Abstract
Gene expression valuated by reverse transcription-quantitative PCR (RT-qPCR) are often applied to study the gene function. To obtain accurate and reliable results, the usage of stable reference genes is essential for RT-qPCR analysis. The traditional southern Chinese medicinal herb, Desmodium styracifolium Merr is well known for its remarkable effect on the treatment of urination disturbance, urolithiasis, edema and jaundice. However, there are no ready-made reference genes identified for D. styracifolium. In this study, 13 novel genes retrieved from transcriptome datasets of four different tissues were reported according to the coefficient of variation (CV) and maximum fold change (MFC) of gene expression. The expression stability of currently used Leguminosae ACT6 was compared to the 13 candidate reference genes in different tissues and 7-day-old seedlings under different experimental conditions, which was evaluated by five statistical algorithms (geNorm/NormFinder/BestKeeper/ΔCT/RefFinder). Our results indicated that the reference gene combinations of PP + UFM1, CCRP4 + BRM and NFD6 + NCLN1 were the most stable reference genes in leaf, stem and root tissues, respectively. The most stable reference gene combination for all tissues was CCRP4 + CUL1. In addition, the most stable reference genes for different experimental conditions were distinct, for instance SMUP1 for MeJA treatment, ERDJ2A + SMUP1 for SA treatment, NCLN1 + ERDJ2A for ABA treatment and SF3B + VAMP721d for salt stress, respectively. Our results lay a foundation for achieving accurate and reliable RT-qPCR results so as to correctly understand the function of genes in D. styracifolium.
Supplementary Information
The online version contains supplementary material available at 10.1007/s13205-021-02954-x.
Keywords: Desmodium styracifolium Merr, Medicinal herb, Reference gene, Tissues
Introduction
Desmodium styracifolium Merr is a traditional southern Chinese medicinal herb, which is mainly distributed in Guangdong, Guangxi and Hainan provinces. It is well known for its remarkable effect on the treatment of urination disturbance, urolithiasis, edema and jaundice, etc. (Xiong et al. 2015; Cheng et al. 2018). It was reported that the main pharmacodynamical ingredients were flavonoids, polysaccharides and triterpenoids (Hirayama et al. 1989a, b. As the major component in 21 flavonoids extracted from D. styracifolium (Guo et al. 2015), the content of schaftoside was the only standard for assessing the quality of D. styracifolium (Committee 2020). It was documented that schaftoside extracted from D. styracifolium can treat cholelithiasis and urolithiasis via inhibiting cholesterol gallstone formation by activation of ileal liver X receptor α and hepatic farnesoid X receptor (Liu et al. 2017). In the kindred plant Desmodium spp., the biosynthetic pathway of schaftoside is predicted (Hamilton et al. 2012). As documented in previous study (Wang et al. 2020b), chalcone synthase (CHS), chalcone isomerase (CHI), flavone synthase II (FNSII) and C-glucosyltransferase (CGT) are involved in schaftoside biosynthesis. However, the exact copy of structural genes, for instance CHS and CGT, responsible for schaftoside biosynthesis in different tissues remains unclear due to lack of the stable reference genes.
To accurately estimate the expression level and/or pattern of gene(s), including novel genes related to schaftoside biosynthesis in D. styracifolium so as to correctly understand its function, RT-qPCR was often applied (Kubista et al. 2006; Kozera and Rapacz 2013; Zhang et al. 2014). When using RT-qPCR for evaluation of gene expression, researchers need to select the stable expression of genes as reference genes. According to our knowledge, there were no ready-made reference genes identified previously in D. styracifolium. Generally, housekeeping genes, for instance actin (ACT), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), glucose-6-phosphate dehydrogenase (G6PDH), elongation factor (EF), 18 S ribosomal RNA (18 S rRNA), ubiquitin (UBQ), tubulin (TUB), eukaryocyte initiation factor (EIF), ubiquitin conjugating enzyme (UBC), cyclophilin (CYP), are highly expressed in plant tissues and are frequently used as reference genes for RT-qPCR analysis in plants (Kozera and Rapacz 2013; Liu et al. 2014; Zhang et al. 2014; Zhou et al. 2016; Li et al. 2017a, b; Joseph et al. 2018). However, increasing number of studies have shown that there are no universal reference genes suitable for all conditions and/or tissues. Gene expression is highly spatiotemporal specific and often vary with the physiological status of the plant or experimental conditions (Nicot et al. 2005; Gutierrez et al. 2008; Hong et al. 2008; Kozera and Rapacz 2013; Gimeno et al. 2014; Li et al. 2017a; Joseph et al. 2018). Therefore, it is necessary to screen and identify the stable reference genes according to specific tissues and experimental conditions.
Recently, increasingly softwares have been exploited to identify the optimal reference gene(s) stably expressed in tissues and specific conditions, including geNorm (Vandesompele et al. 2002), NormFinder (Andersen et al. 2004), BestKeeper (Pfaffl et al. 2004), ΔCt (Silver et al. 2006) and RefFinder (Xie et al. 2012). The first three algorithms were all based on specific software-based approach, in which, geNorm is used to determine the most stable control genes from a panel of candidate reference genes via a stepwise exclusion or ranking process, followed by geometric averaging of a selection of the most stable reference genes (Vandesompele et al. 2002). NormFinder reveals expression variation by calculating the stability value when using reference genes for normalization (Andersen et al. 2004). BestKeeper determines the optimal reference genes on the basis of pair-wise correlation analysis of all pairs of candidate reference genes (Pfaffl et al. 2004). The ΔCt method displays the pairwise comparisons by calculating the standard deviation (SD) of each pair candidate reference genes and the average SD of each gene (Silver et al. 2006). RefFinder is a web-based tool which integrates the four statistical algorithms including geNorm, BestKeeper, NormFinder, and ΔCt, to rank the overall stability of candidate reference genes (Xie et al. 2012). Using these algorithms mentioned above, more and more reference genes have been identified in numerous species, including the common bean (Borges et al. 2012), soybean (Gao et al. 2017; Bansal et al. 2015), and alfalfa (Wang et al. 2015).
In this study, 14 candidate reference genes, including 13 novel genes retrieved from D. styracifolium transcriptome datasets (Wang et al. 2020a) derived from four different tissues (root, stem, leaf and flower) screened as described by previous study, as well as a commonly used housekeeping gene ACT6 in Leguminosae, were assessed by RT-qPCR. Five statistical algorithms, including, geNorm, NormFinder, BestKeeper, ΔCt and RefFinder, were utilized to evaluate the expression stability of these putative reference genes in different tissues and experimental conditions. Furthermore, the key gene CHS essential for synthesizing schaftoside was investigated to validate the suitability of the stable reference genes newly identified in this study. Our results show that the systematical selection and validation of the best stable novel reference genes in this study will facilitate to correctly understand the function of genes in different tissues of D. styracifolium in response to hormone treatments and salt stress.
Materials and methods
Collection of plant materials and hormone treatments
The leaves, stems and roots of 8-month-old plants of D. styracifolium were collected from South China Botanical Garden, CAS. Each sample had at least four biological replicates and each replicate had at least three independent plants.
For stress treatments, the D. styracifolium seedlings were planted at 20–22 °C in a greenhouse with a relative humidity of 50% under long day (16-h light/8-h dark) conditions. Seven-day-old seedlings of the D. styracifolium were used. Seedlings were grown in glass dish containing filter paper irrigating with Hoagland nutrient solution. Elicitors were added in the 7th day of the cultivating for 24 h, including methyl jasmonate (MeJA, 100 μM), abscisic acid (ABA, 100 μM), salicylic acid (SA, 100 μM), salt (NaCl, 100 mM), and the control (CK) was treated with Hoagland nutrient solution. Each treatment has at least three biological replicates, and each biological replicate contains at least 15 seedlings. All samples were frozen immediately in liquid nitrogen after harvest and then stored at − 80 °C for analysis.
RNA extraction and cDNA synthesis
Total RNA was extracted from each sample using the HiPure Plant RNA Mini Kit R4151 (Magen, Guangdong, China) according to the manufacturer’s instructions, followed by PrimeScript™ RT reagent Kit with gDNA Eraser (TaKaRa, Dalian, China) treatment to eliminate DNA contamination. The integrity of RNA was determined by 1.5% agarose gel electrophoresis (Supplementary Fig. S1). The purity and concentration of total RNA was determined using NanoDrop 2000 Spectrophotometer (Thermo Scientific, Waltham, MA, US). The RNAs extracted from tested samples, which displayed two clearly bands (28S and 18S) in the agarose gel electrophoresis, a concentration higher than 60 ng/μL and a ratio of A260/A280 between 1.8 and 2.0 were required for later cDNA preparation. Synthesis of cDNA was conducted in the PrimerScript™ RT cDNA Synthesis Kit (TaKaRa, Dalian, China) using 1.0 μg RNA solution. The resulting cDNA was then diluted with 10 times nuclease-free water to prepare RT-qPCR and stored at − 20 °C for further use.
