Abstract
To gain insight into the biology of Natural Killer (NK) cells, others and we previously identified the NK cell signature, defined as the set of transcripts which expression is highly enriched in these cells compared to other immune subtypes. The transcript encoding the Serine/threonine/ tyrosine kinase 1 (Styk1) is part of this signature. However, the role of Styk1 in the immune system is unknown. Here, we report the generation of a novel transgenic mouse model, in which Styk1 expression is invalidated and replaced by an EGFP reporter cassette. We demonstrated that Styk1 expression is a hallmark of NK cells and other NK1.1 expressing cells such as liver type 1 innate lymphoid cells (ILC1) and NK1.1+ γδ T cells. Styk1 expression is maintained by IL-15 in NK cells and negatively correlates with the expression of educating NK cell receptors. Analysis of phosphorylation levels of mTOR substrates suggested that Styk1 could moderately contribute to the activity of the PI3K/Akt/mTOR pathway. However, Styk1 deficient NK cells develop normally and have normal in vitro and in vivo effector functions. Thus Styk1 expression is a hallmark of NK cells, ILC1 and NK1.1+ T cells but is dispensable for their development and immune functions.
Keywords: Natural Killer cells, receptor tyrosine kinase, development, transcription, innate lymphoid cells
Graphical Abstract
Styk1 is specifically expressed by NK cells and at a lower level by other NK1.1+ cells such as ILC1 and gd T cells. Styk1 expression is not essential for NK cell effector functions.
Introduction
Natural Killer (NK) cells are innate lymphocytes endowed with antitumor and antiviral functions. They have the unique ability to kill cells recognized as targets without prior stimulation. Cytotoxicity is mediated by the oriented release of specialized granules containing perforin, granzymes and other proteins at the synapse with the target cell. NK cells also secrete several cytokines including IFNγ and TNFα that also play different roles in viral or tumor clearance. Recognition of target cells is mediated by an array of cell surface germ-line encoded receptors called the NK cell receptors. NK cell receptor genes are clustered in two important loci, the leukocyte receptor complex and the NK cell receptor complex [1]. These receptors transduce activating or inhibitory signals in response to the engagement by their respective ligands, and the balance between these signals governs NK cell behavior, ie activation or ignorance. NK cells belong to the recently defined family of innate lymphoid cells (ILCs) [2] and are closely related to ILC1s, although the latter are generally less cytotoxic than NK cells and are tissue resident, unlike NK cells that circulate in the body via the blood.
Our understanding of immune cells in general and of NK cells in particular has been largely improved by whole genome analyses of gene expression. For example, we previously identified S1PR5 as an NK-specific transcript using microarray analyses. This led to the discovery that S1PR5 is a chemotactic receptor for sphingosine-1 phosphate essential for NK cell egress from the bone marrow [3], a breakthrough that we recently validated in human as well [4]. Using a similar microarray approach, we also found that NKp46 was a specific marker of NK cells across different mammalian species [5]. Further studies subsequently showed that NKp46 was in fact not totally NK- specific as it was expressed in a fraction of T cells [6],[7] and in NCR1 positive ILC3s in the gut [8],[9]. A selective marker of NK cells is therefore still lacking. The Immgen consortium reported the identification of an NK cell transcriptional signature based on a comprehensive transcriptomic analysis of hundreds of immune subsets in mouse [10]. This signature was composed of 25 transcripts strongly enriched in NK cells compared to all other immune populations, including several of the aforementioned transcripts (S1pr5, Ncr1). Most of these genes were already known in NK cells, many of which encoding NK cell receptors. However, very surprisingly, one of the highest-ranking transcripts defining NK cells was the Serine/threonine/ tyrosine kinase 1 (Styk1) mRNA, whose role in NK cells and more generally in the immune system was unknown.
Styk1 shares homology with platelet-derived growth factor/fibroblast growth factor receptors and has been shown to drive transformation of NIH3T3 and Baf/3 cell lines by regulating cell proliferation and survival through the activation of both MAP kinase and phosphatidylinositol 3′-kinase [11]. Several recent papers confirmed that Styk1 was involved in the activation of this pathway, leading to the induction of aerobic glycolysis, an outcome that may contribute to oncogenesis in cells where Styk1 is overexpressed. Styk1 is overexpressed in many tumors including breast [12], lung [13], acute leukemia [14], prostate [15], ovarian [16] cancers and is associated with progression of renal cell carcinoma [17]. Styk1 may induce epithelial to mesenchymal transition through the PI3K/Akt pathway [18]. Styk1 has a transmembrane domain but no extracellular domain, and only one protein, HSP90AA1, is reported to interact with Styk1 (BioGrid database). How Styk1 is activated under physiological conditions and the pathways this kinase regulates remain unknown. Here, we report the generation of Styk1EGFP/EGFP reporter mice that are also loss-of-function Styk1 mutants. We demonstrate that Styk1 expression is a hallmark of NK cells, ILC1 and NK1.1+ T cells. Styk1 deficient NK cells display a slightly decreased basal level of phosphorylation of mTOR substrates S6 and Akt suggesting that Styk1 may contribute to the PI3K/Akt/mTOR pathway under steady state conditions. However, NK cells develop normally in the absence of Styk1 and Styk1 deficient NK cells have normal in vitro and in vivo effector functions.
Results
Styk1 expression is a hallmark of NK cells, ILC1 and NK1.1+ T cells
A previous report from the Immgen consortium showed that Styk1 mRNA was part of the transcriptional signature of resting mouse NK cells. More specifically, Styk1 was among the 25 genes more highly expressed by NK cells than any other leukocyte populations [10]. Immgen microarray data also predict NK-specific expression of Styk1 in human (Immgen website). Preferential Styk1 expression in mammalian NK cells correlates with the conserved location of the Styk1 gene at the border of the NK-complex on mouse chromosome 6 and human chromosome 12 (Figure 1A). Analyses of genomic data from 28 mouse strains suggest the presence of a functional Styk1 allele in all of them [19]. Similarly, sequence variations of Styk1 predicted to be pathogenic are extremely rare in human, according to the exome aggregation consortium (Exac) database of more than 60 000 exome sequence data [20].
