Abstract
The potential emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike (S) escape mutants is a threat to the efficacy of existing vaccines and neutralizing antibody (nAb) therapies. An understanding of the antibody/S escape mutation landscape is urgently needed to preemptively address this threat. Here we describe a rapid method to identify escape mutants for nAbs targeting the S receptor binding site. We identified escape mutants for five nAbs, including three from the public germline class VH3-53 elicited by natural coronavirus disease 2019 (COVID-19) infection. Escape mutations predominantly mapped to the periphery of the angiotensin-converting enzyme 2 (ACE2) recognition site on the RBD with K417, D420, Y421, F486, and Q493 as notable hotspots. We provide libraries, methods, and software as an openly available community resource to accelerate new therapeutic strategies against SARS-CoV-2.
Keywords: SARS-CoV-2, S RBD, escape mutants, neutralizing antibodies, deep mutational scanning, yeast surface display
Graphical abstract
Francino-Urdaniz et al. describe a deep mutational scanning method to identify escape mutants on SARS-CoV-2 S RBD. Escape mutants were identified for five neutralizing antibodies elicited from natural infection, revealing hotspots at positions K417, D420, Y421, F486, and Q493 at the periphery of the ACE2 recognition site.
Introduction
The type I viral fusion protein spike (S) is a major antigenic determinant of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and is the antigen used in all approved coronavirus disease 2019 (COVID-19) vaccines (Baden et al., 2021; Polack et al., 2020; Voysey et al., 2021). Recently, the B.1.1.7 (N501Y; Alpha), B.1.351 (E484K, N501Y, K417N; Beta), B.1.427 (L452R; Epsilon), B.1.617 (L452R, E484Q; Delta), and C.37 (L452Q, F490S; Lambda) viral lineages have emerged (mutations listed are for S receptor binding domain [RBD] only). Among other mutations on S, all lineages encode single-nucleotide substitutions in the S RBD near the recognition site for its cellular target angiotensin-converting enzyme 2 (ACE2) (Voysey et al., 2021; Wang et al., 2021; Zhang et al., 2021).
Dozens of studies have reported the structural, epitopic, and functional landscape of non-neutralizing monoclonal antibodies and neutralizing antibodies (nAbs) targeting trimeric S (Banach et al., 2021; Rogers et al., 2020; Yuan et al., 2020a). A prophetic understanding of the mutations on S that could evade antibody recognition would enable development of better vaccine boosters and monoclonal antibody therapies. In particular, US Food and Drug Administration (FDA)-approved monoclonal antibody therapies targeting the S RBD developed by Regeneron (Baum et al., 2020) and Lilly (Jones et al., 2020) have shown significantly decreased effectiveness with Beta and Gamma variants (Chen et al., 2021; Greaney et al., 2021) Thus, we sought to develop an S RBD yeast surface display (YSD) platform (Figure S1; Wrapp et al., 2020), because we hypothesized that broad identification of SARS-CoV-2 S escape mutants could be accomplished by integrating high-throughput screening platforms with deep sequencing. Although a similar platform uses the loss of nAb binding to identify escape mutants (Greaney et al., 2021; Starr et al., 2020), we rationalized that a functional screening assay that directly measures the ability of a nAb to compete with ACE2 for S RBD binding would be a comparatively strong predictor of RBD escapability, because it accounts for mutations in RBD that would disrupt S binding to ACE2.
Results
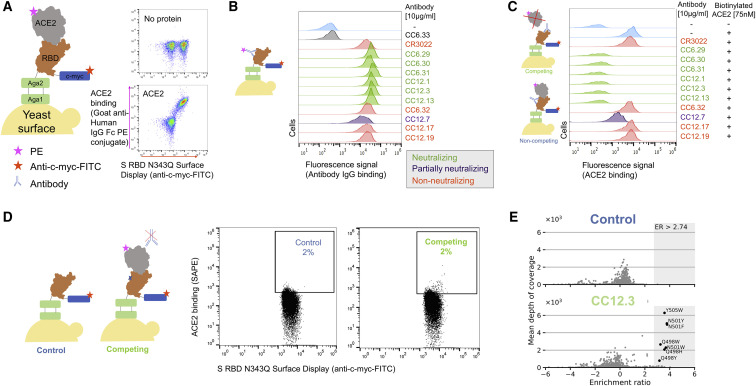
We had previously developed an aglycosylated S-RBD YSD platform (S RBD(333–537)-N343Q) from the original Wuhan-Hu-1 strain (Banach et al., 2021) that can bind specifically to ACE2 (Figure 1 A). This S RBD construct has its one native N-linked glycan removed (N343Q) because the heavy N-linked mannosylation of S. cerevisiae could hamper anti-S RBD mAb recognition (Jigami, 2008). Cell-surface titrations of CR3022 IgG and nAb HKU-910-30 IgG yielded apparent dissociation constants comparable with reported in vitro results (Banach et al., 2021; Yuan et al., 2020a; Figure S2A). We then tested a panel of 11 additional anti-S RBD mAbs for binding to aglycosylated RBD. These mAbs were isolated from convalescent donors infected in late 2019/early 2020 and thus are representative of anti-S mAbs raised during natural infection (Rogers et al., 2020). Ten of the 11 mAbs recognized aglycosylated S RBD (Figure 1B). The one panel member that did not bind, CC6.33, selectively recognizes the S309 epitope on the RBD containing the N-linked glycan at position 343 (Pinto et al., 2020).
Figure 1.
Identification of SARS-CoV-2 S RBD escape mutants using yeast screening
(A) Cartoon of the yeast display construct S-RBD(333–537)-N343Q. Cytograms show specific binding in the presence, but not absence, of ACE2-Fc.
(B) Binding profiles of aglycosylated S RBD labeled with 10 μg/mL of indicated mAb. Antibodies are color coded according to neutralization potency (Rogers et al., 2020).
(C) Competitive binding between IgG and ACE2 was performed by labeled yeast displaying aglycosylated S RBD with 10 μg/mL of indicated mAbs followed by labeling with biotinylated ACE2.
(D) Single-site saturation mutagenesis S RBD libraries were sorted by FACS using a competition experiment. The top cytogram shows the cell population collected for the control population without ACE2 labeling, while the bottom cytogram shows the cell population enriched in mutations able to bind ACE2 in the presence of a competing IgG. The specific cytogram shown is for nAb CC12.3 using the S RBD library corresponding to mutations at positions 437–537.
(E) Per-mutation enrichment ratio (ER) distributions as a function of average depth of coverage control (left) and CC12.3 nAb competing experiment (right).
See also Figure S2.
Next, we evaluated the ability of the mAb panel to competitively inhibit ACE2 binding to aglycosylated S RBD in an assay conceptually similar to the one previously described by Tan et al. (2020). Yeast displaying aglycosylated S RBD was first labeled with a saturating concentration of a given mAb and then co-incubated with biotinylated ACE2. Six mAbs completely ablated ACE2 binding, one mAb partially inhibited ACE2, and the remaining four did not prevent ACE2 binding (Figure 1C). An inverse correlation was observed between the previously determined neutralization potency of the antibody (Banach et al., 2021; Yuan et al., 2020a) and the fluorescence signal increase in the competition assay (Figure 1C). We conclude from these experiments that, excluding the S309 epitope, the aglycosylated S RBD platform faithfully recapitulates binding interactions of nAbs with S RBD (Rogers et al., 2020).
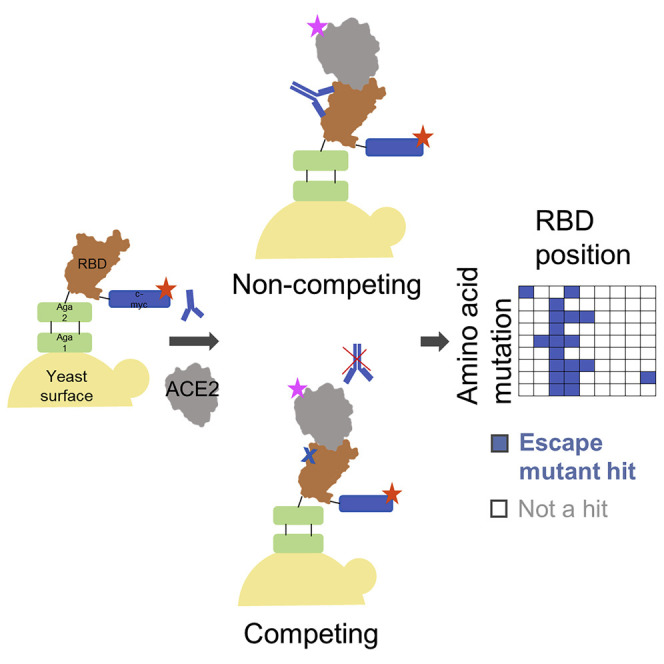
Our strategy for identifying potential S RBD escape mutants was as follows. First, we constructed a saturation mutagenesis library of aglycosylated S RBD containing all possible single missense and nonsense mutations for the 119 surface-exposed positions of the RBD (96% coverage of the 2,380 possible library members; Data S1 contains library coverage statistics) (Wrenbeck et al., 2016). For each codon, mutations were encoded using oligonucleotides containing a degenerate NNK sequence. This degenerate sequence encodes all 20 amino acids plus a stop codon, which is useful as an internal negative control for the assay. We labeled yeast displaying these RBD variants with a saturating concentration of nAb and then co-incubated with a saturating concentration of biotinylated ACE2. We then used fluorescence-activated cell sorting (FACS) to screen for mutants that could bind ACE2, indicating that the RBD mutation allows for evasion of the nAb while not disrupting the ACE2 interaction critical for cell entry (Figure 1D; Figures S2B and S2C). Importantly, a control with no ACE2 labeling was sorted to set an empirical false discovery rate (FDR) for putative escape mutant hits (Figure 1E; Figure S2C). Plasmid DNA from sorted cells was prepped and deep sequenced. We determined the enrichment ratio (ER)—the base-2 logarithm of the ratio of the frequency mutant in the sorted population to its frequency in the reference population—and then used the control population to set the FDR (Figure 1E; Figure S2D). We screened five different nAbs identified earlier as having completely ablated ACE2 binding (CC6.29, CC6.31, CC12.1, CC12.3, CC12.13). In all, we identified a total of 97 S RBD mutants that can escape recognition by at least one nAb (Data S1).
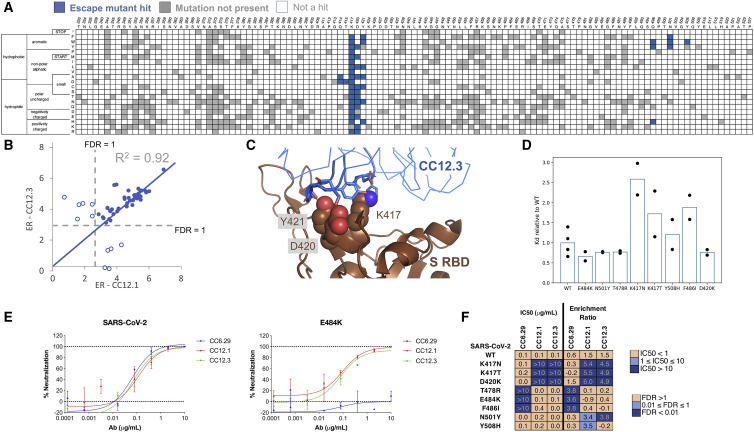
For all five nAbs, the putative escape mutant hits were localized to specific locations within the S RBD primary sequence (Figure 2 A; Data S1). CC12.1 and CC12.3 belong to the public germline class VH3-53 (Banach et al., 2021; Wu et al., 2020b; Yuan et al., 2020a) and are representative of the subset of VH3-53 public antibodies with relatively short CDRH3 regions (Wu et al., 2020a). Strikingly, these two nAbs share over 90% of the same RBD escape mutants (Figure 2B), even though the light chain differs between the nAbs. Structural complexes of antibodies CC12.1 and CC12.3 were previously solved in complex with S RBD (Yuan et al., 2020b), affording a structural explanation for individual escape mutants. Escape mutants for both of the VH3-53 nAbs CC12.1 and CC12.3 clustered at the same location on the S RBD, mainly on the periphery of the ACE2 binding site (Figure 2C; Figure S3A). To confirm that the mutations did not have a large effect on equilibrium binding to ACE2, we determined the dissociation constant of eight single-point mutants using YSD titrations. Binding affinities of each mutant were comparable with the S RBD N343Q dissociation constants and were in agreement with a previous deep mutational scanning study (Starr et al., 2020; Figure 2D). Thus, mutations identified that escape antibody recognition in this assay can still bind ACE2.
Figure 2.
Validation of escape mutants using yeast screening and pseudoneutralization assays
(A) Heatmap showing predicted S RBD escape mutants for CC12.3 in blue. White cells are mutations with a p value for an FDR > 1, while gray cells are mutations not present in the mutational library.
