Abstract
CD8+ T cells play an important role in vaccination and immunity against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Although numerous SARS-CoV-2 CD8+ T cell epitopes have been identified, the molecular basis underpinning T cell receptor (TCR) recognition of SARS-CoV-2-specific T cells remains unknown. The T cell response directed toward SARS-CoV-2 spike protein–derived S269–277 peptide presented by the human leukocyte antigen (HLA)-A∗02:01 allomorph (hereafter the HLA-A2S269–277 epitope) is, to date, the most immunodominant SARS-CoV-2 epitope found in individuals bearing this allele. As HLA-A2S269–277-specific CD8+ T cells utilize biased TRAV12 gene usage within the TCR α-chain, we sought to understand the molecular basis underpinning this TRAV12 dominance. We expressed four TRAV12+ TCRs which bound the HLA-A2S269–277 complex with low micromolar affinity and determined the crystal structure of the HLA-A2S269–277 binary complex, and subsequently a ternary structure of the TRAV12+ TCR complexed to HLA-A2S269–277. We found that the TCR made extensive contacts along the entire length of the S269–277 peptide, suggesting that the TRAV12+ TCRs would be sensitive to sequence variation within this epitope. To examine this, we investigated cross-reactivity toward analogous peptides from existing SARS-CoV-2 variants and closely related coronaviruses. We show via surface plasmon resonance and tetramer studies that the TRAV12+ T cell repertoire cross-reacts poorly with these analogous epitopes. Overall, we defined the structural basis underpinning biased TCR recognition of CD8+ T cells directed at an immunodominant epitope and provide a framework for understanding TCR cross-reactivity toward viral variants within the S269–277 peptide.
Keywords: COVID-19, SARS-CoV-2, HLA-A2, T cell receptor recognition, spike protein
Abbreviations: BSA, buried surface area; HLA, human leukocyte antigen; PBMCs, peripheral blood monocular cells; pHLA, peptide-HLA; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SPR, surface plasmon resonance; TCR, T cell receptor; VDW, van der Waals
The emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and the clinical syndrome associated with the infection (COVID-19) have had unprecedented global impacts on health, lifestyle, and economies. The rapid development of safe and effective COVID-19 vaccines has provided the means to stop the spread of COVID-19 in principle; however, production and administration of vaccines to sufficiently cover the global population poses challenges that will take years to overcome. Meanwhile, global infection rates remain high, which enables SARS-CoV-2 to further adapt to the selection pressures of human immune responses elicited by infection and vaccination. There is a strong correlation between circulating T cell numbers and COVID-19 pathogenesis (1, 2, 3, 4, 5, 6), and evidence demonstrates that asymptomatic individuals mount a highly functional virus-specific cellular immune response (3), whereas acute COVID-19 patients typically exhibit lymphopenia. Notably, the depletion of peripheral T cells has been linked to high viral titers, increased disease severity, and death, whereas recovery of T cell numbers precedes clinical and virological recovery (4). COVID-19 severity has been also linked to CD8+ T cell activation and exhaustion, as well as characteristic changes in CD4+ T cell subsets (5, 6, 7).
As CD8+ T cells specific for SARS-CoV-2-derived epitopes are an important component of the antiviral immune response in COVID-19 infection and vaccination (8, 9, 10, 11, 12, 13, 14, 15, 16, 17), an understanding of the T cell–mediated protective immune responses to SARS-CoV-2 and the potential viral variants is needed to provide insights into the ongoing adaptation of SARS-CoV-2 to human immune responses and our ability to control this evolving pathogen in the long term. A number of SARS-CoV-2 CD8+ T cell epitopes have been defined for high frequency human leukocyte antigen (HLA) class I alleles, including HLA-A∗01:01, A∗02:01, A∗03:01, A∗11:01, A∗24:02, HLA-B∗07:02, B∗27:05, B∗40:01, and B∗44:03 (7, 9, 14, 15, 16, 17, 18). This allowed us to understand the magnitude, phenotype, response kinetics, and immunodominance hierarchy of epitope-specific CD8+ T cells and their origins in COVID-19 (9). However, current knowledge on the underlying SARS-CoV-2-specific T cell receptor (TCR) repertoires and the role of the HLA in providing protective immunity is limited.
We and others investigated the HLA-A2-restricted CD8+ T cell responses to SARS-CoV-2 spike (S) protein and identified an immunodominant CD8+ T cell epitope S269–277 (YLQPRTFLL) (15, 17, 19). Although the relative role of HLA-A2S269–277 CD8+ TCR repertoire in SARS-CoV-2 infection remains unknown, CD8+ T cells directed toward the S269–277 epitope are consistently detected in acute and convalescent HLA-A2+ COVID-19 patients, although their phenotypic profiles seem suboptimal when compared with CD8+ T cell responses directed at the HLA-B7-restricted SARS-CoV-2 nucleoprotein derived epitope N105–113 (HLA-B7N105–113), the most dominant SARS-CoV-2 epitope identified to date (9, 15). The HLA-A2S269–277-restricted CD8+ T cell response in convalescent COVID-19 patients is characterized by a public and diverse TCR repertoire with significant biased TCR gene usage and conserved CDR3 motifs (9, 19). Specifically, unpaired TCR repertoire analysis in 34 convalescent COVID-19 patients showed that 72% of HLA-A2S269–277-specific CD8+ T cells used the TRAV12-1 gene segment and 16% the TRBV7-9 gene segment (19). We recently analyzed the HLA-A2S269–277 tetramer+ T cell repertoire in five convalescent patients using α–β paired single-cell sequencing, which broadly mirrored these observations and additionally provided α–β paired TCR sequences (9). Notably, this revealed two highly prevalent TCR motifs: (i) CDR3α motif CVVNXXXDMRF (where X denotes any amino acid) associated with TRAV12-1 usage and the joining gene segment TRAJ43 (16% of the total TCR repertoire across five patients) and (ii) CDR3α motif CAVNXDDKIIF linked to TRAV12-2 and TRAJ30 (3.4% of the TCR repertoire in 4 of 5 patients). Our previous data revealed that the TRAV12-1 motif was independent of TRBV gene usage, but tightly linked to TRBJ2-2, whereas the TRAV12-2 motif was tightly associated with TRBV7-9, although allowed different joining gene segments. Collectively, the HLA-A2S269–277 TCR repertoire data suggest that TRAV12 germline–encoded elements are directly involved in the T cell selection process and linked to corresponding TCR β gene segments. To investigate the basis of the HLA-A2S269–277-restricted CD8+ T cell repertoire and probe its potential to recognize and cross-react with S269–277 peptide variants, we selected three TRAV12-1 TCRs and one TRAV12-2/TRBV7-9 TCR for further study and determined the ternary crystal structure of the HLA-A2 presented S269–277 epitope and a TRAV12+ TCR bound to HLA-A2S269–277.
