Abstract
The receptor-binding domain (RBD) of SARS-CoV-2 spike (S) protein plays a vital role in binding and internalization through the alpha-helix (AH) of human angiotensin-converting enzyme 2 (hACE2). Thus, it is a potential target for designing and developing antiviral agents. Inhibition of RBD activity of the S protein may be achieved by blocking RBD interaction with hACE2. In this context, inhibitors with large contact surface area are preferable as they can form a potentially stable complex with RBD of S protein and would not allow RBD to come in contact with hACE2. Peptides represent excellent features as potential anti-RBD agents due to better efficacy, safety, and tolerability in humans compared to that of small molecules. The present study has selected 645 antiviral peptides known to inhibit various viruses and computationally screened them against the RBD of SARS-CoV-2 S protein. In primary screening, 27 out of 645 peptides exhibited higher affinity for the RBD of S protein compared to that of AH of the hACE2 receptor. Subsequently, AVP1795 appeared as the most promising candidate that could inhibit hACE2 recognition by SARS-CoV 2 as was predicted by the molecular dynamics simulation. The critical residues in RBD found for protein-peptide interactions are TYR 489, GLY 485, TYR 505, and GLU 484. Peptide-protein interactions were substantially influenced by hydrogen bonding and hydrophobic interactions. This comprehensive computational screening may provide a guideline to design the most effective peptides targeting the spike protein, which could be studied further in vitro and in vivo for assessing their anti-SARS CoV-2 activity.
Keywords: SARS-CoV-2, Antiviral peptide, Molecular dynamics simulation, Receptor-binding domain, Angiotensin converting enzyme 2
Graphical abstract
1. Introduction
A novel coronavirus is causing widespread respiratory tract infections and posing a serious threat to public life and health. Following a devastating first wave, it has emerged as even more dangerous and is causing havoc upon lives around the world. The international committee on taxonomy of viruses (ICTV) officially designated 2019 novel coronavirus (2019-nCov) as SARS-CoV-2 and the disease as COVID-19 [1,2]. This is the third time an animal to human transmission of deadly viruses has been witnessed in the past two decades [3]. The number of affected people and death toll due to COVID-19 are increasing day by day. As of July 26, 2021, 195 million people have been infected and 4.1 million are killed by this deadly virus [4].
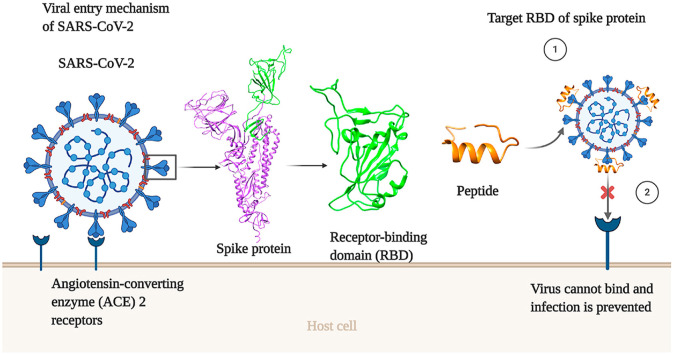
SARS-CoV-2 is a positive sense, single-stranded, enveloped, non-segmented RNA virus belonging to the coronaviridae family [5]. The genome size of SARS-CoV-2 ranges from 29.8 kb to 29.9 kb [6], which encodes four main structural proteins, comprising of spike (S), envelop (E), membrane (M), nucleocapsid (N) proteins, and 16 non-structural proteins (nsp) [5,7]. S protein is a highly N-glycosylated trimeric protein that covers the outer surface of SARS-CoV-2. Each monomer of S protein has a molecular weight of 180 kDa and consists of S1 and S2 subunits [[8], [9], [10]]. The S protein is involved in receptor recognition, membrane fusion, as well as the entry of the virus to host cells. It binds with human angiotensin-converting enzyme 2 (hACE2), which serves as the port of entry for the virus to host lung epithelial cells. hACE2 is found on the surface of many other cell types including epithelial tissues of upper and lower respiratory tracts [11,12]. The receptor-binding-domain (RBD) of S1 subunit binds with an alpha helix of the peptidase domain (PD) of the angiotensin-converting enzyme 2 (ACE2) [13,14], which subsequently triggers the fusion of viral and host cellular membrane by S2 subunit [15]. The RBD of S1 protein contains antiparallel β sheets (β1, β2, β3, β4 and β7) with short joining helices and loops forming the core. The shorter β5 and β6 strands, α4 and α5 helices, and loops are inserted between the β4 and β7 strands [16].
Drug design to counter COVID-19 can be directed towards targeting viral proteins or host-cell proteins. Designing drugs to target viral proteins have several benefits since they could be highly specific against the virus while maintaining minimal detrimental effects on host cells. Among the four structural proteins of SARS-CoV-2, design of inhibitors against S1 protein is an effective choice to arrest viral entry to the host cells, which is a key step in the virus infection cycle [17]. Evidently, SARS-CoV-2 exhibits a high nucleotide sequence similarity with SARS-CoV-1 (79.5%) as well as MERS (50%) [18]. Closest relatives, RaTG13-CoV and RmYN02-CoV, share 96.3% nucleotide identity in the whole genome sequence, ~97% nucleotide identity in the long 1 ab open reading frame (ORF1ab), respectively. Notably, S protein from SARS-CoV-1 and SARS-CoV-2 share critical residues within the RBD of the S1 subunit and bind to the same receptor, hACE2, for internalization [16,19,20]. Given the sequence similarity among different viruses, inhibitors of one virus show great promise as potential therapeutics against others. Moreover, with similarities between critical residues of RBDs, inhibitors known to arrest SARS-CoV-1 entry offer high potential to halt SARS-CoV-2 entry as well.
The binding interface of SARS-CoV-2 and hACE2 shares a high contact surface and contains a hydrogen bonding network as well as a hydrophobic region [21]. Therefore, as potential inhibitors of RBD S1 protein, peptides are superior to small molecules because peptide-protein interaction (PPI) has a large contact surface area [22]. As peptides have a molecular weights in between small molecules (<500 Da) and biologics (up to 150,000 Da), they exhibit some unique chemical features. Peptides are also amenable to chemical adjustment and can bind with PPIs that are therapeutically relevant [23]. This class of therapeutics have many advantages over small-molecule medications because they are highly selective, well-tolerated, have fewer side effects, and go through a shorter clinical development and FDA approval process [27,28]. Despite the challenges of short half-lives, rapid clearance, cost, and intravenous administration, several groups including ours are looking into the possibility of using an antiviral peptide to treat covid-19 [[24], [25], [26], [27]]. In this regard, peptide like molecules can be an ideal solution to inhibit RBD S1 and potentially inhibit RBD-hACE2 interaction. In a previous study, the effectiveness of peptides against the S1 protein of SARS-CoV-1 was established [28]. In another study, a corresponding hexapeptide to the ACE-interacting domain of SARS-CoV-2 (AIDS) has been found to disrupt the association between RBD-ACE2 in mice [29]. Karoyan and colleagues designed peptide-mimics which has been found to inhibit S protein with inhibitory concentration (IC50) in nanomolar range [30]. In a previous work, we have computationally demonstrated that peptides known to inhibit RBD of SARS-CoV-1 S1 protein shows great promise against SARS-CoV-2 [31].
