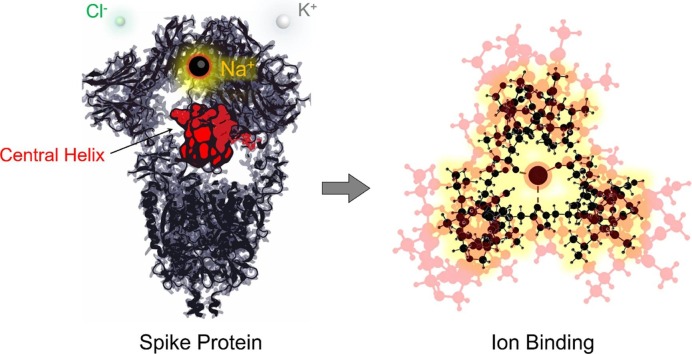
Graphical abstract
Keywords: SARS-CoV, Spike protein, Central helix funnel, Physiological electrolytes, Ion binding, DFT
Abstract
The trimeric spike (S) glycoprotein is the trojan horse and the stronghold of the severe acute respiratory syndrome coronaviruses. Although several structures of the S-protein have been solved, a complete understanding of all its functions is still lacking. Our multi-approach study, based on the combination of structural experimental data and quantum-chemical DFT calculations, led to identify a sequestration site for sodium, potassium and chloride ions within the central cavity of both the SARS-CoV-1 and SARS-CoV-2 spike proteins. The same region was found as strictly conserved, even among the sequences of the bat-respective coronaviruses. Due to the prominent role of the main three electrolytes at many levels, and their possible implication in the molecular mechanisms of COVID-19 disease, our study can take the lead in important discoveries related to the SARS-CoV-2 biology, as well as in the design of novel effective therapeutic strategies.
1. Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) protein is a trimeric class I fusion protein [1]. It triggers the envelope fusion with the human (h) host cell membrane by h-ACE-2 receptor mediated endocytosis [2], [3], [4]. The S-protein represents the trojan horse of coronavirus infection and, consequently, the most promising candidate for the development of vaccines and drugs [5], [6]. Each S-protein monomer weighs ∼180 kDa and is made up of two subunits, S1 and S2 [7]. The S1 contains the N-terminus domain (NTD) and the receptor binding domain (RBD), which interacts with h-ACE-2; the S2 contains the fusion system, constituted by the central helix (CH), the head-repeated region 1 (HR1), the transmembrane head-repeated region 2 (HR2), the fusion peptide (FP), the connecting domains (CD1, CD2), the S1/S2 cleavage site and several glycosylation sites (see Supplementary Information 1.1 (SI-1.1)). Cryo-electron microscopy (cryo-EM) full structures of the 2019-nCoV S-protein have been solved in either its infectious or non-infectious pre-fusion state [1], [8]. However, the underlying structural mechanism of the infection triggering, necessary for h-ACE-2 binding, is not completely understood and many factors could play a significant role in this process. Since 1977, it is well-established that many viruses can take advantage of inorganic cations in the aqueous environment for replication [9]. The intracellular and extracellular concentration of Na+, K+, Ca2+ and Cl− ions is crucial for the survival of human cells, as dictating their physiological Donnan potential [10], [11]. Coronaviruses are enveloped, thus, by logical extension, also SARS-CoV-2 and other enveloped viruses should cope osmolarity [12]. It has been demonstrated that the envelope proteins of SARS-CoV-1 behave as ion channels, showing selectivity towards Na+ over K+ and Cl− ions [13], [14]. Not surprisingly, available commercial drugs like ion channel blockers have been proposed for both the SARS-CoV-1 and SARS-CoV-2 targeting [15], [16]. These findings suggest that the human SARS-related-CoVs could exploit the three major physiological electrolytes (sodium, potassium and chloride ions) not only for adaptation to the aqueous environment, but also to properly engage with the host receptor. In favour of this hypothesis, Duquerroy found in 2005 [17] that the S2 subunit of the SARS-CoV-1 S-protein, as in its post-fusion hairpin conformation (the conformation assumed after fusion between the virus envelope and the host membrane), was stabilized by two chloride atoms, respectively reported as chloride 1 (CL1) and chloride 2 (CL2): CL1 interacted directly by hydrogen bonds (HBs) with a triad of glutamine (Gln/Q) “zippers”, while CL2 interacted indirectly with a triad of asparagine (Asn/N) residues via water molecules (hereinafter, CL1 and CL2 terms will be adopted as referring to the respective chloride atom interaction pattern, direct or indirect). Starting from the structural analysis of the central funnel-shaped domain within available pre-fusion cryo-EM structures of the SARS-CoV-1 and SARS-CoV-2 S-glycoprotein, we identified a conserved Gln-rich structural region, potentially able to bind inorganic ions. The latter were structurally unsolved however, due, probably, to the low resolution of the respective structures (R ≥ 3.0 Å), at which water molecules and ions are generally unobserved. The putative interaction between the experimentally solved protein site and either alkali metal cations or chloride was modelled quantum chemically, using density functional theory (DFT), with the aim to assess if they could be accommodated efficiently despite their lack of resolution.
2. Methods
3D molecular representations were obtained using VMD 1.9.3 [18], Chimera 1.1.2 [19] and ADF 2018.105 software [20] (https://www.scm.com/). Structures were selected from the coronavirus 3D structure database (https://cov3d.ibbr.umd.edu/spike) and retrieved from the Protein Data Bank [21]. S-protein CH domain sequences of human and bat SARS-CoVs were obtained from the Uniprot web server [22].
2.1. Protein cavity analysis
The FTmap software [23] was used to detect interesting small cavities within the CH-funnel of the SARS-CoV-2 spike glycoprotein. Default settings were adopted.