Results
The TRAV12+-restricted CD8+ T cell response toward HLA-A2S269–277 is composed of TCRs with binding affinities typical for antiviral CD8+ T cell responses
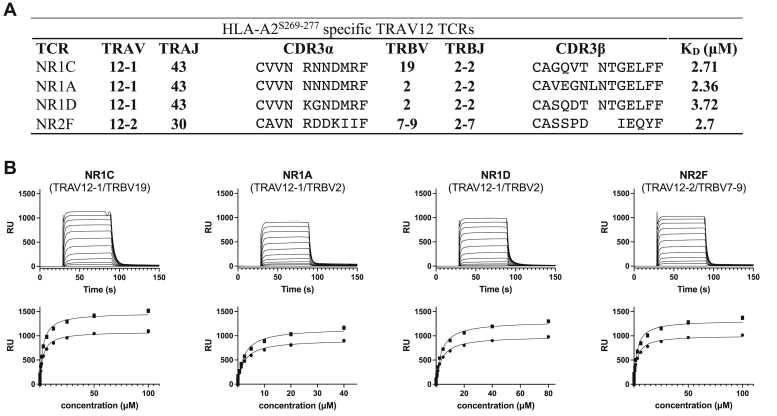
To probe the basis of TRAV12 bias within the immunodominant HLA-A2S269–277-specific CD8+ T cells, we selected three TCRαβ pairs with the prevalent TRAV12-1 motif and one with the TRAV12-2/TRBV7-9 combination. While the three TRAV12-1 TCRs (NR1A, NR1C, and NR1D) used the same joining gene segments TRAJ43 and TRBJ2-2, only two (NR1A and NR1D) used the most prevalent TRBV2 gene and one (NR1C) used the comparatively rare TRBV19 (Fig. 1A). Of note, a previous analysis of the HLA-A2S269–277-restricted CD8+ T cell repertoire encountered TRBV19 in less than 1% of HLA-A2S269–277 reactive T cells (19).
Figure 1.
TRAV12+TCRs bind HLA-A2S269–277with similar affinities.A, TCR αβ gene usage, CDR3 sequences, and equilibrium dissociation constants KD of selected TRAV12+ HLA-A2S269–277-restricted TCRs. B, SPR affinity measurements of TCRs binding to immobilized HLA-A2S269–277. Representative SPR measurements (top) and determination of HLA-A2S269–277 binding affinity (bottom). HLA, human leukocyte antigen; SPR, surface plasmon resonance; TCR, T cell receptor.
TCRs were expressed, refolded, and purified via established procedures (20), and purity (>90%) was assessed by gel filtration chromatography and SDS-PAGE. Biotinylated HLA-A2S269–277 (15) was immobilized, and the equilibrium dissociation constant (KD) for each TCR was determined via surface plasmon resonance (SPR) measurements. The KD values fell within a narrow range of 2.36 to 3.72 μM for all four TCRs (Fig. 1, A and B). These TCR-peptide-HLA (TCR-pHLA) affinities fall into the typical range observed for antiviral CD8+ T cell responses, which shows that the phenotypical profile and suboptimal response of HLA-A2S269–277-restricted CD8+ T cells is not linked to a lack in TCR binding to HLA-A2S269–277. To uncover the basis of the T cell selection bias in the HLA-A2S269–277-restricted T cell repertoire, we next determined the crystal structures of the HLA-A2-presented S269–277 epitope and of the TRAV12-1 TCR NR1C bound to HLA-A2S269–277.
Binary structure of HLA-A2S269–277 reveals a feature-rich epitope
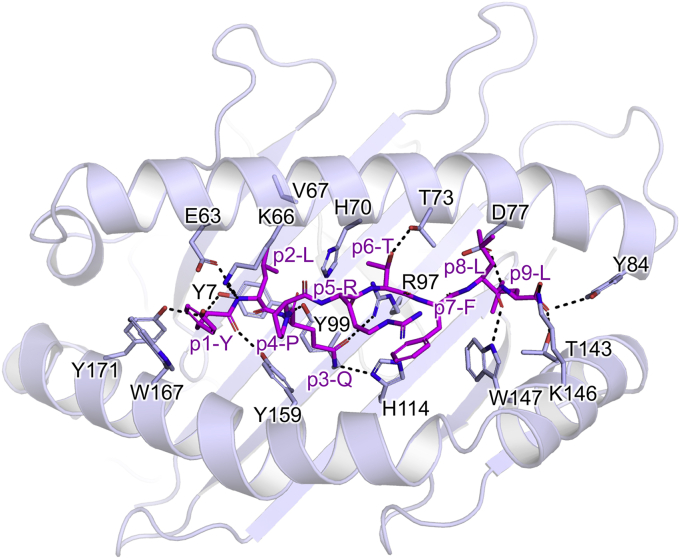
To understand the basis of S269–277 presentation by HLA-A2, we crystallized and solved the structure of HLA-A2S269–277 at a resolution of 2.3 Å (Fig. 2; see Table S1 for data collection and refinement statistics). As expected, the S269–277 peptide (YLQPRTFLL) was bound to HLA-A2 in a canonical conformation, with the side chains of p2-Leu and p9-Leu anchored in the B and F pockets, respectively, and adopting an overall conformation similar to that of the influenza epitope HLA-A2 M158–66 (21). The peptide backbone was bound to HLA-A2 via ten H-bonds and an additional salt bridge between Lys146 and the peptide C terminus (Fig. 2). The side chain of p3-Gln reached into the C-pocket, where it formed H-bonds with R97 and His114, thus acting as ancillary anchor residue. Moreover, the surface-exposed side chain p6-Thr contributed an H-bond with Thr73, and the side chain of p1-Tyr was tightly stacked between Trp167 and Lys66, which partially covered the aromatic ring of p1-Tyr, but exposed its OH group. The side chain of p5-Arg interacted with Gln155, adopting two distinct conformations due to crystal contacts. The p4-Pro and p5-Arg midsection of the peptide was most prominently exposed due do a bulged backbone conformation between p3 and p5 of the peptide, and this arrangement left a narrow opening to the mostly buried side chain of p3-Gln. The side chains of p7-Phe and p8-Leu were nested against the walls of the peptide binding cleft and were also accessible. Thus, while the peptide was tightly bound by HLA-A2, all peptide side chains, with the exception of the canonical anchor residues p2-Leu and p9-Leu, were accessible for potential interactions with the TCR. Therefore, HLA-A2S269–277 provided a relatively feature-rich epitope for potential TCR interaction.
Figure 2.
Structure of HLA-A2S269–277. H-bonding interactions between the S269–277 peptide (purple sticks) and HLA-A2 (cartoon). HLA-A2 contact residues and H-bonds within 3.5 Å of the peptide are shown (light blue sticks and black dashes). HLA, human leukocyte antigen.
Structural overview of the ternary complex between the NR1C TCR and HLA-A2S269–277
To gain insight into the basis of TRAV12-biased gene usage toward HLA-A2S269–277, we chose the NR1C TCR for structural investigation. This TCR contains the TRAV12-1/TRAJ43 TCR motif paired with the canonical TRBJ2-2 segment and the TRBV19 gene. We crystallized and determined the structure of the ternary complex between the NR1C and HLA-A2S269–277 at a resolution of 3.2 Å (Figs. 3 and 4; see Table S1 for data collection and refinement statistics). The model contained two protomers in the asymmetric unit, with an overall Cα RMSD of 0.98 Å, and the electron density at the TCR–pHLA interface region was clear and unambiguous (Fig. S1). Based on the clarity of the electron density maps, we chose protomer 2 (chains D, E, G, H, and F) for our analysis.