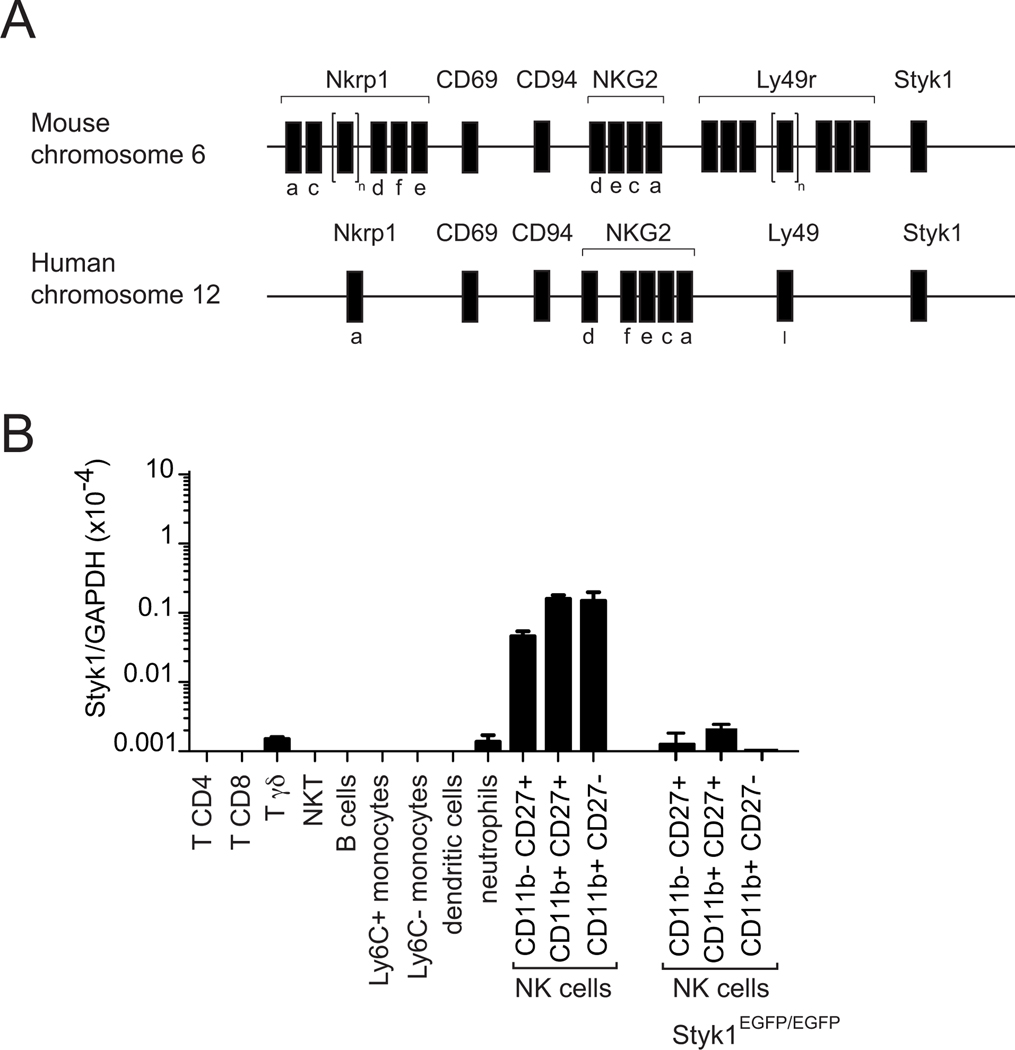
Figure 1: Styk1 mRNA is preferentially expressed in mouse NK cells.
(A) Scheme of the NK complex in mouse and human showing localization of the Styk1 gene adjacent to the complex in both species. (B) RT-QPCR analysis of Styk1 expression in FACS sorted subsets from WT or Styk1EGFP/EGFP mice, as indicated. Immune subsets were defined and gated as shown in Figure S1. Data show representative results from three independent experiments for each species.
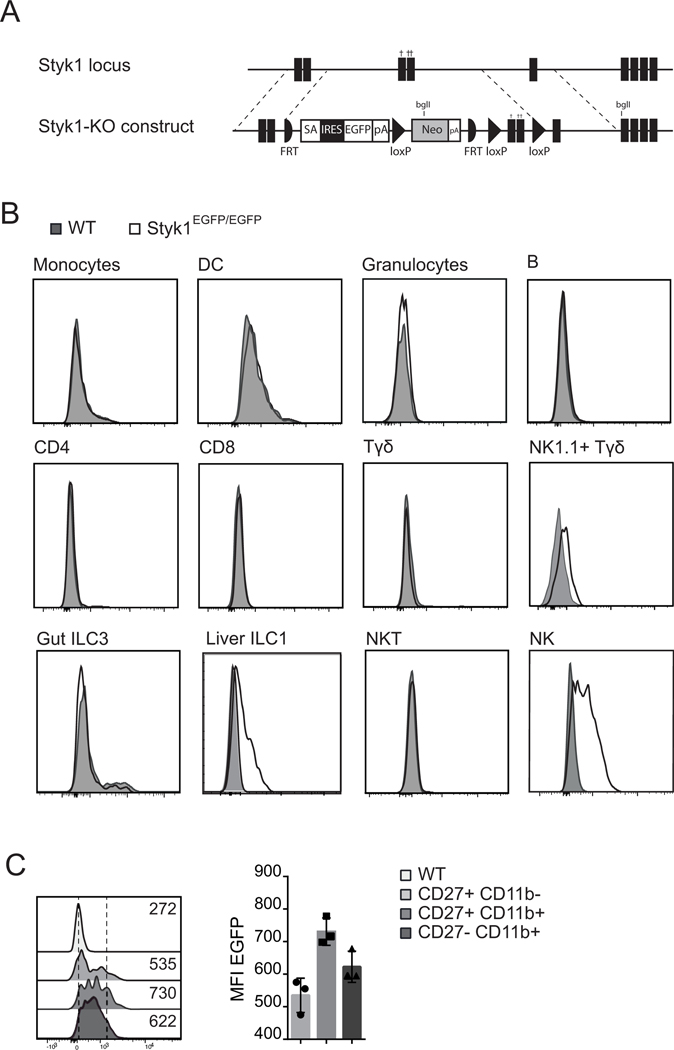
RT-QPCR measurements further supported the preferential expression of Styk1 in mouse NK cells compared to other lymphoid and myeloid cell subsets (Figure 1B, gating of subsets shown in Figure S1). Styk1 mRNA expression was already detectable in immature CD11b-CD27+ NK cells. To further explore the expression of Styk1 in mouse leukocyte subsets, we generated a Styk1-reporter mouse line by inserting an EGFP cassette in the exon 2 of Styk1 that also results in a knockout (Figure 2A). We verified that Styk1 expression was lost in NK cells from these mice (Figure 1B). We then measured EGFP expression in various leukocyte subsets from Styk1EGFP/EGFP mice. As shown in Figure 2B, EGFP expression was only detected in NK cells, and at a weaker level in liver ILC1s and spleen NK1.1+ γδ T cells. EGFP expression was gradually increased during NK cell maturation and maximal in CD27+ CD11b+ NK cells (Figure 2C). We also performed a detailed analysis of EGFP expression in other ILC subsets. EGFP expression was undetectable in gut ILC1, ILC3 and NCR+ILC3s, as defined by gating in figure S2. Thus, high Styk1 expression is a hallmark of the NK cell lineage, and Styk1 is also expressed at lower level in liver ILC1s and spleen NK1.1+ γδ T cells.
