Abstract
Divisions of the genetic material and cytoplasm are coordinated spatially and temporally to ensure genome integrity. This coordination is mediated in part by the major cell cycle regulator cyclin-dependent kinase (Cdk1). Cdk1 activity peaks during mitosis, but during mitotic exit/cytokinesis Cdk1 activity is reduced, and phosphorylation of its substrates is reversed by various phosphatases including Cdc14, PP1, PP2A, and PP2B. Cdk1 is known to phosphorylate several components of the actin- and myosin-based cytokinetic ring (CR) that mediates division of yeast and animal cells. Here we show that Cdk1 also phosphorylates the Schizosaccharomyces pombe CR component paxillin Pxl1. We determined that both the Cdc14 phosphatase Clp1 and the PP1 phosphatase Dis2 contribute to Pxl1 dephosphorylation at mitotic exit, but PP2B/calcineurin does not. Preventing Pxl1 phosphorylation by Cdk1 results in increased Pxl1 levels, precocious Pxl1 recruitment to the division site, and increased duration of CR constriction. In vitro Cdk1-mediated phosphorylation of Pxl1 inhibits its interaction with the F-BAR domain of the cytokinetic scaffold Cdc15, thereby disrupting a major mechanism of Pxl1 recruitment. Thus, Pxl1 is a novel substrate through which S. pombe Cdk1 and opposing phosphatases coordinate mitosis and cytokinesis.
INTRODUCTION
Cytokinesis is the final stage of the cell cycle during which daughter cells physically separate. Yeast and animal cells divide using a conserved actin- and myosin-based contractile apparatus called the cytokinetic ring (CR) (Glotzer, 2017; Mangione and Gould, 2019). Studies of model organisms such as Schizosaccharomyces pombe have provided substantial insight into this process (Bathe and Chang, 2010; Pollard, 2010; Willet et al., 2015b). Forward genetic screens identified the proteins involved in cytokinesis, which include both CR structural components and regulators (Nurse et al., 1976; Minet et al., 1979; Chang et al., 1996; Balasubramanian et al., 1998; Kim et al., 2010; Hayles et al., 2013; Chen et al., 2015, 2016), many of which are conserved in the budding yeast Saccharomyces cerevisiae and metazoan cells (reviewed in Pollard and Wu, 2010). Subsequent fluorescence imaging studies determined the timing of cytokinesis events relative to mitosis in S. pombe (Wu et al., 2003, 2006; Wu and Pollard, 2005). The CR assembles during metaphase from clusters of early cytokinetic proteins called “cytokinesis nodes,” which include the F-BAR protein Cdc15 (Fankhauser et al., 1995; Wachtler et al., 2006) and the formin Cdc12 (Chang et al., 1997). Cdc12 nucleates and elongates F-actin from nodes that eventually coalesce into the CR (Kovar et al., 2003; Vavylonis et al., 2008; Laporte et al., 2011; Zimmermann et al., 2017). The CR then undergoes a maturation period during which it accumulates more components. Then, the CR constricts to bring together opposing membranes, and a cell wall structure called the septum is coordinately deposited (reviewed in Willet et al., 2015b; García Cortés et al., 2016; Perez et al., 2016).
These events are carefully regulated to protect genome integrity and ensure cytokinetic success. This is accomplished in part by the major cell cycle regulator cyclin-dependent kinase (Cdk1). High levels of Cdk1 activity promote mitosis while inhibiting cytokinesis (Wheatley et al., 1997) (reviewed in Wolf et al., 2007; Bohnert and Gould, 2011). Cdk1 inhibition and simultaneous reversal of Cdk1 substrate phosphorylation by opposing phosphatases are required for the events of mitotic exit, which include anaphase and cytokinesis (Stegmeier and Amon, 2004; Clifford et al., 2008a; Wu et al., 2009; Bloom et al., 2011; Wurzenberger and Gerlich, 2011; Grallert et al., 2015; Kuilman et al., 2015). In yeasts, multiple cytokinetic proteins have been identified as Cdk1 substrates, including S. pombe formin Cdc12 (Willet et al., 2018), S. cerevisiae and S. pombe IQGAP proteins Iqg1 and Rng2, respectively (Holt et al., 2009; Chen et al., 2013; Naylor and Morgan, 2014; Morita et al., 2021), and S. cerevisiae C2 domain protein Inn1, the orthologue of S. pombe Fic1 (Palani et al., 2012; Kuilman et al., 2015).
The CR component Pxl1 arrives at the CR during its maturation (Ren et al., 2015). Pxl1 binds the F-BAR domain of the CR scaffold Cdc15, and this interaction is critical for Pxl1 CR localization (Snider et al., 2020). Pxl1 also binds another site on Cdc15 that includes the SH3 domain and part of the intrinsically disordered region (Roberts-Galbraith et al., 2009; Bhattacharjee et al., 2020), and Cdc15’s SH3 domain is important for Pxl1 assembly into the CR throughout cytokinesis (Cortes et al., 2015; Martin-Garcia et al., 2018). While the Pxl1 N-terminus is important for CR localization (Pinar et al., 2008), the C-terminus comprises three tandem LIM (Lin11, Isl-1, and Mec-3) domains that bind mechanically stressed F-actin in vitro and in cultured cells (Sun et al., 2020; Winkelman et al., 2020) and are important for Pxl1 function (Ge and Balasubramanian, 2008; Pinar et al., 2008). pxl1∆ cells have severe cytokinesis defects, displaying CR sliding and splitting during anaphase (Ge and Balasubramanian, 2008; Pinar et al., 2008; Cortes et al., 2015). Additionally, in pxl1∆ cells the phosphatase calcineurin (CN) is not recruited to the CR (Martin-Garcia et al., 2018). Indeed, mutating the Pxl1 binding site on the Cdc15 F-BAR dramatically reduces both Pxl1 and CN localization to the CR, leading to defects in CR nanoscale architecture and constriction (Snider et al., 2020).
Despite its key functions in cytokinesis, the mechanisms that dictate the timing of Pxl1 arrival at the division site are unknown. Potentially relevant to this question, multiple proteome-scale studies determined that Pxl1 is phosphorylated in mitosis (Koch et al., 2011; Carpy et al., 2014; Kettenbach et al., 2015; Swaffer et al., 2016, 2018), and one has provided strong evidence that Pxl1 is a direct Cdk1 substrate (Swaffer et al., 2016). Here we confirmed that Pxl1 is a Cdk1 substrate and determined that Pxl1 is dephosphorylated by both Cdc14 and PP1 phosphatases. We also examined the consequence of Cdk1-mediated Pxl1 phosphorylation to Pxl1 function and to cytokinesis.
RESULTS AND DISCUSSION
Pxl1 is phosphorylated in a cell cycle–dependent manner
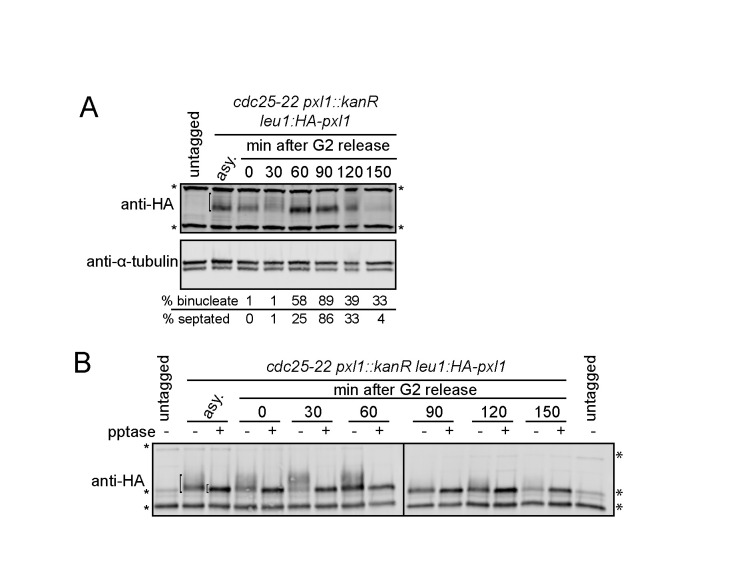
As reported previously, we observed that HA-Pxl1 abundance fluctuated across the cell cycle (Figure 1A) (Ge and Balasubramanian, 2008; Pinar et al., 2008). We also noted that HA-Pxl1 migration on SDS–PAGE changed during mitosis and cytokinesis, with a notable retardation 30 min after release from a G2 arrest that was reversed 60 min after release (Figure 1A). Treatment with lambda protein phosphatase collapsed immunoprecipitated HA-Pxl1 to a single distinct band, indicating that these changes in migration were due to changes in phosphorylation (Figure 1B).
