Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus that has triggered global health and economic crises. Here we report the effects of SARS-CoV-2 infection on peroxisomes of human cell lines Huh-7 and SK-N-SH. Peroxisomes undergo dramatic changes in morphology in SARS-CoV-2-infected cells. Rearrangement of peroxisomal membranes is followed by redistribution of peroxisomal matrix proteins to the cytosol, resulting in a dramatic decrease in the number of mature peroxisomes. The SARS-CoV-2 ORF14 protein was shown to interact physically with human PEX14, a peroxisomal membrane protein required for matrix protein import and peroxisome biogenesis. Given the important roles of peroxisomes in innate immunity, SARS-CoV-2 may directly target peroxisomes, resulting in loss of peroxisome structural integrity, matrix protein content and ability to function in antiviral signaling.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel coronavirus with significant morbidity and mortality. Since the first report of its emergence in late 2019, SARS-CoV-2 has been responsible for a pandemic that has affected a large percentage of the world population, led to severe restrictions on public life, and battered the world economy.
SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus with sequence homology to other coronaviruses including SARS-CoV and MERS-CoV (Sun et al., 2020; Wu et al., 2020). SARS-CoV-2 is highly transmissible and infects primarily the respiratory epithelium, together with tissues in brain, kidney, heart, and liver (Machhi et al., 2020). This can result in mild respiratory distress to severe lung injury, multiorgan failure, and death.
A critical component of SARS-CoV-2 is the viral spike protein (S), which binds its cellular receptor, angiotensin-converting enzyme 2 (ACE2), to gain entry to the host cell (Yan et al., 2020). Binding of S to ACE2 leads to proteolytic cleavage of S by a host cell protease, which results in activation of S and insertion of S into the host cell membrane. This leads to fusion between the SARS-CoV-2 membrane and the plasma membrane, which is followed by virus entry into the cell.
Peroxisomes are ubiquitous organelles with established roles in the metabolism of lipids and reactive oxygen species (Smith and Aitchison, 2013). Notably, peroxisomes also regulate proinflammatory and immune pathways (Wong et al., 2018). Peroxisomes have been linked to the replication and infectivity of a variety of human viruses by acting as reservoirs for viral membrane proteins and by providing specialized lipids to viral envelopes (Lazarow, 2011; Tanner et al., 2014). Peroxisome biogenesis was shown to be essential for the replication of different viruses, including herpes simplex virus (Jean Beltran et al., 2018), Kaposi sarcoma-associated herpesvirus (Sychev et al., 2017), and human cytomegalovirus (Jean Beltran et al., 2018).
Significantly, peroxisomes have been shown to act as platforms for antiviral signaling and to participate in innate immunity (Dixit et al., 2010). Antiviral signaling pathways are activated by the cell’s detection of viral proteins, lipids, or nucleic acids by pathogen recognition receptors, in particular Toll-like receptors (Miyake et al., 2018). Cytosolic RIG-1-like receptors also recognize viral nucleic acids (Kell and Gale, 2015), and these receptors interact with mitochondrial antiviral signaling adaptor (MAVS). MAVS was first found on mitochondria but is now known to reside also at specialized regions of the ER and on the surfaces of peroxisomes (Dixit et al., 2010; Horner et al., 2011). Peroxisome-localized MAVS drives types I and III interferon responses to viral infection (Dixit et al., 2010; Odendall et al., 2014; Wong et al., 2019). Viruses such as dengue, West Nile, and Zika counter this peroxisome-mounted antiviral interferon response by targeting peroxisome biogenic (PEX) proteins for degradation through interaction with viral capsid protein (You et al., 2015; Wong et al., 2019). Other viruses use different strategies to down-regulate the number of peroxisomes and reduce a peroxisome-initiated immune response. For example, HIV inhibits peroxisome biogenesis by up-regulating levels of microRNAs targeting PEX mRNAs (Xu et al., 2017; 2020), while hepatitis C virus inactivates both mitochondrial and peroxisomal MAVS, thereby reducing the different antiviral responses mounted by these organelles (Bender et al., 2015).
As SARS-CoV-2 only recently emerged, our knowledge of how it acts on cells and whether it targets peroxisomes is currently limited. Here we report the effect of SARS-CoV-2 infection on peroxisomes using two human cell lines susceptible to viral infection. We show that infection with SARS-CoV-2 progressively leads to aggregation of peroxisomes, rearrangement of peroxisomal membrane structures, and relocalization of peroxisomal matrix proteins to the cytosol. This represents an extreme example of aberrant peroxisome structure and matrix protein content and of compromised peroxisome populations caused by viral infection.
RESULTS AND DISCUSSION
Human Huh-7 and SK-N-SH cells are permissive to infection by SARS-CoV-2
Huh-7 is a hepatocyte-derived carcinoma cell line used extensively in viral research (Blaney et al., 2010; Barrows et al., 2016). SK-N-SH is a neuroblastoma cell line that has been used for the study of both enveloped and nonenveloped viruses (Zhou and Roizman, 2002; Brown et al., 2004). Both Huh-7 cells and SK-N-SH cells express ACE2, the receptor for SARS-CoV-2, and replication of SARS-CoV-2 in these cell lines has been reported (Appelberg et al., 2020; Jiao et al., 2021; Kumar et al., 2021; Sacramento et al., 2021). These cell lines were chosen to study the effect of SARS-CoV-2 infection on the cellular peroxisome population at a physiological level, that is, without the need to overexpress the ACE2 receptor for virus entry into the host cell.
Nonconfluent Huh-7 or SK-N-SH cells mock-infected or infected at a multiplicity of infection (MOI) of 1 with SARS-CoV-2 were collected at 24 h, 48 h, and 72 h postinfection (hpi).
