Abstract
Neutrophils migrate in response to chemoattractants to mediate host defense. Chemoattractants drive rapid intracellular cytoskeletal rearrangements including the radiation of microtubules from the microtubule-organizing center (MTOC) toward the rear of polarized neutrophils. Microtubules regulate neutrophil polarity and motility, but little is known about the specific role of MTOCs. To characterize the role of MTOCs on neutrophil motility, we depleted centrioles in a well-established neutrophil-like cell line. Surprisingly, both chemical and genetic centriole depletion increased neutrophil speed and chemotactic motility, suggesting an inhibitory role for centrioles during directed migration. We also found that depletion of both centrioles and GM130-mediated Golgi microtubule nucleation did not impair neutrophil directed migration. Taken together, our findings demonstrate an inhibitory role for centrioles and a resilient MTOC system in motile human neutrophil-like cells.
INTRODUCTION
Neutrophils respond rapidly to sites of tissue damage and infection, and are critical for host defense (Borregaard, 2010; de Oliveira, Rosowski et al., 2016). They rapidly integrate a diverse array of signals to mediate efficient directed migration. Although there has been progress in understanding the major mechanisms driving cell migration, the role of the microtubule-organizing center (MTOC) during neutrophil motility remains unclear.
Microtubules regulate neutrophil polarization downstream of chemoattractant stimulation, and regulate front–back polarity (Niggli, 2003; Xu et al., 2005; Yoo et al., 2012). Microtubule networks focus at MTOCs, which facilitate their recruitment and organization. In animal cells, two orthogonal centrioles comprise the core of the centrosome, the main MTOC. Some studies report that centrioles are necessary for directional migration of mesenchymal cells (Xu et al., 2007; Wakida et al., 2010; Cheng et al., 2019), while others suggest they are not required (Xu et al., 2005; Tormanen et al., 2019). Furthermore, these studies were primarily done in mesenchymal and epithelial cells that have adhesion-dependent migration and generally exhibit slower migration speeds. In contrast, neutrophils are amoeboid cells that are generally small, fast, and can rapidly repolarize toward different stimuli (Vorotnikov and Tyurin-Kuzmin, 2014). These intrinsic differences make it likely that the MTOC has distinct functions in neutrophil directed migration.
There is a scarcity of research on the function of centrioles in neutrophils. Early studies used electron microscopy to identify the presence of centrioles and confirm their ability to nucleate microtubules in neutrophils (Schliwa et al., 1982; Euteneuer and Schliwa, 1985). A more recent study suggested the centrosome is required for proper neutrophil polarization and directional migration (Xu et al., 2007). This study targeted an indirect, downstream signaling pathway, used laser ablation to ablate centrioles, and acknowledged that these manipulations may have had effects distinct from centriole depletion. Centrinone, a Polo-like kinase 4 (PLK4) inhibitor, has been shown to be an excellent tool for directly perturbing centrioles and studying their role in various cellular processes (Wong et al., 2015). PLK4 is the central regulator of centriole duplication and recruits downstream effectors, such as SAS6.
We sought to characterize the role of centrioles during neutrophil motility using live-cell imaging. We combined the power of pharmacological and genetic approaches to deplete centrioles in the neutrophil-like PLB-985 cell line. We found that centrioles are not required for efficient neutrophil migration. We also found that Golgi-mediated microtubule nucleation is dispensable for neutrophil directed migration. Our findings suggest that there are multiple sites for microtubule nucleation in amoeboid cells and that the centriole is not required for neutrophil motility and may in fact have inhibitory effects on neutrophil directed migration.
RESULTS AND DISCUSSION
Centrinone partially depletes centrioles and promotes migration of neutrophil-like PLB-985 cells
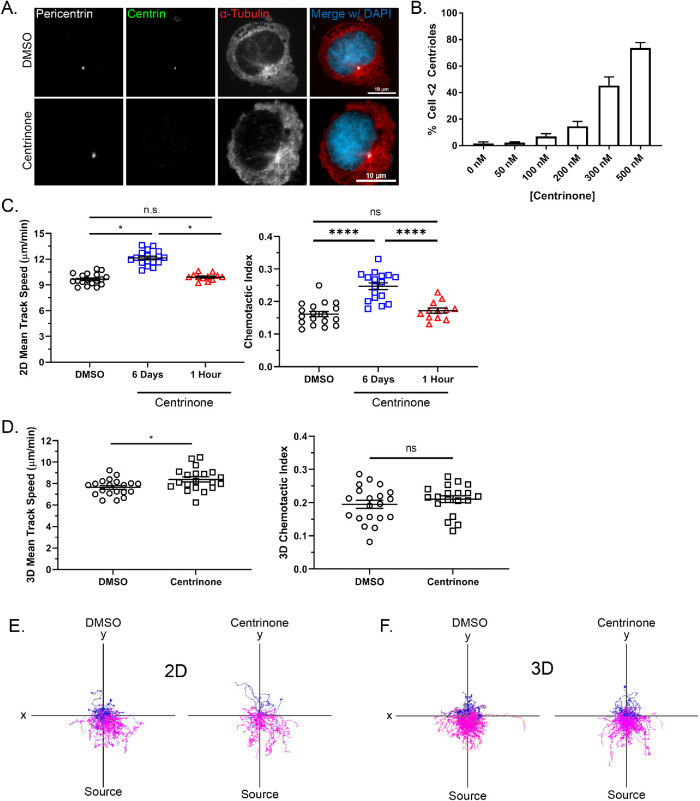
Previous studies have demonstrated that centriole depletion impairs the directed migration of some cell types (Koonce et al., 1984; Wakida et al., 2010; Kazazian et al., 2017). To determine the effects of the MTOC on neutrophil directed migration, we used centrinone, a PLK4 inhibitor, to deplete centrioles in the neutrophil-like PLB-985 cell line (Tucker et al., 1987; Hauert et al., 2002; Pedruzzi et al., 2002). Cells must progress through the cell cycle in the presence of centrinone to lose centrioles (i.e., centrinone cannot eliminate existing centrioles). Centriole depletion was detected using immunofluorescence by staining for centrin and pericentrin to identify centrioles and the pericentriolar material, respectively. We quantified centrin foci colocalizing with pericentrin, and cells with fewer than two centrin-containing foci were considered centriole-depleted. Six days of treatment with 500 nM centrinone depleted centrioles in approximately 70% of cells (Figure 1, A and B).
FIGURE 1:
Centrinone depletes centrioles and promotes migration in neutrophil-like PLB-985 cells. (A) Representative immunofluorescence images showing centriole depletion in centrinone-treated PLB-985 cells and DMSO-treated controls. Scale bars: 10 µm. Cells were treated with centrinone for 6 days and stained for the centriole marker centrin (green) and the pericentriolar matrix marker pericentrin (white). (B) The bar graph depicts quantification of centriole depletion at the indicated concentrations of centrinone for 6 days (data displayed as mean + SD; N = 3 repeats, n = 300 cells). (C) Mean 2D track speed and chemotactic index of 500 nM centrinone- and DMSO-treated PLB-985 cells (data displayed as mean + SEM; N = 3 repeats, n = 12–18 devices, 1547–3560 cells). (D) Mean 3D track speed and chemotactic index of 500 nM centrinone or DMSO-treated dPLB-985 cells (data displayed as mean + SEM; N = 6 repeats, n = 20 devices, 8457–10,809 cells). (E) Representative track plots of cells migrating in 2D on 10 µg/ml fibronectin. Magenta tracks indicate cell tracks with a positive Y displacement toward the fMLP gradient. Blue tracks have a net negative Y displacement away from the gradient. (F) Representative track plots of cells migrating in a 1.5 mg/ml 3D collagen I matrix. Significance for both 2D and 3D migration was determined by mixed-effects REML regression with a Kenward-Rogers degrees of freedom approximation. n.s. = not significant; *, p < 0.05; ****, p < 0.0001.