2.2. DFT calculations
Two S-protein cryo-EM structures were chosen for SARS-CoV-2 and SARS-CoV-1, due to the main Gln rotamers observed as conducive to ion binding in the CH domain (Q1002/Q984): PDB IDs 6VSB (SARS-CoV-2) [1], in open state conformation, being the cationic binding form P+, and 5XLR (SARS-CoV) [24], which represents the anionic binding form P− (see Fig. 4, Fig. 5 in Section 3.1). The X-ray crystal structure of the SARS-CoV-1 spike protein in its post-fusion hairpin conformation (PDB ID 1WYY) [17] was used as a reference for subsequent calculations. Hydrogens were added using the software Chimera 1.1.2 [19] and ionization states checked with the PropKa server tool (https://www.ddl.unimi.it/vegaol/propka.htm). CH residues within 5 Å from the glutamine ion “zipper” in each structure (Q1002, SARS-CoV-2, pre-fusion; Q984, SARS-CoV-1, pre-fusion; Q902, SARS-CoV-1, post-fusion) were selected using the VMD engine [18] (T998 → T1006 and T980 → T988 in SARS-CoV-2 and SARS-CoV-1 post-fusion S-protein, respectively; L898 → A906 in the SARS-CoV-1 post-fusion hairpin conformation). Residues were then extracted from the respective PDB structure. C/N-terminal residues were capped retaining the peptide bond shared with the first adjacent residue to be removed and the α-carbon of the latter (in SARS-CoV-2, I997 and Y1007, respectively). Sidechains extruding outside of the binding cavity were removed and treated as Gly residues, since not participating to the putative ion-protein interaction pattern, preserving however the helical backbone integrity. Missing atoms were added, and all hydrogen atoms were subjected to minimization with the DFT-B forcefield as provided in the ADF package [20]. In presence of ions, pre-fusion and post-fusion models were finally made of 376 and 358 atoms, respectively. All the calculations were performed with the Amsterdam Density Functional 2018.105 (ADF) [20]. The dispersion-corrected BLYP-D3 (BJ) functional was used [25], [26]. The TZ2P basis set was employed along with Becke integration grid [27], [28]. Neither frozen core approximation nor symmetry were adopted [29]. Complexes between cryo-EM pre-fusion structures and ions were optimized in the gas phase constraining proteins at their original conformation, in order to evaluate if the latter may be conducive to ion binding without further refinements. Single point energy calculations on the SARS-CoV-1 spike protein, as in the post-fusion CL1 ion complex (1WYY) [17], were performed in the gas phase for qualitative comparison with the pre-fusion complexes.
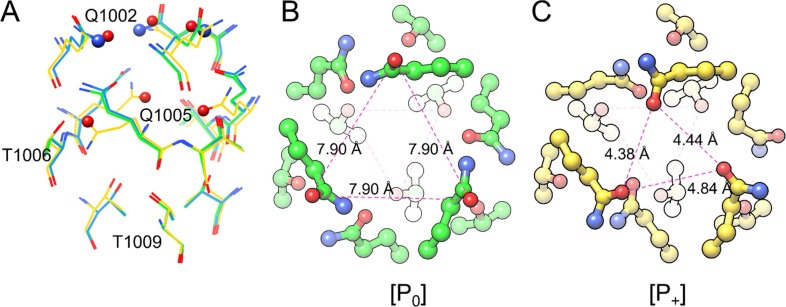
Fig. 4.
S-protein CH site. A) Superposition of the selected structures at the identified region (PDB 6VXX, green, 6VYB, blue, 6VSB, yellow); atoms involved in rotamer conversion are depicted as CPK spheres. B) Top view of the CH site in the closed state of the SARS-CoV-2 S-protein (P0) in which sidechains form triangular planes (pink dot lines); Q1002 amidic oxygen atoms’ distance is reported in angstroms. C) Top view of the protein open state 6VSB (P+). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
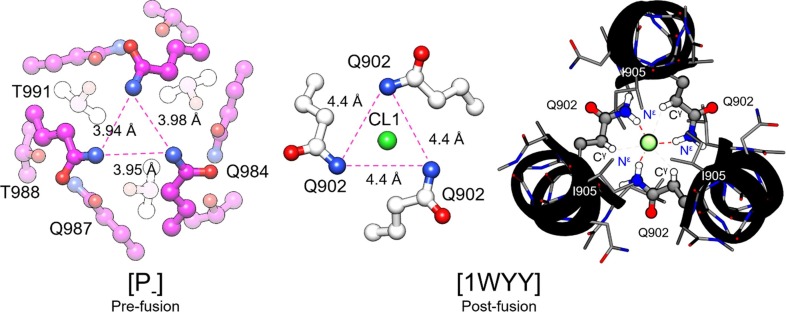
Fig. 5.
Top view of the glutamine (Q) sidechain arrangement in the pre-fusion and post-fusion structure of the SARS-CoV-1 spike protein. Q984 and Q902 sidechains of the SARS-CoV-1 spike protein are shown for the pre-fusion 5XLR19 cryo-EM structure and the post-fusion 1WYY17 X-ray crystal structure, in magenta and white/grey ball-sticks respectively, along with other close residues; the chloride atom bound in the post-fusion state (CL1) is depicted in green; glutamine Nε atoms’ distance is reported in angstroms. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
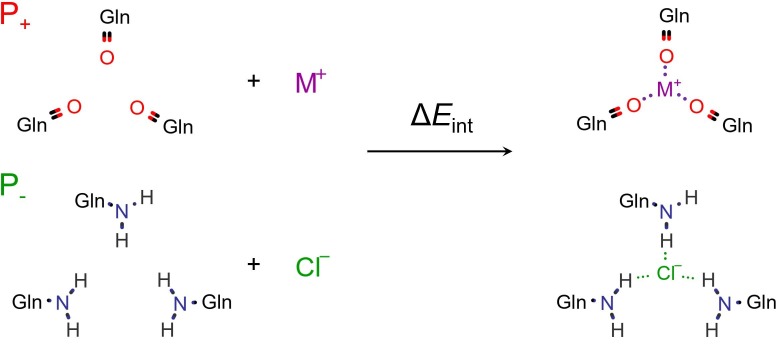
As shown in Fig. 1 , the interaction energy, ΔE int, between the ion (I = Na+, K+, Cl−) and the respective S-protein cavity (P+, P−/P1WYY) was calculated. The interaction energy is calculated as the difference in energy between the optimized complex and the free ligands (protein and ion) in their final state:
Fig. 1.