Figure 2: Styk1 expression marks NK cells and other NK1.1+ cells.
(A) Scheme of the Styk1 locus and of the targeting strategy. (B) Flow cytometry analysis of EGFP expression in various immune subsets from Styk1EGFP/EGFP mice. Immune subsets were defined as shown in Figures S1 (spleen, liver and BM subsets) and S2 (gut subsets) −. (C) Flow cytometry analysis of EGFP expression in gated NK cells of different maturation status as defined by CD27 and CD11b expression. Data in graph are shown as mean/median ± SD. B and C show representative results of at least 3 independent experiments.
Normal NK cell development in Styk1EGFP/EGFP mice
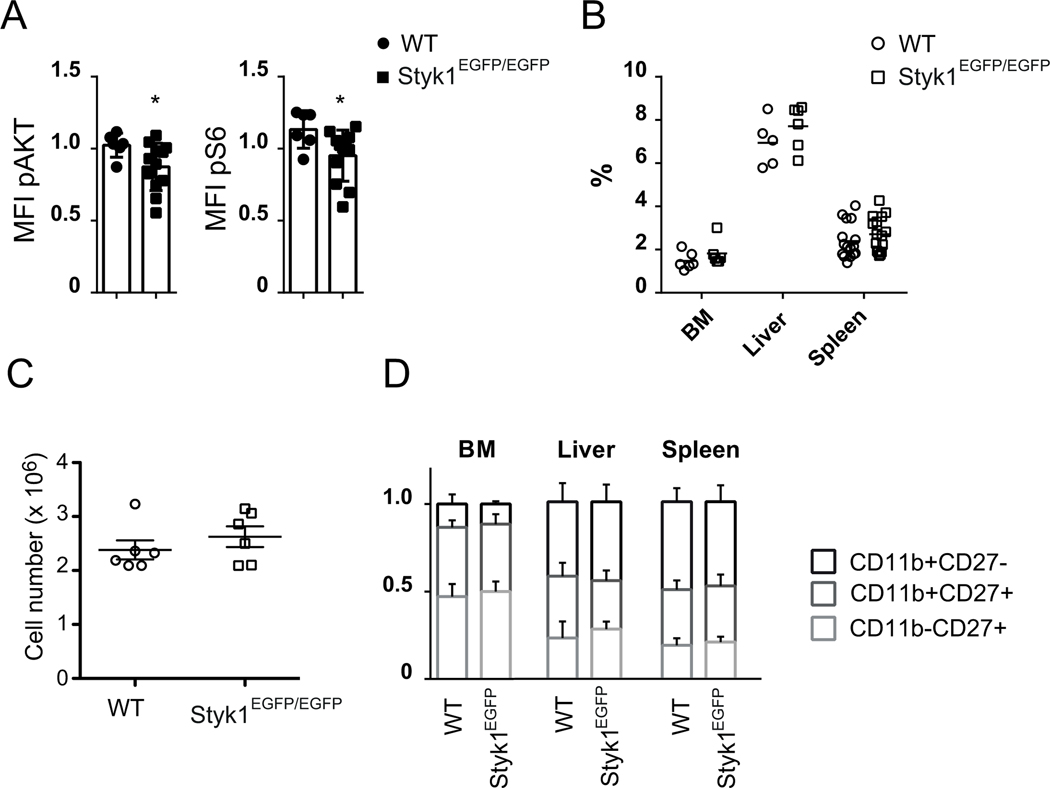
Styk1 has been suggested to regulate cell proliferation and survival by activating both MAP kinase and Akt. Accordingly, we found a slightly but significant decrease in basal level of phosphorylation of Akt and of the mTOR substrate S6 in Styk1EGFP/EGFP NK cells compared to control NK cells (Figure 3A). However, no difference in the level of these phosphorylation events was observed between both types of NK cells following stimulation with anti-NK1.1 antibodies or with IL-15 (data not shown). Given the importance of mTOR in NK cell development and function [21], we explored the NK cell compartment in Styk1EGFP/EGFP mice. As shown in Figure 3B, the percentage of NK cells among leukocytes in different organs was comparable in Styk1EGFP/EGFP versus control mice and the number of spleen NK cells was also comparable in both mouse stains (Figure 3C). The relative proportions of mature and immature cells among NK cells was also normal in the different organs of Styk1EGFP/EGFP mice (Figure 3D), and so were the expression of metabolic markers CD98, CD71 and size/granularity (data not shown), excluding a role for Styk1 in NK cell proliferation, metabolism or maturation. The expression of various receptors from the NK or the leukocyte complex was also comparable in Styk1EGFP/EGFP and control mice (Table I). The targeted Styk1 null allele we generated could be converted into a floxed allele upon Flipase-driven recombination (see figure 2A). Styk1 fl/fl mice were crossed with Ncr1-iCre mice to generate NK-specific Styk1 deletion. NK cells from these mice displayed normal development and distribution (data not shown).
Figure 3: Styk1EGFP/EGFP NK cells have a lower basal mTOR activity than control NK cells but develop normally.