Next, we used microfluidics and live-cell imaging to track and quantify the migration of centrinone-treated PLB-985 cells. We found that centrinone treatment significantly increased both migration speed and chemotactic index of neutrophils responding to f-Met-Leu-Phe (fMLP) on a fibronectin-coated surface (Figure 1C). One-hour centrinone treatment post-differentiation, which does not reduce centriole number, did not affect migration, suggesting that the increased migration phenotype is due to centriole depletion and not acute inhibition of PLK4. Additionally, centrinone-treated cells showed improved directed migration as seen by track plots (Figure 1E and Supplemental Movie S1). Cell speed was also increased with centrinone treatment in 3D collagen, with no effect on directed migration (Figure 1, D and F, and Supplemental Movie S1). Together, these data suggest that centrioles are dispensable for neutrophil migration in both 2D and 3D environments and limit the directed motility of neutrophil-like cells.
Movie S1.
Representative time-lapse imaging of centrinone- vs. DMSO-treated PLB-985 cells, corresponding to Figure 1. Cells are migrating on either 10 μg/mL fibronectin or in a 1.5 mg/mL collagen I matrix towards an fMLP gradient (0.5 μM) in a microfluidic device. The frame rate is 10 frames per second. Images were acquired every 30 seconds for 45 minutes. Cells were labeled with Calcein AM. Scale bar 200 μm.
Effects of centrinone and blebbistatin on polyploidy and cell migration
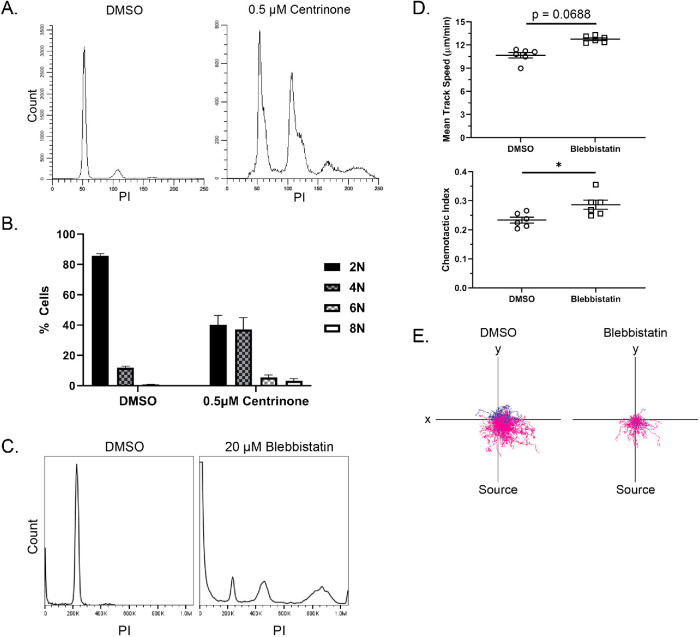
We noticed that centrinone-treated PLB-985 cells were larger and we wondered whether this was due to cytokinesis failure caused by loss of centrioles and/or PLK4 inhibition (Rosario et al., 2010; Press et al., 2019). To characterize the effects of centrinone treatment on polyploidy in differentiated PLB-985 cells, we performed propidium iodide staining and flow cytometry analysis. As expected, centrinone treatment resulted in more polyploidy compared with dimethyl sulfoxide (DMSO)–treated control cells (Figure 2A). The relationship between polyploidy and directed cell migration is unclear, so we used an alternative mechanism to induce polyploidy and probed its effects on the migration of PLB-985 cells. Blebbistatin, a myosin II inhibitor, inhibits cytokinesis thereby inducing polyploidy similar to centrinone treatment (Figure 2, B and C). Treatment with blebbistatin also increased PLB-985 directed motility, although less than what we observed with centrinone treatment (Figure 2, D and E). Although previous reports in the literature suggest centriole manipulation promotes migratory signaling independent of its effects on polyploidy (Godinho et al., 2014), we cannot rule out a possible contribution of polyploidy to the increased directed motility of PLB-985 cells after centrinone treatment.
FIGURE 2:
Effects of centrinone and blebbistatin on polyploidy and cell migration. (A) Representative histograms of propidium iodide (PI) staining for ploidy determination in centrinone-treated compared with DMSO-treated PLB-985 cells. (B) Graph showing quantification of the ploidy (data displayed as mean + SD). (C) Representative histogram of PI staining for ploidy analysis in cells treated with DMSO or 20 µM blebbistatin. Cells were treated for the first 4 days of differentiation and then treatment was washed out; cells were used on day 6. (D) Mean 2D track speed and chemotactic index of blebbistatin and DMSO-treated PLB-985 cells (data displayed as mean + SD; N = 2 repeats, n = 6 devices, 1159–2377 cells). Cells were imaged in a 10-µg/ml fibronectin-coated microfluidic device migrating toward an fMLP gradient every 30 s for 45 min. Significance was determined by mixed-effects REML regression with a Kenward-Rogers degrees of freedom approximation. (D) Representative track plots of blebbistatin and DMSO-treated cells migrating in 2D on 10 µg/ml fibronectin. n.s. = not significant; *, p < 0.05; **, p < 0.01.
The Golgi localizes near the nucleus and MTOC in motile PLB-985 cells
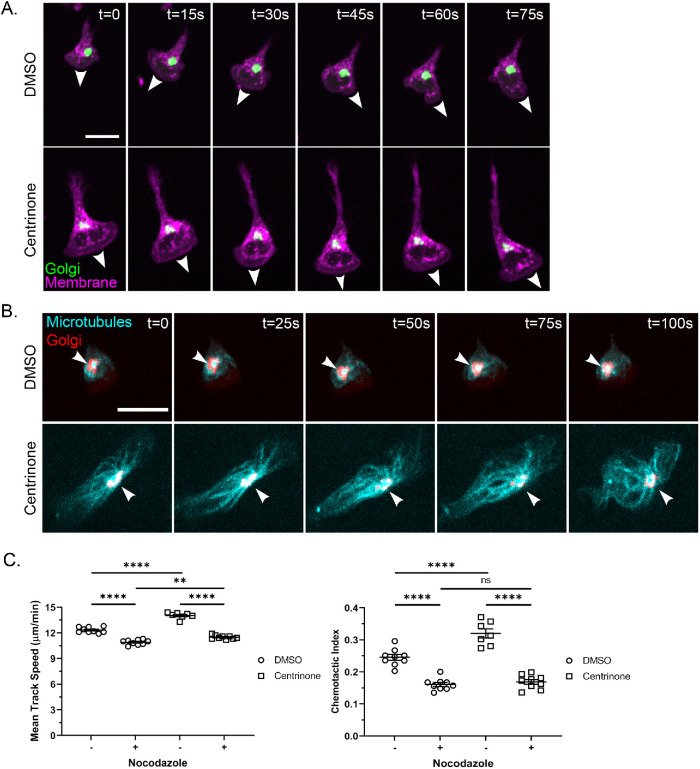
We hypothesized that alternative centers of microtubule nucleation were compensating in centriole-reduced PLB-985 cells. Sites other than the centrosome can serve as centers of microtubule nucleation, including membranous organelles (Sanchez and Feldman, 2017). Gavilan et al. demonstrated an apparent MTOC hierarchy with centrioles being dominant to other sites such as the Golgi (Gavilan et al., 2018). The Golgi is typically closely associated with centrioles in neutrophils (Tang and Clermont, 1989; Olins and Olins, 2005) and is an established noncentrosomal microtubule nucleation center (Chabin-Brion et al., 2001). To address the localization of the Golgi in motile neutrophils, we used a Golgi marker and performed live imaging of PLB-985 responding to a gradient of fMLP. We found that the Golgi localizes near the nucleus and toward the uropod in motile neutrophils, consistent with previous reports in fixed neutrophil-like cells (Figure 3A; Mazaki et al., 2012; Ren et al., 2020). In the presence of centrinone, the cells became more highly polarized without a change in the localization of the Golgi, although the Golgi appeared more fragmented. In addition, the Golgi appeared to nucleate microtubules in centrinone-treated neutrophil-like cells (Figure 3B). These findings suggest that the Golgi can operate as an alternative MTOC in centrinone-treated PLB-985 cells.
