles in our MDCK α-actinin 1/4 double knockout cells contained more uniform distribution of zyxin (Figure 3, A and D).
Using a fine-tip microneedle on the micromanipulator, we carefully placed the tip onto the cell surface to gently sever the actin bundle in α-actinin 1/4-deficient cells expressing both tdDsRed-zyxin and GFP-F-tractin (Figure 3D). Immediately, the zyxin-positive bundle started to shorten (Figure 3D). If the actin bundles are depolymerizing at the severed ends, then F-tractin intensity should decrease at the tip of actin fibers. In contrast, actin fibers are under tension and contracting, then F-tractin intensity should increase at the severed ends. The kymograph along the severed actin fibers shows that the F-tractin intensity increased at the tips of severed fibers (Figure 3D), suggesting that these bundles are indeed contractile. These zyxin-positive, contractile bundles contained both AFAP1 and ARHGAP42 (Figure 3E), further validating the list of force-dependent proteins identified in our proximal biotinylation analysis surrounding zyxin.
Using α-actinin 1/4 knockout cells expressing both tdDsRed-zyxin and GFP-tagged AFAP1 or ARHGAP42, we examined relative dynamics of these proteins along contractile actin bundles to probe potential interactions in cells without external force application. Using a pulse dye laser to selectively photobleach a small region along actin bundles, we simultaneously monitored the fluorescence intensity recovery of both GFP and tdDsRed-tagged proteins (Figure 3F). We quantified the fluorescence intensity recovery based on the mobile fraction (after 5 min) and time to reach 50% of mobile fraction (Figure 3, F and G). Interestingly, AFAP1 recovered much slower than zyxin, suggesting that AFAP1 turnover on the surface of actin bundles is slower (Figure 3, F and G), while ARHGAP42 recovered at a similar rate as zyxin (Figure 3, F and G). These results suggest that, while both AFAP1 and ARHGAP42 are in close proximity to zyxin along the contractile actin bundles, their temporal dynamics are distinct which in turn indicate the different roles played by AFAP1 and ARHGAP42. In fact, AFAP1 is a known actin cross-linker (Qian et al., 2002) and is, thus, thought to play a structural role in organizing actin fibers. Interestingly, both AFAP1 and ARHGAP42 are substrates of Src kinases (Flynn et al., 1993; Luo et al., 2017) whose activation has been shown to depend on physical forces (Wang et al., 2005).
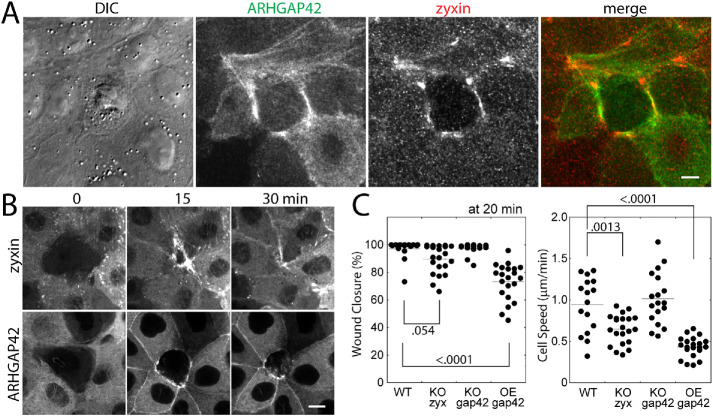
Because of the similar dynamics exhibited by zyxin and ARHGAP42 along the contractile actin bundles, we sought to test how the level of ARHGAP42 may alter zyxin-dependent cell migration. Previously, we have shown that MDCK cells with a reduced level of zyxin are deficient in single cell-sized wound healing (Nguyen et al., 2010). This mode of motility requires the formation of contractile actin bundles (known as a purse-string actin ring), where both zyxin and ARHGAP42 accumulated (Figure 4, A and B; Supplemental Movie S2). The wound closure was slower for the cell lines lacking zyxin and overexpressing GFP-tagged ARHGAP42, but not in ARHGAP42-deficient cells (Figure 4C), suggesting that the excess of ARHGAP42 may be inactivating RhoA (Luo et al., 2017) and inhibiting actin contractility required for apical constriction and wound closure. These data suggest that zyxin and ARHGAP42 are in close proximity at contractile actin bundles to regulate this unique mode of epithelial cell migration.