(A) Flow cytometry analysis of pS6 and pAkt in gated NK cells from freshly isolated spleen cells. Data show mean +/−SD fluorescence intensity of the staining. Each dot corresponds to an individual mouse in a total of 6 (WT) or 11 (KO) mice pooled from 3 experiments. *p <0.05 (Student T-test). (B) Percentage of NK cells in different organs of Styk1EGFP/EGFP and control mice. N=6 mice for each organ (pool of two experiments). (C) Number of NK cells in the spleen of Styk1EGFP/EGFP and control mice. N=6 in each group (pool of two experiments). (D) NK cell maturation as defined by the flow cytometry analysis of CD27/CD11b expression in Styk1EGFP/EGFP and control mice. Data show the mean +/−SD frequency of each subset. N=6 mice (pool of two experiments)
Table I:
Percentage of NK cells expressing the indicated NK cell markers in the spleen of Styk1EGFP/EGFP and control mice.
| CD127 | CXCR3 | CD146 | KLRG1 | Ly49D | Ly49H | NKG2ACE | Ly49E/F | Ly49I | Ly49G2 | |
|---|---|---|---|---|---|---|---|---|---|---|
| WT | 5,7 +/− 2,9 | 54,5 +/− 2,5 | 60,7 +/− 4,4 | 29,2 +/− 15 | 43 +/− 9 | 49 +/− 5 | 53,5 +/− 3 | 8 +/− 2 | 48,5 +/− 5 | 41 +/− 5 |
| Styk1 KO | 3,7 +/− 1,5 | 45,3 +/− 9 | 64,9 +/− 2,9 | 35 +/− 8,5 | 48 +/− 5 | 52,3 +/− 4 | 49 +/− 3 | 8 +/− 2 | 47,6 +/− 4 | 42 +/− 2 |
Styk1 mRNA expression is lower in educated NK cells and is maintained by IL-15
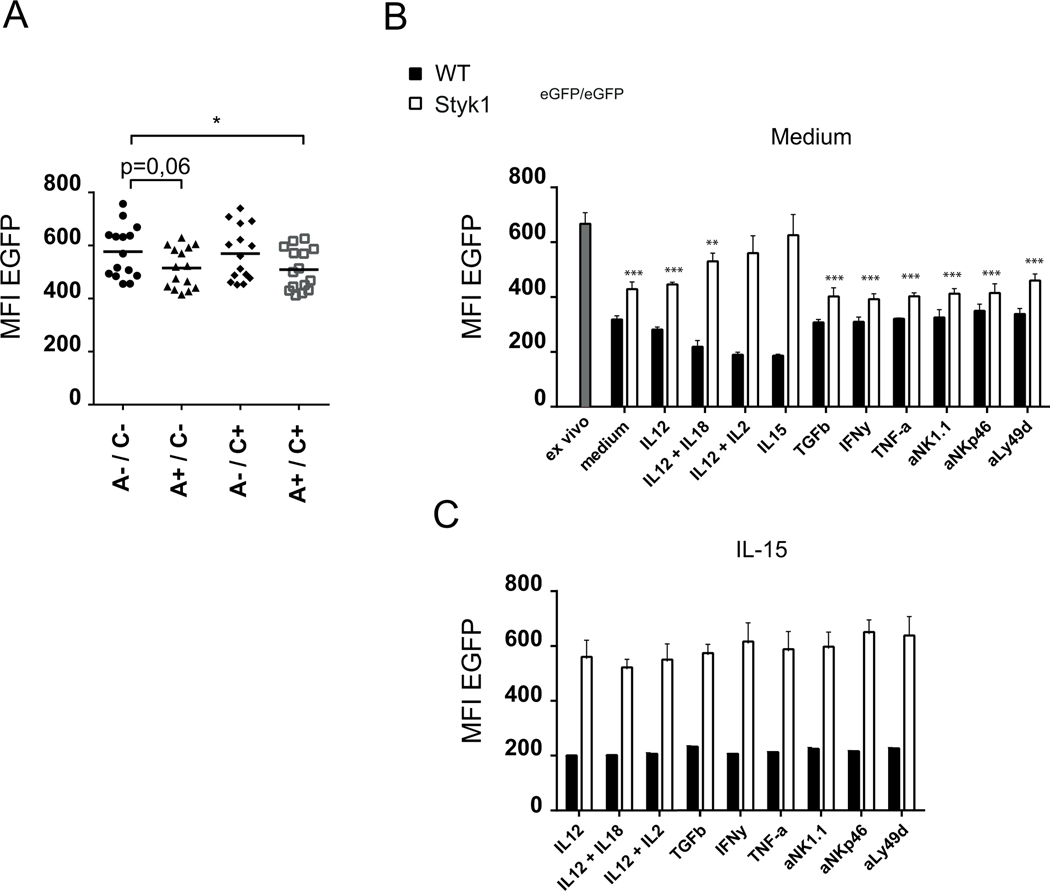
We took advantage of the EGFP reporter allele to study the regulation of Styk1 mRNA in different biological contexts. In C57BL/6 mice, Ly49C and the CD94/NKG2A receptor have been shown to interact with substantial affinity with self-MHC class I molecules, and NK cell subsets expressing these receptors are educated, ie they are more reactive than their non-educated counterparts [22]–[24]. We stained Styk1EGFP/EGFP NK cells with Ly49C and NKG2A antibodies. As shown in Figure 4A, EGFP expression was significantly lower in educated Ly49C+NKG2A+ NK cells compared to uneducated Ly49C-NKG2A- NK cells. Next, we cultured spleen cells from Styk1EGFP/EGFP mice in the presence or absence of different cytokines or plate-bound antibodies directed against diverse activating receptors. In the presence of IL-15, EGFP expression was maintained while all other tested conditions resulted in a decreased EGFP expression (Figure 4B). We then tested if the same stimuli could influence EGFP expression when combined with IL-15. However, as shown in Figure 4C, none of the tested stimuli altered the expression of EGFP when IL-15 was present.
Figure 4: Styk1 expression is maintained by IL-15 in NK cells and is lower in educated NK cells.