FIGURE 3:
The Golgi appears to nucleate microtubules toward the uropod and nocodazole blocks centrinone effects on the motility of neutrophil-like cells. (A) Time lapse of 500 nM centrinone or DMSO-treated dPLB-985 ManII-GFP cells (labeling the Golgi) migrating on 10 µg/ml fibronectin in an fMLP gradient. Cells were stained with CellMask Orange to visualize the membrane (magenta) and Golgi (green) and imaged every 15 s. White arrowheads mark the leading edge and direction of cell migration. Scale bar: 15 µm. (B) Time lapse of representative DMSO- and centrinone-treated cells depicting microtubule convergence at the Golgi. dPLB-985 cells were stained with 1 µM SiR-Tubulin (cyan) for 1 h and then imaged every 5 s to visualize the Golgi (red). White arrowheads indicate the point of microtubule convergence. Cell images are displayed at 25-s intervals. Scale bar: 15 µm. (C) Mean track speed and chemotactic index of nocodazole-treated (10 µM for 1 h before tracking). dPLB-985 cells ± centrinone (data displayed as mean + SEM; N = 3 repeats, n = 9 devices, 3256–4772 cells). Cells were imaged in a 10-µg/ml fibronectin-coated microfluidic device migrating toward an fMLP gradient every 30 s for 45 min. Significance was determined by mixed-effects REML regression with a Satterthwaite degrees of freedom approximation. n.s. = not significant; **, p < 0.01; ****, p < 0.0001.
Centrinone-induced neutrophil directed migration is impaired by nocodazole treatment
To determine whether changes in the microtubule network underlie the improved motility after centrosome depletion, we tested the effect of the microtubule disrupting drug nocodazole. We found that microtubule depletion with nocodazole abrogated both the increased speed and directionality of centrinone-treated PLB-985 cells (Figure 3C). There was a small increase in motility in the presence of both centrinone and nocodazole, suggesting that there may also be microtubule-independent effects of centrinone treatment. Taken together, these results suggest the increased motility induced by centrinone treatment is, at least in part, mediated by effects of the drug on the microtubule network.
CRISPR/Cas9 knockout of SAS6 depletes centrioles and promotes migration in neutrophil-like cells
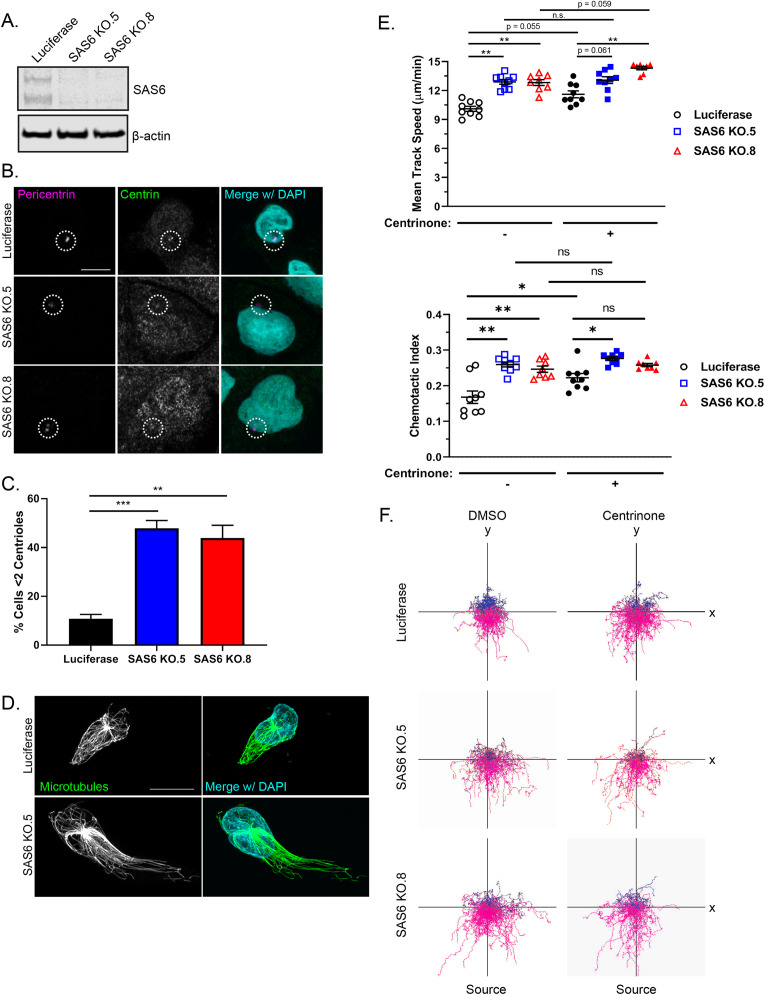
To further evaluate a role for centrioles in limiting neutrophil motility, we took a genetic approach to disrupt centrioles. We knocked out SAS6 centriolar assembly protein, which is necessary for centriole duplication, using CRISPR/Cas9 (Keller et al., 2014; McKinley and Cheeseman, 2017; Yoshiba et al., 2019). We generated multiple SAS6 knockout clones (Figure 4A) and confirmed centriole depletion by immunofluorescence (Figure 4B). Approximately 50% of SAS6 knockout cells were centriole-depleted as compared with 10% of control cells (Figure 4C). SAS6-deficient cells showed a polarized microtubule network (Figure 4D). In addition, SAS6-depleted cells displayed increased directed migration similar to our findings with centrinone treatment (Figure 4, E and F, and Supplemental Movie S3). Centrinone did not significantly improve migration of the SAS6 knockout clones (Figure 4, E and F, and Supplemental Movie S3). These data suggest that centrioles are not necessary for neutrophil directed migration and instead function to limit neutrophil motility.
