Abstract
Objective
Mathematical models are known to help determine potential intervention strategies by providing an approximate idea of the transmission dynamics of infectious diseases. To develop proper responses, not only are more accurate disease spread models needed, but also those that are easy to use.
Materials and methods
As of July 1, 2020, we selected the 20 countries with the highest numbers of COVID-19 cases in the world. Using the Verhulst–Pearl logistic function formula, we calculated estimates for the total number of cases for each country. We compared these estimates to the actual figures given by the WHO on the same dates. Finally, the formula was tested for longer-term reliability at t = 18 and t = 40 weeks.
Results
The Verhulst–Pearl logistic function formula estimated the actual numbers precisely, with only a 0.5% discrepancy on average for the first month. For all countries in the study and the world at large, the estimates for the 40th week were usually overestimated, although the estimates for some countries were still relatively close to the actual numbers in the forecasting long term. The estimated number for the world in general was about 8 times that actually observed for the long term.
Conclusions
The Verhulst–Pearl equation has the advantage of being very straightforward and applicable in clinical use for predicting the demand on hospitals in the short term of 4–6 weeks, which is usually enough time to reschedule elective procedures and free beds for new waves of the pandemic patients.
Keywords: COVID-19, Epidemic forecasting, Mathematical model, Pandemic, SARS-CoV-2
1. Introduction
The COVID-19 pandemic caused enormous changes to the delivery of healthcare across the world, and the routine practice of clinical physicians has been substantially affected.1, 2, 3, 4, 5, 6 Mathematical models are known to help determine potential intervention strategies by providing an approximate idea of the transmission dynamics of infectious diseases.7, 8, 9, 10 To predict the course of epidemics, various complex mathematical concepts and models have been developed, including the following: the epidemic exponential growth rate (mainly the susceptible-infectious-recovered [SIR] model),11, 12, 13 susceptible-exposed-infectious-recovered (SEIR) model,12 , 14, 15, 16, 17 exponential growth rate and basic reproduction number (parametric approach, nonparametric approach), the autoregressive integrated moving average (ARIMA) model,18 least squares estimation, maximum likelihood estimation, and mechanistic and phenomenological models.19 However, working with these models is the purview of highly specialized professionals. To estimate the pandemic growth rate, an adequate mix of medical and advanced mathematical knowledge is required. To develop proper responses, not only are more accurate disease spread models needed, but also those that are easy to use.13 , 20 Few studies have been published in search for a straightforward mathematical model that can be easily used by clinicians to forecast the approximate rate of COVID-19 spread, and unfortunately, none of these few models published are as simple as expected.18 , 20 , 21 Therefore, it may be useful for clinicians and healthcare managers working daily in hospitals to have access to fundamental formulas that can easily predict the course of the epidemic based on its current state of evolution.
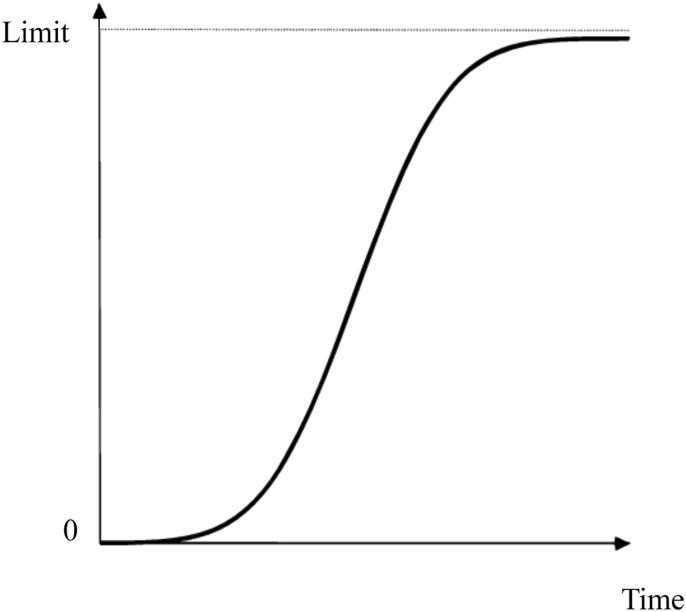
To put it simply, most epidemics are known to grow exponentially in the first stage of an outbreak.22 For this, epidemic curves can be mathematically studied, and the trajectory of the spreading infection can be drawn according to the number of cases in units of time, such as days, weeks, or months. The logistic function curve fits well with the pace of the pandemic over time, both with an early exponential rise and an eventual flattening as the population gains herd immunity.13 , 23, 24, 25 (Fig. 1 ) Pierre François Verhulst (1804–1849), a Belgian mathematician, studied the law of population growth and showed that its curve increases with a convex curvature and then continues to increase towards a limit but with a concave curvature.23 , 25 , 26 Over time, a stable population will reach a saturation level, which is the limit, and the total number of events will not increase higher than this limit.12 Verhulst–Pearl's exponential growth model has formed the basis for other models. Using this formula simply, if the population of a country and the cumulative number of infected cases in two subsequent time units (t0 and t1) in the given country are known, it is possible to estimate the cumulative number of cases at future time points (t2, t3, t4, and so on).
Fig. 1.
The logistic function curve.
We conducted this study to reveal whether Verhulst–Pearl's exponential growth model could be proposed as a simple, reliable formula to predict the cumulative case numbers of the COVID-19 pandemic, and whether it is applicable in neurosurgery for the prediction of the demand on hospitals.
2. Materials and methods
From the situation report dated July 1, 2020, on the World Health Organization's (WHO's) website,27 we chose the 20 countries with the highest numbers of COVID-19 cases in the world ( Table 1 ). We retrieved the latest population figures in millions and the population densities per square meter of these countries from their Wikipedia pages and recorded them (The Wikimedia Foundation, Inc, San Francisco, CA, United States). The motivation behind using Wikipedia as a source for this information was that those pages are updated with the latest data on the countries' populations. We also recorded the world's total population for a comparison. The total number of COVID-19 cases and the total number for these 20 countries and for the world were obtained from the WHO situation report dated July 1, 2010, and recorded as time t = 0.27 Similarly, we found the total numbers of COVID-19 cases from the WHO status report dated July 8, 2020,28 a week later, and recorded them as time t = 1. Then, using the Verhulst–Pearl logistic function formula, we calculated the total case estimates for each country and the world for July 15, July 22, and July 29, 2020 and recorded them as times t = 2, t = 3, and t = 4, respectively. We then compared our estimates with the numbers from the relevant dates of the WHO status reports on July 15, July 22, and July 29, 2020.29, 30, 31 We divided our estimation numbers by actual numbers and examined how much they deviated from the number 1. Finally, the formula was tested for longer-term reliability at t = 18 and t = 40 weeks. The complete data for the actual numbers can be found on the WHO website at https://covid19.who.int/WHO-COVID-19-global-data.csv.32
Table 1.
Basic Information for the calculations.
