Abstract
Unrelated individuals can produce genetically similar clones of antibodies, known as public clonotypes, which have been seen in responses to different infectious diseases, as well as healthy individuals. Here we identify 37 public clonotypes in memory B cells from convalescent survivors of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection or in plasmablasts from an individual after vaccination with mRNA-encoded spike protein. We identify 29 public clonotypes, including clones recognizing the receptor-binding domain (RBD) in the spike protein S1 subunit (including a neutralizing, angiotensin-converting enzyme 2 [ACE2]-blocking clone that protects in vivo) and others recognizing non-RBD epitopes that bind the S2 domain. Germline-revertant forms of some public clonotypes bind efficiently to spike protein, suggesting these common germline-encoded antibodies are preconfigured for avid recognition. Identification of large numbers of public clonotypes provides insight into the molecular basis of efficacy of SARS-CoV-2 vaccines and sheds light on the immune pressures driving the selection of common viral escape mutants.
Keywords: coronavirus, SARS-CoV-2, SARS-CoV, COVID-19, antibodies, monoclonal, human, adaptive immunity, public clonotypes, vaccines
Graphical abstract
Chen et al. describe neutralizing and non-neutralizing public antibodies elicited to SARS-CoV-2 spike in vaccinated and naturally infected individuals. Germline-revertant forms of some public clonotypes bind efficiently to spike protein, suggesting common germline-encoded properties for recognition.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is the causative agent of COVID-19 and the ongoing worldwide pandemic. The spike (S) protein is the principal antigen recognized by the protective antibody response against SARS-CoV-2 (Jiang et al., 2020; Krammer, 2020). The S protein is cleaved into S1, which includes the receptor-binding domain (RBD) and the N-terminal domain (NTD), and S2, contains the fusion peptide and heptad repeats HR1 and HR2 and mediates fusion between virus and host cell membrane (Bosch et al., 2003; Tortorici and Veesler, 2019). SARS-CoV-2 and SARS-CoV-1 share approximately 80% amino acid sequence identity, and both use human angiotensin-converting enzyme 2 (ACE2) as an entry receptor through binding mediated by the RBD (Hoffmann et al., 2020; Li et al., 2003; Wan et al., 2020).
Monoclonal antibodies (mAbs) targeting the SARS-CoV-2 S protein have been a focus for development of medical countermeasures against COVID-19. Many studies have identified antibodies to the S1 and S2 regions on the S protein, with the majority of neutralizing antibodies targeting the RBD in S1 and inhibiting ACE2 binding (Liu et al., 2020; Robbiani et al., 2020; Seydoux et al., 2020; Wec et al., 2020; Zost et al., 2020b). Multiple RBD-specific mAbs have been developed as monotherapies or cocktail therapeutics, and three have received Emergency Use Authorization (EUA) in the United States.
In recent years, public B cell clonotypes have been identified in the human antibody repertoires formed in response to diverse viruses, including Ebola (Cohen-Dvashi et al., 2020; Davis et al., 2019; Ehrhardt et al., 2019), influenza (Joyce et al., 2016; Pappas et al., 2014; Zost et al., 2021; Sui et al., 2009; Wheatley et al., 2015), human immunodeficiency virus 1 (HIV-1) (Setliff et al., 2018; Williams et al., 2015; Wu et al., 2011; Zhou et al., 2015), hepatitis C (Bailey et al., 2017; Giang et al., 2012), SARS-CoV-2 (Dong et al., 2021; Nielsen et al., 2020; Yuan et al., 2020a), respiratory syncytial virus (Mukhamedova et al., 2021), and in healthy individuals (Briney et al., 2019; Soto et al., 2019). These studies reveal a convergence of B cell selection resulting in circulating B cells clones with genetically similar antigen receptor genes in multiple individuals. The selection of public B cell clonotypes often has a structural basis mediated by low-affinity recognition of virus surface antigens by unmutated germline-encoded naive B cell receptors that are preconfigured for binding and cell activation. Public clonotypes are of great interest, because the understanding of viral epitopes that commonly induce antibodies in humans has implications for predicting the most common responses to vaccines in large populations.
Several efforts have characterized public clonotypes in the response to SARS-CoV-2, with most work focused on neutralizing public clonotypes (Dong et al., 2021; Robbiani et al., 2020; Tan et al., 2021; Yuan et al., 2020a) that target the S1 protein RBD and NTD domains. However, it is less clear if public clonotypes are directed to other sites on the S trimer, such as the S2 domain. Epitopes on the S2 domain may be of interest, because these sites may be more conserved than those in RBD in different strains of coronavirus as a result of functional constraints associated with the viral fusion mechanism. In this study, we identified 37 total public clonotypes, 27 of which are shared between vaccinated and convalescent individuals. We found that shared clonotypes comprise a substantial proportion of the elicited human B cell response to the S trimer. We also compared the response following infection or mRNA vaccination to investigate the genetic basis for the efficacy of mRNA vaccines in the population. These data show that many clonotypes are shared between convalescent and vaccinated individuals.
Results
Identification of public clonotypes
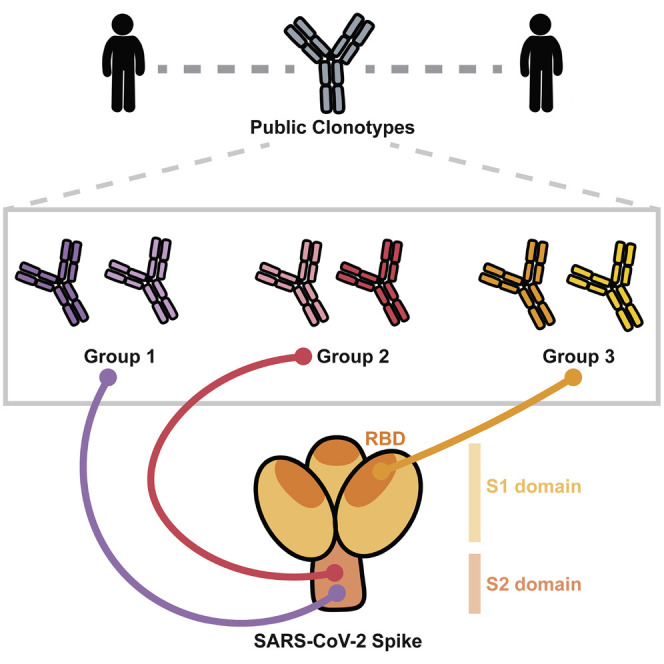
We first collected antibody variable gene sequences for SARS-CoV-2 human mAbs from existing publications that reported mAbs from individuals with a history of SARS-CoV-2 infection (Brouwer et al., 2020; Cao et al., 2020; Kreer et al., 2020; Liu et al., 2020; Robbiani et al., 2020; Rogers et al., 2020; Seydoux et al., 2020; Wec et al., 2020; Zost et al., 2020b). This search identified a panel of 2,865 paired heavy and light chain variable gene sequences. We clustered all sequences by binning the clones based on the inferred immunoglobulin heavy variable (IGHV) gene, immunoglobulin heavy joining (IGHJ) gene, and the amino acid length of the heavy chain complementarity determining region 3 (CDRH3). These sequences then were clustered according to 70% nucleotide sequence identity in the DNA sequence encoding the CDRH3. Next, the sequences were binned further based on the inferred immunoglobulin light variable gene (IGLV or IGKV) and immunoglobulin light joining (IGLJ or IGKJ) genes. Clusters meeting these similarity criteria in both heavy and light chains with sequences originating from two or more individuals were deemed public clonotypes. Eleven public clonotypes were identified in the repertoires of subjects with prior natural infection (Figures 1 A–1C), and these clones are encoded by a variety of heavy and light chain variable genes. Of the 11 public clonotypes identified, 5 of the heavy chain genes have been reported previously to encode potently neutralizing SARS-CoV-2 antibodies that bind to the RBD: IGHV3-53 (Yuan et al., 2020a), IGHV1-58 (Dong et al., 2021), IGHV3-30, IGHV3-30-3 (Robbiani et al., 2020), and IGHV3-66 (Yuan et al., 2020a). Three have not been reported: IGHV1-69, IGHV4-59, and IGHV3-7. IGHV3-53 and IGHV3-66 are commonly observed in antibodies in SARS-CoV-2 patients (Robbiani et al., 2020) because the germline gene segments encode amino acid motifs that are preconfigured for RBD binding (Yuan et al., 2020a). IGHV1-58 also commonly encodes antibodies that neutralize SARS-CoV-2, because this germline gene segment encodes motifs that mediate binding to the S protein (Dong et al., 2021). Notably, IGHV1-58 encodes the mAb COV2-2196, which is the basis for one of the two antibodies in a cocktail currently in phase III clinical trials (Dong et al., 2021; Zost et al., 2020a). Clonally expanded B cell populations containing potently neutralizing antibodies encoded by IGHV3-30 also have been found in multiple individuals (Robbiani et al., 2020). However, the role of IGHV1-69, IGHV4-59, and IGHV3-7 public clonotypes in SARS-CoV-2 responses remains unknown. In this paper, for clarity, we designated public clonotypes incorporating these additional three VH gene segments as members of group 1, 2, or 3 mAbs, respectively (Figures 1C–1E). Group 1 is shared by two donors from the cohort we studied and includes mAbs COV2-2002 and COV2-2333. Group 2 is shared by a donor from our group and a previously described donor IDCnC2 (Kreer et al., 2020) and includes antibodies COV2-2164 and CnC2t1p1_B10. Lastly, group 3 is shared by a donor from our group and a previously described donor COV107 (Robbiani et al., 2020) and includes antibodies COV2-2531 and C126 (Figures 1C–1E). cDNAs for the antibody variable genes encoding each of the six antibodies from the three groups of public clonotypes were synthesized and cloned into an IgG1 expression vector, as previously described (Gilchuk et al., 2020).
Figure 1.
Sequence characteristics of monoclonal antibodies to SARS-CoV-2
(A) Available sequences of mAbs to SARS-CoV-2 from multiple publications were obtained from public databases. Numbers inside each box represent the number of sequences with the indicated gene usage. Colored outlining boxes represent public clonotypes that are shared between the individuals listed in the key to the right side. The heatmap is color coded so that red represents a higher number of sequences using the corresponding genes, and blue represents a lower number of sequences using the corresponding genes.
(B) CDR3 sequences of the heavy and light chains of each of the remaining eight public clonotypes are shown. Dashes represent amino acids that differed in the public clonotype. Each box color correlates to the public clonotypes in (A) and (C).
(C) A Venn diagram illustrating all of the public clonotypes identified between naturally infected individuals. The colored boxes in the Venn diagram overlaps represent the public clonotypes identified in (A). Novel public clonotypes, designated as groups 1, 2, or 3, are highlighted in the purple, pink, or orange overlaps, respectively.
(D) Multiple sequence alignments of the heavy chain sequences for groups 1, 2, or 3 to their respective inferred germline genes IGHV 3-07/IGHJ4, IGHV1-69/IGHJ4, or IGHV4-59/IGHJ3. The CDRH3 sequence is highlighted in dark blue.
(E) Multiple sequence alignments of the light chain sequences for groups 1, 2, or 3 to their respective inferred germline genes IGLV3-01/IGLJ3, IGKV3-11/IGKJ4, or IGHV3-01/IGLJ2. The CDRL3 sequence is highlighted in light blue.
Functional properties of identified public clonotype antibodies
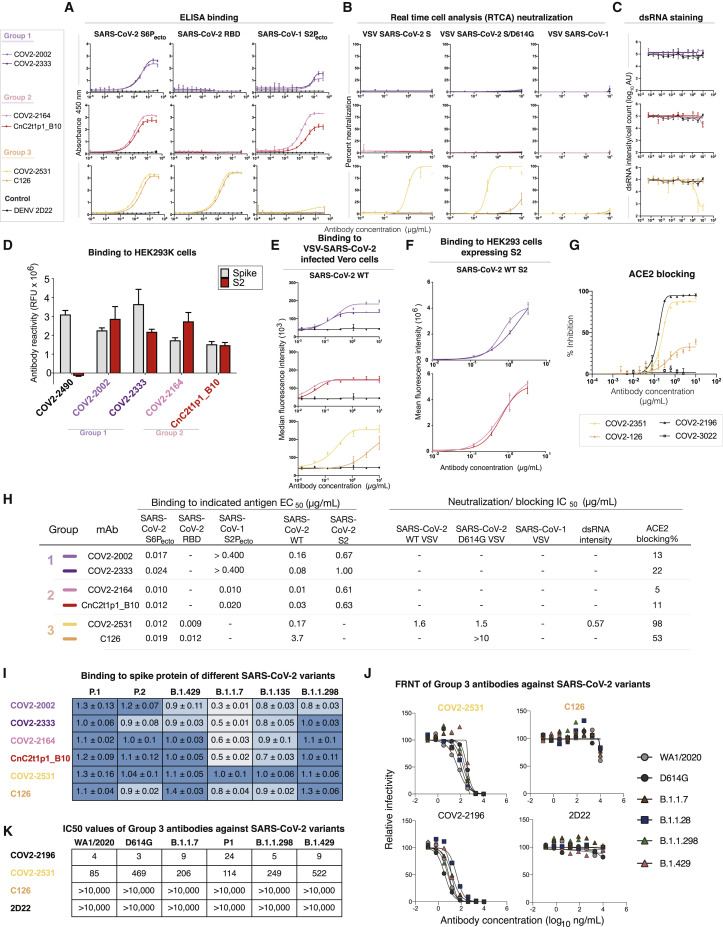
To examine the binding properties of antibodies in these three new SARS-CoV-2 public clonotypes, we tested six recombinant purified antibodies, two for each public clonotype, for binding to recombinant stabilized trimeric prefusion ectodomain of the SARS-CoV-2 S protein (S6Pecto), SARS-CoV-2 RBD, or recombinant stabilized trimeric prefusion ectodomain of the SARS-CoV-1 S protein (S2Pecto) proteins by ELISA (Figure S1). The two group 1 antibodies, COV2-2002 and COV2-2333, did not bind to SARS-CoV-2 RBD, but both bound to SARS-CoV-2 S6Pecto and SARS-CoV-1 S2Pecto proteins. However, they did not saturate in binding to SARS-CoV-1 S2Pecto at the maximum concentration tested (400 ng/mL), indicating relatively weak binding to recombinant SARS-CoV-1 S2Pecto. Group 2 antibodies, which include COV2-2164 and CnC2t1p1_B10, did not bind to SARS-CoV-2 RBD, but both bound to SARS-CoV-2 S6Pecto and SARS-CoV-1 S2Pecto proteins. Group 3 antibodies, which include COV2-2531 and C126, bound to SARS-CoV-2 S6Pecto and SARS-CoV-2 RBD proteins (Figures 2 A and 2H). Antibodies from group 3 did not bind SARS-CoV-1 S2Pecto.
Figure 2.