Selection of candidate reference genes
A candidate reference gene with a small CV and MFC < 2 (MFC, the ratio of the maximum and minimum values among tissues) was defined as a most stable gene. Meanwhile, a mean expression value level lower than the maximum expression level subtracted with twofold SD was a prerequisite for a candidate housekeeping gene reported in previous study (de Jonge et al. 2007). To select the new candidate reference genes of D. styracifolium, we analyzed the transcriptome data derived from the following four tissues of 1-year-old seedlings: roots (RT, four replicates), stems (ST, four replicates), leaves (LF, eight replicates) and flowers (FL, three replicates). To estimate the expression stability of each gene, we analyzed the raw data for all genes as described previously (Wang et al. 2017; Yan et al. 2018): First, FPKM value, the mean expression value (MV) and the SD among all the tested samples were calculated for each gene; Second, CV should be calculated and ranked in order later. In general, the lower the CV value is, the more stable the gene expression is. Based on this principle and the method described by de Jonge et al. (2007), candidate reference genes that met the following requirements were selected: (a) the primary criteria, MFC should be lower than 2 (MFC < 2); (b) the second criteria, MV < MEV–2 × SD (MEV denotes the maximum expression value of each gene among all the tested samples); (c) the third criteria, the CV value of each gene among all the tested samples should be lower than 12% (CV < 12%).
Based on these selection procedures for the transcriptome sequencing data, 13 genes that had a minor variation in expression were selected. These candidate reference genes include vesicle-associated membrane protein 721d (VAMP721d), pinin protein (PP), carbon catabolite repressor protein 4 homolog 1 (CCRP4), splicing factor 3B subunit 2 isoform X2 (SF3B), probable E3 ubiquitin-protein ligase ARI7 (ARI7), ATP-dependent helicase BRM (BRM), probable Ufm1-specific protease isoform X1 (UFM1), protein NUCLEAR FUSION DEFECTIVE 6 (NFD6), unknown protein (UN), suppressor of mec-8 and unc-52 protein homolog 1 (SMUP1), cullin-1 (CUL1), nicalin-1 (NCLN1) and dnaJ protein ERDJ2A (ERDJ2A).
Primer design and RT-qPCR efficiency analysis
The RT-qPCR primers were designed using Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi) based on the sequences retrieved from the RNA-Seq dataset of D. styracifolium. The criteria for primer design were as follows: (a) primer size: 20–23 bp; (b) product size: 80–150 bp (The maximum length should not exceed 300 bp); (c) GC% content: 40–60%, and 45–55% was the best (primers are shown in Table 1). Moreover, primer accuracy and specificity were checked by 2.0% (w/v) agarose gel electrophoresis. The melting curve and no template control (NTC) were prepared to further validate the specificity and absence of primer dimer formation and DNA contamination for every primer pair. A standard curve was established by triplicate repeats of RT-qPCR amplification using serial dilutions (1:1,1:10, 1:100, 1:1000, and 1:10,000) of all tested cDNA sample pools. The correlation coefficient (R2) and amplification efficiency (E) for each gene were calculated based on the standard curve. The amplification efficiency of each gene was calculated using the equation E = (10–1/slope − 1) × 100% (Bustin et al. 2009).
Table 1.
Primer sequences, TM and amplification efficiencies of the candidate reference genes used in this study
| Gene | Gene description | Primer sequences (5′-3′) forward/reverse | Amplicon size (bp) | Tm (°C) | E (%) | R2 |
|---|---|---|---|---|---|---|
| VAMP721d | Vesicle-associated membrane protein 721d | CAGAAGCTTCCTGCCACCAA | 104 | 58 | 98.65 | 0.9999 |
| TCGTCTGCCACGACACAATA | ||||||
| PP | Pinin protein | TTCGTAGAGGTGCGTTGTCC | 111 | 58 | 94.15 | 0.9999 |
| GCTTGGCAGGAGGTTGATCT | ||||||
| CCRP4 | Carbon catabolite repressor protein 4 homolog 1 | CACCCCTACTGCTGATGACG | 132 | 58 | 109.49 | 0.9983 |
| ATAGGTGAGGGAGCGGGAAT | ||||||
| SF3B | Splicing factor 3B subunit 2 isoform X2 | CCCCTGGTGCTAGTTTTGGT | 159 | 58 | 101.27 | 0.9989 |
| AGTCACCCCAGTGTTTGGTC | ||||||
| ARI7 | Probable E3 ubiquitin-protein ligase ARI7 | GACCGAAGACGCTCCTATGG | 89 | 58 | 96.66 | 0.9992 |
| GGACCCGTTCGGAATCATCA | ||||||
| BRM | ATP-dependent helicase BRM | CAGTCCCAGCAGCAACCTAA | 133 | 58 | 95.61 | 0.9996 |
| ACGATGATGCTTGGGCTTGA | ||||||
| UFM1 | Probable Ufm1-specific protease isoform X1 | CGGACGATAGCAATCGCAGA | 70 | 58 | 98.98 | 1.0000 |
| GGGTCGGATCCTTTGGTGTT | ||||||
| NFD6 | Protein NUCLEAR FUSION DEFECTIVE 6 | CTCTCTCGCAGTCCCAATCC | 72 | 58 | 100.45 | 1.0000 |
| ACGGCATCATCGATTCCACA | ||||||
| UN | Unknown protein | GTTGTCTCGGGGACAGATCC | 116 | 58 | 106.52 | 0.9996 |
| AGCCTTGCAAAAGACGTGTG | ||||||
| SMUP1 | Suppressor of mec-8 and unc-52 protein homolog 1 | GGGAGGTGATGCTTCTGTCA | 163 | 58 | 101.64 | 0.9999 |
| AGCTGCAACAAAGTCTCCCC | ||||||
| CUL1 | Cullin-1 | TGAAGTTGGGCTGACATGCT | 174 | 58 | 95.39 | 0.9980 |
| TGATCCATTTGCCCCATTCCA | ||||||
| NCLN1 | Nicalin-1 | TTGGCTCCTGGGAAAACGAA | 172 | 58 | 98.98 | 0.9991 |
| TCATGCTCCCAAGCTACTCG | ||||||
| ERDJ2A | dnaJ protein ERDJ2A | TTCCACATGCGCCCTACTAC | 170 | 58 | 95.31 | 0.9979 |
| TCCCGACCCCTCCATAGATT | ||||||
| ACT6 | Actin-6 | GCGGGAAATTGTAAGGGATGTGAAAG | 141 | 60 | 98.23 | 0.9991 |
| TCCCCAATGGTGATGACCTGACC | ||||||
| DsCHS | Chalcone synthase | TGGCGCTGGAGCAATGATTA | 150 | 60 | 94.23 | 0.9999 |
| AGGTGAAACGTCAGTCCCAC |
E (%) amplification efficiency; R2 correlation coefficient
Data analysis
Five calculation programs, such as ΔCt, BestKeeper, NormFinder, geNorm and RefFinder, a web-based tool (http://www.ciidirsinaloa.com.mx/RefFinder-master/?type=reference#tabs-1), were used to calculate the stability of candidate reference genes. The ΔCt method by calculating the standard deviation (SD) of the candidate reference genes pairwise, and then ranking the candidate reference genes by the average SD values. The lower the average SD is, the more stable the RG performs. The BestKeeper program is an Excel-based tool by calculating the SD, coefficient of correlation (r) and CV of the pairwise reference genes, then ranking by the geomean of the three parameters. The most stable gene expression exhibits the lowest CV ± SD value. NormFinder is a model-based variance estimation approach to identify genes suited for normalization by calculating the stability value (SV). The lower SV is, the higher stability the gene is. The geNorm also is a tool based on Excel table, whose input data should be normalized by the formula, 2ΔCt, ΔCt = min Ct (of each gene) − sample Ct. The expression stability value (M value) and pairwise variation (Vn/Vn+1) were calculated for all candidate genes. The lower M value is, the higher stability the gene is. The Vn/Vn+1 value is used to decide the suitable number of reference genes for normalization. Only if the Vn/Vn+1 value is lower than the cut-off value, an additional reference gene is required. Finally, the RefFinder program is used to obtain a comprehensively value of the front four programs, so as to acquire the optimal ranking for the stability of the candidate reference genes.
Validation of reference genes
To validate the comprehensive ranking of candidate reference genes for stability, the most stable two genes, the least stable gene, and ACT6 were selected to normalize the DsCHS for calculating its relative expression level, using the 2−ΔΔCt method (Livak and Schmittgen 2001). The primer for DsCHS was designed according to the criteria mentioned above. Specificity was checked as described above (primer pairs are shown in Table 1).
