Abstract
Actin-based protrusions vary in morphology, stability, and arrangement on cell surfaces. Microridges are laterally elongated protrusions on mucosal epithelial cells, where they form evenly spaced, mazelike patterns that dynamically remodel by fission and fusion. To characterize how microridges form their highly ordered, subcellular patterns and investigate the mechanisms driving fission and fusion, we imaged microridges in the maturing skin of zebrafish larvae. After their initial development, microridge spacing and alignment became increasingly well ordered. Imaging F-actin and non-muscle myosin II (NMII) revealed that microridge fission and fusion were associated with local NMII activity in the apical cortex. Inhibiting NMII blocked fission and fusion rearrangements, reduced microridge density, and altered microridge spacing. High-resolution imaging allowed us to image individual NMII minifilaments in the apical cortex of cells in live animals, revealing that minifilaments are tethered to protrusions and often connect adjacent microridges. NMII minifilaments connecting the ends of two microridges fused them together, whereas minifilaments oriented perpendicular to microridges severed them or pulled them closer together. These findings demonstrate that as cells mature, cortical NMII activity orchestrates a remodeling process that creates an increasingly orderly microridge arrangement.
INTRODUCTION
Cells create diverse actin-based protrusions to carry out a wide variety of functions. Protrusions not only vary in shape and size but also vary in persistence, plasticity, dynamics, and arrangement on cell surfaces. For example, lamellipodia extend and retract within seconds or minutes (Giannone et al., 2007), whereas invadopodia persist for hours (Murphy and Courtneidge, 2011), and stereocilia are stable throughout an animal’s life (Zhang et al., 2012; Narayanan et al., 2015). Microvilli move rapidly and independently on epithelial cell surfaces (Meenderink et al., 2019), but form stable clusters by establishing protocadherin-based connections at their tips (Crawley et al., 2014; Meenderink et al., 2019). Stereocilia, on the other hand, form highly stable and stereotyped arrangements on epithelial cells, and their orientation is strictly dictated by planar cell polarity (Tarchini and Lu, 2019). Identifying mechanisms regulating the dynamics and arrangement of protrusions is critical to understanding how cell surfaces acquire diverse morphologies.
Microridges are laterally elongated protrusions found on the apical surfaces of mucosal epithelial cells (Depasquale, 2018). Although microridges are less studied than other protrusions, recent work in zebrafish periderm cells, which form the most superficial layer of the skin, has begun to identify mechanisms underlying microridge morphogenesis. Distinct from other protrusions that emerge and extend as unitary structures, microridges form from the coalescence of fingerlike precursor protrusions called pegs (Lam et al., 2015; Pinto et al., 2019; van Loon et al., 2020). Microridge development requires specification of apical-basal cell polarity (Raman et al., 2016; Magre et al., 2019), activity of the branched actin nucleation complex Arp2/3 (Lam et al., 2015; Pinto et al., 2019; van Loon et al., 2020), Plakin cytolinkers (Inaba et al., 2020), keratin filaments (Inaba et al., 2020), and cortical non-muscle myosin II (NMII) contraction, which concomitantly promotes apical constriction (Lam et al., 2015; Pinto et al., 2019; van Loon et al., 2020). Like actin filaments in microvilli (Tyska and Mooseker, 2002), actin networks in microridges constantly turn over (Lam et al., 2015), but the recruitment of keratin filaments by Plakin cytolinkers helps preserve microridge structure (Inaba et al., 2020). Microridges exhibit unusual dynamics, undergoing fission and fusion to form new patterns (Lam et al., 2015). How microridge patterns mature after their initial formation has not been determined, and the molecular mechanisms executing fission and fusion are unknown.
Microridges are arranged in striking, highly ordered patterns: Neighboring microridges are periodically spaced and tend to align parallel to one another, filling cell surfaces in mazelike arrangements that resemble the parallel organization of molecules in liquid crystals, referred to as a “nematic” organization (Needleman and Dogic, 2017). These patterns may optimize cell surface energy and/or the function of microridges in mucus retention. Microridge arrangements are reminiscent of the tissue-level patterns that arise through cell–cell signaling interactions, obeying Alan Turing’s elegant reaction-diffusion model (Turing, 1952; Kondo and Miura, 2010). At the subcellular level, however, periodic patterns often involve the regular arrangement of cytoskeletal elements. For example, axons (Xu et al., 2013), dendrites (Han et al., 2017), cilia (Jia et al., 2019), and the Caenorhabditis elegans syncytial epidermis (Costa et al., 1997) contain periodic membrane-associated cytoskeletal structures. Although reaction-diffusion mechanisms can explain some subcellular phenomena, like waves of actin polymerization in the cortex of dividing oocytes (Bement et al., 2015), regular subcellular patterns are often created by the arrangement of molecules themselves. For example, the spacing of actin rings in neurites is determined by the size of the spectrin tetramers that separate them (Xu et al., 2013). Nothing is known about the mechanisms that create highly ordered, periodic microridge patterns.
The membranes of epithelial cells associate with a thin actomyosin filament network, called the cortex (Kelkar et al., 2020). NMII forms bipolar minifilaments within the cortex, which contract actin filaments to generate forces that regulate membrane tension, cytokinesis, and cellular morphogenesis (Martin and Goldstein, 2014; Kelkar et al., 2020). Both the density and the specific arrangement of NMII minifilaments influence cortical contractility (Kelkar et al., 2020). The cortical network is attached to cell junctions and pulls them to constrict apical surfaces during a variety of morphogenetic events (Martin and Goldstein, 2014). Cortical contraction also regulates protrusion morphogenesis. For example, contraction stimulates actin treadmilling to regulate microvillar length (Chinowsky et al., 2020). In zebrafish periderm cells, pulsatile NMII activity lowers apical membrane tension to permit the formation and elongation of microridges from peg precursors (van Loon et al., 2020). Cortical NMII contraction continues in these cells after microridges have formed (van Loon et al., 2020), but the functional significance of these later contractile events is unknown.
In this study, we characterized microridge dynamics as their patterns mature into an increasingly ordered arrangement and investigated the role of NMII in these processes. We found that after initial development, fission and fusion continuously remodel microridges, but these events dampen as maturation proceeds. High-resolution imaging revealed that cortical NMII minifilaments connect adjacent microridges, and that their specific orientation relative to microridges dictates the nature of rearrangements. These findings demonstrate that cortical NMII minifilaments are not only required for microridge formation but also regulate microridge fission, fusion, and alignment to pattern maturing epithelial cell surfaces.
RESULTS
Microridge patterns mature in larval zebrafish
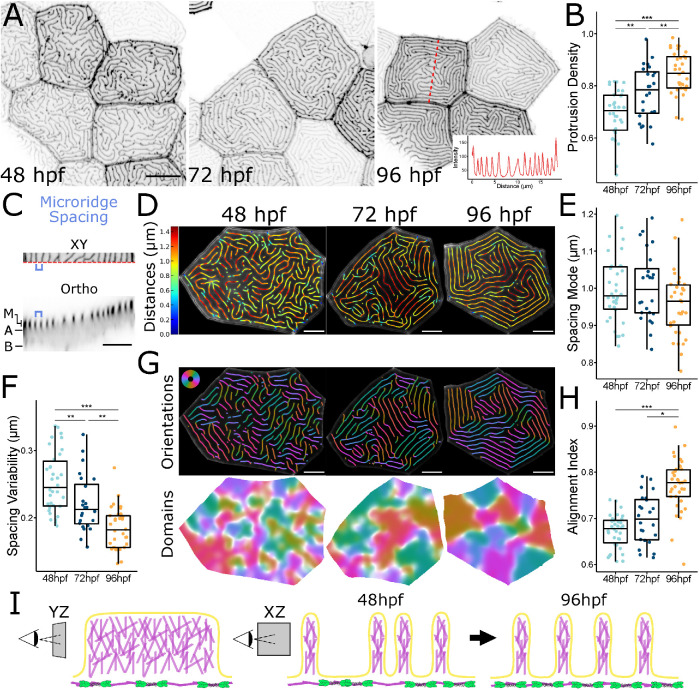
To determine how microridge spacing and patterning change as the developing zebrafish skin matures, we imaged zebrafish periderm cells expressing the F-actin reporter Lifeact-GFP (Riedl et al., 2008) in 48, 72, and 96 h postfertilization (hpf) fish (Figure 1A). Microridges had already formed and elongated by 48 hpf, but became longer on average during this period (Supplemental Figure S1B), likely reflecting a specific reduction in pegs and short microridges (Supplemental Figure S1C). Total microridge density on the apical surface increased between 48 and 96 hpf (Figure 1B), which could result from an increase in microridges or reduced apical area. However, apical cell areas were not reduced, but were in fact slightly larger at 96 hpf than at 48 or 72 hpf (Supplemental Figure S1D). Since microridge development occurs in tandem with apical constriction during early development (van Loon et al., 2020), these observations suggest that changes to microridges after 48 hpf represent a distinct maturation process.