Reactivity and functional activity of group 1, 2, and 3 antibodies
Group 1 antibodies are shown in light or dark purple, group 2 antibodies are in red or pink, and group 3 antibodies are in light or dark orange. mAb DENV 2D22 was used as a negative control antibody, as shown in the lines in black. All experiments are performed in biological replicates and technical triplicates. Biological replicate from representative single experiment is shown.
(A) ELISA binding to SARS-CoV-2 S6Pecto, SARS-CoV-2 RBD, or SARS-CoV-1 S2Pecto was measured by absorbance at 450 nm. Antibody concentrations starting at 0.4 μg/mL were used and titrated 2-fold.
(B) Neutralization activity of antibodies to VSV-SARS-CoV-2, VSV-SARS-CoV-2/D614G, and VSV-SARS-CoV-1 determined by using real-time cell analysis (RTCA) assay. The percent of neutralization is reported. Antibody concentrations started at 10 μg/mL and were titrated 3-fold.
(C) Neutralization activity of antibodies to authentic SARS-CoV-2 (USA-WA1/2020) determined by measuring dsRNA intensity per cell count after Calu3 lung epithelial cells were inoculated with SARS-CoV-2. Antibody concentrations started at 10 μg/mL and were titrated 3-fold.
(D) Antibody binding to full-length S (gray) or S protein C-terminal S2 region (red) expressed on the surface of HEK293T cells that were fixed and permeabilized. Antibodies were screened at 1 μg/mL. Antibody reactivity was measured by flow cytometry, and cellular florescence values were determined. COV2-2490, an NTD-directed antibody, was used as a control.
(E) Binding to VSV-SARS-CoV-2-infected Vero cells (SARS-CoV-2 wild-type [WT]) was measured using flow cytometry, and median florescence intensity values were determined for dose-response binding curves. Antibody was diluted 3-fold starting from 10 μg/mL.
(F) Binding to S protein C-terminal S2 region expressed on HEK293T cells (SARS-CoV-2 WT S2) was measured using flow cytometry, and mean fluorescence intensity values were determined for dose-response binding curves. Antibody was diluted 3-fold starting from 10 μg/mL.
(G) Inhibition of ACE2 binding curves for COV2-2531 or C126. Antibody concentrations started at 10 μg/mL and were titrated 3-fold to identify ACE2 blocking curves. COV2-2531 is shown in light orange, and C126 is shown in dark orange.
(H) The half maximal concentration (EC50) and half maximal inhibitory concentration (IC50) values for each of the assay curves in Figures 3A–3E. All values are denoted as μg/mL. ACE2 blocking was determined by measuring amount of ACE2 with FLAG tag binding in the presence of each antibody, measured by binding of an anti-FLAG antibody. Percent blocking is shown, calculated by using ACE2 binding without antibody as 0% blocking.
(I) Binding of each antibody to several variants of concern spike proteins. Binding of each antibody (at 1 μg/mL) to the SARS-CoV-2 spike variants is shown relative to the antibody’s binding to the WT spike, defined as a value of 1.0. The relative amounts of each variant expressed in cells were estimated using the signal for antibody 1A9, normalizing the average 1A9 binding (at 1 μg/mL) for each variant to the average 1A9 binding with the WT construct. For each public clonotype antibody, the binding values to each variant spike were corrected for spike expression equivalent to that of the WT spike. Darker blue indicates less change to binding of that antibody to the variant. Lighter blue color indicates more change for binding of that antibody to the variant.
(J) Neutralization curves of group 3 antibodies against variant SARS-CoV-2 strains. Antibodies were tested for inhibition of infection of the indicated viruses on Vero-TMPRSS2 cell monolayer cultures using a focus reduction neutralization test.
(K) Antibody neutralization IC50 values for group 3 antibodies against variant SARS-CoV-2 strains. One representative experiment of two performed in duplicate and mean IC50 values (ng/mL) from two independent experiments are shown.
We hypothesized that group 1 and 2 antibodies might bind the S2 domain of the S trimer; SARS-CoV-2 infection can elicit antibodies that recognize cross-reactive epitopes on the S2 domain (Ladner et al., 2021). Antibodies were tested for binding against the S2 domain of SARS-CoV-2 S expressed on HEK293T cells. This experiment showed that group 1 and 2 antibodies bound to S2 in a dose-dependent manner (Figures 2D, 2F, and 2H), and revealed that public clonotypes can be elicited to the S2 domain of the S trimer.
Antibodies from each group then were tested for neutralizing activity using a previously described real-time cell analysis (RTCA) assay that measures cellular impedance (Gilchuk et al., 2020; Zost et al., 2020b). We used recombinant, infectious vesicular stomatitis virus (VSV) expressing the S proteins from SARS-CoV-2 (WA1/2019 strain), SARS-CoV-2/D614G, or SARS-CoV-1 (Urbani strain) (Figure S1). In addition, we used authentic SARS-CoV-2 (WA1/2019) virus and Calu3 cell monolayer cultures, and neutralization was measured by staining for double-stranded RNA, which is produced in the cytoplasm in virus-infected cells (Figure S2). Group 3 mAb COV2-2531 neutralized SARS-CoV-2 (VSV-SARS-CoV-2 and VSV-SARS-CoV-2/D614G) (Figures 2B, 2C, and 2H) and authentic SARS-CoV-2, but not SARS-CoV-1. In contrast, another group 3 mAb, C126, partially neutralized the SARS-CoV-2/D614G variant but did not neutralize the wild-type (WT) VSV-SARS-CoV-2, VSV-SARS-CoV-1, or authentic virus. Group 1 and 2 antibodies did not exhibit neutralizing capacity for any of the viral strains tested.
Because both group 3 antibodies exhibited neutralizing capacity, we considered that they might block virus attachment to ACE2, a principal mechanism of inhibition by RBD-targeted antibodies (Hansen et al., 2020; Zost et al., 2020a). We tested whether each antibody could block binding of soluble trimeric S protein to recombinant human ACE2 protein. Only group 3 antibodies blocked binding to ACE2 (Figure 2H). Similar to the pattern we observed for neutralization, COV2-2531 fully blocked ACE2 binding, whereas C126 partially blocked binding, with less than 50% inhibition at maximal effect (Figure 2G). Therefore, it is likely that COV2-2531 neutralizes virus infection at least in part by blocking binding to ACE2.
We assessed binding of all six antibodies to variants P.1 (Gamma), P.2 (Zeta), B.1.429 (Epsilon), B.1.1.7 (Alpha), B.1.135 (Beta), and B.1.1.298 (Figure S3). Antibody binding of both group 1 and 2 mAbs was largely affected by variant residues in B.1.1.7 or B.1.135. However, group 3 antibody COV-2531 maintained its ability to bind all variants tested. C126, the less mutated counterpart of the group 3 public clonotype, exhibited a decrease in binding to P.2, B.1.1.7, and B.1.135 (Figure 2I).
Because COV2-2531 maintained binding to all variants tested, we tested whether it retained its neutralization capacity. Group 3 antibodies COV2-2531 and C126 were tested against clinical isolates for B.1.1.7 (Alpha), B.1.1.298, and B.1.429 (Epsilon) viruses. The mAbs also were tested against a chimeric WA1/2020 virus encoding the S gene of B.1.1.28 (Gamma, Wash-B.1.1.28). These results were compared with neutralization of WA1/2020 and an isogenic mutant containing the D614G mutation (WA1/2020 D614G) (Figure 2J). C126 lacked inhibitory activity against authentic SARS-CoV-2 viruses. In comparison, COV2-2531 neutralized the panel of variant SARS-CoV-2 viruses comparably with WA1/2020, with IC50 values ranging from 85 to 522 ng/mL (Figure 2K).
Binding sites of identified clonotype antibodies
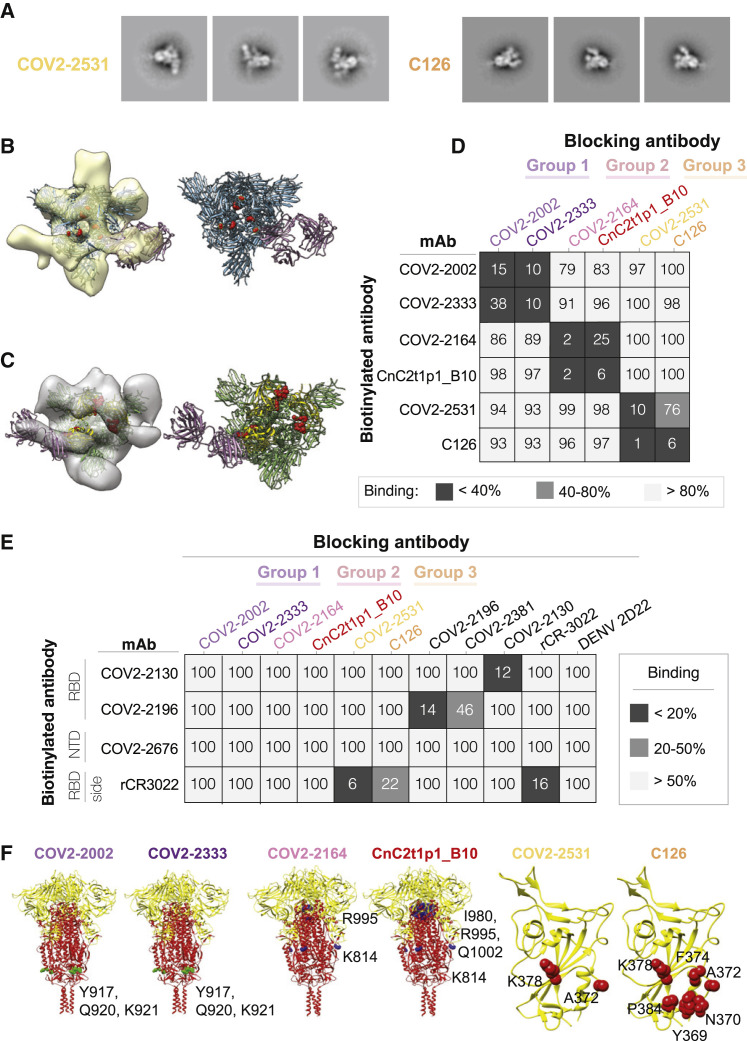
We used negative stain electron microscopy (EM) to image Fab-SARS-CoV-2 S6Pecto complexes. Even though all of the antibodies bound to S protein in ELISA as IgG1, only the group 3 antibody Fabs COV2-2531 and C126 formed complexes visualized on EM grids, suggesting that some antibodies may require an IgG format for strong binding (Figure 3 A; Figure S4). Low-resolution 3D reconstructions for COV2-2531 and C126 showed that these two antibodies bind the side of the RBD and recognize the cryptic face of the RBD that is accessible only in the “open” position of the RBD in the context of the S trimer (Figures 3B and 3C).
Figure 3.
Epitope identification and structural characterization of antibodies
(A) Negative stain EM of SARS-CoV-2 S6Pecto protein in complex with Fab forms of different mAbs. Negative stain 2D classes of SARS-CoV-2 S protein incubated with COV2-2531 or C126. Box size is 128 pix at 4.36 Å/pix.
(B) mAb COV2-2531 3D volume with critical residues 372 and 275 shown in red on the S protein (blue). RBD is in the open position. Density corresponds to three fabs, as we docked a single Fab structure onto the EM density map, shown in magenta.
(C) MAb C126 3D volume with critical binding residues shown in red. The Fab is docked to a protomer of SARS-CoV-2 S protein in the open conformation. Top left is the RBD positioned in open conformation, with the other two protomers in the trimer in closed position. The S protein is shown in green, with the RBD in yellow. The Fab is shown in magenta.
(D) Competition-binding ELISA results for mAbs within each clonotype group. Unlabeled blocking antibodies applied to antigen first are listed across the top, while biotinylated antibodies that are added to antigen-coated wells second are indicated on the left. The number in each box represents percent un-competed binding of the biotinylated antibody in the presence of the indicated competing antibody. Heatmap colors range from dark gray (<40% binding of the biotinylated antibody) to light gray (>80% binding of the biotinylated antibody). Experiment was performed in biological replicate and technical triplicate. Biological replicate from representative single experiment is shown.
(E) Competition-binding ELISA data using group 1, 2, or 3 antibodies against epitope-mapped reference antibodies. Biotinylated antibodies are indicated on the left, and the unlabeled antibodies applied to antigen first are indicated across the top. Heatmap colors range from dark gray (<20% binding of the biotinylated antibody) to light gray (>50% binding of the biotinylated antibody). Experiment was performed in biological replicate and technical triplicates. Biological replicate from representative single experiment shown.
(F) Alanine scanning mutagenesis results for group 1, 2, or 3 antibodies. S2 epitope residues are shown (green spheres or blue spheres) on the S protein structure (PDB: 6XR8), S1 is colored yellow, and S2 is red. RBD epitopes are shown in red on the RBD structure (PDB: 6XR8). Primary data are shown in Figure S5.
We then tested binding of antibodies to infected Vero cells that were inoculated with VSV-SARS-CoV-2 chimeric viruses (Figure S5). Antibodies from each of the groups bound to infected cells dose-dependently, with the group 3 RBD-reactive antibodies exhibiting greater binding than the group 1 or 2 S2-reactive antibodies (Figures 2E and 2H). Binding of the group 3 antibodies correlated with their neutralization capacity, because COV2-2531 showed greater binding than C126. The capacity to bind to infected cells also suggested that these antibodies could act in vivo not only by direct virus neutralization but also through Fc-mediated functions.
We used competition-binding ELISA for pairwise comparison of antibodies binding to the S6Pecto protein (Figure 3D). Members of each public clonotype group clustered with the other member of the same group by competition binding. We also competed the antibodies for binding against a larger group of epitope-mapped antibodies we previously described (Zost et al., 2020b) that cover various sites on the S protein and against rCR3022 (Yuan et al., 2020b), which bind less well to the RBD of SARS-CoV-2 compared with SARS-CoV-1 and do not block ACE2 binding (Figure 2G). Both group 3 antibodies competed with rCR3022, with COV2-2531 exhibiting a higher level of competition than C126. None of the group 1 or 2 antibodies competed with the reference antibodies tested (Figure 3E).
We then determined the critical binding residues at the amino acid level for each of the public clonotype antibodies by screening for binding to alanine scanning mutant libraries of the SARS-CoV-2 S protein. A372 and K378 were critical residues for COV2-2531 binding. For C126, we also identified A372 and K378, but with additional critical residues Y369, N370, F374, and P384 (Figure 3F; Figure S6). The identified residues are consistent with the binding site identified in the negative stain EM analyses and overlap with the epitope of CR3022 (Yuan et al., 2020b). It was curious that several SARS-CoV-2-specific neutralizing antibodies competed with CR3022, which also binds to SARS-CoV-1 but is non-neutralizing. SARS-CoV-1 has an N-glycosylation site at N370 in the binding site for these two mAbs, which SARS-CoV-2 lacks (Yuan et al., 2020b). This difference in glycosylation likely explains why COV2-2531 and C126 do not bind or neutralize SARS-CoV-1, even though they recognize the relatively conserved cryptic face of the RBD (Figure S7). In the alanine scanning libraries, native alanine residues are changed to serine. It is possible that A372 was identified as critical for binding by COV2-2531 and C126 because the A372S mutation results in the introduction of N-linked glycosylation of N370, rather than making direct side-chain contact with the antibodies.