Results
Selection of candidate reference genes based on D. styracifolium transcriptome data
To efficiently identify the stable reference genes, transcriptome data from root, stem, leaf, and flower of D. styracifolium (Wang et al. 2020a) were used to screen the candidate reference genes as described previously (de Jonge et al. 2007). In addition, genes with fragments per kilobase of exon model per million mapped fragments (FPKM) less than 5 were excluded before the stability analyses. In addition, CV value < 12% is considered as a key standard for screening stable candidate reference gene as described previous study (Yan et al. 2018). Consequently, 50 genes with a CV ranging between 8.18 and 11.96% were selected as the rough candidate reference genes (Supplementary Table S1). Subsequently, we randomly selected 13 of the top 25 candidate genes as the final candidate genes for further study (Supplementary Table S2). According to the annotation of the RNA-seq data, they are vesicle-associated membrane protein 721d (VAMP721d), pinin protein (PP), carbon catabolite repressor protein 4 homolog 1 (CCRP4), splicing factor 3B subunit 2 isoform X2 (SF3B), probable E3 ubiquitin-protein ligase ARI7 (ARI7), ATP-dependent helicase BRM (BRM), probable Ufm1-specific protease isoform X1 (UFM1), protein NUCLEAR FUSION DEFECTIVE 6 (NFD6), unknown protein (UN), suppressor of mec-8 and unc-52 protein homolog 1 (SMUP1), cullin-1 (CUL1), nicalin-1 (NCLN1) and dnaJ protein ERDJ2A (ERDJ2A), respectively. In Glycine max, the currently widely used housekeeping gene is ACT6 (NP_001276160.2). In this study, the homologous gene ACT6 in D. styracifolium was chosen to test its usage, although its CV value is 29.0%. Totally, 14 candidate reference genes were selected for further estimation.
Expression profile of candidate reference genes of D. styracifolium
To obtain the gene-specific primers, several selection standards were carried out. First, all of the primers flanking the intron were designed and the resulting PCR amplicon contains the single band with the expected length (Supplementary Fig. S1). Second, the nature of gene-specific primer was further characterized by melting curve analysis with a single peak (Supplementary Fig. S2). The cDNA-free template controls (ddH2O as template) showed no obvious melting curve products and the Ct value higher than 35 (data not shown). These results indicate that the specificity of primer pairs for each candidate reference gene meets the experimental requirements.
Subsequently, with serial dilutions (1:1, 1:10, 1:100, 1:1000, and 1:10,000) of all tested cDNA sample pools, a standard curve was established by triplicate repeats of RT-qPCR amplification, the correlation coefficient (R2) and amplification efficiency (E) for each gene were calculated. As shown in Table 1, the amplification efficiency for primer pairs of all candidate reference genes ranged from 94.15% (PP) to 109.49% (CCRP4), and the R2 values lay between 0.9980 (CUL1) and 1.000 (UFM1 and NFD6).
The expression profiles of the 14 candidate reference genes were shown by the Ct values of RT-qPCR in all experiment situations. The smaller the Ct value of the gene is, the higher transcript abundance of the gene is. As shown in the Fig. 1, the average Ct values ranged from 23.82 to 28, which indicates that the expression level of all candidate reference genes are suitable for the requirement of reference gene expression level (15 < Ct < 30; Wan et al. 2010). The candidate reference gene names, descriptions, primer sequences, Tm values, amplicon lengths, amplification efficiencies and R2 values are listed in Table 1.
Fig. 1.
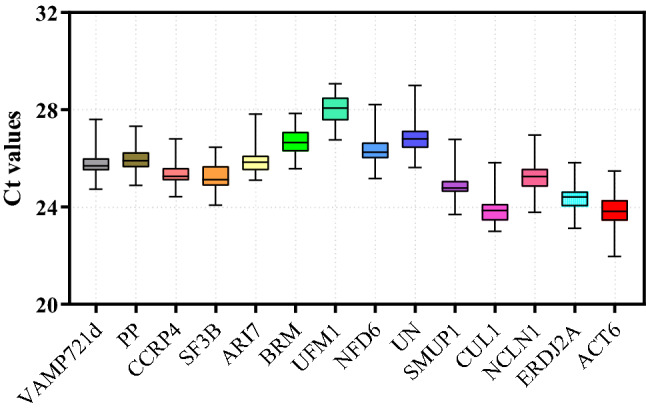
Distribution of Ct values of 14 candidate reference genes in all experimental samples. Expression levels of 14 candidate reference genes used in the study. Expression levels data RT-qPCR quantification computed tomography (Ct) values for each gene in different tissues (root\stem\leaf) and experimental conditions (MeJA\ABA\SA\Salt). The box plot indicates the 25th and 75th percentiles, and whisker caps represent the maximum and minimum values
Expression stability analysis of candidate reference genes of D. styracifolium
To further confirm stable internal reference genes, the tested samples were divided into 7 groups: tissues (leaf, stem, root), MeJA treatment (CK, 100 μM MeJA), ABA treatment (CK, 100 μM ABA), SA treatment (CK, 100 μM SA), salt stress (CK, 100 mM NaCl), total treatment (TREAT) and total tested samples (TOTAL). All the samples were analyzed by the five statistical algorithms: ΔCt, BestKeeper, NormFinder, geNorm and RefFinder.
According to ΔCt analysis method, the ranking order of candidate reference genes was generated by the SD values (Table 2). The lower the mean SD was, the higher the stability of the gene was. NFD6, CCRP4 and PP were the most stable genes. On the contrary, UN was the most least stable gene for Tissues. For the 7-day-old seedlings treatment, SMUP1 and VAMP721d for MeJA, CCRP4 for ABA, TREAT and TOTAL, NCLN1 and ERDJ2A for SA and VAMP721d for salt stress were stable reference genes with the lowest SD means. Interestingly, ACT6, the commonly used housekeeping gene in Glycine max was the most unstable one for MeJA, ABA, TREAT and TOTAL.
