ABSTRACT
Antimicrobial peptides (AMPs) are one of the key immune responses that can eliminate pathogenic bacteria through membrane perturbation. As a successful skin commensal, Staphylococcus epidermidis can sense and respond to AMPs through the GraXRS two-component system and an efflux system comprising the VraG permease and VraF ATPase. GraS is a membrane sensor known to function in AMP resistance through a negatively charged, 9-residue extracellular loop, which is predicted to be linear without any secondary structure. An important question is how GraS can impart effective sensing of AMPs through such a small unstructured sequence. In this study, we verified the role of graS and vraG in AMP sensing in S. epidermidis, as demonstrated by the failure of the ΔgraS or ΔvraG mutants to sense. Deletion of the extracellular loop of VraG did not affect sensing but reduced survival with polymyxin B. Importantly, a specific region within the extracellular loop, termed the guard loop (GL), has inhibitory activity since sensing of polymyxin B was enhanced in the ΔGL mutant, indicating that the GL may act as a gatekeeper for sensing. Bacterial two-hybrid analysis demonstrated that the extracellular regions of GraS and VraG interact, but interaction appears dispensable to sensing activity. Mutation of the extracellular loop of VraG, the GL, and the active site of VraF suggested that an active detoxification function of VraG is necessary for AMP resistance. Altogether, we provide evidence for a unique sensory scheme that relies on the function of a permease to impart effective information processing.
IMPORTANCE Staphylococcus epidermidis has become an important opportunistic pathogen that is responsible for nosocomial and device-related infections that account for considerable morbidity worldwide. A thorough understanding of the mechanisms that enable S. epidermidis to colonize human skin successfully is essential for the development of alternative treatment strategies and prophylaxis. Here, we demonstrate the importance of an AMP response system in a clinically relevant S. epidermidis strain. Furthermore, we provide evidence for a unique sensory scheme that would rely on the detoxification function of a permease to effect information processing.
KEYWORDS: GraRS, VraFG, cationic antimicrobial peptides, Gram-positive bacteria, membrane permease, resistance, sensing, Staphylococcus epidermidis, two-component system
INTRODUCTION
Staphylococcus epidermidis is a coagulase-negative staphylococcus that is ubiquitous on human skin (1). As a commensal, S. epidermidis is a successful colonizer of multiple body sites, including the anterior nares, axillae, and perineum, despite competition from other native flora (1). However, upon breaching of the epithelial barrier with indwelling medical devices or implants, S. epidermidis can gain access to underlying tissue and cause infections that have garnered attention as being especially difficult to treat (1). S. epidermidis infections are often subacute but chronic and are highly recalcitrant to routine antibiotic treatments, often requiring the complete removal of the colonized medical devices and hence increasing morbidity and mortality rates. Therefore, a critical understanding of the basic bacterial mechanisms involved in skin persistence and colonization is necessary to devise strategies to combat opportunistic behaviors like those deployed during S. epidermidis infections.
Antimicrobial peptides (AMPs) are potent and evolutionarily conserved effectors found among almost every class of life. On human skin, AMPs form a critical component of the innate immune response and aid in keeping microbial proliferation in check through their direct bactericidal effects. AMPs are subdivided into groups according to their amino acid composition and structure. One group, the cationic AMPs (CAMPs), are characterized by their cationic and amphipathic properties, which enable efficient interaction with anionic bacterial membranes, the most frequent target of AMPs. CAMPs disrupt membrane stability, often through pore formation in bacterial membranes (2). Respectively, some AMPs are also suggested to inhibit bacterial intracellular processes like cell wall, DNA, and protein synthesis by interfering with lipid II function, indiscriminately associating with DNA, or binding ribosomal proteins directly (3). As a highly successful colonizer, S. epidermidis must have developed means to effectively resist CAMPs in order to survive the hostile human skin environment.
Sensitive and rapid signal transduction pathways play an important part in the resistance of S. epidermidis to CAMPs. These resistance pathways in Gram-positive bacteria with low G+C contents often comprise two families of proteins, i.e., (i) a specialized two-component system (TCS) composed of an intramembrane-sensing histidine kinase (IM-HK) sensory component and a cognate response regulator and (ii) an adjacent ABC transporter that imparts detoxification and/or secondary signaling effects (4). The representative example of this arrangement is the BceRS-BceAB system in Bacillus subtilis, which is responsible primarily for bacitracin resistance. As an IM-HK, BceS lacks any obvious or typical extracellular sensory domain. Catalytic inactivation of the BceA ATPase or deletion of the BceAB transporter system abolished the BceRS-BceAB system’s ability to respond to bacitracin, suggesting that BceS signaling is associated not with direct ligand binding but rather with the transport activity of BceAB (5). Indeed, a membrane-localized BceRS-BceAB sensory complex was detected (6), and mathematical modeling of the system’s response dynamics suggested that BceS kinase activity responds to BceAB-mediated bacitracin flux as part of a “positive flux-sensory” scheme (7). In S. epidermidis, GraXRS, also known as ApsXRS, and the adjacent VraFG form a five-component signal transduction pathway that is thought to be responsible for CAMP resistance, based on its similarity to the BceRS-BceAB system in B. subtilis. GraXRS/ApsXRS (glycopeptide resistance-associated or antimicrobial peptide sensor XRS) represents the TCS that positively regulates the dltXABCD operon and mprF, which catalyze the d-alanylation of teichoic acids and the lysylination of phosphatidylglycerol, respectively (8–12). These cell surface modifications increase the net positive charge of the cell surface and thus electrostatically repulse CAMPs. The genes vraF and vraG (vancomycin resistance-associated F and G), which are located immediately downstream of graXRS, encode the putative ABC transporter of the system that is positively regulated by GraRS (8, 9).
GraR and GraS form the foundation of the TCS pathway, whereby GraS is the IM-HK and GraR is the cognate response regulator. GraS contains a small 9-amino-acid unstructured extracellular loop (EL) in place of a typical extracellular sensing domain, which is framed by two transmembrane segments. Previous work from our laboratory and others has suggested that this small EL, at least in Staphylococcus aureus, confers sensing and selectivity with respect to CAMPs, possibly by an enrichment of negatively charged residues and conformational rigidity in the loop (9, 13, 14). Recently, GraS was found to facilitate resistance to antimicrobial unsaturated free fatty acids in acidic pH through a mechanism unique to CAMP detection (15). Despite sharing considerable predicted membrane topology and primary structure with the bacitracin efflux system VraDE, VraFG is implicated to harbor no CAMP detoxification function in S. aureus, as VraFG alone is not sufficient to confer resistance to colistin in a ΔgraRS background (16). Instead, the VraG permease in S. aureus is thought to be a sensory component that facilitates sensing through GraS, as suggested by their direct interaction and VraFG being necessary for its own synthesis (16). However, the manner in which staphylococci, especially S. epidermidis, remove membrane-associated CAMPs and how VraFG could mediate CAMP signaling to GraS remain unclear, despite its sequence similarities to the B. subtilis Bce system. How this system modulates its own activity in the presence of CAMPs is also poorly understood. The last component of this five-component system, GraX, is thought to be involved in fine-tuning the system’s sensory activity, as evidenced by direct interactions with GraS, GraR, and the ATPase VraF in S. aureus, suggesting that it may act as a scaffold to augment the GraRS signal cascade (16, 17).
In this study, we sought to further characterize the roles of GraS and VraG in signal integration, system regulation, and resistance of the GraXRS-VraFG system in S. epidermidis. We show here that the 9-amino-acid EL of GraS is essential for proper function with VraG, and we suggest that VraG is a secondary sensing component regulating the system activity by a putative “guard loop” (GL) that is thought to function via a gated efflux-sensing mechanism. Together, these results not only further our understanding of how S. epidermidis senses CAMPs but also may provide the mechanistic foundation for how GraXRS-VraFG effects information processing and confers immediate resistance to CAMPs.
RESULTS
Alteration of the GraS EL reduces basal signaling and CAMP resistance.