FIGURE 1:
Microridge patterns mature over time. (A) Representative images of periderm cells expressing Lifeact-GFP in zebrafish larvae at the specified developmental stage. Images were inverted so that high intensity fluorescence appears black and low intensity is white. Inset at 96 hpf is an intensity line profile plot along the dashed red line in the associated image, showing the regular spacing between adjacent microridges along the line. (B) Dot and box-and-whisker plot of microridge density, defined as the sum microridge length (μm) normalized to apical cell area (μm2), on periderm cells at the specified stage; 48 hpf, n = 34 cells from 12 fish; 72 hpf, n = 24 cells from 10 fish; 96 hpf, n = 34 cells from 15 fish. P = 1.87 × 10–9, one-way ANOVA followed by Tukey’s HSD test: 48–72 hpf, P = 3.32 × 10–3; 48–96 hpf, P = 1.17 × 10–9; 72–96 hpf, P = 6.65 × 10–3. (C) Microridge-to-microridge spacing. Top: cropped image of a 96 hpf periderm cell expressing Lifeact-GFP. The blue bracket shows the distance between two adjacent microridges. Bottom: orthogonal optical section from the above periderm cell along the dashed red line at the bottom edge of the XY image. M, microridge protrusions; A, apical; B, basal. Blue bracket shows the distance between the same microridges as above. (D) Visualization of microridge spacing at three developmental stages. Color coding indicates the distance from each point on each microridge to the nearest neighboring microridge. Colors correspond to the distances indicated on the bar to the left. (E) Dot and box-and-whisker plots of the mode distance between neighboring microridges in periderm cells at the specified stage; 48 hpf, n = 34 cells from 12 fish; 72 hpf, n = 24 cells from 10 fish; 96 hpf, n = 34 cells from 15 fish. P = 0.089, one-way ANOVA. (F) Dot and box-and-whisker plot of microridge spacing variability, defined as the interquartile range of distances between neighboring microridges in periderm cells at the specified stages; 48 hpf, n = 34 cells from 12 fish; 72 hpf, n = 24 cells from, 10 fish; 96 hpf n = 34 cells from 15 fish. P = 9.91 × 10–10, one-way ANOVA followed by Tukey’s HSD test: 48–72 hpf, P = 7.72 × 10–3; 48–96 hpf, P = 8.11 × 10–10; 72–96 hpf, P = 1.90 × 10–3. (G) Visualization of microridge orientations at the specified stages. Microridge orientations are color-coded along each microridge (top); colors correspond to the color wheel on the top left. Microridge alignment domains were expanded from microridge orientations (bottom), using the same color wheel and scale as above. See Materials and Methods for details. (H) Dot and box-and-whisker plot of the microridge alignment index for periderm cells at the specified stages; 48 hpf, n = 34 cells from 12 fish; 72 hpf, n = 24 cells from 10 fish; 96 hpf, n = 34 cells from 15 fish. P = 2.96 × 10–12, one-way ANOVA followed by Tukey’s HSD test: 48–72 hpf, P = 0.121; 48–96 hpf, P = 4.02 × 10–10; 72–96 hpf, P = 8.79 × 10–7. (I) Diagram of microridge structure and spacing. (YZ) Branched actin fills microridge protrusions, depicted as a lengthwise section, the apical actomyosin cortex is shown below the protrusions. (XZ) Microridge spacing, depicted as a cross-section, is variable after microridge formation (48 hpf), but gradually matures to a more regularly spaced pattern (96 hpf). Scale bars: 10 μm (A) and 5 μm (C, D, and G). *p ≤ 0.05, **p ≤ 0.01, and ***p ≤ 0.001. For box-and-whisker plots, the middle line is the median, and the bottom and top ends of boxes are 25th and 75th percentiles, respectively.
One of the most striking features of microridges is their regularly spaced and aligned arrangement, reminiscent of the nematic organization of molecules in liquid crystals (Needleman and Dogic, 2017). To investigate how microridge spacing changes as cells mature, we measured the distance between every point on each microridge and the nearest point on a neighboring microridge (Figure 1, C and D). The mode, median, and mean distances between microridges were similar between the three different stages (Figure 1E; Supplemental Figure S1, E and F) and, as expected, corresponded to the orthogonal distance between adjacent microridges (Figure 1, A, C, and D). To measure spacing variability, we compared the interquartile range of distances, revealing that variability decreased over time (Figure 1F). Variability measured as SD or coefficient of variation yielded the same result (not shown). These observations suggest that initially variable microridge spacing matured toward a specific spacing distance (Figure 1I).
To determine how microridge alignment changes as microridge spacing becomes less variable, we developed an “alignment index” measurement. First, we calculated orientation along each microridge, then defined and colored regions containing microridges aligned in a similar orientation (Figure 1G). White regions represent border areas with orientation disorder; in other words, where microridges with different orientations meet. Cells with more white regions (less saturated) have a more disordered microridge arrangement. In our alignment index, a cell with no aligned microridges (all white) would have an alignment index of 0, and a cell with perfectly parallel microridges (no white) would have an index of 1 (Figure 1H). This analysis revealed that the number of domains with aligned microridges decreased (and each domain increased in area) over time (Figure 1, G and H), demonstrating that microridges increasingly align parallel to one another as the skin develops.
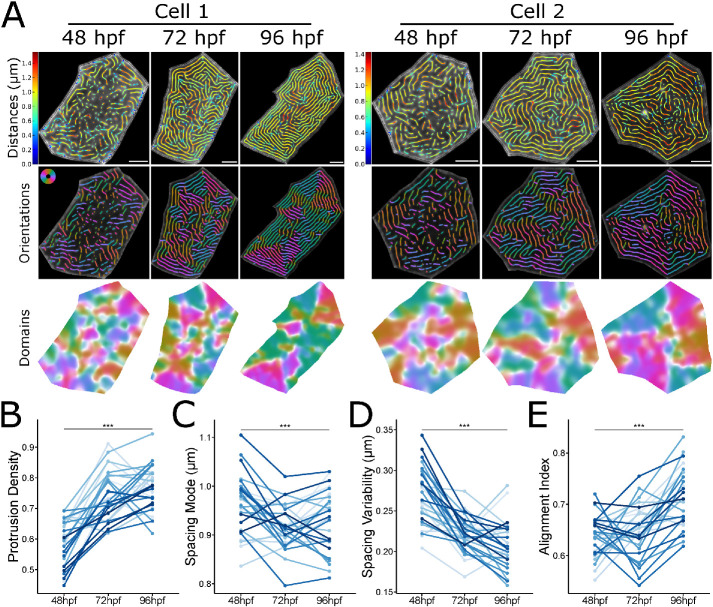
To determine if population-level changes in microridge patterning reflect microridge maturation in individual cells, we scatter-labeled periderm cells with RFP, enabling us to identify the same cells day-to-day and thus track how microridge spacing and alignment change over time. Although each cell behaved differently, on average, microridge density increased, spacing became less variable, and microridges increasingly aligned between 48 and 96 hpf (Figure 2), demonstrating that population-level trends in microridge arrangement reflect the maturation of microridge patterns in individual cells toward a nematic arrangement.
FIGURE 2:
Microridge patterns mature on individual periderm cells. (A) Microridge distances, orientations, and alignment domains in two cells from 48–96 hpf. (B) Line and point plot of microridge density, defined as the sum microridge length (μm) normalized to apical cell area (μm2), in periderm cells over time; n = 28 cells from 9 fish. P = 5.87 × 10–15, one-way repeated measures ANOVA. (C) Line and point plot of the mode distances between neighboring microridges in periderm cells over time. n = 28 cells from 9 fish. P = 4.08 × 10–4, one-way repeated measures ANOVA. (D) Line and point plot of microridge spacing variability (interquartile range of distances) between neighboring microridges in periderm cells over time; n = 28 cells from 9 fish. P = 7.37 × 10–11, one-way repeated measures ANOVA. (E) Line and point plot of microridge alignment index in periderm cells over time; n = 28 cells from 9 fish. P = 5.16 × 10–8, one-way repeated measures ANOVA. Scale bars: 5 μm (A). ***p ≤ 0.001.
Microridges continuously rearrange
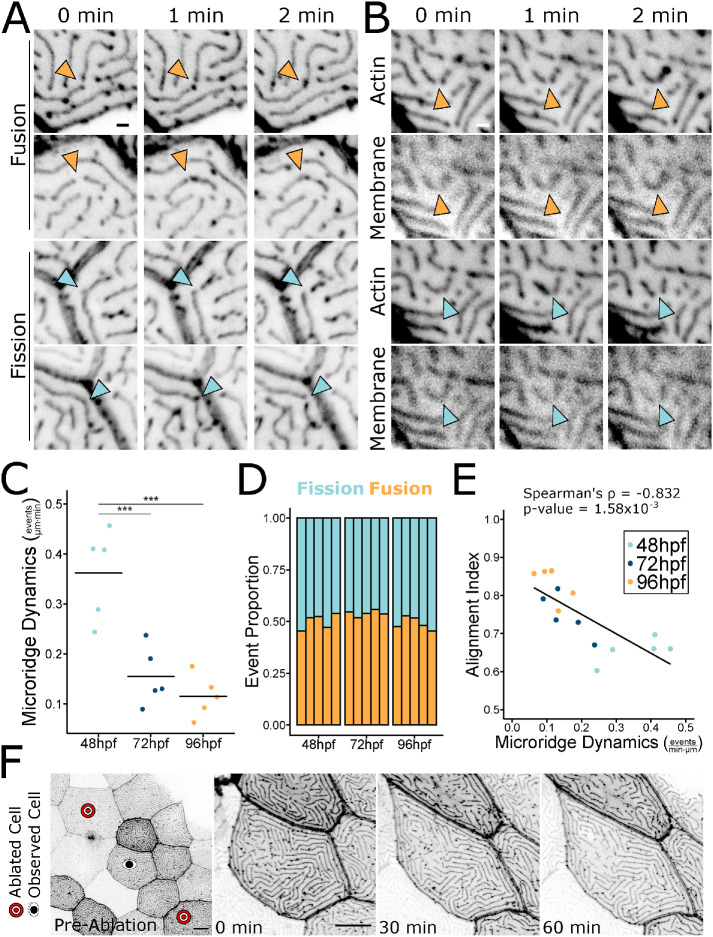
To determine the mechanism by which microridge patterns change over time, we performed time-lapse imaging of periderm cells expressing Lifeact-GFP at 30-s intervals. At each developmental stage, pegs, the fingerlike precursor protrusions that coalesce to form microridges, continued to dynamically appear within and between microridges (Supplemental Video S1), likely contributing to microridge lengthening. As previously observed (Lam et al., 2015), microridges underwent two types of rearrangements that altered their pattern. First, intact microridges sometimes broke apart into two separate microridges; second, two separate microridges sometimes fused end-to-end to form a longer microridge (Figure 3A; Supplemental Videos S1 and S2). Imaging a reporter for the plasma membrane demonstrated that these events reflect fission or fusion of the whole protrusion, not just of its internal actin structure (Figure 3B; Supplemental Video S3). As microridges matured, rearrangement events decreased from 0.362 events/μm⋅min at 48 hpf to 0.155 and 0.115 events/μm⋅min at 72 and 96 hpf, respectively (Figure 3C; Supplemental Video S1). Fission and fusion events occurred with roughly equal frequency. The proportion of fission and fusion events did not change over time (Figure 3D), but the frequency of these rearrangements decreased as the pattern matured (Figure 3E, Spearman Correlation Coefficient = –0.832).
