Abstract
Circular RNAs (circRNAs) generated from back-splicing of exons have been found in a wide range of eukaryotic species and exert a variety of biological functions. Unlike canonical splicing, the mechanism of back-splicing has long remained elusive. We recently determined the cryo-EM structure of the yeast spliceosomal E complex assembled on introns, leading us to hypothesize that the same E complex can assemble across an exon forming the exondefinition complex. This complex, when assembled on long exons, goes through the splicing cycle and catalyzes back-splicing to generate circRNAs. Supporting this hypothesis, we purified the yeast post-catalytic spliceosomal P complex (the best complex in the splicing cycle to trap splicing products and intermediates) and detected canonical and back-splicing products as well as splicing intermediates. Here we describe in detail this procedure, which may be applied to other organisms to facilitate research on the biogenesis and regulation of circRNA.
Keywords: CircRNA, Back-splicing, Spliceosome, P complex
1. Introduction
There are multiple types of circular RNAs [1] and we will mainly focus on the one derived from back-splicing (referred to as circRNA) in this paper. This type of circRNA was first discovered in 1993 when the Bailleul group found circular RNAs containing exons from the ets-1 gene [2]. CircRNA was originally thought to be a rare product of aberrant RNA splicing [2]. However, high throughput sequencing demonstrated that circRNAs are abundant, conserved, and wide-spread in all eukaryotic species investigated including metazoans, plants, fungi, and protists [3]. CircRNAs are naturally stable and resistant to degradation by exonucleases once formed [4, 5]. In some cases, circRNAs accumulate to levels that exceed their associated linear mRNAs. Most circRNAs accumulate in the cytoplasm while a subset remains in the nucleus [6]. Increasing evidence shows that circRNAs play a role in the regulation of microRNA activity, alternative splicing, transcription, and have functional roles in immune response and human diseases [6-12]. In addition, several circRNAs were shown to specifically associate with ribosomes to generate proteins under some circumstances, revealing the potential protein-coding function of circRNAs [13, 14].
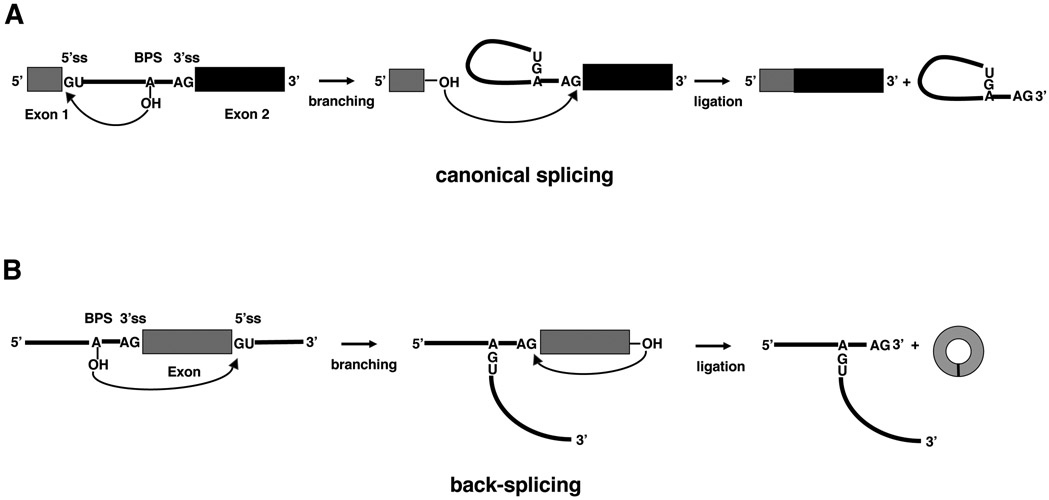
To understand back-splicing and circRNA biogenesis, it is necessary to first introduce canonical splicing. In canonical splicing, intron removal is catalyzed by the spliceosome, a large RNA-protein complex made up of five (U1, U2, U4/U5, U6) small nuclear ribonucleoprotein particles (snRNPs) and numerous non-snRNP factors [15]. The spliceosome undergoes dramatic conformational and compositional changes driven by eight DExH/D-box RNA helicases in the splicing cycle, sequentially forming the E, A, pre-B, B, Bact, B*, C, C*, P, and ILS complexes. Spliceosomal assembly on pre-mRNA is initiated via recognition of intron elements by U1 snRNP and other spliceosomal proteins to form the E complex. Subsequently, the spliceosome transits through the A complex to form the fully assembled pre-B complex, which is then sequentially converted to the B, Bact, and B* complex during the activation stage. The B* complex catalyzes the first transesterification reaction (also called the branching reaction) to form the C complex. In this step, the 2’-OH group of the conserved adenine in the BPS (Branch Point Sequence) attacks the 5’ ss generating a 5’ exon with a free 3’-OH and a lariat intermediate (Fig. 1A). The resulting C complex is converted to C* for the second transesterification reaction in which the newly freed 3’-OH in the 5’ exon attacks the 3’ ss to ligate the two exons and form the P complex. After the release of the ligated exon, the ILS complex forms, which then disassembles to release the intron lariat and recycle the spliceosomal components. However, circRNAs are formed by back-splicing, in which an upstream 3’ ss is joined with a downstream 5’ ss [3, 16, 17] (Fig. 1B). Although back-splicing requires the spliceosome and canonical splice sites [18-20], it is unclear whether the spliceosome that catalyzes back-splicing is the same as the spliceosome that catalyzes the canonical splicing and what triggers an exon to undergo back-splicing instead of canonical splicing.
