Abstract
The Golgi complex is a central hub for intracellular protein trafficking and glycosylation. Steady-state localization of glycosylation enzymes is achieved by a combination of mechanisms involving retention and recycling, but the machinery governing these mechanisms is poorly understood. Herein we show that the Golgi-associated retrograde protein (GARP) complex is a critical component of this machinery. Using multiple human cell lines, we show that depletion of GARP subunits impairs Golgi modification of N- and O-glycans and reduces the stability of glycoproteins and Golgi enzymes. Moreover, GARP-knockout (KO) cells exhibit reduced retention of glycosylation enzymes in the Golgi. A RUSH assay shows that, in GARP-KO cells, the enzyme beta-1,4-galactosyltransferase 1 is not retained at the Golgi complex but instead is missorted to the endolysosomal system. We propose that the endosomal system is part of the trafficking itinerary of Golgi enzymes or their recycling adaptors and that the GARP complex is essential for recycling and stabilization of the Golgi glycosylation machinery.
INTRODUCTION
The Golgi complex plays a central role in the processing, packaging, and sorting of secretory and transmembrane proteins in eukaryotic cells (D’Souza et al., 2020). Although the Golgi complex was discovered more than 120 years ago, the molecular mechanisms of intra-Golgi trafficking and Golgi maintenance are not entirely understood (De Matteis et al., 2019). According to the “cisternal maturation” model, newly synthesized secretory proteins arrive at the cis-Golgi compartment from the ER by vesicular trafficking, and then stay within this compartment while it matures, sequentially changing its identity from cis to medial to trans (Losev et al., 2006; Glick and Nakano, 2009). During this maturation, Golgi-resident glycosylation enzymes are recycled continuously from late to early cisternae by retrograde trafficking, allowing them to maintain their steady-state localization to the Golgi complex (Gleeson, 1998; Ishii et al., 2016). However, the exact trafficking itineraries and molecular machinery involved in the delivery of these enzymes to their corresponding Golgi cisternae are poorly understood. Potential components of this machinery are coat proteins, small GTPases, vesicle tethers, and SNAREs implicated in intercompartmental cargo transport in both anterograde and retrograde pathways (Bonifacino and Hierro, 2011; Cruz and Kim, 2019; D’Souza et al., 2020). Each Golgi cisterna contains different carbohydrate-modifying enzymes. These enzymes catalyze the addition (glycosyltransferases) or removal (glycosidases) of sugars to/from cargo glycoproteins, as well as the addition of sulfate and phosphate groups. Different forms of protein glycosylation are referred to by their sugar–protein linkage. For instance, N-linked glycans are linked to asparagine residues, O-linked glycans to serine, threonine or tyrosine residues, C-linked glycans to tryptophan residues, and glypiation to the C-terminus of proteins (Schjoldager et al., 2020). As a maturing glycoprotein proceeds from cis- to trans-Golgi, several Golgi glycosyltransferases add N-acetylglucosamine, glucose, galactose, mannose, fucose, N-acetylgalactosamine, and sialic acid residues, resulting in complex glycosylation of the protein (Stanley, 2011; Schjoldager et al., 2020).
The conserved oligomeric Golgi (COG) complex is the major Golgi vesicle tethering complex responsible for the maintenance of the Golgi glycosylation machinery (Pokrovskaya et al., 2011; D’Souza et al., 2020). Several lines of evidence indicate that COG and another vesicle tethering complex, the Golgi-associated retrograde protein (GARP) complex, are functionally interconnected (Laufman et al., 2011; D’Souza et al., 2019). In addition, multiple recent CRISPR screens identified both COG and GARP as a requirement for the entry of viruses and bacterial toxins into the host cells (Laufman et al., 2011, 2013; Yamaji et al., 2019). These pathogens interact with glycoproteins or glycolipids on the host plasma membrane, suggesting that complete depletion of COG and GARP complexes may alter not only a particular intracellular trafficking step but also the cellular glycosylation machinery.
GARP is a multisubunit tethering complex located at the trans-Golgi network (TGN) where it functions to tether retrograde transport vesicles derived from endosomes (Siniossoglou and Pelham, 2002; Conibear et al., 2003; Liewen et al., 2005; Pérez-Victoria et al., 2008, 2010a). GARP comprises four subunits named vacuolar protein sorting 51 (VPS51), VPS52, VPS53, and VPS54 (Siniossoglou and Pelham, 2002; Conibear et al., 2003; Liewen et al., 2005; Pérez-Victoria et al., 2008, 2010b). GARP shares its VPS51, VPS52, and VPS53 subunits with another complex known as endosome-associated recycling protein (EARP or GARPII) complex, which has an additional subunit named VPS50 in place of VPS54 (Gillingham et al., 2014; Schindler et al., 2015). GARP complex localization to the TGN is dependent on the small GTPases ARFRP1 and ARL5 (Rosa-Ferreira et al., 2015; Ishida and Bonifacino, 2019). In yeast, mutation of genes encoding any of the GARP subunits inhibits recycling of the cargo receptor Vps10 from endosomes to the TGN, leading to the missorting and secretion of vacuolar hydrolases (Siniossoglou and Pelham, 2002; Conibear et al., 2003). In mammalian cells, GARP participates in retrieval of mannose-6-phosphate receptors (MPRs), the TGN-resident protein TGN46, and the Niemann-Pick C2 protein from endosomes to the TGN (Pérez-Victoria et al., 2008; Wei et al., 2017). Mutation of GARP subunit genes in mammalian cells cause MPR missorting, with consequent secretion of immature cathepsin D and lysosomal dysfunction (Pérez-Victoria et al., 2008). GARP depletion also results in alterations of autophagy (Pérez-Victoria et al., 2010b; Dotiwala et al., 2013), anterograde transport of GPI-anchored and transmembrane proteins (Hirata et al., 2015), and sphingolipid homeostasis (Fröhlich et al., 2015). Furthermore, mutations in GARP subunits have been found to cause neurodevelopmental disorders in humans (Feinstein et al., 2014; Hady-Cohen et al., 2018; Gershlick et al., 2019; Uwineza et al., 2019), and a mutation in VPS54 is the cause of progressive motor neuron death in the wobbler mouse, an animal model for amyotrophic lateral sclerosis (Schmitt-John et al., 2005). Inhibition of sphingolipid synthesis showed improvement in neuropathology and survival in mutant wobbler mice, suggesting that altered sphingolipid homeostasis contributes to the pathogenesis of GARP-deficiency disorders (Petit et al., 2020).
In this study, we have examined if glycosylation is disturbed in human cells depleted of GARP subunits. Using three different cell lines with knockout (KO) of GARP subunits and a combination of microscopy and biochemical approaches, we demonstrate defects in glycosylation and a reduction in the level of Golgi-resident enzymes in GARP-KO cells. We also show that the trans-Golgi enzymes beta-1,4-galactosyltransferase 1 (B4GalT1) and alpha-2,6-sialyltransferase 1 (ST6Gal1) fail to be retained in the Golgi but are instead mislocalized to endolysosomes in GARP-KO cells. These findings indicate that GARP plays a crucial role in normal Golgi glycosylation by mediating the maintenance of the Golgi glycosylation machinery.
RESULTS
GARP KO in RPE1 and HEK293T cells causes N- and O-glycosylation defects
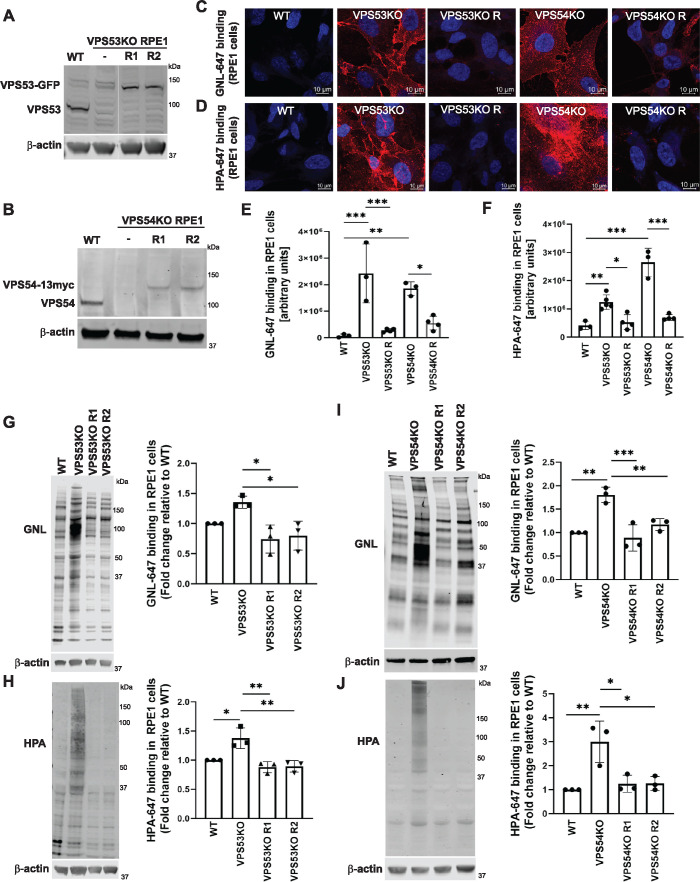
To test if complete depletion of GARP complex activity has an effect on the processing of N- and O-linked oligosaccharides of glycoproteins, we knocked out the VPS53 (Figure 1A) and VPS54 (Figure 1B) subunits of GARP in immortalized retinal pigment epithelial (RPE1) cells. These cells were chosen for their noncancerous origin and superior characteristics for microscopy imaging. To test for protein N-glycosylation defects, nonpermeabilized wild-type (WT) and GARP-KO cells were stained with Galanthus nivalis (GNL) lectin labeled with Alexa 647 (GNL-647). GNL binds terminal 1,3- and 1,6-linked mannose residues on N-linked glycans (Bailey Blackburn et al., 2016) and an increase in binding to the plasma membrane indicates expression of underglycosylated N-linked glycoconjugates (Pokrovskaya et al., 2011) (Figure 1C). To determine O-glycosylation defects, nonpermeabilized WT and GARP-KO cells were stained with Helix pomatia agglutinin (HPA) lectin labeled with Alexa 647 (HPA-647). HPA binds to terminal N-acetylgalactosaminyl residues in O-glycans; therefore, increased binding indicates surface expression of underglycosylated O-linked glycoconjugates (Brooks, 2000) (Figure 1D). GARP-KO cell lines showed a significant increase in binding of both lectins to the plasma membrane (Figure 1, C–F), indicating the presence of altered/immature N- and O-glycosylated proteins. To verify the specificity of the KO and of lectin staining, we rescued KO cells by stably expressing the corresponding GARP subunit under the control of the COG4 promoter. The use of the COG4 promoter (Zinia D’Souza and V.L, unpublished data) allowed expression of GARP subunits at near-endogenous levels (Figure 1, A and B). This expression level was sufficient to completely rescue the Cathepsin D sorting defect in GARP-KO cells (Supplemental Figure S1A). Importantly, in rescued cells, a substantial reduction in lectin binding to the plasma membrane was observed (Figure 1, C–F), confirming that glycosylation defects are directly related to GARP complex malfunction.