It was described previously that S. epidermidis is more resistant to CAMPs than is S. aureus (8, 9). The basis of this resistance has been ascribed to GraS, the sensing component of the GraRS TCS (8, 9). GraS contains a single 9-amino-acid EL framed by two transmembrane segments, and it was suggested that this EL is critical for resistance function and CAMP selectivity (9, 14). Of note, the 9-amino-acid EL of S. epidermidis (DYEISVESV) diverges in sequence from that of S. aureus (DYDFPIDSL); however, its unique contribution to system functionality within S. epidermidis was unclear. In order to uncover the contributions of specific residues within the loop, MIC assays were conducted with polymyxin B sulfate (PMB), a proxy for CAMPs, with a number of key GraS mutants and chimeras (Table 1). As expected, deletion of graS led to an 8-fold reduction in PMB resistance (Table 2). We also examined the three negatively charged residues (D35, E37, and E41) in the EL of GraS in S. epidermidis, because these residues (D35, D37, and D41) were shown to be important for CAMP resistance in S. aureus (14). The conserved aspartic acid residue at position 35 in the EL of GraS in S. epidermidis was mutated to alanine (ΔEL::AYEISVESV graS), resulting in a 4-fold reduction in PMB MIC, analogous to a ΔgraS mutant (Table 2). Similarly, replacing the entire EL of GraS in S. epidermidis with alanines (ΔEL::Ala9 graS) led to a PMB MIC identical to that of a ΔgraS mutant (Table 2). To evaluate the importance of S. aureus EL in a S. epidermidis background, a chimeric GraS was made in strain NIH051475 expressing the EL of S. aureus MW2 (ΔEL::DYDFPIDSL graS). This chimera led to a reduction in PMB resistance similar to that of the other ΔEL mutants (Table 2). A chimera that replaces the two transmembrane segments of GraS in S. epidermidis strain NIH051475 with those of its counterpart from S. aureus MW2 (IMMW2 graS) exhibited resistance to PMB similar to that of the wild-type (WT) strain (Table 2), indicating the importance of the EL rather than the flanking membrane segments in dictating sensitivity to cationic peptides. Crucially, the PMB phenotype of the ΔgraS mutant could not be rescued by overexpressing either vraG (ΔgraS/pEPSA5::vraG) or vraFG (ΔgraS/pEPSA5::vraFG) in a manner similar to that in S. aureus (16), but this could be achieved by chromosomal complementation with the WT graS allele (Table 2). This finding suggests that the detoxification effect of VraFG did not contribute to PMB sensitivity in a graS mutant of S. epidermidis.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant genotype or descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| S. epidermidis | ||
| NIH051475 | 37 | |
| ALC9220 | ΔgraS | This study |
| ALC9221 | ΔgraS WTR | This study |
| ALC9222 | ΔgraS/pEPSA5::vraG; VraG is ectopically overexpressed in ΔgraS background by inducible plasmid pEPSA5 | This study |
| ALC9223 | ΔgraS/pEPSA5::vraFG; VraFG is ectopically overexpressed in ΔgraS background by inducible plasmid pEPSA5 | This study |
| ALC9224 | graS-IMMW2; replacement of strain NIH051475 GraS intramembrane regions with those of S. aureus MW2 over residues 15–34 and 44–64 | This study |
| ALC9225 | ΔEL::Ala9 graS | This study |
| ALC9226 | ΔEL::AYEISVESV graS | This study |
| ALC9227 | ΔEL::DYDFPIDSL graS; replacement of strain NIH051475 9-residue EL with that of S. aureus MW2 | This study |
| ALC9228 | ΔvraG | This study |
| ALC9229 | ΔvraG WT revertant | This study |
| ALC9230 | ΔEL vraG | This study |
| ALC9231 | ΔEL vraG WT revertant | This study |
| ALC9232 | ΔGL vraG | This study |
| ALC9233 | ΔGL vraG WT revertant | This study |
| ALC9234 | ΔGL::Gly10 vraG | This study |
| ALC9235 | vraF-G40A; alanine point mutation of glycine 40 in Walker A motif of VraF | This study |
| ALC9236 | vraF-G40A WT revertant | This study |
| ALC9237 | Strain NIH051475 with C-terminal HA-tagged GraS | This study |
| ALC9238 | Strain NIH051475 with C-terminal HA-tagged EL/DYDFPIDSL GraS | This study |
| ALC9239 | Strain NIH051475 with C-terminal HA-tagged EL/alanine GraS | This study |
| ALC9240 | Strain NIH051475 with N-terminal HA-tagged VraG | This study |
| ALC9241 | Strain NIH051475 with N-terminal HA-tagged ΔEL VraG | This study |
| ALC9242 | Strain NIH051475 with N-terminal HA-tagged ΔGL VraG | This study |
| ALC9243 | Strain NIH051475 with N-terminal HA-tagged GL-Gly10 VraG | This study |
| E. coli | ||
| DC10B | mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG Δdcm | 31 |
| DTH1 | F− cya-854 ilv-691::Tn10 recA1 endA1 gyrA96 (Nalr) thi-1 hsdR17 spoT1 rfbD1 glnV44(AS) | 38 |
| C41(DE3) | F− ompT gal dcm hsdSB(rB− mB−)(DE3) | Lucigen |
| SKC-N05 | DC10BΩPN25-hsdMS of strain NIH051475 integrated between atpI and gidB genes | This study |
| Plasmids | ||
| pSK236 | Shuttle plasmid; ColE1 E. coli origin with Ampr selection and pC194 staphylococcal origin with Cmr selection | 39 |
| pEPSA5 | Xylose inducible vector; p15A E. coli origin with Ampr selection and pC194 staphylococcal origin with Cmr selection | 34 |
| pMAD | Allelic exchange vector; pBR322 E. coli origin with Ampr selection and pE194ts staphylococcal origin with Ermr selection | 33 |
| pKD46 | λ-red recombineering vector for targeted genetic alterations in E. coli; repA101ts origin, Ampr | 40 |
| pKD4 | Template plasmid for FRT-flanked kanamycin cassette; Ampr and Kanr | 40 |
| pKD46′ | pKD46 harboring E. coli K-12 recA coding sequence under native promoter | This study |
| pKT25 | BacTH vector for N-terminal T25 fusion; p15A origin, Kanr | Euromedex |
| pUT18 | BacTH vector for C-terminal T18 fusion; ColE1 origin, Ampr | Euromedex |
| pKT25-zip | Contains T25 fused leucine zipper of GCN4 | Euromedex |
| pUT18C-zip | Contains T18 fused leucine zipper of GCN4 | Euromedex |
Ampr, ampicillin resistance; Cmr, chloramphenicol resistance; Kanr, kanamycin resistance.
TABLE 2.
PMB resistance of different S. epidermidis mutants
| Strain | Relevant genotype | PMB MIC (μg/ml)a |
|---|---|---|
| NIH051475 | 64 | |
| ALC9220 | ΔgraS | 8 |
| ALC9221 | ΔgraS WTR | 64 |
| ALC9222 | ΔgraS/pEPSA5::vraG | 8b |
| ALC9223 | ΔgraS/pEPSA5::vraFG | 4b |
| ALC9224 | IMMW2 graS | 64 |
| ALC9225 | ΔEL::Ala9 graS | 8 |
| ALC9226 | ΔEL::AYEISVESV graS | 16 |
| ALC9227 | ΔEL::DYDFPIDSL graS | 16 |
| ALC9228 | ΔvraG | 16 |
| ALC9229 | ΔvraG WTR | 64 |
| ALC9230 | ΔEL vraG | 16 |
| ALC9231 | ΔEL vraG WTR | 64 |
| ALC9232 | ΔGL vraG | 16 |
| ALC9233 | ΔGL vraG WTR | 64 |
| ALC9234 | ΔGL::Gly10 vraG | 16 |
| ALC9235 | vraF-G40A | 8 |
| ALC9236 | vraF-G40A WTR | 64 |
MICs are representative of a 105 initial inoculum incubated for 48 h at 37°C unless indicated otherwise. All values represent averages of triplicates.
The strain MIC is representative of a 105 initial inoculum with 48 h of incubation at 37°C with 1% xylose induction.
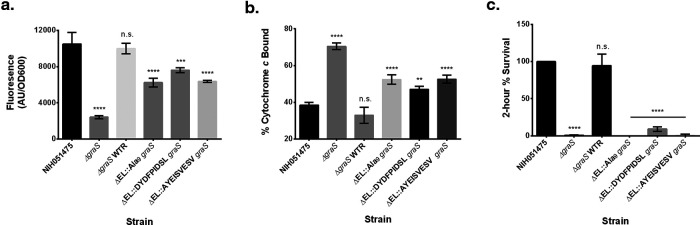
Alterations to the EL of GraS were shown previously to greatly impact the regulation of mprF and the dlt operon, downstream effectors of this TCS (8, 9, 14). To define this effect, a vector-based fluorescent transcriptional reporter was generated by fusing the promoter of mprF to the coding sequence of the mRuby fluorescent protein. Predictably, deletion of graS led to a significant drop in fluorescence (Fig. 1a), but it was restored by reverting to the WT graS allele, suggesting the key role of GraS in signaling of the system. Replacement of the EL with only alanines reduced the mprF promoter activity, as did substitution of the extracellular residues of S. epidermidis with those from S. aureus (DYDFPIDSL), indicating that the EL in S. epidermidis contributed only in part to its sensing activity. Remarkably, replacement of the single aspartic acid residue at position 35 decreased mprF expression to the level of the other two mutants, indicating that this residue, which is directly adjacent to the predicted membrane segment, has significant influence on sensing of PMB. Nevertheless, all of the mutants still displayed fluorescence higher than that of the ΔgraS mutant (Fig. 1a).
FIG 1.
GraS EL is essential for sensing and survival with CAMPs. (a) mprF promoter fusion assays using pSK236::Pmprf-mRuby. Overnight cultures were diluted to an OD600 of 1.6 and incubated for 50 min at 37°C with shaking. Fluorescence was read with an excitation wavelength of 558 nm and an emission wavelength of 592 nm. (b) Cytochrome c binding assays. Overnight cell cultures with an OD650 of 3 were incubated for 10 min with 0.5 mg/ml cytochrome c to assess differences in binding by measuring supernatant A530. (c) Two-hour time-kill assays. Cells were diluted to 106 CFU/ml and incubated with 32 μg/ml PMB at 37°C for 2 h with shaking at 250 rpm. The graph shows the percent difference between colony counts plated at 0 h and 2 h, standardized to 100% colony recovery from strain NIH051475. Each experiment represents means and standard deviations of three biological replicates. Statistical analysis was conducted by multiple-comparison one-way analysis of variance (ANOVA) against strain NIH051475 using the Dunnett post hoc multiple-comparison correction. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., not significant.
