ABSTRACT
Gardnerella spp. in the vaginal microbiome are associated with bacterial vaginosis, in which a lactobacillus-dominated community is replaced with mixed bacteria, including Gardnerella species. Co-occurrence of multiple Gardnerella species in the vaginal environment is common, but different species are dominant in different women. Competition for nutrients, including glycogen, could play an important role in determining the microbial community structure. Digestion of glycogen into products that can be taken up and further processed by bacteria requires the combined activities of several enzymes collectively known as amylases, which belong to glycoside hydrolase family 13 (GH13) within the CAZy classification system. GH13 is a large and diverse family of proteins, making prediction of their activities challenging. SACCHARIS annotation of the GH13 family in Gardnerella resulted in identification of protein domains belonging to eight subfamilies. Phylogenetic analysis of predicted amylase sequences from 26 genomes demonstrated that a putative α-glucosidase-encoding sequence, CG400_06090, was conserved in all Gardnerella spp. The predicted α-glucosidase enzyme was expressed, purified, and functionally characterized. The enzyme was active on a variety of maltooligosaccharides with maximum activity at pH 7. Km, kcat, and kcat/Km values for the substrate 4-nitrophenyl α-d-glucopyranoside were 8.3 μM, 0.96 min−1, and 0.11 μM−1 min−1, respectively. Glucose was released from maltose, maltotriose, maltotetraose, and maltopentaose, but no products were detected when the enzyme was incubated with glycogen. Our findings show that Gardnerella spp. produce an α-glucosidase enzyme that may contribute to the multistep process of glycogen metabolism by releasing glucose from maltooligosaccharides.
IMPORTANCE Increased abundance of Gardnerella spp. is a diagnostic characteristic of bacterial vaginosis, an imbalance in the human vaginal microbiome associated with troubling symptoms, and negative reproductive health outcomes, including increased transmission of sexually transmitted infections and preterm birth. Competition for nutrients is likely an important factor in causing dramatic shifts in the vaginal microbial community but little is known about the contribution of bacterial enzymes to the metabolism of glycogen, a major carbon source available to vaginal bacteria. The significance of our research is characterizing the activity of an enzyme conserved in Gardnerella species that likely contributes to the ability of these bacteria to utilize glycogen.
KEYWORDS: Gardnerella, alpha-glucosidase, glycogen, glycoside hydrolase, vaginal microbiome
INTRODUCTION
Gardnerella spp. in the vaginal microbiome are hallmarks of bacterial vaginosis, a condition characterized by replacement of the lactobacillus-dominated microbial community with mixed aerobic and anaerobic bacteria, including Gardnerella. This change in the microbial community can be associated with increased vaginal pH, malodorous discharge, and the presence of biofilm (1). In addition to troubling symptoms, the presence of non-Lactobacillus-dominated vaginal microbiota is associated with increased risk of HIV transmission and infection with other sexually transmitted pathogens, such as Neisseria gonorrhoeae and Trichomonas spp. (2, 3). Historically, Gardnerella has been considered a single-species genus. Paramel Jayaprakash et al. used cpn60 barcode sequences to divide Gardnerella spp. into four subgroups (A to D) (4), and this framework was supported by whole-genome sequence comparison (5, 6). More recently, Vaneechoutte et al. emended the classification of Gardnerella based on whole-genome sequence comparison, biochemical properties, and matrix-assisted laser desorption ionization–time-of-flight mass spectrometry and proposed the addition of three novel species: Gardnerella leopoldii, Gardnerella piotii, and Gardnerella swidsinskii (7).
Colonization with multiple Gardnerella species is common, and different species are dominant in different women (8). Understanding factors that contribute to differential abundance is important, since the species may differ in virulence (6) and they are variably associated with clinical signs (8, 9). Several factors, including interspecific competition, biofilm formation, and resistance to antimicrobials, could contribute to the differential abundance of the different species. Khan et al. showed that resource-based scramble competition is frequent among Gardnerella subgroups (10). In a scramble competition, one competitor outgrows the others through its superior ability to use shared resources, such as nutrients (11). Unlike the gut microbiome, where members of the microbiota have access to nutrients from digesta as well as host-derived resources, nutrients in the vaginal environment are largely host-derived. Gardnerella spp. have been shown to utilize a variety of carbohydrates and amino acids (12) and to engage in foraging of mucosal sialoglycans (13).
Glycogen is another nutrient available in the vagina, but previous reports on the growth of Gardnerella spp. on glycogen-containing media are inconsistent (14). Species may differ in their ability to digest glycogen and utilize the breakdown products, which may in turn contribute to determining microbial community structure. Glycogen is an energy storage molecule which consists of linear chains of approximately 13 glucose molecules covalently linked with α-1,4 glycosidic linkages, with branches attached through α-1,6 glycosidic bonds (15, 16). A single glycogen molecule consists of approximately 55,000 glucose residues with a molecular mass of ∼107 kDa (17). The size of glycogen particles can vary with source; from 10 to 44 nm in human skeletal muscle to approximately 110 to 290 nm in human liver (18). Glycogen is accumulated in vaginal epithelial cells under the influence of estrogen (19) and is released into the vaginal lumen when superficial vaginal epithelial cells are sloughed off and the cytolytic actions of vaginal bacteria lyse the epithelial cells (20). The concentration of cell-free glycogen in vaginal fluid varies greatly, ranging from 0.1 to 32 μg/μl (21). This long polymeric molecule must be digested into smaller components that can be taken up by bacterial cells and metabolized further (22, 23).
Glycogen digestion is accomplished by the coordinated action of enzymes collectively described as amylases (24). A large majority of these enzymes belong to family 13 within the glycosyl hydrolase class (GH13) of carbohydrate-active enzymes (CAZymes) (25). GH13 enzymes are further classified into more than 43 subfamilies based on structure and activity (26). For example, subfamily 23 is primarily composed of α-glucosidases, which are exo-acting enzymes that act on α-1,4 glycosidic bonds from the nonreducing end to release glucose; subfamily 32 is composed of α-amylases, endo-acting enzymes that cleave α-1,4 glycosidic bonds to produce maltose and α-limit dextrin; and subfamily 14 contains debranching enzymes, such as pullulanase, which target α-1,6 glycosidic bonds (18, 27). Limited information is available on glycogen degradation mechanisms in the vaginal microbiome. Although it has been demonstrated that vaginal secretions exhibit amylase activity that can degrade glycogen (23, 28), the role of bacterial amylase enzymes in this process is not clear, and there is limited information about glycogen metabolism by clinically important bacteria such as Gardnerella spp.