Ion-protein interaction energy scheme: P+ (PDB ID 6VSB) P− (standing also for 1WYY – 5XLR, 1WYY); M+ = Na+, K+.
Implicit solvent effects were not included in the calculation because the pre-fusion putative ion binding site does not result directly exposed to the water solution, being located deep in the CH funnel and surrounded by the HR1 domains. Explicit solvent effects were included in separate geometry optimizations for sodium and potassium ion complexes, to explore the potential coordination of axial water molecules. Interaction energy, ΔE int, was also calculated for the hydrated sodium-protein complex, as the difference in energy between the latter and the free ligands in the gas phase. In the case of chloride, water was not included, as water molecules do not participate to the interaction pattern of the post-fusion chloride experimental complex CL1, used here as a reference, in which the Gln sidechain arrangement correlates specifically with direct binding.
3. Results
3.1. Structural analysis
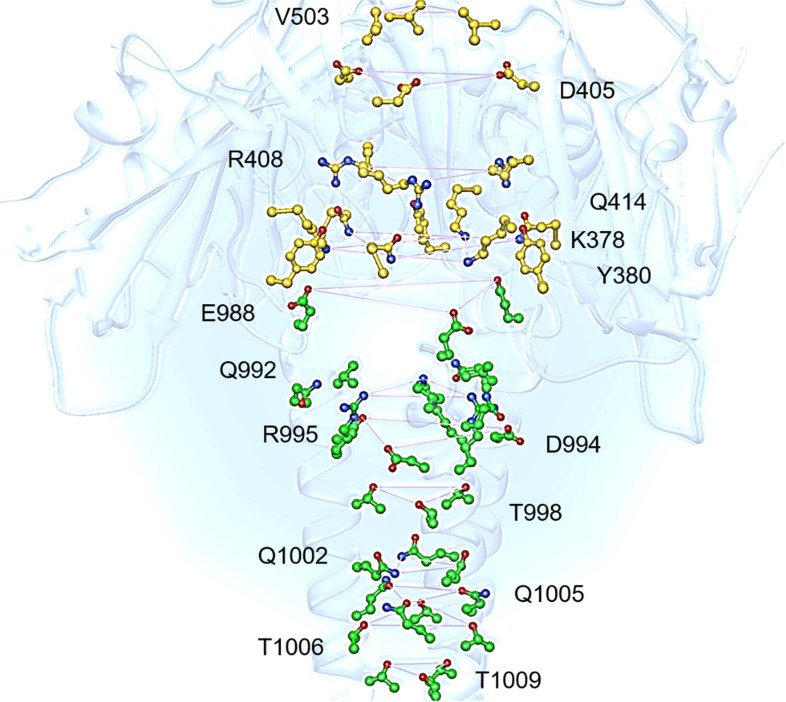
The first solved cryo-EM structure of the SARS-CoV-2 S-protein in its closed state (PDB ID 6VXX, reported hereafter as P0) [8] reveals that RBDs and CHs form together a central tunnel extending axially from S1 to S2. Specific residues shape the tunnel, extruding into the cavity and forming triangular planes by their sidechains (Fig. 2 ).
Fig. 2.
RBD and CH residues extruding into the S-protein central cavity. RBD and CH residues are depicted in yellow and green ball-stick representation, respectively (PDB ID, 6VXX). Triangular planes are shown as pink lines. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
RBD residues exposed are V503, D405, R408, Q414, K378 and Y380. The latter are conserved in SARS-CoV-1 (except V503) as I489, D392, R395, Q401, K365 and Y367, respectively. Helices give rise to a central funnel. CH residues protruding into the cavity by sidechains are: E988, V991, Q992, D994, R995, T998, Q1002, Q1005, T1006 and T1009 (E970, V973, D974, D976, R977, T980, Q984, Q987, T988 and T991 in SARS-CoV-1). Interestingly, the CH domain (45 aa) is shared with 100% of sequence identity by the h-SARS-CoV-1, h-SARS-CoV-2 and bat-SARS-CoVs Rf1, Rp3 and HKU3, as shown in Table 1 .
Table 1.
CH domain sequence of bat and human S-proteins. For each sequence, respective organism and code identifier is reported.
| ORGANISM | SEQUENCE | UNIPROT-ID | ||
|---|---|---|---|---|
| Bat-SARS-CoV Rf1 | 956 | EAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSEC | 1000 | Q0QDZ0 |
| Bat-SARS-like CoV Rp3 | 956 | EAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSEC | 1000 | Q3I5J5 |
| Bat-SARS-like CoV HKU3 | 957 | EAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSEC | 1001 | Q3LZX1 |
| h-SARS-CoV | 970 | EAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSEC | 1025 | P59594 |
| h-SARS-CoV-2 | 988 | EAEVQIDRLITGRLQSLQTYVTQQLIRAAEIRASANLAATKMSEC | 1032 | P0DTC2 |
The CH funnel residues (24 aa) are either charged (10 aa, 22.2%) or polar neutral (14 aa, 31.1%). Importantly, 9 out of the 10 residues that are directly exposed to the inner cavity by their sidechains are polar (90%). The prominent polar composition of the CH funnel suggests that the latter may catalyze ion binding [30]. Unfortunately, the resolution associated to the full cryo-EM structures precludes ions and water molecules from being solved. With the aim to preliminarily identify suitable binding cavities within the CH domain, we docked fragment probes to three representative SARS-CoV-2 structures (PDB IDs 6VXX, P0, in the RBD-closed state, 6VYB [8] and 6VSB [1], P+, in the RBD-open state) (section 2), using the FTmap software [23]. FTmap scans a macromolecule of average size in less than an hour and identifies hot spot regions in it that could serve as ligand binding sites. In practice, it samples 16 small solvent fragments in billions of positions within the target and clusters their populations for each region, offering clues about potential binding site(s). Results disclosed differences between each structure at the bottom of the funnel. This region is characterized by 4 triads of polar residues, namely Q1002, Q1005, T1006 and T1009 (Fig. 2, Fig. 3 ).