Defects in the regulation of mprF and the dlt operon attributable to mutated residues in the EL of GraS were shown previously to impact surface positive charge and to reduce 2-h survival rates with CAMPs, which reflect persistence due to a short-term activation mechanism (14). To decipher the change in surface positive charge, we evaluated binding to cytochrome c, a highly cationic protein; increased binding would indicate reduction in surface positive charge. Predictably, deletion of graS led to increased binding of cytochrome c, compared with the parent (Fig. 1b), coinciding with reduced mprF expression of the mutant in comparison to the parent (Fig. 1a). Similarly, all mutants, including the D35A mutant, displayed comparably greater cytochrome c binding than the parent but not as great as that of the ΔgraS mutant (Fig. 1b), mirroring their reduced expression of mprF (Fig. 1a). The 2-h killing assay revealed that these mutants displayed a defect in immediate survival against PMB that was similar to the level seen with the ΔgraS mutant (Fig. 1c). This defect was consistent with the MICs but was in contrast to the intermediate level of defect seen in the fluorescent reporter and cytochrome c binding assays. All defects reflected by the ΔgraS mutant could be restored to the parental phenotype by reintroducing the WT graS allele (Fig. 1).
VraG is indispensable for CAMP resistance.
The role VraG plays in CAMP resistance in S. epidermidis is not clearly defined (16, 17). Deletion of VraG in S. aureus was shown previously to impart a resistance defect against CAMPs that was indistinguishable from that of a ΔgraS mutant (16, 18). As seen in Table 2, deletion of vraG led to a severe PMB resistance defect that is comparable to that of ΔgraS (Table 2). This defect can be specifically restored by reintroducing the WT vraG allele (Table 2).
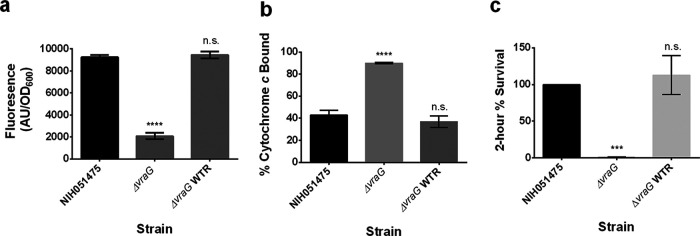
To determine whether VraG is involved in sensing similar to the GraXRS-VraFG system in S. aureus (16, 18, 19), the mprF expression level in the ΔvraG mutant of S. epidermidis strain NIH051475 was queried. This assay showed a severe reduction in fluorescence level in the ΔvraG mutant (Fig. 2a). Parental levels of expression could be restored by reintroducing the WT vraG allele (Fig. 2a).
FIG 2.
VraG plays a critical role in CAMP sensing. (a) mprF promoter fusion assays using pSK236::Pmprf-mRuby. Overnight cultures were diluted to an OD600 of 1.6 and incubated for 50 min at 37°C with shaking. Fluorescence was read with an excitation wavelength of 558 nm and an emission wavelength of 592 nm. (b) Cytochrome c binding assays. Overnight cell cultures with an OD650 value of 3 were incubated for 10 min with 0.5 mg/ml cytochrome c to assess differences in binding by measuring supernatant A530. (c) Two-hour time-kill assays. Cells were diluted to 106 CFU/ml and incubated with 32 μg/ml PMB at 37°C for 2 h with shaking at 250 rpm. The graph shows the percent difference between colony counts plated at 0 h and 2 h, standardized to 100% colony recovery from strain NIH051475. Each experiment represents means and standard deviations of three biological replicates. Statistical analysis was conducted by multiple-comparison one-way ANOVA against strain NIH051475 using the Dunnett post hoc multiple-comparison correction. ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., not significant.
Because deletion of vraG was shown previously to impart a less-positive cell envelope charge and a 2-h survival defect in S. aureus (18), the ΔvraG mutant of S. epidermidis strain NIH051475 was queried by cytochrome c binding and 2-h time-kill assays. Cytochrome c binding of the ΔvraG mutant displayed considerably higher binding levels than the parental control (Fig. 2b), suggesting a less positive surface charge in the mutant. This finding correlated with reduced survival of the ΔvraG mutant in PMB in a 2-h killing assay (Fig. 2c). These defects could be specifically restored to parental levels by reintroducing the WT vraG allele (Fig. 2b and c).
The VraG EL is involved in CAMP resistance.
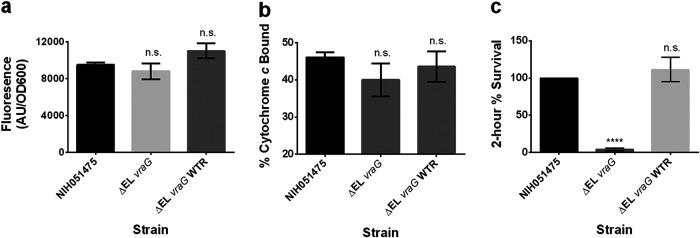
VraG is a membrane permease composed of 10 transmembrane segments and a large EL of some 201 residues. The extracellular domain of VraG has gained attention as possibly the true CAMP sensor in the GraXRS-VraFG system. To examine this possibility, we first deleted the EL of VraG and found a 4-fold reduction in the PMB MIC versus the parent (16 versus 64 μg/ml), analogous to the ΔvraG mutant, but this was restored to the parental level by reintroducing the WT vraG allele (Table 2). We next queried mprF expression in the ΔEL vraG mutant with the mprF reporter. Remarkably, the mprF promoter activity of the ΔEL vraG mutant did not differ from that of the parent (Fig. 3a). In agreement with the mprF data, cytochrome c binding in the ΔEL vraG mutant was indistinguishable from that of the parental strain (Fig. 3b). In contrast to the mprF activity and cytochrome c binding, the survival rate for the ΔEL vraG mutant upon 2-h exposure to PMB was very low, compared to that for the parent, which was set at 100% as a reference, but the rate was restored to the parental level upon reintroduction of the WT vraG allele (Fig. 3c). This result corroborates the increased susceptibility in the MIC assay (Table 2). This divergence between sensing (i.e., mprF activation) and resistance (i.e., MIC) indicates that the EL in its entirety plays a major role in PMB resistance (e.g., maintaining the integrity and efflux of VraG) but not necessarily in sensing.
FIG 3.
The EL of VraG regulates sensing and confers immediate resistance against CAMPs. (a) mprF promoter fusion assays using pSK236::Pmprf-mRuby. Overnight cultures were diluted to an OD600 of 1.6 and incubated for 50 min at 37°C with shaking. Fluorescence was read with an excitation wavelength of 558 nm and an emission wavelength of 592 nm. Statistical analysis was conducted by unpaired t test. n.s., not significant. (b) Cytochrome c binding assays. Overnight cell cultures with an OD650 of 3 were incubated for 10 min with 0.5 mg/ml cytochrome c to assess differences in binding by measuring supernatant A530. (c) Two-hour time-kill assays. Cells were diluted to 106 CFU/ml and incubated with 32 μg/ml PMB at 37°C for 2 h with shaking at 250 rpm. The graph shows the percent difference between colony counts plated at 0 h and 2 h, standardized to 100% colony recovery from strain NIH051475. Each experiment represents means and standard deviations of three biological replicates. Statistical analysis was conducted by multiple-comparison one-way ANOVA against strain NIH051475 using the Dunnett post hoc multiple-comparison correction. ****, P ≤ 0.0001; n.s., not significant.
VraG activity may be modulated by a gated mechanism.
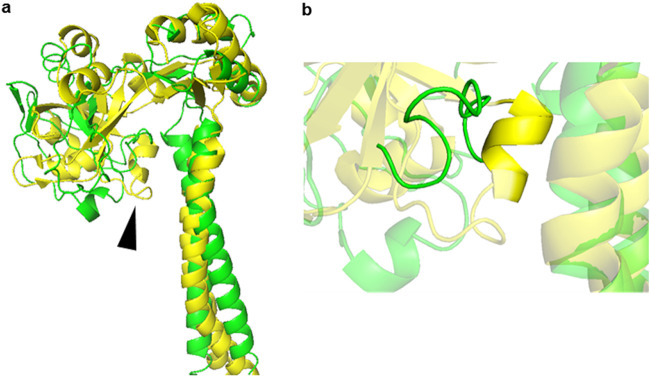
How VraG either senses or coordinates sensing with GraS remains poorly understood. Through threaded template-based modeling (TBM) with I-TASSER (20) and ResNet deep convolutional neural network (DCNN) contact prediction with CASP12- and CASP13-winning method RaptorX-Contact (21), a predicted structure for the EL of VraG and the flanking region was generated (Fig. 4a). TBM indicated that the EL of VraG shares considerable similarity with Spr0695, one component of a MacAB-like AMP efflux system in Streptococcus pneumoniae (22). The efflux function of Spr0695 is thought to rely on an alpha helix of six residues (EAKRSK), termed the guard helix, that controls lateral entrance of the substrate by physically occluding its access to the Spr0693 channel in the resting state (22). Through contact prediction, a putative guard helix of 10 residues from position 432 to position 441 (KVYFMSDVDR) was identified in the EL of VraG, and we tentatively termed it the GL (Fig. 4b).
FIG 4.
Predicted structure of VraG EL and flanking transmembrane regions. (a) RaptorX-predicted structure of VraG EL and flanking transmembrane regions (green) superimposed on a portion of the resolved structure of Spr0695 (yellow). The arrowhead indicates the location of the VraG GL and the Spr0695 guard helix. (b) The putative GL of VraG (green) in reference to the guard helix of Spr0695 (yellow).
