Abstract
FANCI is an essential component of Fanconi anemia pathway, which is responsible for the repair of DNA interstrand cross-links (ICLs). As an evolutionarily related partner of FANCD2, FANCI functions together with FANCD2 downstream of FA core complex. Currently, growing evidences showed that the essential role of FA pathway in male fertility. However, the underlying mechanisms for FANCI in regulating spermatogenesis remain unclear. In the present study, we found that the male Fanci−/− mice were sterile and exhibited abnormal spermatogenesis, including massive germ cell apoptosis in seminiferous tubules and dramatically decreased number of sperms in epididymis. Besides, FANCI deletion impaired maintenance of undifferentiated spermatogonia. Further investigation indicated that FANCI was essential for FANCD2 foci formation and regulated H3K4 and H3K9 methylation on meiotic sex chromosomes. These findings elucidate the role and mechanism of FANCI during spermatogenesis in mice and provide new insights into the etiology and molecular basis of nonobstructive azoospermia.
Subject terms: Cell death, Spermatogenesis
Introduction
Fanconi anemia (FA) is a rare genome instability syndrome characterized by early-onset bone marrow failure, developmental defects, high predisposition to cancers, and reduced fertility [1]. It has been revealed that mutations in FA genes, which are involved in DNA interstrand cross-links (ICLs) repair, usually lead to FA [2–4]. The FA pathway consists of 22 identified FA proteins (FANCA-FANCW) and several FA-associated proteins such as FAAP24 and FAAP100 [1, 5–8]. When DNA ICLs occur, FANCM recognizes the damage sites and recruits FA core complex proteins, which then monoubiquitinates FANCI-FANCD2 heterodimer [9, 10]. The monoubiquitination of the heterodimer is the keynote of FA pathway activation. Monoubiquitinated FANCI and FANCD2 then recruit downstream repair factors such as SLX4 (FANCP), XPF (FANCQ), BRCA1 (FANCS), BRCA2 (FANCD1), and RAD51 (FANCR) to complete the ICLs repair [11–15].
Currently, there are growing evidences for FA gene-associated infertility. It has been reported that mutations in FA genes including FANCA [16], FANCM [17], and FANCU [18] cause isolated non-obstructive azoospermia (NOA) in male patients. Consistently, many FA gene knockout mice showed sterility and hypogonadism in both males and females, and there were severe loss of germ cells and defects in spermatogenesis in male mice [19–23]. Besides, recent studies have reported that a network of FA proteins and DNA damage response (DDR) factors, MDC1 and RNF8, regulates the epigenetic modifications during meiosis, and FANCD2 is reported to be the central component of this network [24, 25]. Among FA proteins, FANCI is an evolutionarily related partner of FANCD2 and functions together with FANCD2 downstream of FA core complex to repair ICLs [26]. However, the exact role and mechanisms of FANCI, which is critical for FA pathway activation, in male meiosis remain unclear.
In the present study, we generated Fanci−/− mice and found that the male Fanci−/− mice were sterile and exhibited aberrant spermatogenesis. Further studies indicated that FANCI was essential for FANCD2 foci formulation and regulated histone methylation during meiosis. Altogether, these findings show that FANCI, an important component of FA pathway, plays an essential role in spermatogenesis.
Materials and methods
Animals
Fanci-flag mice with 3×flag fusing to N-terminus of the FANCI protein were generated using the CRISPR/Cas9 technology. The detailed information of the generation of the Fanci-flag knock-in mouse was shown in Supplementary Fig. 1. Fanci−/− mice were generated by Nanjing Sync Biotech Company using the CRISPR/Cas9 technology. The deletion of 98 bp in exon 5 resulted in frameshift mutation of Fanci gene, which led to a truncated protein of 116 amino acids (Supplementary Fig. 2). Primer sequences were as follows: Fanci-flag, forward: TCTGTTGAATGGATGGTGAAGATGT, reverse: TGGGGCATGTGTACACCGAG; Fanci knockout, forward: CCTAACCTTTGAAGCCCCTCG, reverse: GGGTGTCAATCAACTGCCTC.
Histology, immunohistochemistry, and TUNEL staining
Testes from control and Fanci−/− mice were fixed in 4% PFA overnight, and then dehydrated and embedded in paraffin. For morphological analysis, paraffin sections at 5 μm thick were stained with H&E. For immunohistochemistry, sections were incubated at 4 °C overnight with primary antibodies, after that the sections were incubated with specific secondary antibodies (Zhongshan Biotech) and then stained with DAB Substrate Kit (Vectorlabs). For the TUNEL assay, sections were treated with 15 μg/ml proteinase K for 10 min at 37 °C and then stained with the in situ Cell Death Detection kit (Roche). Images of histological analysis were captured on Olympus BX53 fluorescence microscope.
Meiotic chromosome spread and immunofluorescence microscopy
Meiotic chromosome spread of spermatocytes was performed as previously described [27]. Briefly, seminiferous tubules were incubated in hypotonic buffer (30 mM Tris (pH 7.5), 50 mM sucrose, 17 mM trisodium citrate, 5 mM EDTA, 1 mM PMSF, and 2.5 mM dithiothreitol) for 30 min and then minced in 100 mM sucrose to release spermatocytes. After that, the suspension was spread on slides with 1% PFA and 0.1% Triton X-100. Slides were then incubated in a humid chamber at 4 °C overnight. The following antibodies were used: SOX9 (Sigma, 1:500), DDX4 (Abcam, 1:500), PLZF (Santa Cruz, 1:100), FLAG (Sigma 1:100), FANCD2 (Novus, 1:200), SYCP1 (Abcam, 1:500), SYCP3 (Abcam, 1:500), γH2AX (Cell Signaling Technology, 1:200), RNA Pol II (Sigma, 1:200), RAD51 (Invitrogen, 1:200), MLH1 (BD Biosciences, 1:100), H3K9me2 (Millipore, 1:500), H3K9me3 (Millipore, 1:500), H3K4me2 (Millipore, 1:200) and cleaved PARP1 (Cell Signaling Technology, 1:400). FITC- and TRITC-conjugated secondary antibodies (Invitrogen) were used. Fluorescence images were captured with the Dragonfly spinning disc confocal microscope (ANDOR Technology).
Meiotic stages were determined on the basis of SYCP3 signal as previously described [24]. Briefly, spermatocytes in leptotene have short and unsynapsed SYCP3 signal. In zygotene spermatocytes, homologous chromosomes begin to pair, and the SYCP3 signal on lateral elements becomes longer. At pachytene, homologous chromosomes fully synapse, and the SYCP3 signal is thick and condensed. In the diplotene stage, the SYCP3 signal is compact on XY chromosomes and decreases progressively on autosomes because of the desynapse of homologous chromosomes.
The relative mean fluorescence intensity (RMFI) of histone methylation was quantified using the ImageJ software. Briefly, regions of interest (ROIs) were drawn around XY chromatin, presented as XY in Fig. 8, and the other nuclear area excluding the XY body was presented as Autosome (AU) in Fig. 8. The ROIs of XY chromatin and autosome region were standardized to the background. In this study, we calculated the RMFI of spermatocytes in both pachytene and diplotene stages.