FIGURE 4:
CRISPR/Cas9 knockout of SAS6 depletes centrioles and promotes neutrophil migration. (A) Western blot of two CRISPR/Cas9 SAS6 knockout clones and CRISPR/Cas9 luciferase control. (B) Representative immunofluorescence images showing centriole depletion in SAS6 knockout clones compared with luciferase control. Cells were stained for centrin (green) and pericentrin (magenta). The white dotted circles indicate the location of the centrosome. Scale bar: 5 µm. (C) Quantification of centriole depletion in SAS6 knockout clones compared with luciferase control (data displayed as mean + SEM; N = 3 repeats, n = 230–321 cells). Significance was determined by one-way ANOVA with post hoc Dunnett’s test for multiple comparisons. (D) Representative immunofluorescence images of SAS6 KO cell compared with luciferase controls. Cells were seeded on fibronectin-coated coverslips and stimulated with 100 nM fMLP for 5 min. Cells were fixed with glutaraldehyde and stained for microtubules (green) and nuclei (blue). Scale bar: 10 µm. (E) 2D mean track speed and chemotactic index of SAS6 knockout clones compared with luciferase control ± centrinone (data displayed as mean + SEM; N = 3 repeats, n = 8–9 devices, 3875–5576 cells). Cells were imaged in a 10-µg/ml fibronectin-coated microfluidic device migrating toward an fMLP gradient every 30 s for 45 min. (F) Representative track plots of SAS6 knockout clones or luciferase control ± 500 nM centrinone. Significance was determined by mixed-effects REML regression with a Satterthwaite degrees of freedom approximation. n.s. = not significant; *, p < 0.05; **, p < 0.01; ***, p < 0.001.
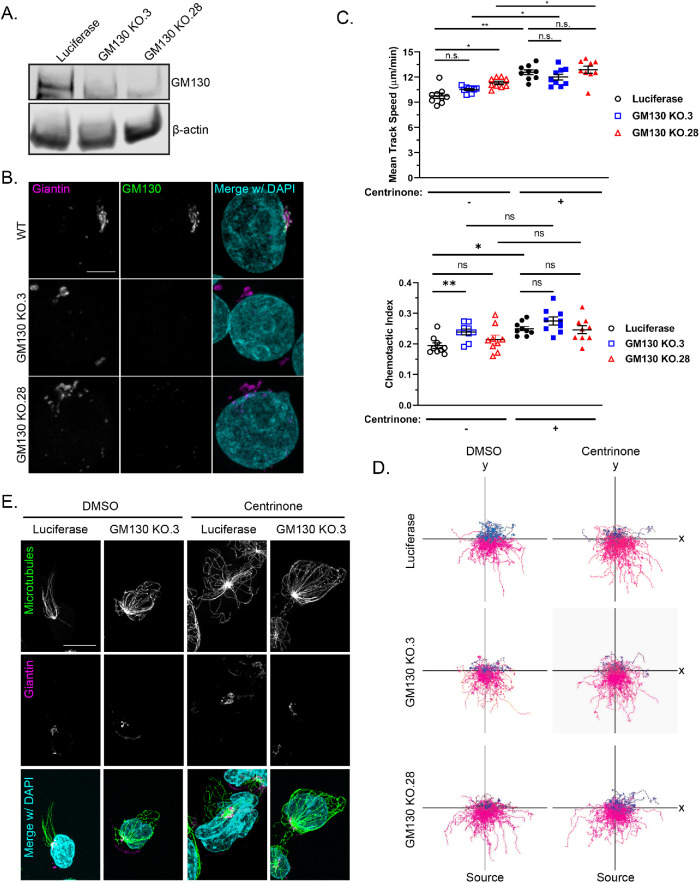
GM130 and Golgi microtubule nucleation are dispensable for neutrophil migration
We next knocked out a key component of the Golgi nucleation apparatus to determine whether the Golgi nucleation center affects neutrophil migration. The Golgi nucleates microtubules in an A-kinase anchoring protein 450 (AKAP450)–dependent manner and GM130 regulates AKAP450 localization to the Golgi (Rivero et al., 2009; Wu et al., 2016). We chose to target GM130 because it is Golgi-specific, whereas AKAP450 also associates with centrosomes (Schmidt et al., 1999; Takahashi et al., 1999; Witczak et al., 1999; Takahashi et al., 2002). We knocked out GM130 using CRISPR/Cas9 (Figure 5, A and B) and found that this had variable effects on the directed migration of PLB-985 cells with a trend toward increased motility (Figure 5, C and D, and Supplemental Movie S2). Centrinone treatment increased the migration speed of GM130 knockout cells similar to centrinone-treated luciferase control cells. There was also a trend toward increased chemotactic index, although this did not reach statistical significance (Figure 5C). In addition, microtubules were organized in centrinone-treated GM130 knockout cells with a clear, intact MTOC (Figure 5E). Surprisingly, we also found that AKAP450 localization was not affected by GM130 depletion (Supplemental Figure S1). Taken together, these findings suggest that there are multiple sites of microtubule nucleation in motile neutrophils, and that directed migration is unaffected with simultaneous loss of centrioles and the key GM130 component of Golgi.
FIGURE 5:
GM130 and Golgi microtubule nucleation are dispensable for neutrophil migration. (A) Western blot of two CRISPR/Cas9 GM130 knockout clones compared with the CRISPR/Cas9 luciferase control. (B) Representative immunofluorescence images showing GM130 depletion in two GM130 knockout clones compared with a wild-type control. Cells were stained for GM130 (green) and the cis-Golgi marker giantin (magenta). Scale bar: 5 µm. (C) 2D mean track speed and chemotactic index of GM130 knockout clones compared with luciferase control ± centrinone (data displayed as mean + SEM; N = 3 repeats, n = 9 devices, 3720–5911 cells). Cells were imaged in a 10-µg/ml fibronectin-coated microfluidic device migrating toward an fMLP gradient every 30 s for 45 min. Significance was determined by mixed-effects REML regression with a Satterthwaite degrees of freedom approximation. (D) Representative track plots of GM130 knockout clones or luciferase control ± centrinone. (E) Representative immunofluorescence images of GM130 KO or luciferase control. Cells were seeded on fibronectin-coated coverslips and stimulated with 100 nM fMLP for 5 min. Cells were stained for microtubules (green) and nuclei (blue). Scale bar: 10 µm. n.s. = not significant; *, p < 0.05; **, p < 0.01.
Movie S2.
Directed cell migration of SAS6 KO PLB-985 cells, corresponds to Figure 4. Luciferase CRISPR control and SAS6 KO PLB-985 cells +/- centrinone migrating on 10 μg/mL fibronectin towards an fMLP gradient (0.5 μM) in a microfluidic device. The frame rate is 10 frames per second. Cells were labeled with Calcein AM. Images were acquired every 30 seconds for 45 minutes.
Movie S3.
Directed cell migration of GM130 KO PLB-985 cells, corresponds to Figure 5. Luciferase CRISPR control and GM130 KO3 PLB-985 cells +/- centrinone migrating on 10 μg/mL fibronectin towards an fMLP gradient (0.5 μM) in a microfluidic device. The frame rate is 10 frames per second. Cells were labeled with Calcein AM. Images were acquired every 30 seconds for 45 minutes.
There have been conflicting reports on the role of the centrosome in cell migration. In general, most studies have shown that the centrosome is important for cell migration (Koonce et al., 1984; Wakida et al., 2010). In this study, we depleted centrioles in neutrophil-like cells and showed that they are not required for neutrophil directed migration. Both chemical and genetic depletion of centrioles increased neutrophil directed migration. The specific PLK4 inhibitor, centrinone, efficiently disrupted centrioles, and under all conditions increased the directed migration of PLB-985 cells. These findings contrast with the effects of laser depletion of centrioles in similar cell types, which can impair migration in newt eosinophils (Koonce et al., 1984) as well as in U2OS and PtK cells. However, photoablation of organelles may damage nearby structures and introduce oxidative or thermal stress. Centrinone impairs directional and invasive cell migration of MDA-MB-231 cancer cells (Kazazian et al., 2017). The differences described here in PLB-985 may be lineage dependent (neutrophil-like) or due to the amoeboid mode of motility. Alternatively, the 6-day exposure used by us would have more effectively depleted centrioles than the 12–24-h exposure in Kazazian et al., differentiating the direct effect of PLK4 inhibition on restraining migration, with the long-term effect of centriole loss on promoting migration. Bolstering this latter view, we find that SAS6-mediated centriole loss can similarly promote migration. Nevertheless, our data do not rule out lineage-specific effects.