Screening the group 1 and 2 antibodies against the SARS-CoV-2 alanine scanning mutation library confirmed that they bound to the S2 domain. For the group 1 antibodies COV2-2002 and COV2-2333, we identified critical residues for both antibodies (Y917, Q920, K921) in the heptad repeat (HR1) region of S2. These residues are conserved between SARS-CoV-2 and SARS-CoV-1. For the group 2 antibodies, screens identified two regions of residues that were critical for binding. For both COV2-2164 and CnC2t1p1 B10, we identified K814 (also conserved between SARS-CoV-2 and SARS-CoV-1) as critical for binding. In addition, for both antibodies, we also identified R995, and for CnC2t1p1_B10, I980 and Q1002 (Figure 3F; Figure S6). K814 is not close to I980, R995, or Q1002 on the S protein structure. However, inspection of the available S protein structures (PDB: 6XR8 and 7C2L) suggested that residues I980, R995, and Q1002 are not readily accessible to antibodies in the full S protein or even in the absence of S1. These residues make interactions that likely help maintain S2 structure, and so their mutation could indirectly affect group 2 antibody binding. We conclude that K814 is an epitope residue for group 2 antibodies COV2-2164 and CnC2t1p1_B10. These results suggest that the mAbs in each public clonotype group have essentially identical critical epitope residues.
Functional properties of germline-revertant forms of antibodies from each identified public clonotype
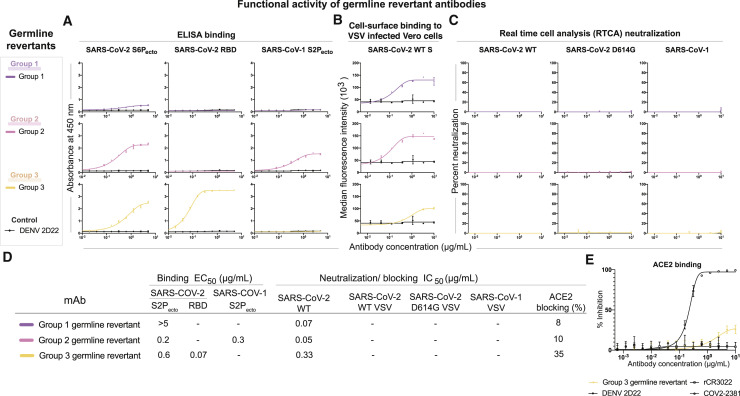
To determine whether the function of each antibody group was due to germline-encoded reactivity or the result of somatic mutations, we investigated the equivalent germline-encoded antibodies for each public clonotype. Heavy and light chain variable region sequences of antibodies COV2-2002, COV2-2164, and COV2-2531 were aligned with the germline sequences of [IGHV3-7/IGHJ4/ + IGLV3-1/IGLJ3], [IGHV1-69/IGHJ4 + IGKV3-11/IGKJ4], or [IGHV4-59/IGHJ3 + IGLV6-57/IGLJ2], respectively. Each residue that differed from the germline gene was reverted back to the inferred germline residue (Figure S8). We then tested whether the germline revertants of the antibodies in each group shared similar functional properties with their somatically mutated counterparts. Each germline-revertant antibody was tested for binding to SARS-CoV-2 S6Pecto, SARS-CoV-2 RBD, or SARS-CoV-1 S2Pecto proteins. The group 1 germline revertant did not bind to SARS-CoV-2 S6Pecto or SARS-CoV-1 S2Pecto. The group 2 germline revertant maintained binding to both SARS-CoV-2 and SARS-CoV-1 proteins but exhibited lower binding avidity (higher EC50 values) than its matured counterparts COV2-2164 or CnC2t1p1_B10. The group 3 germline revertant maintained binding to SARS-CoV-2 S6Pecto and RBD proteins (Figures 4 A and 4D). Each germline revertant also bound to the surface of virus-infected cells (Figures 4B and 4D). Although none of the germline revertants exhibited neutralizing capacity (Figures 4C and 4D), the group 3 germline revertant showed a low level of ACE2 blocking (Figures 4D and 4E).
Figure 4.
Germline-revertant antibody reactivity and functional activity
Group 1, 2, or 3 germline-revertant antibodies are shown in purple, pink, or yellow, respectively. DENV 2D22 was used as a control antibody for all assays, as shown in the lines in black. All experiments were performed in biological replicate and technical triplicate. Biological replicate from representative single experiment is shown, mean ± SD of triplicates is shown.
(A) Binding to SARS-CoV-2 S6Pecto, SARS-CoV-2 RBD, or SARS-CoV-1 S2Pecto was measured by absorbance at 450 nm, as shown in the first three columns.
(B) Binding to Vero cells infected with VSV-SARS-CoV-2, measured by flow cytometric analysis and reported as median florescence intensity.
(C) Results for neutralization curves for replication-competent VSV chimeric viruses in real-time cell analysis (RTCA) are shown in the next three columns, measured by percent neutralization calculated by normalized cell index.
(D) Binding EC50 and neutralization IC50 values for each of the assay curves in Figure 5A. All values are denoted as μg/mL. ACE2 blocking was determined by measuring the amount of ACE2 with FLAG tag binding in the presence of each antibody, measured by binding of an anti-FLAG antibody. Percent blocking is shown, calculated by using ACE2 binding without antibody as 0% blocking.
(E) Inhibition binding curves for the group 3 germline-revertant antibody. The starting antibody concentration used was 10 μg/mL and was titrated 3-fold serially to obtain ACE2-blocking curves.
COV2-2531 confers protection in vivo
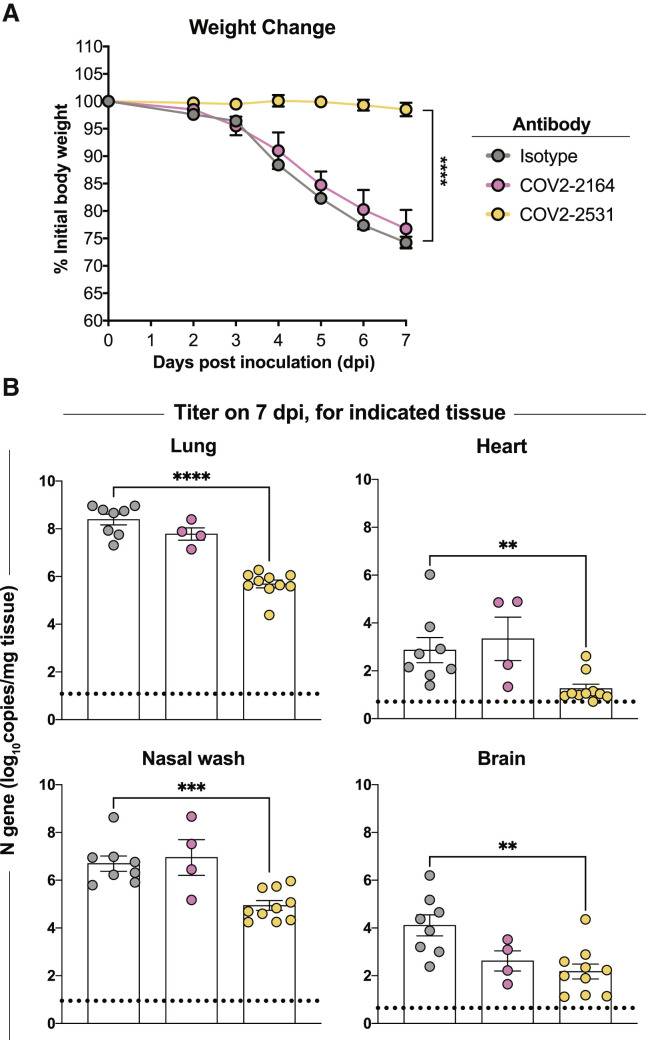
MAbs can act by direct virus inactivation, but binding of some mAbs to the surface of virus-infected cells (Figures 2E and 2H) suggested that these antibodies also might act through Fc-mediated functions. Therefore, it was important to test some public clonotypes in vivo. We tested the efficacy of these antibodies against SARS-CoV-2 in vivo. We used K18-hACE2 transgenic mice, which develop severe lung infection and disease after intranasal inoculation (Golden et al., 2020; Winkler et al., 2020; Zheng et al., 2021). K18-hACE2 transgenic mice received either one antibody from group 2 (COV2-2164), one antibody from group 3 (COV2-2531), or an isotype-control antibody (DENV 2D22) via intraperitoneal injection (200 μg, 10 mg/kg) 1 day prior to intranasal inoculation with 103 plaque-forming units (PFUs) of SARS-CoV-2 (WA1/2020). Mice treated with COV2-2531 were protected completely from weight loss (Figure 5 A) and showed reduced viral infection in the lung, nasal wash, heart, and brain (Figure 5B) compared with the isotype-control antibody-treated group. However, mice treated with COV2-2164 were not protected from weight loss, yet showed a reduction in viral load in the lung and brain but not in the nasal wash and heart (Figure 5B). Thus, antibodies that compete for binding with the SARS-CoV-1 mAb rCR3022 can be elicited after SARS-CoV-2 infection, some of which can confer protection.
Figure 5.
Antibody-mediated protection against SARS-CoV-2 challenge in mice
(A and B) Eight-week-old male K18-hACE2 transgenic mice were inoculated by the intranasal route with 103 PFUs of SARS-CoV-2 (WA1/2020 strain). One day prior to infection, mice were given a single 200-μg dose of COV2-2351 or COV2-2164 by intraperitoneal injection. (A) Weight change. Statistical analysis was performed only between isotype- and COV2-2351-treated groups. For isotype and COV2-2531 (mean ± SEM; n = 8–10, two experiments: unpaired t test of area under the curve; ∗∗∗∗p < 0.0001). For COV2-2164 (mean ± SEM; n = 8, two experiments). (B) Viral RNA levels at 7 days post-infection in the lung, nasal wash, heart, and brain as determined by qRT-PCR. For isotype and COV2-2531 (mean ± SEM; n = 8–10, two experiments: one-way ANOVA with Turkey’s post-test: not significant [ns], ∗p < 0.05, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, comparison with the isotype control mAb-treated group). For COV2-2164 (mean ± SEM; n = 8, two experiments).
Public clonotypes shared between vaccine and convalescent responses to SARS-CoV-2 S protein
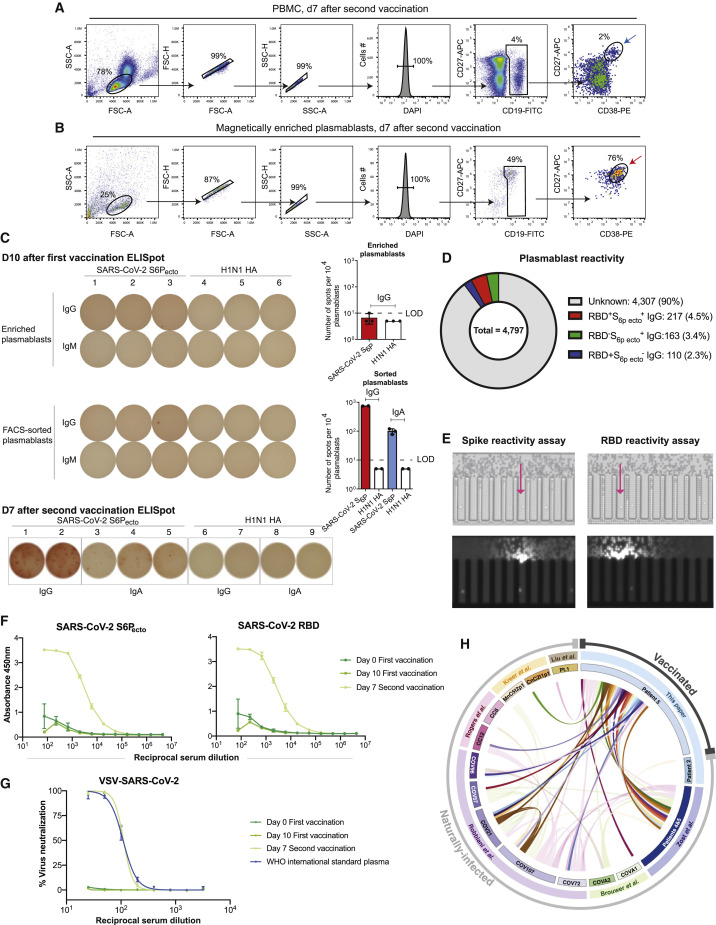
We hypothesized that SARS-CoV-2 mRNA vaccines might induce public clonotypes that are shared with those seen in convalescent individuals after natural infection. We obtained peripheral blood mononuclear cells from a volunteer 10 days after the first vaccine dose and 7 days after second vaccine dose with the Pfizer-BioNTech vaccine. Circulating plasmablasts were enriched directly from blood by negative selection using paramagnetic beads and purified further by flow cytometric sorting (Figures 6 A and 6B). Sorted plasmablasts were loaded on a Beacon microfluidics instrument for single-cell secreted antibody binding screening and antibody gene sequencing or in a Chromium single-cell microfluidics device (10X Genomics) followed by reverse transcription with PCR and sequence analysis to obtain paired antibody sequences, as described previously (Zost et al., 2020b). Enzyme-linked immunospot (ELISpot) assay analysis revealed a large increase in the frequency of S-reactive cells in the enriched plasmablast cell fraction on day 7 after the second vaccination compared with that on day 10 after the first vaccine dose, confirming induction of target-specific responses in this individual. SARS-CoV-2 S6Pecto-specific secreted antibodies were of IgG and IgA classes and accounted for >10% of total plasmablasts (Figure 6C). Further, single-cell antibody secretion analysis of a total of 4,797 purified plasmablasts revealed that a large fraction of SARS-CoV-2-reactive clones (including S6Pecto- and/or RBD-reactive clones) secreted RBD-specific IgG (Figure 6D).
Figure 6.
Analysis of vaccinated donor antibody response
(A and B) Flow cytometric plots showing gating strategy to identify plasmablasts in total PBMC sample collected on day 7 after second vaccine dose (top panel) or identification of plasmablasts after direct enrichment from whole blood at the same time point using negative selection with paramagnetic beads (bottom panel). Blue arrow indicates enriched plasmablasts that were used for ELISpot analysis as in (B), and red arrow indicates plasmablasts (DAPI−CD19loCD27hiCD38hi) that were FACS sorted for single-cell secretion and paired antibody sequencing studies.
