Abstract
Lipid droplets (LDs) are neutral lipid-containing organelles enclosed in a single monolayer of phospholipids. LD formation begins with the accumulation of neutral lipids within the bilayer of the endoplasmic reticulum (ER) membrane. It is not known how the sites of formation of nascent LDs in the ER membrane are determined. Here we show that multiple C2 domain–containing transmembrane proteins, MCTP1 and MCTP2, are at sites of LD formation in specialized ER subdomains. We show that the transmembrane domain (TMD) of these proteins is similar to a reticulon homology domain. Like reticulons, these proteins tubulate the ER membrane and favor highly curved regions of the ER. Our data indicate that the MCTP TMDs promote LD biogenesis, increasing LD number. MCTPs colocalize with seipin, a protein involved in LD biogenesis, but form more stable microdomains in the ER. The MCTP C2 domains bind charged lipids and regulate LD size, likely by mediating ER–LD contact sites. Together, our data indicate that MCTPs form microdomains within ER tubules that regulate LD biogenesis, size, and ER–LD contacts. Interestingly, MCTP punctae colocalized with other organelles as well, suggesting that these proteins may play a general role in linking tubular ER to organelle contact sites.
INTRODUCTION
Lipid droplets (LDs) are highly conserved, dynamic organelles that undergo cycles of growth and consumption in most organisms. LDs store neutral lipids in the form of triglycerides (TAG) and sterol esters (SE) that are excluded from the cytosol by a phospholipid monolayer. In response to stimuli, neutral lipids are mobilized by lipases for membrane synthesis, signal transduction, or energy conversion via fatty acid oxidation. Originally considered as inert cytoplasmic inclusions, LDs have emerged as bona fide organelles that are essential in regulating cellular lipid homeostasis (Walther and Farese, 2012; Krahmer et al., 2013; Cohen, 2018; Henne et al., 2018; Olzmann and Carvalho, 2019). Defects in lipid storage and metabolism lead to a number of human diseases including cardiovascular disease, diabetes, and lipodystrophies (Xu et al., 2018). In eukaryotes, LDs form de novo at the endoplasmic reticulum (ER), where neutral lipids are first synthesized. As neutral lipids accumulate within the ER bilayer, they coalesce into a lens-like structure that grows toward the cytoplasm while maintaining contact with the ER (Choudhary et al., 2015; Thiam and Forêt, 2016; Walther et al., 2017; Olzmann and Carvalho, 2019). How sites of LD formation are determined and the molecular mechanisms of LD biogenesis are unknown, but accumulating evidence suggests that nascent LDs form at discrete ER subdomains (Jacquier et al., 2013; Kassan et al., 2013; Wang et al., 2016, 2018; Joshi et al., 2018; Chung et al., 2019; Choudhary et al., 2020). Proteins associated with these ER subdomains include ACSL3, seipin, and LDAF1 in mammalian cells (Kassan et al., 2013; Salo et al., 2016, 2019; Wang et al., 2016; Chung et al., 2019).
Seipin is an evolutionarily conserved ER membrane protein that is required for formation and maturation of LDs and stabilizes ER–LD connections. Mutations in seipin cause defects in LD storage and lead to Berardinelli–Seip congenital lipodystrophy type 2 in humans (Magré et al., 2001; Szymanski et al., 2007; Fei et al., 2008; Grippa et al., 2015; Wang et al., 2016). Recently, seipin was shown to determine sites of LD formation in complex with LD assembly factor 1 (LDAF1) in mammalian cells by facilitating TAG separation from the ER membrane (Chung et al., 2019). In Saccharomyces cerevisiae, we previously showed that seipin associates at ER sites containing Pex30 (Joshi et al., 2018), an ER-shaping protein with a reticulon homology domain (RHD) that localizes to ER tubules and edges of ER sheets (Joshi et al., 2016). We further found that Pex30-containing subdomains are ER sites at which both nascent LDs and peroxisomes form (Joshi et al., 2017, 2018; Joshi and Cohen, 2019). Cells deficient in both seipin and Pex30 proteins exhibited defective LD formation, suggesting that ER sites containing Pex30 and seipin mediate LD formation by providing a platform for LD-forming protein machinery (Joshi et al., 2018; Wang et al., 2018). A recent report demonstrated the protein machinery at sites of LD biogenesis in yeast. It includes seipin, the protein phosphatase Nem1 (homologous to human protein Dullard), Yft2 (homologous to human FIT2), and Pex30 (Joshi et al., 2018; Choudhary et al., 2020).
Multiple C2 and transmembrane domain–containing proteins (MCTPs) are functional mammalian homologues of Pex30, which is part of the Pex23 family of proteins localized at ER contact sites (David et al., 2013; Joshi et al., 2018; Wu et al., 2020). MCTPs are conserved in higher eukaryotes. Caenorhabditis elegans and Drosophila melanogaster have one MCTP, while humans have two MCTPs, MCTP1 and MCTP2, that are differentially expressed in tissues (Shin et al., 2005). Knockdown of MCTP2 in mammalian cells and genetic ablation of MCTP in C. elegans showed a significant reduction in LD size and number, suggesting that MCTPs play a role in LD biogenesis (Joshi et al., 2018). However, the physiological function of MCTPs and how they affect LD biogenesis are not defined.
In this report, we perform functional characterization of human MCTP1 and MCTP2. We show that MCTP1 and MCTP2 are localized in discrete regions of the ER membrane. Using high-resolution live-cell microscopy, we demonstrate the formation of nascent LDs at MCTP subdomains. Furthermore, we characterize the role of the RHD and C2 domains of MCTPs in LD biogenesis. We show that RHDs promote LD formation, whereas C2 domains regulate LD size, likely by mediating ER–LD contacts.