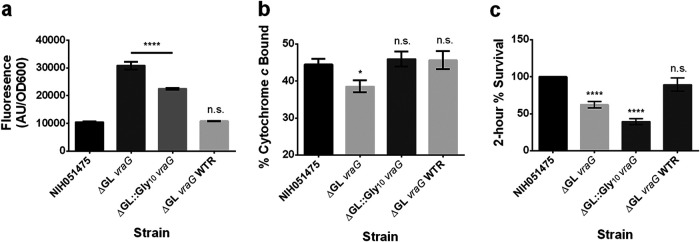
In a previous study, it was shown that mutagenesis of three positively charged residues (Lys207, Arg208, and Lys210) over the guard helix in Spr0695 led to a partial reduction in resistance to LL-37 in S. pneumoniae during antimicrobial susceptibility testing (22). To test the hypothesis that the 10 residues comprising the putative S. epidermidis VraG GL play a role in CAMP resistance, putative GL residues 432 to 441 were either deleted (ΔGL vraG) or substituted with glycines (ΔGL::Gly10 vraG). As seen in Table 2, there was a 4-fold reduction in the PMB MICs for both mutants versus the parental strain, analogous to the ΔvraG and ΔEL vraG mutants, but values were restored to the parental level with introduction of the WT vraG allele (Table 2). Having observed little if any difference in mprF activity between the ΔEL vraG mutant and the parent (Fig. 3a), we assessed whether deletion of GL conferred a difference in sensing of PMB to alter mprF promoter activity. Interestingly, the ΔGL vraG mutant had a significantly higher level of mprF expression than the parent, but it returned to the WT level with vraG restoration. Notably, the mprF activity level of the ΔGL::Gly10 vraG mutant was lower than that of the ΔGL vraG mutant but higher than that of the parent (Fig. 5a). The increased mprF promoter activity in the ΔGL vraG mutant coincided with less binding of cytochrome c (Fig. 5b). The increase in mprF activity in the ΔGL::Gly10 vraG mutant in comparison to the parent did not translate to a reduction in cytochrome c binding; this may be due to the fact that this is a less sensitive assay than the mprF expression assay (Fig. 5b). Surprisingly, both mutants survived significantly less in the 2-h time-kill assay, compared to the parental strain (Fig. 5c), but rates were restored to WT levels upon introduction of the WT vraG allele. Notably, this result coincides with the MIC. Together, we interpret that sensing by VraG requires the GL while resistance to CAMPs involves intact VraG with an intact GL sequence, possibly to maintain the integrity of VraG as a permease.
FIG 5.
The GL of VraG specifically regulates system sensing and may mediate immediate resistance against CAMPs. (a) mprF promoter fusion assays using pSK236::Pmprf-mRuby. Overnight cultures were diluted to an OD600 of 1.6 and incubated for 50 min at 37°C with shaking. Fluorescence was read with an excitation wavelength of 558 nm and an emission wavelength of 592 nm. (b) Cytochrome c binding assays. Overnight cell cultures with an OD650 of 3 were incubated for 10 min with 0.5 mg/ml cytochrome c to assess differences in binding by measuring supernatant A530. (c) Two-hour time-kill assays. Cells were diluted to 106 CFU/ml and incubated with 32 μg/ml PMB at 37°C for 2 h with shaking at 250 rpm. The graph shows the percent difference between colony counts plated at 0 h and 2 h, standardized to 100% colony recovery from strain NIH051475. Each experiment represents means and standard deviations of three biological replicates. Statistical analysis was conducted by multiple-comparison one-way ANOVA against strain NIH051475 using the Dunnett post hoc multiple-comparison correction. *, P ≤ 0.05; ****, P ≤ 0.0001; n.s., not significant.
The EL of VraG associates with GraS.
A direct interaction between VraG and GraS was observed previously in S. aureus (16), but the context for this interaction remained unclear. Here, we sought to examine this interaction through bacterial two-hybrid (BacTH) assays of VraG with mutant GraS proteins and of GraS with mutant VraG proteins. Complementation of native VraG with any mutant GraS proteins, including D35A, ΔEL::DYDFPIDSL (i.e., S. aureus EL), and ΔEL::Ala9, totally abolished the association between VraG and GraS (Table 3). Complementation of WT GraS with ΔEL VraG or ΔGL::Gly10 VraG also disrupted the association between VraG and GraS. Notably complementation of GraS with ΔGL VraG led to a quantifiable but reduced association (Table 3). These data indicated that the ELs of VraG and GraS are required for association with one another in S. epidermidis.
TABLE 3.
BacTH analysis of VraG-GraS and select mutant interactions
| Hybrid protein pair | β-Galactosidase activity (U/mg)a |
|---|---|
| GraS + VraG | 89.48 ± 3.61 |
| ΔEL::DYDFPIDSL GraS + VraG | — |
| ΔEL::AYEISVESV GraS + VraG | — |
| ΔEL::Ala9 GraS + VraG | — |
| GraS + ΔEL VraG | — |
| GraS + ΔGL VraG | 41.60 ± 1.87 |
| GraS + ΔGL::Gly10 VraG | — |
Complementation between hybrid proteins was quantified according to the BacTH quantification described in Materials and Methods. Each interaction represents the average of three biological replicates with standard deviation values. pUT18C-Zip plus PKT25-Zip represented the positive control for this assay and produced an average of approximately 500 U of β-galactosidase activity. Empty pKT25 and pUT18 vectors were used as negative controls and generated less than 5 U of β-galactosidase activity on average. One unit of β-galactosidase activity corresponds to 1 nmol of ONPG hydrolyzed per min at 28°C per mg of bacterial dry weight. Dashes indicate β-galactosidase activity of 5 U or less.
The ATPase VraF is required for proper system function of GraS.
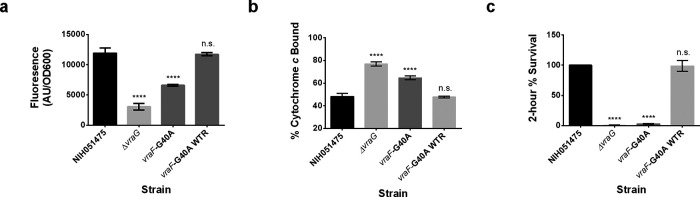
The extracellular region of VraG seems to be required for immediate survival upon 2-h exposure to PMB (Fig. 3c), but the entire EL is not necessarily required for normal sensing (Fig. 3a). The dysregulation of sensing and survival with the GL suggests that VraG is not simply a secondary sensing component. In order to assess a possible detoxification role for VraG, a vraF mutant inactivating the cognate ATPase via a point mutation of the Walker A motif (vraF-G40A) was generated. This mutant harbored impaired resistance to PMB at a level similar to that of the ΔvraG mutant (Table 2), but the level could be restored to the parental level by reintroduction of the WT vraF allele (Table 2). For mprF expression, the vraF-G40A mutant yielded an intermediate level between parental and ΔvraG levels, which could be restored to parental levels by reintroducing the WT vraF allele (Fig. 6a). The vraF-G40A mutant led to an increase in cytochrome c binding that was intermediate between parental and ΔvraG mutant levels (Fig. 6b). This phenotype could be restored to parental levels by introduction of the WT vraF allele (Fig. 6b). Notably, the vraF-G40A mutant survived as poorly as the vraG mutant under short-term acute PMB exposure (Fig. 6c), but levels were restored to parental levels by reintroduction of the WT vraF allele (Fig. 6c).
FIG 6.
Enzymatically competent VraF is required for proper sensing and resistance to PMB, and VraG EL alone is not sufficient to mount effective resistance. (a) mprF promoter fusion assays using pSK236::Pmprf-mRuby. Overnight cultures were diluted to an OD600 of 1.6 and incubated for 50 min at 37°C with shaking. Fluorescence was read with an excitation wavelength of 558 nm and an emission wavelength of 592 nm. (b) Cytochrome c binding assays. Overnight cell cultures with an OD650 of 3 were incubated for 10 min in 0.5 mg/ml cytochrome c to assess differences in binding by measuring supernatant A530. (c) Two-hour time-kill assays. Cells were diluted to 106 CFU/ml and incubated with 32 μg/ml PMB at 37°C for 2 h with shaking at 250 rpm. The graph shows the percent difference between colony counts plated at 0 h and 2 h, standardized to 100% colony recovery from strain NIH051475. Each experiment represents means and standard deviations of three biological replicates. Statistical analysis was conducted by multiple-comparison one-way ANOVA against strain NIH051475 using the Dunnett post hoc multiple-comparison correction. ****, P ≤ 0.0001; n.s., not significant.
Protein expression in key mutant constructs.
Of concern in these assays is whether key mutant proteins were expressed as VraG or GraS protein variants or as degraded proteins. To ascertain this, we cloned in-frame a hemagglutinin (HA) tag to the C terminus of key mutated GraS proteins and the N terminus of VraG proteins expressed in S. epidermidis strain NIH051475. Using equivalent amounts of lysate proteins, the samples were resolved in SDS gels, blotted onto polyvinylidene difluoride (PVDF) membranes, and probed with mouse anti-HA antibody followed by donkey anti-mouse F(ab')2 antibody conjugated to horseradish peroxidase and developing substrate. As seen in Fig. S1 in the supplemental material, all relevant and major mutated proteins, including VraG-HA, ΔGL VraG-HA, ΔGL::Gly10 VraG-HA, ΔEL VraG-HA, GraS-HA, ΔEL::DYDFPIDSL GraS-HA, and ΔEL::Ala9 GraS-HA, were expressed. As expected, ΔEL VraG-HA was expressed at a lower molecular weight versus the native protein (see Fig. S1, left). Also, GraS was detected as a dimer in our membrane extracts (see Fig. S1, right).