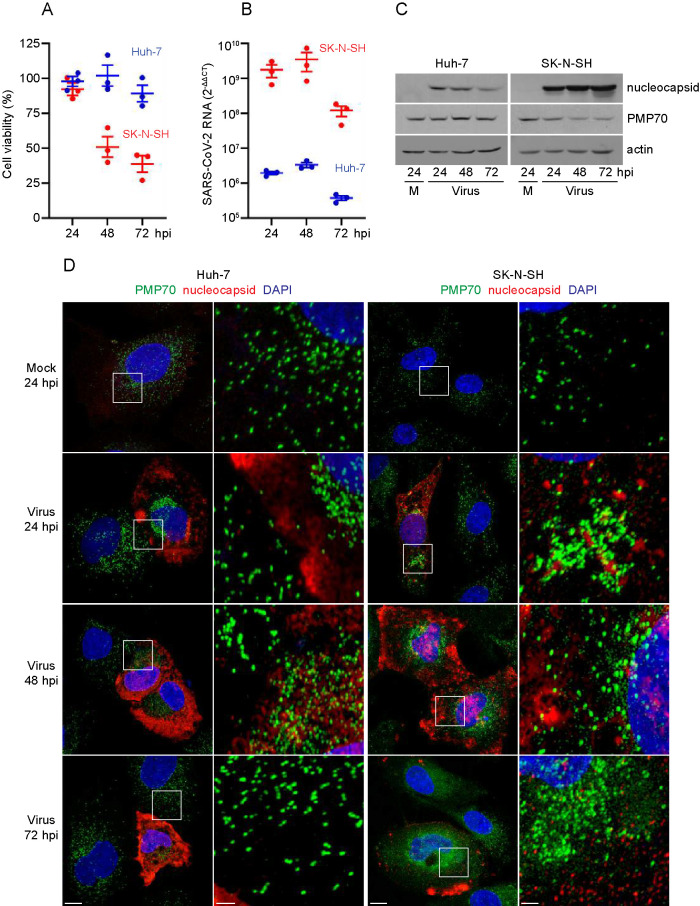
Infected Huh-7 cells were not compromised in viability, as measured by ATP production, at 24 and 48 hpi relative to mock-infected cells, but exhibited a small drop in relative viability to 89% at 72 hpi (Figure 1A). SK-N-SH cells were not compromised in viability at 24 hpi, but exhibited drops to 51% and 39% viability relative to mock-infected cells at 48 hpi and 72 hpi, respectively (Figure 1A).
FIGURE 1:
Huh-7 cells and SK-N-SH cells are permissive to SARS-CoV-2 infection. (A) Huh-7 cells and SK-N-SH cells were seeded into 96-well tissue culture plates and infected with SARS-CoV-2 at a MOI of 1, or were mock-infected. Viability of SARS-CoV-2-infected cells at 24, 48, and 72 hpi was determined using the CellTiter-Glo Luminescent Cell Viability Assay kit and was normalized to the viability of mock-infected cells. The results of three independent experiments are shown. Bars depict mean ± SEM. (B) Huh-7 cells and SK-N-SH cells were infected with SARS-CoV-2 at a MOI of 1 or mock-infected. Total RNA was isolated at 24, 48 and 72 hpi and analyzed by qRT-PCR. Viral S-protein RNA was quantified using the 2-ΔΔCT method (Livak and Schmittgen, 2001); that is, the threshold cycle (CT) values for S-protein RNA were normalized against transcript levels of the human ß-actin gene (ACTB; ΔCT) and further normalized against the ΔCT values for S-protein RNA in mock-infected cells (ΔΔCT). The results of three independent experiments are displayed. Bars depict mean ± SEM. (C) Protein lysates were prepared from mock-infected cells and cells infected with SARS-CoV-2 for 24, 48, and 72 h, separated by SDS-PAGE, and subjected to immunoblotting with antibodies to nucleocapsid, PMP70, and actin. Actin served as a control for protein loading. Representative immunoblots from one of three independent experiments are shown. (D) Huh-7 cells and SK-N-SH cells were grown on coverslips, infected with SARS-CoV-2 at a MOI of 1 or mock-infected, and fixed at 24, 48, and 72 hpi. Cells were immunolabeled with antibodies against nucleocapsid and PMP70. Cell nuclei were stained with DAPI. Cells were observed by confocal fluorescence microscopy. Images presented are maximum-intensity projections and representative of three independent experiments. Squares in panels at left are shown at higher magnification in panels at right. Bars: 10 μm (left panels), 2 μm (right panels).
Total RNA was extracted from cells, and SARS-CoV-2 RNA levels were measured by real-time quantitative reverse-transcriptase PCR (qRT-PCR). We used the 2-ΔΔCT method (Livak and Schmittgen, 2001) to calculate relative changes in viral RNA by normalizing levels of S-protein RNA both to the amount of human ß-actin gene (ACTB) RNA and to the background signal of S-protein RNA in mock-infected cells. Levels of S-protein RNA were abundant in both Huh-7 and SK-SN-SH cells and continued to increase from 24 to 48 hpi, before dropping off at 72 hpi (Figure 1B). The amount of S-protein RNA was approximately 1000-fold greater in SK-N-SH cells than in Huh-7 cells (Figure 1B).
Cell lysates from mock-infected and SARS-CoV-2- infected cells were prepared for SDS-PAGE, and immunoblot analysis revealed that expression of nucleocapsid protein (Figure 1C) paralleled accumulation of viral RNA over time. Nucleocapsid protein was not detected in mock-infected cells. The amount of nucleocapsid protein was greater in infected SK-N-SH cells than Huh-7 cells. In contrast, the peroxisomal membrane protein, PMP70, and actin did not change significantly in amount over the course of infection (Figure 1C).
We next evaluated the effects of SARS-CoV-2 infection on cells by immunofluorescence (IF) confocal microscopy (Figure 1D). Huh-7 and SK-N-SH cells were infected with SARS-CoV-2 and analyzed at 24, 48, and 72 hpi. After fixation, cells were immunolabeled with antibodies against nucleocapsid protein to confirm viral infection and with PMP70 to detect peroxisomes. Nuclei were visualized by DAPI staining. Nucleocapsid protein was not detected in mock-infected cells but 24 hpi and later, the protein was observed throughout the cytosol and sometimes enriched in the perinuclear region. Consistent with viral spread over time, increasing numbers of cells were observed to be positive for staining with antibodies to nucleocapsid.
Cells infected with SARS-CoV-2 displayed changes in morphology, with the cytoplasmic region often appearing more condensed than that of neighboring uninfected cells that were negative for nucleocapsid (Figure 1D). PMP70, which decorates peroxisomal membranes, also was observed to undergo dramatic changes in distribution during infection. In mock-infected cells, PMP70-labeled structures were either punctate or slightly elongated, generally uniform in size, and dispersed throughout the cytoplasm. In contrast, both larger and smaller PMP70-labeled structures were observed in infected cells, suggesting that peroxisomes might undergo coalescence and fragmentation. These observations prompted us to investigate changes to the peroxisome compartment upon infection with virus in more detail.