FIGURE 1:
GARP-KO results in Golgi glycosylation defects in RPE1 cells. (A) WB of RPE1 cell lysates from WT, VPS53-KO, and two VPS53-KO rescued clones expressing VPS53-GFP (R1 and R2) probed with anti-VPS53 antibody. (B) WB of RPE1 cell lysates from WT, VPS54-KO, and two VPS54-KO clones rescued with VPS54-13myc (R1 and R2) probed with anti-VPS54 antibodies. (C) Staining of nonpermeabilized WT, VPS53-KO, and VPS54-KO RPE1 cells, and the corresponding rescued cells with the fluorescently conjugated lectin GNL-647 (specific for terminal α-D-mannosyl residues). (D) Staining of nonpermeabilized WT, VPS53-KO, and VPS54-KO RPE1 cells and the corresponding rescued cells with the fluorescently conjugated lectin HPA-647 (specific for terminal GalNAc residues). (E) Quantification of GNL-647 binding was done on the basis of the total GNL-647 signal per field. Three different fields were used for the quantification of GNL-647 binding and each field included approximately 80 cells. The y-axis in the bar graph represents relative binding of GNL-647 to the surface of WT, GARP-KOs, and rescued RPE1 cells in arbitrary units. Bar graphs with error bars represent mean ± SD. (F) Quantification of HPA-647 binding was done on the basis of the total HPA-647 signal per field. Bar graphs with error bars represent mean ± SD from three different fields. The y-axis in the bar graph represents relative binding of HPA-647 to the surface of WT, GARP-KOs, and rescued RPE1 cells in arbitrary units. (G, H) GNL-647 and HPA-647 staining of total proteins from WT, VPS53-KO, and two rescue clones of RPE1 cells (left panels) and quantification from three independent experiments (right panels). Values in bar graphs represent the mean ± SD from three independent experiments. (I, J) GNL-647 and HPA-647 staining of total proteins from WT, VPS54-KO, and two rescue clones of RPE1 cells (left panels) and quantification from three independent experiments (right panels). Values in bar graphs represent the mean ± SD from three independent experiments. Statistical significance was calculated in GraphPad Prism 8 using one-way ANOVA. ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05.
Lectin blot analysis using GNL-647 and HPA-647 also showed abnormal processing of N- and O-linked oligosaccharides in total cellular glycoproteins (Figure 1, G–J) and secreted glycoproteins (Supplemental Figure S1, B and C) in VPS53- and VPS54-KO RPE1 cells. These glycosylation defects could also be rescued by stable expression of the corresponding genes in the KO cells (Figure 1, E–J). From these experiments we concluded that KO of VPS53 or VPS54 in RPE1 cells caused N- and O-glycosylation defects in plasma membrane, intracellular, and secreted glycoproteins. Similar glycosylation abnormalities were detected in VPS53- and VPS54-KO HEK293T cells (Supplemental Figure S1, D and E), indicating that glycosylation defects in GARP-KO cells are cell line-independent and, hence, that the GARP complex plays a general role in oligosaccharide processing in human cells.
GARP depletion affects the stability and glycosylation of Golgi and lysosomal glycoproteins
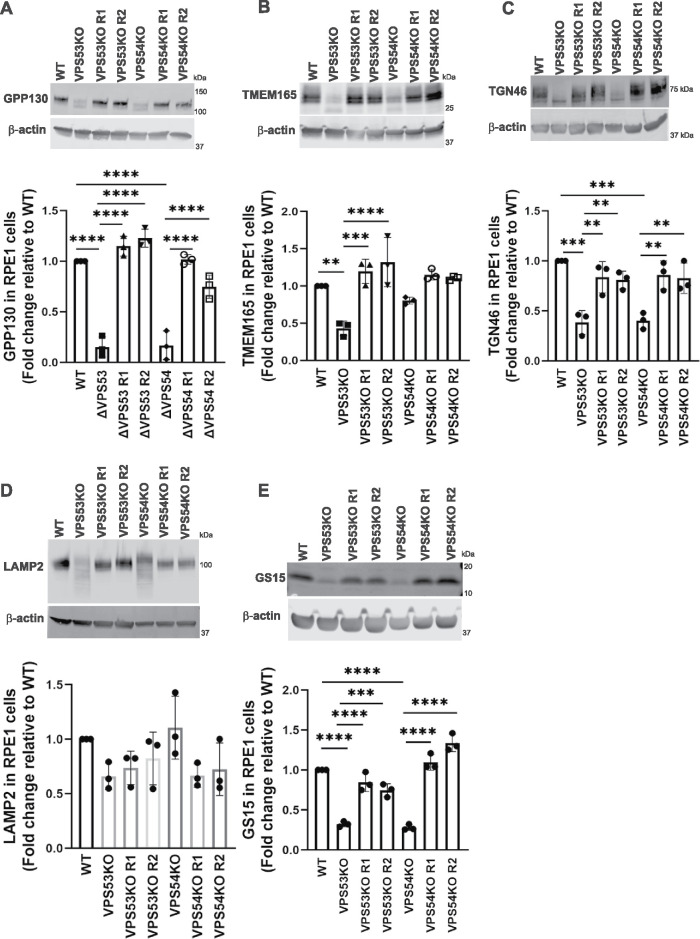
Defective glycosylation influences the stability of glycoproteins (Blackburn et al., 2018), so we reasoned that GARP deficiency might affect the stability of Golgi proteins. Indeed, we found that the levels of cis-Golgi GPP130 (Figure 2A), medial-trans-Golgi TMEM165 (Figure 2B), and TGN-resident TGN46 (Figure 2C) were significantly reduced in GARP-KO cells. Importantly, the levels of all three Golgi proteins were restored in GARP-KO rescued cells. We also detected an electrophoretic mobility shift for all three proteins, indicating problems with their secondary modifications. The total level of the lysosomal protein LAMP2 (Figure 2D) did not significantly change, but the mobility of LAMP2 was increased, indicating altered glycosylation of this protein. A similar pattern was observed in HEK293T cells, which also showed a decrease in the levels and increased electrophoretic mobility of GPP130 (Supplemental Figure S2A), TMEM165 (Supplemental Figure S2B), and TGN46 (Supplemental Figure S2C), and an increase in the mobility of LAMP2 (Supplemental Figure S2D). We concluded that the absence of GARP results in abnormal glycosylation of Golgi and lysosomal proteins, as well as reduced levels of Golgi proteins. To further elucidate if these reductions in protein levels occur only for glycosylated Golgi proteins, we examined the stability of nonglycosylated GS15/Bet1L (Figure 2E). The result showed a significant reduction in the level of GS15 in VPS53-KO and VPS54-KO RPE1 cells, while the rescued cells exhibited normal levels of GS15. In summary, GARP depletion affects the stability of multiple glycosylated and nonglycosylated Golgi proteins.
FIGURE 2:
GARP-KO alters the glycosylation of Golgi and lysosomal glycoproteins in RPE1 cells. (A-E) WB (top panel) and quantification (bottom panel) of (A) GPP130, (B) TMEM165, (C) TGN46, (D) LAMP2, and (E) GS15/Bet1L in WT, VPS53-KO, VPS54-KO, and the corresponding rescued RPE1 cells. Quantification of the blots were done by Image Studio and graphs were prepared in GraphPad Prism 8. Values represent the mean ± SD from three independent experiments. Statistical significance was calculated using one-way ANOVA. ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01.
GARP-KO affects the stability of key glycosylation enzymes in RPE1, HEK293T, and HeLa cells
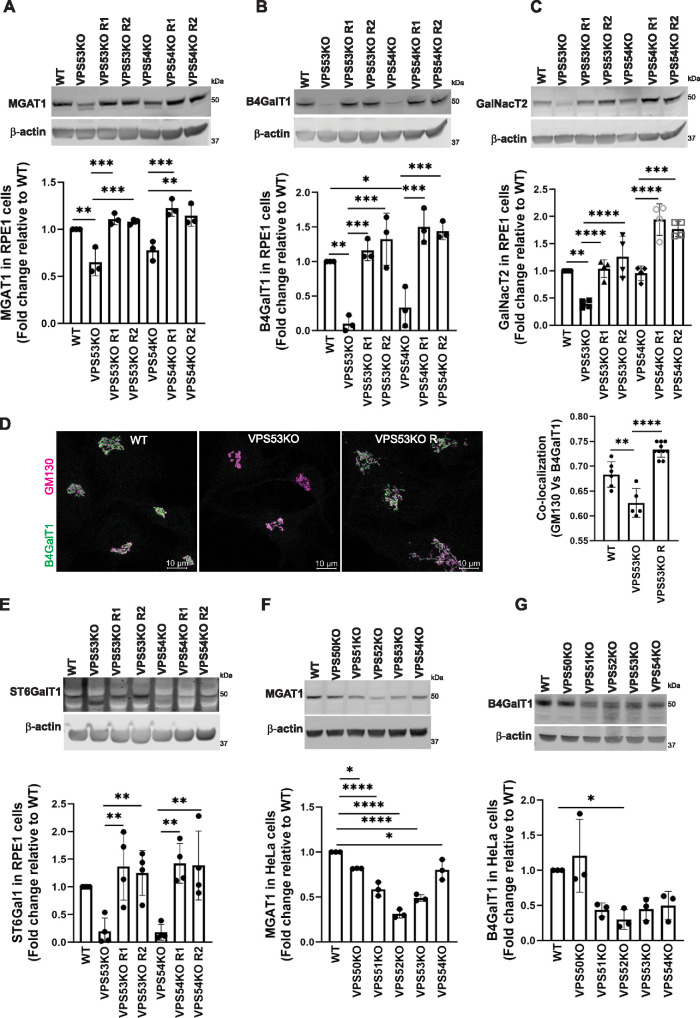
Defective Golgi glycosylation could result from malfunction, mislocalization, and/or destabilization of Golgi enzymes. Indeed, VPS53-KO and VPS54-KO cells showed a reduction in the protein levels of the key N-glycosylation enzymes alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase (MGAT1) (Figure 3A), B4GalT1 (Figure 3B), and beta-galactoside alpha-2,6-sialyltransferase 1 (ST6GalT1) (Figure 3E). We observed an additional low molecular weight band in the ST6Gal1 blot in VPS53-KO RPE1 cells but not in VPS54-KO RPE1 cells; this low molecular weight may correspond to partially degraded or under-glycosylated protein. We also assessed the localization of B4GalT1 in WT, VPS53-KO, and VPS53-KO rescued RPE1 cells by immunofluorescence microscopy (Figure 3D, left panel). We observed that the colocalization of B4GalT1 with the Golgi-marker GM130 was significantly decreased in VPS53-KO (Figure 3D, right panel), indicating that GARP deficiency affects both the stability and the localization of B4GalT1. We also examined polypeptide N-acetylgalactosaminyltransferase 2 (GalNacT2), an O-glycosylation enzyme (Lira-Navarrete et al., 2015), and observed a decrease in the levels of this enzyme in VPS53-KO and VPS54-KO cells (Figure 3C). Rescuing the VPS53-KO and VPS54-KO RPE1 cells by stable expression of the corresponding cDNAs significantly restored the levels and localization of N- and O-glycosylation Golgi enzymes (Figure 3, A–E).