The objective of the current study was to annotate GH13 enzymes in Gardnerella spp. and to characterize the activity of a putative α-glucosidase conserved among all Gardnerella spp.
RESULTS
Identification of putative amylase sequences.
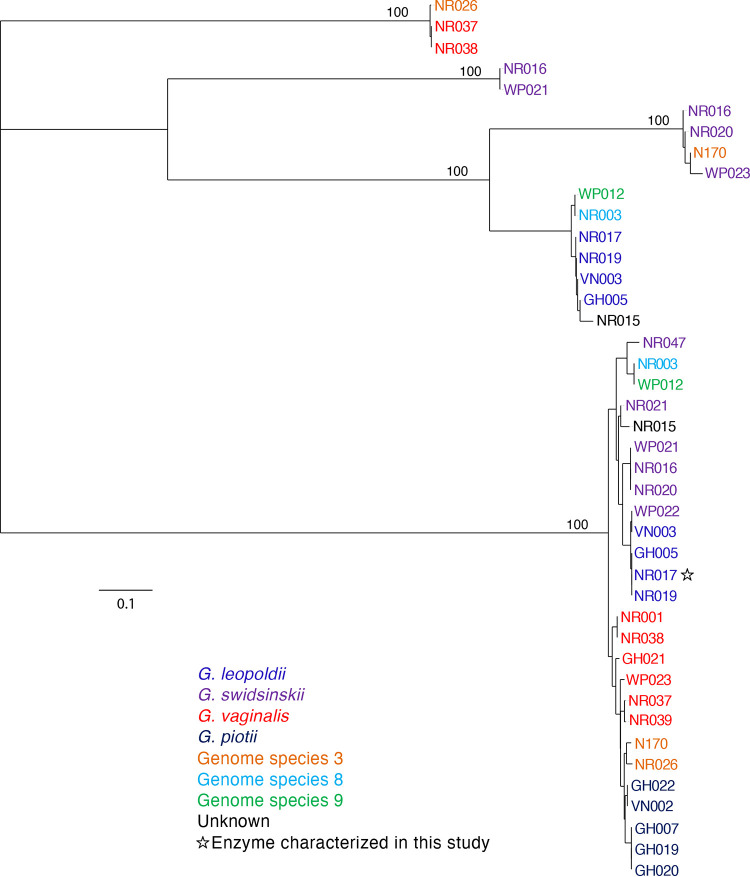
A total of 60 proteins from the 26 study isolates were annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) as amylases. After removal of severely truncated sequences, a multiple-sequence alignment of the remaining 42 sequences was trimmed to the uniform length of 290 amino acids. Phylogenetic analysis of these predicted amylase sequences revealed five clusters, but only one of these clusters contained orthologous sequences from all 26 isolates (Fig. 1). Sequence identities within this cluster were all 91 to 100% identical. This conserved sequence (corresponding to GenBank accession no. WP_004104790) was annotated as an GH13 α-amylase (EC 3.2.1.1) in all genomes, suggesting that it was an endo-acting enzyme that releases maltooligosaccharides from glycogen.
FIG 1.
Phylogenetic analysis of predicted extracellular amylases protein sequences from 26 Gardnerella genomes. Trees are consensus trees of 100 bootstrap iterations. Bootstrap values are shown for major branch points. Species is indicated by label color according to the legend.
The GH13 family in the CAZy database is a large, polyspecific family targeting α-glucosyl linkages. The functional diversity of members in this family can make annotation of new sequences challenging. In order to predict the activity of the conserved protein identified among the study isolates, we employed SACCHARIS, which combines CAZyme family trees generated from biochemically characterized proteins with related sequences of unknown function. Currently, two Gardnerella reference genomes are available in CAZy: Gardnerella vaginalis ATCC 14018 and G. swidsinskii GV37. GH13 family proteins encoded in these Gardnerella genomes were identified, and a family tree including all characterized GH13 sequences in the CAZy database was generated (see Fig. S1 in the supplemental material). SACCHARIS annotation of the GH13 family in Gardnerella resulted in identification of protein domains belonging to eight subfamilies (2, 9, 11, 14, 20, 23, 31, and 32). Alignment with characterized GH13 enzymes from the CAZy database (26) suggested that the conserved amylase identified in the study isolates was an α-glucosidase (EC 3.2.1.20), closely related to subfamilies 23 and 30 of GH13, and not an α-amylase as annotated in GenBank. Subsequent examination of the α-glucosidase with SignalP and SecretomeP indicated that no signal peptide was present and that the protein was not likely secreted through a Sec-independent pathway.
Relationship to other α-glucosidases.