In this research, we extended our search to include 645 peptides, experimentally known to inhibit a wide variety of viruses, and computationally screened them against the RBD of SARS-CoV-2 S1 protein. Only RBD of S1 was chosen as a therapeutic target instead of RBD-hACE2 complex, with the primary hypothesis that peptide inhibitors with stronger affinity for RBD than that of hACE2 would prevent the virus from host cellular entry. We employed state of the art computational tools to screen the selected antiviral peptides, which is to the best of our knowledge, the largest data set screened against SARS-CoV-2 S1 protein. Based on interaction and binding affinity, peptides that exhibited greater binding affinity than that of the alpha-helix of hACE2 peptidase were further investigated by molecular dynamics simulation, principal component analysis, and binding free energy landscape. Our research aims to provide a potential framework for designing and developing promising anti-SARS-CoV-2 peptide therapeutics.
2. Methods
2.1. Molecular docking
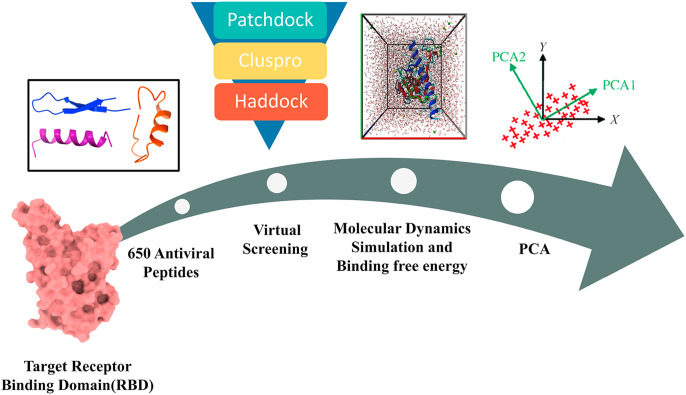
A total of 645 antiviral peptides were taken from the Antiviral Peptide Databases (AVPdb) [32]. These peptides have been experimentally tested against various viruses to interrupt virus cohesion or entry into the host cell. The 3D structure of these peptides were modeled using the PEP-fold 3.5 program [33,34]. The crystal structure of the RBD bound with hACE2 was obtained from the RSCB Protein Data Bank (PDB ID: 6M0J), having a structural resolution of 2.45 Å [16]. The RBD (6M0J) was subjected to molecular docking with the modeled antiviral peptides using Patchdock [35]. The top 1000 RBD-peptide conformers for each complex generated from Patchdock were refined by Firedock [36,37]. In addition, docking was also performed with two other programs, including ClusPro 2.0 [38] and HADDOCK 2.2 [39].
2.2. Molecular dynamics (MD) simulations
150 ns MD simulation was conducted for Apo-RBD, and AH, AVP1795, AVP1775, AVP1784, AVP0673, AVP1869, AVP1871 in complex with RBD by using YASARA Dynamics software. AMBER 14 force field was assigned for the analysis to describe the complexes in the simulated system [40,41]. For automatic parameterization of the force field, YASARA uses AutoSMILES option to incorporate semi-empirical AM1 point charge, general AMBER force field (GAFF) atom types and parameters. The TIP3P model was used to represent the water molecules in the system, and Na+/Cl− were added to it [42]. To carry out the simulation, the condition of periodic boundaries was incorporated, maintaining the cell size of 20 Å larger than the protease size. The box size was maintained at X = 76.5688, Y = 59.6229, Z = 47.1462 Å along three axes. The Particle Mesh Ewald (PME) [43] method was applied to account for the long-range electrostatic interactions at an 8 Å cut off distance. To incorporate the physiological condition of the simulated system at 310 K and pH 7.4, 0.9% NaCl was used [44]. For the initial energy minimization process of each simulation system, a simulated annealing method with the steepest gradient approach was used (5000 cycles). A multi-step algorithm was used to select a simulation time step of 1.25 fs [45]. During the MD simulation of 150 ns for each system, snapshots were saved at every 100 ps. Bond angle, dihedral angle, bond distance, van der Waals and columbic interactions, root-mean-square-deviation (RMSD), root-mean-square-fluctuation (RMSF), solvent-accessible surface area (SASA) and values for alpha carbon, backbone and heavy atoms were calculated from MD trajectories. During 150 ns, MD snapshots were collected to evaluate the interactions between peptide-protein complexes. 150 MD snapshots have been selected for calculating the binding free energy for each complex by the PRODIGY server [46], which predicts the binding affinity in biological complexes.
Principal component analysis (PCA). PCA of MD is a special strategy that can interpret the adjustment of structural qualities of proteins in the presence of peptides in a reduced dimension using different multivariate energy functions [47,48]. Factors assembling the structural and energy information are bond distances, dihedral angles, bond angles, planarity, electrostatic energies, and van der Waals energies. PCA can elicit the concealed structure and energy outline among various groups from an MD trajectory [49,50]. In this study, the last 50 ns of MD trajectory data were considered for PCA analysis. Prior to this analysis, the data were preprocessed by centering and scaling. PCA explains the multivariate report ascertained in an X matrix into a product of two unique matrices, as described in the following equation,
Where, is the matrix of score representing the relation of samples. is the loadings matrix, which contains information about the relationship of variables to one another, k is the number of factors in the model, and E is the unmodeled variance. To execute the calculation, in-house developed codes R [51] and RStudio [52] were applied. For originating plots, the Package Factoextra [53] was used.
2.3. Single residue tracking analysis
The secondary structure elements of highest and lowest frequency residues of the peptides (AH, AVP1795, AVP1775, AVP1784, AVP0673) with RBD during 150 ns MD simulation were analyzed using the STRIDE web interface [54]. For this analysis, a total of 151 MD snapshots for each complex were considered. Briefly, the residues with highest and lowest interaction frequency for AH, AVP1795, AVP1775, AVP1784, and AVP0673 were determined upon analysis of MD snapshots of each of these peptides in the complex with RBD at each time point of the 150 ns MD simulation. Subsequently, the secondary structures of these residues (alpha helix, 310 helix, beta sheet, beta turn, and coil) were determined by using the STRIDE web server and examined over time.