(B) Comparison of ERs for individual hits for CC12.3 versus CC12.1. Closed circles represent escape mutant hits for both nAbs, whereas open circles are escape mutant hits for only one nAb.
(C) Solved structure of nAb CC12.3 in complex with S RBD (PDB: 7KN6).
(D) Dissociation constants of single-point mutants relative to S RBD N343Q (“WT”) determined by yeast surface display titrations. Circles show the relative value for each biological replicate, and the bars represent the mean (n = 2 for each mutant; n = 4 for WT).
(E) Pseudovirus neutralization curves for CC12.1, CC12.3, and CC6.29 on SARS-CoV-2 (left) and SARS-CoV-2 E484K (right).
(F) Pseudovirus IC50 analysis for CC12.1, CC12.3, and CC6.29 on different identified mutations.
See also Figure S3.
Having identified a number of putative escape mutants from the mutagenesis library screening, we sought to determine how this functional screening correlated with the more conventional pseudovirus neutralization assay. A panel of MLV-based SARS-CoV-2 pseudoviruses were generated that contained single mutations predicted by the mutagenesis scanning to allow escape from one of the antibodies screened, as well as several irrelevant control mutations. Antibodies CC12.1, CC12.3, and CC6.29 were screened against the original SARS-CoV-2 pseudovirus, as well as this panel of mutant pseudoviruses in duplicate (Figure 2E), and the resulting IC50 values were compared to calculate the effect on antibody neutralization potency (Figure 2F; Figure S3B). Consistent with the RBD mutagenesis library and structural analysis, CC6.29 failed to neutralize the F486I, E484K, and T478R variants. Additionally, K417N, K417T, and D420K hotspot mutants completely escaped neutralization for both CC12.1 and CC12.3. The only instance we tested where the mutagenesis scanning data differed from the pseudovirus results was at N501Y that was predicted to confer escape from CC12.1 and CC12.3 but had no effect on the in vitro neutralization potency. Although it is unclear why this discrepancy occurred, we note that N501Y significantly increases the affinity of the RBD for ACE2, which could result in ACE2 out-competing bound nAbs.
Finally, we performed biological replicates where the mutagenesis library corresponding to S RBD positions 437–537 was separately transformed into yeast and screened against nAbs CC6.29, CC12.1, and CC12.3. Although the ERs were lower than in the initial experiment, nearly the same set of escape mutants was identified for CC6.29, and escape mutants originally identified for all nAbs had significantly higher ERs than other variants in the replicate (p value range, 4.2e−4 to 1.9e−11, one-sided Welch’s t test) (Figure S3C).
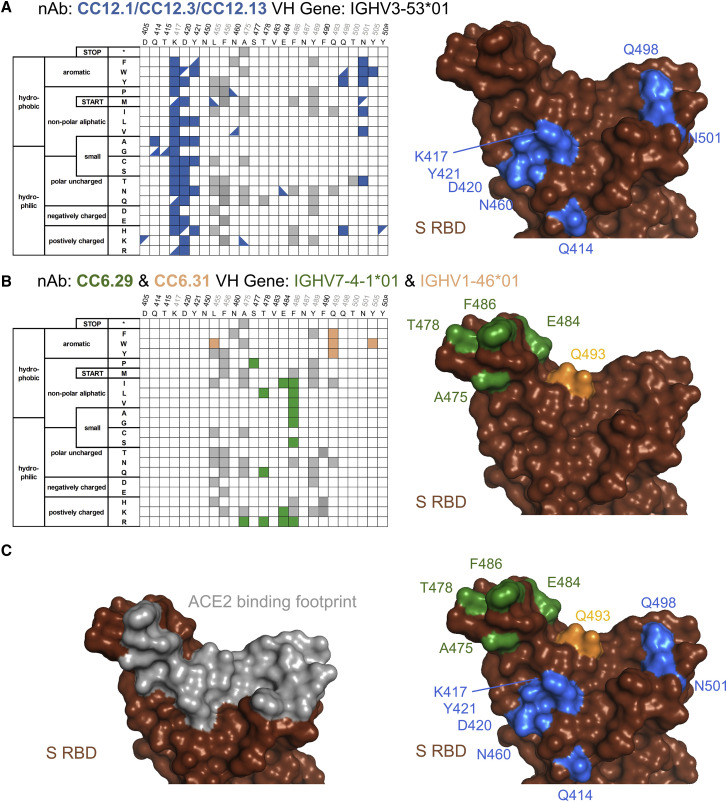
Selected per-position heatmaps and structural mapping of S RBD escape mutants are shown in Figure 3 for all five nAbs. Closer examination of these datasets reveals key features of the RBD escape mutant response. CC12.1 and CC12.3 nAbs share over 90% of the same RBD escape mutants (Figure 2B), including notable hotspot mutations occurring at K417, D420, Y421, and Q498 (Figure 3A). Interestingly, multiple aromatic substitutions at Q498 escape recognition for CC12.1 and CC12.3, even though the antibodies have different light chains and recognition motifs for that position. Introduction of an aromatic residue at Q498 introduces substantial van der Waals clashes that are likely unresolved without antibody loop movement. The other VH3-53 nAb tested, CC12.13, has a 15-amino acid length CDRH3 that likely has a distinct binding mode than that for CC12.1 and CC12.3 (Wu et al., 2020a). Consistent with this, the CC12.13 escape mutants identified are mostly different from those for CC12.1/CC12.3 (Data S1).
Figure 3.
Sequence determinants and structural basis of S RBD escape mutants
Limited per-position heatmap (left) and mutations mapped onto the S RBD-ACE2 structural complex (right; PDB: 6M0J). For clarity, only positions with two or more escape mutations are shown with surface colored. (A) nAbs CC12.1, CC12.3, and CC12.13. Boxes indicate escape mutants for two or more nAbs, while triangles indicate an escape mutant identified for just one nAb (top left: CC12.1, bottom right: CC12.3, bottom left: CC12.13); (B) CC6.29 (green) and CC6.31 (orange); and (C) overlay of escape mutants from all nAbs onto the S RBD-ACE2 structural complex.
Another nAb screened, CC6.29, has a completely different escape mutant profile compared with CC12.1/CC12.3. The 15 potential RBD escape mutants for CC6.29 center around the structural “knob” of positions A475, S477, T478, E484, and F486 (Figure 3B). E484K shared by the B.1.351 and B.1.526 lineages is identified as an escape mutant for this nAb, but the structurally adjacent S477N mutation newly identified in the B.1.526 lineage does not escape CC6.29 neutralization. Intriguingly, S477P is identified as an escape mutant for this nAb. F486 is a mutational hotspot even though that position is involved in the recognition of ACE2. This is consistent with a previous mutational scan of S RBD showing that mutation of F486 does not significantly impact ACE2 binding affinity (Starr et al., 2020). CC6.31 escape mutants partially overlap with CC6.29 but implicate a different set of mutants (Figure 3B). Multiple mutations at Q493 escape CC6.31, including Q493 substitutions to the aromatic amino acids F/W.
In total, the five nAbs map a partially overlapping surface with the ACE2 binding site that is primed for antibody escape. In comparison with the binding footprint of ACE2 (Figure 3C), the escape mutants almost completely map to the outer binding shell and periphery of the interaction surface, akin to an O-ring circumscribing the receptor binding site (RBS). Out of the identified escape mutants, residues K417, F486, Q493, N501, and Y505 are located on the ACE2 footprint (Figure 3C). Although mutations on K417 and F486 do not significantly change the RBD affinity to ACE2, mutations on N501 can increase or decrease affinity depending on the substitution. The Y505W mutant shared by CC6.31, CC12.1, and CC12.3 also increases ACE2 affinity (Starr et al., 2020).
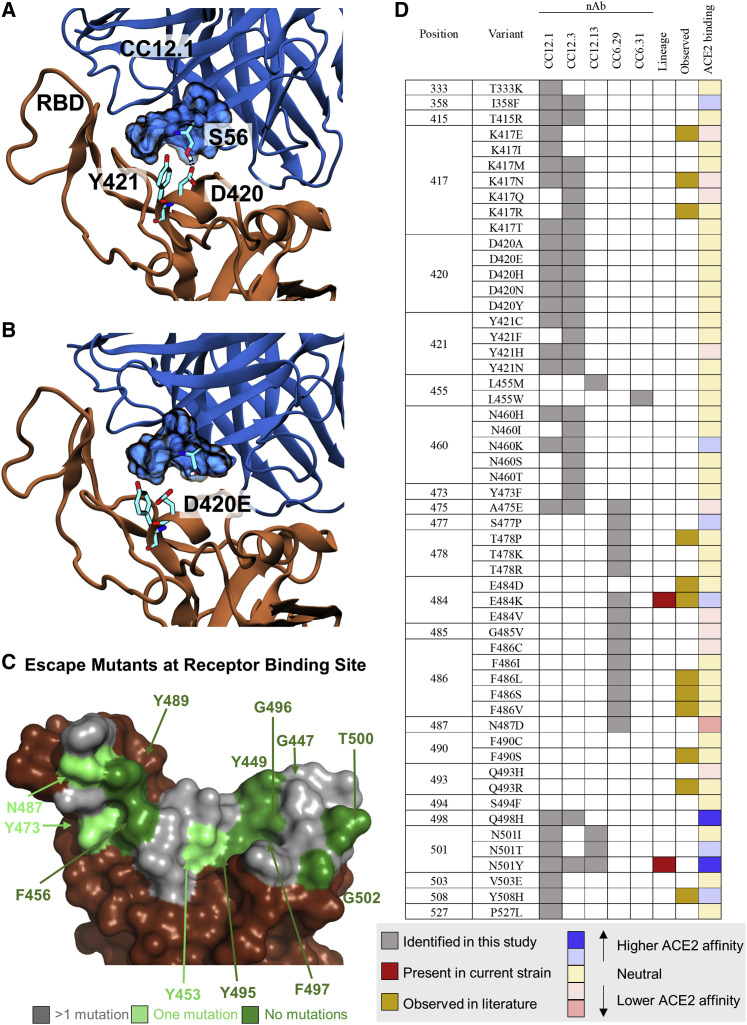
We were puzzled by the fact that the mutations at D420 were so deleterious to the neutralization potency of the VH3-53 nAbs given that this residue is on the outer periphery of the binding epitope. Consequently, we performed 100-ns aqueous molecular dynamics (MD) simulations of CC12.1 and CC12.3 in complex with wild-type S RBD. We also simulated S RBD incorporated with the D420E, D420K, or Y421N mutation (see Supplemental Information for details). In the control simulation with CC12.1, D420 on the RBD and CDRH2 S56 on CC12.1 form persistent hydrogen bonds, and Y421 on the RBD is tightly bound within a pocket of CC12.1 residues (Figure 4 A). With the D420E mutation, the increased length of E420 disrupts its ability to hydrogen bond with S56, requiring it to adopt a bent conformation (Figure 4B). This forces Y421 out of the antibody pocket, causing increased fluctuations in neighboring RBD loops that persist throughout the entire 100-ns production simulation (Figures S4A and S4B). With the D420K mutation, hydrogen bonding with S56 is completely disrupted. With the Y421N mutation, N421 is too short to interact with the antibody pocket (Figure S4C). Similar escape mechanisms are observed for CC12.3 with all three RBD mutations, including increased fluctuations at one of the same key sites (K458) on the RBD in response to the D420E mutation (Figures S4D and S4E).
Figure 4.
Mechanistic, structural, and sequence analysis of SARS-CoV-2 escape mutants
(A and B) Snapshots from MD trajectories showing (A) key interactions in the control simulation of S RBD in complex with CC12.1, and (B) mechanism of escape of S RBD from CC12.1 as a result of the D420E mutation. Images were rendered with Visual Molecular Dynamics (VMD) (Humphrey et al., 1996), and black dotted lines indicate persistent hydrogen bonds.
(C) S RBS positions are colored by the number of escape mutants identified to date. RBS residues involving the S RBD-ACE2 structural complex (PDB: 6M0J) are colored by number of escape mutants identified to date.
(D) Summary of 1-nt escape mutants identified in the present study. Lineage column indicates presence of the given mutation among currently circulating SARS-CoV-2 strains, while the observed column refers to an escape mutant previously identified in literature (Baum et al., 2020; Greaney et al., 2021; Li et al., 2020; Liu et al., 2021; Weisblum et al., 2020). ACE2 binding indicates affinity to ACE2 based on the measurements by Starr et al. (2020).
