Abstract
Lipid droplets (LDs) are dynamic organelles for lipid storage and homeostasis. Cells respond to metabolic changes by regulating the spatial distribution of LDs and enzymes required for LD growth and turnover. The small size of LDs precludes the observation of their associated enzyme densities and dynamics with conventional fluorescence microscopy. Here we employ quantitative photo-activated localization microscopy to study the density of the fatty acid (FA) activating enzyme Faa4 on LDs in live yeast cells with single-molecule sensitivity and 30 nm resolution. During the log phase LDs colocalize with the endoplasmic reticulum (ER) where their emergence and expansion are mediated by the highest observed Faa4 densities. During transition to the stationary phase, LDs with a ∼2-fold increased surface area translocate to the vacuolar surface and lumen and exhibit a ∼2.5-fold increase in Faa4 density. The increased Faa4 density on LDs further suggests its role in LD expansion, is caused by its ∼5-fold increased expression level, and is specific to exogenous FA chain-lengths. When lipolysis is induced by refreshed medium, Faa4 shuttles through ER- and lipophagy to the vacuole, where it may activate FAs for membrane expansion and degrade Faa4 to reset its cellular abundance to levels in the log phase.
INTRODUCTION
Lipid droplets (LDs) are lipid and energy storage organelles of a cell consisting of a neutral lipid core surrounded by a phospholipid monolayer (Tauchi-Sato et al., 2002; Walther and Farese, 2012). Cells maintain their lipid homeostasis and adapt to energy needs by actively regulating the anabolism and catabolism of LDs through regulatory proteins and enzymes (Walther and Farese, 2012; Olzmann and Carvalho, 2019). Fatty acids (FAs) required for the synthesis of neutral lipids such as triacylglycerols (TAGs) as well as FAs released from the breakdown of TAGs by lipases need to be converted into acyl-CoAs by fatty acid activating enzymes (Faergeman et al., 2001). The transition of cells to growth arrest during starvation and back to growth resumption results in major changes in the number, size, and subcellular location of LDs. These transitions are mediated by a change in the composition of regulatory proteins and enzymes on LDs (Kurat et al., 2006; Markgraf et al., 2014; Wang et al., 2014a). Conventional fluorescence microscopy yielded valuable insights into the processes of LD biogenesis, breakdown, and overall colocalization with regulatory enzymes (Kurat et al., 2006; Wang et al., 2016; Seo et al., 2017). However, since conventional fluorescence microscopy cannot resolve LDs and their subcellular location below the optical diffraction limit, little is known about the relation between the exact size of LDs and the density of LD-localized regulatory enzymes. Likewise, it is not known how this relation changes during metabolic shifts. Here we expand our recently developed live-cell single-molecule localization microscopy approach (Adhikari et al., 2019, 2020) by resolving LDs at different metabolic states in living yeast cells. For the first time, we gain quantitative insights into the relation between the size of LDs and the density of the endogenously tagged fatty acid activation enzyme Faa4 on LDs during metabolic transitions. We simultaneously monitor the subcellular localization and redistribution of LDs and Faa4, quantifying their mobility in different metabolic states.
LDs emerge from the ER and maintain a close association with the ER and other organelles for lipid exchange (Jacquier et al., 2011; Schuldiner and Bohnert, 2017). Most of the LD-resident proteins are targeted to LDs during their biogenesis in the ER membrane (Jacquier et al., 2011; Bersuker and Olzmann, 2017). In yeast, the fatty acid activation protein 4 (Faa4) is one of the fatty acyl CoA-synthetases responsible for import and activation of FAs for the synthesis of TAG (Faergeman et al., 2001; Black and DiRusso, 2007). Faa4 as well as the diacylglycerol acyltransferase Dga1, which produces TAGs, are localized to the ER membrane where LDs emerge (Jacquier et al., 2011) and to matured LDs (Kurat et al., 2006; Markgraf et al., 2014).
During nutrient deprivation when yeast cells reach a stationary growth phase, Dga1 and Faa4 dynamically relocalize from the ER to the surface of LDs. In addition, Faa4 exhibits increased expression during the stationary phase, which is also crucial for stationary phase survival (Ashrafi et al., 1998; Kurat et al., 2006). The increased amount of Dga1 and Faa4 on LDs causes an increase in local TAG synthesis on LDs and results in their expansion (Markgraf et al., 2014). This expansion of LDs is thought to occur since cells don’t need lipids for membrane expansion during the absence of their growth. In addition, LD-mediated buffering of FAs released during lipolysis reduces lipotoxicity of free FAs (Listenberger et al., 2003). While increased amounts of Faa4 have been observed with conventional fluorescence microscopy on enlarged LDs during the stationary phase, it is unknown if and how cells dynamically control the density of regulatory proteins between the ER and the LDs based on metabolic changes.
During the stationary growth phase, some LDs are degraded in the vacuole through microautophagy, commonly referred to as microlipophagy (van Zutphen et al., 2014; Wang et al., 2014a; Seo et al., 2017). The resulting FAs need to be reactivated by Faa4, which has been shown to enter the vacuole along with LDs during microlipophagy (van Zutphen et al., 2014; Wang et al., 2014a). At the same time, LDs at nuclear vacuolar junctions increase their TAG content using the activated FAs (Hariri et al., 2018, 2019). During growth resumption from the stationary phase, TAGs stored in LDs are rapidly broken down by LD-resident lipases to release and channel DAGs and FAs toward the ER and the vacuole for membrane proliferation (Kurat et al., 2006; Markgraf et al., 2014; Ouahoud et al., 2018; Ganesan et al., 2019). Again, Faa4 is needed to reactivate the released FAs for membrane synthesis.
Yeast cells (S. cerevisiae) can import and utilize exogenously supplied FAs for intracellular lipid storage as well as for the synthesis of various membrane lipids. Yeast cells contain four fatty acyl-CoA synthetases (Faa1, Faa2, Faa3, Faa4) and Fat1 for the import and the activation of fatty acids (Black and DiRusso, 2007). The catalytic action of Faa4/Faa1 is essential for the activation and the utilization of exogenously added long-chain (C12–C18) fatty acids, whereas Fat1 is essential for the activation of very long-chain (C22–C26) fatty acids (Watkins et al., 1998; Faergeman et al., 2001). LD proliferation as well as the overall redistribution of LD-associated proteins have been shown in earlier studies when growth medium is supplemented with external FAs like oleic acid (C18) (Jacquier et al., 2011). However, these studies lack the quantitative information of such redistribution of LD-associated enzymes like Faa4 as well as their density changes on LDs when exogenous fatty acids of various chain lengths are added.
Previous studies using conventional fluorescence microscopy lack the ability to accurately quantify the spatial distribution of Faa4 and the size of LDs below the optical diffraction limit of ∼ (250–300) nm. The accurate and simultaneous quantification of LD size, molecular composition, and mobility requires superresolution microscopy techniques such as photo-activated localization microscopy (PALM) (Betzig et al., 2006). In this study, we employ correlative PALM and conventional fluorescence microscopy to study, for the first time, the nanoscopic spatial distribution and the density of endogenously tagged Faa4 on the ER and LDs and inside the vacuole during transitions of living yeast cells to different metabolic states. As cells transition to the stationary phase, we find that LDs grow in size with increased Faa4 density on them and get immobilized at the vacuole. Some LDs along with their surface localized Faa4 are eventually taken up by the vacuole where they freely diffuse. When cells in the stationary phase are diluted back to fresh medium for 2 h, a few LDs localize again to the ER while Faa4 is predominantly localized to the vacuolar lumen, suggesting a mechanism for FA activation inside the vacuole and for degrading excess Faa4. When cells in the log phase are supplied with exogenous oleic acid (C18) that is preferentially activated by Faa4, the number of LDs per cell as well as the Faa4 density on them increase due to the relocalization of Faa4 from ER to LDs. In contrast, the addition of lignoceric acid (C24) does not result in a significant relocalization of Faa4 from the ER to LDs. The observed relocalization of Faa4 to LDs is therefore specific to the catalytic activity of Faa4 to activate imported oleic acid prior to their conversion to TAGs. Our approach to resolve and quantify the density of regulatory proteins on LDs in living cells may help in future studies to better understand how cells differently regulate their enzyme densities on LDs to maintain lipid and energy homeostasis based on metabolic needs.
RESULTS
Live-cell PALM resolves the subcellular localization and dynamics of Faa4 and LDs with ∼30 nm resolution
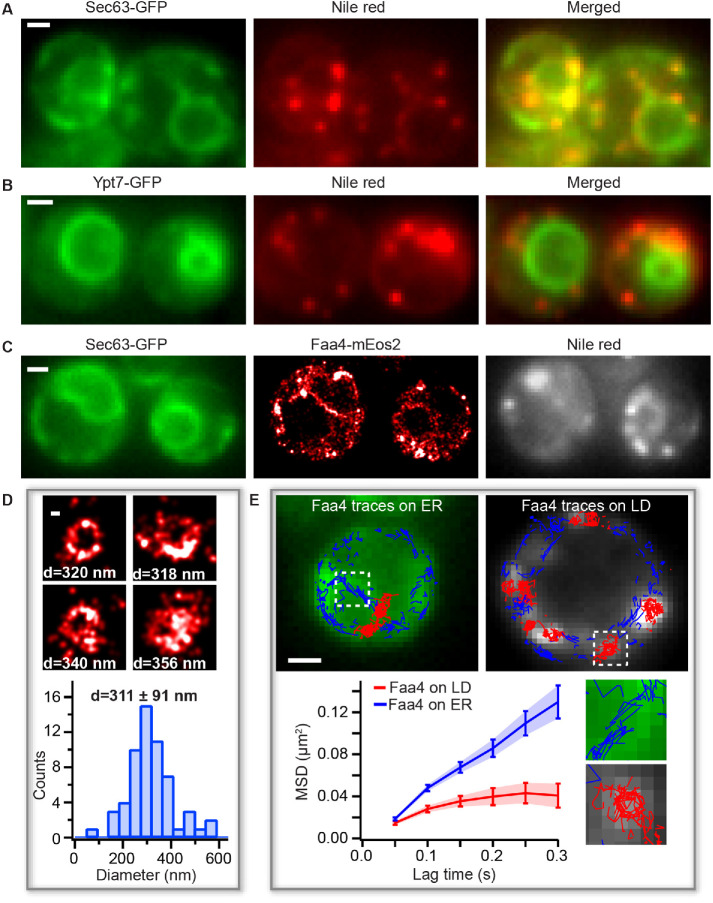
Yeast cells actively regulate the spatial distribution, number, and size of LDs based on their metabolic state (van Zutphen et al., 2014; Wang et al., 2014a). During the log phase, LDs emerge from and remain closely associated with the ER, where they take up neutral lipids for their expansion. The newly formed LDs are diffraction limited in size and most of the regulatory proteins are targeted to the surface of LDs during their biogenesis (Kory et al., 2016). Faa4 is one of the fatty acid activating proteins in yeast that has been shown to localize to the ER and to LDs (Natter et al., 2005; Kurat et al., 2006), where its catalytic activity is required to form neutral lipids, phospholipids, and protein myristoylation (Ashrafi et al., 1998; Black and DiRusso, 2007). While previous studies using conventional fluorescence microscopy yielded valuable information about the cellular distribution of Faa4, they lacked an accurate quantification of the subcellular localization of Faa4 and of the size of LDs below the optical diffraction limit. To overcome these limitations, we employed correlative live-cell PALM and conventional fluorescence microscopy. This approach allows us to study the nanoscopic localization and the single-molecule dynamics of Faa4 along with a simultaneous quantification of LD sizes and their motion in living yeast cells. First, we endogenously tagged Faa4 with the photo-switchable fluorescent protein mEos2 in a Saccharomyces cerevisiae strain (W303), which additionally expressed the ER marker Sec63-GFP for correlative conventional fluorescence and PALM imaging. mEos2 has been extensively used in the past for quantitative PALM studies of cellular organelles and protein clusters (McKinney et al., 2009; Lee et al., 2012; Puchner et al., 2013). We then recorded a repetitive sequence with one frame of conventional GFP fluorescence at low 488 nm excitation power in the green channel followed by nine frames with high 561 nm laser power and sparse 405 nm photo activation to detect the single-molecule fluorescence of Faa4-mEos2 in the red channel (Supplemental Movie S1). As expected, LDs in the log phase were localized to the ER but not to the vacuole in colocalization experiments with Sec63-GFP (ER marker) and Ypt7-GFP (vacuole marker) (Figure 1, A and B). Conventional fluorescence images of Sec63-GFP showed the expected outline of the ER around the nucleus and in proximity to the plasma membrane (Figure 1C, left). The reconstructed PALM images of single Faa4-mEos2 molecules revealed their localization to the ER as well as to LDs (Figure 1C, middle), which is consistent with previous studies (Natter et al., 2005; Kurat et al., 2006). The localization of Faa4 to LDs was further confirmed by staining LDs with Nile red (Greenspan et al., 1985) (Figure 1C, right). While conventional fluorescence images exhibit low resolution and out-of-focus fluorescence, the highly specific staining of LDs together with the overlapping Faa4 localizations that form dense superresolved clusters or donut shapes allow to identify LD-localized Faa4. Furthermore, the high ∼30 nm precision of individual localizations in PALM images estimated with the Thompson formula (Thompson et al., 2002) resolves the size of LDs with a diameter ranging from 100 to 600 nm (Figure 1D).