FIGURE 3:
Microridges dynamically rearrange. A) Stills from time-lapse movies of 48 hpf periderm cells expressing Lifeact-GFP, showing microridges undergoing fission or fusion. Orange arrowheads indicate fusion; blue arrowheads indicate fission. Images were inverted, so that high intensity fluorescence appears black and low intensity is white. Images are still frames from Supplemental Video S2. (B) Stills from time-lapse movies of 48 hpf periderm cells expressing Lifeact-GFP (actin) and mRuby-PH-PLC (membrane), showing microridges undergoing fission or fusion. Orange arrowheads indicate fusion; blue arrowheads indicate fission. Images were inverted, so that high intensity fluorescence appears black and low intensity is white. Images are still frames from Supplemental Video S3. (C) Jittered dot plot of the sum of fission and fusion events in each cell, normalized to cell apical area, over a 9.5-min period (events/μm⋅min) at the specified stage. Middle bar represents the mean. n = 5 cells from 5 fish at all stages. P = 1.62 × 10–4, one-way ANOVA followed by Tukey’s HSD test: 48–72 hpf, P = 9.75 × 10–4; 48–96 hpf, P = 2.17 × 10–4; 72–96 hpf, P = 0.629. (D) Stacked bar plot of the proportion of fission and fusion events indicated stages. Each bar represents one cell, n = 5 cells from 5 fish at all developmental stages. Fusion events were used in a Test of Equal Proportions (P = 0.224) and fusion event proportion estimates were 0.501, 0.543, and 0.488 for 48, 72, and 96 hpf, respectively. (E) Scatter plot of microridge dynamics (events/μm⋅min) over a 9.5-min period vs. the microridge alignment index at the start of the 9.5-min period; n = 5 cells from 5 fish at all developmental stages. P = 1.16 × 10–4, Spearman’s rank correlation ρ = –0.832. (F) Stills from a time-lapse movie of Lifeact-GFP-expressing periderm cells stretching in response to neighbor cell ablation in a 72 hpf zebrafish. Preablation image shows the cell of interest between the ablated cells and the images of the cell of interest immediately after ablation (0 min) and at 30-min intervals after ablation. The cell stretched dramatically, but microridge rearrangements did not appreciably increase. Microridge rearrangements occurred at a rate of 0.00393 events/μm⋅min over the course of the video (compare to rates in C). Images were inverted, so that high intensity fluorescence appears black and low intensity is white. Images are still frames from Supplemental Video S4. Scale bars: 1 μm (A and B) and 10 μm (E). ***p ≤ 0.001.
Movie S1.
Microridge fusion and fission diminish as microridge patterns mature. 9.5-minute time-lapse videos with 30-second intervals of periderm cells expressing Lifeact-GFP in zebrafish at the indicated developmental stage. Microridge fusion and fission attenuate as microridges become longer and more aligned at each stage. Orange circles show locations of microridge fusions. Blue circles show locations of microridge fissions. Scale bar: 10μm.
Movie S2.
Microridges fuse and fission. 4.5-minute time-lapse videos with 30-second intervals of periderm cells expressing Lifeact-GFP in 48hpf zebrafish. White arrowheads show locations of microridge fusion and fission events. Scale bar: 1μm.
Movie S3.
Microridge fusion and fission reflect fusion and fission of the plasma membrane. 9.5-minute time-lapse video with 30-second intervals of periderm cells expressing fluorescent reporters for actin (Lifeact-GFP) and membrane (mRuby-PH-PLC) on 48hpf zebrafish. Microridge fusion (yellow arrowhead) and fission (white arrowhead) in the actin channel are mimicked by fission and fusion of projections in the membrane channel. Time-lapse frames are sum projection images. Scale bar: 1μm.
Cell stretching does not induce microridge rearrangement
Periderm cells are constantly pushed and pulled by neighboring cells as the epidermis grows. We therefore speculated that microridge fission and fusion may be induced by forces associated with cell shape distortion. To test this idea, we ablated periderm cells on either side of an observed cell using a laser on a 2-photon microscope (O’Brien et al., 2009b; van Loon et al., 2020). This procedure caused the central cell to stretch between the two wounds and often pucker or bulge in the orthogonal axis. Surprisingly, cell elongation did not increase microridge fission or fusion, but simply distorted microridges to accommodate the cells’ new shapes (Figure 3F; Supplemental Video S4). This observation suggests that microridges do not undergo fission or fusion simply as a result of cellular distortion, thus implying that remodeling events are actively regulated.
Movie S4.
Rapid cell shape changes do not induce microridge fusion and fission. 60-minute time-lapse video with 1-minute intervals of periderm cells expressing Lifeact-GFP on 72hpf zebrafish. Time-lapse begins immediately after laser ablation of periderm cells on either side of the cell of interest. The cell of interest rapidly elongates between the two wounds, but does not increase fusion and fission events. Orange circles show locations of microridge fusions. Blue circles show locations of microridge fissions. Microridge rearrangements occurred at a rate of 0.00393 events/μm·min over the course of the video. Scale bar: 10μm.
Microridge rearrangements require cortical NMII contraction
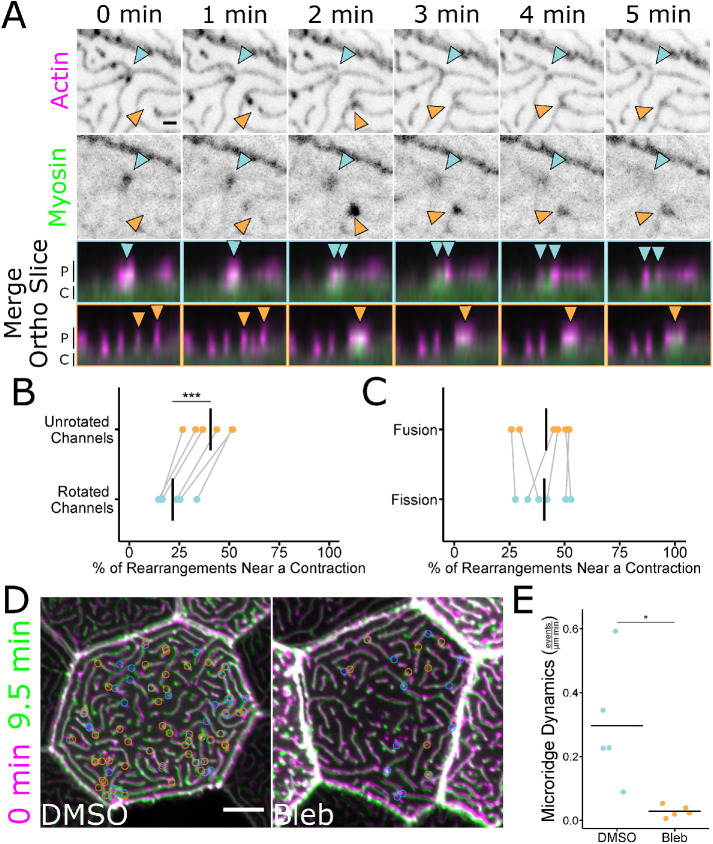
The apical cortex of periderm cells experiences pulsatile NMII-based contractions through at least 48 hpf (van Loon et al., 2020). These contractions are required for apical constriction and the coalescence of peg precursors to form and elongate microridges during initial stages of microridge morphogenesis (van Loon et al., 2020), but later functions have not been described. To test if cortical contraction affects microridge fission or fusion events, we made time-lapse videos of periderm cells expressing fluorescent reporters for both F-actin (Lifeact-Ruby) and NMII (Myl12.1-EGFP) (Maître et al., 2012; van Loon et al., 2020). At 48 hpf, periderm cells displayed local pulses of NMII reporter fluorescence in the apical cortex (Figure 4A; Supplemental Video S5), which we previously found to reflect NMII contraction (van Loon et al., 2020). Many of these contraction events correlated spatially and temporally with microridge rearrangements (Figure 4A). To quantify this correlation, we measured the distance between microridge rearrangement events and the nearest detectable NMII contractile pulse in the same frame. On average, 41% of microridge rearrangements occurred within 1 μm of an NMII contraction (Figure 4B). By contrast, when the NMII reporter channel was rotated 90°, only 22% occurred within 1 μm of a contraction (Figure 4B), indicating that the coincidence between contraction and rearrangement events did not occur by chance. These observations likely underestimate the number of rearrangement events associated with contraction, since contractions may be shorter-lasting or dimmer than we can detect with our reporter. NMII contractions equally correlated with fission and fusion events (Figure 4C).