FIGURE 4:
The roles of zyxin and ARHGAP42 along contractile actin bundles during wound healing of cell monolayer. (A) Relative localization of zyxin and GFP-ARHGAP42 during the wound healing. Scale bar, 10 μm. (B) Time-lapse montages of zyxin or GFP-ARHGAP42 expressing cells undergoing wound closure. Scale bar, 10 μm. (C) Quantification of wound closure at 20 min after ablation (left) and the cell speed of wound closure (right) of wildtype (WT), knockout (KO) of zyxin and ARHGAP42, and ARHGAP42 over-expressing (OE) cells. The p values are calculated using one-way ANOVA.
Movie S2.
Single cell wound healing of zyxin‐GFP (left) and GFP‐ARHGAP42 (right) expressing cells. See also Figure 4B.
Using a promiscuous biotin ligase to label the proximal proteins in situ, we have identified a unique set of force-sensitive proximal proteins surrounding zyxin. Our approach also suggests the presence of the VASP-zyxin complex irrespective of mechanical stimuli and thus providing additional insights into the known protein interactions in cells. Interestingly, AFAP1 is a known binding partner of Src (Baisden et al., 2001) and ARHGAP42 is a substrate of Src (Luo et al., 2017), suggesting that zyxin may be a key mediator of ARHGAP42 activation at the site of force-bearing fibers. Further analysis on other candidates will shed new insights into how zyxin is recruited to force-bearing actin filaments and how zyxin initiates or activates signaling pathways under tension (e.g., via ARHGAP42). Therefore, elucidating the force-dependent protein interactome should provide mechanistic details into functions regulated by zyxin and other force-sensitive proteins.
MATERIALS AND METHODS
Cell lines and reagents
MDCK GII cells were cultured in DMEM (Life Technologies) supplemented with 10% fetal bovine serum (Atlanta Biological) and antibiotics. The cells were treated with the mycoplasma removal agent (Bio-Rad), tested with the universal mycoplasma detection kit (ATCC), and verified to be free of mycoplasma. Cells were either transiently or stably transfected with plasmids using Lipofectamine 3000 (Invitrogen) or jetOPTIMUS (Polyplus) according to the manufacturer’s protocol. The stably transfected cells were selected using G418 (Invitrogen). ACTN1 and ACTN4 cDNA were purchased from Harvard PlasmID Database, and subcloned into pEGFP vector (Invitrogen) using a Gibson assembly kit (NEB). GFP-tagged F-tractin (58473) was purchased from Addgene. GFP-tagged zyxin was a gift from Masahiro Sokabe (Nagoya University, Japan), GFP-tagged VASP was a gift from Frank Gertler (MIT), GFP-tagged ARHGAP42 was a gift from Daniel Rosel (Charles University, Czech Republic) (Luo et al., 2017), and GFP-tagged AFAP1 was a gift from Mingyao Liu (University of Toronto) (Han et al., 2004). The antibodies used in this study were anti-v5 (SV5-Pk1, Bio-Rad), anti-zyxin (EPR4302, Abcam), anti-α-tubulin (DM1A, Cell Signaling), anti-VASP (43, BD Biosciences), anti-α-actinin 1 (23, BD Biosciences), anti-α-actinin 4 (D7U5A, Cell Signaling), anti-AFAP1 (21093, Proteintech), anti-β-actin (8H10D10, Cell Signaling), anti-γ-actin (2A3, Millipore), anti-ABLIM1 (15129, Proteintech), anti-ABLIM3 (27981, Proteintech), anti-striatin 3 (23966, Proteintech), anti-β-catenin (14, BD Biosciences), and anti-SVIL (AF7338, R&D Systems). Alexa 568-tagged streptavidin (Invitrogen) was used for detection of biotinylated proteins.