FIGURE 3:
GARP-KO affects the stability of key N- and O- Golgi glycosylation enzymes in RPE1 and HeLa cells. (A–C) WB (top panel) and quantification (bottom panels) of MGAT1 (A), B4GalT1 (B), and GalNacT2 (C) in WT, VPS53-KO, VPS54-KO, and the corresponding rescued RPE1 cells. (D) WT, VPS53-KO, and VPS53-KO rescued RPE1 cells were co-stained for endogenous B4GalT1 (green) and GM130 (magenta), and images were taken (left panel). Colocalization of B4GalT1 with GM130 was determined by calculation of the Pearson’s correlation coefficient (right panel). At least 30 cells were imaged per sample for the quantification. Each dot in the bar graph (right panel) represents the colocalization of GM130 and B4GalT1 in several (1 to 10) cells imaged per field. (E) WB (top panel) and quantification (bottom panels) of ST6Gal1 in WT, VPS53-KO, VPS54-KO, and the corresponding rescued RPE1 cells. For quantification of ST6Gal1 blot, the additional low molecular weight band in VPS53-KO cells was not included. (F, G) WB (top panel) and quantification (bottom panels) of MGAT1 (F) and B4GalT1 (G) in WT, VPS50-, VPS51-, VPS52-, VPS53-, and VPS54-KO HeLa cells. Values in bar graphs represent the mean ± SD from three independent experiments. Statistical significance was calculated using one-way ANOVA. ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, *P ≤ 0.05.
In line with these findings, KO of any of the GARP subunits (VPS51, VPS52, VPS53, and VPS54) in HeLa cells (Ishida and Bonifacino, 2019) also caused a significant reduction of MGAT1 (Figure 3F) and B4GalT1 protein levels (Figure 3G). In contrast, KO of the VPS50 subunit of EARP had little or no effect on the protein levels of MGAT1 (Figure 3F) and B4GalT1 (Figure 3G), indicating that glycosylation defects are due to a malfunction of the GARP and not the EARP complex. Interestingly, GalNacT2 (Supplemental Figure S3D) and ST6Gal1 (Supplemental Figure S3E) did not show reduced stability in HeLa GARP-KO cells, possibly indicating altered Golgi physiology in cancer cells. We also observed reduced protein levels of MGAT1 (Supplemental Figure S3A) and B4GalT1 (Supplemental Figure S3B) in VPS53-KO and VPS54-KO HEK293T cells. In contrast, the protein levels of ST6Gal1 were not reduced (Supplemental Figure S3C), but its electrophoretic motility in VPS53- and VPS54-KO HEK293T cells was altered, consistent with potential defects in ST6Gal1 modification and/or with partial degradation.
The defects in Golgi enzyme stability observed in GARP-KO cells are similar to those previously described in COG-KO cells (Blackburn et al., 2018; D’Souza et al., 2019), so we tested if GARP depletion affects COG localization and/or stability. We found that this was not the case, as both localization (Supplemental Figure S4A) and levels (Supplemental Figure S4B) of COG subunits were unaltered in GARP-KO cells. We concluded that depletion of the GARP complex specifically affects the stability of key glycosylation enzymes in all tested human cell lines.
ST6Gal1 is not retained in the Golgi complex in GARP-KO cells
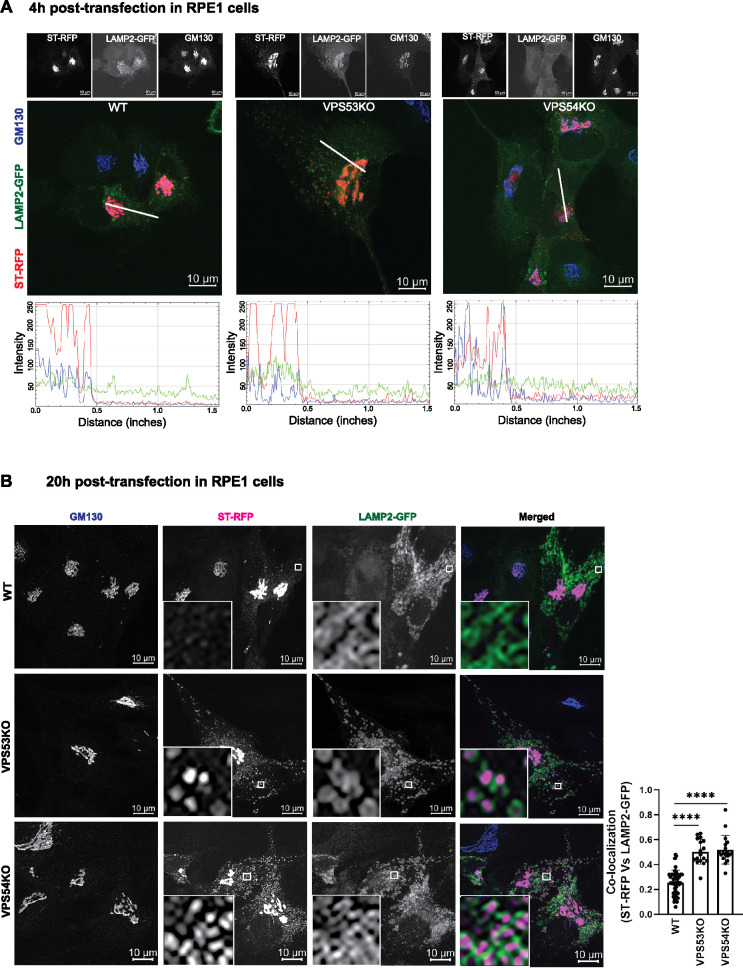
The GARP complex is localized to the TGN (Bonifacino and Hierro, 2011); therefore, we hypothesized that in GARP-deficient cells, newly synthesized glycosylation enzymes may not be efficiently retained in and/or recycled to the Golgi cisternae and may instead be targeted to endolysosomes. To test this hypothesis, we transfected WT and VPS53-KO and VPS54-KO RPE1 cells with plasmids encoding RFP-tagged ST6Gal1 (ST-RFP) and GFP-tagged LAMP2 (LAMP2-GFP) and examined the localization of both proteins relative to the Golgi marker GM130 at 4 and 20 h after transfection. As expected, 4 h was sufficient for ST-RFP to become localized to the Golgi complex in both WT and GARP-KO cells (Figure 4A). At 20 h after transfection, most of the ST-RFP remained localized to the trans-Golgi in WT cells (Figure 4B). In contrast, in VPS53-KO and VPS54-KO cells, ST-RFP exhibited partial localization to cytoplasmic puncta, the majority of which colocalized with LAMP2-GFP (Figure 4B). This result revealed that ST-RFP is not efficiently retained in the Golgi complex in GARP-KO cells but is instead partially mistargeted to the endolysosomal compartment for degradation.
FIGURE 4:
ST-RFP is not retained in the Golgi in GARP-deficient RPE1 cells. (A) WT, VPS53-KO, and VPS54-KO RPE1 cells were co-transfected with plasmids encoding ST-RFP and LAMP2-GFP for 4 h, followed by staining for the Golgi marker GM130. Microscopic images in A show individual channels in grayscale on top of each merged image. Bottom panels in A are the line scan plots of relative intensity over distance that demonstrates the overlap between the channels. Line scan analysis was done using ImageJ. (B) WT, VPS53-KO and VPS54-KO RPE1 cells were co-transfected with plasmids encoding ST-RFP and LAMP2-GFP for 20 h, followed by staining for the Golgi marker GM130. Insets are zoomed in views of the small white-boxed areas (10× inset). Colocalization of ST-RFP with LAMP2-GFP was determined by calculation of the Pearson’s correlation coefficient (right panel) in approximately 20 cells. Statistical significance was calculated using one-way ANOVA. ****P ≤ 0.0001.
RUSH assay reveals mislocalization of B4GalT1 in GARP-KO cells
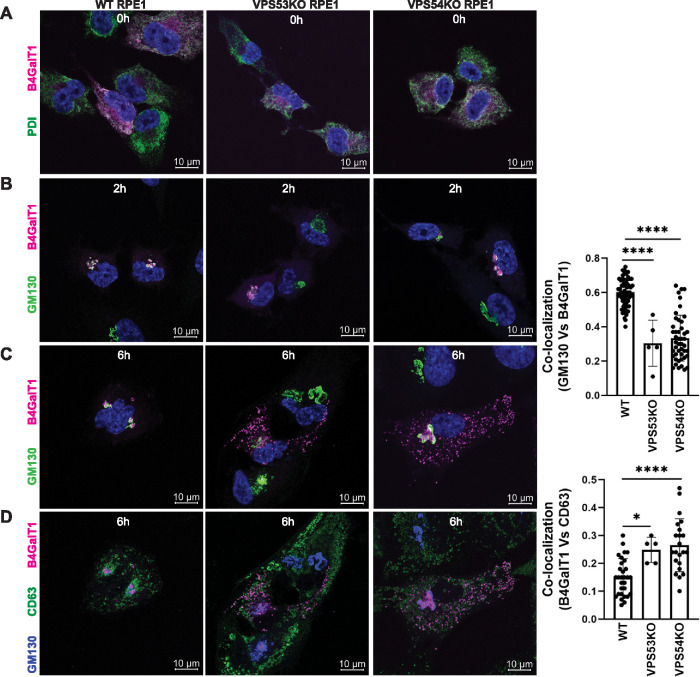
We next employed the Retention Using Selective Hooks (RUSH) system (Boncompain et al., 2012) to examine the transport of the Golgi enzyme B4GalT1 in WT and GARP-KO cells. RPE1 cells were transfected with B4GalT1 isoform 1 (Str-KDEL_flB4GalT1-SBP-mCherry) to express streptavidin-KDEL as a hook and B4GalT1-SBP-mCherry as a reporter. Following overnight incubation, biotin and cycloheximide were added to release B4GalT1-SBP-mCherry from ER retention. Samples were analyzed at 0, 2, and 6 h after ER release to compare the intracellular trafficking of B4GalT1 in WT and GARP-KO cells. Staining for the ER marker PDI (shown in green) revealed, as expected, that B4GalT1 (shown in magenta) was localized to the ER in the absence of biotin (0 h) in WT, VPS53-KO, and VPS54-KO cells (Figure 5A). Two hours after the addition of biotin, B4GalT1 moved to the perinuclear area, where it colocalized with the cis-Golgi marker GM130 (Figure 5B) in both WT and GARP-KO cells, indicating that ER-Golgi traffic is unaltered in these cells. Six hours after release from the ER (Figure 5C, left panel), B4GalT1 was localized to the Golgi area in WT cells. In contrast, VPS53-KO and VPS54-KO cells showed a dramatic decrease in B4GalT1 colocalization with GM130 (Figure 5C, right panel) and the appearance of B4GalT1 in multiple puncta that partially colocalized with the endolysosomal marker CD63 (Figure 5D). We also confirmed that the colocalization of B4GalT1 and GM130 was similar in cells with different expression levels of B4GalT1-mCherry (Supplemental Figure S5A). Similarly, B4GalT1/MAN2A RUSH assay for 1 h in HeLa WT and VPS54-KO cells showed Golgi localization of enzymes (Supplemental Figure S5B). However, 6 h after ER release, both B4GalT1-mCherry and MAN2A-GFP were mislocalized to off-Golgi puncta-like structures in VPS54-KO cells (Supplemental Figure S5C).