FIGURE 1:
Pxl1 phosphorylation changes throughout the cell cycle. Cells were grown to mid–log phase, shifted to 36°C for 4 h, and then released to permissive temperature (25°C). Samples were collected at the indicated times. Denatured lysates (A) and IP-phosphatase (pptase) samples (B) were separated by SDS–PAGE and analyzed by immunoblot (IB) with HA and tubulin antibodies. Asterisks indicate background bands, and the bracket in asynchronous (asy.) samples indicates HA-Pxl1.
Pxl1 is phosphorylated by Cdk1
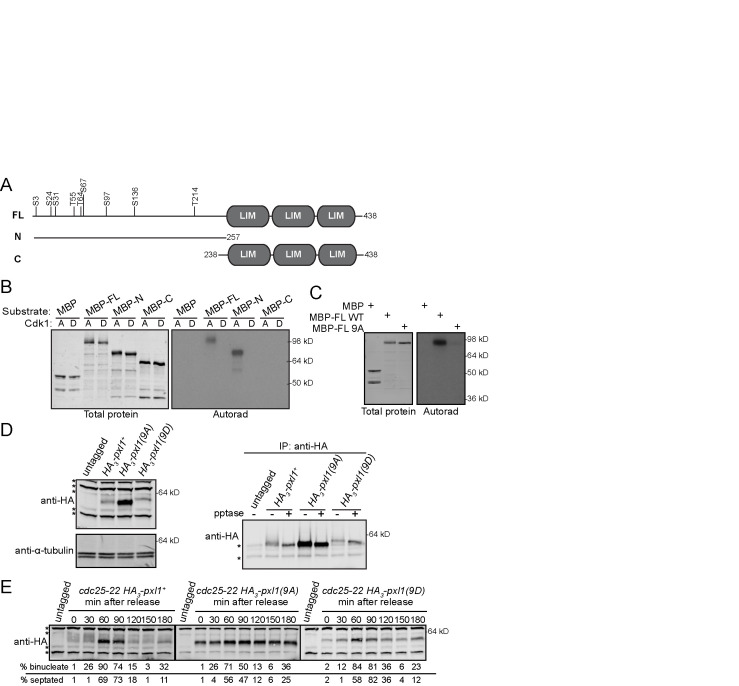
Previous studies (Koch et al., 2011; Carpy et al., 2014; Kettenbach et al., 2015; Swaffer et al., 2016, 2018) and our own analysis of green fluorescent protein (GFP)-Pxl1 purifications identified multiple phosphorylation sites on Pxl1, many of which matched the Cdk1 minimal consensus sequence of [S/T]P (Supplemental Figure S1). Therefore, we tested whether Cdc2-Cdc13 (active Cdk1) could phosphorylate recombinant MBP-Pxl1. Cdk1 phosphorylated both an N-terminal fragment (N) and full-length Pxl1 (FL), but not a C-terminal fragment (C) (Figure 2, A and B). These results were expected because all of the [S/T]P phosphorylation sites identified in Pxl1 from cell lysates or purifications reside in the N-terminus. Phosphoamino acid analysis indicated that Cdk1 phosphorylates both serines and threonines in MBP-FL and MBP-N (Supplemental Figure S2A), and comparison of the tryptic phosphopeptide maps of MBP-FL and MBP-N confirmed that the vast majority of Cdk1 phosphorylation sites reside in the N-terminus of Pxl1 (Supplemental Figure S2B).
FIGURE 2:
Pxl1 is phosphorylated by Cdk1. (A) To-scale schematics of Pxl1 full-length (FL) and N- and C-terminal fragments. Numbers indicate amino acid positions. Black vertical lines indicate sites that match the [S/T]P consensus sequence and are mutated in Pxl1(9A) or Pxl1(9D). (B) In vitro kinase assays (IVKA) using active (A) or dead (D) Cdk1, the indicated substrate, and radiolabeled ATP were separated by SDS–PAGE, transferred to PVDF, stained with Revert Total Protein Stain, and then exposed to film (Autorad). (C) IVKA using active Cdk1, the indicated substrate, and radiolabeled ATP were separated by SDS–PAGE, stained with Coomassie, and then exposed to film (Autorad). (D) Lysates from the indicated strains (left) and subsequent IP-pptase (right) were separated by SDS–PAGE and IB for indicated proteins. (E) Cells were grown to mid–log phase, shifted to 36°C for 4 h, and then released to permissive temperature (25°C). Samples were collected at the indicated times. Denatured lysates were separated by SDS–PAGE and analyzed by IB with HA antibody. (B–E) Positions of molecular weight markers indicated on the right. (D, E) Asterisks indicate background bands.
Mutating all nine potential Cdk1 sites in the N-terminus of Pxl1 to alanine abolished Pxl1 phosphorylation by Cdk1 in vitro (Figure 2, A and C). To determine the consequence of Pxl1 phosphorylation on these sites, we replaced pxl1+ with alleles encoding HA-tagged pxl1 or pxl1 phosphomutants: all nine residues matching the [S/T]P consensus sequence were mutated to alanine (9A) or aspartic acid (9D) to eliminate or potentially mimic phosphorylation, respectively. Unlike HA-Pxl1, HA-Pxl1(9A) and HA-Pxl1(9D) migrated as single bands on SDS–PAGE. HA-Pxl1(9D) migrated more slowly than HA-Pxl1(9A), which comigrated with dephosphorylated HA-Pxl1 (Figure 2D). HA-Pxl1(9A) also appeared to be more abundant than HA-Pxl1 or HA-Pxl1(9D) (Figure 2, D and E), which is investigated below. While phosphatase treatment of HA-Pxl1 collapsed the multiple phospho-species to a single band, the SDS–PAGE mobilities of HA-Pxl1(9A) and HA-Pxl1(9D) were largely unaltered by the treatment. To substantiate this observation, we also monitored the SDS–PAGE mobilities of the phosphomutants throughout the cell cycle (Figure 2E) and did not observe any changes as the cells progressed from G2 through mitosis and cytokinesis. Together these data support that we identified the major sites of Pxl1 phosphorylation.
Cdc14 phosphatase Clp1 and PP1 phosphatase Dis2, but not CN, regulate Pxl1 dephosphorylation
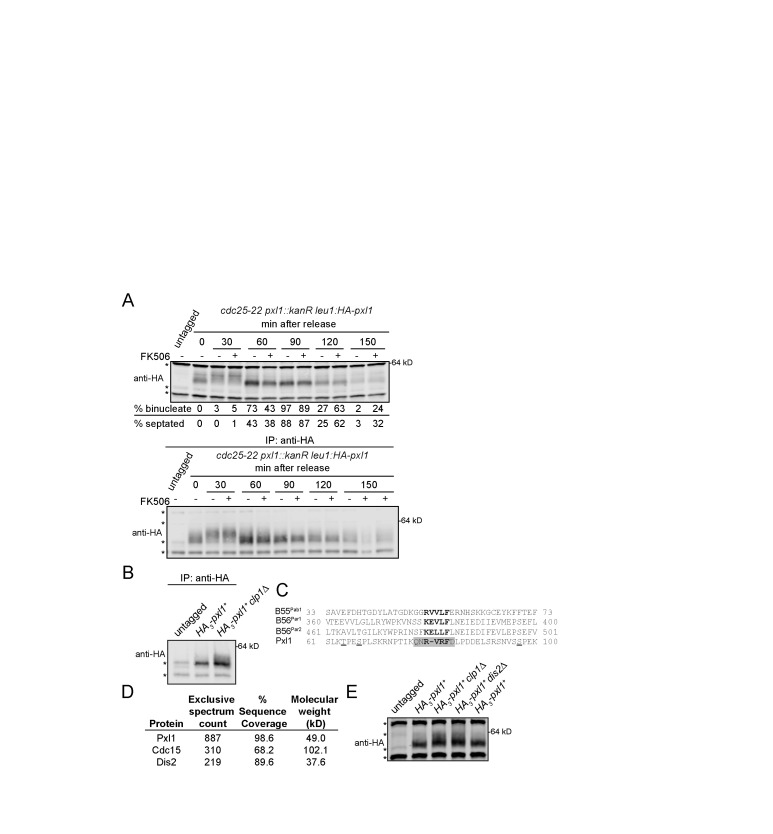
Pxl1 is dynamically phosphorylated as cells enter mitosis and dephosphorylated during anaphase (Figures 1 and 2E). Therefore, we asked which phosphatase(s) mediates Pxl1 dephosphorylation. Because Pxl1 recruits CN to the mature CR (Martin-Garcia et al., 2018), we first tested whether Pxl1 is a CN substrate. We released cells from a G2 arrest for 30 min to allow mitotic onset and then treated them with the CN inhibitor FK506 to test whether this prevented Pxl1 dephosphorylation (Figure 3A). We observed no change in Pxl1 dephosphorylation, so although CN depends on Pxl1 for its CR localization, Pxl1 is not a CN substrate.