There have been a number of recent approaches to identify specific S escape mutants (summarized in Data S2; Baum et al., 2020; Greaney et al., 2021; Li et al., 2020; Liu et al., 2021; Weisblum et al., 2020). A survey of the existing escape mutant literature, along with escape mutants identified in the present work, allows us to identify the absolute and near-absolute escape-resistant ACE2 RBS residues in the context of the original lineage (Figure 4C). One resistant patch is found around F456/Y473/N487/Y489, while other residues are discontinuous patches on the remainder of the RBS. We note that many of these same resistant residues are identical to those from SARS-CoV (Y449, N487, Y489, G496, T500, and G502). The lack of a contiguous surface at the RBS that is conserved makes it highly unlikely that one could identify a naive nAb targeting the RBS that is completely resistant to escape.
A major near-term concern with public health implications is identification of the set of single-nucleotide polymorphisms that encode for escape mutants on the S RBD. A summary of 1-nt escape mutants identified in the present work is shown in Figure 4D. To our knowledge, 40/54 (74%) of 1-nt escape mutants identified from this nAb panel have not previously been identified, including hotspot positions D420 and Y421 that escape recognition by the abundant VH3-53 nAbs. Other notable residues identified here include S477, Q498, and Y501, because these positions lie directly on the RBS and all have been shown to slightly increase binding affinity to ACE2 (Starr et al., 2020). Mutants K417N, E484K, and N501Y in currently circulating lineages escape some, but not all, of the nAbs on the panel.
The current study has been performed in the context of the original Wuhan-Hu-1 strain. Nonetheless, new variants are emerging, and further research should be conducted to gain insight on the escape mutants in the presence of multiple mutations. To that end, we have constructed new libraries containing a constant mutation to E484K and N501Y present in the Alpha, Beta, and Iota variants of concern (88.7% and 91.8% library coverage, respectively; Data S1 contains library coverage statistics).
Discussion
We have developed a yeast platform that allows for the rapid identification of SARS-CoV-2 S RBD escape mutants for a given nAb. Although other platforms to identify escape mutants have recently been described, key advantages of the approach presented here include: (1) screening by competitive binding against ACE2, which more precisely mimics how actual viral infection can still persist despite antibody binding; (2) a robust and rigorous hit identification algorithm; (3) a safe working environment, because it does not use live virus; and (4) a relatively fast identification, because the RBD library can be screened against a given nAb and analyzed in under a week.
Limitations of study
There also exist drawbacks. First, the present method is limited to mapping escape mutants for anti-S-RBD nAbs that directly compete with ACE2 for binding. Many nAbs neutralize by targeting S epitopes across protomers (Barnes et al., 2020) or on the N-terminal domain (Chi et al., 2020), and a robust platform for S ectodomain display would enable more comprehensive studies. We attempted to develop a YSD platform for the full S ectodomain but were unsuccessful: we screened media composition, expression temperature, protein orientation (Figures S1 and S5), and mutations (1,909 mutants screened with only two potential hits) (Data S3; Figure S5). Second, the presented assay measures the ability of a given mutant to escape nAb blockade of ACE2. Although from all available data the assay appears to correlate well in the context of pseudo-virus, each mutation is pleiotropic with unknown fitness effects beyond escape for a given nAb; the true RBS escape mutants that do not appreciably impede viral fitness will be a subset of the mutations identified here.
Still, using this method, we were able to identify specific failure mechanisms for five different nAbs. This tool can be easily adapted and contribute to developing the next generation of broad nAbs against SARS-CoV-2, as well as suggest mutations to include for the next generation of vaccines. The two major prospective applications for this tool then are for monoclonal antibody therapy and universal vaccine design against SARS-CoV-2 (i.e., generating vaccine-elicited antibodies that are resistant to viral escape). The rationale for using this tool in the context of monoclonal antibody therapy is arguably stronger, because FDA-approved therapies such as bamlanivimab (Jones et al., 2020), among others, are not as effective against currently circulating variants. The antibodies used here are from convalescent patients and represent antibodies raised during natural infection. Although some FDA-approved antibodies were not derived from convalescent patients, in principle, any nAb that directly competes with ACE2 binding should be amenable to this technique. We have developed mutagenesis libraries in three different RBD backgrounds, and new libraries could be developed to match genotypes for future variants of concern.
In contrast, it remains to be seen whether this yeast platform could be used in the context of universal vaccine design, because individual nAbs, or combinations thereof, are often not representative of bulk sera. Thus, it would be interesting to see whether our yeast platform presented here is robust enough to identify escape mutants from bulk sera from convalescent or vaccinated individuals.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-c-myc FITC | Miltenyi Biotec | Cat# 130-116-485 |
| Goat anti-Human IgG Fc Secondary antibody, PE, eBiosciences | Invitrogen | Cat# 12-4998-82 |
| CR3022 | Institute of Protein Design, Laboratory of Prof. Neil King | AB_2848080 |
| CC12.1 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC12.3 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC12.13 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC12.7 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC12.17 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC12.19 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC6.29 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC6.30 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC6.31 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC6.32 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| CC6.33 | The Scripps Research Institute, Laboratory of Prof. Dennis Burton | Rogers et al., 2020 |
| Bacterial and virus strains | ||
| E. coli XL1-Blue Competent cells | Agilent | Cat# 200228 |
| E. coli Mach1™ | Thermo Scientific | Cat# C862003 |
| Chemicals, peptides, and recombinant proteins | ||
| ACE2-Fc | Institute of Protein Design, Laboratory of Prof. Neil King | Walls et al., 2020 |
| NotI-HF | NEB | Cat# R3189 |
| BsaI-HFv2 | NEB | Cat# R3733 |
| CutSmart® Buffer | NEB | Cat# B7204S |
| Nuclease Free Water | IDT | Cat# 11-05-01-14 |
| Ultrapure Agarose | Invitrogen | Cat# 16500-500 |
| TAE Buffer (Tris-acetate-EDTA) (50X) | Thermo Scientific | Cat# B49 |
| SYBR Safe DNA gel stain | Invitrogen | Cat# S33102 |
| Gel loading dye, Purple | NEB | Cat# B7024 |
| UltraPure Salmon Sperm DNA Solution | Invitrogen | Cat# 15632011 |
| Glycerol | Macron™ Chemicals | Cat# 5092-16 |
| HEPES free acid | Millipore | Cat# 391338 |
| HEPES sodium salt | Amresco | Cat# 0485 |
| PEG 3350 | Spectrum | Cat #P0125 |
| Lithium acetate dihydrate | Sigma-Aldrich | Cat# L6883 |
| PBS - Phosphate-Buffered Saline (10X) pH 7.4 | Invitrogen | Cat# AM9624 |
| Streptavidin phycoerythrin (SAPE) | Invitrogen | Cat# S866 |
| Bovine Serum Albumina (BSA), Fraction V, Fatty acid free | VWR | Cat# 7907-25 |
| EZ-Link NHS-Biotin | ThermoFisher | Cat# 20217 |
| Zymolyase | Zymo Research | Cat# E1005 |
| DMSO, Amine-free, Sequencing grade | Thermo Scientific | Cat# 20688 |
| Sodium Chloride | Sigma-Aldrich | Cat# 746398 |
| Difico yeast nitrogen base without amino acids | Sigma-Aldrich | Cat#Y026 |
| Bacto casamino acids, technical grade | Fisher | Cat# 223120 |
| Sodium Phosphate dibasic anhydrous | Fisher Chemical | Cat# BP3321 |
| Sodium Phosphate monobasic monohydrate | Fisher Chemical | Cat# S369-500 |
| D-Galactose | Fisher Bioreagents | Cat# BP656-500 |
| Dextrose | Fisher Chemical | Cat# D19212 |
| Pen/Strep | Fisher | Cat# 15140-122 |
| Kanamycin | GoldBio | Cat# K-120-25 |
| Exonuclease I | NEB | Cat# M0293S |
| Lambda exonuclease | NEB | Cat# M0262S |
| Lambda exonuclease reaction buffer 10X | NEB | Cat# B0262S |
| Q5 HotStart 2X MasterMix | NEB | Cat# M0494L |
| rSAP | NEB | Cat# M0371L |
| 70% v/v Denatured ethanol solution | Fisher Bioreagents | Cat# BP82031GAL |
| IDTE pH 8.0 (1X TE Solution) | IDT | Cat# 11-05-01-13 |
| Quant-iT PicoGreen dsDNA Reagent | Thermo Scientific | Cat# T7581 |
| Lambda DNA | Thermo Scientific | Cat# SD0011 |
| Potassium hydroxide | VWR | Cat# MK698412 |
| Sodium hydroxide | VWR | Cat# MK77081 |
| MES acid | GoldBio | Cat# M-090-1 |
| Phosphoric acid | Sigma-Aldrich | Cat# 345245 |
| Citric acid | Sigma-Aldrich | Cat# 791725 |