Fig. 1.
A schematic of canonical splicing and back-splicing. (A) In canonical splicing, the 2’-OH group of the conserved adenine in the BPS attacks the 5’ ss, generating an exon 1 with a free 3’-OH and a lariat intermediate in the first transesterification reaction (branching). In the second transesterification reaction (ligation), the free 3’ OH of exon 1 attacks the 3’ ss, generating a ligated mRNA and intron lariat. (B) In back-splicing, the 2’-OH group of the conserved adenine in the BPS attacks the downstream 5’ ss. Then the free 3’ OH of the exon attacks the upstream 3’ ss, forming an intronic T-branch structure and circRNA consisting of the exon.
To answer these questions, it is important to understand how introns and exons are recognized and defined in the long pre-mRNA sequence. In lower eukaryotes such as yeast, which often contain single or small introns, the intron definition model prevails where the spliceosome initially recognizes and assembles across an intron [21, 22]. Vertebrate genes, on the other hand, typically contain much longer introns separated by short exons [23]. The splicing machinery is more likely to recognize and assemble across an exon through the exon definition model [24]. Most biochemical and structural studies have used model substrates consisting of two exons and an intron, and biochemical and structural analyses of the exon definition process were limited.
We recently determined the structure of the yeast E complex assembled on introns using cryo-EM [17]. Based on the relative positions between the U1 snRNP that recognizes the 5’ ss and the BBP/Mud2 heterodimer that recognizes the BPS, we proposed that the same E complex can form across an exon forming the exon definition complex without the need of any structural rearrangements or additional components. We use the exon definition complex to represent any spliceosomal complexes (including the E complex) assembled across an exon. It is worth noting that the exon definition E complex is likely fluid and commitment to splicing site pairing may not occur until the A complex stage [25, 26]. If the exon on which the spliceosome assembles is short, the exon definition complex will be stalled at the pre-B complex stage and be prompted to undergo remodeling to become intron-spanning and catalyze the canonical splicing reaction. If this exon is long, the exon definition complex catalyzes back-splicing generating a circRNA and a unique T-branch structure (previously referred to as a Y-shaped intermediate) through the same transesterification reactions used by canonical splicing [17].
As a first step to test this hypothesis, we purified spliceosomes stalled at the P complex (which increases the chance of detecting splicing products and intermediates before their release) and evaluated whether the complexes contained both the canonical and back-splicing products and intermediates. There are three approaches to stall the spliceosome at a particular stage in the splicing cycle. One is to use a mutant pre-mRNA substrate that cannot proceed to the next step of the splicing cycle. For example, a U to A mutation (UACUAAC to UACAAAC) that is two nucleotides upstream of the branch point A in the pre-mRNA was successfully used to stall the spliceosome at the A complex stage [27]. The second approach is to adjust the ATP concentration of the reaction, which is required by the DExD/H-box RNA helicases to drive splicing cycle progression. For example, splicing reactions containing 50 μM ATP stall at the B complex stage [28]. The third approach is to use mutant helicases. For example, mutations in helicases Prp2 and Prp22 allow the enrichment of the Bact and P complex since Prp2 and Prp22 are essential for the transition from Bact to B* and from P to ILS complex, respectively [29-32]. Stalling the spliceosome at defined stages combined with tandem affinity purification (TAP) allows for purification of the relevant complexes for subsequent biochemical and structural analyses [33]. Here we describe how we purified the yeast spliceosomal P complex using tandem affinity tags from a yeast strain carrying the Prp22H606A mutant, followed by reverse transcription polymerase chain reaction (RT-PCR) to detect the canonical and back-splicing products and intermediates.
