Abstract
The endoplasmic reticulum (ER) is composed of a controlled ratio of sheets and tubules, which are maintained by several proteins with multiple functions. Reticulons (RTNs), especially RTN4, and DP1/Yop1p family members are known to induce ER membrane curvature. RTN4B is the main RTN4 isoform expressed in nonneuronal cells. In this study, we identified FAM134C as a RTN4B interacting protein in mammalian, nonneuronal cells. FAM134C localized specifically to the ER tubules and sheet edges. Ultrastructural analysis revealed that overexpression of FAM134C induced the formation of unbranched, long tubules or dense globular structures composed of heavily branched narrow tubules. In both cases, tubules were nonmotile. ER tubulation was dependent on the reticulon homology domain (RHD) close to the N-terminus. FAM134C plays a role in the autophagy pathway as its level elevated significantly upon amino acid starvation but not during ER stress. Moreover, FAM134C depletion reduced the number and size of autophagic structures and the amount of ER as a cargo within autophagic structures under starvation conditions. Dominant-negative expression of FAM134C forms with mutated RHD or LC3 interacting region also led to a reduced number of autophagic structures. Our results suggest that FAM134C provides a link between regulation of ER architecture and ER turnover by promoting ER tubulation required for subsequent ER fragmentation and engulfment into autophagosomes.
INTRODUCTION
The endoplasmic reticulum (ER) is a continuous membrane system of nuclear envelope and peripheral ER comprising flat sheets and tubules (Baumann and Walz, 2001; Lynes and Simmen, 2011), each carrying out specific vital cellular processes (Baumann and Walz, 2001; Shibata et al., 2006; Hu et al., 2011; Goyal and Blackstone, 2013). Multiple factors are responsible for maintaining the ER shape and size. It is not known how all these factors compete with or complement each other in their functioning.
Reticulons (RTNs) and DP1/Yop1p family members regulate the morphology of the tubular ER. Mammals have four reticulon genes (RTN1, RTN2, RTN3, and NOGO-A/RTN4), each of which gives rise to a range of alternatively spliced transcript variants that encode different isoforms due to differential splicing and promoter usage (Oertle and Schwab, 2003). The RTN family is characteristic for its highly conserved C-terminal reticulon homology domain (RHD) of 150–200 amino acids (aa) containing two hydrophobic stretches separated by a 66-aa hydrophilic loop and followed by a short C-terminal tail (Yang and Strittmatter, 2007). No sequence homology can be observed in the N-terminus of the variants. NOGO-A/RTN4A together with DP1 is sufficient to induce ER tubulation (Voeltz et al., 2006). Similar results were reported for NOGO-B/RTN4B (hereafter called RTN4B) (Rämö et al., 2016). Overexpression of NOGO-A/RTN4A and RTN4B or the yeast reticulon orthologue Rtn1p generates long unbranched tubules in mammalian (Cos-7 and Huh-7) cells and yeast Saccharomyces cerevisiae, whereas their deletion leads to loss of tubular ER (Voeltz et al., 2006; Rämö et al., 2016). RTNs are reported to induce such structural conformations by forming homo- and hetero-oligomers that form arc-like scaffolds on and around membranes to give rise to a stable ER tubule (Shibata et al., 2008). Interaction of RTNs with other proteins also is known to complement RTN function in a sound maintenance of ER structure (Christodoulou et al., 2016). The formation of a reticular ER network of tubules requires interplay between several other proteins. REEPs belong to such a protein family of membrane-associated proteins that have been shown to affect ER structure (Park et al., 2010; Kumar et al., 2019).
ER sheets on the other hand are stabilized by membrane-bound poly/ribosomes and some specific factors like kinectin, CLIMP-63, and p180 (Shibata et al., 2006; Puhka et al., 2007). The consistent thickness of the ER sheets is maintained by the coiled-coil domain of CLIMP-63, which forms intraluminal bridges (Shibata et al., 2010). The RTNs are known to localize also to ER sheet edges, thereby maintaining ER morphology (Shibata et al., 2010) as well. FAM134B (also known as RETREG1) has been characterized as an RHD-containing protein having a role in ER structural maintenance and ER-phagy (autophagy of the ER) (Bernales et al., 2007; Khaminets et al., 2015). The protein has been reported to localize to the ER membrane, specifically to tubules and sheet edges (Khaminets et al., 2015; Mo et al., 2020), and in another study, the protein has been shown to primarily localize to the Golgi apparatus (Kurth et al., 2009). The RHD of FAM134B is required for membrane fragmentation in vitro and ER-phagy in vivo under ER stress (Bhaskara et al., 2019; D’Eletto et al., 2020; Jiang et al., 2020).
We screened for RTN4B interacting partners by using two complementary mass spectrometry–based approaches, affinity purification coupled with mass spectrometry (AP-MS) to detect stable partners and biotin identification (BioID) to also identify transient proximal interactors (Liu et al., 2018). Among the hits was a FAM134 family member, FAM134C/RETREG3. We showed that FAM134C localizes exclusively to ER tubules and sheet edges. Overexpression of FAM134C induced a massive sheet-to-tubule transition and dramatically hampered ER dynamics. Similarly to RTN4B, the RHD of FAM134C is responsible for promoting membrane curvature. FAM134C plays a role in the autophagy pathway as its level rose specifically upon amino acid starvation but not during ER stress. Furthermore, the depletion of FAM134C led to a reduced number of autophagosomes. The remaining autophagosomes were smaller, and their content was altered. FAM134C’s function in autophagosome formation is dependent on intact RHD and LC3 interacting region (LIR) domains.
RESULTS
FAM134C interacts with RTN4B in Huh-7 cells
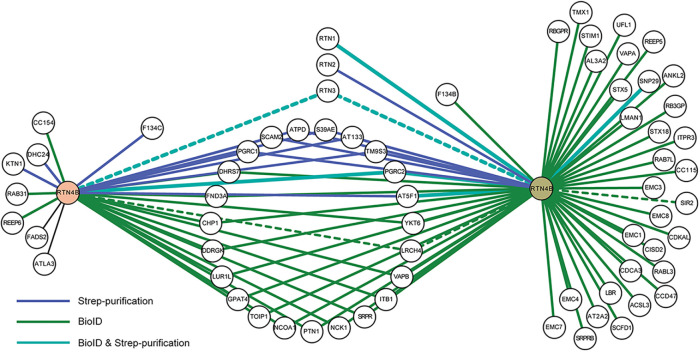
Although several RTN4 interacting partners, such as RTN3, have already been identified (Qi et al., 2003; Dodd et al., 2005; Voeltz et al., 2006), we aimed to complement those studies and find novel partners for RTN4B, which we had identified as the major RTN4 isoform in human hepatoma (Huh-7) cells (Rämö et al., 2016). For this, we used Huh-7 and human embryonic kidney (HEK-293) cell lines. Both cell lines expressing RTN4B constructs transiently (in Huh-7) or stably (in HEK-293) were tagged either with streptavidin (strep) for affinity purification (Varjosalo et al., 2013) or with BirA for proximity-dependent BioID (Roux et al., 2012). BioID is suitable for identifying dynamic interactions, whereas the strep purification tends to detect more stable interactions. The proteomics analysis disclosed many novel interacting partners from the two cell lines (Figure 1 and Supplemental Figure S1). Analysis gave 68 hits, which ranged from proteins belonging to metabolic processes (17 hits) to catalytic activity, ranging from hydrolase to transferase activity (17 hits). Around 35 hits were ER membrane–associated proteins. Hits also comprised membrane trafficking proteins (12 hits), transporter proteins (10 hits), and proteins with calcium ion and calcium-dependent phospholipid binding functions (five hits). The Huh-7 run revealed FAM134C as a significant and novel interactor. FAM134B on the other hand was among the transient interacting hits in HEK-293 cells (BioID run).
FIGURE 1:
RTN4B interaction partners in Huh-7 and HEK-293 cells. The protein–protein interaction map of RTN4B generated with Cytoscape represents interacting proteins from Huh-7 (on the left) and HEK-293 (on the right) cell lines using two purification methods. Blue lines represent experimentally validated interaction by strep purification, green lines for validated interactions from BioID purification, and overlap of two purification methods is shown with the light blue color. Dotted lines represent known interactions derived from the protein interaction network analysis (PINA2.0) platform.
We used a bimolecular fluorescence complementation assay (BiFC) (Kaddoum et al., 2010) to confirm the interaction of FAM134C with RTN4B (Supplemental Figure S2). RTN4B has been shown to form homo-oligomers (Shibata et al., 2008) and was used as a positive control. We have shown earlier that myosin 1C associates with ER sheets and has no effect on ER tubules (Joensuu et al., 2014). Myosin 1C was not among the mass spectrometry hits and, thus, served as a negative control. FAM134C/RTN4B and RTN4B/RTN4B pairs gave positive BiFC signals, while the myosin 1C/FAM134C pair gave a negative signal. These results confirmed our mass spectrometry findings that FAM134C is an interacting partner of RTN4B.