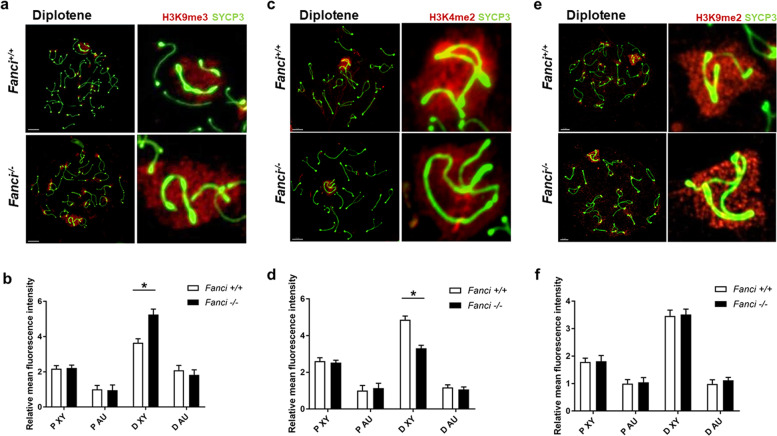
Fig. 8. FANCI regulates epigenetic modification on sex chromosomes during meiosis.
a, c, e Immunofluorescence analysis of meiotic chromosome spreads from wild type and Fanci-/- mice with indicated antibodies. Areas surrounding sex chromosomes are magnified. Scale bars, 5 μm. Quantification of H3K9me3 (b), H3K4me2 (d), and H3K9me2 (f) relative mean fluorescence intensity (RMFI) on autosome regions (Au) and sex chromosomes (XY) in pachytene (P) and diplotene (D) spermatocytes. For each panel, 30 cells were analyzed for each group. Data are presented as mean ± SD. *, P < 0.05. Student’s t-test.
Protein extraction and western blotting
Total protein of testes was extracted with the Total Protein Extraction Kit (Invent) and the protein concentration was examined by BCA method (Thermo). Protein samples were separated by SDS-PAGE and then transferred to PVDF membrane (Millipore). After blocking in 5% milk, the membranes were incubated with indicated primary antibodies at 4 °C overnight. The following antibodies were used: FANCI (Novus, 1:5000), FLAG (Sigma, 1:5000), and α-Tubulin (Proteintech, 1:3000). After being incubated with secondary antibodies, the membranes were detected with an ECL system (Millipore). The ChemiDoc MP System (Bio-Rad) was used to capture images.
Quantitative RT-PCR analysis
Total RNA from tissues was extracted with Trizol reagent (Thermo) and then reverse-transcribed using a PrimeScript RT Reagent Kit (TAKARA). The mRNA level of Fanci was quantitated by real-time PCR using the LightCycler 480 SYBR Green I Master Mix (Roche). Gapdh was used as the internal control. Primer sequences were as follows: Fanci, forward: CATCTTGATGGATTCCTATGGGC, reverse: GTTCGCGTAACAACTCTGTTGA; Gapdh, forward: AATGGATTTGGACGCATTGGT, reverse: TTTGCACTGGTACGTGTTGAT. Independent experiments were conducted three times.
mRNA probe analysis
Paraffin testicular sections were pretreated and analyzed with Fanci BaseScope™ Probe (ACD), which was a method of in situ hybridization (ISH) to visualize mRNA. The RNA-specific probe was designed to detect the 98 bp which was deleted in Fanci−/− mice. According to the manufacturer’s instructions, testicular sections were first pretreated with reagents such as hydrogen peroxide and protease, and then incubated with RNA-specific probe. After that, the sections were incubated with amplification reagents to enhance signals, and the signals were further detected using a red chromogenic substrate. Images were acquired with the Olympus BX53 microscope.
Statistical analysis
Data in this study were analyzed using the SPSS version 21.0. Numeric variables were compared by Student’s t-test. For categorical variables, the Chi-square test (Fisher’s exact test) was used to compare the differences. P < 0.05 was considered statistically significant.
Results
Expression pattern of FANCI during meiotic prophase I
Measurement of mRNA levels by real-time PCR showed that Fanci was differentially expressed in various mouse tissues, with much higher level in testis than other tissues (Fig. 1a).
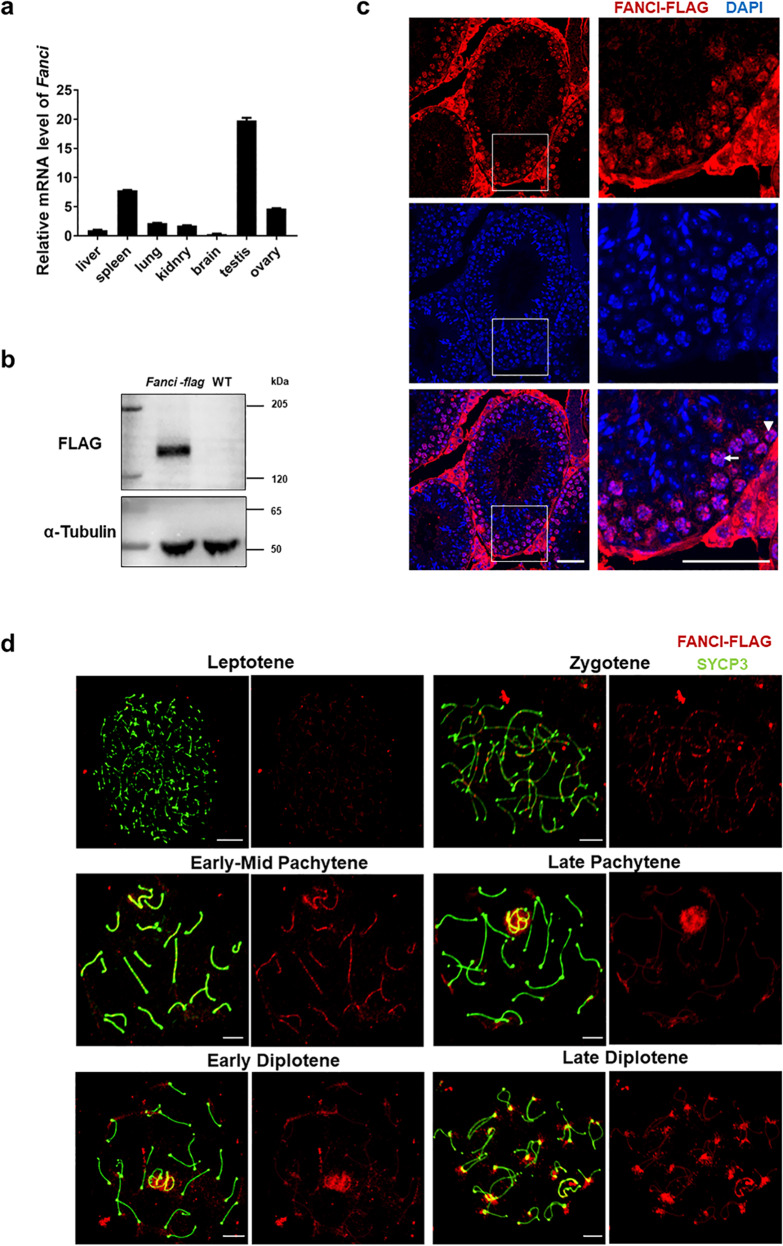
Fig. 1. Temporal and spatial expression of FANCI during meiotic prophase I.