Peroxisomal membranes undergo dramatic restructuring after SARS-CoV-2 infection
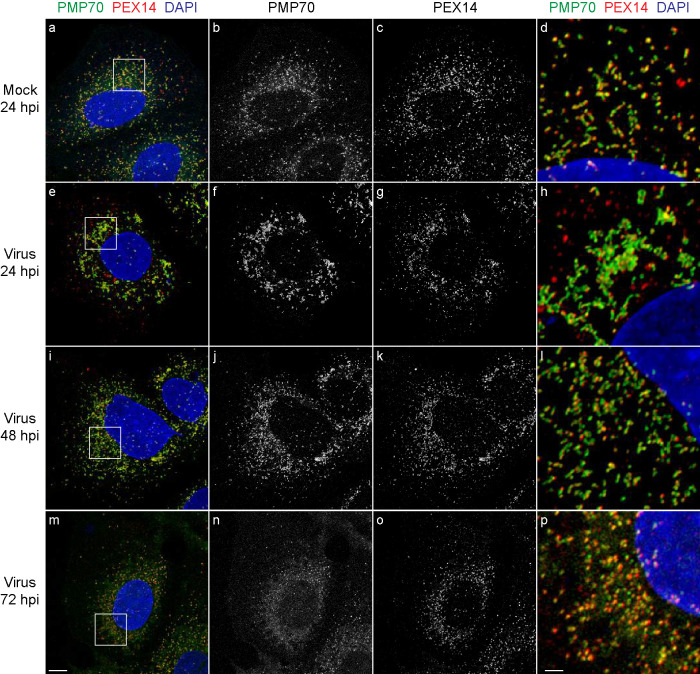
We analyzed the intracellular localization of two peroxisomal membrane proteins in response to SARS-CoV-2 infection (Figure 2). PMP70 is an ATP-binding cassette (ABC) protein that is involved in the transport of acyl-CoAs into the peroxisome (Imanaka et al., 2000). PMP70 may also enrich at the interface between peroxisomes and the ER where synthesis and transport of lipids occur (Olzmann and Carvalho, 2019). PEX14 is a peroxisomal membrane protein that is involved in the import of peroxisomal matrix proteins as part of the docking complex for PEX5, the shuttling receptor for protein import from the cytosol into the peroxisomal matrix (Meinecke et al., 2016).
FIGURE 2:
Peroxisomal membranes undergo rearrangement in SARS-CoV-2-infected Huh-7 cells. Cells were grown on coverslips, infected with SARS-CoV-2 at a MOI of 1 or mock-infected, and fixed at 24, 48, and 72 hpi. Cells were labeled with antibodies against PMP70 and PEX14. Cell nuclei were stained with DAPI. Cells were observed by IF confocal microscopy. Images are maximum intensity projections. Black and white images show single labeling by PMP70 or PEX14 antibodies. Squares in a, e, I, and m are at higher magnification in d, h, l, and p, respectively. Bars: 10 μm (a–c, e–g, i–k, m–o), 2 μm (d, h, l, p). Also see Supplemental Video 1.
In mock-infected cells, PMP70-labeled structures were seen to be dispersed throughout the cytosol and to exhibit both a punctate morphology characteristic of peroxisomes and an elongated morphology suggestive of localization to another compartment such as the ER (Figure 2, a, b, and d). PEX14 was confined to punctate or slightly elongated structures in close proximity to, but not fully overlapping with, PMP70 (Figure 2, a, c, and d). At higher magnification (D), PMP70 and PEX14 could be seen in close proximity and appeared to interdigitate along tubular structures. PEX14 and PMP70 were seldomly found isolated from each other in mock-infected cells.
At 24 hpi, PMP70-labeled tubules were observed to coalesce and exhibited significant clumping, often in the perinuclear region (Figure 2, e and f). PMP70 localized almost exclusively to these tubules, which were elongated compared with the PMP70-decorated structures in mock-infected cells, and sometimes exhibited multiple branchings (Figure 2h). PEX14 also exhibited coalescence and clumping similar to those for PMP70 at 24 hpi (Figure 2, e and g). Notably, PMP70 and PEX14 were segregated from each other by 24 hpi; that is, a large number of PEX14-positive structures did not have any PMP70-associated signals in their vicinity (Figure 2, e and h). At 48 hpi, PMP70-labeled elongated tubular elements appeared to have broken down into more numerous, shorter tubular structures, leaving the impression that peroxisome numbers might be increased in response to virus infection (Figure 2, i, j, and l). However, by 72 hpi, many of the peroxisomal membranes appeared to have disintegrated, as PMP70 was largely dispersed through the cell and not confined to defined structural elements (Figure 2, m, n, and p). Supplemental Video 1 shows the morphological changes of peroxisome structures decorated with PMP70 over the time course of virus infection.
Movie S1.
3D reconstruction of membrane surfaces in Huh-7 cells labeled by anti-PMP70 antibody. The images presented in Figure 2 were processed with Imaris software using automatic threshold settings of the “surpass” and “surface” modules. Rotational models of the surface renderings were recorded as MP4 files and combined in FiJI/ImageJ. (A) mock-infected cell. Cells infected with SARS-CoV-2 at (B) 24 hpi, (C) 48 hpi and (D) 72 hpi.
SARS-CoV-2 infection results in redistribution of peroxisomal matrix proteins to the cytosol and depletion of functional peroxisomes
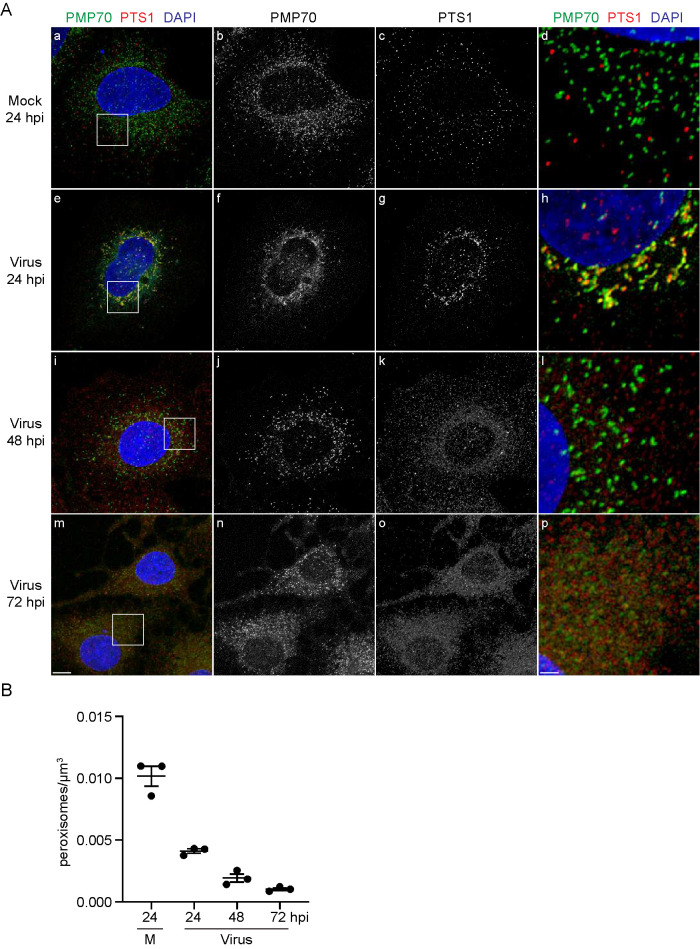
We performed IF confocal microscopy on cells double-labeled with antibodies to PMP70 and antibodies against the peroxisome targeting signal type 1 (PTS1) (Figure 3A). Many peroxisomal matrix proteins have PTS1s at their C-termini. PEX5 interacts with PTS1-containing matrix proteins in the cytosol to form receptor–cargo complexes that translocate to the peroxisomal membrane, bind to the protein import docking machinery, and initiate the formation of transient pores in the peroxisomal membrane for entry of cargo proteins into the matrix (Walter and Erdmann, 2019). Peroxisomes become metabolically active only after importation of matrix proteins. By evaluating the intracellular localization of PTS1-containing matrix proteins, we sought to determine if there was a change in the number of functionally active peroxisomes in SARS-CoV-2-infected cells.