Generation of knockout cell lines
Knockout cell lines were generated by transfecting cells with a plasmid encoding Cas9 and the gRNA (Ran et al., 2013) modified to be compatible with the PiggyBac Transposon System. The gRNA sequences were selected based on ECRISP (Heigwer et al., 2014). The cells were then screened for the absence of the target protein via Western blots (Figure 2C and Supplemental Figure S1E) and immunofluorescence labeling, and the mutations were verified by sequencing the target sequences using TOPO cloning or deep sequencing of amplicons (Genewiz). See Supplemental Table S1 for the gRNA sequences and resulting mutations.
Stretch-dependent proximal biotinylation analysis
MDCK cells were cotransfected with v5-TurboID-tagged zyxin in Piggybac transposon containing plasmid and transposase-containing plasmid (System Biosciences). The transfected cells were selected using G418 (Invitrogen), and the protein levels were analyzed in Western blot (Figure 1A). For the small-scale cell stretch analysis (Figure 1A), the cells were plated on commercially available, collagen-coated PDMS chamber (Strex), and the cells were stretched by inserting a laser-cut acrylic frame with its outer length matching the final stretch dimension of the chamber. Cells were stretched for 10 min in the presence of 50 μM biotin, then stained with anti-v5 antibody and streptavidin (Figure 1A). For the large-scale analysis for mass spectrometry analysis, the cells were plated on the thin collagen-coated PDMS membranes attached to custom acrylic plate and inserted into a square cell culture dish (see details in Renner et al., 2017). After 24 h, the membranes were uniaxially stretched to 60% for 30 min in the presence of 50 μM biotin (Sigma-Aldrich). The control membranes were not stretched, but incubated with or without 50 μM biotin (Sigma-Aldrich). The biotinylated proteins were then harvested as previously described (Cheah and Yamada, 2017). Briefly, the cells were washed with phosphate-buffered saline (PBS) thrice, scraped in PBS, centrifuged, and frozen until purification day. The cells were then lysed, frozen in dry ice/ethanol solution, rapidly thawed at 37°C in a water bath, then incubated for 30 min at 4°C (Hesketh et al., 2017). The lysates were sonicated with Sonifier 250 (Branson) at 30% duty cycle and output at 3 for 1 min and then centrifuged for 20 min. The lysate concentrations were measured with DC protein assay kit (Bio-Rad). Equal amounts of cell lysates (2 mg/ml, 4 ml total) were added to 500 μl of Dynabead streptavidin C1 (Invitrogen) and incubated overnight at 4°C. The beads were washed with the lysis buffer thrice and 2% SDS once, then washed with the lysis buffer two more times. The biotinylated proteins were eluted by 25 mM biotin and heated at 95°C for 5 min. This approach elutes 40–60% of bound proteins from streptavidin beads (Cheah and Yamada, 2017). The concentration of eluted biotinylated proteins was 300–350 µg/ml for control and stretch conditions, and ∼150 µg/ml for no biotin control based on Bradford assay (Bio-Rad). Eluted samples were embedded into SDS–PAGE gel and the gel-cutouts containing all eluted proteins were submitted to Taplin Mass Spectrometry Facility (Harvard Medical School) for in-gel digest of the samples, followed by microcapillary LC/MS/MS analysis, protein database searching.
