Abstract
Background and Aim
The apolipoprotein C3 (APOC3) polymorphism has been reported to predispose to non-alcoholic fatty liver disease (NAFLD). However, the results remain inconclusive. This meta-analysis aimed to provide insights into the association between APOC3 polymorphisms and NAFLD risk.
Methods
Studies with terms “NALFD” and “APOC3” were retrieved from PubMed, Web of Science, CNKI and Wanfang databases up to August 1, 2019. Pooled odds ratio (OR) and 95% confidence interval (95% CI) for the association of APOC3 polymorphisms and NAFLD risk were calculated using fixed and random-effects models.
Results
A total of twelve studies from eleven articles were included. Of them, eight studies (1750 cases and 2181 controls) reported the strong association of variant rs2854116 with NAFLD and six studies (1523 cases and 1568 controls) found the association of rs2854117 polymorphism with NAFLD. Overall, a statistically significant association between rs2854116 polymorphism of APOC3 gene and NAFLD risk was found only under dominant model. However, association of rs2854117 polymorphism with NAFLD risk was not detected under all four genetic models. In sub-group analysis of NAFLD subjects based on country, no association among them in China was detected. Besides, four studies analyze the association between the two polymorphisms and clinical characteristics in all subjects or NAFLD patients, and we also failed detect any association between the wild carriers and variant carriers.
Conclusion
The meta-analyses suggests that the rs2854116 polymorphism but not rs2854117 polymorphism in APOC3 gene might be a risk factor for NAFLD among Asians. That is, individuals with CT+CC genotype have higher risk of developing NAFLD. However, studies with sufficient sample size are needed for the further validation.
Keywords: Apolipoprotein C3, polymorphism, non-alcoholic fatty liver disease, meta-analysis
Introduction
Non-alcoholic fatty liver disease (NAFLD) is one of the most common cause of liver disease worldwide with prevalence varying from 15% to 40% in general population depending on the demographic area 1. Currently, the population prevalence in Asia is around 25%. Among more affluent regions of China, the community prevalence is about 15% 2,3. The spectrum of this disease ranges from bland steatosis, non-alcoholic steatohepatitis (NASH), fibrosis, cirrhosis, and occasionally to hepatocellular carcinoma 4 . NAFLD is considered to be the hepatic manifestation of the metabolic syndrome (MS), which is characterized by obesity, type 2 diabetes, dyslipidemia and hypertension with insulin resistance being the main mechanisms 5. Although the exact etiology of NAFLD is not well delineated, it is a complex metabolic condition in which both lifestyle and genetic factors play a pathogenic role 6.
The apolipoprotein C3 gene (APOC3) is a member of the APOA1/C3/A4/A5 gene cluster and located on chromosome 11q23, an area in strong linkage with lipid metabolism 7. APOC3 is a 79-amino-acid glycoprotein and a major component of TG-rich lipoproteins (TRLs). Indeed, APOC3 impairs the lipolysis of TRLs by inhibiting lipoprotein lipase and the hepatic uptake of TRLs by remnant receptors 8,9. A few genetic studies suggest that the single nucleotide polymorphisms (SNPs) in the APOC3 gene may have implications for hypertriglyceridemia 10. Homozygotes for the C-482T and T-455C variants are resistant to insulin-mediated down-regulation of APOC3 gene transcription, which results in high TG levels 11. Recently, several studies have investigated the association between single nucleotide polymorphisms (SNPs) in the APOC3 gene and NAFLD risk. Of those SNPs, two were most commonly investigated: rs2854116 (T455C in the promoter region), and rs2854117 (C482T in the promoter region). However, the results have been inconsistent 12–25. In this study, we conducted a meta-analysis in order to get a robust conclusion about the association between the two polymorphisms in the APOC3 gene and NAFLD susceptibility.
Materials and Methods
Search strategy
We searched the PubMed, Web of Science, CNKI and Wanfang database before August 1, 2019, by using the key subjects “apolipoprotein C3 or APOC3” AND “genetic polymorphism or polymorphisms or variant” AND “non-alcoholic fatty liver disease or NAFLD”. Studies were restricted to human populations and studies written in other languages except for English were also considered. Additional studies were identified by a hand search of references of original or review articles on this topic.
Inclusion criteria and exclusion criteria
Studies were included according to the following criteria: (1) studies with clear diagnostic criteria, (2) studies that evaluatd the association between the APOC3 polymorphisms and NAFLD, (3) studies in a case-control design, and (4) studies with detailed genotype frequency of cases and controls or providing the necessary data to calculate genotype frequency. Studies were excluded when they were: (1) case-only study, case reports, and review articles, (2) studies without the raw data of the APOC3 genotype, and (3) repetitive publications.