| Yeast nitrogen base with ammonium sulfate | Sigma-Aldrich | Cat# Y0626 |
| DMEM | Corning | Cat# 15/013/CV |
| L-Glutamine | Corning | Cat# 25-005-CL |
| Critical commercial assays | ||
| Monarch PCR & DNA Cleanup kit | NEB | Cat# T1030 |
| Monarch DNA Gel Extraction kit | NEB | Cat# T1020 |
| Monarch Plasmid Miniprep kit | NEB | Cat# T1010 |
| Zymoprep Yeast Plasmid miniprep II | Zymo Research | Cat# D2004 |
| Agencourt AMPure XP | Beckman Coulter | Cat# A63881 |
| PhiX | Illumina | Cat# FC-110-3001 |
| Deposited data | ||
| Raw sequencing data | This study | SAMN18250431-SAMN18250483 and SAMN20095117-SAMN20095120 |
| Experimental models: Cell lines | ||
| HEK293T | ATCC | CRL-3216 |
| HeLa-hACE2 | Rogers et al., 2020 | N/A |
| Vero-E6 | ATCC | CRL-1586 |
| Experimental models: Organisms/strains | ||
| Saccharomyces cerevisiae EBY100 | ATCC | MYA-4941™ |
| Oligonucleotides | ||
| Down c-myc Secondary primer for nicking mutagenesis CAAGTCCTCTTCAGAAATAAGCTTTG | Kowalsky et al., 2015 | N/A |
| M13F Secondary primer for oligo pool mutagenesis TGTAAAACGACGGCCAGT | IDT | N/A |
| MBK-175 KanR-fwd CAATAATATTGAAAAAGGAAGAGT | This study | N/A |
| MBK-176 KanR-rev ATGAGTAAACTTGGTCTGACAGTT | This study | N/A |
| MBK-177 A-pUC19-fwd AACTGTCAGACCAAGTTTACTCAT | This study | N/A |
| MBK-178 A-pUC19-rev ACTCTTCCTTTTTCAATATTATTG | This study | N/A |
| MBK-180 DS-tile1-fwd GTTCAGAGTTCTACAGTCCGACGATCACACG TGGTGTTTATTACCCT |
This study | N/A |
| MBK-181 DS-tile1-rev CCTTGGCACCCGAGAATTCCACATAAGA AAAGGCTGAGAGACATA |
This study | N/A |
| MBK-301 DS-tile2-fwd GTTCAGAGTTCTACAGTCCGACGATCCTTAG GGAATTTGTGTTTAAG |
This study | N/A |
| MBK-302 DS-tile2-rev CCTTGGCACCCGAGAATTCCAAACTTCACC AAAAGGGCACAA |
This study | N/A |
| MBK-303 DS-tile3-fwd GTTCAGAGTTCTACAGTCCGACGA TCAGGAAGAGAATCAGCAACTGT |
This study | N/A |
| MBK-304 DS-tile3-rev CCTTGGCACCCGAGAATTCCAATGATTGTAAA GGAAAGTAACA |
This study | N/A |
| MBK-305 DS-tile4-fwd GTTCAGAGTTCTACAGTCCGACGATCAGTAGTAGT ACTTTCTTTTGAACTT |
This study | N/A |
| MBK-306 DS-tile4-rev CCTTGGCACCCGAGAATTCCAGGTGTAATGTC AAGAATCTCAAG |
This study | N/A |
| MBK-307 DS-tile5-fwd GTTCAGAGTTCTACAGTCCGACGATCGAC TCACTTTCTTCCACAGCA |
This study | N/A |
| MBK-308 DS-tile5-rev CCTTGGCACCCGAGAATTCCAAGCTCTGA TTTCTGCAGCTCT |
This study | N/A |
| PJS-P2192 pETCON-NK-BsaI-C-term-fwd TGTTATGGAGCGGGTCTCAGGGGGCGGATCCGAA | This study | N/A |
| PJS-P2193 pETCON-NK-BsaI-C-term-rev ACGTTCAGTGATGGTCTCTACTAGCCTGCAGAGC | This study | N/A |
| PJS-P2194 pETCON-NK-BsaI-N-term-fwd TGTTATGGAGCGGGTCTCACAGGAACTGACA ACTATATGC |
This study | N/A |
| PJS-P2195 pETCON-NK-BsaI-N-term-rev ACGTTCAGTGATGGTCTCTGAAAATATTGAA AAACAGCGAAGTAA |
This study | N/A |
| IFU-128 N501Y fwd CCAACCCACTtatGGTGTTGGTT | This study | N/A |
| IFU-129 N501Y rev AAACCATATGATTGTAAAGGAAAGTAAC | This study | N/A |
| IFU-124 E484K fwd TAATGGTGTTaaaGGTTTTAATTGTTACTTTCCTTTAC | This study | N/A |
| IFU-125 E484K rev CAAGGTGTGCTACCGGCC | This study | N/A |
| IFU-116 K417N fwd GCAAACTGGAaatATTGCTGATTATAATT ATAAATTAC |
This study | N/A |
| IFU-117 K417N rev CCTGGAGCGATTTGTCTG | This study | N/A |
| IFU-118 K417T fwd GCAAACTGGAacgATTGCTGATTATAATTATAAAT | This study | N/A |
| IFU-119 K417T rev CCTGGAGCGATTTGTCTG | This study | N/A |
| IFU-120 D420K fwd AAAGATTGCTaaaTATAATTATAAATTAC CAGATGATTTTACAGGC |
This study | N/A |
| IFU-121 D420K rev CCAGTTTGCCCTGGAGCG | This study | N/A |
| IFU-122 T478R fwd GGCCGGTAGCagaCCTTGTAATG | This study | N/A |
| IFU-123 T478R rev TGATAGATTTCAGTTGAAATATCTCTCTCAAAAG | This study | N/A |
| IFU-126 F486I fwd TGTTGAAGGTattAATTGTTACTTTC | This study | N/A |
| IFU-127 F486I rev CCATTACAAGGTGTGCTAC | This study | N/A |
| IFU-130 Y508H fwd TTACCAACCAcacAGAGTAGTAG | This study | N/A |
| IFU-131 Y508H rev CCAACACCATTAGTGGGTTG | This study | N/A |
| RBD_1 T333-NNK TGGAGGCGGTAGCGGAGGCGGAGG GTCGNNKAACTTGTGCCCTTTTGGTGAAGTTTTTC |
This study | N/A |
| RBD_2 N334-NNK AGGCGGTAGCGGAGGCGGAGGGTCGA CANNKTTGTGCCCTTTTGGTGAAGTTTTTCAAG |
This study | N/A |
| RBD_3 L335-NNK CGGTAGCGGAGGCGGAGGGTCGAC AAACNNKTGCCCTTTTGGTGAAGTTTTTCAAGCCA |
This study | N/A |
| RBD_4 G339-NNK CGGAGGGTCGACAAACTTGTGCCCTTTTN NKGAAGTTTTTCAAGCCACCAGATTTGCAT |
This study | N/A |
| RBD_5 E340-NNK AGGGTCGACAAACTTGTGCCCTTTTGG TNNKGTTTTTCAAGCCACCAGATTTGCATCTG |
This study | N/A |
| RBD_6 A344-NNK CTTGTGCCCTTTTGGTGAAGTTTTTCAAN NKACCAGATTTGCATCTGTTTATGCTTGGA |
This study | N/A |
| RBD_7 T345-NNK GTGCCCTTTTGGTGAAGTTTTTCAAGCCN NKAGATTTGCATCTGTTTATGCTTGGAACA |
This study | N/A |
| RBD_8 R346-NNK CCCTTTTGGTGAAGTTTTTCAAGCCACCN NKTTTGCATCTGTTTATGCTTGGAACAGGA |
This study | N/A |
| RBD_9 S349-NNK TGAAGTTTTTCAAGCCACCAGATTTGCA NNKGTTTATGCTTGGAACAGGAAGAGAATCA |
This study | N/A |
| RBD_10 Y351-NNK TTTTCAAGCCACCAGATTTGCATCTGT TNNKGCTTGGAACAGGAAGAGAATCAGCAACT |
This study | N/A |
| RBD_11 A352-NNK TCAAGCCACCAGATTTGCATCTGTTT ATNNKTGGAACAGGAAGAGAATCAGCAACTGTG |
This study | N/A |
| RBD_12 N354-NNK CACCAGATTTGCATCTGTTTATGCTT GGNNKAGGAAGAGAATCAGCAACTGTGTTGCTG |
This study | N/A |
| RBD_13 K356-NNK ATTTGCATCTGTTTATGCTTGGAACAGG NNKAGAATCAGCAACTGTGTTGCTGATTATT |
This study | N/A |
| RBD_14 R357-NNK TGCATCTGTTTATGCTTGGAACAGGAAGN NKATCAGCAACTGTGTTGCTGATTATTCTG |
This study | N/A |
| RBD_15 I358-NNK ATCTGTTTATGCTTGGAACAGGAAGAGAN NKAGCAACTGTGTTGCTGATTATTCTGTCC |
This study | N/A |
| RBD_16 S359-NNK TGTTTATGCTTGGAACAGGAAGAGAATCN NKAACTGTGTTGCTGATTATTCTGTCCTAT |
This study | N/A |
| RBD_17 N360-NNK TTATGCTTGGAACAGGAAGAGAATCAGC NNKTGTGTTGCTGATTATTCTGTCCTATATA |
This study | N/A |
| RBD_18 V362-NNK TTGGAACAGGAAGAGAATCAGCAACTGTN NKGCTGATTATTCTGTCCTATATAATTCCG |
This study | N/A |
| RBD_19 A363-NNK GAACAGGAAGAGAATCAGCAACTGTGTT NNKGATTATTCTGTCCTATATAATTCCGCAT |
This study | N/A |
| RBD_20 D364-NNK CAGGAAGAGAATCAGCAACTGTGTTGCTNN KTATTCTGTCCTATATAATTCCGCATCAT |
This study | N/A |
| RBD_21 S366-NNK GAGAATCAGCAACTGTGTTGCTGATTATN NKGTCCTATATAATTCCGCATCATTTTCCA |
This study | N/A |
| RBD_22 V367-NNK AATCAGCAACTGTGTTGCTGATTATTCTNN KCTATATAATTCCGCATCATTTTCCACTT |
This study | N/A |
| RBD_23 N370-NNK CTGTGTTGCTGATTATTCTGTCCTATATNN KTCCGCATCATTTTCCACTTTTAAGTGTT |
This study | N/A |
| RBD_24 A372-NNK TGCTGATTATTCTGTCCTATATAATTCCN NKTCATTTTCCACTTTTAAGTGTTATGGAG |
This study | N/A |
| RBD_25 S373-NNK TGATTATTCTGTCCTATATAATTCCGCAN NKTTTTCCACTTTTAAGTGTTATGGAGTGT |
This study | N/A |
| RBD_26 S375-NNK TTCTGTCCTATATAATTCCGCATCATTTNN KACTTTTAAGTGTTATGGAGTGTCTCCTA |
This study | N/A |
| RBD_27 T376-NNK TGTCCTATATAATTCCGCATCATTTTCCN NKTTTAAGTGTTATGGAGTGTCTCCTACTA |
This study | N/A |
| RBD_28 F377-NNK CCTATATAATTCCGCATCATT TTCCACTNNKAAGTGTTATGGAGTGT CTCCTACTAAAT |
This study | N/A |
| RBD_29 K378-NNK ATATAATTCCGCATCATTTTCCACTTTTNN KTGTTATGGAGTGTCTCCTACTAAATTAA |
This study | N/A |
| RBD_30 Y380-NNK TTCCGCATCATTTTCCACTTTTAAGTGT NNKGGAGTGTCTCCTACTAAATTAAATGATC |
This study | N/A |
| RBD_31 G381-NNK CGCATCATTTTCCACTTTTAAGTGTTATN NKGTGTCTCCTACTAAATTAAATGATCTCT |
This study | N/A |
| RBD_32 V382-NNK ATCATTTTCCACTTTTAAGTGTTATGGA NNKTCTCCTACTAAATTAAATGATCTCTGCT |
This study | N/A |
| RBD_33 S383-NNK ATTTTCCACTTTTAAGTGTTATGGAGTGN NKCCTACTAAATTAAATGATCTCTGCTTTA |
This study | N/A |
| RBD_34 P384-NNK TTCCACTTTTAAGTGTTATGGAGTGTCTN NKACTAAATTAAATGATCTCTGCTTTACTA |
This study | N/A |
| RBD_35 T385-NNK CACTTTTAAGTGTTATGGAGTGTCTCCTN NKAAATTAAATGATCTCTGCTTTACTAATG |
This study | N/A |
| RBD_36 K386-NNK TTTTAAGTGTTATGGAGTGTCTCCTAC TNNKTTAAATGATCTCTGCTTTACTAATGTCT |
This study | N/A |
| RBD_37 N388-NNK GTGTTATGGAGTGTCTCCTACTAAATTAN NKGATCTCTGCTTTACTAATGTCTATGCAG |
This study | N/A |
| RBD_38 D389-NNK TTATGGAGTGTCTCCTACTAAATTAAATNN KCTCTGCTTTACTAATGTCTATGCAGATT |
This study | N/A |
| RBD_39 L390-NNK TGGAGTGTCTCCTACTAAATTAAATGAT NNKTGCTTTACTAATGTCTATGCAGATTCAT |
This study | N/A |
| RBD_40 N394-NNK TACTAAATTAAATGATCTCTGCTTTACTNN KGTCTATGCAGATTCATTTGTAATTAGAG |
This study | N/A |
| RBD_41 Y396-NNK ATTAAATGATCTCTGCTTTACTAATGTCNN KGCAGATTCATTTGTAATTAGAGGTGATG |
This study | N/A |
| RBD_42 D405-NNK CTATGCAGATTCATTTGTAATTAGAGGT NNKGAAGTCAGACAAATCGCTCCAGGGCAAA |
This study | N/A |
| RBD_43 R408-NNK TTCATTTGTAATTAGAGGTGATGAAGTCN NKCAAATCGCTCCAGGGCAAACTGGAAAGA |
This study | N/A |
| RBD_44 A411-NNK AATTAGAGGTGATGAAGTCAGACAAATCN NKCCAGGGCAAACTGGAAAGATTGCTGATT |
This study | N/A |
| RBD_45 P412-NNK TAGAGGTGATGAAGTCAGACAAATCGCT NNKGGGCAAACTGGAAAGATTGCTGATTATA |
This study | N/A |
| RBD_46 G413-NNK AGGTGATGAAGTCAGACAAATCGCTCC ANNKCAAACTGGAAAGATTGCTGATTATAATT |
This study | N/A |
| RBD_47 Q414-NNK TGATGAAGTCAGACAAATCGCTCCAG GGNNKACTGGAAAGATTGCTGATTATAATTATA |
This study | N/A |
| RBD_48 T415-NNK TGAAGTCAGACAAATCGCTCCAGGGC AANNKGGAAAGATTGCTGATTATAATTATAAAT |
This study | N/A |
| RBD_49 K417-NNK CAGACAAATCGCTCCAGGGCAAACTGGA NNKATTGCTGATTATAATTATAAATTACCAG |
This study | N/A |
| RBD_50 D420-NNK CGCTCCAGGGCAAACTGGAAAGATT GCTNNKTATAATTATAAATTACCAGATGATTTTA |
This study | N/A |
| RBD_51 Y421-NNK TCCAGGGCAAACTGGAAAGATTGCT GATNNKAATTATAAATTACCAGATGATTTTACAG |
This study | N/A |
| RBD_52 K424-NNK AACTGGAAAGATTGCTGATTATAATTA TNNKTTACCAGATGATTTTACAGGCTGCGTTA |
This study | N/A |
| RBD_53 P426-NNK AAAGATTGCTGATTATAATTATAAATTAN NKGATGATTTTACAGGCTGCGTTATAGCTT |
This study | N/A |
| RBD_54 D427-NNK GATTGCTGATTATAATTATAAATTACCAN NKGATTTTACAGGCTGCGTTATAGCTTGGA |
This study | N/A |
| RBD_55 D428-NNK TGCTGATTATAATTATAAATTACCAGAT NNKTTTACAGGCTGCGTTATAGCTTGGAATT |