Surprisingly, loss of both centrosomes and GM130-dependent MTOCs did not impair the directed migration of PLB-985 cells. However, one limitation of our data is that we did not identify the fraction of cells that experienced simultaneous loss of both organelles. Although it is possible that the experimental system or lethality precluded simultaneous loss of both MTOCs, our findings also may suggest alternative sites of microtubule nucleation in neutrophil-like cells. In the absence of centrosomal and GM130-dependent microtubule nucleation, pericentrin-containing MTOCs can still form in the cytoplasm (Gavilan et al., 2018). It is possible that neutrophils are following this compensatory MTOC hierarchy. As depletion of pericentrin abolishes these cytoplasmic MTOCs (Gavilan et al., 2018), a neutrophil model of triple MTOC depletion is key to determining the necessity of any given MTOC. Further work is necessary to clearly delineate the roles of individual MTOCs and establish a hierarchy in neutrophil migration.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Antibodies and reagents
| Antibodies, Reagent, or Chemical | Source | Identifier | |
|---|---|---|---|
| Antibodies | Dilutions | ||
| Mouse anti-human α-Tubulin | Cell Signaling | 3873S | 1:500 (IF) |
| Rabbit anti-human β-Actin | Sigma-Aldrich | SAB5500001 | 1:1000 (WB) |
| Mouse anti-human Centrin | EMD-Millipore | 04-1624 | 1:200 (IF) |
| Rabbit anti-human Giantin | Atlas Antibodies | HPA011008 | 1:250 (IF) |
| Mouse anti-human GM130 | BD Transduction Laboratories | 610823 | 1:250 (IF), 1:500 (WB) |
| Rabbit anti-human Pericentrin | Abcam | 4448 | 1:1000 (IF) |
| Mouse anti-human SAS6 | Santa Cruz Biotechnology | sc-81431 | 1:500 (WB) |
| Mouse anti-human AKAP450 | BD Transduction Laboratories | 611518 | 1:250 (IF) |
| Goat anti-mouse IgG2a Alexa Fluor 488 | Invitrogen | A-21131 | 1 µg/ml (IF) |
| Goat anti-mouse Alexa Fluor 488 | Invitrogen | A-11001 | 1 µg/ml (IF) |
| Goat anti-mouse Alexa Fluor 555 | Invitrogen | A32727 | 1 µg/ml (IF) |
| Goat anti-mouse Alexa Fluor 680 | Invitrogen | A-21057 | 1:10,000 (WB) |
| Goat anti-mouse Alexa Fluor 790 | Invitrogen | A11357 | 1:10,000 (WB) |
| Goat anti-mouse IgG (H&L) Antibody Dylight 800 Conjugated | Rockland Immunochemicals | 610-140-002-0.5 | 1:10,000 (WB) |
| Goat anti-rabbit IgG Alexa Fluor 568 | Invitrogen | A-11011 | 1 µg/ml (IF) |
| Goat anti-rabbit IgG Alexa Fluor 647 | Invitrogen | A-21244 | 1 µg/ml (IF) |
| Goat anti-rabbit Alexa Fluor 680 | Invitrogen | A-21076 | 1:10,000 (WB) |
| Goat anti-rabbit Alexa Fluor 790 | Invitrogen | A11369 | 1:10,000 (WB) |
| Goat ant-Rabbit IgG (H&L) Antibody Dylight 800 Conjugated | Rockland Immunochemicals | 611-145-002-0.5 | 1:10,000 (WB) |
| Reagents and Chemicals | |||
| Bovine Serum Albumin | Sigma-Aldrich | A7906 | |
| D-PBS | Corning | 21-031-CV | |
| HEPES | Corning | 25060CI | |
| Human Serum Albumin | Lee BioSolutions | 101-15-100 | |
| Nocodazole | Sigma-Aldrich | M1404 | 10 µM |
| Taxol | Tocris Bioscience | 10-917-0 | 10 µM |
| Y-27632 (ROCK inhibitor) | Tocris Bioscience | 1254 | 10 µM |
Cell culture and differentiation
PLB-985 cells were maintained in RPMI 1640 1X with l-glutamine and 25 mM HEPES (Corning 10-041-CV) supplemented with 10% heat-inactivated fetal bovine serum (FBS; HyClone, SH30071.03) and 1X penicillin–streptomycin (Corning; 30-002-CI) at 37°C, 5% CO2.
All PLB-985 cells were differentiated in RPMI 1640 complete media supplemented with 1.3% DMSO (Sigma-Aldrich; D2650) at a density of (3–4) × 105 cells/ml in 10 ml for 6 d at 37°C, 5% CO2. For centrinone experiments, cells were treated with either DMSO or 500 nM centrinone B (Tocris; 5690) in addition to normal differentiation media for the duration of differentiation. For blebbistatin (Sigma-Aldrich; B0560) experiments, cells were treated with DMSO or 20 µM blebbistatin for 4 days before differentiation media was replaced with drug-free media. Cells were harvested and all experiments were done on day 6.
HEK293T cells were maintained in DMEM 1X (Corning; 10-013-CV) supplemented with 10% heat-inactivated Fetal Clone III (Hyclone; SH30609.03) and 1X penicillin–streptomycin (Corning; 30-002-CI) at 37°C, 5% CO2.
Retrovirus production and transduction
For retrovirus production, 3 × 106 HEK293T cells were seeded and grown to 70% confluency overnight. Cells were transfected with pBABE-Puro-ManII-eGFP (kindly provided by Anjon Audhya, University of Wisconsin-Madison), pUMVC (Addgene; 8449), and VSV-G (Addgene; 8454) at a 10:10:1 µg ratio using TransIT-LT1 Transfection Reagent (Mirus; MIR2300) according to the manufacturer’s instructions. Cell supernatants were harvested after 72 h and centrifuged to remove cell debris. Virus was concentrated in the cleared supernatants using Retro-X Concentrator (Clontech; PT5063-2) according to the manufacturer’s instructions. Concentrated virus was resuspended in RPMI 1640 and stored at −80°C.
PLB-985 cells were transduced with pBABE-Puro-ManII-eGFP virus in RPMI complete media containing 8 µg/ml polybrene (Sigma-Aldrich; 107689) for 48 h. Cells were selected with 1 µg/ml puromycin (Sigma-Aldrich; P7255) for at least 72 h until all nontransduced control cells were dead.