RESULTS AND DISCUSSION
MCTP1 and MCTP2 are ER membrane tubulating proteins
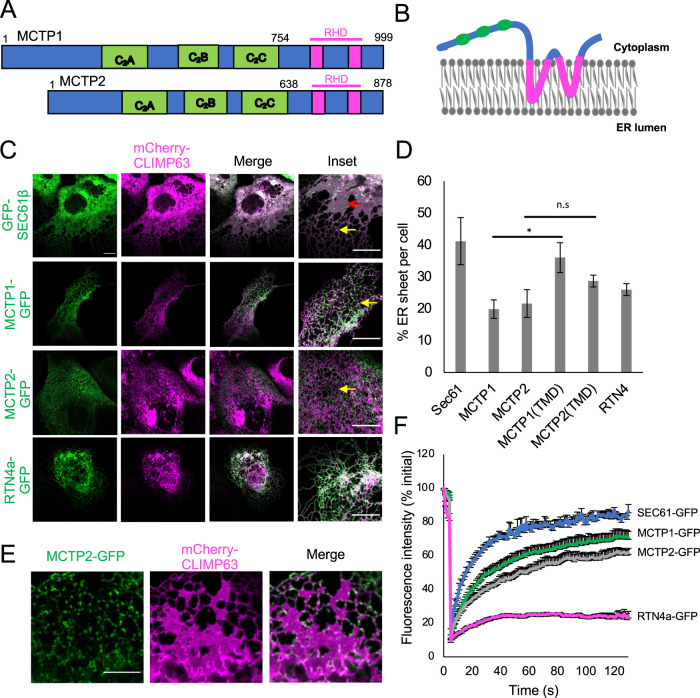
MCTPs are part of an evolutionarily conserved group of proteins that include synaptotagmins, ferlins, and extended synaptotagmins (E-Syts) (Shin et al., 2005). These proteins contain at least one transmembrane domain (TMD) at either the C- or N-terminus and more than one C2 domain. C2 domains bind membrane phospholipids in a calcium-dependent or -independent manner (Lemmon, 2008; Elíes et al., 2020). MCTPs contain TMDs at the C-erminus and three C2 domains (C2A, C2B, and C2C) at the N-terminus (Figure 1A). Using HHpred, we found that the TMDs of MCTPs are similar to the RHDs of several reticulon and reticulon-like proteins in humans (Supplemental Figure S1A) (Alva et al., 2016). Similar to RHDs, the TMDs of MCTPs have two hydrophobic regions 30–37 amino acids long (Figure 1A) (Shin et al., 2005). The RHDs are proposed to form two hairpins in the outer leaflet of the ER membrane (Voeltz et al., 2006). Both the N- and C-termini are proposed to face the cytosolic side (Figure 1B). We hypothesized that if MCTPs have RHDs, then these proteins might tubulate the ER membrane in cells. Overexpression of the ER membrane protein CLIMP63 shifts the balance of ER morphology from tubules to sheets (Shibata et al., 2010). In COS-7 cells, overexpression of the control membrane protein GFP-SEC61β and mCherry-CLIMP63 led to prominent sheet-like structures in the ER membrane (Figure 1C). However, when we overexpressed MCTP1-GFP, MCTP2-GFP, or RTN4a-GFP along with mCherry-CLIMP63, the ER membrane was mostly tubulated. This suggests that like RTN4a, MCTPs can tubulate the ER membrane. ER membrane was also tubulated when we overexpressed only the reticulon-like TMD of MCTP2, MCTP2(TMD)-GFP (640–878 amino acids), along with mCherry-CLIMP63 (Figure 1D and Supplemental Figure S1B). Thus, the TMD of MCTP2 was sufficient to tubulate the ER membrane. However, we found that the TMD of MCTP1, MCTP1(TMD)-GFP (757–999 amino acids), was not sufficient to stabilize the ER tubules (Figure 1D and Supplemental Figure S1B). We observed that MCTP1(TMD)-GFP was not uniformly distributed in the ER membrane upon overexpression and generated bright punctae in some cells (Supplemental Figure S1B), which may explain why MCTP1(TMD)-GFP is less efficient in tubulating the ER membrane. Both MCTP1(C2)-GFP (Supplemental Figure S3D) and MCTP2(C2)-GFP (unpublished data) were localized in the cytoplasm when overexpressed, confirming that TMDs of both MCTP1 and MCTP2 are required for targeting of MCTPs to the ER membrane.
FIGURE 1:
MCTP1 and MCTP2 are ER membrane tubulating proteins. (A) Domains in MCTP1 and MCTP2. The green blocks represent the three C2 domains, whereas the magenta blocks are the TMDs, which are similar to RHDs. (B) Predicted topology of MCTPs. Both N- and C-terminus are in the cytoplasm. The two inverted hairpins in the outer leaflet of the ER membrane are the TMDs, which are like RHDs. (C) Confocal images of live COS-7 cells expressing mCherry-CLIMP63 with GFP-SEC61β, MCTP1-GFP, MCTP2-GFP, or RTN4a-GFP. Red arrows indicate ER sheet, whereas yellow arrows indicate ER tubules. Scale bar, 10 µm. (D) Quantification of percent ER sheet per cell. Error bars indicate mean ± SE, n = 7–10 cells/condition, *p < 0.05, n.s, not significant, Student’s t test. (E) Airyscan images of live COS-7 cells expressing mCherry-CLIMP63 and MCTP2-GFP. Scale bar, 5 µm. (F) Mean fluorescence intensities normalized to initial value plotted over time of FRAP analysis on MCTP1-GFP, MCTP2-GFP, GFP-SEC61β, and RTN4a-GFP. Error bars indicate ± SE, n = 10 cells.
We next determined whether MCTPs, similar to reticulons, localize to highly curved regions such as tubules and edges of ER sheets in the ER membrane (Shibata et al., 2008). Overexpressed full-length MCTP1-GFP and MCTP2-GFP localized to tubules and edges of ER sheets (Figure 1C). Airyscan imaging of COS-7 cells with lower expression of full-length MCTP2-GFP revealed that MCTP2 is localized as discrete punctae on ER tubules and at the edges of ER sheets (Figure 1E). Cells with lower expression of MCTP2-GFP had more sheets than cells with higher levels of MCTP2-GFP, supporting the previous observation that the expression of reticulons and reticulon-like proteins regulates ER morphology (Schuck et al., 2009; Shibata et al., 2010; Joshi et al., 2016). To test whether MCTPs are regulated by changes in ER morphology, we next examined the number of MCTP2 punctae in cells overexpressing CLIMP63 or RTN4a using immunofluorescence. We found that changes in ER morphology did not significantly change the number of endogenous MCTP2 punctae, suggesting that MCTPs are not regulated by changes in ER morphology (Supplemental Figure S1C). Taken together, the results show that MCTPs affect ER morphology when overexpressed (Figure 1, C and E). We found that MCTPs form punctae upon low expression in live cells and when endogenously localized using immunofluorescence. MCTP punctae localized to ER subdomains on ER tubules and edges of ER sheets, suggesting that these proteins, like reticulons, have high affinity for highly curved regions of the ER. Whether MCTPs affect the ER morphology at these ER subdomains is not known.
Previous reports have demonstrated that reticulons diffuse slowly in the ER membrane (Shibata et al., 2008). To test whether this applied to MCTPs, we determined the relative rate at which MCTPs diffuse in the ER using fluorescence recovery after photobleaching (FRAP) on cells overexpressing MCTP1-GFP or MCTP2-GFP. RTN4a-GFP and GFP-SEC61β were used as controls. We selected small regions of the ER in the periphery of the cell where a single layer of the continuous tubular network can be visualized to analyze the diffusion mobility of MCTPs. Upon photobleaching, both MCTP1-GFP and MCTP2-GFP diffused relatively slowly into the bleached area compared with the control membrane protein GFP-SEC61β, which showed maximal recovery (Figure 1F). However, the recovery of MCTP1-GFP and MCTP2-GFP was greater than that of RTN4a-GFP, suggesting that the diffusion mobility of MCTPs is faster than that of RTN4a-GFP but slower than that of GFP-SEC61β. Further, we checked the motility of MCTP punctae in cells with low expression of MCTP1-GFP or MCTP2-GFP and found that a significant fraction of large punctae were immobile, whereas smaller punctae were mobile in the ER membrane (Supplemental Video 1).
Movie S1.
Associated with Figure 1. Dynamics of MCTP punctae on the ER membrane. Airyscan images of cells expressing SEC61-mApple and MCTP1-GFP (left) or MCTP2-GFP (right) were taken for 2 minutes with a time interval of 5 seconds. Scale bar, 5 μm.