FIGURE 1:
PALM reveals LD sizes and Faa4 dynamics in living cells. (A) The conventional fluorescence images of Sec63-GFP (left), Nile red (middle), and merged (right) show that LDs are colocalized with the ER in the log phase. (B) The conventional fluorescence image of Ypt7-GFP (left), Nile red (middle), and merged (right) indicate that LDs are not localized to the vacuole. (C) Conventional image of Sec63-GFP (left), PALM image of Faa4-mEos2 (middle), and conventional image of LDs stained with Nile red (right) visualize Faa4-mEos2 localizations at the ER and in dense spots that colocalize with LDs. (D) Representative images of superresolved LDs and histogram of LD diameters with a mean of 311 ± 91 nm (N = 58 LDs from 25 cells). Error represents the SD of all LD sizes. (E) Single-molecule tracking of Faa4-mEos2 superimposed on Sec63-GFP (left) and Nile red (right) reveals traces on the ER (blue) and on LDs (red). The MSD of Faa4 traces on the ER (N = 3 cells,1270 traces longer than three frames) and on LDs (N = 8 LDs from three cells, 280 traces longer than three frames) exhibit free diffusion of Faa4 at shorter lag times but confinement on LDs at longer lag times. Scale bars: 1 μm; D: 100 nm.
Movie S1.
Faa4-mEos2 single molecule signal (left) under 561 nm excitation shows blinking mEos2 molecules. Sec63-GFP (right, average of 50 frames) under 488 nm excitation shows ER around the cell periphery and the nucleus. Scale bar: 1 μm.
To gain additional insights into the dynamics of single Faa4 enzymes, we quantified their motion in the ER and on LDs using single-molecule tracking (Manley et al., 2008). The single-molecule tracking of Faa4 revealed its motion along the ER membrane and on the surface of LDs (Figure 1E, top). While we observed a few Faa4-mEos2 traces (∼1%) moving from the ER to LDs, an accurate quantification of this flux is too challenging due to the low number and short duration of these traces. The calculated mean-squared displacement (MSD) curves showed that Faa4 freely diffuses on ER and on the surface of LDs, but exhibited confinement at longer lag times on LDs (Figure 1E, bottom). The mean diffusion coefficient of all Faa4 enzymes was determined to be D = 0.061 ± 0.014 μm2/s (Supplemental Figure S1C), which is comparable to other membrane-localized proteins (Wu et al., 2015). Since LDs in the log phase are immobile, these results show that Faa4 does not undergo any significant immobilization on LDs compared with the ER.
These results demonstrate that our correlative live-cell PALM approach reveals the subcellular localization and the dynamics of Faa4 at the nanoscopic length scale and allows us to quantify the size of LDs in living cells.
In the stationary phase, Faa4 and enlarged LDs redistribute to the vacuole
When yeast cells exhaust the nutrients in their medium, they enter the stationary growth phase which is characterized by cell cycle arrest along with LD expansion to buffer excess FAs (Wang et al., 2014a; Hariri et al., 2018). During the transition to the stationary phase, LDs have been shown to redistribute to vacuolar microdomains followed by entry into the vacuole in a process called microlipophagy (van Zutphen et al., 2014; Wang et al., 2014a; Seo et al., 2017).
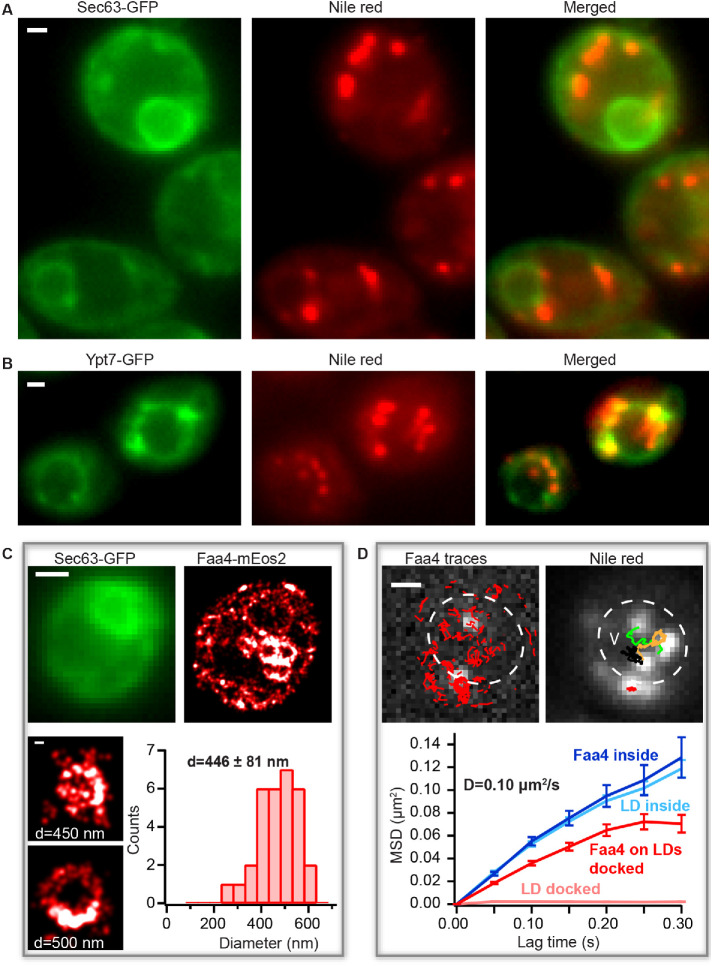
To obtain high-resolution insights in the distribution and dynamics of Faa4 and LDs during the transition to the stationary phase, we grew a yeast culture for ∼30 h without exchange of medium. Colocalization experiments of Nile red with Sec63-GFP and Ypt7-GFP confirmed that LDs redistributed from the ER to either the vacuolar surface or to the vacuolar lumen in these conditions (Figure 2, A and B). To measure the nanoscopic spatial distribution and the dynamics of Faa4 in this growth phase, we again performed live-cell PALM imaging of Faa4-mEos2. The superresolved images revealed that Faa4 is localized to bigger LDs in clusters (Figure 2C, top). Consistent with previous reports (Wang et al., 2014a), the quantified LD sizes exhibited an increased diameter (446 ± 81 nm) compared with the log phase (Figure 2C, bottom). In some cells, a fraction of Nile red-stained LDs already randomly diffused inside the vacuole (Figure 2D, top right, Supplemental Movie S2). Likewise, the single-molecule localizations and traces from Faa4-mEos2 were distributed throughout the vacuolar lumen (Figure 2D, top left). To test if Faa4 was still localized to LDs inside the vacuole, we quantified single-molecule tracking of Faa4-mEos2 and single-particle tracking (SPT) of LDs stained with Nile red (Figure 2D, left and right). The MSD of the traces from LDs and Faa4-mEos2 inside the vacuole revealed a similar diffusion coefficient of D = 0.10 µm2 /s (Figure 2D, bottom). Since freely diffusing Faa4 would have a diffusion coefficient larger than ∼2 orders of magnitude (Swaminathan et al., 1997) and would not be detectable with the used frame rates (Supplemental Figure S4A, right), these data indicate that Faa4 is still localized to LDs inside the vacuole.
FIGURE 2:
Faa4 redistribution and LD dynamics during the transition to the stationary phase. (A) The conventional fluorescence images of Sec63-GFP (left), Nile red (middle), and merged (right) show LDs not colocalized with the ER during the stationary phase. (B) Conventional fluorescence images of Ypt7-GFP (left), Nile red (middle), and merged (right) reveal LDs either docked to or inside the vacuole. (C) Sec63-GFP (top, left) visualizes the ER. PALM image of Faa4-mEos2 (top, right) reveals Faa4 localized to the ER and to LDs. Representative superresolved images (bottom) of LDs with an increased mean diameter of 446 ± 81 nm (N = 31 LDs from 10 cells). Error is calculated from the SD of all LD diameters. (D) The single-molecule traces (red) superimposed on single-molecule signals (white) of Faa4-mEos2 show confined diffusion along the surface of immobile LDs at the vacuole and free diffusion on diffusing LDs inside the vacuole (left). SPT of LDs with the conventional fluorescence signal from Nile red reveals immobile LDs at the vacuole and free diffusion of LDs inside (right). The MSD of Faa4-mEos2 (N = 140 traces from 3 LDs) and LDs (N = 425 traces from 3 LDs) exhibit a similar slope with D = 0.10 µm2/s inside the vacuole (bottom). LDs on the vacuole are immobile while the MSD of Faa4 indicates free diffusion with confinement at longer lag times on the surface of LDs as in the log phase (bottom). Scale bars: 1 μm; zoom in C: 100 nm.