FAM134C membrane insertion and topology
The FAM134/RETREG family contains three known proteins, namely FAM134A (RETREG2; Q8NC44, UniProtKB), FAM134B (Q9H6L5), and FAM134C (Q86VR2). FAM134C is a conserved protein found in many eukaryotes (Supplemental Figure S3A) but has not been functionally characterized yet. Currently, two alternatively spliced isoforms of human FAM134C, Q86VR2-1 and Q86VR2-2 (UniProtKB), corresponding to 466 and 271 aa, respectively, have been described. According to Western blotting, FAM134C corresponds to ∼65 kDa in Huh-7 (see Figure 5C later in this article).
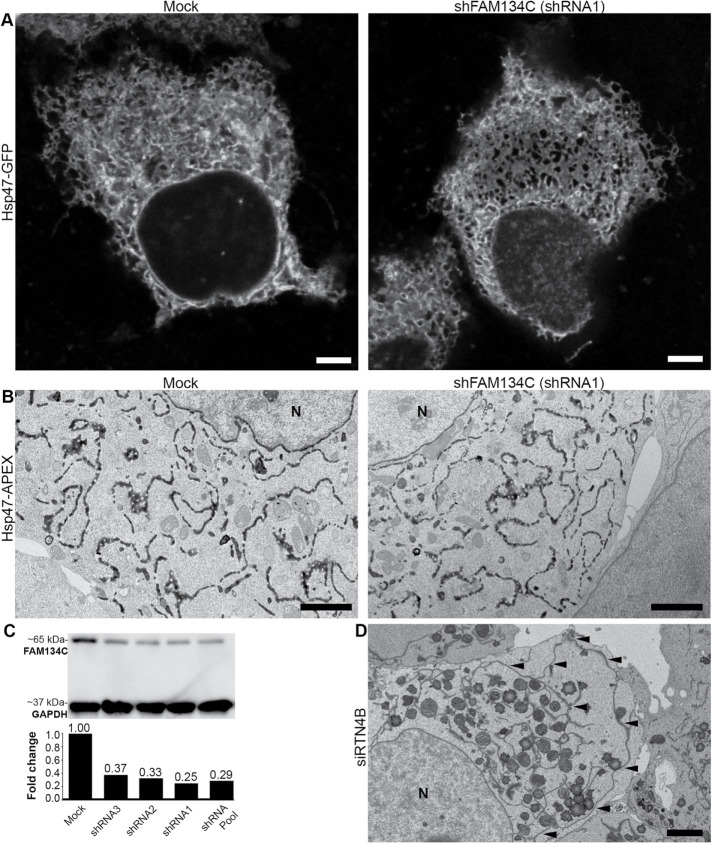
FIGURE 5:
Depletion of FAM134C does not affect ER morphology. (A) Confocal LM images show FAM134C depleted (shRNA1) and mock-treated Huh-7 cells expressing ER marker Hsp47-GFP. (B) TEM micrographs showing Huh-7 cells expressing ER marker Hsp47-APEX with mock and FAM134C silenced conditions (shRNA1). (C) Huh-7 cell lysates from mock and FAM134C silenced cells (shRNAs 1–3 and pool of three) were analyzed by Western blotting. GAPDH was used as the loading control. (D) RTN4B silencing induces formation of large ER sheets. Black arrowheads indicate the long ER sheet. N denotes the nucleus. Scale bars: 5 μm (A), 2 μm (B), and 2 μm (D).
FAM134C lacks a conventional signal sequence but has been predicted to have several transmembrane (TM) domains according to UniProtKB, Protter (Omasits et al., 2014), and TMHMM (Krogh et al., 2001). Both NCBI’s conserved domains tool and HHpred (Soding et al., 2005) predict that FAM134C possesses an RHD. The RHD corresponded to 60–210 aa of the FAM134C, where all predicted TMs of FAM134C fall as well (Supplemental Figure S3B). BLAST (basic local alignment search tool; NCBI) between FAM134C and RTN4B gave an overall sequence similarity of around 26%, where the RHD of RTN4B and some of the TM domains of FAM134C showed clear overlap.
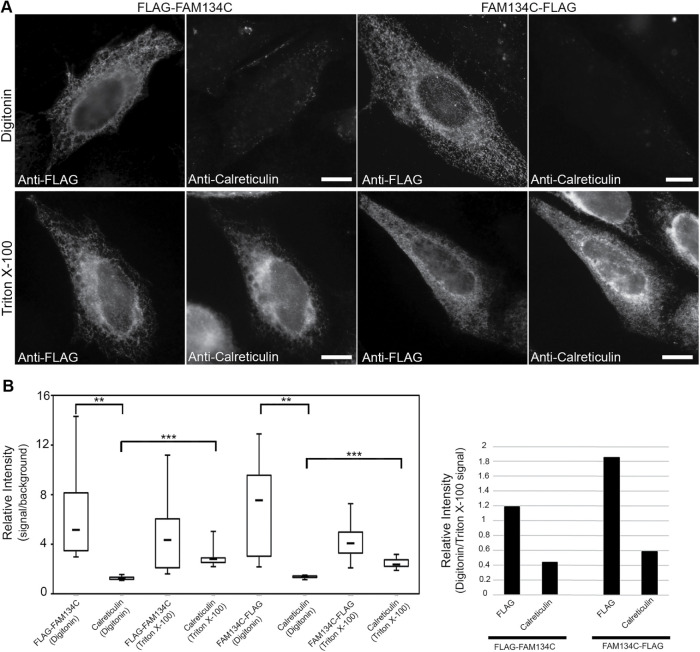
Protter and TMHMM software predict that only the N-terminus of FAM134C is cytosolic. To test this, HeLa cells mildly overexpressing N- or C-terminally tagged FAM134C (FLAG-FAM134C and FAM134C-FLAG, respectively) were fixed and permeabilized with Triton X-100 or digitonin. Cells were then immunostained with antibodies against FLAG and the ER luminal protein calreticulin. Wide-field imaging revealed that in digitonin-permeabilized cells both N- and C-terminal FLAG-tag gave a significant signal, while calreticulin gave a significant signal only in Triton X-100–permeabilized cells (Figure 2). Therefore, we concluded that both the N- and C-terminus of FAM134C face the cytoplasm.
FIGURE 2:
Membrane topology of FAM134C. (A) LM images and quantification of relative intensities (>10 cells) of HeLa cells mildly expressing FLAG-FAM134C and FAM134C-FLAG. Cell were fixed and treated with either digitonin or Triton X-100 and immunolabeled with anti-FLAG antibody and calreticulin. FLAG-FAM134C and FAM134C-FLAG were accessible to the anti-FLAG antibody in both digitonin and Triton X-100–permeabilized cells, whereas calreticulin was recognized only upon Triton X-100 permeabilization. (B) The left plot shows the relative intensity calculated by dividing the mean intensity of the cytoplasmic area by the background signal, and the right plot shows the normalized median of signals under digitonin permeabilization with respect to Triton X-100 permeabilization. Error bars indicate ±SD. The statistical significance of the differences was estimated by Student’s t test, and ** indicates p < 0.005 and ***p < 0.0005. Scale bars: 10 μm.
FAM134C localizes to the ER, and overexpression induces extensive ER tubulation via RHD
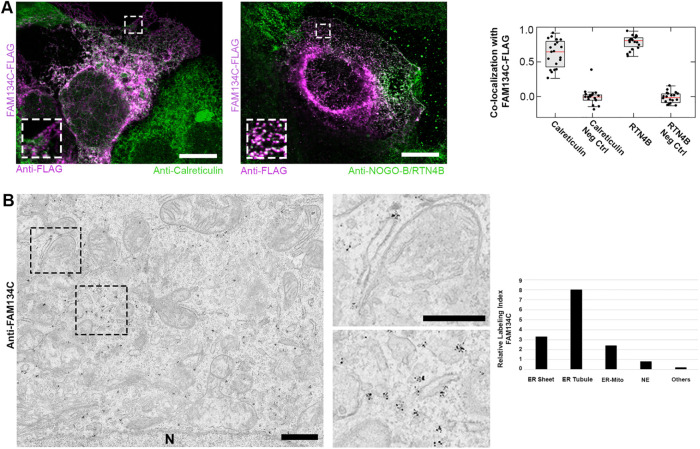
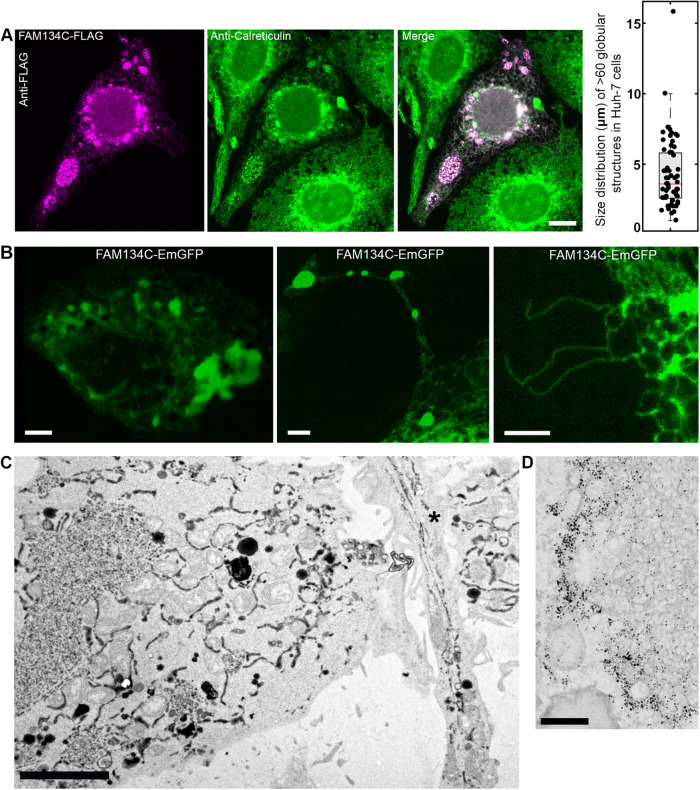
Huh-7 cells overexpressing FAM134C-FLAG were used for localization studies. Confocal microscopy revealed clear colocalization of FAM134C-FLAG with ER markers calreticulin and RTN4B (Figure 3A). RTN4B was shown to localize exclusively to ER tubules and sheet edges (Rämö et al., 2016). FAM134C gave a higher colocalization with RTN4B than with the general luminal calreticulin. Compared with calreticulin staining, FAM134C seemed to localize preferably to ER tubules and sheet edges (Figure 3A, insets). Pearson’s coefficient analysis clearly showed no significant colocalization of FAM134C with the intermediate compartment marker ERGIC53 (Hauri and Schweizer, 1992; Kappeler et al., 1997) or the Golgi marker GM130 (Nakamura et al., 1995) (Supplemental Figure S4). Immunoelectron microscopy (immuno-EM) against the endogenous FAM134C revealed a clear preference to tubules and sheet edges in human bone osteosarcoma (U2-OS) cells. Occasionally, the labeling was observed at ER–mitochondria contact sites (Figure 3B).