| Country | Population | Density/km2 | 1 July 2020 Total Case Number t = 0 |
8 July 2020 Total Case Number t = 1 |
b | e-c | |
|---|---|---|---|---|---|---|---|
| 1 | USA | 328,239,523 | 33.6 | 2,573,393 | 2,923,432 | 127 | 0.879 |
| 2 | Brazil | 210,147,125 | 25 | 1,368,195 | 1,623,284 | 152 | 0.842 |
| 3 | Russian Federation | 146,748,590 | 8.4 | 654,405 | 700,792 | 223 | 0.933 |
| 4 | India | 1352642280 | 407 | 585,493 | 742,417 | 2309 | 0.788 |
| 5 | United Kingdom | 67,886,004 | 270.7 | 312,658 | 286,353 | 216 | 1.092 |
| 6 | Peru | 32,824,358 | 23 | 282,365 | 305,703 | 115 | 0.924 |
| 7 | Chile | 17,574,003 | 24 | 279,393 | 301,019 | 62 | 0.927 |
| 8 | Spain | 47,431,256 | 94 | 249,271 | 252,130 | 189 | 0.989 |
| 9 | Italy | 60,317,116 | 201.3 | 240,578 | 241,956 | 250 | 0.994 |
| 10 | Iran | 83,183,741 | 48 | 227,662 | 245,688 | 364 | 0.926 |
| 11 | Mexico | 128,649,565 | 61 | 220,657 | 261,750 | 582 | 0.842 |
| 12 | Pakistan | 212,228,286 | 244.4 | 213,470 | 237,489 | 993 | 0.898 |
| 13 | Turkey | 83,154,997 | 105 | 199,906 | 207,897 | 415 | 0.961 |
| 14 | Germany | 83,166,711 | 232 | 194,725 | 197,341 | 426 | 0.986 |
| 15 | Saudi Arabia | 34,218,169 | 15 | 190,823 | 217,108 | 178 | 0.878 |
| 16 | France | 67,081,000 | 104 | 157,194 | 159,909 | 426 | 0.983 |
| 17 | South Africa | 59,622,350 | 42.4 | 151,209 | 215,855 | 393 | 0.699 |
| 18 | Bangladesh | 161,376,708 | 1,106 | 145,483 | 168,645 | 1108 | 0.862 |
| 19 | Canada | 37,971,020 | 3.92 | 103,918 | 105,935 | 354 | 0.981 |
| 20 | Qatar | 2,795,484 | 176 | 96,088 | 100,945 | 28 | 0.953 |
| World | 7,800,000,000 | 14.7 | 10,357,662 | 11,669,259 | 752 | 0.887 |
2.1. Mathematical model
The single, simple formula we used in this study is an application of the Verhulst–Pearl logistic function:26
(* world population, if calculations are made for the whole world).where N represents the cumulative number of infected cases at time t, e represents a mathematical constant approximately equal to 2.71828, c is the constant of integration, and b is the exponential function base (the rate of change). The result is defined as a logistic curve, which is S-shaped (Fig. 1).
We can explain this formula using an example. Let the cumulative number of infected cases at time t0 be 600 and the size of population be 90,000. If the cumulative number of infected cases reaches 1,800 one week later (t1), the cumulative number of infected cases at t4 (forth week) can be estimated as follows:
When t = 0 then thus b=149
When t = 1 hence e -c =
When t = 4 ≈ 32,000
This means that after four weeks, the total number infected people will reach approximately 32,000.
In the classical population growth scenario, the Verhulst-Pearl logistic equation comprises three assumptions.33 First, all individuals are equivalent, i.e., the addition of every new individual reduces the actual rate of increase by the same fraction at every density. Second, reproductive rate and carrying capacity are unchangeable constants. Third, no time lag exists in the response of the actual rate of increase per individual to changes in population density. In the pandemic scenario, herd immunity is an assumption, i.e., it is assumed that each individual can be infected only once, then becomes immune afterwards. It also is assumed that population size does not change and that no migrations, deaths, or births occur. In addition, it is assumed that no change in the reproduction rate occurs regardless of virus mutations or protective measures taken.
2.2. Data collection and statistical analysis
The Statistical Package for Social Sciences (SPSS v.20 IBM, Armonk, New York) was utilized to record and analyze the data. Variables included patient demographics: cumulative case numbers, ratio of the estimated values/actual numbers for cumulative cases, and country populations. Descriptive analyses were done to determine frequencies, mean, mode, maximum and minimum values, standard deviation, and standard error of the mean.
3. Results
On July 1, 2020, the countries that had the highest numbers of cumulative COVID-19 cases were the following, in descending order: the USA, Brazil, the Russian Federation, India, the United Kingdom, Peru, Chile, Spain, Italy, Iran, Mexico, Pakistan, Turkey, Germany, Saudi Arabia, France, South Africa, Bangladesh, Canada, and Qatar (cumulative case numbers: maximum = 2,573,393, minimum = 96,088, mean = 422,344) (Table 1). The actual cumulative case numbers on July 1 and July 8, which were the bases of the estimates of the following weeks, are shown in Table 1.
3.1. Short-term estimates
When the estimates for July 15, 2020 (t = 2) were calculated using the data from July 1 (t = 0) and July 8 (t = 1) and divided by the actual numbers of July 15, 2020, the mean was 1.005 (range: 0.899–1.039) (Table 2 ). This shows that the formula estimated the actual numbers with only a 0.5% discrepancy on average. We noticed that the total number of cases in the United Kingdom suddenly decreased from 313,487 on July 2, 2020 to 283,761 on July 3, 2020. This change occurred because the United Kingdom revised its historical data, leading to a negative number of new cases and an overall decrease in cases for the country. When we removed the United Kingdom from the list, the mean ratio of the estimated number/actual number was 1.01 for July 15, 2020. Then the result showed, on average, only a 1% discrepancy between the estimated and observed numbers. The estimates for two countries, Italy and Peru, deviated from the actual numbers by less than 0.1%. The estimates for July 15 for three countries, Pakistan, Saudi Arabia, and South Africa, were less accurate. However, the discrepancies in the estimates for those three countries were still minimal and were equal to or less than 3.9%.
Table 2.
The actual-estimate cumulative case numbers and their ratios for July 15, 2020 in the 20 countries.
| Country | 15 July 2020 Total Case Number t = 2 |
15 July 2020 Estimated Total Case Number by the Formula |
15 July 2020 Estimated/Actual Ratio |
|
|---|---|---|---|---|
| 1 | USA | 3344783 | 3320581 | 0,992 |
| 2 | Brazil | 1884967 | 1925835 | 1,021 |
| 3 | Russian Federation | 746369 | 750862 | 1,006 |
| 4 | India | 936181 | 942804 | 1,007 |
| 5 | The United Kingdom | 291377 | 262108 | 0,899 |
| 6 | Peru | 330123 | 330258 | 1 |
| 7 | Chile | 319493 | 324303 | 1,015 |
| 8 | Spain | 256619 | 255006 | 0,993 |
| 9 | Italy | 243344 | 243410 | 1 |
| 10 | Iran | 262173 | 265085 | 1,011 |
| 11 | Mexico | 304435 | 310522 | 1,019 |
| 12 | Pakistan | 255769 | 264162 | 1,032 |
| 13 | Turkey | 214993 | 216549 | 1,007 |
| 14 | Germany | 199726 | 199963 | 1,001 |
| 15 | Saudi Arabia | 237803 | 247241 | 1,039 |
| 16 | France | 162390 | 162660 | 1,001 |
| 17 | South Africa | 298292 | 308125 | 1,032 |
| 18 | Bangladesh | 190057 | 195465 | 1,028 |
| 19 | Canada | 108155 | 105945 | 0,979 |
| 20 | Qatar | 104533 | 105889 | 1,012 |
When the same calculations were performed for July 22, 2020 (t = 3), the mean ratio of the estimated cumulative case number divided by the actual cumulative case number was found to be 1.017 (1.065 excluding the United Kingdom), and the range was 0.813–1.154 (Table 3 ). This means that the formula estimated the actual numbers with only a 1.7% error on average. The mean value came closer to 1 when the UK was included, but this was a paradox and resulted from the buffering effect of the UK on excessively diverging values in the opposite direction from those of other countries, as the UK revised and reduced the number of historical cases. The estimates for three countries, Italy, Peru, and Germany, deviated from the actual cumulative case numbers by less than 0.1%, which showed greater certainty. The formula estimated the case numbers of five countries (the United Kingdom, Pakistan, Saudi Arabia, Bangladesh, and Brazil) for July 22 less accurately than it did for other countries.
Table 3.
The actual-estimate cumulative case numbers and their ratios for July 22, 2020 in the 20 countries.
| Country | 22 July 2020 Total Case Number t = 3 |
22 July 2020 Estimated Total Case Number by the Formula |
22 July 2020 Estimated/Actual Ratio |
|
|---|---|---|---|---|
| 1 | USA | 3805524 | 3777209 | 0,992 |
| 2 | Brazil | 2118646 | 2284207 | 1,078 |
| 3 | Russian Federation | 789190 | 804984 | 1,020 |
| 4 | India | 1192915 | 1196181 | 1,002 |
| 5 | The United Kingdom | 295821 | 240730 | 0,813 |
| 6 | Peru | 357681 | 357953 | 1 |
| 7 | Chile | 334683 | 349383 | 1,043 |
| 8 | Spain | 266194 | 257778 | 0,968 |
| 9 | Italy | 244752 | 245191 | 1.001 |
| 10 | Iran | 278827 | 280642 | 1,006 |
| 11 | Mexico | 349396 | 334676 | 0,957 |
| 12 | Pakistan | 267428 | 294761 | 1,102 |
| 13 | Turkey | 221500 | 225169 | 1,016 |
| 14 | Germany | 202799 | 203192 | 1,001 |
| 15 | Saudi Arabia | 255825 | 281399 | 1,099 |
| 16 | France | 166511 | 165509 | 0,993 |
| 17 | South Africa | 381798 | 440667 | 1,154 |
| 18 | Bangladesh | 210510 | 227099 | 1,078 |
| 19 | Canada | 111124 | 110092 | 0,990 |
| 20 | Qatar | 104533 | 110931 | 1,032 |
When the same calculations were performed for July 29, 2020 (t = 4), the mean ratio of the estimated cumulative case number/actual cumulative case number was found to be 1.068 (range: 732–1.367) (Table 4 ).