Data extraction
For each study, the following data were extracted independently by two investigators: the first author's name, year of publication, country of origin, age, gender, study design, diagnostic criteria of NAFLD, number of cases and controls, and HWE in controls (P value) and analysis were carried on independently. The results obtained by those two investigators were compared, and disagreements were discussed among all authors and resolved with consensus.
Statistical analysis
The risk of NAFLD associated with the two polymorphisms of the APOC3 gene was estimated for each study by odds ratio (OR) and 95% confidence interval (95%CI). A χ2-test-based Q statistic test was performed to assess the between-study heterogeneity 26. We also quantified the effect of heterogeneity by I2 test. When a significant Q test (P≤ 0.05) or I2 >50% indicated heterogeneity across studies, the random effects model was used 27. Otherwise, the fixed effects model was adopted 28. Before estimating the effect of APOC3 polymorphisms on NAFLD, we tested whether genotype frequencies of controls were in HWE using χ2 test. We first estimated the risks of the heterozygote and variant homozygote compared with the wild-type homozygote, respectively, and then evaluated the risks of the combined variant homozygote and heterozygote versus the wild-type homozygote, and the variant homozygote versus the combined heterozygote and wildtype homozygote, assuming dominant and recessive effects of the variant allele, respectively. We performed stratification analyses on country. Analysis of sensitivity was performed to evaluate the stability of the results. Finally, potential publication bias was investigated using Begg' funnel plot and Egger's regression test 29,30. P<0.05 was regarded as statistically significant.
All statistical analyses were performed using software Cochrane Collaboration RevMan 5.2 and STATA package version 12.0 (Stata Corporation, College Station, Texas).
Results
Study characteristics
After an initial search, a total of 118 published articles relevant to the topic were identified. According to the inclusion criteria, 15 studies with full-text were included in this meta-analysis and 103 studies were excluded. Because the study by Cai et al 12 included two populations, we treated them separately in this meta-analysis. We excluded three studies 23–25 due to the absence of detailed genotyping information. The process of study selection is briefly summarized in Figure 1. As shown in Table 1, there were 8 case-control studies with 1750 NAFLD cases and 2181 controls concerning rs2854116 polymorphism and 6 case-control studies with 1523 cases and 1568 controls concerning rs2854117 polymorphism. Four studies with 2384 subjects reported the clinical and biochemical parameters in the study grops. The distribution of genotypes in the controls was consistent with the Hardy-Weinberg equilibrium for almost all selected studies except one study19.
Figure 1.
Flow chart showing study selection procedure.
Table 1.
Characteristics of studies included in the meta-analysis
| Study | Year | Country | Age, mean ± SD, year | Gender(male/female) | Diagnostic criteria | Genotype (case/control) | PHWE | ||||
| case | control | case | control | WT Ho | Ht | VR Ho | |||||
| rs2854116 | TT | TC | CC | ||||||||
| Cai et al. | 2013 | China (Han) |
43.68±10.72 | 42.88±10.0 | NA | NA | The criteria of NAFLD designed by the Chinese Liver Disease Association (2010) |
51/70 | 109/103 | 33/38 | 0.992 |
| Cai et al. | 2013 | China (Uyghur) |
42.39±8.79 | 41.59±9.07 | NA | NA | The criteria of NAFLD designed by the Chinese Liver Disease Association (2010) |
56/54 | 115/94 | 32/40 | 0.939 |