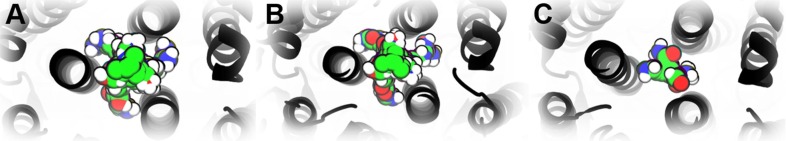
Fig. 3.
Probe populations at the CH site. A) Closed state structure 6VXX, P0. B) Open state structure 6VYB. C) Open state structure 6VSB, P+. Fragment probes are shown in green van der Waals sphere representation, the CH domains and protein in black ribbons. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In the S-protein closed state 6VXX (P0) and in the open state 6VYB the identified site was populated most (Fig. 3A-B), while in the open state structure 6VSB (P+) it appeared highly less occupied (Fig. 3C) (see SI-1.2, Supplementary Table 1, for cluster populations). The different fragment probe populations suggest the presence of meaningful structural rearrangements in the middle of the CH domain. The determined protein site as in the closed state 6VXX (P0) was therefore subjected to comparison with the two open states 6VSB (P+) and 6VYB (Fig. 4 A).
In the closed state, P0, the following HBs were observed: Q1002-Q1005 (inter-helix, sidechain-sidechain, N—O distance ∼2.88 Å), Q1002-T1006 (intra-helix, backbone-sidechain, O—O distance ∼3.05 Å), Q1005-T1009 (intra-helix, backbone-sidechain; O—O distance ∼2.95 Å). T1009 residues define the bottom of the funnel, forming a tight equilateral triangular plane, in which γ carbons are equidistant from each other by only 3.92 Å. A wider equilateral triangular plane is formed by the above located Q1002 amidic oxygen atoms (O—O distance ∼7.90 Å). The same pattern was observed for the related open state structure 6VYB (Fig. 4B), but not for the first solved open state 6VSB (Fig. 4C, P+). We report the following differences for 6VSB: 1) the inter-helix Q1002-Q1005 HBs are disrupted; 2) Q1002 sidechains are largely rotated and the respective amidic oxygens are coplanar, oriented towards each other by an average distance of 4.5 ± 0.2 Å. The second observation is surprising from a chemical point of view, in fact, this kind of arrangement could be ideal for alkali metal cation binding. After inspection of 69 available SARS-CoV-2 full spike protein cryo-EM structures (SI-1.3, Supplementary Table 2), 10 reproduced a similar situation to 6VSB (P+, Fig. 4C), 52 showed a Gln sidechain arrangement comparable to the closed state 6VXX (P0, Fig. 4B), not ideal for direct ion binding, while 7 structures showed another rotamer state, which may be conducive to direct chloride binding (P−, Fig. 5 ).
The P− rotamer state shown in Fig. 5 seems to reproduce well the CL1 Gln arrangement described by Duquerroy for the hairpin post-fusion conformation of the SARS-CoV-1 spike protein, showing a chloride atom bound directly to trimeric Gln sidechains (PDB 1WYY) [17]. Since the SARS-CoV-1 and SARS-CoV-2 S-proteins share the 100% of sequence identity at the CH, we decided to check in the former the presence of the peculiar sidechain arrangements herein described. 13 out of the 18 available SARS-CoV-1 full S-protein cryo-EM structures did not suggest direct ion binding; none showed the P+ pattern of Fig. 4C, potentially able to bind cations, while 5 structures revealed their respective conserved Q984 residues in the CL1 pattern (Q1002 in SARS-CoV-2), P− (see SI-1.2, Supplementary Table 2, for the full list of respective PDB IDs). The intrinsic limitations of the available resolution techniques, as specifically related to the amidic sidechain rotamer assignments and the B-factors’ optimization process, however, warn about the actual reliability of these structural observations. Consequently, for the most interesting cryo-EM structures under study, 6VSB (P+) and 5XLR (P−), isothermal factors of Gln Nε and Oε atoms were inspected. For each structure, B-factors result in a low relative range with respect to the maximum B values registered (∼72 vs 304.6 Å2 and ∼60 vs 286.43 Å2, in P+ and P−, respectively). Thus, it is reasonable to assume that the selected amide rotamers lie on a region of enough low thermal motion to be considered structurally reliable for further considerations. In light of these findings, we decided to assess the ability of the CH-site to bind physiological alkali cations (Na+ and K+) and chloride (Cl−) in the representative pre-fusion conformations of the SARS-CoV-2 and SARS-CoV-1 S-protein, by means of Density Functional Theory (DFT) [31].
3.2. Ion-S-protein interaction
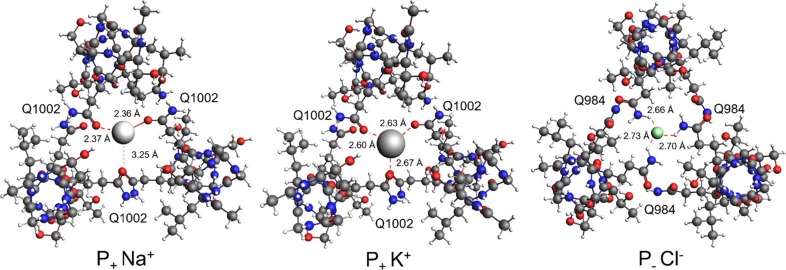
Complexes with sodium (6VSB, P+Na+), potassium (6VSB, P+K+) and chloride (5XLR, P−Cl−) ions were optimized quantum-chemically. The optimized complexes are shown in Fig. 6 (see SI-2 for structure coordinates).
Fig. 6.
Top view of ion-S-protein modelled complexes in the pre-fusion state. Coordinated glutamine (Q) residues are highlighted along with the interacting atom pair distance in angstroms (red dot lines; P+, 6VSB, SARS-CoV-2; P−, 5XLR, SARS-CoV-1). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Sodium cation interacts preferentially with two Gln oxygens by distance values lower than 3 Å and gives rise to a distorted trigonal planar complex. Potassium, having a bigger radius than sodium, accommodates in the CH site interacting almost equally with all three Q1002 amidic oxygen atoms and forms a quite regular trigonal planar complex. Chloride is complexed directly by the Gln Nε atoms, leading to a tetrahedral coordination geometry, which resembles the CL1 interaction state described by Duquerroy in 2005 [17]. The fourth coordination ligand is absent also in the present case, being replaced by a triad of threonine alkyl groups (T998 and T980, in SARS-CoV-2 and SARS-CoV-1, respectively).