FIGURE 3:
FAM134C localizes to the ER. (A) Confocal LM images of Huh-7 cells overexpressing FAM134C-FLAG (magenta) show colocalization between FAM134C and the general ER marker calreticulin and tubular ER marker RTN4B (green). Insets show higher magnification of boxed areas. Colocalization was quantified using Pearson’s correlation coefficient (∼20 cells). For the negative control (Neg Ctrl) one of the channels from the same ROI was rotated by 90°. Error bars indicate ±SD. (B) Immuno-EM micrographs of U2-OS cells revealed localization of endogenous FAM134C to tubules and sheet edges (insets show higher magnification of boxed area). Scale bars: 10 μm (A), 1 μm (B), and 200 nm (B inset).
Higher overexpression of FAM134C resulted in clear morphological changes on the ER structure as large globular structures randomly distributed throughout the cell could be detected (Figure 4A). The size of the globules varied greatly from cell to cell, ranging on average from 3 to 7 μm in diameter. The globules were positive for calreticulin, verifying their ER origin. Cells overexpressing FAM134C also displayed long protrusions extending from the plasma membrane (Figure 4B). Some of the protrusions contained globules that had similar size distribution as those found from cell cytoplasm, whereas some protrusions comprised long and unbranched ER tubules only. Confocal live cell imaging revealed that the ER globules in the cell cytoplasm and in protrusions as well as the unbranched long tubules were nondynamic (Supplemental Movies 1–3).
FIGURE 4:
FAM134C overexpression induces extensive ER tubulation. (A) LM images of Huh-7 cells overexpressing FAM134C-FLAG (magenta) costained with the general ER marker calreticulin (green) reveal large globular structures colocalizing with calreticulin in addition to normal-looking ER. The size distribution for the globular structures was measured from >20 cells. Error bars indicate ±SD; the number of structures analyzed was 60. (B) Confocal LM images depicting three prominent phenotypes from FAM134C-EmGFP overexpression (green): large globular structures, long protrusions from the cell edge comprising of smaller globular structures and long tubular unbranched profiles extending from the cell edge; also see Supplemental Figure S5 and Supplemental Movies 1–3. (C) TEM micrograph of Huh-7 cells overexpressing FAM134C-EmGFP (positive for transfection marker ssHRP-KDEL) showing the dense globular structures of highly branched tubular ER and otherwise normal-looking ER network. A protrusion containing long ER profiles is indicated with an asterisk (*). (D) Immuno-EM micrograph of FAM134C-EmGFP overexpression induced heavily tubular and branched ER tubules that are positive for GFP labeling. Scale bars: 10 μm (A), 5 μm (B, C), and 200 nm (D).
Movie S1.
Live cell imaging of FAM134C overexpression -induced large globular structures in Huh-7. Structures vary greatly in size, are randomly distributed throughout the cell, and are non-dynamic in nature. The display rate is 3.14 fps.
Movie S2.
Live cell imaging of FAM134C overexpression -induced long protrusions in Huh-7. Small globular structures can be seen at random locations within the protrusions, and both protrusions and globular structures within them are non-dynamic in nature. The display rate is 3.14 fps.
Movie S3.
Live cell imaging of FAM134C overexpression -induced long unbranched tubules in Huh-7. These long tubular structures are non-dynamic in nature. The display rate is 3.14 fps.
Transmission electron microscopy (TEM) was used to analyze the ER globules and cell protrusions in more detail. For this, Huh-7 cells were cotransfected with FAM134C-FLAG and ER marker pssHRP-KDEL (Connolly et al., 1994). FAM134C overexpressing cells revealed large and smaller-size clusters of highly branched ER tubules (Figure 4C). The size of these profiles matched the size of globules seen in light microscopy (LM). These structures were found only in overexpressing cells, and they were composed of a dense network of short ribosome-free tubules that were connected to normal sheets at the immediate vicinity of the cluster. The diameter of the tubules ranged between 40 and 50 nm. Electron tomography confirmed the continuity of the tubules and their connections to normal ER sheets surrounding the globules (Supplemental Figure S5B and Supplemental Movie 4). Besides these globular clusters, the rest of the ER network looked normal and continuous. Immuno-EM studies on FAM134C-EmGFP overexpressing Huh-7 cells showed that the clustered tubules were positive for the fusion protein. The labeling was heavy at the periphery of the globular cluster and milder at the central parts of the cluster (Figure 4D).
Movie S4.
FAM134C overexpression induced formation of dense network of ER tubules. Models generated from serial electron tomograms revealing normal ER (depicted in yellow) connected to heavily branched and tubulated ER (depicted in green) in a Huh-7 cell co-expressing FAM134C-EmGFP and ER marker Hsp47-APEX. Voxel size: 2.28 × 2.28 × 2.28 nm.
To elucidate the role of FAM134C’s RHD in promoting the membrane curvature, we created a FAM134C construct where the RHD was extended (TM-mutFAM134C-GFP) (Supplemental Figure S6A). The length of four helixes in RHD was extended by hydrophobic alpha helixes without altering the position of the arginine in hairpin inversion (Supplemental Figure S6A). Analogous mutations where the loop tip between two transmembrane helixes was extended have been shown to abolish the curvature-promoting properties of the RHD of Reticulon 4 and REEP4 (Zurek et al., 2011; Kumar et al., 2019). The TM-mutFAM134C’s C-terminus was facing the cytoplasm; thus, the extension of RHD did not affect the protein topology (Supplemental Figure S6B). Wide-field LM analysis revealed that overexpression of TM-mutFAM134C did not induce ER tubulation or formation of tubular clusters seen during the overexpression of the wtFAM134C (Supplemental Figure S6C). Seventy percent of the analyzed TM-mut overexpressing cells (n = 85 cells) had normal-looking ER in comparison to only 16% of wild-type overexpressing cells (n = 106 cells). In 30% of cells overexpressing TM-mut, we found in addition to reticular, normal-looking ER staining bright blobs ranging on average from 1 to 3 μm in diameter, with occasional individual larger blobs up to 8 µm. TEM analysis of Huh-7 and U2-OS cells overexpressing the TM-mut revealed normal-looking ER in most of the cells (n = 23 Huh-7 and 10 U2-OS cells), and we could not find any cells with tubular clusters (Supplemental Figure S6D). We found a few cells having large tightly packed ER sheets that recall phenotypic changes typically connected to overexpression of ER membrane proteins. The severity of the ER phenotype in both samples was directly connected to the expression level. The transfection efficiency with both constructs was around 30–40%. From this we concluded that FAM134C thus brings about ER tubulation via its RHD upon overexpression.
Short hairpin RNA (shRNA)-mediated silencing was used to deplete FAM134C in Huh-7 cells. To exclude the possibility of off-target effects, we used three different shRNAs in our silencing experiments. According to Western blotting, 48 h silencing resulted in around 75% efficiency with each construct and pool of three (Figure 5C). Confocal microscopy and TEM revealed that silencing of FAM134C in Huh-7 cells did not induce clear structural changes on the ER (Figure 5, A and B). As a control, we silenced RTN4B, where exceptionally large ER sheets were found (Figure 5D) (Rämö et al., 2016).