Table 2.
Expression stability of 14 candidate reference genes calculated by Delta Ct, BestKeeper, NormFinder, geNorm and RefFinder
| Group | Rank | Delta Ct | BestKeeper | NormFinder | GeNorm | RefFinder | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SD | Gene | geomean | Gene | SV | Gene | SV | Gene | SV | ||
| Tissues | 1 | NFD6 | 0.35 | SF3B | 0.41 | PP | 0.128 | NFD6 | 0.346 | CCRP4 | 1.86 |
| 2 | CCRP4 | 0.35 | CCRP4 | 0.54 | NFD6 | 0.152 | CCRP4 | 0.347 | NFD6 | 2.55 | |
| 3 | PP | 0.35 | BRM | 0.56 | CCRP4 | 0.163 | PP | 0.350 | CUL1 | 3.36 | |
| 4 | CUL1 | 0.37 | ARI7 | 0.57 | CUL1 | 0.195 | CUL1 | 0.367 | PP | 3.41 | |
| 5 | BRM | 0.38 | UFM1 | 0.57 | BRM | 0.216 | BRM | 0.381 | BRM | 4.61 | |
| 6 | UFM1 | 0.40 | ERDJ2A | 0.60 | ERDJ2A | 0.241 | UFM1 | 0.398 | UFM1 | 5.26 | |
| 7 | ERDJ2A | 0.40 | NFD6 | 0.60 | VAMP721d | 0.245 | ERDJ2A | 0.399 | SF3B | 5.57 | |
| 8 | VAMP721d | 0.41 | CUL1 | 0.64 | UFM1 | 0.270 | VAMP721d | 0.407 | ERDJ2A | 6.48 | |
| 9 | SMUP1 | 0.47 | PP | 0.66 | SMUP1 | 0.348 | SMUP1 | 0.471 | VAMP721d | 8.82 | |
| 10 | SF3B | 0.47 | SMUP1 | 0.69 | NCLN1 | 0.359 | SF3B | 0.471 | SMUP1 | 9.49 | |
| 11 | NCLN1 | 0.47 | ACT6 | 0.78 | ACT6 | 0.363 | NCLN1 | 0.473 | ARI7 | 10.24 | |
| 12 | ACT6 | 0.47 | VAMP721d | 0.79 | SF3B | 0.379 | ACT6 | 0.474 | ACT6 | 11.24 | |
| 13 | ARI7 | 0.55 | NCLN1 | 0.82 | ARI7 | 0.493 | ARI7 | 0.553 | NCLN1 | 11.45 | |
| 14 | UN | 0.73 | UN | 1.06 | UN | 0.681 | UN | 0.728 | UN | 14.00 | |
| MeJA | 1 | SMUP1 | 0.22 | SMUP1 | 0.13 | VAMP721d | 0.036 | SMUP1 | 0.216 | SMUP1 | 1.19 |
| 2 | VAMP721d | 0.22 | ERDJ2A | 0.16 | SMUP1 | 0.040 | VAMP721d | 0.223 | VAMP721d | 1.57 | |
| 3 | CCRP4 | 0.24 | VAMP721d | 0.18 | CCRP4 | 0.138 | CCRP4 | 0.240 | CCRP4 | 3.57 | |
| 4 | ERDJ2A | 0.25 | NCLN1 | 0.18 | ERDJ2A | 0.144 | ERDJ2A | 0.255 | ERDJ2A | 4.00 | |
| 5 | UFM1 | 0.26 | UN | 0.19 | UFM1 | 0.161 | UFM1 | 0.265 | UFM1 | 6.30 | |
| 6 | NFD6 | 0.27 | CCRP4 | 0.21 | NFD6 | 0.173 | NFD6 | 0.272 | ARI7 | 6.88 | |
| 7 | ARI7 | 0.27 | NFD6 | 0.21 | UN | 0.198 | ARI7 | 0.273 | NFD6 | 6.90 | |
| 8 | UN | 0.28 | PP | 0.22 | ARI7 | 0.199 | UN | 0.283 | UN | 7.27 | |
| 9 | SF3B | 0.28 | UFM1 | 0.22 | SF3B | 0.221 | SF3B | 0.285 | SF3B | 8.17 | |
| 10 | CUL1 | 0.30 | ARI7 | 0.25 | NCLN1 | 0.232 | CUL1 | 0.297 | NCLN1 | 8.34 | |
| 11 | NCLN1 | 0.30 | SF3B | 0.29 | PP | 0.241 | NCLN1 | 0.303 | CUL1 | 9.64 | |
| 12 | PP | 0.32 | CUL1 | 0.29 | CUL1 | 0.243 | PP | 0.321 | PP | 10.61 | |
| 13 | BRM | 0.35 | BRM | 0.36 | BRM | 0.310 | BRM | 0.350 | BRM | 13.00 | |
| 14 | ACT6 | 0.52 | ACT6 | 0.39 | ACT6 | 0.510 | ACT6 | 0.523 | ACT6 | 14.00 | |
| ABA | 1 | CCRP4 | 0.25 | UFM1 | 0.26 | CCRP4 | 0.103 | CCRP4 | 0.252 | ERDJ2A | 2.28 |
| 2 | SMUP1 | 0.26 | SMUP1 | 0.27 | SMUP1 | 0.122 | SMUP1 | 0.258 | SMUP1 | 2.74 | |
| 3 | ERDJ2A | 0.26 | ERDJ2A | 0.30 | ERDJ2A | 0.125 | ERDJ2A | 0.263 | CCRP4 | 2.85 | |
| 4 | NFD6 | 0.26 | BRM | 0.31 | NFD6 | 0.125 | NFD6 | 0.264 | NFD6 | 4.23 | |
| 5 | UN | 0.29 | NFD6 | 0.32 | UN | 0.178 | UN | 0.288 | NCLN1 | 4.60 | |
| 6 | ARI7 | 0.29 | SF3B | 0.32 | ARI7 | 0.183 | ARI7 | 0.289 | UFM1 | 5.20 | |
| 7 | CUL1 | 0.29 | CUL1 | 0.32 | NCLN1 | 0.188 | CUL1 | 0.293 | UN | 5.48 | |
| 8 | NCLN1 | 0.29 | NCLN1 | 0.33 | VAMP721d | 0.189 | NCLN1 | 0.293 | ARI7 | 7.82 | |
| 9 | UFM1 | 0.29 | PP | 0.33 | UFM1 | 0.202 | UFM1 | 0.295 | VAMP721d | 7.95 | |
| 10 | VAMP721d | 0.30 | VAMP721d | 0.35 | CUL1 | 0.212 | VAMP721d | 0.296 | CUL1 | 8.37 | |
| 11 | SF3B | 0.31 | CCRP4 | 0.35 | SF3B | 0.234 | SF3B | 0.311 | SF3B | 9.45 | |
| 12 | PP | 0.37 | UN | 0.38 | PP | 0.303 | PP | 0.370 | BRM | 9.49 | |
| 13 | BRM | 0.37 | ARI7 | 0.39 | BRM | 0.325 | BRM | 0.371 | PP | 11.39 | |
| 14 | ACT6 | 0.58 | ACT6 | 0.55 | ACT6 | 0.559 | ACT6 | 0.577 | ACT6 | 14.00 | |
| SA | 1 | NCLN1 | 0.18 | ARI7 | 0.26 | NCLN1 | 0.064 | NCLN1 | 0.181 | NCLN1 | 1.57 |
| 2 | ERDJ2A | 0.18 | UN | 0.27 | CUL1 | 0.068 | ERDJ2A | 0.181 | ERDJ2A | 2.34 | |
| 3 | CUL1 | 0.19 | CUL1 | 0.27 | ERDJ2A | 0.073 | CUL1 | 0.187 | CUL1 | 3.46 | |
| 4 | VAMP721d | 0.19 | NFD6 | 0.27 | VAMP721d | 0.122 | VAMP721d | 0.195 | VAMP721d | 4.28 | |
| 5 | NFD6 | 0.21 | ERDJ2A | 0.28 | NFD6 | 0.139 | NFD6 | 0.205 | ARI7 | 4.41 | |
| 6 | ACT6 | 0.22 | NCLN1 | 0.28 | ARI7 | 0.148 | ACT6 | 0.222 | NFD6 | 4.47 | |
| 7 | ARI7 | 0.22 | VAMP721d | 0.29 | CCRP4 | 0.159 | ARI7 | 0.223 | UN | 6.03 | |
| 8 | CCRP4 | 0.23 | ACT6 | 0.30 | ACT6 | 0.171 | CCRP4 | 0.229 | ACT6 | 6.62 | |
| 9 | SMUP1 | 0.24 | UFM1 | 0.30 | SMUP1 | 0.181 | SMUP1 | 0.236 | SMUP1 | 8.68 | |
| 10 | UN | 0.24 | SMUP1 | 0.33 | UFM1 | 0.182 | UN | 0.239 | CCRP4 | 9.24 | |
| 11 | UFM1 | 0.25 | PP | 0.34 | UN | 0.191 | UFM1 | 0.247 | UFM1 | 10.22 | |
| 12 | BRM | 0.28 | BRM | 0.37 | BRM | 0.240 | BRM | 0.285 | BRM | 12.00 | |
| 13 | PP | 0.30 | CCRP4 | 0.38 | PP | 0.249 | PP | 0.298 | PP | 12.47 | |
| 14 | SF3B | 0.31 | SF3B | 0.49 | SF3B | 0.274 | SF3B | 0.313 | SF3B | 14.00 | |
| Salt | 1 | VAMP721d | 0.21 | SF3B | 0.15 | SF3B | 0.088 | VAMP721d | 0.206 | SF3B | 1.97 |
| 2 | SMUP1 | 0.21 | CUL1 | 0.17 | VAMP721d | 0.089 | SMUP1 | 0.209 | VAMP721d | 2.00 | |
| 3 | SF3B | 0.21 | SMUP1 | 0.18 | SMUP1 | 0.107 | SF3B | 0.213 | SMUP1 | 2.06 | |