DISCUSSION
The innate barrier formed by human skin is accentuated by the production of potent CAMPs, which help regulate microbial proliferation and maintain a healthy skin microbiome (1). As a ubiquitous and highly successful colonizer of human skin, S. epidermidis resists these dangerous antimicrobials primarily through charge modifications of its cell wall and membrane. GraRS is a TCS that confers detection and response to CAMPs in both S. aureus and S. epidermidis (8, 9, 13, 14, 18, 19, 23, 24). Once CAMPs are detected by GraS, the cognate response regulator GraR positively regulates mprF and the dltXABCD operon. MprF, a bifunctional membrane protein, catalyzes the synthesis and translocation of lysyl-phosphatidylglycerol within the cell membrane (10, 11). The dltXABCD operon encodes enzymes involved with the addition of d-alanine to wall teichoic acids (12). These cell envelope modifications increase the net positive charge of the cell surface and help resist CAMPs by electrostatic repulsion. Additionally, the genes encoding the VraFG ABC transporter, lying immediately downstream of the graXRS operon, are positively regulated by GraR and are known to play an essential role in CAMP resistance by an unknown mechanism (16, 18).
A unique feature of the GraXRS system is that the GraS sensor-histidine kinase consists of two transmembrane segments framing a very small 9-residue EL, which lacks any recognizable extracellular sensory domain. This is a characteristic of the IM-HK subfamily within the TCSs (25). These 9 residues are known to be critical for efficient sensing and selectivity of CAMPs. In S. aureus, substitution of the first aspartic acid in the GraS loop results in system dysregulation similar to that of the ΔgraS mutant. Work done by Otto and co-workers showed that heterologous complementation in S. aureus with the S. epidermidis graS or the native graS expressing the EL derived from S. epidermidis was able to alter the S. aureus CAMP induction profile (9). Thus, loop charge and/or structural rigidity may facilitate CAMP sensing, but the exact mechanism of GraS-mediated signal transmission remained unclear.
The genes encoding the putative ABC transporter VraFG are adjacent to graXRS and are genetically regulated by GraRS. This TCS-ABC transporter arrangement places GraXRS-VraFG into a burgeoning Firmicutes group of transporter-based information processors (4, 26). The loss of VraG is known to nullify the activation of the CAMP resistance system in S. aureus, leading to a reduction in the expression of GraR-regulated targets downstream (16, 19). Functional consequences of deletion of VraG involve the failure to properly modify the cell envelope, a decrease in surface positive charge, and a reduction in 2-h survival with CAMPs. Work done by Msadek and coworkers with chimeric VraG expressing the EL of VraE, a homologous protein involved with bacitracin efflux, suggested that the EL of VraG is what confers specificity of substrate recognition during CAMP sensing (16, 27). Furthermore, failed attempts to restore parental levels of colistin resistance by overexpression of vraFG in a S. aureus ΔgraRS background suggested that VraFG may not be a stand-alone detoxification module (16). This finding, combined with the physical interaction observed between GraS and VraG (16), suggests that VraG acts via an unknown mechanism as either the true sensor of this system or a secondary sensory component.
Here, we corroborated and confirmed in S. epidermidis what had been demonstrated in GraS of S. aureus. Any change imposed here on the 9-residue EL of GraS led to defects in the parameters we tested, including mprF expression, changes in surface charge, and 2-h survival rates with PMB. Interestingly, there is a divergence in phenotypic effects between MICs (Table 2) and mprF signaling/cytochrome c binding in EL replacement and D35A mutants of GraS (Fig. 1a and b). This result may be explained by the transient effect of signaling, while resistance to PMB is linked to a longer period that involves active detoxification and lingering transient system activation. Exemplifying these observations was the chimeric substitution of the S. epidermidis strain NIH051475 EL with that of S. aureus MW2 (ΔEL::DYDFPIDSL graS), resulting in a MIC close to that of the graS mutant (Table 2). Indeed, the MIC effect was likely due to the EL, as substitution of the two membrane segments of GraS in S. epidermidis strain NIH051475 with their counterparts from S. aureus MW2 (IMMW2 graS) did not change the PMB MICs (Table 2). Importantly, alterations to the EL of GraS led to the abolishment of interaction with VraG in the BacTH assay (Table 3), thus implying that the EL of GraS likely facilitates the interaction between GraS and VraG. However, the possibility of altering the EL of GraS to influence another transmembrane conformation necessary for the GraS-VraG interaction cannot be ruled out. Further biochemical studies are required in order to determine the interaction interface between GraS and VraG.
The role of VraG in the GraXRS-VraFG system has been the source of much contention regarding whether it is a secondary or the primary CAMP sensor and whether it even retains a discrete detoxification function at all. Indeed, our initial results for the ΔvraG mutant suggest that it is a sensor because mprF expression in this mutant was disrupted to a magnitude similar to that in ΔgraS (Fig. 2a). Concurrently, the functional consequences of lacking vraG mimic those seen in a ΔgraS mutant, as mprF activation, cytochrome c binding, and 2-h time-kill assay results were similar to those for a ΔgraS mutant (Table 2 and Fig. 2b and c). As seen previously in S. aureus with colistin resistance (16), attempts to rescue PMB resistance in a ΔgraS background of S. epidermidis strain NIH051475 by overexpressing either VraG (ΔgraS/pEPSA5::vraG) or VraFG (ΔgraS/pEPSA5::vraFG) failed (Table 2). These results together support the view that VraG is required for signaling and survival advantages as a result of activation of mprF and the dltXABCD operon but is not fully sufficient for increasing MICs in the absence of GraS. In contrast to S. aureus mutants lacking the EL of VraG (ΔEL vraG), in which signaling is enhanced (28), the mprF promoter activity and cytochrome c binding of the ΔEL vraG mutant of S. epidermidis did not change and were similar to those of the parent (Fig. 3a and b). Furthermore, the ΔEL vraG mutant failed to associate with WT GraS during BacTH complementation (Table 3). Combined, these results suggest that (i) the EL of VraG is dispensable for mprF activation of the GraRS system, (ii) GraS activity is mediated by an interaction with the EL of VraG, and (iii) the EL of VraG is essential for conferring survival against CAMPs.
Through threaded protein folding prediction (20) and de novo protein folding (21), it was determined that the EL of VraG shares considerable predicted structure with Spr0695 (Fig. 4a), a component of a MacAB-like cationic peptide efflux system in Streptococcus pneumoniae. Spr0695 employs a gated mechanism via a 6-residue guard helix (Fig. 4b) that modulates lateral entry of AMPs for entrance into the Spr0693 channel. Through structure prediction, we were able to identify a putative 10-residue guard helix in VraG that we termed the GL (Fig. 4b). Deletion of the GL (ΔGL vraG) or its substitution with glycines (ΔGL::Gly10 vraG) led to a dramatic increase in mprF activity; however, only the ΔGL vraG mutant displayed a significant reduction in cytochrome c binding (Fig. 5a and b). Interestingly, these observations did not translate to enhanced survival in 2-h time-kill assays (Fig. 5c). BacTH analysis revealed that the ΔGL vraG mutant protein led to a reduction (∼50%) in the association between VraG and GraS, whereas all other mutant proteins, including that with GL substitution with glycine (ΔGL::Gly10 VraG protein) exhibited no association with GraS (Table 3). Nevertheless, these results suggest that (i) the EL of VraG has both an inhibitory element (GL) and an unknown stimulatory element for GraS or, alternatively, the GL in S. epidermidis requires the EL of VraG to be functional and (ii) the GL is not necessarily the interaction interface between GraS and VraG, since deletion of GL still allows VraG-GraS interaction (Table 3).
In order to further characterize VraG, we employed genetic studies to assess functional effects of the Walker A motif of the VraF ATPase (vraF-G40A). In the vraF-G40A mutant, the mprF promoter activity revealed expression intermediate between parental and ΔvraG levels, accompanied by an increase in cytochrome c binding versus the parent that was less than that of the ΔvraG mutant (Fig. 6a and b). Because the vraF-G40A mutation would render the efflux pump VraG ineffective, it is not surprising that the vraF-G40A mutant phenocopied the ΔvraG mutant in both the MIC and 2-h time-kill assays (Table 2). These results suggest that VraG may be a discrete detoxification module but sensory function can be maintained at a reduced level (e.g., vraF-G40A mutation) even when the permease becomes inactivated, as was seen previously with the Walker A region inactivation of the BceA ATPase in the closely related bacitracin resistance BceRS-BceAB system (5).
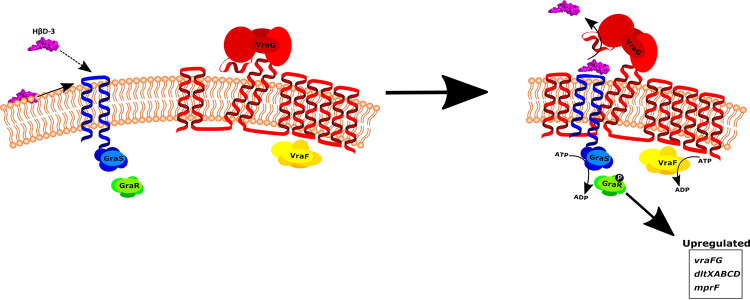
Now, a clearer model for the GraXRS-VraFG system in S. epidermidis can be derived, considering all of the evidence discovered here (Fig. 7). In our model, the dedicated sensory function of GraS is tied to the activity of VraG in much the same fashion as the positive flux-sensory scheme found in the BceRS-BceAB bacitracin resistance system (7). We propose that VraG functions both as a CAMP detoxification module and as a secondary sensory component. The efflux activity of VraG would rely on lateral substrate entrance and displacement of the GL. This displacement would presumably activate the VraF-dependent efflux function of VraG through a currently unknown mechanism. GraS, with the negatively charged EL imbedded in the membrane, may selectively associate with certain CAMPs via this loop, as suggested by both its assumed direct association with CAMPs and its capacity to specifically select CAMPs (8, 9, 14), and present these CAMPs as substrates for VraG. The efflux activity of VraG could then transmit a conformational change in GraS, enabling autophosphorylation and positive regulation of the downstream effectors of the system. Indeed, GraS may be absolutely necessary for proper VraG efflux functionality, as evidenced by the inability to restore resistance to PMB by overexpression of VraG or VraFG in a ΔgraS mutant background (Table 2). Because alterations to the membrane can lead to various deleterious effects, tying the efflux function of a membrane stress-related transporter to the activation of a membrane-based resistance mechanism by a flux-sensory arrangement offers extraordinarily rapid and tight control. Interestingly, although there is a high degree of similarity between the GraXRS-VraFG systems of S. epidermidis and S. aureus, previous work from our laboratory suggests that S. aureus may not follow the same proposed model of flux-sensory regulation as described here (28). More work is necessary to thoroughly understand the differences that may exist in the regulation of the systems within the two bacteria. Furthermore, our proposed mechanism is specific for the detection of CAMPs and may be independent of the mechanism that activates this system at acidic pH (15). Indeed, under low-pH conditions, the assumed biochemical contributions of the GraS EL were determined to be dispensable for system activation in S. aureus (15). Additionally, we presume that our flux-sensory model would not apply, because a lowered pH would reduce the charge on the GraS EL acidic residues, rendering our proposed electrochemical association between CAMPs and the GraS EL ineffective. Therefore, we surmise that activation of this system at acidic pH may bypass VraFG entirely and rely solely on GraS-mediated detection of membrane perturbation. System activation may occur through detection of membrane fluidity, because a lower pH would reduce the charge on polar lipid head groups and result in the tightening of phospholipid lateral packing, which may be detected through subsequent physical reorganization of the GraS intramembrane regions (15, 29).