Table 4.
The actual-estimate cumulative case numbers and their ratios for July 29, 2020 in the 20 countries.
| Country | 29 July 2020 Total case Number t =4 |
29 July 2020 Estimated Total case Number by the Formula |
29 July 2020 Estimated/Actual Ratio |
|
|---|---|---|---|---|
| 1 | USA | 4263531 | 4288470 | 1,005 |
| 2 | Brazil | 2442375 | 2706337 | 1,108 |
| 3 | Russian Federation | 828990 | 862414 | 1,040 |
| 4 | India | 1531669 | 1517640 | 0,990 |
| 5 | The United Kingdom | 300696 | 220308 | 0,732 |
| 6 | Peru | 389717 | 387079 | 0,993 |
| 7 | Chile | 349800 | 376316 | 1,075 |
| 8 | Spain | 280610 | 260611 | 0,928 |
| 9 | Italy | 246488 | 246494 | 1,000 |
| 10 | Iran | 296273 | 328064 | 1,107 |
| 11 | Mexico | 395489 | 438329 | 1,108 |
| 12 | Pakistan | 276288 | 328171 | 1,187 |
| 13 | Turkey | 227982 | 234305 | 1,027 |
| 14 | Germany | 206926 | 206062 | 0,995 |
| 15 | Saudi Arabia | 270831 | 320095 | 1,181 |
| 16 | France | 172148 | 168545 | 0,979 |
| 17 | South Africa | 459761 | 628927 | 1,367 |
| 18 | Bangladesh | 229185 | 263386 | 1,149 |
| 19 | Canada | 114597 | 112340 | 0,980 |
| 20 | Qatar | 109880 | 116478 | 1,060 |
The estimation for the total world numbers was nearly exact for July 15, and the estimated numbers differed very little in the short term in general. The estimated/actual ratios were 0.08% for cumulative cases (Table 5 ).
Table 5.
The actual and estimate cumulative case and death numbers in the World in general for short-term.
| World Population = 7,800,000,000 Density = 14.7 | |||||||
|---|---|---|---|---|---|---|---|
| Actual Total Case Number |
Actual Total Death Number |
Estimated Total Case Number |
Estimated Total Death Number |
Ratio (Estimated /Actual) (Case) |
Ratio (Estimated/Actual) (Death) |
||
| 1 July 2020 | (t = 0) | 10357662 | 508055 | – | – | – | – |
| 8 July 2020 | (t = 1) | 11669259 | 539906 | – | – | – | – |
| 15 July 2020 | (t = 2) | 13150645 | 574464 | 13162335 | 573740 | 1.0008 | 0.9987 |
| 22 July 2020 | (t = 3) | 14765256 | 612054 | 14854313 | 609756 | 1.0060 | 0.9962 |
| 29 July 2020 | (t = 4) | 16558289 | 656093 | 16738197 | 648001 | 1.0100 | 0,9870 |
3.2. Longer-term estimates at t = 18 weeks
Four months after the first estimation week t = 2, we tested the formula's precision for the worldwide cumulative case number estimation for t = 18 weeks (November 5, 2020) and found a number 80% greater than the actual number (the estimate is higher than the actual number). This may suggest that the estimates for the long term may be misleading. However, it may also suggest that fewer cases have been diagnosed worldwide than estimated even though the second wave of the pandemic has continued in Europe since the summer months. The cumulative case number estimates for t = 18 weeks were more accurate for Turkey, Peru, and the Russian Federation (5%, 25%, and 30% discrepancy, respectively) than the total case estimates for all countries. It is worth noting that Turkey revised the number of cases in November 2020. Turkey did not report the number of asymptomatic cases after July 29, 2020 until revision. As of November 25, 2020, those who were positive for the polymerase chain reaction (PCR) test were added to the number of patients even though they were asymptomatic. Asymptomatic cases from July 29 to December 10 were also added to the number of patients and updated in Turkey.
3.3. Longer-term estimates at t = 40 weeks
For all countries in the study and the world at large, the estimates for the 40th week were not as accurate as in the first month (Table 6 ). The estimated number for the world in general was about 8 times that actually observed. In twelve countries, as in the world in general, the estimated numbers were higher than the observed. However, in some countries this difference was relatively close to the respective estimates (Russian Federation, Iran, Qatar, and Peru). In contrast, the difference was much pronounced in some countries (India, Bangladesh, Saudi Arabia, South Africa, and Mexico). These countries had significantly less case observations than anticipated at the longer term. Eight countries had many more case observations than anticipated. Of these, Italy, France, and the UK were the most prominent. (However, it is necessary to remember that the UK revised the case numbers at the time we obtained the reference values). In Fig. 2 , the curves of the estimated and observed cumulative case numbers in selected 6 countries in short- and long-term are demonstrated as an overview. The complete data of this chart are shown in Table 7 .
Table 6.
The actual-estimate cumulative case numbers and their ratios for April 07, 2021 in the 20 countries.
| Country | 07 April 2021 Total case Number t = 40 |
07 April 2021 Estimated Total case Number by the Formula |
07 April 2021 Estimated/Actual Ratio |
|
|---|---|---|---|---|
| 1 | USA | 30475874 | 190011326 | 6,234 |
| 2 | Brazil | 13013601 | 182736630 | 14,041 |
| 3 | Russian Federation | 4606162 | 9827278 | 2,133 |
| 4 | India | 12801785 | 1158404820 | 90,48 |
| 5 | The United Kingdom | 4358882 | 9303 | 0,002 |
| 6 | Peru | 1590209 | 5591373 | 3,516 |
| 7 | Chile | 1037780 | 4415578 | 4,254 |
| 8 | Spain | 3325600 | 387193 | 0,116 |
| 9 | Italy | 3686707 | 305745 | 0,082 |
| 10 | Iran | 1963394 | 4673243 | 2,380 |
| 11 | Mexico | 2251705 | 80911676 | 35,933 |
| 12 | Pakistan | 696184 | 14748317 | 21,184 |
| 13 | Turkey | 3579185 | 972292 | 0,271 |
| 14 | Germany | 2910445 | 341855 | 0,117 |
| 15 | Saudi Arabia | 394169 | 17369642 | 44,066 |
| 16 | France | 4807569 | 311714 | 0,064 |
| 17 | South Africa | 1552853 | 59622350 | 38,395 |
| 18 | Bangladesh | 651652 | 41293937 | 63,368 |
| 19 | Canada | 1014373 | 223385 | 0,220 |
| 20 | Qatar | 185261 | 551377 | 2,976 |
Fig. 2.
Estimated and observed cumulative case numbers in selected 6 countries in short- and long-term. In the short-term of about 2–3 months, estimated numbers were close to the observed numbers in all of the countries. In the longer-term (up to ten months), an overestimation was observed in some countries such as India, Peru, Russian federation and USA, and an underestimation was observed in some countries such as Italy and Turkey. However, in some countries such as Russian Federation and Peru, error in the prediction was relatively low even in the longer-term. (The countries in the chart were listed in alphabetical order.)
Table 7.