FAM134C expression elevates upon amino acid starvation and is needed for autophagosome formation
FAM134C is predicted to have a LIR motif at 445–450 aa (Q86VR2; UniProtKB) within the protein’s intrinsically unstructured region (220–466 aa) (Supplemental Figure S3B), and it has been shown to interact with LC3 in mammalian cells (Khaminets et al., 2015). Amino acid starvation in U2-OS, Huh-7, and HeLa cells elevated FAM134C expression levels (Figure 6A and Supplemental Figure S7, A and B). In U2-OS cells, the basal level is higher in general and the elevation was prominent when cells were grown at high confluency of more than 90% (Figure 6A).
FIGURE 6:
FAM134C expression elevates upon starvation. (A) Western blots and quantifications showing the changes in FAM134C levels at normal growth conditions (DMEM) and upon amino acid starvation (EBSS) with/without bafilomycin A1 in U2-OS cells at both high and low cell confluency, silencing of FAM134C in Huh-7 cells (B), and upon DTT-induced ER stress in Huh-7 cells (C). LC3II was used as a general reporter for autophagy and Phospho-elF2α as a marker for ER stress. GAPDH and α-tubulin were used as loading controls. For quantification, the relative average signal was obtained by normalizing for the signal from cells after a 1 h incubation in DMEM. Error bars indicate ±SEM.
Induction of autophagy upon amino acid starvation was confirmed with autophagy marker LC3 (microtubule-associated protein 1A/1B-light chain 3) (Kabeya et al., 2000). LC3II protein levels did not change upon silencing of FAM134C (shRNA1) (Figure 6B). No elevation in FAM134C level was observed in Huh-7 cells upon induction of ER stress using dithiothreitol (DTT) in comparison to the ER stress marker Phospho-eIF2α (Figure 6C) (Kaufman, 1999). Collectively, the data suggested FAM134C to play a role in the regulation of autophagosome formation by elevating its level specifically upon amino acid starvation and not during ER stress. Interestingly, bafilomycin A1 treatment did not have a similar impact on FAM134C levels compared with LC3 levels, suggesting that FAM134C is not targeted to lysosomal degradation (Figure 6A). Immuno-EM in HeLa cells supported this finding as the endogenous FAM134C labeling was detected in the ER and not in autophagosomal structures (five cells and 10 autophagic structures analyzed; Supplemental Figure S7C).
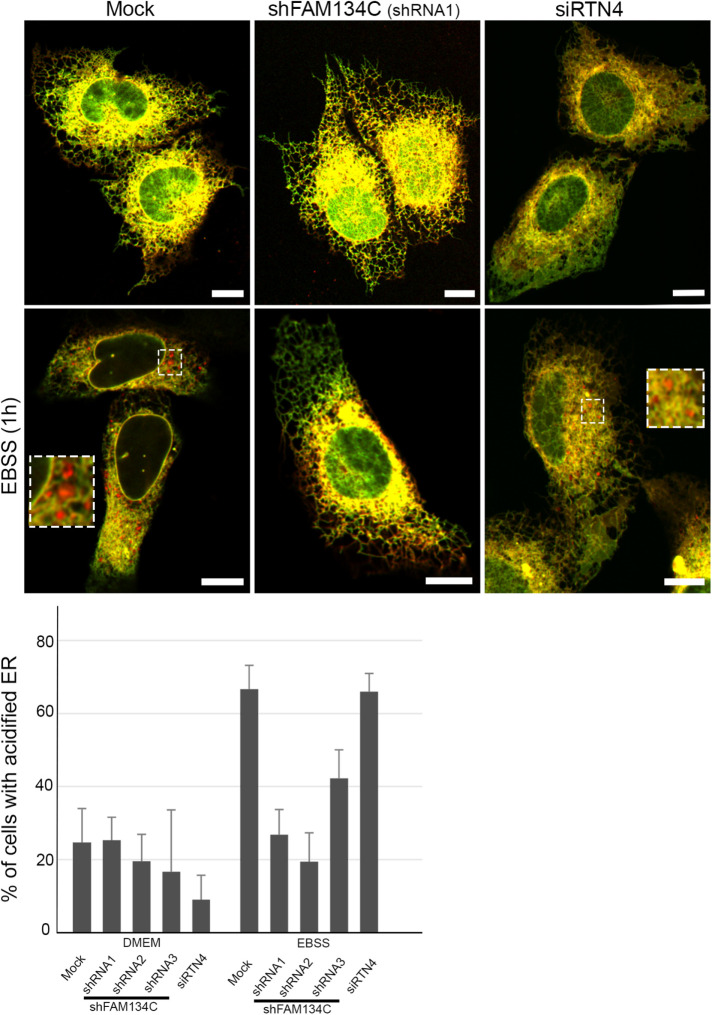
To test the role of FAM134C in ER-phagy, we used the EATR assay to monitor the lysosomal degradation of the autophagocytozed ER (Liang et al., 2018). This assay is based on the more pH-tolerant nature of mCherry compared with enhanced green fluorescent protein (eGFP), which loses its fluorescence in the acidic pH of the lysosome. U2-OS cells were transiently cotransfected with mock, shFAM134C (shRNAs 1–3), or siRTN4 and ER translocon complex subunit RAMP fused with eGFP and mCherry. Upon amino acid starvation for 1 h, FAM134C silenced cells failed to efficiently bring about ER-phagy, as red fluorescent puncta indicating acidified ER inside lysosomes could be detected less frequently (Figure 7). Upon starvation, in FAM134C silenced cells the percentage of cells with acidified ER remained at the same level compared with the nonstarved condition with two different silencing constructs (shRNA1 and 2), in comparison to mock cells, where the amount of positive cells increased 2.7 times. Silencing with shRNA3 had only a mild effect. On the other hand, RTN4 depleted cells containing large peripheral ER sheets were able to bring about ER-phagy efficiently upon amino acid starvation (Figure 7).
FIGURE 7:
FAM134C is required for amino acid starvation–induced ER-phagy. Confocal LM images of U2-OS cells expressing RAMP-eGFP-mCherry and mock, FAM134C shRNAs 1–3, or RTN4 siRNA at basal or amino acid starvation (EBSS) conditions. Red puncta represent acidified ER. Insets show higher magnification of boxed areas. The percentage of cells having acidified ER was calculated from three to 10 experimental replicates. Error bars indicate ±SEM. Total number of cells analyzed per condition was 176, 88, 26, 19, 119, 202, 74, 34, 24, and 120 according to the order in the graph. Scale bars: 10 µm.
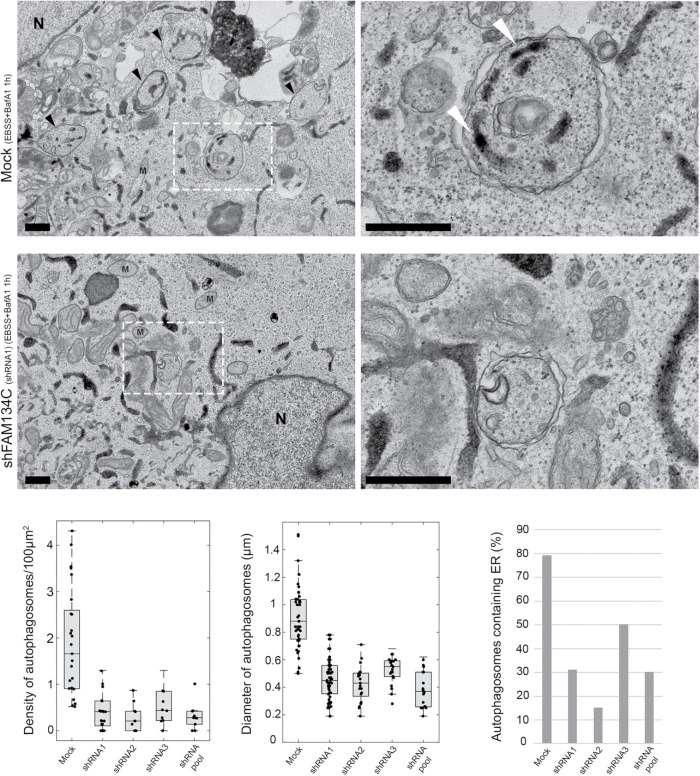
Next, we examined the role of FAM134C in autophagosome formation. U2-OS were transiently cotransfected with mock, shFAM134C (shRNAs 1–3, or the pool of three constructs), and general ER marker Hsp47-APEX for 48 h. Upon amino acid starvation together with bafilomycin A1 treatment for 1 h, FAM134C silenced cells showed a significant reduction in the number and the size of autophagosomes, as well as changes in autophagosome cargo (Figure 8). The average number of autophagosomes per 100 µm2 of cytoplasmic area was reduced from 1.84 (n = 25 cells) to 0.38 (shRNA1; n = 25 cells), 0.29 (shRNA2; n = 10 cells), 0.52 (shRNA3; n = 10 cells), and 0.32 (pool; n = 10 cells). The average diameter of autophagosomes was reduced from 0.92 µm in mock structures (n = 50 structures) to 0.46 µm (shRNA1; n = 50 structures), 0.43 µm (shRNA2; n = 20 structures), 0.52 µm (shRNA3; n = 20 structures), and 0.39 µm (pool; n = 20 structures). Furthermore, the percentage of autophagosomes having ER as cargo in these structures was reduced from 79% to 31/15/50/30%, respectively. In TEM images of the control cells, the mean area of autophagosomes was 876 nm2, of which the ER occupied 15% (133 nm2). In FAM134C silenced cells, the mean area of autophagosomes was 77.5% smaller (197 nm2; averaged from samples using three shRNAs individually), suggesting that the decrease in autophagosome size upon FAM134C silencing could not be explained only by the decrease in be the amount of ER as cargo (Supplemental Figure S7D). These results were confirmed in Huh-7 cells using shRNA1 (Supplemental Figure S8). The amount of bulk ER showed a slight increase in Huh-7 silenced cells.