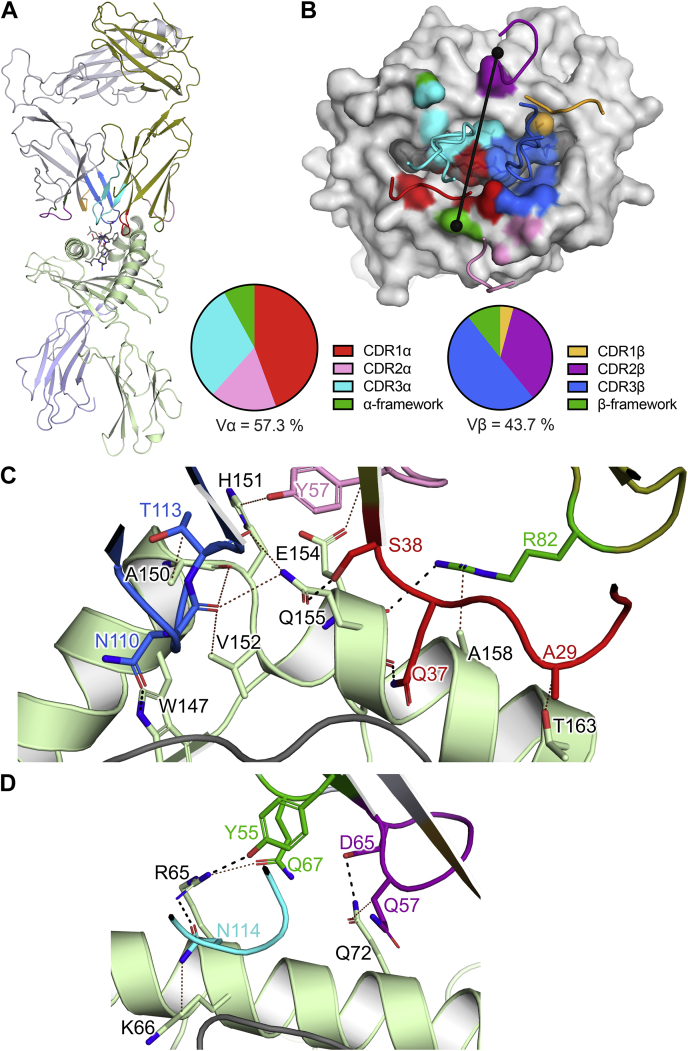
Figure 3.
Structure of the NR1C TCR–HLA-A2S269–277ternary complex.A, overview of the ternary complex, with NR1C (top) and HLA-A2S269–277(bottom) shown in cartoon representation. B, TCR footprint of NR1C on HLA-A2S269–277. NR1C CDR loops, HLA-A2, and peptide are shown as tubes, light surface, and dark surface, respectively, with surface contacts (top) and partial BSA contributions (bottom) colored according to each CDR loop. TCR Vα and Vβ center of mass positions and docking angle are shown as black spheres and a line, respectively. C and D, interactions between the NR1C TCR and HLA-A2 with interface residues shown as sticks. H-bonds are shown as black dashed lines and VDW interactions as brown dotted lines, and a distance cut-off of 3.5 Å and 4.0 Å was applied for H-bonds and VDW interactions, respectively. C, interactions between the NR1C TCR and HLA-A2 α2-domain. D, interactions between the NR1C TCR and HLA-A2 α1-domain. Colors: light green for HLA-A2 α-chain, olive for TCR α-chain, red, pink, and cyan for CDR1α, CDR2α, and CDR3α, orange, purple, and blue for CDR1β, CDR2β, and CDR3β, and green for α/β-framework contact residues, respectively. BSA, buried surface area; HLA, human leukocyte antigen; TCR, T cell receptor; VDW, multiple van der Waals.
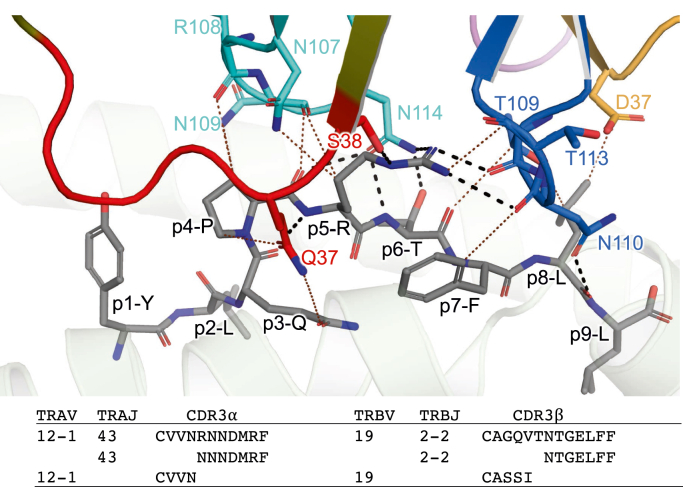
Figure 4.
Interactions between NR1C and the peptide NR1C TCR–HLA-A2S269–277ternary complex. The S269–277 peptide and TCR residues interacting with the peptide are shown sticks. The peptide is colored gray and TCR contact residues are colored red, cyan, orange, and blue for CDR1α CDR3α, CDR1β, and CDR3β, respectively. H-bonds are shown as black dashed lines and VDW interactions as brown dotted lines, and a distance cut-off of 3.5 Å and 4.0 Å was applied for H-bonds and VDW interactions, respectively. CDR3 sequences and germline-encoded elements are listed below the graphic. HLA, human leukocyte antigen; TCR, T cell receptor.
The structure of the NR1C TCR–HLA-A2S269–277 ternary complex revealed that NR1C TCR engaged HLA-A2S269–277 in a canonical docking orientation at an angle of 85° with respect to the peptide, and an overall buried surface area (BSA) of 2013 Å2. The partial contributions of the TCR α and β chains to the BSA on HLA-A2S269–277 were 57.3% and 42.7%, respectively, whereas the partial contributions of the peptide and the HLA to the BSA of the TCR were 34.9% and 65.1%, respectively (Fig. 3, A and B). Compared with the binary HLA-A2S269–277 complex, the HLA-A2S269–277 structure within the NR1C ternary complex was largely unchanged (RMSD for HLA α chain residues 1 to 180 was between 1.0 and −1.3 Å for all protomers). The near-orthogonal docking angle of the NR1C TCR allowed both the CDR1α and CDR3α loops to adopt backbone orientations alongside the peptide, such that the CDR1α loop occupied the space between p1 to p5 of the peptide and the HLA-A2 α2 helix, while the CDR3α loop occupied the opposing gap between the HLA-A2 α1 helix and p4 to p5 of the peptide. As with CDR1α, the CDR3β loop was wedged between the p5 to p6 of the peptide and the HLA-A2 α2 helix while CDR2α sat atop the HLA-A2 α2 helix. CDR1β and CDR2β interacted with p8 of the peptide and the HLA-A2 α1 helix, respectively (Fig. 3B).