Forty-week estimated and observed cumulative case numbers of selected (as example) 6 countries, of which diagrammatic representations are shown in Fig. 2.
| Week |
Date |
Countrya |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| India |
Italy |
Peru |
Russian F. |
Turkey |
USA |
||||||||
| Estimate | Observed | Estimate | Observed | Estimate | Observed | Estimate | Observed | Estimate | Observed | Estimate | Observed | ||
| 0 | 1-Jul-20 | – | 585493 | – | 240578 | – | 282365 | – | 654405 | – | 199906 | – | 2573393 |
| 1 | 8-Jul-20 | – | 742417 | – | 241956 | – | 305703 | – | 700792 | – | 207897 | – | 2923432 |
| 2 | 15-Jul-20 | 942804 | 936181 | 243410 | 243344 | 330258 | 330123 | 750862 | 746369 | 216549 | 214993 | 3320581 | 3344783 |
| 3 | 22-Jul-20 | 1196181 | 1192915 | 245191 | 244752 | 357953 | 357681 | 804984 | 789190 | 225169 | 221500 | 3777209 | 3805524 |
| 4 | 29-Jul-20 | 1517640 | 1531669 | 246494 | 246488 | 387079 | 389717 | 862414 | 828990 | 234305 | 234712 | 4288470 | 4263531 |
| 5 | 5-Aug-20 | 1925348 | 1908254 | 247914 | 248419 | 418376 | 433100 | 923954 | 866627 | 243752 | 288774 | 4869597 | 4678610 |
| 6 | 12-Aug-20 | 2442400 | 2329638 | 249404 | 251237 | 452314 | 483133 | 989857 | 902701 | 253614 | 344130 | 5528638 | 5039709 |
| 7 | 19-Aug-20 | 3097987 | 2767273 | 250903 | 254636 | 488963 | 541493 | 1060426 | 937321 | 263873 | 399865 | 6275141 | 5393138 |
| 8 | 26-Aug-20 | 3929035 | 3234474 | 252411 | 261174 | 528533 | 600438 | 1135998 | 970865 | 274517 | 456364 | 7120217 | 5682811 |
| 9 | 2-Sep-20 | 4982192 | 3769523 | 253929 | 270189 | 571249 | 652037 | 1216888 | 1005000 | 285650 | 513985 | 8076245 | 5968380 |
| 10 | 9-Sep-20 | 6316319 | 4370128 | 255455 | 280153 | 617351 | 691575 | 1303498 | 1041007 | 297201 | 572660 | 9156977 | 6248989 |
| 11 | 16-Sep-20 | 8005576 | 5020359 | 256990 | 289990 | 667097 | 733860 | 1396214 | 1079519 | 309218 | 631120 | 10377641 | 6496246 |
| 12 | 23-Sep-20 | 10143210 | 5646010 | 258535 | 300897 | 720762 | 772896 | 1495456 | 1122241 | 321718 | 689912 | 11755031 | 6779609 |
| 13 | 30-Sep-20 | 12846178 | 6225763 | 260089 | 313011 | 778639 | 808714 | 1601675 | 1176286 | 334722 | 747992 | 13307582 | 7077015 |
| 15 | 7-Oct-20 | 16260709 | 6757131 | 261652 | 330263 | 841042 | 829999 | 1715349 | 1248619 | 348249 | 805387 | 15055434 | 7380326 |
| 15 | 14-Oct-20 | 20568894 | 7239389 | 263224 | 365467 | 908305 | 851171 | 1836988 | 1340409 | 362320 | 863719 | 17020445 | 7728436 |
| 16 | 21-Oct-20 | 25996305 | 7651107 | 264806 | 434449 | 980781 | 870876 | 1967137 | 1447335 | 376957 | 923463 | 19226183 | 8124633 |
| 17 | 28-Oct-20 | 32820534 | 7990322 | 266398 | 564778 | 1058850 | 890574 | 2106372 | 1563976 | 392183 | 985368 | 21697840 | 8611256 |
| 18 | 4-Nov-20 | 41380298 | 8313876 | 267998 | 759829 | 1142909 | 906545 | 2255308 | 1693454 | 408021 | 1048388 | 24462092 | 9193765 |
| 19 | 11-Nov-20 | 52084394 | 8591730 | 269609 | 995463 | 1233382 | 923527 | 2414600 | 1836960 | 424495 | 1112740 | 27546857 | 9990620 |
| 20 | 18-Nov-20 | 65419243 | 8912907 | 271229 | 1238072 | 1330716 | 938268 | 2584942 | 1991998 | 441631 | 1181903 | 30980950 | 11085184 |
| 21 | 25-Nov-20 | 81953001 | 9222216 | 272859 | 1455022 | 1435383 | 950557 | 2767070 | 2162503 | 459454 | 1268523 | 34793613 | 12276834 |
| 22 | 2-Dec-20 | 102333224 | 9499413 | 274498 | 1620901 | 1547877 | 963605 | 2961767 | 2347401 | 477993 | 1461758 | 39013899 | 13385755 |
| 23 | 9-Dec-20 | 127274001 | 9735850 | 276148 | 1757394 | 1668719 | 975116 | 3169861 | 2541199 | 497275 | 1686431 | 43669905 | 14755996 |
| 24 | 16-Dec-20 | 157527526 | 9932547 | 277807 | 1870576 | 1798453 | 986130 | 3392231 | 2734454 | 517331 | 1898447 | 48787838 | 16245376 |
| 25 | 23-Dec-20 | 193834864 | 10099066 | 279476 | 1977370 | 1937646 | 998475 | 3629806 | 2933753 | 538189 | 2062960 | 54390930 | 17895109 |
| 26 | 30-Dec-20 | 236851968 | 10244852 | 281155 | 2067487 | 2086887 | 1008908 | 3883570 | 3131550 | 559884 | 2178580 | 60498205 | 19147627 |
| 27 | 6-Jan-21 | 287050889 | 10374932 | 282844 | 2181619 | 2246787 | 1021058 | 4154559 | 3308601 | 582446 | 2270101 | 67123149 | 20643544 |
| 28 | 13-Jan-21 | 344603224 | 10495147 | 284514 | 2303263 | 2417974 | 1037350 | 4443869 | 3471053 | 605911 | 2346285 | 74272319 | 22428591 |
| 29 | 20-Jan-21 | 409262743 | 10595660 | 286253 | 2400598 | 2601095 | 1068802 | 4752654 | 3633952 | 630314 | 2399781 | 81943976 | 23884299 |
| 30 | 27-Jan-21 | 480274236 | 10689527 | 287973 | 2485956 | 2796809 | 1102795 | 5082129 | 3774672 | 655693 | 2442350 | 90126837 | 25050308 |
| 31 | 3-Feb-21 | 556340752 | 10777284 | 289703 | 2570608 | 3005784 | 1142716 | 5433571 | 3901204 | 682084 | 2492977 | 98799047 | 26055512 |
| 32 | 10-Feb-21 | 635676017 | 10858371 | 291443 | 2655319 | 3228695 | 1191221 | 5808319 | 4012710 | 709529 | 2548195 | 107927488 | 26832826 |
| 33 | 17-Feb-21 | 716150026 | 10937320 | 293194 | 2739591 | 3466216 | 1238501 | 6207778 | 4112151 | 738068 | 2602034 | 117467536 | 27433718 |
| 34 | 24-Feb-21 | 795508001 | 11030176 | 294955 | 2832162 | 3719014 | 1286757 | 6633416 | 4200902 | 767744 | 2655633 | 127363358 | 27883560 |
| 35 | 3-Mar-21 | 871617473 | 11139516 | 296726 | 2955434 | 3987745 | 1332939 | 7086767 | 4278750 | 798602 | 2723316 | 137548796 | 28345585 |
| 36 | 10-Mar-21 | 942687791 | 11262707 | 298509 | 3101093 | 4273043 | 1374467 | 7569427 | 4351553 | 830088 | 2807387 | 147948852 | 28827195 |
| 37 | 17-Mar-21 | 1007416606 | 11438734 | 300301 | 3258770 | 4575514 | 1418974 | 8083057 | 4418436 | 864049 | 2911642 | 158481728 | 29208890 |
| 38 | 24-Mar-21 | 1065043237 | 11734058 | 302105 | 3419616 | 4895724 | 1472790 | 8629380 | 4483471 | 898735 | 3061520 | 169061308 | 29594849 |
| 39 | 31-Mar-21 | 1115316668 | 12149335 | 303919 | 3561012 | 5234191 | 1533121 | 9210175 | 4545095 | 934799 | 3277880 | 179599942 | 30033063 |
| 40 | 7-Apr-21 | 1158404820 | 12801785 | 305745 | 3686707 | 5591373 | 1590209 | 9827278 | 4606162 | 972292 | 3579185 | 190011326 | 30475874 |
Countries were listed alphabetically.