Movie S2.
Faa4-mEos2 single molecule signal (left) under 561 nm excitation from cells grown to stationary phase. Nile red signal (right) under 488 nm excitation visualizes a fraction of LDs moving randomly inside vacuole and the other fraction immobile. Faa4-mEos2 signal is also visible from LDs moving randomly inside (top cell). Scale bar: 1 μm.
Since LDs transition from immobility on the vacuole to random diffusion inside the vacuole (Supplemental Movie S3), the average size measurement before and the diffusion after the vacuolar entry can be used to calculate the vacuolar viscosity. By using the Stokes-Einstein equation (Saks et al., 2008) with the average diameter of LDs before their entry into the vacuole (446 ± 81 nm, Figure 2C) as well as the average diffusion coefficient of D = 0.15 ± 0.084 μm2/s inside the vacuole (Supplemental Figure S2A), the vacuolar viscosity resulted in a value of 6.6 ± 3.9 cP, in agreement with previous studies (Puchkov, 2010). In summary, our correlative conventional fluorescence and PALM approach revealed an increase in LD size during transition to the stationary growth phase followed by an entry into the vacuole where LDs freely diffuse while Faa4 still localizes to their surface.
Movie S3.
BODIPY-NL signal under 488 nm excitation from a cell grown to stationary phase visualizes a transition of a LD from immobility to random diffusion after it enters the vacuole. Scale bar: 1 μm.
Quantification of live-cell PALM reveals a ∼2.5-fold density increase of Faa4 on LDs for their expansion during the stationary phase
During the transition to the stationary phase, yeast cells stop expanding their membranes for cell division and instead increase their TAG content in LDs of increasing size through local synthesis of TAG (Kurat et al., 2006; Hariri et al., 2018). Dga1 catalyzes the final step in the conversion of DAG to TAG with FAs activated by Faa4/Faa1. Consequently, Dga1 relocalizes from the ER to LDs where its activity is needed (Markgraf et al., 2014). During the stationary phase the fatty acid-activating enzyme Faa4 has increased expression levels and is also required for stationary phase survival (Ashrafi et al., 1998). When cells are diluted with fresh medium to resume growth (lag phase), TAGs stored in LDs are broken down through lipolysis to release DAG and FAs for the synthesis of other membrane lipids (Kurat et al., 2006; Ouahoud et al., 2018). The catalytic action of Faa4 is required to reactivate FAs released during lipolysis for TAG synthesis on LDs during the stationary phase and for membrane lipid synthesis during the lag phase. During both the stationary and the lag phase, the futile cycle of TAG synthesis and breakdown on LDs has to be broken for a net synthesis or the breakdown of TAGs. One way to break this futile cycle on LDs is a change in density of specific proteins required for TAG synthesis or the breakdown. Hence, we sought to investigate how the density of Faa4 on LDs changes during these metabolic transitions using quantitative PALM.
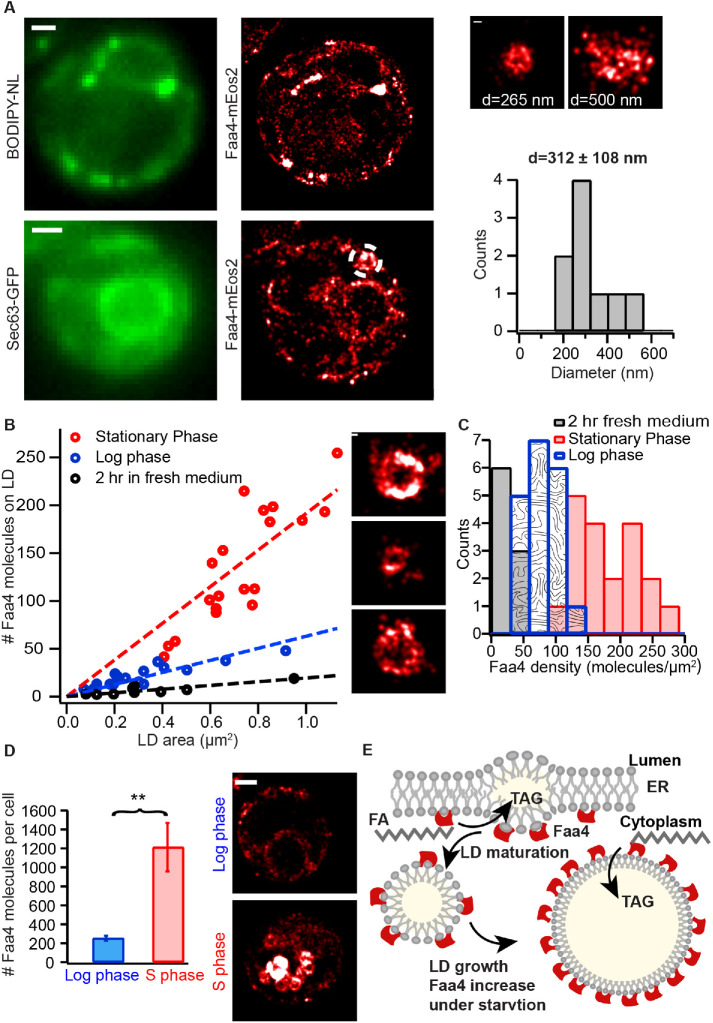
To establish the conditions of cells in the lag phase, we exchanged the medium of yeast cells in the stationary phase with fresh medium ∼2 h prior to imaging. The conventional fluorescence and PALM imaging of LDs and Faa4-mEos2 as well as Sec63-GFP showed a subcellular distribution of LDs and Faa4 that resembled cells in the log phase. LDs again localized to the ER with a mean diameter of ∼312 nm and single Faa4-mEos2 localizations were predominantly found along the ER and on the surface of LDs (Figure 3A). To measure the Faa4 density on LDs, we quantified the surface area and the number of Faa4-mEos2 molecules on individual LDs from the log, stationary, and lag phases using their superresolved images. Counting the number of molecules in protein complexes or organelles with PALM has been a major development in the field and requires methods to correct for blinking artifacts by combining fluorescent bursts emitted from the same fluorophore within a spatiotemporal threshold (Annibale et al., 2011; Puchner et al., 2013; Deschout et al., 2014). However, the application of these methods has so far been limited to fixed cells due to the potential motion of protein complexes or organelles during the data acquisition. Here we apply molecule counting to live-cell PALM with the following rationales: first, we selected multiple immobile LDs with varying numbers of mEos2 localizations at low 405 nm photo-activation power to avoid spatial overlap of multiple emitters (Supplemental Figure S3A). For each individual LDs, the number of apparent blink corrected molecules at various allowed dark times (tdark) was determined by grouping the localizations that appear on the LD within the allowed dark time. This results in a double exponential decrease in the number of apparent molecules at shorter dark times due to blinking followed by linear decrease at longer dark times due to false linking (Supplemental Figure S3B). Previous studies with mEos2 have also shown the double exponential distribution of dark times due to the presence of two dark states (Annibale et al., 2010; Lee et al., 2012). By fitting the number of apparent molecules versus dark time curve with the sum of a double exponential and linear function, we determine the optimal choice for the dark time that balances overcounting due to blinking and undercounting due to false linking (Supplemental Figure S3, B–D). Using this procedure with multiple LDs allows us to determine the average number of localizations from single mEos2 molecules (Supplemental Figure S3C) as in previous studies (Lee et al., 2012) and to convert detected localizations to molecules with an error of 17%. Although there may be an exchange of bleached and preactivated mEos2 on LDs with their surroundings, the employed widefield excitation and photo-switching results in the same fraction of fluorophores that are photo activated and bleached on LDs and their surroundings. Therefore, any bias in over- or underestimating the number of Faa4 molecules due to exchange cancels out. The high degree of correlation (ρ = 0.89) between the number of molecules and their localizations from multiple LDs (Supplemental Figure S3E) confirms the accuracy of this approach in quantifying protein numbers using detected localizations.
FIGURE 3:
Quantitative live-cell PALM reveals a ∼2.5-fold density increase of Faa4 on LDs for their expansion during the stationary phase. (A) In the lag phase single Faa4-mEos2 molecules are localized to LDs visualized with BODIPY-NL (top) and to the ER visualized with Sec63-GFP (bottom). The diameter of LDs with a mean of 312 ± 108 nm (N = 9 LDs from four cells) is similar to LDs in the log phase (right). (B) The quantified number of Faa4 molecules on individual LDs linearly correlates with their surface area for each growth phase. The larger slope of LDs in the stationary phase compared with log and lag phases indicates an increase in Faa4 density on LDs during the stationary phase. The insets show PALM images of LDs in the stationary (top), log (middle), and lag phase (bottom). (C) The histograms of Faa4 densities on LDs shows a roughly 2.5-fold increase of Faa4 density on LDs in the stationary phase compared with the log phase. (D) The total number of Faa4 molecules per cell in the stationary phase is roughly fivefold higher (p = 0.0033, t test) compared with the lag phase and explains the increased Faa4 density on LDs. Error bars are the SE of the mean calculated using N = 9 cells for each case. The representative superresolution images of Faa4-mEos2 exhibit dense localizations on LDs in the stationary phase and overall much less localizations of Faa4 on the ER and on few emerging LDs in the log phase. (E) A model for LD expansion mediated by a higher density of Faa4 on the LD surface. Scale bars: 1 μm; zooms: 100 nm. Asterisks indicate statistical significance from t test (**p ≤ 0.005).
We applied this quantitative PALM approach in each metabolic phase in order to determine the density of Faa4 on individual LDs. The number of Faa4 molecules depended linearly on the surface area of LDs for each growth phase (Figure 3B). Therefore, the density of Faa4 on LDs in each phase is roughly constant regardless of their surface area. However, the slope of Faa4 molecules versus LD surface area was different in different growth conditions as seen in the histograms of the respective Faa4 density distributions (Figure 3C). Log phase cells exhibited a tight distribution with a mean of 78.5 ± 21 Faa4 molecules/µm2, whereas cells in the stationary phase contained on average a ∼2.5-fold higher density of Faa4 (180 ± 50 molecules/µm2) and a ∼4-fold higher number of molecules per LD. The variability of the Faa4 density on LDs in the stationary phase was increased, which indicates a more heterogeneous population of LDs compared with the log phase. When cells in the stationary phase were diluted with fresh medium for ∼2 h and entered the lag phase, the Faa4 density on LDs decreased to values that were lower but similar to cells in the log phase. This observation is consistent with the overall distribution of LDs and Faa4 (Figure 3A) that resembles the one of log phase cells.