3. Results and discussion
3.1. Peptide binding affinity and interactions in peptide-protein docking
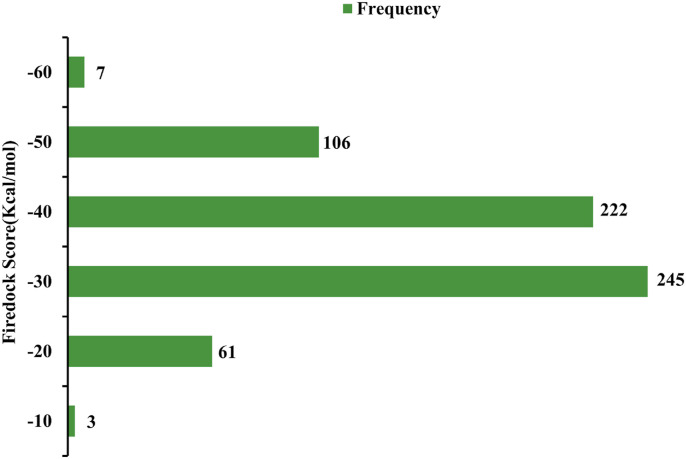
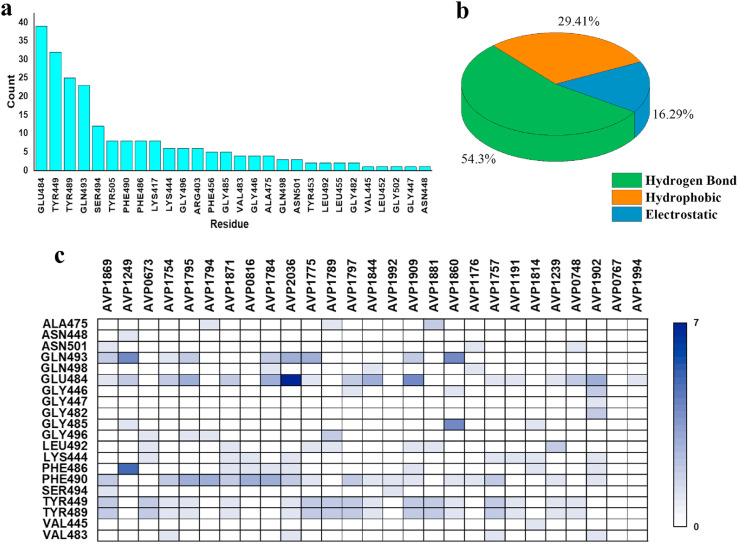
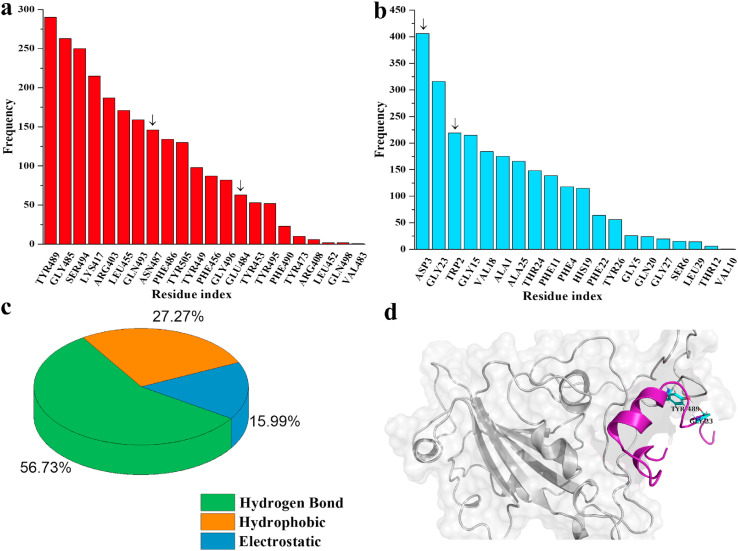
Initially, 645 peptides that are known to inhibit several viruses were screened against the binding pocket of receptor-binding domain (RBD) of SARS CoV-2 S1 protein using Patchdock and Firedock servers (Table S1). Fig. 1 summarizes the entire workflow, tools, and software used for screening the antiviral peptides. The amino acid sequence, length, and efficacy of the selected peptides against corresponding cell lines, retrieved from Antiviral Peptide Databases (AVPdb), are also presented in Table S1. The alpha helix (AH) of hACE2 peptidase domain served as a control peptide. The distribution of binding affinities of 645 peptides are summarized in Fig. 2 . The binding affinity and non-covalent interactions of 27 peptides out of 645 that exhibited substantially higher binding affinity than that of AH of the hACE2 (−36.74 kcal/mol) were further verified using ClusPro 2.0 and HADDOCK 2.2 docking protocols (Table 1 ). Upon analysis of binding pose and non-bonding interactions, AVP1795, AVP1775, AVP1871, AVP1869, AVP1784, and AVP0673 were revealed as the most promising inhibitors of RBD of S1 protein (Fig. 3 ). In RBD interactions with the 27 superior binding peptides, residues, such as GLU 484, TYR 449, TYR 489, GLN 493, and SER 494 of RBD were the key contributors of multiple non-covalent interactions (Fig. 4 a and c) and the types of interactions were predominantly hydrogen bonds (54.3%) (Fig. 4b).
Fig. 1.
Flowchart for methodology.
Fig. 2.
Frequency distribution of all antiviral peptides over a range of docking scores.
Table 1.
Docking result of the 27 selected peptides that showed higher affinity towards RBD of SARS CoV-2 spike protein.
| Peptide ID | FireDock Score | ClusPro 2.0 Score | HADDOCK Score |
|---|---|---|---|
| AVP1249 | −68.08 | −925.6 | −94.7 ± 3.4 |
| AVP1757 | −66.89 | −810.4 | −70.1 ± 2.3 |
| AVP1239 | −65.64 | −751.6 | −64.8 ± 4.5 |
| AVP0816 | −65.03 | −776.1 | −79.1 ± 2.5 |
| AVP1191 | −63.58 | −694 | −66.2 ± 5.3 |
| AVP1795 | −62.94 | −755.9 | −81.5 ± 12.8 |
| AVP1794 | −60.02 | −768.9 | −79.6 ± 6.1 |
| AVP1909 | −59.9 | −843 | −74.5 ± 3.9 |
| AVP1871 | −59.72 | −972.3 | −79.3 ± 2.7 |
| AVP1869 | −59.66 | −702.3 | −99.8 ± 4.3 |
| AVP1994 | −59.56 | −659.1 | −49.5 ± 6.9 |
| AVP1754 | −59.48 | −805.1 | −89.4 ± 3.6 |
| AVP1814 | −59.14 | −674.3 | −65.9 ± 4.0 |
| AVP1775 | −58.87 | −677.5 | −78.2 ± 7.7 |
| AVP1860 | −58.65 | −809.5 | −71.5 ± 6.3 |
| AVP1881 | −58.37 | −798.1 | −73.1 ± 6.1 |
| AVP2036 | −58.34 | N | −78.3 ± 5.9 |
| AVP1902 | −58.17 | N | −58.2 ± 8.6 |
| AVP1789 | −57.92 | −899.2 | −77.8 ± 5.7 |
| AVP0767 | −57.77 | −588.2 | −54.3 ± 5.5 |
| AVP0673 | −57.74 | −883.4 | −94.1 ± 3.5 |
| AVP1176 | −57.73 | −816.1 | −71.3 ± 7.9 |
| AVP1844 | −57.63 | −769.5 | −80.0 ± 1.8 |
| AVP1992 | −57.6 | −784.3 | −77.3 ± 2.8 |
| AVP1797 | −57.35 | −659.8 | −77.6 ± 3.1 |
| AVP0748 | −57.33 | −662.3 | −64.4 ± 1.6 |
| AVP1784 | −57.12 | −863.6 | −82.9 ± 3.8 |
| α1 helix of peptidase domain of RBD | −36.74 | −749.6 | −70.2±11.0 |
N = No desired binding interaction is obtained.