(C) ELISpot analysis of SARS-CoV-2 S6Pecto-specific antibody secretion using enriched plasmablasts from blood collected on day 10 after the first vaccine dose (IgG) and day 7 after the second vaccine dose (IgG and IgA). A/Darwin/42/2020 H1N1 influenza virus hemagglutinin (HA) was used as a control for specificity of the plasmablast response. Wells with spots (left) and number of SARS-CoV-2 S6Pecto-specific responses detected (right) are shown. Dotted line indicates values below limit of the detection (LOD = 10 spots per 104 cells), which were set up to 5 spots per 104 cells.
(D) Pie chart representation showing frequency of RBD and SARS-COV-2 S6Pecto reactive (red), SARS-COV-2 S6Pecto reactive only (green), or RBD reactive only (blue) plasmablasts identified as in (C). Fraction of cells that did not react to either SARS-COV-2 RBD or S6Pecto is shown in gray.
(E) Flow-cytometry-sorted plasmablasts were loaded on a Beacon instrument and assessed for binding to S6Pecto or RBD-coated beads using single-cell antibody secretion analysis. Bright-field images of the Beacon instrument chip with individual plasmablasts loaded into the pens of the chip are shown for the selected fields of view for each screening condition (top). False-color fluorescent images from the same fields of view (bottom) showing binding of the detection anti-human Alexa Fluor 568-labeled antibody to the S6Pecto or RBD-coated beads that captured human antibodies secreted by single plasmablasts (visualized as a plume from the beads that loaded into the channel of the chip). Arrow indicates cells secreting antigen-reactive IgG antibodies.
(F) ELISA binding to SARS-CoV-2 S6Pecto of serum from patient 5 at day 0 of first vaccination, day 10 after first vaccination, or day 7 after second vaccination were measured by absorbance at 450 nm. Serum was diluted 1:75 and then diluted serially 3-fold. Experiment was performed in biological replicate and technical triplicate. Biological replicate from representative single experiment is shown, mean ± SD of triplicates is shown.
(G) Neutralization curves of serum from patient 5 at day 0 of first vaccination, day 10 after first vaccination, or day 7 after second vaccination. A World Health Organization (WHO) International standard for anti-SARS-CoV-2 human immunoglobulin was used as the positive control. Serum was diluted starting at a 1:25 dilution, then diluted serially 2-fold. Experiment was performed in technical triplicate.
(H) Circos plot indicating public clonotypes identified in this paper. The more opaque ribbons within the circle represent public clonotypes that are shared between the vaccinated donor and convalescent donors after natural infection. Translucent ribbons indicate public clonotypes shared between convalescent infection individuals. The individuals from whom sequences were derived are indicated on the inner circle. The published sources from which the sequences were obtained are shown on the second circle. The outside circle indicates whether the individuals were naturally infected or vaccinated.
We also analyzed antibody reactivity and neutralization in serum collected on the day before vaccination (day 0), on day 10 after the first vaccine dose, on day 7 after the second vaccine dose, and on day 28 after the second vaccine dose. The reactivity of serum antibodies to both SARS-CoV-2 S6Pecto and SARS-CoV-2 RBD was measured by ELISA for binding (Figure 6F) and by VSV-SARS-CoV-2 neutralizing assay (Figure 6G). Binding and neutralizing activities steadily increased over time, with maximal activity detected on day 28 after the second vaccine dose.
We obtained 725 paired heavy and light chain sequences from plasmablasts following primary immunization and 8,298 paired sequences from plasmablasts following the second dose of vaccine. The same procedure was carried out on a sample collected 35 days after onset of symptoms from a convalescent individual with confirmed SARS-CoV-2 infection. This individual’s serum had been determined previously to contain neutralizing antibodies (Zost et al., 2020b). Single-cell antibody secretion analysis revealed that a minor fraction (0.5%) of total plasmablasts produced S-protein-reactive antibodies. We identified 1,883 paired heavy and light chain antibody sequences from this specimen.
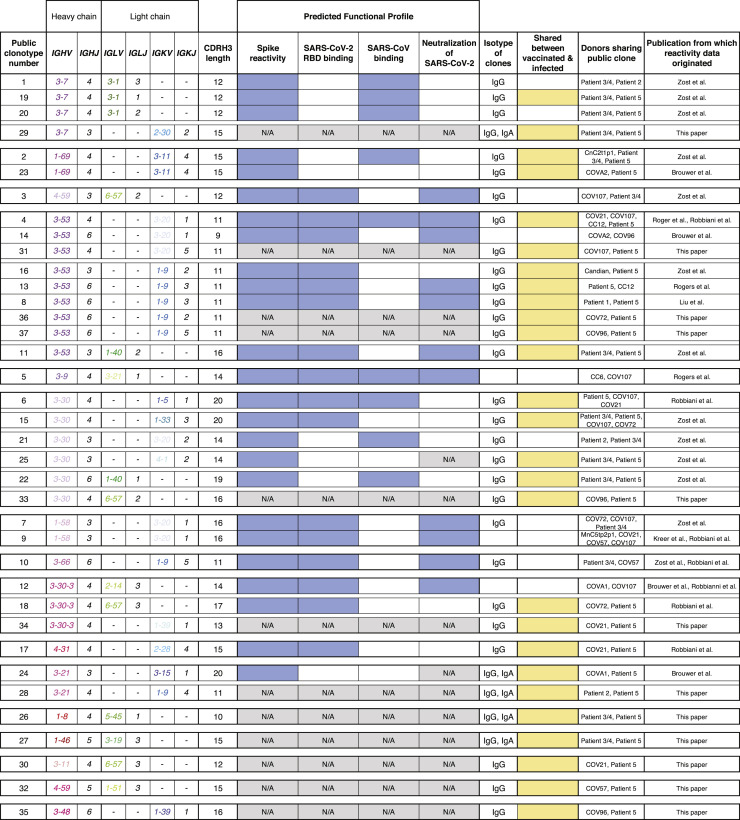
Antibody sequences identified in these new studies and sequences we collected from previous SARS-CoV-2 antibody discovery studies were clustered as described in Figure 1. We identified a total of 37 public clonotypes, 26 of which represented clonotypes shared between antibodies isolated from the vaccinee and individuals with exposure history to natural SARS-CoV-2 infection (Figure 6H). The antigen-binding specificity of each group was inferred through review of data in each respective publication in which the antibodies were reported. Fourteen of the 26 newly identified shared clonotypes encoded antibodies specific to the SARS-CoV-2 S protein. Within that panel of mAbs, 8 of 26 clonotypes reacted with SARS-CoV-2 RBD protein, and 6 of the 26 public clonotypes cross-reacted with both SARS-CoV-1 and SARS-CoV-2 (Figure 7 ). Most antibodies shared in public clonotypes were IgG, with a subset of IgA noted. This finding shows that the Pfizer-BioNTech vaccine induces many antibodies that are genetically similar to ones elicited through natural SARS-CoV-2 infection, including multiple public clonotypes in convalescent donors encoded by commonly used VH genes, such as IGHV3-53, IGHV3-66, IGHV1-58, IGHV3-30, and IGHV3-30-3. Additionally, of the 37 total public clonotypes, 16 bound to RBD, and of these, 11 of 16 were neutralizing. All neutralizing public clonotypes recognized RBD. However, of the 37 public clonotypes identified, 21 are directed to antigenic sites other than the RBD, including ones described here directed to the S2 domain. Overall, these results suggest that many of the public clonotypes observed in previously infected individuals likely are found in vaccinated individuals.
Figure 7.
Identification of public clonotypes shared between naturally infected individuals and a vaccinated donor
Table showing all public clonotypes identified. Gene usage for each clone or CDRH3 length is shown in columns 2 or 3. Reactivity profiles obtained from published sources are shown for comparative purposes. Blue indicates positive reactivity, while white indicates that binding reactivity or neutralization was not detected. Gray indicates reactivity profile was not found in either publication and therefore is unknown. Isotypes of antibodies in each group are listed in the eighth column. If the group contained sequences from both vaccinated and infected individuals, it was denoted in yellow. White was used for clonotypes that were shared only between convalescent individuals following natural infection.
Discussion
The high number of identified public B cell clonotypes in the response to SARS-CoV-2 infection or vaccination is striking. Many public clonotypes are shared between both infected and vaccinated individuals. Public clonotypes were induced by each of the currently known antigenic sites on the S protein and are found in both the neutralizing and non-neutralizing repertoires. Some clonotypes in the shared SARS-CoV-2 response appear preconfigured in the germline state to recognize particular S epitopes, and this recognition likely is driven by particular structural features on S. In this study, we compared the sequences of more than 14,000 paired B cell sequences encoding antibodies to S protein of SARS-CoV-2. Likely, the availability of large numbers of paired antibody gene sequences from a multitude of donors contributed to our ability to identify such a high number of paired-sequence public clonotypes.
Several neutralizing public clonotypes have been identified previously, most commonly clonotypes encoded by the closely related heavy chain genes IGHV3-53, IGHV3-66 (Tan et al., 2021; Yuan et al., 2020a), IGHV1-2 (Rapp et al., 2021), and IGHV3-30 (Robbiani et al., 2020). Structural features of these public clonotypes likely drive the frequent selection of such clones, such as the canonical configuration of aromatic residues in the public clonotype IGHV1-58 + IGHJ3 and IGKV3-20 + IGKJ1 that engages the SARS-CoV-2 RBD F486 residue (Dong et al., 2021). Members of this public clonotype, such as COV2-2196, engage the RBD using predominantly germline-encoded residues in both the heavy and light chain (Dong et al., 2021; Kreer et al., 2020; Nielsen et al., 2020; Robbiani et al., 2020; Tortorici et al., 2020). Identification of public clonotypes from multiple donors suggests these antibodies could contribute to humoral responses that mediate protection if they appear not only in memory B cells but also as antibodies from plasma cells secreted into the serum (Voss et al., 2020). The high prevalence of public clonotypes elicited to the SARS-CoV-2 S trimer may contribute to the high efficacy of S-encoding mRNA vaccines in large populations.
If diverse individuals independently make the same antibody in response to an antigen, there could be a constant and collective selective pressure on that epitope, resulting in a high potential for escape variants at that site. For example, although IGHV3-53- and IGHV3-66-encoded public clonotypes have been described in numerous individuals, neutralization of these antibodies is impacted adversely by the K417N or K417T substitutions present in the B.1.351 or P.1 SARS-CoV-2 variants of concern, respectively (Yuan et al., 2021). A similar case was described for IGHV1-2-encoded antibodies that target the RBD and IGHV1-24-encoded antibodies that target the NTD. These antibodies are found in the serum of convalescent individuals (Voss et al., 2020), but neutralization of these antibodies is negatively affected for 501Y.V2 variant viruses (Wibmer et al., 2021). A possible explanation for the selective pressure that led to the emergence and propagation of these variants is the humoral immunity mediated by these public clonotypes.
The new group 3 public clonotype neutralizing and protective antibodies described here bind to the cryptic face of the RBD and compete with the SARS-CoV-2 non-neutralizing mAb CR3022. Neutralizing antibodies that bind to the more conserved base of the RBD are of interest, because these sites are largely unaffected by common mutations in the variants of concern, such as E484K, K417N, and N501Y (Yuan et al., 2021). Importantly, recent work has identified a B.1.1.7 variant with a deletion of RBD residues 375–377. This deletion disrupts the epitope of CR3022, yet appears to be functionally tolerated (Rennick et al., 2021). Because group 3 antibodies share a similar epitope, with critical residues of COV2-2531 and C126 being K378 and A372, but with additional critical residues of Y369, N370, F374, and P384 identified for C126, this deletion might abrogate binding of antibodies from this public clonotype. Group 3 antibody characterization reveals additional insights on antibodies binding to epitopes similar to that of CR3022. Recently published literature suggests that there is an avidity threshold that affects antibody neutralization at the cryptic face of the RBD (Wu et al., 2020). We hypothesize that affinity improvements via somatic hypermutation play an important role in the ability of COV2-2531 to neutralize, particularly because neither C126 nor the germline revertant of group 3 neutralizes virus.
To our knowledge, public clonotypes specific to the S2 domain have not been described. In this study, we identified two public clonotypes that target the S2 domain of the S trimer. These mAbs do not neutralize, but they react with S proteins of both SARS-CoV-2 and SARS-CoV-1. It is likely that these S2 epitopes are the target of non-neutralizing antibodies in multiple individuals following infection or vaccination. Previous studies have identified broadly immunogenic epitopes that are conserved in the functional domains of the SARS-CoV-2 S trimer S2 domain, including cross-reactivity to endemic coronaviruses, and therefore these findings have important implications for antibody and vaccine design (Ladner et al., 2021). The S2 region of the S trimer may be more capable of recruiting preexisting memory B cells for diverse coronaviruses, because the S2 domain is more conserved for functionally important sites such as the heptad repeat regions and fusion loop (Anderson et al., 2021).
We propose that there are essentially four classes of public clonotypes: (1) neutralizing public clonotypes that bind to relatively invariant sites on S, (2) neutralizing public clonotypes that bind to sites that tolerate high sequence variability, (3) non-neutralizing public clonotypes that target relatively invariant sites, and (4) non-neutralizing public antibodies that target variable sites. The first class of antibodies is likely the most protective class in a population, because these mAbs neutralize and recognize residues unlikely to be sustained with mutations because of loss of viral fitness. An example of this class would be IGHV1-58-encoded antibodies as described previously (Dong et al., 2021). Many public clones currently identified for SARS-CoV-2 are categorized in the second class. While these clones initially offer protection, this property could be lost as widespread selective pressure on the virus is exerted on a region with genetic and structural plasticity. Examples of this group were discussed here, such as IGHV3-53- and IGHV3-66-encoded antibodies that target the RBD (Yuan et al., 2021). Here, we described three new public clonotypes following natural infection (groups 1, 2, and 3) and a total of 29 new clonotypes after mRNA vaccination. Public clonotype groups 1 and 2 fall into the third class of antibodies described here (non-neutralizing antibodies that target invariant sites), and public clonotype group 3 antibodies fall into the second class (neutralizing public clonotypes that bind to variable sites). Future public clonotypes to SARS-CoV-2 could be binned with this four-quadrant scheme to better understand how public clonotypes contribute to humoral immunity against COVID-19.