The interaction energy between each ion and the S-protein pre-fusion CH site, ΔE int, was calculated (see Section 2, Fig. 1). The same was done also for the X-ray crystal structure of the SARS-CoV-1 post-fusion ion complex, as a reference for our predictions (1WYY-Cl−). Results are summarized in Table 2 (see the SI-3 for total energies and basis set superposition errors, BSSE). Counterpoise corrected interaction energies, calculated for the modelled pre-fusion ion complexes and the reference complex 1WYY-Cl− are of the same order of magnitude, which thing allows to consider our models as generally reliable in terms of predicted interaction pattern. All the ions interact favourably with the S-protein, suggesting the ion binding nature of this site. The ΔE int trend for the pre-fusion S-protein ion complexes is in the order Na+ > K+ > Cl−, making sodium the best putative bound ion. With specific regards to the interaction energy of the CL1 pre-fusion (P−Cl−) and post-fusion complex (1WYY-Cl−), the latter results more favourable. This can be related to the tighter HBs formed with the chloride ion in the post-fusion structure (Nε-H∙∙∙Cl−, 2.17 Å vs 2.66 Å), the involvement of Gln902 γ-carbons in the interaction pattern of the latter, not observed in the P− pre-fusion complex, and probably a better dispersion exerted by the fourth coordination ligand, isoleucine (I905, Fig. 5), more hydrophobic than threonine.
Table 2.
Calculated ion-protein interaction energies. Interaction energy values for each complex (1WYY, post-fusion; P+, pre-fusion; P−, pre-fusion) are reported along with the counterpoise corrected energy in parentheses, considering the error resulting from the basis set overlap of individual ligands as they approach to each other to form a complex (basis set superposition error, BSSE); correction is applied performing single point energy calculations with the mixed basis set. Details related to each experimental structure and the shortest bond distance between the main interacting glutamine atoms (X = Nε-H in 1WYY-Cl− and P−; X = O in P+) and the respective complexed ion (I = Cl−, Na+, K+).
| PDB ID | Resolution (Å) | Complex | ΔEInt (kcal∙mol−1)a | X∙∙∙I (Å) | |||
|---|---|---|---|---|---|---|---|
| Reference | 1WYY | (X-Ray) | 2.20 | 1WYY-Cl− | −108.1 | (−100.4) | 2.17 |
| 6VSB | (Cryo-EM) | 3.46 | P+Na+ | −91.9 | (−91.6) | 2.36 | |
| P+K+ | −83.1 | (−82.7) | 2.6 | ||||
| 5XLR | (Cryo-EM) | 3.8 | P−Cl− | −78.3 | (−74.2) | 2.66 | |
Energies and optimized geometries were computed at the ZORA-BLYP-D3(BJ)/TZ2P level of theory.
3.3. Water coordination in cation complexes
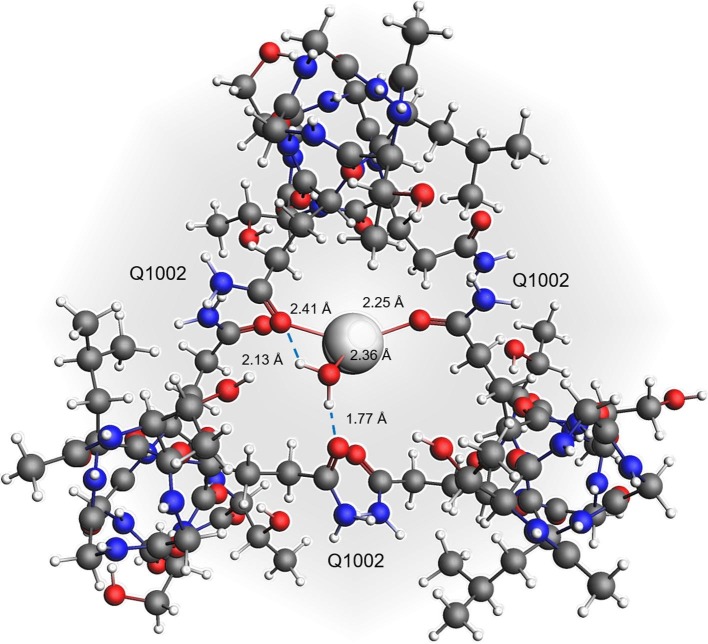
The P+Na+ complex of Fig. 6 was further optimized after inclusion of a water molecule, above the trigonal sodium-protein coordination plane, yielding a distorted tetrahedral geometry (Fig. 7 ).
Fig. 7.
Top view of the monohydrated sodium-protein modelled complex (P+Na+∙H2O) in the pre-fusion state. Water-mediated hydrogen bonds are shown as blue dot lines along with atom pair distances in angstroms for any interaction. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The insertion of another water ligand below the same plane, conversely, did not lead to any minimum energetic state. As referring to the P+K+ complex, no axial coordination was predicted. In the P+Na+∙H2O model, the water molecule interacts with Na+ and engages HBs with two pre-coordinated oxygen atoms, increasing the stability of the complex. Indeed, the ΔE int calculated for the hydrated complex is −110.9 kcal∙mol−1 (BSSE = −0.9 kcal∙mol−1), corresponding to a gain in stabilization equal to −19 kcal∙mol−1 after inclusion of a single water molecule (P+Na+, ΔE int −91.9 kcal∙mol−1). Water coordination is sterically allowed because the sodium cation coordinates preferentially with two out of three oxygen atoms, i.e., it interacts asymmetrically with the protein and leaves enough space for the accommodation of a water molecule in proximity of the third (weakly bound) atom. Consistently, upon hydration, Na+ is predicted to coordinate one amidic oxygen more strongly than the other two (Na+∙∙O distance decreases from 2.36 to 2.25 Å in presence of water; Fig. 6, Fig. 7) and to loosen further the weakest pre-existing interaction (Na+∙∙O distance increases from 3.25 to 3.39 Å in presence of water). Similar considerations suggest that the K+-water coordination is impossible because the symmetric interaction pattern of potassium within the cavity and its radius, bigger than sodium, are not sterically in favour of it.