| Li M et al. | 2014 | China | 40.7±9.7 | 39.5±9.1 | 200/100 | 200/100 | Ultrasonography | 94/134 | 131/123 | 75/43 | 0.093 |
| Niu et al. | 2014 | China | 49.76±16.17 | 47.69±15.86 | 175/215 | 198/211 | The criteria of NAFLD designed by the AASID |
102/104 | 180/195 | 108/110 | 0.350 |
| Niu et al. | 2014 | China | 49.96±16.20 | 47.81±16.19 | 132/155 | 146/164 | The criteria of NAFLD designed by the Chinese Liver Disease Association (2010) |
81/80 | 139/147 | 67/83 | 0.364 |
| Puppala et al. |
2014 | India | 44.1±12.1 | 42.6±10.6 | 92/58 | 92/58 | Ultrasonography | 44/60 | 75/81 | 31/9 | 0.007 |
| Song et al. | 2017 | China | NA | NA | 39/93 | 94/158 | The criteria of NAFLD designed by the Chinese Liver Disease Association (2010) |
44/88 | 63/117 | 23/46 | 0.517 |
| Yang et al. |
2018 | China | 70.95±4.73 | 72.53±5.67 | 25/72 | 133/229 | Guidelines for management of nonalcoholic fatty liver disease: an updated and revised edition |
39/135 | 46/177 | 12/50 | 0.506 |
| rs2854117 | |||||||||||
| Cai et al. | 2013 | China (Han) |
43.68±10.72 | 42.88±10.0 | NA | NA | The criteria of NAFLD designed by the Chinese Liver Disease Association (2010) |
52/71 | 106/99 | 35/41 | 0.539 |
| Cai et al. | 2013 | China (Uyghur) |
42.39±8.79 | 41.59±9.07 | NA | NA | The criteria of NAFLD designed by the Chinese Liver Disease Association (2010) |
73/71 | 105/86 | 25/31 | 0.567 |
| Li et al. | 2014 | China | 40.7±9.7 | 39.5±9.1 | 200/100 | 200/100 | The criteria of NAFLD designed by the Chinese Liver Disease Association (2010) |
108/126 | 144/127 | 48/47 | 0.118 |
| Niu et al. | 2014 | China | 49.76±16.17 | 47.69±15.86 | 175/215 | 198/211 | The criteria of NAFLD designed by the AASID |
107/104 | 176/203 | 107/102 | 0.882 |
| Niu et al. | 2014 | China | 49.96±16.20 | 47.81±16.19 | 132/155 | 146/164 | The criteria of NAFLD designed by the Chinese Liver Disease Association (2010) |
81/79 | 132/156 | 74/75 | 0.907 |
| Puppala et al. |
2014 | India | 44.1±12.1 | 42.6±10.6 | 92/58 | 92/58 | Ultrasonography | 55/62 | 57/46 | 38/42 | 0.000 |
HWE, Hardy-Weinberg equilibrium; PHWE was calculated by goodness-of fit χ2-test, PHWE < 0.05 was considered statistically significant; NA, not available. Ht, heterozygote; VR Ho, variant homozygote; WT Ho, wide-type homozygote.
Quantitative synthesis
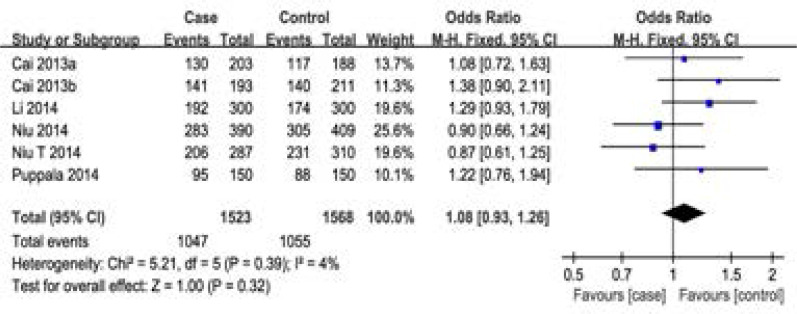
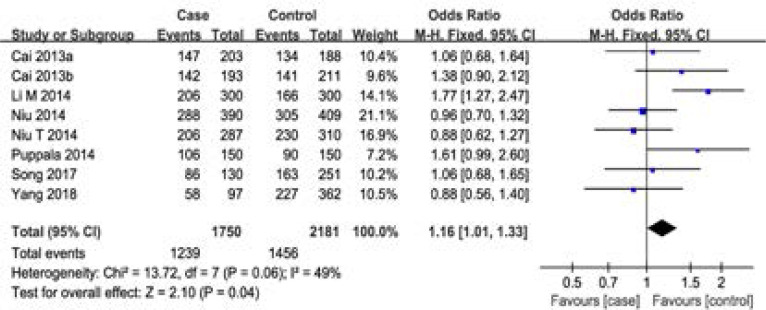
For rs2854116 polymorphism, 8 case-control studies with 1750 cases and 2181 controls were identified. Overall, a significant association was detected under dominant model (OR=1.16, 95%CI: 1.01–1.33, P=0.04), however, there was no significant difference in APOC3 genotype distribution between NAFLD and control (OR=1.14, 95%CI: 0.82–1.57, P=0.44 for recessive model, OR=1.13, 95%CI: 0.98–1.31, P=0.10 for CT vs TT, OR=1.24, 95%CI: 0.84–1.82, P=0.28 for CC vs TT) (Figure 2, Table 2). That is to say, individuals with TC or CC genotype had a higher risk of developing NAFLD. In the sub-group analysis by country, no significant association was found in Chinese populations. Although only one study focused on Indian population, our result suggested that the APOC3 gene polymorphism T-455C (rs2854116) was significantly associated with NAFLD. For rs2854117 polymorphism, 6 case-control studies with 1523 cases and 1568 controls were identified. Overall, no significant association between the polymorphism of APOC3 gene and NAFLD risk was detected (OR=1.08, 95%CI: 0.93–1.26 for dominant model, OR=1.00, 95%CI: 0.84–1.19 for recessive model, OR=1.10, 95%CI: 0.93–1.29 for CT vs CC, OR=1.02, 95%CI: 0.84–1.25 for TT vs CC) (Figure 3, Table 2). No correlation was detected between groups stratified on country among Chinese populations. Furthermore, one study indicated that the C-482T polymorphism was not a risk indicator of NAFLD for Indian.