a Relative mRNA level of Fanci in various tissues. Data are presented as mean ± SD. b Western blot analysis of the wild type and Fanci-flag testes (8 weeks old). α-Tubulin was used as a loading control. c Expression and localization of FANCI in adult testes. Immunofluorescence analysis of FLAG was performed on paraffin sections from Fanci-flag testes. The nuclei were stained with DAPI. Arrowhead represents spermatogonia. Arrow represents spermatocyte. Scale bars, 20 μm. d Localization of FANCI in chromosome spread of Fanci-flag spermatocytes. Meiotic chromosome spreads were immunolabeled with anti-FLAG and anti-SYCP3 antibodies. Scale bars, 5 μm.
We next detected the localization and expression of FANCI by generating a Fanci-Flag mouse model (Fig. 1b, Supplementary Fig. 1). The Fanci-Flag mice were normal in appearance. The body weight and the testis/body ratio of adult mice were comparable between wild-type and Fanci-Flag mice. Besides, both Fanci-Flag males and females were fertile (Supplementary Fig. 1d−g). Immunofluorescence staining of testicular sections from Fanci-Flag mice showed that FANCI was localized in the nuclei of spermatogonia and spermatocytes (Fig. 1c). Consistent with our results, a single-cell RNA-seq analysis also indicated that Fanci had relatively high expression levels in spermatogonia and spermatocyte (Supplementary Fig. 3) [28]. Immunofluorescence microscopy of meiotic chromosome spreads from Fanci-Flag mice was performed and each meiotic stage was determined on the appearance pattern of chromosome axes. Results showed that FANCI was not detected in the leptotene stage. Its accumulation on the chromosome axes began in the zygotene stage and increased through the mid-pachytene stage. After mid-pachytene stage, FANCI signal on the autosome region gradually decreased. Meanwhile, FANCI signal on the sex chromosomes progressively increased and spread onto the entire XY domain in the early diplotene stage. After that FANCI signal decreased on the XY domain and diffused throughout the entire chromatin in the late diplotene stage (Fig. 1c). These results indicated that FANCI was expressed in a temporal and spatial manner during meiotic prophase I.
FANCI deletion results in male sterility and causes germ cell loss in mice
To explore the function of FANCI in male fertility, we generated Fanci−/− mice by targeted deletion of 98 bp in exon 5 using the CRISPR/Cas9 technology (Supplementary Fig. S2). FANCI was not detected by western blotting in Fanci−/− testes (Fig. 2a). Furthermore, the results of mRNA probe did not show any signal in the testis of Fanci−/− mice (Fig. 2b).
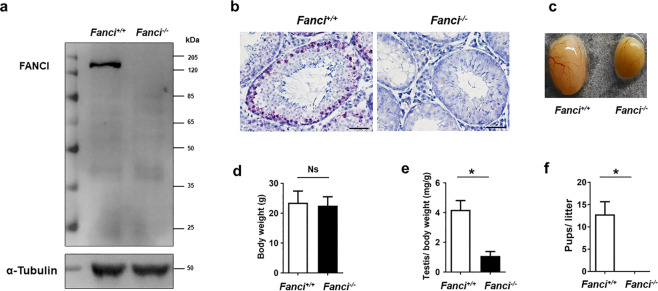
Fig. 2. FANCI deletion results in atrophytestes and male sterility.
a Western blot analysis of the wild type and Fanci−/− testes (8 weeks old). α-Tubulin was used as a loading control. b The mRNA probe detection of wild type and Fanci−/− testes. Scale bars, 20 μm. c Representative images of the testes of wild-type and Fanci−/− mice. d Body weights of wild-type and Fanci−/− mice at 8 weeks after birth. Six mice were analyzed for each group, and data are presented as mean ± SD. *, P < 0.05. Student’s t-test. e Mean ratio of testis/body weight from 8 weeks old wild type and Fanci−/− mice. Six mice were analyzed for each group, and data are presented as mean ± SD. *, P < 0.05. Student’s t-test. f Fertility test result of the mean ratio of pups/litter from wild type and Fanci−/− mice. Six wild-type mice: 34 litters and 12.8 pups/litter. Six Fanci−/− mice: 0 litter. Data are presented as mean ± SD. *, P < 0.05. Student’s t-test.
Genotyping of offspring from heterozygous mating showed a normal Mendelian distribution (Supplementary Fig. 2d). Besides, Fanci−/− mice appeared to be normal without microphthalmia or skeletal malformations (Supplementary Fig. 2e−g). The body weight of adult mice was comparable between wild-type and Fanci−/− mice (Fig. 2d). However, the Fanci−/− testes were smaller than those of littermate controls, with the ratio of the testis to body weight decreased significantly (Fig. 2c, e). Furthermore, both Fanci−/− males (Fig. 2f) and females were infertile.
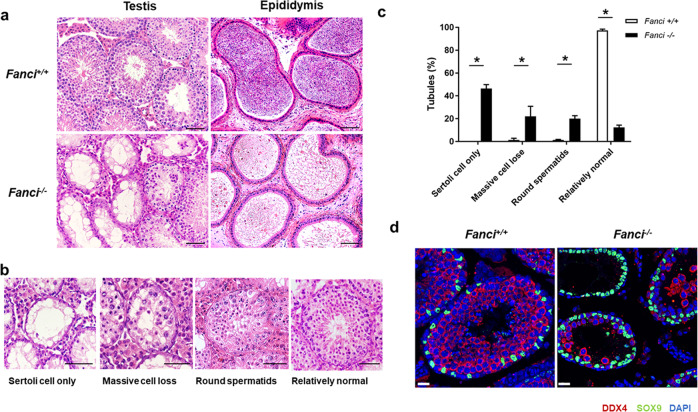
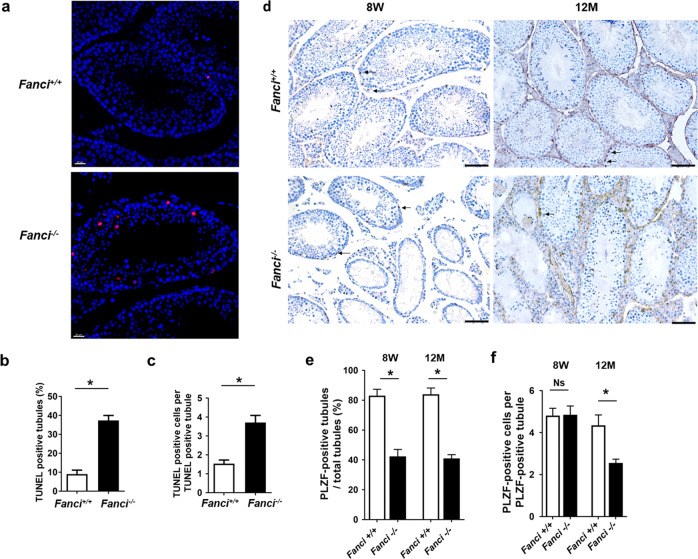
To further explore the functions of FANCI, we compared testis sections from 8 weeks old wild-type and Fanci−/− mice using H&E staining. Seminiferous tubules from wild-type mice had normal architecture and contained germ cells at all stages. In contrast, seminiferous tubules from Fanci−/− testes were predominantly degenerated and exhibited a significant decrease in the number of cells. Consistent with the histologic analysis of testis sections, sperms in epididymides of Fanci−/− mice were much fewer than controls (Fig. 3a). Additionally, in Fanci−/− testes, 46.3% of seminiferous tubules only contained Sertoli cells, and 22% of seminiferous tubules had massive cell loss. Moreover, 20% of Fanci−/− tubules had round spermatids as the most advanced spermatogenic cells, and only 12.3% tubules in Fanci−/− testes had relatively normal morphology (Fig. 3b, c). Furthermore, immunofluorescence staining of testicular sections for DDX4 (a germ cells marker) and SOX9 (a marker for Sertoli cells) indicated massive germ cell loss in Fanci−/− testes (Fig. 3d), suggesting that FANCI deletion resulted in the dramatically decrease of germ cells and relative enrichment of Sertoli cells. In addition, massive loss of germ cells was found in Fanci−/− testes at 12 months, which was more severe than that at 8 weeks (Supplementary Fig. 4a, c). Exactly, 60.7% of seminiferous tubules only contained Sertoli cells, and only 9.0% tubules had relatively normal morphology (Supplementary Fig. 4b). Collectively, our results indicate that FANCI is required for male fertility.