Structural basis of TRAV12-1 bias and CDR3α motif selection
While the overall binding parameters of the NR1C TCR–HLA-A2S269–277 complex were well within the boundaries of typical TCR–pHLA interactions, the structure stood out in that the interface between the TRAV12–1-encoded residues and HLA-A2S269–277 was much more involved than the interface formed by TCR β germline–encoded residues. As it is likely that the structural conservation of extensive interactions between TRAV12-1 germline–encoded residues and HLA-A2S269–277 across different TCRs with the same TRAV is directly linked to the T cell selection bias, we calculated and compared the partial contributions of the different CDR loops to the overall BSA of HLA-A2S269–277. Within the overall dominant interface of the TCR α-chain, CDR1α, CDR2α, CDR3α, and α-framework residues contributed 44.4%, 17.2%, 30.4%, and 8% to the TCR α footprint, respectively (Fig. 3B). Thus, CDR1α residues were likely involved in TRAV12-1 selection. On the other hand, the BSA contributions of CDR1β, CDR2β, CDR3β, and β-framework residues of 4%, 33.7%, 48.6%, and 13.8%, respectively, were clearly dominated by CDR3β, which implied that TRBV germline–encoded interactions were less likely to be involved in selection of NR1C (Fig. 3B). To gain an understanding of the molecular basis for the TRAV12-1 selection bias, we next analyzed the interactions of the CDR loops with HLA-A2S269–277.
Interactions of NR1C with the HLA
In accordance with the large TCR-α footprint, NR1C formed an extended interface with HLA-A2 α2 domain which involved residues from the CDR1α, CDR2α, α-framework, and CDR3β regions. While CDR2α Tyr57 and α-framework residue Arg82 interacted with the ridge of HLA-A2 α2 domain helix (Fig. 3C), the CDR1α and CDR3β loops both interacted with the inner wall of the peptide-binding cleft via four H-bonds as well as multiple van der Waals (VDW) interactions. Specifically, CDR1α Gln37 and Ser38 both formed two H-bonds with the backbone and side chain of Gln155, and CDR3β formed an H-bond with Trp147 as well as a VDW interface involving Ala150, Val152, and Gln155 (Fig. 3C). By comparison, the interactions of NR1C with the HLA-A2 α1 domain appeared more incidental, with the residues CDR2β Asp65 and CDR3α Asn109 and the β-framework Tyr55 forming two H-bonds with HLA residue Arg65, and one with Gln72 (Fig. 3D).
Interactions of NR1C with the peptide
The interface between the NR1C TCR and the peptide was formed by CDR1α, CDR3α, CDR1β, and CDR3β and overall comprised nine H-bonds (four with the side chain of p5-Arg, one with the side chain of p6-Thr, and four with the backbone of p5, p6, and p8), as well as a substantial number of VDW interactions (Fig. 4). The TCR effectively engaged the side chains of peptide residues p3-p8, with the prominent p5-Arg forming a central interaction hub surrounded by CDR1α, CDR3α, and CDR3β (Fig. 4). The backbone of CDR3β was looped around the p5-Arg side chain which also interacted with CDR1α and CDR3α and formed an extensive H-bond network that interconnected the peptide with the HLA-A2 α2 helix. Notably, the CDR1α residues Gln37 and Ser38, together with CDR3β residue Asn110, formed three bridging H-bond interactions that interlinked p5-Arg and p8-Leu of the peptide (Fig. 4) with the HLA-A2 α2 residues Gln155 and Trp147 (Fig. 3C), and these bridging H-bonds were further supported by two H-bonds between the backbone of CDR3β Thr109 and Asn110 p5-Arg (Fig. 4). This intricate network of H-bonds was further bolstered by VDW contacts between CDR1α Gln37, CDR3α Asn107, and Asn109 and CDR3β Thr109, Asn110, and Thr113, with the side chains of p3-Gln, p4-Pro, p5-Arg, p7-Phe, and p9-Leu (Fig. 4). The side of the peptide near the HLA-A2 α1 helix, was solely engaged by CDR3α, which formed overall four H-bonds with the peptide, namely between CDR3α Asn114, p5-Arg and p6-Thr, and one between the backbones of CDR3α Asn109 and p4-Pro (Fig. 4).
Our analysis revealed that the H-bonding network formed by CDR1α and CDR3β effectively anchored each loop in the narrow gap between the peptide and the HLA-A2 α2 helix, which suggests that the main contributors to this network, namely CDR1α residues Gln37 and Ser38 and CDR3β residue Asn110, were important for the selection of NR1C. Crucially, a comparison of TRAV gene segments revealed that CDR1α residues Gln37 and Ser38 are exclusive to both TRAV12-1 and TRAV12-2, which were predominantly selected by HLA-A2S269–277, whereas the closely related TRAV12-3 gene segment, encoding Tyr38, was absent from the HLA-A2S269–277-restricted T cell repertoire. Regarding CDR3 motif selection, the CDR3α residue Asn114 encoded by TRAJ43 also appeared to play key role, as it was both part of the H-bonding network and precisely positioned to form H-bonds with p5-Arg and p7-Thr (Fig. 4). Closer inspection of the CDR3α motifs associated with TRAV12-1 CVVNXXXDMRF revealed that CDR3α residue 114 was interchangeably selected as Asn or Asp (i.e., CVVNXX(N/D)DMRF), while the conserved TRAV12–1-encoded CDR3α motif (Val106, Asn107, and Arg117) directly interacted with CDR1α and thereby contributed to the stable juxtaposition of CDR1α and CDR3α. Of note, the CDR3β residue Asn110 encoded by frequently selected junctional element TRBJ2-2 is also part of a conserved motif CDR3β associated with TRAV12-1 selection (15).
Accordingly, our data provide a basis for the T cell recognition of HLA-A2S269–277 by linking the observed T cell repertoire bias and CDR motifs to structurally conserved interactions between HLA-A2S269–277 and unique TRAV12-1 and TRAV12-2 germline–encoded residues together with compatible junctional sequences.
SARS-CoV-2 HLA-A2S269–277-specific TRAV12 TCRs do not cross-react with viral variants
As CD8+ T cells can provide immune protection against viruses containing structurally similar immunogenic peptides, we first investigated the cross-reactive potential of SARS-CoV-2 HLA-A2S269–277-specific TCRs containing the dominant TRAV12-1 and TRAV12-2 gene segments. To define the potential cross-reactive capacity of the HLA-A2S269–277 CD8+ T cells, we searched available SARS-CoV-2 sequencing data in the GISAID database (22) for variants with mutations within the S269–277 epitope. The three most frequently changed amino acids were Pro272, Arg273, and Thr274, which were detected in 8000, 100, and 80 samples, respectively. Based on relative frequencies, we selected the S269–277 peptide variants P272L, R273S, and T274I for testing. To further investigate the possibility of heterologous CD8+ T cell immunity associated with the HLA-A2S269–277 epitope, we selected highly homologous peptides from related coronavirus species (SARS, MERS, PNRL, and BTRL; accession numbers YP_009825051.1, ABD72984.1, QIQ54048.1, QSQ01650.1, respectively), as well as the homologous self-derived peptide PAR1 (Table 1). We produced HLA-A2 complexes presenting each of the homologous peptides and measured binding of the TRAV12-1 TCR NR1C and the TRAV12-2 TCR NR2F via SPR (Table 1 and Fig. S2). In addition, we measured binding of the two remaining TRAV12-1 TCRs NR1A and NR1D to the S269–277 variants P272L, and R273S, that is, variants that, based on the structural data, were expected to directly impact on CDR1α binding. Our SPR measurements demonstrated that the TRAV12+ TCRs were all extremely sensitive to substitutions in the HLA-A2S269–277 epitope, and changes in HLA-A2S269–277 either completely abrogated TCR binding (S269–277R273S, MERS, SARS) or had a strongly negative impact (S269–277P272L, S269–277T274I, BTRL), with a >10-fold reduction in affinity. A notable exception was a peptide derived from a pangolin coronavirus (PNRL), which showed a >10-fold reduction in binding of the TRAV12-1 TCR NR1C, but only had minor (>2 fold) impact on binding of the TRAV12-2 TCR NR2F.