2. Description of the method
2.1. Purification of Yeast Spliceosomal Post-Catalytic P Complex
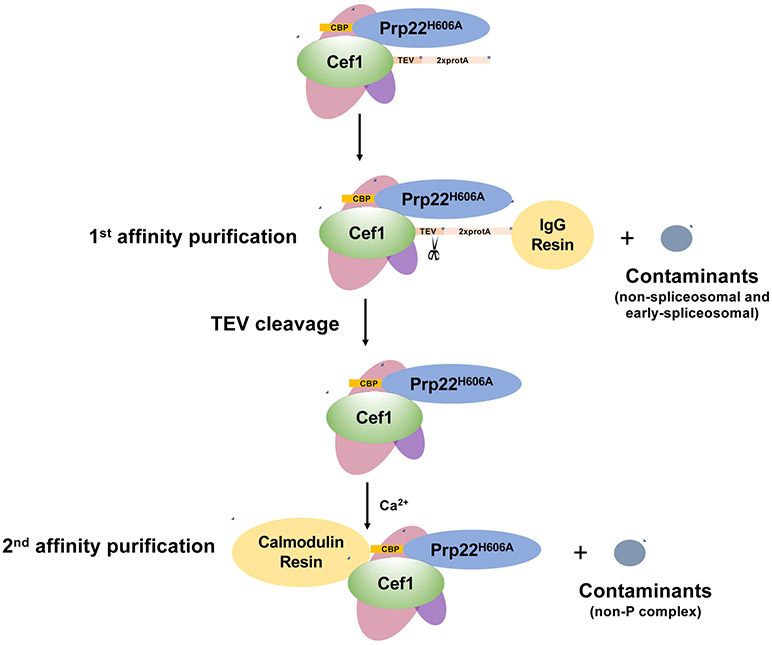
The tandem-affinity method was used to purify the yeast spliceosomal post-catalytic complex through a protein A tag on the C-terminus of Cef1 and a CBP tag on the N-terminus of Prp22H606A from yeast strain yXL352 (Fig. 2). The first affinity purification was carried out using IgG resin utilizing the protein A tag on Cef1. Cef1 belongs to the Nineteen Complex (NTC) which is recruited to the activated spliceosome at the Bact complex stage [34, 35]. Spliceosomes purified using the protein A tag on Cef1 will contain complexes at or after the Bact stage in the splicing cycle. Calmodulin resin was used for a second affinity purification through the CBP tag on Prp22H606A. The essential splicing factor Prp22 catalyzes the release of mRNA from the spliceosome [36]. The H606A in Prp22 leads to defective exon release, and will greatly enrich the P complex and RNA products at this step [32]. We will describe below in detail the steps involved in purifying the P complex.
Fig. 2.
A schematic of yeast spliceosomal post-catalytic P Complex purification using tandem-affinity-purification (TAP). The first affinity purification was carried out with the 2xprotA tag on Ceflwith IgG resin. The second affinity purification was carried out with the CBP tag on Prp22H606A using Calmodulin resin, followed by elution using buffer supplemented with EGTA.
2.1.1. Generation of the Cef1 and Prp22 double tagged yeast strain
To generate the Cef1-protein A and CBP-Prp22H606A tagged yeast strain, the plasmid p358-H606A carrying Prp22H606A and yeast strain YBST1 (Mata ura3-52 trp1-63 his3-Δ200 leu2Δ1 ade2-101 lys2-801 prp22::LEU2 [p360-Prp22 (URA3 CEN)]) were obtained from Dr. Beate Schwer [31, 37]. Cef1 in YBST1 was C-terminal tagged by 2xProtein A tag using PCR-based method [38], resulting in yeast strain yXL332 (Mata ura3-52 trp1-63 his3-Δ200 leu2Δ1 ade2-101 lys2-801 prp22::LEU2 Cef1-2protA::HIS3 [p360-Prp22 (URA3 CEN)]). The coding sequence of the calmodulin-binding peptide (CBP) tag was introduced into p358-H606A using PCR and NEBuilder HiFi DNA Assembly Kit (New England BioLabs). p358-CBP-H606A plasmid was then transformed into yXL332 yeast strain. The resulting Prp22H606 yeast strain yXL352 (Mata ura3-52 trp1-63 his3-Δ200 leu2Δ1 ade2-101 lys2-801 prp22::LEU2 Cef1-2xprotA::HIS3 [p358-CBP-H606A (TRP1 CEN)]) was obtained after shuffling out the wild-type Prp22 plasmid on a FOA plate [39].
2.1.2. Yeast culture
50 ml of starter culture was grown in YPD medium from a large colony on a streaked plate (less than one month old) by shaking at 200 rpm overnight at 30 °C. The doubling time was calculated by checking the OD600 of the yeast strain to estimate how much starter culture to use to inoculate each liter of media to reach OD600 of around 2 after overnight (~16 hr) growth. Three liters of cells were grown another 2hr at 16 °C after OD600 reaches 2. The OD600 of the culture should be 3-4 at the time of harvesting. Cells were then harvested by centrifugation at 5,500 rpm using a F6-6x1000y rotor (Fiberlite) for 8 min at 6 °C. Cell pellet was washed once with 500 ml of cold double-distilled water to remove the YPD medium and weighed. Typically, about 4-5 grams of wet yeast cells can be obtained from one liter of culture.
2.1.3. Cell lysis
The yeast pellet was resuspended in lysis buffer containing 50 mM Tris, pH 8.0, 150 mM NaCl, 0.1% NP-40, 1.5 mM MgCl2, 0.5 mMDTT and 1mM PMSF. Typically, 1 ml lysis buffer was added per 3 g of wet cell pellet. The mixture was vortexed to obtain a thick cell suspension. The cells were flash-frozen in liquid nitrogen to form yeast ‘popcorns’ and cryogenically ground using a SPEX 6870 Freezer/Mill at a rate of 8 cps, 5 min precool time and 2 min run time for 6 cycles. The powder generated can be stored at −80 °C or used immediately in the next purification step.
2.1.4. Preparation of lysate supernatant
The frozen cell powder was thawed on ice and re-suspended in lysis buffer containing 1 mM benzamidine, 1 μM leupeptin, and 1 μg/ml pepstatin A as protease inhibitors. An additional 4.5 ml lysis buffer was added per 3 g of original wet cell pellet at this step. The cell lysate was first centrifuged at 17,000 rpm for 1 hr at 4 °C using a SS34 rotor (Sorvall) and then the supernatant was further centrifuged at 40,000 rpm for 1.5 hr at 4 °C in a 55.2 Ti rotor (Beckman) to obtain cleaner cell lysate.