FIGURE 3:
Pxl1 is regulated by Cdc14 and PP1 phosphatases, but not by calcineurin. (A) Cells were grown to mid–log phase, shifted to 36°C for 4 h, and then released to permissive temperature (25°C). The CN inhibitor FK506 (10 µg/ml) or DMSO was added to cell cultures at 30 min after release from G2 arrest. Samples were collected at the indicated times. Denatured lysates (top) and IP samples (bottom) were separated by SDS–PAGE and analyzed by IB with HA antibody. (B) IP samples separated by SDS–PAGE and IB for the indicated proteins. (C) Alignment of PP1 docking motifs in S. pombe proteins. Cdk1 phosphorylation sites in Pxl1 are underlined. PP1 docking motif in Pxl1 is highlighted in gray. (D) Selected proteins pulled down by GFP-Pxl1 from prometaphase-arrested cells. (E) Denatured lysates separated by SDS–PAGE and IB for the indicated proteins. (A, B, E) Asterisks indicate background bands. Positions of molecular weight markers indicated on the right.
The Cdc14 phosphatase Clp1 opposes Cdk1 activity in S. pombe (Clifford et al., 2008b; Chen et al., 2013), so we next tested whether it has a role in Pxl1 dephosphorylation. The slower-migrating bands corresponding to phosphorylated Pxl1 were increased in clp1∆ compared to clp1+ (Figure 3B), consistent with Clp1 contributing to Pxl1 dephosphorylation. However, not all of the rapidly migrating Pxl1 species was abolished, indicating that another phosphatase may also dephosphorylate Pxl1. We searched the Eukaryotic Linear Motif (ELM) database (Gouw et al., 2018) for linear motifs in Pxl1 that could provide insight into its regulation and/or function, and we identified a protein phosphatase 1 (PP1) catalytic subunit interacting motif (Egloff et al., 1997; Aggen et al., 2000) at amino acids 78–84 (Figure 3C). Consistent with the possibility that PP1 dephosphorylates Pxl1, the catalytic subunit of S. pombe PP1 Dis2 was identified in a purification of GFP-Pxl1 from cells blocked in prometaphase by a nda3-KM311 arrest (Figure 3D). Pxl1 phosphorylation was increased in lysates from dis2∆ cells, similar to what was observed in clp1∆ cells (Figure 3E), and Pxl1 phosphorylation in a double phosphatase deletion was similar compared with both single mutants (Supplemental Figure S2C). Thus, two phosphatases previously implicated in mitotic exit and reversal of CDK phosphorylation (Clifford et al., 2008; Chen et al., 2013; Heim et al., 2015; Qian et al., 2015; Martín et al., 2020), Clp1/Cdc14 and Dis2/PP1, contribute to Pxl1 dephosphorylation.
Phosphorylation regulates the timing of Pxl1 localization to the CR
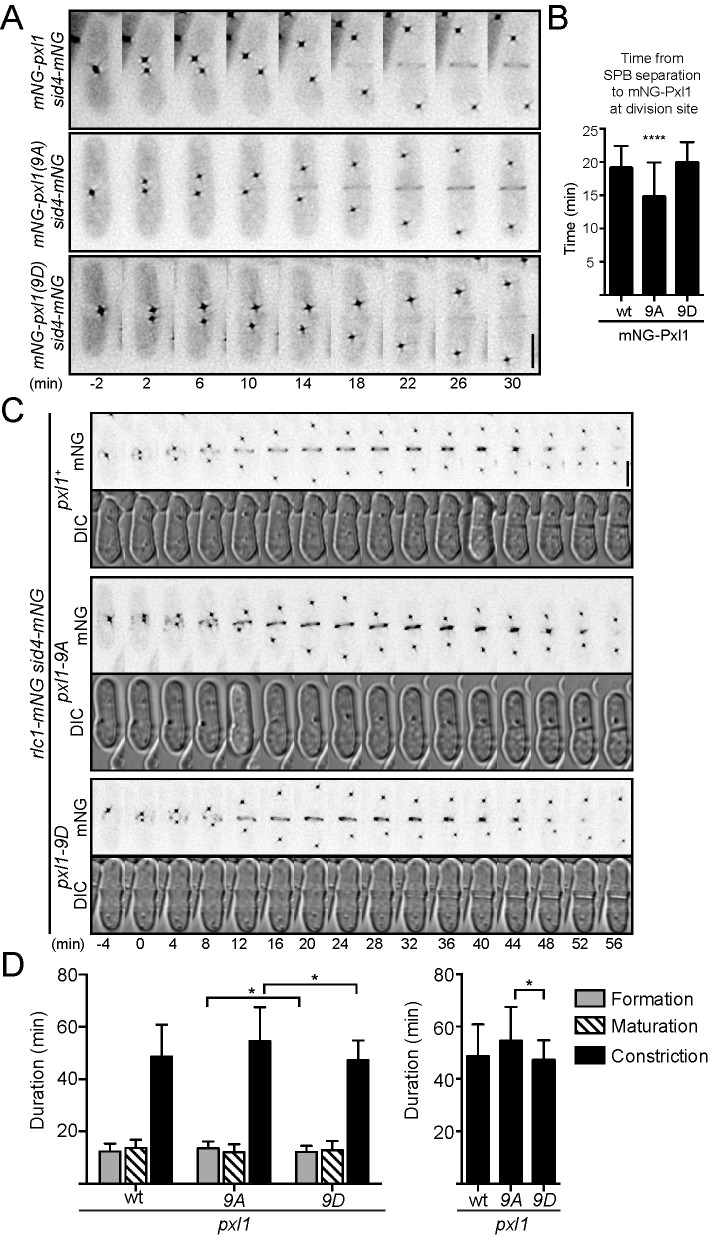
Cdk1 phosphorylation of S. pombe formin Cdc12 (Willet et al., 2018), S. cerevisiae and S. pombe IQGAP Iqg1/Rng2 (Naylor and Morgan, 2014; Morita et al., 2021), and S. cerevisiae C2 domain protein Inn1 (Palani et al., 2012; Kuilman et al., 2015) temporally regulates their recruitment to the CR. Therefore, we performed time-lapse imaging of mNeonGreen (mNG)-Pxl1, mNG-Pxl1(9A), and mNG-Pxl1(9D) in cells also producing Sid4-mNG, a spindle pole body (SPB) marker for monitoring progression through mitosis. Relative to SPB separation, mNG-Pxl1(9A) arrived at the division site earlier (14.7 ± 5.2 min; 47 cells) than either mNG-Pxl1 (19.0 ± 3.4 min; 31 cells) or mNG-Pxl1(9D) (19.9 ± 3.1 min; 27 cells) (Figure 4, A and B). Thus, abolishing phosphorylation on Pxl1 results in its early CR recruitment.
FIGURE 4:
Loss of Cdk1 phosphorylation leads to early recruitment of Pxl1 to the CR and increased duration of constriction. (A) Representative montages taken from time-lapse fluorescence imaging; deconvolved sum projections are shown. Images were acquired every 2 min, and every other time point is shown. Time 0 is set as the first frame with two SPBs. (B) Average time between SPB separation to division site localization of the indicated Pxl1 proteins. ****p < 0.0001, ordinary one-way analysis of variance (ANOVA) with Dunnett’s test for multiple comparisons to wild type (wt). (C) Representative fluorescence (mNG) or differential interference contrast (DIC) montages taken from time-lapse imaging; fluorescence images are deconvolved, max intensity projections, and DIC images are sum projections. Images were acquired every 2 min, and every other time point is shown. Time 0 is set as the first frame with two SPBs. (D) Average duration of the cytokinesis stages (left); duration of constriction shown alone on the right. *p < 0.05, Welch’s one-way ANOVA with Dunnett’s T3 test for multiple comparisons. (A, C) Scale bars are 5 µm. (B, D) Error bars are SD.