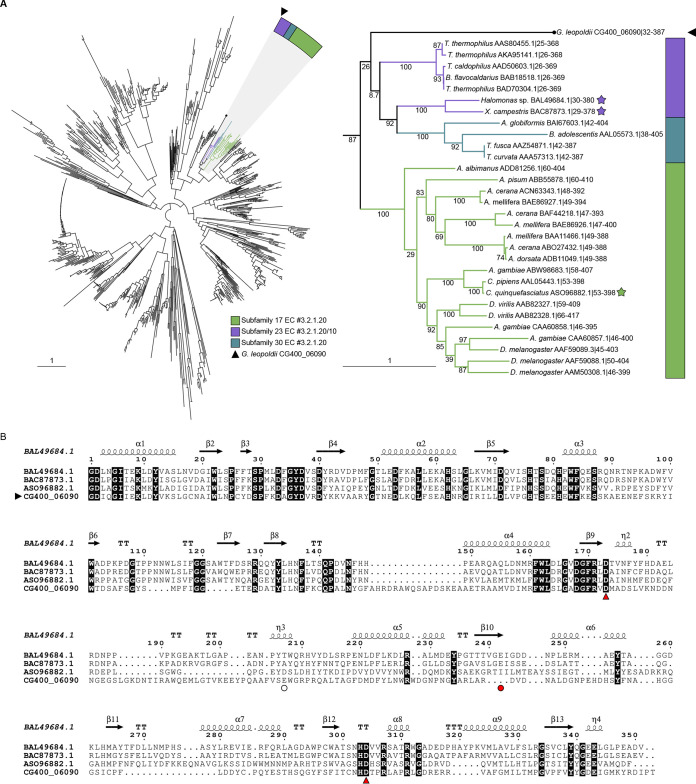
A representative sequence of the predicted α-glucosidase was selected from G. leopoldii (NR017). This sequence (CG400_06090) was combined with functionally characterized members of the GH13 CAZy database using SACCHARIS (Fig. 2A). CG400_06090 partitioned with members of GH13 subfamilies 17, 23, and 30, of which the prominent activity is α-glucosidase. This clade was expanded to clarify the relationship of CG400_06090 with related characterized sequences as an apparently deep-branching member of subfamily 23, although with low bootstrap support (Fig. 2A). When additional, uncharacterized sequences from the CAZy database were included, CG400_06090 clustered within subfamily 23 (Fig. S2).
FIG 2.
(A) (Left) Phylogenetic tree of characterized GH13 members in the CAZy database and predicted G. leopoldii NR017 GH13 sequence CG400_06090. (Right) Expanded tree of subfamilies 17, 23, and 30. Stars indicate structurally characterized sequences, and bootstrap values are included. (B) CLUSTAL alignment of the catalytic domain of CG400_06090 with catalytic domains of GH13 subfamily 23 proteins from Halomonas sp. (BAL49684.1) and Xanthomonas campestris (BAC87873.1) and subfamily 17 protein from Culex quinquefasciatus (ASO96882.1). Red arrowheads indicate the conserved members of the catalytic triad, and the red circle indicates the acid/base (Glu242) in BAL49684.1 and BAC87873.1. The white circle indicates the predicted acid/base in CG400_06090. Invariant sequences are highlighted in black.
To identify conserved catalytic residues between CG400_06090 and characterized members of GH13, CG400_06090 was aligned with structurally characterized members of subfamily 23 (BAL49684.1 and BAC87873.1) and subfamily 17 (ASO96882.1) (Fig. 2B). Of the GH13 catalytic triad, the aspartate nucleophile and the aspartate that stabilizes the transition state are conserved among the four sequences (29). The glutamate general acid/base, however, does not appear to be sequence conserved between CG400_06090 and the GH13 subfamily 23 members. Upon modeling of CG400_06090 in PHYRE2 and aligning with BAL49684.1 (PDB ID 3WY1), CG400_06090 Glu256 did appear to be spatially conserved with the general acid/base (data not shown).
Expression and purification of α-glucosidase protein.
The G. leopoldii NR017 CG400_06090 open reading frame comprises 1,701 bp encoding a 567-amino-acid protein with a predicted mass of 62 kDa. The full-length open reading frame encoding amino acids 2 to 567 was PCR amplified and ligated into vector pQE-80L for expression in Escherichia coli as an N-terminal hexahistidine-tagged protein. A distinct protein band in SDS-PAGE between 50 kDa and 75 kDa was obtained from IPTG (isopropyl-β-d-thiogalactopyranoside)-induced E. coli cells compared with noninduced cells, indicating the expression of the α-glucosidase protein (Fig. 3A). The recombinant protein was soluble and was purified using a nickel-nitrilotriacetic acid (Ni-NTA) spin column (Fig. 3B). From a 50-ml broth culture, approximately 700 μl of purified protein at 4.0 mg/ml was obtained.
FIG 3.
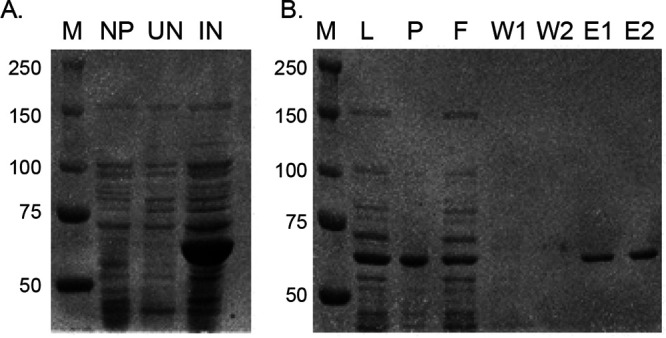
Production and purification of α-glucosidase protein. (A) A protein with a predicted mass of 62 kDa was observed in induced cultures. M, size marker; NP, E. coli with no vector; UN, uninduced culture; IN, induced with IPTG. Numbers on the left are marker sizes, in kilodaltons. (B) Fractions from Ni-NTA affinity purification of His-tagged recombinant protein. M, size marker; L, lysate; P, pellet; F, flowthrough; W1, first wash; W2, second wash; E1, first elution; E2, second elution.
Effect of pH on enzyme activity.
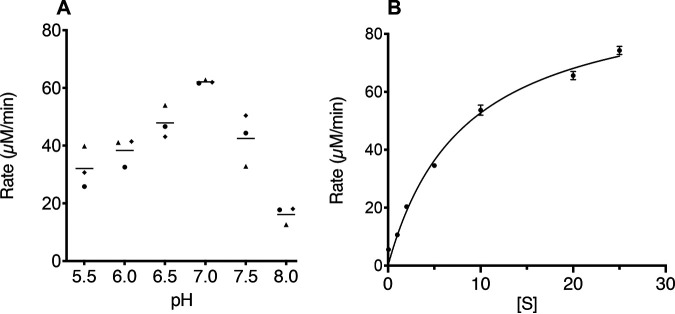
α-Glucosidase activity of the purified protein was demonstrated by release of 4-nitrophenol from the chromogenic substrate 4-nitrophenyl α-d-glucopyranoside. A preliminary analysis of enzyme activity over pH 3 to 8 showed that the product was produced over a broad pH range from 4.0 to 8.0 (Fig. S3). To more precisely examine effects of pH on activity, a pH rate profile was determined, and the maximum rate was observed at pH 7.0 (Fig. 4A). The dependence of rate on substrate concentration fit the Michaelis-Menten equation (Fig. 4B), with a Michaelis constant (Km) of 8.3 μM and a Vmax of 96 μM/min, corresponding to a kcat value of 0.96 (± 0.01) min−1 and a kcat/Km of 0.11 μM−1 min−1.