Microscopy and image analysis
Cells were imaged using a Zeiss AxioObserver equipped with a Yokogawa CSU-10 spinning disk confocal system, 40 or 63× objective, 488- and 561-nm solid-state lasers, and a Photometrics CoolSNAP HQ2 camera. The microscope system was controlled by Slidebook software (Intelligent Imaging Innovations). For live-cell imaging, the temperature was set to 37°C by a custom microscope heating chamber and the media were supplemented with 25 mM HEPES (Invitrogen). Fluorescence Recovery After Photobleaching experiments were also conducted on the same confocal system. The photobleaching system (Intelligent Imaging Innovation) is equipped with a pulsed nitrogen-pumped tunable dye laser combined with a Micropoint system with x-y axis galvomirrors. Coumarin 440 dye was used to photobleach both GFP and DsRed signals. The same laser system (set to a higher laser power) was used to ablate a single cell in cell monolayers for wound healing analysis. All images were analyzed using ImageJ and Microsoft Excel and graphed using KaleidaGraph.
Microneedle cell stretching analysis
Custom-drawn needles were fabricated using P-97 Micropipette puller (Sutter Instrument). To maximize the microneedle’s contact with adherent cells, the tip of microneedle was placed on the cells at a shallow angle, while the rest of the microneedle was carefully bent at two locations at 90° angles in opposite directions to clear the sidewall of a p35 dish. The microneedle was attached to a 3D micromanipulator (Physik Instrumente) controlled with a gaming joystick and a custom-written software. The microneedle was carefully placed on neighboring sacrificial cells, then slowly moved away from the cells of interest (denoted by yellow arrows in the figures) to apply mechanical strain across cell–cell contacts. Note that none of the cells analyzed came in direct contact with the microneedle. Since the intensity of zyxin accumulation along the fibers often exceeded the cytoplasmic zyxin intensity, the thresholding based on fluorescence intensity was used to define the areas of zyxin accumulation after the Background Subtraction command in ImageJ was used to minimize the background intensity fluctuations. The difference in the zyxin area from prestretch and at maximum stretch (normalized to total cell area) was used as a proxy for the extent of zyxin accumulation along the force-bearing actin filaments (Figures 2D and 3C). Colocalization with zyxin and other proteins was quantified based on the relative intensities of the respective proteins at sites of zyxin accumulation and cytoplasm (Figure 2B).
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health R03 EB021636 (S.Y.), National Science Foundation 1562095 (SY, REU supplement to JC), UC CRCC C21CR2138 (S.Y., Diversity supplement to T.W.L.), and University of California, Davis Provost’s Undergraduate Fellowships (to J.S.C., K.A.J., T.W.L., R.C., and J.L.Y.). J.S.C. is a recipient of the Beckman Scholars Award.
Abbreviations used:
- BioID
BIOtin Identification
- MDCK
Madin-Darby Canine Kidney
- PBS
phosphate-buffered saline
- SVIL
supervillain
- tdDsRed
tandem dimer DsRed
- TurboID-LIM
TurboID-tagged zyxin LIM.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E19-10-0568) on April 28, 2021.