FIGURE 4:
Microridge rearrangements spatially and temporally correlate with NMII contraction. (A) Stills from a time-lapse movie of 48 hpf zebrafish periderm cells expressing Lifeact-mRuby (actin) and Myl12.1-EGFP (myosin). Microridge fission occurred as a myosin contraction event dissipated (blue arrowheads). Microridges fused as a myosin contraction intensified (orange arrowheads). Merged images show the microridge protrusions (P) and apical cortex (C) of the above fission (blue arrowheads and borders) and fusion (orange arrowheads and borders) events from an orthogonal view. Single-channel images were inverted, so that high intensity fluorescence appears black and low intensity is white. Images are still frames from Supplemental Video S5. (B) Dot plot of the percentage of microridge fission and fusion events within 1 μm of an NMII contraction over a 9.5-min period. The graph compares unrotated channels to data analyzed after rotating the NMII fluorescence channel 90° relative to the actin fluorescence channel. The gray lines connect the unrotated samples to their rotated counterparts; n = 6 cells from 6 fish, including 3 cells from 3 fish at 24 hpf and 3 cells from 3 fish at 48 hpf. P = 2.27 × 10–4, paired t test. (C) Dot plot of the percentage of microridge fission and fusion events within 1 μm of an NMII contraction event over a 9.5-min period. The graph compares contraction-associated fusion events to contraction-associated fission events in the same cells. The gray lines connect points from the same cell; n = 6 cells from 6 fish, including 3 cells from 3 fish at 24 hpf and 3 cells from 3 fish at 48 hpf. P = 0.778, paired t test. (D) Overlap frames from 9.5-min time-lapse movies of 49 hpf zebrafish periderm cells expressing Lifeact-GFP after 1-h exposure to 1% DMSO (vehicle control) or 50 μM blebbistatin. Circles indicate the locations where fission (blue) and fusion (orange) events were detected over the course of the 9.5-min movies (frames were collected at 30-s intervals). Overlapped images are from Supplemental Video S6. (E) Jittered dot plot of the sum of fission and fusion events in each cell, normalized to cell apical area, over a 9.5-min period (events/μm⋅min) in cells after 1-h exposure to 1% DMSO (vehicle control) or 50 μM blebbistatin; n = 5 cells from 5 fish for control and treatment. P = 0.033, unpaired t test. Scale bars: 1 μm (A) and 5 μm (D). *p ≤ 0.05 and ***p ≤ 0.001. Bars in dot plots represent the mean.
Movie S5.
NMII contractions correlate with microridge fusion and fission. 9.5-minute time-lapse video with 30-second intervals of periderm cells expressing fluorescent reporters for actin (Lifeact-mRuby) and NMII (Myl12.1-EGFP) on 48hpf zebrafish. Microridges fuse near sites of intensifying myosin fluorescence signal (yellow arrowheads) and fission near sites of diminishing myosin fluorescence signal (white arrowhead). Scale bar: 1μm.
To directly test if NMII contraction is required for microridge rearrangements, we treated 48 hpf fish with the specific NMII inhibitor blebbistatin (Straight et al., 2003) for 1 h, then made 9.5-min videos of periderm cells expressing Lifeact-GFP. NMII inhibition dramatically reduced fission and fusion compared with controls (Figure 4, D and E; Supplemental Video S6), demonstrating that NMII activity is required for microridge remodeling.
Movie S6.
Short-term NMII inhibition reduces microridge fusion and fission. 9.5-minute time-lapse video with 30-second intervals of periderm cells expressing Lifeact-GFP on 49hpf zebrafish after 1-hour treatment with 1% DMSO or 50μM blebbistatin. Microridge fusion and fission decrease in periderm cells after 1-hour treatment with blebbistatin. Orange circles show locations of microridge fusions. Blue circles show locations of microridge fissions. Scale bar: 5μm.
NMII contraction regulates microridge density and spacing
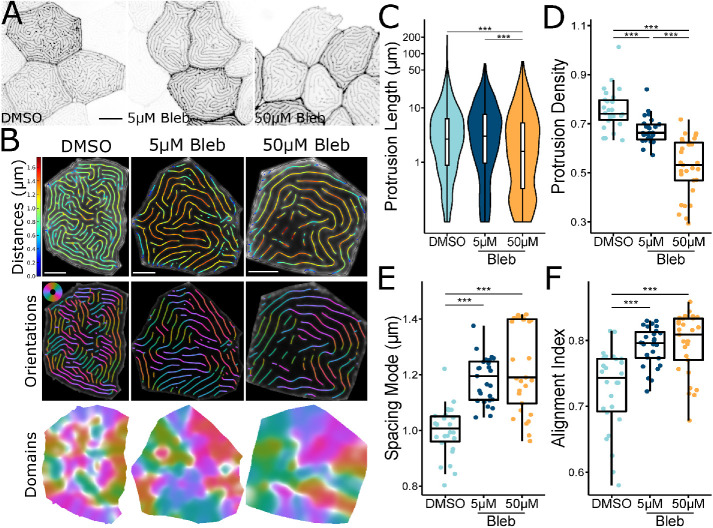
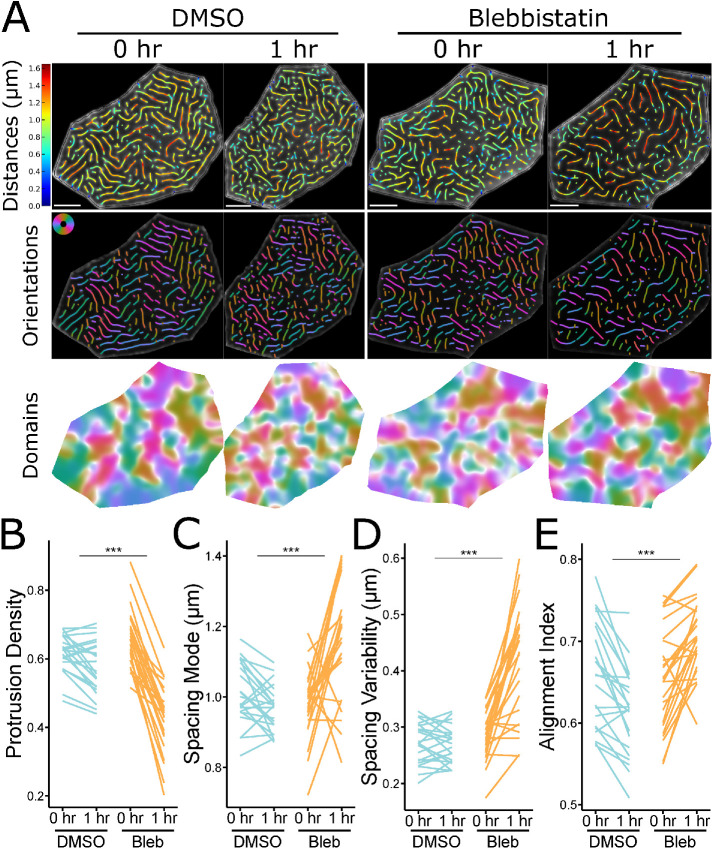
Given that NMII contraction promotes microridge rearrangements, and that these dynamic events negatively correlate with microridge alignment, we hypothesized that inhibiting NMII contraction may disrupt microridge maturation. To determine the long-term consequences of suppressing NMII activity, we treated zebrafish with blebbistatin for 24 h, starting at 48 hpf. Compared to controls, microridges in blebbistatin-treated animals were shorter, distributed less densely, and spaced more widely (Figure 5, A–E). These observations indicate that microridges must be actively maintained by contraction, which can facilitate the incorporation of new pegs into established microridges. Blebbistatin also increased microridge alignment (Figure 5F). This effect on alignment may be a consequence of the lower microridge density, since our alignment index measures the number of domains containing aligned microridges (Figure 5, B, D, and F), but could also indicate that suppressing contraction allows the system to settle into a local energy minimum (see Discussion). Since long-term NMII inhibition can have deleterious, indirect effects on cells, we compared microridges on individual cells before and after 1-h blebbistatin treatment. Similar to 24-h treatment, 1-h exposure to blebbistatin disrupted microridge spacing, decreasing density and increasing the microridge alignment index (Figure 6).
FIGURE 5:
Inhibiting NMII changes microridge patterns. (A) Representative images of periderm cells expressing Lifeact-GFP on 72 hpf zebrafish after 24-h exposure to the specified concentration of blebbistatin or vehicle control (DMSO). Images were inverted, so that high intensity fluorescence appears black and low intensity is white. (B) Visualizations of microridge distances, orientations, and alignment domains from periderm cells at 72 hpf, after 24-h exposure to the specified concentration of blebbistatin or vehicle control (DMSO). (C) Violin and box-and-whisker plot of projection length for periderm cells in 72 hpf zebrafish, after 24-h exposure to the specified concentration of blebbistatin or vehicle control (DMSO). DMSO, n = 26 cells from 9 fish; 5 μM blebbistatin, n = 27 cells from 9 fish; 50 μM blebbistatin, n = 29 cells from 9 fish. P < 2.2 × 10–16, Kruskal–Wallis test followed by Dunn’s test with Benjamini-Hochberg p value adjustment: DMSO-5 μM blebbistatin, P = 0.173; DMSO-50 μM blebbistatin, P = 2.51 × 10–13; 5 μM blebbistatin-50 μM blebbistatin, P = 3.79 × 10–17. (D) Dot and box-and-whisker plot of microridge density, defined as the sum microridge length (μm) normalized to apical cell area (μm2), for periderm cells in 72 hpf zebrafish after 24-h exposure to the specified concentration of blebbistatin or vehicle control (DMSO). DMSO, n = 26 cells from 9 fish; 5 μM blebbistatin, n = 27 cells from 9 fish; 50 μM blebbistatin, n = 29 cells from 9 fish. P = 2.80 × 10–14, one-way ANOVA followed by Tukey’s HSD test: DMSO-5 μM blebbistatin, P = 3.07 × 10–3; DMSO-50 μM blebbistatin, P <2 × 10–16; 5 μM blebbistatin-50 μM blebbistatin, P = 7.70 × 10–8. (E) Dot and box-and-whisker plot of the mode distance between neighboring microridges in periderm cells in 72 hpf zebrafish after 24-h exposure to the specified concentration of blebbistatin or vehicle control (DMSO). DMSO, n = 26 cells from 9 fish; 5 μM blebbistatin, n = 27 cells from 9 fish; 50 μM blebbistatin, n = 29 cells from 9 fish. P = 0.318, one-way ANOVA. (F) Dot and box-and-whisker plot of the alignment index on periderm cells in 72 hpf zebrafish after 24-h exposure to the specified concentration of blebbistatin or vehicle control (DMSO). DMSO, n = 26 cells from 9 fish; 5 μM blebbistatin, n = 27 cells from 9 fish; 50 μM blebbistatin, n = 29 cells from 9 fish. P = 4.56 × 10–7, one-way ANOVA followed by Tukey’s HSD test: DMSO-5 μM blebbistatin, P = 1.11 × 10–5; DMSO-50 μM blebbistatin, P = 2.38 × 10–6; 5 μM blebbistatin-50 μM blebbistatin, P = 0.951. Scale bars: 10 µm (A) and 5 μm (B). **p ≤ 0.01 and ***p ≤ 0.001. For box-and-whisker plots, the middle line is the median, and bottom and top ends of boxes are 25th and 75th percentiles, respectively.