To test if the increased Faa4 density on LDs during the stationary phase is caused by an increased expression of Faa4 or by a regulated increase in its affinity to LDs, we measured the total number of Faa4 molecules per cell in the stationary and the log phase. Our results indeed show that stationary phase cells have a roughly fivefold higher number of Faa4 molecules per cell compared with log phase cells (Figure 3D). Taken together, our quantitative PALM approach reveals a ∼5-fold higher expression level of Faa4 during the stationary phase compared with the log phase, which is the predominant mechanism for the measured ∼2.5-fold increase in Faa4 density of LDs and its associated increase in catalytic activity for buffering FAs in expanding LDs (Figure 3E).
Subcellular localization and density of Faa4 during metabolic transitions is dynamically regulated among the ER, LDs, and the vacuole for its enzymatic activity
Having gained quantitative insights in the growth phase-dependent Faa4 density on LDs, we next quantified the distribution of Faa4 enzymes on the three major organelles where Faa4 is found: the ER, LDs, and the vacuole. While a lower pH of the vacuole could affect mEos2 photophysics inside the vacuole, previous studies found no significant change in cytoplasmic pH in log, stationary, and lag phases, which allows us to reliably compare Faa4 densities on ER and LDs (Cimprich et al., 1995). This measurement of Faa4 redistribution in different growth phases can shed light on where its catalytic activity is needed and how the high expression level in the stationary phase is reset to the default values in the log phase.
During the stationary phase, LDs expand at the vacuole through local synthesis of TAGs. Consequently, proteins like Dga1 and Faa4, which are required for TAG and SE synthesis, relocalize from the ER to LDs (Kurat et al., 2006; Markgraf et al., 2014) through the cytosol or ER-LD bridges (Kory et al., 2016). The change in the relative abundance of proteins on the ER and LDs has therefore been proposed as a regulatory strategy to control their local activity under different metabolic states (Natter et al., 2005). On growth resumption from the stationary phase, LDs are rapidly consumed through degradation of TAG into DAG and FAs to meet the lipid needs of growing cells (Kurat et al., 2006; Ouahoud et al., 2018). This degradation has been shown to require functional interaction of LDs with the vacuole. The reactivation of released FAs for membrane lipid synthesis is dependent on the enzymatic action of Faa4 (Kiegerl et al., 2019). Therefore, the redistribution of Faa4 among the ER, LDs, and the vacuole can be a strategy to direct its catalytic activity to organelles within a cell where it is needed in different metabolic states.
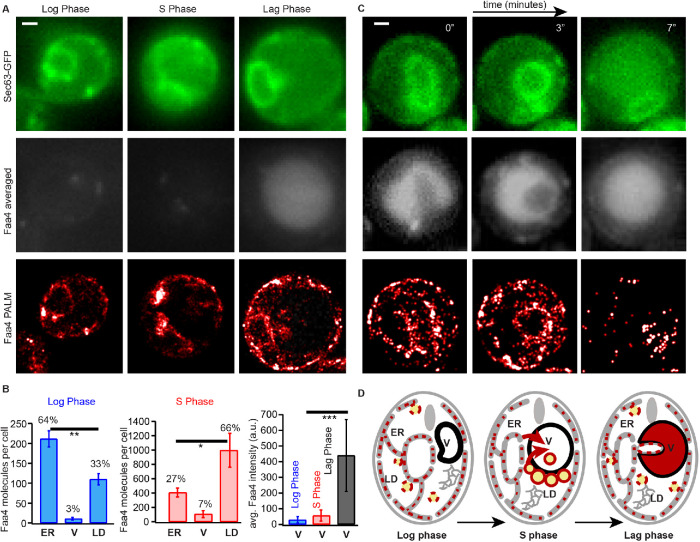
To gain insights in the redistribution of Faa4, we quantified the number and the percentage of Faa4 molecules per cell that are localized to the ER, LDs, and the vacuole in the log, stationary, and lag phase. Overall, Faa4 was predominately localized to the ER (64%) in the log phase but to LDs (66%) in the stationary phase (Figure 4, A and B). This redistribution was accompanied by a dramatic 10-fold increase in the number of Faa4 on LDs compared with the log phase (Figure 4, A and B). The number of Faa4 molecules on LDs in the lag phase was roughly twofold lower compared with the log phase and roughly 15-fold lower compared with the stationary phase (Supplemental Figure S4D). In the stationary phase, the number and the percentage of Faa4 localizations in the vacuole were slightly increased compared with the log phase. However, when stationary phase cells were diluted with fresh medium for ∼2 h (lag phase), the Faa4-mEos2 signal showed an intense and diffuse fluorescence throughout the vacuole, which was identified by its dark appearance in transmitted light images (Supplemental Figure S4, A–C). This diffuse signal is likely due to the fast diffusion of Faa4 molecules in the vacuole and prevents the detection of single-molecule localizations. In addition, vacuolar Faa4-mEos2 is exposed to a more acidic pH compared with LD- and ER-localized Faa4, which might influence the photophysics of mEos2 and prevent an absolute comparison of molecule numbers. We therefore quantified the average conventional fluorescence intensity of mEos2 and obtained a relative comparison of vacuolar Faa4 abundance in the different growth phases (Figure 4B, right). These data indicate a strong accumulation of fast-diffusing Faa4-mEos2 inside the vacuole where rarely LDs were detected (Figure 4A, right, and Supplemental Figure S4C).
FIGURE 4:
Subcellular Faa4 distribution on the ER, LDs, and the vacuole is coupled to the growth phase. (A) Sec63-GFP fluorescence in the log (top, left), stationary (top, middle) and lag phase (top, right) visualizes the ER. The averaged Faa4-mEos2 signal in the log (middle, left), stationary (middle, middle), and lag (middle, right) phases reveals an intense and diffuse Faa4 signal from the circular vacuolar region only in the lag phase. Faa4-mEos2 is similarly localized to the ER and the LDs in the log and lag phase (bottom) but mostly forms dense coats on LDs in the stationary phase. (B) The quantification of the absolute number and the percentage of Faa4 molecules that colocalized with ER, vacuole and LDs in the log (N = 14 cells) and stationary phase (N = 7 cells). Error bars represent the SE of the mean. The quantification of the average intensity counts per pixel inside the vacuole reveal that Faa4 is predominately localized to the vacuole after 2 h of dilution in fresh medium (lag phase) (N = 5 cells). (C) Time lapse shows how a portion of the Sec63-GFP signal is progressively taken inside the averaged Faa4-mEos2 signal, which indicates that a part of the ER is taken inside vacuole after 30 min of dilution in fresh medium. (D) Schematics showing localization and redistribution of Faa4 among the ER, LDs, and the vacuole in the log, stationary and lag phase. Scale bars: 1 μm. Asterisks indicate statistical significance from t test (*p ≤ 0.05, **p ≤ 0.005, ***p ≤ 0.0005).
The accumulation of Faa4 in the vacuole during the lag phase is likely the mechanism by which cells down-regulate their high Faa4 expression level of the stationary phase to the values observed in the lag phase (Figure 3D and Supplemental Figure S4D). So far, our data demonstrate entry of Faa4 into the vacuole at high densities on LDs undergoing lipophagy (Figure 2D). However, other routes may exist; recently, piecemeal microER-phagy has been shown to maintain proper ER size and protein homeostasis during ER stress recovery in an ESCRT-dependent manner (Loi et al., 2019; Schäfer et al., 2019). To test if ER-phagy contributes to the Faa4 relocalization to the vacuole on growth resumption, we imaged the Faa4 distribution ∼30 min after stationary phase cells were diluted in fresh medium. Indeed, we found a significant number of cells that were in the process of or already had taken up parts of the ER into the vacuole, as confirmed by the Sec63-GFP signal (Figure 4C; Supplemental Figure S4E; Supplemental Movie S4). Since single-molecule localizations of Faa4-mEos2 were detected on the ER inside the vacuole, our data suggest that ER-phagy contributes to Faa4 relocalization into the vacuole on growth resumption.
Movie S4.
Time lapse of averaged Faa4-mEos2 signal (left) and averaged Sec63-GFP signal (right) imaged after adding fresh media for 30 minutes to stationary phase cells. A part of ER (Sec63-GFP) is progressively internalized inside vacuole (Faa4-mEos2) revealing ER-Phagy. Scale bar: 1 μm
Subcellular redistribution of Faa4 to LDs is dependent on the carbon chain length of exogenously added fatty acids
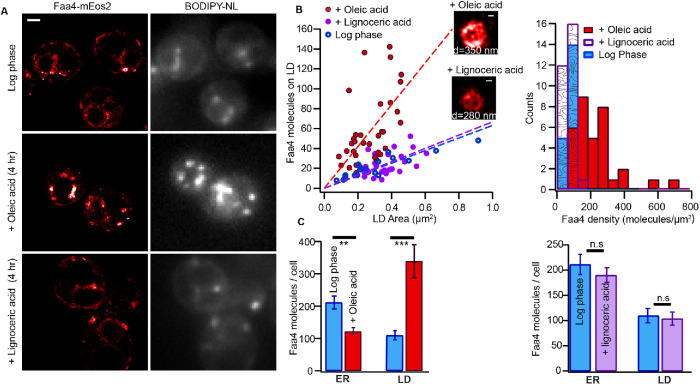
Exogenous fatty acids have to be activated to acyl-CoAs by fatty acyl-CoA synthetases prior to their metabolic utilization. In yeast, Faa4 preferentially activates long-chain fatty acids (C12:C18), whereas Fat1 is responsible for the activation of very long-chain fatty acids (C22:C26) (Faergeman et al., 2001; Johnson et al., 1994; Watkins et al., 1998). Since Faa4 preferentially activates fatty acids of length C12–C18 for their conversion to neutral lipids and storage in LDs, we investigated how the subcellular distribution and density of Faa4 on LDs are affected by exogenously added oleic acid (C18) and lignoceric acid (C24). When yeast cells in the log phase were incubated with 3.5 mM oleic acid for 4 h, we observed a significantly higher number of LDs per cell that often appeared in cluster compared with log phase cells without added oleic acid (Supplemental Figure S6A). In contrast, the number of LDs per cell and their intracellular distribution in the presence of 3.5 mM lignoceric acid resembled that of cells in the log phase (Supplemental Figure S6A). Faa4-mEos2 PALM images revealed that Faa4 localized to the ER and LDs in log phase cells in the presence and absence of lignoceric acid but densely accumulated on LDs in the presence of oleic acid (Figure 5A). The calculated size distribution of individual LDs in the presence of oleic acid (d = 297 ± 65 nm) was similar to one in the presence of lignoceric acid (d = 333 ± 48 nm) (Supplemental Figure S6B). However, the slope of the number of molecules on individual LDs versus area revealed a roughly threefold higher density of Faa4 on LDs in the presence of oleic acid (Figure 5B, left). The higher density of Faa4 in the presence of oleic acid is also apparent in the Faa4 density histograms with a mean density of 229 molecules/µm2 for oleic acid and 70 molecules/µm2 for lignoceric acid-treated cells (Figure 5B, right). In contrast, the Faa4 density distribution on LDs in the presence of lignoceric was similar to cells in log phase.