FIG 4.
(A) pH-rate profile of α-glucosidase. Each point represents the average of two technical replicates for each of three independent experiments as indicated by different shapes. Horizontal lines indicate means. (B) Michaelis-Menten plot. Results shown are the averages from three independent experiments, with error bars indicating standard deviations. The line represents the fit to the Michaelis-Menten equation.
Analysis of substrate hydrolysis.
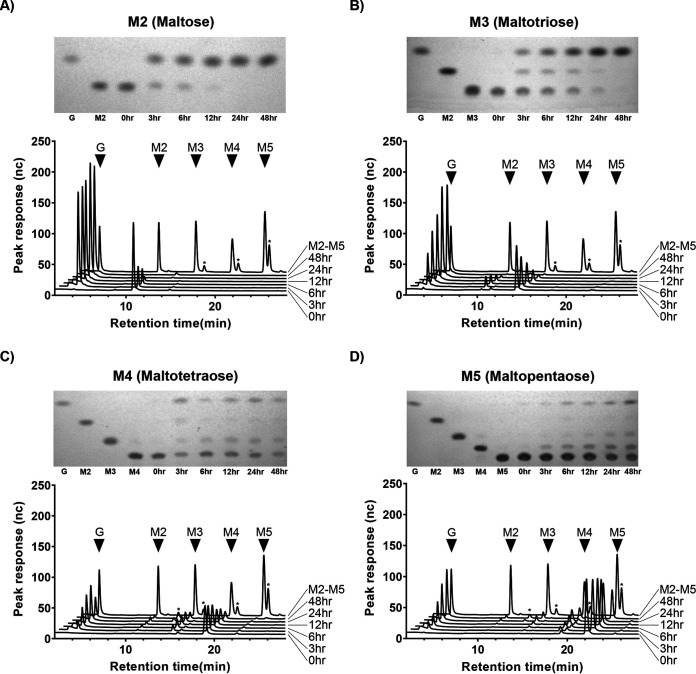
Production of glucose was detected when the purified α-glucosidase was incubated with maltose (M2), maltotriose (M3), maltotetraose (M4), and maltopentaose (M5) (Fig. 5). Samples analyzed by high-performance anion exchange chromatography with pulsed amperometric detection (HPAEC-PAD) contained large peaks corresponding to maltooligosaccharides ranging from maltose to maltopentaose; minor peaks, especially in the maltotetraose and maltopentaose samples, resolved products not observed in the thin-layer chromatography (TLC) analysis. These peaks did not align to isomaltotriose, panose, or 6-o-α-d-glucosyl-maltose standards in HPAEC-PAD (data not shown) and likely represent larger, mixed linkage products. No appreciable activity was detected on maltodextrins (MD 4 to 7, MD 13 to 17, and MD 16.5 to 19.5) or glycogen (Fig. S4).
FIG 5.
α-Glucosidase CG400_06090 digests of maltose to maltopentaose. Each panel shows a TLC plate and HPAEC-PAD trace of CG400_06090 digests of M2 to M5 between 0 h and 48 h. Major peaks from M2 to M5 standards are indicated with black triangles, while asterisks represent unique, undefined oligosaccharides.
DISCUSSION
Glycogen is a major nutrient available to members of the vaginal microbiota, and its utilization likely plays an important role in the survival and success of Gardnerella spp. in the vaginal microbiome. The ability of Gardnerella spp. to utilize glycogen has been reported in previous studies. Dunkelberg et al. described the growth of four strains of what was then known as Haemophilus vaginalis in buffered peptone water with 1% glycogen, suggesting their ability to ferment glycogen, but no additional details were provided (30). Similarly, Edmunds reported the glycogen-fermenting ability of 14 of 15 Haemophilus vaginalis isolates, although serum was included in the media (31). Piot et al. examined the starch fermentation ability of 175 Gardnerella isolates on Mueller-Hinton agar supplemented with horse serum and found that all Gardnerella strains were capable of hydrolyzing starch (32). They did not, however, assess glycogen degradation, and further, it is not clear if serum amylase contained in the medium contributed to the observed amylase activity. Taylor-Robinson, in his review, reported that starch can be utilized by Gardnerella (33) while Catlin, in another review, reported that glycogen utilization is inconsistent in Gardnerella (14).
Glycogen is digested into smaller products, such as maltose and maltodextrins, by a group of enzymes collectively known as amylases. Amylases that contribute to glycogen utilization in the vaginal environment are not well studied. Spear et al. (23) suggested a role for a human amylase enzyme in the breakdown of vaginal glycogen. They detected host-derived pancreatic α-amylase and acidic α-glucosidase in genital fluid samples collected from women and showed that genital fluid containing these enzymes can degrade the glycogen. The presence of these enzyme activities alone in vaginal fluid, however, was not correlated with glycogen degradation, suggesting contributions of activity from the resident microbiota. Many vaginal bacteria likely produce amylases that contribute to this activity, as various anaerobic taxa are known to encode amylases and digest glycogen (34); however, the contributions of the vaginal microbiota to this process and the importance of these processes to vaginal microbial community dynamics are not yet well understood. Regardless of the source, amylases digest glycogen into maltose and or maltooligosaccharides, which are transported inside bacteria via ABC transporter systems for further processing (35).
Protein sequences belonging to eight different GH13 subfamilies that could have roles in glycogen degradation were identified in G. vaginalis ATCC 14018 and G. swidsinskii GV37 (Fig. S1), and multiple amylase-like proteins were detected in the proteomes of additional Gardnerella spp. (Fig. 1), all of which were annotated as “alpha-amylase” in the GenBank records. Our results demonstrate the value of using more nuanced analyses, including dbCAN and SACCHARIS, to guide discovery of CAZymes based on comparison at a catalytic-domain level to functionally characterized enzymes. Further investigation will be required to demonstrate the actual functions of these proteins and how they contribute to carbohydrate-degrading activities of Gardnerella spp.