Immunoblotting suggested that HA-Pxl1(9A) protein levels were increased compared with those in HA-Pxl1 and HA-Pxl1(9D) (Figure 2D). Therefore, we compared the whole cell fluorescence intensities of mNG-Pxl1, mNG-Pxl1(9A), and mNG-Pxl1(9D). mNG-Pxl1(9A) whole cell fluorescence intensity was increased approximately twofold compared with those of mNG-Pxl1 and mNG-Pxl1(9D), with a proportional increase in the fluorescence intensity of mNG-Pxl1(9A) in the CR (Supplemental Figure S3, A–C). There was also a trend toward increased mNG-Pxl1 in both clp1∆ and dis2∆ (Supplemental Figure S3, D–F), which could represent an intermediate phenotype given that some Pxl1 phosphorylation remains in clp1∆ and dis2∆ (Figure 3, B and E, and Supplemental Figure S2C). We hypothesized that increased CR Pxl1-9A would recruit more of its binding partner calcineurin (Martin-Garcia et al., 2018; Snider et al., 2020). To test this we examined the localization of Cnb1-mNG, the regulatory subunit of calcineurin (Supplemental Figure S3, G–I) but did not see a similar twofold increase in Cnb1 localization in Pxl1-9A. The whole cell intensity of Cnb1-mNG was reduced in Pxl1-9A and Pxl1-9D compared with that of wild type (Supplemental Figure S3, G and H), which increased the ratio of CR:whole cell intensity in both the Pxl1-9A and Pxl1-9D (Supplemental Figure S3I). These results suggest that Pxl1 localization alone is not sufficient for Cnb1 recruitment and that there are other factors regulating their interaction.
Loss of Pxl1 phosphorylation alters cytokinesis timing
We next asked whether early CR recruitment and an increased amount of Pxl1 at the CR affected cytokinesis dynamics. To determine this, we performed time-lapse imaging of cells producing Sid4-mNG and the CR marker Rlc1-mNG in pxl1+(wt), pxl1(9A), or pxl1(9D) (Figure 4C). Then, we quantified the duration of CR formation (i.e., the period from SPB separation to CR formation), maturation (i.e., the period between formation and constriction), and constriction (Figure 4D, left). Although there was a small difference in CR formation between pxl1(9A) (13.6 ± 2.5; 33 cells) and pxl1(9D) (12.2 ± 2.3 min; 41 cells), formation was similar in pxl1(9D) and pxl1+ (12.4 ± 3.0; 32 cells), and there were no significant differences in the durations of CR maturation. Constriction took longer in pxl1(9A) (54.5 ± 13.0 min; 33 cells) compared with pxl1+(48.7 ± 12.2 min; 32 cells) and pxl1(9D) (47.3 ± 7.5; 41 cells) (Figure 4D, right). These results suggest that phosphorylation delays Pxl1 CR recruitment and that early recruitment and/or increased levels of Pxl1 has negative downstream consequences on the efficiency of cytokinesis.
Cdk1-mediated phosphorylation inhibits Pxl1 binding to Cdc15’s N-terminus but not its C-terminus
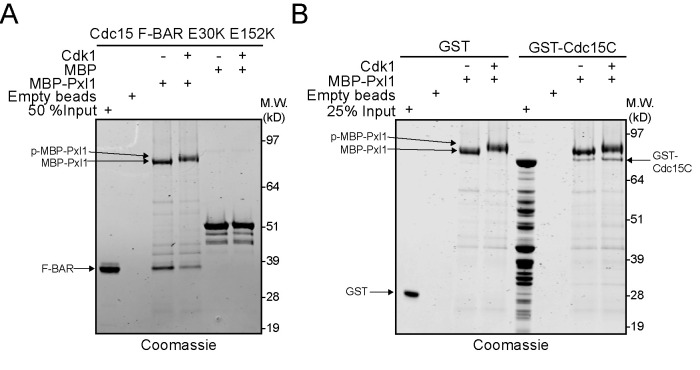
The N-terminus of Pxl1 is the site of Cdk1 phosphorylation and contains CR targeting information (Pinar et al., 2008). Pxl1 interacts directly with the F-BAR protein Cdc15 (Roberts-Galbraith et al., 2009; Bhattacharjee et al., 2020; Snider et al., 2020). Because Cdk1 phosphorylation of the formin Cdc12 inhibits its interaction with Cdc15 (Willet et al., 2018), we tested whether Cdk1 phosphorylation inhibits Pxl1 binding to Cdc15 similarly. However, unlike Cdc12 (Willet et al., 2015a), Cdc15 has two known binding sites for Pxl1: one in the N-terminal F-BAR domain (Snider et al., 2020) and one made up of the C-terminal intrinsically disordered region and SH3 domain (Cdc15C) (Bhattacharjee et al., 2020). We tested whether either of these Pxl1-Cdc15 interactions was modulated by Pxl1 phosphostate using sequential in vitro phosphorylation and binding assays. Cdk1 phosphorylation of Pxl1 reduced binding to the F-BAR domain of Cdc15 (Figure 5A), but not to Cdc15C (Figure 5B).
FIGURE 5:
Phosphorylation of MBP-Pxl1 inhibits interaction with Cdc15 F-BAR domain but not C-terminus. (A) Coomassie-stained SDS–PAGE of recombinant Cdc15 F-BAR (amino acids 19–312) that was input or pulled down by empty amylose beads, bead-bound MBP, bead-bound MBP-Pxl1, or bead-bound Cdk1-phosphorylated MBP-Pxl1. (B) Coomassie-stained SDS–PAGE of recombinant GST and GST-Cdc15(amino acids 441–927) that was input or pulled down by empty beads, bead-bound MBP-Pxl1, or bead-bound Cdk1-phosphorylated MBP-Pxl1.
Summary
Pxl1 is an important contributor to CR stability. Here we showed that Pxl1 is phosphorylated by Cdk1, and we identified two phosphatases that regulate Pxl1 dephosphorylation: the Cdc14 phosphatase Clp1 and the PP1 phosphatase Dis2, supporting their function to reverse Cdk1 phosphorylation and promote mitotic exit (Stegmeier and Amon, 2004; Clifford et al., 2008a, b; De Wulf et al., 2009; Wu et al., 2009; Mocciaro and Schiebel, 2010; Bloom et al., 2011; Bouchoux and Uhlmann, 2011; Wurzenberger and Gerlich, 2011; Palani et al., 2012; Grallert et al., 2015; Kuilman et al., 2015; Orii et al., 2016). Inconveniently for these studies, we suspect that the pxl1(9D) is not a true phosphomimetic because Pxl1(9D) is not reduced at the division site and the expected cytokinetic defects from Pxl1 loss are not observed. The deficiencies of aspartic acid as a replacement for phosphorylated serine or threonine are well-known (Dephoure et al., 2013), and therefore other molecular manipulation is required to determine the effect of persistent phospho-Pxl1 and whether both phosphatases simultaneously, sequentially, or redundantly dephosphorylate Pxl1.
Interestingly, our data showed that CN does not impact the Pxl1 phosphostate, even though Pxl1 is necessary for CN localization to the CR (Martin-Garcia et al., 2018) and CN/PP2B has been implicated in mitotic exit in S. pombe and metazoans (Yoshida et al., 1994; Mochida and Hunt, 2007; Nishiyama et al., 2007; Martin-Garcia et al., 2018). Combined with the finding that Pxl1 CR localization is not sufficient to recruit CN in a cdc15 mutant lacking a segment of its intrinsically disordered region (Mangione et al., 2019), it is clear that more investigation is necessary to understand CN recruitment to the CR and its downstream targets in cytokinesis.
We found that Pxl1 phosphorylation reduces its ability to bind the F-BAR domain of Cdc15 in vitro, and because Pxl1-Cdc15 F-BAR domain interaction is the major mechanism of Pxl1 CR recruitment (Snider et al., 2020), our results provide a mechanistic explanation for the phosphorylation-mediated change we observe in Pxl1 localization in vivo. It also seems possible, however, that phosphorylation regulates Pxl1 protein levels given that we observed an approximately twofold increase in the abundance of Pxl1(9A) using tags for Pxl1 detection. Increased levels may have allowed us to detect Pxl1(9A) CR arrival earlier, and the increased Pxl1 levels in the CR may have perturbed cytokinesis. Pxl1 abundance seems to be regulated at least in part by transcription, with transcript levels peaking during M phase (Rustici et al., 2004). Although the products of other genes with this transcriptional profile are subsequently eliminated by phosphorylation-directed ubiquitin-mediated proteolysis, it seems unlikely that phosphorylation triggers Pxl1 degradation because Pxl1 persists well after the peak of its phosphorylation, through cytokinesis and into the next cell cycle. It is also present at normal levels in phosphatase mutants where it remains hyperphosphorylated. Perhaps the binding interactions in the CR that accompany Pxl1 dephosphorylation protect it from degradation. Future studies to map and then perturb the Cdc15 binding site(s) on Pxl1 could test this hypothesis.