Figure 2.
Meta-analysis of the association between rs2854116 polymorphism and susceptibility to NAFLD under dominant model.
Table 2.
Summary of ORs of the APOC3 polymorphisms and NAFLD risk
| SNP | N | Dominant model | Recessive model | Ht vs. WT Ho | VR Ho vs. WT Ho | ||||||||
| OR(95% CI) | Pa | I2 | OR(95% CI) | Pa | I2 | OR(95% CI) | Pa | I2 | OR(95% CI) | Pa | I2 | ||
| rs2854116 | 8 | 1.16(1.01,1.33) | 0.06 | 49% | 1.14(0.82,1.57) | 0.0007 | 72% | 1.13(0.98,1.31) | 0.44 | 0% | 1.24(0.84,1.82) | 0.0003 | 74% |
| rs2854117 | 6 | 1.08(0.93,1.26) | 0.39 | 4% | 1.00(0.84,1.19) | 0.76 | 0% | 1.10(0.93,1.29) | 0.16 | 38% | 1.02(0.84,1.25) | 0.93 | 0% |
N: number of studies
Test for heterogeneity.
# Random-effect model was used when the P for heterogeneity test was ≤. 0.05, otherwise the fixed-effect model was used. CI, confidence interval; OR, odds ratio; SNP, single-nucleotide polymorphism; Ht+VR Ho vs. WT Ho, dominant model; VR Ho vs. Ht+WT Ho, recessive model.
Figure 3.
Meta-analysis of the association between rs2854117 polymorphism and susceptibility to NAFLD under dominant model.
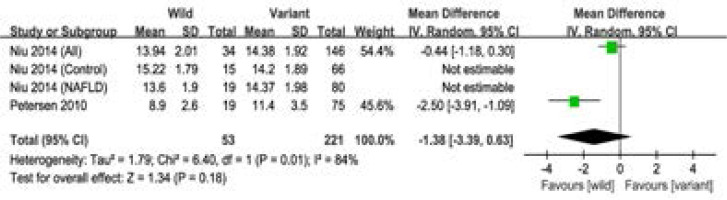
In addition, four studies with 2384 subjects reported the clinical characteristics in the study groups. No significant association was observed between the polymorphism and clinical characteristics (BMI, Waist, APOC3, AST, ALT, FINS, HOMA-IR, FPG, TG, HDL, LDL, TC) (Figure 4, Table 3). Moreover, three studies with 1148 subjects reported the clinical characteristics in the NAFLD group, and Our results suggested the polymorphism was not associated with the clinical characteristics (BMI, HOMA-IR, FPG, TG, HDL, LDL) (Table 3).
Figure 4.
Meta-analysis of the association between the two polymorphisms and the APOC3 level.
Table 3.
Summary of SMD of the clinical characteristics between the wild carriers and variant carriers
| All subjects | NAFLD group | |||||||
| N | SMD(95% CI) | Pa | I2 | N | SMD(95% CI) | Pa | I2 | |
| BMI | 5 | 0.16(-0.25,0.58) | 0.86 | 0 | 3 | 0.48(-0.04,0.99) | 0.83 | 0 |
| Waist | 2 | -0.09(-3.48,3.30) | 0.16 | 50 | ||||
| FPG | 5 | -1.18(-2.94,0.58) | 0.42 | 0 | 3 | -0.49(-4,27,3.29) | 0.19 | 40 |
| HOMA-IR | 4 | -0.06(-0.13,0.00) | 0.29 | 19 | 3 | -0.11(-0.22,0.01) | 0.60 | 0 |
| TG | 6 | -4.50(-19.36,10.35) | 0.005 | 70 | 3 | 2.69(-0.97,15.10) | 0.20 | 38 |
| HDL | 6 | 0.83(-0.68,2.34) | 0.19 | 33 | 3 | -3.09(-8.17,1.99) | 0.003 | 83 |
| LDL | 6 | 1.81(-6.09,9.71) | 0.004 | 72 | 3 | 4.02(-2.39,10.43) | 0.57 | 0 |
| TC | 3 | 1.37(-11.98,14.73) | 0.01 | 77 | ||||
| FINS | 2 | -0.06(-0.26,0.13) | 0.42 | 0 | ||||
| APOC3 | 2 | -1.38(-3.39,0.63) | 0.01 | 84 | ||||
| ALT | 2 | -0.01(-4.88,4.85) | 0.99 | 0 | ||||
| AST | 2 | -2.13(-5.65,1.38) | 0.47 | 0 | ||||
N: number of studies
Test for heterogeneity
BMI, body mass index; FPG, fasting blood glucose; HOMA-IR, homeostatic model assessment of insulin resistance; TG, triglyceride; HDL, high-density lipoprotein; LDL, low-density lipoprotein; TC, total cholesterol; FINS, fasting blood insulin; APOC3, apolipoprotein C3; ALT, alanine aminotransferase; AST, aspartate aminotransferase; NAFLD, nonalcoholic fatty liver disease; SMD, standardized mean difference
Heterogeneity and sensitivity analysis
For rs2854116 polymorphism, significant heterogeneity between studies was observed in overall comparisons under recessive and CC versus TT models (I2=72%, Pheterogeneity=0.0007, I2=74%, Pheterogeneity= 0.0003, respectively). For rs2854117 polymorphism, no significant heterogeneity between studies was observed in overall comparisons. Then, sensitivity analysis, after removing one study at a time, was performed to evaluate the stability of the results. The results was not changed when any single study was omitted, confirming the stability of the results.