FIGURE 6:
Short-term inhibition of NMII contractility alters microridge patterns in individual cells. (A) Representative visualizations of microridge distances, orientations, and alignment domains in periderm cells expressing Lifeact-GFP before (48 hpf, 0 h) and after (49 hpf, 1 h) 1-h treatment with 50 μM blebbistatin or vehicle (DMSO). (B) Line plot of microridge density, defined as the sum microridge length (μm) normalized to apical cell area (μm2), from periderm cells before (48 hpf, 0 h) and after (49 hpf, 1 h) 1-h treatment with 50 μM blebbistatin or vehicle control (DMSO). DMSO, n = 22 cells from 4 fish; 50 μM blebbistatin, n = 25 cells from 4 fish. P = 4.09 × 10–11, one-way repeated measures ANOVA. (C) Line plot of microridge spacing mode from periderm cells before (48 hpf, 0 h) and after (49 hpf, 1 h) 1-h treatment with 50 μM blebbistatin or vehicle control (DMSO). DMSO, n = 22 cells from 4 fish; 50 μM blebbistatin, n = 25 cells from 4 fish. P = 7.76 × 10–6, one-way repeated measures ANOVA. (D) Line plot of microridge spacing variability (interquartile range of distances) between neighboring microridges in periderm cells before (48 hpf, 0 h) and after (49 hpf, 1 h) 1-h treatment with 50 μM blebbistatin or vehicle control (DMSO). DMSO, n = 22 cells from 4 fish; 50 μM blebbistatin, n = 25 cells from 4 fish. P < 2 × 10–16, one-way repeated measures ANOVA. (E) Line plot of the alignment index in periderm cells before (48 hpf, 0 h) and after (49 hpf, 1 h) 1-h treatment with 50 μM blebbistatin or vehicle control (DMSO). Note that control treatment with DMSO decreased alignment, likely reflecting disruption of the pattern by the mounting and unmounting procedure required for this experiment, but treatment with blebbistatin increased alignment, emphasizing the role of NMII in this process. DMSO, n = 22 cells from 4 fish; 50 μM blebbistatin, n = 25 cells from 4 fish. P = 1.02 × 10–8, one-way repeated measures ANOVA. Scale bars: 5 μm (A). ***p ≤ 0.001.
High-resolution imaging reveals individual NMII minifilaments in the cortex
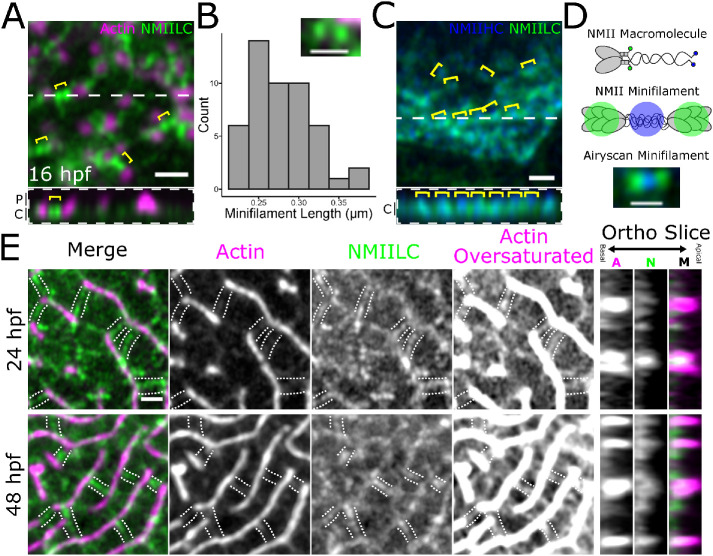
Since NMII inhibition experiments could not disambiguate NMII’s role in regulating microridge fission and fusion, length maintenance, and spacing, we addressed these questions by imaging NMII organization and activity in the periderm cortex directly. To image NMII and F-actin with improved spatial resolution, we used Airyscan microscopy (Weisshart, 2014). Using this approach, the NMII reporter often appeared as pairs of puncta (Figure 7A). The Myl12.1-EGFP NMII reporter is a fusion of EGFP to a myosin regulatory light chain (Maître et al., 2012; van Loon et al., 2020), which binds near myosin heads at opposing ends of NMII minifilaments. We thus speculated that puncta pairs represent ends of single bipolar minifilaments. Consistent with this possibility, the median distance between intensity maxima of NMII reporter doublets was 281 nm (Figure 7B), similar to the reported length of bipolar minifilaments assembled in vitro (∼300 nm in length; Billington et al., 2013). To further test if these structures are individual minifilaments, we imaged periderm cells expressing reporters for both NMII light chain (Myl12.1-Ruby) and a C-terminally tagged NMII heavy chain (Myh9a-EGFP). A fluorophore at the C-terminus of NMII heavy chains should localize to the middle of minifilaments, between NMII heavy chain heads (Figure 7, C and D). Puncta in periderm cells expressing both reporters were arranged in the expected alternating pattern (Figure 7, C and D). Thus, our imaging system allows us to distinguish individual NMII minifilaments within the plane of the apical cortex in cells of living animals.
FIGURE 7:
NMII minifilaments connect adjacent pegs and microridges. (A) Airyscan image of a 16 hpf zebrafish periderm cell expressing fluorescent reporters for actin (Lifeact-Ruby) and NMII light chain (Myl12.1-GFP). Pairs of green puncta (yellow brackets) appear in the cortex between adjacent pegs (magenta puncta). Below is an orthogonal view of the peg protrusions (P) and apical cortex (C) along the dashed white line in the top image. (B) Histogram of distances between the intensity maxima of presumptive NMII minifilaments. Inset is a representative image showing GFP signal at opposing ends of a presumptive NMII minifilament in a periderm cell expressing reporters for actin (Lifeact-Ruby) and NMII light chain (Myl12.1-GFP); n = 49 minifilaments from 4 cells on 4 fish. (C) Airyscan image of a 24 hpf zebrafish periderm cell expressing fluorescent reporters for NMII heavy chain (NMIIHC, Myh9a-mCherry) and NMII light chain (NMIILC, Myl12.1-GFP). NMIIHC channel was pseudo-colored blue. Yellow brackets show examples of GFP-mCherry-GFP fluorescence patterns. Below is an orthogonal view of apical cortex (C) along the dashed white line in the top image. (D) Diagram of NMII fluorescent protein fusion design and expected NMII minifilament fluorescence pattern. The top graphic shows an NMII macromolecule, composed of two heavy chains, two essential light chains, and two regulatory light chains. GFP was fused to the regulatory light chains (Myl12.1-GFP), while mCherry was fused to the tail of the heavy chains (Myh9a-mCherry; represented in blue). The middle graphic shows the expected fluorescence pattern when multiple NMII macromolecules, labeled like the one in the top graphic, assemble into an NMII minifilament. The bottom Airyscan image shows an NMII minifilament in the cortex of a 24 hpf zebrafish periderm cell expressing Myl12.1-GFP and Myh9a-mCherry. (E) Airyscan images showing NMII minifilaments connecting adjacent microridges side-to-side and end-to-end during (24 hpf) and after (48 hpf) microridge formation in periderm cells expressing reporters for actin (Lifeact-Ruby) and NMII light chain (Myl12.1-GFP). The oversaturated images reveal actin filaments in the cortex. The panels to the right show actin (C), NMII (N),s and merged channels (M) in an orthogonal section. The dotted lines track along NMII minifilament “bridges” and F-actin in the apical cortex. Scale bars: 1 μm (A, C, and E) and 500 nm (B and D)
Cortical NMII minifilaments associate with pegs and microridges
To determine how NMII minifilaments are arranged relative to cell protrusions, we imaged them, along with F-actin, at several developmental stages. Prior to microridge formation (16 hpf), NMII minifilaments in the apical cortex were closely associated with microridge peg precursors (Figure 7A) and continued to associate with protrusions as pegs coalesced to form microridges. Rotating the NMII channel relative to the actin channel significantly reduced the proximity between actin and NMII fluorescence signals, demonstrating that the association of minifilaments with protrusions did not occur by chance (Supplemental Figure S2, A and B). At 24 hpf, NMII minifilaments were often attached to two separate microridges, bridging them end-to-end or side-to-side (Figure 7E). This organization was maintained as microridges matured: At 48 hpf many cortical NMII “bridges” formed perpendicular connections between adjacent microridges, often appearing to consist of two end-to-end minifilaments (Figure 7E).
NMII minifilaments orchestrate microridge rearrangement and spacing
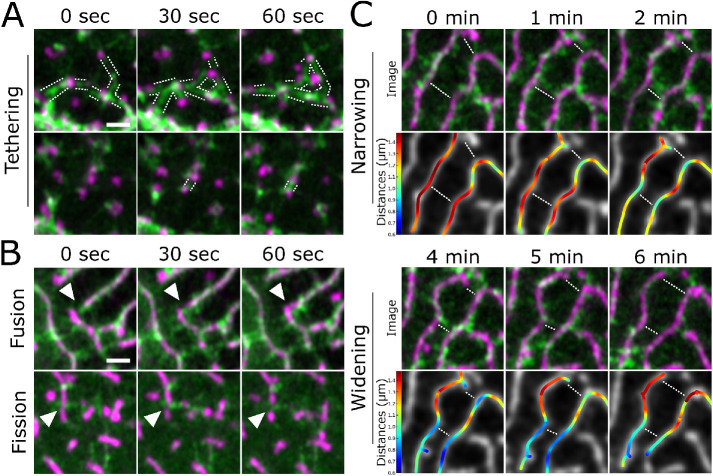
To observe how the organization of NMII minifilaments in the cortex relates to protrusion dynamics, we made high-resolution videos of periderm cells expressing Lifeact-Ruby and Myl12.1-EGFP. During early morphogenesis, appearance and disappearance of pegs often correlated with appearance and disappearance of NMII reporter signal, and movement of pegs was associated with a corresponding movement of the NMII reporter (Figure 8A; Supplemental Video S7), confirming that NMII minifilaments are tethered to protrusions. At later stages, when microridges remodel, the orientation of NMII minifilaments correlated with the type of microridge rearrangement observed. Minifilaments connecting the ends of two microridges appeared to pull them together, fusing them into a longer microridge (Figure 8B; Supplemental Figure S2C; Supplemental Video S7). By contrast, minifilaments oriented perpendicular to microridges were often associated with fission events, which occurred at the point where microridges attached to the minifilaments (Figure 8B; Supplemental Figure S2D; Supplemental Video S7). To quantify these observations, we identified 30 fusion and 30 fission events by examining the actin channel, then examined the NMII channel to score the orientation of minifilaments relative to those events. Remarkably, in all cases of fusion (30/30), a minifilament appeared between the ends of the microridges that were about to fuse. Similarly, in all cases of fission (30/30), a minifilament was attached to the parent microridge prior to fission.