In Fig. 3 , when examining the curves of the worldwide observed numbers and estimated cumulative case numbers during the 40 weeks between July 2020 and April 2021 it can be easily seen that the formula quite accurately predicted the observed numbers for the first four months (from July to October 2020). However, the estimates for November and December 2020 were slightly above the observed figures. This overestimation has become increasingly apparent after January 2021. This may not be surprising, as this period coincides with the start and gradual acceleration of vaccination around the world.
Fig. 3.
Global estimated and observed cumulative case numbers.
4. Discussion
Our model using the simple Verhulst–Pearl logistic function equation proves to be in perfect agreement with the real data for forecasting the pandemic progress in the short but not long term, probably because vaccination had begun or perhaps because of the assumptions of the model which may not fit well with the COVID-19 pandemic. Several other authors also reported using the Verhulst–Pearl logistic function in predicting cumulative case numbers from the COVID-19 pandemic.13 , 34, 35, 36 Mahanty et al.36 compared two nonlinear growth models (Verhulst and Gompertz) and the susceptible-infectious-recovered model (SIR) to predict active COVID-19 cases in India, Pakistan, Italy, Germany, Brazil, and Myanmar, and found that the Verhulst model's fitting effect is better than that of the Gompertz and SIR models. Niazkar et al.37 used the three mathematical prediction models (a recursive-based method, the Boltzmann function-based model, and Beesham's prediction model) to forecast COVID-19 case and death numbers in Iran and Turkey. They concluded that these models failed to predict the first 10–20 days of data, although the recursive-based method was more effective than the others. Postnikov13 reported that the SIR model, being sequentially reduced to the Verhulst equation, could provide an accurate description of the COVID-19 epidemic for a period of about three months in various countries, including Italy, the U.S., China, and Russia.
Whether static, deterministic, or dynamic stochastic models are used, studies generally covered one- to three-month periods. Therefore, we were unable to make comparisons with other studies in the literature in the longer term, although our study covers a 10-month period. However, for the short term, the model we used provided estimation results that were similar to those of the dynamic stochastic complex models. Three main parameters limit the ability to predict the long-term growth of the COVID-19 pandemic.38 First, the extent of protective immunity is still unclear. Second, the extent of transmission and immunity among asymptomatic or minimally symptomatic individuals, as it is more common in children, is unknown. Third, it is nearly impossible to measure and model contact rates between susceptible and contagious individuals in various lockdown or reopening scenarios. It is possible that the Verhulst-Pearl model does not work in the longer term just because of its inherent assumptions, which may not fit with the COVID-19 pandemic. However, short-term forecasts of a few weeks based on the logistics growth model can give very reliable results. Due to changes in the external environment, such as modifications in government control policies and new treatment methods, the mathematical models will not necessarily be accurate for long-term estimates, and therefore, the model parameters should be updated to provide reliable long-term forecasts.22 This can be explained by the S-shape of the logistic function curve, which is convex in the early stages of a pandemic unlike the concave curvature seen in the later periods. In other words, when the curve approaches the limit (the saturation of transmission), the change over time will be less. Some published research suggests that mathematical models provide more accurate results at the early exponential growth stage of outbreaks, perhaps because any precautions have not yet been taken.13 Since we have been interested in the long-term course of the pandemic leading to a potential herd immunity rather than the very short-term course, we used “week” instead of “day” for the time t-value in the formula, although we were aware that mathematical models were more accurate for short term daily forecasts and weekly forecasts are more prone to error as Arora et al. noted.39
To get an idea of whether the simple model we used in this study is as reliable as the complex models, we compared it with two published studies, one using the Susceptible, Infected and Recovered (SIR) model40 and the other using recurrent neural networks, and long short-term memory (LSTM) based model.39 In the first of these studies, using the SIR model with correction factor, Malavika et al.40 estimated the cumulative numbers of cases in India as 58912, 81709 and 102974 for 8, 15 and 22 May 2020, respectively. The cumulative numbers of cases reported by WHO for India on the same dates are 56342, 81970 and 118447, respectively, and when we compare them with their respective estimates, we get 1.045, 0.996, and 0.869, respectively. In the other compared study, Arora et al.39 conducted a research on predicting the number of new coronavirus positive cases in India for one day to one week beforehand, using recurrent neural networks and long short-term memory based model. They reported that their model was quite accurate for 1–3 day forecasts, but the error increased for weekly forecasts. The prediction/actual ratios for 7 and 14 May 2020 in their study were 1.005 and 1.002 respectively. In our study, the prediction/actual ratios we found for India on 15, 22 and 29 July are 1,007 (936181/942804), 1,002 (1192915/1196181), and 0,990 (1531669/1517640) respectively. Thus, our forecasting seems to be as reliable as that of Malakavi et al. and Arora et al. Both of these studies were conducted in the pre-vaccination period covering a 2 to 3-week period using daily initial references. In contrast, our study provides estimates for a longer period of 40 weeks, including the vaccination period, using weekly rather than daily initial references for practical purposes.
Appropriately measuring the transmission dynamics of infectious diseases is a function of the calendar period.41 The most commonly used determinant of the transmission potential is the basic reproduction number, which is defined as the expected number of secondary cases arising from a typical primary case throughout its entire course of infection in a fully susceptible population.42, 43, 44 One of the methods to estimate the reproduction number is to use the growth rate of the cumulative incidence of cases during the exponential growth phase of the pandemic.44 As can be expected, an exponential growth model estimates parameters more precisely during a pandemic's exponential growth phase.45
All mathematical models are a simplifications of reality.46 However, when these simplifications have little effect on the characteristics of the epidemic of interest, it can lead to a satisfactory result. Therefore, the Verhulst–Pearl equation is valid and useful in solving a simple optimal growth problem.25 , 26 , 47 Of course, it is not flawless; the Verhulst model is a deterministic model (not a process wherein the random variable exists) and does not include any stochastic (randomly determined) components. Stochastic effects make the invasion threshold challenging to discern.48 For example, the number of individuals susceptible to infection and the reproduction number decreases as the outbreak progresses.49 The fluctuations on the level of the limit (in other words, endemic disease die-outs or the population saturation point or carrying capacity, similar to that of the increase of a country's population, is not unlimited in terms of herd immunity) cannot be explained by the Verhulst–Pearl model.50 However, while adding more complexity to a mathematical model increases the realism, it often makes analysis difficult for an ordinary clinician, and more parameters increase the uncertainty.
Using the mathematical formula in this study, we can predict when the pandemic will end around the world. For herd immunity in the community, typically 60–90% of the population must be immunized via infection or vaccination.51 , 52 This percentage is called the herd immunity threshold. The basic reproduction number (R0) is the most important parameter in calculating the herd immunity threshold and refers to the average number of infected people caused by a single infectious person.53 , 54 Although it is not yet known what this rate is for COVID-19, it is estimated to be between 50 and 83%; deducted from that, the R0 value has been reported to be between 2 and 6 in COVID-19.51 , 52 , 54 , 55 This means that when 50–83% of the population becomes immune to the COVID-19 disease, herd immunity will be theoretically achieved (calculated from the formula: herd immunity threshold R = 1- 1/R0).53 , 54 , 56 Hence, assuming there are no reinfections in COVID-19 disease and a lifelong immunity is obtained, and the R0 value is an average of 4 (i.e., 75% of the population must be infected for herd immunity), we found that 75% (approximately 5.8 billion) of the world population will be infected and immunized 64 weeks (1.10.2021) after t0 time (1.7.2020) in our study, and thus, herd immunity will be reached. This finding supports claims that herd immunity against COVID-19 cannot be achieved without unacceptably high case fatality rates, even if we assume that the death rate is 2%.52 If the immunity gained by ongoing vaccination is considered, it may be possible to achieve herd immunity earlier. These estimates are for the whole world and, because there is heterogeneity in transmission, much better scenarios can be expected for an individual country based on the individual transmission dynamics including their specific R0 value and mortality rate, which are themselves dependent on many other factors, including age, genetic factors, socioeconomic factors, healthcare system, behavioral factors, use of personal protective equipment, and the spatial distribution of people.53
As herd immunity and a constant force of infection are assumptions in the Verhulst-Pearl exponential growth model we used, this static model has some weaknesses in forecasting the COVID-19 pandemic in the longer term. With the COVID-19 pandemic, the herd immunity threshold seems highly unlikely due to factors such as the emergence of new variants, vaccine hesitancy, limited use of vaccines for children, and uncertainty about the extent to which vaccinated people are infected and spread the virus.57 In the model used in our study, it was assumed that each individual could have only one COVID-19 infection, regardless of age, gender, or any other variable. In reality, we now know that it is possible to become infected more than once with COVID-19 and even that the virus might have a potential to reactivate after a latent period,58 but we also know that this is not a very common situation, at least not during our study period. Therefore, we think that the herd immunity assumption may not have affected our study's overall results too much.