| 4 | CCRP4 | 0.23 | NCLN1 | 0.19 | CCRP4 | 0.152 | CCRP4 | 0.230 | CCRP4 | 4.43 | |
| 5 | NFD6 | 0.24 | ERDJ2A | 0.19 | NFD6 | 0.176 | NFD6 | 0.243 | NFD6 | 5.36 | |
| 6 | NCLN1 | 0.26 | CCRP4 | 0.19 | CUL1 | 0.189 | NCLN1 | 0.259 | CUL1 | 5.38 | |
| 7 | CUL1 | 0.27 | UFM1 | 0.20 | NCLN1 | 0.194 | CUL1 | 0.265 | NCLN1 | 5.63 | |
| 8 | UN | 0.27 | VAMP721d | 0.20 | BRM | 0.207 | UN | 0.269 | UFM1 | 8.15 | |
| 9 | UFM1 | 0.27 | BRM | 0.21 | UN | 0.213 | UFM1 | 0.275 | ERDJ2A | 8.59 | |
| 10 | BRM | 0.28 | PP | 0.24 | UFM1 | 0.216 | BRM | 0.278 | BRM | 9.43 | |
| 11 | ERDJ2A | 0.28 | NFD6 | 0.25 | ERDJ2A | 0.230 | ERDJ2A | 0.281 | UN | 10.49 | |
| 12 | ARI7 | 0.29 | ARI7 | 0.25 | PP | 0.232 | ARI7 | 0.286 | ACT6 | 11.73 | |
| 13 | ACT6 | 0.29 | ACT6 | 0.25 | ARI7 | 0.241 | ACT6 | 0.289 | PP | 12.38 | |
| 14 | PP | 0.30 | UN | 0.25 | ACT6 | 0.243 | PP | 0.296 | ARI7 | 12.49 | |
| TREAT | 1 | CCRP4 | 0.27 | SMUP1 | 0.26 | CCRP4 | 0.129 | CCRP4 | 0.274 | VAMP721d | 2.45 |
| 2 | VAMP721d | 0.29 | PP | 0.27 | VAMP721d | 0.157 | VAMP721d | 0.286 | ERDJ2A | 2.71 | |
| 3 | ERDJ2A | 0.29 | VAMP721d | 0.29 | ERDJ2A | 0.162 | ERDJ2A | 0.290 | CCRP4 | 2.72 | |
| 4 | SMUP1 | 0.29 | ARI7 | 0.30 | SMUP1 | 0.166 | SMUP1 | 0.291 | SMUP1 | 2.83 | |
| 5 | NCLN1 | 0.31 | NFD6 | 0.30 | NCLN1 | 0.195 | NCLN1 | 0.307 | NCLN1 | 3.98 | |
| 6 | PP | 0.31 | ERDJ2A | 0.31 | PP | 0.202 | PP | 0.311 | PP | 4.74 | |
| 7 | NFD6 | 0.32 | UFM1 | 0.32 | NFD6 | 0.222 | NFD6 | 0.318 | NFD6 | 6.19 | |
| 8 | ARI7 | 0.32 | CUL1 | 0.33 | ARI7 | 0.225 | ARI7 | 0.323 | ARI7 | 6.73 | |
| 9 | SF3B | 0.33 | BRM | 0.33 | SF3B | 0.236 | SF3B | 0.328 | CUL1 | 9.46 | |
| 10 | CUL1 | 0.34 | NCLN1 | 0.33 | CUL1 | 0.256 | CUL1 | 0.335 | UFM1 | 9.82 | |
| 11 | UFM1 | 0.35 | CCRP4 | 0.33 | UFM1 | 0.265 | UFM1 | 0.345 | SF3B | 9.87 | |
| 12 | UN | 0.37 | UN | 0.33 | UN | 0.296 | UN | 0.367 | BRM | 11.62 | |
| 13 | BRM | 0.37 | SF3B | 0.39 | BRM | 0.309 | BRM | 0.371 | UN | 12.24 | |
| 14 | ACT6 | 0.47 | ACT6 | 0.45 | ACT6 | 0.427 | ACT6 | 0.469 | ACT6 | 14.00 | |
| TOTAL | 1 | CCRP4 | 0.38 | ARI7 | 0.41 | CCRP4 | 0.154 | CCRP4 | 0.383 | CCRP4 | 2.06 |
| 2 | PP | 0.39 | PP | 0.42 | PP | 0.166 | PP | 0.392 | PP | 2.38 | |
| 3 | VAMP721d | 0.41 | CCRP4 | 0.43 | VAMP721d | 0.221 | VAMP721d | 0.410 | ERDJ2A | 3.16 | |
| 4 | CUL1 | 0.43 | ERDJ2A | 0.43 | CUL1 | 0.255 | CUL1 | 0.426 | VAMP721d | 4.05 | |
| 5 | ERDJ2A | 0.43 | CUL1 | 0.43 | ERDJ2A | 0.259 | ERDJ2A | 0.427 | NCLN1 | 4.46 | |
| 6 | NCLN1 | 0.44 | SF3B | 0.43 | NCLN1 | 0.280 | NCLN1 | 0.441 | CUL1 | 4.47 | |
| 7 | NFD6 | 0.46 | NFD6 | 0.45 | SF3B | 0.315 | NFD6 | 0.457 | ARI7 | 5.20 | |
| 8 | SF3B | 0.46 | SMUP1 | 0.46 | NFD6 | 0.317 | SF3B | 0.460 | SF3B | 7.20 | |
| 9 | ARI7 | 0.48 | UFM1 | 0.46 | ARI7 | 0.348 | ARI7 | 0.482 | NFD6 | 7.24 | |
| 10 | BRM | 0.52 | BRM | 0.47 | BRM | 0.409 | BRM | 0.518 | SMUP1 | 9.92 | |
| 11 | SMUP1 | 0.54 | VAMP721d | 0.48 | SMUP1 | 0.448 | SMUP1 | 0.543 | BRM | 9.97 | |
| 12 | UN | 0.59 | NCLN1 | 0.53 | UN | 0.498 | UN | 0.589 | UN | 12.24 | |
| 13 | UFM1 | 0.59 | UN | 0.61 | ACT6 | 0.502 | UFM1 | 0.589 | UFM1 | 12.98 | |
| 14 | ACT6 | 0.60 | ACT6 | 0.63 | UFM1 | 0.510 | ACT6 | 0.601 | ACT6 | 13.74 | |
SD standard deviation of each gene among the tested samples; geomean comprehensive index value of each gene among the tested samples; SV stability value of each gene among the tested samples
The ranking order estimated by BestKeeper was determined by the coefficient of determination (r), SD and CV of each gene (Pfaffl et al. 2004). The most stable reference genes must have the highest r, lowest SD and CV. As shown in Table 2, the geomean value of each candidate reference gene was calculated and ranking. The ranking order showed the optimum reference gene in different situations. For example, SF3B was the optimum one for TOTAL and salt stress, SMUP1 was the best one for MeJA and TREAT, UFM1 for ABA, ARI7 for SA and TOTAL. The stability of ACT6 was the most unstable reference gene in four groups (MeJA, ABA, TREAT and TOTAL), which is consistent with ΔCt results.
The NormFinder analysis shown that PP was the most suitable gene for tissues and the UN was the most unsuitable one. For MeJA treatment, VAMP721d was the most stable one. For ABA treatment, TREAT and TOTAL, the most stable reference gene was CCRP4. Under salt stress, the best stable reference gene was SF3B. Interestingly, SF3B was the worst one for SA treatment. Additionally, ACT6 performed worst in MeJA, ABA, salt stress and TREAT.
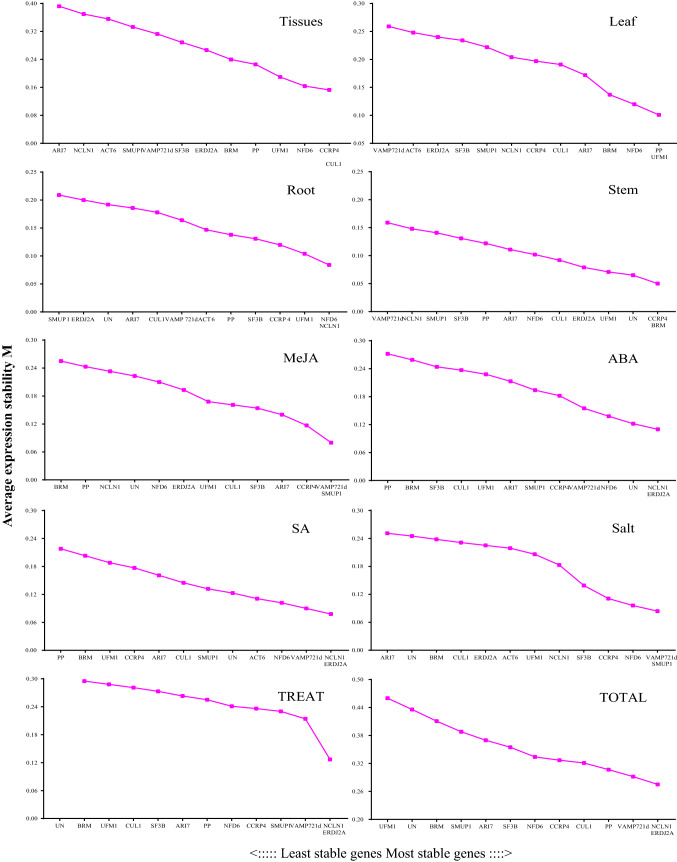
According to the manual of the geNorm, the M value of each candidate reference gene was calculated. As shown in Table 2, M value of all tested samples are smaller than 1.5, indicating that all selected reference genes were relatively stable. After stepwise exclusion of the least stable reference gene, the two most stable genes were obtained finally. As shown in Fig. 2, CCRP4 and CUL1 were the best stable reference gene for Tissues and VAMP721d and SMUP1 were the optimal reference gene for MeJA and salt stress. NCLN1 and ERDJ2A were most stable genes for ABA, SA, TREAT and TOTAL. Noticeably, ACT6 was the most unstable reference gene in MeJA, ABA, TREAT and TOTAL. UN and SF3B expressed most unstable in tissues and SA treatment, respectively.
Fig. 2.