FIGURE 8:
FAM134C is needed for normal autophagosome formation upon amino acid starvation. TEM micrographs of mock and FAM134C silenced (shRNA1) U2-OS cells expressing Hsp47-APEX upon amino acid starvation and bafilomycin A1 incubation. Boxed areas are shown at higher magnification on the right. Density of autophagosomes/100 µm2 of cytoplasmic area, the average diameter of autophagosomes, and the percentage of autophagosomes containing ER as cargo were calculated from mock and shRNAs 1–3 treated cells and are presented in the accompanying graphs. See Supplemental Figure S8 for similar analysis done in Huh-7 cells. In TEM images, black arrowheads indicate autophagosomes that have ER as cargo (indicated with white arrowheads); mitochondria are marked with M and nucleus with N. Scale bars: 500 nm.
Next, we created a LIR-mutFAM134C construct, where the LIR motif (DDFELL) was replaced with six alanines, and studied the effect of dominant-negative expression of the mutated FAM134C construct on autophagosome formation. Upon 2 h of amino acid starvation together with bafilomycin A1, the density of autophagosomes in mock was 3.3/100 µm2 of cytoplasmic area and in wild-type FAM134C overexpressing cells 2.5/100 µm2 of cytoplasmic area, whereas overexpression of LIR-mutFAM134C or TM-mutFAM134C in similar conditions led to a reduced amount of autophagosomes (0.9/100 µm2 of cytoplasmic area for both). RTN4 overexpression did not reduce autophagosome formation in the starved condition in a same way, although it clearly induced ER sheet-to-tubule transformation (Supplemental Figure S9). Moreover, overexpression of wild-type FAM134C increased the amount of captured ER by autophagocytosis to late endocytic organelles compared with control (ER occupied 9.5% of the area of endocytic organelles in overexpressing cells and 5.7% in mock calculated from thin-section TEM images, n = 48 organelles from 25 cells and 25 organelles from 28 cells, respectively). Overexpression of mutated FAM134C caused a reduction in autophagosome number, indicating the importance both LC3 binding and membrane bending capacity in the formation of those structures.
Overall, these observations suggested an indispensable role for FAM134C in autophagosome formation, along with its presumable function in ER-phagy, in accordance with the role of FAM134B in selective ER-phagy (Jiang et al., 2020).
DISCUSSION
The reticulon family of proteins have been the hallmarks of positively curved membranes of ER (Voeltz et al., 2006). The proteomics screen using RTN4B as bait in two human cell lines gave a list of proteins belonging to many known families of associated proteins of ER. Hits revealed proteins involved in membrane traffic, like vesicle formation/movement and fusion (the RAB and VAMP family, respectively), thus showing the extent of the possible functions of RTN4B. Among the novel interactors were FAM134C (in Huh-7 cells) and FAM134B (in HEK-293 cells), and we focused our attention on the less studied RHD family protein and FAM134 member, FAM134C.
FAM134C is not as highly expressed as RTN4B (Rämö et al., 2016), but its RNA expression is substantially higher than that of FAM134B in the U2-OS, Huh-7, or HeLa cells (37-, 44-, and 1.7-fold, respectively) according to the Human Protein Atlas (https://www.proteinatlas.org/cell). In healthy human tissues, FAM134C protein was detected in all 29 organs analyzed (Wang et al., 2019), and it was the most abundant protein of the FAM134 family, with the highest levels in bone marrow, duodenum, and testes, tissues where cell division and maturation is abundant. From the other two FAM134 family members, only low FAM134B expression was observed in nine organs mainly connected to the endocrine system, and the amount of FAM134B exceeded FAM134C expression only in the pancreas, whereas FAM134A was moderately expressed in 26 organs and was the most abundant FAM134 form in the brain and testicles. At the RNA level, the GTEx Consortium (2015) studied 54 body sites and discovered that FAM134B expression was higher than that of FAM134C only in part of the spinal cord, substantia nigra, left ventricle, and skeletal muscles. In summary, FAM134C is more abundantly represented than FAM134B at RNA and protein levels in most cell types and organs studied so far. Disorders of FAM134C or B production in the brain tissues have caused phenotypic changes: FAM134C deficiency in hippocampal nerves reduced the dendritic number and axon length (Wang et al., 2013), a nonsense mutation in FAM134B caused hereditary sensory autonomic neuropathy (Kurth et al., 2009), and knockout mice had defects in peripheral sensory neurons (Khaminets et al., 2015), indicating that both proteins are important for the normal human lifespan.
In this study, LM revealed that overexpression of FAM134C induced either immobile globular tubular clusters or long, unbranched protrusions of the ER membrane. These tightly packed tubular structures were unorganized in orientation and FAM134C positive, as confirmed by electron tomography and immuno-EM, respectively. Similar kinds of structures were observed under RTN4B overexpression (Rämö et al., 2016) and ATP depletion (Lingwood et al., 2009) conditions. We detected FAM134C to have both its N- and C-terminus facing the cytoplasm, like FAM134B (Khaminets et al., 2015; Bhaskara et al., 2019), thus allowing a similar kind of fold predictions of RHD for FAM134C. The RHD present in the N-terminal part of FAM134C has 30–50% sequence identity to the corresponding part in FAM134B. When we extended FAM134C membrane-embedded α-helixes from 15 to 19 aa, one full turn in the α-helix, we lost the striking overexpression phenotype on the ER and instead found occasional cells with phenotypic changes typically connected to overexpression of ER membrane proteins. Thus, the RHD of the FAM134C is most probably responsible for the membrane curvature by partial membrane insertion. This concept was previously described for RTN4 and for FAM134B in silico simulations that were proven in vitro (Shibata et al., 2006; Bhaskara et al., 2019).
Depletion of RTN4B induces striking structural changes in the ER morphology including formation of large peripheral ER sheets in Huh-7 cells (Rämö et al., 2016). Also, Khaminets et al. (2015) reported especially peripheral CLIMP-63–positive ER expansion after FAM134B silencing in U2-OS cells, and knockout studies in mouse embryonic fibroblasts support the finding. We did not observe the distinctive ER phenotype upon FAM134C silencing under normal growth conditions, suggesting that substantial membrane bending or obvious ER sheet removal are not the main functions of FAM134C or are compensated by the more highly expressed RTN4B and other RHD proteins. However, we found a significant increase in FAM134C protein expression due to amino acid starvation for several hours in three cell lines studied. The cells lacking FAM134C formed fewer autophagosomes compared with the control, and the existing autophagosomes were strikingly smaller in diameter. Comparable results were observed with three different shRNA constructs, ruling out potential off-target effects. The majority of autophagosomes in control U2-OS cells were harboring ER as cargo for lysosomal degradation, while in FAM134C silenced cells ER-phagy remained a rare event. Furthermore, FAM134C was not destined for lysosomal degradation under these conditions, because its amount was not increased upon inhibition of lysosomal function with bafilomycin A1. These results differ from those obtained for FAM134B, where no or very small differences in autophagosome number or autophagic marker LC3 amount were reported (Khaminets et al., 2015). In this study, the authors came to the conclusion that FAM134B is not required for bulk autophagy. Our results point in the other direction, as FAM134C is universally expressed and in FAM134C silenced cells, autophagosomes are small and rare and the LC3 amount increases.