In contrast to the Verhulst-Pearl exponential growth model's static nature, dynamic models simulate changes in infectious strength and the infectious pathogen's dynamics.57 , 59 Dynamic models are highly sophisticated and complex models, but they need detailed knowledge of transmission routes and infectiousness. Therefore, these models' results still might contain much uncertainty. However, the Verhulst-Pearl exponential growth model is deterministic, i.e., if the state of the system at a certain point in time is known, all future states can be determined by solving the relevant model.59 Thus, when a rapid investigation is required, analysis using the Verhulst-Pearl exponential growth model, without taking potential stochastic effects into account, may be an option that provides initial results and an idea of the general sequence of events.57 , 60 Therefore, large populations in stable environments often are modelled with deterministic models, while stochastic models are more useful in demonstrating the system's inherent variability for small population sizes.59 , 60 Thinking in deterministic terms is a way of generating concepts and trying to determine the most probable course of events, even if it excludes stochastic dynamics from the model.60
Where does our investigation leave us in terms of the allocation of medical resources in general and neurosurgical planning in particular? The COVID-19 pandemic prompted the application of new priority guidelines for patients requiring neurosurgery interventions. As a result, most elective surgeries have been delayed. Furthermore, patients who need surgery have been avoiding admission to hospitals or have been delaying their treatment as much as possible because they are afraid of being infected by SARS-CoV-2.1, 2, 3 , 24 , 61 , 62
A European survey from the EANS Ethico-Legal Committee highlighted how the COVID-19 pandemic led to a rationing of neurosurgical care in 80% of the responding countries.3 While the study demonstrated a correlation between the resources available before the pandemic and the ability to uphold neurosurgical services, it also highlighted the collateral damage that prioritization and rationing may have on the population served by the neurosurgical centers participating in the survey. When forced to ration resources due to a pandemic event, an accurate estimation of its evolution would be extremely useful. COVID-19 is expected to evolve into multiple waves, and forecasts on bed availability might mean the difference between being offered surgical or conservative treatment for individual neurosurgical patients. Neurosurgery is under a higher economic burden than many other medical branches in terms of both the cost of patient treatment and the training of residents and medical students interested in neurosurgery63, 64, 65, 66, 67, 68, 69, 70, 71, 72, 73 The fair and effective use of resources has become even more important during the pandemic, which seems to be lasting longer than initially thought. Therefore, it is imperative that neurosurgery clinics and neurosurgical societies organize longer-term work patterns and training programs, including professional congresses.
As well as the direct effects of the epidemic, attempts to control it have profound economic consequences.55 , 70 , 71 This continuing situation, the length of which is unknown, has caused many government to make economic decisions. Economic decisions have a tremendous influence on the character of life in a society.74 Therefore, a reliable estimation of the course of the pandemic will help physicians design more effective plans and arrangements for their patients and the training of residents.
4.1. Limitations
Despite numerous speculations about the epidemiological course of the pandemic, identifying the correct prediction is difficult. We made this study based on figures we obtained from the WHO's sources. However, the WHO warned that the completeness of the indicators varies by countries and regions, and that the decrease in cases shown over time should be interpreted with the trend in country participation, that delays in reporting may be observed, and that the number of cases depends on detection and testing strategies that vary between countries and over time. Therefore, due to variable information collection and reporting, it is more appropriate to analyze each country/region independently.
The benefit of this algorithm, although limited to a short time horizon, is its relative simplicity. Essentially, methods for modeling viral disease dynamics are often more complex, including intrahost, interhost, and environmental factors and involving a large number of stochastic processes. The simple design of the formula we used has not been validated for long time periods. It is just a way of projecting current trends into the near future. However, although this mathematical approach is not complex, its application still requires a basic knowledge of mathematics.
5. Conclusions
The Verhulst–Pearl equation is a valid and useful tool to solve a simple optimal growth problem. Unlike complex mathematical models that increase accuracy, this equation makes the analysis easy for an ordinary clinician. When forced to ration resources due to a pandemic event, an accurate estimation of its evolution would be extremely useful. This model has the advantage of being very simple and applicable in neurosurgery for predicting the demand on hospitals in the short term of 4–6 weeks, which is usually enough time to reschedule elective procedures and free beds for new waves of the pandemic patients. However, the reliability of the formula in long-term estimates is uncertain.
Conflict-of-interest disclosure
The authors declare no competing financial interests and no sources of funding and support, including any for equipment and medications.
Ethical issues
The ethical issues for this study involving human subjects have been carefully considered in line with the Declaration of Helsinki (1964).
Financial support
None
Authors’ contributions
Naci Balak: Conception of the idea, data collection, analysis design and performance results interpretation, writing the text, literature search, text revision
Deniz Inandenizlukuslu@marmara.edu.tr Analysis performance, literature search, text revision, critical review of the intellectual content,
Mario GanauMario.Ganau@alumni.harvard.edu Data analysis and design, interpretation of the results, literature search, text revision, critical review of the intellectual content,
Cesare Zoiagioiaoffice@gmail.com Data analysis and design, interpretation of the results, literature search, text revision, critical review of the intellectual content,
Sinan Sönmezssonmez@marmara.edu.tr Literature search, text revision, critical review of the intellectual content,
Batuhan Kurtbatuhan@marmara.edu.tr Literature search, text revision, critical review of the intellectual content,
Ahmet Akgülahmetakgul@marmara.edu.tr Literature search, Text revision, critical review of the intellectual content
Müjgan Tezmtez@marmara.edu.tr Data analysis, literature search, text revision, critical review of the intellectual content, supervision of the course of the project and the article
Acknowledgments
We would like to express our gratitude to Prof. Mehmet Oktav for his exceptional support, without which the completion of this article would not have been possible. We also thank Mrs. Ann Hazinedar for her help in proofreading.