Nascent LD formation occurs at ER subdomains containing MCTPs
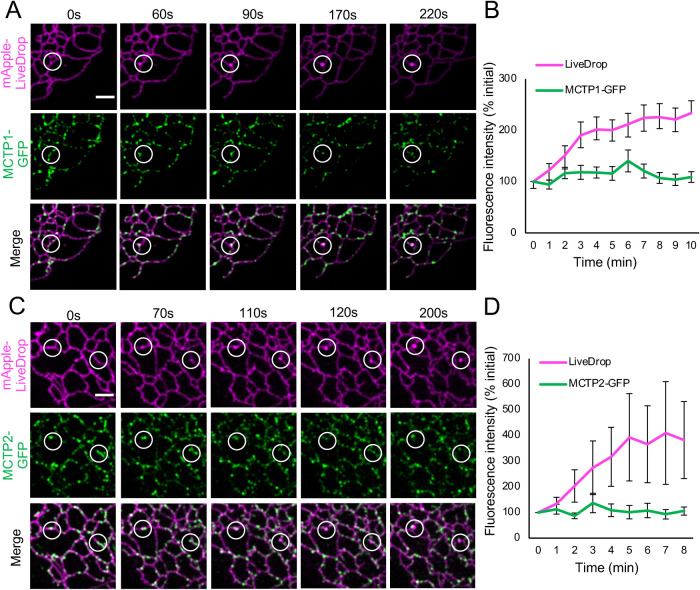
LiveDrop is a fusion protein that targets both nascent LDs forming in the ER and mature LDs (Wang et al., 2016). Previous reports showed that MCTP2 punctae in ER subdomains colocalize with LiveDrop under basal conditions, suggesting that MCTP2 is at nascent LD formation sites (Joshi et al., 2018). However, the spatiotemporal dynamics of MCTPs at LD formation sites were not determined. Here, we induced formation of new LDs by adding 400 µM oleic acid (OA) to cells grown under serum-free starvation conditions for 16–20 h. This allowed us to test whether nascent LDs form at MCTP subdomains in the ER. Airyscan time-lapse imaging indicated that new LDs form at sites in the ER membrane that contain MCTP1-GFP (Figure 2A) or MCTP2-GFP (Figure 2C). Upon quantification, we found that the fluorescence intensities of MCTP1-GFP or MCTP2-GFP punctae did not change over time, whereas the intensity of mApple-LiveDrop increased as it accumulated at LD biogenesis sites after LD induction (Figure 2, B and D). This suggests that MCTP1-GFP and MCTP2-GFP form stable ER subdomains, whereas mApple-LiveDrop is recruited to these sites under conditions of neutral lipid synthesis and TAG accumulation. MCTPs remained in the ER after the formation of LDs (Figure 2, A and C, and Supplemental Video 2). A recent report showed that endogenously tagged Plin3, a cytosolic protein that is recruited to the LD surface, was recruited to nascent LDs before LiveDrop (Chung et al., 2019). Therefore, we expressed MCTP1-GFP or MCTP2-GFP along with mApple-LiveDrop and Halo-Plin3 (Supplemental Figure S2). Cells with lower expression of Halo-Plin3 were selected as overexpression of Plin3 is known to inhibit LD biogenesis. Upon LD induction, we found that both Halo-Plin3 and mApple-LiveDrop are recruited to MCTP1-GFP (Supplemental Figure S2) or MCTP2-GFP (unpublished data) punctae. Using Plin3 as an additional marker for nascent LDs suggested that accumulation of LiveDrop at MCTP1 and MCTP2 subdomains as observed in Figure 2, A and C, is not an artifact of membrane deformation but is due to accumulation of neutral lipids. Our findings indicate that ER subdomains that contain MCTPs could be predetermined sites where new LDs form.
FIGURE 2:
Nascent LDs form at ER subdomains containing MCTPs. (A, C) Airyscan images of live COS-7 cells expressing mApple-LiveDrop together with MCTP1-GFP or MCTP2-GFP. White circles indicate sites of nascent LD formation. Cells were incubated in starvation medium for 16–20 h. Images were taken immediately after addition of 400 µM OA at the interval of 5 s for at least 10 min. Scale bar, 2 µm. (B, D) Quantification of fluorescence intensity of mApple-LiveDrop and MCTP1-GFP or MCTP2-GFP at 25–30 sites of new LD formation from three independent experiments. Error bars indicate mean ± SE.
Movie S2.
Associated with Figure 2. Formation of nascent LDs at MCTP ER subdomains. Associated with Figures 2A and 2C. Airyscan images of live COS7 cells expressing mApple-LiveDrop together MCTP2-GFP. Cells were incubated in starvation medium for 16-20 hours. Images were taken immediately after addition of 400 μM oleic acid at the interval of 5 secs for at least 10 minutes. Scale bar, 2 μm.
MCTP2 is associated with a subpopulation of LDs
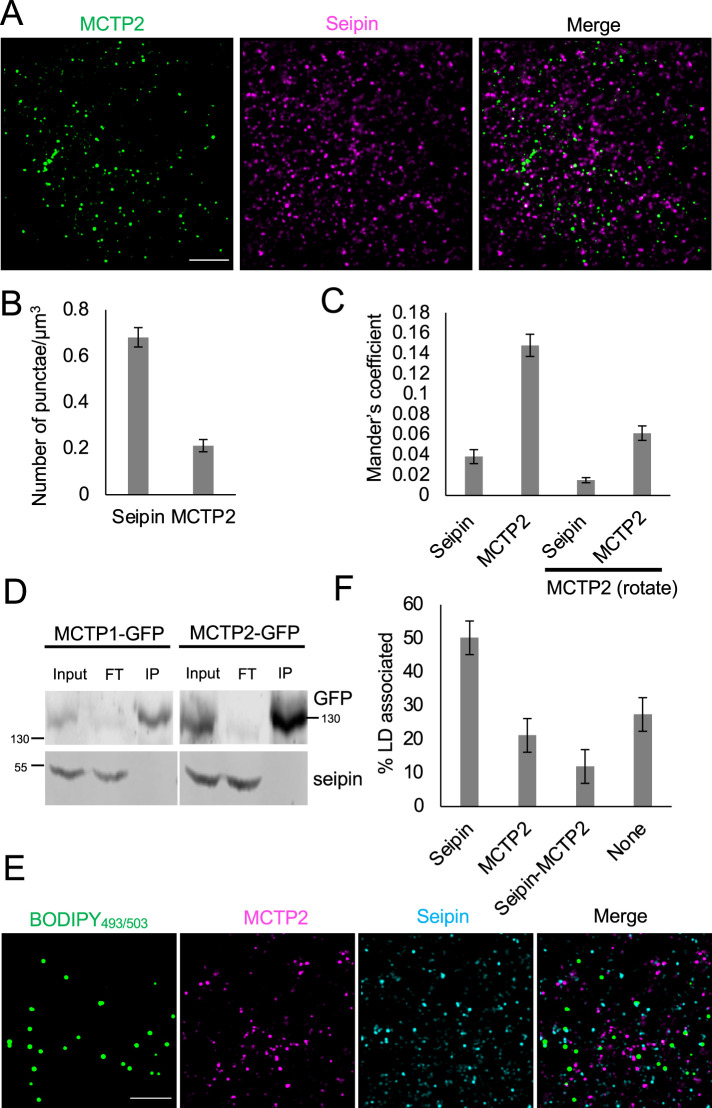
Next, we explored the involvement of MCTP2 in LD biogenesis by analyzing the localization of endogenous MCTP2 and seipin. Seipin is a well-established ER membrane protein localized at ER–LD contact sites, so we sought to determine whether MCTP2 colocalized with seipin under basal conditions using immunofluorescence. We validated the antibodies by staining for MCTP2 and seipin in cells that overexpressed MCTP2-GFP and seipin-mApple, respectively, and observed complete colocalization (unpublished data). We found that seipin punctae are three times more abundant than MCTP2 punctae (Figure 3, A and B) and that MCTP2 punctae colocalized with seipin with a Mander’s coefficient of 0.14. This colocalization was not random as clockwise rotation of the MCTP2 image by 90° caused the Mander’s coefficient to decrease to 0.06 (Figure 3C). Conversely, only a small fraction of seipin colocalized with MCTP2 (Mander’s coefficient 0.038; Figure 3, A and C). We then investigated whether seipin and MCTPs physically interact by performing immunoprecipitation of proteins interacting with MCTP1-GFP or MCTP2-GFP. The immunoblots did not detect seipin in the immuno-pulldown fractions from cells expressing either MCTP1-GFP or MCTP2-GFP (Figure 3D). Our findings suggest that even though seipin and MCTP2 colocalize, they do not exhibit stable interaction under basal conditions. We then determined colocalization between seipin and MCTP2 relative to LDs. We stained LDs with BODIPY493/503 in cells that were labeled for endogenous seipin and MCTP2 (Figure 3E). We found that 50% of LDs were associated with seipin, consistent with previous findings (Salo et al., 2016). About 21% of LDs were associated with MCTP2 punctae and 12% of LDs were associated with both seipin and MCTP2 (Figure 3F). Our findings suggest that MCTP2 punctae are associated with only a small fraction of seipin-positive LDs under basal conditions.