FIGURE 5:
RUSH assay reveals mislocalization of B4GalT1 to endolysosomes in GARP-KO RPE1 cells. RPE1 cells were transfected with plasmids encoding Str-KDEL_flB4GalT1-SBP-mCherry in biotin-free medium. Following overnight incubation, biotin (40 µM) and cycloheximide (50 µM) were added for 0, 2, and 6 h. (A) Colocalization of B4GalT1 with ER marker PDI at time 0. (B, C) Colocalization of B4GalT1 with Golgi protein GM130 at 2 h (B) and 6 h (C). The graph on the right side of C shows the quantification of B4GalT1 and GM130 colocalization at 6 h of biotin/cycloheximide addition. Values in the bar graph represent the mean ± SD of the colocalization between B4GalT1 and GM130 in approximately 40 cells in WT and VPS54-KOs. Colocalization in VPS53-KO cells was measured using each field in approximately 20 cells. Statistical significance was calculated using one-way ANOVA. ****P ≤ 0.0001. (D) Colocalization of B4GalT1, GM130 and the endolysosomal marker CD63 after addition of biotin/cycloheximide mix for 6 h. Quantification of the colocalization between CD63 and B4GalT1 was done by Pearson’s correlation coefficient analysis in approximately 25 cells. Statistical significance was calculated using one-way ANOVA. ****P ≤ 0.0001, *P ≤ 0.05.
Taken together, these results indicate that GARP is essential for proper localization of Golgi enzymes in different cell types.
RUSH assay reveals mislocalization of B4GalT1 in GARP (VPS54 KO), but not in EARP (VPS50 KO) HeLa cells
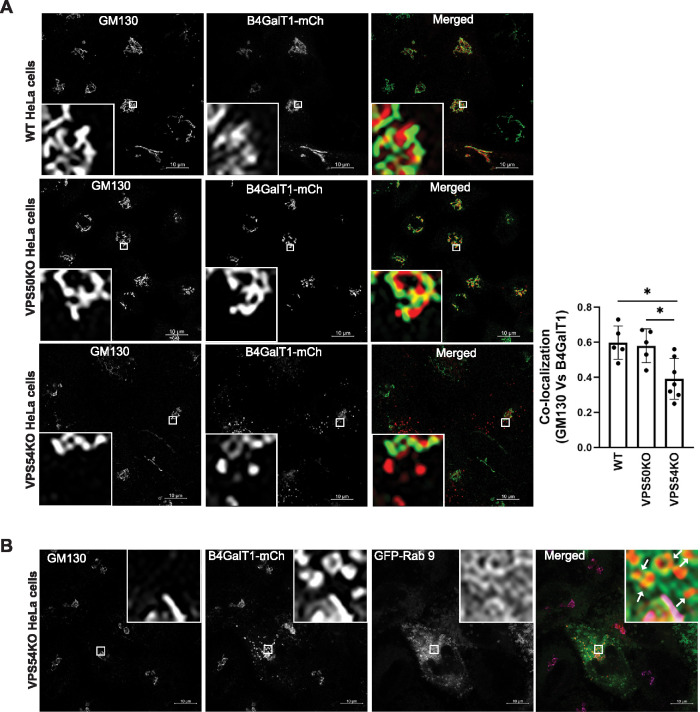
To further verify that the retrieval of Golgi enzymes is GARP but not EARP dependent, we performed a RUSH assay in HeLa cells lacking unique GARP (i.e., VPS54) and EARP (i.e., VPS50) subunits. HeLa WT, VPS50-KO, and VPS54-KO cells were transfected to express B4GalT1 isoform 1 (Str-KDEL_flB4GalT1-SBP-mCherry). Following overnight incubation in biotin-free medium, biotin (40 µM) and cycloheximide (50 µM) were added for 6 h to release B4GalT1-SBP-mCherry from ER retention and to inhibit new protein synthesis, respectively. The cells were then stained for GM130 as a marker of the Golgi complex. WT and VPS50-KO HeLa cells showed striking colocalization of GM130 and B4GalT1 in the Golgi region (Figure 6A, top and middle panels). However, in VPS54-KO HeLa cells, like in VPS54-KO RPE1 cells (Figure 5C), a fraction of B4GalT1 was distributed in punctate structures throughout the cell (Figure 6A, bottom panel). Consequently, the colocalization of B4GalT1 with GM130 was significantly decreased in VPS54-KO cells compared with WT and VPS50-KO HeLa cells (Figure 6A, right panel). To determine the identity of B4GalT1-bearing punctate structures, transient expression of different GFP-tagged endosomal Rabs (Rab7, Rab9, and Rab11) was done and the data revealed that B4GalT1 puncta are GFP-Rab9 positive (Figure 6B and unpublished data), indicating relocalization to a late endosomal compartment. Taken together, our data indicate that Golgi enzymes are not retained in the Golgi complex in GARP-deficient cells but are partially relocalized to a late endosomal compartment.
FIGURE 6:
RUSH assay reveals mislocalization of B4GalT1 in GARP- (VPS54-KOs), but not in EARP- (VPS50-KO)-deficient HeLa cells. (A) HeLa WT, VPS50-KO, and VPS54-KO cells were transfected with plasmids encoding Str-KDEL_flB4GalT1-SBP-mCherry. Following overnight incubation, the cells were chased with biotin/cycloheximide mix for 6 h. Cells were stained with GM130 as a Golgi marker. Quantification of colocalization of GM130 with B4GalT1 was done using Pearson’s correlation coefficient (right panel). Each dot on the bar graph indicates the colocalization in cells per field (bottom left corner; 10× inset). Statistical analysis was done using GraphPad Prism one way ANOVA software in 30 WT, VPS50-KO, and VPS54-KO cells. *P ≤ 0.05. (B) HeLa VPS54-KO cells were cotransfected with plasmids encoding Str-KDEL_flB4GalT1-SBP-mCherry and GFP-Rab 9. After overnight incubation, the cells were chased with biotin/cycloheximide mix for 6 h. Cells were stained with GM130 as a Golgi marker. Insets with white arrows indicate the endocytic GFP-Rab9-positive vesicles filled with B4GalT1-mCherry in VPS54-KO cells (top right corner; 10× inset).
Endogenous B4GalT1 can recycle via an endosomal compartment
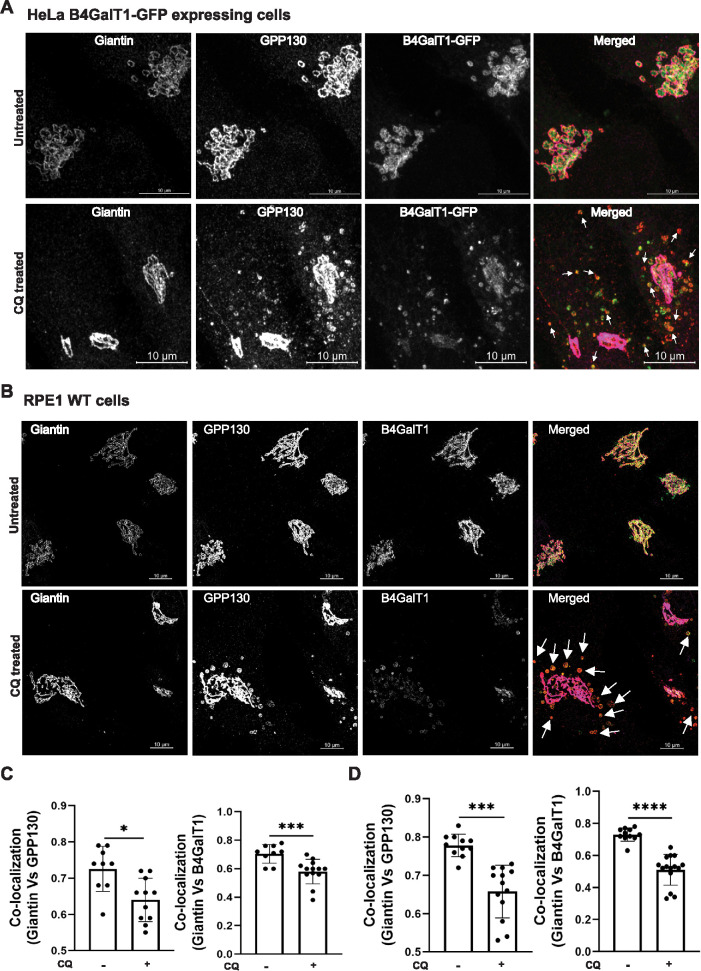
Several proteins, including GPP130, TGN46, and MPR, are known to cycle between endosomes and the Golgi complex in a manner dependent on the GARP complex (Pérez-Victoria et al., 2008). To test if Golgi enzymes can cycle via an endosomal compartment, we employed a chloroquine (CQ) treatment procedure. CQ prevents the acidification of endolysosomal compartments and blocks transport out of late endosomes. The cis–Golgi-resident protein GPP130 was used as a positive control, as it actively redistributes to endosomes after CQ treatment (Linstedt et al., 1997). This experiment was performed in HeLa B4GalT1-GFP and RPE1 cells. In both cell lines, giantin colocalizes with B4GalT1 and GPP130 in control conditions (Figure 7, A and B, top panels). Following 3 h of CQ treatment, a fraction of the B4GalT1 colocalized with GPP130 in endosomal punctate structures away from the Golgi complex (Figure 7, A and B, bottom panels). Colocalization of giantin with GPP130 (Figure 7C, left panel) or B4GalT1 (Figure 7C, right panel) in HeLa cells was significantly decreased in CQ-treated cells, consistent with the decrease in colocalization between giantin with GPP130 (Figure 7D, left panel) or B4GalT1 (Figure 7D, right panel) in RPE1 cells.
FIGURE 7:
B4GalT1 is efficiently relocalized to an endosomal compartment in response to CQ treatment. (A) HeLa cells expressing endogenously tagged B4GalT1-GFP were left untreated or treated for 3 h with 0.1 mM CQ and stained for giantin and GPP130. (B) RPE1 WT cells were left untreated or treated for 3 h with 0.1 mM CQ and stained for giantin, GPP130, and B4GalT1. Arrows in the merged image indicate putative endocytic vesicles. (C) Quantification of colocalization of GPP130 (left panel) and B4GalT1-GFP (right panel) with giantin in 40 cells using Pearson’s correlation coefficient (HeLa cells). (D) Quantification of colocalization of GPP130 (left panel) and B4GalT1 (right panel) with giantin in 40 cells using Pearson’s correlation coefficient (RPE1 cells). Statistical analysis was done using GraphPad Prism (paired t test). Each dot on the bar graph indicates the colocalization in cells per field. ****P ≤ 0.0001, ***P ≤ 0.001, *P ≤ 0.05.