This study | N/A |
| RBD_56 T430-NNK TTATAATTATAAATTACCAGATGATTTTN NKGGCTGCGTTATAGCTTGGAATTCTAACA |
This study | N/A |
| RBD_57 N437-NNK TGATTTTACAGGCTGCGTTATAGC TTGGNNKTCTAACAATCTTGATTCTAAGGTTGGTG |
This study | N/A |
| RBD_58 N439-NNK TACAGGCTGCGTTATAGCTTGGAATTC TNNKAATCTTGATTCTAAGGTTGGTGGTAATT |
This study | N/A |
| RBD_59 N440-NNK AGGCTGCGTTATAGCTTGGAATTCTAACN NKCTTGATTCTAAGGTTGGTGGTAATTATA |
This study | N/A |
| RBD_60 L441-NNK CTGCGTTATAGCTTGGAATTCTAACAA TNNKGATTCTAAGGTTGGTGGTAATTATAATT |
This study | N/A |
| RBD_61 K444-NNK AGCTTGGAATTCTAACAATCTTGATTCTN NKGTTGGTGGTAATTATAATTACCTGTATA |
This study | N/A |
| RBD_62 V445-NNK TTGGAATTCTAACAATCTTGATTCTAAGN NKGGTGGTAATTATAATTACCTGTATAGAT |
This study | N/A |
| RBD_63 G446-NNK GAATTCTAACAATCTTGATTCTAAGGTT NNKGGTAATTATAATTACCTGTATAGATTGT |
This study | N/A |
| RBD_64 G447-NNK TTCTAACAATCTTGATTCTAAGGTT GGTNNKAATTATAATTACCTGTATAGATTGTTTA |
This study | N/A |
| RBD_65 N448-NNK TAACAATCTTGATTCTAAGGTTGG TGGTNNKTATAATTACCTGTATAGATTGTTTAGGA |
This study | N/A |
| RBD_66 Y449-NNK CAATCTTGATTCTAAGGTTGG TGGTAATNNKAATTACCTGTATAG ATTGTTTAGGAAGT |
This study | N/A |
| RBD_67 N450-NNK TCTTGATTCTAAGGTTGGTGGTAATTATN NKTACCTGTATAGATTGTTTAGGAAGTCTA |
This study | N/A |
| RBD_68 L452-NNK TTCTAAGGTTGGTGGTAATTATAATTACNNKTAT AGATTGTTTAGGAAGTCTAATCTCA |
This study | N/A |
| RBD_69 L455-NNK TGGTGGTAATTATAATTACCTGTATAGANNK TTTAGGAAGTCTAATCTCAAACCTTTTG |
This study | N/A |
| RBD_70 F456-NNK TGGTAATTATAATTACCTGTATAGATTGN NKAGGAAGTCTAATCTCAAACCTTTTGAGA |
This study | N/A |
| RBD_71 R457-NNK TAATTATAATTACCTGTATAGATTGTTTNNKAA GTCTAATCTCAAACCTTTTGAGAGAG |
This study | N/A |
| RBD_72 K458-NNK TTATAATTACCTGTATAGATTGTTTAGG NNKTCTAATCTCAAACCTTTTGAGAGAGATA |
This study | N/A |
| RBD_73 S459-NNK TAATTACCTGTATAGATTGTTTAGGAAGNNKA ATCTCAAACCTTTTGAGAGAGATATTT |
This study | N/A |
| RBD_74 N460-NNK TTACCTGTATAGATTGTTTAGGAAGTCTNN KCTCAAACCTTTTGAGAGAGATATTTCAA |
This study | N/A |
| RBD_75 K462-NNK GTATAGATTGTTTAGGAAGTCTAATCTCN NKCCTTTTGAGAGAGATATTTCAACTGAAA |
This study | N/A |
| RBD_76 P463-NNK TAGATTGTTTAGGAAGTCTAATCTCAA ANNKTTTGAGAGAGATATTTCAACTGAAATCT |
This study | N/A |
| RBD_77 F464-NNK ATTGTTTAGGAAGTCTAATCTCAAAC CTNNKGAGAGAGATATTTCAACTGAAATCTATC |
This study | N/A |
| RBD_78 E465-NNK GTTTAGGAAGTCTAATCTCAAACCTT TTNNKAGAGATATTTCAACTGAAATCTATCAGG |
This study | N/A |
| RBD_79 R466-NNK TAGGAAGTCTAATCTCAAACCTTTTGAG NNKGATATTTCAACTGAAATCTATCAGGCCG |
This study | N/A |
| RBD_80 I468-NNK GTCTAATCTCAAACCTTTTGAGAGAGATN NKTCAACTGAAATCTATCAGGCCGGTAGCA |
This study | N/A |
| RBD_81 S469-NNK TAATCTCAAACCTTTTGAGAGAGATATTN NKACTGAAATCTATCAGGCCGGTAGCACAC |
This study | N/A |
| RBD_82 T470-NNK TCTCAAACCTTTTGAGAGAGATATTTC ANNKGAAATCTATCAGGCCGGTAGCACACCTT |
This study | N/A |
| RBD_83 E471-NNK CAAACCTTTTGAGAGAGATATTTCAACTN NKATCTATCAGGCCGGTAGCACACCTTGTA |
This study | N/A |
| RBD_84 Y473-NNK TTTTGAGAGAGATATTTCAACTGAAATCNN KCAGGCCGGTAGCACACCTTGTAATGGTG |
This study | N/A |
| RBD_85 Q474-NNK TGAGAGAGATATTTCAACTGAAATCTATN NKGCCGGTAGCACACCTTGTAATGGTGTTG |
This study | N/A |
| RBD_86 A475-NNK GAGAGATATTTCAACTGAAATCTATCAGN NKGGTAGCACACCTTGTAATGGTGTTGAAG |
This study | N/A |
| RBD_87 S477-NNK TATTTCAACTGAAATCTATCAGGCCGGTN NKACACCTTGTAATGGTGTTGAAGGTTTTA |
This study | N/A |
| RBD_88 T478-NNK TTCAACTGAAATCTATCAGGCCGGTAGC NNKCCTTGTAATGGTGTTGAAGGTTTTAATT |
This study | N/A |
| RBD_89 P479-NNK AACTGAAATCTATCAGGCCGGTAGCACAN NKTGTAATGGTGTTGAAGGTTTTAATTGTT |
This study | N/A |
| RBD_90 N481-NNK AATCTATCAGGCCGGTAGCACACCTTG TNNKGGTGTTGAAGGTTTTAATTGTTACTTTC |
This study | N/A |
| RBD_91 G482-NNK CTATCAGGCCGGTAGCACACCTTGTAAT NNKGTTGAAGGTTTTAATTGTTACTTTCCTT |
This study | N/A |
| RBD_92 V483-NNK TCAGGCCGGTAGCACACCTTGTAATGGT NNKGAAGGTTTTAATTGTTACTTTCCTTTAC |
This study | N/A |
| RBD_93 E484-NNK GGCCGGTAGCACACCTTGTAATGGTGTTN NKGGTTTTAATTGTTACTTTCCTTTACAAT |
This study | N/A |
| RBD_94 G485-NNK CGGTAGCACACCTTGTAATGGTGTTGAANN KTTTAATTGTTACTTTCCTTTACAATCAT |
This study | N/A |
| RBD_95 F486-NNK TAGCACACCTTGTAATGGTGTTGAAGGTN NKAATTGTTACTTTCCTTTACAATCATATG |
This study | N/A |
| RBD_96 N487-NNK CACACCTTGTAATGGTGTTGAAGGTTTTN NKTGTTACTTTCCTTTACAATCATATGGTT |
This study | N/A |
| RBD_97 Y489-NNK TTGTAATGGTGTTGAAGGTTTTAATTG TNNKTTTCCTTTACAATCATATGGTTTCCAAC |
This study | N/A |
| RBD_98 F490-NNK TAATGGTGTTGAAGGTTTTAATTGTTACN NKCCTTTACAATCATATGGTTTCCAACCCA |
This study | N/A |
| RBD_99 L492-NNK TGTTGAAGGTTTTAATTGTTACTTTCCTNN KCAATCATATGGTTTCCAACCCACTAATG |
This study | N/A |
| RBD_100 Q493-NNK TGAAGGTTTTAATTGTTACTTTCCTTTA NNKTCATATGGTTTCCAACCCACTAATGGTG |
This study | N/A |
| RBD_101 S494-NNK AGGTTTTAATTGTTACTTTCCTTTACAANN KTATGGTTTCCAACCCACTAATGGTGTTG |
This study | N/A |
| RBD_102 Q498-NNK TTACTTTCCTTTACAATCATATGGTTTCNN KCCCACTAATGGTGTTGGTTACCAACCAT |
This study | N/A |
| RBD_103 P499-NNK CTTTCCTTTACAATCATATGGTTTCCAANN KACTAATGGTGTTGGTTACCAACCATACA |
This study | N/A |
| RBD_104 T500-NNK TCCTTTACAATCATATGGTTTCCAACCCN NKAATGGTGTTGGTTACCAACCATACAGAG |
This study | N/A |
| RBD_105 N501-NNK TTTACAATCATATGGTTTCCAACCCACTNN KGGTGTTGGTTACCAACCATACAGAGTAG |
This study | N/A |
| RBD_106 V503-NNK ATCATATGGTTTCCAACCCACTAATGGT NNKGGTTACCAACCATACAGAGTAGTAGTAC |
This study | N/A |
| RBD_107 G504-NNK ATATGGTTTCCAACCCACTAATGGTGT TNNKTACCAACCATACAGAGTAGTAGTACTTT |
This study | N/A |
| RBD_108 Y505-NNK TGGTTTCCAACCCACTAATGGTGTTGGT NNKCAACCATACAGAGTAGTAGTACTTTCTT |
This study | N/A |
| RBD_109 Q506-NNK TTTCCAACCCACTAATGGTGTTGGTT ACNNKCCATACAGAGTAGTAGTACTTTCTTTTG |
This study | N/A |
| RBD_110 Y508-NNK ACCCACTAATGGTGTTGGTTACCAACCA NNKAGAGTAGTAGTACTTTCTTTTGAACTTC |
This study | N/A |
| RBD_111 E516-NNK ACCATACAGAGTAGTAGTACTTTCTTTT NNKCTTCTACATGCACCAGCAACTGTTTGTG |
This study | N/A |
| RBD_112 L517-NNK ATACAGAGTAGTAGTACTTTCTTTTGAANN KCTACATGCACCAGCAACTGTTTGTGGAC |
This study | N/A |
| RBD_113 L518-NNK CAGAGTAGTAGTACTTTCTTTTGAACTTN NKCATGCACCAGCAACTGTTTGTGGACCTA |
This study | N/A |
| RBD_114 H519-NNK AGTAGTAGTACTTTCTTTTGAACTTCTAN NKGCACCAGCAACTGTTTGTGGACCTAAAA |
This study | N/A |
| RBD_115 A520-NNK AGTAGTACTTTCTTTTGAACTTCTACA TNNKCCAGCAACTGTTTGTGGACCTAAAAAGT |
This study | N/A |
| RBD_116 P521-NNK AGTACTTTCTTTTGAACTTCTACATGCA NNKGCAACTGTTTGTGGACCTAAAAAGTCTA |
This study | N/A |
| RBD_117 A522-NNK ACTTTCTTTTGAACTTCTACATGCACCA NNKACTGTTTGTGGACCTAAAAAGTCTACTA |
This study | N/A |
| RBD_118 T523-NNK TTCTTTTGAACTTCTACATGCACCAGC ANNKGTTTGTGGACCTAAAAAGTCTACTAATT |
This study | N/A |
| RBD_119 P527-NNK TCTACATGCACCAGCAACTGTTTGTGG ANNKAAAAAGTCTACTAATTTGGTTAAAAACA |
This study | N/A |
| IFU-104 L1_Inner_FWD gttcagagttctacagtccgacgatcTGGAGGAGGCTCTGG | This study | N/A |
| IFU-105 L1_Inner_REV ccttggcacccgagaattccaCCAAGCTATAACGCAGCC | This study | N/A |
| IFU-106 L2_Inner_FWD gttcagagttctacagtccgacgatcGGCTGCG TTATAGCTTGG |
This study | N/A |
| IFU-107 L2_Inner_REV ccttggcacccgagaattccaGCCCCCTTTG TTTTTAACCAA |
This study | N/A |
| Forward Outer Primer AATGATACGGCGACCACCGAGATCTACAC GTTCAGAGTTCTACAGTCCGACGATC |
Kowalsky et al., 2015 | N/A |
| Reverse Outer Primer CAAGCAGAAGAC GGCATACGAGATNNNNNNGTGACTGGAGTTCCTT GGCACCCGAGAATTCCA “NNNNNN” is the barcode. |
Kowalsky et al., 2015 | N/A |
| Recombinant DNA | ||
| pUC19-S-ecto-B S ectodomain fragment positions 501-814 with BsaI sites for assembly and BbvCI site | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pUC19-S-ecto-C-Nterm S ectodomain fragment positions 815-1198 with a C-terminal T4 fibritin trimerization domain with BsaI sites for assembly and BbvCI site | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pUC19-S-ecto-A-Nterm-KanR S ectodomain fragment positions 13-500 with BsaI sites for assembly and BbvCI site | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pUC19-S-ecto-Nterm S ectodomain for N-terminal YSD | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pJS697 YSD vector backbone (C-terminal fusion) for in vivo HR | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pJS698 YSD vector backbone (N-terminal fusion) for in vivo HR | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pJS699 S-RBD(333-537)-N343Q | Banach et al., 2021 | https://www.addgene.org/Timothy_Whitehead/ |
| pIFU001 S-RBD(333-537)-N343Q-E484K | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pIFU002 S-RBD(333-537)-N343Q-N501Y | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pIFU003 S-RBD(333-537)-N343Q-T478R | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pIFU004 S-RBD(333-537)-N343Q-K417N | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pIFU005 S-RBD(333-537)-N343Q-K417T | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pIFU006 S-RBD(333-537)-N343Q-Y508H | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pIFU007 S-RBD(333-537)-N343Q-F486I | This study | https://www.addgene.org/Timothy_Whitehead/ |
| pIFU008 S-RBD(333-537)-N343Q-D420K | This study | https://www.addgene.org/Timothy_Whitehead/ |
| Software and algorithms | ||
| Python software (dms and analysis modules) | This paper | https://github.com/WhiteheadGroup/SpikeRBDStabilization |
| GraphPad | N/A | https://www.graphpad.com/ |
| Python3 | N/A | https://www.python.org/ |
| Benchling | N/A | https://www.benchling.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Timothy A. Whitehead (timothy.whitehead@colorado.edu).
