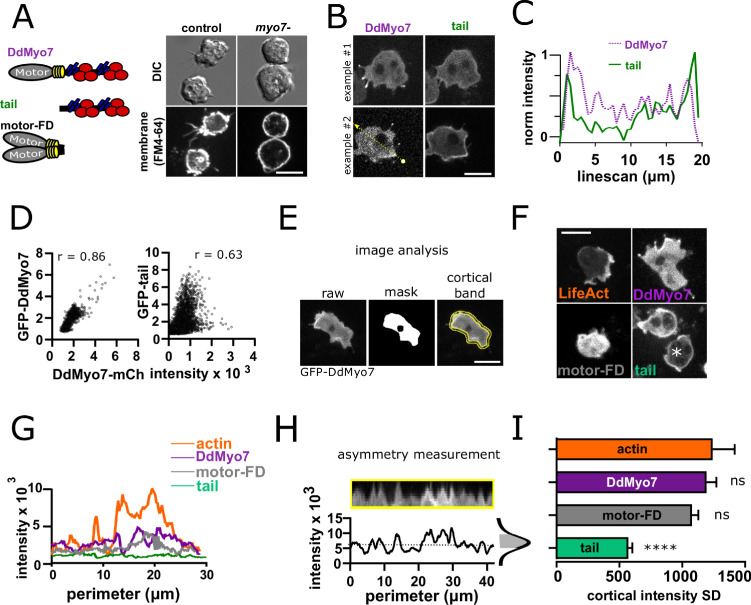
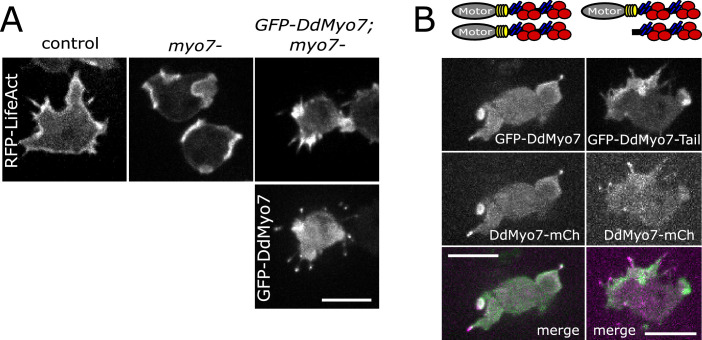
Figure 1. DdMyo7 has a distinct cortical localization from its tail domain.
(A) (left) Schematic of DdMyo7 illustrating its motor domain (gray), 4 IQ domains (yellow) and tandem MyTH4-FERM domains (blue-MyTH4, red-FERM) in the tail, the tail fragment, and a motor forced dimer (motor-FD); (right) Dictyostelium control, or myo7 null cells visualized with DIC and the membrane dye FM4-64 showing DdMyo7 is critical for filopodia formation. (B) Confocal images showing two examples of wild-type cells co-expressing DdMyo7-mCherry and GFP-DdMyo7-tail. The localization of DdMyo7-mCherry is at the cortex and in filopodia tips, and GFP-tail fragment localized around cortex. (C) Line intensity profile along the line shown in panel B. (D) Cytofluorograms of a representative field of cells comparing the colocalization between DdMyo7-mCherry intensity (x-axis) and GFP-DdMyo7 or GFP-DdMyo7 tail intensity (y-axis). (E) Analysis strategy for measuring entire cell peripheral intensity. (F) Micrographs of cells expressing RFP-Lifeact, GFP-DdMyo7, GFP-Tail, or GFP-Motor-Forced Dimer (FD) asterisks (*) on GFP-tail indicates the cell analyzed in G. (A,B,E,F) Scale bars are 10 µm. (G) Peripheral line scan intensity of cells from F. (H). Sample cortical band intensity showing the mean and variation of intensities around the periphery (asymmetry measurement), shaded region of the intensity distribution represents the standard deviation. (I). Cortical band standard deviation (SD; n > 93 cells from three experiments for each group) (see also Figure 1—source data 1). A higher SD indicates asymmetric localization. One-way ANOVA with multiple comparison correction compared to actin, ****p<0.001, ns not significant (see also Figure 1—source data 2).