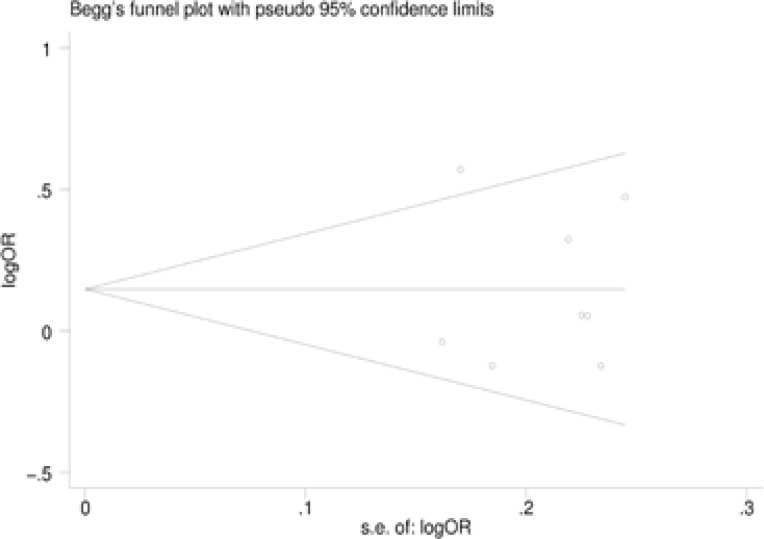
Publication bias
Begg's funnel plot and Egger's test were performed to assess the potential publication bias in the available literature. The shape of funnel plots did not reveal any evidence of funnel plot asymmetry (Figure 5). Egger's test also showed that there was no statistical significance for the evaluation of publication bias under dominant model (rs2854116: P=0.987, rs2854117: P=0.383).
Figure 5.
Begg's funnel plot for publication bias under dominant model. a rs2854116. b rs2854117.
Discussion
NAFLD is one of the most common liver disorders31,32. Although the pathogenesis of NAFLD remains largely unknown, insulin resistance, oxidative stress and inflammation play important roles in the development and progression of NAFLD 33. Many candidate genes were reported to be associated with NAFLD risk, such as TNF-α, PNPLA3, APOC3, PPAR-γ and adiponectin34–37. Our previous study38 performed a meta-analysis to evaluate the association between adiponectin polymorphisms and NAFLD susceptibility and the result suggested that adiponectin +45T>G and −11377C>G polymorphisms might be a risk factor for NAFLD, and +276G>T polymorphism may be a protective factor for NAFLD among Asians. As for APOC3, it is a glycoprotein synthesized mainly in the liver and the intestinal, and plays an essential role in regulating the serum triglyceride levels. Two polymorphisms, T-455C and C-482T, were located in the 5'promoter region and were in a strong linkage disequilibrium with each other. This polymorphism leads to a roughly 30% higher plasma concentration of ApoC3, and postprandial hypertriglyceridaemia. As a result, individuals with carrier of those polymorphisms can take up increased amounts of lipid from the chylomicron remnant, leading to NAFLD and hepatic insulin resistance 39. Transgenic mice overexpressing human APOC3 were predisposed to hepatic steatosis, indicating that APOC3 might play an important role in the development of NAFLD 40,41. However, the results remain controversial. Although Petersen et al. have recently reported that the polymorphisms C-482T and T-455C in APOC3 are associatd with nonalcoholic fatty liver disease and insulin resistance, the association of APOC3 variants and NAFLD risk has not yet been validated by others so far.
Recently, the APOC3 polymorphisms have been investigated the association with many diseases, such as metabolic syndrome 42, type 2 diabetes 43, coronary heart disease 44 and plasma APOC3 and lipid levels 45. As for NAFLD, a previous meta-analysis conducted by Zhang et al. 46, evaluated the association between APOC3 polymorphisms and risk of NAFLD based on 7 studies and reported that the APOC3 gene polymorphism is not a genetic risk factor for NAFLD. Ithis study, we conducted a comprehensive literature search in different databases and included several additional studies, which allowed for a larger number of subjects (12 studies) and more precise risk estimation. Besides, we further analyze the association between the two polymorphisms and clinical characteristics in all subjects or NAFLD patients. A significant association between the rs2854116 polymorphism of APOC3 gene and NAFLD risk were found under dominant model. However, no association of rs2854117 polymorphism with NAFLS was detected. When stratified by country, no association among them in China was detected. Interestingly, Puppala et al suggested that the APOC3 gene polymorphism T-455C (rs2854116) was significantly associated with NAFLD in Southern Indian population. These inconsistent results may be attributed to differences in genetic backgrounds, environmental factors, and other factors, such as small sample size or inadequate adjustment for confounding factors.