Materials availability
Pooled libraries and plasmids from this study will be available at Addgene: https://www.addgene.org/Timothy_Whitehead/.
Experimental model and subject details
Cell lines
Saccharomyces cerevisiae strain EBY100 (ATCC MYA-4941™) was cultured at 30°C for cell growth, and at 22°C for cell induction in flasks while shaking at 300rpm. Cells were incubated in 6.7g/L Difco yeast nitrogen base, 5g/L Bacto casamino acids, 5.4g/L Na2HPO4, and 8.56 g/L NaH2PO4·H2O and 20 g/L carbon source (dextrose for cell growth and galactose for cell induction). HEK293T (ATCC CRL-3219) were cultured in DMEM (Corning 15-013-CV) with 10% heat-inactivated FBS, 2 mM L-glutamine and 1X PenStrep at 37°C in a humidified 5% CO2 incubator. Vero-E6 cells (ATCC CRL-1586) were plated in a T225 flask with complete DMEM (Corning 15-013-CV) containing 10% FBS, 1X PenStrep, 2 mM L- Glutamine (Corning 25-005-CL) overnight at 37°C and 5% CO2. HeLa-ACE2 cells were seeded in 12 μL complete DMEM at a density of 2x103 cells per well.
Bacterial strains
Escherichia coli strain XL1-Blue and Mach1™ were incubated in LB media at 37°C and 300rpm in culture tubs.
Method details
Plasmid constructs
All plasmids and primers used for this work are listed in the Key resources table. All plasmids were verified by Sanger sequencing. Yeast display constructs for SARS-CoV-2 spike protein ectodomain (GenBank MN908947 with a GSAS substitution at the furin cleavage site (682-685) and proline substitutions at positions 986 and 987 (Wrapp et al., 2020), and a C-terminal T4 fibritin trimerization domain), as shown in Figure S1, were constructed as follows. Spike was codon optimized for Saccharomyces cerevisiae with Benchling software using default options, split into three gene blocks (hereafter labeled A, B, and C) each encoded with BsaI restriction sites with overhangs (Potapov et al., 2018), synthesized as gBlocks (IDT), and cloned into pUC19 (Addgene: #50005) using SalI/KpnI restriction sites. This yielded the spike fragment entry plasmids pUC19-S-ecto-B and pUC19-S-ecto-C-Nterm. To construct pUC19-S-ecto-A-Nterm-KanR (the spike fragment destination plasmid), PCR was used to amplify both the kanamycin resistance gene from pETconNK (Addgene: #81169) with primers MBK-175 and MBK-176, and the pUC19-S-ecto-A-Nterm plasmid with primers MBK-177 and MBK-178. NEBuilder HiFi DNA Assembly protocol (NEB) was used to insert the kanamycin resistance gene into the plasmid. pUC19-S-ecto-Nterm was constructed by Golden Gate cloning (Engler and Marillonnet, 2014) using pUC19-S-ecto-A-Nterm-KanR, pUC19-S-ecto-B, and pUC19-S-ecto-C-Nterm.
To construct pJS698 (N-terminal fusion Spike ectodomain YSD backbone), pETconNK-Nterm-Aga2p was first constructed by inserted a gene block with a multiple cloning site between the AGA2 signal peptide and the remainder of the AGA2 coding sequence following standard restriction enzyme cloning practices. pETconNK-Nterm-Aga2p was amplified with primers PJS-P2194 and PJS-P2195 using KAPA HiFi HotStart Readymix (Kapa Biosystems). The reaction was fractionated by agarose gel electrophoresis and the 6062 bp band excised and purified using a Monarch DNA Gel Extraction kit (NEB). The fragment (40 ng) was circularized using the Q5® Site-Directed Mutagenesis Kit (NEB) in a 10 μL reaction and transformed into E. coli Mach1 chemically competent cells (Invitrogen).
To construct pJS697 (C-terminal fusion RBD YSD backbone), pETconNK (Addgene: #81169) was amplified with primers PJS-P2192 and PJS-P2193 using KAPA HiFi HotStart Readymix (Kapa Biosystems). The reaction was fractionated by agarose gel electrophoresis and the 6084 bp band excised and purifed using a Monarch DNA Gel Extraction kit (NEB). The fragment (40 ng) was circularized using the Q5® Site-Directed Mutagenesis Kit (NEB) in a 10 μL reaction and transformed into E. coli Mach1 chemically competent cells (Invitrogen).
pJS699 (Wuhan-Hu-1 S-RBD(333-537)-N343Q for fusion to the C terminus of AGA2) was previously described (Banach et al., 2021). S RBD Single point mutants were introduced following the Kapa HiFi Hotstart Readymix protocol (Cat# 7958927001) with the following protocol:
-
•
3 min @ 95°C
-
•
25 cycles of:
-
•
20 s @ 98°C
-
•
15 s @ melting temperature of each primer
-
•
2:40 min @ 72°C
-
•
3:40 min @ 72°C
-
•
Hold @ 4°C
Amplicons were fractionated by agarose gel electrophoresis and purified using a Monarch DNA Gel Extraction Kit (NEB). Further ligation of the purified amplicons was performed using T4 ligase and PEG. Finally, the plasmids were transformed into E. coli Mach1 cells and incubated overnight. On the following day the DNA was extracted using an NEB Miniprep Kit. pIFU001 - pIFU008 contain the single mutants E484K, N501Y, T478R, K417N. K417T, Y508H, F486I and D420K respectively.
Recombinant protein production, purification, and preparation
ACE2-Fc was produced and purified following Walls et al. (2020). CR3022 (ter Meulen et al., 2006) was expressed by transient transfection in Expi293F cells and purified by protein A affinity chromatography and SEC using a Superdex 200 10/300 GL. Specificity was verified by measuring binding to SARS-CoV-2 RBD and irrelevant antigen. The anti-SARS-CoV-2 RBD antibody panel used (CC6.29, CC6.30, CC6.31, CC6.32, CC6.33, CC12.1, CC12.3, CC12.7, CC12.13, CC12.17, CC12.19) was a kind gift from Dennis Burton’s lab at Scripps and were produced and purified according to Rogers et al. (2020).
All proteins that were chemically biotinylated were prepared at a 20:1 molar ratio of biotin to protein using EZ-Link NHS-Biotin (Thermo Scientific) according to the manufacturer’s protocol. All proteins were stored at 4°C in phosphate buffered saline (8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, 0.24 g/L KH2PO4) pH 7.4.
Preparation of Mutagenic Libraries
All 119 surface exposed positions on S RBD (positions 333-537) were mutated to every other amino acid plus stop codon using NNK primers using comprehensive nicking mutagenesis exactly as described (Wrenbeck et al., 2016). For compatibility with Illumina sequencing, two tiles were made: tile 1 encompassed positions 333-436, while tile 2 encompassed positions 437-527 containing the critical receptor binding site. Serial dilutions were plated to calculate the transformation efficiency (Data S1).
To create the display construct of S-RBD(333-537)-N343Q fused to the C terminus of Aga2p, pJS697 was digested with BsaI-HFv2 (NEB) and purified using a Monarch PCR & DNA Cleanup Kit (NEB). Each mutated pJS699 library was digested with NotI-HF (NEB), the reaction fractionated by agarose gel electrophoresis, and the band corresponding to S-RBD (0.83kb) excised and purified using a Monarch DNA Gel Extraction Kit (NEB). Yeast transformation was performed exactly as described (Medina-Cucurella and Whitehead, 2018). For each library, the two fragments were co-transformed (in a 3:1 molar ratio of S-RBD to backbone) into chemically competent S. cerevisiae EBY100 (Boder and Wittrup, 1997). Serial dilutions were plated on SDCAA and incubated 3 days to calculate the efficiency of the transformation (Data S1). Biological replicates were made on a different day by co-transforming each tile into EBY100 exactly as described. Yeast stocks for each transformation were stored in yeast storage buffer (20 w/v % glycerol, 200 mM NaCl, 20 mM HEPES pH 7.5) at −80°C.
Mutagenic libraries for the N-terminal spike orientation were constructed following oligo pool mutagenesis exactly as described (Medina-Cucurella et al., 2019; Wrenbeck et al., 2016) using pUC19-S-ecto-A-Nterm-KanR, pUC19-S-ecto-B, and pUC19-S-ecto-C-Nterm as templates. For the oligo pool we computationally selected 1,909 mutations hypothesized to either destabilize the ‘down’ conformation, stabilize the ‘up’ conformation, or both (Data S3). The majority of these mutations targeted S1 (94%, 1793/1909) at the NTD, RBD, SD1, and SD2 domains, with the remainder mapping to the boundary between the HR1 and CH domains on S2. After mutagenesis, the mutational libraries were digested with BsaI-HFv2, fractionated by agarose gel electrophoresis, and gel excised and purified with Monarch Gel Extraction kit (NEB). 40 fmol of pUC19-S-ecto-A-NSM-Nterm-KanR, pUC19-S-ecto-B-NSM, and pUC19-S-ecto-C-NSM-Nterm were ligated together with T4 DNA Ligase (NEB), cleaned up and concentrated each to a final volume of 6 μL with Monarch PCR & DNA Cleanup kit (NEB), and transformed into chemically competent E.coli Mach1 cells (Invitrogen cat. #C862003). The resulting library had on average 3 mutations per spike protein per plasmid. Library statistics were determined post sequencing.
To construct the surface display library in yeast, the spike plasmid library was digested with NotI-HF (NEB) and the S coding region was gel purified. The YSD vector pJS698 was digested with BsaI-HFv2 and column purified. 1.3 μg of insert (S coding region) and 1.7 μg of vector were electroporated into 400 μL EBY100 using the method of Benatuil et al. (2010) as written, except that electroporation was performed at 2 kV rather than 2.5 kV. Serial dilutions were plated on SDCAA Agar to calculate the complexity of the library. After recovery, the cells were transferred to 50 mL SDCAA (20 g/L dextrose, 6.7g/L Difco yeast nitrogen base, 5g/L Bacto casamino acids, 5.4g/L Na2HPO4, and 8.56 g/L NaH2PO4·H2O) and grown at 30°C for two days to saturation. The cultures were passaged twice in medium M37D (diluted to OD600 = 0.05 in 120 ml, then to OD600 = 0.4 in 50 ml) and stocks prepared at OD600 = 1 as in Whitehead et al. (2012). The final composition of M37 is 20 g L-1 dextrose or galactose (for M37D, M37G respectively), 5 g L-1 casamino acids, 6.7 g L-1 yeast nitrogen base with ammonium sulfate, 50 mM citric acid, 50 mM phosphoric acid, 80 mM MES acid, neutralized with 90% sodium hydroxide / 10% potassium hydroxide to pH 7. Both media should be prepared by dissolving all reagents except yeast nitrogen base into MilliQ water, adjusting the pH to 7.0 with freshly prepared sodium hydroxide / potassium hydroxide mixture, and adjusting the volume to 9/10th of the final desired volume. Pass the solution through a 0.22 μm filter, both for sterility and to remove particulates that would nucleate struvite. Finish the media by addition of 1/10th volume of 10x filtered yeast nitrogen base.