(A) EGFP expression in educated vs uneducated NK cells defined as NK cells co-expressing NKG2A and Ly49C and NK cells negative for both receptors, respectively. (B-C) Spleen cells from Styk1EGFP/EGFP and control mice were cultured in the indicated conditions for 24h. EGFP expression was then measured in gated NK cells by flow cytometry. Histograms show mean +/−SD fluorescence intensity of EGFP fluorescence. The blue bar indicated EGFP levels in freshly isolated NK cells. Data are pooled from the analysis of N=3 mice in each group in a single experiment and are representative of three independent experiments. In (B) one way Anova statistical analysis was performed, with a post-test to compare all groups with the IL-15 group.
Taken together, these results show that Styk1 expression is maintained by IL-15 and suggest that Styk1 expression could be regulated during NK cell education.
Redundant role of Styk1 in NK cell function
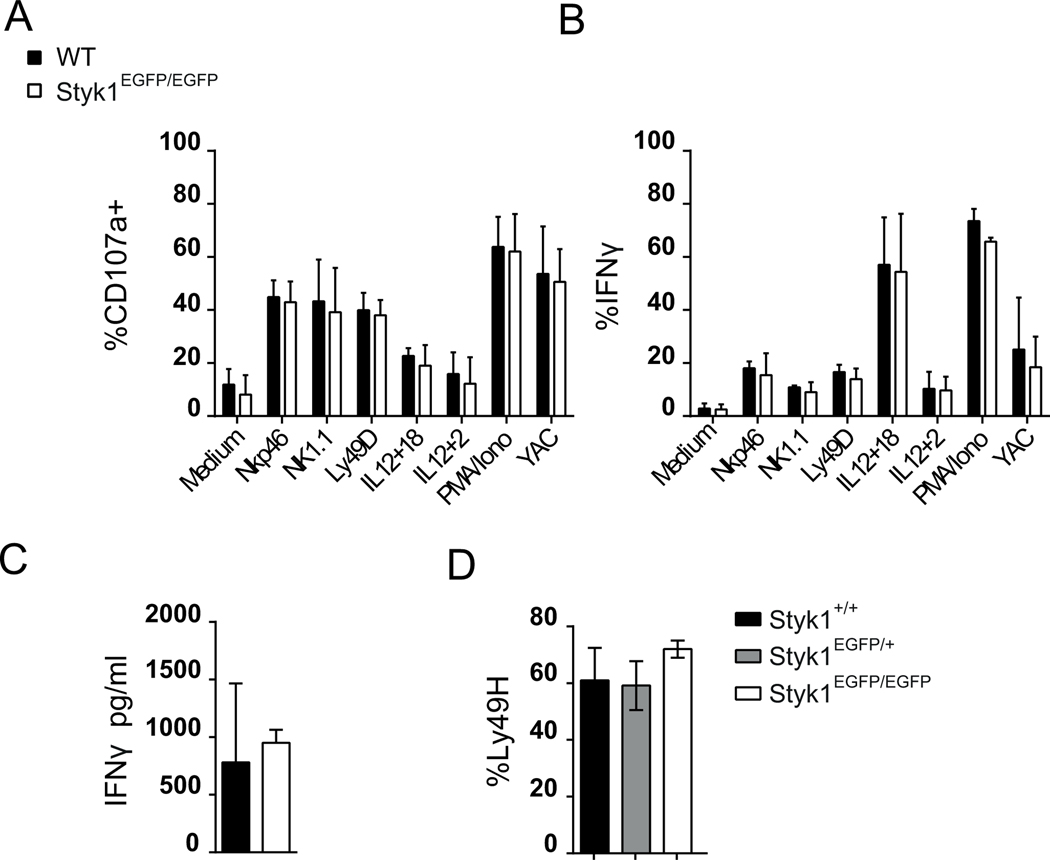
To test the role of Styk1 in NK cell effector functions, we stimulated Styk1EGFP/EGFP and control NK cells with cytokine cocktails, plate-bound antibodies against activating NK cell receptors, YAC1 tumor cells or PMA / ionomycin for 4 hours and measured their degranulation by staining for surface CD107a and their capacity to secrete IFNγ using intracellular staining. As shown in Figure 5A–B, all these stimuli triggered both NK cell functions at different levels, but no difference was observed between Styk1EGFP/EGFP and control NK cells. No difference was observed either in the ability of Styk1EGFP/EGFP and control NK cells to kill YAC1 cells in a standard cytotoxicity assay (data not shown). In these various experiments, control mice were from the C57BL/6J line while Styk1EGFP/EGFP had a C57BL/6N background. Our own unpublished analyses failed to detect any differences in terms of NK cell development between these strains. Yet, subtle differences were previously noted in NK cell response to cytokines between these strains [25]. To exclude that the mouse background did not bias our functional results, we backcrossed Styk1EGFP/EGFP mice six times with C57BL/6J mice. Resultant Styk1EGFP/+ mice were intercrossed to obtain Styk1+/+, Styk1EGFP/+ and Styk1EGFP/EGFP littermates, whose NK cells were assayed for functional responses as in Figure 5A–B. However, no difference was observed between the groups (data not shown).
Figure 5: Normal in vitro and in vivo function of Styk1 deficient NK cells.
(A-B) Spleen cells from Styk1EGFP/EGFP and control mice were stimulated in the indicated conditions for 4 hours. NK cell degranulation was measured by staining for surface CD107 and IFNγ secretion was measured by intracellular staining. Results show mean +/−SD and are pooled from 6 mice in 3 independent experiments. (C) IFNγ levels measured by Elisa in the serum of the indicated mice 2 days post infection with MCMV. Each dot represents an individual mouse and results show mean+/−SD of a pool of 3 independent experiments. (D) The percentage of Ly49H positive NK cells was measured by flow cytometry in gated NK cells from the spleen of infected mice of the indicated strain 7 days post infection. Results show the mean+/−SD (pool of 3 independent experiments).
Next, to further test the role of Styk1 in NK cell function, we used Styk1+/+, Styk1EGFP/+ and Styk1EGFP/EGFP littermate mice, all on a C57BL/6J background that we challenged with MCMV Smith strain. All mice in each group survived the infection and recovered in terms of weight (data not shown). We monitored IFNγ expression at day +2. At this time point, IFNγ production is mediated by NK cells in response to an array of innate cytokines produced by myeloid cells. Despite significant inter individual variation, no significant difference between groups was noted (Figure 5C). Ly49H positive NK cells expand in the spleen and in the liver during MCMV infection, following the recognition of the viral epitope m157 on infected cells. This proliferative response peaks seven days after infection. We monitored the frequency of Ly49H positive NK cells in the spleen of mice at day-7 post infection. This frequency was similar in all mouse groups (Figure 5D).