4. Discussion
The structural analysis of the SARS-CoV-2 S-protein in its first solved cryo-EM structures led to the identification of a highly polar region in the central helix domain. The latter is characterized by a triad of Gln amidic oxygen atoms arranged in peculiar geometries, potentially able to form alkali metal and anion complexes, respectively (P+, P−) [1], [17]. Among them, the P− Gln arrangement highly resembles the CL1 interaction pattern described by Duquerroy in 2005 for the post-fusion ion complex of the SARS-CoV-1 spike protein, solved by X-ray diffraction technique (Fig. 5) [17]. Interestingly, the identified protein sequence was found as fully conserved among either the human or bat SARS-CoVs (Table 1). In keeping with this, the CL1 Gln arrangement P− has been detected for the cryo-EM structures of both the SARS-CoV-2 and SARS-CoV-1 S-protein, while, on the other hand, the SARS-CoV-2 cation-binding conformation P+ has not been observed in SARS-CoV-1. Although apparently inconsistent, we expect that such result may be related to the higher number of available (full) S-protein structures in SARS-CoV-2, i.e., to a larger conformational sampling of the latter as compared to SARS-CoV-1 (69 vs 18 solved structures, respectively), and to different settings used for either sample preparation or model refinement. Unfortunately, ions are experimentally unobserved in the pre-fusion states of both SARS-CoV-1 and SARS-CoV-2 spike protein, probably because of the low structural resolution of the cryo-EM structures, lying under the detection limit required for water molecules and ions (generally lower than 3 Å). Thus, we adopted quantum-chemical DFT calculations to assess the possibility of ion binding in the chosen representative pre-fusion structures of the SARS-CoV-1 and SARS-CoV-2 spike proteins. Results were subsequentially compared to the SARS-CoV-1 post-fusion complex reference (PDB ID 1WYY) [17], in which the interacting ion (chloride) is solved at high resolution (X-ray) and interacts in a similar way to what preliminarily expected for our modelled complexes (see Fig. 5, Fig. 6 for comparison). Using this approach, in other words, we considered whether ions may be present according to both chemical intuition and accurate quantum chemistry rules, while experimentally unsolved as a drawback of the cryo-EM technique resolution. Indeed, the calculated ion-protein interaction energies and binding geometries indicate that Na+, K+ and Cl− atoms can interact efficiently with the conserved CH domain of the h-SARS-CoV spike proteins as in their cryo-EM conformation (Table 2). Furthermore, the resulting complex geometries and energies are qualitatively consistent with the interaction pattern observed in the experimental X-ray reference structure (for which ions have been solved). We also explored the axial coordination of explicit water molecules by cations, and this resulted favourable only for the top-ranked sodium ion complex, giving further validity to our assumptions. Indeed, the better ΔE int of sodium ion compared to potassium and chloride in the pre-fusion S-protein structures (Table 2), along with the prediction of a stable water complex only for Na+, can be linked to the respective concentration of these electrolytes in the extracellular solution. In fact, the sodium ion is more concentrated than potassium and chloride out of the cell membrane, while the opposite is observed in the intracellular solution, due to the Donnan potential. This result is of special importance, since it is consistent with our hypothesis, considering SARS-CoVs as able to take advantage from the extracellular media to assault the host by their spike proteins, as well as with the experimental evidence of chloride sequestration in the intracellular environment (where the anion is more present) after membrane fusion. Accordingly, the post-fusion chloride complex (1WYY-Cl−), is more stable than the one predicted for the pre-fusion state (P−Cl−). In other words, the SARS-CoV spike proteins may exploit both the extracellular and intracellular solution by cation and anion binding, respectively, in order to drive the first stages of the infection process.
5. Conclusions
Basing on the present study, the existence of an ion binding site shared by SARS-CoVs is reasonable and highly expected. Although further investigations are required to confirm the potential role of physiological electrolytes in spike proteins, our results suggest that they may represent a term of structural stabilization for the central helix trimer before the infection triggering, in line with the evidence of such role as exerted by chloride atoms on the helical structure of the SARS-CoV-1 spike protein, after fusion with the host cell. Subsequent computational and experimental studies may, therefore, give precious insights about the observed high conservation of the CH domain in bat and human SARS-CoVs, in terms of evolutionary biofunction. Nevertheless, small molecules could also be designed with the property to specifically bind and interfere with the CH funnel, preventing membrane fusion and infection to occur. Indeed, other investigations are ongoing in our laboratories for this purpose.
From a biochemical and physio-pathological perspective, the favourable interaction of the three major electrolytes with the S-protein, in either the pre-fusion or post-fusion state, correlates well with their presence in the extracellular solution, enforcing the idea that SARS-CoV-2 (and, in general, SARS-CoVs) may realistically sequestrate electrolytes from physiological body fluids and take advantage from them in order to succeed, depriving the human cells of the salt balance required for their homeostasis. Basing on our results, the main three electrolytes should therefore represent the landmark for translational investigations on the SARS-CoV-2 spike glycoprotein structure functions and on the way the latter can be impaired by ad hoc therapeutic strategies, aiding finally the clinical practice in treating efficiently SARS-related coronavirus diseases.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank the Netherlands Organization for Scientific Research (NWO) for financial support. We also thank Prof. Paolo Ruggerone and Dr. Giuliano Malloci from the University of Cagliari for insightful discussions about the web server algorithms adopted in the present study.