FIGURE 5:
The subcellular distribution of Faa4 and density on LDs is dependent on the carbon chain length of exogenously added fatty acids. (A) Faa4-mEos2 PALM and BODIPY-NL image of yeast cells grown to log phase (top). Faa4 is localized to the ER and to a few LDs as confirmed by the BODIPY-NL signal. In the presence of oleic acid (C18), a higher number of LDs per cell is observed and Faa4 predominately localizes to LDs (middle). The Faa4 and LD distribution in the presence of lignoceric acid (C24) resembles that of the log phase (bottom). (B) Area of individual LDs vs. the number of Faa4 molecules on LDs surface from cells in the log phase and in the presence of oleic acid and lignoceric acid (left). Histogram of surface densities of Faa4 on individual LDs (right). (C) Quantification of the number of Faa4 molecules per cell localized to the ER and LDs in the log phase (N = 14 cells) and in the presence of oleic acid (left) (N = 20 cells) and in the presence of lignoceric acid (right) (N = 17 cells). Error bars represent the SE of the mean. Scale bar: 1 μm; zooms: 100 nm. Asterisks indicate statistical significance from t test (**p ≤ 0.005, ***p ≤ 0.0005).
To test if the increased density of Faa4 on LDs in the presence of oleic is caused by subcellular redistribution of Faa4, we next quantified the number of Faa4 molecules localized to the ER and LDs. In the presence of oleic acid, the majority of Faa4 molecules localized to LDs, whereas in the absence of oleic acid, the majority of Faa4 localized to the ER (Figure 5C, left). The number of Faa4 molecules on LD increased roughly 3.5-fold in the presence of oleic acid (Figure 5C, left). This increased Faa4 density on LDs is also partly explained by the increase in overall Faa4 expression in the presence of oleic acid (Supplemental Figure S6C). In contrast, there was no significant difference in the subcellular distribution of Faa4 molecules between the cells in the log in the presence and absence of lignoceric acid but a slight increase in overall Faa4 expression level in the presence of lignoceric acid (Figure 5C, right, and Supplemental Figure S6C). These results demonstrate that the increased expression of Faa4 and its redistribution to LDs are specific to the chain length of exogenously added fatty acids and correlates with the catalytic activity of Faa4.
DISCUSSION
LDs are increasingly recognized as actively regulated organelles that store neutral lipids (TAGs and SEs) in their hydrophobic core (Romanauska and Köhler, 2018; Mejhert et al., 2020). Especially during metabolic transitions of cells, the enzymatic activity of proteins involved in the expansion and breakdown of LDs needs to be actively controlled. Since the density of enzymes is the most fundamental way of tuning the overall catalytic activity, a quantification of LD size and the number of specific proteins on them are key in understanding the mechanisms of LD metabolism. While previous studies using conventional fluorescence microscopy yielded valuable insights into the overall distribution of regulatory proteins on LDs and the dynamics of LDs, they cannot resolve LDs below the diffraction limit or count the number of regulatory proteins they contain. To be able to measure protein densities on LDs, quantitative superresolution microscopy techniques are required that resolve LDs below the optical diffraction limit and enable specific protein counting. In the past, most quantitative superresolution microscopy experiments were performed in fixed cells to avoid movement of the structures under investigation during the long data acquisition time. These experiments employed low 405 nm photo-activation rates to carefully characterize the photophysical properties of irreversibly bleaching photoactivatable fluorescent proteins such as mEos2. Using this characterization, the fluorescent bursts originating from the same fluorophores were grouped to a photon-weighted position with increased precision, and the number of molecules were counted (Puchner et al., 2013; Durisic et al., 2014; Fricke et al., 2015).
Here we transferred this approach to live-cell PALM in order to quantify the density of proteins on LDs and to capture their dynamics while cells are transitioning to different metabolic states. Quantitative live-cell PALM is based on two requirements. First, the structures under investigation should not extensively move and should be trackable in the conventional fluorescence channel during data acquisition. This requirement enables grouping of fluorescent bursts from the same fluorophore and the correct assignment of molecules to a structure. Second, the concentration of a protein on a structure must be in equilibrium and not change during the data acquisition time. We note that the binding and unbinding of a protein will not affect the outcome of threshold-based counting techniques; the same fraction of bound and unbound fluorophores is photo-activated and bleached in a given time. For instance, when 50% of fluorophores on a LD have been imaged and bleached, 50% of unbound fluorophores will have been bleached as well. Therefore, binding and unbinding will not alter the fraction of unbleached fluorophores and the quantification of their number. The linear relation (ρ = 0.89) between the number of localizations and the number of blink-corrected molecules on individual LDs further demonstrate that the number of detected localizations is an accurate measure for quantifying protein numbers as previously demonstrated in other systems (Lee et al., 2012) and confirmed with intracellular calibration standards (Puchner et al., 2013). In this study, only LDs that freely diffused inside the vacuole during starvation violated the first requirement of immobility. All other LDs were immobile to determine their size and the number of associated proteins. Since the transition of cells to different metabolic states and the associated change in density of regulatory proteins take place on the timescale of hours (Brauer et al., 2008), potential density changes during the data acquisition time of several minutes can also be considered insignificant.
In this study we applied quantitative live-cell PALM to Faa4, one of the fatty-acyl CoA synthetases required to activate FAs for the expansion of LDs (synthesis of neutral lipids and other complex lipids) as well as for their breakdown (beta-oxidation). Previous studies using conventional bulk fluorescence of Faa4-GFP showed that Faa4 dynamically localizes among the ER, LDs, and the vacuole based on the nutritional state of a yeast cell (Natter et al., 2005; Kurat et al., 2006; van Zutphen et al., 2014; Wang et al., 2014a). However, these studies lacked an accurate quantification of Faa4 abundance, LD size, and Faa4 densities on LDs, a fundamental quantity in determining catalytic activity.
In the log phase, we observed exclusively immobile LDs colocalized with the ER where they emerge and expand, consistent with previous studies. The superresolved images of LDs with Faa4-mEos2 further revealed the size distribution of LDs with a mean diameter of 311 nm, which agrees with previous EM size measurements (Wang et al., 2014b). Most importantly, our quantitative live-cell PALM approach enabled us to quantify the number of Faa4 enzymes and their diffusion on the ER, LDs, and the vacuole. In the log phase, more than 60% of Faa4 localizes to the ER and about 34% to LDs, which are the subcellular locations where the catalytic activity of Faa4 is needed for the activation of FAs and their subsequent synthesis into neutral lipids. The average diffusion coefficient of Faa4 on the ER of 0.061 ± 0.014 μm2/s agrees with the published diffusion coefficient of FAs measured using BODIPY-C12 (Adhikari et al., 2019) and indicates that Faa4 freely diffuses at the ER. Similarly, we found that Faa4 freely diffuses on the surface of LDs as also observed with the LD-localized protein Dga1 using FLIP (Jacquier et al., 2011). These results suggest that Faa4 is homogeneously distributed in the ER membrane and targeted to emerging LDs through passive diffusion from the ER membrane.
During starvation and the transition to the stationary phase, LDs have been shown to expand by accumulating neutral lipids followed by lipophagic degradation in the vacuole (Markgraf et al., 2014; van Zutphen et al., 2014; Wang et al., 2014a). Consistent with these findings, our data showed LDs either docked to the vacuole or diffusing inside the vacuole. Notably, our quantified average diameter of LDs (446 nm) in the stationary phase was significantly larger compared with the 311 nm in the log phase and corresponds to a ∼2-fold increase in surface area and a ∼3-fold expansion of their volume. The unique capability of PALM to accurately measure the size and to estimate the number of Faa4 molecules on LDs further revealed a ∼2.5-fold increase in Faa4 density on LDs compared with the log phase. This density increase of Faa4 might therefore serve to activate FAs for LD expansion in the stationary phase. In fact, Dga1, responsible for converting DAG to TAG by adding one fatty acid activated by Faa4, has been shown to exclusively localize to LDs during the stationary phase (Markgraf et al., 2014). The linear correlation of the Faa4 numbers with the surface area of LDs and the overall increase in Faa4 expression level suggest that the Faa4 density is passively regulated through its abundance rather than through changes in its affinity to LDs. This suggestion agrees with a previous study about the role of an increased Faa4 expression level for cell survival during the stationary phase (Ashrafi et al., 1998).
Our tracking data revealed that LDs docked to the vacuole are immobile while single Faa4 molecules still freely diffuse on the surface of LDs. However, LDs exhibit random diffusion inside the vacuole with an average diffusion coefficient of D = 0.15 µm2/s. The similar diffusion coefficient of Faa4 indicates that Faa4 is still localized to LDs that entered the vacuole through lipophagy (Wang et al., 2014a; Tsuji et al., 2017). The viscosity inside the vacuole estimated using Stokes-Einstein with the determined diffusion coefficient of LDs (D = 0.15 µm2/s) and their average diameter (446 nm) was 6.6 ± 3.9 cP, which is in agreement with previous measurements (Puchkov, 2010, 2012).
Previous studies identified the specificity of Faa4 and Faa1 for activating long-chain fatty acids (C12–C18), whereas Fat1 was shown to activate very long-chain fatty acids (C22–C26) in yeast (Watkins et al., 1998; Choi and Martin, 1999; Faergeman et al., 2001). In this work, we observed a larger number of LDs per cell in the presence of exogenous oleic acid (C18), consistent with earlier studies (van Zutphen et al., 2014) (Supplemental Figure S6A). The higher number of LDs per cell indicates the conversion of imported oleic acid to its activated form by Faa1/Faa4 and then to neutral lipids. Most importantly, our quantitative PALM approach enabled us to quantify the size of individual LDs and the number of Faa4 enzymes on each LDs to determine the density of Faa4 on LDs. LDs in the presence of oleic acid have a ∼3-fold higher number of Faa4 molecules on their LDs as in the absence of oleic acid but exhibit a similar size (Figure 5B; Supplemental Figure S6B). Interestingly, the measured mean density of Faa4 on LDs in the presence of oleic acid is similar to LDs in the stationary phase (Figures 3C and 5B). The increased density of Faa4 on LDs in the presence of oleic acid reflects the higher enzymatic activity of Faa4 needed to activate imported oleic acid for the local synthesis of TAGs for LD expansion. In addition, the Faa4-mediated conversion of oleic acid to TAGs on LDs and the subsequent LD expansion could prevent the toxic accumulation of free oleic acids in the ER and the cytoplasm. This notion is based on previous studies demonstrating that the inability of yeast to convert imported oleic acid to TAGs in various mutants induces lipotoxicity (Garbarino et al., 2009; Petschnigg et al., 2009). Earlier studies also revealed the relocalization of Dga1, which is involved in the terminal step of TAG synthesis from ER to LDs on oleic acid addition (Jacquier et al., 2011; Markgraf et al., 2014). A similar future study quantifying the density of Dga1 on LDs and correlating the Dga1 density on LDs with the Faa4 density in the presence of oleic acid could further elucidate the role of the increased Faa4 density on LDs. Our work also revealed that the addition of very long-chain fatty acids such as lignoceric acid (C24), which is not a substrate of Faa4, does not significantly increase in the number of LDs or the density of Faa4 on individual LDs. First of all, this result suggests that imported oleic acids are preferentially converted to neutral lipids and stored in LDs compared with lignoceric acid, which is consistent with a previous study (Petschnigg et al., 2009). Second, this result implies that the increase of Faa4 density on LDs is specific to the preferred fatty acid substrate of Faa4 and indicates a higher enzymatic action of LD-localized Faa4. Our correlative PALM and conventional fluorescence microscopy approach further enabled us to quantify the number of Faa4 molecules localized to ER and LDs under each condition. The increased number of Faa4 molecules on LDs in the presence of oleic acid is explained by the relocalization of Faa4 from ER to LDs and by an increased expression level.