Fig. 3. FANCI deletion causes germ cell loss in mice.
a H&E staining of testes and epididymides from 8 weeks old wild type and Fanci−/− mice. Scale bars, 50 μm. b Representative images of H&E stained testicular sections showing various seminiferous tubules in 8 weeks old Fanci−/− mice, including Sertoli cell-only tubules, tubules with massive cell loss, tubules with round spermatids as the most advanced spermatogenic cells, and relatively normal tubules. Scale bars, 50 μm. c Quantification of different types of seminiferous tubules in 8 weeks old wild type and Fanci−/− mice. Six wild-type mice and six Fanci−/− mice were analyzed. Data are presented as mean ± SD. *, P < 0.05. Chi-square test (Fisher’s exact test). d Immunofluorescence staining for DDX4 (a germ cell marker) and SOX9 (a Sertoli cell marker) in wild type and Fanci−/− mice testes of 8-week-old mice. Scale bars, 20 μm.
FANCI deletion increases apoptosis of germ cells and affects the maintenance of undifferentiated spermatogonia
Consistent with massive germ cell loss during spermatogenesis, TUNEL assay and immunofluorescence staining for cleaved PARP1 showed that apoptosis was dramatically increased in Fanci−/− testes (Fig. 4a−c and Supplementary Fig. 5a−c), suggesting that Fanci−/− germ cells were eliminated by increased apoptosis. Furthermore, we found that most cleaved PARP1 signals were localized in SYCP3-positive cells in Fanci−/− testes, indicating that most apoptotic cells were spermatocytes. Only a few apoptotic cells were undifferentiated spermatogonia or spermatids in Fanci−/− testes (Supplementary Fig. 5d, e).
Fig. 4. FANCI deletion increases apoptosis of germ cells and affects the maintenance of undifferentiated spermatogonia.
a TUNEL assay of testes from 8-week-old wild-type and Fanci−/− mice. Cells stained red are TUNEL-positive. Scale bars, 20 μm. b Frequencies of TUNEL-positive tubules in wild type and Fanci−/− mice. In each group, six mice were analyzed. Data are presented as mean ± SD. *, Chi-square test (Fisher’s exact test). c Numbers of TUNEL-positive cells per TUNEL-positive tubule in wild type and Fanci−/− mice. Six wild-type mice and six Fanci−/− mice were analyzed. Data are presented as mean ± SD. *, P < 0.05. Student’s t-test. d Immunohistochemistry staining against PLZF (an undifferentiated spermatogonia marker) on wild type and Fanci−/− testes at 8 weeks old and at 12 months old. Arrows represent the undifferentiated spermatogonia. Scale bars, 50 μm. Scoring of the percentage of PLZF-positive tubules per total tubule (e) and scoring of the number of PLZF-positive cells per PLZF-positive tubule (f). A total of 100 tubules from three independent mice were examined at 8 weeks and 12 months, both for wild type and Fanci−/− mice. In each group, six mice were analyzed. Data are presented as mean ± SD. *, P < 0.05. Ns no statistical significant difference. Frequencies were compared by the Chi-square test (Fisher’s exact test). Numbers were compared by Student’s t-test.
As the maintenance of undifferentiated spermatogonia is required for continuous spermatogenesis, we next investigated whether the number of undifferentiated spermatogonia was altered in Fanci−/− testes. Immunohistochemistry staining utilizing PLZF (an undifferentiated spermatogonia marker) showed that percentage of PLZF-positive tubules were significantly decreased in Fanci−/− mice compared with controls at both 8 weeks and 12 months (Fig. 4d, e). Despite the number of PLZF-positive cells per PLZF-positive tubules was similar between Fanci−/− and wild-type mice at 8 weeks, the average number of PLZF-positive cells decreased dramatically in Fanci−/− testes at 12 months compared with wild type (Fig. 4d, f). These results suggest that FANCI plays an important role in the maintenance of undifferentiated spermatogonia.
FANCI colocalizes with FANCD2 on sex chromosomes and is required for FANCD2 foci formation in spermatocytes
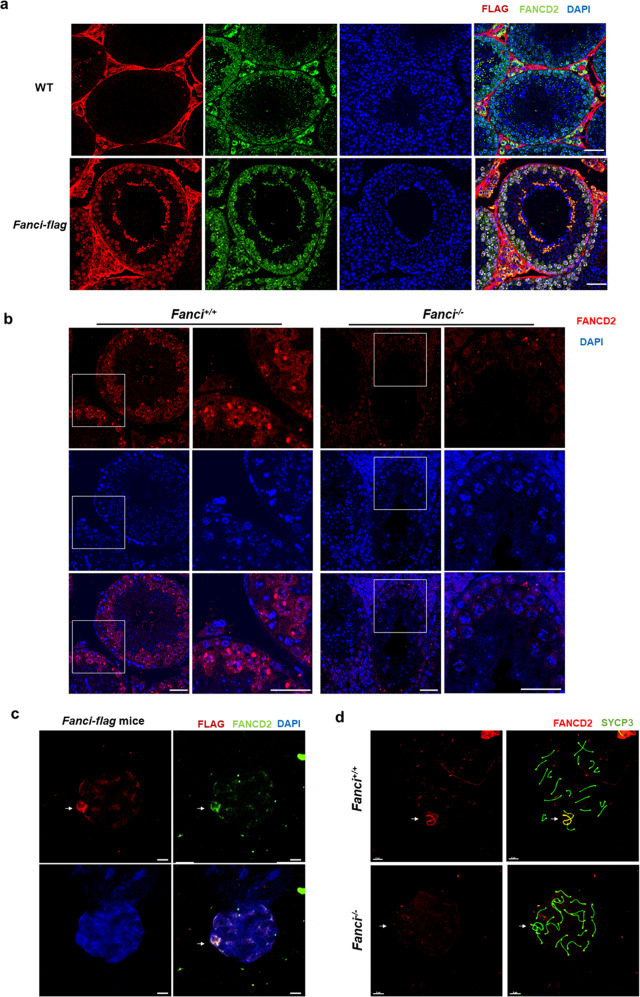
It is well-known that FANCI and FANCD2 form a heterodimer and play a critical role in FA pathway activation. Thus, we firstly investigated the localization of FANCD2 and FANCI in Fanci-Flag mice, and found that FANCD2 and FANCI were mainly colocalized in spermatogonia and spermatocytes (Fig. 5a). Besides, nuclear spread analysis of spermatocytes from Fanci-Flag mice indicated that FANCD2 and FANCI were colocalized on sex chromosomes (Fig. 5c).