To compare B4GalT1 and GPP130 recycling rates, we shortened CQ treatment to 90 min (Supplemental Figure S6, A and B). The short CQ treatment was sufficient to mislocalize a significant fraction of B4GalT1 away from the Golgi (Supplemental Figure S6, C, D, left panel), while GPP130 relocation from the Golgi was visible, but not statistically significant (Supplemental Figure S6, C, D, right panel), indicating that B4GalT1 Golgi-endosomal cycling rates are faster or similar to the cycling rates of GPP130.
Furthermore, a 3 h washout after CQ treatment resulted in relocalization of both B4GalT1 and GPP130 back to the Golgi (Supplemental Figure S7, A and B, bottom panels). As a result, colocalization of giantin with B4GalT1 after CQ washout was significantly restored both in HeLa (Supplemental Figure S7C) and in RPE1 (Supplemental Figure S7D) cells. These data indicate that the intracellular trafficking of B4GalT1 is similar to that of GPP130 and that a trans-Golgi enzyme can be relocalized to the endosomal compartment and possesses information for its return to the Golgi.
DISCUSSION
In this study, we have demonstrated that the GARP complex is important for the proper modification of oligosaccharide chains of glycoproteins in the Golgi complex. Using multiple cell lines, we discovered that complete depletion of two GARP subunits (VPS53 and VPS54) is detrimental for the modification of both N- and O-linked oligosaccharides and the stability of glycoproteins and components of the Golgi glycosylation machinery.
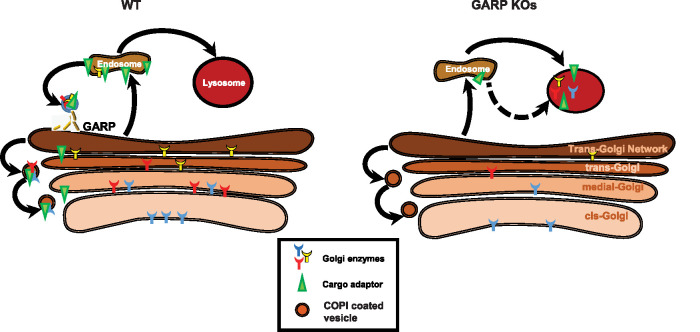
In addition to the Golgi glycosylation defect, our results demonstrated a reduction in the levels of several Golgi-glycosylated and -nonglycosylated proteins and an increase in electrophoretic mobility of LAMP2 in GARP-KO cells. Two of the tested glycoproteins (TGN46 and GPP130) are known to cycle via endosomal compartments (Natarajan and Linstedt, 2004; Pérez-Victoria et al., 2008) and their retrieval to the Golgi complex is likely enabled by the GARP complex. We also observed alterations in the localization and stability of multiple Golgi glycosyltransferases in GARP-KO cells. On the basis of these results, we propose that glycosylation defects in GARP-KO cells are directly linked to altered stability of the glycosylation machinery due to mislocalization of Golgi glycosyltransferases (Figure 8). The stability and localization of Golgi enzymes was unaltered in VPS50-KO cells, indicating that only GARP (VPS51-54), and not EARP (VPS50-53), is needed for the maintenance of the Golgi glycosylation machinery. There might be several reasons for the reduction in stability of Golgi-resident glycosyltransferases in GARP-KO cells. One reason might be a failure in the retrieval of glycosyltransferases to their corresponding Golgi cisternae. As a consequence, these Golgi enzymes would be unable to maintain their proper steady-state localization and meet their substrates. This would be similar to COG subunit mutants, which exhibit a reduction in the Golgi localization of B4GalT1 and MGAT1 (Foulquier et al., 2006; Blackburn et al., 2018). Another reason might be the mislocalization of Golgi-resident enzymes away from the Golgi to other organelles. This would be similar to observations in COG4-KO cells, which showed mislocalization of the Golgi enzyme ST6Gal1 tagged with RFP to enlarged endolysosomes (EELs), indicating that these Golgi-resident enzymes can be degraded in EELs (D’Souza et al., 2019). Our results showed increased colocalization of ST6Gal1 with LAMP2 (Figure 4B) and B4GalT1 with CD63 and GFP-Rab9 (Figures 5D and 6B). Glycosylation abnormalities were previously described in cells depleted for another Golgi vesicle-tethering complex, COG (Pokrovskaya et al., 2011), and GARP and COG were shown to functionally interact via VPS51 (Luo et al., 2011) and a STX16-containing SNARE complex (Pérez-Victoria and Bonifacino, 2009; Willett et al., 2013). Therefore, one possibility was that GARP depletion affects COG complex function. However, both the localization and the stability of COG complex subunits were unaltered in GARP-KO cells, indicating that the glycosylation defects observed in this study are directly related to GARP. Side-by-side comparison of glycosylation defects in HEK293T and RPE1 cells depleted for multiple COG and GARP subunits showed that stability of the tested Golgi enzymes was altered to a comparable degree, but the N- and O-glycosylation defects were less severe in GARP-KO cells (A.K., T.K, V.V.L., unpublished data), indicating that COG deficiency affects either a wider range or a different subset of Golgi glycosylation machinery.
FIGURE 8:
Proposed model of altered Golgi trafficking in GARP-KO cells. The GARP complex plays a key role in recycling of Golgi enzymes and/or enzyme adaptors from endosomes to the Golgi complex. In WT cells (left), the Golgi enzymes that modify newly synthesized glycoproteins are constantly recycled back to their corresponding Golgi compartments, allowing them to maintain their proper steady-state localization. In contrast, in GARP-KO cells (right) Golgi enzymes and/or Golgi enzyme adaptors fail to recycle back to the Golgi complex and are instead delivered to lysosomes for degradation. Consequently, there are decreased levels of enzymes in the Golgi and carbohydrate modification of glycoproteins is hindered.
The effect of GARP depletion on the Golgi glycosylation machinery was unexpected, since current models for trafficking and retention of Golgi enzymes include only intra-Golgi and Golgi-ER cycling pathways (Storrie et al., 1998; Pérez-Victoria et al., 2008; Martínez-Menárguez, 2013). The GARP complex is predicted to work as a vesicular tether for docking and fusion at the trans-Golgi/TGN compartment (Pérez-Victoria and Bonifacino, 2009). If this prediction is correct, one would expect accumulation of nontethered vesicles in cells deficient for GARP complex subunits. Our results did not reveal accumulation of endogenous or newly synthesized Golgi enzymes in vesicle-like structures. The likely explanation for the lack of Golgi enzyme-carrying GARP-dependent vesicles is the use of cells completely lacking GARP function (GARP KO cells). Our previously published work (Willett et al., 2014) that investigated cells depleted for another Golgi vesicular tether, COG complex, demonstrated that CCD (COG-complex dependent) vesicle accumulation occurred transiently only after the acute (2–6 d) but not prolonged (9 d) depletion of COG complex subunits, indicating a transient nature of vesicle accumulation.
Golgi-resident proteins are concentrated in “proper” Golgi subcompartments by a combination of retention and retrieval mechanisms (Banfield, 2011), but for the majority of Golgi enzymes the localization mechanisms are still enigmatic. One model proposed that the thickness of the lipid bilayer is important for the proper localization and retention of transmembrane proteins (Welch and Munro, 2019). Other models rely on efficient intra-Golgi and Golgi-ER recycling (Mironov and Beznoussenko, 2012; Sengupta et al., 2015). Recycling is achieved by packaging of Golgi-resident proteins into COPI vesicles (Gaynor et al., 1998), although COPI-independent Golgi-ER recycling was also described (Storrie et al., 2000). A small subset of enzymes depends on direct interaction with COPI (Liu et al., 2018), while another subset uses the GOLPH3 adaptor for interaction with COPI (Eckert et al., 2014). There are multiple scenarios for how GARP deficiency may affect the maintenance of Golgi enzymes. In the “indirect” models, GARP deficiency may affect the thickness of TGN membranes (by somehow altering lipid balance in this compartment), Golgi pH, ion homeostasis, or proper retrieval of enzyme adaptors. Our preliminary analysis indicates that the cholesterol content of the Golgi and localization of COPI coat are not severely altered in GARP-KO cells and that glycosylation defects observed in GOLPH3/GOLPH3L double-KO cells are less severe than in GARP-depleted cells (T.K., V.V.L. unpublished observation). In the “direct “model, GARP deficiency may affect recycling of a subset Golgi enzymes and enzyme receptors via the endosomal compartment (Figure 8). One possibility is that Golgi enzymes could bind and follow secretory proteins during the glycosylation process and, therefore, have to be retrieved from post-Golgi compartments. Another possibility is that the Golgi enzymes pass the endosomes as a part of their quality control mechanisms.
It is important to note that GARP deficiency affects not only enzymes located at the trans-Golgi (B4GalT1 and ST6Gal1) but also enzymes located in earlier Golgi compartments (MGAT1, MAN2A, and GalNacT2). The loss of MGAT1 activity in GARP-deficient cells is also supported by GNL binding data (Figure 1C). GNL binds to immature terminal mannose residues in N-glycans, and MGAT1 activity is primarily responsible for the addition of GlcNAc to the growing N-glycan chain (Chen and Stanley, 2003). The lack of MGAT1 activity in both VPS54-KO and VPS53-KO cells results in higher binding of GNL-647 to the plasma membrane of GARP-deficient cells. Although both MGAT1 and B4GalT1 were found in COPI vesicles, these enzymes do not bind the COPI coat directly and the nature of a possible adaptor is still unknown.
Interestingly, live-cell imaging of WT HeLa cells co-expressing endogenously tagged B4GalT1-GFP and exogenous ST-RFP revealed that both enzymes appeared not only in the Golgi cisternae and multiple vesicular structures, but also in the tubular sorting/late endosomelike compartment (Supplemental Movie S1), indicating that this compartment could be a part of the normal trafficking itinerary for Golgi enzymes, and that defects in GARP-mediated retrieval from this compartment may cause mislocalization and degradation of the enzymes, causing numerous glycosylation defects (Figure 8). Importantly, endogenous B4GalT1 can be redistributed to an endosomal compartment in CQ-treated cells and returned back to the Golgi after CQ washout, indicating that at least some Golgi enzymes can travel to endosomal compartments and possess information for their return to the Golgi complex, likely in a GARP-dependent manner. Our result is in agreement with an earlier study where exposure of cells to CQ for 24 h resulted in mislocalization of B4GalT1 (Rivinoja et al., 2009). While we propose that GARP regulates stability and proper localization of Golgi enzymes by recycling Golgi enzymes or associated adaptors from endosomes to the Golgi complex, another recent study indicates that four Golgi enzymes (MGAT2, B4GalT7, B3GalT6, and POMGNT1) are retrieved back to the Golgi complex from the plasma membrane (Sun et al., 2021). These studies and our data indicate that a subset of Golgi enzymes possess signal(s) for their recycling from post-Golgi compartments. Future experiments should test if the GARP complex could directly tether vesicles that recycle Golgi enzymes.
