Abstract
Motivation
One major goal of single-cell RNA sequencing (scRNAseq) experiments is to identify novel cell types. With increasingly large scRNAseq datasets, unsupervised clustering methods can now produce detailed catalogues of transcriptionally distinct groups of cells in a sample. However, the interpretation of these clusters is challenging for both technical and biological reasons. Popular clustering algorithms are sensitive to parameter choices, and can produce different clustering solutions with even small changes in the number of principal components used, the k nearest neighbor and the resolution parameters, among others.
Results
Here, we present a set of tools to evaluate cluster stability by subsampling, which can guide parameter choice and aid in biological interpretation. The R package scclusteval and the accompanying Snakemake workflow implement all steps of the pipeline: subsampling the cells, repeating the clustering with Seurat and estimation of cluster stability using the Jaccard similarity index and providing rich visualizations.
Availabilityand implementation
R package scclusteval: https://github.com/crazyhottommy/scclusteval Snakemake workflow: https://github.com/crazyhottommy/pyflow_seuratv3_parameter Tutorial: https://crazyhottommy.github.io/EvaluateSingleCellClustering/.
1 Introduction
One of the most powerful applications of single-cell RNAseq is to define cell types based on the transcriptional profiles of the cells. A number of tools such as Seurat (Macosko et al., 2015), scanpy (Wolf et al., 2018) and SINCERA (Guo et al., 2015) have implemented unsupervised clustering methods for single-cell RNAseq data. Although benchmarking studies have examined the performance of different clustering algorithms (Duò et al., 2018), less attention has been given to optimizing clustering algorithms for a particular dataset.
Two main questions exist in this perspective. First, given a set of selected parameters, how robust is each cluster? Second, what is the best way to select parameters for a specific dataset? A partial way to address this problem is clustree (Zappia and Oshlack, 2018), which plots the clusters as a tree structure to visualize the relationship among clusters with different resolutions and aid in the determination of cluster numbers and appropriate parameters. However, when the cluster number is large, the tree becomes hard to interpret. Furthermore, it does not provide any quantitative assessment and requires manual inspections of the trees. While methods have begun to approach the problem, there remains an urgent need for a data-driven evaluation of the cluster stability.
2 Materials and methods
To address part of the challenges of evaluating the clustering results, we deployed a re-sampling method in which we re-sample a subset of the cells from the population and repeat clustering. The cell identity is recorded for each re-sampling, and for each cluster, a Jaccard index is calculated to evaluate cluster similarity before and after re-clustering. We then repeat the re-clustering for a number of times and use the mean or median of the Jaccard indices as a metric to evaluate the stability of the cluster. While the theory behind this method has been extensively developed (Hennig, 2007; Lun, 2019), practical implementations are lacking.
We implemented a Snakemake (Köster and Rahmann, 2012) workflow to perform the subsampling and re-clustering steps while taking advantage of multiple CPUs available in a high-performance computing cluster. In addition, we developed a new R package scclusteval to aid the analysis and visualization of the output from the Snakemake workflow. To facilitate the reproducibility of the workflow, we have created a Docker container for the Snakemake workflow which includes support for Seurat V3. The Snakemake workflow generates two rds objects: one contains the cell identity (cluster id) information before and after the reclustering for the subsampled data and the other contains the cell identity information for the full dataset for various combinations of parameters.
The accompanying R package relies extensively on functions from the tidyverse packages. The fundamental object the scclusteval package interacts with is a tibble in a tidy format. The input of the R package is obtained directly from the Snakemake pipeline output.