Understanding the antibody response that is shared between convalescent and vaccinated individuals also will be of continued interest as the percentage of vaccinated individuals increases in the face of the emergence of new viral variants of concern. The understanding of viral epitopes that induce protective antibodies in multiple individuals has implications for predicting the most common responses to new vaccines in large populations. The emergence of SARS-CoV-2 variants with acquired mutations in epitopes for neutralizing antibodies, including antibody regimens currently authorized for EUA, is a cause for concern (Collier et al., 2021; Tegally et al., 2020; Wang et al., 2021a, 2021b). Our analyses of public clonotypes after natural infection and vaccination and their shared epitope targets may predict sites of future major antigenic changes in the S trimer.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| COV2-2196 | Zost et al., 2020a | N/A |
| COV2-2130 | Zost et al., 2020a | N/A |
| COV2-2490 | Zost et al., 2020b | N/A |
| COV2-2676 | Zost et al., 2020b | N/A |
| CR3022 | Dingens et al., 2020 | N/A |
| r2D22 | Smith et al., 2012 | N/A |
| COV2-2002 | This paper | N/A |
| COV2-2333 | This paper | N/A |
| COV2-2164 | This paper | N/A |
| CnC2t1p1_B10 | Kreer et al., 2020 | N/A |
| COV2-2531 | This paper | N/A |
| C126 | Robbiani et al., 2020 | N/A |
| Goat anti-human IgG Fc, Multispecies ads-HRP | Southern Biotech | Cat#: 2014-05 |
| Goat anti-human IgG Alexa Fluor 647 | Invitrogen | Cat#: A-21445 |
| Angiotensin Converting Enzyme-2, ACE-2 | Sigma-Aldrich | Cat#: SAE0064 |
| Monoclonal ANTI-FLAG M2-Peroxidase HRP | Sigma-Aldrich | Cat#: A8592 |
| Anti-S2 antibody 1A9 | Genetex | Cat#: GTX632604 |
| Bacterial and virus strains | ||
| VSV-SARS-CoV-2 | Case et al., 2020b | N/A |
| VSV-SARS-CoV-2 D614G | Case et al., 2020b | N/A |
| VSV-SARS-CoV-1 | Case et al., 2020b | N/A |
| SARS-CoV-2 D614G | Plante et al., 2021 | N/A |
| SARS-CoV-2 (strain 2019 n-CoV/USA_WA1/2020) | CDC/BEI Resources | NR52281 |
| SARS-CoV-2 (isolate B.1.1.7) | Chen et al., 2021 | N/A |
| SARS-CoV-2 (isolate B.1.429) | Chen et al., 2021 | N/A |
| SARS-CoV-2 (isolate B.1.1.298) | Chen et al., 2021 | N/A |
| SARS-CoV-2 (isolate B.1.1.28) | Chen et al., 2021 | N/A |
| Biological samples | ||
| Plasmablasts from SARS-CoV-2 vaccinee | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| SARS-CoV-2 S6Pecto | Greaney et al., 2021 | N/A |
| Avidin–Peroxidase | Sigma | Cat#: A3151 |
| 1-step Ultra TMB-ELISA substrate solution | Thermo Fisher Scientific | Cat#: 34029 |
| ExpiCHO Expression Medium | Thermo Fisher Scientific | Cat#: A2910001 |
| FreeStyle 293 expression medium | Thermo Fisher Scientific | Cat#: 1238002 |
| Fetal Bovine Serum, ultra-low IgG | Thermo Fisher Scientific | Cat#: 16250078 |
| EZ-Link NHS-PEG4-Biotin, No-Weigh Format | Thermo Fisher Scientific | Cat#: A39259 |
| FabALACTICA® Fab kit | Genovis | Cat#: A2-AFK-025 |
| BioLock biotin blocking solution | IBA Lifescience | Cat#: 2-0205-050 |
| Uranyl Formate | EMS | Cat#: CF400-CU-50 |
| Deposited data | ||
| Sequence Read Archive deposition for the public clonotypes identified | This paper | NCBI: PRJNA511481 |
| Paired heavy and light chain plasmablast sequences, that were identified as public clonotypes, following mRNA vaccination | This paper | GenBank accession numbers MZ555491 through MZ555628 |
| Sequences of COV2-2164 | This paper | GenBank accession numbers MZ555629 and MZ555630 |
| Experimental models: Cell lines | ||
| Monkey: Vero E6 | American Type Culture Collection | CRL-1586, RRID: CVCL_0574 |
| Monkey: Vero E6 +TMPRSS2 | Diamond laboratory | N/A |
| Monkey: Vero E6 + ACE2 + TMPRS22 | A. Creanga and B. Graham (Vaccine Research Center, NIH) | N/A |
| Monkey: Vero CCL-81 | American Type Culture Collection | CCL-81; RRID: CVCL_0059 |
| Monkey: Vero Furin | Mukherjee et al., 2016 | N/A |
| Monkey: MA104 | American Type Culture Collection | CRL-2378.1 |
| Calu-3 | Thorne laboratory | N/A |
| Hamster: ExpiCHO-S | Thermo Fisher Scientific | Cat# A29127, RRID: CVCL_5J31 |
| Human: FreeStyle 293-F | Thermo Fisher Scientific | Cat# R79007, RRID: CVCL_D603 |
| Human: HEK293T | American Type Culture Collection | CRL-3216 |
| Human: HEK293T+ACE2 | Diamond laboratory | N/A |
| Human: HEK293 | American Type Culture Collection | CRL-1473 |
| Experimental models: Organisms/Strains | ||
| Mouse: B6.Cg-Tg(K18-ACE2)2Prlmn/J | Jackson Laboratory | Cat# 034860; RRID: IMSR_JAX:03486 |
| Recombinant DNA | ||
| Plasmid: pTwist-CMV S6Pecto | Greaney et al., 2021 | N/A |
| Plasmid: pTwist-mCis_G1 COV2-2196 | Zost et al., 2020a | N/A |
| Plasmid: pTwist-mCis_G1 COV2-2130 | Zost et al., 2020a | N/A |
| Plasmid: pTwist-mCis_G1 COV2-2490 | Zost et al., 2020b | N/A |
| Plasmid: pTwist-mCis_G1 COV2-2676 | Zost et al., 2020b | N/A |
| Plasmid: pTwist-mCis_G1 rCR3022 | Dingens et al., 2020 | N/A |
| Plasmid: pTwist-mCis_G1 r2D22 | Smith et al., 2012 | N/A |
| Plasmid: pTwist-mCis_G1 COV2-2002 | This paper | N/A |
| Plasmid: pTwist-mCis_G1 COV2-2333 | This paper | N/A |
| Plasmid: pTwist-mCis_G1 COV2-2164 | This paper | N/A |
| Plasmid: pTwist-mCis_G1 CnC2t1p1_B10 | Kreer et al., 2020 | N/A |
| Plasmid: pTwist-mCis_G1 COV2-2531 | This paper | N/A |
| Plasmid: pTwist-mCis_G1 C126 | Robbiani et al., 2020 | N/A |
| Software and algorithms | ||
| RTCA version 2.1.0 | Acea Biosciences, Inc. | RTCA Software, RRID: SCR_014821 |
| Serial EM 3.7 | D.N. Mastronarde | SerialEM, RRID: SCR_017293 |
| Topaz 0.2.3 | Bepler et al., 2019, Bepler et al., 2020 | Topaz |
| UCSF Chimera 1.14 | Pettersen et al., 2004 | UCSF Chimera, RRID: SCR_004097 |
| Cryosparc 3.0.0 | Punjani et al., 2017 | RRID: SCR_016501 |
| FlowJo | FlowJo, LLC | v10 |
| Computational pipeline for the clustering analysis | This paper | Zenodo; DOI 10.5281/zenodo.5150784 |
| Script to create heatmaps | This paper | Zenodo; DOI 10.5281/zenodo.5150784 |
| PyIR script used to determine sequence characteristics of each antibody | This paper | Zenodo; DOI 10.5281/zenodo.5150784 |
| GraphPad Prism | GraphPad | v 9.0.0 |
| iQue® Forecyt® Software | Sartorius | N/A |
| Nanozoomer Digital Pathology | Hamamatsu | v2 |
| Other | ||
| xCELLigence RTCA MP analyzer | Acea Biosciences, Inc. | N/A |
| xCELLigence E-Plate 96 PET cell culture plates | Acea Biosciences, Inc. | Cat# 300601010 |
| ÄKTA pure chromatography system | GE Healthcare Life Sciences | N/A |
| Biostack microplate stacker | BioTek | N/A |
| FEI TF20 electron microscope with Gatan US4000 4k × 4k CCD camera | Thermo Fisher Scientific | N/A |
| StrepTrap HP column | GE Healthcare Life Sciences | Cat# 28-9075-48 |
| 400 mesh copper EM grids | Electron Microscopy Services | Cat# 22451 |
| EL406 washer dispenser | BioTek | N/A |
| HisTrap Excel column | GE Healthcare Life Sciences | Cat# 17-3712-06 |
| HiTrap MabSelect SuRe 5 mL column | GE Healthcare Life Sciences | Cat# 29-0491-04 |
| Intellicyt iQue Screener Plus | Sartorius | N/A |
| ImmunoSpot S6 Plate Imager | Cellular Technology Limited | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, James. E. Crowe, Jr. (james.crowe@vumc.org)
Materials availability
Materials described in this paper are available for distribution for nonprofit use using templated documents from Association of University Technology Managers “Toolkit MTAs,” available at: https://autm.net/surveys-and-tools/agreements/material-transfer-agreements/mta-toolkit.
Experimental model and subject details
Research participants
We studied three patients (patient 2 [52-year-old female], 3 [56-year-old male] and 4 [56-year-old female]) in North America with laboratory-confirmed symptomatic SARS-CoV-2 infections that we have described previously (Zost et al., 2020b). We studied one patient (59-year-old male) who received Pfizer-BioNTech vaccine. The studies were approved by the Institutional Review Board of Vanderbilt University Medical Center.
Cell lines
Vero E6 (ATCC, CRL-1586) cells were maintained at 37°C in 5% CO2 in Dulbecco’s minimal essential medium (DMEM) containing 10% heat inactivated fetal bovine serum (FBS), 10 mM HEPES pH 73, 1 mM sodium pyruvate, 1 × non-essential amino acids, and 100 U/mL of penicillin-streptomycin. ExpiCHO cells (Thermo Fisher Scientific, A29127) were maintained at 37°C in 8% CO2 in ExpiCHO Expression Medium (Thermo Fisher Scientific, A2910002). Mycoplasma testing of cell lines was performed on monthly basis using a PCR-based mycoplasma detection kit (ATCC, 30-1012K), with negative results at each testing. Calu-3 (ATCC, HTB-55) cells were maintained at 37°C in 5% CO2 in DMEM with high glucose and L-glutamine (GIBCO 11965092), containing 10% heat inactivated fetal bovine serum (FBS), and 100 U/mL of penicillin-streptomycin. Vero-TMPRSS2 cells (Zang et al., 2020) were cultured at 37°C in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mM HEPES pH 7.3, 1 mM sodium pyruvate, 1 × non-essential amino acids, 100 U/mL of penicillin–streptomycin, and 5 μg/mL of blasticidin.
Viruses
The generation of a replication-competent VSV expressing SARS-CoV-2 S protein with a 21 amino acid C-terminal deletion that replaces the VSV G protein (VSV-SARS-CoV-2) was described previously (Case et al., 2020b). The S protein-expressing VSV virus was propagated in MA104 cell culture monolayers (African green monkey, ATCC CRL-2378.1) (Case et al., 2020b). Viral stocks were titrated on Vero E6 cell monolayer cultures by visualizing VSV plaques using neutral red staining. VSV-SARS-CoV-2/D614G was introduced by site directed mutagenesis. The 2019n-CoV/USA_WA1/2019 isolate of SARS-CoV-2 was obtained from the US Centers for Disease Control (CDC). Infectious stocks were propagated by inoculating Vero CCL81 cells. Supernatant was aliquoted and stored at −80°C. The University of Arizona group obtained the USA-WA1/2020 isolate of SARS-CoV-2 from WRCEVA. Early passage virus stock was generated by a single passage on Vero CCL81 for 48 h. Infected cell lysate and culture supernatant was combined, subjected to one freeze-thaw, and then centrifuged to pellet cell debris. The stock was titered to ∼3 x106 PFU/mL by standard plaque assay on Vero CCL81 cells. Nanopore sequencing of these early passages confirmed the genome sequence was identical to the GenBank WA1/2020 sequence (MN985325.1), with no mutations in the spike furin cleavage site. All work with infectious SARS-CoV-2 was performed in Institutional Biosafety Committee-approved BSL3 or A-BSL3 facilities at Washington University School of Medicine or University of Arizona, using appropriate positive pressure air respirators and protective equipment. The WA1/2020 recombinant strains with substitutions (D614G or N501Y/D614G) or a chimeric spike gene (B.1.1.28) were obtained from an infectious cDNA clone of the 2019n-CoV/USA_WA1/2020 strain, as previously described (Plante et al., 2021). The B.1.1.7, B.1.429, and B.1.1.298 isolates were obtained from nasopharyngeal swabs, and all viruses were passaged once in Vero-TMPRSS2 cells and subjected to next-generation sequencing as previously described (Chen et al., 2021) to confirm the introduction and stability of substitutions. Substitutions for each variant were as follows: B.1.1.7: deletion of 69 and 70 and 144 and 145, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H; B.1.1.28: L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y, D614G, H655Y, and T1027I; B.1.429: S13I, W152C, L452R, and D614G; B.1.1.298: deletion of 69 and 70, Y453F, D614G, I692V, and M1229I. All virus experiments were performed in an approved biosafety level 3 facility.
Method details
Clustering for identification of public clonotypes
Publicly available paired sequence sets of antibody genes were obtained (Brouwer et al., 2020; Kreer et al., 2020; Liu et al., 2020; Rogers et al., 2020; Seydoux et al., 2020; Wec et al., 2020; Zost et al., 2020b). Together with sequences derived from this paper, public clonotypes were determined by genetic similarities of antibody sequences using the following clustering scheme. The sequences were first binned by the same heavy chain V and J genes. Following sequences then were clustered according to 70% sequence similarity on their CDRH3 nucleotide sequence. Lastly, sequences then were binned together again if they used the same light chain V and J genes. Clusters of sequences containing sequences from two or more donors were determined to be public clonotypes. This clustering allowed us to identify a total of 11 public clonotypes, three of which had not been previously described. Below are the number of sequences we derived from each publication.
| Publication | Number of sequences identified | Number of public clonotypes that are SARS-CoV-reactive |
|---|---|---|
| Liu et al., 2020, Nature | 19 | |
| Kreer et al., 2020, Cell | 18 | |
| Robbiani et al., 2020, Nature | 533 | 2 |
| Rogers et al., 2020Science | 33 | 1 |
| Seydoux et al., 2020, Immunity | 32 | |
| Brouwer et al., 2020, Science | 84 | |
| Hansen et al., 2020, Science | 8 | |
| Wec et al., 2020, Science | 54 | |
| Zost et al., 2020b, Nature Medicine | 389 | 4 |
Heatmap generation
All sequences that were identified to be public clonotypes were analyzed with PyIR (Soto et al., 2020) to identify the V and J genes. The number of sequences with corresponding V and J genes on the heavy and light chains were counted. These frequency counts then were plotted onto the heatmap using Python Seaborn Library.