Yeast Display Titrations and Competition Binding
For cell surface titrations, EBY100 harboring the RBD display plasmid was grown in 1 mL M19D (5 g/l casamino acids, 40 g/l dextrose, 80 mM MES free acid, 50 mM citric acid, 50 mM phosphoric acid, 6.7 g/l yeast nitrogen base, adjusted to pH7 with 9M NaOH, 1M KOH) overnight at 30°C. Expression was induced by resuspending the M19D culture to OD600 = 1 in M19G (5 g/l casamino acids, 40 g/l galactose, 80 mM MES free acid, 50 mM citric acid, 50 mM phosphoric acid, 6.7 g/l yeast nitrogen base, adjusted to pH7 with 9M NaOH, 1M KOH) and growing 22 h at 22°C with shaking at 300 rpm. For CR3022 IgG, yeast surface display titrations were performed as described by Chao et al. (2006) with an incubation time of 4h at room temperature and using secondary labels anti-c-myc-FITC (Miltenyi Biotec) and Goat anti-Human IgG Fc PE conjugate (Invitrogen Catalog # 12-4998-82). Titrations were performed in biological replicates and technical triplicates (n = 6). The levels of display and binding were assessed by fluorescence measurements for FITC and SAPE using the Sony SH800 cell sorter equipped with a 70 μm sorting chip and 488 nm laser.
To test the individual antibody panel binding to S RBD, EBY100 harboring the RBD display plasmid was grown from −80°C cell stocks in 1 mL SDCAA for 4h at 30°C. Expression was induced by resuspending the SDCAA culture to OD600 = 1 in SGCAA and growing at 22h at 22°C with shaking at 300rpm. 1x105 yeast cells were labeled with 10 μg/ml antibody IgG for 30 min at room temperature in PBSF (PBS containing 1g/l BSA). The cells were centrifuged and washed with 200 μL PBSF. They were labeled with 0.6 μL FITC (Miltenyi Biotec), 0.25 μL Goat anti-Human IgG Fc PE (ThermoFisher Scientific) and 49.15 μL PBSF for 10min at 4°C. Cells were then centrifuged, washed with PBSF, and read on a flow cytometer to measure binding of the ACE2. Experiments were performed at least in biological duplicate.
Competitive binding assays on the yeast surface were performed between a free antibody and biotinylated ACE2. S. cerevisiae EBY100 harboring the RBD display plasmid was grown from −80°C cell stocks in 1 mL SDCAA for 4h at 30°C. Expression was induced by resuspending the SDCAA culture to OD600 = 1 in SGCAA and growing at 22h at 22°C with shaking at 300rpm. 1x105 yeast cells were labeled with 10 μg/ml antibody IgG for 30 min at room temperature in PBSF (PBS containing 1g/l BSA). The same cells were labeled with 30nM chemically biotinylated hACE2, in the same tube without washing, for 30min at room temperature in PBSF. The cells were centrifuged and washed with 200 μL PBSF. They were labeled with 0.6 μL FITC (Miltenyi Biotec), 0.25 μL SAPE (Invitrogen) and 49.15 μL PBSF for 10min at 4°C. Cells were then centrifuged, washed with PBSF. The pellet was resuspended in 100 μL and read on a flow cytometer to measure binding of the hACE2.
Yeast Display Screening of S and S RBD libraries
For full-length S ectodomain screening, pUC19-S-ecto-Nterm and pJS698 were independently linearized via digest with restriction enzymes at 37°C for 1 hour, and gel extracted based on size using Monarch DNA Gel Extraction Kit. The linearized regions were co-transformed in a molar ratio of 3:1 insert to vector into chemically competent EBY100 following published protocols (Medina-Cucurella and Whitehead, 2018). EBY100 cells were recovered in nuclease free water for 5 minutes and then plated on two different yeast media agar plates: SDCAA and M37D. Cells were incubated at 30°C for 3 days. After initial growth, colonies from each plate were selected and grown up at 30°C and 250 rpm overnight in the respective dextrose media: SDCAA, M37D. Cells were then induced in respective galactose media at an OD600 = 1 at three different temperatures, 18°C, 22°C, and 30°C for 20 hours.
Induced EBY100 cells were washed with PBSF (8 g/L NaCl, 0.2 g/L KCl, 1.44 g/L Na2HPO4, 0.24 g/L KH2PO4, and 1g/L bovine serum albumin, pH to 7.4 and filter sterilized) and resuspended in PBSF at an OD600 = 10. The cells were then incubated with either 500nM of the biotinylated ACE2-Fc or 500nM of the biotinylated CR3022 for 1 hour at room temperature. The cells were then washed with PBSF and labeled with anti-c-myc fluorescein isothiocyanate (FITC) (Miltenyi Biotec) and streptavidin–R-phycoerythrin (SAPE) (Invitrogen) and incubated on ice for 10 minutes.
The Spike mutagenic library was labeled with CR3022 and, separately, ACE2-Fc under the optimal conditions were screened. Approximately 108 yeast cells were sorted using fluorescence activated cell sorting (FACS), and the top 1% of cells by fluorescence were collected. The two resulting sorted libraries were expanded and sorted in a second round, again screening 108 cells and collecting the top 1% by fluorescence intensity. The selected populations were amplified and purified based on tile, deep sequenced, and count data compared with a reference population.
For the escape mutant screening of the S RBD, 3x107 induced EBY100 yeast cells displaying S RBD were labeled with 10 μg/ml antibody IgG for 30 min at room temperature with mixing by pipetting every 10 min in PBSF (PBS containing 1g/l BSA). The same cells were labeled with 75nM chemically biotinylated ACE2, in the same tube, for 30min at room temperature in PBSF with mixing by pipetting every 10 min. The cells were centrifuged and washed with 1mL PBSF. Cells were then labeled with 1.2 μL FITC, 0.5 μL SAPE and 98.3 μL PBSF for 10min at 4°C. Cells were centrifuged, washed with 1mL PBSF, resuspended to 1 mL PBSF and sorted using FACS. Multiple gates were used for sorting as shown in Figure S3, including an FSC/SSC+ gate for isolation of yeast cells, FSC-H/FSC-A gate to discriminate single cells, a FSC-A/FITC+ gate selects the cells displaying the RBD on their surface and from this last gate, the top 2% by a PE+/FITC+ is collected. At least 2.0x105 cells were collected and were recovered in SDCAA with 50 μg/mL Kanamycin and 1x PenStrep for 30h. For the biological replicates (Figure S3) the ACE2 concentration was 30nM but all other conditions were identical.
Deep Sequencing Preparation
Libraries were prepared for deep sequencing following the “Method B” protocol from Kowalsky et al. (2015) exactly as described for the spike ectodomain libraries and with a few changes for the RBD libraries. A Monarch PCR & DNA Cleanup kit was used. PCR of extracted and cleaned-up yeast plasmid DNA was performed using 2xQ5 HotStart Master Mix (NEB) and the following protocol:
-
•
1 min @ 98°C
-
•
25 cycles of:
-
•
10 s @ 98°C
-
•
20 s @ 64°C
-
•
30 s (replicate 1) or 1 min (replicate 2) @ 72°C
-
•
2 min @ 72°C
-
•
Hold @ 4°C
Primers used in library prep are given in Key resources table. Amplicons were fractionated by agarose gel electrophoresis and purified using a Monarch DNA Gel Extraction Kit (NEB). Samples were then further purified using Agencourt Ampure XP beads (Beckman Coulter), quantified using PicoGreen (ThermoFisher), pooled, and sequenced on an Illumina MiSeq using 2 × 250 bp paired-end reads at the BioFrontiers Sequencing Core (University of Colorado, Boulder, CO).
Molecular Dynamics Simulations
GROMACS 2018.3 (Abraham et al., 2015) was employed for all molecular dynamics (MD) simulations along with the TIP3P (Jorgensen et al., 1983) water model and Amber99SB-ILDN (Lindorff-Larsen et al., 2010) force field to model the receptor binding domain (RBD) of the spike (S) protein of SARS-CoV-2 and neutralizing antibodies CC12.1 and CC12.3. Simulations were initiated from crystal structures of the RBD in complex with CC12.1 (PDB code 6XC2; Yuan et al., 2020b) and CC12.3 (PDB code 6XC4; Yuan et al., 2020b). All systems containing a positive charge were neutralized by the addition of Cl− ions, also modeled with the Amber99SB-ILDN force field. Each simulation consisted of approximately 192,000 atoms.
A steepest descent energy minimization of the initial coordinates for each system was carried out for 5,000 steps. NVT equilibration simulations were then performed for 0.5 ns at 310 K with the Bussi−Donadio−Parrinello (Bussi et al., 2007) thermostat. Subsequent NPT equilibration simulations were performed for 1 ns at 310 K and 1.0 bar, using the same thermostat and Berendsen (Berendsen et al., 1984) barostat. The time constant for coupling in both the NVT and NPT simulations was 0.1 ps. Production simulations in the NPT ensemble were then carried out at 310 K and 1.0 bar with the Bussi−Donadio−Parrinello thermostat and Parrinello–Rahman (Parrinello and Rahman, 1981) barostat. Long-range electrostatic interactions were calculated using particle mesh Ewald summations and a cutoff of 1.0 nm, and Lennard Jones interactions were calculated over 1.0 nm and shifted beyond this distance. Neighbor lists were updated every 10 steps with a cutoff of 1.0 nm. Bonds between hydrogen and heavy atoms were constrained with the LINCS (Hess et al., 1997) algorithm. Furthermore, periodic boundary conditions were used in all simulations in all directions. Production simulations were carried out for 100 ns, leading to a total of 0.8 microseconds of simulation time across the eight simulations.
Pseudo Neutralization Assays
SARS-CoV-2 pseudovirus neutralization assays were performed as previously described (Rogers et al., 2020). Briefly, pseudovirus was generated by cotransfecting MLV-gag/pol and MLV-CMV-Luciferase plasmids with truncated wild-type SARS-CoV-2 or mutant SARS-CoV-2 plasmid respectively onto HEK293T cells. After 48h or 72h of transfection, supernatants containing pseudovirus were collected and frozen at −80°C. Neutralization assay was performed as follows. First, monoclonal antibodies were serially diluted into half-area 96-well plates (Corning, 3688) and incubated with pseudovirus at 37°C for 1 h. Next, HeLa-hACE2 cells were transferred in the 96-well plates at 10,000 cells/well. After 48h of incubation, supernatants were removed, cells were lysed with 1x luciferase lysis buffer (25 mM Gly-Gly pH 7.8, 15 mM MgSO4, 4 mM EGTA, 1% Triton X-100). Finally, Bright-Glo (Promega, PR-E2620) was added onto 96-well plates according to manufacturer’s instructions. Neutralization IC50s were calculated using “One-Site LogIC50” regression in GraphPad Prism 8.0. Pseudovirus mutant constructs were generated by amplifying two overlapped fragments of SARS-CoV-2 mutant sequences with Q5 enzyme (NEB, M0492) following manufacturer’s instructions. Two fragments were then joint into one fragment by bridge PCR, and gibson cloned into digested pcDNA3.3 backbone.
Quantification and statistical analysis
Dissociation constants
The dissociation constants on Figure 2D represent the mean of the replicates (values show as open circles). There are two replicates for each single point mutant and 4 for the wild-type.
Deep Sequencing Analysis
All deep sequencing data analysis was performed by scripts written in Python, available at GitHub (https://github.com/WhiteheadGroup/SpikeRBDStabilization).
Because all sequenced samples were PCR amplicons of known length, paired-end reads were merged by aligning at the known overlap. Mismatches in overlapping regions were resolved by selecting the base pair with the higher quality score and assigning it a quality score given by the absolute difference of the quality scores at the mismatch. Paired reads with more than 10 mismatches in the overlapping region and merged reads containing any quality score less than 10 were discarded. The total number of retained reads in each sample was recorded as , the number of reads in sample .
Each read was compared to the wild-type sequence to identify all mutations. Counts for synonymous single mutations were combined to give , the number of reads in sample encoding the single amino acid mutation . Reads including multiple mutations or mutations not encoded in the library oligos were not analyzed further. The frequency of single mutant in sample was calculated as .
Each experiment consisted of two samples: a reference sample and a selected sample . For each experiment, the risk ratio of variant was calculated as i.e., the ratio of the variant’s frequency in the selected population to its frequency in the reference population. Enrichment ratios were calculated as the binary logarithm of the risk ratio: . Variants with five or fewer counts in the reference population were not analyzed further. Variants with at least five counts in the reference population but no counts in the selected population were given a pseudocount of one.