Funding
The present research did not receive any specific grant.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.comptc.2021.113392.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. doi: 10.1126/science:abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3(1):237–261. doi: 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang T., Bidon M., Jaimes J.A., Whittaker G.R., Daniel S. Coronavirus membrane fusion mechanism offers a potential target for antiviral development. Antiviral Res. 2020;178:104792. doi: 10.1016/j.antiviral.2020.104792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A., Dold C., Faust S.N., Finn A., Flaxman A.L., Hallis B., Heath P., Jenkin D., Lazarus R., Makinson R., Minassian A.M., Pollock K.M., Ramasamy M., Robinson H., Snape M., Tarrant R., Voysey M., Green C., Douglas A.D., Hill A.V.S., Lambe T., Gilbert S.C., Pollard A.J., Aboagye J., Adams K., Ali A., Allen E., Allison J.L., Anslow R., Arbe-Barnes E.H., Babbage G., Baillie K., Baker M., Baker N., Baker P., Baleanu I., Ballaminut J., Barnes E., Barrett J., Bates L., Batten A., Beadon K., Beckley R., Berrie E., Berry L., Beveridge A., Bewley K.R., Bijker E.M., Bingham T., Blackwell L., Blundell C.L., Bolam E., Boland E., Borthwick N., Bower T., Boyd A., Brenner T., Bright P.D., Brown-O'Sullivan C., Brunt E., Burbage J., Burge S., Buttigieg K.R., Byard N., Cabera Puig I., Calvert A., Camara S., Cao M., Cappuccini F., Carr M., Carroll M.W., Carter V., Cathie K., Challis R.J., Charlton S., Chelysheva I., Cho J.-S., Cicconi P., Cifuentes L., Clark H., Clark E., Cole T., Colin-Jones R., Conlon C.P., Cook A., Coombes N.S., Cooper R., Cosgrove C.A., Coy K., Crocker W.E.M., Cunningham C.J., Damratoski B.E., Dando L., Datoo M.S., Davies H., De Graaf H., Demissie T., Di Maso C., Dietrich I., Dong T., Donnellan F.R., Douglas N., Downing C., Drake J., Drake-Brockman R., Drury R.E., Dunachie S.J., Edwards N.J., Edwards F.D.L., Edwards C.J., Elias S.C., Elmore M.J., Emary K.R.W., English M.R., Fagerbrink S., Felle S., Feng S., Field S., Fixmer C., Fletcher C., Ford K.J., Fowler J., Fox P., Francis E., Frater J., Furze J., Fuskova M., Galiza E., Gbesemete D., Gilbride C., Godwin K., Gorini G., Goulston L., Grabau C., Gracie L., Gray Z., Guthrie L.B., Hackett M., Halwe S., Hamilton E., Hamlyn J., Hanumunthadu B., Harding I., Harris S.A., Harris A., Harrison D., Harrison C., Hart T.C., Haskell L., Hawkins S., Head I., Henry J.A., Hill J., Hodgson S.H.C., Hou M.M., Howe E., Howell N., Hutlin C., Ikram S., Isitt C., Iveson P., Jackson S., Jackson F., James S.W., Jenkins M., Jones E., Jones K., Jones C.E., Jones B., Kailath R., Karampatsas K., Keen J., Kelly S., Kelly D., Kerr D., Kerridge S., Khan L., Khan U., Killen A., Kinch J., King T.B., King L., King J., Kingham-Page L., Klenerman P., Knapper F., Knight J.C., Knott D., Koleva S., Kupke A., Larkworthy C.W., Larwood J.P.J., Laskey A., Lawrie A.M., Lee A., Ngan Lee K.Y., Lees E.A., Legge H., Lelliott A., Lemm N.-M., Lias A.M., Linder A., Lipworth S., Liu X., Liu S., Lopez Ramon R., Lwin M., Mabesa F., Madhavan M., Mallett G., Mansatta K., Marcal I., Marinou S., Marlow E., Marshall J.L., Martin J., McEwan J., McInroy L., Meddaugh G., Mentzer A.J., Mirtorabi N., Moore M., Moran E., Morey E., Morgan V., Morris S.J., Morrison H., Morshead G., Morter R., Mujadidi Y.F., Muller J., Munera-Huertas T., Munro C., Munro A., Murphy S., Munster V.J., Mweu P., Noé A., Nugent F.L., Nuthall E., O'Brien K., O'Connor D., Oguti B., Oliver J.L., Oliveira C., O'Reilly P.J., Osborn M., Osborne P., Owen C., Owens D., Owino N., Pacurar M., Parker K., Parracho H., Patrick-Smith M., Payne V., Pearce J., Peng Y., Peralta Alvarez M.P., Perring J., Pfafferott K., Pipini D., Plested E., Pluess-Hall H., Pollock K., Poulton I., Presland L., Provstgaard-Morys S., Pulido D., Radia K., Ramos Lopez F., Rand J., Ratcliffe H., Rawlinson T., Rhead S., Riddell A., Ritchie A.J., Roberts H., Robson J., Roche S., Rohde C., Rollier C.S., Romani R., Rudiansyah I., Saich S., Sajjad S., Salvador S., Sanchez Riera L., Sanders H., Sanders K., Sapaun S., Sayce C., Schofield E., Screaton G., Selby B., Semple C., Sharpe H.R., Shaik I., Shea A., Shelton H., Silk S., Silva-Reyes L., Skelly D.T., Smee H., Smith C.C., Smith D.J., Song R., Spencer A.J., Stafford E., Steele A., Stefanova E., Stockdale L., Szigeti A., Tahiri-Alaoui A., Tait M., Talbot H., Tanner R., Taylor I.J., Taylor V., Te Water Naude R., Thakur N., Themistocleous Y., Themistocleous A., Thomas M., Thomas T.M., Thompson A., Thomson-Hill S., Tomlins J., Tonks S., Towner J., Tran N., Tree J.A., Truby A., Turkentine K., Turner C., Turner N., Turner S., Tuthill T., Ulaszewska M., Varughese R., Van Doremalen N., Veighey K., Verheul M.K., Vichos I., Vitale E., Walker L., Watson M.E.E., Welham B., Wheat J., White C., White R., Worth A.T., Wright D., Wright S., Yao X.L., Yau Y. Oxford COVID Vaccine Trial Group, Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Robson B. COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles’ heel conserved region to minimize probability of escape mutations and drug resistance. Comput. Biol. Med. 2020;121:103749. doi: 10.1016/j.compbiomed.2020.103749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrasco L. The inhibition of cell functions after viral infection A proposed general mechanism. FEBS Lett. 1977;76:11–15. doi: 10.1016/0014-5793(77)80110-5. [DOI] [PubMed] [Google Scholar]