In the phylogenetic analysis of putative amylase sequences, one cluster contained sequences common to all 26 isolates (Fig. 1). Although this protein sequence was annotated as an α-amylase, suggesting that it would produce maltooligosaccharides from glycogen, SACCHARIS predicted it to be closely related to α-glucosidases, which cleave glucose from the nonreducing end of its substrate. Automated annotation of sequences deposited in primary sequence databases such as GenBank has resulted in significant problems with functional misannotation (36). This problem is exacerbated by the increasing rate at which whole-genome sequences are being generated and deposited. The genome sequences of the study isolates were annotated using the NCBI Prokaryotic Genome Annotation Pipeline, which identifies genes and annotates largely based on sequence similarity to previously annotated sequences, thus potentially propagating incorrect functional annotations.
Phylogenetic analysis of GH13 family enzymes showed that the conserved α-glucosidase from Gardnerella spp. appears to be most closely related to GH13 subfamily 23 (Fig. S2). Primary sequences of GH13 members of amylases have seven highly conserved regions and three amino acids that form the catalytic triad (27). Comparison of the primary sequence and three-dimensional model of CG400_06090 with characterized α-glucosidases from GH13 subfamily 17 and 23 did show sequence conservation of nucleophile and spatial conservation of the catalytic triad.
Our findings suggest that, although α-glucosidase does not hydrolyze glycogen, it can digest smaller maltooligosaccharides. The purified α-glucosidase enzyme was able to completely digest maltose and maltotriose to glucose, but digestion of maltotetraose and maltopentaose was incomplete, suggesting a preference for smaller oligosaccharides (Fig. 5). A previously characterized α-glucosidase (algB) from Bifidobacterium adolescentis DSM20083 also showed activity against maltose and maltotriose, while no activity against maltotetraose and maltopentaose was reported (37). α-Glucosidases can have different substrate specificities due to the variable affinity of the substrate binding site for particular substrates (38).
Most secreted proteins have a short N-terminal signal peptide to guide for extracellular translocation of the newly synthesized protein (39); however, bacterial proteins can be also secreted via nonclassical secretion pathways in a Sec-independent manner, without having a signal peptide (40). Analysis of the Gardnerella α-glucosidase sequence with SignalP and SecretomeP showed that the protein is unlikely to be secreted by either route and thus likely acts on intracellular substrates, including products of extracellular glycogen degradation transported into the cell. Many free-living and host-associated bacteria synthesize glycogen as a storage molecule (41), but the extent to which this occurs in Gardnerella is not known. The possibility that products of digestion of intracellular glycogen stores are substrates for the α-glucosidase characterized in this study remains a question for future study. Purified α-glucosidase from G. leopoldii NR017 was active across a broad pH range of 4.0 to 8.0 (Fig. S3), with the highest rate detected at pH 7.0 (Fig. 4A). This is not unexpected for an intracellular enzyme in a host-associated bacterium and is consistent with at least one other report of an α-glucosidase from a commensal bacterium, Bifidobacterium adolescentis (37).
Our findings show that Gardnerella spp. have an α-glucosidase enzyme that likely contributes to the complex and multistep process of glycogen utilization by releasing glucose from maltooligosaccharides. Identification and biochemical characterization of additional enzymes involved in glycogen metabolism will provide insight into whether utilization of this abundant carbon source is an important factor in population dynamics and competition among Gardnerella spp. The functional annotation strategy demonstrated here provides a powerful approach to guide future experiments aimed at determining enzyme substrates and activities.
MATERIALS AND METHODS
Gardnerella isolates.
Isolates of Gardnerella used in this study were from a previously described culture collection (6) and included representatives of cpn60 subgroup A (G. leopoldii [n = 4], G. swidsinskii [n = 6], and other [n = 1]), subgroup B [G. piotii [n = 5] and genome species 3 [n = 2]), subgroup C (G. vaginalis [n = 6]), and subgroup D (genome species 8 [n = 1] and genome species 9 [n = 1]). Whole-genome sequences of these isolates had been determined previously (BioProject accession no. PRJNA394757) and annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) (42).
Identification and functional annotation of putative amylase sequences.
Putative amylase sequences were identified in proteomes of 26 Gardnerella isolates used in this study based on the PGAP annotations. Multiple-sequence alignments of predicted amylase sequences were performed using CLUSTALW, with results viewed and edited in AliView (version 1.18) (43) prior to phylogenetic tree building using PHYLIP (44). SignalP v 5.0 (45) and SecretomeP 2.0 (40) were used to identify signal peptides.
Putative amylolytic enzymes in G. vaginalis ATCC 14018 and G. swidsinskii GV37 (genomes available in the CAZy database) were identified using dbCAN, and predicted sequences were run through the SACCHARIS pipeline. SACCHARIS combines user sequences with CAZy-derived sequences and trims sequences to the catalytic domain using dbCAN2 (46). Domain sequences were aligned with MUSCLE (47), and a best-fit model was generated with ProtTest (48). Final trees were generated with FastTree (49) and visualized with iTOL (50). Alignment of CG400_06090 from G. leopoldii NR017 to orthologues from Halomonas sp. (BAL49684.1), Xanthomonas campestris (BAC87873.1), and Culex quinquefasciatus (ASO96882.1) was done using CLUSTALW and visualized in ESPript (51). A predicted structure model of CG400_06090 was aligned with Halomonas sp. HaG (BAL49684.1, PDB accession no. 3WY1) (52) to identify putative catalytic residues using PHYRE2 (53). BLASTp (54) alignment of CAZy-derived sequences from reference genomes to a database of the predicted proteomes of 26 study isolates was conducted to identify orthologs.
Expression and purification of the CG400_06090 gene product.