Table 1.
Summary of SPR affinity measurements
| Epitope | Sequence | NR1C |
NR2F |
NR1A |
NR1D |
|---|---|---|---|---|---|
| KD (μΜ) 95% CI | |||||
| S269–277 | YLQPRTFLL | 2.71 | 2.75 | 2.36 | 3.72 |
| 2.30–3.18 | 2.21–3.40 | 1.92–2.89 | 3.33–4.15 | ||
| S269–277P272L | YLQLRTFLL | 272.4 | 170.2 | 157.5 | 163.1 |
| 235.5–320.5 | 160.1–181.5 | 140.9–178.0 | 155.4–171.3 | ||
| S269–277R273S | YLQPSTFLL | N.B. | N.B. | N.B. | N.B. |
| S269–277T274I | YLQPRIFLL | 109.3 | 360.3 | ND | ND |
| 104.2–114.7 | 270.6–520.8 | ||||
| PNRL | YLQQRTFLL | 77.6 | 5.87 | ND | ND |
| 74.9–80.6 | 5.17–6.67 | ||||
| BTRL | YLKPRTFML | >100 | >100 | ND | ND |
| SARS | YLKPTTFML | N.B. | N.B. | ND | ND |
| MERS | KQLPLTFLL | N.B. | N.B. | ND | ND |
| PAR1 | TLDPRSFLL | N.B. | N.B. | ND | ND |
TCR binding to immobilized HLA-A2S269–277 and homologous epitopes (sequence differences to the SARS-CoV-2 epitope are highlighted in bold) were measured in two independent experiments with two replicates. Dissociation constant (KD) (in bold) and confidence interval (95% CI) (in italics) were calculated from all data using a single-site-binding model with KD as a shared variable.
Abbreviations: N.B., no binding; ND, not determined.
Lack of variant-reactive HLA-A2S269–277+CD8+ T cells in COVID-19 convalescent individuals
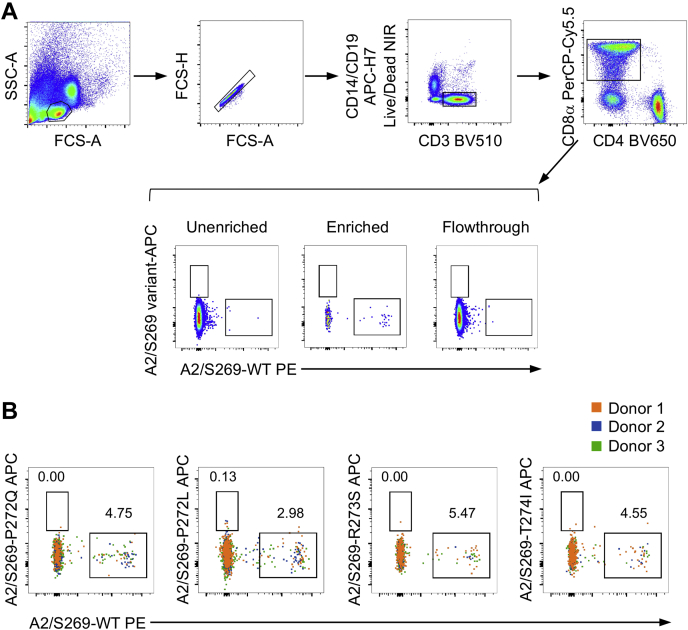
Although the TRAV12-1 and TRAV12-2 TCRs we tested via SPR represent the prominent TCR α sequences within the dominant TRAV12 gene segment, HLA-A2S269–277+CD8+ TCR repertoire also comprises non-TRAV12 TCRs. Thus, we further investigated a potential of HLA-A2S269–277+CD8+ T cell cross-reactivity toward the viral variants in peripheral blood monocular cells (PBMCs) obtained from COVID-19 convalescent individuals directly ex vivo. We generated tetramer reagents with HLA-A2S269–277 and each the single substituted S269–277 variants (P272L, pangolin (P272Q), R273S, and T274I) and performed ex vivo T cell tetramer staining of PBMCs from convalescent COVID-19 patients not to skew the data by in vitro manipulations (Fig. 5). In line with our SPR measurements and the broad structural conservation in the TCRαβ repertoire indicated by our structural data, we found no HLA-A2S269–277+CD8+ T cell cross-reactivity between the WT S269–277 and the variant peptides by ex vivo dual tetramer staining. In contrast, we detected only CD8+ T cell responses directed at the WT HLA-A2S269–277 epitope. Overall, our data demonstrate a lack of cross-reactive HLA-A2S269–277+CD8+ T cell responses between the epitopes encompassing the WT and closely related S269–277 immunogenic peptides.
Figure 5.
Screening of cross-reactivity between HLA-A2S269–277WT and variant epitopes.A, gating strategy of dual-tetramer enrichment with representative fluorescence-activated cell sorting plots of the unenriched, enriched, and flowthrough fractions of tetramer+ populations (donor 2), gated on live/CD14−/CD19−CD3+/CD8+ T cells. B, dual-TAME-enriched WT HLA-A2S269–277–WT-tetramer+ populations against the different variant–tetramer+ populations from three donors as concatenated fluorescence-activated cell sorting plots. HLA, human leukocyte antigen.
Discussion
The robustness of the CD8+ T cell responses to SARS-CoV-2 infection has been inversely linked to COVID-19 severity, with asymptomatic individuals eliciting highly functional and prominent SARS-CoV-2-specific CD8+ T cells (3). We and others have shown that HLA-A2S269–277 is the most immunodominant CD8+ T cell epitope restricted by the common HLA-A2 allele expressed in ∼40% of the global population. Moreover, our data suggested that, despite being immunodominant, the HLA-A2S269–277-restricted CD8+ T cell response appeared to be suboptimally activated as indicated by their phenotypic and activation profiles (15). Furthermore, TCR repertoire diversity of HLA-A2S269–277+CD8+ T cells was limited, with a strong bias in the TRAV12 gene segment. Here, we investigated the structural basis for the presentation of S269–277 by HLA-A2 and for the recognition of the HLA-A2S269–277 complex by CD8+ T cells. Presentation of the S269–277 peptide by HLA-A2S269–277 was overall similar to that of the immunodominant influenza epitope M158–66, suggesting that HLA presentation of this epitope is not linked to suboptimal activation of HLA-A2S269–277+CD8+ T cells. We further determined the structure of the NR1C–HLA-A2S269–277 complex, representing the first ternary structure of a TCR–peptide–HLA complex with SARS-CoV2-reactive T cells and provided a basis for understanding biased T cell recognition of the HLA-A2S269–277 epitope.