Fig. 5. FANCI colocalizes with FANCD2 on sex chromosomes and is required for FANCD2 foci formation in spermatocytes.
a Expression and localization of FANCI and FANCD2 in adult testes. Immunofluorescence analysis of FLAG and FANCD2 was performed on paraffin sections from wild type and Fanci-flag testes. The nuclei were stained with DAPI. Scale bars, 20 μm. b Immunostaining of paraffin sections from wild-type testes and Fanci−/− testes with FANCD2. DNA was stained with DAPI. Scale bars, 20 μm. c Expression and localization of FANCI and FANCD2 in spermatocytes. Immunofluorescence analysis of FLAG and FANCD2 was performed on chromosome spread from Fanci-flag testes. The nuclei were stained with DAPI. Arrows represent the sex chromosomes. Scale bars, 5 μm. d Immunostaining of chromosome spreads from wild type and Fanci−/− spermatocytes with FANCD2 and SYCP3. SYCP3 immunolabeling was used to distinguish meiotic stages. Scale bars, 5 μm.
Next, we examined the expression of FANCD2 in wild type and Fanci−/− testes. Results showed that FANCD2 expression was dramatically decreased in Fanci−/− testes compared with wild-type testes, and the FANCD2 signal was attenuated in both spermatogonia and meiotic cells (Fig. 5b). Furthermore, immunofluorescence microscopy of meiotic chromosome spreads revealed that FANCD2 foci was eliminated in Fanci−/− cells (Fig. 5d), suggesting that FANCI is required for FANCD2 foci formation in spermatocytes.
Meiotic prophase I stage progression and meiotic recombination remain unaffected in Fanci−/− spermatocytes
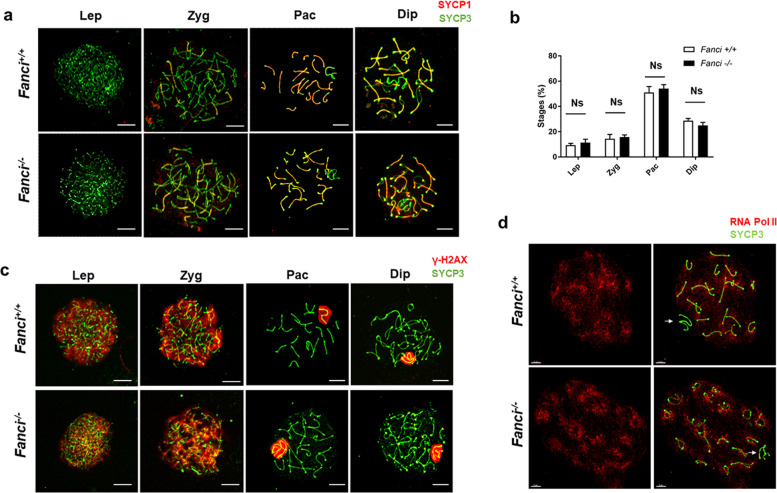
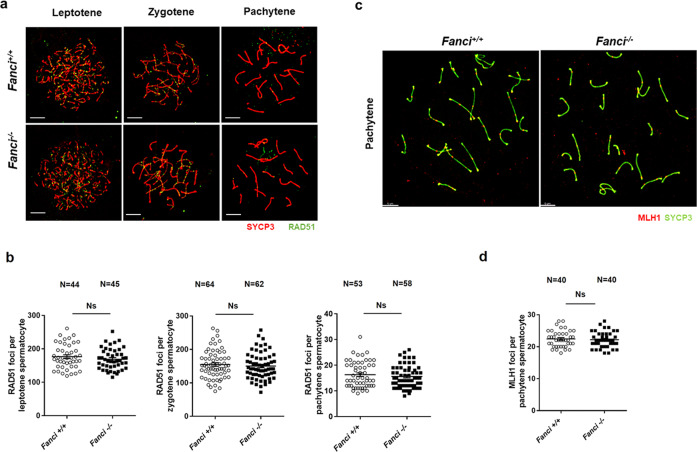
To investigate the roles of FANCI in meiotic prophase I, we analyzed multiple meiosis-related markers in wild type and Fanci−/− spermatocytes by immunofluorescence staining on meiotic chromosome spreads. As shown in Fig. 6a,b, normal chromosome synapsis and meiotic progression were observed in Fanci−/− spermatocytes. Besides, a normal phosphorylated H2AX (γH2AX) signal was detected in all stages of meiotic prophase I in both wild-type and Fanci−/− mice (Fig. 6c). To determine whether meiotic recombination was disturbed by FANCI deletion, we examined the accumulation of RAD51 on chromosome axes in Fanci−/− spermatocytes. Results showed that the number of RAD51 foci were comparable between wild type and Fanci−/− spermatocytes at both early stages (leptotene and zygotene) and later stage (pachytene) (Fig. 7a, b). Furthermore, we examined the accumulation of MLH1, which is the marker for meiotic crossover. We found that the number of MLH1 foci on chromosome axes were also comparable in both groups in pachytene spermatocytes (Fig. 7c, d), suggesting that crossover formation was unaffected in Fanci−/− spermatocytes. Collectively, these results reflect that FANCI is dispensable for meiotic progression and meiotic recombination.
Fig. 6. Meiotic prophase I stage progression and synapsis in Fanci−/− spermatocytes.
a Wild type and Fanci−/− spermatocytes were immunolabeled with SYCP1 and SYCP3 in leptotene (Lep), zygotene (Zyg), pachytene (Pac), and diplotene (Dip) spermatocytes. Meiotic stages were determined on the basis of SYCP3 signals. b Percentage of each stage of meiotic prophase as determined by SYCP3 immunostaining. In each group, six mice were analyzed. Data are presented as mean ± SD. Ns no statistical significant difference. Chi-square test (Fisher’s exact test). c Wild type and Fanci−/− spermatocytes were immunolabeled with γ-H2AX and SYCP3 at leptotene (Lep), zygotene (Zyg), pachytene (Pac), and diplotene (Dip) spermatocytes. d Spermatocytes immunolabeled for RNA Polymerase II (RNA Pol II) and SYCP3 at diplotene spermatocytes. Arrows represent the sex chromosomes. Scale bars, 5 μm.