Ultimately, although pxl1 phosphoablating mutants had only mild effects on cytokinesis, our data are consistent with the concept that Cdk1 controls cytokinesis by phosphorylating many substrates and that multiple redundant mechanisms exist to regulate the timing of cytokinesis.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Experimental model and subject details
Supplemental Table S1 lists S. pombe strains used in this study. Cells were cultured in rich media (5 g/l yeast extract, 30 g/l glucose, 225 mg/l adenine) with supplements (YES) or Edinburgh minimal medium (3 g/l potassium hydrogen phthalate, 20 g/l glucose, 2.2 g/l disodium phosphate) plus selective supplements.
Fusion proteins were produced in Escherichia coli Rosetta2(DE3) pLysS cells. Bacteria were grown in Terrific Broth medium (12 g/l tryptone, 24 g/l yeast extract, 4 ml/l glycerol) with antibiotics. For MBP-Pxl1, 150 µM ZnCl2 was added to the culture media.
Strain construction
For integration of pxl1 wild type and phosphomutant alleles at the endogenous locus, pxl1::ura4+(KGY16851) was transformed with linear DNA consisting of 500 base pairs of the pxl1 5′ noncoding region, then sequences encoding 3xHA or mNG (Roberts-Galbraith et al., 2009; Shaner et al., 2013; Willet et al., 2015a) where indicated, followed by the desired pxl1 allele, a kanR cassette including the ADH1 terminator sequences, and 500 base pairs of the pxl1 3′ noncoding region. Integration was verified by growth on selective media containing 5-fluorourotic acid and G418, then PCR and/or microscopy, and finally sequencing. These were the pxl1 versions used throughout the paper except in Figure 1.
Introduction of pxl1 (and mutants thereof) into other genetic backgrounds was accomplished using standard S. pombe mating, sporulation, and tetrad dissection techniques, except for sid4, which was tagged at the 3′ end of its open reading frame with mNG:hygR using a pFA6 cassette as previously described (Bahler et al., 1998; Willet et al., 2015a). Integration of the tag was verified using whole cell PCR and/or microscopy.
Transformation of yeast with plasmid or linear DNA was accomplished using lithium acetate methods (Keeney and Boeke, 1994).
Molecular biology methods
Expression and integration vectors were constructed using standard molecular biology techniques. Mutations were made with a QuikChange Multi-lightning mutagenesis kit (Agilent Technologies). All constructs were sequenced for verification.
cdc25-22 block-and-release and calcineurin inhibition
Cells were arrested in G2 by being shifted to the restrictive temperature (36°C) for 3.5–4 h and then released back into the cell cycle by shifting to the permissive temperature (25°C). Twenty-five OD cell pellets were collected at the time of release (0 min) and at subsequent time points as indicated. Additionally, cells were fixed in ice-cold 70% ethanol for staining with 4’,6-diamidino-2-phenylindole (DAPI) and methyl blue to visualize nuclei and septa, respectively (see Microscopy and image analysis section).
For testing whether calcineurin regulates Pxl1 in Figure 3A, a cell culture was blocked-and-released as described. After the time 0 pellet was collected, the culture was split into two cultures. At 30 min after release, one culture was treated with dimethyl sulfoxide (DMSO) as a vehicle control and the other with FK506 at final concentration 10 µg/ml (LC Laboratories Cat No F-4900). Immediately after addition of drug or vehicle, the 30-min pellet was collected.
Denatured lysis and immunoprecipitation-phosphatase assays for detecting Pxl1 phosphorylation status
Twenty-five OD pellets were snap-frozen in dry ice-ethanol baths and then lysed by bead disruption in EDTA-free NP-40 lysis buffer (6 mM Na2HPO4, 4 mM NaH2PO4, 1% Nonidet P-40, 150 mM NaCl, 50 mM NaF, 0.1 mM Na3VO4) modified from Gould et al. (1991) with the addition of 0.5 mM diisopropyl fluorophosphate (Sigma-Aldrich), benzamidine, and phenylmethylsulfonyl fluoride. Lysates were denatured by boiling at 95°C for 1 min in EDTA-free SDS lysis buffer (10 mM NaPO4, pH 7.0, 0.5% SDS, 1 mM dithiothreitol [DTT], 50 mM NaF, 1 mM Na3VO4, 4 µg/ml leupeptin). Then lysates were cleared by spinning at 10,000 rpm for 6 min at 4°C. Cleared lysates were normalized by Pierce(TM) BCA protein assay kit assay. A lysate sample was collected at this point by adding to sample buffer and boiling for 1 min.
For immunoprecipitation, Pxl1 was immunoprecipitated from lysates using 4 µg anti-HA antibody (12CA5) and 30 µl protein A sepharose (GE Healthcare; 17-5280-04) for 1 h at 4°C. Then sepharose was washed three times with EDTA-free NP-40 lysis buffer. If proceeding directly to immunoblotting, beads were added to sample buffer and boiled for 1 min.
If proceeding to phosphatase assay, the sepharose was also washed two times with 1× phosphatase buffer (50 mM HEPES, pH 7.4, 150 mM NaCl). Samples were treated with lambda protein phosphatase (New England Biolabs; P0753) according to the manufacturer’s protocol, and reactions were stopped by the addition of sample buffer.
Immunoblot analysis was performed as previously described (Roberts-Galbraith et al., 2009). Briefly, to best visualize Pxl1 phospho-species, proteins were resolved by SDS–PAGE using freshly poured (within 24 h) 6% Tris-glycine gels at 150 V for 2 h. Then, protein samples were transferred to polyvinylidene fluoride (PVDF) membrane (Immobilon FL; EMD Millipore) at 30 V for 1.5 h. Anti-HA antibody (12CA5) was used as the primary antibody. Secondary antibodies were conjugated to IRDye 680RD or IRDye 800CW (LI-COR Biosciences) and visualized using an Odyssey machine (LI-COR Biosciences).
In vitro kinase assays
Kinase reactions were performed in protein kinase buffer (10 mM Tris, pH 7.4, 10 mM MgCl2, and 1 mM DTT) with 10 μM cold ATP, 1 μCi of [γ32P]ATP, and 100 ng of kinase-active or kinase-dead insect cell–produced Cdk1-Cdc13 in 20 μl reactions that were incubated at 30°C for 30 min with shaking. MBP was used as a negative control substrate for Cdk1-Cdc13. Reactions were quenched by the addition of 5 μl of 5× SDS sample buffer. Proteins were separated by SDS–PAGE. The acrylamide gels were either stained with Coomassie blue and then dried or transferred to PVDF membranes (Immobilon P; EMD Millipore) and stained with REVERT Total Protein stain (LI-COR Biosciences). Phosphorylated proteins were visualized by autoradiography.
In vitro phosphorylation of recombinant proteins used in in vitro binding assays was performed via identical kinase assays, except that radioactive [γ32P]ATP was eliminated, and the final concentration of unlabeled ATP in reactions was increased to 2 mM.
Phosphoamino acid analysis
PVDF pieces containing 32P-labeled Pxl1 were subjected to partial acid hydrolysis using boiling 6 M HCl for 1 h. Hydrolyzed amino acids were separated by two-dimensional thin-layer electrophoresis (Boyle et al., 1991).
Tryptic phosphopeptide mapping
Phosphopeptide mapping was performed as described previously (Boyle et al., 1991; McCollum et al., 1999). Pieces of PVDF membrane containing 32P-labeled Pxl1 were pretreated with methanol for 30 s and then incubated at 37°C for 30 min with 0.1% Tween 20 in 50 mM ammonium bicarbonate, pH 8.0. After three short washes with 50 mM ammonium bicarbonate, phosphopeptides were released from the membrane with two 3-h incubations at 37°C in 50 mM ammonium bicarbonate, pH 8.0, with 10 μg of N-tosyl-l-phenylalanine chloromethyl ketone-trypsin added for each incubation. After lyophilization, the phosphopeptides were separated in two dimensions with electrophoresis at pH 1.9. Tryptic phosphopeptides were visualized by autoradiography.