FIGURE 3:
MCTP2 is associated with a subpopulation of LDs. (A) Airyscan images of a fixed cell stained for endogenous MCTP2 and seipin. Scale bar, 5 µm. (B) Quantification of number of MCTP2 and seipin punctae shown in A. Error bars indicate mean ± SE, n = 17 fields of view from 17 cells. (C) Quantification of colocalization of seipin and MCTP2 punctae by Mander’s colocalization coefficient. Error bars indicate mean ± SE, n = 17 fields of view from 17 cells. (D) Immunoblots of anti-GFP immunoprecipitates from COS-7 cells expressing either MCTP1-GFP or MCTP2-GFP. FT, flowthrough; IP, immuno-pulldown. (E) Airyscan images of fixed cells stained for MCTP2 and seipin. LDs were labeled with BODIPY493/503 Scale bar, 5 µm. (F) Quantification of the percent of LDs associated with seipin, MCTP2, seipin and MCTP2, or none. Error bars indicate mean ± SE, n = 15 fields of view from 15 cells.
RHD and C2 domains of MCTPs regulate LD biogenesis and size
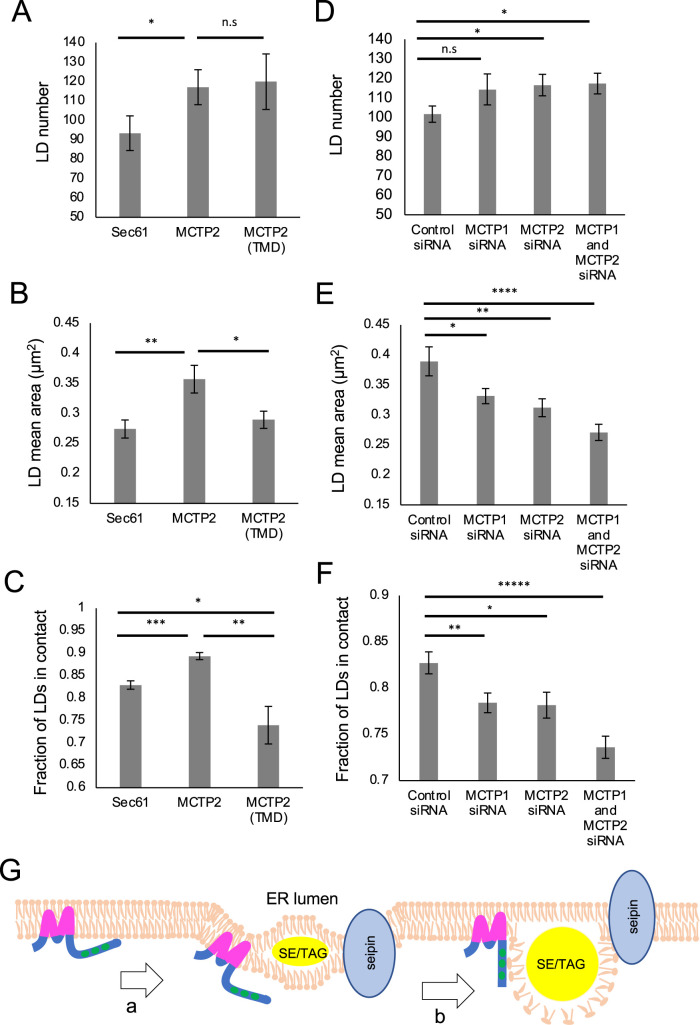
Depletion of MCTP2 in HeLa cells and MCTP in C. elegans leads to a decrease in LD number and size (Joshi et al., 2018). To characterize the function of the RHD and C2 domains of MCTPs, we overexpressed full-length MCTP2 or a truncated version that consists of the TMD tagged with green fluorescent protein (GFP) at the C-terminus, and labeled LDs with BODIPY665/676. We found that the number of LDs was significantly increased in cells expressing MCTP2-GFP or MCTP2(TMD)-GFP compared with GFP-SEC61β (Figure 4A). As shown above, both MCTP2-GFP as well as MCTP2(TMD)-GFP were able to tubulate the ER membrane (Figure 1D). Thus, it is possible that membrane curvature generated by these proteins affects LD number. Our observations are consistent with recent findings demonstrating that overexpression of reticulon protein increases the number of LDs (Santinho et al., 2020). Overexpression of full-length MCTP1-GFP but not MCTP1(TMD)-GFP also increased LD number (Supplemental Figure S3A). Expression of MCTP1(TMD)-GFP did not increase LD number, likely because the TMD of MCTP1 is less efficient at tubulating the ER membrane than the TMD of MCTP2 (Figure 1D).
FIGURE 4:
RHD and C2 domains of MCTP2 regulate LD biogenesis and size. (A–C) Overexpression of full-length or truncated MCTP2. (D–F) Knockdown of MCTP1, MCTP2, or MCTP1 and MCTP2. (A–F) Quantification of number of LDs per cell (A and D); median LD size µm2 (B and E); and fraction of LDs in contact with Sec61, MCTP2, or MCTP2(TMD) (C) or SEC61ß-GFP (F) per cell. Error bars indicate mean ± SE, n = 30 cells from three independent experiments. *p < 0.05, **p < 0.005, ***p < 0.0005, ****p < 0.00005, *****p < 0.000005, n.s, not significant, Student’s t test. (G) Proposed model for role of MCTPs in LD biogenesis. (a) MCTPs contain RHDs that generate membrane bending at discrete ER subdomains, which facilitates accumulation of neutral lipids after LD induction. (b) RHDs are responsible for modulating LD formation, whereas the C2 domains might generate contact via interaction with phospholipids on LD surface to regulate LD size.
In addition to affecting LD number, overexpression of full-length MCTP1-GFP or MCTP2-GFP increased LD size compared with control GFP-SEC61β. However, overexpression of MCTP1(TMD)-GFP or MCTP2(TMD)-GFP did not affect the size of LDs (Figure 4B; Supplemental Figure S3B). This suggests that MCTP C2 domains play a role in regulating the size of LDs, possibly by regulating the contact between ER and LDs to promote ER-to-LD lipid transfer. To test this, we measured contacts between LDs and the ER upon overexpression of full-length or truncated MCTPs. We found that LD–ER contact was significantly increased in cells overexpressing full-length MCTP1-GFP or MCTP2-GFP but not in cells overexpressing MCTP1(TMD)-GFP or MCTP2(TMD)-GFP (Figure 4C; Supplemental Figure S3C). The expression of MCTP TMDs significantly decreased ER–LD contact compared with the control, suggesting a dominant negative effect of overexpression of MCTP TMDs on ER–LD contact. In addition, we found that the soluble C2 domains of MCTP1 localized to LDs when cells were supplemented with OA (Supplemental Figure S3D), consistent with a role of C2 domains in generating ER–LD contact.