We propose that cells need FAM134C in the early stages of autophagy to facilitate ER remodeling, which is required during autophagosome formation and cargo loading. During amino acid starvation, FAM134C’s LIR domain is involved in the initial cargo selection pathway of ER-phagy and RHD in ER remodeling by increasing positive membrane curvature.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture, constructs, overexpression, and silencing
HeLa (CCL-2; American Type Culture Collection [ATCC], LGC Standards GmbH, Germany), HEK-293 (Flp-In T-REx 293; R78007; Thermo Fisher Scientific, Waltham, MA), and U2-OS (HTB-96; ATCC, Manassas, VA) were cultured in DMEM (BioWhittaker, Lonza, Verviers, Belgium) and Huh-7 (JCRB0403; Japanese Collection of Research Bioresources Cell Bank, Osaka, Japan) in Eagle’s Minimum Essential Medium (EMEM) (Lonza), containing 5 or 10% fetal bovine serum (Life Technologies, Thermo Fisher Scientific) and other supplements (Lonza). When indicated, for low and high confluency, 100,000 cells/ml and 500,000 cells/ml, respectively, were plated. Amino acid starvation was induced by replacing growth medium with Earle’s balanced salt solution (EBSS; Life Technologies) for 1–4 h. For the BiCF assay, FAM134C, MYO1C, and RTN4B, in a pENTR221 Gateway–compatible vector (with/without stop codon) from the Orfeome library (Genome Biology Unit, University of Helsinki) were taken through the LR reaction (Gateway Cloning, Life Technologies, Thermo Fisher Scientific) into pDEST-GFP-1-10-N3, pDEST-GFP-11-N3, pDEST-GFP-1-10-C1, or pDEST-GFP-11-C1 (Viita et al., 2019). pcDNA6.2/C-EmGFP-DEST and pDEST-FLAG-N3 Gateway–compatible vectors (Genome Biology Unit) were used to create FAM134C-EmGFP, FLAG-FAM134C, and FAM134C-FLAG. TM-mutFAM134C-GFP was cloned using a stepwise strategy, with overlapping PCR fragments produced in multiple PCRs with oligonucleotides, where nucleotides encoding the helical extension are indicated in bold (CAGGTTGAGGCGGCGc,
GCCGCGTTCAGAGCAATGTTCAGCCCCAGGCACCa,
ATTGCTCTGAACGCGGCTTTC TGGTTTTTTG, AATGCAAGCAGGAACACAAGACGAAGAGATGTCAGGG, AATGCAAGCAGGAACACAAGACaag,
GTGTTCCTGCTTGCATTTTTACTTGCATTT GGCTTGATGA, AAGAACTGCCAGGACAGCCAAAAAGGTCAGTATCC,
AAGAACTGCCAGGACAGCCaaa,
CTGTCCTGGCAGTTCTTTTGGGCCGCTACGTCCC,
CTGTCCTGGCAGTTCTTTTGG, gcatTGCAAGCAGGAACAAGTAGGACAGCAGAAGCCCA,
TTCCTGCTTGCAATGCTT GTCACTGTCATGATGTGG,
ACACTGAAGTCTAGCCGCTGCA) and overexpression construct plasmid FAM134C described above as a template in the Phusion-polymerase (Thermo Fisher Scientific) catalyzed reaction. Finally, the resulting fragment was ligated between Apa1 and Pst1 restiction enzyme sites of the wild-type FAM134C construct. The plasmid-encoded mutant protein of FAM134C is illustrated in the Supplemental Material. TetOn-mCherry-eGFP-RAMP4 was from Addgene (Watertown, MA). DNA transfections were done using Fugene HD (NGF; Promega, Madison, WI) according to the manufacturer’s instructions. Hsp47-GFP and Hsp47-APEX were described earlier (Kano et al., 2005; Kumar et al., 2019). For DNA and small interfering RNA (siRNA) cotransfections jetPRIME (Polyplus, New York, NY) was used according to the manufacturer’s instructions. For cotransfections, a weight ratio of 1:3 or 0.5:0.5:4 was used for two or three plasmids, respectively. We have earlier shown that the cotransfection efficiency is more than 95% (http://urn.fi/URN:ISBN:978-952-10-9877-2). FAM134C silencing was done for 48 h using shRNA from the Biomedicum Functional Genomics Unit (University of Helsinki); shRNA was cloned into the pGEM vector backbone. The following sequences were used:
shRNA1: CCGGCAGCTGGACGATTCTACTGTTCTCGAGAACAGTAGAATCGTCCAGCTGTTTTTTG; shRNA2: CCGGCCTGTACAGACAATGGCACATCTCGAGATGTGCCATTGTCTGTACAGGTTTTTTG and shRNA3: CCGGCCTGACATCTCTTCGTCTTGTCTCGAGACAAGACGAAGAGATGTCAGGTTTTTTG. The silencing efficiency of the shRNA was quantified using Western blotting. The pGEM vector with nonsense control DNA (Agilent Technologies, Espoo, Finland) was used in transfections as a control, and pssHRP-KDEL was used for cytochemical staining of the ER in EM studies (Connolly et al., 1994).
Antibodies
Antibodies against FAM134C (A304-684A; Bethyl Laboratories, Montgomery, TX, for Western blotting; and HPA16492, Atlas Antibodies, Sweden; for immuno-EM), RTN4B (AB-163; Kinasource Ltd, Dundee, UK), LC3 (L7543; Sigma-Aldrich, Germany), GM130 (610823; BD Biosciences, Franklin Lakes, NJ), ERGIC-53 (ALX-804-602; Enzo Life Sciences, Farmingdale, NY), HA (MMS-101R-50; Covance, Princeton, NJ), FLAG (F7425; Sigma-Aldrich), calreticulin (2679S; Cell Signaling Technologies), green fluorescent protein (GFP) (ab290; abcam, Cambridge, UK), and glyseraldehyde-3-phosphate dehydrogenase (GAPDH;MAB374; Merck Millipore, Billerica, MA) were used as primary antibodies. When indicated, rabbit anti-sheep bridging antibody (313-001-003; Jackson ImmunoResearch Labs, West Grove, PA) was used. Secondary antibodies were conjugated with Rhodamine Red-X (016-290-084; Jackson ImmunoResearch), Alexa 647 (A31571; Life Technologies, Thermo Fisher Scientific) and Alexa 488 (A-11008; Life Technologies). Nanogold-conjugated anti-rabbit secondary antibody (1.4 nm) was used in immuno-EM (Nanoprobes, Stony Brook, NY).
Purification, mass spectrometry, and data processing
For affinity purification, tetracycline-inducible Flp-In T-REx 293 cell lines expressing RTN4B or GFP were generated as described earlier (Varjosalo et al., 2013). Affinity purification for Strep/HA-tagged proteins was performed as described earlier (Varjosalo et al., 2013). LR recombination was performed between the RTN4B in the pENTR221 Gateway–compatible vector and the in-house–designed destination vector; the C-terminal tagging vector pcDNA5/FRT/TO/BirA*/Myc/GW (Yadav et al., 2017) or pcDNA5/FRT/TO/StrepII/HA/GW (Varjosalo et al., 2013) was used for all the experiments. Huh-7 cells were made to mildly express RTN4B or GFP using the above-mentioned destination vectors and harvested 16 h posttransfection for lysis and affinity purification.
For BioID purification, ∼5 × 107 cells in two biological replicates were induced with 2 µg/ml doxycycline and 50 μM biotin 24 h before harvesting. Cells were washed with 0.1 mM MgCl2, 0.1 mM CaCl2 in phosphate-buffered saline (PBS) and harvested in 1 mM EDTA-PBS. Purification of the in vivo biotinylated proteins and mass spectrometry were performed as explained earlier (Heikkinen et al., 2017).
For protein identification, Thermo RAW files were uploaded into Proteome Discoverer 1.4 (Thermo Scientific) and searched against the Sequest search engine of the selected human part of the UniProtKB/SwissProt database (http://www.uniprot.org, version 2015-01). The following parameters were applied: Trypsin was selected as the enzyme, and a maximum of two miss cleavages was permitted, precursor mass tolerance at ±15 ppm, and fragment mass tolerance at 0.05 Da. Carbamidomethylation of cysteine were defined as static modifications; oxidation of methionine and biotinylation of lysine was specified as variable modifications. All reported data were based on high-confidence peptides assigned in Proteome Discoverer with a 0.05% false discovery rate (FDR) by Percolator.
The high-confidence protein–protein interactions were identified using stringent filtering against the control contaminant database. The high-confidence protein–protein interaction data were imported into Cytoscape 3.2.1 for visualization.
Immunofluorescence staining and Western blotting
For RTN4B labeling, cells were fixed with –20°C methanol, blocked with 10% goat serum (Life Technologies) and 1% bovine serum albumin, labeled with the indicated antibodies, and mounted in Mowiol (Hoechst, Frankfurt, Germany) supplemented with Dabco (Sigma-Aldrich). All the other samples were fixed with 4% formaldehyde (Electron Microscopy Sciences, Hatfield, PA), 0.1 mM MgCl2, and 0.1 mM CaCl2 in PBS, quenched with 50 mM NH4Cl, permeabilized with 0.1% Triton X-100, and then blocked with 0.2% bovine serum albumin in Dulbecco PBS (Lonza). When appropriate, the cells were incubated consecutively with primary and secondary antibodies, diluted in blocking solution. Samples were mounted in Mowiol supplemented with Dabco. For determining the protein topology, HeLa cells expressing FAM134C–N-Term-FLAG and FAM134C-C-Term-FLAG were fixed with 4% paraformaldehyde, permeabilized with 30 μg/ml digitonin in PBS or 0.1% Triton-X 100 in PBS on ice for 5 min, washed with PBS, and processed for immunofluorescence. Western blotting was done with the indicated antibodies according to the manufacturer’s instructions and by using standard protocols.