Antibody production and purification
Sequences of mAbs were synthesized using a rapid high-throughput cDNA synthesis platform (Twist Bioscience) and subsequently cloned into an IgG1 monocistronic expression vector (designated as pTwist-mCis_G1) for mAb secretion from mammalian cell culture. This vector contains an enhanced 2A sequence and GSG linker that allows simultaneous expression of mAb heavy- and light-chain genes from a single construct upon transfection (Chng et al., 2015). We performed transfections of ExpiCHO cell cultures using the GIBCO ExpiCHO Expression System and protocol for 50mL mini bioreactor tubes (Corning) as described by the vendor. Culture supernatants were purified using HiTrap MabSelect SuRe (Cytiva, formerly GE Healthcare Life Sciences) on a 24-column parallel protein chromatography system (Protein Biosolutions). Purified monoclonal antibodies were buffer exchanged into PBS, concentrated using Amicon Ultra-4 50-kDa centrifugal filter units (Millipore Sigma) and stored at 4°c until use.
Expression and purification of recombinant receptor binding domain (RBD) of SARS-CoV-2 S protein
For electron microscopy imaging of S protein in complex with Fab forms of human mAbs, we expressed a variant of S6Pecto protein containing a C-terminal Twin-Strep-tag, similar to that described previously (Zost et al., 2020b). Expressed protein was incubated with BioLock (IBA Lifesciences) and then isolated by Strep-tag affinity chromatography on StrepTrap HP columns (GE Healthcare), followed by size-exclusion chromatography on TSKgel G4000SWXL (TOSOH) if needed.
ELISA binding assays
Wells of 384-well microtiter plates were coated with purified recombinant SARS-CoV-2 S6Pecto, SARS-CoV-2 RBD, or SARS-CoV S2Pecto at 4°C overnight. Plates were blocked with 2% non-fat dry milk and 2% normal goat serum in DPBS containing 0.05% Tween-20 for 1 h. All antibodies were diluted to a concentration of either 0.4 μg/mL for the matured antibodies or 5 μg/mL for the germline-revertant antibodies. Antibodies were diluted in two-fold dilutions until binding was no longer detected. Bound antibodies were detected using goat anti-human IgG conjugated with horseradish peroxidase and TMB substrate. The reaction was quenched with 1N hydrochloric acid once color was developed. The absorbance was measured at 450 nm using a spectrophotometer (Biotek).
Cell-surface antigen-display assay
Vero cell monolayers were monitored until 80% confluent and then inoculated with VSV-SARS-CoV-2 V (WA1/2020 strain) at an MOI of 0.5 in culture medium (DMEM with 2% FBS). For a T-225 flask, 10 mL of diluted VSV-SARS-CoV-2 virus was added to the monolayer, then incubated for 40 min. During the incubation, the flask was gently rocked back and forth every 10 min to ensure even infection. Following, the incubation the flask volume was topped off to 30 mL with 2% FBS containing DMEM and incubated for 14 h. Cells were monitored for CPE under a microscope, were trypsinized and washed in fluorescence activated cell sorting (FACS) buffer. 100,000 infected cells were seeded per well to stain with respective antibodies. All antibody was diluted to 10 μg/mL in FACS buffer, and then serially diluted 3-fold 7 times to stain for antibodies that react to cell-surface-displayed S protein. Infected cells then were resuspended in 50 μL of diluted antibody. Antibody binding was detected with anti-IgG Alexa Fluor-647-labeled secondary antibodies. Cells were analyzed on an iQue cytometer for staining first by gating to identify infected cells as indicated by GFP-positive cells, and then gated for secondary antibody binding.
Real-time cell analysis (RTCA) neutralization assay
To determine neutralizing activity of purified antibodies or human serum, we used real-time cell analysis (RTCA) assay on an xCELLigence RTCA MP Analyzer (ACEA Biosciences Inc.) that measures virus-induced cytopathic effect (CPE) (Gilchuk et al., 2020; Suryadevara et al., 2021; Zost et al., 2020b). Briefly, 50 μL of cell culture medium (DMEM supplemented with 2% FBS) was added to each well of a 96-well E-plate to obtain background reading. A suspension of 18,000 Vero cells in 50 μL of cell culture medium was seeded in each well, and the plate was placed on the analyzer. Measurements were taken automatically every 15 min, and the sensograms were visualized using RTCA software version 2.1.0 (ACEA Biosciences Inc). SARS-CoV-2 S VSV, SARS-CoV-2 S D614G VSV, or SARS-CoV-1 (∼0.02 MOI, ∼120 PFU per well) was mixed 1:1 with a respective dilution of mAb or heat-inactivated human serum in a total volume of 100 μL using DMEM supplemented with 2% FBS as a diluent and incubated for 1 h at 37°C in 5% CO2. At 16 h after seeding the cells, the virus-mAb mixtures were added in replicates to the cells in 96-well E-plates. Triplicate wells containing virus only (maximal CPE in the absence of mAb) and wells containing only Vero cells in medium (no-CPE wells) were included as controls. Plates were measured continuously (every 15 min) for 48 h to assess virus neutralization. Normalized cellular index (CI) values at the endpoint (48 h after incubation with the virus) were determined using the RTCA software version 2.1.0 (ACEA Biosciences Inc.). Results are expressed as percent neutralization in a presence of respective mAb relative to control wells with no CPE minus CI values from control wells with maximum CPE. RTCA IC50 values were determined by nonlinear regression analysis using Prism software.
Competition-binding ELISA
Wells of 384-well microtiter plates were coated with purified recombinant SARS-CoV-2 S6Pecto protein at 4°C overnight. Plates were blocked with 2% bovine serum albumin (BSA) in DPBS containing 0.05% Tween-20 for 1 h. Each antibody was diluted to a concentration of 10 μg/mL. Next, biotinylated antibodies were diluted to 2.5 μg/mL and added to the primary antibody solution without washing the plate to a final concentration of 0.5μg/mL. Biotinylated antibody binding was detected with horseradish peroxidase-conjugated avidin (Sigma) and developed with TMB. The reaction was quenched with 1N hydrochloric acid once color was developed. Absorbance was measured at 450 nm using a spectrophotometer.
ACE2 blocking assay
Wells of 384-well microtiter plates were coated with purified recombinant SARS-CoV-2 S6Pecto protein at 4°C overnight. Plates were blocked with 2% nonfat dry milk in DPBS containing 0.05% Tween-20 for 1 h. Each antibody was diluted to a concentration of 10 μg/mL. Next, recombinant human ACE2 protein with a C-terminal FLAG tag was diluted to 2 μg/mL and added to the antibody solution without washing the plate to a final concentration of ACE2 of 0.4 μg/mL. ACE2 binding was detected using HRP-conjugated anti-FLAG antibodies and developed with TMB substrate. The reaction was quenched with 1 N hydrochloric acid once color was developed. Absorbance was measured at 450 nm using a spectrophotometer.
dsRNA staining neutralization assay
Calu-3 cells were seeded at 5,000 cells per well in SCREENSTAR 384-well black plates (Greiner) and allowed to adhere overnight. The cells then were treated with antibodies in 12 concentrations spanning from 5.65 × 10−5 μg/mL to 10 μg/mL and immediately transferred to a BSL-3 facility where they were inoculated with SARS-CoV-2 at an approximate MOI of 1 PFU/cell in 50 μL medium, and incubated for 48 h. At the end of the incubation, plates were submerged in PBS with 4% paraformaldehyde and 4% sucrose solution for 30 minutes to fix. Cells then were permeabilized with 0.2% Triton X-100/PBS for 10 min and blocked with 5% BSA/PBS for 1 h. Primary J2 anti-dsRNA (Scicons #10010500) antibody solution at a 1:1,000 dilution was placed on the cells overnight at 4°C. Cells were washed with 0.1% Tween-20/PBS (PBST) three times and plates were incubated with secondary goat anti-mouse Alexa Fluor-546-labeled antibody at 1:1,000 dilution (Thermo Fisher Scientific) for 2 h at room temperature in the dark. Plates were washed three times with PBST and incubated with DAPI for 30 min at room temperature in the dark. Plates were then imaged with fluorescent microscopy on a Nikon Eclipse TI2 automated microscopy system with a 20 × objective. Six frames per well were imaged and sum dsRNA fluorescence intensity, normalized to cell count by DAPI, was measured by Nikon Elements imaging software.
Focus reduction neutralization test
Serial dilutions of mAbs (starting at 10 μg/mL dilution) were incubated with 100 FFU of different SARS-CoV-2 strains for 1 h at 37°C. Antibody–virus complexes were added to Vero-TMPRSS2 cell monolayers in 96-well plates and incubated at 37°C for 1 h. Subsequently, cells were overlaid with 1% (w/v) methylcellulose in MEM. Plates were collected 30 h later by removing overlays and fixed with 4% PFA in PBS for 20 min at room temperature. Plates were washed and sequentially incubated with an oligoclonal pool of SARS2-2, SARS2-11, SARS2-16, SARS2-31, SARS2-38, SARS2-57 and SARS2-71 (Case et al., 2020a) anti-S antibodies and HRP-conjugated goat anti-mouse IgG (Sigma, 12-349) in PBS supplemented with 0.1% saponin and 0.1% bovine serum albumin. SARS-CoV-2-infected cell foci were visualized using TrueBlue peroxidase substrate (KPL) and quantitated on an ImmunoSpot microanalyzer (Cellular Technologies).
Mouse experiments
Animal studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocols were approved by the Institutional Animal Care and Use Committee at the Washington University School of Medicine (assurance number A3381–01). Virus inoculations were performed under anesthesia that was induced and maintained with ketamine hydrochloride and xylazine, and all efforts were made to minimize animal suffering. Heterozygous K18-hACE c57BL/6J mice (strain: 2B6.Cg-Tg(K18-ACE2)2Prlmn/J) were obtained from The Jackson Laboratory. Animals were housed in groups and fed standard chow diets. One day prior to infection, mice were given a single 200 μg dose of COV2-2351 or COV2-2164 by intraperitoneal injection. Eight- to nine-week-old mice were administered 103 PFU of SARS-CoV-2 by intranasal administration.
Measurement of viral burden in mouse tissues
Tissues were weighed and homogenized with zirconia beads in a MagNA Lyser instrument (Roche Life Science) in 1,000 μL of DMEM medium supplemented with 2% heat-inactivated FBS. Tissue homogenates were clarified by centrifugation at 10,000 rpm for 5 min and stored at −80°C. RNA was extracted using the MagMax mirVana Total RNA isolation kit (Thermo Fisher Scientific) on the Kingfisher Flex extraction robot (Thermo Fisher Scientific). RNA was reverse transcribed and amplified using the TaqMan RNA-to-CT 1-Step Kit (Thermo Fisher). Reverse transcription was carried out at 48°C for 15 min followed by 2 min at 95°C. Amplification was accomplished over 50 cycles as follows: 95°C for 15 s and 60°C for 1 min. The number of copies of SARS-CoV-2 N gene RNA in samples was determined using a previously published assay (Case et al., 2020a). Briefly, a TaqMan assay was designed to target a highly conserved region of the N gene (forward primer: ATGCTGCAATCGTGCTACAA; Reverse primer:m GACTGCCGCCTCTGCTC; Probe: /56-FAM/TCAAGGAAC/ZEN/AACATTGCCAA/3IABkFQ/). This region was included in an RNA standard to allow for copy number determination down to 10 copies per reaction. The reaction mixture contained final concentrations of primers or probe of 500 or 100 nM, respectively.
Electron microscopy sample and grid preparation, imaging and processing of S6Pecto–Fab complexes
Fabs were produced by digesting recombinant chromatography-purified IgGs using resin-immobilized cysteine protease enzyme (FabALACTICA, Genovis). The digestion occurred in 100 mM sodium phosphate and 150 mM NaCl pH 7.2 (PBS) for around 16 h at ambient temperature. To remove cleaved Fc from intact IgG, the digestion mix was incubated with CaptureSelect Fc resin (Genovis) for 30 min at ambient temperature in PBS buffer.
For screening and imaging of negatively-stained SARS-CoV-2 S6Pecto protein in complex with human Fabs, the proteins were incubated at a Fab:S molar ratio of 4:1 for about 1 h at ambient temperature or overnight at 4°C. Approximately 3 μL of the sample at concentrations of about 10 to 15 μg/mL was applied to a glow-discharged grid with continuous carbon film on 400 square mesh copper electron microscopy grids (Electron Microscopy Sciences). The grids were stained with 0.75% uranyl formate (Ohi et al., 2004). Images were recorded on a Gatan US4000 4k × 4k CCD camera using an FEI TF20 (TFS) transmission electron microscope operated at 200 keV and control with Serial EM. All images were taken at 50,000 × magnification with a pixel size of 2.18 Å per pixel in low-dose mode at a defocus of 1.5 to 1.8 μm. The total dose for the micrographs was around 30 e− per Å2. Image processing was performed using the cryoSPARC software package. Images were imported, CTF-estimated, and particles were picked. The particles were extracted with a box size of 256 pixels and binned to 128 pixels (pixel size of 4.36 Å/pix) and 2D class averages were performed.
Epitope mapping of antibodies by alanine scanning
Epitope mapping was performed essentially as described previously (Davidson and Doranz, 2014) using SARS-CoV-2 (Wuhan-Hu-1 strain) S protein RBD and S2 shotgun mutagenesis mutation libraries, made using a full-length expression construct for S protein. 184 residues of the RBD (between S residues 335 and 526), and 513 S2 residues (between residues 689 −1247) were mutated individually to alanine, and alanine residues to serine. Mutations were confirmed by DNA sequencing, and clones arrayed in a 384-well plate, one mutant per well. Binding of mAbs to each mutant clone in the alanine scanning library was determined, in duplicate, by high-throughput flow cytometry. A plasmid encoding cDNA for each S protein mutant was transfected into HEK293T cells and allowed to express for 22 h. Cells were fixed in 4% (v/v) paraformaldehyde (Electron Microscopy Sciences), and permeabilized with 0.1% (w/v) saponin (Sigma-Aldrich) in PBS plus calcium and magnesium (PBS++) before incubation with mAbs diluted in PBS++, 10% normal goat serum (Sigma), and 0.1% saponin. mAb screening concentrations were determined using an independent immunofluorescence titration curve against cells expressing wild-type S protein to ensure that signals were within the linear range of detection. Antibodies were detected using 3.75 μg/mL of Alexa Fluor-488-labeled secondary antibodies (Jackson ImmunoResearch Laboratories) in 10% normal goat serum with 0.1% saponin. Cells were washed three times with PBS++/0.1% saponin followed by two washes in PBS, and mean cellular fluorescence was detected using a high-throughput Intellicyte iQue flow cytometer (Sartorius). Antibody reactivity against each mutant S protein clone was calculated relative to wild-type S protein reactivity by subtracting the signal from mock-transfected controls and normalizing to the signal from wild-type S-transfected controls. Mutations within clones were identified as critical to the mAb epitope if they did not support reactivity of the test mAb but supported reactivity of other SARS-CoV-2 antibodies. This counter-screen strategy facilitates the exclusion of S protein mutants that are locally misfolded or have an expression defect.