References
- 1.Ganau M., Netuka D., Broekman M., et al. Neurosurgeons and the fight with COVID-19: a position statement from the EANS individual membership committee. Acta Neurochir. 2020;162(8):1777–1782. doi: 10.1007/s00701-020-04360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hulsbergen A.F.C., Eijkholt M.M., Balak N., et al. Ethical triage during the COVID-19 pandemic: a toolkit for neurosurgical resource allocation. Acta Neurochir. 2020;162(7):1485–1490. doi: 10.1007/s00701-020-04375-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mathiesen T., Arraez M., Asser T., et al. A snapshot of European neurosurgery December 2019 vs. March 2020: just before and during the Covid-19 pandemic. Acta Neurochir. 2020;162(9):2221–2233. doi: 10.1007/s00701-020-04482-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Montemurro N., Perrini P. Will COVID-19 change neurosurgical clinical practice? Br J Neurosurg. 2020;(Jun 1):1–2. doi: 10.1080/02688697.2020.1773399. [DOI] [PubMed] [Google Scholar]
- 5.Leong A.Z., Lim J.X., Tan C.H., et al. COVID-19 response measures - a Singapore Neurosurgical Academic Medical Centre experience segregated team model to maintain tertiary level neurosurgical care during the COVID-19 outbreak. Br J Neurosurg. 2020:1–6. doi: 10.1080/02688697.2020.1758629. [DOI] [PubMed] [Google Scholar]
- 6.Eijkholt M., Broekman M., Balak N., Mathiesen T., committee EE-l. Three pitfalls of accountable healthcare rationing. J Med Ethics. 2021 doi: 10.1136/medethics-2020-106943. [DOI] [PubMed] [Google Scholar]
- 7.Chowell G., Sattenspiel L., Bansal S., Viboud C. Mathematical models to characterize early epidemic growth: a review. Phys Life Rev. 2016;18:66–97. doi: 10.1016/j.plrev.2016.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarnaglia A.J.Q., Zamprogno B., Fajardo Molinares F.A., de Godoi L.G., Jimenez Monroy N.A. Correcting notification delay and forecasting of COVID-19 data. J Math Anal Appl. 2021;(125202):1–15. doi: 10.1016/j.jmaa.2021.125202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva C.J., Cantin G., Cruz C., et al. Complex network model for COVID-19: human behavior, pseudo-periodic solutions and multiple epidemic waves. J Math Anal Appl. 2021;(125171):1–25. doi: 10.1016/j.jmaa.2021.125171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shah N.H., Sheoran N., Jayswal E., et al. Modelling COVID-19 transmission in the United States through interstate and foreign travels and evaluating impact of governmental public health interventions. J Math Anal Appl. 2020:124896. doi: 10.1016/j.jmaa.2020.124896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chowell G., Viboud C., Simonsen L., Moghadas S.M. Characterizing the reproduction number of epidemics with early subexponential growth dynamics. J R Soc Interface. 2016;13(123):1–12. doi: 10.1098/rsif.2016.0659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ivorra B., Ferrandez M.R., Vela-Perez M., Ramos A.M. Mathematical modeling of the spread of the coronavirus disease 2019 (COVID-19) taking into account the undetected infections. The case of China. Commun Nonlinear Sci Numer Simulat. 2020;88(105303):1–21. doi: 10.1016/j.cnsns.2020.105303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postnikov E.B. Estimation of COVID-19 dynamics "on a back-of-envelope": does the simplest SIR model provide quantitative parameters and predictions? Chaos, Solit Fractals. 2020;135(109841):1–6. doi: 10.1016/j.chaos.2020.109841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chowell G., Nishiura H., Bettencourt L.M. Comparative estimation of the reproduction number for pandemic influenza from daily case notification data. J R Soc Interface. 2007;4(12):155–166. doi: 10.1098/rsif.2006.0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowell G., Simonsen L., Towers S., Miller M.A., Viboud C. Transmission potential of influenza A/H7N9, february to may 2013, China. BMC Med. 2013;11(214):1–13. doi: 10.1186/1741-7015-11-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Distante C., Piscitelli P., Miani A. Covid-19 outbreak progression in Italian regions: approaching the peak by the end of march in northern Italy and first week of April in southern Italy. Int J Environ Res Publ Health. 2020;17(3025):1–9. doi: 10.3390/ijerph17093025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smirnova A., deCamp L., Chowell G. Forecasting epidemics through nonparametric estimation of time-dependent transmission rates using the SEIR model. Bull Math Biol. 2019;81(11):4343–4365. doi: 10.1007/s11538-017-0284-3. [DOI] [PubMed] [Google Scholar]
- 18.de Lima C.L., da Silva C.C., da Silva A.C.G., et al. COVID-SGIS: a smart tool for dynamic monitoring and temporal forecasting of covid-19. Front Public Health. 2020;8(580815):1–21. doi: 10.3389/fpubh.2020.580815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma J. Estimating epidemic exponential growth rate and basic reproduction number. Infect Dis Model. 2020;5:129–141. doi: 10.1016/j.idm.2019.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brunner N., Kuhleitner M. Bertalanffy-Putter models for the first wave of the COVID-19 outbreak. Infect Dis Model. 2021;6:532–544. doi: 10.1016/j.idm.2021.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blanco N., Stafford K.A., Lavoie M.C., Brandenburg A., Gorna M.W., Merski M. A simple model for the total number of SARS-CoV-2 infections on a national level. Epidemiol Infect. 2021;149:e80. doi: 10.1017/S0950268821000649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shang C., Yang Y., Chen G.Y., Shang X.D. A simple transmission dynamics model for predicting the evolution of COVID-19 under control measures in China. Epidemiol Infect. 2021;149:e43. doi: 10.1017/S0950268821000339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bacaër N. In: A Short History of Mathematical Population Dynamics. Bacaër N., editor. Springer; London: 2011. Verhulst and the logistic equation (1838) pp. 35–39. [Google Scholar]
- 24.Chibbaro S., Ganau M., Todeschi J., Proust F., Cebula H. How SARS-CoV-2 is forcing us to reconsider and reorganize our daily neurosurgical practice. Neurochirurgie. 2020;66(4):189–191. doi: 10.1016/j.neuchi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsoularis A., Wallace J. Analysis of logistic growth models. Math Biosci. 2002;179(1):21–55. doi: 10.1016/s0025-5564(02)00096-2. [DOI] [PubMed] [Google Scholar]
- 26.Haeussler E., Paul R., Wood R. twelfth ed. ed. Pearson Education, Inc.; Upper Saddle River, NJ: 2008. Introductory Mathematical Analysis. For Business, Economics, and the Life and Social Sciences. [Google Scholar]
- 27.Coronavirus disease (COVID-19) Situation Report 163. Data as received by WHO from national authorities by 10:00 CEST, 1 July 2020. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200701-covid-19-sitrep-163.pdf?sfvrsn=c202f05b_2 Last accessed on 20.07.2021.
- 28.Coronavirus disease (COVID-19) Situation Report 170. Data as received by WHO from national authorities by 10:00 CEST, 8 July 2020. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200708-covid-19-sitrep-170.pdf?sfvrsn=bca86036_2 Last accessed on 20.07.2021.
- 29.Coronavirus disease (COVID-19) Situation Report 177. Data as received by WHO from national authorities by 10:00 CEST, 15 July 2020. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200715-covid-19-sitrep-177.pdf?sfvrsn=b1a193f3_2 Last accessed on 20.07.2021.
- 30.Coronavirus disease (COVID-19) Situation Report 184. Data as received by WHO from national authorities by 10:00 CEST, 22 July 2020. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200722-covid-19-sitrep-184.pdf?sfvrsn=7680210a_2 Last accessed on 20.07.2021.
- 31.Coronavirus disease (COVID-19) Situation Report 191. Data as received by WHO from national authorities by 10:00 CEST, 29 July 2020. 2020. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200729-covid-19-sitrep-191.pdf?sfvrsn=2c327e9e_2 Last accessed on 20.07.2021.
- 32.WHO-COVID-19-global-data. 2021. https://covid19.who.int/WHO-COVID-19-global-data.csv 2021 Last accessed on 20.07.2021.