Movie S1.
Two-color super-resolution Airyscan live cell imaging of cells expressing B4GalT1-GFP and ST-RFP. HeLa cells were CRISPR knock-in with GFP to express endogenous B4GalT1-GFP and were transiently transfected with plasmid expressing ST-RFP. Live cell imaging of cells was done for 7 minutes with time interval of 3 seconds. Time lapse corresponds to Figure 8. Note the appearance of both enzymes in the Golgi membranes, rapidly moving vesicles and endosome-like tubular compartment. The time stamp represents seconds.
Our findings may be relevant to the pathogenesis of GARP-deficiency syndrome in humans (Feinstein et al., 2014; Hady-Cohen et al., 2018; Gershlick et al., 2019; Uwineza et al., 2019). Indeed, a patient with a neurodevelopmental disorder caused by mutations in the VPS51 subunit of GARP and EARP was shown to exhibit abnormal glycosylation of cellular and serum glycoproteins, reminiscent of congenital disorders of glycosylation (Gershlick et al., 2019). These defects could result from the loss of glycosylation enzymes from the Golgi complex described here, thus shedding light on potential mechanisms for this syndrome.
In summary, we have demonstrated that the GARP-KO human cells have defects in Golgi processing of N- and O-linked oligosaccharides. Moreover, GARP-KO cells are unable to retain the tested Golgi glycosylation enzymes. Hence, the GARP complex is a new component of the cellular machinery that regulates the proper localization and abundance of Golgi enzymes.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Cell culture
hTERT RPE1 and HEK293T cells used for all experiments were purchased from ATCC. HeLa-KO used in the study were previously described (Ishida and Bonifacino, 2019). RPE1, HEK293T, and HeLa cells were cultured in DMEM containing Nutrient mixture F-12 (Corning) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher). Cells were incubated in a 37°C incubator with 5% CO2 and 90% humidity.
Creation of VPS53- and VPS54-KO stable cell lines
To create RPE1 VPS53 and VPS54 stable KOs, dual gRNAs were purchased from Transomic with the following target sequences:
For VPS53-KO
transEDIT-dual CRISPR for VPS53 (TEDH-1088057)
grna-a: GCATTGAAATCTGCTCGATCTAGAGGGTCC
grna-b: TGACAATATTCGAACTGTTGTAAGAGGTCA
transEDIT-dual CRISPR for VPS53 (TEDH-1088055)
grna-a: ATGCACCACTCACGTGGAAACATGCGGCCG
grna-b: CTGGAGCACTTCCACAAGTATATGGGGATT
transEDIT-dual CRISPR for VPS53 (TEDH- 1088056)
grna-a: CCAAGATTATGCGTACCAGAGCGAAGGAAA
grna-b: CCGCAGATCCGGCAGCTTTCCGAAAGGTAA
For VPS54-KO
transEDIT-dual CRISPR for VPS54 (TEDHG1001)
grna-a: ACAAATATTCCTGAAACAGGCAGAAGGAAC
grna-b: ATCTAGAAAGTGTTATGAATTCCATGGAAT
transEDIT-dual CRISPR for VPS54 (TEDHG1001)
grna-a: CAAAAGATAATTCACTGGACACAGAGGTGG
grna-b: CATTCTACCTCCCACAGATCAGCAAGGAAC
transEDIT-dual CRISPR for VPS54 (TEDHG1001)
grna-a: CTTAACTCTGTAGCCACAGAAGAAAGGAAA
grna-b: GTAAGCATGTCAGTAGTAACAGATGGGATG
An RPE1-Cas9 stable cell line was created by lentiviral transduction with a plasmid encoding FLAG-tagged Cas9. HEK293FT cells were used for the generation of lentiviral particles. Equal amounts of the three lentiviral packaging plasmids pMD2.G, pRSV-Rev, and pMDLg/pRRE, plus destination plasmid pLenti Cas9-Blast, were used. Briefly, transfected HEK293FT cells were placed in serum-reduced Opti-MEM with 25 μm CQ and GlutaMAX. The next day, the medium was changed to Opti-MEM supplemented with GlutaMAX. At 72 h after transfection, the medium was collected, and cell debris was removed by centrifugation at 600 × g for 10 min. The supernatant was passed through a 0.45-μm polyethersulfone membrane filter and lentiviral medium was stored at 4°C overnight; 1 ml of this lentiviral medium was used to transduce RPE1 cells on a 10-cm dish along with 50 mM sodium butyrate. After 24 h, the medium was changed to DMEM/F12 supplemented with 10% FBS and 10 μg/ml blasticidin.
After the creation of RPE1-Cas9 stable cell line, these cells were transfected with a cocktail of three dual gRNAs specific for VPS53 and VPS54. Neon electroporation (Thermo Fisher) was used for transfecting cells according to the manufacturer’s protocol. TransEDIT-dual CRISPR plasmids express GFP; therefore, the efficiency of transfection was estimated by counting GFP-positive cells. At 48 h post-transfection, cells were spun down at 600 × g and resuspended in cell-sorting medium (phosphate-buffered saline [PBS], 25 mM HEPES, pH 7.0, 2% FBS [heat-inactivated], 1 mM EDTA. 0.2 µm sterile-filtered). Cell sorting was based on high-GFP-Alexa Flour 488 fluorescence; cells were sorted into a 96-well plate containing culture medium using a BD FACS Aria IIIu cell sorter.
At 10–14 d after sorting, the 96-well plates were examined for colonies. Wells with colonies were marked and allowed to grow for 1 wk before expanding. After 14 d, colonies were expanded from 96-well to 12-well plates using trypsin detachment. Cells were maintained in DMEM/F12 medium supplemented with 10% FBS and allowed to grow further. KO of VPS53 and VPS54 was confirmed by genome sequencing and Western blot (WB) analysis for absence of the targeted protein. All the antibodies used for the western blot analysis in the study are listed in Table 1.
TABLE 1:
List of antibodies used.
| Antibody | Source/Catalog # | Species | WB dilution | IF dilution |
|---|---|---|---|---|
| COG3 | Lupashin lab | Mouse | 1:1000 | 1:1000 |
| COG8 | Sigma SAB4200427 | Rabbit | 1:1000 | 1:500 |
| Giantin | Covance PRB-114C | Rabbit | – | 1:1000 |
| GM130 | BD Biosciences, 610823 | Mouse | – | 1:500 |
| GM130 | CalBiochem, CB1008 | Rabbit | – | 1:300 |
| β-actin | Sigma, A5441 | Mouse | 1:1000 | – |
| TGN46 | Bio-Rad, AHP500G | Sheep | 1:2000 | – |
| CD63 | DSHB, H5C6-C | Mouse | – | 1:200 |
| PDI | Affinity Bio Reagents, MA3-019 | Rabbit | – | 1:1000 |
| LAMP2 | DSHB, H4B4 | Mouse | 1:500 | – |
| Cathepsin D | Sigma, C0715 | Mouse | 1:500 | – |
| TMEM165 | Sigma, HPA038299 | Rabbit | 1:500 | – |
| MGAT1 | Abcam, ab180578 | Rabbit | 1:500 | – |
| B4GalT1 | R&D Systems, AF-3609 | Goat | 1:500 | 1:300 |
| ST6Gal1 | R&D Systems, AF-5924 | Goat | 1:500 | – |
| GPP130 | Covance, PRB-144C | Rabbit | 1:1000 | – |
| VPS53 | Thermo Fisher, PA520548 | Rabbit | 1:1000 | – |
| VPS54 | St John’s lab, STJ115181 | Rabbit | 1:1000 | – |
| GALNT2 (GalNacT2) | Thermo Fisher, PA521541 | Rabbit | 1:1000 | – |
| IRDye 680 anti-Mouse | LiCOR/926-68170 | Goat | 1:40000 | – |
| IRDye 800 anti-Rabbit | LiCOR/926-32211 | Goat | 1:40000 | – |
| IRDye 800 anti-Goat | LiCOR/926-32214 | Donkey | 1:40000 | – |
| Alexa Fluor 647 anti-Rabbit | Jackson Immuno Research/711-605-152 | Donkey | 1:500 | 1:1000 |
| Alexa Fluor 647 anti-Mouse | Jackson Immuno Research/715-605-151 | Donkey | 1:500 | 1:1000 |
| Alexa Fluor 647 anti-Goat | Jackson Immuno Research/ 705-605-147 | Donkey | – | 1:1000 |
| DyLight 647 anti-Sheep | Jackson Immuno Research/713-605-147 | Donkey | – | 1:1000 |
| anti-Rabbit Cy3 | Jackson Immuno Research/711-165-152 | Donkey | – | 1:1000 |
| anti-Mouse Cy3 | Jackson Immuno Research/715-165-151 | Donkey | 1:500 | 1:1000 |
| Alexa Fluor 488 anti-Rabbit | Jackson Immuno Research/711-545-152 | Donkey | 1:500 | 1:1000 |
| Alexa Fluor 488 anti-Mouse | Jackson Immuno Research/715-545-151 | Donkey | 1:500 | 1:1000 |
Plasmid preparation, generation of lentiviral particles, and stable cell lines
All constructs were generated using standard molecular biology techniques and are listed in Table 2.
TABLE 2:
List of plasmids used.
| Plasmid name | Source | Citation |
|---|---|---|
| Lenti Cas-9 blast | Feng Zhang, Addgene #52962 | Sanjana et al., 2014 |
| hVPS53-GFP | Origene | |
| pCl-neo-VPS54-13myc | Juan Bonifacino | Ishida and Bonifacino, 2019 |
| pENTR1A no ccDB (w48-1) | Eric Campeau and Paul Kaufman, Addgene #17398 | Campeau et al., 2009 |
| pLenti CMV Neo DEST (705-1) | Eric Campeau and Paul Kaufman, Addgene #17392 | Campeau et al., 2009 |
| pMD2.G | Didier Trono, Addgene #12259 | Dull et al., 1998 |
| pRSV-Rev | Didier Trono, Addgene #12253 | Dull et al., 1998 |
| pMDLg/pRRE | Didier Trono, Addgene #12251 | Dull et al., 1998 |
| hVps53-GFP pLenti COG4 promoter Neo DEST | This study | This study |
| mVps54-13myc pLenti COG4 promoter Neo DEST | This study | This study |
| pLenti COG4 promoter Neo DEST (705-1) | This study | This study |
| LAMP2-GFP | Santiago Di Pietro | Ambrosio et al., 2012 |
| ST-RFP | James Rothman | Lavieu et al., 2013 |
| Str-KDEL_flB4GalT1-SBP-mCherry | Franck Perez, Addgene #65273 | Boncompain et al., 2012 |
To generate pLenti COG4 Neo DEST construct, a chromosomal DNA fragment encoding the COG4 promoter region was amplified from human genomic DNA by PCR using the following forward and reverse primers:
Forward: GCTTATCGATTTCCCCCACGTCTGTTTACCA
Reverse: GAATTCTAGACTTGGTCCCCATTCGGCACTT
The amplified fragment was cloned into pLenti CMV Neo DEST (705-1) using XbalI and ClaI as restriction sites. Lentiviral particles were generated using the HEK293FT cell line as described above.