We further tested the roles of MCTP RHD and C2 domains in LD biogenesis and growth by generating three chimeric proteins (Supplemental Table 1). We 1) replaced the RHD of MCTP2 with the RHD of RTN4a; 2) replaced the RHD of MCTP2 with the TMD of SEC61β, which does not generate ER tubules; or 3) replaced the C2 domains of MCTP2 with the LD-binding C-nexin (CN) domain of SNX14 (Supplemental Figure S4A). SNX14 was recently shown to promote LD growth at ER–LD contacts via a CN domain that is essential for binding to the LDs (Datta et al., 2019). Upon overexpression of these chimeric proteins, similar to full-length MCTP2, we observed a significant increase in LD number (Supplemental Figure S4B) and size (Supplemental Figure S4C) when the RHD of MCTP2 was replaced with the RHD of RTN4a. In contrast, replacing the RHD of MCTP2 with the TMD of SEC61β resulted in only a moderate increase in LD number (Supplemental Figure S4B). We also observed an increase in LD size and ER–LD contact upon overexpression of the chimera protein where the C2 domains of MCTP2 were replaced with the CN domain of SNX14 (Supplemental Figure S4, C and D). These results are consistent with a model in which the MCTP2 RHD domain increases LD number by tubulating the ER membrane, while the C2 domains increase LD size by promoting ER–LD tethering.
Finally, we determined the effect of depleting MCTPs on LD number, size, and contact with the ER (Figure 4, D–F). As expected, knockdown of either MCTP1 or MCTP2 decreased LD size and ER–LD contact (Figure 4, E and F). This suggests nonredundant roles of MCTPs in regulating LD size and tethering to the ER. Depletion of both MCTP1 and MCTP2 led to even more dramatic decreases in the size of LDs and in ER–LD contact. Surprisingly, we found that LD number increased upon depletion of MCTP2 or MCTP1 and MCTP2 (Figure 4D). This increase in LD number could be to compensate for the decrease in LD size. Our results suggest that depletion of MCTPs does not affect total neutral lipid synthesis, so that as LD size decreases with MCTP depletion, LD number increases.
Proposed model for the role of MCTPs in LD biogenesis
In this report, we show that MCTP1 and MCTP2 are ER membrane proteins that have the ability to tubulate the ER membrane in mammalian cells. Both MCTP1-GFP and MCTP2-GFP formed punctae in the ER membrane at lower expression (Figures 1E and 2, A and C) and at endogenous levels (Figure 3, A and E). We show that these punctae are at sites in the ER membrane where new LDs form (Figure 2, A and C, and Supplemental Figure S2). We also show that overexpression of these proteins leads to an increase in LD number and size (Figure 4, A and B; Supplemental Figure S3, A and B), which is consistent with the previous finding that depletion of MCTP2 in HeLa cells and MCTP in C. elegans leads to a decrease in the number and size of LDs (Joshi et al., 2018). In fact, depletion of both MCTP1 and MCTP2 has an additive effect on LD size, as smaller LDs were observed in these cells along with decreased ER–LD contact (Figure 4, E and F).
How do MCTPs regulate formation and growth of new LDs? We propose a model in which MCTPs localize as punctae in discrete regions of the ER membrane. The localized membrane bending by the RHD of MCTPs facilitates accumulation of TAG as determined by LiveDrop and Plin3, the earliest markers of LD formation (Figure 2, A and C, and Supplemental Figure S2) (Chung et al., 2019). The ER membrane curvature facilitates accumulation of TAG and modulates LD number (Santinho et al., 2020). Accumulation of TAG within the bilayer of the ER membrane is the initiation step for LD formation. Once LDs are formed, the C2 domains keep the ER in contact with LDs by interacting with phospholipids on the LD surface. This may facilitate lipid transfer from the ER, where lipids are synthesized, to LDs, promoting LD growth (Figure 4G).
MCTP2 is at ER contact sites with multiple organelles
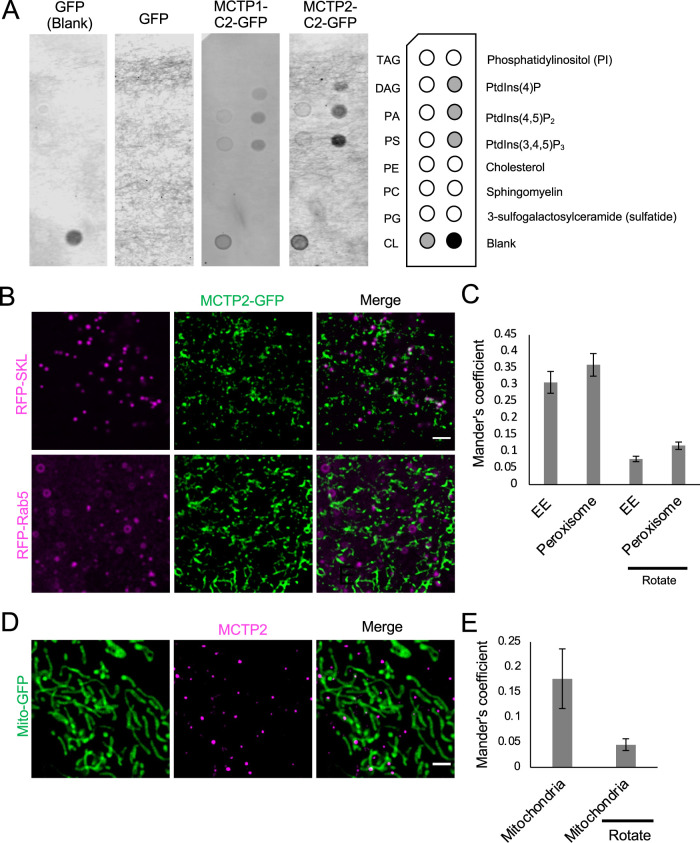
The C2 domains of other proteins such as E-Syts and dysferlin have been reported to bind to charged phospholipids (Therrien et al., 2009; Giordano et al., 2013). The C2 domains of MCTPs do not bind to major phospholipids; however, whether they bind to different species of phosphoinositides was not investigated (Shin et al., 2005). We used a lipid-protein overlay assay to check the binding of C2 domains to different lipids. We found that the C2 domains of MCTP1 and MCTP2 can bind to phosphatidylinositol-4-phosphate (PI4P), phosphatidylinositol-4,5-bisphosphate (PI4,5P2), phosphatidylinositol-3,4,5-trisphosphate (PI3,4,5P3), and the mitochondrial phospholipid cardiolipin (CL) (Figure 5A). Our results are consistent with a recent report suggesting that Arabidopsis thaliana MCTP4 binds to PI4P in the plasma membrane via its C2 domains (Brault et al., 2019). Moreover, the presence of PI4P on the LDs was recently reported (Du et al., 2020). Therefore, we propose that the C2 domains of MCTP1 and MCTP2 proteins could bind PI4P on the surface of LDs via its C2 domains.
FIGURE 5:
MCTP2 is at ER contact sites with multiple organelles. (A) Lipid binding of C2 domains of MCTPs. List of lipids found on the Echelon lipid strip (on the right). Lysate of cells expressing GFP only or GFP-tagged C2 domains of MCTP1 or MCTP2. Lipid-protein interactions were revealed with anti-GFP antibody. (B) Airyscan images of live cells transfected with mCherry-Rab5 or RFP-SKL and MCTP2-GFP. Scale bar, 2 µm. (C) Quantification of colocalization of mCherry-Rab5 or RFP-SKL and MCTP2-GFP by Mander’s colocalization coefficient. Error bars indicate mean ± SE, n = 22 and 28 fields of view for RFP-SKL and mCherry-Rab5, respectively. (D) Airyscan images of fixed cells transfected with Mito-GFP and stained for MCTP2. Scale bar, 2 µm. (E) Quantification of colocalization of MCTP2 punctae and Mito-GFP by Mander’s colocalization coefficient. Error bars indicate mean ± SE, n = 23 fields of view.