Fig. 7. Meiotic recombination and crossover in Fanci−/− spermatocytes.
a Spermatocytes immunolabeled for RAD51 at leptotene, zygotene, and pachytene spermatocytes. b Quantification of the number of RAD51 foci per cell at leptotene, zygotene, and pachytene in wild type and Fanci−/− spermatocytes. Total numbers of analyzed spermatocytes are indicated. Data are presented as mean ± SD. Ns no statistical significant difference. Student’s t-test. c Spermatocytes immunolabeled for MLH1 in Wild type and Fanci−/− spermatocytes. d Quantification of the number of MLH1 foci per cell in wild type and Fanci−/− spermatocytes. Total numbers of analyzed spermatocytes are indicated. Data are presented as mean ± SD. Ns no statistical significant difference. Student’s t-test.
FANCI regulates H3K4 and H3K9 methylation on meiotic sex chromosomes
It has been reported that FA core proteins and FANCD2 regulate H3K4 and H3K9 methylation on meiotic sex chromosomes [24, 25]. To determine whether FANCI is involved in such histone methylation, we performed immunostaining for H3K4me2, H3K9me2, and H3K9me3 on chromosome spreads from wild type and Fanci−/− spermatocytes and then quantified the RMFI both on the autosomes regions and XY body. Consistent with previous studies, the accumulation of H3K9me3 on sex chromosomes was increased in Fanci−/− spermatocytes in the diplotene stage (Fig. 8a), indicating that FANCI was also involved in the regulation of H3K9 methylation. Besides, in Fanci−/− spermatocytes, H3K4me2 intensity was decreased significantly on diplotene sex chromosomes (Fig. 8c). Furthermore, H3K9me2 on XY chromatin was unaffected during the pachytene-to-diplotene transition in Fanci−/− spermatocytes (Fig. 8e). These results suggest that FANCI negatively regulates H3K9me3 and positively regulates H3K4me2 in the diplotene stage. To determine whether the changes in histone modifications in Fanci−/− sex chromosomes are associated with transcriptional changes, we examined the distribution of RNA polymerase II (RNA Pol II) in diplotene spermatocytes. Surprisingly, RNA Pol II signal was excluded from the sex body in diplotene spermatocytes in both wild type and Fanci−/− mice (Fig. 6d), suggesting a normal sex chromosome transcriptional inactivation in Fanci−/− spermatocytes.
Discussion
In this study, we elucidated temporal and spatial expression of FANCI during meiotic prophase I. Besides, FANCI deletion eliminated FANCD2 foci and altered histone methylation during meiosis, which contributed to the spermatogenetic failure in Fanci−/− mice. Collectively, our results elucidate the involvement and mechanism of FANCI during spermatogenesis.
During spermatogenesis, FANCI was reported to localize in spermatocytes and spermatids [29]. However, the exact expression patterns and localization of FANCI during meiotic prophase I are unclear. In the present study, we revealed the exact expression patterns and localization of FANCI from leptotene to late diplotene for the first time. We found that FANCI localized on the chromosome axes from zygotene to mid pachytene stage, after that FANCI signal accumulated on the XY region from late pachytene to early diplotene stage, and then it diffused throughout the entire chromatin in the late diplotene stage. These findings indicated the temporal and spatial manner of FANCI expression and may provide information for future functional studies.
Importantly, we demonstrated that Fanci−/− mice exhibited germline defects including massive germ cell loss and increased apoptosis in seminiferous tubules. The germline phenotypes and compromised fertility in Fanci−/− mice in our study are consistent with many previous FA mouse models [22, 23, 25], indicating that FANCI, as an important component of FA pathway, also plays essential roles in the male germline. Previous studies have revealed that FA proteins were involved in the maintenance of hematopoietic stem cells and human pluripotent stem cells [30, 31], and it is reasonable that FA proteins are associated with undifferentiated spermatogonia. Kato et al. [25] indicated that FANCB regulated maintenance of undifferentiated spermatogonia. Consistent with this study, our findings also suggest an important role for FANCI in the maintenance of undifferentiated spermatogonia, but the underlying mechanism is uncertain and requires further exploration.
It is well-known that FA pathway is a DDR pathway that repairs DNA ICLs in the genome [1]. During meiosis, one important DDR event is meiotic recombination [32]. However, our results demonstrated that FANCI deletion did not affect the number of RAD51 foci on chromosome axes from leptotene to pachytene, suggesting an unessential role of FANCI in RAD51 recruitment and exclusion during meiosis. Similar results were also observed in Fancb mutant mice [25]. Despite the unessential role of FANCI in RAD51 accumulation during meiosis, many studies have revealed that FANCI is involved in the regulation of RAD51 functions during DDR. Studies indicated that FANCI-FANCD2 complex colocalized with RAD51 at the stalled replication forks and stabilizes the RAD51-DNA complex [33–35]. Besides, Sato et al. indicated that FANCI, but not FANCD2, was essential for the FANCI-FANCD2 complex-mediated RAD51-DNA stabilization [36]. Furthermore, a recent study also claimed that FANCI interacted directly with RAD51 and stimulated RAD51-mediated D-loop formation [29]. The underlying mechanism behind this difference is unclear and requires further exploration. Additionally, we also found the normal γ-H2AX and MLH1 signal in Fanci−/− spermatocytes from leptotene to pachytene, which is consistent with the observation in Fancb mutant mice [25]. Collectively, our results revealed the normal programmed DSB repair and meiotic recombination in Fanci−/− mice, suggesting a dispensable role of FANCI in these events.
Among the FA proteins, FANCI and FANCD2 form a protein complex (ID2 complex), and the monoubiquitination of ID2 complex is a critical step in FA pathway activation during ICLs repair [37]. When it comes to spermatogenesis, our results indicated that the global FANCD2 level was decreased in Fanci−/− testes. One possible reason for the global decreased FANCD2 is that FANCI and FANCD2 are partially interdependent for their protein stability [38]. In line with our findings, a previous study has also reported that meiotic FANCD2 chromosomal localization was dependent on FANCI in C. elegans, suggesting the important role of FANCI during meiosis in different species [39]. When it comes to spermatogenesis, recent studies reported that FANCD2 foci on chromosome axes were abolished in mutant spermatocytes deficient for FANCA, FANCB, and FANCC [24, 25], indicating the similar function of FA core complex in regulating FANCD2 foci in both somatic DNA damage response DDR and meiosis. In line with these studies, our results showed that FANCD2 foci were eliminated in Fanci−/− spermatocytes, indicating that FA pathway activation is indispensable during spermatogenesis.