CRISPR/Cas9-mediated genome editing
Guide RNAs (gRNAs) were designed against early exons of target genes using the Crispor tool (Haeussler et al., 2016; http://crispor.tefor.net/). The gRNAs with highest specificity and lowest off-target likelihood were chosen and synthesized by IDT as Alt-R synthetic single guide RNA (sgRNA). For generation of SAS-6 knockout cell lines, three individual gRNAs were simultaneously transfected. For GM130, two gRNAs targeting the same exon about 100 base pairs apart were simultaneously transfected.
| Gene | Exon | Sequence | Source |
|---|---|---|---|
| LUC1 | 6 | ACAACTTTACCGACCGCGCC | Hart, Chandrashekhar et al., 2015 |
| SAS6 | 1 | CCACCAACTAGTCCCGTTGC | McKinley and Cheeseman, 2017 |
| 1 | CCTGCAACGGGACTAGTTGG | McKinley and Cheeseman, 2017 | |
| 3 | TATTCGTCTGACTGATGACA | Designed with CRISPOR | |
| GM130 | 1 | CCATGGTCCTGCCATCGGAGTGG | Designed with CRISPOR |
| 1 | TCTTCCGACATCGCGGGGCGGGG | Designed with CRISPOR |
Ribonucleoprotein (RNP) complexes were made by incubating Alt-R sgRNA with Alt-R S.p. HiFi Cas9 Nuclease V3 (IDT; 1081060) at a 2:1 M ratio (240 pmol:120 pmol) for 20 min at room temperature (RT). Cells (2 × 106) were resuspended in 100 µl Nucleofector Solution V from the Cell Line Nucleofector Kit V (Lonza; VCA-1003). RNPs and 1 µM Alt-R Cas9 Electroporation Enhancer (IDT; 1075916) were added to the cells and then added to electroporation cuvettes. Cells were electroporated using program C-023 on the Nucleofector 2b device (Lonza; AAB-1001). After electroporation, 500 µl of prewarmed RPMI complete medium was added to the cuvettes and then transferred into 2 ml of prewarmed complete medium in a six-well plate using thin transfer pipettes. Cells recovered for 48 h before single-cell sorting clones at the UWCCC Flow Cytometry Core on a BD FACS AriaII BSL-2 Cell Sorter. The PLB-985 CRISPR Luciferase line was not single-cell sorted as it was used as a pooled control line. Cell lines were expanded and screened for target depletion via Western blot and/or immunofluorescence.
Microfluidics and live-cell imaging
Microfluidic devices were fabricated as previously described (Berthier et al., 2010; Cavnar et al., 2012). Gold Seal coverslips 48 × 60 mm (Thermo Scientific; 3334) and devices were plasma treated for 2 min and devices adhered to the coverslips. Microfluidic chambers were coated with 10 μg/ml fibronectin (Sigma-Aldrich; F1141) in 1X phosphate-buffered saline (PBS) for 30 min at 37°C. Fibronectin solution was removed and the devices were blocked with 5% bovine serum albumin (BSA) in 1X PBS for 30 min at 37°C and then washed twice with mHBSS (0.5% HSA, 20 mM HEPES). For inhibitor studies, devices were washed with mHBSS containing the final concentration of inhibitor.
Cells were stained with Calcein AM (Invitrogen; C3100MP; 1:1000) in 1X PBS for 10 min and then resuspended in mHBSS at a concentration of 3 × 106 cells/ml. Cells were loaded and allowed to adhere for 30 min at 37°C, 5% CO2. For inhibitor studies, cells were seeded in microfluidic devices in mHBSS containing inhibitor for 30 min. Cells were treated with the following inhibitor: nocodazole (10 µM). The gradient was generated by adding 0.5 μM fMLP (Sigma-Aldrich; 47729) to the source port and allowing it to set up and equilibrate for 30 min before imaging. Time-lapse imaging was performed on a Nikon Eclipse TE300 inverted epifluorescent microscope equipped with a CoolSNAP ES2 CCD camera (Teledyne Photometrics), motorized stage (Ludl Electronic Products), and light guide. Images were captured using a 10×, NA 0.45 ∞/0.17 objective every 30 s for 45 min in MetaMorph imaging software v7.8 (Molecular Devices).
For Golgi visualization, PLB-985 ManII-eGFP cells were stained with CellMask Orange (Invitrogen; C10045; 1:1000) in 1X PBS for 10 min and resuspended in mHBSS. Cells were loaded into devices prepared as above with one exception: µ-Slide two-well chambered coverslips (Ibidi; 80286) were used instead of 48 × 60-mm coverslips. Cells were imaged on a Zeiss inverted spinning-disk confocal microscope equipped with a motorized stage, Evolve camera (Photometrics), and a Pecon Incubator PS1 stage-top incubator. Images were taken with a 40× air, NA 0.75 ∞/0.17 objective every 15–30 s at 37°C for 10 min using ZenBlue software (Zeiss).
For microtubule imaging, cells were stained with 1 µM SiR-Tubulin (Spirochrome-Cytoskeleton; CY-SC002) in 1X PBS for 1 h. Cells were prepared as above using the µ-Slide two-well chambered coverslip setup. Imaging was performed on an Opterra Sweptfield Laser Scanning Confocal microscope (Bruker) equipped with a motorized stage and EMCCD camera–based detector. Images were acquired using a Nikon Plan Apo VC 60×/1.40 oil ∞/0.17 DIC N2 objective combined with a 60-µm pinhole aperture plate and 488 and 640 nm lasers. Full cell volumes were taken at the fastest acquisition speed for 15–30 min at RT using Prairie View software (Bruker).
Cell-tracking analysis
Cell-tracking analysis was done in Imaris v9.5 (Bitplane) using the Spots function. Prior to analysis, exclusion criteria were applied to prevent improper analysis: movies in which cells drifted due to no attachment, movies in which a majority of the cells appeared dead or unhealthy, or movies in which the device did not adhere properly resulting in an incorrect gradient.
Spots were thresholded at 12 µm and the histogram was adjusted to include only single cells. Tracks were generated using the autoregressive motion algorithm with a maximum distance value of 20 µm and five frames. Tracks were thresholded to only include those ≥100 µm with a track time of at least 480 s to exclude dead and/or stationary cells. The means of all individual tracks were calculated and used for statistical analysis. Imaris-generated cell tracks were imported into MATLAB for the calculation of chemotactic index (CI) using previously described methods (Kim and Haynes, 2013). Briefly, for each cell track, the net displacement in the direction of the gradient was divided by the total migration distance.
Western blotting
Cell pellets were collected and lysed in 1X RIPA buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) with 1X Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific; 78840) for 30 min on ice. Lysates were sonicated for three cycles at 20% amplitude (5 s on, 10 s off) and cleared by centrifugation at 15,000 × g, 4°C for 15 min. Protein concentrations were determined using the Pierce BCA Protein Assay (Thermo Scientific; 23225) and samples stored at −80°C. Protein (20–30 µg) was heated in 4X Bolt LDS Sample Buffer (Invitrogen; B0007) with 10X Bolt Sample Reducing Agent (Invitrogen; B0004) for 10 min at 70°C. SeeBlue Plus2 Pre-stained Protein Standard (Invitrogen; LC5925) and samples were loaded into precast Bolt 4–12% Bis-Tris Plus Gels (Invitrogen) and run for 35 min at 200 V in 1X Bolt MES SDS Running Buffer (Invitrogen; B0002). Proteins were transferred onto nitrocellulose membranes for 60 min at 90 V. Membranes were stained with Revert 700 Total Protein Stain Kit (Licor; 926-11010) according to the manufacturer’s instructions, scanned and destained, and then blocked with 5% milk in TBS-0.1% Tween 20 (TBS-T) for 1 h at RT. Membranes were washed and inverted onto primary antibody in 5% milk TBS-T overnight at 4°C. After washing, the membranes were incubated in secondary antibody for 60 min at RT. Membranes were washed and scanned on an Odyssey scanner (LICOR) at 700 and 800 nm wavelengths. Images were analyzed in Image Studio software (LICOR).