Fig. 3.
Structure and sequences of the six best peptides obtained from docking protocols.
Fig. 4.
Binding interaction. (A) Interacting residues of RBD; (B) Distribution of non-covalent interactions; (C) Residue-residue Contact of the peptide-RBD docked complexes.
3.2. Structural and energy parameters of peptides-RBD complexes in molecular dynamics (MD) simulation
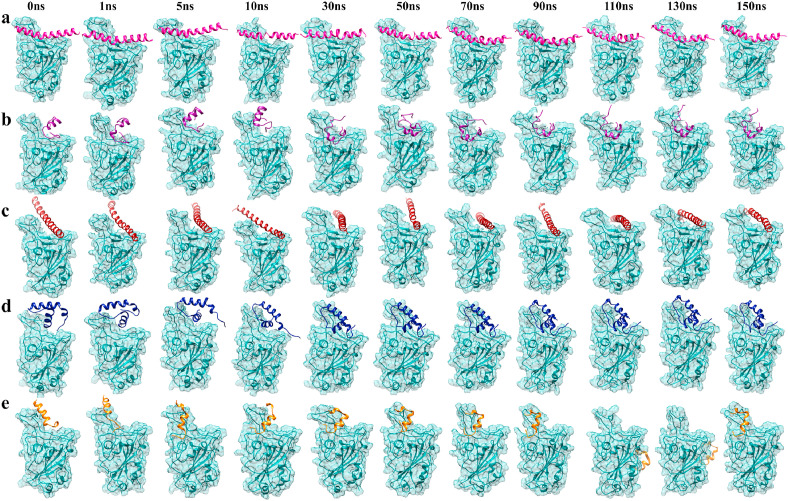
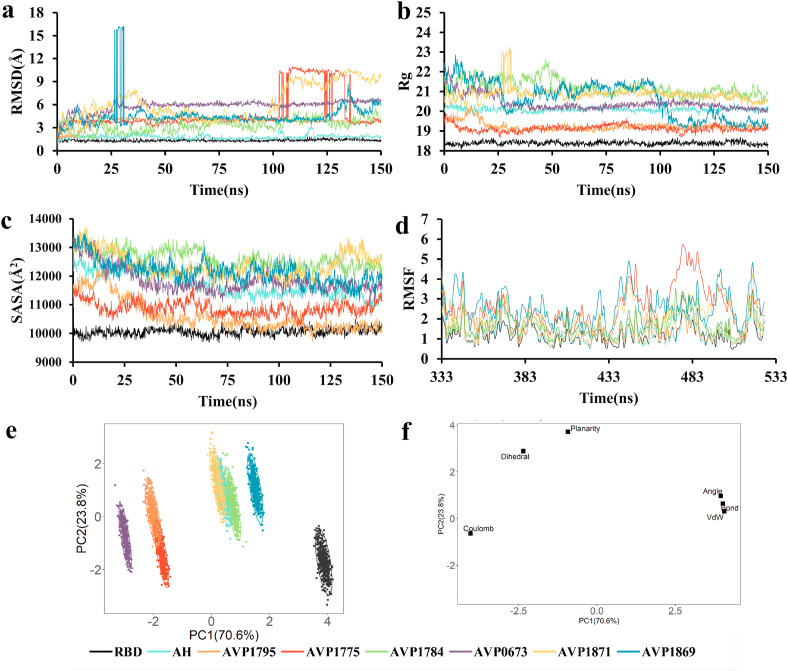
The peptides-RBD complexes of the six best peptides (AVP1795, AVP1775, AVP1871, AVP1869, AVP1784, and AVP0673) from molecular docking were examined during 150 ns of MD simulation. Several structural parameters including RMSD, Rg, SASA, bond, angle, dihedral, planarity and RMSF as well as energy parameters such as coulomb, and van der Waals energies of the complexes relative to AH-RBD and apo-RBD were calculated from MD trajectories. RMSD, Rg, SASA of all the complexes including apo-RBD underwent an initial hike at time 0, which was due to equilibration of the macromolecular complexes in a simulated physiological environment. Subsequently, the complexes took different times to attain an average conformation, and with time showed major or minor upward or downward shifts based on the complex stability. Upon RMSD analysis for all the 7 complexes and apo-RBD, it was observed that apo-RBD quickly reached a stable conformation and maintained it during the entire simulation period. AH-RBD attained its average conformation at around 5 ns with an average RMSD a little higher (~1.6 Å) than that of apo-RBD (~1.3 Å). Unlike, apo-RBD, RMSD values of AH-RBD showed minor upward shifts at different time points. Among the test complexes, AVP1871-RBD never attained a stable conformation, continuously showing major upward shifts. On the contrary, AVP1869-RBD reached a pseudo stable conformation with RMSD around 4 Å at 50 ns, which continued until ~130 ns only to again become unstable. AVP1784-RBD was the closest of all to AH-RBD in terms of RMSD; however, it showed continuous minor fluctuations around 3 Å during the full course of simulation. AVP1775-RBD reached an average conformation rather quickly (at 5 ns); however, during the simulation period it experienced fluctuations at different time points and was dislocated finally from the binding pocket at around 100 ns (Fig. 6e). AVP1795-RBD and AVP0673-RBD showed similar stabilization times (~20 ns) and maintained the average structures at ~6 Å and ~4 Å, respectively. AVP1795-RBD showed minimum fluctuations once it attained stability when compared to that of the other complexes (Fig. 5 a).
Fig. 6.
Representative snapshots (a) AH-RBD; (b) AVP1795-RBD; (c) AVP1784-RBD; (d) AVP0673-RBD; and (e) AVP1775-RBD over the course of 150 ns simulations. RBD is shown in cyan color.
Fig. 5.
Molecular dynamics simulation (a) Root-mean-square deviation (RMSD); (b) Radius of gyration (Rg); (c) Solvent accessible surface area (SASA); and (d) Root-mean-square fluctuation (RMSF); (e) Score plot and (f) Loading plot of top eight peptide.