Genomic DNA from G. leopoldii NR017 was extracted using a modified salting-out procedure, and the CG400_06090 open reading frame (locus tag CG400_06090 in GenBank accession no. NNRZ01000007, protein accession no. RFT33048) was PCR amplified with primers JH0729-F (5’ATG CAT GCG CAT TAT ACG ATC ATG CTC-3′) and JH0730-R (5’ATG GTA CCT TAC ATT CCA AAC ACT GCA-3′); underlining indicates SphI and KpnI restriction sites. PCR utilized 1×PCR buffer (0.2 M Tris-HCl [pH 8.4], 0.5 M KCl), a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 2.5 mM MgCl2, a 400 nM concentration of each primer, 1 U/reaction of Platinum Taq DNA polymerase high-fidelity (5 U/μl in 50% glycerol, 20 mM Tris-HCl, 40 mM NaCl, 0.1 mM EDTA, and stabilizers) (Life Technologies), and 2 μl of template DNA, in a final volume of 50 μl. PCR was performed with following parameters: initial denaturation at 94°C for 3 min, 35 cycles of denaturation at 94°C for 15 s, annealing at 60°C for 15 s, and extension at 72°C for 2 min, and final extension at 72°C for 5 min. Purified PCR products were digested with KpnI and SphI and ligated into expression vector pQE-80L (Qiagen, Mississauga, Ontario, Canada) digested with same restriction endonucleases. The resulting recombinant plasmid was used to transform One Shot TOP10 chemically competent E. coli cells (Invitrogen, Carlsbad, CA). Colony PCR was performed to identify transformants containing the vector with the insert. Insertion of the putative amylase gene in frame with the N-terminal 6× histidine tag was confirmed by sequencing of the purified plasmid.
E. coli cells containing the plasmid were grown overnight at 37°C in LB medium with 100 μg/ml ampicillin. Overnight culture was diluted 1:60 in fresh medium to a final volume of 50 ml, and the culture was incubated at 37°C with shaking at 225 rpm until it reached an OD600 of 0.6. At this point, expression was induced with 0.1 mM IPTG, and a further 4 h of incubation was done at 37°C at 225 rpm. Cells were harvested by centrifugation at 10,000 × g for 30 min, and the pellet was resuspended in lysis buffer (50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole [pH 8.0]). Lysozyme was added to 1 mg/ml, and cells were lysed by sonication (8 min total run time with 15 s on and 30 s off). The lysate was clarified by centrifugation at 10,000 × g for 30 min at 4°C, and the supernatant was applied to an Ni-NTA affinity column according to the manufacturer’s instructions (Qiagen, Germany). Bound proteins were washed twice with wash buffer (50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 8.0) and eluted with elution buffer (50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole [pH 8.0]). Eluted protein was buffer exchanged with the same elution buffer but without imidazole using a protein concentrator with a 30-kDa molecular weight cutoff (MWCO) (Thermo Fisher Scientific). Final protein concentration was measured by spectrophotometry.
Enzyme assay and kinetics.
Purified protein was tested for α-glucosidase activity by measuring the release of 4-nitrophenol from a chromogenic substrate 4-nitrophenyl α-d-glucopyranoside as described elsewhere (55). Briefly, 200 μl of enzyme solution (50 nM) was added to 200 μl of 20 mM 4-nitrophenyl α-d-glucopyranoside substrate, and the reaction mixture was incubated at 37°C. A 50-μl aliquot of the reaction mixture was removed and added to 100 μl of 1 M Na2CO3 solution every 90 s up to 7.5 min (total of 6 different time points). The absorbance of this solution was measured at 420 nm, and the amount of 4-nitrophenol released was calculated from a standard curve. To determine activity at different pH values, the substrate was prepared in 50 mM sodium phosphate buffer (pH 4.0 to 8.0). Kinetic constants were determined for 4-nitrophenyl α-d-glucopyranoside by measuring the rate of reaction of enzyme (50 nM) with substrate concentrations of 0.05, 1, 2, 5, 10, 20, and 25 mM at pH 7. Values were fitted to the Michaelis-Menten equation, v = kcat [E]0[S]/(Km + [S]), where v is the observed rate of reaction, [E]0 is the initial enzyme concentration, [S] is the substrate concentration, Km is the Michaelis constant, and kcat is the turnover number. Kinetics calculations were performed in GraphPad Prism 8.
Enzymatic activity on different substrates.
The activity of the purified enzyme was examined with different substrates. Briefly, 750 μl of enzyme solution (50 nM) was added to 750 μl of 3 mM maltose, maltotriose, maltotetraose, or maltopentaose or 0.1% (wt/vol) maltodextrins (MD 4 to 7, MD 13 to 17, and MD 16.5 to 19.5) or bovine liver glycogen in 50 mM sodium phosphate buffer, pH 6.0, and the reaction mixture was incubated aerobically at 37°C. Aliquots of the reaction mixture (250 μl) were removed at various time intervals of incubation and were heat inactivated at 93°C for 1 min. The reaction mixtures were centrifuged at 10,000 × g for 1 min, and supernatant was stored at −20°C and analyzed by thin-layer chromatography (TLC) and high-performance anion-exchange chromatography (HPAEC) with pulsed amperometric detection (PAD). TLC was performed on silica plates in a 1-butanol–acetic acid–distilled water (2:1:1 [vol/vol/vol]) mobile phase and stained using ethanol-sulfuric acid (70:3 [vol/vol]) solution containing 1% (wt/vol) orcinol monohydrate (Sigma). HPAEC-PAD samples were diluted to 50 μl and resolved on a Dionex PA20 column in a mobile phase of 30 mM NaOH and gradient of 10 mM to 120 mM sodium acetate over 40 min at a flow rate of 0.5 ml min−1. Data were analyzed using Chromeleon v6.80 chromatography data system software. Maltooligosaccharide standards ranging from maltose to maltopentaose (Carbosynth) were used for both TLC and HPAEC-PAD analysis, whereas isomaltotriose, d-panose, and 6-o-α-d-glucosyl-maltose (Carbosynth) were used to identify minor peaks resolved by HPAEC-PAD.
ACKNOWLEDGMENTS
This research was supported by a Natural Sciences and Engineering Research Council of Canada Discovery grant (J.E.H.) and Agriculture and Agri-Food Canada project number J-001589 (J.P.T. and D.W.A.). P.B. is supported by a Devolved Scholarship from the University of Saskatchewan.