REFERENCES
- Baisden JM, Qian Y, Zot HM, Flynn DC (2001). The actin filament-associated protein AFAP-110 is an adaptor protein that modulates changes in actin filament integrity. Oncogene 20, 6435–6447. [DOI] [PubMed] [Google Scholar]
- Branon TC, Bosch JA, Sanchez AD, Udeshi ND, Svinkina T, Carr SA, Feldman JL, Perrimon N, Ting AY (2018). Efficient proximity labeling in living cells and organisms with TurboID. Nat Biotechnol 36, 880–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah JS, Jacobs KA, Heinrich V, Lo SH, Yamada S (2019). Force-induced recruitment of cten along keratin network in epithelial cells. Proc Natl Acad Sci USA 116, 19799–19801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheah JS, Yamada S (2017). A simple elution strategy for biotinylated proteins bound to streptavidin conjugated beads using excess biotin and heat. Biochem Biophys Res Commun 493, 1522–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombelli J, Besser A, Kress H, Reynaud EG, Girard P, Caussinus E, Haselmann U, Small JV, Schwarz US, Stelzer EH (2009). Mechanosensing in actin stress fibers revealed by a close correlation between force and protein localization. J Cell Sci 122, 1665–1679. [DOI] [PubMed] [Google Scholar]
- Crawford AW, Michelsen JW, Beckerle MC (1992). An interaction between zyxin and alpha-actinin. J Cell Biol 116, 1381–1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drees B, Friederich E, Fradelizi J, Louvard D, Beckerle MC, Golsteyn RM (2000). Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J Biol Chem 275, 22503–22511. [DOI] [PubMed] [Google Scholar]
- Flynn DC, Leu TH, Reynolds AB, Parsons JT (1993). Identification and sequence analysis of cDNAs encoding a 110-kilodalton actin filament-associated pp60src substrate. Mol Cell Biol 13, 7892–7900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto M, Kurisu S, Yamada T, Takenawa T (2015). alpha-Actinin-4 enhances colorectal cancer cell invasion by suppressing focal adhesion maturation. PLoS One 10, e0120616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Egelman EH (2012). Actin filaments as tension sensors. Curr Biol 22, R96–R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Bai XH, Lodyga M, Xu J, Yang BB, Keshavjee S, Post M, Liu M (2004). Conversion of mechanical force into biochemical signaling. J Biol Chem 279, 54793–54801. [DOI] [PubMed] [Google Scholar]
- Harris AR, Jreij P, Fletcher DA (2018). Mechanotransduction by the Actin Cytoskeleton: Converting Mechanical Stimuli into Biochemical Signals. Annual Review of Biophysics 47, 617–631. [Google Scholar]
- Heigwer F, Kerr G, Boutros M (2014). E-CRISP: fast CRISPR target site identification. Nat Methods 11, 122–123. [DOI] [PubMed] [Google Scholar]
- Hesketh GG, Youn JY, Samavarchi-Tehrani P, Raught B, Gingras AC (2017). Parallel exploration of interaction space by BioID and affinity purification coupled to mass spectrometry. Methods Mol Biol 1550, 115–136. [DOI] [PubMed] [Google Scholar]
- Hirata H, Tatsumi H, Sokabe M (2008). Mechanical forces facilitate actin polymerization at focal adhesions in a zyxin-dependent manner. J Cell Sci 121, 2795–2804. [DOI] [PubMed] [Google Scholar]
- Hoffman LM, Jensen CC, Chaturvedi A, Yoshigi M, Beckerle MC (2012). Stretch-induced actin remodeling requires targeting of zyxin to stress fibers and recruitment of actin regulators. Mol Biol Cell 23, 1846–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Dasbiswas K, Guo Z, Tee YH, Thiagarajan V, Hersen P, Chew TL, Safran SA, Zaidel-Bar R, Bershadsky AD (2017). Long-range self-organization of cytoskeletal myosin II filament stacks. Nat Cell Biol 19, 133–141. [DOI] [PubMed] [Google Scholar]
- Hu S, Grobe H, Guo Z, Wang YH, Doss BL, Pan M, Ladoux B, Bershadsky AD, Zaidel-Bar R (2019). Reciprocal regulation of actomyosin organization and contractility in nonmuscle cells by tropomyosins and alpha-actinins. Mol Biol Cell 30, 2025–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp JP Jr, Brieher WM (2018). The actin filament bundling protein alpha-actinin-4 actually suppresses actin stress fibers by permitting actin turnover. J Biol Chem 293, 14520–14533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Roux KJ (2016). Filling the Void: Proximity-Based Labeling of Proteins in Living Cells. Trends Cell Biol 26, 804–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo JC, Han X, Hsiao CT, Yates JR 3rd, Waterman CM (2011). Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for beta-Pix in negative regulation of focal adhesion maturation. Nat Cell Biol 13, 383–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCroix AS, Rothenberg KE, Hoffman BD (2015). Molecular-Scale Tools for Studying Mechanotransduction. Annu Rev Biomed Eng 17, 287–316. [DOI] [PubMed] [Google Scholar]