Our structural data revealed a significant involvement of the TCR with the peptide and the HLA-A2 α2 domain, whereas the interface with the HLA-A2 α1 helix was comparatively sparse. The TCR–pHLA interface was characterized by an extensive, peptide-centric H-bonding network involving CDR1α, CDR3α, and CDR3β. Specifically, CDR1α engaged HLA-A2S269–277 by inserting the key residues Gln37 and Ser38 into the narrow space between the p3-p5 of the peptide and HLA-A2 α2 helix, which is highly significant in the context of the available TRAV-encoded sequence space for CDR1α. Namely, the two most frequently selected gene segments (TRAV12-1 and TRAV12-2) in the HLA-A2S269–277-restricted CD8+ T cell repertoire are unique in that they both carry CDR1α Gln37 and Ser38, whereas the closely related TRAV12-3 featuring Gln37 and Tyr38 was absent from the HLA-A2S269–277-restricted repertoire. Moreover, the bridging interactions of CDR1α were mirrored by CDR3β Asn110, which H-bonded to both p5, p7, and p8 of the peptide and to the HLA-A2 α2 domain. Thus, CDR3β Asn110 can also be considered to play a central role for HLA-A2S269–277 recognition by NR1C, which provides a basis for the consistent coselection of the TRBJ2-2 gene segment in TRAV12-1+ TCRs. The general lack of TRBV selectivity in TRAV12-1+ TCRs can be explained by the observation that TRBV germline encoded residues only contributed isolated bonds to the interface because such sparse bonding can likely be realized by a number of TRBV gene segments. Interestingly, the coselection of TRBV7-9 in TRAV12-2+ TCRs was associated with more diverse TRBJ gene usage (9), which indicates that important interactions of the TCR β-domain are mediated by residues encoded by TRBV7-9 rather than selected TRBJ elements.
Based on the involvement of the CDR1α motif, it can be further hypothesized that HLA-A2S269–277-restricted TRAV12-2 TCRs engage HLA-A2S269–277 in similar fashion, that is, with analogous CDR1α and CDR2α interactions. However, the observation that the selection of TRAV12-2 TCRs is strongly linked to the TRBV7-9 gene segment but rather indiscriminate in TRBJ selection is unclear. It is possible that this bimodal coselection of TCR α and TCR β germline elements by HLA-A2S269–277 is linked to differences in CDR3α–CDR3β interactions that determine the precise geometry requirements for CDR3β motif selection and thereby shift the TRAV12-1 and TRAV12-2 associated coselection of TCR β germline elements from a TRBJ encoded to a TRBV-encoded element.
In general, T cells can crossreact with a substantial number of epitopes, including closely related epitope variants that differ from the canonical epitope at amino acids, which are not critical for TCR recognition. Thus, it is possible that homologous epitopes stemming from other coronaviruses can prime T cell responses to SARS-CoV-2 infection, which raises the possibility that different circulating coronavirus strains may provide some level of pre-existing cross-protective T cell immunity. However, the role of such heterologous T cell immune responses in COVID-19 remains to be established. This can be exemplified by SARS-CoV-2-specific CD8+ T cells directed at the immunodominant epitope HLA-B7N105–113 with a limited capacity to cross-recognize epitopes from related coronaviruses (23). Notably, the SARS-CoV-2 HLA-B7N105–113-restricted CD8+ T cell response is highly diverse and CD8+ T cells that specifically recognize the HLA-B7N105–113 epitope have been shown to be present in prepandemic controls (9). However, the naïve phenotype of these pre-existing CD8+ T cells suggests that they are not derived from prior exposure to related viruses (9).
Here, our data revealed that the HLA-A2S269–277-restricted CD8+ TCR repertoire can be characterized by a high level of structural conservation that governs antigen recognition despite significant sequence diversity in the selected TCR (9). We further demonstrated that the HLA-A2S269–277-restricted TCR repertoire is highly sensitive to peptide variations and therefore unlikely to cross-recognize homologous epitopes from other coronaviruses circulating in the human population. Using SPR measurements, we provided evidence that frequently selected HLA-A2S269–277-restricted TCRs bind to the HLA-A2S269–277 epitope with affinities in the typical range for viral epitopes, but alterations of the HLA-A2S269–277 epitope either had a strong negative impact on TCR binding or completely abrogated interactions. Together with our ex vivo tetramer staining data, our study thus showed that the HLA-A2S269–277-restricted TCR repertoire is unable to effectively cross-react with the SARS-CoV-2 S269–277 epitope variants or closely related homologous epitopes from other beta-coronaviruses, providing important insights into potential viral escape from immunodominant SARS-CoV-2-specific CD8+ T cell responses.
Experimental procedures
Study participants and ethics statement
Convalescent COVID-19 individuals were recruited via the Alfred Hospital or University of Melbourne. Peripheral blood was collected in heparinized tubes, and PBMCs were isolated via Ficoll-Paque separation. Experiments conformed to the Declaration of Helsinki principles and the Australian National Health and Medical Research Council Code of Practice. Written informed consents were obtained from all blood donors before the study. The study was approved by the Alfred Hospital (#280/14) and the University of Melbourne (#2057366.1, #2056901.1, #2056689, #2056761, #1442952, #1955465, and #1443389).
Ex vivo dual tetramer–associated magnetic enrichment
PBMCs (2.4–10 × 106) were stained with WT HLA-A2S269–277-PE and variant HLA-A2S269–277-APC tetramers at room temperature for 1 h in MACS buffer (PBS with 0.5% BSA and 2 mM EDTA). Cells were then incubated with anti-PE and anti-APC microbeads (Miltenyi Biotec), and tetramer+ cells were enriched using magnetic separation (9). Lymphocytes were stained with anti-CD71-BV421 (#562995), anti-CD4-BV650 (#563875), anti-CD27-BV711 (#563167), anti-CD38-BV786 (#563964), anti-CCR7-AF700 (#561143), anti-CD14-APC-H7 (#560180), anti-CD19-APC-H7 (#560177), anti-CD45RA-FITC (#555488), anti-CD8-PerCP-Cy5.5 (#565310), anti-CD95-PE-CF594 (#562395), anti-PD1-PE-Cy7 (#561272) (BD Biosciences), anti-CD3-BV510 (#317332), anti-HLA-DR-BV605 (#307640) (BioLegend), and LIVE/DEAD near-infrared stain (#L10119, Invitrogen) for 30 min, fixed with 1% PFA before acquiring data on an LSRII Fortessa (BD Bioscience). FCS files were analyzed using FlowJo v10 software.