- 10.Blaustein M.P., Kao J.P.Y., Matteson D.R. Elsevier Health Sciences; 2011. Cellular Physiology and Neurophysiology E-Book: Mosby Physiology Monograph Series. [Google Scholar]
- 11.Khitrin A.K., Khitrin K.A., Model M.A. A model for membrane potential and intracellular ion distribution. Chem. Phys. Lipids. 2014;184:76–81. doi: 10.1016/j.chemphyslip.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 12.Choi H.-J., Song J.-M., Bondy B.J., Compans R.W., Kang S.-M., Prausnitz M.R., Krammer F. Effect of osmotic pressure on the stability of whole inactivated influenza vaccine for coating on microneedles. PLoS ONE. 2015;10(7):e0134431. doi: 10.1371/journal.pone.0134431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson L., Mckinlay C., Gage P., Ewart G. SARS coronavirus E protein forms cation-selective ion channels. Virology. 2004;330(1):322–331. doi: 10.1016/j.virol.2004.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verdiá-Báguena C., Nieto-Torres J.L., Alcaraz A., DeDiego M.L., Enjuanes L., Aguilella V.M. Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim. Biophys. Acta. 2013;1828(9):2026–2031. doi: 10.1016/j.bbamem.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson L., Gage P., Ewart G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology. 2006;353(2):294–306. doi: 10.1016/j.virol.2006.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.A. Chernyshev, Pharmaceutical Targeting the Envelope Protein of SARS-CoV-2: the Screening for Inhibitors in Approved Drugs (2020). 10.26434/chemrxiv.12286421.v1. [DOI]
- 17.Duquerroy S., Vigouroux A., Rottier P.J.M., Rey F.A., Jan Bosch B. Central ions and lateral asparagine/glutamine zippers stabilize the post-fusion hairpin conformation of the SARS coronavirus spike glycoprotein. Virology. 2005;335(2):276–285. doi: 10.1016/j.virol.2005.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 19.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25(13):1605–1612. doi: 10.1002/(ISSN)1096-987X10.1002/jcc.v25:1310.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 20.te Velde G., Bickelhaupt F.M., Baerends E.J., Fonseca Guerra C., van Gisbergen S.J.A., Snijders J.G., Ziegler T. Chemistry with ADF. J. Comput. Chem. 2001;22(9):931–967. doi: 10.1002/(ISSN)1096-987X10.1002/jcc.v22:910.1002/jcc.1056. [DOI] [Google Scholar]
- 21.Burley S.K., Bhikadiya C., Bi C., Bittrich S., Chen L., Crichlow G.V., Christie C.H., Dalenberg K., Di Costanzo L., Duarte J.M., Dutta S., Feng Z., Ganesan S., Goodsell D.S., Ghosh S., Green R.K., Guranović V., Guzenko D., Hudson B.P., Lawson C.L., Liang Y., Lowe R., Namkoong H., Peisach E., Persikova I., Randle C., Rose A., Rose Y., Sali A., Segura J., Sekharan M., Shao C., Tao Y.-P., Voigt M., Westbrook J.D., Young J.Y., Zardecki C., Zhuravleva M. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2021;49:D437–D451. doi: 10.1093/nar/gkaa1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The UniProt Consortium, UniProt: the universal protein knowledgebase in 2021, Nucleic Acids Res. 49 (2021) D480–D489. 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed]
- 23.Kozakov D., Grove L.E., Hall D.R., Bohnuud T., Mottarella S.E., Luo L., Xia B., Beglov D., Vajda S. The FTMap family of web servers for determining and characterizing ligand binding hot spots of proteins. Nat Protoc. 2015;10(5):733–755. doi: 10.1038/nprot.2015.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gui M., Song W., Zhou H., Xu J., Chen S., Xiang Y.e., Wang X. Cryo-electron microscopy structures of the SARS-CoV spike glycoprotein reveal a prerequisite conformational state for receptor binding. Cell Res. 2017;27(1):119–129. doi: 10.1038/cr.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Becke A.D. Density-functional exchange-energy approximation with correct asymptotic behavior. Phys. Rev. A. 1988;38(6):3098–3100. doi: 10.1103/PhysRevA.38.3098. [DOI] [PubMed] [Google Scholar]
- 26.Grimme S., Antony J., Ehrlich S., Krieg H. A consistent and accurate ab initio parametrization of density functional dispersion correction (DFT-D) for the 94 elements H-Pu. J. Chem. Phys. 2010;132(15):154104. doi: 10.1063/1.3382344. [DOI] [PubMed] [Google Scholar]
- 27.Brauer B., Kesharwani M.K., Kozuch S., Martin J.M.L. The S66x8 benchmark for noncovalent interactions revisited: explicitly correlated ab initio methods and density functional theory. PCCP. 2016;18(31):20905–20925. doi: 10.1039/C6CP00688D. [DOI] [PubMed] [Google Scholar]
- 28.Johnson E.R., Becke A.D. A post-Hartree–Fock model of intermolecular interactions. J. Chem. Phys. 2005;123(2):024101. doi: 10.1063/1.1949201. [DOI] [PubMed] [Google Scholar]
- 29.van der Wijst T., Lippert B., Swart M., Fonseca Guerra C., Bickelhaupt F.M. Differential stabilization of adenine quartets by anions and cations. J. Biol. Inorg. Chem. 2010;15(3):387–397. doi: 10.1007/s00775-009-0611-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Laio A., Torre V. Physical origin of selectivity in ionic channels of biological membranes. Biophys. J. 1999;76(1):129–148. doi: 10.1016/S0006-3495(99)77184-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fonseca Guerra C., Snijders J.G., te Velde G., Baerends E.J. Towards an order- N DFT method. Theor. Chem. Accounts: Theory, Comput., Model. (Theoretica Chimica Acta) 1998;99:391–403. doi: 10.1007/s002140050353. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.