FIG 7.
Flux-sensory model of the S. epidermidis five-component CAMP resistance system. In this model, GraS loads CAMPs onto the negatively charged 9-residue EL either directly from the membrane (solid arrow) or from the environment (dashed arrow). GraS would then interact with VraG, and the presence of the CAMP on GraS would dislodge the GL of VraG, leading to activation of the VraG efflux function. Presumably, once efflux is activated, this conformational change in VraG may be transmitted to GraS and lead to autophosphorylation of GraS. The activation of GraS then leads to phosphorylation of GraR and upregulation of downstream targets necessary for cell envelope modifications and CAMP resistance. Therefore, the primary functions of GraS and VraG are dependent on and inseparable from each other. GraS is the true sensor of the system but relies on the efflux function of VraG to activate, while concurrently VraG relies on GraS to efficiently present substrate for efflux. In such an arrangement, GraS and VraG can tightly regulate potentially deleterious modifications to the cell envelope in situations where it is absolutely essential for survival.
In this study, we sought to further characterize how S. epidermidis regulates CAMP detection through the unique GraXRS-VraFG system. Here, our work suggests that VraG may couple CAMP detoxification to sensing through GraS by a novel gated mechanism. This work may offer the mechanistic foundation for how S. epidermidis detects CAMPs, as well as one way in which coupled TCS-ABC transporters process information in Firmicutes. Further work is necessary to define how VraG and GraS interact, the exact mechanism of gated VraG efflux, and how efflux can lead to activation of GraS.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. A detailed list of all constructs can be found in Table S2 in the supplemental material. Escherichia coli was routinely grown in Luria-Bertani (LB) broth (Difco), while S. epidermidis was maintained in either Trypticase soy broth (TSB) (Difco) or Mueller-Hinton broth (MHB) (Difco). Media were sterilized by autoclaving except for MHB, which was filter sterilized. For plasmid and integration selection, antibiotic concentrations of 100 μg/ml ampicillin, 50 μg/ml kanamycin, 10 μg/ml chloramphenicol, and/or 2.5 μg/ml erythromycin were used when necessary. Bacteria were routinely grown at 37°C with shaking at 250 rpm. E. coli and S. epidermidis strains were transformed chemically or through electroporation, respectively, and transformants were selected on LB agar (LBA) or Trypticase soy agar (TSA) (Difco) with appropriate antibiotics.
Generation of competent E. coli or S. epidermidis cells and transformation procedures.
Chemically competent E. coli was generated as described previously (30). Briefly, 1 ml of overnight culture was diluted 1:100 in fresh LB broth and incubated at 37°C with shaking to an optical density at 600 nm (OD600) of 0.375. After chilling on ice for 10 min, the culture was centrifuged at 3,200 rpm for 7 min at 4°C, with all subsequent steps performed at 4°C or on ice. The pellet was resuspended in 20 ml of CaCl2 solution [60 mM CaCl2, 15% glycerol, and 10 mM piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES; pH 7)], centrifuged for 5 min at 3,200 rpm, resuspended in 5 ml CaCl2 solution, and rested on ice for 30 min. After a final centrifugation at 3,200 rpm for 5 min, the pellet was resuspended in 1 ml of CaCl2 solution, and 50-μl aliquots were stored at −80°C until use. Transformation was carried out by standard heat shock procedures.
Electrocompetent S. epidermidis strain NIH051475 was generated as described previously (31). Briefly, an overnight culture was diluted to an OD578 of 0.5 in TSB and then incubated for 30 min at 37°C with shaking. The culture was chilled on ice for 10 min, collected by centrifugation at 4,000 rpm for 10 min at 4°C, washed, and centrifuged serially with 1 volume, 1/10 volume, and 1/25 volume of ice-cold autoclaved distilled water. After the final wash, cells were resuspended in a final volume of 1/200 volume of cold 10% glycerol, divided into 50-μl aliquots, and stored at −80°C until use. Electroporation of S. epidermidis was performed as described previously (31, 32) with some modification. Briefly, competent cells were thawed first on ice for 5 min and then at room temperature for 5 min. Cells were then pelleted by centrifugation at 5,000 × g for 1 min and resuspended in 50 μl of 10% glycerol supplemented with 500 mM sucrose. Plasmid DNA was added, and the cell-DNA mixture was transferred to a 1-mm electroporation cuvette (Bulldog-Bio) and pulsed three times, in succession, at 2.1 kV with a truncated time constant of 1.1 ms. Cells were then immediately resuspended in 1 ml of recovery medium (TSB supplemented with 500 mM sucrose) and allowed to recover for 1 h at either 30°C or 37°C with shaking. The cells were then centrifuged at 5,000 × g for 3 min, resuspended in 50 to 100 μl of recovery medium, plated in their entirety on TSA with appropriate antibiotics, and incubated at either 30°C or 37°C until colonies developed.
DNA manipulation.
Oligonucleotides used in this study were synthesized by Integrated DNA Technologies, and their sequences are listed in Table S1. S. epidermidis genomic DNA was isolated with the GenElute bacterial genomic DNA kit (Sigma-Aldrich). Plasmids were purified using the E.Z.N.A. plasmid DNA minikit (Omega Bio-Tek). PCR products were purified by either the GeneJET gel extraction kit or the GeneJET gel extraction and DNA cleanup microkit (Thermo Fisher Scientific). Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific), restriction enzymes, Antarctic phosphatase, NEBuilder HiFi DNA assembly kit, and T4 DNA ligase (New England Biolabs) were used according to the manufacturers’ specifications. Sequencing of plasmid constructs and mutants was conducted by the Molecular Shared Resources Lab at Dartmouth College.
Plasmid construction and mutant generation.
Markerless gene deletions, chimeras, chromosomal epitope tagging, and site-directed mutagenesis were achieved by allelic exchange with the thermosensitive vector pMAD (33). DNA fragments of approximately 1,000 bp, corresponding to homologous regions flanking the site of interest, were amplified and conjoined with strand overlap extension (SOEing) PCR, digested with BamHI and SalI or BamHI and XmaI, and then introduced into similarly digested pMAD with T4 DNA ligase. Nucleotide sequences were confirmed by DNA sequencing, and plasmid DNA was amplified in the restriction bypass E. coli strain SKC-N05, purified, and then electroporated into S. epidermidis strain NIH051475. Selection of desired mutants and modifications was achieved as described previously (32). Briefly, transformed strain NIH051475 cells were plated on TSA with erythromycin and 80 μg/ml 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (GoldBio) at 30°C to select for transformants containing the recombinant pMAD. One blue colony was selected and cultured overnight at 30°C in TSB supplemented with erythromycin. The culture was then diluted 1:100 in 5 ml of fresh TSB without antibiotic and incubated overnight at 43°C, a nonpermissive temperature for pMAD replication, to promote single crossover. This procedure was repeated, and dilutions of this culture were plated at 43°C for single colonies on TSA supplemented with erythromycin and 80 μg/ml X-Gal. A light blue colony, signifying a single-crossover event, was selected and grown overnight at 30°C in TSB without antibiotic, with shaking. Dilutions of this overnight culture were then plated on TSA supplemented with 80 μg/ml X-Gal and were incubated overnight at 37°C. Resulting white colonies were then patched onto TSA plates containing either 80 μg/ml X-Gal or erythromycin. The colonies that remained white on the X-Gal plate but failed to grow on the erythromycin plate were then validated for the desired mutation or modification by sequencing.
Plasmid complementation constructs were made using the complementation vector pEPSA5 (34). Coding sequences of vraG and vraFG were introduced into pEPSA5 via Gibson assembly (see Table S1).
The E. coli-staphylococcus shuttle vector pSK236 was used to monitor gene expression in S. epidermidis from a mprF promoter fused to the fluorescent mRuby gene. The promoter of mprF was amplified by PCR and fused to the coding sequence of mRuby by SOEing (see Table S1). The fusion was then introduced into pSK236 using BamHI and SalI sites, yielding plasmid pSK236::Pmprf-mRuby.