The radius of gyration (Rg) exhibits the overall dimension of a peptide-protein conformation, which is also a measure of complex compactness. During the simulation, the AVP1784-RBD displayed fluctuation as well as the highest Rg (Fig. 5b). With 41 residues, a larger RMSD is justified for AVP1784, however it was not the highest one. AVP0673 consisting of 45 residues, showed comparable Rg values to that of AH-RBD (33 residues). The folded structure of AVP0673 might help with better placement in the binding groove of RBD as opposed to the linear helical structure of AVP1784 (Fig. 6c and d). Interestingly, AVP1795-RBD and AVP1775-RBD showed lower Rg than that of apo-RBD, indicating better compaction of these complexes compared to that of AH-RBD. When AVP1795-RBD and AVP1775-RBD were compared in terms of Rg, AVP1795-RBD was better once the complex attained a stable conformation at around 20 ns (Fig. 5b). Since AVP1775 is dislocated from the binding pocket at around 100 ns, a lower observed Rg in its case does not make it a better inhibitor of RBD. Although the residue count in AVP1795 (29 residues) was close to that of AH (33 residues), it showed substantially lower Rg than that of AH, which could be due to a better shape complementary between AVP1795 and RBD than that of AH and RBD (Fig. 3, Fig. 6b).
The protein flexibility for the solvent accessible surface area (SASA) was calculated to clarify the deviation imposed in RBD structure by the peptides binding. The highest amount of SASA was created upon 1784 binding to RBD, which was higher than that of AVP0673, although AVP0673 has a greater residue count (45 residues) than that of AVP1784 (41 residues) (Fig. 5c). AVP0673-RBD and AH-RBD showed a comparable SASA profile. However, an area of attenuation was observed in case of AVP0673-RBD compared to that of AH-RBD. With 45 residues, AVP0673 was presumed to create more SASA than that of AH (33 residues) upon binding to RBD. The folded shape of AVP0673 as opposed to the linear helical shape of AH might explain the observed difference (Fig. 3, Fig. 6d). AVP1795-RBD and AVP1775-RBD showed lower newly created SASA than that of AH-RBD. Unlike AVP1775-RBD, AVP1795-RBD mimicked the SASA profile of apo-RBD once it attained stability, indicating substantial attenuation in complex volume (Fig. 5c).
Root-mean-square fluctuation (RMSF) demonstrates how each amino acid residue present in a protein structure, or a complex fluctuates over the simulation. In the apo-RBD structure, regions such as 366–374, 382–388, 476–487 and 517–519 were noted to be zones with the greatest fluctuation. Upon AH or AVP1795 binding to RBD, the fluctuations in the 366–374 region substantially increased. AVP0673-RBD and AVP1784-RBD showed a similar pattern in fluctuation to that of AH-RBD or AVP1795-RBD; however, with higher fluctuations in each residue. AVP1775-RBD showed a strikingly different RMSF profile than that of the rest (Fig. 5d).
A PCA model containing eight training sets (Apo-RBD, AH-RBD, and six peptide-RBD complexes) was developed to unveil dissimilarities in energy landscape among Apo-RBD and different peptide-RBD complexes during the MD simulation. The generated PCA model reliably explained the variance among the eight training sets, as it altogether explained 94.4% of total variance (PC1 70.6% and PC2 23.8%). With a PCA scores plot, the complexes were clustered to visualize their energy landscape, where each dot represents one time point. A PCA loading plot explained the shifts of clusters along PC1 or PC2 in the scores plot. Looking at the PCA scores plot, a pronounced shift was observed in clusters of the peptide-protein complexes relative to apo-RBD along both PC1 and PC2 (Fig. 5e). The clustering of AVP1795-RBD at the far right indicates the highest change in coulomb energy upon binding of AVP1795 to RBD. On the contrary, this complex experienced minimal change in dihedral and planarity energies, indicating minimal structural change upon the peptide binding. Changes in coulomb energy were relatively less in case of AVP0673-RBD and AVP1775-RBD, which also showed a leftward shift along PC1 compared to that of apo-RBD. The other complexes including AH-RBD showed both leftward and upward shifts relative to apo-RBD, indicating changes in coulomb, dihedral and planarity energies upon peptides binding (Fig. 5f). Notably, change in coulomb energy was substantially lower in AH-RBD compared to that of AVP1795-RBD, which indicates stronger coulombic interactions in AVP1795-RBD complex. This observation further substantiates the lower Rg and SASA values of the AVP1795-RBD complex.
3.3. Binding free energy and interaction frequency
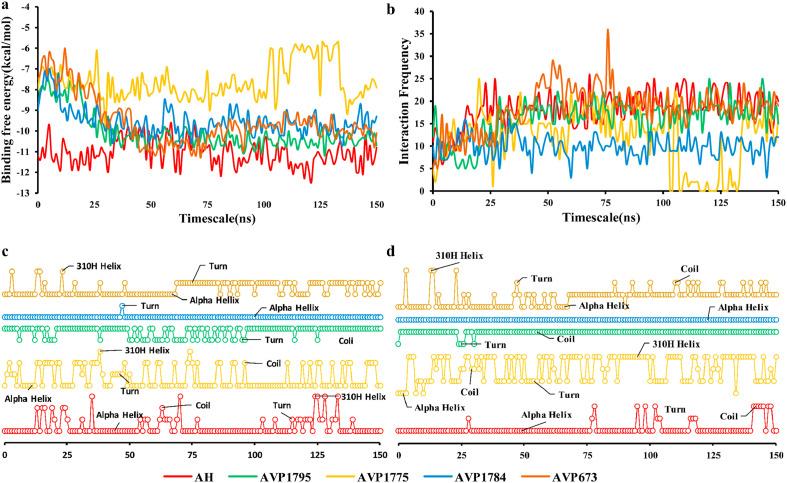
The binding free energy was calculated for four peptide-protein complexes (AVP1795-RBD, AVP1784-RBD, AVP1775-RBD, and AVP0673-RBD) including AH-RBD. The binding free energy of the forerunning peptide, AVP1795, was close to that of AH (Fig. 7 a). The average binding affinities of AVP1795-RBD (−10.04 ± 0.87) were found to be better than those of AVP0673-RBD, AVP1784-RBD, and AVP1775-RBD (Fig. S1). Interaction counts of AH, AVP1795, AVP1784, AVP1775 and AVP0673 were analyzed, and those of AVP1795 and AVP0673 closely matched that of AH. AVP1784 and AVP1775 showed lower interaction counts than the others (Fig. 7b).
Fig. 7.
Binding free energies, interactions, and secondary structure. (a) Distribution of binding free energies (Kcal/mol); (b) frequency of interaction (hydrogen, hydrophobic, electrostatic); Secondary structure of the (c) highest and (d) lowest frequency peptide residues.