FIGURE 8:
NMII minifilaments dynamically connect pegs and organize microridge rearrangements. A) Airyscan time-lapse images of NMII minifilaments dynamically connecting pegs as they emerge in the cortex of a periderm cell expressing fluorescent reporters for actin (Lifeact-mRuby) and NMII (Myl12.1-GFP). The dotted lines track along NMII minifilament “bridges.” Images are still frames from Supplemental Video S7. (B) Airyscan time-lapse images of microridge rearrangements (white arrowheads) in periderm cells expressing fluorescent reporters for actin (Lifeact-mRuby) and NMII (Myl12.1-GFP). In the top panels, an NMII minifilament connects the ends of adjacent microridges, fusing them together. In the bottom panels, NMII minifilaments oriented perpendicular to a microridge appear to sever it. Images are still frames from Supplemental Video S7. (C) Stills from an Airyscan time-lapse movie showing the spacing between microridges narrowing (first 2 min; top) and then widening (last 2 min; bottom) in a periderm cell expressing fluorescent reporters for actin (Lifeact-mRuby) and NMII (Myl12.1-GFP). Top rows show Airyscan images; bottom rows show color-coded distances. The top two rows show NMII minifilaments connecting adjacent microridges and apparently pulling them together. The bottom two rows show the NMII minifilament bridge between microridges dissipating as the adjacent microridges move further apart. The dotted lines highlight narrowing and widening regions. Distance map colors correspond to color bars on the left. Images are still frames from Supplemental Video S7. Scale bars: 1 μm (A and B).
Movie S7.
NMII minifilaments coordinate peg dynamics and microridge fusion, fission, and spacing. 9-minute time-lapse videos with 1-minute intervals of periderm cells expressing fluorescent reporters for actin (Lifeact-mRuby) and NMII (Myl12.1-EGFP). NMII minifilaments appear as two green puncta. Different NMII-mediated events indicated by title cards. "Bridges" of one or two NMII minifilaments attach to pegs as they appear, and occasionally pull them toward one another. NMII minifilaments connect two microridge ends and fuse them into a longer microridge. NMII minifilaments oriented perpendicular to a microridge in the x-y plane sever a microridge. Finally, NMII minifilament "bridges" connecting two adjacent microridges contract to pull the microridges closer together, and allow microridges to drift further apart as they disappear. Scale bar: 1μm.
Minifilaments arranged perpendicular to microridges also appeared to regulate microridge spacing: the attachment of minifilaments to two parallel microridges brought them closer together, whereas their disappearance or detachment allowed the two microridges to drift apart (Figure 8C; Supplemental Figure S2E; Supplemental Video S7). These observations suggest that attachment to cortical NMII minifilaments allows microridges to sample different spacing arrangements on the apical surface.
DISCUSSION
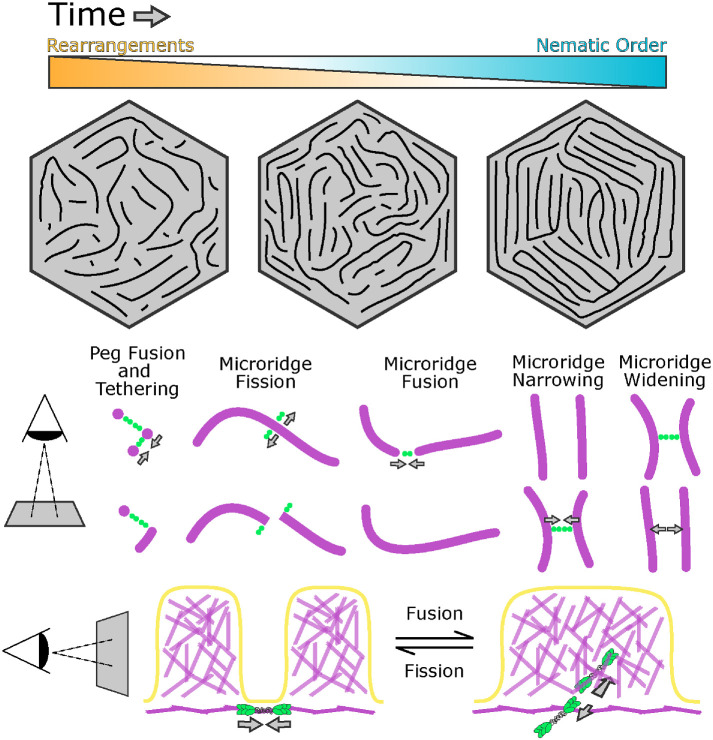
Our study reveals that cortical NMII orchestrates a unique process for the patterning and maturation of microridges. Cells retain microridges on their surfaces for days, and likely even weeks, but, unlike extremely stable stereocilia, microridges continuously remodel through an NMII-mediated “recombination” process of fission and fusion as they mature toward a more ordered, nematic arrangement (Figure 9). Thus, at least during the first week of development, microridges are not permanent cell identifiers, like a fingerprint, but evolving structures that form new patterns over time.
FIGURE 9:
Model for microridge maturation and minifilament-mediated rearrangements. Top: the nematic order of microridge patterns increases as rearrangements decrease in frequency. Bottom: the orientation of NMII minifilaments determines the outcome of rearrangement events and regulates spacing (see Discussion).
Microridge pattern maturation minimizes surface energy
The increasing nematic order of maturing microridge patterns suggests that they are governed by an energy minimization principle, which can be explained with concepts defined by physics. Optimal parallel packing of microridges likely minimizes the bending and stretching energy of the lipid bilayer that is coupled to the underlying cortex. Consistent with this idea, we found that the size of local alignment domains increased, and their number decreased, as microridge patterns matured (Figures 1 and 2). Inevitably, initial disorder in the emerging pattern brings about sharp boundaries between the domains of local alignment. These boundaries are defects in the nematic order and thus associated with an energy penalty, a phenomenon well known in liquid crystals (Needleman and Dogic, 2017). The global energy minimum likely corresponds to concentric microridges arranged in parallel rings, like a target. Our observations show that microridge patterns, which are initially in states with many alignment domains, progress toward this well-ordered global minimum over time, a process that requires crossing energy barriers associated with fission and fusion of preexisting microridges.
Our results suggest that myosin activity facilitates energy barrier crossing, promoting fission and fusion and thus rearranging microridge patterns. The fact that myosin activity leads to microridge fission and fusion with approximately equal probability (Figure 4C) suggests that it does not increase their order or disorder per se, but rather provides quantal “kicks” that locally alter pattern topology. Thus, myosin activity is analogous to the thermodynamic temperature of the pattern—by randomly breaking and fusing individual microridges, myosin allows the pattern to cross energy barriers separating local energy minima. Following this thermodynamic analogy, the decrease in microridge rearrangement events over time corresponds to slowly lowering the temperature, or annealing, which is well known in physics to help systems reach lower energy states on complex energy landscapes with multiple minima (van Laarhoven and Aarts, 1987). Blebbistatin may represent rapid quenching (a sharp temperature drop) that allows the system to descend to the closest energy minimum, perhaps explaining why blebbistatin in our experiments increased the alignment index.
Microridges are modular protrusions
Both the initial formation and the remodeling of microridges demonstrate that they are modular structures: individual units (pegs) assemble into longer structures (microridges); once assembled, microridges can be broken at any point and attached to other microridges. This modular nature distinguishes microridges from other protrusions. However, the apparent simplicity of this process elides the complexity of rearrangement events at the molecular level, which likely involve multiple, locally regulated activities. Fission requires not just severing actin filaments but also locally disassembling a supramolecular network of F-actin, keratin filaments, and actin-binding proteins (Pinto et al., 2019), as well as membrane remodeling. Microridge remodeling events require NMII activity, but fission is likely instigated by upstream regulators that coordinate multiple biochemical activities. Such roles could be played by Rho family GTPases, which can regulate both F-actin stability and NMII contraction (Ridley, 2015; Kelkar et al., 2020), or Aurora B kinase, which promotes NMII activity (Minoshima et al., 2003; Touré et al., 2008) and disassembly of actin and keratin filaments (Field et al., 2019) at the cytokinetic furrow. Fusion likely requires local F-actin polymerization, the activity of F-actin cross-linking proteins that connect the cytoskeletal networks of the two parent microridges, and the reintegration of keratin filaments, which have the potential to connect with themselves end-to-end (Çolakog˘lu and Brown, 2009). Identifying the molecular mechanisms that execute fission and fusion would make it possible to test how each type of event separately contributes to the patterning process.