It has been reported that FA proteins regulate epigenetic programming during meiosis. Alavattam et al. found that FA core proteins (FANCA and FANCC) and FANCD2 regulated both H3K9me2 and H3K9me3, while FANCD2 regulated H3K4me2 independently of FA core complex [24]. Besides, FANCB was reported to regulate H3K9me2 and H3K9me3 on the sex chromosomes during meiosis [25]. Therefore, in the present study, we detected the epigenetic markers H3K9me2, H3K9me3, and H3K4me2 in Fanci−/− spermatocytes. Despite the unaffected H3K9me2 in Fanci−/− spermatocytes, our results clearly indicated that FANCI negatively regulated H3K9me3 and positively regulated H3K4me2 on XY chromatin in diplotene stage during meiosis. Notably, recent studies have indicated that the BRCA1-MDC1-RNF8 axis is upstream of the FA pathway in histone modification [40, 41]. Consistent with our results, FANCD2 was also reported to regulate RNF8-dependent histone modification H3K4me2, suggesting that RNF8 integrates the FA pathway and the BRCA1-MDC1-RNF8 axis [24]. Thus, our findings reinforce the intermediate role of RNF8 in this FA-DDR network, and further explorations are required to clarify the whole FA-DDR system during meiosis.
Despite the altered modification of H3K9me3 and H3K4me2 in diplotene, the meiotic progression was not arrested in Fanci−/− mice. Besides, we observed the normal sex chromosome transcriptional inactivation in Fanci−/− spermatocytes, indicating that the altered epigenetic modification did not result in transcriptional changes. It is possible that the varied epigenetic state in Fanci−/− spermatocytes is associated with increased germ cell apoptosis.
Recently, it was reported that a Fanci knockout model in C57BL/6J mouse strain exhibited severe hypogonadism without any spermatocytes [29]. Our Fanci−/− mice showed milder phenotypes by crossing C57BL/6J Fanci−/− mice to ICR strains. Apart from germline defects, the mixed background Fanci−/− mice in our study appeared to be healthy without any other known FA phenotypes. Notably, the difference between our study and the previous Fanci knockout model indicates the strain-dependent phenotypes. Previous studies have also revealed that FA gene knockout mice in mixed backgrounds exhibit much milder phenotypes than those in pure C57BL/6J strain [20, 21]. Our study also supports the point that strain background affects the phenotypic profiles in FA mouse models.
In summary, we have elucidated the important role and mechanism of FANCI in spermatogenesis. Our study provides new insights into the interaction between the FA pathway and meiotic epigenetic modification underlying spermatogenesis, which will reinforce the notion that DNA repair pathways play important roles in spermatogenesis and suggest the FANCI gene may be a candidate gene for human NOA.
Supplementary information
Acknowledgements
The authors thank all of their colleagues for technical support.
Author contributions
LX performed most of the experiments, treated data, and wrote the paper. WX, DL, and XY performed mouse treatment experiments. YY and SZ participated in research conception and design and revised paper. YQ and FG reviewed the paper.
Funding
This study was supported by grants from the National Key Research and Developmental Program of China (2018YFC1003800 and 2017YFC1001100), the National Natural Science Foundation of China (81873823 and 81771541), Taishan Scholars Program for Young Experts of Shandong Province (tsqn20161069), Young Scholars Program of Shandong University (2016WLJH26) and The Fundamental Research Funds of Shandong University.
Ethics
Mice in this study were maintained and used for experimentation according to the protocol approved by the Animal Care and Research Committee of Shandong University.
Competing interests
The authors declare no competing interests.
Footnotes
Edited by M. Agostini
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
8/26/2021
A Correction to this paper has been published: 10.1038/s41419-021-04096-7
Contributor Information
Yajuan Yang, Email: yangyajuan1991@163.com.
Shidou Zhao, Email: shidouzhao@sdu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41419-021-04034-7.
References
- 1.Ceccaldi R, Sarangi P, D’Andrea AD. The Fanconi anaemia pathway: new players and new functions. Nat Rev Mol Cell Biol. 2016;17:337–49. doi: 10.1038/nrm.2016.48. [DOI] [PubMed] [Google Scholar]
- 2.Wang AT, Smogorzewska A. SnapShot: Fanconi anemia and associated proteins. Cell. 2015;160:354–e1. doi: 10.1016/j.cell.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 3.Kottemann MC, Smogorzewska A. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 2013;493:356–63. doi: 10.1038/nature11863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan W, Deans AJ. A defined role for multiple Fanconi anemia gene products in DNA-damage-associated ubiquitination. Exp Hematol. 2017;50:27–32. doi: 10.1016/j.exphem.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 5.Lopez-Martinez D, Liang CC, Cohn MA. Cellular response to DNA interstrand crosslinks: the Fanconi anemia pathway. Cell Mol Life Sci. 2016;73:3097–114. doi: 10.1007/s00018-016-2218-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Knipscheer P, Raschle M, Smogorzewska A, Enoiu M, Ho TV, Scharer OD, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326:1698–701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sawyer SL, Tian L, Kahkonen M, Schwartzentruber J, Kircher M, University of Washington Centre for Mendelian G, et al. Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Disco. 2015;5:135–42. doi: 10.1158/2159-8290.CD-14-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moldovan GL, D’Andrea AD. How the Fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–49. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Twest S, Murphy VJ, Hodson C, Tan W, Swuec P, O’Rourke JJ, et al. Mechanism of ubiquitination and deubiquitination in the Fanconi anemia pathway. Mol Cell. 2017;65:247–59. doi: 10.1016/j.molcel.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Rajendra E, Oestergaard VH, Langevin F, Wang M, Dornan GL, Patel KJ, et al. The genetic and biochemical basis of FANCD2 monoubiquitination. Mol Cell. 2014;54:858–69. doi: 10.1016/j.molcel.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deans AJ, West SC. DNA interstrand crosslink repair and cancer. Nat Rev Cancer. 2011;11:467–80. doi: 10.1038/nrc3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park JY, Virts EL, Jankowska A, Wiek C, Othman M, Chakraborty SC, et al. Complementation of hypersensitivity to DNA interstrand crosslinking agents demonstrates that XRCC2 is a Fanconi anaemia gene. J Med Genet. 2016;53:672–80. doi: 10.1136/jmedgenet-2016-103847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumaraswamy E, Shiekhattar R. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol Cell Biol. 2007;27:6733–41. doi: 10.1128/MCB.00961-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park JY, Singh TR, Nassar N, Zhang F, Freund M, Hanenberg H, et al. Breast cancer-associated missense mutants of the PALB2 WD40 domain, which directly binds RAD51C, RAD51, and BRCA2, disrupt DNA repair. Oncogene. 2014;33:4803–12. doi: 10.1038/onc.2013.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamamoto KN, Kobayashi S, Tsuda M, Kurumizaka H, Takata M, Kono K, et al. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proc Natl Acad Sci USA. 2011;108:6492–6. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krausz C, Riera-Escamilla A, Chianese C, Moreno-Mendoza D, Ars E, Rajmil O, et al. From exome analysis in idiopathic azoospermia to the identification of a high-risk subgroup for occult Fanconi anemia. Genet Med. 2019;21:189–94. doi: 10.1038/s41436-018-0037-1. [DOI] [PubMed] [Google Scholar]