Average expression stability values (M) and ranking of the candidate reference genes calculated using geNorm. A lower value of the average expression stability indicates more stable expression. The stable genes are on the right side, while the least stable genes on the left side
Moreover, the average expression stability value M of candidate reference genes in leaf, stem and root was also calculated, respectively. As shown in Fig. 2, NFD6 + NCLN1 was the best combination of stable reference genes for root, CCRP4 + BRM for stem and PP + UFM1for leaf.
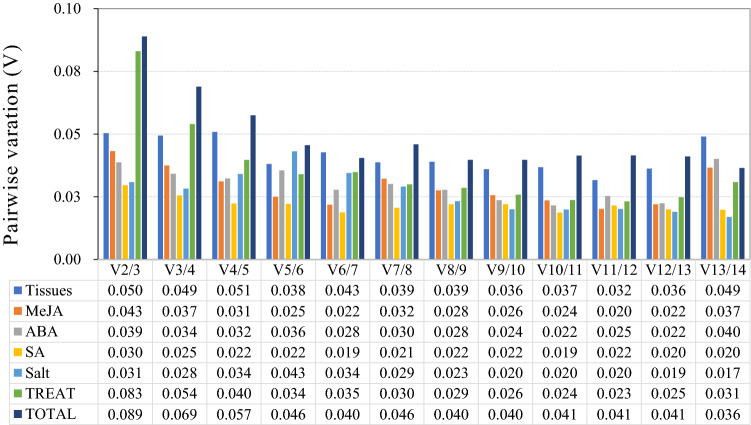
The geNorm software was also used to determine the optimal number of reference genes for normalization of gene expression level by the values of pairwise variation (Vn/n+1). The value should be below 0.15, or an additional reference gene was required (Stanton et al. 2017). As shown in Fig. 3, all the values of V2/3 were smaller than 0.1 and far less than default threshold 0.15, which indicating that the two most stable reference genes were sufficient to normalize expression data in these experimental situations.
Fig. 3.
Pairwise variation (V) analysis of 14 selected reference genes using geNorm software. The pairwise variations Vn/Vn + 1 were calculated by geNorm in different tissues and under hormone and salt stress treatment samples. There was a cut-off value 0.15, only when the value was below this, an additional reference gene was required
RefFinder was finally used to comprehensively determine the stability of the candidate reference genes. According to the Table 2, SMUP1, ERDJ2A, NCLN1, SF3B and VAMP721d were the most suitable reference genes for MeJA, ABA, SA, Salt and TREAT, respectively. CCRP4 was best one for tissues and TOTAL. ACT6 was the worst one for MeJA, ABA, TREAT and TOTAL.
Validating the stability of reference genes
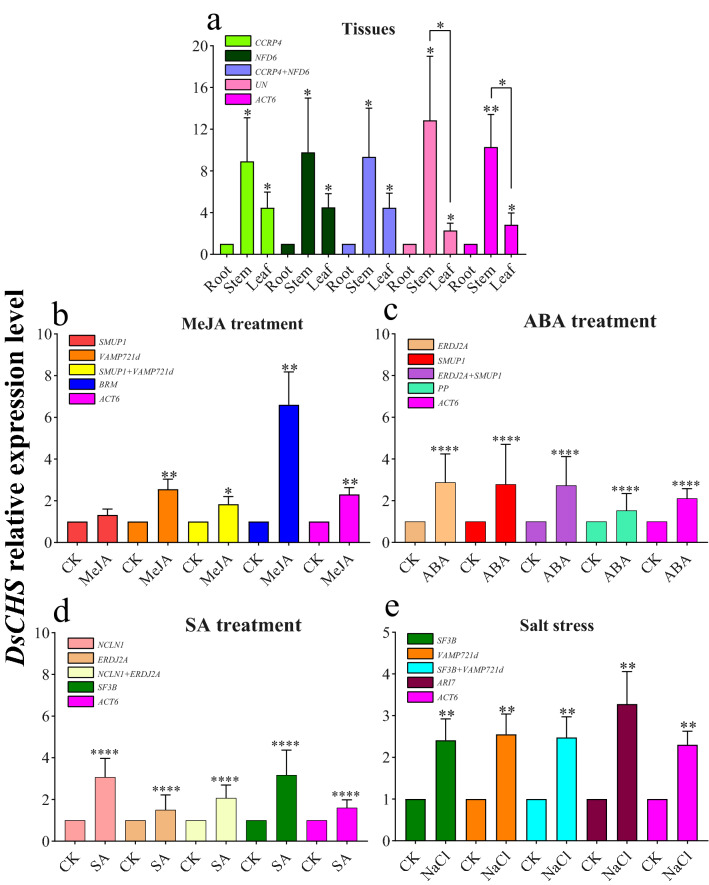
To validate the identified stable reference genes, DsCHS, a key gene committed in flavonoid and schaftoside biosynthesis, was chosen to validate the stability of reference genes in Tissues, and ABA, MeJA, SA treatment and salt stress.
For different tissues, root, stem and leaf, the most stable reference genes CCRP4 and NFD6, and the combination of them (CCRP4 + NFD6), were all used to normalize the RT-qPCR data. As shown in Fig. 4a, the expression level of DsCHS normalized by the combination of CCRP4 + NFD6 provides more accurate estimation than those normalized by single reference gene. When the most unstable reference gene UN and ACT6 were used to calculate the relative expression level of DsCHS, it had a same trend with the most stable genes, highly expressed in stem, followed by leaf and root. But the relative expression of DsCHS was obviously over-estimated in tested stems, and under-estimated in tested roots when UN and ACT6 were used, respectively. Moreover, there were no significant difference between expression level of DsCHS in the tested stems and leaves normalized by the first two stable reference genes alone or combination, but it showed significant difference after normalized by the worst stable reference genes. The effect of the five kinds of normalization was the same as that of comprehensive ranking in Tissues.
Fig. 4.
Relative expression levels of DsCHS normalized by a validated reference gene alone or combination in Tissues a MeJA treatment b ABA treatment c SA treatment d and salt stress e of D. styracifolium. Bars indicate standard deviation calculated from three biological replicates. Asterisk indicates significance at P < 0.05 (*), P < 0.01 (**), P < 0.001 (***) and P < 0.0001 (****) using T test by GraphPad Prism
For MeJA treatment, the average expression level of DsCHS was 1.3, 2.6, 1.8 times higher than CK when normalized by the most stable reference genes (SMUP1, VAMP721d) and their combination (SMUP1 + VAMP721d), respectively (Fig. 4b). The result normalized by the single SMUP1 (the most stable one) showed no significant changes after MeJA treatment. However, the result normalized by the single VAMP721d (the top 2 stable one) showed more significant effect by the MeJA treatment, the combination of them showed the significant result, it seemed that the final result was neutralized by the combination of the former two. But when normalized by the least stable reference gene BRM, it was 6.6 times higher than CK in average relative expression level. When ACT6 was used as reference gene, the relative expression level was slightly higher than those normalized by the combination of SMUP1 + VAMP721d. In sum, the expression level of DsCHS normalized by the second stable reference gene (VAMP721d), the least stable reference gene (BRM) and ACT6, was significantly enhanced by MeJA treatment, except the most stable reference gene (SMUP1). Therefore, the best normalization choice for the most stable reference gene was the SMUP1 for MeJA treatment.
For ABA treatment, the expression level of DsCHS showed slight difference normalized by the most stable two reference genes alone or combination and was under-estimated when normalized by the worst unstable reference gene (PP) and ACT6 (Fig. 4c). Noticeably, the transcript level of DsCHS normalized by ERDJ2A and/or SMUP1 was slightly higher than that of DsCHS normalized by PP or ACT6. As for SA treatment, it seems that the stability of the best candidate gene NCLN1 was similar to the worst SF3B and the second-best candidate gene ERDJ2A similar to ACT6, suggesting that the stability of these candidate reference genes identified by our strategy using RNA-seq transcriptome was similar. Meanwhile, the expression level of DsCHS normalized by NCLN1 + ERDJ2A was slightly distinct when compared to single reference genes including ACT6 (Fig. 4d). According to the geNorm, the combination of NCLN1 + ERDJ2A was the best stable reference gene and was the best choice for using as stable reference gene in SA treatment. In salt stress case, the top 2 best stable reference genes and their combination show similar stability to ACT6 and better stability than the worst reference gene ARI7 (Fig. 4d). Consequently, ACT6, and SF3B and/or VAMP721d can be used as stable reference genes in salt stress.