The orientation of cortical NMII minifilaments determines the nature of microridge rearrangements
Visualizing individual NMII minifilaments in the cortex of living cells provided insight into how they execute microridge rearrangements, as well as evidence that they play a direct role in microridge spacing (Figure 9). From the earliest steps of microridge morphogenesis, cortical minifilaments associate with protrusions (Supplemental Figure S2, A and B). This observation suggests that the ends of individual minifilaments are biochemically tethered to the base of pegs and microridges, orthogonal to actin filaments in these protrusions. When two pegs are tethered to opposite ends of a minifilament, contraction brings them closer together, providing an opportunity for them to fuse into a nascent microridge. Similarly, contraction of peg-to-microridge minifilament bridges may contribute to microridge elongation, and contraction of minifilament bridges connecting two microridge ends may promote microridge fusion. The recruitment of keratin filaments into these growing protrusions likely helps stabilize them (Inaba et al., 2020), preventing fusion events from reversing. By contrast, minifilaments tethered to the sides of microridges sometimes promoted fission, suggesting that minifilaments may pull on microridges to facilitate their local disassembly. If minifilaments bridged two parallel microridges, they often appeared to pull them closer together without severing them, providing direct evidence that NMII can regulate microridge spacing. At later stages, microridges were often linked by a bridge of two minifilaments aligned end-to-end. This arrangement raises the intriguing possibility that minifilaments could serve as molecular spacers for aligned microridges, similar to how spectrin tetramers determine the spacing of actin rings in axons (Xu et al., 2013). However, our observation with time-lapse imaging that cortical NMII minifilaments turn over within minutes makes them unable to serve as stable spacers. It is more likely that spacing length is determined by another factor, such as the minimization of membrane bending energy, and that minifilament-mediated rearrangements allow cells to sample patterns to achieve that minimal energy state.
Our findings demonstrate a surprisingly direct role for individual cortical minifilaments in physically pulling protrusions to rupture them, fuse them together, or alter their relative spacing. In a previous study, we identified a mechanistically distinct role for NMII activity during the earlier morphogenetic step of peg coalescence to form microridges (van Loon et al., 2020). Computational modeling suggested that cortical NMII activity reduced surface energy to allow peg coalescence and, indeed, experimentally reducing surface energy by treating animals with hyperosmolar media was sufficient to induce the spontaneous formation of microridges from pegs. Moreover, stretching cells by laser ablation induced cortical flow that promoted oriented peg coalescence. The distinct NMII-mediated mechanisms that we have identified in these two studies—reducing surface tension and directly pulling on microridges—act over different spatial scales, but likely both contribute to peg coalescence, since in this study we observed that two pegs are often connected by a minifilament. Conversely, the reduction of surface tension by cortical NMII contraction may also contribute to fission, fusion, and spacing events, alongside the direct pulling mechanism that we identified here. However, altering surface tension is not alone sufficient to cause fission or fusion, since stretching cells did not appreciably increase the frequency of these events.
Minifilaments are typically thought to be isotropically oriented in the cortex of interphase cells (Kelkar et al., 2020), but our findings suggest that their association with microridges causes them to adopt a highly organized arrangement in the cortex of periderm cells. In mature periderm cells, since microridges align with cell borders and with one another, their arrangement approximates an ideal targetlike concentric pattern; because minifilaments form perpendicular bridges between adjacent microridges, they are predominantly arranged radially toward the center of cells. Since contractility is influenced not just by minifilament density but also by the relative arrangement of NMII in the cortex (Kelkar et al., 2020), this unusual radial minifilament organization likely endows periderm cells with unique contractile properties. Collectively, our observations reveal a surprisingly organized arrangement of cortical minifilaments, indicating that understanding how cortical contraction executes specific biological processes will require a better understanding of cortical minifilament architecture.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Zebrafish
Zebrafish (Danio rerio) were raised at 28.5°C on a 14-h/10-h light/dark cycle. Embryos were raised at 28.5°C in embryo water composed of 0.3 g/l Instant Ocean salt (Spectrum Brands) and 0.1% methylene blue. Previously characterized zebrafish lines in this paper include AB wild-type fish (ZFIN: ZDB-GENO-960809-7), Tg(krt5:Gal4) (Rasmussen et al., 2015), Tg(UAS:Lifeact-GFP) (Helker et al., 2013), Tg(krt5:Lifeact-Ruby), and Tg(krt5:Myl12.1-EGFP) (van Loon et al., 2020). Tg(krt5:Gal4/+;UAS:Lifeact-GFP/+) zebrafish were incrossed or outcrossed to wild type and screened for brightness on the day of imaging using a fluorescence dissecting microscope. For Airyscan microscopy, Tg(krt5:Myl12.1-EGFP) zebrafish were incrossed and injected with krt5:Lifeact-Ruby and krt5:Myl12.1-EGFP plasmids to improve brightness. All experimental procedures were approved by the Chancellor’s Animal Research Care Committee at the University of California, Los Angeles (UCLA).
Plasmids
Previously characterized plasmids in this paper include krt5:Myl12.1-EGFP (van Loon et al., 2020) and UAS:mRuby-PH-PLC (Jiang et al., 2019). The krt5-Myh9a-mCherry was constructed using the Gateway-based Tol2-kit (Kwan et al., 2007). The following vectors used to construct krt5-Myh9a-mCherry have previously been described: p5E-krt5 (Rasmussen et al., 2015), p3E-mCherrypA (Kwan et al., 2007), and pDestTol2pA2 (Kwan et al., 2007). The Myh9a coding sequence was cloned from a cDNA library of 5dpf zebrafish larvae using the following primers: Forward: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTATATGTCAGACGCAGAGAAGTTC-3′; Reverse: 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTCTTACTCAGGAGTTGGCTCG-3′.
For transient transgene expression, ∼5 nl plasmid (∼25 ng/μl) was injected into single-cell zebrafish embryos.
Microscopy
Live fluorescent images and videos of periderm cells were acquired on a Zeiss LSM800 confocal microscope. Images were acquired with Zeiss Zen Blue software using an EC Plan-Neofluar 40×/1.30 oil DIC M27 objective with 2–3× digital zoom. Optimal resolution and Z-stack intervals were set using Zen software, except for videos for which a Z-stack interval of 0.75 μm was used to improve imaging speed. During imaging, zebrafish slide chambers were mounted on a heated stage set to 28°C. The x-y position and z-stack were occasionally adjusted during time-lapse imaging to keep the cells of interest in the frame. For longitudinal experiments, between 48 and 96 hpf, zebrafish were rescued from mounting agarose each day after imaging using forceps, then placed in separate petri dishes for mounting and imaging on subsequent days.
Airyscan microscopy was performed on a Zeiss LSM 880 confocal microscope with Airyscan in the Broad Stem Cell Institute Research Center/Molecular, Cell and Developmental Biology microscopy core at UCLA. Images were acquired with Zeiss Zen Black software using an Plan-Apochromat 63×/1.4 oil DIC M27 objective with 2–5× digital zoom. After acquisition, Airyscan processing was performed with the default settings on Zen Black.
To ablate periderm cells expressing Lifeact-GFP, we adapted a previously described method (O’Brien et al., 2009a; van Loon et al., 2020). Videos of cell stretching by periderm cell ablation were acquired using Zeiss Zen Black Software on a Zeiss LSM 880 multiphoton microscope using an EC Plan-Neofluar 40×/1.30 oil DIC M27 objective and a Coherent Chameleon Ultra II laser at a wavelength of 813 nm. A 488-nm laser was used to find and focus on the cell surface at 250× digital zoom, and the cell was then exposed to 813-nm laser illumination for 3–4 s at 5–6% laser power using “live” scanning.
Drug treatment
The (-)-Blebbistatin (Cayman Chemical) was dissolved in DMSO (Fisher Scientific). Treatment solutions were made with Ringer’s Solution and included the inhibitor, or equivalent concentration of DMSO (≤ 1%), as well as up to 0.4 mg/ml MS-222 (Sigma). Zebrafish larvae were exposed to the treatment solution for the specified period of time, then mounted in agarose and immersed in the same solution. For treatments longer than 2 h, larvae were initially exposed to a treatment solution without MS-222 and then transferred to a similar solution containing up to 0.4 mg/ml MS-222 ≥ 30 min prior to imaging. For longitudinal experiments with blebbistatin, fish were first mounted in agarose and imaged, then rescued from agarose using forceps and exposed to treatment solutions. Approximately 30 min after exposure to treatment solutions, zebrafish were again mounted in agarose and slide chambers were filled with treatment solution. Zebrafish were imaged again after 1-h exposure to treatment solutions.
Image analysis and statistics
All statistical testing was performed using RStudio (RStudio). Data distributions were assessed for normality using the Shapiro–Wilk test and visually inspected using Q-Q plots. The appropriate parametric or nonparametric tests were then selected based on the normality of the data distributions being compared.
Microridge analysis was performed using a custom Python script. Images of periderm cells were sum-projected and smoothened with a Gaussian filter. Pixel intensities were then normalized based on the modality of their intensity distribution. Unimodal distributions were normalized to the full width at the half maximum, while bimodal distributions were normalized to values between both maxima. Images were then processed with a Hessian filter, thresholded, and skeletonized. Vectorized skeletons were smoothened and fitted to a normalized cell image to produce vectorized microridge lines. Distances between microridges and microridge orientations were then calculated. Microridge alignment domains were calculated by interpolating Q-tensor (Q = v ⊗ v – ½I, where v is a unit tangent vector and I is a unit tensor).
Image management for presentation was performed using FIJI (Schindelin et al., 2012). The brightness and contrast of images were adjusted for the purpose of presentation. All movies were stabilized for presentation and analysis purposes using the Image Stabilizer FIJI plugin (Kang Li@CMU - Image Stabilizer Plugin for ImageJ).
Microridge fusion and fission events were identified manually using the FIJI Multi-point tool. To measure distances from NMII contractions to fusion and fission events, NMII images were smoothened and contractions were automatically thresholded in FIJI with the Triangle method, then distances were measured between microridge rearrangement events and the edge of the nearest contraction using “rgeos” and “sp” R packages.
Supplementary Material
Acknowledgments
We thank Sally Horne-Badovinac, Yasuko Inaba, and Kadidia Pemba Adula for comments on the manuscript; Son Giang and Linda Dong for excellent fish care; and Nat Prunet in the BSCRC/MCDB microscopy core for help with Airyscan microscopy. This work was funded by National Institutes of Health Grant R01GM122901 to A.S. and by the Biotechnology and Biological Sciences Research Council of UK Grants BB/P01190X and BB/P006507 to A.B.G. A.P.v.L. was supported by the Ruth L. Kirschstein National Research Service Award (GM007185).