When cells in the stationary phase are diluted back to fresh medium, LDs are rapidly consumed for membrane expansion during cell proliferation. Previous studies have shown that LD-resident lipases (Tgl3) are required for the lipolysis of TAGs to generate DAG and FAs (Kurat et al., 2006; Ouahoud et al., 2018; Ganesan et al., 2019). The resulting FAs in turn need to be activated by fatty acid-activating enzymes to serve together with DAG as building blocks for the synthesis of membrane phospholipids. In our study, we observed almost no LDs around or inside the vacuole, indicating their rapid degradation on growth resumption. Therefore, our detected dramatic increase of Faa4 inside the vacuole may serve the activation of FAs released from LD breakdown in the vacuole. Since cells in the log phase have a fivefold lower expression level of Faa4, the localization of Faa4 to the vacuole could therefore in addition contribute to down-regulating Faa4 expression levels after growth resumption (Figure 4D). Our data support two mechanisms for how Faa4 enters the vacuole. The first mechanism is the dramatic 10-fold increase of Faa4 on LDs in the stationary phase and its subsequent uptake into the vacuole during lipophagy. The second mechanism is ER-phagy, which may account for the twofold decrease of ER-localized Faa4 in the log phase compared with the stationary phase.
In summary, we presented the first study that employs PALM in living cells to quantify the size of LDs and their associated Faa4 numbers and mobility throughout different metabolic states. Our results demonstrate that LD expansion and breakdown during metabolic transitions is coupled with density changes of Faa4 on LDs. This is consistent with the required catalytic activity of Faa4 on LDs in each metabolic state. The 2.5-fold increase in Faa4 density on LDs in the stationary phase is functionally correlated to their increased size and caused by an increased expression level. In future studies our quantitative live-cell PALM approach can be transferred to numerous other regulatory proteins. The resulting high-resolution data may give new insights into the regulation of LD biogenesis, growth, and mobilization in a cell’s normal physiological condition and how such regulation might be altered in metabolic disorders.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Yeast strains
The endogenous tagging of Faa4 with mEos2 in a W303 S. cerevisiae strain as well as the chromosomal integration of Sec63-GFP was described previously in Adhikari et al. (2019). The W303 yeast strain with integrated Ypt7-GFP was described in Puchner et al. (2013).
Sample preparation
Yeast strains were grown overnight in synthetic complete dextrose (SCD) medium with shaking (270 rpm) at 30°C. The next morning, cells were diluted 1:50 in SCD to an optical density (OD) of ∼0.15 and allowed to grow for 4 h to the log phase with and OD of 0.5. For cells in the late diauxic-shift/stationary phase, cells were grown for ∼30 h without exchange of medium. To study the effect of exogenously added long-chain fatty acids, cells in the log phase were incubated in medium with 0.1% (3.5 mM) oleic acid (C18) and 0.05% Tween 80 (Sigma-Aldrich) for 4 h. To study the effect of very long-chain fatty acids, cells were incubated with 3.5 mM lignoceric acid (C24) and 1% brij58 (Sigma-Aldrich) for 4 h. To prepare cells in the lag phase, stationary phase cells (∼30 h in same medium) were diluted with fresh SCD to an OD of ∼0.2 and imaged after ∼2 h. Cells in Figure 4C and Supplemental Figure S4E were imaged after diluting stationary phase cells in fresh media for ∼30 min to observe the Faa4 distribution at a shorter lag time. To adhere cells for imaging, the chambered borosilicate coverglasses (8-well, Lab-Tek II; Sigma-Aldrich) were incubated with ∼75 μl sterile filtered Concanavalin A (Sigma -Aldrich) at a concentration of 0.8 mg/ml in diH2O for 30 min. After washing the coverglasses three times with deionized water (Millipore), 300 µl of yeast cells were incubated on the coverglass at an OD of ∼0.12 and allowed to settle for ∼30 min. For LD staining with Nile red (Thermo Fisher) or BODIPY-NL (Thermo Fisher), dyes were directly added at a concentration of 100 nM and imaged after ∼10 min.
Experimental setup and data acquisition
All conventional fluorescence and PALM data were acquired on a Nikon Ti-E inverted microscope with a Perfect Focus System. Four excitation lasers (405, 488, and 561 nm, OBIS-CW; Coherent) were combined using dichroic mirrors. The laser beams were aligned, expanded, and focused to the back focal plane of the objective (Nikon-CFI Apo 100× Oil immersion N.A 1.49). The power and shutters of lasers were computer controlled using the Hal4000 software (Zhuang lab, Harvard). A quad-band dichroic mirror (zt405/488/561/640rdc; Chroma) was used to separate the fluorescence emission from the excitation light. For two-color imaging the red (595 nm) and green (525 nm) fluorescence were split by a dichroic longpass beam-splitter (T562lpxr BS; Chroma) followed by bandpass filters ET525/50 (Chroma) in the green channel and ET595/50 (Chroma) in the red channel, respectively. The fluorescence emission was recorded at the frame rate of 20 Hz on an electron multiplying CCD camera (Ixon89Ultra DU-897; Andor) cooled to –68°C and with a gain and preamp gain of 30 and 5.1, respectively.
The 405-nm laser was used to photo-activate mEos2 molecules at a power density of 0.1–1 Wcm–2, whereas the 488-nm laser was used to excite GFP, BODIPY-NL (493/503), and Nile red molecules at a power density between 0.07 and 0.7 Wcm–2 for conventional fluorescence imaging. The excitation of single photo-activated mEos2 molecules for PALM imaging and single-molecule tracking was performed with a 561-nm laser at a power density of 0.8–1 kWcm–2. Since Nile red can also be excited at 561 nm, we added Nile red after imaging mEos2 in the Sec63-GFP+Faa4-mEos2 yeast strain. We note that preactivated mEos2 has been shown to have a 10-fold lower fluorescence intensity compared with GFP (Puchner et al., 2013) and accordingly we did not observe any significant leaking into the GFP channel. For the simultaneous imaging of GFP and Nile red, both fluorophores were excited with the 488-nm laser and the fluorescence of GFP was detected in the 525 nm emission channel and Nile red in the 595 nm channel. BODIPY-NL was added and imaged after Sec63-GFP imaging or imaged at a ∼10-fold lower 488 nm excitation intensity compared with GFP imaging to avoid potential cross-talk with the GFP signal.
Camera calibration
The EMCCD camera was calibrated to obtain to gain (e/ADU) and the camera offset. For the calibration, camera pixels were exposed to varying intensities of light with an EMCCD gain of 30. The slope of the mean intensity versus variance graph accounting for the extra noise factor (F2) was used to calculate the gain. The calculated gain of 0.166 e/ADU and the offset of 200 counts matched the manufacturer’s specification at EMCCD gain 30 and preamp gain 5.1.
Conventional fluorescence image analysis
The conventional fluorescence images of Sec63-GFP, Ypt7-GFP, BODIPY-NL and Nile red were generated by averaging (50–100) image frames. The conventional fluorescence images of Faa4-mEos2 were obtained by averaging (20–60) 561-nm excitation frames.
Superresolution and single-molecule tracking data analysis
PALM data analysis was performed using the INSIGHT software (Zhuang lab, Harvard) that uses a Gaussian PSF model (Gaussian heights ≥ 50 photons, width (260–650) nm; ROI: 7 × 7 pixels with pixel size 160 nm) to fit the single-molecule signal. The obtained molecule list consisting of single-molecule co-ordinates, frames of appearance, widths, heights and total counts were exported for the further analysis described below. Superresolution images were generated by rendering single-molecule localizations as 2D Gaussians whose widths are weighted by the inverse square root of the detected number of photons.
For single-molecule tracking analysis, the single-molecule localizations that appeared within 0.48 µm in consecutive frames (50 ms exposure) were linked to a trace. The average distance between activated mEos2 molecules in a frame was kept low by adjusting the photo-activation density so that different molecules are not accidentally linked. Only single-molecules traces lasting for at least three frames (20 Hz) were extracted for further analysis. From each trace, the MSD for a particular lag time Δt was calculated by averaging the squared displacements over all time intervals of length Δt with a custom-written Igor-pro program. The mean diffusion coefficient (D) was then calculated by linear fitting the averaged MSD versus Δt curve from more than 100 traces with MSD = 4D Δt +2σ2 and localization precision σ.
SPT of LDs
The fluorescence signals from the neutral lipid core of LDs stained with Nile red or BODIPY-NL were detected with INSIGHT as diffraction-limited blobs and fitted with Gaussians to determine the centroids. The obtained localizations were linked to traces and further quantified as described in the previous section about single-molecule tracking.
Calibration of Z with PSF width
To avoid errors in measuring Faa4 numbers on LDs that are not fully within the detectable z-range, we performed a PSF width calibration experiment at different z positions using fluorescent TetraSpeck microspheres (Invitrogen T7279) (Supplemental Figure S5A). The width of the PSF is obtained from the BSF (Bead Spread Function) using Huygens PSF distiller (Scientific Volume Imaging, Netherlands, http://svi.nl) that accounts for the bead size (d = 200 nm), numerical aperture (N.A 1.49) of objective, and emission wavelength. As expected, the width of the PSF is dependent on the distance in z away from the focal plane (Supplemental Figure S5B). Using this calibration curve, the width of mEos2 localizations from individual LDs can be converted to their distance in z from the focal plane. From the width distribution of mEos2 localizations, we then calculated the median and the mean as well as the percentage of localizations with a width greater than 550 nm (max allowed = 650 nm), which corresponds to a z-distance range of ±500 nm (Supplemental Figure S5C). Only 1% of localizations had widths greater than 550 nm, which is well below our width cutoff of 650 nm and ensures that LDs included in the analysis were within the detectable z-range.