- 17.Yin H, Ma H, Hussain S, Zhang H, Xie X, Jiang L, et al. A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet Med. 2019;21:62–70. doi: 10.1038/s41436-018-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang Y, Guo J, Dai L, Zhu Y, Hu H, Tan L, et al. XRCC2 mutation causes meiotic arrest, azoospermia, and infertility. J Med Genet. 2018;55:628–36. doi: 10.1136/jmedgenet-2017-105145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng NC, van de Vrugt HJ, van der Valk MA, Oostra AB, Krimpenfort P, de Vries Y, et al. Mice with a targeted disruption of the Fanconi anemia homolog Fanca. Hum Mol Genet. 2000;9:1805–11. doi: 10.1093/hmg/9.12.1805. [DOI] [PubMed] [Google Scholar]
- 20.Wong JC, Alon N, McKerlie C, Huang JR, Meyn MS, Buchwald M. Targeted disruption of exons 1 to 6 of the Fanconi Anemia group A gene leads to growth retardation, strain-specific microphthalmia, meiotic defects, and primordial germ cell hypoplasia. Hum Mol Genet. 2003;12:2063–76. doi: 10.1093/hmg/ddg219. [DOI] [PubMed] [Google Scholar]
- 21.Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–35. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang Q, Xie H, Zhong Y, Li D, Ke X, Ying H, et al. Severe Fanconi anemia phenotypes in Fancd2 depletion mice. Biochem Biophys Res Commun. 2019;514:713–9. doi: 10.1016/j.bbrc.2019.04.201. [DOI] [PubMed] [Google Scholar]
- 23.Tsui V, Crismani W. The Fanconi anemia pathway and fertility. Trends Genet. 2019;35:199–214. doi: 10.1016/j.tig.2018.12.007. [DOI] [PubMed] [Google Scholar]
- 24.Alavattam KG, Kato Y, Sin HS, Maezawa S, Kowalski IJ, Zhang F, et al. Elucidation of the Fanconi anemia protein network in meiosis and its function in the regulation of histone modifications. Cell Rep. 2016;17:1141–57. doi: 10.1016/j.celrep.2016.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kato Y, Alavattam KG, Sin HS, Meetei AR, Pang Q, Andreassen PR, et al. FANCB is essential in the male germline and regulates H3K9 methylation on the sex chromosomes during meiosis. Hum Mol Genet. 2015;24:5234–49. doi: 10.1093/hmg/ddv244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sims AE, Spiteri E, Sims RJ, 3rd, Arita AG, Lach FP, Landers T, et al. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat Struct Mol Biol. 2007;14:564–7. doi: 10.1038/nsmb1252. [DOI] [PubMed] [Google Scholar]
- 27.Jiang H, Wang L, Cui Y, Xu Z, Guo T, Cheng D, et al. Meiotic chromosome behavior in a human male t(8;15) carrier. J Genet Genomics. 2014;41:177–85. doi: 10.1016/j.jgg.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 28.Lukassen S, Bosch E, Ekici AB, Winterpacht A. Single-cell RNA sequencing of adult mouse testes. Sci Data. 2018;5:180192. doi: 10.1038/sdata.2018.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubois EL, Guitton-Sert L, Beliveau M, Parmar K, Chagraoui J, Vignard J, et al. A Fanci knockout mouse model reveals common and distinct functions for FANCI and FANCD2. Nucleic Acids Res. 2019;47:7532–47. doi: 10.1093/nar/gkz514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haneline LS, Gobbett TA, Ramani R, Carreau M, Buchwald M, Yoder MC, et al. Loss of FancC function results in decreased hematopoietic stem cell repopulating ability. Blood. 1999;94:1–8. [PubMed] [Google Scholar]
- 31.Yung SK, Tilgner K, Ledran MH, Habibollah S, Neganova I, Singhapol C, et al. Brief report: human pluripotent stem cell models of Fanconi anemia deficiency reveal an important role for Fanconi anemia proteins in cellular reprogramming and survival of hematopoietic progenitors. Stem Cells. 2013;31:1022–9. doi: 10.1002/stem.1308. [DOI] [PubMed] [Google Scholar]
- 32.Amunugama R, Fishel R. Homologous recombination in eukaryotes. Prog Mol Biol Transl Sci. 2012;110:155–206. doi: 10.1016/B978-0-12-387665-2.00007-9. [DOI] [PubMed] [Google Scholar]
- 33.Hussain S, Wilson JB, Medhurst AL, Hejna J, Witt E, Ananth S, et al. Direct interaction of FANCD2 with BRCA2 in DNA damage response pathways. Hum Mol Genet. 2004;13:1241–8. doi: 10.1093/hmg/ddh135. [DOI] [PubMed] [Google Scholar]
- 34.Schlacher K, Wu H, Jasin M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 2012;22:106–16. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lossaint G, Larroque M, Ribeyre C, Bec N, Larroque C, Decaillet C, et al. FANCD2 binds MCM proteins and controls replisome function upon activation of s phase checkpoint signaling. Mol Cell. 2013;51:678–90. doi: 10.1016/j.molcel.2013.07.023. [DOI] [PubMed] [Google Scholar]
- 36.Sato K, Shimomuki M, Katsuki Y, Takahashi D, Kobayashi W, Ishiai M, et al. FANCI-FANCD2 stabilizes the RAD51-DNA complex by binding RAD51 and protects the 5’-DNA end. Nucleic Acids Res. 2016;44:10758–71. doi: 10.1093/nar/gkw876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song IY, Palle K, Gurkar A, Tateishi S, Kupfer GM, Vaziri C. Rad18-mediated translesion synthesis of bulky DNA adducts is coupled to activation of the Fanconi anemia DNA repair pathway. J Biol Chem. 2010;285:31525–36. doi: 10.1074/jbc.M110.138206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thompson EL, Yeo JE, Lee EA, Kan Y, Raghunandan M, Wiek C, et al. FANCI and FANCD2 have common as well as independent functions during the cellular replication stress response. Nucleic Acids Res. 2017;45:11837–57. doi: 10.1093/nar/gkx847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Germoglio M, Valenti A, Gallo I, Forenza C, Santonicola P, Silva N, et al. In vivo analysis of FANCD2 recruitment at meiotic DNA breaks in Caenorhabditis elegans. Sci Rep. 2020;10:103. doi: 10.1038/s41598-019-57096-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yan Z, Guo R, Paramasivam M, Shen W, Wang WJMC. A Ubiquitin-binding protein, FAAP20, links RNF8-mediated ubiquitination to the Fanconi anemia DNA repair network. Mol Cell. 2012;47:61–75. doi: 10.1016/j.molcel.2012.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sin HS, Barski A, Zhang F, Kartashov AV, Nussenzweig A, Chen J, et al. RNF8 regulates active epigenetic modifications and escape gene activation from inactive sex chromosomes in post-meiotic spermatids. Genes Dev. 2012;26:2737–48. doi: 10.1101/gad.202713.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.