We show that a fraction of endogenous MCTP2 is associated with seipin and LDs (Figure 3F). However, there are also MCTP2 punctae that are not associated with LDs. A recent study in A. thaliana has reported atMCTP4 as a molecular tether between the ER and plasma membrane (Brault et al., 2019). Moreover, our finding that MCTP C2 domains bind to several species of PIs and CL suggested that MCTPs may localize to other organelles in addition to LDs. We tested whether MCTP2 punctae at ER subdomains contact peroxisomes, endosomes, or mitochondria. Using live-cell imaging, we found that MCTP2-GFP colocalized with peroxisomes (RFP-SKL) and early endosomes (mCherry-Rab5) with Mander’s coefficients of 0.36 and 0.30, respectively (Figure 5, B and C). We confirmed that this colocalization was not random as the Mander’s coefficient decreased to 0.07 and 0.11 upon clockwise rotation of peroxisome and early endosome images (Figure 5C). Using immunofluorescence, we found that endogenous MCTP2 formed punctae that colocalized with mitochondria with a Mander’s coefficient of 0.17 (Figure 5, D and E). This colocalization was not random as the Mander’s coefficient decreased to 0.04 upon clockwise rotation of mitochondria images (Figure 5E). Our results suggest that MCTP2 is localized at ER contact sites with multiple organelles, including LDs, peroxisome, early endosomes, and mitochondria. This observation raises the intriguing possibility that MCTPs function more broadly to link membrane tubulation at ER subdomains with organelle contact sites. These subdomains function in the biogenesis of LDs and may also function in the biogenesis of other organelles such as peroxisomes (Joshi et al., 2018). In addition, MCTP subdomains could play roles in organelle fission and fusion, as the ER has been implicated in the fission of mitochondria and endosomes, as well as mitochondrial fusion (Friedman et al., 2011; Rowland et al., 2014; Hoyer et al., 2018; Abrisch et al., 2020). High curvature of the ER membrane may be necessary for budding of new organelles, while ER tubules wrap around endosomes and mitochondria at sites of division. If MCTPs are present at multiple ER–organelle contact sites, this could provide a mechanism for coordinated regulation of organelle metabolism, biogenesis, and division.
MATERIALS AND METHODS
Cell culture and reagents
COS-7 and HeLa cells (UNC Lineberger Tissue Culture Facility) were maintained at 37°C with 5% CO2 in culture media (CM) DMEM (Corning 15-013-CV) containing 10% fetal bovine serum (FBS; VWR97068-085), 2 mM l-glutamine (Corning 25005CI), and 1× penicillin/streptomycin (Corning; 30-002-CI). For LD induction in Figure 2, A and C, and Supplemental Figure S2, cells were incubated in starvation media containing DMEM, l-glutamine, pyruvate, and nonessential amino acids but in the absence of serum for 16–20 h. This was followed by the addition of 400 µM of sodium-oleate (Sigma; O7501) in imaging media (IM; same as CM except that the DMEM has no phenol red [Corning 17-205-CV]) (Kassan et al., 2013; Li et al., 2019). For staining of Halo tags, cells were incubated with 50 nM of Janelia Fluor dye (JF646, kind gift from Luke Lavis, Janelia Research Campus) for 30 min followed by one wash with phosphate-buffered saline (PBS) (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH adjusted to 7.4) and replacement with appropriate media. For LD staining, BODIPY493/503 (Invitrogen; D3922) and BODIPY665/676 (Invitrogen; B3932) were used.
Plasmids
Plasmids used are listed in Supplemental Table 1. Full-length MCTP1 and MCTP2 were tagged with GFP at the C-terminus by cloning in the pYEMc-eGFP vector. MCTP1 and MCTP2 were inserted in the BamHI- and KpnI-digested pYEMc-eGFP plasmid by targeted homologous recombination using budding yeast. All other plasmids were generated using HiFi DNA Assembly Master mix (E2621; New England Biolabs). For plasmid construction, all PCRs were performed using Q5 High Fidelity DNA polymerase (M0419; New England Biolabs) and restriction enzymes were from New England Biolabs. The following plasmids were kind gifts: pYEMc-eGFP from Alexandre Toulmay (NIH), mCherry-Rab5 (Addgene #49201), mCherry-CLIMP63 and RTN4a-GFP (Addgene #61807) from Gia Voeltz (University of Colorado, Boulder), mApple-LiveDrop from Chi-Lun Chang (St. Jude Children’s Research Hospital), RFP-SKL from Peter Kim (The Hospital for Sick Children, University of Toronto), and HaloTag-C1 from Chris Obara (Janelia Research Campus).
Generation of MCTP2 antibody
Rabbit polyclonal MCTP2 antibody was generated by Yenzyme Antibodies. Rabbits were immunized with a peptide consisting of 18 amino acids (CQGDFKRHRWSNRKRLSAS-amide) of MCTP2 (317–334) with an additional cysteine at the N-terminus to generate polyclonal antibodies.
Transfection and immunofluorescence
For live-cell imaging, COS-7 cells were cultured in an eight-well chambered cover glass (Cellvis, Mountain View, CA) coated with 10 μg/ml fibronectin (Millipore, Burlingon, MA). Cells were transfected using Lipofectamine 2000 with appropriate plasmids (100–150 ng) and incubated with BODIPY665/676 (100 ng/ml; Life Technologies) for 16 h to label LDs. For immunofluorescence microscopy, COS-7 cells were cultured on #1.5, 12-mm-diameter coverslips (Electron Microscopy Sciences; 7223001) coated with 10 μg/ml poly-d-lysine (Millipore A-003-E). Cells were fixed 24 h posttransfection with 4% paraformaldehyde (Electron Microscopy Sciences; 15710) in PBS for 15 min and permeabilized and blocked in 10% normal donkey serum, 1% bovine serum albumin (BSA, Fisher Scientific, BP9703), and 0.1% saponin in PBS for 45 min at room temperature. Cells were washed with 0.1% saponin in PBS and probed with rabbit anti-MCTP2 (1:1000) and mouse anti-BSCL2 (1:2000; Abnova; H00026580-A02) overnight at 4°C in 1% BSA and 0.1% saponin in PBS. Cells were washed with 0.1% saponin in PBS and incubated with fluorescently labeled secondary antibodies for 1 h at room temperature in 1% BSA and 0.1% saponin in PBS. For Figure 3A, we used donkey anti-rabbit Alexa Flour Plus 488 (1:2000; Abcam; ab150073) and donkey anti-mouse Alexa Fluor Plus 647 (1:1000; Invitrogen; A32787) to stain MCTP2 and seipin, respectively. For Figure 3E, cells were incubated for 1 h with 50 ng/ml BODIPY493/503 (Invitrogen; D3922), concomitant with incubation with secondary antibodies. The secondary antibodies used were donkey anti-rabbit Alexa Fluor 568 (1:500; Invitrogen; A10042) for staining MCTP2 and donkey anti-mouse Alexa Fluor Plus 647 (1:1000; Invitrogen; A32787) to stain seipin. Cells were then washed with PBS and mounted onto slides using SlowFade Diamond Antifade Mountant (Invitrogen).
Image acquisition
Images were acquired on an inverted Zeiss 800/Airyscan laser scanning confocal microscope equipped with 405, 488, 561, and 647 nm diode lasers and galium arsenide phosphid (GaAsP) and Airyscan detectors. Confocal images were acquired using a 63×/1.4 NA objective lens, at 37°C and 5% CO2 (Carl Zeiss, Oberkochen, Germany). For Figures 1E, 2, A and C, 3, A and E, and 5, B and D, and Supplemental Figure S2, Airyscan images were taken using 3.5× magnification. These images were processed by Airyscan processing using the Zeiss ZEN software package.