We are grateful to Josseline Ramos-Figueroa, Douglas Fansher, and Natasha Vetter (Department of Chemistry, University of Saskatchewan) for guidance with thin-layer chromatography and Champika Fernando for technical support.
Footnotes
Supplemental material is available online only.
Contributor Information
Janet E. Hill, Email: Janet.Hill@usask.ca.
Michael Y. Galperin, NCBI, NLM, National Institutes of Health
REFERENCES
- 1.Nugent RP, Krohn MA, Hillier SL. 1991. Reliability of diagnosing bacterial vaginosis is improved by a standardized method of gram stain interpretation. J Clin Microbiol 29:297–301. 10.1128/jcm.29.2.297-301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin HL, Richardson BA, Nyange PM, Lavreys L, Hillier SL, Chohan B, Mandaliya K, Ndinya‐Achola JO, Bwayo J, Kreiss J. 1999. Vaginal Lactobacilli, microbial flora, and risk of human immunodeficiency virus type 1 and sexually transmitted disease acquisition. J Infect Dis 180:1863–1868. 10.1086/315127. [DOI] [PubMed] [Google Scholar]
- 3.Taha TE, Hoover DR, Dallabetta GA, Kumwenda NI, Mtimavalye LAR, Yang L-P, Liomba GN, Broadhead RL, Chiphangwi JD, Miotti PG. 1998. Bacterial vaginosis and disturbances of vaginal flora: association with increased acquisition of HIV. AIDS 12:1699–1706. 10.1097/00002030-199813000-00019. [DOI] [PubMed] [Google Scholar]
- 4.Paramel Jayaprakash T, Schellenberg JJ, Hill JE. 2012. Resolution and characterization of distinct cpn60-based subgroups of Gardnerella vaginalis in the vaginal microbiota. PLoS One 7:e43009. 10.1371/journal.pone.0043009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmed A, Earl J, Retchless A, Hillier SL, Rabe LK, Cherpes TL, Powell E, Janto B, Eutsey R, Hiller NL, Boissy R, Dahlgren ME, Hall BG, Costerton JW, Post JC, Hu FZ, Ehrlich GD. 2012. Comparative genomic analyses of 17 clinical isolates of Gardnerella vaginalis provide evidence of multiple genetically isolated clades consistent with subspeciation into genovars. J Bacteriol 194:3922–3937. 10.1128/JB.00056-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schellenberg JJ, Paramel Jayaprakash T, Withana Gamage N, Patterson MH, Vaneechoutte M, Hill JE. 2016. Gardnerella vaginalis subgroups defined by cpn60 sequencing and sialidase activity in isolates from Canada, Belgium and Kenya. PLoS One 11:e0146510. 10.1371/journal.pone.0146510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vaneechoutte M, Guschin A, Van Simaey L, Gansemans Y, Van Nieuwerburgh F, Cools P. 2019. Emended description of Gardnerella vaginalis and description of Gardnerella leopoldii sp. nov., Gardnerella piotii sp. nov. and Gardnerella swidsinskii sp. nov., with delineation of 13 genomic species within the genus Gardnerella. Int J Syst Evol Microbiol 69:679–687. 10.1099/ijsem.0.003200. [DOI] [PubMed] [Google Scholar]
- 8.Hill JE, Albert AYK, the VOGUE Research Group.2019. Resolution and cooccurrence patterns of Gardnerella leopoldii, G. swidsinskii, G. piotii, and G. vaginalis within the vaginal microbiome. Infect Immun 87:e00532-19. 10.1128/IAI.00532-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Balashov SV, Mordechai E, Adelson ME, Gygax SE. 2014. Identification, quantification and subtyping of Gardnerella vaginalis in noncultured clinical vaginal samples by quantitative PCR. J Med Microbiol 63:162–175. 10.1099/jmm.0.066407-0. [DOI] [PubMed] [Google Scholar]
- 10.Khan S, Voordouw MJ, Hill JE. 2019. Competition among Gardnerella subgroups from the human vaginal microbiome. Front Cell Infect Microbiol 9:374. 10.3389/fcimb.2019.00374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. 2010. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 8:15–25. 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khan S, Vancuren SJ, Hill JE. 2020. A generalist lifestyle allows rare Gardnerella spp. to persist at low levels in the vaginal microbiome. Microb Ecol 10.1007/s00248-020-01643-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewis WG, Robinson LS, Gilbert NM, Perry JC, Lewis AL. 2013. Degradation, foraging, and depletion of mucus sialoglycans by the vagina-adapted actinobacterium Gardnerella vaginalis. J Biol Chem 288:12067–12079. 10.1074/jbc.M113.453654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Catlin BW. 1992. Gardnerella vaginalis: characteristics, clinical considerations, and controversies. Clin Microbiol Rev 5:213–237. 10.1128/CMR.5.3.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meléndez-Hevia E, Waddell TG, Shelton ED. 1993. Optimization of molecular design in the evolution of metabolism: the glycogen molecule. Biochem J 295:477–483. 10.1042/bj2950477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng B, Sullivan MA, Chen C, Li J, Powell PO, Hu Z, Gilbert RG. 2016. Molecular structure of human-liver glycogen. PLoS One 11:e0150540. 10.1371/journal.pone.0150540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roach PJ, Depaoli-Roach AA, Hurley TD, Tagliabracci VS. 2012. Glycogen and its metabolism: some new developments and old themes. Biochem J 441:763–787. 10.1042/BJ20111416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Adeva-Andany MM, González-Lucán M, Donapetry-García C, Fernández-Fernández C, Ameneiros-Rodríguez E. 2016. Glycogen metabolism in humans. BBA Clin 5:85–100. 10.1016/j.bbacli.2016.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cruickshank R, Sharman A. 1934. The biology of the vagina in the human subject. BJOG 41:208–226. 10.1111/j.1471-0528.1934.tb08759.x. [DOI] [Google Scholar]
- 20.Lundstrom P. 1960. Glycogen content of the female vaginal mucosa and starch degradation activity associated with the mucosa. Acta Soc Med Ups 65:30–48. [PubMed] [Google Scholar]