Cell-surface binding to full- length S protein, variant S proteins, or S2 domain protein
A plasmid encoding the S protein C terminus S2 region (starting at residue S685) was transfected into HEK293T cells arrayed in a 384-well plate and allowed to express for 22 h. Cells transfected with vector alone acted as negative controls. MAbs were screened over a range of concentrations, 4 replicates for each mAb concentration, as described for epitope mapping. Fluorescence values were background subtracted.
ELISA binding assay for serum analysis
To assess serum reactivity, 384-well microtiter plates were coated with purified recombinant SARS-CoV-2 S6Pecto at 4°c overnight. Plates were blocked with blocking buffer (2% non-fat dry milk and 2% normal goat serum in DPBS containing 0.05% Tween-20) for 1 h. Serum was diluted 1:75 in blocking buffer, and then diluted three-fold serially 15 times, and added to wells. Binding was detected with goat anti-human IgG conjugated with horseradish peroxidase and TMB substrate. The reaction was quenched with 1N hydrochloric acid once color was developed. The absorbance was measured at 450 nm using a spectrophotometer (Biotek).
Plasmablasts isolation and flow cytometric analysis
Blood was collected into tubes containing heparin. To assess plasmablasts frequency in PBMCs for analytical flow cytometric studies, PBMCs were enriched from whole blood (day 10 after first, and day 7 after second vaccination) using direct PBMCs isolation kit (StemCell Technologies). For singe-cell antibody secretion and paired antibody sequencing studies, plasmablasts were enriched from the whole blood (day 7 after second vaccination) by negative selection using custom direct human plasmablasts isolation kit containing paramagnetic beads and antibodies for negative selection (StemCell Technologies). Enriched cells were stained 30 min on ice in a RoboSep buffer (StemCell Technologies) containing following phenotyping antibodies; anti-CD19-FITC (1:20 dilution, eBioscience), anti-CD27-APC (1:20 dilution), and anti-CD38-PE (1:25 dilution, BD Biosciences), and then analyzed by flow cytometry using an SH800 cell sorter (Sony). A DAPI stain was used as a viability dye to exclude dead cells. Plasmablasts were identified as DAPI-CD19loCD27hiCD38hi cells. Approximately 40,000 and ∼6,000 plasmablasts were FACS-sorted in a bulk for paired antibody sequencing and single-cell antibody secretion studies, respectively.
Generation of antibody variable-gene libraries from single plasmablasts
For paired antibody sequencing, cells were resuspended into DPBS containing 0.04% non-acetylated BSA, split into four replicates, and separately added to 50 μL of RT Reagent Mix, 5.9 μL of Poly-dt RT Primer, 2.4 μL of Additive A and 10 μL of RT Enzyme Mix B to complete the Reaction Mix as per the vendor’s protocol. The reactions then were loaded onto a Chromium chip (10x Genomics). Chromium Single Cell V(D)J B Cell-enriched libraries were generated, quantified, normalized and sequenced according to the User Guide for Chromium Single Cell V(D)J Reagents kits (CG000086_REV C). Amplicons were sequenced on an Illumina Novaseq 6000, and data were processed using the CellRanger software v3.1.0 (10X Genomics).
Single-cell antibody secretion analysis using Beacon instrument
FACS-purified plasmablasts were resuspended in plasmablast survival medium that promotes antibody secretion and assessed for reactivity of secreted antibodies using the 11k chip on Beacon optofluidic instrument (Berkley Lights) as previously described (Zost et al., 2020b). Single cell-antibody secretion binding assay was performed as previously described (Zost et al., 2020b) using SARS-CoV-2 S6Pecto- and SARS-CoV-2 RBD-coated beads.
ELISpot assay
Direct enzyme-linked immunosorbent spot (ELISpot) assay was performed to enumerate plasmablasts present in the PBMC samples secreting IgG, IgM, or IgA antibodies reacting with either SARS-CoV-2-S6Pecto protein or influenza A/Darwin/42/2020 H1N1 hemagglutinin protein (as a negative control). Briefly, 96-well ELISpot MSIP plates (Millipore) were activated with 100 μL 100% methanol/well for 10 s, washed three times with 1 × DPBS, coated overnight either with 100 μL of 2 μg/mL of SARS-CoV-2-S6Pecto or influenza HA protein in PBS overnight at 4°C. Plates were washed three times with 1 × DPBS and blocked by incubation with RPMI containing 10% FCS at 37°C for 2 h. Enriched plasmablasts or FACS-sorted plasmablasts were added to the plates and incubated 18-24 h at 37°C. Plates were washed with PBS and then PBS containing 0.05% Tween, and then incubated with either goat anti-human IgG-HRP conjugated antibodies (Southern Biotech), goat anti-human IgA-HRP conjugated antibodies (Southern Biotech), or goat anti-human IgM-HRP conjugated antibodies (Southern Biotech) for 2 h at room temperature. After washing three times with PBS containing 0.05% Tween/1% BSA, plates were developed using 3-amino-9-ethyl-carbazole (AEC) substrate (Sigma). The developed plates were scanned, and spots were analyzed using an automated ELISpot counter (Cellular Technologies Ltd.). Plasmablasts or sorted plasmablasts from PBMCs were added to the plates and incubated 18-24 h at 37°C. Plates were washed with PBS and then PBS containing 0.05% Tween, and then incubated with either goat anti-human IgG-HRP conjugated antibodies (Southern Biotech, catalog no. 2040-05), goat anti-human IgA-HRP conjugated antibodies (Southern Biotech, catalog no. 2050-05), or goat anti-human IgM-HRP conjugated antibodies (Southern Biotech, catalog no. 2020-05) for 2 h at room temperature. After washing three times with PBS containing 0.05% Tween/1% BSA, plates were developed using 3-amino-9-ethyl-carbazole (AEC) substrate (Sigma). The developed plates were scanned and spots were analyzed using an automated ELISpot counter (Cellular Technologies Ltd.).
Quantification and statistical analysis
The descriptive statistics mean ± SEM or mean ± SD were determined for continuous variables as noted. Virus titers in the tissues were compared using one-way ANOVA with Turkey’s post-test. Curves for antibody binding and neutralization were fitted after log transformation of antibody concentrations using non-linear regression analysis. Technical and biological replicates are indicated in the figure legends. Statistical analyses were performed using Prism v8.4.3 (GraphPad).
Acknowledgments
EM data collection was collected at the Center for Structural Biology Cryo-EM facility at Vanderbilt University. We thank Cinque Soto for technical advice on study of public clonotypes. We thank Rachel Nargi, Joseph Reidy, Erica Armstrong, and Christopher Gainza for support in antibody purifications. We also thank Berkeley Lights and Jonathan Didier for expert technical support, and STEMCELL Technologies and Aida Mayhew for supplying crucial B cell enrichment reagents. We thank Jem Uhrlaub, Brendan Larson, and Dr. Mike Worobey (University of Arizona) for preparation and sequencing of early-passage SARS-CoV-2 stocks. We thank Eun-Hyung Lee, Doan C. Nguyen, and Ignacio Sanz from Emory University for sharing the plasmablast survival medium that promotes antibody secretion. We thank the anonymous donors of the plasma samples for their consent that allowed the WHO International standard for anti-SARS-CoV-2 human immunoglobulin to be prepared; we express our gratitude to those who have coordinated the collection of the convalescent plasma: Malcom Semple (University of Liverpool, UK), Lance Turtle (University of Liverpool, UK), Peter Openshaw (Imperial College London, UK), and Kenneth Baillie (University of Edinburgh) on behalf of the ISARIC4C Investigators; and Heli Harvala Simmonds and David Roberts (National Health Service Blood and Transplant, UK). We also thank NIBSC Standards Production and Development staff for the formulation and distribution of materials. We thank Chris Selverian for assistance in epitope mapping and Mallorie Fouch for S2 binding studies. This work was supported by Defense Advanced Research Projects Agency (DARPA) grants HR0011-18-2-0001 and HR0011-18-3-0001; US NIH contracts 75N93019C00074, 75N93019C00062, and 75N93019C00073 (to B.J.D.); NIH grants AI150739, AI130591, R35 HL145242, AI157155, AI141707, AI12893, AI083203, AI149928, AI095202, AI083203, GM136853, DK103126, and UL1 TR001439; the Dolly Parton COVID-19 Research Fund at Vanderbilt; RII COVID-19 Seed Grant 002196 from University of Arizona; a grant from Fast Grants, Mercatus Center, George Mason University; and funding from AstraZeneca. E.C.C. was supported by NIH grant T32 AI138932, and E.S.W. was supported by NIH grant F30 AI152327. J.E.C. is a recipient of the 2019 Future Insight Prize from Merck KGaA, which supported this work with a grant.
Author contributions
Conceptualization, E.C.C. and J.E.C.; investigation, E.C.C., P.G., S.J.Z., N.S., M.S.D., E.S.W., C.R.C., C.A.T., S.L., S.K.C., E.B., J.K.W., S.D., L.M., A.T., J.R., R.E.S., E.D., and R.E.C.; writing first draft: E.C.C. and J.E.C.; all authors edited the manuscript and approved the final submission; supervision, B.J.D., R.H.C., C.A.T., M.S.D., and J.E.C.; funding acquisition, M.S.D. and J.E.C.
Declaration of interests
E.D., J.K.W., and B.J.D. are employees of Integral Molecular, and B.J.D. is a shareholder in that company. M.S.D. is a consultant for Inbios, Vir Biotechnology, NGM Biopharmaceuticals, and Carnival Corporation and is on the Scientific Advisory Boards of Moderna and Immunome. The Diamond laboratory has received funding support in sponsored research agreements from Moderna, Vir Biotechnology, and Emergent BioSolutions. J.E.C. has served as a consultant for Luna Biologics, is a member of the Scientific Advisory Board of Meissa Vaccines and is Founder of IDBiologics. The Crowe laboratory has received funding support in sponsored research agreements from AstraZeneca, IDBiologics, and Takeda.
Published: August 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109604.
Supplemental information
Data and code availability
Sequence Read Archive deposition for the public clonotypes identified is deposited at the NCBI: PRJNA511481. Deposition for the paired heavy and light chain plasmablast sequences, that were identified as public clonotypes, following mRNA vaccination is deposited in GenBank with accession numbers MZ555491 through MZ555628. The sequences of COV2-2164 are deposited in GenBank with accession numbers MZ555629 and MZ555630. All other data are available in the main text or the supplementary materials. The computational pipeline for the clustering analysis, the script to create heatmaps, and the PyIR script used to determine sequence characteristics of each antibody are available as listed in the Key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Anderson E.M., Goodwin E.C., Verma A., Arevalo C.P., Bolton M.J., Weirick M.E., Gouma S., McAllister C.M., Christensen S.R., Weaver J., UPenn COVID Processing Unit Seasonal human coronavirus antibodies are boosted upon SARS-CoV-2 infection but not associated with protection. Cell. 2021;184:1858–1864.e10. doi: 10.1016/j.cell.2021.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey J.R., Flyak A.I., Cohen V.J., Li H., Wasilewski L.N., Snider A.E., Wang S., Learn G.H., Kose N., Loerinc L. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight. 2017;2:92872. doi: 10.1172/jci.insight.92872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bepler T., Kelley K., Noble A.J., Berger B. Topaz-Denoise: gen- eral deep denoising models for cryoEM and cryoET. Nat. Commun. 2020;11:5208. doi: 10.1038/s41467-020-18952-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bepler T., Morin A., Rapp M., Brasch J., Shapiro L., Noble A.J., Berger B. Positive-unlabeled convolutional neural networks for particle picking in cryo-electron micrographs. Nat. Methods. 2019;16:1153–1160. doi: 10.1038/s41592-019-0575-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosch B.J., van der Zee R., de Haan C.A., Rottier P.J. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J. Virol. 2003;77:8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briney B., Inderbitzin A., Joyce C., Burton D.R. Commonality despite exceptional diversity in the baseline human antibody repertoire. Nature. 2019;566:393–397. doi: 10.1038/s41586-019-0879-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer P.J.M., Caniels T.G., van der Straten K., Snitselaar J.L., Aldon Y., Bangaru S., Torres J.L., Okba N.M.A., Claireaux M., Kerster G. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Su B., Guo X., Sun W., Deng Y., Bao L., Zhu Q., Zhang X., Zheng Y., Geng C. Potent neutralizing antibodies against SARS-CoV-2 identified by high-throughput single-cell sequencing of convalescent patients’ B cells. Cell. 2020;182:73–84.e16. doi: 10.1016/j.cell.2020.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Bailey A.L., Kim A.S., Chen R.E., Diamond M.S. Growth, detection, quantification, and inactivation of SARS-CoV-2. Virology. 2020;548:39–48. doi: 10.1016/j.virol.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case J.B., Rothlauf P.W., Chen R.E., Liu Z., Zhao H., Kim A.S., Bloyet L.M., Zeng Q., Tahan S., Droit L. Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe. 2020;28:475–485.e5. doi: 10.1016/j.chom.2020.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chng J., Wang T., Nian R., Lau A., Hoi K.M., Ho S.C., Gagnon P., Bi X., Yang Y. Cleavage efficient 2A peptides for high level monoclonal antibody expression in CHO cells. MAbs. 2015;7:403–412. doi: 10.1080/19420862.2015.1008351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Dvashi H., Zehner M., Ehrhardt S., Katz M., Elad N., Klein F., Diskin R. Structural basis for a convergent immune response against Ebola virus. Cell Host Microbe. 2020;27:418–427.e4. doi: 10.1016/j.chom.2020.01.007. [DOI] [PubMed] [Google Scholar]
- Collier D.A., De Marco A., Ferreira I.A.T.M., Meng B., Datir R.P., Walls A.C., Kemp S.A., Bassi J., Pinto D., Silacci-Fregni C. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature. 2021;593:136–141. doi: 10.1038/s41586-021-03412-7. [DOI] [PubMed] [Google Scholar]
- Davidson E., Doranz B.J. A high-throughput shotgun mutagenesis approach to mapping B-cell antibody epitopes. Immunology. 2014;143:13–20. doi: 10.1111/imm.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis C.W., Jackson K.J.L., McElroy A.K., Halfmann P., Huang J., Chennareddy C., Piper A.E., Leung Y., Albarino C.G., Crozier I. Longitudinal analysis of the human B cell response to Ebola virus infection. Cell. 2019;177:1566–1582.e17. doi: 10.1016/j.cell.2019.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingens A.S., Crawford K.H.D., Adler A., Steele S.L., Lacombe K., Eguia R., Amanat F., Walls A.C., Wolf C.R., Murphy M. Serological identification of SARS-CoV-2 infections among children visiting a hospital during the initial Seattle outbreak. Nat Commun. 2020;11:4378. doi: 10.1038/s41467-020-18178-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J., Zost S.J., Greaney A.J., Starr T.N., Dingens A.S., Chen E.C., Chen R.E., Case J.B., Sutton R.E., Gilchuk P. Genetic and structural basis for recognition of SARS-CoV-2 spike protein by a two-antibody cocktail. bioRxiv. 2021 doi: 10.1101/2021.01.27.428529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt S.A., Zehner M., Krähling V., Cohen-Dvashi H., Kreer C., Elad N., Gruell H., Ercanoglu M.S., Schommers P., Gieselmann L. Polyclonal and convergent antibody response to Ebola virus vaccine rVSV-ZEBOV. Nat. Med. 2019;25:1589–1600. doi: 10.1038/s41591-019-0602-4. [DOI] [PubMed] [Google Scholar]