Immunofluorescence and confocal microscopy
Acid-washed 22-mm glass circle coverslips were coated with 10 µg/ml fibronectin for at least 1 h at 37°C and then blocked for 30 min with 5% BSA-PBS. Coverslips were washed twice with mHBSS. Cells (0.8 × 106) in 500 µl were seeded per coverslip in a 12-well plate (one coverslip per well) and allowed to adhere for 30 min. A 2X solution of fMLP-mHBSS was added for a final concentration of 100 nM fMLP and cells allowed to migrate for 5 min at 37°C, 5% CO2. Media was aspirated and fixation was performed as follows: 1 ml of 37°C preheated 0.1% glutaraldehyde (Sigma-Aldrich, G6257) in cytoskeletal buffer (10 mM MES, pH 6.1, 150 mM NaCl, 5 mM EGTA, pH 8.0, 5 mM MgCl2, 5 mM glucose) with 0.25% Triton X-100 was added for a 45-s extraction step. This was aspirated and 0.5% glutaraldehyde–0.25% Triton X-100 in cytoskeletal buffer was added for 10 min at 37°C. Coverslips were washed three times with PBS, blocked in 10% FBS-PBS for 60 min at RT, washed, and incubated in primary antibody solution overnight at 4°C. Coverslips were washed and incubated in secondary antibody solution for 60 min at RT. Autofluorescence from glutaraldehyde fixation was quenched for 2 min with the TrueVIEW Autofluorescence Quenching Kit (Vector Laboratories; SP-8400-15) according to the manufacturer’s instructions. Coverslips were washed, counterstained with DAPI (Invitrogen; D1306; IF 10 µg/ml) for 5–10 min, washed with ddH2O, and mounted on Rite-On Frosted Slides (Fisher Scientific; 3050-002) with ProLong Gold Antifade Mountant (Invitrogen; P36930).
For Golgi staining, undifferentiated PLB-985 cell lines were cytospun onto acid-washed 22-mm glass circle coverslips with a Shandon Cytospin 3 centrifuge (Thermo Scientific). Cells were fixed and permeabilized with 4% paraformaldehyde (Electron Microscopy Sciences; 15710) + 0.25% Triton X-100 in cytoskeletal buffer for 10 min at 37°C and processed as described above for staining.
For centriole quantification, cells were cytospun onto coverslips and then fixed in ice-cold 100% methanol for 10 min at −20°C. Coverslips were washed and stained as described above. Centrin puncta were counted in FIJI. Cells with less than two centrin puncta were considered centriole-depleted.
All immunofluorescence slides were imaged on an upright Zeiss LSM 800 Laser Scanning Confocal Microscope equipped with a motorized stage and Airyscan module. Images were acquired with a 60× oil, NA 1.40 objective using the Airyscan acquisition mode. Images were processed in ZenBlue software (Zeiss) using the Airyscan processing method and maximum intensity projections generated for further analysis.
Polyploidy analysis
Cells were fixed by adding 70% EtOH dropwise and resting at 4°C overnight. Cells were centrifuged for 10 min at 500 × g and then rehydrated in 1X PBS for 1 min before repeating centrifugation. Cells were stained overnight in 50 µg/ml propidium iodide (Biotium; 40017) in 1X PBS at 4°C. Cells were washed, resuspended in 1X PBS, and data were collected with an Attune NxT Flow Cytometer (Invitrogen) using Attune NxT software (Invitrogen). Data were analyzed with FlowJo v10 (BD).
Statistical analysis
Graphs were made with GraphPad Prism 8 (GraphPad Software) and statistical analyses were performed in either GraphPad Prism 8 or Stata 16 SE (StataCorp). For cell-tracking experiments, significance was determined by mixed-effects REML regression with either a Kenward-Rogers or Satterthwaite degrees of freedom approximation. Significance for SAS6 centriole depletion was determined by one-way ANOVA with a post hoc Dunnett’s test for multiple comparisons. Significance was denoted according to the following: n.s. = not significant (p > 0.05); *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
Supplementary Material
Acknowledgments
This work was supported by funding from National Institutes of Health (NIH) Grant no. AI-134749 to A.H., NIH Grant no. F30 CA-203271 to R.D., NIH Grant no. R01 CA-234904 to M.B., Molecular and Cellular Pharmacology Training Grant no. T32 GM 8688-17 to L.C.K., and University of Wisconsin Carbone Cancer Center Support Grant no. P30 CA014520. We would like to thank members of the Huttenlocher lab for providing insightful discussion. We would like to thank Michael Lasarev for his expert assistance in performing statistical analysis.
Abbreviations used:
- AKAP450
A-kinase anchoring protein 450
- Arp2/3
actin-related protein 2,3 complex
- DMSO
dimethyl sulfoxide
- fMLP
f-Met-Leu-Phe
- MTOC
microtubule-organizing center
- Rac1
Rac family small GTPase 1
- SAS6
SAS6 centriolar assembly protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-02-0060) on June 30, 2021.
REFERENCES
- Berthier E, Surfus J, Verbsky J, Huttenlocher A, Beebe D (2010). An arrayed high-content chemotaxis assay for patient diagnosis. Integr Biol (Camb) 2, 630–638. [DOI] [PubMed] [Google Scholar]
- Borregaard N (2010). Neutrophils, from marrow to microbes. Immunity 33, 657–670. [DOI] [PubMed] [Google Scholar]
- Cavnar PJ, Mogen K, Berthier E, Beebe DJ, Huttenlocher A (2012). The actin regulatory protein HS1 interacts with Arp2/3 and mediates efficient neutrophil chemotaxis. J Biol Chem 287, 25466–25477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabin-Brion K, Marceiller J, Perez F, Settegrana C, Drechou A, Durand G, Pous C (2001). The Golgi complex is a microtubule-organizing organelle. Mol Biol Cell 12, 2047–2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HW, Hsiao CT, Chen YQ, Huang CM, Chan SI, Chiou A, Kuo JC (2019). Centrosome guides spatial activation of Rac to control cell polarization and directed cell migration. Life Sci Alliance 2, e201800135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Oliveira S, Rosowski EE, Huttenlocher A (2016). Neutrophil migration in infection and wound repair: going forward in reverse. Nat Rev Immunol 16, 378–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Euteneuer U, Schliwa M (1985). Evidence for an involvement of actin in the positioning and motility of centrosomes. J Cell Biol 101, 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavilan MP, Gandolfo P, Balestra FR, Arias F, Bornens M, Rios RM (2018). The dual role of the centrosome in organizing the microtubule network in interphase. EMBO Rep 19, e45942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Picone R, Burute M, Dagher R, Su Y, Leung CT, Polyak K, Brugge JS, Théry M, Pellman D (2014). Oncogene-like induction of cellular invasion from centrosome amplification. Nature 510, 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeussler M, Schonig K, Eckert H, Eschstruth A, Mianne J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. (2016). Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol 17, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, Mis M, Zimmermann M, Fradet-Turcotte A, Sun S, et al. (2015). High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 163, 1515–1526. [DOI] [PubMed] [Google Scholar]
- Hauert AB, Martinelli S, Marone C, Niggli V (2002). Differentiated HL-60 cells are a valid model system for the analysis of human neutrophil migration and chemotaxis. Int J Biochem Cell Biol 34, 838–854. [DOI] [PubMed] [Google Scholar]