FRAP
FRAP experiments were conducted on a Zeiss LSM 800 laser scanning inverted microscope using Plan Apo63×/1.4 oil objective with laser line 488 nm. LSM software ZEN was used for image acquisition and analysis. For photobleaching, the tubular ER was magnified using the 3.5× zoom function so that individual tubules could be seen clearly. Images with the region of interest of 11 × 11 pixels were bleached at 100% laser power until the intensity reached 50% of original intensity. After photobleaching, images were taken at 1 s intervals for 130–150 s during recovery. For FRAP of MCTPs, cells that exhibited uniform distribution of MCTPs in the ER membrane were selected.
Image analysis
For image analysis in Figure 4 and Supplemental Figures S3 and S4, we used a custom Matlab pipeline to quantify LD number, size, and ER contacts as previously described (Valm et al., 2017; Arribat et al., 2020). Matlab code is available at https://github.com/TimXQi/Cohen-Lab. This pipeline was used to generate cell and organelle masks through median or Gaussian blurring, Otsu thresholding, and watershed segmentation when appropriate. Organelle number and size were calculated from the corresponding organelle masks. To quantify LD–ER contacts, LDs were dilated by one pixel and an overlap image between LDs and ER was generated. This image was used to calculate the total number of LD–ER contacts/cell, as well as the fraction of LDs in contact with the ER. ImageJ was used for image analysis in all other figures. For Figure 1D, we used ImageJ to quantify % ER sheet as previously described (English and Voeltz, 2013). For Figures 3C and 5, C and E, we used JACoP to measure colocalization (Bolte and Cordelières, 2006).
Immunoprecipitation and immunoblotting
For immunoprecipitation in Figure 3D, we used a protocol provided by Chromotek for GFP-trap Dynabeads with some modifications. Briefly, ∼106–107 cells transfected with 3 µg of MCTP1-GFP or MCTP2-GFP were lysed in 500 µl ice-cold lysis buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 1% digitonin, and cOmplete protease Inhibitor Cocktail tablet, EDTA-Free). After solubilization of membrane at 4°C for 1.5 h with lysate rocking, the cell lysates were centrifuged at 17,000 × g for 10 min at 4°C, and the supernatants were collected. Fifty microliters of the supernatant was used as input fraction. The remaining supernatant was incubated with 25 µl of GFP-trap Dynabeads (ChromoTek; gtd-10) in a cold room for 1.5 h. Dynabeads were separated using magnet until the supernatant was clear. The supernatant was stored as flow through (FT). The bead-bound materials were washed with 500 µl of washing buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% digitonin) three times and were eluted with Laemmli sample buffer for 5 min at 95°C before SDS–PAGE. For immunoblotting analysis, proteins were separated by 8% SDS–PAGE gels, transferred to 0.2-micron nitrocellulose membrane (Bio-Rad; 1620112), and analyzed using primary antibodies GFP (1:1000; Fisher; A10262) and BSCL2 (1:2000; Abnova; H00026580-A02). Proteins were analyzed using IRDye secondary antibody (1:10,000; LI-COR Biosciences) followed by detection using the Odyssey CLx system.
Knockdown of MCTP1 and MCTP2
For knockdown, sets of four individual small interfering RNA (siRNA) for MCTP1 (MQ-016557-01-0002) and MCTP2 (MQ-020810-01-0002), as well as the nontargeting siRNA (D-001206-14-050), were purchased from Horizon Discovery. For siRNA transfection, 1 × 104 HeLa cells were cultured in an eight-chambered cover glass dish (Cellvis; C8-1.5H-N) coated with 10 µg/ml fibronectin (Millipore, Burlington, MA) and left overnight at 37°C with 5% CO2. Cells were transfected according to the manufacturer’s instructions with 5 pmol control siRNA, 5 pmol MCTP1 or MCTP2 pooled siRNA, or both (10 pmol total) for double knockdown using the DharmaFECT 1 transfection reagent (Horizon Discovery; T-2001-01). The knockdown of MCTP1 and MCTP2 was determined by checking the expression of MCTP1-GFP and MCTP2-GFP using fluorescence microscopy.
Protein-lipid overlay assay
Lipid-binding specificity was assessed with protein-lipid overlay assays using commercially available lipid strips blotted with 100 pmol of biologically relevant lipids (Echelon Biosciences; P-6002). Cells were lysed using lysis buffer (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, 0.5% NP-40 and EDTA-Free cOmplete protease inhibitor Cocktail tablet [Thermo Scientific; A32955]). Lipid strips were blocked with 1% nonfat milk in PBS for 45 min followed by a 1 h incubation at room temperature with 1 mg of lysate of cells expressing only GFP or C2 domains of either MCTP1 or MCTP2 proteins tagged with GFP at the C-terminus. Next, the blots were washed three times with PBS containing 0.01% Tween-20 (PBST) for 5 min followed by incubation with a chicken anti-GFP antibody (1:5000; Invitrogen; A10262) in PBST overnight at 4°C. The membranes were washed with PBST three times and incubated using IRDye secondary antibody (1:10,000; LI-COR Biosciences) followed by detection using the Odyssey CLx system. The protein-lipid interactions were detected using the Odyssey CLx system.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under award number R35 GM133460 (S.C.) and by the intramural research program of the National Institute of Diabetes and Digestive and Kidney Diseases (W.A.P.).
Abbreviations used:
- ER
endoplasmic reticulum
- LDs
lipid droplets
- OA
oleic acid
- PIPs
phosphoinositides
- RHD
reticulon homology domain
- TMD
transmembrane domain.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-09-0590) on April 7, 2021.