Taken together, these results suggest that Styk1 is dispensable for mouse NK cell effector functions, in particular in the context of virus infections.
Discussion
Here, we show that Styk1 is a specific marker of NK cells and some NK1.1 expressing cells such as liver ILC1 and NK1.1+ γδ T cells. Styk1 is not expressed by other ILC subsets such as gut ILC1s and NCR+ILC3s. In the present manuscript, we also describe an EGFP reporter mouse, Styk1EGFP/EGFP that can be used to track mouse NK cells and could be useful for NK cell developmental studies.
Cell specific transcripts usually encode for important cellular functions. For instance, S1pr5 is essential to promote trafficking of NK cells throughout the body [3], and is likely essential for their anti-viral and anti-tumor functions; Ly49H encodes an activating NK cell receptor specific to NK cells, which is essential to promote activation and expansion of NK cells in response to MCMV infection [26]–[28]. A previous comprehensive transcriptomic study reported the identification of a set of transcripts that were enriched in NK cells compared to many other leukocyte populations [10]. Among the genes whose expression was uniquely increased in NK cells, several of them had not reported function in NK cells. These data suggested that a rich biology related to NK cells remained undiscovered. As our own unpublished analyses had pointed to Styk1 as being specifically expressed in NK cells, we decided to generate a Styk1 loss-of-function mouse model.
Styk1 expression was regulated during NK cell maturation and was maximal in mature NK cells, starting at the CD11b+ CD27+ stage. The observation that Styk1 mRNA expression was dependent on IL-15 in vitro fits with the recent report that Styk1 is down regulated in Runx3 deficient NK cells [29]. Indeed, this study showed that Runx3 cooperates with ETS and T-box transcription factors to drive the interleukin-15-mediated transcription program during activation of NK cells. Educating receptors negatively correlated with Styk1 expression, which together with the localization of the Styk1 gene next to the NK complex constitutes an interesting link between Styk1 and NK cell receptors. However, Styk1 does not influence expression of NK cell receptors as Styk1EGFP/EGFP NK cells normally expressed all the NK cell receptors tested.
A pioneer study showed in stably expressing cell lines that Styk1 could concomitantly activate both MAP kinase and PI3K pathways [11]. This initial report was followed by other observations that confirmed activation of PI3K/Akt by Styk1 in other cell lines [18],[30],[31]. Styk1 overexpression leads to an increased glycolytic capacity, and decreases oxidative phosphorylation via PI3K/Akt induction [32]. These data were of particular interest when put in the context of our own previous observations of the importance of the Akt/mTOR pathway in the control of NK cell development, bioenergetic metabolism and IL-15 [21] or NK cell receptor-mediated activation [33]. This prompted us to assess the activity of the mTOR pathway and the effector functions of Styk1 deficient NK cells. We consistently observed a slight decrease in basal level of pS6 and pAkt in freshly isolated Styk1EGFP/EGFP NK cells compared to control NK cells, suggesting that Styk1 could play a role in the basal regulation of the Akt/mTOR pathway in NK cells. However, we failed to detect any developmental, differentiation, metabolic or functional defect in NK cells in the absence of Styk1, which suggests that the role of Styk1 in the Akt/mTOR pathway in NK cells is minor. Styk1 might therefore function in some specific contexts that were not tested in our study. Van Roosmalen and colleagues previously identified Styk1 as a kinase promoting tumor cell migration in a large siRNA-based screen for kinases and other molecules involved in this cellular process [34]. However, the normal distribution of NK cells in Styk1EGFP/EGFP mice does not support a role for Styk1 in NK cells migration.
In conclusion, we show here that high Styk1 expression is a hallmark of NK cells and that Styk1 is not essential for NK cell development, activation and antiviral functions. Further studies are required to understand the role of Styk1 in NK cells but Styk1EGFP/EGFP mice represent a novel and interesting model for developmental studies of mouse NK cells.
Material and Methods
Generation of Styk1 deficient mice
A plasmid “KO-first” designed and made by the European Conditional mouse mutagenesis program (EUCOMM) and targeting the mouse Styk1 gene was purchased (Clone PRPGS00082_B_F08). This plasmid contains 5’ and 3’ homology arms, and a gene trap cassette that allows the expression of the LacZ reporter cassette in place of Styk1. It also contains FRT and LoxP sites that allow Cre-mediated conditional targeting of Styk1 following Flp-mediated recombination. We modified this vector using E/T cloning (Genebridges) and replaced the LacZ coding sequence with the EGFP coding sequence (obtained from the Clontech plasmid pEGFP.N1). JM8.A3 ES cells (C57BL/6N) were transfected with this construct and G418-resistant clones were obtained. We checked for correct homologous recombination by PCR followed by southern blot using different probes. Chimeric mice were obtained following microinjection of ES cells into C57BL/6 blastocysts and germline transmission was monitored by PCR using different sets of primers encompassing different parts of the targeted locus. Plasmid and PCR primer sequences are available on request.
Animals
This study was carried in strict accordance with the French recommendation in the Guide for the ethical evaluation of experiments using laboratory animals and the European guidelines 86/609/CEE. Procedures including animals were approved by local ethics review board under ENS2015–014 agreement. All mouse strains were maintained in our animal facility.
Virus production and infection
MCMV-smith was propagated by infecting 3 weeks old Balb/c mice with 50 PFU of MCMV cultured until passage 2 on primary BALB/c mouse whole fetus cells. Briefly, salivary glands were collected on ice in 3% FCS DMEM, homogenized with an organ dissociator and centrifuged at 800g for 5 min at 4°C. Supernatant was collected and stored at −80°C until use. Titers were determined by plaque assay using NIH-3T3 cells. Mice were infected intraperitoneally with 1 × 105 PFU of MCMV.
