Author response image 6. Integration of nuclei from R1/R2/R3 snRNA-seq with cells from Tepe et al.
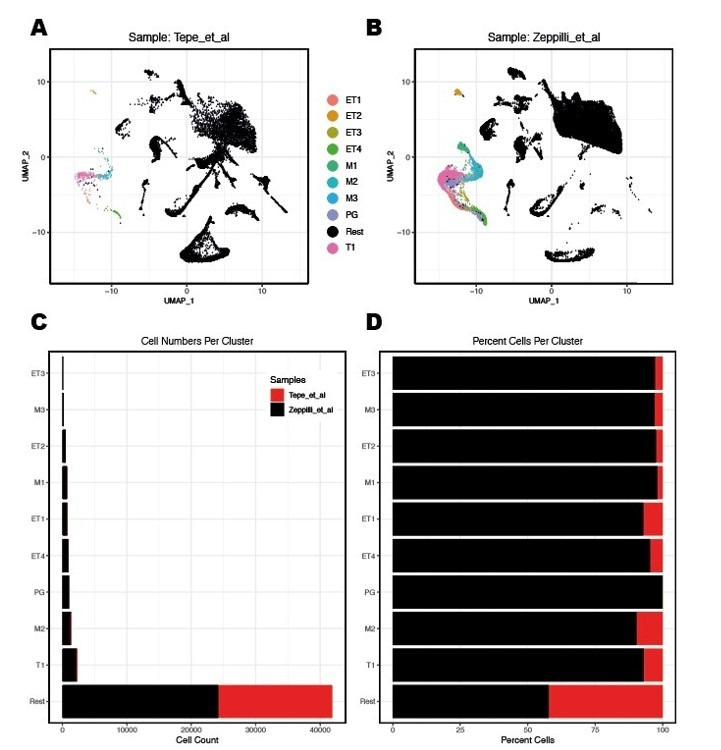
2018 dataset.(A,B) Integration of WT replicates from the Tepe et al. study with the integrated R1/R2/R3 snRNAseq dataset of the present study. Count data for samples WT1 and WT2 were extracted from file GSE121891_OB_6_runs.raw.dge.csv.gz. All WT1 and WT2 cells listed in the GSE121891_OB_metaData_seurat.csv.gz metadata file were used for the integration with the QCfiltered nuclei from the R1/R2/R3 snRNAseq dataset. Data integration between the two full datasets and label transfer of only projection neuron clusters (Figure 2D in the manuscript) to both full datasets were performed using the Seurat 3 R-package. MC/TC clusters are color-coded as indicated.(C,D) Quantification of cells/nuclei labelled as MC/TC clusters in the two datasets.