2.1.5. Purification using IgG resin
The clear supernatant was transferred to pre-cooled 50 ml conical tubes containing IgG Sepharose-6 Fast Flow resin (GE Healthcare) equilibrated with lysis buffer. Typically, 1 ml of dry resin was used for lysate from every 4 liters of yeast culture. The supernatant and the resin were incubated at 4 °C overnight while rotating. After IgG resin was spun down at 1,000 g for 10 min, the supernatant was removed. The resin was combined into a new 15 ml conical tube and washed with 8 x 5 ml of IgG washing buffer containing 20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1.5 mM MgCl2, 0.05% NP40, and 0.5 mM DTT. Then one more wash was performed with 5 ml TEV cutting buffer containing 20 mM Tris-HCl, pH 8.0, 120 mM NaCl, 1.5 mM MgCl2, 0.01% NP40, and 0.5 mM DTT and residual buffer was carefully aspirated. 5 μl of “pre-cut” resin as sample A was taken for SDS-PAGE analysis later.
2.1.6. TEV cleavage
3/5 resin volume of TEV cutting buffer and 1/100 resin volume of TEV protease (Invitrogen) were added to the washed IgG resin with fresh DTT to cut the complex off the resin. The mixture was rotated at 4 °C overnight to release the complex from the IgG resin. The resin was spun down at 1,000g for 10 min the next morning. The supernatant was transferred to a new 1.5 ml microcentrifuge tube as elution 1 (E1) which contains most of the spliceosome complex. The IgG resin was further eluted with 3/5 resin volume of TEV cutting Buffer five times as E2 to E6. 5 μl of “post-cut” resin as sample B was taken. 10 μl of sample from each elution together with Sample A and B was analyzed on SDS-PAGE.
2.1.7. Purification using Calmodulin resin
Eluants containing the P complex were combined in a 15 ml conical tube and supplemented with 2.5 mM CaCl2 and incubated with Calmodulin affinity resin (Agilent) equilibrated with Cal-Washing buffer containing 20 mM Hepes 7.9, 120 mM NaCl, 2 mM CaCl2, ImM MgCl2 and 0.01% NP-40. Typically, 10 μl of dry resin was used for eluants from every 2 liters of original culture. The mixture was rotated for 3 hr at 4 °C. After the resin was spun down and the supernatant discarded, the resin was transferred to a low-adhesion 1.5 ml microfuge tube and washed with 5 x 0.5 ml Cal-washing buffer. After the last wash, the residual washing buffer was removed as thoroughly as possible. The resin was eluted 8 times with 30-50 μl of Cal-Elution buffer containing 20 mM Hepes 7.9, 120 mM NaCl, 2 mM EGTA, and 1 mM MgCl2. These eluted samples can be used to analyze the protein components and RNA products if desired.
2.2. Canonical and Back-Splicing Product and Intermediates Detection
2.2.1. RNA isolation from the P complex
RNA from the purified P complex was isolated using the phenol-chloroform extraction method [40]. Phenol-chloroform (1:1) was added into an equal volume of the P complex elution. The content was mixed thoroughly until an emulsion was formed. The mixture was centrifuged at 18,000 g for 5 min at room temperature. The aqueous phase was transferred into a fresh tube. An equal volume of chloroform was added followed by another round of mixing and centrifugation. The top phase was transferred to a fresh tube with 1/9 volume of 3 M NaOAc (pH 5.2), and 2.5 volumes of 100% ethanol, and 0.5 μl of GlycoBlue (Invitrogen). The sample was frozen in −80 °C for 2-3 hours to allow the RNA to precipitate.
2.2.2. Reverse transcription of RNA into cDNA
RT-PCR can be used to detect ligated exons in purified spliceosomes. In addition, the branched nucleotide can be read through by several reverse transcriptases (such as the AMV and MMLV reverse transcriptases), making it possible to also detect RNA containing the unique lariat and T-branch structure using RT-PCR [41, 42]. The extracted RNA stored in - 80 °C was spun down at 18,000 g for 30 min at 4 °C. A small blue pellet was visible at the bottom of the tube. The pellet was rinsed using cold 70% ethanol followed by centrifugation at 18,000 g for 5 min and air-dried at room temperature. Then the pellet was dissolved in 2.5 μl RNase-free water containing 0.05 μl DNase I (Roche) and 0.1 μl RNase inhibitor (NEB), and was incubated at 37 °C for 20 min to remove DNA contaminants. circRNAs are naturally resistant to digestion by RNA exonucleases due to their covalently closed structure and 0.2 μl RNase R (Lucigen) can therefore be added at this step to digest linear RNA and enrich for circular RNA, if desired. For inactivation of DNase I and RNase R, 0.5 μl of 20 mM EDTA was added and the samples were heated at 75 °C for 10 min. The resulting RNAs were reverse transcribed into cDNAs using ProtoScript II First Strand cDNA Synthesis Kit (NEB) according to the manufacturer’s instruction. The products were used as templates for the following PCRs to detect the presence of the lariat, ligated exon, T-branch, and circRNA using specific primers as described below.