Flow cytometry
Blood was collected in 0.5M EDTA and red blood cells were lysed with ACK lysis buffer. Mononuclear cells were isolated from spleen and liver injected with complete RPMI 1640 5% FCS 0.4 mg/ml Collagenase IV (Serlabo Technologies, Vedène, France) and 0.1mg/ml DNase I (Roche), cut into small pieces and incubated at 37°C with 150RPM shaking for 30 min. After incubation, organs were homogenized and cells from the liver were purified using Percoll (GE Healthcare) density gradient separation. Gut cells were prepared as previously described [8]. Flow cytometry was carried out on a FACS Fortessa (Becton-Dickinson). Data were analysed using FlowJo (V10, Treestar). The following antibodies from eBioscience or BD-biosciences or Biolegend were used: anti-CD19 (ebio1D3), anti-CD3 (145–2C11), anti-NK1.1 (PK136), anti NKp46 (29A1.4), anti-CD11b (M1/70), anti-CD27 (LG.7F9), anti-CD122 (5H4 or TMb1), anti-CD127 (A7R34), anti-CXCR3 (CXCR3–173), anti-CCR6 (HM-CCR6), anti-Ly49D (4e5), Ly49E (CM4), anti-Ly49G2 (4D11), anti-Ly49H (3D10), anti-Ly49I (YLI-90), anti-NKG2ACE (20d5), anti-NKG2D (CX5), anti-KLRG1 (2F1), anti-CD146 (Me9F1), anti-Eomes (Dan11mag), anti- IFN-γ (XMG1), anti-CD107a (1D4B), anti-GR1 (RB6.8), anti-Ly6C (AL21), anti-TCRγδ (GL3), anti-CD4 (GK1.5), anti-CD8 (XMG), anti-CD11c (N418), anti-RORγt, anti-T-bet (ebio4B10), and relevant isotype controls. Antibodies against phosphorylated proteins were from BD-biosciences: pAkt S473 (M89–61), or Cell Signaling Technologies: pS6 S235/236 (5316).
Functional analysis of NK cells
Splenic lymphocytes were prepared and cultured with cytokines (rmIL-15 100ng/ml; rmIL-12 25ng/ml from Peprotech and rmIL-18 5ng/ml from PBL), or on antibody coated plates (anti-NKp46, anti-NK1.1, anti-Ly49D all at 10μg/ml on Immulon 2HB plates) and Golgi-stop (4μl in 6ml; BD-Biosciences) in the presence of anti-CD107a for 4h. Surface and intracellular stainings were then performed and IFN-γ production as well as CD107a exposure was measured by flow cytometry.
Semi-quantitative RT-PCR
We used High capacity RNA-to-cDNA kit (applied biosystem, Carlsbad, USA) to generate cDNA for RT-PCR. PCR was carried out with a SybrGreen-based kit (FastStart Universal SYBR Green Master, Roche, Basel, Switzerland) or SensiFast SYBR No-ROX kit (Bioline) on a StepOne plus instrument (Applied biosystems, Carlsbad, USA) or a LightCycler 480 system (Roche). Primers were designed using the Roche software. The following primers were used for QPCR: Styk1 F TGGAAAGCAGATCCTTTTGG, Styk1 R CCACATCCCCATGAAACAG. These primers amplify a cDNA region spanning exons 6 and 7.
Measurement of serum IFNγ
IFNy level was measured in the serum using a commercial Elisa (DuoSet, RnD)
Statistical analysis
Data were analyzed with GraphPad Prism 6 software, using standard T-tests or One-way Anova, as explained. Differences were considered to be statistically significant when p < .05 ( * if p < .05, ** < .01, *** < .001, **** < .0001).
Supplementary Material
Acknowledgements
The authors thank Drs Marc Dalod and Elena Tomasello for providing the MCMV virus and for technical advice. The authors also thank core facilities of the SFR Biosciences, and in particular the flow cytometry and animal experimentation facilities. The TW lab is funded by the Agence Nationale pour la Recherche (ANR), by the Association pour la Recherche contre le Cancer (ARC, équipe labellisée), by the Institut National du Cancer (INCA) and by institutional grants from INSERM, CNRS, Université de Lyon and Ecole Normale Supérieure de Lyon.
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest
References
- 1.Lanier LL. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 2008; 9:495–502.DOI: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, et al. Innate lymphoid cells--a proposal for uniform nomenclature. Nat. Rev. Immunol. 2013; 13:145–149.DOI: 10.1038/nri3365. [DOI] [PubMed] [Google Scholar]
- 3.Walzer T, Chiossone L, Chaix J, Calver A, Carozzo C, Garrigue-Antar L, Jacques Y, et al. Natural killer cell trafficking in vivo requires a dedicated sphingosine 1-phosphate receptor. Nat. Immunol. 2007; 8:1337–1344.DOI: 10.1038/ni1523. [DOI] [PubMed] [Google Scholar]
- 4.Drouillard A, Mathieu A-L, Marçais A, Belot A, Viel S, Mingueneau M, Guckian K, et al. S1PR5 is essential for human natural killer cell migration toward sphingosine-1 phosphate. J. Allergy Clin. Immunol. 2017.DOI: 10.1016/j.jaci.2017.11.022. [DOI] [PubMed] [Google Scholar]
- 5.Walzer T, Bléry M, Chaix J, Fuseri N, Chasson L, Robbins SH, Jaeger S, et al. Identification, Activation, and Selective in Vivo Ablation of Mouse NK Cells Via NKp46. Proc. Natl. Acad. Sci. 2007; 104:3384–3389.DOI: 10.1073/pnas.0609692104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yu J, Mitsui T, Wei M, Mao H, Butchar JP, Shah MV, Zhang J, et al. NKp46 identifies an NKT cell subset susceptible to leukemic transformation in mouse and human. J. Clin. Invest. 2011; 121:1456–1470.DOI: 10.1172/JCI43242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, Gregoire C, et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. U. S. A. 2011; 108:18324–18329.DOI: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Luci C, Reynders A, Ivanov II, Cognet C, Chiche L, Chasson L, Hardwigsen J, et al. Influence of the transcription factor RORgammat on the development of NKp46+ cell populations in gut and skin. Nat Immunol. 2009; 10:82. [DOI] [PubMed] [Google Scholar]