- Giang E., Dorner M., Prentoe J.C., Dreux M., Evans M.J., Bukh J., Rice C.M., Ploss A., Burton D.R., Law M. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc. Natl. Acad. Sci. USA. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchuk P., Bombardi R.G., Erasmus J.H., Tan Q., Nargi R., Soto C., Abbink P., Suscovich T.J., Durnell L.A., Khandhar A. Integrated pipeline for the accelerated discovery of antiviral antibody therapeutics. Nat. Biomed. Eng. 2020;4:1030–1043. doi: 10.1038/s41551-020-0594-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden J.W., Cline C.R., Zeng X., Garrison A.R., Carey B.D., Mucker E.M., White L.E., Shamblin J.D., Brocato R.L., Liu J. Human angiotensin-converting enzyme 2 transgenic mice infected with SARS-CoV-2 develop severe and fatal respiratory disease. JCI Insight. 2020;5:142032. doi: 10.1172/jci.insight.142032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hil- ton S.K., Huddleston J., Eguia R., Crawford K.H.D. Complete mapping of mutations to the SARS-CoV-2 spike receptor-binding domain that escape antibody recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Science. 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Hillyer C., Du L. Neutralizing antibodies against SARS-CoV-2 and other human coronaviruses. Trends Immunol. 2020;41:355–359. doi: 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce M.G., Wheatley A.K., Thomas P.V., Chuang G.Y., Soto C., Bailer R.T., Druz A., Georgiev I.S., Gillespie R.A., Kanekiyo M., NISC Comparative Sequencing Program Vaccine-induced antibodies that neutralize Group 1 and Group 2 influenza A viruses. Cell. 2016;166:609–623. doi: 10.1016/j.cell.2016.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- Kreer C., Zehner M., Weber T., Ercanoglu M.S., Gieselmann L., Rohde C., Halwe S., Korenkov M., Schommers P., Vanshylla K. Longitudinal isolation of potent near-germline SARS-CoV-2-neutralizing antibodies from COVID-19 patients. Cell. 2020;182:843–854.e12. doi: 10.1016/j.cell.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladner J.T., Henson S.N., Boyle A.S., Engelbrektson A.L., Fink Z.W., Rahee F., D’ambrozio J., Schaecher K.E., Stone M., Dong W. Epitope-resolved profiling of the SARS-CoV-2 antibody response identifies cross-reactivity with endemic human coronaviruses. Cell Rep. Med. 2021;2:100189. doi: 10.1016/j.xcrm.2020.100189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Wang P., Nair M.S., Yu J., Rapp M., Wang Q., Luo Y., Chan J.F., Sahi V., Figueroa A. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature. 2020;584:450–456. doi: 10.1038/s41586-020-2571-7. [DOI] [PubMed] [Google Scholar]
- Mukhamedova M., Wrapp D., Shen C.H., Gilman M.S.A., Ruckwardt T.J., Schramm C.A., Ault L., Chang L., Derrien-Colemyn A., Lucas S.A.M. Vaccination with prefusion-stabilized respiratory syncytial virus fusion protein induces genetically and antigenically diverse antibody responses. Immunity. 2021;54:769–780.e6. doi: 10.1016/j.immuni.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Sirohi D., Dowd K.A., Chen Z., Diamond M.S., Kuhn R.J., Pierson T.C. Enhancing dengue virus maturation using a stable furin over-expressing cell line. Virology. 2016;497:33–40. doi: 10.1016/j.virol.2016.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen S.C.A., Yang F., Hoh R.A., Jackson K.J.L., Roeltgen K., Lee J.Y., Rustagi A., Rogers A.J., Powell A.E., Kim P.S. B cell clonal expansion and convergent antibody responses to SARS-CoV-2. Res Sq. 2020 doi: 10.21203/rs.3.rs-27220/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi M., Li Y., Cheng Y., Walz T. Negative staining and image classification – Powerful tools in modern electron microscopy. Biol. Proced. Online. 2004;6:23–34. doi: 10.1251/bpo70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas L., Foglierini M., Piccoli L., Kallewaard N.L., Turrini F., Silacci C., Fernandez-Rodriguez B., Agatic G., Giacchetto-Sasselli I., Pellicciotta G. Rapid development of broadly influenza neutralizing antibodies through redundant mutations. Nature. 2014;516:418–422. doi: 10.1038/nature13764. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Plante J.A., Liu Y., Liu J., Xia H., Johnson B.A., Lokugamage K.G., Zhang X., Muruato A.E., Zou J., Fontes-Garfias C.R. Spike mutation D614G alters SARS-CoV-2 fitness. Nature. 2021;592:116–121. doi: 10.1038/s41586-020-2895-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punjani A., Rubinstein J.L., Fleet D.J., Brubaker M.A. cryo- SPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- Rapp M., Guo Y., Reddum E.R., Liu L., Wang P., Yu J., Cerutti G., Bimela J., Bahna F., Mannepalli S. Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class. bioRxiv. 2021 doi: 10.1101/2021.01.11.426218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rennick L.J., Robinson-McCarthy L.R., Nambulli S., Duprex W.P., McCarthy K.R. Deletion disrupts a conserved antibody epitope in a SARS-CoV-2 variant of concern. bioRxiv. 2021 doi: 10.1101/2021.03.05.434168. [DOI] [Google Scholar]
- Robbiani D.F., Gaebler C., Muecksch F., Lorenzi J.C.C., Wang Z., Cho A., Agudelo M., Barnes C.O., Gazumyan A., Finkin S. Convergent antibody responses to SARS-CoV-2 infection in convalescent individuals. bioRxiv. 2020 doi: 10.1101/2020.05.13.092619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setliff I., McDonnell W.J., Raju N., Bombardi R.G., Murji A.A., Scheepers C., Ziki R., Mynhardt C., Shepherd B.E., Mamchak A.A. Multi-donor longitudinal antibody repertoire sequencing reveals the existence of public antibody clonotypes in HIV-1 infection. Cell Host Microbe. 2018;23:845–854.e6. doi: 10.1016/j.chom.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux E., Homad L.J., MacCamy A.J., Parks K.R., Hurlburt N.K., Jennewein M.F., Akins N.R., Stuart A.B., Wan Y.H., Feng J. Characterization of neutralizing antibodies from a SARS-CoV-2 infected individual. bioRxiv. 2020 doi: 10.1101/2020.05.12.091298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S.A., Zhou Y., Olivarez N.P., Broadwater A.H., de Silva A.M., Crowe J.E., Jr. Persistence of circulating B memory cell clones with potential for dengue virus disease enhancement for decades following infec- tion. J. Virol. 2012;86:2665–2675. doi: 10.1128/JVI.06335-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C., Bombardi R.G., Branchizio A., Kose N., Matta P., Sevy A.M., Sinkovits R.S., Gilchuk P., Finn J.A., Crowe J.E., Jr. High frequency of shared clonotypes in human B cell receptor repertoires. Nature. 2019;566:398–402. doi: 10.1038/s41586-019-0934-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto C., Finn J.A., Willis J.R., Day S.B., Sinkovits R.S., Jones T., Schmitz S., Meiler J., Branchizio A., Crowe J.E., Jr. PyIR: a scalable wrapper for processing billions of immunoglobulin and T cell receptor sequences using IgBLAST. BMC Bioinformatics. 2020;21:314. doi: 10.1186/s12859-020-03649-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J., Hwang W.C., Perez S., Wei G., Aird D., Chen L.M., Santelli E., Stec B., Cadwell G., Ali M. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009;16:265–273. doi: 10.1038/nsmb.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryadevara N., Shrihari S., Gilchuk P., VanBlargan L.A., Binshtein E., Zost S.J., Nargi R.S., Sutton R.E., Winkler E.S., Chen E.C. Neutralizing and protective human monoclonal antibodies recognizing the N-terminal domain of the SARS-CoV-2 spike protein. bioRxiv. 2021 doi: 10.1101/2021.01.19.427324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan T.J.C., Yuan M., Kuzelka K., Padron G.C., Beal J.R., Chen X., Wang Y., Rivera-Cardona J., Zhu X., Stadtmueller B.M. Sequence signatures of two IGHV3-53/3-66 public clonotypes to SARS-CoV-2 receptor binding domain. bioRxiv. 2021 doi: 10.1101/2021.01.26.428356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorici M.A., Veesler D. Structural insights into coronavirus entry. Adv. Virus Res. 2019;105:93–116. doi: 10.1016/bs.aivir.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegally H., Wilkinson E., Giovanetti M., Iranzadeh A., Fonseca V., Giandhari J., Doolabh D., Pillay S., San E.J., Msomi N. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv. 2020;(Published online December 22, 2020) doi: 10.1101/2020.12.21.20248640. [DOI] [Google Scholar]
- Tortorici M.A., Beltramello M., Lempp F.A., Pinto D., Dang H.V., Rosen L.E., McCallum M., Bowen J., Minola A., Jaconi S. Ultrapotent human antibodies protect against SARS-CoV-2 challenge via multiple mechanisms. Science. 2020;370:950–957. doi: 10.1126/science.abe3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss W.N., Hou Y.J., Johnson N.V., Kim J.E., Delidakis G., Horton A.P., Bartzoka F., Paresi C.J., Tanno Y., Abbasi S.A. Prevalent, protective, and convergent IgG recognition of SARS-CoV-2 non-RBD spike epitopes in COVID-19 convalescent plasma. bioRxiv. 2020 doi: 10.1101/2020.12.20.423708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J. Virol. 2020;94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021;593:130–135. doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- Wang Z., Schmidt F., Weisblum Y., Muecksch F., Barnes C.O., Finkin S., Schaefer-Babajew D., Cipolla M., Gaebler C., Lieberman J.A. mRNA vaccine-elicited antibodies to SARS-CoV-2 and circulating variants. Nature. 2021;592:616–622. doi: 10.1038/s41586-021-03324-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wec A.Z., Wrapp D., Herbert A.S., Maurer D.P., Haslwanter D., Sakharkar M., Jangra R.K., Dieterle M.E., Lilov A., Huang D. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science. 2020;369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheatley A.K., Whittle J.R., Lingwood D., Kanekiyo M., Yassine H.M., Ma S.S., Narpala S.R., Prabhakaran M.S., Matus-Nicodemos R.A., Bailer R.T. H5N1 vaccine-elicited memory B cells are genetically constrained by the IGHV locus in the recognition of a neutralizing epitope in the hemagglutinin stem. J. Immunol. 2015;195:602–610. doi: 10.4049/jimmunol.1402835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wibmer C.K., Ayres F., Hermanus T., Madzivhandila M., Kgagudi P., Oosthuysen B., Lambson B.E., de Oliveira T., Vermeulen M., van der Berg K. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. bioRxiv. 2021 doi: 10.1101/2021.01.18.427166. [DOI] [PubMed] [Google Scholar]
- Williams W.B., Liao H.X., Moody M.A., Kepler T.B., Alam S.M., Gao F., Wiehe K., Trama A.M., Jones K., Zhang R. HIV-1 VACCINES. Diversion of HIV-1 vaccine-induced immunity by gp41-microbiota cross-reactive antibodies. Science. 2015;349:aab1253. doi: 10.1126/science.aab1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., Fox J.M., Chen R.E., Earnest J.T., Keeler S.P. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat. Immunol. 2020;21:1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X., Zhou T., Zhu J., Zhang B., Georgiev I., Wang C., Chen X., Longo N.S., Louder M., McKee K., NISC Comparative Sequencing Program Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing. Science. 2011;333:1593–1602. doi: 10.1126/science.1207532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Bangaru S., Huang D., Zhu X., Lee C.D., Turner H.L., Peng L., Yang L., Burton D.R. A natural mutation between SARS-CoV-2 and SARS-CoV determines neutralization by a cross-reactive antibody. PLoS Pathog. 2020;16:e1009089. doi: 10.1371/journal.ppat.1009089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Huang D., Lee C.C.D., Wu N.C., Jackson A.M., Zhu X., Liu H., Peng L., van Gils M.J., Sanders R.W. Structural and functional ramifications of antigenic drift in recent SARS-CoV-2 variants. bioRxiv. 2021 doi: 10.1101/2021.02.16.430500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang R., Gomez Castro M.F., McCune B.T., Zeng Q., Rothlauf P.W., Sonnek N.M., Liu Z., Brulois K.F., Wang X., Greenberg H.B. TMPRSS2 and TMPRSS4 promote SARS-CoV-2 infection of human small intestinal enterocytes. Sci. Immunol. 2020;5:eabc3582. doi: 10.1126/sciimmunol.abc3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Wong L.R., Li K., Verma A.K., Ortiz M.E., Wohlford-Lenane C., Leidinger M.R., Knudson C.M., Meyerholz D.K., McCray P.B., Jr., Perlman S. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589:603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T., Lynch R.M., Chen L., Acharya P., Wu X., Doria-Rose N.A., Joyce M.G., Lingwood D., Soto C., Bailer R.T., NISC Comparative Sequencing Program Structural repertoire of HIV-1-neutralizing antibodies targeting the CD4 supersite in 14 donors. Cell. 2015;161:1280–1292. doi: 10.1016/j.cell.2015.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Case J.B., Binshtein E., Chen R.E., Nkolola J.P., Schäfer A., Reidy J.X., Trivette A., Nargi R.S. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–449. doi: 10.1038/s41586-020-2548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Gilchuk P., Chen R.E., Case J.B., Reidy J.X., Trivette A., Nargi R.S., Sutton R.E., Suryadevara N., Chen E.C. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat. Med. 2020;26:1422–1427. doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost S.J., Dong J., Gilchuk I.M., Gilchuk P., Thornburg N.J., Bangaru S., Kose N., Finn J.A., Bombardi R., Soto C. Canonical features of human antibodies recognizing the influenza hemagglutinin trimer interface. J. Clin. Invest. 2021;131:e146791. doi: 10.1172/JCI146791. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence Read Archive deposition for the public clonotypes identified is deposited at the NCBI: PRJNA511481. Deposition for the paired heavy and light chain plasmablast sequences, that were identified as public clonotypes, following mRNA vaccination is deposited in GenBank with accession numbers MZ555491 through MZ555628. The sequences of COV2-2164 are deposited in GenBank with accession numbers MZ555629 and MZ555630. All other data are available in the main text or the supplementary materials. The computational pipeline for the clustering analysis, the script to create heatmaps, and the PyIR script used to determine sequence characteristics of each antibody are available as listed in the Key resources table. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.