2.2.3. PCR Primer design
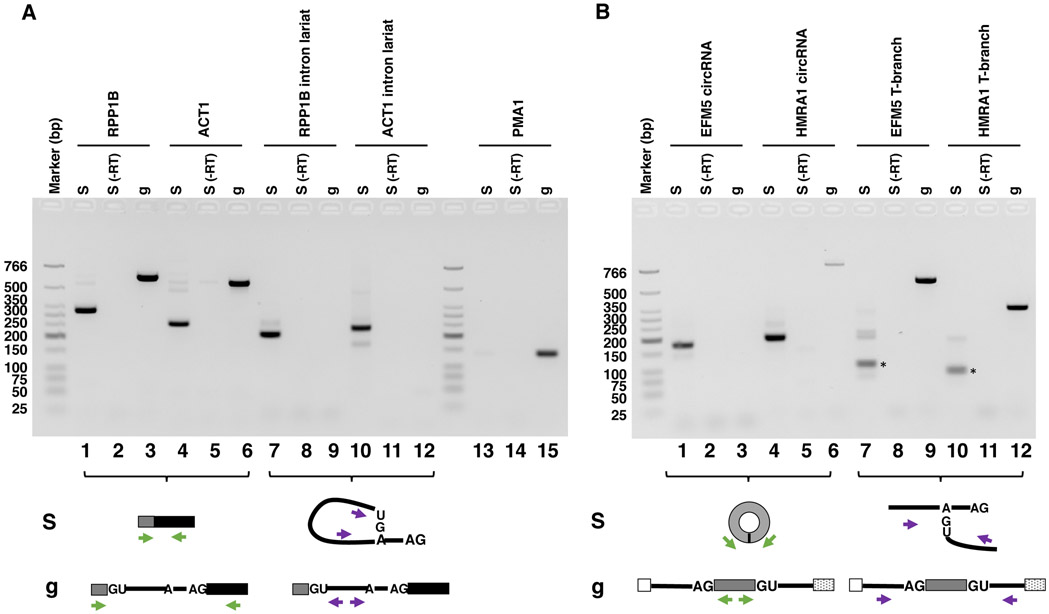
Here single-intronic gene RPP1B was used as an example to illustrate the process of primer design and canonical splicing product detection. After splicing, exons 1 and 2 are ligated together and can be detected using an inward-facing primer pair (Fig. 3A, green). The forward primer is located in exon 1 with the sequence 5’-TTGCAGTTTCAGTTAATCTTGGAC-3’ and the reverse primer is located in exon 2 with the sequence 5’-CAGCAGCTTCTTCTTCTTTTTCTT-3’. A 334 bp PCR product is expected for the ligated exon. The intron lariat can be detected by the purple outward-facing primer pair across the branch site with the sequences of 5’-AACAATCGTCCCGAGAAATG-3’ for the forward primer and 5’-GTTTTAGGTTGCATTTTTGTACATAC/TTAG-3’ for the reverse primer (Fig. 3A, purple, “/” denotes the 2’-5’ linkage at the branch point), similar to the primer design used to detect the ACT1 intron lariat [43, 44]. Note that the reverse primer does not need to cross the branch point for the PCR to be successful. A 210 bp PCR product is expected for the intron lariat. Another single-intronic gene ACT1 was also evaluated using the following primer pairs to detect the ligated mRNA and intron lariat: forward primer for the ligated ACT1 mRNA: 5’-CTACTCAAACCAAGAAGAAAAAGA-3’; reverse primer for the ligated ACT1mRNA: 5’-TGATACCTTGGTGTCTTGGTCT-3’; forward primer for the ACT1 intron lariat: 5’-CACTCTCCCATAACCTCCTA-3’; reverse primer for the ACT1 intron lariat: 5’-GCAAGCGCTAGAACATACTTAG-3’. The expected PCR products length for ligated ACT1 mRNA and intron lariat are 261 and 236 bp, respectively. In addition, PCR amplification from intronless gene PMA1 using primers in the gene body (forward primer: 5’-GGCTGGTGTCGAAATCTTGT-3’ and reverse primer: 5’-CTTTCTGGAAGCAGCCAAAC-3’) was used as a negative control. No PCR product is expected to be generated using RNA extracted from the purified spliceosome (Fig. 3A, lane 13).
Fig. 3.

Detection of canonical and back-splicing products and intermediates in purified spliceosomal P complex. RT-PCR products generated from cDNA isolated from the purified spliceosomal P complex (S) or PCR products from yeast genomic DNA (g, as controls) were analyzed on an agarose gel and stained with EtBr. Primer positions are indicated as arrows in the schematic diagrams below. (−RT) designates negative controls for RT-PCR reactions without reverse transcriptases. (A) Purified spliceosomal P complex contains spliced mRNAs (lanes 1 and 4) and intron lariats (lanes 7 and 10) for single intronic genes RPP1B and ACT1. PCR product from intronless gene PMA1 using primers in the gene body serves as a negative control (lanes 13). (B) Purified spliceosomal P complex contains circRNAs (lanes 1 and 4) and T-branch (lanes 7 and 10) for multi-intronic genes EFM5 and HMRA1. The * in lanes 7 and 10 indicates PCR bands at the expected size. (C) Sanger sequencing of RT-PCR products in lanes 1, 4, 7, and 10 of panel B, confirming that these products were derived from circRNAs and T-branches of EFM5 and HMRA1. “/” indicates where two ends of exon 2 are ligated. “∣” indicates where the 5’ ss of intron 2 is ligated to the BP of intron 1. The 5’ ss and BPS are in bold. “-” represents deletions in the BPS due to errors caused by reverse transcriptase reading through the branch.