In this(our) study, the potential correlation was also analyzed between the two polymorphisms and clinical characteristics. Four studies were enrolled and the combined results showed that there was no significant difference between the wide-type genotype and the variant genotypes in either all study groups or NAFLD group. Since only few studies reported the available data, this result should be interpreted with caution, and more studies are needed.
Heterogeneity between studies is very common in the meta-analysis of genetic association studies 47. In this(our) study, heterogeneity was found under recessive model and CC versus TT model for rs2854116 polymorphism. So stratified analysis by country was performed and the heterogeneity also existed in Asian population. Then sensitivity analyses were conducted by successively excluding one study, and the estimated pooled OR changed quite little, strengthening the conclusions from this meta-analysis. Besides, no publication bias was not observed, backuping the validity and generalization of our conclusions.
Some limitations of this meta-analysis should be addressed. First, due to incomplete raw data or publication limitations, some relevant studies could not be included in our analysis. Second, the number of published studies, especially for Indian population was not sufficiently large for a comprehensive analysis, and some studies with small sample size may not have enough statistical power to explore the real association. Third, our results were based on unadjusted estimates, and lacking of the necessary information (such as age, gender, family history and other risk factors) for the date analysis may cause serious confounding bias.
In summary, this meta-analysis suggested that APOC3 rs2854116 polymorphism is associated with NAFLD risk. Individuals with CT+CC genotype had an increased risk of NAFLD. Studies with large sample zise are needed to validate our findings.
Conflict of interest
None declared.
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-Meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology. 2016;64:73–84. doi: 10.1002/hep.28431. PubMed. [DOI] [PubMed] [Google Scholar]
- 2.Fan JG, Kim SU, Wong VW. New trends on obesity and NAFLD in Asia. J Hepatol. 2017;67(4):862–873. doi: 10.1016/j.jhep.2017.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Fan JG, Farrell GC. Epidemiology of non-alcoholic fatty liver disease in China. J Hepatol. 2009;50(1):204–210. doi: 10.1016/j.jhep.2008.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313:2263–2273. doi: 10.1001/jama.2015.5370. PubMed. [DOI] [PubMed] [Google Scholar]
- 5.Mendez-Sanchez N, Arrese M, Zamora-Valdes D, Uribe M. Current concepts in the pathogenesis of nonalcoholic fatty liver disease. Liver Int. 2007;27:423–433. doi: 10.1111/j.1478-3231.2007.01483.x. [DOI] [PubMed] [Google Scholar]
- 6.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 7.Bossé Y, Chagnon YC, Després JP, Rice T, Rao DC, Bouchard C, et al. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Quebec Family Study. J Lipid Res. 2004;45:419–426. doi: 10.1194/jlr.M300401-JLR200. [DOI] [PubMed] [Google Scholar]
- 8.Ekman R, Nilsson-Ehle P. Effects of apolipoproteins on lipoprotein lipase activity of human adipose tissue. Clin Chim Acta. 1975;63:29–35. doi: 10.1016/0009-8981(75)90374-5. [DOI] [PubMed] [Google Scholar]
- 9.Quarfordt SH, Michalopoulos G, Schirmer B. The effect of human C apolipoproteins on the in vitro hepatic metabolism of triglyceride emulsions in the rat. J Biol Chem. 1982;257:14642–14647. [PubMed] [Google Scholar]
- 10.Talmud PJ, Humphries SE. Apolipoprotein C-III gene variation and dyslipidaemia. Curr Opin Lipidol. 1997;8:154–158. doi: 10.1097/00041433-199706000-00005. [DOI] [PubMed] [Google Scholar]
- 11.Li WW, Dammerman MM, Smith JD, Metzger S, Breslow JL, Leff T. Common genetic variation in the promoter of the human apo CIII gene abolishes regulation by insulin and may contribute to hypertriglyceridemia. J Clin Invest. 1995;96:2601–2605. doi: 10.1172/JCI118324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cai W, Yao H. The study on the correlation of nonalcoholic fatty liver disease with polymorphism gene of lipid metabolism and cytokines. Wulumuqi, People's Republic of China: XinJiang Medical University; 2013. PhD thesis. [Google Scholar]