- Kazazian K, Go C, Wu H, Brashavitskaya O, Xu R, Dennis JW, Gingras AC, Swallow CJ (2017). Plk4 promotes cancer invasion and metastasis through Arp2/3 complex regulation of the actin cytoskeleton. Cancer Res 77, 434–447. [DOI] [PubMed] [Google Scholar]
- Keller D, Orpinell M, Olivier N, Wachsmuth M, Mahen R, Wyss R, Hachet V, Ellenberg J, Manley S, Gonczy P (2014). Mechanisms of HsSAS-6 assembly promoting centriole formation in human cells. J Cell Biol 204, 697–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Haynes CL (2013). On-chip evaluation of neutrophil activation and neutrophil-endothelial cell interaction during neutrophil chemotaxis. Anal Chem 85, 10787–10796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonce MP, Cloney RA, Berns MW (1984). Laser irradiation of centrosomes in newt eosinophils: evidence of centriole role in motility. J Cell Biol 98, 1999–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazaki Y, Nishimura Y, Sabe H (2012). GBF1 bears a novel phosphatidylinositol-phosphate binding module, BP3K, to link PI3Kγ activity with Arf1 activation involved in GPCR-mediated neutrophil chemotaxis and superoxide production. Mol Biol Cell 23, 2457–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley KL, Cheeseman IM (2017). Large-Scale Analysis of CRISPR/Cas9 Cell-Cycle Knockouts Reveals the Diversity of p53-Dependent Responses to Cell-Cycle Defects. Dev Cell 40, 405-420.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V (2003). Microtubule-disruption-induced and chemotactic-peptide-induced migration of human neutrophils: implications for differential sets of signalling pathways. J Cell Sci 116, 813–822. [DOI] [PubMed] [Google Scholar]
- Olins AL, Olins DE (2005). The mechanism of granulocyte nuclear shape determination: possible involvement of the centrosome. Eur J Cell Biol 84, 181–188. [DOI] [PubMed] [Google Scholar]
- Pedruzzi E, Fay M, Elbim C, Gaudry M, Gougerot-Pocidalo MA (2002). Differentiation of PLB-985 myeloid cells into mature neutrophils, shown by degranulation of terminally differentiated compartments in response to N-formyl peptide and priming of superoxide anion production by granulocyte-macrophage colony-stimulating factor. Br J Haematol 117, 719–726. [DOI] [PubMed] [Google Scholar]
- Press MF, Xie B, Davenport S, Zhou Y, Guzman R, Nolan GP, O’Brien N, Palazzolo M, Mak TW, Brugge JS, Slamon DJ (2019). Role for polo-like kinase 4 in mediation of cytokinesis. Proc Natl Acad Sci USA 116, 11309–11318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C, Yuan Q, Jian X, Randazzo PA, Tang W, Wu D (2020). Small GTPase ARF6 is a coincidence-detection code for RPH3A polarization in neutrophil polarization. J Immunol 204, 1012–1021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivero S, Cardenas J, Bornens M, Rios RM (2009). Microtubule nucleation at the cis-side of the Golgi apparatus requires AKAP450 and GM130. EMBO J 28, 1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosario CO, Ko MA, Haffani YZ, Gladdy RA, Paderova J, Pollett A, Squire JA, Dennis JW, Swallow CJ (2010). Plk4 is required for cytokinesis and maintenance of chromosomal stability. Proc Natl Acad Sci USA 107, 6888–6893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AD, Feldman JL (2017). Microtubule-organizing centers: from the centrosome to non-centrosomal sites. Curr Opin Cell Biol 44, 93–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliwa M, Pryzwansky KB, Euteneuer U (1982). Centrosome splitting in neutrophils: an unusual phenomenon related to cell activation and motility. Cell 31, 705–717. [DOI] [PubMed] [Google Scholar]
- Schmidt PH, Dransfield DT, Claudio JO, Hawley RG, Trotter KW, Milgram SL, Goldenring JR (1999). AKAP350, a multiply spliced protein kinase A-anchoring protein associated with centrosomes. J Biol Chem 274, 3055–3066. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Shibata H, Shimakawa M, Miyamoto M, Mukai H, Ono Y (1999). Characterization of a novel giant scaffolding protein, CG-NAP, that anchors multiple signaling enzymes to centrosome and the Golgi apparatus. J Biol Chem 274, 17267–17274. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Yamagiwa A, Nishimura T, Mukai H, Ono Y (2002). Centrosomal proteins CG-NAP and kendrin provide microtubule nucleation sites by anchoring gamma-tubulin ring complex. Mol Biol Cell 13, 3235–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang XM, Clermont Y (1989). Granule formation and polarity of the Golgi apparatus in neutrophil granulocytes of the rat. Anat Rec 223, 128–138. [DOI] [PubMed] [Google Scholar]
- Tormanen K, Ton C, Waring BM, Wang K, Sutterlin C (2019). Function of Golgi-centrosome proximity in RPE-1 cells. PLoS One 14, e0215215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker KA, Lilly MB, Heck L Jr, Rado TA (1987). Characterization of a new human diploid myeloid leukemia cell line (PLB-985) with granulocytic and monocytic differentiating capacity. Blood 70, 372–378. [PubMed] [Google Scholar]
- Vorotnikov AV, Tyurin-Kuzmin PA (2014). Chemotactic signaling in mesenchymal cells compared to amoeboid cells. Genes Dis 1, 162–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakida NM, Botvinick EL, Lin J, Berns MW (2010). An intact centrosome is required for the maintenance of polarization during directional cell migration. PLoS One 5, e15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witczak O, Skalhegg BS, Keryer G, Bornens M, Tasken K, Jahnsen T, Orstavik S (1999). Cloning and characterization of a cDNA encoding an A-kinase anchoring protein located in the centrosome, AKAP450. EMBO J 18, 1858–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong YL, Anzola JV, Davis RL, Yoon M, Motamedi A, Kroll A, Seo CP, Hsia JE, Kim SK, Mitchell JW, et al. (2015). Cell biology. Reversible centriole depletion with an inhibitor of Polo-like kinase 4. Science 348, 1155–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, de Heus C, Liu Q, Bouchet BP, Noordstra I, Jiang K, Hua S, Martin M, Yang C, Grigoriev I, et al. (2016). Molecular pathway of microtubule organization at the Golgi apparatus. Dev Cell 39, 44–60. [DOI] [PubMed] [Google Scholar]
- Xu J, Van Keymeulen A, Wakida NM, Carlton P, Berns MW, Bourne HR (2007). Polarity reveals intrinsic cell chirality. Proc Natl Acad Sci USA 104, 9296–9300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wang F, Van Keymeulen A, Rentel M, Bourne HR (2005). Neutrophil microtubules suppress polarity and enhance directional migration. Proc Natl Acad Sci USA 102, 6884–6889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SK, Lam PY, Eichelberg MR, Zasadil L, Bement WM, Huttenlocher A (2012). The role of microtubules in neutrophil polarity and migration in live zebrafish. J Cell Sci 125, 5702–5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshiba S, Tsuchiya Y, Ohta M, Gupta A, Shiratsuchi G, Nozaki Y, Ashikawa T, Fujiwara T, Natsume T, Kanemaki MT, Kitagawa D (2019). HsSAS-6-dependent cartwheel assembly ensures stabilization of centriole intermediates. J Cell Sci 132, jcs217521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.