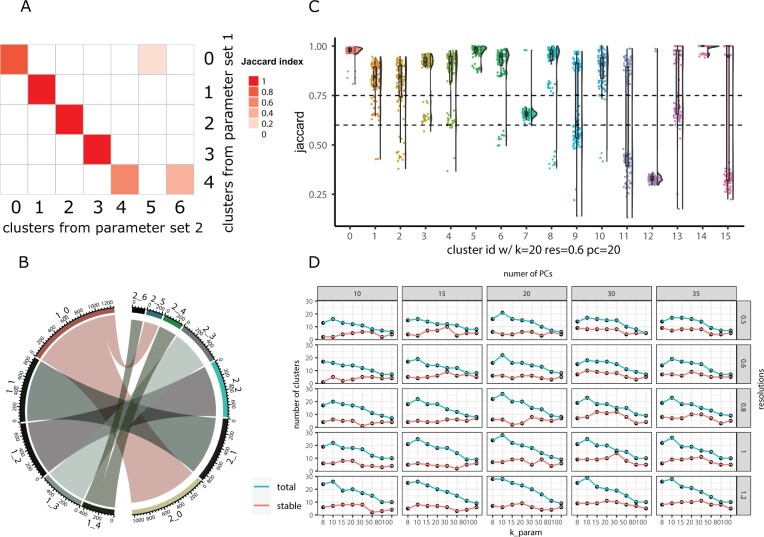
To explore the cell identity changes across different parameters for the full dataset, one can use the PairWiseJaccardSetsHeatmap function to visualize the pairwise Jaccard index across clusters (Fig. 1A). Alternatively, one can use the ClusterIdentityChordPlot function to visualize how the cells switch from one cluster to a different cluster (Fig. 1B).
Fig. 1.
Visualizations methods from the scclusteval R package. (A) A pairwise Jaccard index heatmap to visualize the clusters’ relationship between two sets of different clustering parameters for the full dataset. X-axis represents the clusters from parameter set 2, y-axis represents the clusters from parameter set 1. In this example, cluster 0 in the y-axis split into cluster 0 and 5 in the x-axis; cluster 4 in the y-axis split into cluster 4 and 6 in the x-axis. (B) A cluster chord diagram showing cell identity switching between two different clustering parameters with additional information of the cluster size compared to (A). (C) A Jaccard Raincloud plot showing the stability of each cluster. A boxplot with a half-side violin plot showing the distribution of the Jaccard indices (highest Jaccard index used for matching clusters for each subsample) before and after re-clustering across 100 subsamples. The dotted lines are Jaccard cutoffs of 0.6 and 0.75. (D) A line plot showing the relationship between different parameters, and the total number of clusters (blue line) and number of stable clusters (red line). The x-axis represents the k parameter, the y-axis represents the number of clusters. The columns are split by the number of PCs and rows are split by different resolutions.
As a rule of thumb, clusters with a mean/median stability score less than 0.6 should be considered unstable. Scores between 0.6 and 0.75 indicate that the cluster is measuring a pattern in the data. Clusters with stability scores greater than 0.85 are highly stable (Zumel and Mount, 2014). For each subset of cells, we calculate pairwise Jaccard index of each cluster before and after reclustering and assign the highest Jaccard as the stability score for each cluster. The distribution of the Jaccard indices across subsamples measures the robustness of the cluster. If a cluster is robust and stable, random subsetting and reclustering will keep the cell identities within the same cluster. The heart of the visualization is the raincloud plot (Allen et al., 2019). The plot can be created using the JaccardRainCloudPlot function. The raincloud plot gives an intuitive sense of the stability of clusters (Fig. 1C). Because increasing resolution always generates more clusters, we also use the percentage of cells in the stable clusters to evaluate a particular clustering. We want to maximize the number of clusters but also want the majority of the cells to be in stable clusters. The CalculatePercentCellInStable function can be used to calculate the percentage of cells in the stable clusters. Finally, a scatter plot to explore the relationship between the clustering parameters and the number of total clusters, total number of stable clusters and percentage of cells in stable clusters can be made using the ParameterSetScatterPlot function (Fig. 1D).
To demonstrate the usage of the scclusteval package, we analyzed two example public datasets: a mixture control dataset (Tian et al., 2019) and a 5k PBMC dataset. These analyses are available at website https://crazyhottommy.github.io/EvaluateSingleCellClustering/index.htm, powered by the workflowr (Blischak et al., 2019) R package. All the processed datasets can be downloaded from https://osf.io/rfbcg/.