To increase the efficiency of pKD46 λ-red recombineering in DC10B (recA1), the complete coding sequence and native promoter of recA from E. coli K-12 were integrated into pKD46. The coding sequence and promoter of recA were amplified from K-12 genomic DNA and introduced into pKD46 via Gibson assembly (see Table S1) between the oriR101 replication origin and the ampicillin resistance cassette, yielding plasmid pKD46′.
Vectors for BacTH complementation assays were constructed by amplifying the coding sequence of the gene of interest (see Table S1) using genomic DNA from the corresponding mutant of interest. DNA fragments were digested with BamHI and EcoRI and ligated into similarly digested pKT25 or pUT18. Vectors resulting from pKT25 integration expressed a hybrid protein in which the T25 fragment of adenylate cyclase was fused to the N terminus of VraG (see Table S2). Conversely, integration into pUT18 yielded a hybrid protein in which the T18 fragment of adenylate cyclase was fused to the C terminus of GraS, lacking a stop codon (see Table S2).
Construction of the NIH051475 restriction bypass E. coli strain.
Most S. epidermidis strains present a formidable restriction barrier to genetic manipulation. In a previous study, we constructed a plasmid-based artificial modification (PAM) system to bypass these restriction barriers. However, transformation with PAM remained difficult, and efficiency was low. To circumvent the issues of plasmid size and instability, a stable chromosomally integrated restriction bypass tool for S. epidermidis strain NIH051475 was made in DC10B, a restriction-modification-deficient E. coli strain. Oligonucleotides SC37 and SC38 (see Table S1) were used to amplify the upstream 50-bp homologous region required for integration and also to introduce the pN25 promoter. Oligonucleotides SC39 and SC40 amplified the coding sequence of the host specificity determinant modification and specificity (hsdMS) genes while providing homologous overlap regions upstream for fusion to the pN25 promoter and downstream for fusion to a flippase recognition target (FRT)-flanked kanamycin cassette, which was amplified with the oligonucleotide pair SC41 and SC42. Oligonucleotides SC41 and SC43 were then used to fuse the kanamycin cassette to the downstream 50-bp homologous region via SOEing. The upstream homologous site and promoter were fused via SOEing to the strain NIH051475 hsdMS coding sequence using oligonucleotides SC37 and SC40. Lastly, the two fragments were fused via SOEing with oligonucleotides SC44 and SC45 to generate the final amplicon for λ-red recombineering. This final amplicon targets integration at a neutral site between atpI and gidB on the DC10B chromosome, as was done previously (35).
For recombineering in DC10B, separate electrocompetent E. coli preparation and transformation procedures were carried out. Briefly, DC10B harboring pKD46′ through chemical transformation was first grown overnight at 30°C with shaking in LB broth supplemented with ampicillin. The overnight culture was then diluted 1:1,000 in super optimal broth (SOB) and allowed to grow with shaking at 30°C with ampicillin until the OD600 reached 0.4. After the addition of l-arabinose to a concentration of 0.2%, the culture was allowed to incubate for another 1 h at 30°C with shaking to enable expression of the recombinase machinery. The cells were then centrifuged at 5,000 rpm for 10 min at 4°C and resuspended in an equal volume of ice-cold 10% glycerol, followed by another washing step with 10% glycerol. The competent cells were then resuspended in 6% of the starting culture volume of 10% glycerol, centrifuged once at 5,000 rpm for 10 min at 4°C, resuspended in 10% glycerol according to 2 ml/liter of starting culture, and then stored at −80°C in 100-μl aliquots until use. Transformation was accomplished with 30 μl of thawed competent cells and 1 μg of target amplicon in a 1-mm electroporation cuvette pulsed once at 1.8 kV with 25 μF, followed by transfer of 1 ml of SOB with catabolite repression (SOC) into the cuvette to allow recovery at room temperature overnight. The cells were then pelleted at 5,000 × g for 3 min, resuspended in 50 to 100 μl SOC recovery medium, plated in their entirety on kanamycin-containing LBA, and incubated overnight at 37°C. pKD46′ was then cured from the strain by growing one colony overnight at 43°C in LB broth without antibiotic. Dilutions of this culture were plated on LBA with kanamycin and grown overnight at 43°C. Colonies were then patched onto LBA with either kanamycin or ampicillin, and those that grew on kanamycin but not ampicillin were selected and validated for integration by sequencing.
MIC testing.
MICs were determined with PMB according to Clinical and Laboratory Standards Institute standards (36). Briefly, strains were grown in TSB overnight and then diluted to 106 CFU/ml in MHB. One hundred microliters of the diluted cells was added to a 96-well round-bottomed plate. Two-fold serial dilutions of PMB beginning at 128 μg/ml were then prepared and added to the cells. Plates were incubated at 37°C for 48 h, and MICs were assessed.
mRuby fluorescence reporter assay.
Strains carrying pSK236::Pmprf-mRuby were grown overnight in MHB with chloramphenicol selection. Cultures were diluted to an OD600 of 1.6 on a Biophotometer Plus (Eppendorf), and 200 μl was then added to a 96-well flat-bottomed plate. Following a 50-min incubation at 37°C with shaking, fluorescence was read with an excitation wavelength of 558 nm and an emission wavelength of 592 nm.
Cytochrome c binding assay.
Cytochrome c binding was performed similarly to what was described previously (18). Briefly, strains were grown overnight in TSB at 37°C with shaking, and 2 ml of cells was pelleted at 5,000 × g for 2 min. The pellet was washed twice with 1 ml of 20 mM morpholinepropanesulfonic acid (MOPS) (pH 7) and then resuspended in 1 ml of MOPS. Cell densities were evaluated by OD650 values and adjusted to an OD650 of 3. The aliquots were then centrifuged for 2 min at 5,000 × g, resuspended in 300 μl of 0.5 mg/ml cytochrome c dissolved in MOPS, incubated for 10 min at room temperature, and centrifuged for 2 min at 5,000 × g. The absorbance of 200 μl of supernatant was then read at 530 nm in a 96-well flat-bottomed plate.
Two-hour killing assay.
Strains were grown overnight in TSB at 37°C with shaking and then diluted to 106 CFU/ml in TSB supplemented with 32 μg/ml PMB. A 5-ml culture was incubated at 37°C with shaking for 2 h, and a 100-μl aliquot was diluted to ∼103 CFU/ml in 0.85% NaCl, followed by plating (45 μl) on TSA plates for enumeration the following day. For comparison, samples were taken at 0 h., diluted, and plated on TSA plates.
BacTH quantification.
Chemically competent DHT1 cells were cotransformed with either pKT25-vraG and an assortment of mutated graS in pUT18 or pUT18-graS and mutated vraG in pKT25 (see Table S2). Transformants were recovered at 30°C for 1 h in LB broth with shaking, and then 50 μl was plated on LBA plates with kanamycin, ampicillin, 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG), and 40 μg/ml X-Gal. After 24 to 48 h at 30°C, individual colonies were picked and inoculated in LB broth with kanamycin, ampicillin, and 0.5 mM IPTG for overnight growth at 30°C with shaking. Overnight cultures were diluted 1:5 in M63 medium, and the OD600 was recorded. A 1.25-ml aliquot of diluted cells was then permeabilized with 25 μl of 0.1% SDS and 25 μl of chloroform by vortex-mixing and then shaking at room temperature for 30 min. Thirty-two microliters of permeabilized cells was mixed with 128 μl of PM2 buffer (70 mM Na2HPO4·12H2O, 30 mM NaH2PO4·H2O, 1 mM MgSO4, and 0.2 mM MnSO4 [pH 7.0]) containing 100 mM β-mercaptoethanol, followed by addition of 40 μl of PM2 buffer with 0.4% o-nitrophenyl-β-galactoside (ONPG) to each well of a 96-well round-bottomed plate. The plate was incubated at room temperature for 10 min with shaking, after which the reaction was stopped with 80 μl of Na2CO3. Two hundred microliters of the reaction mixture was transferred to a new 96-well plate for measurement of A405. β-Galactosidase activity was then calculated with the following equation: A = [1,000 × (OD405 of sample − OD405 of control/OD600 of sample)]/time (in minutes).
Validation of mutant protein expression.
S. epidermidis strains expressing C-terminal GraS or N-terminal VraG with an in-frame HA tag were grown overnight at 30°C in 100 ml of TSB with shaking, harvested by centrifugation at 4,000 rpm for 15 min, washed with 50 mM Tris-HCl (pH 7.4), and recentrifuged. Pelleted cells were resuspended in 2 ml of lysis buffer (50 mM Tris-HCl [pH 7.5], 50 mM NaCl, and 1 tablet of cOmplete mini protease inhibitor cocktail [Roche]) supplemented with 20 μl of 2.5 mg/ml lysostaphin and 1 μl of 250 U/μl benzonase (Millipore) and were incubated for 30 min on ice. Cells were then lysed with a BeadBeater (BioSpec, Bartlesville, OK) following the manufacturer’s protocol. After removal of unbroken cells and large cellular debris by centrifugation at 5,000 rpm for 15 min at 4°C, the supernatant was subjected to ultracentrifugation at 72,000 rpm for 30 min at 4°C to collect the membrane fraction, which was resuspended in 100 μl of 1% SDS. After protein determination with the Bradford assay, 32 μg of each protein sample was separated by SDS-PAGE and then transferred to a PVDF membrane using an iBlot 2 gel transfer device (Thermo Fisher Scientific). The membrane was blocked with 5% skim milk, washed, and then probed with a 1:1,000 dilution of primary mouse anti-HA antibody (Invitrogen) followed by a 1:5,000 dilution of donkey anti-mouse F(ab′)2 antibody conjugated to horseradish peroxidase (Jackson ImmunoResearch). The membrane was then developed with the enhanced chemiluminescence (ECL) Western blotting substrate kit (Pierce).