FIGURE 3:
SARS-CoV-2-infected Huh-7 cells exhibit a redistribution of peroxisomal matrix proteins to the cytosol and a reduction in peroxisome numbers. (A) Cells were grown on coverslips, infected with SARS-CoV-2 at a MOI of 1 or mock-infected, and fixed at 24, 48, or 72 hpi. Cells were labeled with antibodies against PMP70 and PTS1. Cell nuclei were stained with DAPI. Cells were observed by IF confocal microscopy. Images are maximum-intensity projections. Black and white images show single labeling by PMP70 or PTS1 antibodies. Squares in a, e, I, and m are at higher magnification in d, h, l, and p, respectively. Bars: 10 μm (a–c, e–g, i–k, m–o), 2 μm (d, h, l, p). (B) Mature peroxisomes defined as “PTS1-labeled puncta” were quantified using the Spot function of Imaris software. Points show peroxisome counts per μm3 of cytoplasm from three independent experiments, in which a minimum of 50 cells were analyzed. Bars depict mean ± SEM. Peroxisome counts were significantly different in SARS-CoV-2-infected cells from those in mock-treated cells, as determined by one-way ANOVA (p < 0.0001).
In mock-infected cells, anti-PMP70 antibody labeled punctate or slightly elongated structures (Figure 3A, a, b and d), as was observed in mock-infected cells double-labeled with antibodies to PMP70 and PEX14 (Figure 2, a, b, and d). PTS1 signal was confined to punctate elements found dispersed in the cell (Figure 3A, a, c, and d). PMP70-labeled and PTS1-labeled structures showed little overlap (Figure 3A, a–d), suggesting that PMP70-labeled and PTS1-labeled structures marked different stages of peroxisome maturation.
At 24 hpi, both PMP70-labeled and PTS1-labeled structures aggregated in the perinuclear region (Figure 3A, e–g). Both markers also colocalized to a greater degree at this stage after infection (Figure 3A, a-h), which may indicate that peroxisome maturation was affected by this time. By 48 hpi, most of the PTS1 signal no longer was associated with punctate structures but was instead dispersed in the cytosol (Figure 3A; i and k), suggesting breakdown and loss of functional peroxisomes. Also at 48 hpi (Figure 3A, i, j, and l), PMP70-labeled elements were smaller structures, suggesting that the larger PMP70-labeled structures seen at 24 hpi (Figure 3A, e, f, and h; Figure 2, e, f, and h) either broke down or disaggregated. By 72 hpi, both PMP70 and PTS1 signals were scattered throughout the cell (Figure 3A, m–p), and in many areas the two signals no longer localized to distinct organellar structures but rather appeared diffused in the cytosol (Figure 3A, m–p). Similar changes in the morphologies of peroxisomal membranes decorated with anti-PMP70 and anti-PEX14 antibodies and in the localization of matrix proteins decorated with anti-PTS1 antibodies were seen in SARS-CoV-2-infected SK-N-SH cells (Supplemental Figures S1 and S2). Quantification of PTS1-labeled puncta suggested that the numbers of metabolically active peroxisomes in virus-infected Huh-7 cells decreased by more than 50% in the first 24 hpi compared with those in mock-infected cells (Figure 3B). The numbers of peroxisomes continued to decline with time after infection until at 72 hpi, only 10–15% of PTS1-labeled peroxisomes remained as compared with mock-infected cells.
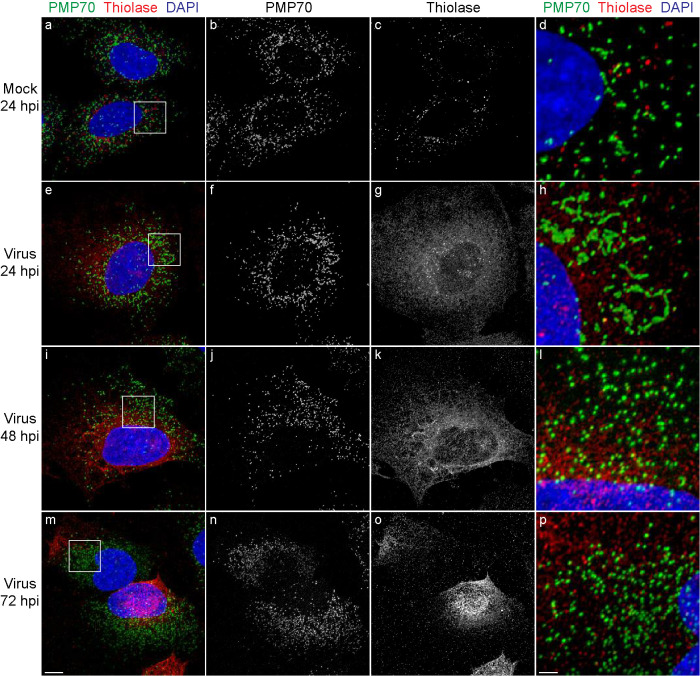
3-Ketoacyl-CoA thiolase (hereafter referred to as “thiolase”) is a matrix enzyme of the peroxisomal fatty-acid β-oxidation pathway (Poirier et al., 2006). Thiolase is targeted to the peroxisome matrix by an N-terminal peroxisome targeting signal type 2 (PTS2), which is recognized in the cytosol by the PTS2 receptor PEX7 (Walter and Erdmann, 2019). We also evaluated what effect SARS-CoV-2 infection of cells might have on the localization of a PTS2-containing peroxisomal matrix protein. We found by IF confocal microscopy that anti-PMP70 and anti-thiolase antibodies decorated largely different structures in mock-infected Huh-7 cells (Figure 4, a–d). Whereas the PMP70 signal was confined to punctate or slightly elongated structures, thiolase-positive structures were predominantly punctate. Although often found in proximity to one another, thiolase-positive structures and PMP70-positive structures generally did not show overlap. Peroxisomes underwent similar changes in morphology after infection of Huh-7 cells with SARS-CoV-2, as was observed by IF confocal microscopy analysis of Huh-7 cells stained with anti-PMP70 antibodies and anti-PTS1 antibodies. At 24 hpi, PMP70-positive membrane elements appeared elongated and sometimes appeared to branch (Figure 4 h). At 48 hpi and 72 hpi, the PMP70-positive structures appeared to have disintegrated, leaving many small PMP70-positive fragments scattered throughout the cell (Figure 4, l and p). Akin to the dispersed cytosolic localization of the PTS1-signal in SARS-CoV-2 infected cells, we found that the thiolase signal was predominantly distributed to the cytosol at 24 hpi (Figure 4g). At later times of virus infection, the thiolase signal was often observed to be distributed unevenly in a cell (Figure 4, m and o). Overall, the fragmented appearance of peroxisomal membranes, as decorated by anti-PMP70 antibody, and the cytosolic localization of PTS1-containing and PTS2-containing peroxisomal matrix proteins, as decorated by anti-PTS1 and anti-thiolase antibodies, suggest that metabolically active peroxisomes are greatly reduced in number in Huh-7 cells at later times after SARS-CoV-2 infection.