Determining hits from yeast display screens
For each escape mutant screen, we collected the top 2% (PE channel) of the population of FITC+ (RBD displaying) cells. This population was not labeled with biotinylated ACE2 and so serves as a null experiment where the observed enrichment ratios are due to other sources of variance and not to differential nAb binding. We fit the distribution of enrichment ratios for each of these control samples using kernel density estimation (KDE) (SciPy’s scipy.stats.gaussian_kde with default parameters) (Shalloo et al., 2020). We then treated this distribution estimate as an empirical null hypothesis. Under this null hypothesis, we expect false positives, where N is the number of variants tested, F is the cumulative distribution function (CDF) of the control ER KDE, and ERt is a threshold. Therefore, for a target false discovery rate (FDR), we chose , where F-1 is the inverse CDF of the KDE. In data from samples labeled with nAbs, we then tested the hypothesis that each observed ER was greater than the associated ERt using an one-sided exact Poisson rate ratio test (statsmodels.stats.rates.test_poisson_2indep from the Python library statsmodels) (Seabold and Perktold, 2010). For these tests, the null ratio was . The counts were given by the number of reads for the variant in the selected and reference populations, respectively, and the exposures were given by the total number of reads in the reference and selected populations, respectively. For this analysis, we identified hits for replicate 1 (tiles 1 & 2 for nAbs CC6.29, CC12.1, and CC12.3) using a target FDR of 1 and a Poisson rate ratio test significance level of 0.01. For replicate 2 (tile 2 for nAbs CC6.31, CC12.13) escape mutant hits were identified using a target FDR of 1.
For the full-length S ectodomain screen, our null experiment was the collected reference populations without selections for each of the ACE2-Fc and CR3022 experiments. These reference populations were passaged, sorted, and amplified identically to the sorted libraries except that no screen was employed. We fit the distribution of enrichment ratios for these control samples using a logistic CDF (custom MATLAB script), and the empirical FDR was calculated exactly as above.
Acknowledgments
We thank scientists K. Jackson and A. Scott at the BioFrontiers sequencing core for technical guidance and members of the Whitehead lab for intellectual and material support, including Matt Bedewitz, for reagent prep. The anti-SARS-CoV-2 RBD antibody panel used was a kind gift from Dennis Burton’s lab at Scripps, and the ACE2-Fc and CR3022 were kind gifts from Neil King’s lab at the University of Washington. Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R01AI141452 to T.A.W. and the NIH/CU Molecular Biophysics Program and NIH Biophysics Training Grant T32 GM-065103 to A.C.L. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. This research was also supported by a Balsells Fellowship to I.M.F.-U. and a CU Boulder BSI fellowship to C.M.H.
Author contributions
Designed experiments: I.M.F.-U., P.J.S., M.B.K., E.R.R., K.G.S., J.G.J., and T.A.W.; performed experiments: I.M.F.-U., P.J.S., M.B.K., S.B., L.P., F.Z., E.R.R., and A.C.L.; performed simulations: E.R.R. and K.G.S.; developed algorithms and software: P.J.S., C.M.H., and T.A.W.; wrote paper: I.M.F.-U., P.J.S., M.B.K., K.G.S., and T.A.W.
Declaration of interests
I.M.F.-U., P.J.S., M.B.K., C.M.H., J.G.J., and T.A.W. have a US provisional patent application “IDENTIFICATION OF SARS-COV-2 S RBD ESCAPE MUTANTS” covering some of the mutants identified in the present study.
Published: August 10, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109627.
Supplemental information
Processed Spike ectodomain library results for sorting against ACE2, CR3022.
Data and code availability
-
•
Raw sequencing reads for this work have been deposited in the SRA (Accession #s SAMN18250431-SAMN18250483 for the original Wuhan-Hu-1 S RBD and S ectodomain and Accession #s SAMN20095117-SAMN20095120 for the S RBD E484K and N501Y variants).
-
•
All scripts used to process and analyze deep sequencing data are freely available on Github (https://github.com/WhiteheadGroup/SpikeRBDStabilization).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abraham M.J., Murtola T., Schulz R., Páll S., Smith J.C., Hess B., Lindahl E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. [Google Scholar]
- Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., COVE Study Group Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N. Engl. J. Med. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banach B.B., Cerutti G., Fahad A.S., Shen C.-H., Olivera de Souza M., Katsamba P.S., Tsybovsky Y., Wang P., Nair M.S., Huang Y. Paired Heavy and Light Chain Signatures Contribute to Potent SARS-CoV-2 Neutralization in Public Antibody Responses. bioRxiv. 2021 doi: 10.1101/2020.12.31.424987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C.O., Jette C.A., Abernathy M.E., Dam K.A., Esswein S.R., Gristick H.B., Malyutin A.G., Sharaf N.G., Huey-Tubman K.E., Lee Y.E. SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies. Nature. 2020;588:682–687. doi: 10.1038/s41586-020-2852-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Science. 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benatuil L., Perez J.M., Belk J., Hsieh C.-M. An improved yeast transformation method for the generation of very large human antibody libraries. Protein Eng. Des. Sel. 2010;23:155–159. doi: 10.1093/protein/gzq002. [DOI] [PubMed] [Google Scholar]
- Berendsen H.J.C., Postma J.P.M., van Gunsteren W.F., DiNola A., Haak J.R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- Boder E.T., Wittrup K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997;15:553–557. doi: 10.1038/nbt0697-553. [DOI] [PubMed] [Google Scholar]
- Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- Chao G., Lau W.L., Hackel B.J., Sazinsky S.L., Lippow S.M., Wittrup K.D. Isolating and engineering human antibodies using yeast surface display. Nat. Protoc. 2006;1:755–768. doi: 10.1038/nprot.2006.94. [DOI] [PubMed] [Google Scholar]
- Chen R.E., Zhang X., Case J.B., Winkler E.S., Liu Y., VanBlargan L.A., Liu J., Errico J.M., Xie X., Suryadevara N. Resistance of SARS-CoV-2 variants to neutralization by monoclonal and serum-derived polyclonal antibodies. Nat. Med. 2021;27:717–726. doi: 10.1038/s41591-021-01294-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Yan R., Zhang J., Zhang G., Zhang Y., Hao M., Zhang Z., Fan P., Dong Y., Yang Y. A neutralizing human antibody binds to the N-terminal domain of the Spike protein of SARS-CoV-2. Science. 2020;369:650–655. doi: 10.1126/science.abc6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C., Marillonnet S. In: Valla S., Lale R., editors. Humana Press; 2014. Golden gate cloning; pp. 119–131. (DNA Cloning and Assembly Methods). [Google Scholar]
- Greaney A.J., Starr T.N., Gilchuk P., Zost S.J., Binshtein E., Loes A.N., Hilton S.K., Huddleston J., Eguia R., Crawford K.H.D. Complete Mapping of Mutations to the SARS-CoV-2 Spike Receptor-Binding Domain that Escape Antibody Recognition. Cell Host Microbe. 2021;29:44–57.e9. doi: 10.1016/j.chom.2020.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Bekker H., Berendsen H.J.C., Fraaije J.G.E.M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1997;18:1463–1472. [Google Scholar]
- Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38, 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jigami Y. Yeast Glycobiology and Its Application. Biosci. Biotechnol. Biochem. 2008;72:637–648. doi: 10.1271/bbb.70725. [DOI] [PubMed] [Google Scholar]
- Jones A.B.E., Brown-Augsburger P.L., Corbett K.S., Westendorf K., Davies J., Cujec T.P., Wiethoff C.M., Blackbourne, Heinz B.A., Foster B. LY-CoV555, a rapidly isolated potent neutralizing antibody, provides protection in a non-human primate model of SARS-CoV-2 infection. bioRxiv. 2020 doi: 10.1101/2020.09.30.318972. [DOI] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- Kowalsky C.A., Klesmith J.R., Stapleton J.A., Kelly V., Reichkitzer N., Whitehead T.A. High-resolution sequence-function mapping of full-length proteins. PLoS ONE. 2015;10:e0118193. doi: 10.1371/journal.pone.0118193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., Zhao C., Zhang Q., Liu H., Nie L. The Impact of Mutations in SARS-CoV-2 Spike on Viral Infectivity and Antigenicity. Cell. 2020;182:1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindorff-Larsen K., Piana S., Palmo K., Maragakis P., Klepeis J.L., Dror R.O., Shaw D.E. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., VanBlargan L.A., Bloyet L.M., Rothlauf P.W., Chen R.E., Stumpf S., Zhao H., Errico J.M., Theel E.S., Liebeskind M.J. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe. 2021;29:477–488.e4. doi: 10.1016/j.chom.2021.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Cucurella A.V., Whitehead T.A. Characterizing protein-protein interactions using deep sequencing coupled to yeast surface display. Methods Mol. Biol. 2018;1764:101–121. doi: 10.1007/978-1-4939-7759-8_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Cucurella A.V., Steiner P.J., Faber M.S., Beltrán J., Borelli A.N., Kirby M.B., Cutler S.R., Whitehead T.A. User-defined single pot mutagenesis using unamplified oligo pools. Protein Eng. Des. Sel. 2019;32:41–45. doi: 10.1093/protein/gzz013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M., Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Physiol. 1981;52:7182–7190. [Google Scholar]
- Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., C4591001 Clinical Trial Group Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov V., Ong J.L., Kucera R.B., Langhorst B.W., Bilotti K., Pryor J.M., Cantor E.J., Canton B., Knight T.F., Evans T.C., Jr., Lohman G.J.S. Comprehensive Profiling of Four Base Overhang Ligation Fidelity by T4 DNA Ligase and Application to DNA Assembly. ACS Synth. Biol. 2018;7:2665–2674. doi: 10.1021/acssynbio.8b00333. [DOI] [PubMed] [Google Scholar]
- Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seabold S., Perktold J. Statsmodels: Econometric and Statistical Modeling with Python. Proc. 9th Python Sci. Conf. 2010;2010:57–61. [Google Scholar]
- Shalloo R.J., Dann S.J.D., Gruse J.-N., Underwood C.I.D., Antoine A.F., Arran C., Backhouse M., Baird C.D., Balcazar M.D., Bourgeois N. Automation and control of laser wakefield accelerators using Bayesian optimization. Nat. Commun. 2020;11:6355. doi: 10.1038/s41467-020-20245-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310.e20. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan C.W., Chia W.N., Qin X., Liu P., Chen M.I.-C., Tiu C., Hu Z., Chen V.C.-W., Young B.E., Sia W.R. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat. Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- ter Meulen J., van den Brink E.N., Poon L.L.M., Marissen W.E., Leung C.S.W., Cox F., Cheung C.Y., Bakker A.Q., Bogaards J.A., van Deventer E. Human monoclonal antibody combination against SARS coronavirus: synergy and coverage of escape mutants. PLoS Med. 2006;3:e237. doi: 10.1371/journal.pmed.0030237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., Angus B., Baillie V.L., Barnabas S.L., Bhorat Q.E., Oxford COVID Vaccine Trial Group Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Liu L., Iketani S., Luo Y., Guo Y., Wang M., Yu J., Zhang B., Kwong P.D., Graham B.S. Increased Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7 to Antibody Neutralization. bioRxiv. 2021 doi: 10.1038/s41586-021-03398-2. 2021.01.25.428137. [DOI] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. eLife. 2020;9:e61312. doi: 10.7554/eLife.61312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead T.A., Chevalier A., Song Y., Dreyfus C., Fleishman S.J., De Mattos C., Myers C.A., Kamisetty H., Blair P., Wilson I.A., Baker D. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat. Biotechnol. 2012;30:543–548. doi: 10.1038/nbt.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrenbeck E.E., Klesmith J.R., Stapleton J.A., Adeniran A., Tyo K.E.J., Whitehead T.A. Plasmid-based one-pot saturation mutagenesis. Nat. Methods. 2016;13:928–930. doi: 10.1038/nmeth.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N.C., Yuan M., Liu H., Lee C.D., Zhu X., Bangaru S., Torres J.L., Caniels T.G., Brouwer P.J.M., van Gils M.J. An Alternative Binding Mode of IGHV3-53 Antibodies to the SARS-CoV-2 Receptor Binding Domain. Cell Rep. 2020;33:108274. doi: 10.1016/j.celrep.2020.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Wang F., Shen C., Peng W., Li D., Zhao C., Li Z., Li S., Bi Y., Yang Y. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science. 2020;368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Wu N.C., Zhu X., Lee C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan M., Liu H., Wu N.C., Lee C.D., Zhu X., Zhao F., Huang D., Yu W., Hua Y., Tien H. Structural basis of a shared antibody response to SARS-CoV-2. Science. 2020;369:1119–1123. doi: 10.1126/science.abd2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W., Davis B.D., Chen S.S., Martinez J.M.S., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 strain in Southern California, USA. medRxiv. 2021 doi: 10.1001/jama.2021.1612. 2021.01.18.21249786. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Processed Spike ectodomain library results for sorting against ACE2, CR3022.
Data Availability Statement
-
•
Raw sequencing reads for this work have been deposited in the SRA (Accession #s SAMN18250431-SAMN18250483 for the original Wuhan-Hu-1 S RBD and S ectodomain and Accession #s SAMN20095117-SAMN20095120 for the S RBD E484K and N501Y variants).
-
•
All scripts used to process and analyze deep sequencing data are freely available on Github (https://github.com/WhiteheadGroup/SpikeRBDStabilization).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.