Purification of GFP-Pxl1 from prometaphase-arrested cells
GFP-Pxl1 was purified using a nanobody-based immunoprecipitation (IP) (Rothbauer et al., 2008) that was performed as previously described (Chen et al., 2016) except that EDTA was removed from all buffers. A 2 l culture of GFP-pxl1 nda3-KM311 (KGY18174) was grown in 4× YES media at 32°C to OD 0.3 and then shifted to 18°C for 6.5 h. Cells were pelleted and then lysed under native conditions in a glass bead beater with NP-40 buffer (10 mM sodium phosphate, pH 7.0, 0.15 M NaCl, 1% Nonidet P40, 50 mM sodium fluoride, 100 μM sodium orthovanadate, 5 µg/ml leupeptin) plus Protease Inhibitors (Roche). The lysate was cleared at low speed (ca. 3000 rpm) on a tabletop centrifuge. The supernatant was transferred to a clean Falcon tube and GFP-TRAP magnetic agarose beads (30 μl) (GFP-Trap_MA; ChromoTek) prewashed with NP-40 buffer were added and nutated at 4°C for 1–1.5 h. The beads were then washed with NP-40 buffer two times (5 ml) and then once with 5 ml Low-NP-40 buffer (0.02% NP-40) before elution from beads with 100 μl of 200 mM glycine twice. The eluate was trichloroacetic acid (TCA) precipitated (25% TCA on ice), and proteins were identified using LC-mass spectrometry (MS) as detailed below. A control purification was also performed from untagged yeast, and proteins identified by MS were subtracted from the list of potential interactors.
Mass spectrometry analysis
GFP-Pxl1 purification was analyzed by mass spectrometry as previously described (Elmore et al., 2014) with the following modification: a newer version of Scaffold (v4.8.4; Proteome Software, Portland, OR) was used, and the minimum peptide identification probability was changed to 95.0%. Phosphorylation sites were annotated using Scaffold PTM (v3.0.0; Proteome Software, Portland, OR).
Microscopy and image analysis
Microscope.
All images were acquired using a Personal DeltaVision microscope system (GE Healthcare, Issaquah, WA), which includes an Olympus IX71 microscope, 60×/NA 1.42 PlanApo and 100×/NA 1.40 UPlanSApo objectives, fixed- and live-cell filter wheels, a Photometrics CoolSnap HQ2 camera, and softWoRx imaging software.
Sample preparation.
For live-cell fluorescence imaging, strains were grown overnight to log phase in YES media at 25°C in a shaking water bath. Cells were imaged in YES media at 23–29°C.
Yeast cells were fixed by adding ice-cold 70% ethanol while vortexing and then incubating at 4°C for at least 15 min. Cells were fixed at a ratio of 0.5 OD per 1 ml 70% ethanol.
For visualizing nuclei and cell wall, approximately 0.5 OD fixed cells were washed once with phosphate-buffered saline, pH 7.5, and then resuspended in 50 µl of 1 mg/ml methyl blue (Sigma; M6900) and incubated at room temperature for 30 min. Then, the cells were pelleted and mixed 1:1 with 5 µg/ml DAPI (Sigma; D9542) before imaging immediately.
Time-lapse imaging.
Time-lapse imaging was performed using an ONIX microfluidics perfusion system (CellASIC ONIX; EMD Millipore). Fifty microliters of a 40 × 106 cells/ml YES suspension was loaded into Y04C plates for 5 s at 8 psi. YES medium flowed into the chamber at 5 psi throughout imaging. Time-lapse images were obtained at 2-min intervals. Ten Z-series optical sections were taken at 0.5-µm spacing. The stage temperature was maintained at 29°C. Images were deconvolved with 10 iterations using softWoRx, and maximum intensity Z-projections were generated using ImageJ (Schindelin et al., 2012). Cytokinesis stages were defined as follows: formation is the time from SPB separation to Rlc1 coalescence into a ring; maturation is the period between formation and constriction; constriction is the time from first decrease in Rlc1 ring diameter to a single point.
Quantifying fluorescence intensity.
For all intensity measurements, the background was subtracted by creating a region of interest (ROI) in the same image where there were no cells. The raw intensity of the background was divided by the area of the background, which was multiplied by the area of the ROI. This number was subtracted from the raw integrated intensity of that ROI. For CR intensity quantification, an ROI was drawn around the CR and measured for raw integrated density. Three biological replicates were performed. All images for intensity analyses of the CR and whole cell were not deconvolved, and the z-slices were sum projected.
Recombinant protein purification
Pxl1 was expressed as an MBP fusion as previously described (Bhattacharjee et al., 2020) in the pMAL-c2 vector in Rosetta2(DE3)pLysS cells, which were grown to log phase in Terrific Broth + 150 μM ZnCl2 (to increase production), induced overnight at 17°C with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) (Fisher Scientific; BP1755). Cells were harvested, flash frozen, and lysed by sonication in 20 mM Tris, pH 7.4, 150 mM NaCl, 0.1% NP-40, and cOmplete EDTA-free protease inhibitor cocktail (Roche). Lysate was cleared at high speed and then incubated with amylose resin (New England BioLabs) for 2 h at 4°C. Resin was washed thoroughly in lysis buffer. After washing, buffer was added to the resin to create a 1:1 slurry of buffer and resin-bound protein. The concentration of MBP-Pxl1 fusion on resin was calculated by running a sample on SDS–PAGE alongside bovine serum albumin (BSA) standards and staining with Coomassie blue.
Cdc15 F-BAR domain (residues 19–312) was produced as previously described (Snider et al., 2020). Cdc15 F-BAR domains were expressed as a 6xHis fusion in E. coli Rosetta2(DE3)pLysS cells grown to log phase in Terrific Broth, induced overnight at 17°C with 0.1 mM IPTG (Fisher Scientific; BP1755). Cells were lysed by sonication and F-BAR domains were purified on cOmplete His-Tag resin (Roche) according to the manufacturer’s protocol. His tags were then cleaved by thrombin digestion, and F-BARs were further purified on a HiTrap Q SP anion exchange column (Cytiva Life Sciences) and concentrated with Amicon Ultra Centrifugal Filters (EMD-Millipore).
GST-Cdc15C fusion proteins were produced as previously described (Bhattacharjee et al., 2020). Bacteria were grown in Terrific Broth media with antibiotics to log phase (OD595 1–1.5) at 36°C. Then, induction was initiated by incubation for 15 min on ice before the addition of 0.4 mM IPTG (Fisher Scientific; BP1755). Protein was produced for 16–18 h at 18°C.Frozen cell pellets were lysed in GST buffer (4.3 mM NaHPO4, 137 mM NaCl, 2.7 mM KCl, 1 mM DTT) with the addition of 200 µg/ml lysozyme (Sigma-Aldrich; L6876), cOmplete EDTA-free protease inhibitor cocktail (Roche; Cat#05056489001), and 0.1% NP-40 (US Biologicals; N3500). Continuous agitation on ice for 20 min was used to suspend the cell pellet. Then, lysates were sonicated three times for 30 s, with at least a 30 s pause between sonications (Sonic Dismembrator Model F60; Fisher Scientific; power 15 W). Lysates were cleared for 15–30 min at 10,000–13,000 rpm. Cleared lysate was then used in a batch purification protocol by adding GST-bind (EMD Millipore; 70541) resin for 2 h at 4°C. Then, resin was washed three times for 5 min at 4°C with the appropriate buffer. To elute proteins from beads, dry beads were resuspended in an equal volume of GST elution buffer (50 mM Tris-HCl, pH 8, 10 mM glutathione [Sigma-Aldrich; G4251]) and nutated for 30 min at 4°C. The supernatant was separated from the resin to a fresh tube. NaCl (100 mM) was added to GST fusion proteins. Eluted fusion proteins were then aliquoted, snap frozen, and stored at –80°C.
Protein concentration was calculated from Coomassie Brilliant Blue G (Sigma-Aldrich; B0770) staining of SDS–PAGE-separated purified proteins and BSA standards (Sigma-Aldrich).
In vitro binding assays
After the kinase assay, 1 µM recombinant MBP-Pxl1 immobilized with amylose resin was incubated with 1 µM binding partners in a 50-µl binding reaction. Binding buffer was 20 mM Tris, pH 7.0, 150 mM NaCl, 0.1% NP-40. After nutating for 1 h at 4°C, beads were rinsed 2× with 0.5 ml binding buffer and then boiled with 30 µl 2× sample buffer. Proteins were resolved in SDS–PAGE for Coomassie staining analysis.