FIGURE 4:
SARS-CoV-2-infected cells show a redistribution of the PTS2-containing peroxisomal matrix protein thiolase to the cytosol. Cells were grown on coverslips, infected with SARS-CoV-2 at a MOI of 1 or mock-infected, and fixed at 24, 48, or 72 hpi. Cells were labeled with antibodies against PMP70 and thiolase. Cell nuclei were stained with DAPI. Cells were observed by IF confocal microscopy. Images are maximum-intensity projections. Black and white images show single labeling by PMP70 or thiolase antibodies. Squares in a, e, I, and m are at higher magnification in d, h, l, and p, respectively. Bars: 10 μm (a–c, e–g, i–k, m–o), 2 μm (d, h, l, p).
Reduced numbers of peroxisomes have been reported for cells infected by dengue, West Nile, and Zika viruses (You et al., 2015; Wong et al., 2019); however, no disruption of the peroxisome compartment during viral infection of cells such as we have observed has been reported. Therefore, infection of cells by SARS-CoV-2 may more negatively affect the structure of peroxisomes and their functionality than does infection by other viruses, although demonstration of this awaits future directly comparative studies.
SARS-CoV-2 ORF14 protein interacts physically with human PEX14
A recent analysis of the interactome of SARS-CoV-2 proteins with human host cell proteins identified the 73–amino acid protein encoded by ORF14 (also designated as ORF9c) as a putative interaction partner of various PEX proteins, including PEX3, an early acting peroxisome biogenesis factor; PEX11β, a peroxisome division factor; and PMP70 (Gordon et al., 2020). Because our data show that peroxisomal matrix proteins are mostly cytosolic in SARS-CoV-2-infected cells, we investigated whether ORF14 protein also interacts with human PEX14, a component of the docking complex mediating matrix protein import into the peroxisome (Meinecke et al., 2016). Recombinant ORF14 and PEX14 proteins were made as maltose binding protein (MBP) or glutathione-S-transferase (GST) fusion proteins respectively in E. coli. In vitro binding showed that strong binding of GST-PEX14 to immobilized MBP–ORF14 but not MBP alone (Figure 5).
FIGURE 5:
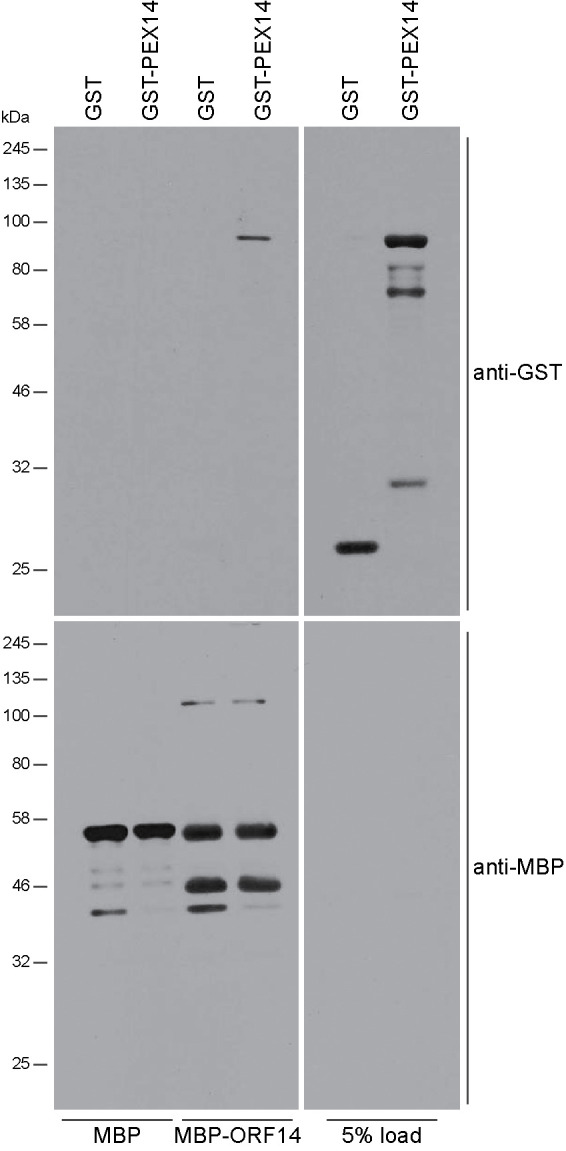
SARS-CoV-2 ORF14 protein binds human PEX14. MBP alone or an MBP–ORF14 protein fusion made in E. coli was immobilized on amylose resin and incubated with E. coli extracts containing GST alone or a GST–PEX14 protein fusion. Bound protein was detected by immunoblotting with anti-GST antibody. Total MBP and MBP–ORF14 were detected by immunoblotting with anti-MBP antibody. Numbers at left denote migrations of molecular mass markers. One of five independent experiments is presented.
Since the start of the COVID-19 pandemic, there has been a massive international effort to produce vaccines against SARS-CoV-2 in record time. Many initial studies focused on analysis and interrogation of the structural components of the virus to identify surface epitopes on the virion that could be used to produce neutralizing antibodies (Wrapp et al., 2020) or on the development of nucleic acid vaccines that exploit the virus’s use of S protein to gain entry to the host cell (Fuller and Berglund, 2020; Jackson et al., 2020). Other studies have dissected the interactome between SARS-CoV-2 proteins and host cell proteins as a means to identify potential molecular targets for the development of therapeutics against the virus (Gordon et al., 2020). However, comparatively little was known about how SARS-CoV-2 affects the structure, integrity, and functionality of the host cell and its organelles.