- 21.Mirmonsef P, Hotton AL, Gilbert D, Gioia CJ, Maric D, Hope TJ, Landay AL, Spear GT. 2016. Glycogen levels in undiluted genital fluid and their relationship to vaginal pH, estrogen, and progesterone. PLoS One 11:e0153553. 10.1371/journal.pone.0153553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spear GT, French AL, Gilbert D, Zariffard MR, Mirmonsef P, Sullivan TH, Spear WW, Landay A, Micci S, Lee BH, Hamaker BR. 2014. Human alpha-amylase present in lower-genital-tract mucosal fluid processes glycogen to support vaginal colonization by Lactobacillus. J Infect Dis 210:1019–1028. 10.1093/infdis/jiu231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berg JM, Tymoczko JL, Stryer L. 2002. Glycogen breakdown requires the interplay of several enzymes.Biochemistry, 5th ed. WH Freeman, New York, NY. [Google Scholar]
- 25.Cantarel BL, Coutinho PM, Rancurel C, Bernard T, Lombard V, Henrissat B. 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res 37:D233–D238. 10.1093/nar/gkn663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. 2014. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res 42:D490–D495. 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehta D, Satyanarayana T. 2016. Bacterial and archaeal α-amylases: diversity and amelioration of the desirable characteristics for industrial applications. Front Microbiol 7:1129. 10.3389/fmicb.2016.01129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nunn KL, Clair GC, Adkins JN, Engbrecht K, Fillmore T, Forney LJ. 2020. Amylases in the human vagina. mSphere 5:e00943-20. 10.1128/mSphere.00943-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uitdehaag JCM, Mosi R, Kalk KH, van der Veen BA, Dijkhuizen L, Withers SG, Dijkstra BW. 1999. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α-amylase family. Nat Struct Biol 6:432–436. 10.1038/8235. [DOI] [PubMed] [Google Scholar]
- 30.Dunkelberg WE, McVeigh I. 1969. Growth requirements of Haemophilus vaginalis. Antonie Van Leeuwenhoek 35:129–145. 10.1007/BF02219124. [DOI] [PubMed] [Google Scholar]
- 31.Edmunds PN. 1962. The biochemical, serological and haemagglutinating reactions of Haemophilus vaginalis. J Pathol Bacteriol 83:411–422. 10.1002/path.1700830211. [DOI] [PubMed] [Google Scholar]
- 32.Piot P, Van Dyck E, Totten PA, Holmes KK. 1982. Identification of Gardnerella (Haemophilus) vaginalis. J Clin Microbiol 15:19–24. 10.1128/jcm.15.1.19-24.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor-Robinson D. 1984. The bacteriology of Gardnerella vaginalis. Scand J Urol Nephrol Suppl 86:41–55. [PubMed] [Google Scholar]
- 34.Nunn KL, Forney LJ. 2016. Unraveling the dynamics of the human vaginal microbiome. Yale J Biol Med 89:331–337. [PMC free article] [PubMed] [Google Scholar]
- 35.Boos W, Shuman H. 1998. Maltose/Maltodextrin System of Escherichia coli: transport, Metabolism, and Regulation. Microbiol Mol Biol Rev 62:204–229. 10.1128/MMBR.62.1.204-229.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schnoes AM, Brown SD, Dodevski I, Babbitt PC. 2009. Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol 5:e1000605. 10.1371/journal.pcbi.1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van den Broek L, Struijs K, Verdoes J, Beldman G, Voragen A. 2003. Cloning and characterization of two α-glucosidases from Bifidobacterium adolescentis DSM20083. Appl Microbiol Biotechnol 61:55–60. 10.1007/s00253-002-1179-1. [DOI] [PubMed] [Google Scholar]
- 38.Okuyama M, Saburi W, Mori H, Kimura A. 2016. α-Glucosidases and α-1,4-glucan lyases: structures, functions, and physiological actions. Cell Mol Life Sci 73:2727–2751. 10.1007/s00018-016-2247-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Owji H, Nezafat N, Negahdaripour M, Hajiebrahimi A, Ghasemi Y. 2018. A comprehensive review of signal peptides: structure, roles, and applications. Eur J Cell Biol 97:422–441. 10.1016/j.ejcb.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 40.Bendtsen JD, Kiemer L, Fausbøll A, Brunak S. 2005. Non-classical protein secretion in bacteria. BMC Microbiol 5:58. 10.1186/1471-2180-5-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Preiss J. 1984. Bacterial glycogen synthesis and its regulation. Annu Rev Microbiol 38:419–458. 10.1146/annurev.mi.38.100184.002223. [DOI] [PubMed] [Google Scholar]
- 42.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsson A. 2014. AliView: a fast and lightweight alignment viewer and editor for large datasets. Bioinformatics 30:3276–3278. 10.1093/bioinformatics/btu531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Felsenstein J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164–166. [Google Scholar]
- 45.Almagro Armenteros JJ, Tsirigos KD, Sønderby CK, Petersen TN, Winther O, Brunak S, von Heijne G, Nielsen H. 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat Biotechnol 37:420–423. 10.1038/s41587-019-0036-z. [DOI] [PubMed] [Google Scholar]
- 46.Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z, Busk PK, Xu Y, Yin Y. 2018. dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46:W95–W101. 10.1093/nar/gky418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Letunic I, Bork P. 2019. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robert X, Gouet P. 2014. Deciphering key features in protein structures with the new ENDscript server. Nucleic Acids Res 42:W320–W324. 10.1093/nar/gku316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shen X, Saburi W, Gai Z, Kato K, Ojima-Kato T, Yu J, Komoda K, Kido Y, Matsui H, Mori H, Yao M. 2015. Structural analysis of the α-glucosidase HaG provides new insights into substrate specificity and catalytic mechanism. Acta Crystallogr D Biol Crystallogr 71:1382–1391. 10.1107/S139900471500721X. [DOI] [PubMed] [Google Scholar]
- 53.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. 2015. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Angelov A, Putyrski M, Liebl W. 2006. Molecular and biochemical characterization of alpha-glucosidase and alpha-mannosidase and their clustered genes from the thermoacidophilic archaeon Picrophilus torridus. J Bacteriol 188:7123–7131. 10.1128/JB.00757-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download JB.00213-21-s0001.pdf, PDF file, 1.8 MB (1.8MB, pdf)