- 13.Hyysalo J, Stojkovic I, Kotronen A, Hakkarainen A, Sevastianova K, Makkonen J, et al. Genetic variation in PNPLA3 but not APOC3 influences liver fat in non-alcoholic fatty liver disease. J Gastroenterol Hepatol. 2012;27:951–956. doi: 10.1111/j.1440-1746.2011.07045.x. [DOI] [PubMed] [Google Scholar]
- 14.Li MR, Zhang SH, Liao XH, Zhong BH. Relation between apolipoproteinC3 (-482C>T) polymorphism and nonalcoholic fatty liver disease in the Han Chinese population. The Journal of Practical Medicine. 2014;30:2566–2549. [Google Scholar]
- 15.Li MR, Zhang SH, Chao K, Liao XH, Yao JY, Chen MH, et al. Apolipoprotein C3 (-455T>C) polymorphism confers susceptibility to nonalcoholic fatty liver disease in the Southern Han Chinese population. World J Gastroenterol. 2014;20:14010–14017. doi: 10.3748/wjg.v20.i38.14010. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niu TH, Jiang M, Xin YN, Jiang XJ, Lin ZH, Xuan SY. Lack of association between apolipoprotein C3 gene polymorphisms and risk of nonalcoholic fatty liver disease in a Chinese Han population. World J Gastroenterol. 2014;20:3655–3662. doi: 10.3748/wjg.v20.i13.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Niu T, Jiang M, Liu H, Jiang X, Lin Z, Zhang M, et al. Association between APOC3 promoter region polymorphisms and non-alcoholic fatty liver disease. Zhonghua Gan Zang Bing Za Zhi. 2014;22:374–379. doi: 10.3760/cma.j.issn.1007-3418.2014.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Petersen KF, Dufour S, Hariri A, Nelson-Williams C, Foo JN, Zhang XM, et al. Apolipoprotein C3 gene variants in nonalcoholic fatty liver disease. N Engl J Med. 2010;362:1082–1089. doi: 10.1056/NEJMoa0907295. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Puppala J, Bhrugumalla S, Kumar A, Siddapuram SP, Viswa PD, Kondawar M, et al. Apolipoprotein C3 gene polymorphisms in Southern Indian patients with nonalcoholic fatty liver disease. Indian J Gastroenterol. 2014;33:524–529. doi: 10.1007/s12664-014-0504-9. [DOI] [PubMed] [Google Scholar]
- 20.Song X, Song C, Fan L, Ma Q, Mao J, Xu W, Li X. Association of single nucleotide polymorphism of SIRT1 and APOC3 with nonalcoholic fatty liver disease. Wei Sheng Yan Jiu. 2017;46(4):527–532. PubMed. [PubMed] [Google Scholar]
- 21.Valenti L, Nobili V, Al-Serri A, Rametta R, Leathart JB, Zappa MA, et al. The APOC3 T-455C and C-482T promoter region polymorphisms are not associated with the severity of liver damage independently of PNPLA3 I148M genotype in patients with nonalcoholic fatty liver. J Hepatol. 2011;55:1409–1414. doi: 10.1016/j.jhep.2011.03.035. PubMed. [DOI] [PubMed] [Google Scholar]
- 22.Yang H, Chen G, Song C, Li D, Ma Q, Chen G, Li X. A novel index including SNPs for the screening of nonalcoholic fatty liver disease among elder Chinese: A population-based study. Medicine (Baltimore) 2018;97(13):e0272. doi: 10.1097/MD.0000000000010272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kozlitina J, Boerwinkle E, Cohen JC, Hobbs HH. Dissociation between APOC3 variants, hepatic triglyceride content and insulin resistance. Hepatology. 2011;53:467–474. doi: 10.1002/hep.24072. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sentinelli F, Romeo S, Maglio C, Incani M, Burza MA, Scano F, et al. Lack of effect of apolipoprotein C3 polymorphisms on indices of liver steatosis, lipid profile and insulin resistance in obese Southern Europeans. Lipids Health Dis. 2011;10:93. doi: 10.1186/1476-511X-10-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang RN, Zheng RD, Mi YQ, Zhou D, Shen F, Chen GY, et al. APOC3 rs2070666 Is Associated with the Hepatic Steatosis Independently of PNPLA3 rs738409 in Chinese Han Patients with Nonalcoholic Fatty Liver Diseases. Dig Dis Sci. 2016;61:2284–2293. doi: 10.1007/s10620-016-4120-7. PubMed. [DOI] [PubMed] [Google Scholar]
- 26.Lau J, Ioannidis JP, Schmid CH. Quantitative synthesis in systematic reviews. Ann Intern Med. 1997;127:820–826. doi: 10.7326/0003-4819-127-9-199711010-00008. [DOI] [PubMed] [Google Scholar]
- 27.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 28.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. PubMed. [DOI] [PubMed] [Google Scholar]