3.4. Secondary structure analyses
Secondary structural elements of representative residues (i.e., residues that showed the highest and lowest interaction counts during the 150 ns MD simulation) were analyzed for AVP1795, AVP0673, AVP1784 and AVP1775 compared to that of AH. In single residue tracking, both the highest and lowest frequency residues of AVP1775 (LEU 5 and ASP14) and AVP0673 (GLU 11 and PHE 12) experienced very frequent shifting between different secondary structures (alpha helix, 310 helices, coil, turn), unlike that of AH, AVP1795, and AVP1784. The frequent shifts in secondary structural elements of representative residues of these peptides structures is a mark of their inherent instability in those structures (Fig. 7c and d). This was further substantiated by the observed overall secondary structural changes of AVP1775-RBD and AVP0673-RBD complexes (Fig. S2). On the other hand, both the highest and lowest frequency residues of AVP1784 experienced the least secondary structural flipping during the simulation period, supporting its lower interaction frequency with RBD (Fig. 7b) and reduced binding free energy compared to that of AH and AVP1795 (Fig. 7a).
3.5. Analysis of non-covalent interactions in peptides-RBD complexes obtained from MD simulation
Conformers were generated for four peptide-RBD complexes (AVP1795-RBD, AVP0673-RBD, AVP1784-RBD, and AVP1775-RBD) and AH-RBD at a 1 ns interval during the 150 ns MD simulation. Precise analysis of these conformers revealed participating RBD and peptide residues, types of interactions (hydrogen, hydrophobic, electrostatic), and the percent participation of a residue in its corresponding bond at each time point.
Analysis of AVP1795-RBD complex showed that TYR 489, GLY 485, and SER 494 are the major interacting residues of RBD, whereas ASN 487 and GLU 484 (participation in bond formation for ≥ 90% time of the MD simulation) tied the peptide in its binding groove. On the contrary, residues such as ASP 3 and TRP 2 (participation in bond formation for ≥ 90% time of the MD simulation) of AVP1795 were its major anchoring points. Several other residues demonstrated flexible interactions within the binding pocket (Fig. 8 a and b). As a result, predominance of both hydrogen bond (56.73%) and hydrophobic interactions (27.27%) was observed for AVP1795-RBD (Fig. 8c). A representative hydrogen bond interaction between RBD and AVP1795 has been shown between TYR 489 and GLY 23 in Fig. 8d.
Fig. 8.
RBD-AVP 1795. (a) Interacting RBD residues; (b) Interacting AVP 1795 residues; (c) Distribution of non-covalent interactions; (d) Representative snapshot (at 150 ns); AVP 1795 (purple) and RBD (grey) over 150 ns MD simulation. Arrows mark ≥90% time participation in bond formation during MD simulation.
In the case of AVP1784-RBD complex, TYR 449, GLY 496, PHE 490 or RBD formed multiple bonds with the peptide, whereas TYR 505, GLU 484 (participation in bond formation for ≥ 90% time of the MD simulation) served as the anchoring points to hold AVP1784 in the binding groove (Fig. S3a). On the other hand, ARG 19, PHE 8, CYS 4, and ALA 11 were found to be the key residues of AVP1784, although only ARG 19 showed more than a 90% participation time in bond formation (Fig. S3b). The dominance of hydrogen bonding (42.67%) was maintained. Electrostatic interactions (31.37%) and hydrophobic interactions (25.96%) were also notable (Fig. S3c). An important hydrophobic interaction between TYR 505 and PHE 8 of RBD and AVP1784, respectively, is shown in Fig. S3d.
The major RBD residues in AVP1775-RBD complex were LYS 417, ARG 403, ASN 487, and TYR 489) (Fig. S4a). On the contrary, ASP 14, HIS 3, LEU 13 of AVP 1775 were key to its affinity towards RBD ((Fig. S4b). None of the peptide or RBD residues served as anchoring points, rather the participating residues showed flexible interactions. Hydrogen bond (56.73%), hydrophobic (27.27%), as well as electrostatic (15.99%) interactions were observed (Fig. S4c). A representative hydrogen bond between LYS 417 and LEU 13 of RBD and AVP1775 is shown in Fig. S4d.
In the case of AVP0673-RBD, ASN 487, TYR 489, and PHE486 participated in bond formation for more than 90% time of the MD simulation period (Fig. S5a). GLU11 of AVP0673 showed multiple interactions with RBD residues, whereas ARG16, LEU18, and VAL17 (participation in bond formation for ≥ 90% time of the MD simulation) were its true anchoring points, (Fig. S5b). Hydrogen bond (52.72%), hydrophobic (27.73%) and electrostatic (19.55%) interactions were observed (Fig. S5c). Representative hydrogen bonds between ASN487 and LEU18 of RBD and AVP0673, respectively, are shown in Fig. S5d.
In the AH-RBD complex, RBD residues such as ARG 403 and LYS417 (participation in bond formation for ≥ 90% time of the MD simulation) served as anchoring points and were its principal interaction points. Residues, such as GLN 493, ASN 501, TYR 505, and TYR 489 showed substantial interactions with the peptide (Fig. S6a). On the other hand, GLU 18 and GLN 23 (participation in bond formation for ≥ 90% time of the MD simulation) of AH remained it in its binding groove. Among the other important AH residues were HIS15, LYS12, and GLN5 (Fig. S6b). Non-covalent interactions were predominantly hydrogen bonds (58.34%), whereas hydrophobic (18.8%) as well as electrostatic (22.86%) interactions were also observed (Fig. S6c). Hydrogen bonding is shown between ARG403 and GLU18 of AH-RBD in Fig. S6d.
4. Conclusions
Since peptides have excellent object specificity and selectivity, they can also provide a promising therapeutic strategy for COVID-19 treatment via the development of high-affinity antiviral peptides. To block the SARS-CoV-2 RBD interaction with hACE2 and subsequent viral entry to the host cells, developing peptide inhibitors could be considered an initial step. Our comprehensive computational screening showed that 27 antiviral peptides exhibited better binding affinity than that of the alpha-helix of hACE2. Molecular dynamics simulations and the binding free energy landscape indicate that AVP1795 has the most promising features as a SARS-CoV-2 inhibitor, and AVP0673 also exhibited better results. Upon analysis of non-covalent interactions, we also identified critical RBD and peptide residues that can spearhead identifying a peptide inhibitor with excellent selectivity and specificity. As a future direction, these peptides will be synthesized using the standard Fmoc-based synthesis protocol, purified by RP-HPLC, and analyzed with mass spectrometry. To assess the inhibition efficiency of these antiviral peptides, high-throughput screening (HTS) will be performed. Our comprehensive computational screening may help other researchers design effective antiviral peptides against SARS-CoV-2.
Conflict of interest and authorship conformation form
Please check the following as appropriate.
•All authors have participated in (a) conception and design, or analysis and interpretation of the data; (b) drafting the article or revising it critically for important intellectual content; and (c) approval of the final version.
•This manuscript has not been submitted to, nor is under review at, another journal or other publishing venue.