For detection of the back-splicing products, multi-intronic genes EFM5 and HMRA1 were used as examples. To detect circRNA, outward-facing primers in the middle exon were designed with the sequences of 5’- AACCACTGGACTTCAGTGATGA-3’ and 5’- CGTCGTAAAGCTTTTGAAAGGCTTC-3’ for EFM5 which generates a 179 bp PCR fragment [3] (Fig. 3B, green), as well as 5’-GAAAGCAAAGCCTTAATTCCAAGG-3’ and 5’ TTTCCCTTTGGGCTCTTCTCT 3’ for HMRA1 which generates a 196 bp PCR fragment. To detect T-branch for EFM5, a forward primer located in intron 1 with the sequence 5’-TTTTCAACACAGTAACGTAGAATTAC-3’ and the reverse primer located in intron 2 with the sequence 5’-AACAGTTAGTAAGATGAAAAGATACTGG-3’ were used, which generates a 105 bp PCR product (Fig. 3B, purple). For HMRA1, a forward primer located in intron 1 with the sequence 5’-GTATGTTTTCATTTCAAGGATAG-3’ and a reverse primer located in intron 2 with the sequence 5’-TGTTAGTATAGGATATATTTAAGTTTGA-3’ were designed to detect the T-branch which generates an 85 bp PCR product. The identities of these PCR products were confirmed through Sanger sequencing after cloning the PCR product on a pminiT 2.0 vector using the PCR Cloning Kit (NEB) (Fig. 3C).
2.2.4. Canonical and back-splicing products and intermediates detection using PCR
Using cDNA from the post-catalytic spliceosomal P complex purified from the yeast strain yXL352 or yeast genomic DNA, PCR was performed using primers described above to detect the splicing products and intermediates. PCR reactions were carried out in a total volume of 20 μl containing 0.2 μl Q5 High-Fidelity DNA Polymerase (New England Biolabs), 0.2 mM dNTPs, 0.5 μM each primer and 1 μl reverse transcription product (or 100 ng genomic DNA), in a C1000 Thermal Cycler (Bio-Rad). An initial denaturation step was performed at 98 °C for 1 min, followed by 32-40 cycles of 98 °C for 15 sec and 58-62 °C (depending on primers) for 30 sec, with a final extension step at 72°C for 30 sec.
For the single-intronic gene RPP1B and ACT1, the ligated exon can be detected in cDNA from purified P complex (Fig. 3A, lanes 1 and 4) and the gene containing the intron can be detected in genomic DNA (Fig. 3A, lanes 3 and 6) using the primers colored in green. The lariat product produced from canonical splicing can be detected in cDNA from purified P complex using the primers colored in purple (Fig. 3A, lanes 7 and 10) and no product can be detected from genomic DNA using the same primers (Fig. 3A, lanes 9 and 12). As expected, CircRNA can be detected in cDNA from purified P complex using primers colored in green both for EFM5 (Fig. 3B, lane 1) and HMRA1 (Fig. 3B, lanes 4). The T-branch products can be detected using the primers colored in purple (Fig. 3B, lanes 7 and 10). These results show that purified post-catalytic P complexes contain both canonical and back-splicing products that can be detected using RT-PCR.
3. Concluding Remarks
Here we described a detailed protocol for purifying the yeast spliceosomal post-catalytic P complex and detecting both canonical and back-splicing products and intermediates in the purified complex. The results support our model based on the recently determined cryo-EM structure of the yeast spliceosomal E complex that the same spliceosome can carry out both canonical as well as back-splicing to generate circRNAs.
Furthermore, back-splicing occurs in much lower frequency than canonical splicing and circRNA can be difficult to detect in total RNAs. Purified spliceosomes may offer an advantage in detecting the low abundance circRNAs. However, ligation products including circRNA and intermediates such as lariat or T-branch are normally quickly released from the spliceosome. Our approach of stalling and purifying the P complex enriches these products and intermediates, making them easier to detect. In addition to our strategy of replacing the endogenous Prp22 with the Prp22H606A mutant, Prp22 ATPase/helicase mutants act in a dominant negative fashion [37]. Expressing Prp22K512A in the presence of WT Prp22 in yeast [45], or supplementing yeast or HeLa splicing extract with recombinant yeast Prp22S635A or human Prp22K594A (or Prp22S717A) [46, 47] also successfully stalls the spliceosomal P complex. The latter approach is particularly attractive in organisms where genetic manipulation may not be feasible. Taken together, similar to what we have done with the yeast circRNA, stalling and purifying the spliceosomal P complex may be a useful approach for studying circRNA in other species.