Coronaviruses replicate in the cytoplasm of their host cells, a process that entails remodeling of the host cell endomembrane system. Electron tomography revealed that organelles involved in coronavirus replication are derived predominantly from the ER (Knoops et al., 2008; Klein et al., 2021). The ER plays a central role in the biogenesis of other membrane organelles, including peroxisomes, and has been shown to make dynamic contacts with different organelles to coordinate a cell’s responses to ever changing environments (Prinz et al., 2020). Given these links between the ER and organelles and the ER’s role in coronavirus replication, changes to the structure, integrity, and functionality of host cell organelles, including peroxisomes, could be expected upon infection with SARS-CoV-2. Indeed, a recent study demonstrated gross morphological changes in the Golgi apparatus, mitochondrial network, and cytoskeleton in cells infected with SARS-CoV-2 (Cortese et al., 2020). This study concluded that peroxisomes increase in abundance in SARS-CoV-2-infected cells, although the structures observed may not be functional mature peroxisomes but rather peroxisomal membrane fragments that contain PMP70, as this study has shown. Moreover, our demonstration of extensive relocalization of peroxisomal matrix proteins to the cytosol in SARS-CoV-2-infected cells is concordant with virus-induced loss of peroxisome function. Recent findings that treatment of COVID-19 patients with type III interferon led to accelerated viral decline (Feld et al., 2021) are consistent with the requirement for functional peroxisomes to mount a type III interferon response to viral infection (Dixit et al., 2010; Odendall et al., 2014; Wong et al., 2019) and our demonstration of the disruption of the peroxisome compartment by SARS-CoV-2 infection.
How might SARS-CoV-2 infection cause a loss of peroxisome integrity and function? We observed that the small viral protein encoded by ORF14 binds the critical peroxisome biogenesis factor PEX14. It is tempting to speculate that this interaction could sequester or inactivate PEX14 during SARS-CoV-2 infection, thus rendering PEX14 unable to perform its role in matrix protein import. This scenario is consistent with the mislocalization of matrix proteins to the cytosol in SARS-CoV-2-infected cells that we observed. Future work will be required to elucidate whether the observed effects of SARS-CoV-2 infection on the peroxisome compartment are due to generic changes in the endomembrane system of the host cell or are the result of more direct effects of viral proteins on the peroxisome biogenesis machinery. Either way, inactivation of peroxisome-mediated antiviral signaling through disruption of the peroxisome compartment may be a key strategy by which SARS-CoV-2 circumvents one of the primary defense mechanisms mounted against it.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture and virus infection
Vero E6 cells (American Type Culture Collection) used for the production of SARS-CoV-2 virus, Huh-7 cells (Thermo-Fisher), and SK-N-SH cells (Millipore-Sigma) were cultured in DMEM (Life Technologies) supplemented with 100 U penicillin/mL, 100 U streptomycin/mL, 1 mM HEPES (Life Technologies), 2 mM glutamine (Life Technologies), and 10% heat-inactivated fetal bovine serum (FBS; Life Technologies) at 37°C in 5% CO2. Cells were seeded the day before infection with SARS-CoV-2 and were infected at a MOI of 1.
Cell lines were limited to 20 passages, during which time they were monitored for aberrant morphology. After 20 passages, aliquots of new cells were revived. Cell lines upon receipt or when revived were tested for the presence of mycoplasma by PCR using primers specific for the 16S rRNA gene of Mycoplasma hyorhinis (Eldering et al., 2004). Cell cultures were tested for mycoplasma every 6–8 wk, and on the rare occasion that a positive test arose, cultures were discarded and the cell line was revived.
Cell viability
The viability of mock-infected and SARS-CoV-2-infected Huh-7 and SK-N-SH cells was measured using the CellTiter-Glo Luminescent Cell Viability Assay kit (Promega). Luminescence was measured using a Synergy HTX multimode microplate reader (BioTek).
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from cells using the RNA NucleoSpin Kit (Machery Nagel) and reverse-transcribed using random primers (Invitrogen) and Improm-II reverse transcriptase (Promega) at 42°C for 1.5 h to produce cDNAs. cDNAs were diluted 1:5 with water, and equal-volume samples were mixed with the appropriate primers (Integrated DNA Technologies) and PerfecTa SYBR green SuperMix with Low ROX (Quanta Biosciences) and amplified for 40 cycles (30 s at 94°C, 40 s at 55°C, 20 s at 68°C) in a Bio-Rad CFX96 qRT-PCR machine. CT values were normalized to a human ACTB mRNA control. ΔΔCT values were determined using control samples as the reference value. Relative levels of mRNAs were calculated using the formula 2(−ΔΔCT). Oligonucleotide sequences were 5′-CCTACTAAATTAAATGATCTCTGCTTTACT (forward) and 5′-CAAGCTATAACGCAGCCTGTA (reverse) for sequence encoding SARS-CoV-2 S protein, and 5′-CCTGGCACCCAGCACAAT (forward) and 5′-GCCGATCCACACGGAGTACT (reverse) for ACTB.
Assay for protein–protein binding
Binding between human PEX14 and SARS-CoV-2 ORF14 protein was assayed essentially as described by Knoblach et al. (2013). GST fusion to human PEX14 was constructed in pGEX4T-1 (GE Healthcare). MBP fusion to SARS-CoV-2 ORF14 was constructed in pMAL-c2 (New England Biolabs). All constructs were verified by sequencing. Recombinant proteins were expressed in E. coli strain BL21 (Invitrogen) or strain Rosetta DE3 (Sigma-Aldrich; GST-PEX14 only). MBP alone or MBP-SARS-CoV-2 ORF14 protein fusion was immobilized on amylose beads and incubated with E. coli lysates containing GST alone or GST-PEX14 in binding buffer (20 mM Tris-HCl, pH 7.5, 100 mM KCl, 5 mM MgCl2, 0.5% [vol/vol] Triton X-100). Unbound proteins were removed by washing five times in binding buffer. Immobilized proteins were eluted in sample buffer (50 mM Tris-HCl, pH 6.8, 2% SDS, 5% [vol/vol] glycerol, 0.002% bromophenol blue, 100 mM 2-mercaptoethanol) and subjected to SDS–PAGE and immunoblotting.