Abbreviations used:
- dpf
days post-fertilization
- hpf
hours post-fertilization
- Myl12.1
Myosin light chain 12.1
- NMII
non-muscle myosin II.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-05-0258) on June 3, 2021.
REFERENCES
- Bement WM, Leda M, Moe AM, Kita AM, Larson ME, Golding AE, Pfeuti C, Su K-C, Miller AL, Goryachev AB, von Dassow G (2015). Activator-inhibitor coupling between Rho signalling and actin assembly makes the cell cortex an excitable medium. Nat Cell Biol 17, 1471–1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billington N, Wang A, Mao J, Adelstein RS, Sellers JR (2013). Characterization of three full-length human nonmuscle myosin II paralogs. J Biol Chem 288, 33398–33410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinowsky CR, Pinette JA, Meenderink LM, Lau KS, Tyska MJ (2020). Non-muscle myosin-2 contractility-dependent actin turnover limits the length of epithelial microvilli. Mol Biol Cell mbcE20090582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Çolakog˘lu G, Brown A (2009). Intermediate filaments exchange subunits along their length and elongate by end-to-end annealing. J Cell Biol 185, 769–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M, Draper BW, Priess JR (1997). The role of actin filaments in patterning the Caenorhabditis elegans cuticle. Dev Biol 184, 373–384. [DOI] [PubMed] [Google Scholar]
- Crawley SW, Shifrin Jr DA, Grega-Larson NE, McConnell RE, Benesh AE, Mao S, Zheng Y, Zheng QY, Nam KT, Millis BA, et al. (2014). Intestinal brush border assembly driven by protocadherin-based intermicrovillar adhesion. Cell 157, 433–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depasquale JA (2018). Actin Microridges: Actin microridges in epithelium. Anat Rec 31, 81. [DOI] [PubMed] [Google Scholar]
- Field CM, Pelletier JF, Mitchison TJ (2019). Disassembly of actin and keratin networks by aurora b kinase at the midplane of cleaving Xenopus laevis eggs. Curr Biol 29, 1999–2008.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G, Dubin-Thaler BJ, Rossier O, Cai Y, Chaga O, Jiang G, Beaver W, Döbereiner H-G, Freund Y, Borisy G, Sheetz MP (2007). Lamellipodial actin mechanically links myosin activity with adhesion-site formation. Cell 128, 561–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han B, Zhou R, Xia C, Zhuang X (2017). Structural organization of the actin-spectrin-based membrane skeleton in dendrites and soma of neurons. Proc Natl Acad Sci USA 114, E6678–E6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helker CSM, Schuermann A, Karpanen T, Zeuschner D, Belting H-G, Affolter M, Schulte-Merker S, Herzog W (2013). The zebrafish common cardinal veins develop by a novel mechanism: lumen ensheathment. Development 140, 2776–2786. [DOI] [PubMed] [Google Scholar]
- Inaba Y, Chauhan V, van Loon AP, Choudhury LS, Sagasti A (2020). Keratins and Plakin family cytolinker proteins control the length of epithelial microridge protrusions. Elife 9. 10.7554/eLife.58149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N, Rasmussen JP, Clanton JA, Rosenberg MF, Luedke KP, Cronan MR, Parker ED, Kim H-J, Vaughan JC, Sagasti A, Parrish JZ (2019). A conserved morphogenetic mechanism for epidermal ensheathment of nociceptive sensory neurites. Elife 8. 10.7554/eLife.42455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia R, Li D, Li M, Chai Y, Liu Y, Xie Z, Shao W, Xie C, Li L, Huang X, et al. (2019). Spectrin-based membrane skeleton supports ciliogenesis. PLoS Biol 17, e3000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K@CMU - Image Stabilizer Plugin for ImageJ.
- Kelkar M, Bohec P, Charras G (2020). Mechanics of the cellular actin cortex: From signalling to shape change. Curr Opin Cell Biol 66, 69–78. [DOI] [PubMed] [Google Scholar]
- Kondo S, Miura T (2010). Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616–1620. [DOI] [PubMed] [Google Scholar]
- Kwan KM, Fujimoto E, Grabher C, Mangum BD, Hardy ME, Campbell DS, Parant JM, Yost HJ, Kanki JP, Chien C-B (2007). The Tol2kit: a multisite gateway-based construction kit for Tol2 transposon transgenesis constructs. Dev Dyn 236, 3088–3099. [DOI] [PubMed] [Google Scholar]
- van Laarhoven PJM, Aarts EHL (1987). Simulated annealing. In: Simulated Annealing: Theory and Applications, ed. van Laarhoven PJM, Aarts EHL, Springer Netherlands, Dordrecht. 7–15. [Google Scholar]
- Lam P-Y, Mangos S, Green JM, Reiser J, Huttenlocher A (2015). In vivo imaging and characterization of actin microridges. PLoS One 10:e0115639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Loon AP, Erofeev IS, Maryshev IV, Goryachev AB, Sagasti A (2020). Cortical contraction drives the 3D patterning of epithelial cell surfaces. J Cell Biol 219. 10.1083/jcb.201904144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magre I, Fandade V, Damle I, Banerjee P, Yadav SK, Sonawane M, Joseph J (2019). Nup358 regulates microridge length by controlling SUMOylation-dependent activity of aPKC in zebrafish epidermis. J Cell Sci 132. 10.1242/jcs.224501. [DOI] [PubMed] [Google Scholar]
- Maître J-L, Berthoumieux H, Krens SFG, Salbreux G, Jülicher F, Paluch E, Heisenberg C-P (2012). Adhesion functions in cell sorting by mechanically coupling the cortices of adhering cells. Science 338, 253–256. [DOI] [PubMed] [Google Scholar]
- Martin AC, Goldstein B (2014). Apical constriction: themes and variations on a cellular mechanism driving morphogenesis. Development 141, 1987–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meenderink LM, Gaeta IM, Postema MM, Cencer CS, Chinowsky CR, Krystofiak ES, Millis BA, Tyska MJ (2019). Actin dynamics drive microvillar motility and clustering during brush border assembly. Dev Cell 50, 545–556.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoshima Y, Kawashima T, Hirose K, Tonozuka Y, Kawajiri A, Bao YC, Deng X, Tatsuka M, Narumiya S, May WS Jr, et al. (2003). Phosphorylation by aurora B converts MgcRacGAP to a RhoGAP during cytokinesis. Dev Cell 4, 549–560. [DOI] [PubMed] [Google Scholar]
- Murphy DA, Courtneidge SA (2011). The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat Rev Mol Cell Biol 12, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan P, Chatterton P, Ikeda A, Ikeda S, Corey DP, Ervasti JM, Perrin BJ (2015). Length regulation of mechanosensitive stereocilia depends on very slow actin dynamics and filament-severing proteins. Nat Commun 6, 6855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman D, Dogic Z (2017). Active matter at the interface between materials science and cell biology. Nature Reviews Materials 2, 17048. [Google Scholar]
- O’Brien GS, Martin SM, Söllner C, Wright GJ, Becker CG, Portera-Cailliau C, Sagasti A (2009a). Developmentally regulated impediments to skin reinnervation by injured peripheral sensory axon terminals. Curr Biol 19, 2086–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien GS, Rieger S, Martin SM, Cavanaugh AM, Portera-Cailliau C, Sagasti A (2009b). Two-photon axotomy and time-lapse confocal imaging in live zebrafish embryos. J Vis Exp 10.3791/1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto CS, Khandekar A, Bhavna R, Kiesel P, Pigino G, Sonawane M (2019). Microridges are apical epithelial projections formed of F-actin networks that organize the glycan layer. Sci Rep 9, 12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman R, Damle I, Rote R, Banerjee S, Dingare C, Sonawane M (2016). aPKC regulates apical localization of Lgl to restrict elongation of microridges in developing zebrafish epidermis. Nat Commun 7, 11643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen JP, Sack GS, Martin SM, Sagasti A (2015). Vertebrate epidermal cells are broad-specificity phagocytes that clear sensory axon debris. J Neurosci 35, 559–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ (2015). Rho GTPase signalling in cell migration. Curr Opin Cell Biol 36, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, et al. (2008). Lifeact: a versatile marker to visualize F-actin. Nat Methods 5, 605–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Cheung A, Limouze J, Chen I, Westwood NJ, Sellers JR, Mitchison TJ (2003). Dissecting temporal and spatial control of cytokinesis with a myosin II Inhibitor. Science 299, 1743–1747. [DOI] [PubMed] [Google Scholar]
- Tarchini B, Lu X (2019). New insights into regulation and function of planar polarity in the inner ear. Neurosci Lett 709, 134373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touré A, Mzali R, Liot C, Seguin L, Morin L, Crouin C, Chen-Yang I, Tsay Y-G, Dorseuil O, Gacon G, Bertoglio J (2008). Phosphoregulation of MgcRacGAP in mitosis involves Aurora B and Cdk1 protein kinases and the PP2A phosphatase. FEBS Lett 582, 1182–1188. [DOI] [PubMed] [Google Scholar]
- Turing A (1952). The chemical basis of morphogenesis. Phil Trans R Soc Lond B 237, 37–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, Mooseker MS (2002). MYO1A (brush border myosin I) dynamics in the brush border of LLC-PK1-CL4 cells. Biophys J 82,1869–1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisshart K (2014). The basic principle of airyscanning. Zeiss Technology Note 22, https://www.embl.de/services/core_facilities/almf/events_ext/2017/EN_wp_LSM-880_Basic-Principle-Airyscan.pdf. [Google Scholar]
- Xu K, Zhong G, Zhuang X (2013). Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science 339, 452–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D-S, Piazza V, Perrin BJ, Rzadzinska AK, Poczatek JC, Wang M, Prosser HM, Ervasti JM, Corey DP, Lechene CP (2012). Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature 481, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.