Light microscopy and image quantitation
Wide-field images of fixed cells were taken with a Zeiss AxioImager M2 482 epifluorescence microscope equipped with a 63×/Plan-Apochromat/1.40 oil/M27 and 483 AxioCam HRm camera (Zeiss, Oberkochen, Germany) or with a Leica DM6000B upright fluorescence wide-field microscope equipped with 40×/1.25-0.75 HCX PL APO CS oil objective, Hamamatsu Orca-Flash4.0 V2 sCMOS camera (Wetzlar, Germany). Images were acquired with AxioVision4 (Zeiss) or LAS X software (Leica). Confocal images of fixed cells were taken with a LSM880 confocal laser scanning microscope (Zeiss) with a 63× plan-apochromat (NA = 1.40) oil objective, GaAsP detector, and ZEN 2 software (Zeiss). Live Huh-7 cells were imaged at 37°C and 5% CO2 on glass-bottom dishes (MatTek, Ashland, MA). Live cell imaging was done using a 3I Marianas (3I Intelligent Imaging Innovations, Denver, CO) on a Zeiss Axio Observer Z1 microscope equipped with a 63×/1.2 W C-Apochromat Corr (working distance [WD] = 0.28 M27) objective, spinning-disk confocal, LED lasers, and Andor Neo sCMOS camera (Andor, Oxford Instruments, Abingdon, UK). Pearson’s correlation coefficients were calculated using the Microscopy Image Browser (MIB) (Belevich et al., 2016) for square regions of interest (ROIs) placed with the cell area (n > 20). As a negative control, the Pearson’s correlation coefficient was also calculated for the same ROIs after rotating one of the two channels by 90°.
For BiFC analysis, the average intensity of the BiFC signal was calculated from the cytoplasmic area (excluding the nucleus) of at least 10 transfected and nontransfected cells per condition. The signal was normalized by subtracting the average background intensity, and the relative BiFC signal was calculated as the ratio of intensities between transfected and nontransfected cells. The analysis was automatized using the Triple Area Intensity plug-in of the MIB (Belevich et al., 2016). The relative BiFC signal values were plotted using the box plot function in Matlab.
For the EATR assay, U2-OS cells were transiently cotransfected with TetOn-mCherry-eGFP-RAMP4 and mock, siRTN4, or FAM134C shRNAs 1–3 for 48 h. Doxycycline hyclate (D9891; Sigma-Aldrich, Saint Louis, MO) was added 16 h before imaging to a final concentration of 4 µg/ml. Three to 10 experimental replicates were imaged live with a Leica TCS SP5 II or SP8 equipped with a HC PL APO 63×/1,2 W Corr/0,17 CS2 (water) objective, HeNe 633 nm/12 mW and DPSS 561 nm/20 mW laser, three photomultiplier tube (PMT) and two HyD detectors for fluorescence with adjustable detection range and LAS AF 2.7.7 12.4.2018 software. For analysis, the images from all the different conditions were pooled together, shuffled, and anonymized using MIB (with plug-ins called Rename and shuffle and Restore). The anonymized images were manually categorized based on the presence or absence of acidified ER puncta within the cytoplasm by three experts. The percentage of cells with acidified ER was calculated.
Transmission electron microscopy and image quantitation
Cells grown on glass coverslips were cytochemically stained and flat embedded as described previously (Jokitalo et al., 2001). Cells were fixed with 2% glutaraldehyde (Sigma-Aldrich) and 1.5% formaldehyde in 0.1 M sodium cacodylate buffer, pH 7.4, for 20 min at room temperature (RT). Cytochemical staining with 3,3′-diaminobenzidine (TAAB, Berks, UK) was done for pssHRP-KDEL or pssHsp47-APEX transfected cells. Cells were then postfixed with 1% reduced osmium tetroxide in sodium cacodylate buffer for 1 h on ice, dehydrated through series of ethanols and acetone, and infiltrated with Epon (TAAB 812) for 2 h before14 h polymerization at 60°C. Thin sections (60 or 100 nm) were cut and poststained with uranyl acetate and lead citrate. For immuno-EM, the cells were fixed with paraformaldehyde–lysine–periodate fixative (Mclean and Nakane, 1974). Cells were permeabilized with 0.01% saponin (Sigma-Aldrich) and immunolabeled with anti-FAM134C primary antibody and 1.4 nm nanogold-conjugated anti-rabbit secondary antibody, silver enhanced with the HQ Silver kit (Nanoprobes, Stony Brook, NY), and gold toned with 0.05% gold chloride. Finally, cells were processed for osmication, dehydration, Epon embedding, and sectioning as described above. TEM imaging was done at 80 or 120 kV with FEI-Tecnai 12 (Thermo Fisher Scientific, Waltham, MA) or with Jeol JEM-1400 microscopes equipped with Gatan Orius SC 1000B bottom-mounted charge-coupled device (CCD) cameras (Gatan, AMETEK). The specificity of the FAM134C antibody in immuno-EM was verified as abolished labeling in U2-OS cells transfected with FAM134C shRNA1 for 48 h.
Electron tomography was done on serial 230-nm-thick sections as previously described (Puhka et al., 2007), except that the tilt series images between ±60° were acquired with an UltraScan 4000 CCD camera, 4 k × 4 k (Gatan) at nominal magnification of 9.600×. Dual axis tilt series were acquired using SerialEM software running on a FEI-Tecnai FEG 20 microscope (Thermo Fisher Scientific) operating at 200 kV. Gold particles on tomograms were quantified by manual tagging on separately modeled ER structures. Reconstructions were done using IMOD software (Kremer et al., 1996) followed by visualization and modeling using MIB and Amira (Thermo Fisher Scientific).
Images were taken at 500× (10 or 25 cells per condition), where Hsp47-APEX staining was indicating positive transfection. Autophagosome density was calculated by dividing the number of autophagosomes by the area occupied by the ER per cell profile in section using MIB software. For autophagosome size determination, images taken at 5000× and were analyzed using MIB by taking the mean of the longest distance from end to end within an autophagosome membrane and the distance subtended by the nearest autophagosome membrane to the middle of the longest distance falling perpendicular to the same. The results were visualized using a customized box plot function of Matlab (MathWorks, Natick, MA).
The distribution of the gold labeling was quantified as described in Mayhew (2011), using MIB segmentation software (Belevich et al., 2016). Ten cells were randomly selected at low magnification, and 8000× magnification images were acquired at the north and south positions relative to the nucleus from these cells. The images were annotated in MIB, and gold particles were assigned to the following categories: ER sheet, ER tubule, nuclear envelope, ER-mitochondria contact site, and others. Segmentation of the cellular compartments was done in MIB. To estimate the area of cellular compartments per image, a grid of 180 nm between branch points was placed on each image and the number of intersections falling on top of an organelle was calculated. Nuclei and extracellular area were excluded. The relative labeling index (RLI) was calculated based on the area of the respective cellular organelle and the attribution of the counted gold particles and the expected number of gold particles per organelle (Mayhew, 2011). The RLI shows whether the distribution pattern of the protein is random. RLI = 1 indicates that the distribution of the protein is random, whereas RLI > 1 states the significance of the distribution of the protein in the respective cellular organelle.
Supplementary Material
Acknowledgments
We acknowledge the Electron Microscopy Unit (EMBI) and Light Microscopy Unit of the Institute of Biotechnology, University of Helsinki, for excellent support during this project. B.L. and D.K. were graduate students in the Doctoral Programme in Integrative Life Science, University of Helsinki. This work was funded by the Academy of Finland (projects 1287975 and 1331998, E.J.), the Sigrid Jusélius Foundation (E.J.), the Biocenter Finland (E.J., and I.B.), and the Helsinki Institute of Life Science Fellow’s program (E.J.).
Abbreviations used:
- aa
amino acid
- BiFC
bimolecular fluorescence complementation assay
- DTT
dithiothreitol
- EBSS
Earle’s balanced salt solution
- ER
endoplasmic reticulum
- GAPDH
glyseraldehyde-3-phosphate dehydrogenase
- immuno-EM
immunoelectron microscopy
- LIR
LC3 interacting region
- LM
light microscopy
- MIB
Microscopy Image Browser
- PBS
phosphate-buffered saline
- RHD
reticulon homology domain
- RTN
reticulon
- TEM
transmission electron microscopy
- TM
transmembrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-06-0409) on April 7, 2021.