- 9.Satoh-Takayama N, Vosshenrich CAJ, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention J-J, et al. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008; 29:958–970.DOI: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 10.Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, Best JA, et al. Molecular definition of the identity and activation of natural killer cells. Nat. Immunol. 2012; 13:1000–1009.DOI: 10.1038/ni.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu L, Yu X-Z, Li T-S, Song L-X, Chen P-L, Suo T-L, Li Y-H, et al. A novel protein tyrosine kinase NOK that shares homology with platelet- derived growth factor/fibroblast growth factor receptors induces tumorigenesis and metastasis in nude mice. Cancer Res. 2004; 64:3491–3499.DOI: 10.1158/0008-5472.CAN-03-2106. [DOI] [PubMed] [Google Scholar]
- 12.Moriai R, Kobayashi D, Amachika T, Tsuji N, Watanabe N. Diagnostic relevance of overexpressed NOK mRNA in breast cancer. Anticancer Res. 2006; 26:4969–4973. [PubMed] [Google Scholar]
- 13.Amachika T, Kobayashi D, Moriai R, Tsuji N, Watanabe N. Diagnostic relevance of overexpressed mRNA of novel oncogene with kinase-domain (NOK) in lung cancers. Lung Cancer Amst. Neth. 2007; 56:337–340.DOI: 10.1016/j.lungcan.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Kondoh T, Kobayashi D, Tsuji N, Kuribayashi K, Watanabe N. Overexpression of serine threonine tyrosine kinase 1/novel oncogene with kinase domain mRNA in patients with acute leukemia. Exp. Hematol. 2009; 37:824–830.DOI: 10.1016/j.exphem.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 15.Chung S, Tamura K, Furihata M, Uemura M, Daigo Y, Nasu Y, Miki T, et al. Overexpression of the potential kinase serine/ threonine/tyrosine kinase 1 (STYK 1) in castration-resistant prostate cancer. Cancer Sci. 2009; 100:2109–2114.DOI: 10.1111/j.1349-7006.2009.01277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson KA, Oprea G, Handy J, Kimbro KS. Aberrant STYK1 expression in ovarian cancer tissues and cell lines. J. Ovarian Res. 2009; 2:15.DOI: 10.1186/1757-2215-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao Q, Chen M, Li Z, Huang W, Jin Y, Ye X, Tong M. High Novel Oncogene with Kinase-Domain (NOK) Gene Expression is Associated with the Progression of Renal Cell Carcinoma. Clin. Lab. 2016; 62:179–186. [DOI] [PubMed] [Google Scholar]
- 18.Wang Z, Qu L, Deng B, Sun X, Wu S, Liao J, Fan J, et al. STYK1 promotes epithelial-mesenchymal transition and tumor metastasis in human hepatocellular carcinoma through MEK/ERK and PI3K/AKT signaling. Sci. Rep. 2016; 6:33205.DOI: 10.1038/srep33205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adams DJ, Doran AG, Lilue J, Keane TM. The Mouse Genomes Project: a repository of inbred laboratory mouse strain genomes. Mamm. Genome Off. J. Int. Mamm. Genome Soc. 2015; 26:403–412.DOI: 10.1007/s00335-015-9579-6. [DOI] [PubMed] [Google Scholar]
- 20.Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016; 536:285–291.DOI: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marçais A, Cherfils-Vicini J, Viant C, Degouve S, Viel S, Fenis A, Rabilloud J, et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nat. Immunol. 2014; 15:749–757.DOI: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez NC. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005; 105:4416–4423.DOI: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joncker NT, Shifrin N, Delebecque F, Raulet DH. Mature natural killer cells reset their responsiveness when exposed to an altered MHC environment. J. Exp. Med. 2010; 207:2065–2072.DOI: 10.1084/jem.20100570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song Y-J, Yang L, French AR, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005; 436:709–713.DOI: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 25.Simon MM, Greenaway S, White JK, Fuchs H, Gailus-Durner V, Wells S, Sorg T, et al. A comparative phenotypic and genomic analysis of C57BL/6J and C57BL/6N mouse strains. Genome Biol. 2013; 14:R82.DOI: 10.1186/gb-2013-14-7-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith HRC, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl. Acad. Sci. U. S. A. 2002; 99:8826–8831.DOI: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002; 296:6. [DOI] [PubMed] [Google Scholar]
- 28.Daniels KA, Devora G, Lai WC, O’Donnell CL, Bennett M, Welsh RM. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J Exp Med. 2001; 194:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levanon D, Negreanu V, Lotem J, Bone KR, Brenner O, Leshkowitz D, Groner Y. Transcription factor Runx3 regulates interleukin-15-dependent natural killer cell activation. Mol. Cell. Biol. 2014; 34:1158–1169.DOI: 10.1128/MCB.01202-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li J, Wu F, Sheng F, Li Y-J, Jin D, Ding X, Zhang S. NOK/STYK1 interacts with GSK-3β and mediates Ser9 phosphorylation through activated Akt. FEBS Lett. 2012; 586:3787–3792.DOI: 10.1016/j.febslet.2012.09.011. [DOI] [PubMed] [Google Scholar]
- 31.Zhao Y, Yang L, He J, Yang H. STYK1 promotes Warburg effect through PI3K/AKT signaling and predicts a poor prognosis in nasopharyngeal carcinoma. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2017; 39:1010428317711644.DOI: 10.1177/1010428317711644. [DOI] [PubMed] [Google Scholar]
- 32.Shi W-Y, Yang X, Huang B, Shen WH, Liu L. NOK mediates glycolysis and nuclear PDC associated histone acetylation. Front. Biosci. Landmark Ed. 2017; 22:1792–1804. [DOI] [PubMed] [Google Scholar]
- 33.Marçais A, Marotel M, Degouve S, Koenig A, Fauteux-Daniel S, Drouillard A, Schlums H, et al. High mTOR activity is a hallmark of reactive natural killer cells and amplifies early signaling through activating receptors. eLife. 2017; 6.DOI: 10.7554/eLife.26423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van Roosmalen W, Le Dévédec SE, Golani O, Smid M, Pulyakhina I, Timmermans AM, Look MP, et al. Tumor cell migration screen identifies SRPK1 as breast cancer metastasis determinant. J. Clin. Invest. 2015; 125:1648–1664.DOI: 10.1172/JCI74440. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.