- 33.Pianka E. seventh ed. Eric R. Pianka; USA: 2011. Evolutionary Ecology. [Google Scholar]
- 34.Abusam A., Abusam R., Al-Anzi B. Adequacy of Logistic models for describing the dynamics of COVID-19 pandemic. Infect Dis Model. 2020;5:536–542. doi: 10.1016/j.idm.2020.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vandamme L., Rocha P. Analysis and simulation of epidemic COVID-19 curves with the verhulst model applied to statistical inhomogeneous age groups. Appl Sci. 2021;11(4159):1–16. [Google Scholar]
- 36.Mahanty C., Kumar R., Mishra B., Hemanth D., Gupta D., Khanna A. Prediction of COVID‐19 active cases using exponential and non‐linear growth models. Expet Syst. 2020;(e12648):1–22. [Google Scholar]
- 37.Niazkar M, Eryilmaz Turkkan G, Niazkar HR, Turkkan YA. Assessment of three mathematical prediction models for forecasting the COVID-19 outbreak in Iran and Turkey. Comput Math Methods Med 2020; 2020(7056285): 1-13 DOI: 0.1155/2020/7056285. [DOI] [PMC free article] [PubMed]
- 38.Holmdahl I., Buckee C. Wrong but useful - what covid-19 epidemiologic models can and cannot tell us. N Engl J Med. 2020;383(4):303–305. doi: 10.1056/NEJMp2016822. [DOI] [PubMed] [Google Scholar]
- 39.Arora P., Kumar H., Panigrahi B.K. Prediction and analysis of COVID-19 positive cases using deep learning models: a descriptive case study of India. Chaos, Solit Fractals. 2020;139(110017):1–9. doi: 10.1016/j.chaos.2020.110017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malavika B., Marimuthu S., Joy M., Nadaraj A., Asirvatham E.S., Jeyaseelan L. Forecasting COVID-19 epidemic in India and high incidence states using SIR and logistic growth models. Clin Epidemiol Glob Health. 2021;9:26–33. doi: 10.1016/j.cegh.2020.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishiura H. Time variations in the generation time of an infectious disease: implications for sampling to appropriately quantify transmission potential. Math Biosci Eng. 2010;7(4):851–869. doi: 10.3934/mbe.2010.7.851. [DOI] [PubMed] [Google Scholar]
- 42.de Silva E., Ferguson N.M., Fraser C. Inferring pandemic growth rates from sequence data. J R Soc Interface. 2012;9(73):1797–1808. doi: 10.1098/rsif.2011.0850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Roberts M.G., Nishiura H. Early estimation of the reproduction number in the presence of imported cases: pandemic influenza H1N1-2009 in New Zealand. PloS One. 2011;6(5) doi: 10.1371/journal.pone.0017835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon Mendez L., de Mateo Ontanon S., Larrauri Camara A., et al. [Transmissibility and severity of the pandemic influenza A (H1N1) 2009 virus in Spain] Gac Sanit. 2011;25(4):296–302. doi: 10.1016/j.gaceta.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 45.Hedge J., Lycett S.J., Rambaut A. Real-time characterization of the molecular epidemiology of an influenza pandemic. Biol Lett. 2013;9(20130331):1–5. doi: 10.1098/rsbl.2013.0331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Britton T., Lindenstrand D. Epidemic modelling: aspects where stochasticity matters. Math Biosci. 2009;222(2):109–116. doi: 10.1016/j.mbs.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 47.Gatto M., Muratori S., Rinaldi S. A functional interpretation of the logistic equation. Ecol Model. 1988;42(2):155–159. [Google Scholar]
- 48.Hu H., Nigmatulina K., Eckhoff P. The scaling of contact rates with population density for the infectious disease models. Math Biosci. 2013;244(2):125–134. doi: 10.1016/j.mbs.2013.04.013. [DOI] [PubMed] [Google Scholar]
- 49.Chowell G., Nishiura H. Quantifying the transmission potential of pandemic influenza. Phys Life Rev. 2008;5(1):50–77. [Google Scholar]
- 50.Hillen T. Applications and limitations of the verhulst model for populations. Math Biol, University of Ualberta. 2003;6:19–20. [Google Scholar]
- 51.GAO Science & Tech Spotlight; 2020. Herd immunity for COVID-19.https://www.gao.gov/assets/710/707975.pdf GAO-20-646SP. 2020. Last accessed on 08.09.2020. [Google Scholar]
- 52.Britton T., Ball F., Trapman P. A mathematical model reveals the influence of population heterogeneity on herd immunity to SARS-CoV-2. Science. 2020;369(6505):846–849. doi: 10.1126/science.abc6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Anderson R.M., May R.M. Vaccination and herd immunity to infectious diseases. Nature. 1985;318(6044):323–329. doi: 10.1038/318323a0. [DOI] [PubMed] [Google Scholar]
- 54.Randolph H.E., Barreiro L.B. Herd immunity: understanding COVID-19. Immunity. 2020;52(5):737–741. doi: 10.1016/j.immuni.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitchell P. What could coronavirus teach us? Br J Neurosurg. 2020;34(2):117–118. doi: 10.1080/02688697.2020.1756417. [DOI] [PubMed] [Google Scholar]
- 56.Bouter L., Zielhuis G., Zeegers M., editors. Textbook of Epidemiology. Bohn Stafleu van Loghum; Houten: 2018. [Google Scholar]
- 57.Krämer A., Kretzschmar M., Krickeberg K. Springer Science+Business Media, LLC; New York Dordrecht Heidelberg London: 2010. Modern Infectious Disease Epidemiology. Concepts, Methods, Mathematical Models, and Public Health. [Google Scholar]
- 58.Bonomo G., Caldiroli D., Bonomo R., Pugliese R., DiMeco F., Zoia C. Reactivation of COVID-19 in a neurosurgical patient with early neuropsychiatric presentation. Does seroconversion mean immunity? Surg Neurol Int. 2021;12(166):1–5. doi: 10.25259/SNI_831_2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Vries G., Hillen T., Lewis M., Müller J., Schönfisch B. Society for Industrial and Applied Mathematics; Philadelphia: 2006. A Course in Mathematical Biology: Quantitative Modeling with Mathematical and Computational Methods. [Google Scholar]
- 60.Smith F. Experimental methods in population dynamics: a critique. Ecology. 1952;33(4):441–450. [Google Scholar]
- 61.Zoia C., Bongetta D., Veiceschi P., et al. Neurosurgery during the COVID-19 pandemic: update from Lombardy, northern Italy. Acta Neurochir. 2020;162(6):1221–1222. doi: 10.1007/s00701-020-04305-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zoia C., Raffa G., Somma T., et al. COVID-19 and neurosurgical training and education: an Italian perspective. Acta Neurochir. 2020;162(8):1789–1794. doi: 10.1007/s00701-020-04460-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Balak N., Broekman M.L.D., Mathiesen T. Ethics in contemporary health care management and medical education. J Eval Clin Pract. 2020;26(3):699–706. doi: 10.1111/jep.13352. [DOI] [PubMed] [Google Scholar]
- 64.Balak N., Elmaci I. Costs of disorders of the brain in Europe. Eur J Neurol. 2007;14(2):e9. doi: 10.1111/j.1468-1331.2006.01570.x. [DOI] [PubMed] [Google Scholar]
- 65.Balak N., Tisell M. In: Ethics in Neurosurgical Practice. Honeybul S., editor. Cambridge University Press; Cambridge: 2020. Healthcare economics; pp. 73–84. [Google Scholar]
- 66.Castlen J.P., Cote D.J., Moojen W.A., et al. The changing health care landscape and implications of organizational ethics on modern medical practice. World Neurosurg. 2017;102:420–424. doi: 10.1016/j.wneu.2017.03.073. [DOI] [PubMed] [Google Scholar]
- 67.Tisell M., Balak N. In: Ethics in Neurosurgical Practice. Honeybul S., editor. Cambridge University Press; Cambridge: 2020. Surgical training; pp. 54–63. [Google Scholar]
- 68.Zaed I. COVID-19 consequences on medical students interested in neurosurgery: an Italian perspective. Br J Neurosurg. 2020:1–2. doi: 10.1080/02688697.2020.1777260. [DOI] [PubMed] [Google Scholar]
- 69.Balak N. Letter: length of stay beyond medical readiness in a neurosurgical patient population and associated healthcare costs. Neurosurgery. 2021 doi: 10.1093/neuros/nyab198. [DOI] [PubMed] [Google Scholar]
- 70.Dannhoff G., Cebula H., Chibbaro S., et al. Investigating the real impact of COVID-19 pandemic on the daily neurosurgical practice? Neurochirurgie. 2021;67(2):99–103. doi: 10.1016/j.neuchi.2021.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Minhas Z., Ganau M., Thakar C., et al. COVID-19: new challenges, risks, and the future provision of care in spinal services. Bone Joint Lett J. 2020;102-B(6):655–657. doi: 10.1302/0301-620X.102B6.BJJ-2020-0686. [DOI] [PubMed] [Google Scholar]
- 72.Finn R., Ganau M., Jenkinson M.D., Plaha P. COVID-legal study: neurosurgeon experience in Britain during the first phase of the COVID-19 pandemic - medico-legal considerations. Br J Neurosurg. 2021:1–4. doi: 10.1080/02688697.2021.1902475. [DOI] [PubMed] [Google Scholar]
- 73.Rubulotta F., Soliman-Aboumarie H., Filbey K., et al. Technologies to optimize the care of severe COVID-19 patients for health care providers challenged by limited resources. Anesth Analg. 2020;131(2):351–364. doi: 10.1213/ANE.0000000000004985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Case K., Fair R., Oster S. 13 ed. Pearson Education Limited; Harlow: 2020. Principles of Economics. [Google Scholar]