REFERENCES
- Baumann O, Walz B (2001). Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int Rev Cytol 205, 149–214. [DOI] [PubMed] [Google Scholar]
- Belevich I, Joensuu M, Kumar D, Vihinen H, Jokitalo E (2016). Microscopy Image Browser: a platform for segmentation and analysis of multidimensional datasets. PloS Biol 14, e1002340.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernales S, Schuck S, Walter P (2007). ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy 3, 285–287. [DOI] [PubMed] [Google Scholar]
- Bhaskara RM, Grumati P, Garcia-Pardo J, Kalayil S, Covarrubias-Pinto A, Chen W, Kudryashev M, Dikic I, Hummer G (2019). Curvature induction and membrane remodeling by FAM134B reticulon homology domain assist selective ER-phagy. Nat Commun 10, 2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulou A, Santarella-Mellwig R, Santama N, Mattaj IW (2016). Transmembrane protein TMEM170A is a newly discovered regulator of ER and nuclear envelope morphogenesis in human cells. J Cell Sci 129, 1552–1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly CN, Futter CE, Gibson A, Hopkins CR, Cutler DF (1994). Transport into and out of the Golgi complex studied by transfecting cells with cDNAs encoding horseradish peroxidase. J Cell Biol 127, 641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Eletto M, Oliverio S, Di Sano F (2020). Reticulon homology domain-containing proteins and ER-phagy. Front Cell Dev Biol 8, 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd DA, Niederoest B, Bloechlinger S, Dupuis L, Loeffler JP, Schwab ME (2005). Nogo-A, -B, and -C are found on the cell surface and interact together in many different cell types. J Biol Chem 280, 12494–12502. [DOI] [PubMed] [Google Scholar]
- Goyal U, Blackstone C (2013). Untangling the web: mechanisms underlying ER network formation. Biochim Biophys Acta 1833, 2492–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauri HP, Schweizer A (1992). The endoplasmic reticulum-Golgi intermediate compartment. Curr Opin Cell Biol 4, 600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkinen T, Kampjarvi K, Keskitalo S, von Nandelstadh P, Liu X, Rantanen V, Pitkanen E, Kinnunen M, Kuusanmaki H, Kontro M, et al. (2017). Somatic MED12 nonsense mutation escapes mRNA decay and reveals a motif required for nuclear entry. Hum Mutat 38, 269–274. [DOI] [PubMed] [Google Scholar]
- Hu J, Prinz WA, Rapoport TA (2011). Weaving the web of ER tubules. Cell 147, 1226–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Wang X, Ding X, Du M, Li B, Weng X, Zhang J, Li L, Tian R, Zhu Q, et al. (2020). FAM134B oligomerization drives endoplasmic reticulum membrane scission for ER-phagy. EMBO J 39, e102608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joensuu M, Belevich I, Rämö O, Nevzorov I, Vihinen H, Puhka M, Witkos TM, Lowe M, Vartiainen MK, Jokitalo E (2014). ER sheet persistence is coupled to myosin 1c-regulated dynamic actin filament arrays. Mol Biol Cell 25, 1111–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jokitalo E, Cabrera-Poch N, Warren G, Shima DT (2001). Golgi clusters and vesicles mediate mitotic inheritance independently of the endoplasmic reticulum. J Cell Biol 154, 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, Kominami E, Ohsumi Y, Yoshimori T (2000). LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J 19, 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaddoum L, Magdeleine E, Waldo GS, Joly E, Cabantous S (2010). One-step split GFP staining for sensitive protein detection and localization in mammalian cells. Biotechniques 49, 727–+. [DOI] [PubMed] [Google Scholar]
- Kano F, Kondo H, Yamamoto A, Kaneko Y, Uchiyama K, Hosokawa N, Nagata K, Murata M (2005). NSF/SNAPs and p97/p47/VCIP135 are sequentially required for cell cycle-dependent reformation of the ER network. Genes Cells 10, 989–999. [DOI] [PubMed] [Google Scholar]
- Kappeler F, Klopfenstein DR, Foguet M, Paccaud JP, Hauri HP (1997). The recycling of ERGIC-53 in the early secretory pathway. ERGIC-53 carries a cytosolic endoplasmic reticulum-exit determinant interacting with COPII. J Biol Chem 272, 31801–31808. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ (1999). Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes Dev 13, 1211–1233. [DOI] [PubMed] [Google Scholar]
- Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, et al. (2015). Regulation of endoplasmic reticulum turnover by selective autophagy. Nature 522, 354–358. [DOI] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996). Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116, 71–76. [DOI] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580. [DOI] [PubMed] [Google Scholar]
- Kumar D, Golchoubian B, Belevich I, Jokitalo E, Schlaitz A-L (2019). REEP3 and REEP4 determine the tubular morphology of the endoplasmic reticulum during mitosis. Mol Biol Cell 30, 1377–1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurth I, Pamminger T, Hennings JC, Soehendra D, Huebner AK, Rotthier A, Baets J, Senderek J, Topaloglu H, Farrell SA, et al. (2009). Mutations in FAM134B, encoding a newly identified Golgi protein, cause severe sensory and autonomic neuropathy. Nat Genet 41, 1179–1181. [DOI] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Ahmed S, Corn JE (2018). Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Biol 217, 3354–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingwood D, Schuck S, Ferguson C, Gerl MJ, Simons K (2009). Generation of cubic membranes by controlled homotypic interaction of membrane proteins in the endoplasmic reticulum. J Biol Chem 284, 12041–12048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Salokas K, Tamene F, Jiu Y, Weldatsadik RG, Ohman T, Varjosalo M (2018). An AP-MS- and BioID-compatible MAC-tag enables comprehensive mapping of protein interactions and subcellular localizations. Nat Commun 9, 1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynes EM, Simmen T (2011). Urban planning of the endoplasmic reticulum (ER): how diverse mechanisms segregate the many functions of the ER. Biochim Biophys Acta 1813, 1893–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayhew TM (2011). Mapping the distributions and quantifying the labelling intensities of cell compartments by immunoelectron microscopy: progress towards a coherent set of methods. J Anat 219, 647–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclean IW, Nakane PK (1974). Periodate-lysine-paraformaldehyde fixative—new fixative for immunoelectron microscopy. J Histochem Cytochem 22, 1077–1083. [DOI] [PubMed] [Google Scholar]
- Mo J, Chen J, Zhang B (2020). Critical roles of FAM134B in ER-phagy and diseases. Cell Death Dis 11, 983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Rabouille C, Watson R, Nilsson T, Hui N, Slusarewicz P, Kreis TE, Warren G (1995). Characterization of a cis-Golgi matrix protein, GM130. J Cell Biol 131, 1715–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oertle T, Schwab ME (2003). Nogo and its paRTNers. Trends Cell Biol 13, 187–194. [DOI] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Muller S, Wollscheid B (2014). Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics 30, 884–886. [DOI] [PubMed] [Google Scholar]
- Park SH, Zhu PP, Parker RL, Blackstone C (2010). Hereditary spastic paraplegia proteins REEP1, spastin, and atlastin-1 coordinate microtubule interactions with the tubular ER network. J Clin Invest 120, 1097–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puhka M, Vihinen H, Joensuu M, Jokitalo E (2007). Endoplasmic reticulum remains continuous and undergoes sheet-to-tubule transformation during cell division in mammalian cells. J Cell Biol 179, 895–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi B, Qi Y, Watari A, Yoshioka N, Inoue H, Minemoto Y, Yamashita K, Sasagawa T, Yutsudo M (2003). Pro-apoptotic ASY/nogo-B protein associates with ASYIP. J Cell Physiol 196, 312–318. [DOI] [PubMed] [Google Scholar]
- Rämö O, Kumar D, Gucciardo E, Joensuu M, Saarekas M, Vihinen H, Belevich I, Smolander OP, Qian K, Auvinen P, Jokitalo E (2016). NOGO-A/RTN4A and NOGO-B/RTN4B are simultaneously expressed in epithelial, fibroblast and neuronal cells and maintain ER morphology. Sci Rep , 6, 35969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA (2010). Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voeltz GK, Rapoport TA (2006). Rough sheets and smooth tubules. Cell 126, 435–439. [DOI] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK (2008). The reticulon and DP1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem 283, 18892–18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soding J, Biegert A, Lupas AN (2005). The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33, W244–W248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varjosalo M, Sacco R, Stukalov A, van Drogen A, Planyavsky M, Hauri S, Aebersold R, Bennett KL, Colinge J, Gstaiger M, Superti-Furga G (2013). Interlaboratory reproducibility of large-scale human protein-complex analysis by standardized AP-MS. Nat Methods 10, 307–314. [DOI] [PubMed] [Google Scholar]
- Viita T, Kyheroinen S, Prajapati B, Virtanen J, Frilander MJ, Varjosalo M, Vartiainen MK (2019). Nuclear actin interactome analysis links actin to KAT14 histone acetyl transferase and mRNA splicing. J Cell Sci 132, jcs226582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586. [DOI] [PubMed] [Google Scholar]
- Wang DX, Eraslan B, Wieland T, Hallstrom B, Hopf T, Zolg DP, Zecha J, Asplund A, Li LH, Meng C, et al. (2019). A deep proteome and transcriptome abundance atlas of 29 healthy human tissues. Mol Systems Biol 15, e8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JL, Tong CW, Chang WT, Huang AM (2013). Novel genes FAM134C, C3orf10 and ENOX1 are regulated by NRF-1 and differentially regulate neurite outgrowth in neuroblastoma cells and hippocampal neurons. Gene 529, 7–15. [DOI] [PubMed] [Google Scholar]
- Yadav L, Tamene F, Goos H, van Drogen A, Katainen R, Aebersold R, Gstaiger M, Varjosalo M (2017). Systematic analysis of human protein phosphatase interactions and dynamics. Cell Syst 4, 430–444.e5. [DOI] [PubMed] [Google Scholar]
- Yang YS, Strittmatter SM (2007). The reticulons: a family of proteins with diverse functions. Genome Biol 8, 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek N, Sparks L, Voeltz G (2011). Reticulon short hairpin transmembrane domains are used to shape ER tubules. Traffic 12, 28–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.