- 29.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. PubMed. [PubMed] [Google Scholar]
- 30.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018 Jan;67(1):328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 32.Younossi ZM. Non-alcoholic fatty liver disease - A global public health perspective. J Hepatol. 2018 Nov 9; doi: 10.1016/j.jhep.2018.10.033. pii: S0168-8278(18)32517-0. [DOI] [PubMed] [Google Scholar]
- 33.Obika M, Noguchi H. Diagnosis and evaluation of nonalcoholic fatty liver disease. Exp Diabetes Res. 2012;2012:145754. doi: 10.1155/2012/145754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Di Rosa M, Malaguarnera L. Genetic variants in candidate genes influencing NAFLD progression. J Mol Med (Berl) 2012;90:105–118. doi: 10.1007/s00109-011-0803-x. [DOI] [PubMed] [Google Scholar]
- 35.Wang JK, Feng ZW, Li YC, Li QY, Tao XY. Association of tumor necrosis factor-α gene promoter polymorphism at sites -308 and -238 with non-alcoholic fatty liver disease: a meta-analysis. J Gastroenterol Hepatol. 2012;27:670–676. doi: 10.1111/j.1440-1746.2011.06978.x. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, You W, Zhang H, Peng R, Zhu Q, Yao A, et al. PNPLA3 polymorphisms (rs738409) and non-alcoholic fatty liver disease risk and related phenotypes: a meta-analysis. J Gastroenterol Hepatol. 2015;30:821–829. doi: 10.1111/jgh.12889. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Guo X, Wu P, Song J, Ye C, Yu S, et al. Association between the Pro12Ala polymorphism of PPAR-γ gene and the non-alcoholic fatty liver disease: a meta-analysis. Gene. 2013;528:328–334. doi: 10.1016/j.gene.2013.07.014. PubMed. [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Guo XF, Yu SJ, Song J, Zhang JX, Cao Z, et al. Adiponectin polymorphisms and non-alcoholic fatty liver disease risk: a meta-analysis. J Gastroenterol Hepatol. 2014;29:1396–1405. doi: 10.1111/jgh.12562. [DOI] [PubMed] [Google Scholar]
- 39.Samuel VT, Petersen KF, Shulman GI. Lipid-induced insulin resistance: unravelling the mechanism. Lancet. 2010;375:2267–2277. doi: 10.1016/S0140-6736(10)60408-4. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ito Y, Azrolan N, O'Connell A, Walsh A, Breslow JL. Hypertriglyceridemia as a result of human apo CIII gene expression in transgenic mice. Science. 1990;249:790–793. doi: 10.1126/science.2167514. PubMed. [DOI] [PubMed] [Google Scholar]
- 41.Lee HY, Birkenfeld AL, Jornayvaz FR, Jurczak MJ, Kanda S, Popov V, et al. Apolipoprotein CIII overexpressing mice are predisposed to diet-induced hepatic steatosis and hepatic insulin resistance. Hepatology. 2011;54:1650–1660. doi: 10.1002/hep.24571. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Povel CM, Boer JM, Reiling E, Feskens EJ. Genetic variants and the metabolic syndrome: a systematic review. Obes Rev. 2011;12:952–967. doi: 10.1111/j.1467-789X.2011.00907.x. PubMed. [DOI] [PubMed] [Google Scholar]
- 43.van Hoek M, van Herpt TW, Dehghan A, Hofman A, Lieverse AG, van Duijn CM, et al. Association of an APOC3 promoter variant with type 2 diabetes risk and need for insulin treatment in lean persons. Diabetologia. 2011;54:1360–1367. doi: 10.1007/s00125-011-2092-x. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang JZ, Xie X, Ma YT, Zheng YY, Yang YN, Li XM, et al. Association between Apolipoprotein C-III Gene Polymorphisms and Coronary Heart Disease: A Meta-analysis. Aging Dis. 2016;7:36–44. doi: 10.14336/AD.2015.0709. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Song Y, Zhu L, Richa M, Li P, Yang Y, Li S. Associations of the APOC3 rs5128 polymorphism with plasma APOC3 and lipid levels: a meta-analysis. Lipids Health Dis. 2015;14:32. doi: 10.1186/s12944-015-0027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Chen L, Xin Y, Lou Y, Liu Y, Xuan S. Apolipoprotein c3 gene polymorphisms are not a risk factor for developing non-alcoholic Fatty liver disease: a meta-analysis. Hepat Mon. 2014;14(10):e23100. doi: 10.5812/hepatmon.23100. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boccia S, De Feo E, Gallı P, Gianfagna F, Amore R, Ricciardi G. A systematic review evaluating the methodological aspects of meta-analyses of genetic association studies in cancer research. Eur J Epidemiol. 2010;25:765–775. doi: 10.1007/s10654-010-9503-z. [DOI] [PubMed] [Google Scholar]