Highlights.
Detailed protocol for purifying yeast post-catalytic spliceosomal P complex
Method for detecting circRNA in purified spliceosomal P complex
Acknowledgements
We thank Dr. Joseph Giovinazzo for critical reading of the manuscript. This work was supported by NIH grant GM130673 (R.Z.).
Abbreviations used in the text:
- circRNAs
circular RNAs
- snRNPs
small nuclear ribonucleoproteins
- ss
splice site
- BPS
Branch Point Sequence
- TAP
tandem affinity purification
- RT-PCR
reverse transcription polymerase chain reaction
- NTC
Nineteen Complex
- CBP
calmodulin-binding peptide
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Lasda E, Parker R, Circular RNAs: diversity of form and function, RNA 20(12) (2014) 1829–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Cocquerelle C, Mascrez B, Hetuin D, Bailleul B, Mis-splicing yields circular RNA molecules, FASEB J 7(1) (1993) 155–60. [DOI] [PubMed] [Google Scholar]
- [3].Wang PL, Bao Y, Yee MC, Barrett SP, Hogan GJ, Olsen MN, Dinneny JR, Brown PO, Salzman J, Circular RNA Is Expressed across the Eukaryotic Tree of Life, Plos One 9(3) (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Suzuki H, Tsukahara T, A view of pre-mRNA splicing from RNase R resistant RNAs, Int J Mol Sci 15(6) (2014) 9331–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Xiao MS, Wilusz JE, An improved method for circular RNA purification using RNase R that efficiently removes linear RNAs containing G-quadruplexes or structured 3' ends, Nucleic Acids Res 47(16) (2019) 8755–8769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, Zhu P, Chang Z, Wu Q, Zhao Y, Jia Y, Xu P, Liu H, Shan G, Exon-intron circular RNAs regulate transcription in the nucleus, Nat Struct Mol Biol 22(3) (2015) 256–64. [DOI] [PubMed] [Google Scholar]
- [7].Piwecka M, Glazar P, Hernandez-Miranda LR, Memczak S, Wolf SA, Rybak-Wolf A, Filipchyk A, Klironomos F, Cerda Jara CA, Fenske P, Trimbuch T, Zywitza V, Plass M, Schreyer L, Ayoub S, Kocks C, Kuhn R, Rosenmund C, Birchmeier C, Rajewsky N, Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function, Science 357(6357) (2017). [DOI] [PubMed] [Google Scholar]
- [8].Conn VM, Hugouvieux V, Nayak A, Conos SA, Capovilla G, Cildir G, Jourdain A, Tergaonkar V, Schmid M, Zubieta C, Conn SJ, A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation, Nat Plants 3 (2017) 17053. [DOI] [PubMed] [Google Scholar]
- [9].Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M, Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1, RNA Biol 14(3) (2017) 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H, Qu B, Zhou Z, Shen N, Yang L, Chen LL, Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity, Cell 177(4) (2019) 865–880 e21. [DOI] [PubMed] [Google Scholar]
- [11].Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, Robinson DR, Nesvizhskii AI, Chinnaiyan AM, The Landscape of Circular RNA in Cancer, Cell 176(4) (2019) 869–881 e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen LL, The expanding regulatory mechanisms and cellular functions of circular RNAs, Nat Rev Mol Cell Biol 21(8) (2020) 475–490. [DOI] [PubMed] [Google Scholar]
- [13].Legnini I, Di Timoteo G, Rossi F, Morlando M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade M, Laneve P, Rajewsky N, Bozzoni I, Circ-ZNF609 Is a Circular RNA that Can Be Translated and Functions in Myogenesis, Molecular Cell 66(1) (2017) 22-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E, Perez-Hernandez D, Ramberger E, Shenzis S, Samson M, Dittmar G, Landthaler M, Chekulaeva M, Rajewsky N, Kadener S, Translation of CircRNAs, Mol Cell 66(1) (2017) 9–21 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Jurica MS, Moore MJ, Pre-mRNA splicing: awash in a sea of proteins, Mol Cell 12(1) (2003) 5–14. [DOI] [PubMed] [Google Scholar]
- [16].Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. June 4;73(5) (1993) 1019–30. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- [17].Li X, Liu S, Zhang L, Issaian A, Hill RC, Espinosa S, Shi S, Cui Y, Kappel K, Das R, Hansen KC, Zhou ZH, Zhao R, A unified mechanism for intron and exon definition and back-splicing, Nature 573(7774) (2019) 375–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Starke S, Jost I, Rossbach O, Schneider T, Schreiner S, Hung LH, Bindereif A, Exon circularization requires canonical splice signals, Cell Rep 10(1) (2015) 103–11. [DOI] [PubMed] [Google Scholar]
- [19].Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S, circRNA biogenesis competes with pre-mRNA splicing, Mol Cell 56(1) (2014) 55–66. [DOI] [PubMed] [Google Scholar]