Protein production and purification
Peptides used in this study were >85% pure and purchased from Mimotopes Mulgrave and GL Biochem. Human β2m micro-globulin and HLA-A2 heavy chain with and without C-terminal BirA tag were expressed as inclusion bodies in E. coli BL21(DE3) and refolded and purified as previously described (9). Briefly, 60 mg HLA-A2 or HLA-A2BirA heavy chain, 20 mg β2m, and 6 mg peptide were incubated overnight at 4 °C in 500 ml refolding buffer containing 3 M Urea, 100 mM Tris (pH 8.0), 400 mM arginine, 2 mM EDTA, 0.1 mM PMSF, one complete protease inhibitor tablet (Roche), and 0.5 mM oxidized and 5 mM reduced glutathione (for HLA-A2BirA refolding, 1 mM oxidized and 10 mM reduced glutathione were used). Refolded HLA-A2–peptide complex was purified with diethylaminoethyl anion-exchange chromatography and S200 size-exclusion chromatography. HLA-A2BirA refolded with WT, mutant, and variant peptides were biotinylated using BirA ligase before tetramer staining and SPR experiments. TCR variable domain sequences were obtained as E. coli codon-optimized synthetic DNA and cloned into pET30 expression vectors containing TCR α and β constant domains with an engineered interchain disulfide linkage. TCR α and β chains were expressed as inclusion bodies in E. coli and refolded and purified as previously described (24). Briefly, TCRs were refolded by incubating 60 mg each of TCR α- and β-chain in 1 l refolding buffer containing 5 M urea at 4 °C for 2 days. Refolded TCRs were purified via diethylaminoethyl anion-exchange, S200 size-exclusion, hydrophobic-interaction (HiTrap-Phenyl), and strong anion-exchange (HiTrap-Q) chromatography.
Crystallization, data collection, and structure determination
Crystals of the HLA-A2S269–277 binary and NR1C–HLA-A2S269–277 ternary complex were grown using the hanging-drop vapor diffusion method at 20 °C using a 1:1 ratio protein and reservoir solution. Binary complex crystals were obtained using a protein concentration of 5.0 mg/ml and a reservoir solution containing 0.2 M sodium thiocyanate and 20% PEG 3350. Ternary complex crystals were obtained using a protein concentration of 12 mg/ml and a reservoir solution containing 0.1 M Tris (pH 8.0), 0.2 M sodium thiocyanate, and 20% PEG 3350. Before data collection, crystals of the binary and ternary complexes were gradually transferred into the mother liquor supplemented with 20% (w/v) cryoprotectant ethylene glycol or PEG-400, respectively, and flash-frozen in liquid nitrogen. X-ray diffraction data were collected at MX-2 beamline of Australian Synchrotron on a Dectris Eiger detector. Data were processed using XDS (25) and Aimless from the CCP4 program suite (26). The structures were solved via molecular replacement in Phaser (27), and structure models were built via iterative rounds of model building in Coot (28) and restrained refinement in Phenix (29). TCR variable domains were numbered according to the IMGT system (30), and graphical representations were produced in PyMOL (version 2.2, Schrodinger, LCC).
SPR
SPR measurements were performed on a Biacore T200 instrument (GE Healthcare) at 25 °C in 10 mM Hepes (pH 7.5), 150 mM NaCl, 1 mM EDTA, and 0.005% surfactant P20. Biotinylated HLA-A2BirA complexes of HLA-A2S269–277 and variants and HLA-A2PB1 (31) as negative control were immobilized at a flow rate of 5 μl/min on a streptavidin sensor chip (GE Healthcare) to a surface loading of 1500 to 2000 response units, followed by injection of 200 μM biotin. TCR binding to HLA-A2S269–277 and variants was determined by passing serial dilutions of purified TCRs over the chip at a flow rate of 10 μl/min for 60 s followed by 120-s dissociation time. Two independent experiments with two replicates were performed for each TCR, and equilibrium dissociation constants KD were determined by fitting a single-site-binding model to the data.
Data availability
The structures were deposited in the PDB database (PDB codes 7N6D for the HLA-A2S269–277 binary and 7N6E for the NR1C-HLA-A2S269–277 ternary complexes).
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We thank all the participants involved in the study, Joseph Toressi, Allen Cheng, Robyn Esterbauer, Hannah Kelly, Jane Batten, and Helen Kent for support with the cohorts. We thank Jill Garlick, Janine Roney, Anne Paterson, and the research nurses at the Alfred Hospital. We thank the Monash Molecular Crystallisation Facility for assistance with crystallization. The protein crystal X-ray diffraction data were collected on MX2 beamline at the Australian Synchrotron facility, Melbourne, Australia.
Author contributions
P. C., K. K., J. R., and J. P. conceptualization; P. C., T. H. O. N., L. C. R., J. A. J., K. K., and J. P. formal analysis; P. C., J. R., and J. P. supervision; P. C., S. J. K., K. K., and J. R. funding acquisition; P. C., T. H. O. N., L. C. R., J. A. J., and J. P. investigation; P. C. and T. H. O. N. methodology; P. C., K. K., J. R., and J. P. writing–original draft; P. C., K. K., J. R., and J. P. project administration; P. C., K. K., J. R., and J. P. writing–review and editing; J. A. J., A. K. W., S. J. K., and J. R. resources; J. P. validation.
Funding and additional information
This work was supported by the NHMRC Leadership Investigator Grant to K. K. (1173871), NHMRC Emerging Leadership Level 1 Investigator Grant to T. H. O. N. (#1194036), Research Grants Council of the Hong Kong Special Administrative Region, China (#T11-712/19-N) to K. K., and MRFF Award (#2005544) to K. K., S. J. K., A. K. W., and J. A. J. A. K. W. are supported by Emerging Leadership 1 Investigator Grant (#1173433), J. A. J. by an NHMRC Early Career Fellowship (ECF) (#1123673), and S. J. K. by NHMRC Senior Principal Research Fellowship (#1136322). J. R. is supported by an ARC Laureate fellowship.
Edited by Peter Cresswell
Contributor Information
Katherine Kedzierska, Email: kkedz@unimelb.edu.au.
Jamie Rossjohn, Email: Jamie.rossjohn@monash.edu.
Jan Petersen, Email: jan.petersen@monash.edu.