Blink correction and estimate of Faa4 molecules
To convert the number of localizations to the number of molecules, we employed a spatiotemporal threshold method for grouping localizations from the same mEos2 molecule that has been applied and confirmed in fixed cells (Puchner et al., 2013; Durisic et al., 2014). These methods are based on the fact that mEos2, once photoactivated, blinks for a variable time with various dark times between fluorescent bursts until it irreversibly bleaches (Supplemental Figure S3A). The low 405 nm photoactivation power used throughout PALM experiments results in significantly longer times between the photoactivation of nearby mEos2 molecules compared with the time between fluorescent bursts from the same fluorophore. Therefore, fluorescent bursts appearing within the spatial resolution and a maximum dark time can be grouped to determine the blink-corrected number of molecules.
To determine the optimal dark time in this study, localizations from individual LDs at low 405 nm photo-activation power were selected for further analysis (Supplemental Figure S3A, right). The distribution of widths from localizations on LDs ensured that LDs within our detectable z range are included (Supplemental Figure S5). For each individual LD, the number of apparent blink-corrected molecules at various allowed dark times td was determined by grouping localizations appearing within 650 nm and the allowed td (Supplemental Figure 3B). The spatial threshold of 650 nm is the maximum diameter of LDs and accounts for diffusion of photoactivated mEos2 along the surface of LDs. At td = 0 s localizations that appear in consecutive frames are grouped and the number of apparent molecules therefore corresponds to the number of fluorescent bursts. As td increases, more and more fluorescent bursts from the same mEos2 molecules are grouped, which results in a double exponential decay of the apparent number of molecules. This double exponential decay is caused by two previously identified dark states with different decay times (Lee et al., 2012; De Zitter et al., 2019). At longer dark times when most fluorescent bursts are incorrectly grouped, the probability for falsely grouping two mEos2 molecules keeps increasing and results in a dominating linear decrease in the number of apparent molecules. Therefore, the obtained apparent molecules versus td curve for each individual LD was fitted with the sum of a double exponential and linear function (Supplemental Figure 3B). For each LD, the optimal td was determined to be where the fraction of overcounting balances the fraction of undercounting due to false grouping (Supplemental Figure S3, B–D). The average number of molecules per localizations of 0.107 ± 0.04 from N = 9 different LDs (Supplemental Figure S3C) was then used for all PALM data to convert localizations to molecules and to quantify the Faa4 density on LDs (Figure 3B and C, and Figure 5, B and C), Faa4 molecules per cell (Figure 3D; Supplemental Figure S6C) and Faa4 molecules on the ER, LDs, and in the vacuole (Figure 4B) with an error of 17%. While the number of Faa4 molecules on immobile LDs can directly be determined by blink correction, the free diffusion of Faa4-mEos2 on the ER and in the vacuole prevents reliable blink correction and makes a localization-based approach more comparable and reliable in different cellular compartments. We note that previous reports in fixed cells found that only ∼60% of mEos2 molecules are detected in PALM experiments (Puchner et al., 2013; Durisic et al., 2014). While this observable fraction has not been determined so far in living cells due to the experimental challenges, the true number of molecules is likely larger than the number of detected molecules. However, both uncertainties for converting localizations to the number of molecules only affect the absolute numbers and do not bias the relative densities or abundances that are relevant for this study.
405 nm photo-activation energy correction
We found that the expression level of Faa4-mEos2 is ∼5-fold higher in the stationary phase compared with the log phase (Figure 3D). To maintain a constant and nonoverlapping detection of mEos2 molecules in each data acquisition frame, this difference in expression levels makes it necessary to acquire PALM data with different 405 nm photo-activation energy densities. As a result, different fractions of total mEos2 molecules can be activated in different movies based on the delivered 405 nm energy. Therefore, it is necessary to verify that the fraction of activated molecules only depends on the delivered 405 nm energy but is independent of acquisition time and mEos2 expression level. Once a calibration curve relating the fraction of activated mEos2 molecules and the delivered 405 nm is obtained, the difference in the fraction of activated mEos2 can be adjusted to compare the localizations across different movies using the delivered 405 nm energy.
To verify that the fraction of activated mEos2 molecules only depends on the delivered 405 nm energy, we analyzed PALM data from cells in the log and stationary phases that were imaged with different 405 nm powers and data acquisition times. From the obtained mEos2 localizations for both the log and the stationary phases, the fraction of activated mEos2 molecules was calculated as a function of data acquisition time. As expected, the fraction of activated mEos2 molecules is lower for stationary phase cells at a particular data acquisition time since a longer total data acquisition time is required for 100% activation (Supplemental Figure S3F). Next, we calculated the cumulative mEos2 localizations versus delivered 405 nm energy using the 405 nm power and exposure time (Supplemental Figure S3G). The resulting cumulative fraction of activated mEos2 molecules versus 405 nm energy overlapped for both log and stationary phase cells and verifies that the fraction of detected mEos2 molecules only depends on the delivered 405 nm energy (Supplemental Figure S3H).
To quantify Faa4 densities on LDs in Figure 3, B and C recorded at different 405 nm energies, the delivered 405 nm energy was calculated by multiplying the 405 nm power with the exposure time of each frame and integrating over time. For LDs from movies with a total delivered energy less than ∼5 mJ (95% photo activation), the number of localizations was adjusted to 100% activation using the percentage activation versus energy calibration curve (Supplemental Figure S3H). For the quantification of Faa4 molecules in Figures 3D, 4B, and 5, B and C, the energy delivered in each movie corresponded to more than ∼95% activation. Therefore, no adjustments for the number of localizations were used.
Diameter and Faa4 density of LDs
To determine the diameter of LDs, molecule lists of Faa4-mEos2 belonging to individual LDs were first extracted. The radius r of LDs was then approximated by computing the average SD in x and y assuming LDs are spherical in 3D. This approach is more robust and easier to automate compared with fit-based approaches. For instance, fits do not converge well for LDs that do not exhibit a clear donut shape. We validated this by correlating the LD sizes obtained using SD with sizes obtained using double gaussian fitting of x and y projections (Supplemental Figure S1, A and B). The diameter (d = 2r) of LDs was obtained by multiplying the obtained radius by 2. The surface density of Faa4 on LDs was then obtained by calculating the ratio of the number of Faa4 molecules and the surface area of a sphere of diameter d.
Subcellular localizations of Faa4
To quantify the subcellular localizations of Faa4 on the ER, LDs, and the vacuole in different metabolic states, Faa4-mEos2 localizations from superresolution images were first transformed to and superimposed on the conventional (GFP) channel in the INSIGHT software. This third-order polynomial transformation was determined and verified by localizing and mapping TetraSpeck microspheres (Invitrogen T7279) which fluoresce in both channels. LDs in PALM images are identifiable as dense circular/donut-shaped puncta which are separable from ER localizations. The number of Faa4 localizations that colocalized with the bright Nile red/BODIPY-NL puncta (LD marker) and Sec63-GFP (ER marker) was determined from superimposed superresolution and conventional images in INSIGHT. In the superimposed images, the dense circular Faa4-mEos2 structures always colocalized with the Nile red signal. To further increase the statistics, dense circular structures in superresolution images of Faa4-mEos2 in cells with no Nile red/BODIPY-NL staining were therefore also considered to be from LDs and included. The remaining localizations in each cell that did not colocalize with the ER marker Sec63-GFP were considered to be from the vacuole. While vacuolar Faa4-mEos2 was detected as single-molecule localizations in the log and stationary phase due to its slow diffusion, the fast vacuolar diffusion of Faa4-mEos2 in the lag phase prevented the detection of single molecules. However, the signal of the freely diffusing Faa4-mEos2 in the vacuolar lumen was evident from a homogenous and diffuse florescence. To compare the relative abundance of Faa4 in the vacuole in different growth conditions (Figure 4A), we averaged ∼20 frames of the 561 nm channel and used the CCD counts per pixel as a metric for its relative abundance.
Supplementary Material
Acknowledgments
We thank Doug Mashek and his lab as well as Christer Ejsing and Florian Froehlich for helpful discussions. From the Puchner lab, we thank Maria-Paz Ramirez Lopez for making the Sec63 yeast strain. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award number R21GM127965.
Abbreviations used:
- FA
fatty acid
- Faa4
fatty acid activation protein 4
- LD
lipid droplet
- MSD
mean-squared displacement
- OD
optical density
- PALM
photo-activated localization microscopy
- SCD
synthetic complete dextrose
- SMLM
single-molecule localization microscopy
- SPT
single-particle tracking
- TAG
triacylglycerol.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E20-11-0695) on June 23, 2021.