Discussion
It is very important for correctly and concisely evaluating the expression of gene normalized by stable reference gene. Currently, there are no stable reference genes previously reported in D. styracifolium. In this study, 14 candidate reference genes were systematically evaluated, including 13 new candidate reference genes selected form the RNA-Seq data of D. styracifolium and 1 commonly used housekeeping gene (ACT6) in Leguminosae, by five different statistical algorithms (GeNorm, NormFinder, BestKeeper, ΔCt and RefFinder). As shown in Fig. 1, all of these 14 candidate reference genes had a low or medium expression level and met the selection criteria of reference genes (Wan et al. 2010; Yan et al. 2018). The stability ranking of candidate reference genes by different programs highly was consistent with each other, which could easily exclude the least stable reference gene. For example, the least stable reference genes evaluated by five algorithms was UN in Tissues, SF3B in SA, ACT6 in MeJA, ABA, TREAT and TOTAL. Furthermore, almost all algorithms ranked SMUP1 as the most stable reference genes in MeJA treatment, and CCRP4 was ranked top 3 in different tissues. Noticeably, ranking order of gene stability by five algorithms had slight difference due to their distinct principle for evaluating reference gene. BestKeeper sometimes gave a higher-ranking order to a certain gene when compared to the other algorithms in some cases. For example, in different tissues of Santalum album, the stability ranking of ACT ranked first using BestKeeper method while ranked the middle using GeNorm and NormFinder (Yan et al. 2018). In Iris germanica case, BestKeeper ranked IgACT6 as top 2 stable genes while its ranking order was in the middle or bottom position using GeNorm and NormFinder methods (Wang et al. 2021). The same thing also happened in our study, SF3B was ranked first in the BestKeeper result. However, it was ranked 10th in ΔCt, 12th in NormFinder, 10th in geNorm, and 7th in RefFinder. In our study, CCRP4, ERDJ2A, SMUP1, NCLN1 and SF3B were the most stable reference gene in three tissues, ABA, MeJA, SA and salt stress treatment, respectively. Meanwhile, CCRP4 and VAMP721d were the most stable reference gene for TOTAL and TREAT, respectively.
To study the difference between the SD/CV of the RNA-seq data and the ranking order of five algorithms, the original RNA-seq data were also studied. Our results indicated that the stability ranking of reference genes was not similar to the real result. For example, VAMP721d had the lowest CV value in FPKM. However, it is ranked as the most unstable reference gene in Tissues. Therefore, CV values did not determine the final stability order.
Lines of studies showed that the geometric mean of multiple carefully selected reference genes was validated as an accurate normalization factor (Vandesompele et al. 2002; Kozera and Rapacz 2013). In this study, to verify the accuracy and reliability of the results normalized by the most stable reference genes under various conditions, the expression level of DSCHs was evaluated by different candidate reference genes and their combinations. Our studies showed that the combination of top 2 stable reference genes (CCRP4 and NFD6) could be more accurate to reflect the expression level of DsCHS in root, stem and leaf of D. styracifolium when compared with the single reference gene. The similar situation also happened in ABA, SA, salt stress treatment, although there was no significant difference between the expression level of DsCHS normalized by the first two stable reference genes single or combination and ACT6. However, there was an exception in MeJA treatment. The expression level of DsCHS normalized by the first stable gene SMUP1 was different from the second stable gene VAMP721d and their combination (SMUP1 + VAMP721d). The former one showed no significant change after MeJA treatment while the latter two showed a contrary result compared to SMUP1. As shown in Table 2, SMUP1 was always ranked top1 by all methods except for NormFinder. These results indicate that accurate estimation of gene expression using one or two stable reference genes should be careful and dependent on the exact experimental condition.
Increasing evidences documented that identification of the stable reference genes screening by the transcriptome dataset is feasible and efficient (Yan et al. 2018; Liang et al. 2020; Zhang et al. 2020). Moreover, high-throughput omics methods provide us a chance to find new and stable reference genes. Our study also verified that several novel reference genes (CCRP4, NFD6, SMUP1, etc.), screened from our previous transcriptome dataset of D. styracifolium (Wang et al. 2020a), performed better than the traditional housekeeper gene ACT6 in some case (Fig. 4). It is undeniable that using transcriptome dataset to screen and identify stable reference genes is an efficient strategy for less-studied plants. Due to the sensitivity of RT- qPCR and the importance of an accurate reference gene for the final normalization results, the further validation after screening from the transcriptome dataset should be performed.
Conclusion
In conclusion, our study systematically evaluated candidate reference genes in D. styracifolium using transcriptome dataset and validated these putative reference genes in Tissues (leaf, stem and root), and seedlings under hormone treatments and salt stress. A total of 14 candidate reference genes were selected and statistically ranked by five statistics methods, including geNorm, NormFinder, BestKeeper, ΔCt and RefFinder. The resulting stable reference genes were further validated using DsCHS. Our results indicated that the reference gene combination of PP + UFM1, CCRP4 + BRM and NFD6 + NCLN1 were the most stable reference genes in leaf, stem and root tissues, respectively. The most stable reference gene combination for all tissues was CCRP4 + CUL1. In addition, the most stable reference genes for different experimental conditions were distinct, for instance SMUP1 for MeJA treatment, ERDJ2A + SMUP1 for SA treatment, NCLN1 + ERDJ2A for ABA treatment and SF3B + VAMP721d for salt stress, respectively. Our results lay a foundation for achieving accurate and reliable RT-qPCR results, so as to correctly understand the function of genes in D. styracifolium.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1: Supplementary tables. Table S1 The rough candidate genes whose CV was lower than 12%. Table S2 The candidate reference genes selected according to RNA-seq data. Supplementary figures. Figure S1 Electrophoresis analysis of the specificity of primer pairs for RT-PCR amplification (a) and the RNA quality (b). In the Fig. S1(a), ‘1–16’ represent VAMP721d, PP, CCRP4, SF3B, ARI7, BRM, UFM1, NFD6, UN, SMUP1, CUL1, NCLN1, ERDJ2A, ACT6, DsCHS and 2000 bp marker, respectively. In the Fig. S1(b), ‘0’ represent the 1kb ladder marker, and ‘1–21’ represent the RNA from tissues (root, stem and leaf), MeJA, ABA, SA and salt treatment, each sample has three biological repeats. Figure S2 Dissociation curves for the reference genes tested in this study. The dissociation curves for each candidate gene showed a single peak indicated that the primers of each tested gene were specific. (DOCX 297 KB)
Author contributions
This research was designed by FX, SZ and YW; ZW, FY and DS carried out the experiments; ZW performed data analysis; ZW prepared the hormonal treatment and salt stress samples; ZW drafted the manuscript; FX, SZ and YW revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was partially supported by Grants from Key Area R&D Project of Guangdong Province (2020B020221001), Guangdong Provincial Key Laboratory of Applied Botany (AB2018017), Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams, China (NO.2019KJ148), Youth Innovation Promotion Association CAS (2015286).
Declarations
Conflict of interest
The authors declare no conflict of interest.
Contributor Information
Feng Xu, Email: xufeng198@126.com.
Shaohua Zeng, Email: shhzeng@scbg.ac.cn.