Quantification and statistical analysis
No statistical methods were used to predetermine sample size. Details of statistical tests performed are described in the relevant figures and legends.
Supplementary Material
Acknowledgments
We are grateful to Chloe Snider and Rahul Bhattacharjee for assistance with recombinant protein production, to Liping Ren for strain construction, and to Jitender Singh Bisht, Sierra Cullati, Chloe Snider, and Alaina Willet for constructive comments on the manuscript. M.C.M. was supported by National Institutes of Health (NIH) Grants F31GM119252 and T32GM007347. This work was supported by NIH Grant R35GM131799 to K.L.G.
Abbreviations used:
- CN
calcineurin
- Cdk1
cyclin-dependent kinase
- CR
cytokinetic ring
- mNG
mNeonGreen
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-12-0807) on June 16, 2021.
REFERENCES
- Aggen JB, Nairn AC, Chamberlin R (2000). Regulation of protein phosphatase-1. Chem Biol 7, R13–R23. [DOI] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A 3rd, Steever AB, Wach A, Philippsen P, Pringle JR (1998). Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14, 943–951. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Chang L, Wong KC, Naqvi NI, He X, Sazer S, Gould KL (1998). Isolation and characterization of new fission yeast cytokinesis mutants. Genetics 149, 1265–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathe M, Chang F (2010). Cytokinesis and the contractile ring in fission yeast: towards a systems-level understanding. Trends Microbiol 18, 38–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharjee R, Mangione MC, Wos M, Chen JS, Snider CE, Roberts-Galbraith RH, McDonald NA, Presti LL, Martin SG, Gould KL (2020). DYRK kinase Pom1 drives F-BAR protein Cdc15 from the membrane to promote medial division. Mol Biol Cell 31, 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom J, Cristea IM, Procko AL, Lubkov V, Chait BT, Snyder M, Cross FR (2011). Global analysis of Cdc14 phosphatase reveals diverse roles in mitotic processes. J Biol Chem 286, 5434–5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Gould KL (2011). On the cutting edge: post-translational modifications in cytokinesis. Trends Cell Biol 21, 283–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchoux C, Uhlmann F (2011). A quantitative model for ordered Cdk substrate dephosphorylation during mitotic exit. Cell 147, 803–814. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, van der Geer P, Hunter T (1991). Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol 201, 110–149. [DOI] [PubMed] [Google Scholar]
- Carpy A, Krug K, Graf S, Koch A, Popic S, Hauf S, Macek B (2014). Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (fission yeast). Mol Cell Proteomics 13, 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Drubin D, Nurse P (1997). cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J Cell Biol 137, 169–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F, Woollard A, Nurse P (1996). Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci 109 (Pt 1), 131–142. [DOI] [PubMed] [Google Scholar]
- Chen JS, Beckley JR, McDonald NA, Ren L, Mangione M, Jang SJ, Elmore ZC, Rachfall N, Feoktistova A, Jones CM, et al. (2015). Identification of new players in cell division, DNA damage response, and morphogenesis through construction of Schizosaccharomyces pombe deletion strains. G3 (Bethesda) 5, 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Beckley JR, Ren L, Feoktistova A, Jensen MA, Rhind N, Gould KL (2016). Discovery of genes involved in mitosis, cell division, cell wall integrity and chromosome segregation through construction of Schizosaccharomyces pombe deletion strains. Yeast 33, 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JS, Broadus MR, McLean JR, Feoktistova A, Ren L, Gould KL (2013). Comprehensive proteomics analysis reveals new substrates and regulators of the fission yeast clp1/cdc14 phosphatase. Mol Cell Proteomics 12, 1074–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clifford DM, Chen CT, Roberts RH, Feoktistova A, Wolfe BA, Chen JS, McCollum D, Gould KL (2008a). The role of Cdc14 phosphatases in the control of cell division. Biochem Soc Trans 36, 436–438. [DOI] [PubMed] [Google Scholar]
- Clifford DM, Wolfe BA, Roberts-Galbraith RH, McDonald WH, Yates JR 3rd, Gould KL (2008b). The Clp1/Cdc14 phosphatase contributes to the robustness of cytokinesis by association with anillin-related Mid1. J Cell Biol 181, 79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes JC, Pujol N, Sato M, Pinar M, Ramos M, Moreno B, Osumi M, Ribas JC, Perez P (2015). Cooperation between paxillin-like protein Pxl1 and glucan synthase Bgs1 is essential for actomyosin ring stability and septum formation in fission yeast. PLoS Genet 11, e1005358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dephoure N, Gould KL, Gygi SP, Kellogg DR (2013). Mapping and analysis of phosphorylation sites: a quick guide for cell biologists. Mol Biol Cell 24, 535–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Wulf P, Montani F, Visintin R (2009). Protein phosphatases take the mitotic stage. Curr Opin Cell Biol 21, 806–815. [DOI] [PubMed] [Google Scholar]
- Egloff M-P, Johnson DF, Moorhead G, Cohen PTW, Cohen P, Barford D (1997). Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J 16, 1876–1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore ZC, Beckley JR, Chen J-S, Gould KL (2014). Histone H2B ubiquitination promotes the function of the anaphase-promoting complex/cyclosome in Schizosaccharomyces pombe. G3 (Bethesda) 4, 1529–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Reymond A, Cerutti L, Utzig S, Hofmann K, Simanis V (1995). The S. pombe cdc15 gene is a key element in the reorganization of F-actin at mitosis. Cell 82, 435–444. [DOI] [PubMed] [Google Scholar]
- García Cortés JC, Ramos M, Osumi M, Pérez P, Ribas JC (2016). The cell biology of fission yeast septation. Microbiol Mol Biol Rev 80, 779–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge W, Balasubramanian MK (2008). Pxl1p, a paxillin-related protein, stabilizes the actomyosin ring during cytokinesis in fission yeast. Mol Biol Cell 19, 1680–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M (2017). Cytokinesis in metazoa and fungi. Cold Spring Harb Perspect Biol 9, a022343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KL, Moreno S, Owen DJ, Sazer S, Nurse P (1991). Phosphorylation at Thr167 is required for Schizosaccharomyces pombe p34cdc2 function. EMBO J 10, 3297–3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouw M, Michael S, Samano-Sanchez H, Kumar M, Zeke A, Lang B, Bely B, Chemes LB, Davey NE, Deng Z, et al. (2018). The eukaryotic linear motif resource—2018 update. Nucleic Acids Res 46, D428–D434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grallert A, Boke E, Hagting A, Hodgson B, Connolly Y, Griffiths JR, Smith DL, Pines J, Hagan IM (2015). A PP1-PP2A phosphatase relay controls mitotic progression. Nature 517, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayles J, Wood V, Jeffery L, Hoe KL, Kim DU, Park HO, Salas-Pino S, Heichinger C, Nurse P (2013). A genome-wide resource of cell cycle and cell shape genes of fission yeast. Open Biol 3, 130053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heim A, Konietzny A, Mayer TU (2015). Protein phosphatase 1 is essential for Greatwall inactivation at mitotic exit. EMBO Rep 16, 1501–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO (2009). Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science 325, 1682–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney JB, Boeke JD (1994). Efficient targeted integration at leu1-32 and ura4-294 in Schizosaccharomyces pombe. Genetics 136, 849–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettenbach AN, Deng L, Wu Y, Baldissard S, Adamo ME, Gerber SA, Moseley JB (2015). Quantitative phosphoproteomics reveals pathways for coordination of cell growth and division by the conserved fission yeast kinase pom1. Mol Cell Proteomics 14, 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. (2010). Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28, 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Krug K, Pengelley S, Macek B, Hauf S (2011). Mitotic substrates of the kinase aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci Signal 4, rs6. [DOI] [PubMed] [Google Scholar]