Acknowledgments
We are grateful to our donors who supported the build of the computational platform (http://grc-bd.org/donate/). The authors acknowledge the World Academy of Science (TWAS) for the purchase of the High-Performance Computer for performing molecular dynamics simulations. We are grateful to Suprio Kamal for helping in the percent time interaction calculation. BioRender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104759.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
M.A.Halim conceived the idea. M.M.H.S., A.A.N., M.T.I., M.A.R.U., M.S.I., F.F.B.H., M.I.A., M.S.B., T.H., U.S.T., S.I.S., N.R.J., M.A., M.A.I. and M.M.H. performed molecular docking of 645 peptides and analyzed data. M.A.A. conducted the MD simulations. R.I. performed the principal component analysis on MD data. M.M.H.S. and A.A.N. draft the manuscript. M.A.S. M. A. Hossain and M.A.Halim revised the manuscript. M.A.Hossain and M. A. Halim supervised the project. All authors approved the manuscript in its final form.
Supporting information
The Supporting Information available free of charge are sequence, length, inhibition efficiency, binding affinity, secondary structures percentage, interaction residues, distribution of non-covalent interactions best peptides.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Gorbalenya A.E., Baker S.C., Baric R.S., de Groot R.J., Drosten C., Gulyaeva A.A., Haagmans B.L., Lauber C., Leontovich A.M., Neuman B.W., Penzar D., Perlman S., Poon L.L.M., Samborskiy D.V., Sidorov I.A., Sola I., Ziebuhr J. The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.-M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.-C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H.-D., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348 doi: 10.1056/nejmoa030747. 1967–1976. [DOI] [PubMed] [Google Scholar]
- 3.Zaki A.M., van Boheemen S., Bestebroer T.M., Osterhaus A.D.M.E., Fouchier R.A.M. Isolation of a novel coronavirus from a man with pneumonia in Saudi arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/nejmoa1211721. [DOI] [PubMed] [Google Scholar]
- 4.W.H. Organisation, WHO coronavirus disease (COVID-19) dashboard | WHO coronavirus disease (COVID-19) dashboard, who.Int. (202AD) 20–26. htttps://covid19.who.int/.%0Ahttps://covid19.who.int/?gclid=CjwKCAjwnK36BRBVEiwAsMT8WJ3y00_BUzvrLsvbl3uthuoTH_Occ45gyEUbpYRyEqAzll3aZB6TYxoCcM0QAvD_BwE (accessed October 23, 2020).
- 5.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., Mclellan J.S. Vol. 367. Science; 2020. Cryo-EM Structure of the 2019-nCoV Spike in the Prefusion Conformation; pp. 1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Khailany R.A., Safdar M., Ozaslan M. Genomic characterization of a novel SARS-CoV-2. Gene Reports. 2020;19:100682. doi: 10.1016/j.genrep.2020.100682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hassan S.S., Aljabali A.A.A., Panda P.K., Ghosh S., Attrish D., Choudhury P.P., Seyran M., Pizzol D., Adadi P., Abd El-Aziz T.M., Soares A., Kandimalla R., Lundstrom K., Lal A., Azad G.K., Uversky V.N., Sherchan S.P., Baetas-da-Cruz W., Uhal B.D., Rezaei N., Chauhan G., Barh D., Redwan E.M., Dayhoff G.W., Bazan N.G., Serrano-Aroca Á., El-Demerdash A., Mishra Y.K., Palu G., Takayama K., Brufsky A.M., Tambuwala M.M. A unique view of SARS-CoV-2 through the lens of ORF8 protein. Comput. Biol. Med. 2021;133:104380. doi: 10.1016/j.compbiomed.2021.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alejandra Tortorici M., Walls A.C., Lang Y., Wang C., Li Z., Koerhuis D., Boons G.J., Bosch B.J., Rey F.A., de Groot R.J., Veesler D. Structural basis for human coronavirus attachment to sialic acid receptors. Nat. Struct. Mol. Biol. 2019;26:481–489. doi: 10.1038/s41594-019-0233-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ou X., Liu Y., Lei X., Li P., Mi D., Ren L., Guo L., Guo R., Chen T., Hu J., Xiang Z., Mu Z., Chen X., Chen J., Hu K., Jin Q., Wang J., Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat. Commun. 2020;11:1–12. doi: 10.1038/s41467-020-15562-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar S., Maurya V.K., Prasad A.K., Bhatt M.L.B., Saxena S.K. Structural, glycosylation and antigenic variation between 2019 novel coronavirus (2019-nCoV) and SARS coronavirus (SARS-CoV) VirusDisease. 2020;31:13–21. doi: 10.1007/s13337-020-00571-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dejnirattisai W., Zhou D., Ginn H.M., Duyvesteyn H.M.E., Supasa P., Case J.B., Zhao Y., Walter T.S., Mentzer A.J., Liu C., Wang B., Paesen G.C., Slon-Campos J., López-Camacho C., Kafai N.M., Bailey A.L., Chen R.E., Ying B., Thompson C., Bolton J., Fyfe A., Gupta S., Tan T.K., Gilbert-Jaramillo J., James W., Knight M., Carroll M.W., Skelly D., Dold C., Peng Y., Levin R., Dong T., Pollard A.J., Knight J.C., Klenerman P., Temperton N., Hall D.R., Williams M.A., Paterson N.G., Bertram F.K.R., Siebert C.A., Clare D.K., Howe A., Radecke J., Song Y., Townsend A.R., Huang K.Y.A., Fry E.E., Mongkolsapaya J., Diamond M.S., Ren J., Stuart D.I., Screaton G.R. The antigenic anatomy of SARS-CoV-2 receptor binding domain. Cell. 2021;184:2183–2200. doi: 10.1016/j.cell.2021.02.032. e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boopathi S., Poma A.B., Kolandaivel P. Novel 2019 coronavirus structure, mechanism of action, antiviral drug promises and rule out against its treatment. J. Biomol. Struct. Dyn. 2020:1–10. doi: 10.1080/07391102.2020.1758788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panda P.K., Arul M.N., Patel P., Verma S.K., Luo W., Rubahn H.-G., Mishra Y.K., Suar M., Ahuja R. Structure-based drug designing and immunoinformatics approach for SARS-CoV-2. Sci. Adv. 2020;6 doi: 10.1126/sciadv.abb8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shang J., Wan Y., Luo C., Ye G., Geng Q., Auerbach A., Li F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. U.S.A. 2020;117:11727–11734. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 17.Huang Y., Yang C., feng Xu X., Xu W., wen Liu S. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41:1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 20.Wrobel A.G., Benton D.J., Xu P., Roustan C., Martin S.R., Rosenthal P.B., Skehel J.J., Gamblin S.J. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat. Struct. Mol. Biol. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Liu M., Gao J. Enhanced receptor binding of SARS-CoV-2 through networks of hydrogen-bonding and hydrophobic interactions. Proc. Natl. Acad. Sci. U.S.A. 2020;117 doi: 10.1073/pnas.2008209117. 13967–13974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith M.C., Gestwicki J.E. Features of protein-protein interactions that translate into potent inhibitors: topology, surface area and affinity. Expet Rev. Mol. Med. 2012;14:1–20. doi: 10.1017/erm.