References
- Bansal R, Mittapelly P, Cassone BJ, Mamidala P, Redinbaugh MG, Michel A. Recommended reference genes for quantitative PCR analysis in soybean have variable stabilities during diverse biotic stresses. PLoS ONE. 2015;10(8):e0134890. doi: 10.1371/journal.pone.0134890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges A, Tsai SM, Caldas DG. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 2012;31(5):827–838. doi: 10.1007/s00299-011-1204-x. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, Vandesompele J, Wittwer CT. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cheng X-x, Tang X-m, Guo C-c, Zhang C-r, Yang Q. New flavonol glycosides from the seeds of Desmodiumstyracifolium. Chem Nat Compd. 2018;54(5):846–850. doi: 10.1007/s10600-018-2496-7. [DOI] [Google Scholar]
- Andersen CL, Jensen JL, Ørntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Can Res. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Committee CP. Pharmacopoeia of the people’s Republic of China Part I. Beijing: Chemical Industry Press; 2020. p. 46. [Google Scholar]
- de Jonge HJ, Fehrmann RS, de Bont ES, Hofstra RM, Gerbens F, Kamps WA, de Vries EG, van der Zee AG, te Meerman GJ, ter Elst A. Evidence based selection of housekeeping genes. PLoS ONE. 2007;2(9):e898. doi: 10.1371/journal.pone.0000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao M, Liu Y, Ma X, Shuai Q, Gai J, Li Y. Evaluation of reference genes for normalization of gene expression using quantitative RT-PCR under aluminum, cadmium, and heat stresses in soybean. PLoS ONE. 2017;12(1):e0168965. doi: 10.1371/journal.pone.0168965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno J, Eattock N, Van Deynze A, Blumwald E. Selection and validation of reference genes for gene expression analysis in switchgrass (Panicum virgatum) using quantitative real-time RT-PCR. PLoS ONE. 2014;9(3):e91474. doi: 10.1371/journal.pone.0091474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P, Yan W, Han Q, Wang C, Zhang Z. Simultaneous quantification of 25 active constituents in the total flavonoids extract from Herba Desmodii Styracifolii by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J Sep Sci. 2015;38(7):1156–1163. doi: 10.1002/jssc.201401360. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mauriat M, Guenin S, Pelloux J, Lefebvre JF, Louvet R, Rusterucci C, Moritz T, Guerineau F, Bellini C, Van Wuytswinkel O. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J. 2008;6(6):609–618. doi: 10.1111/j.1467-7652.2008.00346.x. [DOI] [PubMed] [Google Scholar]
- Hamilton ML, Kuate SP, Brazier-Hicks M, Caulfield JC, Rose R, Edwards R, Torto B, Pickett JA, Hooper AM. Elucidation of the biosynthesis of the di-C-glycosylflavone isoschaftoside, an allelopathic component from Desmodium spp. that inhibits Striga spp. development. Phytochemistry. 2012;84:169–176. doi: 10.1016/j.phytochem.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Hirayama H, Ikegami K, Irino N, Kajimoto T, Kubo T, Nohara T, Ou S (1989) Preventive of kidney stone containing flavonoid as active ingredient and production thereof. Japan patent JAH01305080(A)
- Hirayama H, Ikegami K, Irino N, Kajimoto T, Kubo T, Nohara T, Ou S (1989) Triterpenoidalsaponin, production thereof and preventive for renal litiasis containing above-mentioned saponin derivative as active ingredient. Japan patent JPH63131390(A)
- Hong SY, Seo PJ, Yang MS, Xiang F, Park CM. Exploring valid reference genes for gene expression studies in Brachypodium distachyon by real-time PCR. BMC Plant Biol. 2008;8:112. doi: 10.1186/1471-2229-8-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele Jo, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):00341–003411. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph JT, Poolakkalody NJ, Shah JM. Plant reference genes for development and stress response studies. J Biosci. 2018;43(1):173–187. doi: 10.1007/s12038-017-9728-z. [DOI] [PubMed] [Google Scholar]
- Kozera B, Rapacz M. Reference genes in real-time PCR. J Appl Genet. 2013;54(4):391–406. doi: 10.1007/s13353-013-0173-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonak J, Lind K, Sindelka R, Sjoback R, Sjogreen B, Strombom L, Stahlberg A, Zoric N. The real-time polymerase chain reaction. Mol Aspects Med. 2006;27(2–3):95–125. doi: 10.1016/j.mam.2005.12.007. [DOI] [PubMed] [Google Scholar]
- Li D, Hu B, WangWu QW. The research on reference genes in medicinal plant. Mol Plant Breed. 2017;15(3):903–910. doi: 10.13271/j.mpb.015.000903. [DOI] [Google Scholar]
- Li T, Wang J, Lu M, Zhang T, Qu X, Wang Z. Selection and validation of appropriate reference genes for qRT-PCR analysis in Isatisindigotica Fort. Front Plant Sci. 2017;8:1139. doi: 10.3389/fpls.2017.01139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, He Z, Yu H, Wang E, Zhang X, Zhang B, Zhang C, Liang Z. Selection and validation of reference genes for gene expression studies in Codonopsispilosula based on transcriptome sequence data. Sci Rep. 2020;10(1):1362. doi: 10.1038/s41598-020-58328-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wang Q, Sun M, Zhu L, Yang M, Zhao Y. Selection of reference genes for quantitative real-time PCR normalization in Panaxginseng at different stages of growth and in different organs. PLoS ONE. 2014;9(11):e112177. doi: 10.1371/journal.pone.0112177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Liu C, Chen H, Huang X, Zeng X, Zhou J, Mi S. Prevention of cholesterol gallstone disease by schaftoside in lithogenic diet-induced C57BL/6 mouse model. Eur J Pharmacol. 2017;815:1–9. doi: 10.1016/j.ejphar.2017.10.003. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2[-Delta Delta C(T)] method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Nicot N, Hausman JF, Hoffmann L, Evers D. Housekeeping gene selection for real-time RT-PCR normalization in potato during biotic and abiotic stress. J Exp Bot. 2005;56(421):2907–2914. doi: 10.1093/jxb/eri285. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity, BestKeeper-Excel-based tool using pair-wise correlations. Biotech Lett. 2004;26(6):509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- Silver N, Best S, Jiang J, Thein SL. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol Biol. 2006;7:33. doi: 10.1186/1471-2199-7-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton KA, Edger PP, Puzey JR, Kinser T, Cheng P, Vernon DM, Forsthoefel NR, Cooley AM. A whole-transcriptome approach to evaluating reference genes for quantitative gene expression studies: a case study in mimulus. G3. 2017;7(4):1085–1095. doi: 10.1534/g3.116.038075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J. Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem. 2010;399(2):257–261. doi: 10.1016/j.ab.2009.12.008. [DOI] [PubMed] [Google Scholar]
- Wang H, Zhang X, Liu Q, Liu X, Ding S. Selection and evaluation of new reference genes for RT-qPCR analysis in Epinephelusakaara based on transcriptome data. PLoS ONE. 2017;12(2):e0171646. doi: 10.1371/journal.pone.0171646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Fu Y, Ban L, Wang Z, Feng G, Li J, Gao H. Selection of reliable reference genes for quantitative real-time RT-PCR in alfalfa. Genes Genet Syst. 2015;90(3):175–180. doi: 10.1266/ggs.90.175. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Y, Liu Q, Tong H, Zhang T, Gu C, Liu L, Huang S, Yuan H. Selection and validation of appropriate reference genes for RT-qPCR analysis of flowering stages and different genotypes of Irisgermanica L. Sci Rep. 2021;11(1):9901. doi: 10.1038/s41598-021-89100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gong H, Xu X, Wei X, Wang Y, Zeng S. Transcriptome and small RNAome facilitate to study schaftoside in Desmodiumstyracifolium Merr. Ind Crops Prod. 2020 doi: 10.1016/j.indcrop.2020.112352. [DOI] [Google Scholar]
- Wang ZL, Gao HM, Wang S, Zhang M, Chen K, Zhang YQ, Wang HD, Han BY, Xu LL, Song TQ, Yun CH, Qiao X, Ye M. Dissection of the general two-step di-C-glycosylation pathway for the biosynthesis of (iso)schaftosides in higher plants. Proc Natl Acad Sci USA. 2020;117(48):30816–30823. doi: 10.1073/pnas.2012745117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie F, Xiao P, Chen D, Xu L, Zhang B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol Biol. 2012 doi: 10.1007/s11103-012-9885-2. [DOI] [PubMed] [Google Scholar]
- Xiong Y, Wang J, Deng J. Comparison between LysimachiaeHerba and DesmodiiStyracifoliiHerba in pharmacological activities. China J Chin Materia Med. 2015;40(11):2106–2111. [PubMed] [Google Scholar]
- Yan H, Zhang Y, Xiong Y, Chen Q, Liang H, Niu M, Guo B, Li M, Zhang X, Li Y, Teixeira da Silva JA, Ma G. Selection and validation of novel RT-qPCR reference genes under hormonal stimuli and in different tissues of Santalum album. Sci Rep. 2018;8(1):17511. doi: 10.1038/s41598-018-35883-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Fan W, Chen D, Jiang L, Li Y, Yao Z, Yang Y, Qiu D. Selection and validation of reference genes for quantitative gene expression normalization in Taxus spp. Sci Rep. 2020;10(1):22205. doi: 10.1038/s41598-020-79213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhao L, Zeng Y. Selection and application of reference genes for gene expression studies. Plant Physiol J. 2014 doi: 10.13592/j.cnki.ppj.2014.0201. [DOI] [Google Scholar]
- Zhou X, Wang J, Shi H, Xu H. The exploration of 18S rRNA for quantitative RT-PCR as reference gene. Jilin Normal University. 2016;37(2):115–119. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file 1: Supplementary tables. Table S1 The rough candidate genes whose CV was lower than 12%. Table S2 The candidate reference genes selected according to RNA-seq data. Supplementary figures. Figure S1 Electrophoresis analysis of the specificity of primer pairs for RT-PCR amplification (a) and the RNA quality (b). In the Fig. S1(a), ‘1–16’ represent VAMP721d, PP, CCRP4, SF3B, ARI7, BRM, UFM1, NFD6, UN, SMUP1, CUL1, NCLN1, ERDJ2A, ACT6, DsCHS and 2000 bp marker, respectively. In the Fig. S1(b), ‘0’ represent the 1kb ladder marker, and ‘1–21’ represent the RNA from tissues (root, stem and leaf), MeJA, ABA, SA and salt treatment, each sample has three biological repeats. Figure S2 Dissociation curves for the reference genes tested in this study. The dissociation curves for each candidate gene showed a single peak indicated that the primers of each tested gene were specific. (DOCX 297 KB)