Antibodies
Anti-PMP70 mouse monoclonal antibody clone 70-18 was from Sigma-Aldrich (SAB4200181). Anti-PEX14 antibody (rabbit) was from Novus Biologicals (NBP1-71841). Anti-SARS-CoV-2-nucleocapsid antibody (rabbit) was from GeneTex (GTX135357). Anti-actin antibody (rabbit) was from Sigma-Aldrich (A2066). Anti-PTS1 antiserum raised in rabbit (Aitchison et al., 1992) and anti-peroxisomal thiolase antiserum raised in rabbit (Bodnar and Rachubinski, 1990; Di Cara et al., 2019) have been described. Anti-MBP mouse monoclonal antibody was from New England Biolabs (E8032). GST was detected with mouse monoclonal antibody GST-2 (Sigma-Aldrich, SAB4200692). Donkey–anti rabbit Cy3-conjugates and donkey–anti mouse Cy2-conjugates were from Jackson Immunoresearch Laboratories. Horseradish peroxidase-conjugated anti-mouse and anti-rabbit antibodies were from GE Healthcare. SDS–PAGE and immunoblot analyses were carried out according to standard protocols.
Confocal fluorescence microscopy
Huh-7 cells and SK-N-SH cells grown on coverslips were washed with PBS, fixed for 10 min at room temperature with 4% electron microscopy–grade paraformaldehyde (Electron Microscope Sciences), and again washed with PBS. Cells were first incubated in PBG (PBS, 0.5% BSA, 0.5% fish gelatin) to prevent nonspecific binding of antibodies and then incubated with primary antibody in PBG overnight at room temperature. All primary antibodies were applied at 1:1,000 dilution of stock solution. Cells were washed in PBG and then incubated with secondary antibody in PBG for 1 h. Cells were washed in PBS, stained for nuclei with DAPI (Hoechst) for 30 s, and mounted in 50% glycerol/n-propyl gallate.
Images were acquired with an LSM710 confocal fluorescence microscope (Carl Zeiss) equipped with a 63 × 1.4 NA Plan Apochromat oil immersion objective and a piezoelectric stage to allow rapid acquisition of z-stacks. Images were collected with a z-resolution of 0.1 μm to a total stack height of 8 μm. Cy2 was excited with a 488-nm laser, and its emission was collected within a range of 493–561 nm, while Cy3 was excited with a 561-nm laser and its emission was collected between 596 and 684 nm. DAPI was excited with a 408-nm laser and its emission was collected between 410 and 497 nm.
Acquired fluorescence images were deconvolved using algorithms provided by Huygens Professional Software (Scientific Volume Imaging BV). Three-dimensional data sets were processed to remove noise and reassign blur through iterative classic maximum likelihood estimation and an experimentally derived point spread function. Imaris software (Bitplane) was then used to display the deconvolved 3D data set and to prepare the image files before final figure assembly in Adobe Photoshop and Adobe Illustrator (Adobe Systems).
Supplementary Material
Acknowledgments
The authors thank Darryl Falzarano, Vaccine and Infectious Disease Organization, University of Saskatchewan, for kindly providing SARS-CoV-2 (strain CANADA/VIDO01/2020). The authors also thank Valeria Mancinelli, Eileen Reklow, and Kelly Tedrick for technical support, and Guobin Sun and Xuejun Sun for help with confocal microscopy. This work was funded by grants from the Canadian Institutes of Health Research to T.C.H. (OV3-172302) and R.A.R. (FDN-143289) and the Li Ka Shing Institute of Virology to T.C.H.
Abbreviations used:
- GST
glutathione-S-transferase
- hpi
hours postinfection
- IF
immunofluorescence
- MBP
maltose binding protein
- MOI
multiplicity of infection
- PEX
protein required for peroxisome assembly
- PTS1
peroxisome targeting signal type 1
- PTS2
peroxisome targeting signal type 2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-02-0074) on May 19, 2021.
REFERENCES
- Aitchison JD, Szilard RK, Nuttley WM, Rachubinski RA (1992). Antibodies directed against a yeast carboxyl-terminal peroxisomal targeting signal specifically recognize peroxisomal proteins from various yeasts. Yeast 8, 721–734. [DOI] [PubMed] [Google Scholar]
- Appelberg S, Gupta S, Svensson Akusjärvi S, Ambikan AT, Mikaeloff F, Saccon E, Á V, Benfeitas R, Sperk R, Ståhlberg M, et al. (2020). Dysregulation in Akt/mTOR/HIF-1 signaling identified by proteo-transcriptomics of SARS-CoV-2 infected cells. Emerg Microbes Infect 9, 1748–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows NJ, Campos RK, Powell ST, Prasanth KR, Schott-Lerner G, Soto-Acosta R, Galarza-Muños G, McGrath E, Urrabaz-Garza R, Gao J, et al. (2016). A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe 20, 259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender S, Reuter A, Eberle F, Einhorn E, Binder M, Bartenschlager R (2015). Activation of type I and III interferon response by mitochondrial and peroxisomal MAVS and inhibition by hepatitis C virus. PLoS Pathog. 11, e1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaney JE, Drubin AP, Murphy BR, Whitehead SS (2010). Targeted mutagenesis as a rational approach to dengue virus vaccine development. In: Current Topics in Microbiology and Immunology, ed. Rothman A., Berlin, Heidelberg: Springer, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodnar AG, Rachubinski RA (1990). Cloning and sequence determination of cDNA encoding a second rat liver peroxisomal 3-ketoacyl-CoA thiolase. Gene 91, 193–199. [DOI] [PubMed] [Google Scholar]
- Brown DM, Kauder SE, Cornell CT, Jang GM, Racaniello VR, Semler BL (2004). Cell-dependent role for the poliovirus 3′ noncoding region in positive-strand RNA synthesis. J Virol 78, 1344–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M, Lee J-Y, Cerikan B, Neufeldt CJ, Oorschot VMJ, Köhrer S, Hennies S, Schieber NL, Ronchi P, Mizzon G, et al. (2020). Integrative imaging reveals SARS-CoV-2-induced reshaping of subcellular morphologies. Cell Host Microbe 18, 853–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cara F, Rachubinski RA, Simmonds AJ (2019). Distinct roles for peroxisomal targeting signal receptors Pex5 and Pex7 in Drosophila. Genetics 211, 141–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. (2010). Peroxisomes are signaling platforms for antiviral innate immunity. Cell 141, 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldering JA, Felten C, Veilleux CA, Potts BJ (2004). Development of a PCR method for mycoplasma testing of Chinese hamster ovary cell cultures used in the manufacture of recombinant therapeutic proteins. Biologicals 32, 183–193. [DOI] [PubMed] [Google Scholar]
- Feld JJ, Kandel C, Biondi MJ, Kozak RA, Zahoor MA, Lemieux C, Borgia SM, Boggild AK, Powis J, McCready J, et al. (2021). Peginterferon lambda for the treatment of outpatients with COVID-19: a phase 2, placebo-controlled randomised trial. Lancet Respir Med https//.org/10.1016/S-2213-2600(20)30566-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DH, Berglund P (2020). Amplifying RNA vaccine development. N Engl J Med 382, 2469–2471. [DOI] [PubMed] [Google Scholar]
- Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O’Meara MJ, Razelj VV, Guo JZ, Swaney DL, et al. (2020). A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 583, 459–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Liu HM, Park HS, Briley J, Gale M Jr (2011). Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci USA 108, 14590–14595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imanaka T, Aihara K, Suzuki Y, Yokota S, Osumi T (2000). The 70-kDa peroxisomal membrane protein (PMP70), an ATP-binding cassette transporter. Cell Biochem Biophys 32, 131–138. [DOI] [PubMed] [Google Scholar]
- Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, McCullough MP, Chappell JD, Denison MR, Stevens LJ, et al. (2020). An mRNA vaccine against SARS-CoV-2—preliminary report. 2020. N Engl J Med 383, 1920–1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jean Beltran PM, Cook KC, Hashimoto Y, Galitzine C, Murray LA, Vitek O, Cristea IM (2018). Infection-induced peroxisome biogenesis is a metabolic strategy for herpesvirus replication. Cell Host Microbe 24, 526–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao L, Yang Y, Yu W, Zhao Y, Long H, Gao J, Ding K, Ma C, Li J, Zhao S, et al. (2021). The olfactory route is a potential way for SARS-CoV-2 to invade the central nervous system of rhesus monkeys. Signal Transduct Target Ther 6, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kell AM, Gale M Jr (2015). RIG-I in RNA virus recognition. Virology 479-480, 110–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P (2021). SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. 11, 5885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblach B, Sun X, Coquelle N, Fagarasanu A, Poirier RL, Rachubinski RA (2013). An ER–peroxisome tether exerts peroxisome population control in yeast. EMBO J 32, 2439–2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K, Kikkert M, van den Worm SHE, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder E (2008). SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol 6, e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Sarma P, Kaur H, Prajapat M, Bhattacharyya A, Avti P, Sehkhar N, Kaur H, Bansal S, Mahendiratta S, et al. (2021). Clinically relevant cell culture models and their significance in isolation, pathogenesis, vaccine development, repurposing and screening of new drugs for SARS-CoV-2: a systematic review. Tissue Cell 70, 101497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow PB (2011). Viruses exploiting peroxisomes. Curr Opin Microbiol 14, 458–469. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Machhi J, Herskovitz J, Senan AM, Dutta D, Nath B, Oleynikov MD, Blomberg WR, Meigs DD, Hasan M, Patel M, et al. (2020). The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J Neuroimmune Pharmacol 15, 359–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinecke M, Bartsch P, Wagner R (2016). Peroxisomal protein import pores. Biochim Biophys Acta 1863, 821–827. [DOI] [PubMed] [Google Scholar]
- Miyake K, Sibata T, Ohto U, Shimizu T, Saitoh SI, Fukui R, Murakami Y (2018). Mechanisms controlling nucleic acid-sensing Toll-like receptors. Int Immunol 30, 43–51. [DOI] [PubMed] [Google Scholar]
- Odendall C, Dixit E, Stavru F, Bierne H, Franz KM, Durbin AF, Boulant S, Gehrke L, Cossart P, Kagan JC (2014). Diverse intracellular pathogens activate type III interferon expression from peroxisomes. Nat Immunol 15, 717–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olzmann JA, Carvalho P (2019). Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 20, 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirier Y, Antonenkov VD, Glumoff T, Hiltunen JK (2006). Peroxisomal ß-oxidation—A metabolic pathway with multiple functions. Biochim Biophys Acta 1763, 1413–1426. [DOI] [PubMed] [Google Scholar]
- Prinz WA, Toulmay A, Balla T (2020). The functional universe of membrane contact sites. Nat Rev Mol Cell Biol 21, 7–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacramento CQ, Fintelman-Rodrigues N, Temerozo JR, de Paula Da Silva A, da Silva Gomes Dias S, dos Santos da Silva C, Ferreira AC, Mattos M, Pão CRR, de Freitas CS, et al. (2021). In vitro antiviral activity of the anti-HCV drugs daclatasvir and sofosbuvir against SARS-CoV-2, the aetiological agent of COVID-19. J Antimicrobiol Chemother, dkab072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Aitchison JD (2013). Peroxisomes take shape. Nat Rev Mol Cell Biol 14, 803–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, He W, Wang L, Lai A, Ji X, Zhai X, Li G, Suchard MA, Tian J, Zhou J, et al. (2020). COVID-19: epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol Med 26, 483–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sychev ZE, Hu A, DiMaio TA, Gitter A, Camp ND, Noble WS, Wolf-Yadlin A, Lagunoff M (2017). Integrated systems biology analysis of KSHV latent infection reveals viral induction and reliance on peroxisome mediated lipid metabolism. PLoS Pathog 13, e1006256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner LB, Chng C, Guan XL, Lei Z, Rozen SG, Wenk MR (2014). Lipidomics identifies a requirement for peroxisomal function during influenza virus replication. J Lipid Res 55, 1357–1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter T, Erdmann R (2019). Current advances in protein import into peroxisomes. Protein J 38, 351–362. [DOI] [PubMed] [Google Scholar]
- Wong CP, Xu Z, Hou S, Limonta D, Kumar A, Power C, Hobman TC (2019). Interplay between Zika virus and peroxisomes during infection. Cells 8, 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CP, Xu Z, Power C, Hobman TC (2018). Targeted elimination of peroxisomes during viral infection: lessons from HIV and other viruses. DNA Cell Biol 37, 417–421. [DOI] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS (2020). Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367, 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Peng Y, Huang B, Ding X, Wang X, Niu P, Meng J, Zhu Z, Zhang Z, Wang J, et al. (2020). Genome composition and divergence of novel coronavirus (2019-nCoV) originating in China. Cell Host Microbe 27, 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Asahchop EL, Branton WG, Gelman BB, Power C, Hobman TC (2017). MicroRNAs upregulated during HIV infection target peroxisome biogenesis factors: implications for virus biology, disease mechanisms and neuropathology. PLoS Pathog 13, e1006360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Lodge R, Power C, Cohen EA, Hobman TC (2020). The HIV-1 accessory protein Vpu downregulates peroxisome biogenesis. mBio 11, e03395–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q (2020). Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367, 1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You J, Hou S, Malik-Soni N, Xu Z, Kumar A, Rachubinski RA, Frappier L, Hobman TC (2015). Flavivirus infection impairs peroxisome biogenesis and early antiviral signaling. J Virol 89, 12349–12361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G, Roizman B (2002). Cation-independent mannose-6-phosphate receptor blocks apoptosis induced by herpes simplex virus 1 mutants lacking glycoprotein D and is likely the target of antiapoptotic activity of the glycoprotein. J Virol 76, 6197–6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.