REFERENCES
- Abrisch RG, Gumbin SC, Wisniewski BT, Lackner LL, Voeltz GK (2020). Fission and fusion machineries converge at ER contact sites to regulate mitochondrial morphology. J Cell Biol 219, e201911122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva V, Nam S-Z, Söding J, Lupas AN (2016). The MPI bioinformatics Toolkit as an integrative platform for advanced protein sequence and structure analysis. Nucleic Acids Res 44, W410–W415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arribat Y, Grepper D, Lagarrigue S, Qi T, Cohen S, Amati F (2020). Spastin mutations impair coordination between lipid droplet dispersion and reticulum. PLoS Genet 16, e1008665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte S, Cordelières FP (2006). A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224, 213–232. [DOI] [PubMed] [Google Scholar]
- Brault ML, Petit JD, Immel F, Nicolas WJ, Glavier M, Brocard L, Gaston A, Fouché M, Hawkins TJ, Crowet J, et al. (2019). Multiple C2 domains and transmembrane region proteins (MCTPs) tether membranes at plasmodesmata. EMBO Rep 20, e47182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V, El Atab O, Mizzon G, Prinz WA, Schneiter R (2020). Seipin and Nem1 establish discrete ER subdomains to initiate yeast lipid droplet biogenesis. J Cell Biol 219, e201910177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary V, Ojha N, Golden A, Prinz WA (2015). A conserved family of proteins facilitates nascent lipid droplet budding from the ER. J Cell Biol 211, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Wu X, Lambert TJ, Lai ZW, Walther TC, Farese RV (2019). LDAF1 and seipin form a lipid droplet assembly complex. Dev Cell 51, 551–563.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S (2018). Lipid droplets as organelles. Int Rev Cell Mol Biol 337, 83–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Liu Y, Hariri H, Bowerman J, Henne WM (2019). Cerebellar ataxia disease–associated Snx14 promotes lipid droplet growth at ER–droplet contacts. J Cell Biol 218, 1335–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C, Koch J, Oeljeklaus S, Laernsack A, Melchior S, Wiese S, Schummer A, Erdmann R, Warscheid B, Brocard C (2013). A combined approach of quantitative interaction proteomics and live-cell imaging reveals a regulatory role for endoplasmic reticulum (ER) reticulon homology proteins in peroxisome biogenesis. Mol Cell Proteomics 12, 2408–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du X, Zhou L, Aw YC, Mak HY, Xu Y, Rae J, Wang W, Zadoorian A, Hancock SE, Osborne B, et al. (2020). ORP5 localizes to ERlipid droplet contacts and regulates the level of PI(4)P on lipid droplets. J Cell Biol 219, e201905162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elíes J, Yáñez M, Pereira TMC, Gil-Longo J, MacDougall DA, Campos-Toimil M (2020). An update to calcium binding proteins. Adv Exp Med Biol 1131, 183–213. [DOI] [PubMed] [Google Scholar]
- English AR, Voeltz GK (2013). Rab10 GTPase regulates ER dynamics and morphology. Nat Cell Biol 15, 169–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, Yang H (2008). Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J Cell Biol 180, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK (2011). ER tubules mark sites of mitochondrial division. Science 334, 358–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P (2013). PI(4,5)P2-dependent and Ca2+-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153, 1494–1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grippa A, Buxó L, Mora G, Funaya C, Idrissi F-Z, Mancuso F, Gomez R, Muntanyà J, Sabidó E, Carvalho P (2015). The seipin complex Fld1/Ldb16 stabilizes ER–lipid droplet contact sites. J Cell Biol 211, 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Reese ML, Goodman JM (2018). The assembly of lipid droplets and their roles in challenged cells. EMBO J 37, e98947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyer MJ, Chitwood PJ, Ebmeier CC, Striepen JF, Qi RZ, Old WM, Voeltz GK (2018). A novel class of ER membrane proteins regulates ER-associated endosome fission. Cell 175, 254–265.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Mishra S, Choudhary V, Schneiter R (2013). Expression of oleosin and perilipins in yeast promotes formation of lipid droplets from the endoplasmic reticulum. J Cell Sci 126, 5198–5209. [DOI] [PubMed] [Google Scholar]
- Joshi AS, Cohen S (2019). Lipid droplet and peroxisome biogenesis: do they go hand-in-hand? Front Cell Dev Biol 7, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Huang X, Choudhary V, Levine TP, Hu J, Prinz WA (2016). A family of membrane-shaping proteins at ER subdomains regulates pre-peroxisomal vesicle biogenesis. J Cell Biol 215, 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Nebenfuehr B, Choudhary V, Satpute-Krishnan P, Levine TP, Golden A, Prinz WA (2018). Lipid droplet and peroxisome biogenesis occur at the same ER subdomains. Nat Commun 9, 2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Zhang H, Prinz WA (2017). Organelle biogenesis in the endoplasmic reticulum. Nat Cell Biol 19, 876–882. [DOI] [PubMed] [Google Scholar]
- Kassan A, Herms A, Fernández-Vidal A, Bosch M, Schieber NL, Reddy B, Fajardo A, Gelabert-Baldrich M, Tebar F, Enrich C, et al. (2013). Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol 203, 985–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Farese RV, Walther TC (2013). Balancing the fat: lipid droplets and human disease. EMBO Mol Med 5, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon MA (2008). Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 9, 99–111. [DOI] [PubMed] [Google Scholar]
- Li D, Zhao YG, Li D, Zhao H, Huang J, Miao G, Feng D, Liu P, Li D, Zhang H (2019). The ER-localized protein DFCP1 modulates ER-lipid droplet contact formation. Cell Rep 27, 343–358.e5. [DOI] [PubMed] [Google Scholar]
- Magré J, Delépine M, Khallouf E, Gedde-Dahl Jr T, Van Maldergem L, Sobel E, Papp J, Meier M, Mégarbané A, BSCL Working Group, et al. (2001). Identification of the gene altered in Berardinelli-Seip congenital lipodystrophy on chromosome 11q13. Nat Genet 28, 365–370. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Carvalho P (2019). Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 20, 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowland AA, Chitwood PJ, Phillips MJ, Voeltz GK (2014). ER contact sites define the position and timing of endosome fission. Cell 159, 1027–1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Belevich I, Li S, Karhinen L, Vihinen H, Vigouroux C, Magré J, Thiele C, Hölttä-Vuori M, Jokitalo E, et al. (2016). Seipin regulates ER–lipid droplet contacts and cargo delivery. EMBO J 35, 2699–2716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salo VT, Li S, Vihinen H, Hölttä-Vuori M, Szkalisity A, Horvath P, Belevich I, Peränen J, Thiele C, Somerharju P, et al. (2019). Seipin facilitates triglyceride flow to lipid droplet and counteracts droplet ripening via endoplasmic reticulum contact. Dev Cell 50, 478–493.e9. [DOI] [PubMed] [Google Scholar]
- Santinho A, Salo VT, Chorlay A, Li S, Zhou X, Omrane M, Ikonen E, Thiam AR (2020). Membrane curvature catalyzes lipid droplet assembly. Curr Biol 30, 2481–2494.e6. [DOI] [PubMed] [Google Scholar]
- Schuck S, Prinz WA, Thorn KS, Voss C, Walter P (2009). Membrane expansion alleviates endoplasmic reticulum stress independently of the unfolded protein response. J Cell Biol 187, 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Shemesh T, Prinz WA, Palazzo AF, Kozlov MM, Rapoport TA (2010). Mechanisms determining the morphology of the peripheral ER. Cell 143, 774–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Voss C, Rist JM, Hu J, Rapoport TA, Prinz WA, Voeltz GK (2008). The reticulon and Dp1/Yop1p proteins form immobile oligomers in the tubular endoplasmic reticulum. J Biol Chem 283, 18892–18904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin O-H, Han W, Wang Y, Südhof TC (2005). Evolutionarily conserved multiple C 2 domain proteins with two transmembrane regions (MCTPs) and unusual Ca 2+ binding properties. J Biol Chem 280, 1641–1651. [DOI] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li WP, Agarwal AK, Garg A, Anderson RGW, Goodman JM (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc Natl Acad Sci USA 104, 20890–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therrien C, Di Fulvio S, Pickles S, Sinnreich M (2009). Characterization of lipid binding specificities of dysferlin C2 domains reveals novel interactions with phosphoinositides. Biochemistry 48, 2377–2384. [DOI] [PubMed] [Google Scholar]
- Thiam AR, Forêt L (2016). The physics of lipid droplet nucleation, growth and budding. Biochim Biophys Acta 1861, 715–722. [DOI] [PubMed] [Google Scholar]
- Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott-Schwartz J (2017). Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 546, 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA (2006). A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell 124, 573–586. [DOI] [PubMed] [Google Scholar]
- Walther TC, Chung J, Farese RV (2017). Lipid droplet biogenesis. Annu Rev Cell Dev Biol 33, 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV (2012). Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81, 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, et al. (2016). Seipin is required for converting nascent to mature lipid droplets. eLife 5, e16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Idrissi F-Z, Hermansson M, Grippa A, Ejsing CS, Carvalho P (2018). Seipin and the membrane-shaping protein Pex30 cooperate in organelle budding from the endoplasmic reticulum. Nat Commun 9, 2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, de Boer R, Krikken AM, Aks˛it A, Bordin N, Devos DP, van der Klei IJ (2020). Pex24 and Pex32 are required to tether peroxisomes to the ER for organelle biogenesis, positioning and segregation in yeast. J Cell Sci 133, jcs246983. [DOI] [PubMed] [Google Scholar]
- Xu S, Zhang X, Liu P (2018). Lipid droplet proteins and metabolic diseases. Biochim Biophys Acta 1864, 1968–1983. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.