To generate lentiviruses encoding GARP subunits, hVPS53 (GFP-tagged), transcript variant 1 (NM_001128159) purchased from Origene or mVps54-13myc (Ishida and Bonifacino, 2019) were subcloned into the pENTRA 1A no ccDB (w48-1) entry vector and recombined into pLenti COG4 promoter Neo DEST (705-1) destination vector using Gateway LR Clonase II Enzyme Mix (Thermo Fisher) according to the manufacturer’s instructions.
Lentiviral medium (100 µl) with polybrene (10 µg /ml) was used to transduce RPE1 cell lines in a 6-well dish. At 24 h after transduction, the medium was replaced with 10% FBS without antibiotic in DMEM/F12. At 48 h after transduction, the medium was replaced with 600 µg/ml G418 selection medium and incubated for 72 h. Then, cells were single-sorted into a 96-well plate to obtain clonal populations. At 10–14 d after sorting, the 96-well plates were examined for colonies. Wells with colonies were marked and allowed to grow for 1 wk more before expanding. Cells were maintained in DMEM/F12 medium with 10% FBS and allowed to grow further. Once colonies were split onto 10-cm dishes, aliquots were cryopreserved in freezing medium (90% FBS plus 10% DMSO).
Preparation of cell lysates and Western blot analysis
For preparation of cell lysates, cells grown on tissue culture dishes were washed twice with PBS and lysed in 2% SDS that was heated for 5 min at 70°C. Total protein concentration in the cell lysates was measured using the BCA protein assay (Pierce). The protein samples were prepared in 6× SDS sample buffer containing beta-mercaptoethanol and denatured by incubation at 70°C for 10 min; 10–30 µg of protein samples were loaded onto Bio-Rad (4–15%) gradient gels or Genescript (8–16%) gradient gels. Gels were transferred onto nitrocellulose membranes using the Thermo Scientific Pierce G2 Fast Blotter. Membranes were rinsed in PBS, blocked in Odyssey blocking buffer (LI-COR) for 20 min, and incubated with primary antibodies overnight at 4°C. Membranes were washed with PBS and incubated with secondary fluorescently tagged antibodies diluted in Odyssey blocking buffer for 60 min. All the primary and secondary antibodies used in the study are listed in Table 1. Blots were then washed and imaged using the Odyssey Imaging System. Images were processed using the LI-COR Image Studio software.
Lectin blotting
To perform blots with fluorescent lectins, 10 µg of cell lysates were loaded onto Bio-Rad (4–15%) gradient gels and run at 160V. Next, proteins were transferred to nitrocellulose membrane using the Thermo Scientific Pierce G2 Fast Blotter. The nitrocellulose membrane was blocked with 3% bovine serum albumin (BSA) for 30 min. The lectins HPA or GNL conjugated to Alexa 647 fluorophore were diluted 1:1000 in 3% BSA from their stock concentration of 1 and 5 µg/µl, respectively. Blots were incubated with lectin solutions for 30 min and then washed in PBS four times for 4 min each and imaged using the Odyssey Imaging System.
Secretion assay
Cells were plated in three 6-cm dishes and grown to 90–100% confluency. Cells were then rinsed 3× with PBS and placed in 2 ml serum-free, chemically defined medium (BioWhittaker Pro293a-CDM, Lonza) with 1× GlutaMAX (100× stock, Life Technologies) added per well. After 36 h, the collected medium was spun down at 3000 × g to remove floating cells. The supernatant was concentrated using a 10k concentrator (Amicon Ultra 10k, Millipore); final concentration was 24× that of cell lysates.
Immunofluorescence microscopy
Cells were plated on glass coverslips to 80–90% confluency and fixed with 4% paraformaldehyde (PFA) (freshly made from 16% stock solution) in PBS for 15 min at room temperature. Cells were then permeabilized with 0.1% Triton X-100 for 1 min followed by treatment with 50 mM ammonium chloride for 5 min, treated with 6 M urea for 2 min (only for COG3 staining), and washed with PBS. After washing and blocking twice with 1% BSA, 0.1% saponin in PBS for 10 min, cells were incubated with primary antibody (diluted in 1% cold fish gelatin, 0.1% saponin in PBS) for 40 min, washed, and incubated with fluorescently conjugated secondary antibodies for 30 min. Cells were washed four times with PBS, then coverslips were dipped in PBS and water 10 times each and mounted on glass microscope slides using Prolong Gold antifade reagent (Life Technologies). Cells were imaged with a 63× oil 1.4 NA objective of a LSM880 Zeiss Laser inverted microscope and Airyscan superresolution microscope using ZEN software. Quantitative analysis was performed using single-slice confocal images. All the microscopic images shown are Z-stacked maximum intensity projection images.
RUSH assay
The RUSH construct Str-KDEL_flB4GalT1-SBP-mCherry (isoform 1 of B4GalT1) was transfected into cells using an electroporation-based system or Lipofectamine 3000 reagent protocol. Cells were plated on glass coverslips in the presence of biotin-free medium containing avidin (100 μg/ml) to prevent biotin in the medium from interfering with the RUSH reporter. At 16 h post-transfection, the medium was changed with biotin-free medium supplemented with biotin (40 μm) and cycloheximide (50 μm) for 2 and 6 h, respectively, followed by fixing of cells with 4% PFA in PBS. Cells were stained for the ER maker PDI, Golgi marker GM130, endolysosomal marker CD63, and microscopic images of cells with moderate expression levels of FP-tagged proteins were taken.
Statistical analysis
All results are representative of at least three independent experiments. WB images are representative from three repeats. WBs were quantified by densitometry using the LI-COR Image Studio software. Error bars for all graphs represent SD. Statistical analysis was done using one-way ANOVA, two-way ANOVA, or paired t test using GraphPad Prism software.
Supplementary Material
Acknowledgments
We are thankful to Eric Campeau, Rainer Duden, Wei Guo, Paul Kaufman, Taroh Kinoshita, James Rothman, Frank Perez, Santiago M. Di Pietro, Didier Trono, and others who provided reagents and cell lines. We also thank the UAMS sequencing, flow cytometry and digital microscopy core facilities for the use of their equipment and expertise. We are grateful to all members of Lupashin’s lab for comments on the paper. This work was supported by the National Institutes of Health (R01GM083144), by UAMS Easy Win Early Victory grant program (V L), and by the Intramural Program of NICHD (ZIA HD001607).
Abbreviations used:
- B4GalT1
beta-1,4-galactosyltransferase 1
- BSA
bovine serum albumin
- COG
conserved oligomeric golgi
- CQ
chloroquine
- EEL
enlarged endolysosome
- FBS
fetal bovine serum
- GalNacT2
N-acetyl galactosaminyltransferase 2
- GARP
Golgi-associated retrograde protein
- GNL
Galanthus nivalis lectin
- KO
knockout
- LAMP2-GFP
GFP-tagged LAMP2
- MGAT1
alpha-1,3-mannosyl-glycoprotein 2-beta-n-acetylglucosaminyltransferase
- PBS
phosphate-buffered saline
- PFA
paraformaldehyde
- RPE1
retinal pigment epithelial
- RUSH
Retention Using Selective Hooks
- ST-RFP
RFP-tagged ST6Gal1
- TGN
trans-Golgi network
- VPS
vacuolar protein sorting
- WB
Western blot
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E21-04-0169) on June 23, 2021.
REFERENCES
- Ambrosio AL, Boyle JA, Di Pietro SM (2012). Mechanism of platelet dense granule biogenesis: study of cargo transport and function of Rab32 and Rab38 in a model system. Blood 120, 4072–4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey Blackburn J, Pokrovskaya I, Fisher P, Ungar D, Lupashin VV (2016). COG complex complexities: detailed characterization of a complete set of HEK293T cells lacking individual COG subunits. Front Cell Dev Biol 4, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banfield DK (2011). Mechanisms of protein retention in the Golgi. Cold Spring Harb Perspect Biol 3, a005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn JB, Kudlyk T, Pokrovskaya I, Lupashin VV (2018). More than just sugars: Conserved oligomeric Golgi complex deficiency causes glycosylation-independent cellular defects. Traffic 19, 463–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boncompain G, Divoux S, Gareil N, De Forges H, Lescure A, Latreche L, Mercanti V, Jollivet F, Raposo G, Perez F (2012). Synchronization of secretory protein traffic in populations of cells. Nature Methods 9, 493–498. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Hierro A (2011). Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol 21, 159–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks SA (2000). The involvement of Helix pomatia lectin (HPA) binding N-acetylgalactosamine glycans in cancer progression. Histol Histopathol 15, 143–158. [DOI] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD (2009). A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One 4, e6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Stanley P (2003). Five Lec1 CHO cell mutants have distinct Mgat1 gene mutations that encode truncated N-acetylglucosaminyltransferase I. Glycobiology 13, 43–50. [DOI] [PubMed] [Google Scholar]
- Conibear E, Cleck JN, Stevens TH (2003). Vps51p mediates the association of the GARP (Vps52/53/54) complex with the late Golgi t-SNARE Tlg1p. Mol Biol Cell 14, 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruz MD, Kim K (2019). The inner workings of intracellular heterotypic and homotypic membrane fusion mechanisms. J Biosci 44, 91. [PubMed] [Google Scholar]
- D’Souza Z, Taher FS, Lupashin VV (2020). Golgi inCOGnito: From vesicle tethering to human disease. Biochim Biophys Acta Gen Subj 1864, 129694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza Z, Blackburn JB, Kudlyk T, Pokrovskaya I, Lupashin V. (2019). Defects in COG-mediated Golgi trafficking alter endo-lysosomal system in human cells. Front Cell Dev Biol 7, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Matteis MA, Corda D, Luini A (2019). The Golgi complex: 120 years and it doesn’t show. FEBS Lett 593, 2277–2279. [DOI] [PubMed] [Google Scholar]
- Dotiwala F, Eapen VV, Harrison JC, Arbel-Eden A, Ranade V, Yoshida S, Haber JE (2013). DNA damage checkpoint triggers autophagy to regulate the initiation of anaphase. Proc Natl Acad Sci USA 110, E41–E49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dull T, Zufferey R, Kelly M, Mandel R, Nguyen M, Trono D, Naldini L (1998). A third-generation lentivirus vector with a conditional packaging system. J Virol 72, 8463–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert ES, Reckmann I, Hellwig A, Röhling S, El-Battari A, Wieland FT, Popoff V (2014). Golgi phosphoprotein 3 triggers signal-mediated incorporation of glycosyltransferases into coatomer-coated (COPI) vesicles. J Biol Chem 289, 31319–31329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstein M, Flusser H, Lerman-Sagie T, Ben-Zeev B, Lev D, Agamy O, Cohen I, Kadir R, Sivan S, Leshinsky-Silver E (2014). VPS53 mutations cause progressive cerebello-cerebral atrophy type 2 (PCCA2). J Med Genet 51, 303–308. [DOI] [PubMed] [Google Scholar]