- [20].Wilusz Jeremy E. A 360° view of circular RNAs: From biogenesis to functions. Wiley Interdiscip Rev RNA. July;9(4) (2018) e1478. doi: 10.1002/wrna.1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Romfo CM, Alvarez CJ, van Heeckeren WJ, Webb CJ, Wise JA, Evidence for splice site pairing via intron definition in Schizosaccharomyces pombe, Mol Cell Biol 20(21) (2000) 7955–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].De Conti L, Baralle M, Buratti E, Exon and intron definition in pre-mRNA splicing, Wiley Interdiscip Rev RNA 4(1) (2013) 49–60. [DOI] [PubMed] [Google Scholar]
- [23].Hawkins JD, A survey on intron and exon lengths, Nucleic Acids Res 16(21) (1988) 9893–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Berget SM, Exon recognition in vertebrate splicing, J Biol Chem 270(6) (1995) 2411–4. [DOI] [PubMed] [Google Scholar]
- [25].Lim SR, Hertel KJ, Commitment to splice site pairing coincides with A complex formation, Mol Cell 15(3) (2004) 477–83. [DOI] [PubMed] [Google Scholar]
- [26].Kotlajich MV, Hertel KJ, Death by splicing: tumor suppressor RBM5 freezes splicesite pairing, Mol Cell 32(2) (2008) 162–4. [DOI] [PubMed] [Google Scholar]
- [27].Plaschka C, Lin PC, Charenton C, Nagai K, Prespliceosome structure provides insights into spliceosome assembly and regulation, Nature 559(7714) (2018) 419–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Luhrmann R, The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome, Mol Cell 36(4) (2009) 593–608. [DOI] [PubMed] [Google Scholar]
- [29].Warkocki Z, Odenwalder P, Schmitzova J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Luhrmann R, Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components, Nat Struct Mol Biol 16(12) (2009) 1237–43. [DOI] [PubMed] [Google Scholar]
- [30].Wan R, Bai R, Yan C, Lei J, Shi Y, Structures of the Catalytically Activated Yeast Spliceosome Reveal the Mechanism of Branching, Cell 177(2) (2019) 339–351 e13. [DOI] [PubMed] [Google Scholar]
- [31].Campodonico E, Schwer B, ATP-dependent remodeling of the spliceosome: intragenic suppressors of release-defective mutants of Saccharomyces cerevisiae Prp22, Genetics 160(2) (2002) 407–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Liu SH, Li XN, Zhang LD, Jiang JS, Hill RC, Cui YX, Hansen KC, Zhou ZH, Zhao R, Structure of the yeast spliceosomal postcatalytic P complex, Science 358(6368) (2017) 1278–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, Wilm M, Seraphin B, The tandem affinity purification (TAP) method: a general procedure of protein complex purification, Methods 24(3) (2001) 218–29. [DOI] [PubMed] [Google Scholar]
- [34].Hogg R, McGrail JC, O'Keefe RT, The function of the NineTeen Complex (NTC) in regulating spliceosome conformations and fidelity during pre-mRNA splicing, Biochem Soc Trans 38(4) (2010) 1110–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chan SP, Kao DI, Tsai WY, Cheng SC, The Prp19p-associated complex in spliceosome activation, Science 302(5643) (2003) 279–82. [DOI] [PubMed] [Google Scholar]
- [36].Schwer B, Gross CH, Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing, EMBO J 17(7) (1998) 2086–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Schwer B, Meszaros T, RNA helicase dynamics in pre-mRNA splicing, EMBO J 19(23) (2000) 6582–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E, Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines, Yeast 15(10B) (1999) 963–72. [DOI] [PubMed] [Google Scholar]
- [39].Widlund PO, Davis TN, A high-efficiency method to replace essential genes with mutant alleles in yeast, Yeast 22(10) (2005) 769–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Green MR, Sambrook J, Isolation of High-Molecular-Weight DNA Using Organic Solvents, Cold Spring Harb Protoc 2017(4) (2017) pdb prot093450. [DOI] [PubMed] [Google Scholar]
- [41].Vogel J, Hess WR, Borner T, Precise branch point mapping and quantification of splicing intermediates, Nucleic Acids Res 25(10) (1997) 2030–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Lorsch JR, Bartel DP, Szostak JW, Reverse transcriptase reads through a 2'-5'linkage and a 2'-thiophosphate in a template, Nucleic Acids Res 23(15) (1995) 2811–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Cheng Z, Menees TM, RNA branching and debranching in the yeast retrovirus-like element Ty1, Science 303(5655) (2004) 240–3. [DOI] [PubMed] [Google Scholar]
- [44].Menees TM, Methods for analyzing lariat rna, U.S. Patent 20130150251A1 (2013).
- [45].Bai R, Yan C, Wan R, Lei J, Shi Y, Structure of the Post-catalytic Spliceosome from Saccharomyces cerevisiae, Cell 171(7) (2017) 1589–1598 e8. [DOI] [PubMed] [Google Scholar]
- [46].Wilkinson ME, Fica SM, Galej WP, Norman CM, Newman AJ, Nagai K, Postcatalytic spliceosome structure reveals mechanism of 3'-splice site selection, Science 358(6368) (2017) 1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Fica SM, Oubridge C, Wilkinson ME, Newman AJ, Nagai K, A human postcatalytic spliceosome structure reveals essential roles of metazoan factors for exon ligation, Science 363(6428) (2019) 710–714. [DOI] [PMC free article] [PubMed] [Google Scholar]