3. Discussion
Identifying cell clusters in a single-cell experiments is challenging because there typically is not one correct clustering for any dataset, and no principled way to select a single best clustering. There are usually cells in different cell cycle stages in typical cell cultures, and cells with different ploidies in cancer cell lines. Moreover, the concept of cell identity is evolving with the advance of the scRNAseq technology (Morris, 2019; Xia and Yanai, 2019). The end cluster results need to be confirmed by our understanding of the biology, and making sense of the novel clusters/cell-types/cell states is important. Nevertheless, our new Snakemake workflow and R package provide valuable guidance in choosing parameters for clustering and facilitate the biological interpretation of the clusters derived from scRNAseq data.
Acknowledgements
The authors thank 10x genomics and Luyi Tian to have their single-cell RNAseq data publicly available. They also thank the Harvard FAS research computing center to provide the computing resource.
Funding
This work was supported by grants to Catherine Dulac from the NIH [1U19MH114830 and 1U19MH114821] and the Howard Hughes Medical Institute.
Conflict of Interest: none declared.
Contributor Information
Ming Tang, FAS Informatics Group, Harvard University, Cambridge, MA, USA; Department of Molecular and Cellular Biology, Center for Brain Science, Harvard University, Cambridge, MA, USA; Howard Hughes Medical Institute, Cambridge, MA, USA.
Yasin Kaymaz, FAS Informatics Group, Harvard University, Cambridge, MA, USA.
Brandon L Logeman, Department of Molecular and Cellular Biology, Center for Brain Science, Harvard University, Cambridge, MA, USA; Howard Hughes Medical Institute, Cambridge, MA, USA.
Stephen Eichhorn, Department of Chemistry, Harvard University, Cambridge, MA, USA.
Zhengzheng S Liang, Department of Molecular and Cellular Biology, Center for Brain Science, Harvard University, Cambridge, MA, USA; Howard Hughes Medical Institute, Cambridge, MA, USA.
Catherine Dulac, Department of Molecular and Cellular Biology, Center for Brain Science, Harvard University, Cambridge, MA, USA; Howard Hughes Medical Institute, Cambridge, MA, USA.
Timothy B Sackton, FAS Informatics Group, Harvard University, Cambridge, MA, USA.
References
- Allen M. et al. (2019) Raincloud plots: a multi-platform tool for robust data visualization. Wellcome Open Res., 4, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blischak J. et al. (2019) Creating and sharing reproducible research code the workflowr way [version 1; peer review: 3 approved]. F1000Research, 8, 1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duò,A.et al. (2018) A systematic performance evaluation of clustering methods for single-cell RNA-seq data. F1000Res., 7, 1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M. et al. (2015) SINCERA: a pipeline for single-cell RNA-seq profiling analysis. PLoS Comput. Biol., 11, e1004575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig C. (2007) Cluster-wise assessment of cluster stability. Comput. Stat. Data Anal., 52, 258–271. [Google Scholar]
- Köster J., Rahmann S. (2012) Snakemake—a scalable bioinformatics workflow engine. Bioinformatics, 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Lun A. (2019) Bootstrapping for cluster stability. https://ltla.github.io/SingleCellThoughts/general/bootstrapping.html.
- Macosko E.Z. et al. (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell, 161, 1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S.A. (2019) The evolving concept of cell identity in the single cell era. Development, 146, dev169748. [DOI] [PubMed] [Google Scholar]
- Tian L. et al. (2019) Benchmarking single cell RNA-sequencing analysis pipelines using mixture control experiments. Nat. Methods, 16, 479–487. [DOI] [PubMed] [Google Scholar]
- Wolf F.A. et al. (2018) SCANPY: large-scale single-cell gene expression data analysis. Genome Biol., 19, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B., Yanai I. (2019) A periodic table of cell types. Development, 146, dev169854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappia,L. Oshlack,A. (2018) Clustering trees: a visualization for evaluating clusterings at multiple resolutions. GigaScience, 7, giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zumel N., Mount J. (2014) Practical Data Science with R. 1st edn. Manning Publications Co., Greenwich, CT, USA. [Google Scholar]