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quartararo A.J., Gates Z.P., Somsen B.A., Hartrampf N., Ye X., Shimada A., Kajihara Y., Ottmann C., Pentelute B.L. Ultra-large chemical libraries for the discovery of high-affinity peptide binders. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-16920-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marqus S., Pirogova E., Piva T.J. Evaluation of the use of therapeutic peptides for cancer treatment. J. Biomed. Sci. 2017;24:1–15. doi: 10.1186/s12929-017-0328-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.G. (MIT) Zhang, S. (MIT) Pomplun, A.R. (MIT) Loftis, A. (MIT) Loas, B.L. (MIT) Pentelute The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han Y., Král P. Computational design of ACE2-based peptide inhibitors of SARS-CoV-2. ACS Nano. 2020;14:5143–5147. doi: 10.1021/acsnano.0c02857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schütz D., Ruiz-Blanco Y.B., Münch J., Kirchhoff F., Sanchez-Garcia E., Müller J.A. Peptide and peptide-based inhibitors of SARS-CoV-2 entry, Adv. Drug Deliv. Rev. E. 2020;167:47–65. doi: 10.1016/j.addr.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ho T.Y., Wu S.L., Chen J.C., Wei Y.C., Cheng S.E., Chang Y.H., Liu H.J., Hsiang C.Y. Design and biological activities of novel inhibitory peptides for SARS-CoV spike protein and angiotensin-converting enzyme 2 interaction. Antivir. Res. 2006;69:70–76. doi: 10.1016/j.antiviral.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paidi R.K., Jana M., Mishra R.K., Dutta D., Raha S., Pahan K. ACE-2-interacting domain of SARS-CoV-2 (AIDS) peptide suppresses inflammation to reduce fever and protect lungs and heart in mice: implications for COVID-19 therapy. J. Neuroimmune Pharmacol. 2021;16:59–70. doi: 10.1007/s11481-020-09979-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karoyan P., Vieillard V., Gómez-Morales L., Odile E., Guihot A., Luyt C.E., Denis A., Grondin P., Lequin O. Human ACE2 peptide-mimics block SARS-CoV-2 pulmonary cells infection. Commun. Biol. 2021;4:1–9. doi: 10.1038/s42003-021-01736-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chowdhury S.M., Talukder S.A., Khan A.M., Afrin N., Ali M.A., Islam R., Parves R., Al Mamun A., Sufian M.A., Hossain M.N., Hossain M.A., Halim M.A. Antiviral peptides as promising therapeutics against SARS-CoV-2. J. Phys. Chem. B. 2020;124:9785–9792. doi: 10.1021/acs.jpcb.0c05621. [DOI] [PubMed] [Google Scholar]
- 32.Qureshi A., Thakur N., Tandon H., Kumar M. AVPdb: a database of experimentally validated antiviral peptides targeting medically important viruses. Nucleic Acids Res. 2014;42:D1147–D1153. doi: 10.1093/nar/gkt1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maupetit J., Derreumaux P., Tuffery P. PEP-FOLD: an online resource for de novo peptide structure prediction. Nucleic Acids Res. 2009;37:W498–W503. doi: 10.1093/nar/gkp323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shen Y., Maupetit J., Derreumaux P., Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theor. Comput. 2014;10:4745–4758. doi: 10.1021/ct500592m. [DOI] [PubMed] [Google Scholar]
- 35.Schneidman-Duhovny D., Inbar Y., Nussinov R., Wolfson H.J. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:363–367. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mashiach E., Schneidman-Duhovny D., Andrusier N., Nussinov R., Wolfson H.J. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36:229–232. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrusier N., Nussinov R., Wolfson H.J. FireDock: fast interaction refinement in molecular docking. Proteins Struct. Funct. Genet. 2007;69:139–159. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 38.Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., Van Dijk M., De Vries S.J., Bonvin A.M.J.J. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 40.Krieger E., Darden T., Nabuurs S.B., Finkelstein A., Vriend G. Making optimal use of empirical energy functions: force-field parameterization in crystal space. Proteins Struct. Funct. Genet. 2004;57:678–683. doi: 10.1002/prot.20251. [DOI] [PubMed] [Google Scholar]
- 41.Maier J.A., Martinez C., Kasavajhala K., Wickstrom L., Hauser K.E., Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J. Chem. Theor. Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. 2001;105:9954–9960. doi: 10.1021/jp003020w. [DOI] [Google Scholar]
- 43.Darden T., York D., Pedersen L. Particle mesh Ewald: an N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 44.Krieger E., Nielsen J.E., Spronk C.A.E.M., Vriend G. Fast empirical pKa prediction by Ewald summation. J. Mol. Graph. Model. 2006;25:481–486. doi: 10.1016/j.jmgm.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 45.Krieger E., Vriend G. New ways to boost molecular dynamics simulations. J. Comput. Chem. 2015;36:996–1007. doi: 10.1002/jcc.23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32:3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 47.De Jong S., calibration Multivariate, Martens H., Naes T. Vol. 4. Wiley; New York: 1989. p. 441. Price: £75.00, US$138.00. No. of pages: 504, J. Chemom. 1990. [DOI] [Google Scholar]
- 48.Wold S., Esbensen K., Geladi P. Principal component analysis. Chemometr. Intell. Lab. Syst. 1987;2:37–52. doi: 10.1016/0169-7439(87)80084-9. [DOI] [Google Scholar]
- 49.Islam R., Parves M.R., Paul A.S., Uddin N., Rahman M.S., Al Mamun A., Hossain M.N., Ali M.A., Halim M.A. A molecular modeling approach to identify effective antiviral phytochemicals against the main protease of SARS-CoV-2. J. Biomol. Struct. Dyn. 2021;39:3213–3224. doi: 10.1080/07391102.2020.1761883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmed S., Mahtarin R., Ahmed S.S., Akter S., Islam M.S., Al Mamun A., Islam R., Hossain M.N., Ali M.A., Sultana M.U.C., Parves M.R., Ullah M.O., Halim M.A. Investigating the binding affinity, interaction, and structure-activity-relationship of 76 prescription antiviral drugs targeting RdRp and Mpro of SARS-CoV-2. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1796804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.R Core Development Team, R Core Team . 2014. R: a language and environment for statistical computing. R Found. Stat. Comput. Vienna, Austria. URL Http//Www.R-Project.Org/. (2014) [Google Scholar]
- 52.RStudio Team Integrated development for R., RStudio. 2020;42:14. https://rstudio.com [Google Scholar]
- 53.Kassambara A., Mundt F. Factoextra: extract and visualize the results of multivariate data analyses. R package. https://cran.r-project.org/package=factoextrahttps://cran.r-project.org/package=factoextra%0Ahttps://rpkgs.datanovia.com/factoextra/index.html Retrieved from, 1 (2020) 1–76. factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R package version 1.0.7.
- 54.Heinig M., Frishman D., STRIDE A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32 doi: 10.1093/nar/gkh429. W500–W502. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.