Supporting information
References
- 1.Sattler A., Angermair S., Stockmann H., Heim K.M., Khadzhynov D., Treskatsch S., Halleck F., Kreis M.E., Kotsch K. SARS-CoV-2-specific T cell responses and correlations with COVID-19 patient predisposition. J. Clin. Invest. 2020;130:6477–6489. doi: 10.1172/JCI140965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bonifacius A., Tischer-Zimmermann S., Dragon A.C., Gussarow D., Vogel A., Krettek U., Godecke N., Yilmaz M., Kraft A.R.M., Hoeper M.M., Pink I., Schmidt J.J., Li Y., Welte T., Maecker-Kolhoff B. COVID-19 immune signatures reveal stable antiviral T cell function despite declining humoral responses. Immunity. 2021;54:340–354.e346. doi: 10.1016/j.immuni.2021.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.A., Shankar N., Lim J.M.E., Sun L.J., Zahari M., Tun Z.M., Kumar V. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adamo S., Chevrier S., Cervia C., Zurbuchen Y., Raeber M.E., Yang L., Sivapatham S., Jacobs A., Bächli E., Rudiger A., Stüssi-Helbling M., Huber L.C., Schaer D.J., Bodenmiller B., Boyman O. Lymphopenia-induced T cell proliferation is a hallmark of severe COVID-19. bioRxiv. 2020 doi: 10.1101/2020.08.04.236521. [preprint] [DOI] [Google Scholar]
- 5.Zheng H.Y., Zhang M., Yang C.X., Zhang N., Wang X.C., Yang X.P., Dong X.Q., Zheng Y.T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell Mol. Immunol. 2020;17:541–543. doi: 10.1038/s41423-020-0401-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diao B., Wang C., Tan Y., Chen X., Liu Y., Ning L., Chen L., Li M., Liu Y., Wang G., Yuan Z., Feng Z., Zhang Y., Wu Y., Chen Y. Reduction and functional exhaustion of T cells in patients with coronavirus disease 2019 (COVID-19) Front. Immunol. 2020;11:827. doi: 10.3389/fimmu.2020.00827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Amaya Hernandez S.P., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8(+) T cell activation in COVID-19 patients. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thevarajan I., Nguyen T.H.O., Koutsakos M., Druce J., Caly L., van de Sandt C.E., Jia X., Nicholson S., Catton M., Cowie B., Tong S.Y.C., Lewin S.R., Kedzierska K. Breadth of concomitant immune responses prior to patient recovery: A case report of non-severe COVID-19. Nat. Med. 2020;26:453–455. doi: 10.1038/s41591-020-0819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen T.H.O., Rowntree L.C., Petersen J., Chua B.Y., Hensen L., Kedzierski L., van de Sandt C.E., Chaurasia P., Tan H.X., Habel J.R., Zhang W., Allen L.F., Earnest L., Mak K.Y., Juno J.A. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity. Immunity. 2021;54:1066–1082.e5. doi: 10.1016/j.immuni.2021.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koutsakos M., Rowntree L.C., Hensen L., Chua B.Y., van de Sandt C.E., Habel J.R., Zhang W., Jia X., Kedzierski L., Ashhurst T.M., Putri G.H., Marsh-Wakefield F., Read M.N., Edwards D.N., Clemens E.B. Integrated immune dynamics define correlates of COVID-19 severity and antibody responses. Cell Rep. Med. 2021;2:100208. doi: 10.1016/j.xcrm.2021.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., Nakao C., Rayaprolu V., Rawlings S.A., Peters B., Krammer F. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., Marrama D., de Silva A.M., Frazier A., Carlin A.F., Greenbaum J.A. Targets of T cell responses to SARS-CoV-2 coronavirus in humans with COVID-19 disease and unexposed individuals. Cell. 2020;181:1489–1501.e1415. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weiskopf D., Schmitz K.S., Raadsen M.P., Grifoni A., Okba N.M.A., Endeman H., van den Akker J.P.C., Molenkamp R., Koopmans M.P.G., van Gorp E.C.M., Haagmans B.L., de Swart R.L., Sette A., de Vries R.D. Phenotype and kinetics of SARS-CoV-2-specific T cells in COVID-19 patients with acute respiratory distress syndrome. Sci. Immunol. 2020;5 doi: 10.1126/sciimmunol.abd2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W., Whitton H.J., Virbasius A., Olivier K.J., Jr., Buckner L.R., Alistar A.T. Unbiased screens show CD8(+) T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53:1095–1107.e1093. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., Koutsakos M., Hensen L., Jia X., Chua B., Zhang W., Tan H.-X., Flanagan K.L., Doolan D.L., Torresi J. Suboptimal SARS-CoV-2-specific CD8(+) T cell response associated with the prominent HLA-A∗02:01 phenotype. Proc. Natl. Acad. Sci. U. S. A. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., Lopez-Camacho C., Slon-Campos J., Zhao Y., Stuart D.I., Paesen G.C. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar, Daul F., Salvat Lago M., Decker A., Luxenburger H., Binder B., Bettinger D., Sogukpinar O., Rieg S. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 18.Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P., Benner S.E., Patel E.U., Littlefield K., Laeyendecker O., Shoham S. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Invest. 2021;131 doi: 10.1172/JCI145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shomuradova A.S., Vagida M.S., Sheetikov S.A., Zornikova K.V., Kiryukhin D., Titov A., Peshkova I.O., Khmelevskaya A., Dianov D.V., Malasheva M., Shmelev A., Serdyuk Y., Bagaev D.V., Pivnyuk A., Shcherbinin D.S. SARS-CoV-2 epitopes are recognized by a public and diverse repertoire of human T cell receptors. Immunity. 2020;53:1245–1257.e1245. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Petersen J., Ciacchi L., Tran M.T., Loh K.L., Kooy-Winkelaar Y., Croft N.P., Hardy M.Y., Chen Z., McCluskey J., Anderson R.P., Purcell A.W., Tye-Din J.A., Koning F., Reid H.H., Rossjohn J. T cell receptor cross-reactivity between gliadin and bacterial peptides in celiac disease. Nat. Struct. Mol. Biol. 2020;27:49–61. doi: 10.1038/s41594-019-0353-4. [DOI] [PubMed] [Google Scholar]
- 21.Valkenburg S.A., Josephs T.M., Clemens E.B., Grant E.J., Nguyen T.H., Wang G.C., Price D.A., Miller A., Tong S.Y., Thomas P.G., Doherty P.C., Rossjohn J., Gras S., Kedzierska K. Molecular basis for universal HLA-A∗0201-restricted CD8+ T-cell immunity against influenza viruses. Proc. Natl. Acad. Sci. U. S. A. 2016;113:4440–4445. doi: 10.1073/pnas.1603106113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu Y., McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lineburg K.E., Grant E.J., Swaminathan S., Chatzileontiadou D.S.M., Szeto C., Sloane H., Panikkar A., Raju J., Crooks P., Rehan S., Nguyen A.T., Lekieffre L., Neller M.A., Tong Z.W.M., Jayasinghe D. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity. 2021;54:1055–1065.e1055. doi: 10.1016/j.immuni.2021.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clements C.S., Kjer-Nielsen L., MacDonald W.A., Brooks A.G., Purcell A.W., McCluskey J., Rossjohn J. The production, purification and crystallization of a soluble heterodimeric form of a highly selected T-cell receptor in its unliganded and liganded state. Acta Crystallogr. D Biol. Crystallogr. 2002;58:2131–2134. doi: 10.1107/s0907444902015482. [DOI] [PubMed] [Google Scholar]
- 25.Kabsch W. XDS. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., Keegan R.M., Krissinel E.B., Leslie A.G., McCoy A., McNicholas S.J., Murshudov G.N., Pannu N.S., Potterton E.A., Powell H.R. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P., Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., McCoy A.J., Moriarty N.W., Oeffner R., Read R.J., Richardson D.C. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lefranc M.P., Giudicelli V., Ginestoux C., Jabado-Michaloud J., Folch G., Bellahcene F., Wu Y., Gemrot E., Brochet X., Lane J., Regnier L., Ehrenmann F., Lefranc G., Duroux P. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koutsakos M., Illing P.T., Nguyen T.H.O., Mifsud N.A., Crawford J.C., Rizzetto S., Eltahla A.A., Clemens E.B., Sant S., Chua B.Y., Wong C.Y., Allen E.K., Teng D., Dash P., Boyd D.F. Human CD8(+) T cell cross-reactivity across influenza A, B and C viruses. Nat. Immunol. 2019;20:613–625. doi: 10.1038/s41590-019-0320-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The structures were deposited in the PDB database (PDB codes 7N6D for the HLA-A2S269–277 binary and 7N6E for the NR1C-HLA-A2S269–277 ternary complexes).