Statistical analyses.
Statistical analyses were performed using GraphPad Prism version 6.0c, with detailed statistical methods described in the figure legends.
ACKNOWLEDGMENTS
We sincerely thank Scot Ouellette from the University of Nebraska Medical Center for sharing with us the DHT1 E. coli cells used in our BacTH assays.
This project was supported in part by NIH grant R21 AI127922.
Footnotes
Supplemental material is available online only.
Contributor Information
Ambrose L. Cheung, Email: ambrose.cheung@dartmouth.edu.
Michael J. Federle, University of Illinois at Chicago
REFERENCES
- 1.Otto M. 2012. Molecular basis of Staphylococcus epidermidis infections. Semin Immunopathol 34:201–214. 10.1007/s00281-011-0296-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bechinger B, Gorr S-U. 2017. Antimicrobial peptides: mechanisms of action and resistance. J Dent Res 96:254–260. 10.1177/0022034516679973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scocchi M, Mardirossian M, Runti G, Benincasa M. 2016. Non-membrane permeabilizing modes of action of antimicrobial peptides on bacteria. Curr Top Med Chem 16:76–88. 10.2174/1568026615666150703121009. [DOI] [PubMed] [Google Scholar]
- 4.Piepenbreier H, Fritz G, Gebhard S. 2017. Transporters as information processors in bacterial signalling pathways. Mol Microbiol 104:1–15. 10.1111/mmi.13633. [DOI] [PubMed] [Google Scholar]
- 5.Rietkötter E, Hoyer D, Mascher T. 2008. Bacitracin sensing in Bacillus subtilis. Mol Microbiol 68:768–785. 10.1111/j.1365-2958.2008.06194.x. [DOI] [PubMed] [Google Scholar]
- 6.Dintner S, Staron A, Berchtold E, Petri T, Mascher T, Gebhard S. 2011. Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes bacteria. J Bacteriol 193:3851–3862. 10.1128/JB.05175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fritz G, Dintner S, Treichel NS, Radeck J, Gerland U, Mascher T, Gebhard S. 2015. A new way of sensing: need-based activation of antibiotic resistance by a flux-sensing mechanism. mBio 6:e00975-15. 10.1128/mBio.00975-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A 104:9469–9474. 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol 66:1136–1147. 10.1111/j.1365-2958.2007.05986.x. [DOI] [PubMed] [Google Scholar]
- 10.Oku Y, Kurokawa K, Ichihashi N, Sekimizu K. 2004. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology (Reading) 150:45–51. 10.1099/mic.0.26706-0. [DOI] [PubMed] [Google Scholar]
- 11.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KPM, van Strijp JAG. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor Mprf is based on modification of membrane lipids with l-lysine. J Exp Med 193:1067–1076. 10.1084/jem.193.9.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J Biol Chem 274:8405–8410. 10.1074/jbc.274.13.8405. [DOI] [PubMed] [Google Scholar]
- 13.Chaili S, Cheung AL, Bayer AS, Xiong YQ, Waring AJ, Memmi G, Donegan N, Yang S-J, Yeaman MR. 2016. The GraS sensor in Staphylococcus aureus mediates resistance to host defense peptides differing in mechanisms of action. Infect Immun 84:459–466. 10.1128/IAI.01030-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheung AL, Bayer AS, Yeaman MR, Xiong YQ, Waring AJ, Memmi G, Donegan N, Chaili S, Yang S-J. 2014. Site-specific mutation of the sensor kinase GraS in Staphylococcus aureus alters the adaptive response to distinct cationic antimicrobial peptides. Infect Immun 82:5336–5345. 10.1128/IAI.02480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuiack RC, Veldhuizen RAW, McGavin MJ. 2020. Novel functions and signaling specificity for the GraS sensor kinase of Staphylococcus aureus in response to acidic pH. J Bacteriol 202:e00219-20. 10.1128/JB.00219-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Falord M, Karimova G, Hiron A, Msadek T. 2012. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob Agents Chemother 56:1047–1058. 10.1128/AAC.05054-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muzamal U, Gomez D, Kapadia F, Golemi-Kotra D. 2014. Diversity of two-component systems: insights into the signal transduction mechanism by the Staphylococcus aureus two-component system GraSR. F1000Res 3:252. 10.12688/f1000research.5512.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meehl M, Herbert S, Götz F, Cheung A. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother 51:2679–2689. 10.1128/AAC.00209-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang S-J, Bayer AS, Mishra NN, Meehl M, Ledala N, Yeaman MR, Xiong YQ, Cheung AL. 2012. The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect Immun 80:74–81. 10.1128/IAI.05669-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roy A, Kucukural A, Zhang Y. 2010. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc 5:725–738. 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Källberg M, Wang H, Wang S, Peng J, Wang Z, Lu H, Xu J. 2012. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7:1511–1522. 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang H-B, Hou W-T, Cheng M-T, Jiang Y-L, Chen Y, Zhou C-Z. 2018. Structure of a MacAB-like efflux pump from Streptococcus pneumoniae. Nat Commun 9:196. 10.1038/s41467-017-02741-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kraus D, Herbert S, Kristian SA, Khosravi A, Nizet V, Götz F, Peschel A. 2008. The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol 8:85. 10.1186/1471-2180-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falord M, Mäder U, Hiron A, Débarbouillé M, Msadek T. 2011. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One 6:e21323. 10.1371/journal.pone.0021323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mascher T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol Lett 264:133–144. 10.1111/j.1574-6968.2006.00444.x. [DOI] [PubMed] [Google Scholar]
- 26.Coumes-Florens S, Brochier-Armanet C, Guiseppi A, Denizot F, Foglino M. 2011. A new highly conserved antibiotic sensing/resistance pathway in Firmicutes involves an ABC transporter interplaying with a signal transduction system. PLoS One 6:e15951. 10.1371/journal.pone.0015951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiron A, Falord M, Valle J, Débarbouillé M, Msadek T. 2011. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol Microbiol 81:602–622. 10.1111/j.1365-2958.2011.07735.x. [DOI] [PubMed] [Google Scholar]
- 28.Cho J, Costa SK, Wierzbicki RM, Rigby WFC, Cheung AL. 2021. The extracellular loop of the membrane permease VraG interacts with GraS to sense cationic antimicrobial peptides in Staphylococcus aureus. PLoS Pathog 17:e1009338. 10.1371/journal.ppat.1009338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lähdesmäki K, Ollila OHS, Koivuniemi A, Kovanen PT, Hyvönen MT. 2010. Membrane simulations mimicking acidic pH reveal increased thickness and negative curvature in a bilayer consisting of lysophosphatidylcholines and free fatty acids. Biochim Biophys Acta 1798:938–946. 10.1016/j.bbamem.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 30.Seidman CE, Struhl K, Sheen J, Jessen T. 1997. Introduction of plasmid DNA into cells. Curr Protoc Mol Biol 32:1.8.4–1.8.5. [DOI] [PubMed] [Google Scholar]
- 31.Monk IR, Shah IM, Xu M, Tan M-W, Foster TJ. 2012. Transforming the untransformable: application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. mBio 3:e00277-11. 10.1128/mBio.00277-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costa SK, Donegan NP, Corvaglia A-R, François P, Cheung AL. 2017. Bypassing the restriction system to improve transformation of Staphylococcus epidermidis. J Bacteriol 199:e00271-17. 10.1128/JB.00271-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnaud M, Chastanet A, Débarbouillé M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, Gram-positive bacteria. Appl Environ Microbiol 70:6887–6891. 10.1128/AEM.70.11.6887-6891.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Forsyth RA, Haselbeck RJ, Ohlsen KL, Yamamoto RT, Xu H, Trawick JD, Wall D, Wang L, Brown-Driver V, Froelich JM, C KG, King P, McCarthy M, Malone C, Misiner B, Robbins D, Tan Z, Zhu Z, Carr G, Mosca DA, Zamudio C, Foulkes JG, Zyskind JW. 2002. A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol Microbiol 43:1387–1400. 10.1046/j.1365-2958.2002.02832.x. [DOI] [PubMed] [Google Scholar]
- 35.Monk IR, Tree JJ, Howden BP, Stinear TP, Foster TJ. 2015. Complete bypass of restriction systems for major Staphylococcus aureus lineages. mBio 6:e00308-15. 10.1128/mBio.00308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clinical and Laboratory Standards Institute. 2015. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard—10th ed. M07-A10: Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 37.Conlan S, Mijares LA, Becker J, Blakesley RW, Bouffard GG, Brooks S, Coleman H, Gupta J, Gurson N, Park M, Schmidt B, Thomas PJ, Otto M, Kong HH, Murray PR, Segre JA, NISC Comparative Sequencing Program. 2012. Staphylococcus epidermidis pan-genome sequence analysis reveals diversity of skin commensal and hospital infection-associated isolates. Genome Biol 13:R64. 10.1186/gb-2012-13-7-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dautin N, Karimova G, Ullmann A, Ladant D. 2000. Sensitive genetic screen for protease activity based on a cyclic AMP signaling cascade in Escherichia coli. J Bacteriol 182:7060–7066. 10.1128/JB.182.24.7060-7066.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahmood R, Khan S. 1990. Role of upstream sequences in the expression of the staphylococcal enterotoxin B gene. J Biol Chem 265:4652–4656. 10.1016/S0021-9258(19)39612-7. [DOI] [PubMed] [Google Scholar]
- 40.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2 and Fig. S1. Download JB.00178-21-s0001.pdf, PDF file, 0.2 MB (255.7KB, pdf)