- Foulquier F, Vasile E, Schollen E, Callewaert N, Raemaekers T, Quelhas D, Jaeken J, Mills P, Winchester B, Krieger M (2006). Conserved oligomeric Golgi complex subunit 1 deficiency reveals a previously uncharacterized congenital disorder of glycosylation type II. Proc Natl Acad Sci USA 103, 3764–3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fröhlich F, Petit C, Kory N, Christiano R, Hannibal-Bach H-K, Graham M, Liu X, Ejsing CS, Farese RV Jr, Walther TC (2015). The GARP complex is required for cellular sphingolipid homeostasis. Elife 4, e08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaynor EC, Graham TR, Emr SD (1998). COPI in ER/Golgi and intra-Golgi transport: do yeast COPI mutants point the way? Biochimica et Biophysica Acta Biochim Biophys Acta Mol Cell Res 1404, 33–51. [DOI] [PubMed] [Google Scholar]
- Gershlick DC, Ishida M, Jones JR, Bellomo A, Bonifacino JS, Everman DB (2019). A neurodevelopmental disorder caused by mutations in the VPS51 subunit of the GARP and EARP complexes. Hum Mol Genet 28, 1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillingham AK, Sinka R, Torres IL, Lilley KS, Munro S (2014). Toward a comprehensive map of the effectors of rab GTPases. Dev Cell 31, 358–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson PA (1998). Targeting of proteins to the Golgi apparatus. Histochem Cell Biol 109, 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Nakano A (2009). Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev 25, 113–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hady-Cohen R, Ben-Pazi H, Adir V, Yosovich K, Blumkin L, Lerman-Sagie T, Lev D (2018). Progressive cerebello-cerebral atrophy and progressive encephalopathy with edema, hypsarrhythmia and optic atrophy may be allelic syndromes. Eur J Paediatr Neurol 22, 1133–1138. [DOI] [PubMed] [Google Scholar]
- Hirata T, Fujita M, Nakamura S, Gotoh K, Motooka D, Murakami Y, Maeda Y, Kinoshita T (2015). Post-Golgi anterograde transport requires GARP-dependent endosome-to-TGN retrograde transport. Mol Biol Cell 26, 3071–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida M, Bonifacino JS (2019). ARFRP1 functions upstream of ARL1 and ARL5 to coordinate recruitment of distinct tethering factors to the trans-Golgi network. J Cell Biol 218, 3681–3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M, Suda Y, Kurokawa K, Nakano A (2016). COPI is essential for Golgi cisternal maturation and dynamics. J Cell Sci 129, 3251–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufman O, Hong W, Lev S (2011). The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J Cell Biol 194, 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laufman O, Hong W, Lev S (2013). The COG complex interacts with multiple Golgi SNAREs and enhances fusogenic assembly of SNARE complexes. J Cell Sci 126, 1506–1516. [DOI] [PubMed] [Google Scholar]
- Lavieu G, Zheng H, Rothman JE (2013). Stapled Golgi cisternae remain in place as cargo passes through the stack. Elife 2, e00558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liewen H, Meinhold-Heerlein I, Oliveira V, Schwarzenbacher R, Luo G, Wadle A, Jung M, Pfreundschuh M, Stenner-Liewen F (2005). Characterization of the human GARP (Golgi associated retrograde protein) complex. Exp Cell Res 306, 24–34. [DOI] [PubMed] [Google Scholar]
- Linstedt A, Mehta A, Suhan J, Reggio H, Hauri H (1997). Sequence and overexpression of GPP130/GIMPc: evidence for saturable pH-sensitive targeting of a type II early Golgi membrane protein. Mol Biol Cell 8, 1073–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira-Navarrete E, de Las Rivas M, Compañón I, Pallarés MC, Kong Y, Iglesias-Fernández J, Bernardes GJ, Peregrina JM, Rovira C, Bernadó P (2015). Dynamic interplay between catalytic and lectin domains of GalNAc-transferases modulates protein O-glycosylation. Nat Commun 6, 6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Doray B, Kornfeld S (2018). Recycling of Golgi glycosyltransferases requires direct binding to coatomer. Proc Natl Acad Sci USA 115, 8984–8989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losev E, Reinke CA, Jellen J, Strongin DE, Bevis BJ, Glick BS (2006). Golgi maturation visualized in living yeast. Nature 441, 1002–1006. [DOI] [PubMed] [Google Scholar]
- Luo L, Hannemann M, Koenig S, Hegermann J, Ailion M, Cho M-K, Sasidharan N, Zweckstetter M, Rensing S, Eimer S (2011). The Caenorhabditis elegans GARP complex contains the conserved Vps51 subunit and is required to maintain lysosomal morphology. Mol Biol Cell 22, 2564–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Menárguez JA (2013). Intra-Golgi transport: roles for vesicles, tubules, and cisternae. ISRN Cell Biol 2013, 126731. [Google Scholar]
- Mironov AA, Beznoussenko GV (2012). The kiss-and-run model of intra-Golgi transport. Int J Mol Sci 13, 6800–6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Natarajan R, Linstedt AD (2004). A cycling cis-Golgi protein mediates endosome-to-Golgi traffic. Mol Biol Cell 15, 4798–4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Abascal-Palacios G, Tascón I, Kajava A, Magadán JG, Pioro EP, Bonifacino JS, Hierro A (2010a). Structural basis for the wobbler mouse neurodegenerative disorder caused by mutation in the Vps54 subunit of the GARP complex. Proc Natl Acad Sci USA 107, 12860–12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Bonifacino JS (2009). Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-golgi network. Mol Cell Biol 29, 5251–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Mardones GA, Bonifacino JS (2008). Requirement of the human GARP complex for mannose 6-phosphate-receptor-dependent sorting of cathepsin D to lysosomes. Mol Biol Cell 19, 2350–2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Victoria FJ, Schindler C, Magadán JG, Mardones GA, Delevoye C, Romao M, Raposo G, Bonifacino JS (2010b). Ang2/fat-free is a conserved subunit of the Golgi-associated retrograde protein complex. Mol Biol Cell 21, 3386–3395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CS, Lee JJ, Boland S, Swarup S, Christiano R, Lai ZW, Mejhert N, Elliott SD, McFall D, Haque S (2020). Inhibition of sphingolipid synthesis improves outcomes and survival in GARP mutant wobbler mice, a model of motor neuron degeneration. Proc Natl Acad Sci USA 117, 10565–10574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrovskaya ID, Willett R, Smith RD, Morelle W, Kudlyk T, Lupashin VV (2011). Conserved oligomeric Golgi complex specifically regulates the maintenance of Golgi glycosylation machinery. Glycobiology 21, 1554–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivinoja A, Hassinen A, Kokkonen N, Kauppila A, Kellokumpu S (2009). Elevated Golgi pH impairs terminal N-glycosylation by inducing mislocalization of Golgi glycosyltransferases. J Cell Physiol 220, 144–154. [DOI] [PubMed] [Google Scholar]
- Rosa-Ferreira C, Christis C, Torres IL, Munro S (2015). The small G protein Arl5 contributes to endosome-to-Golgi traffic by aiding the recruitment of the GARP complex to the Golgi. Biol Open 4, 474–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjana NE, Shalem O, Zhang F (2014). Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11, 783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindler C, Chen Y, Pu J, Guo X, Bonifacino JS (2015). EARP is a multisubunit tethering complex involved in endocytic recycling. Nat Cell Biol 17, 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schjoldager KT, Narimatsu Y, Joshi HJ, Clausen H (2020). Global view of human protein glycosylation pathways and functions. Nat Rev Mol Cell Biol 21, 729–749. [DOI] [PubMed] [Google Scholar]
- Schmitt-John T, Drepper C, Mußmann A, Hahn P, Kuhlmann M, Thiel C, Hafner M, Lengeling A, Heimann P, Jones JM (2005). Mutation of Vps54 causes motor neuron disease and defective spermiogenesis in the wobbler mouse. Nat Genet 37, 1213–1215. [DOI] [PubMed] [Google Scholar]
- Sengupta P, Satpute-Krishnan P, Seo AY, Burnette DT, Patterson GH, Lippincott-Schwartz J (2015). ER trapping reveals Golgi enzymes continually revisit the ER through a recycling pathway that controls Golgi organization. Proc Natl Acad Sci USA 112, E6752–E6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Pelham HR (2002). Vps51p links the VFT complex to the SNARE Tlg1p. J Biol Chem 277, 48318–48324. [DOI] [PubMed] [Google Scholar]
- Stanley P (2011). Golgi glycosylation. Cold Spring Harb Perspect Biol 3, a005199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storrie B, Pepperkok R, Nilsson T (2000). Breaking the COPI monopoly on Golgi recycling. Trends Cell Biol 10, 385–390. [DOI] [PubMed] [Google Scholar]
- Storrie B, White J, Röttger S, Stelzer EH, Suganuma T, Nilsson T (1998). Recycling of Golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J Cell Biol 143, 1505–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Chen B, Song Z, Lu L (2021). A quantitative study of the Golgi retention of glycosyltransferases. bioRxiv, https//.org/10.1101/2021.02.15.431224. [DOI] [PubMed] [Google Scholar]
- Uwineza A, Caberg J-H, Hitayezu J, Wenric S, Mutesa L, Vial Y, Drunat S, Passemard S, Verloes A, El Ghouzzi V (2019). VPS51 biallelic variants cause microcephaly with brain malformations: A confirmatory report. Eur J Med Genet 62, 103704. [DOI] [PubMed] [Google Scholar]
- Wei J, Zhang Y-Y, Luo J, Wang J-Q, Zhou Y-X, Miao H-H, Shi X-J, Qu Y-X, Xu J, Li B-L (2017). The GARP complex is involved in intracellular cholesterol transport via targeting NPC2 to lysosomes. Cell Rep 19, 2823–2835. [DOI] [PubMed] [Google Scholar]
- Welch LG, Munro S (2019). A tale of short tails, through thick and thin: investigating the sorting mechanisms of Golgi enzymes. FEBS Lett 593, 2452–2465. [DOI] [PubMed] [Google Scholar]
- Willett R, Pokrovskaya I, Kudlyk T, Lupashin V (2014). Multipronged interaction of the COG complex with intracellular membranes. Cell Logist 4, e27888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willett R, Ungar D, Lupashin V (2013). The Golgi puppet master: COG complex at center stage of membrane trafficking interactions. Histochem Cell Biol 140, 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaji T, Hanamatsu H, Sekizuka T, Kuroda M, Iwasaki N, Ohnishi M, Furukawa J-i, Yahiro K, Hanada K (2019). A CRISPR Screen using Subtilase cytotoxin identifies SLC39A9 as a glycan-regulating factor. Iscience 15, 407–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.