REFERENCES
- Adhikari S, Moscatelli J, Smith EM, Banerjee C, Puchner EM (2019). Single-molecule localization microscopy and tracking with red-shifted states of conventional BODIPY conjugates in living cells. Nat Commun 10, 3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adhikari S, Banerjee C, Moscatelli J, Puchner EM (2020). Conventional BODIPY conjugates for live-cell super-resolution microscopy and single-molecule tracking. J Vis Exp JoVE 160, e60590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annibale P, Scarselli M, Kodiyan A, Radenovic A (2010). Photoactivatable fluorescent protein mEos2 displays repeated photoactivation after a long-lived dark state in the red photoconverted form. J Phys Chem Lett 1, 1506–1510. [Google Scholar]
- Annibale P, Vanni S, Scarselli M, Rothlisberger U, Radenovic A (2011). Quantitative photo activated localization microscopy: unraveling the effects of photoblinking. PLoS One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashrafi K, Farazi TA, Gordon JI (1998). A role for Saccharomycescerevisiae fatty acid activation protein 4 in regulating protein N-myristoylation during entry into stationary phase. J Biol Chem 273, 25864–25874. [DOI] [PubMed] [Google Scholar]
- Bersuker K, Olzmann JA (2017). Establishing the lipid droplet proteome: Mechanisms of lipid droplet protein targeting and degradation. Biochim Biophys Acta 1862, 1166–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645. [DOI] [PubMed] [Google Scholar]
- Black PN, DiRusso CC (2007). Yeast acyl-CoA synthetases at the crossroads of fatty acid metabolism and regulation. Biochim Biophys Acta 1771, 286–298. [DOI] [PubMed] [Google Scholar]
- Brauer MJ, Huttenhower C, Airoldi EM, Rosenstein R, Matese JC, Gresham D, Boer VM, Troyanskaya OG, Botstein D (2008). Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol Biol Cell 19, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Martin CE (1999). The Saccharomyces cerevisiae FAT1 gene encodes an acyl-CoA synthetase that is required for maintenance of very long chain fatty acid levels. J Biol Chem 274, 4671–4683. [DOI] [PubMed] [Google Scholar]
- Cimprich P, Slavík J, Kotyk A (1995). Distribution of individual cytoplasmic pH values in a population of the yeast Saccharomyces cerevisiae. FEMS Microbiol Lett 130, 245–251. [DOI] [PubMed] [Google Scholar]
- De Zitter E, Thédié D, Mönkemöller V, Hugelier S, Beaudouin J, Adam V, Byrdin M, Van Meervelt L, Dedecker P, Bourgeois D (2019). Mechanistic investigation of mEos4b reveals a strategy to reduce track interruptions in sptPALM. Nat Methods 16, 707–710. [DOI] [PubMed] [Google Scholar]
- Deschout H, Shivanandan A, Annibale P, Scarselli M, Radenovic A (2014). Progress in quantitative single-molecule localization microscopy. Histochem Cell Biol 142, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durisic N, Laparra-Cuervo L, Sandoval-Álvarez A, Borbely JS, Lakadamyali M (2014). Single-molecule evaluation of fluorescent protein photoactivation efficiency using an in vivo nanotemplate. Nat Methods 11, 156–162. [DOI] [PubMed] [Google Scholar]
- Faergeman NJ, Black PN, Zhao XD, Knudsen J, DiRusso CC (2001). The Acyl-CoA synthetases encoded within FAA1 and FAA4 in Saccharomyces cerevisiae function as components of the fatty acid transport system linking import, activation, and intracellular Utilization. J Biol Chem 276, 37051–37059. [DOI] [PubMed] [Google Scholar]
- Fricke F, Beaudouin J, Eils R, Heilemann M (2015). One, two or three? Probing the stoichiometry of membrane proteins by single-molecule localization microscopy. Sci Rep 5, 14072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan S, Sosa Ponce ML, Tavassoli M, Shabits BN, Mahadeo M, Prenner EJ, Terebiznik MR, Zaremberg V (2019). Metabolic control of cytosolic-facing pools of diacylglycerol in budding yeast. Traffic Cph Den 20, 226–245. [DOI] [PubMed] [Google Scholar]
- Garbarino J, Padamsee M, Wilcox L, Oelkers PM, D’Ambrosio D, Ruggles KV, Ramsey N, Jabado O, Turkish A, Sturley SL (2009). Sterol and diacylglycerol acyltransferase deficiency triggers fatty acid-mediated cell death. J Biol Chem 284, 30994–31005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan P, Mayer EP, Fowler SD (1985). Nile red: a selective fluorescent stain for intracellular lipid droplets. J Cell Biol 100, 965–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri H, Rogers S, Ugrankar R, Liu YL, Feathers JR, Henne WM (2018). Lipid droplet biogenesis is spatially coordinated at ER-vacuole contacts under nutritional stress. EMBO Rep. 19, 57–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hariri H, Speer N, Bowerman J, Rogers S, Fu G, Reetz E, Datta S, Feathers JR, Ugrankar R, Nicastro D, et al. (2019). Mdm1 maintains endoplasmic reticulum homeostasis by spatially regulating lipid droplet biogenesis. J Cell Biol 218, 1319–1334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacquier N, Choudhary V, Mari M, Toulmay A, Reggiori F, Schneiter R (2011). Lipid droplets are functionally connected to the endoplasmic reticulum in Saccharomyces cerevisiae. J Cell Sci 124, 2424–2437. [DOI] [PubMed] [Google Scholar]
- Johnson DR, Knoll LJ, Levin DE, Gordon JI (1994). Saccharomyces cerevisiae contains four fatty acid activation (FAA) genes: an assessment of their role in regulating protein N-myristoylation and cellular lipid metabolism. J Cell Biol 127, 751–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiegerl B, Tavassoli M, Smart H, Shabits BN, Zaremberg V, Athenstaedt K (2019). Phosphorylation of the lipid droplet localized glycerol‑3‑phosphate acyltransferase Gpt2 prevents a futile triacylglycerol cycle in yeast. Biochim Biophys Acta Mol Cell Biol Lipids 1864, 158509. [DOI] [PubMed] [Google Scholar]
- Kory N, Farese RV, Walther TC (2016). Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biol 26, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurat CF, Natter K, Petschnigg J, Wolinski H, Scheuringer K, Scholz H, Zimmermann R, Leber R, Zechner R, Kohlwein SD (2006). Obese yeast: triglyceride lipolysis is functionally conserved from mammals to yeast. J Biol Chem 281, 491–500. [DOI] [PubMed] [Google Scholar]
- Lee S-H, Shin JY, Lee A, Bustamante C (2012). Counting single photoactivatable fluorescent molecules by photoactivated localization microscopy (PALM). Proc Natl Acad Sci USA 109, 17436–17441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Ory DS, Schaffer JE (2003). Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc Natl Acad Sci USA 100, 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loi M, Raimondi A, Morone D, Molinari M (2019). ESCRT-III-driven piecemeal micro-ER-phagy remodels the ER during recovery from ER stress. Nat Commun 10, 5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley S, Gillette JM, Patterson GH, Shroff H, Hess HF, Betzig E, Lippincott-Schwartz J (2008). High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods 5, 155–157. [DOI] [PubMed] [Google Scholar]
- Markgraf DF, Klemm RW, Junker M, Hannibal-Bach HK, Ejsing CS, Rapoport TA (2014). An ER protein functionally couples neutral lipid metabolism on lipid droplets to membrane lipid synthesis in the ER. Cell Rep. 6, 44–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL (2009). A bright and photostable photoconvertible fluorescent protein. Nat Methods 6, 131–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mejhert N, Kuruvilla L, Gabriel KR, Elliott SD, Guie M-A, Wang H, Lai ZW, Lane EA, Christiano R, Danial NN, et al. (2020). Partitioning of MLX-family transcription factors to lipid droplets regulates metabolic gene expression. Mol Cell 77, 1251–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natter K, Leitner P, Faschinger A, Wolinski H, McCraith S, Fields S, Kohlwein SD (2005). The spatial organization of lipid synthesis in the yeast Saccharomyces cerevisiae derived from large scale green fluorescent protein tagging and high resolution microscopy. Mol Cell Proteomics MCP 4, 662–672. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, Carvalho P (2019). Dynamics and functions of lipid droplets. Nat Rev Mol Cell Biol 20, 137–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouahoud S, Fiet MD, Martínez-Montañés F, Ejsing CS, Kuss O, Roden M, Markgraf DF (2018). Lipid droplet consumption is functionally coupled to vacuole homeostasis independent of lipophagy. J Cell Sci 131, 213876. [DOI] [PubMed] [Google Scholar]
- Petschnigg J, Wolinski H, Kolb D, Zellnig G, Kurat CF, Natter K, Kohlwein SD (2009). Good fat, essential cellular requirements for triacylglycerol synthesis to maintain membrane homeostasis in yeast. J Biol Chem 284, 30981–30993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puchkov EO (2010). Brownian motion of polyphosphate complexes in yeast vacuoles: characterization by fluorescence microscopy with image analysis. Yeast Chichester Engl 27, 309–315. [DOI] [PubMed] [Google Scholar]
- Puchkov EO (2012). Single yeast cell vacuolar milieu viscosity assessment by fluorescence polarization microscopy with computer image analysis. Yeast Chichester Engl 29, 185–190. [DOI] [PubMed] [Google Scholar]
- Puchner EM, Walter JM, Kasper R, Huang B, Lim WA (2013). Counting molecules in single organelles with superresolution microscopy allows tracking of the endosome maturation trajectory. Proc Natl Acad Sci USA 110, 16015–16020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanauska A, Köhler A (2018). The Inner Nuclear Membrane Is a Metabolically Active Territory that Generates Nuclear Lipid Droplets. Cell 174, 700–715.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saks V, Beraud N, Wallimann T (2008). Metabolic Compartmentation – A System Level Property of Muscle Cells. Int J Mol Sci 9, 751–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer JA, Schessner JP, Bircham PW, Tsuji T, Funaya C, Pajonk O, Schaeff K, Ruffini G, Papagiannidis D, Knop M, et al. (2019). ESCRT machinery mediates selective microautophagy of endoplasmic reticulum in yeast. EMBO J e102586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Bohnert M (2017). A different kind of love - lipid droplet contact sites. Biochim Biophys Acta Mol Cell Biol Lipids 1862, 1188–1196. [DOI] [PubMed] [Google Scholar]
- Seo AY, Lau P-W, Feliciano D, Sengupta P, Gros MAL, Cinquin B, Larabell CA, Lippincott-Schwartz J (2017). AMPK and vacuole-associated Atg14p orchestrate μ-lipophagy for energy production and long-term survival under glucose starvation. ELife 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swaminathan R, Hoang CP, Verkman AS (1997). Photobleaching recovery and anisotropy decay of green fluorescent protein GFP-S65T in solution and cells: cytoplasmic viscosity probed by green fluorescent protein translational and rotational diffusion. Biophys J 72, 1900–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauchi-Sato K, Ozeki S, Houjou T, Taguchi R, Fujimoto T (2002). The surface of lipid droplets is a phospholipid monolayer with a unique Fatty Acid composition. J Biol Chem 277, 44507–44512. [DOI] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW (2002). Precise nanometer localization analysis for individual fluorescent probes. Biophys J 82, 2775–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T, Fujimoto M, Tatematsu T, Cheng J, Orii M, Takatori S, Fujimoto T (2017). Niemann-Pick type C proteins promote microautophagy by expanding raft-like membrane domains in the yeast vacuole. ELife 6, e25960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Farese RV (2012). Lipid droplets and cellular lipid metabolism. Annu Rev Biochem 81, 687–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-W, Miao Y-H, Chang Y-S (2014a). A sterol-enriched vacuolar microdomain mediates stationary phase lipophagy in budding yeast. J Cell Biol 206, 357–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C-W, Miao Y-H, Chang Y-S (2014b). Control of lipid droplet size in budding yeast requires the collaboration between Fld1 and Ldb16. J Cell Sci 127, 1214–1228. [DOI] [PubMed] [Google Scholar]
- Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, et al. (2016). Seipin is required for converting nascent to mature lipid droplets. ELife 5, e16582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins PA, Lu JF, Steinberg SJ, Gould SJ, Smith KD, Braiterman LT (1998). Disruption of the Saccharomycescerevisiae FAT1 gene decreases very long-chain fatty acyl-CoA synthetase activity and elevates intracellular very long-chain fatty acid concentrations. J Biol Chem 273, 18210–18219. [DOI] [PubMed] [Google Scholar]
- Wu C-Y, Roybal KT, Puchner EM, Onuffer J, Lim WA (2015). Remote control of therapeutic T cells through a small molecule-gated chimeric receptor. Science 350, aab4077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Zutphen T, Todde V, de Boer R, Kreim M, Hofbauer HF, Wolinski H, Veenhuis M, van der Klei IJ, Kohlwein SD (2014). Lipid droplet autophagy in the yeast Saccharomyces cerevisiae. Mol Biol Cell 25, 290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.