- Lele TP, Pendse J, Kumar S, Salanga M, Karavitis J, Ingber DE (2006). Mechanical forces alter zyxin unbinding kinetics within focal adhesions of living cells. J Cell Physiol 207, 187–194. [DOI] [PubMed] [Google Scholar]
- Li B, Trueb B (2001). Analysis of the alpha-actinin/zyxin interaction. J Biol Chem 276, 33328–33335. [DOI] [PubMed] [Google Scholar]
- Luo W, Janostiak R, Tolde O, Ryzhova LM, Koudelkova L, Dibus M, Brabek J, Hanks SK, Rosel D (2017). ARHGAP42 is activated by Src-mediated tyrosine phosphorylation to promote cell motility. J Cell Sci 130, 2382–2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TN, Uemura A, Shih W, Yamada S (2010). Zyxin-mediated actin assembly is required for efficient wound closure. J Biol Chem 285, 35439–35445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakes PW, Wagner E, Brand CA, Probst D, Linke M, Schwarz US, Glotzer M, Gardel ML (2017). Optogenetic control of RhoA reveals zyxin-mediated elasticity of stress fibres. Nat Commun 8, 15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Baisden JM, Cherezova L, Summy JM, Guappone-Koay A, Shi X, Mast T, Pustula J, Zot HG, Mazloum N, et al. (2002). PC phosphorylation increases the ability of AFAP-110 to cross-link actin filaments. Mol Biol Cell 13, 2311–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan M, Majzoub K, Rao DS, Neela PH, Zarnegar BJ, Mondal S, Roth JG, Gai H, Kovalski JR, Siprashvili Z, et al. (2018). RNA-protein interaction detection in living cells. Nat Methods 15, 207–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner DJ, Ewald ML, Kim T, Yamada S (2017). Biochemical analysis of force-sensitive responses using a large-scale cell stretch device. Cell Adh Migr 11, 504–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller HB, Friedel CC, Boulegue C, Fassler R (2011). Quantitative proteomics of the integrin adhesome show a myosin II-dependent recruitment of LIM domain proteins. EMBO Rep 12, 259–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Blankman E, Deakin NO, Hoffman LM, Jensen CC, Turner CE, Beckerle MC (2013). LIM domains target actin regulators paxillin and zyxin to sites of stress fiber strain. PLoS One 8, e69378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Phua DYZ, Axiotakis L Jr, Smith MA, Blankman E, Gong R, Cail RC, Espinosa de Los Reyes S, Beckerle MC, Waterman CM, Alushin GM (2020). Mechanosensing through direct binding of tensed F-actin by LIM domains. Dev Cell 55, 468–482.e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda S, Blee AM, Macway KG, Renner DJ, Yamada S (2015). Force dependent biotinylation of myosin IIA by alpha-catenin tagged with a promiscuous biotin ligase. PLoS One 10, e0122886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Botvinick EL, Zhao Y, Berns MW, Usami S, Tsien RY, Chien S (2005). Visualizing the mechanical activation of Src. Nature 434, 1040–1045. [DOI] [PubMed] [Google Scholar]
- Watanabe-Nakayama T, Saito M, Machida S, Kishimoto K, Afrin R, Ikai A (2013). Requirement of LIM domains for the transient accumulation of paxillin at damaged stress fibres. Biol Open 2, 667–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JD, Anderson CA, Suarez C, Kovar DR, Gardel ML (2020). Evolutionarily diverse LIM domain-containing proteins bind stressed actin filaments through a conserved mechanism. Proc Natl Acad Sci USA 117, 25532–25542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshigi M, Hoffman LM, Jensen CC, Yost HJ, Beckerle MC (2005). Mechanical force mobilizes zyxin from focal adhesions to actin filaments and regulates cytoskeletal reinforcement. J Cell Biol 171, 209–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.