- Kovar DR, Kuhn JR, Tichy AL, Pollard TD (2003). The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J Cell Biol 161, 875–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Maiolica A, Godfrey M, Scheidel N, Aebersold R, Uhlmann F (2015). Identification of Cdk targets that control cytokinesis. EMBO J 34, 81–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Coffman VC, Lee IJ, Wu JQ (2011). Assembly and architecture of precursor nodes during fission yeast cytokinesis. J Cell Biol 192, 1005–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangione MC, Gould KL (2019). Molecular form and function of the cytokinetic ring. J Cell Sci 132, jcs226928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangione MC, Snider CE, Gould KL (2019). The intrinsically disordered region of the cytokinetic F-BAR protein Cdc15 performs a unique essential function in maintenance of cytokinetic ring integrity. Mol Biol Cell 30, 2790–2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín R, Stonyte V, Lopez-Aviles S (2020). Protein phosphatases in G1 regulation. Int J Mol Sci 21, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Garcia R, Arribas V, Coll PM, Pinar M, Viana RA, Rincon SA, Correa-Bordes J, Ribas JC, Perez P (2018). Paxillin-mediated recruitment of calcineurin to the contractile ring is required for the correct progression of cytokinesis in fission yeast. Cell Rep 25, 772–783.e774. [DOI] [PubMed] [Google Scholar]
- McCollum D, Feoktistova A, Gould KL (1999). Phosphorylation of the myosin-II light chain does not regulate the timing of cytokinesis in fission yeast. J Biol Chem 274, 17691–17695. [DOI] [PubMed] [Google Scholar]
- Minet M, Nurse P, Thuriaux P, Mitchison JM (1979). Uncontrolled septation in a cell division cycle mutant of the fission yeast Schizosaccharomyces pombe. J Bacteriol 137, 440–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mocciaro A, Schiebel E (2010). Cdc14: a highly conserved family of phosphatases with non-conserved functions? J Cell Sci 123, 2867–2876. [DOI] [PubMed] [Google Scholar]
- Mochida S, Hunt T (2007). Calcineurin is required to release Xenopus egg extracts from meiotic M phase. Nature 449, 336–340. [DOI] [PubMed] [Google Scholar]
- Morita R, Numata O, Nakano K, Takaine M (2021). Cell cycle-dependent phosphorylation of IQGAP is involved in assembly and stability of the contractile ring in fission yeast. Biochem Biophys Res Commun 534, 1026–1032. [DOI] [PubMed] [Google Scholar]
- Naylor SG, Morgan DO (2014). Cdk1-dependent phosphorylation of Iqg1 governs actomyosin ring assembly prior to cytokinesis. J Cell Sci 127, 1128–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama T, Yoshizaki N, Kishimoto T, Ohsumi K (2007). Transient activation of calcineurin is essential to initiate embryonic development in Xenopus laevis. Nature 449, 341–345. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K (1976). Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet 146, 167–178. [DOI] [PubMed] [Google Scholar]
- Orii M, Kono K, Wen H-I, Nakanishi M (2016). PP1-dependent formin Bnr1 dephosphorylation and delocalization from a cell division site. PloS One 11, e0146941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palani S, Meitinger F, Boehm ME, Lehmann WD, Pereira G (2012). Cdc14-dependent dephosphorylation of Inn1 contributes to Inn1-Cyk3 complex formation. J Cell Sci 125, 3091–3096. [DOI] [PubMed] [Google Scholar]
- Perez P, Cortes JC, Martin-Garcia R, Ribas JC (2016). Overview of fission yeast septation. Cell Microbiol 18, 1201–1207. [DOI] [PubMed] [Google Scholar]
- Pinar M, Coll PM, Rincon SA, Perez P (2008). Schizosaccharomyces pombe Pxl1 is a paxillin homologue that modulates Rho1 activity and participates in cytokinesis. Mol Biol Cell 19, 1727–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD (2010). Mechanics of cytokinesis in eukaryotes. Curr Opin Cell Biol 22, 50–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Wu JQ (2010). Understanding cytokinesis: lessons from fission yeast. Nat Rev Mol Cell Biol 11, 149–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian J, Beullens M, Huang J, De Munter S, Lesage B, Bollen M (2015). Cdk1 orders mitotic events through coordination of a chromosome-associated phosphatase switch. Nat Commun 6, 10215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren L, Willet AH, Roberts-Galbraith RH, McDonald NA, Feoktistova A, Chen JS, Huang H, Guillen R, Boone C, Sidhu SS, et al. (2015). The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol Biol Cell 26, 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Galbraith RH, Chen JS, Wang J, Gould KL (2009). The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J Cell Biol 184, 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H (2008). A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics 7, 282–289. [DOI] [PubMed] [Google Scholar]
- Rustici G, Mata J, Kivinen K, Lió P, Penkett CJ, Burns G, Hayles J, Brazma A, Nurse P, Bähler J (2004). Periodic gene expression program of the fission yeast cell cycle. Nat Genet 36, 809–817. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, et al. (2013). A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat Methods 10, 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snider CE, Chandra M, McDonald NA, Willet AH, Collier SE, Ohi MD, Jackson LP, Gould KL (2020). Opposite surfaces of the Cdc15 F-BAR domain create a membrane platform that coordinates cytoskeletal and signaling components for cytokinesis. Cell Rep 33, 108526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F, Amon A (2004). Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Ann Rev Genet 38, 203–232. [DOI] [PubMed] [Google Scholar]
- Sun X, Phua DYZ, Axiotakis L Jr, Smith MA, Blankman E, Gong R, Cail RC, Espinosa de Los Reyes S, Beckerle MC, Waterman CM, Alushin GM (2020). Mechanosensing through direct binding of tensed F-actin by LIM domains. Dev Cell 55, 468–482.e467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P (2016). CDK substrate phosphorylation and ordering the cell cycle. Cell 167, 1750–1761.e1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaffer MP, Jones AW, Flynn HR, Snijders AP, Nurse P (2018). Quantitative phosphoproteomics reveals the signaling dynamics of cell-cycle kinases in the fission yeast Schizosaccharomyces pombe. Cell Rep 24, 503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD (2008). Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science 319, 97–100. [DOI] [PubMed] [Google Scholar]
- Wachtler V, Huang Y, Karagiannis J, Balasubramanian MK (2006). Cell cycle-dependent roles for the FCH-domain protein Cdc15p in formation of the actomyosin ring in Schizosaccharomyces pombe. Mol Biol Cell 17, 3254–3266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley SP, Hinchcliffe EH, Glotzer M, Hyman AA, Sluder G, Wang Y (1997). CDK1 inactivation regulates anaphase spindle dynamics and cytokinesis in vivo. J Cell Biol 138, 385–393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet AH, Bohnert KA, Gould KL (2018). Cdk1-dependent phosphoinhibition of a formin-F-BAR interaction opposes cytokinetic contractile ring formation. Mol Biol Cell 29, 713–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet AH, McDonald NA, Bohnert KA, Baird MA, Allen JR, Davidson MW, Gould KL (2015a). The F-BAR Cdc15 promotes contractile ring formation through the direct recruitment of the formin Cdc12. J Cell Biol 208, 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willet AH, McDonald NA, Gould KL (2015b). Regulation of contractile ring formation and septation in Schizosaccharomyces pombe. Curr Opin Microbiol 28, 46–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JD, Anderson CA, Suarez C, Kovar DR, Gardel ML (2020). Evolutionarily diverse LIM domain-containing proteins bind stressed actin filaments through a conserved mechanism. Proc Nat Acad Sci USA 117, 25532–25542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F, Sigl R, Geley S (2007). “… The end of the beginning”: cdk1 thresholds and exit from mitosis. Cell Cycle 6, 1408–1411. [PubMed] [Google Scholar]
- Wu JQ, Guo JY, Tang W, Yang CS, Freel CD, Chen C, Nairn AC, Kornbluth S (2009). PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat Cell Biol 11, 644–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Kuhn JR, Kovar DR, Pollard TD (2003). Spatial and temporal pathway for assembly and constriction of the contractile ring in fission yeast cytokinesis. Dev Cell 5, 723–734. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD (2005). Counting cytokinesis proteins globally and locally in fission yeast. Science 310, 310–314. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD (2006). Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol 174, 391–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurzenberger C, Gerlich DW (2011). Phosphatases: providing safe passage through mitotic exit. Nat Rev Mol Cell Biol 12, 469–482. [DOI] [PubMed] [Google Scholar]
- Yoshida T, Toda T, Yanagida M (1994). A calcineurin-like gene ppb1+ in fission yeast: mutant defects in cytokinesis, cell polarity, mating and spindle pole body positioning. J Cell Sci 107, 1725–1735. [DOI] [PubMed] [Google Scholar]
- Zimmermann D, Homa KE, Hocky GM, Pollard LW, De La Cruz EM, Voth GA, Trybus KM, Kovar DR (2017). Mechanoregulated inhibition of formin facilitates contractile actomyosin ring assembly. Nat Commun 8, 703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.